Abstract
Understanding how pathogens defend themselves against host defence mechanisms, such as hydrogen peroxide (H2O2) production, is crucial for comprehending fungal infections. H2O2 poses a significant threat to invading fungi due to its potent oxidizing properties. Our research focuses on the hemibiotrophic fungal wheat pathogen Zymoseptoria tritici, enabling us to investigate host–pathogen interactions. We examined two catalase‐peroxidase (CP) genes, ZtCpx1 and ZtCpx2, to elucidate how Z. tritici deals with host‐generated H2O2 during infection. Our analysis revealed that ZtCpx1 was up‐regulated during biotrophic growth and asexual spore formation in vitro, while ZtCpx2 showed increased expression during the transition from biotrophic to necrotrophic growth and in‐vitro vegetative growth. Deleting ZtCpx1 increased the mutant's sensitivity to exogenously added H2O2 and significantly reduced virulence, as evidenced by decreased Septoria tritici blotch symptom severity and fungal biomass production. Reintroducing the wild‐type ZtCpx1 allele with its native promoter into the mutant strain restored the observed phenotypes. While ZtCpx2 was not essential for full virulence, the ZtCpx2 mutants exhibited reduced fungal biomass development during the transition from biotrophic to necrotrophic growth. Moreover, both CP genes act synergistically, as the double knock‐out mutant displayed a more pronounced reduced virulence compared to ΔZtCpx1. Microscopic analysis using fluorescent proteins revealed that ZtCpx1 was localized in the peroxisome, indicating its potential role in managing host‐generated reactive oxygen species during infection. In conclusion, our research sheds light on the crucial roles of CP genes ZtCpx1 and ZtCpx2 in the defence mechanism of Z. tritici against host‐generated hydrogen peroxide.
Keywords: catalase peroxidase, defence mechanisms, hydrogen peroxide, reactive oxygen species, Zymoseptoria tritici
Zymoseptoria tritici uses catalase‐peroxidase genes, such as ZtCpx1 and ZtCpx2, to detoxify host‐generated reactive oxygen species, with their synergistic action highlighted by the double knock‐out mutant's significantly reduced virulence.
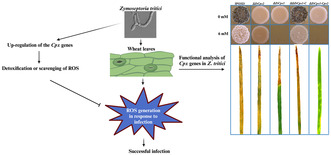
1. INTRODUCTION
Plants have developed various basal defence mechanisms in response to pathogen attacks, including the generation of reactive oxygen species (ROS), such as superoxide anion radicals (O2 −), hydroxyl radicals (HO˙), and hydrogen peroxide (H2O2) at the site of penetration, a phenomenon known as the oxidative burst (Doke et al., 1996; Singh et al., 2021; Wojtaszek, 1997). In general, high levels of ROS can disrupt the balance between radical‐generating and radical‐scavenging systems, resulting in oxidative stress, which is detrimental for living cells due to oxidation of DNA, proteins and lipids, leading to cell damage and malfunction. During plant infection, elevated levels of ROS stimulate fungal pathogens to develop infection structures (Heller & Tudzynski, 2011). Among ROS, H2O2 is the most stable and amenable for experimental studies compared to O2 − and HO˙, which both have a very short half‐life and are highly toxic (Costet et al., 2002). Notably, H2O2 has both antifungal properties and functions as a signalling molecule in numerous biological processes, including phytoalexin production, activation of defence‐related genes and programmed cell death (Apostol et al., 1989; Gadjev et al., 2008; Joseph et al., 1998). There is growing evidence that H2O2 exposure has dual effects on plant‐pathogenic fungi. It can reduce tissue colonization by biotrophic and hemibiotrophic fungal pathogens, but conversely, it can enhance the ramification of host tissue by necrotrophs (Govrin & Levine, 2000; Mellersh et al., 2002; Shetty et al., 2007). To counteract the lethal effects of ROS, plant pathogens produce small molecules such as glutathione and employ antioxidant enzymes like catalase and catalase‐peroxidases (CPs) (Lehmann et al., 2015; Nanda et al., 2010). For instance, MoHYR1 in Magnaporthe oryzae encodes a glutathione peroxidase (GSHPx) that detoxifies host‐derived ROS and is crucial for full virulence (Huang et al., 2011). Catalases and peroxidase are well‐known enzymes that protect cells from oxidative stress by catalysing the decomposition of H2O2 (Vidossich et al., 2012). MoPRX1 in M. oryzae, encoding a thioredoxin peroxidase (TPx), modulates host‐derived H2O2 during early colonization, contributing to virulence (Mir et al., 2015). CPs are unique bifunctional enzymes in the class I peroxidases, exhibiting both catalase and peroxidase activities. Phylogenetic analysis suggests that plant‐pathogenic fungi acquired CP genes through lateral gene transfer (from Negibacteria), followed by gene duplication and diversification (Passardi et al., 2007). Currently, CPs are classified in two distinct clades: one containing cytoplasmic enzymes found in saphrophytic and plant‐pathogenic fungi, and another containing extracellular enzymes predominantly identified in plant‐pathogenic fungi, which differ in location, structure and function (Zámocký et al., 2012, 2020).
Biotrophic fungal pathogens obtain nutrients from living cells and require robust antioxidant mechanisms to overcome oxidative stress imposed by host defences during early infection stages. For example, BghCatB of Blumeria graminis f. sp. hordei, which encodes a secreted catalase, is speculated to play a role in virulence by detoxifying H2O2 produced at the sites of invasion (Skamnioti et al., 2007). Conversely, deletion mutants of the gene encoding secreted catalase CpCAT1 from Claviceps purpurea did not show a significant reduction in virulence, indicating it is not required for pathogenicity (Garre et al., 1998). Functional analysis of CfCAT2, a cytoplasmic catalase of the biotrophic tomato pathogen Cladosporium fulvum demonstrated its dispensability for virulence, despite its preferential expression in response to exogenously applied H2O2.
Unlike biotrophs, necrotrophic fungal pathogens kill host cells, obtain nutrition from decaying host tissue and should have the ability to handle rising ROS levels during oxidative stress situations following infection establishment. Indeed, the growth of necrotrophic plant pathogens is stimulated in the presence of H2O2 (Govrin & Levine, 2000). Recent studies have shown that deletion mutants of genes encoding both secreted and cytoplasmic catalases in Cochliobolus heterostrophus (ChCAT3), Sclerotinia sclerotiorum (Scat1) and Botrytis cinerea (BcCAT2) lose their pathogenicity on their respective hosts and exhibit increased sensitivity to exogenously added H2O2 during in‐vitro mycelial growth (Robbertse et al., 2003; Schouten et al., 2002; Yarden et al., 2014).
Hemibiotrophic pathogens initially grow intercellularly on host plants in a biotrophic phase, followed by a necrotrophic phase at later infection stages where they need to neutralize host‐generated H2O2. In M. oryzae, MgCATB, a secreted catalase plays a central role in the onset of infection by maintaining the integrity of fungal cell walls and regulating appressorium function (Skamnioti et al., 2007). However, functional analysis of secreted fungal CPs, such as MgCPXB of M. oryzae, has demonstrated their importance primarily during the early stages of infection but not in advanced stages (Tanabe et al., 2011).
Zymoseptoria tritici (Quaedvlieg et al., 2011) is a significant foliar pathogen of bread and durum wheat, causing Septoria tritici blotch (STB) in wheat‐growing areas worldwide (Eyal, 1999; Fones & Gurr, 2015). As a hemibiotroph, Z. tritici shows two distinct colonization phases: an initial biotrophic stage and an advanced necrotrophic phase (Kema et al., 1996). Yang et al. (2013) showed that incompatible Z. tritici–wheat interactions were associated with a rapid and substantial early accumulation of H2O2, whereas in compatible interactions, significantly lower levels of H2O2 accumulated during the initial biotrophic phase (Yang et al., 2013). Moreover, a substantial accumulation of H2O2 was detected during the switch to necrotrophy in compatible interactions, coinciding with the occurrence of severe disease symptoms. This suggests that Z. tritici is able to cope with different levels of H2O2 during the two phases of infection. Recently, a forward genetic approach was employed to explore the genetic factors contributing to oxidative stress tolerance in Z. tritici and to identify candidate genes influencing this trait. This investigation revealed a specific quantitative trait locus with a high LOD score of 15.8 on the chromosome 8 associated with oxidative stress that encompasses the functional ZtCpx1 gene (Zhong et al., 2021).
In this study, we demonstrate that Z. tritici produces a cytoplasmic and an extracellular catalase peroxidase, encoded by CP genes ZtCpx1 and ZtCpx2, respectively, and show that both are crucial for virulence.
