Abstract
We reviewed all diagnoses of Shigella species notified to the UK Health Security Agency from January 2016 to March 2023. An overall increase in notifications of shigellosis was seen between 2016 (n = 415/quarter) and 2023 (n = 1 029/quarter). However, notifications dramatically declined between March 2020 and September 2021 during the COVID-19 pandemic (n = 208/quarter) highlighting the impact of travel and social distancing restrictions on transmission. S. sonnei diagnoses were more affected by lockdown restrictions than S. flexneri, most likely due to a combination of species-specific characteristics and host attributes. Azithromycin resistance continued to be associated with epidemics of sexually transmissible S. flexneri (adult males = 45.6% vs. adult females = 8.7%) and S. sonnei (adult males = 59.5% vs. adult females = 14.6%). We detected resistance to ciprofloxacin in S. sonnei from adult male cases not reporting travel at a higher frequency (79.4%) than in travel-associated cases (61.7%). Extensively drug-resistant Shigella species associated with sexual transmission among men almost exclusively had ESBL encoded by bla CTX-M-27, whereas those associated with returning travellers had bla CTX-M-15. Given the increasing incidence of infections and AMR, we recommend that enhanced surveillance is used to better understand the impact of travel and sexual transmission on the acquisition and spread of MDR and XDR Shigella species.
Keywords: antimicrobial resistance, gastrointestinal illness, impact of COVID, microbial genomics, sexual transmission, shigellosis, surveillance trends, travel
Key results
Except for the years covered by the COVID-19 pandemic, there was a steady increase in shigellosis notifications from January 2016 to March 2023.
During the pandemic, notifications dramatically declined and then gradually increased as lockdown restrictions were relaxed.
S. sonnei diagnoses were more influenced by lockdown restrictions than S. flexneri, suggesting that returning travellers may seed domestic transmission of S. sonnei.
Azithromycin resistance continued to be a factor in epidemics of sexually transmissible S. flexneri and S. sonnei.
Resistance to ciprofloxacin in S. sonnei from adult male cases not reporting travel was higher than from travel-associated cases.
Extensively-drug resistant Shigella species associated with sexual transmission among men almost exclusively had bla CTX-M-27, whereas those associated with returning travellers had bla CTX-M-15.
Enhanced collection of travel and sexual transmission data will improve understanding of this clinically significant pathogen.
Introduction
Shigellosis, previously known as bacillary dysentery, is a gastrointestinal illness characterized by diarrhoea containing blood and/or mucus, and sometimes stomach cramps, fever, and vomiting. Although most people recover without treatment within a few days, others require hospitalization to treat dehydration, blood stream infections, and/or other complications [1]. Globally, most cases of shigellosis are caused by Shigella sonnei or S. flexneri, although S. boydii and S. dysenteriae also contribute to the burden of gastrointestinal disease in endemic regions and may cause infectious gastrointestinal disease in travellers returning from the Indian sub-continent and Africa [2, 3].
Shigella species are patho-adapted to infect the colonic mucosa of the human gut, and there is no animal reservoir. The infectious dose is low (10–100 bacteria) and the incubation period is usually 24–48 hours. Due to the low infectious dose, shigellosis is highly transmissible, and transmission is maintained primarily by direct person to person contact but can occur indirectly via the ingestion of food or water contaminated by human faeces. There is evidence that some individuals continue to be carriers for weeks or even months after their symptoms have resolved [4]. Historically in England, sporadic cases of shigellosis were associated with travellers returning from endemic regions (e.g., Africa, Asia, and Latin America) [5–7]. Outbreaks mostly occurred amongst children in school and nursery school settings, although foodborne outbreaks have been described [8–10]. More recently, domestic transmission has been sustained by sexual transmission among gay, bisexual, and other men who have sex with men (GBMSM) [3, 11, 12].
Sexually transmissible shigellosis among GBMSM is a public health concern. Surveillance data indicate that the burden of disease is high, clinical outcomes can be severe, and outbreaks have been caused by multidrug resistant (MDR) and extensively drug resistant (XDR) strains of S. flexneri and S. sonnei [13–15]. However, not all MDR and XDR cases of shigellosis are sexually transmitted, and previous studies in the UK and elsewhere have identified MDR and XDR Shigella in travellers returning from endemic regions [16, 17]. We reviewed all diagnoses of Shigella species notified to the UK Health Security Agency (UKHSA) from January 2016 to March 2023. The aim of this study was to describe the epidemiology, microbiology, and antimicrobial resistance profiles of travel-associated and sexually transmitted shigellosis in England.