2. RESULTS
2.1. Identification and characterization of Z. tritici catalases and catalase‐peroxidases
In the Z. tritici genome (Goodwin et al., 2011), two bifunctional CPs were identified: CP A (ZtCpx1; Protein ID: 105409) and CP B (ZtCpx2; Protein ID: 67250), along with two monofunctional catalase‐encoding genes, ZtCat1 (Protein ID: 85387) and ZtCat2 (Protein ID: 98331). In‐silico analysis (SignalP) revealed that only ZtCpx2 possesses a signal peptide, with a cleavage site located between positions 22 and 23, suggesting its secretion. These enzymes were found both in apoplastic fluids extracted from compatible and incompatible interactions between Z. tritici and wheat (M'Barek et al., 2015), motivating further characterization. ZtCpx1 has a 2508 bp open reading frame (ORF) with one intron, encoding a 752 amino acid (aa) protein, while ZtCpx2 has a 2636 bp ORF with four introns, encoding a 797 aa secreted protein. Sequence analysis revealed that both ZtCpx1 and ZtCpx2 encode two‐domain peroxidases (PF00141), with domains positioned between aa stretches 119–449/454–762 and 61–410/415–716, respectively. A phylogenetic tree constructed by comparing catalases and CPs from other plant pathogens showed two distinct clades, one containing the two catalases and the other containing the two CPs. ZtCPX1 clustered with cytoplasmic CPs (Figure 1), closely related to CPEA1 and CPEA2 of Verticillium longisporum (Singh et al., 2012). ZtCPX2 aligned with the group of secreted CPs that have been predicted in other fungal plant pathogens (Figure 1), closely related to CP MgCPXB of Magnaporthe grisea, and CPX2 of Verticillium dahliae (Tanabe et al., 2011; Tran et al., 2014). In‐silico analysis using the peroxisomal targeting signal‐1 (PTS1) predictor tool (https://mendel.imp.ac.at/pts1/) indicated that only ZtCpx1 possesses a PTS1 signal located at its C‐terminus (DLKAGQVSSAKL). This signal is highly likely to be functional in directing it to the peroxisomes within fungal cells.
FIGURE 1.
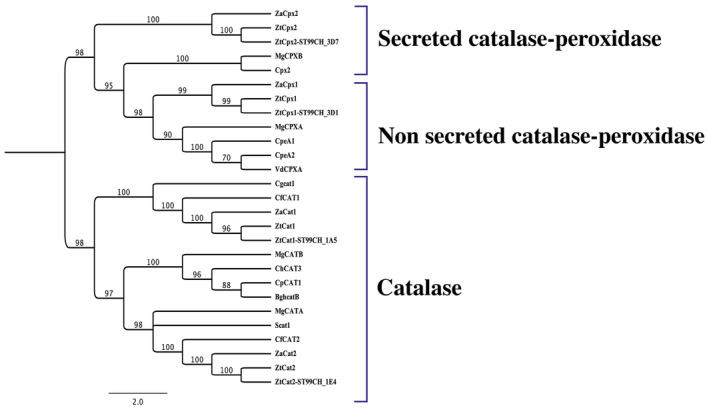
Phylogenetic analysis of Zymoseptoria tritici IPO323 catalases (ZtCat1 and ZtCat2) and catalase‐peroxidases (ZtCpx1 and ZtCpx2), Z. tritici ST99CH_3D7 (ZtCpx2‐ ST99CH_3D7), Z. tritici ST99CH_3D1 (ZtCpx1‐ ST99CH_3D1), Z. tritici ST99CH_1A5 (ZtCat1‐ ST99CH_1A5), Z. tritici ST99CH_1E4 (ZtCat2‐ ST99CH_1E4), Z. ardabiliae (ZaCpx1, ZaCpx2, ZaCat1, and ZaCat2) and their homologues in Verticillium longisporum (CPEA1‐2), Verticillium dahliae (VdCPXA and Cpx2), Magnaporthe grisea (MgCPXA, MgCPXB, MgCATA and MgCATB), Cladosporium fulvum (CfCAT1‐2), Colletotrichum gloeosporioides (Cgcat1), Sclerotinia sclerotiorium (Scat1), Cochliobolus heterostrophus (ChCAT3), Blumeria graminis f. sp. hordei (BghcatB) and Claviceps purpurea (CAT1). The phylogenetic tree was constructed using the CLC software, applying the unweighted pair group method with arithmetic mean (UPGMA), and it was subjected to 1000 bootstrap replicates for robust statistical support. The statistical confidence values for the branching points on the tree are indicated at the nodes.
2.2. Disruption and complementation of ZtCpx1 and ZtCpx2
To assess the potential role of ZtCpx1 in virulence, we created gene disruption and complementation mutants through homologous recombination. We obtained three independent transformants, showing similar morphological phenotypes (Figure S1a). One of these (ΔZtCpx1) was chosen for complementation with the ZtCpx1 wild‐type allele, which resulted in ZtCpx1‐C and homologous recombination was validated via PCR (Figure S1a). The knock‐out construct for ZtCpx2 was generated through the user‐friendly protocol as described previously (Mirzadi Gohari et al., 2014) and eventually one transformant, ΔZtCpx2, was obtained (Figure S1b). Subsequently, the ΔZtCpx2 strain was used to delete ZtCpx1, yielding in the double knock‐out strain ΔΔZtCpx1‐Cpx2 (Figure S1c). We used quantitative real‐time PCR (qPCR) to count the number of hph gene copies in the genomes of mutants with one (ΔZtCpx1 and ZtCpx2) and two (ΔΔZtCpx1‐Cpx2) gene deletions to check for additional ectopic insertions. We used Z. tritici β‐tubulin as a control because it has a single copy in the wild‐type IPO323 genome. All the mutants showed a single copy of the transgene, indicating a single T‐DNA insertion, which is typical for Agrobacterium tumefaciens‐mediated transformation (ATMT) (Figure S2).
2.3. ΔZtCpx2 and ΔΔZtCpx1 ‐Cpx2 are sensitive to exogenously added H2O2
To investigate the roles of ZtCpx1 and ZtCpx2 in the tolerance of Z. tritici to H2O2, we exposed single and double knock‐out strains to various conditions, including continuous exposure to H2O2 for 14 days. The ΔZtCpx1, ΔZtCpx2 and ΔΔZtCpx1‐Cpx2 strains and both controls, including the Z. tritici IPO323 wild type (WT) and complemented strain (ΔZtCpx1‐C) were plated on potato dextrose agar (PDA) supplemented with 0 and 6 mM H2O2. The most distinct phenotypes were observed at 6 mM H2O2, as shown in Figure 2. Both control strains as well as the ΔZtCpx2 strain were able to grow under these conditions similar to the WT, but the ΔZtCpx1 and ΔΔZtCpx1‐Cpx2 strains were clearly affected and unable to grow (Figure 2), indicating that ZtCpx1 is essential for H2O2 tolerance or degradation under in vitro conditions.
FIGURE 2.
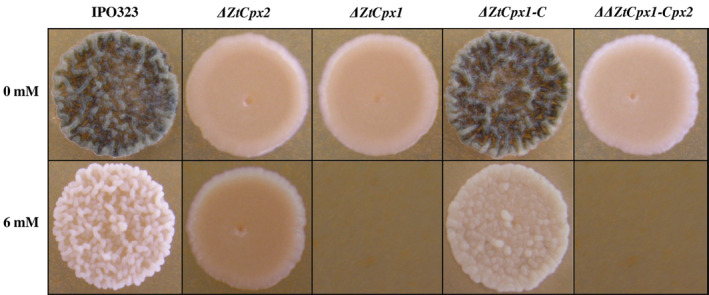
Sensitivity assay of Zymoseptoria tritici strains to H2O2. The spore suspensions of Z. tritici IPO323 (wild type, WT), disruptant strains ΔZtCpx1, ΔZtCpx2, ΔΔZtCpx1‐Cpx2 and the complementation strain ΔZtCpx1‐C were plated on potato dextrose agar amended with 6 mM H2O2 and subsequently incubated at 18°C. Pictures were taken 14 days post‐incubation. It is noteworthy that both ΔZtCpx1 and ΔΔZtCpx1‐Cpx‐2 strains displayed an inability to thrive in the presence of 6 mM H2O2.
2.4. Reduced spore germination of ΔZtCpx1 and ΔZtCpx2 strains
All mutant strains and the controls were inoculated on PDA amended with different H2O2 concentrations, and the spore germination frequency was recorded at 24 and 48 hours after inoculation (hai). At 24 hai in PDA with 4 mM H2O2, spore germination of the ΔZtCpx1 and ΔΔZtCpx1‐Cpx2 strains was decreased to 34% and 4%, respectively, whereas the germination of ΔZtCpx2 was similar to the control strain (94%). At increased H2O2 concentrations (6 mM), however, none of the ΔZtCpx1 and ΔΔZtCpx1‐Cpx2 spores germinated, whereas the germination frequency of the ΔZtCpx2 strain was decreased to 30% (Figure 3A), with the control (IPO323) showing a germination frequency of approximately 83% under the same conditions. At 48 hai, nearly all spores of all strains germinated equally on PDA without H2O2, indicating that H2O2 inhibits spore germination of the ΔZtCpx1 and ΔΔZtCpx1‐Cpx2 strains (Figure 3B).
FIGURE 3.
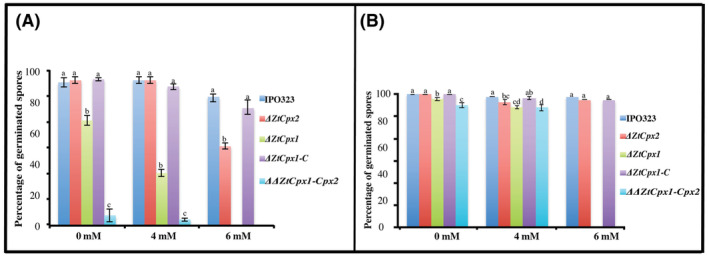
(A) Spore germination frequencies of Zymoseptoria tritici IPO323 (wild type, WT) after treatment with H2O2. Spores of the disruptant strains ΔZtCpx2 and ΔZtCpx1 and the double disruptant ΔΔZtCpx1‐Cpx2 and the complemented strain ZtCpx1‐C were plated on potato dextrose agar amended with 4 and 6 mM H2O2. Spore germination was assessed at two time points: 24 h (A) and 48 h after treatment (B). This analysis involved the examination of 50 spores for each strain across three independent biological replicates. Duncan's test was employed to compare the means at a 5% probability level, and columns with the same letters are not significantly different from each other.
2.5. Expression profiling of the Z. tritici catalase and catalase‐peroxidase genes
To study the in‐planta expression levels of ZtCpx1, ZtCpx2, ZtCat1 and ZtCat2, we inoculated the susceptible wheat cv. Taichung 29 with the WT and collected samples every 4 days after inoculation up to 20 days post‐inoculation (dpi). We compared this with in‐vitro conditions under both nutrient‐rich (yeast glucose at 18°C) and nutrient‐poor (minimal medium at 18°C) growing conditions using reverse transcription (RT)‐qPCR. ZtCpx1 exhibited a bimodal pattern with peaks at 4 dpi and 16/20 dpi, aligning with the initial biotrophic stage and the late necrotrophic stage. In contrast, ZtCpx2 peaked at 8 dpi and gradually decreased, suggesting a relationship between ZtCpx2 expression and to the transition from biotrophic to necrotrophic growth, coinciding with the onset of H2O2 production, its accumulation and subsequent decrease due to cell death (Figure 4A). The expression levels of ZtCpx1 and ZtCpx2 were also analysed under in‐vitro conditions during vegetative growth and asexual spore formation. As previously noted, the production of yeast‐like spores and mycelium was induced at two distinct temperatures: 18°C for yeast‐like spores and 25°C for mycelium (Figure 4B). ZtCpx1 was primarily expressed in spores, whereas ZtCpx2 was exclusively expressed in vegetative mycelium, consistent with the significant increase in fungal biomass in planta at 8 dpi and onwards (Figure 4B) (Kema et al., 1996).
FIGURE 4.
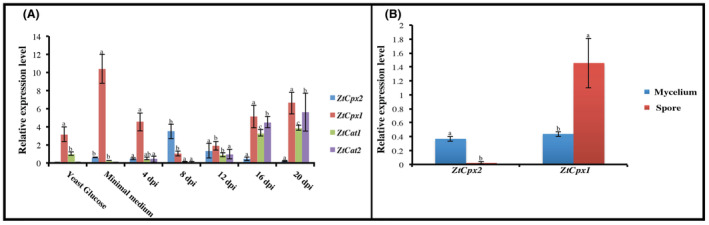
(A) Expression profiling of the Zymoseptoria tritici catalase genes ZtCat1 and ZtCat2, and catalase‐peroxidase genes ZtCpx1 and ZtCpx2 in yeast extract‐glucose (YG) medium, minimal medium mimicking in‐planta conditions, and actual in‐planta conditions. Leaves of cv. Taichung 29 were inoculated with the wild‐type (WT) strain and harvested 4, 8, 12, 16 and 20 days post‐inoculation (dpi). (B) Under in‐vitro growth conditions, expression of ZtCpx1 and ZtCpx2 was profiled in mycelium and spore statuses. Data were normalized using the constitutively expressed Z. tritici β‐tubulin gene. Duncan's test was employed to compare the means at a 5% probability level, and columns with the same letters are not significantly different from each other.
2.6. Loss of function of ZtCpx1 modulates expression patterns of related antioxidant genes
Disruption of ZtCpx1 affected the transcription of ZtCpx2, ZtCat1 and ZtCat2 during infection. Profiling of these genes in the ΔZtCpx1 strain showed that the expression of ZtCpx2 was delayed (12 dpi) compared to the WT (8 dpi). ZtCat1 was specifically up‐regulated at 4 dpi, different from its WT expression pattern, whereas ZtCat2 showed a similar expression profile in both ΔZtCpx1 and WT strains (Figure 5A). Finally, we monitored the relative expression levels of ZtCat1 and ZtCat2 in the ΔΔZtCpx1‐Cpx2 double mutant and showed that the former was only expressed at 12 dpi, while the latter had a variable expression pattern over the entire time course (Figure 5B).
FIGURE 5.
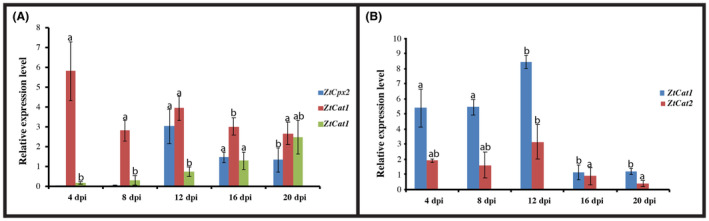
Relative in‐planta transcription of ZtCpx2, ZtCat1 and ZtCat2 in the ΔZtCpx1 disruptant background (A) and of ZtCat1 and ZtCat2 in the ΔΔZtCpx1‐Cpx2 double disruptant background (B). Leaves of cv. Taichung 29 were inoculated with the wild‐type (WT) strain and RNA was isolated from inoculated leaves at 4, 8, 12, 16 and 20 days post‐inoculation (dpi). Duncan's test was employed to compare the means at a 5% probability level, and columns with the same letters are not significantly different from each other.
2.7. ZtCpx1 and ZtCpx2 are required for full virulence
In addition, we investigated the effect of individual and combined disruption of ZtCpx1 and ZtCpx2 on virulence. The WT and all mutants (ΔZtCpx1, ΔZtCpx2 and ΔΔZtCpx1‐Cpx2) as well as the complemented strain ΔZtCpx1‐C were used to inoculate cv. Taichung 29 and assayed as described before (Mehrabi et al., 2006). The WT and ΔZtCpx2 strains produced typical symptoms with small chlorotic flecks around 7–8 dpi that expanded into larger chlorotic lesions at 10–12 dpi and eventually coalesced into typical necrotic STB blotches bearing numerous pycnidia at 14–16 dpi (Figure 6), indicating that ZtCpx2 is dispensable for full virulence. In contrast, the expression of disease symptoms was significantly delayed and not uniformly distributed over the inoculated leaf area after inoculation with the ΔZtCpx1 strain, whereas the WT phenotype was completely restored in the complemented ΔZtCpx1‐C strain. Finally, the double mutant ΔΔZtCpx1‐Cpx2 showed severely attenuated symptoms and only caused a limited number of necrotic lesions with few pycnidia at 21 dpi (Figure 6). To ensure consistency and observe variations among the tested mutant strains, the full leaves of cv. Taichung 29 were inoculated using a hand sprayer, and STB disease development was documented at 20 dpi (Figure S3). Additionally, the percentage of necrotic area formed on the inoculated leaves is presented (Figure S4). To further validate these observations, we quantified fungal biomass in all above‐mentioned interactions using a TaqMan assay (Figure 7). The fungal biomass of the WT and ΔZtCpx1‐C strains began to increase at 8 dpi, which significantly differed from the ΔZtCpx2, ΔZtCpx1 and ΔΔZtCpx1‐Cpx2 strains. Fungal biomass increase of the ΔZtCpx2 strain was delayed and at 12 dpi eventually reached a level comparable with that of WT at 8dpi, indicating that ZtCpx2 is dispensable for virulence but may play a role in the switch from biotrophic to necrotrophic growth, possibly involving the modulation of host‐derived H2O2 levels. Fungal biomass of the ΔZtCpx1 strain developed slower and remained significantly lower during the entire infection process and, as expected, biomass of the ΔΔZtCpx1‐Cpx2 strain hardly developed throughout the infection process (Figure 7).
FIGURE 6.
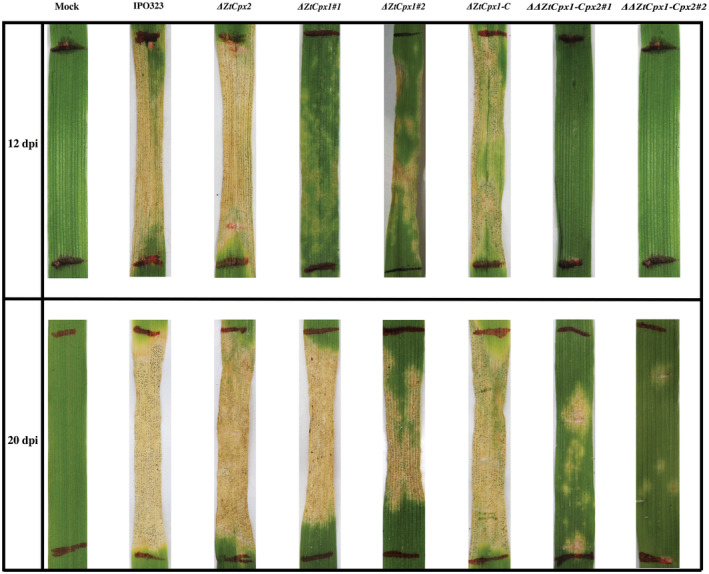
The effect of ZtCpx1 and ZtCpx2 disruption on pathogenicity of Zymoseptoria tritici. Leaves of susceptible wheat cv. Taichung 29 were inoculated between the marked lines with Z. tritici IPO323 (wild type, WT), the single disruptants ΔZtCpx2, ΔZtCpx1#1‐2, the double disruptant ΔΔZtCpx1‐Cpx2#1‐2 and the complemented strain ZtCpx1‐C. Photographs were taken at 12 and 20 days post‐inoculation (dpi).
FIGURE 7.
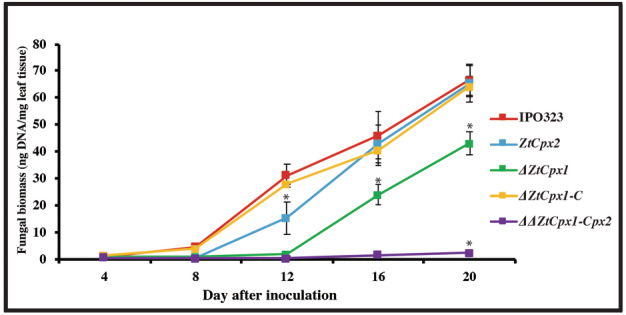
Fungal biomass quantifications in the susceptible wheat cv. Taichung 29 at 4, 8, 12, 16 and 20 days post‐inoculation (dpi). Zymoseptoria tritici IPO323 (wild type, WT), the single disruptant strains ΔZtCpx2, ΔZtCpx1, the double disruptant ΔΔZtCpx1‐Cpx2 and the complemented strain ΔZtCpx1‐C were inoculated on leaves of cv. Taichung 29 and DNA was isolated from inoculated leaves at 4, 8, 12, 16 and 20 dpi. Duncan's test was employed to compare the means at a 5% probability level. * indicates significant difference at α = 0.05.
2.8. ZtCpx1 is localized in the peroxisome
As only ZtCpx1 carries a peroxisomal targeting signal‐1 (PTS1), as shown by our in‐silico analysis of the four studied genes here using the PTS1 predictor tool, the subcellular localization of ZtCpx1 was investigated by tagging ZtCpx1 to green fluorescent protein (GFP) and stimulating peroxisome activity with 1% oleic acid. Our findings indicated that ZtGFP‐ZtCpx1 was indeed localized within the peroxisomes. This conclusion was supported by the significant increase in the intensity of fluorescent signals following treatment with 1% oleic acid. The quantified data, derived from three independent experiments (n = 30/29 cells), demonstrated this increase with high statistical significance (p = 0.001, Student's t test). The combination of fluorescence and differential interference contrast (DIC) microscopy along with statistical analysis provides robust evidence for the localization of ZtCpx1 in the peroxisomes of Z. tritici cells (Figure 8).
FIGURE 8.
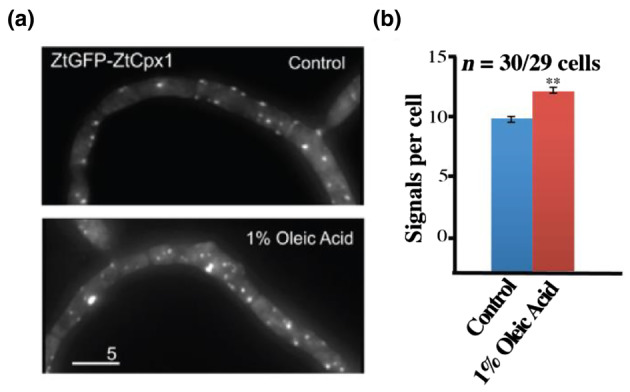
ZtCpx1 is localized to peroxisomes. (a) Zymoseptoria tritici cells expressing ZtGFP‐ZtCpx1 after integration of pCZtGFPCpx1 in to the succinate dehydrogenase locus. 2D‐deconvolved maximum projection of a z‐axis stack, adjusted in brightness, contrast and gamma settings. Bar: 5 μm. (b) Quantitative analysis of peroxisome number per cell. Cells were treated with 1% oleic acid for 1.5 h. Data are from three independent experiments (n = 30/29 cells) and are presented as mean ± SEM. ** indicates significant difference at α = 0.001, Student's t test. Treatment with 1% oleic acid increased the number of fluorescent signals. This strongly indicates that ZtGFP‐ZtCpx1 is localized in the peroxisomes.
3. DISCUSSION
Reactive oxygen species (ROS) serve as a sophisticated defence mechanism employed by plants to impede fungal attacks (Heller & Tudzynski, 2011). Nevertheless, maintaining a delicate balance between ROS generation and breakdown is crucial for proper plant cell function, preventing detrimental effects on plant cells (Nanda et al., 2010). During pathogen infection, various families of enzymes participate in both the production and detoxification of ROS within plant cells. NADPH oxidases, present in all biological kingdoms (NOx/RBOH), are associated with ROS generation (Sumimoto, 2008). Conversely, enzymes like glutathione peroxidases (Bela et al., 2022) and peroxiredoxins (Tripathi et al., 2009) play a pivotal role in scavenging and detoxifying ROS. The interaction between host plants and fungal pathogens triggers a substantial increase in ROS levels as an early defence response. In response, fungal pathogens are equipped with several ROS detoxification enzymes, such as CPs, to counteract the harmful effects of ROS (Tanabe et al., 2011).
Numerous studies have endeavoured to shed light on the intricate roles of detoxifying enzymes in both necrotrophic and biotrophic pathosystems. It has been proposed that ROS‐scavenging enzymes could wield substantial influence in the establishment of diseases by biotrophic pathogens (Bussink & Oliver, 2001; Garre et al., 1998), while necrotrophic pathogens are reported to benefit from ROS production (Govrin & Levine, 2000). However, our understanding remains limited regarding how hemibiotrophic pathogens deal with elevated ROS levels during their interactions with host organisms and the precise mechanisms by which fungal detoxifying enzymes facilitate host infection.
To address this question, we identified and functionally analysed the biological role of two CP genes (designated ZtCpx1 and ZtCpx2) in Z. tritici, a fungal wheat pathogen exhibiting a distinctive hemibiotrophic lifestyle. Phylogenetic analysis revealed that ZtCpx1 grouped with CpeA1‐2 of V. longisporum (Singh et al., 2012), which points at a presumed role of ZtCpx1 in protecting the fungus against oxidative stress generated by the host plant. Additionally, ZtCpx2 clustered closely with M. oryzae CPXB (Tanabe et al., 2011) and V. dahliae Cpx2 (Tran et al., 2014), suggesting a potential role of ZtCpx2 in the scavenging or detoxification of host‐derived H2O2. Our findings showed that deletion of the ZtCpx1 gene resulted in enhanced sensitivity of the mutant to H2O2 and significantly reduced its virulence. Conversely, ZtCpx2 was dispensable for full virulence of Z. tritici, albeit that disruption significantly reduced fungal biomass development during the transition from biotrophic to necrotrophic growth. Intriguingly, the generation of double mutants for both genes demonstrated a synergistic action of both CPs in facilitating wheat infection. Similar to our findings, deletion of MgCat2, encoding a secreted catalase, in the hemibiotroph M. grisea severely affected virulence by partly impaired appressorium formation and reduced sporulation. Additionally, conidial melanization was impaired, which is an important metabolic process to fortify fungal cell walls (Skamnioti et al., 2007). Additionally, M. oryzae MgCPXB, which encodes a secreted CP, is required for neutralizing host‐produced H2O2 during initial colonization, but not for full virulence (Tanabe et al., 2011). Singh et al. (2012) reported that CpeA1‐2 from V. longisporum, which encodes a cytoplasmic CP, plays an important role during late phases but not during the initial phases of infection of oilseed rape (Singh et al., 2012). Recent research by Tran et al. (2014) highlighted the significance of Vta2, a transcription activator of adhesion in V. dahliae, and the secreted CP Cpx2 in detoxifying extracellular ROS (Tran et al., 2014). These collective findings underscore the necessity for hemibiotrophic fungi to deploy mechanisms for coping with host‐derived H2O2 throughout the entire infection cycle. When a pathogen enters leaves and colonizes the apoplast surrounding the host mesophyll cells or enters the cells, the host responds with an oxidative burst as shown in several pathosystems (Ghiasi Noei et al., 2022; Shetty et al., 2003). At this critical stage, the neutralization of ROS is paramount to initiate biotrophy successfully. Following this stage, a transition from biotrophy to necrotrophy occurs, leading to host cell collapse and triggering a second wave of defensive responses, characterized by a substantial generation of ROS. Thus, hemibiotrophic fungi rely on scavenging mechanisms to counteract host‐generated ROS during both phases of the infection process.
Hemibiotrophic fungi, such as Z. tritici, may exhibit behaviour similar to biotrophic fungal pathogens. Biotrophic fungi rely on nutrients released from living plant cells, necessitating a robust antioxidant defence mechanism to counteract oxidative stress when invading and colonizing plants. However, contrary to this hypothesis, targeted gene replacement experiments involving both secreted and cytoplasmic catalase genes in biotrophic fungi like C. purpurea and C. fulvum have demonstrated that these genes are dispensable for virulence (Bussink & Oliver, 2001; Garre et al., 1998). One plausible explanation for these unexpected findings could be the existence of functional redundancy among catalases or the activation of other related antioxidant enzymes that compensate for the disruption of these catalase genes. Notably, upon genome analysis of C. fulvum and C. purpurea (Amselem et al., 2011; De Wit et al., 2012), it became evident that both fungi contain four genes encoding catalases, suggesting that the effect of disruption of single catalase genes might be masked by the enzymes encoded by the other genes. Nevertheless, our previous study demonstrated that deletion of ZtCat1 in Z. tritici had no impact on virulence (our unpublished data). Additionally, we speculate that ZtCat2 has no or limited impact on the modulation of host‐derived H2O2, considering its low expression levels in the WT strain and minimal change in the ΔΔZtCpx1‐Cpx2. Collectively, it can be concluded that both catalases could not compensate the loss of function of either or both investigated CPs.
Nevertheless, in necrotrophs, ROS‐degrading enzymes do not seem to play a critical role in virulence (Robbertse et al., 2003; Schouten et al., 2002; Yarden et al., 2014). For instance, in the case of C. heterostrophus, when all monofunctional catalase‐encoding genes were deleted, it became evident that only ChCAT3, which encodes a secreted catalase, played a crucial role in safeguarding the fungus from oxidative stress during its vegetative growth phase but had no significant impact on virulence (Robbertse et al., 2003). Similarly, BcCAT2‐deficient mutants of B. cinerea were hypersensitive to extracellular H2O2 but were unaffected in virulence on tomato (Schouten et al., 2002).
One of the technical limitations in detecting slightly reduced virulence in mutants compared to the WT strain may arise from the predominantly quantitative nature of symptom expression, which can lead to minor variations being easily overlooked. As a result, it has been proposed that the use of more sensitive monitoring tools is necessary to detect subtle changes in virulence within the disrupted strains (Robbertse et al., 2003; Singh et al., 2012). Quantification of fungal DNA during the infection process was therefore suggested to reveal such slight changes. In this study, we used qPCR in order to detect minor changes in fungal biomass within infected leaf tissues at different time points. Our results demonstrated the sensitivity and reliably of this technique, enabling the precise and distinctive patterns of fungal biomass between the ΔZtCpx2 mutant and the WT strain, even at very early infection stages when no discernible disease symptoms were observed. The growth of the ΔZtCpx2 mutant exhibited a slower progression, and by 12 dpi, it ultimately reached a level comparable to that of the WT at 8 dpi. This suggests that ZtCpx2 may not be essential for infection but may have a role in the transition from biotrophic to necrotrophic growth. Similarly, Singh et al. (2012) were able to show that CpeA1‐2 of V. longisporum was not involved in launching the initial phase of plant infection while the examined CP played an important role in advanced stages of infection, but the overall V. longisporum DNA content in plants infected by the CpeA1‐2 mutant was significantly lower than that of the WT at 35 dpi (Singh et al., 2012).
Our findings provided compelling evidence that ZtCpx1 is indeed localized within the peroxisomes of Z. tritici cells. This conclusion was supported by several key observations. First, ZtGFP‐ZtCpx1 fusion proteins displayed a significant increase in fluorescence following treatment with 1% oleic acid, which is known to induce peroxisome proliferation. This increase in fluorescence signals strongly indicated the presence of ZtCpx1 within the peroxisomes. The quantified data, derived from three independent experiments (n = 30/29 cells), demonstrated this increase with high statistical significance (p = 0.001, Student's t test). This rigorous statistical analysis further bolstered our confidence in the peroxisomal localization of ZtCpx1. The significance of localizing ZtCpx1 within the peroxisomes aligns with its potential role in managing ROS generated by the host during infection. Peroxisomes are known to be key players in ROS catabolism, and their efficient functioning is critical for the success of fungal pathogens. Our study not only elucidates the subcellular localization of ZtCpx1 but also underscores its importance in the context of fungal–plant interactions.
Zymoseptoria tritici is commonly classified as a hemibiotrophic fungus, exhibiting an extended symptomless phase that is typically regarded as biotrophic (Kema et al., 1996). Notably, the penetration of Z. tritici into wheat plants triggers the accumulation of H2O2, which has been documented to have adverse effects (Shetty et al., 2003). Infiltrating wheat leaves with H2O2 has been shown to extend the latency period while simultaneously reducing stomatal penetration and mesophyll colonization, indicating the harmful nature of H2O2 during this covert colonization phase (Shetty et al., 2007). Therefore, Z. tritici requires specific genes enabling it to overcome stressful condition caused by H2O2 to efficiently initiate biotrophic growth. In this study, we elucidated the crucial role of ZtCpx1, the sole cytoplasmic CP, in protecting against host‐generated H2O2 during initial colonization. Up‐regulation of ZtCpx1 at 4 dpi and during spore production indicated its vital role in establishing the biotrophic phase. Loss of ZtCpx1 resulted in reduced virulence, evidenced by significantly decreased fungal biomass in the ΔZtCpx1 strain throughout infection. Overall, ZtCpx1 plays a pivotal role in managing host‐generated H2O2 during both initial and final colonization phases, enabling Z. tritici to complete its life cycle and sporulate within infected tissues. It is important to note that after 8–10 dpi, a transition from biotrophic to necrotrophic growth takes place (Kema et al., 1996), which likely necessitates the activation of additional mechanisms to effectively respond to the fluctuating waves of ROS during infection (Shetty et al., 2003; Yang et al., 2013).
Initially, it was hypothesized that Z. tritici would leverage plant defence responses similar to those employed by necrotrophic pathogens like B. cinerea to enhance its ability to colonize the host (Govrin & Levine, 2000). In contrast, Shetty et al. (2007) demonstrated that Z. tritici would benefit from scavenging H2O2 during the transition from biotrophy to necrotrophy, enabling the pathogen to promote colonization and intensify symptom expression. In line with these findings, the functional characterization of ZtCpx2, the sole secreted CP in the Z. tritici genome, revealed its role in regulating H2O2 levels during the transition from biotrophy to necrotrophy, as evidenced by the failure of ZtCpx2 mutants to increase fungal biomass compared to the WT strain. This observation is further supported by in‐planta expression analyses showing that ZtCpx2 expression peaked at 8 dpi, coinciding with the transition from biotrophy to necrotrophy. Furthermore, a comprehensive analysis of the double knock‐out strain, ΔΔZtCpx1‐Cpx2, demonstrated the additive contributions of both ZtCpx1 and ZtCpx2 to pathogenicity. This was evident through reduced symptom development and significantly less fungal biomass accumulation in the double mutant compared to individual mutants and the WT strain. These findings suggest that ZtCpx1 and ZtCpx2 collaboratively and dynamically contribute to scavenging H2O2 throughout the entire infection process. In summary, our study collectively provides robust evidence of how specific genes contribute to a plant pathogen's adaptability in response to ROS challenges in planta and environmental conditions. To our knowledge, this is the first demonstration of two CPs working in synergy to enhance pathogenicity in a fungal plant pathogen.
4. EXPERIMENTAL PROCEDURES
4.1. Strains, media and growth conditions
The fully sequenced Z. tritici reference strain IPO323 (Goodwin et al., 2011), known for its high pathogenicity on wheat cultivar Taichung 29, served as the WT strain and recipient strain for gene replacement experiments. Yeast‐like spores were produced in yeast glucose broth (YGB) medium (yeast extract 10 g/L, glucose 30 g/L) subjecting the cultures to orbital shaking using an Innova 4430 shaker (New Brunswick Scientific) at 18°C. To stimulate mycelial growth, all Z. tritici strains were grown under the same conditions, but at 25°C. Aspergillus nidulans minimal medium (MM) was used for in‐vitro expression analyses (Barratt et al., 1965). Escherichia coli DH5α and DH10β were used for general plasmid transformation and Agrobacterium tumefaciens AGL‐1 was used for fungal transformation procedure as described previously.
4.2. Phylogenetic tree construction
Phylogenetic analyses of Z. tritici catalases and CP enzymes with their homologues from other fungal plant pathogens were conducted using the CLC Genomics Workbench package. Fungal proteins were retrieved from public databases and aligned using CLC software, with gap opening costs and gap extension penalties set at 10 and 1, respectively. The phylogenetic tree was constructed based on the unweighted pair group method with arithmetic mean (UPGMA) algorithm, and the statistical robustness of the tree was assessed through bootstrap analysis with 1000 repetitions.
4.3. Generation of gene replacement and complementation constructs
A 2.5‐kb full‐length cDNA clone, ZtEST2P12K00276, containing the ZtCpx1 gene (referred to as pSport1‐ZtCpx1), was identified in Z. tritici IPO323 cDNA libraries (Kema et al., 2008). To enable genetic manipulations, the ZtCpx1 insert was excised from pSport1‐ZtCpx1 using KpnI/XbaI restriction enzymes and ligated into the binary vector pCGN1589, creating pCGNZtCpx1. For ZtCpx1 gene disruption, the GPS‐Mutagenesis system from New England Biolabs was employed. We used a customized donor construct known as pGPS3HygKan for transposition (Mehrabi et al., 2006). The target construct, pCGNZtCpx1, underwent transposition using pGPS3HygKan, and the resulting mixture was introduced into E. coli DH10β. Colony PCR identified clones with transposon insertion within the ZtCpx1. A disruption construct, named pCGNΔZtCpx1, was chosen, featuring transposon insertion near the middle of the ZtCpx1 ORF. This construct was used to disrupt the ZtCpx1 in Z. tritici IPO323 through ATMT, following established procedures (Zwiers & De Waard, 2001).
For the ZtCpx1 complementation construct (pZtCpx1com), we used the Invitrogen multisite Gateway three‐fragment vector construction kit. This kit facilitated the cloning of the complete ZtCpx1 ORF, including 994 bp upstream (including its promoter) and 498 bp downstream (as a terminator), into pDONRP221. The resulting plasmid, p221‐ZtCpx1com, was then combined with two Entry vectors (pRM245 and pRM234) and used to clone these three fragments into the Destination vector, pPm43GW, via the LR reaction (Mehrabi et al., 2015). For the ZtCpx2 deletion construct, approximately 2 kb of upstream and downstream sequences of ZtCpx2 were cloned into pDONRP4‐P1R and pDONRP2R‐P3, respectively. These constructs, along with pRM250 (containing the hygromycin phosphotransferase [hph] gene as a selection marker) (Mehrabi et al., 2015), were then assembled into the Destination vector, pPm43GW, using the LR reaction. For the double knock‐out construct, pZtCpx1‐2, approximately 1.2 kb of sequences upstream and downstream of the ZtCpx1 gene were cloned into pDONRP4‐P1R and pDONRP2R‐P3, respectively. These Entry vectors, along with pRM251 (containing geneticin as a selectable marker) (Mehrabi et al., 2015), were used to clone the three fragments into the Destination vector pPm43GW. Primer details can be found in Table 1.
TABLE 1.
Primers used in this study.
| Name | Sequence (5′–3′) | Location |
|---|---|---|
| PrimerE | ATGTCTGCAAACGGTTGCCCAA | ZtCpx1 |
| PrimerF | CTACAACTTCGCGCTGCTGAC | ZtCpx1 |
| PrimerG | ATGAAGGGTTGTCTCAATCATCTG | ZtCpx2 |
| PrimerH | CTACTGGACATCGTTCTGAGGA | ZtCpx2 |
| PrimerK | GTGCTCACCGCCTGGACGACTAAAC | Middle of hph gene |
| PrimerL | GATGAGACCCGGCGACAAGT | Downstream of ZtCpx2 |
| PrimerM | TTCGACCGTGGCTTGACACC | Upstream of ZtCpx1 |
| PrimerN | TCCACCCAAGCGGCCGGA | Beginning of geneticin gene |
| hph‐F | CTCGATGAGCTGATGCTTTG | hph gene |
| hph‐R | GTCAATGACCGCTGTTATGC | hph gene |
| ZtβTUB‐F | GCCCAGACAACTTCGTGTTC | β‐tubulin gene |
| ZtβTUB‐R | ACGACATCGAGAACCTGGTC | β‐tubulin gene |
| Q‐ZtCpx1‐F | ACAACGCCAATCTCGACAAG | ZtCpx1 |
| Q‐ZtCpx1‐R | GACTCAATGGCGACATTTCC | ZtCpx1 |
| Q‐ZtCpx2‐F | TCCTAAATCCGAGCCTTTCC | ZtCpx2 |
| Q‐ZtCpx2‐R | TCAACCCACTGCCAAGAATC | ZtCpx2 |
| Q‐ZtCat1‐F | GATGCACATGAAGAGCAAGC | ZtCat1 |
| Q‐ZtCat1‐F | GAAACTCGCCCTTCTCAATG | ZtCat1 |
| Q‐ZtCat2‐F | ACTTTGGCGTTCAGGTCATC | ZtCat2 |
| Q‐ZtCat2‐F | ACTCCGACGTTCTGAATTGG | ZtCat2 |
| SK‐Sep‐348 | ATCACCCTCGGCATGGACGAGCTCTACAAGATGTCTGCAAACGGTTGCCCAATC | ZtCpx1 |
| SK‐Sep‐349 | CCACAAGATCCTGTCCTCGTCCGTCGTCGCCTACAACTTCGCGCTGCTGACC | ZtCpx1 |
4.4. Generation of vector pCZtGFPCpx1
The vector pCZtGFPCpx1 was constructed through in‐vivo recombination within the yeast Saccharomyces cerevisiae DS94 (MATa, ura3‐52, trp1‐1, leu2‐3, his3‐111, lys2‐801) following established protocols (Tang et al., 1996), as described in published procedures (Kilaru & Steinberg, 2015; Raymond et al., 1999).Vector pCZtGFPCpx1 comprises a fusion of ZtGFP with the full‐length ZtCpx1 gene, both of which are regulated by the constitutive ZtTub2 promoter and terminated by specific terminator sequences. This vector was designed for precise integration into the sdi1 locus of Z. tritici, with carboxin serving as the selection agent. To construct pCZtGFPCpx1, a 16,597‐bp fragment of pCZtGFPSpa2 (Guo et al., 2015) was digested with PshAI and ligated with a 2319‐bp full‐length ZtCpx1 gene, which was amplified using primers SK‐Sep‐348 and SK‐Sep‐349 (Table 1). Subsequently, pCZtGFPCpx1 was introduced into A. tumefaciens EHA105 using the heat shock method (Holsters et al., 1978). A selection agent, carboxin at a concentration of 40 μg/mL, was employed during the transformation process. Finally, transformation of the pCZtGFPCpx1 vector into Z. tritici IPO323 resulted in the generation of IPO323_CZtGFPCpx1 transformant strain.
4.5. Fungal transformation
All transformations were carried out using the ATMT procedure, following previously established protocol (Zwiers & De Waard, 2001). Genomic DNA from stable transformants was extracted following standard procedures (Sambrook et al., 1989). For complementation and double knock‐out strategies, the same method was employed with minor adjustments, including the use of 250 μg/mL geneticin for mutant selection.
4.6. Hph copy number determination
ZtβTUB‐F/R and hph‐F/R primers were used in qPCR to determine the copy number of the hph resistance marker gene. Primers were tested with a series of 10× dilutions of genomic DNA from the wild‐type strain IPO323 (500 ng/μL) and the double‐gene deletion strain ΔΔZtCpx1‐Cpx2 (350 ng/μL). The qPCR tests were run at an annealing temperature of 60°C, where both primer pairs' efficiencies were nearly 100%. The C t value of hph was normalized to the single‐copy β‐tubulin gene and calculated using the E−∆Ct method as described previously (Tian et al., 2021).
4.7. In‐vitro oxidative stress assays
To assess the sensitivity of Z. tritici strains to continuous exposure to H2O2, we conducted experiments using PDA supplemented with varying concentrations of H2O2. Autoclaved PDA was cooled to 40°C, and H2O2 (Sigma) was added to reach final concentrations of 6, 8 and 10 mM. The PDA plates were inoculated with 5 μL of a spore suspension from each strain, with a concentration of 108 spores/mL, incubated in a 20°C incubator for 7 days, and photographic documentation was performed using an Olympus camera.
4.8. Germination frequency assays
For germination frequency assays, we prepared PDA plates supplemented with 0, 4 and 6 mM H2O2. These plates were cut into 1 cm2 plugs, which were then placed on glass slides. Subsequently, each plug was inoculated with 10 μL of a yeast‐like spore suspension containing 104 spores/mL. A coverslip was used to enclose the inoculated area, and the samples were placed in Petri plates containing dampened cotton wool to maintain high relative humidity. Incubation was carried out at 20°C for 2 days. The germination frequency of each strain was determined by counting the number of spores that exhibited initial hyphal growth out of 50 randomly selected spores, using a light microscope (Zeiss) at 40× magnification. In this study, spores were considered germinated when blastospores began growing as hyphae. The experiments were conducted in triplicate, and the percentage of germinated spores was recorded, as presented in Figure 3.
4.9. RNA isolation, and RT‐qPCR
For RT‐qPCR analysis of in‐vitro and in‐planta expression profiles of ZtCpx1, ZtCpx1, ZtCat1 and ZtCat2, the following procedure was adopted: For in‐planta analyses, susceptible wheat cv. Taichung 29 was inoculated with the WT strain, as previously described (Mehrabi et al., 2006). This assay was conducted using three biological samples, with each replicate consisting of three separate seedlings, each carrying one fully unfolded leaf. At 2, 4, 8, 12, 16 and 20 dpi, three infected leaves were harvested from each replicate, flash‐frozen, and ground into powder using a mortar and pestle cooled with liquid nitrogen. Total RNA was extracted either from ground leaves or fungal biomass produced in yeast glucose broth (YGB) using the RNeasy plant mini kit from Qiagen. Subsequently, DNA contamination was eliminated using the DNA‐free kit (Ambion Inc.). First‐strand cDNA was synthesized from approximately 2 μg of total RNA, primed with oligo(dT), using the SuperScript III kit, following the manufacturer's instructions. One microlitre of the resulting cDNA was used in a 25 μL PCR employing a QuantiTect SYBR Green PCR Kit. The reaction was run and analysed using an ABI 7500 Real‐Time PCR System. The relative expression of each gene was initially normalized using the constitutively expressed Z. tritici β‐tubulin gene (Keon et al., 2007) and subsequently calculated using the comparative C t method, as outlined previously (Schmittgen & Livak, 2008).
4.10. Pathogenicity assays
Pathogenicity assays were conducted on wheat cv. Taichung 29 in a greenhouse. Plants used in this study were cultivated in a greenhouse under controlled conditions, maintained at a temperature of 18/16°C (day/night) and a relative humidity (RH) of 70%. They were grown until reaching the stage where the first leaves were fully unfolded. For each pot, three to five seeds were initially sown, and after 7 days, three seedlings were selected in each pot, each with their first fully unfolded leaves. Inoculum for all strains was produced in YGB at 18°C for 7 days using an orbital shaker. Yeast‐like spores were obtained after centrifugation and washing, and spore concentrations were adjusted to 107 spores/mL with 0.15% Tween 20 as a surfactant. A marked 5 cm leaf segment was inoculated with spores using a cotton swab. Inoculated plants were enclosed in transparent plastic bags for 48 h to maintain high humidity before being transferred to a greenhouse compartment (22°C, >90% relative humidity, 16 h photoperiod). Disease development was monitored and recorded every 3 days. This assay was conducted using three biological samples, with each replicate consisting of three separate seedlings, each carrying one fully unfolded leaf. Fungal DNA quantification was performed on leaves of cv. Taichung 29 harvested at 4, 8, 12, 16 and 20 dpi. Genomic DNA was extracted from approximately 100 mg of infected leaves using standard phenol/chloroform DNA extraction, as detailed by Sambrook et al. (1989). For each biological replicate, three inoculated leaves were pooled together, ground into a fine powder, and then 100 mg was weighed out from the resulting mixture. The TaqMan assay was conducted to quantify the fungal biomass in infected leaf tissues, following the method described previously (Waalwijk et al., 2002; Ware, 2006).
4.11. Epifluorescence microscopy
Fluorescence microscopy was conducted following established procedures (Kilaru et al., 2015). In brief, IPO323_CZtGFPCpx1 cells were cultured in YG medium, with growth induced at 18°C (200 rpm) for yeast‐like growth or 24°C (100 rpm) for hyphal growth. Subsequently, cells were placed on a 2% agar cushion for direct observation using an inverted microscope (IX81; Olympus) with a PlanApo 100_/1.45 Oil TIRF objective. Fluorescent tagging (excited at 488 nm/75 mW) captured single images or z‐stacks (6 μm depth, 0.2 μm z resolution) using an objective piezo (Piezosystem Jena GmbH) and a 150 ms exposure time. A DIC image was acquired for each cell using a CoolSNAP HQ2 camera (Photometrics/Roper Scientific). Fluorescent and DIC image overlays were created using MetaMorph software (Molecular Devices). The entire system was controlled using the MetaMorph software package.
4.12. Induction of peroxisome proliferation by oleic acid
Peroxisome proliferation was induced by oleic acid, following the established procedure (Camões et al., 2015). IPO323_CZtGFPCpx1 cells were cultured for 24 h in YGB at 18°C with 200 rpm. Oleic acid (Merck) was added to a final concentration of 1% (vol/vol) and incubated for 1.5 h at 18°C with 200 rpm. The numbers of peroxisomes were determined using laser‐based epifluorescence microscopy. To achieve this, z‐stacks were captured at a 200 nm step size with a 150 ms exposure time, using a piezo drive (Piezosystem Jena GmbH). The data was then analysed as a maximum projection using MetaMorph software.
4.13. Statistical analysis
The statistical analysis employed in this study used a factorial design conducted in a randomized design with three replications, aiming to ensure the robustness and reliability of the results. Statistical analyses were carried out using SPSS 26 software (IBM SPSS). Mean differences were further scrutinized using Duncan's test at a significance level of 5% to compare the levels of the factors. This approach facilitated a comprehensive assessment of the data, enhancing the validity and interpretability of the findings.
Supporting information
Figure S1.
Figure S2.
Figure S3.
Figure S4.
ACKNOWLEDGEMENTS
A. Mirzadi Gohari was financially supported by the Ministry of Research and Technology of Iran. We would like to thank Bertus vander Laan at Unifarm of Wageningen University for maintaining the glasshouse in an excellent condition.
Mirzadi Gohari, A. , Mehrabi, R. , Kilaru, S. , Schuster, M. , Steinberg, G. , de Wit, P.P.J.G.M. et al. (2024) Functional characterization of extracellular and intracellular catalase‐peroxidases involved in virulence of the fungal wheat pathogen Zymoseptoria tritici . Molecular Plant Pathology, 25, e70009. Available from: 10.1111/mpp.70009
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Amselem, J. , Cuomo, C.A. , van Kan, J.A. , Viaud, M. , Benito, E.P. , Couloux, A. et al. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genetics, 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostol, I. , Heinstein, P.F. & Low, P.S. (1989) Rapid stimulation of an oxidative burst during elicitation of cultured plant cells: role in defense and signal transduction. Plant Physiology, 90, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt, R. , Johnson, G.B. & Ogata, W. (1965) Wild‐type and mutant stocks of Aspergillus nidulans . Genetics, 52, 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bela, K. , Riyazuddin, R. & Csiszár, J. (2022) Plant glutathione peroxidases: non‐heme peroxidases with large functional flexibility as a core component of ROS‐processing mechanisms and signalling. Antioxidants, 11, 1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussink, H.J. & Oliver, R. (2001) Identification of two highly divergent catalase genes in the fungal tomato pathogen, Cladosporium fulvum . European Journal of Biochemistry, 268, 15–24. [DOI] [PubMed] [Google Scholar]
- Camões, F. , Islinger, M. , Guimarães, S.C. , Kilaru, S. , Schuster, M. , Godinho, L.F. et al. (2015) New insights into the peroxisomal protein inventory: acyl‐CoA oxidases and dehydrogenases are an ancient feature of peroxisomes. Biochimica et Biophysica Acta (BBA) ‐ Molecular Cell Research, 1853, 111–125. [DOI] [PubMed] [Google Scholar]
- Costet, L. , Dorey, S. , Fritig, B. & Kauffmann, S. (2002) A pharmacological approach to test the diffusible signal activity of reactive oxygen intermediates in elicitor‐treated tobacco leaves. Plant and Cell Physiology, 43, 91–98. [DOI] [PubMed] [Google Scholar]
- De Wit, P.J. , Van Der Burgt, A. , Ökmen, B. , Stergiopoulos, I. , Abd‐Elsalam, K.A. , Aerts, A.L. et al. (2012) The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genetics, 8, e1003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke, N. , Miura, Y. , Sanchez, L. , Park, H. , Noritake, T. , Yoshioka, H. et al. (1996) The oxidative burst protects plants against pathogen attack: mechanism and role as an emergency signal for plant bio‐defence—a review. Gene, 179, 45–51. [DOI] [PubMed] [Google Scholar]
- Eyal, Z. (1999) The Septoria tritici and Stagonospora nodorum blotch diseases of wheat. European Journal of Plant Pathology, 105, 629–641. [Google Scholar]
- Fones, H. & Gurr, S. (2015) The impact of Septoria tritici blotch disease on wheat: an EU perspective. Fungal Genetics and Biology, 79, 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev, I. , Stone, J.M. & Gechev, T.S. (2008) Programmed cell death in plants: new insights into redox regulation and the role of hydrogen peroxide. International Review of Cell and Molecular Biology, 270, 87–144. [DOI] [PubMed] [Google Scholar]
- Garre, V. , Müller, U. & Tudzynski, P. (1998) Cloning, characterization, and targeted disruption of cpcat1, coding for an in planta secreted catalase of Claviceps purpurea . Molecular Plant–Microbe Interactions, 11, 772–783. [DOI] [PubMed] [Google Scholar]
- Ghiasi Noei, F. , Imami, M. , Didaran, F. , Ghanbari, M.A. , Zamani, E. , Ebrahimi, A. et al. (2022) Stb6 mediates stomatal immunity, photosynthetic functionality, and the antioxidant system during the Zymoseptoria tritici–wheat interaction. Frontiers in Plant Science, 13, 1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, S.B. , Ben M'Barek, S. , Dhillon, B. , Wittenberg, A.H. , Crane, C.F. , Hane, J.K. et al. (2011) Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genetics, 7, e1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govrin, E.M. & Levine, A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea . Current Biology, 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Guo, M. , Kilaru, S. , Schuster, M. , Latz, M. & Steinberg, G. (2015) Fluorescent markers for the Spitzenkörper and exocytosis in Zymoseptoria tritici . Fungal Genetics and Biology, 79, 158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller, J. & Tudzynski, P. (2011) Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annual Review of Phytopathology, 49, 369–390. [DOI] [PubMed] [Google Scholar]
- Holsters, M. , De Waele, D. , Depicker, A. , Messens, E. , Van Montagu, M. & Schell, J. (1978) Transfection and transformation of Agrobacterium tumefaciens . Molecular and General Genetics, 163, 181–187. [DOI] [PubMed] [Google Scholar]
- Huang, K. , Czymmek, K.J. , Caplan, J.L. , Sweigard, J.A. & Donofrio, N.M. (2011) HYR1‐mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathogens, 7, e1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, L.M. , Koon, T.T. & Man, W.S. (1998) Antifungal effects of hydrogen peroxide and peroxidase on spore germination and mycelial growth of Pseudocercospora species. Canadian Journal of Botany, 76, 2119–2124. [Google Scholar]
- Kema, G.H. , van der Lee, T.A. , Mendes, O. , Verstappen, E.C. , Lankhorst, R.K. , Sandbrink, H. et al. (2008) Large‐scale gene discovery in the Septoria tritici blotch fungus Mycosphaerella graminicola with a focus on in planta expression. Molecular Plant–Microbe Interactions, 21, 1249–1260. [DOI] [PubMed] [Google Scholar]
- Kema, G.H. , Yu, D. , Rijkenberg, F.H. , Shaw, M.W. & Baayen, R.P. (1996) Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology, 86, 777–786. [Google Scholar]
- Keon, J. , Antoniw, J. , Carzaniga, R. , Deller, S. , Ward, J.L. , Baker, J.M. et al. (2007) Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Molecular Plant–Microbe Interactions, 20, 178–193. [DOI] [PubMed] [Google Scholar]
- Kilaru, S. , Schuster, M. , Latz, M. , Guo, M. & Steinberg, G. (2015) Fluorescent markers of the endocytic pathway in Zymoseptoria tritici . Fungal Genetics and Biology, 79, 150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilaru, S. & Steinberg, G. (2015) Yeast recombination‐based cloning as an efficient way of constructing vectors for Zymoseptoria tritici . Fungal Genetics and Biology, 79, 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, S. , Serrano, M. , L'Haridon, F. , Tjamos, S.E. & Metraux, J.‐P. (2015) Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry, 112, 54–62. [DOI] [PubMed] [Google Scholar]
- M'Barek, S.B. , Cordewener, J.H. , Ghaffary, S.M.T. , van der Lee, T.A. , Liu, Z. , Gohari, A.M. et al. (2015) FPLC and liquid‐chromatography mass spectrometry identify candidate necrosis‐inducing proteins from culture filtrates of the fungal wheat pathogen Zymoseptoria tritici . Fungal Genetics and Biology, 79, 54–62. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , Gohari, A.M. , da Silva, G.F. , Steinberg, G. , Kema, G.H. & de Wit, P.J. (2015) Flexible Gateway constructs for functional analyses of genes in plant‐pathogenic fungi. Fungal Genetics and Biology, 79, 186–192. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , Zwiers, L.‐H. , de Waard, M.A. & Kema, G.H. (2006) MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola . Molecular Plant–Microbe Interactions, 19, 1262–1269. [DOI] [PubMed] [Google Scholar]
- Mellersh, D.G. , Foulds, I.V. , Higgins, V.J. & Heath, M.C. (2002) H2O2 plays different roles in determining penetration failure in three diverse plant–fungal interactions. The Plant Journal, 29, 257–268. [DOI] [PubMed] [Google Scholar]
- Mir, A.A. , Park, S.‐Y. , Sadat, M.A. , Kim, S. , Choi, J. , Jeon, J. et al. (2015) Systematic characterization of the peroxidase gene family provides new insights into fungal pathogenicity in Magnaporthe oryzae . Scientific Reports, 5, 11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadi Gohari, A. , Mehrabi, R. , Robert, O. , Ince, I.A. , Boeren, S. , Schuster, M. et al. (2014) Molecular characterization and functional analyses of ZtWor1, a transcriptional regulator of the fungal wheat pathogen Zymoseptoria tritici . Molecular Plant Pathology, 15, 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, A.K. , Andrio, E. , Marino, D. , Pauly, N. & Dunand, C. (2010) Reactive oxygen species during plant–microorganism early interactions. Journal of Integrative Plant Biology, 52, 195–204. [DOI] [PubMed] [Google Scholar]
- Passardi, F. , Zamocky, M. , Favet, J. , Jakopitsch, C. , Penel, C. , Obinger, C. et al. (2007) Phylogenetic distribution of catalase‐peroxidases: are there patches of order in chaos? Gene, 397, 101–113. [DOI] [PubMed] [Google Scholar]
- Quaedvlieg, W. , Kema, G. , Groenewald, J. , Verkley, G. , Seifbarghi, S. , Razavi, M. et al. (2011) Zymoseptoria gen. nov.: a new genus to accommodate Septoria‐like species occurring on graminicolous hosts. Persoonia, 26, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, C.K. , Pownder, T.A. & Sexson, S.L. (1999) General method for plasmid construction using homologous recombination. BioTechniques, 26, 134–141. [DOI] [PubMed] [Google Scholar]
- Robbertse, B. , Yoder, O. , Nguyen, A. , Schoch, C.L. & Turgeon, B.G. (2003) Deletion of all Cochliobolus heterostrophus monofunctional catalase‐encoding genes reveals a role for one in sensitivity to oxidative stress but none with a role in virulence. Molecular Plant–Microbe Interactions, 16, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. & Maniatis, T. (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schmittgen, T.D. & Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative Ct method. Nature Protocols, 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schouten, A. , Tenberge, K.B. , Vermeer, J. , Stewart, J. , Wagemakers, L. , Williamson, B. et al. (2002) Functional analysis of an extracellular catalase of Botrytis cinerea . Molecular Plant Pathology, 3, 227–238. [DOI] [PubMed] [Google Scholar]
- Shetty, N. , Kristensen, B. , Newman, M.‐A. , Møller, K. , Gregersen, P.L. & Jørgensen, H.L. (2003) Association of hydrogen peroxide with restriction of Septoria tritici in resistant wheat. Physiological and Molecular Plant Pathology, 62, 333–346. [Google Scholar]
- Shetty, N.P. , Mehrabi, R. , Lütken, H. , Haldrup, A. , Kema, G.H. , Collinge, D.B. et al. (2007) Role of hydrogen peroxide during the interaction between the hemibiotrophic fungal pathogen Septoria tritici and wheat. New Phytologist, 174, 637–647. [DOI] [PubMed] [Google Scholar]
- Singh, S. , Braus‐Stromeyer, S.A. , Timpner, C. , Valerius, O. , von Tiedemann, A. , Karlovsky, P. et al. (2012) The plant host Brassica napus induces in the pathogen Verticillium longisporum the expression of functional catalase peroxidase which is required for the late phase of disease. Molecular Plant–Microbe Interactions, 25, 569–581. [DOI] [PubMed] [Google Scholar]
- Singh, Y. , Nair, A.M. & Verma, P.K. (2021) Surviving the odds: from perception to survival of fungal phytopathogens under host‐generated oxidative burst. Plant Communications, 2, 100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skamnioti, P. , Henderson, C. , Zhang, Z. , Robinson, Z. & Gurr, S.J. (2007) A novel role for catalase B in the maintenance of fungal cell‐wall integrity during host invasion in the rice blast fungus Magnaporthe grisea . Molecular Plant–Microbe Interactions, 20, 568–580. [DOI] [PubMed] [Google Scholar]
- Sumimoto, H. (2008) Structure, regulation and evolution of Nox‐family NADPH oxidases that produce reactive oxygen species. The FEBS Journal, 275, 3249–3277. [DOI] [PubMed] [Google Scholar]
- Tanabe, S. , Ishii‐Minami, N. , Saitoh, K.‐I. , Otake, Y. , Kaku, H. , Shibuya, N. et al. (2011) The role of catalase‐peroxidase secreted by Magnaporthe oryzae during early infection of rice cells. Molecular Plant–Microbe Interactions, 24, 163–171. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Halleck, M.S. , Schlegel, R.A. & Williamson, P. (1996) A subfamily of P‐type ATPases with aminophospholipid transporting activity. Science, 272, 1495–1497. [DOI] [PubMed] [Google Scholar]
- Tian, H. , MacKenzie, C.I. , Rodriguez‐Moreno, L. , van den Berg, G.C. , Chen, H. , Rudd, J.J. et al. (2021) Three LysM effectors of Zymoseptoria tritici collectively disarm chitin‐triggered plant immunity. Molecular Plant Pathology, 22, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, V.T. , Braus‐Stromeyer, S.A. , Kusch, H. , Reusche, M. , Kaever, A. , Kühn, A. et al. (2014) Verticillium transcription activator of adhesion Vta2 suppresses microsclerotia formation and is required for systemic infection of plant roots. New Phytologist, 202, 565–581. [DOI] [PubMed] [Google Scholar]
- Tripathi, B.N. , Bhatt, I. & Dietz, K.‐J. (2009) Peroxiredoxins: a less studied component of hydrogen peroxide detoxification in photosynthetic organisms. Protoplasma, 235, 3–15. [DOI] [PubMed] [Google Scholar]
- Vidossich, P. , Alfonso‐Prieto, M. & Rovira, C. (2012) Catalases versus peroxidases: DFT investigation of H2O2 oxidation in models systems and implications for heme protein engineering. Journal of Inorganic Biochemistry, 117, 292–297. [DOI] [PubMed] [Google Scholar]
- Waalwijk, C. , Mendes, O. , Verstappen, E.C. , de Waard, M.A. & Kema, G.H. (2002) Isolation and characterization of the mating‐type idiomorphs from the wheat septoria leaf blotch fungus Mycosphaerella graminicola . Fungal Genetics and Biology, 35, 277–286. [DOI] [PubMed] [Google Scholar]
- Ware, S.B. (2006) Aspects of sexual reproduction in Mycosphaerella species on wheat and barley: genetic studies on specificity, mapping, and fungicide resistance [Thesis]. Wageningen: Wageningen University and Research.
- Wojtaszek, P. (1997) Oxidative burst: an early plant response to pathogen infection. Biochemical Journal, 322, 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, F. , Melo‐Braga, M.N. , Larsen, M.R. , Jørgensen, H.J. & Palmisano, G. (2013) Battle through signaling between wheat and the fungal pathogen Septoria tritici revealed by proteomics and phosphoproteomics. Molecular and Cellular Proteomics, 12, 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden, O. , Veluchamy, S. , Dickman, M.B. & Kabbage, M. (2014) Sclerotinia sclerotiorum catalase SCAT1 affects oxidative stress tolerance, regulates ergosterol levels and controls pathogenic development. Physiological and Molecular Plant Pathology, 85, 34–41. [Google Scholar]
- Zámocký, M. , Gasselhuber, B. , Furtmüller, P.G. & Obinger, C. (2012) Molecular evolution of hydrogen peroxide degrading enzymes. Archives of Biochemistry and Biophysics, 525, 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zámocký, M. , Kamlárová, A. , Maresch, D. , Chovanová, K. , Harichová, J. & Furtmüller, P.G. (2020) Hybrid heme peroxidases from rice blast fungus Magnaporthe oryzae involved in defence against oxidative stress. Antioxidants, 9, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Z. , McDonald, B.A. & Palma‐Guerrero, J. (2021) Tolerance to oxidative stress is associated with both oxidative stress response and inherent growth in a fungal wheat pathogen. Genetics, 217, iyaa022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiers, L.‐H. & De Waard, M.A. (2001) Efficient Agrobacterium tumefaciens‐mediated gene disruption in the phytopathogen Mycosphaerella graminicola . Current Genetics, 39, 388–393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