Methods
Data collection
Faecal specimens from hospitalized patients and individuals in the community with gastrointestinal symptoms are sent to local hospital, private or regional laboratories in England for culture (https://www.gov.uk/government/publications/smi-b-30-investigation-of-faecal-specimens-for-enteric-pathogens). All Shigella isolated at these diagnostic laboratories are captured via the Second Generation Surveillance System (SGSS) (https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/739854/PHE_Laboratory_Reporting_Guidelines.pdf). This dataset comprises comprehensive, national coverage of primary laboratory-based notifications of shigellosis.
UKHSA guidelines recommend that diagnostic laboratories submit isolates of presumptive Shigella to the Gastrointestinal Bacterial Reference Unit (GBRU) at UKHSA for confirmation by PCR and typing by whole genome sequencing (WGS). PCR and sequencing results are stored in the Gastro Data Warehouse (GDW) and linked to patient demographic data, including age, sex, area of residence, and travel history. Not all isolates of Shigella are referred to the GBRU, and therefore the data in GDW are a subset (approximately two-thirds) of diagnoses reported in SGSS. However, the sequencing data in GDW were used in this analysis for detailed analyses of molecular typing, strain differentiation and relatedness, and AMR profiling.
Data analysis
Microbiological typing data from all isolates of Shigella species submitted to the GBRU between January 2016 and March 2023 were extracted and analyzed (Supplementary Table). Travel-associated cases were defined as those reporting recent foreign travel to any country seven days prior to the onset of symptoms, based on information from laboratory reports. Travel history is poorly captured for cases of Shigella species; if no travel history is reported, it cannot be inferred that the case did not travel. Laboratory surveillance data lack information on patient sexual orientation. However, in line with previous work, we used a proxy indicator of cases that may be attributed to sexual transmission among GBMSM, defined as cases among adult males without a history of travel [18]. Patients aged <16 years were classified as children.
The analysis was undertaken in three stages. First, we analyzed the demographic data available in SGSS, specifically, age, sex, and travel history, over time. We then analyzed the demographic data available in GDW, specifically age, sex, and travel history over time, linked to the reference laboratory-confirmed cases of shigellosis in England for all Shigella species and then for each individual species of Shigella. Finally, we analyzed the sequencing data for genomic markers of resistance to azithromycin (defined here as the presence of ermB and/or mphA), ciprofloxacin (defined here as the presence of mutations in gyrA, parC and/or the presence of qnr), and the third generation cephalosporins (defined here by the presence any bla CTX-M variant) for all Shigella species and for each individual species of Shigella. We defined extensively-drug resistant isolates as those expressing genomic markers of resistance to azithromycin, ciprofloxacin and third-generation cephalosporins.
Data are presented quarterly; Quarter 1 (Q1) from January to March, Q2 from April to June, Q3 from July to September and Q4 from October to December.
Whole-genome sequencing
Since August 2015, microbiological typing, including confirmation of the species and the serotype, has been performed at UKHSA using WGS [6, 7]. DNA was extracted for sequencing on an Illumina HiSeq 2500 instrument. Identification to the species level was based on kmer identification [19]. Genome-derived serotyping and AMR determinant profiling were performed using the GeneFinder tool (https://github.com/phe-bioinformatics/gene_finder), run using the default parameters. For serotyping, a reference database containing the gene sequences encoding the 12 O-antigen synthesis or modification genes was constructed and then used to determine serotype as previously described [7].
For AMR determinant profiling, genes were defined as present if they represented 100% of the reference sequence with greater than 90% nucleotide identity [17]. The reference database for AMR determinants can also be found in the GeneFinder github repository (https://github.com/phe-bioinformatics/gene_finder/tree/master/refs). The prevalence of resistance to streptomycin, tetracycline, sulphonamides, and trimethoprim in Shigella species is high, and the trends have been consistent for many years [20–22]. We, therefore, focused our AMR analysis on the presence of genomic makers of resistance to azithromycin, ciprofloxacin, and the third generation cephalosporins as the trends in resistance to these clinically relevant classes of antimicrobials fluctuated during the study period.
Results
Overview of local and regional diagnostic laboratory data from SGSS
Between Q1 2016 and Q1 2020, notifications of shigellosis showed an increasing trend (Figure 1). However, following the implementation of the first lockdown restrictions in Q2 2020 in response to the COVID-19 pandemic, reported diagnoses more than halved (Figure 1). From Q1 2016 to Q1 2020, on average there were 581 diagnoses of shigellosis reported to SGSS each quarter, whereas from Q2 2020 to Q3 2021, there was an average of 208 diagnoses of shigellosis recorded in SGSS each quarter (Figure 1). Following the relaxation of the lockdown restrictions in Q3 2021, the number of notifications among all persons returned to, and then exceeded, pre-pandemic levels, with an average of 785 reported diagnoses per quarter (Figure 1 and Table 1). The increase in diagnoses following the final removal of lockdown restrictions occurred among both adults and children and in both males and females (Table 1 and Figure 2). The largest proportional increases were among adult males who did not travel or where travel history was not reported (Figure 2).
Figure 1.
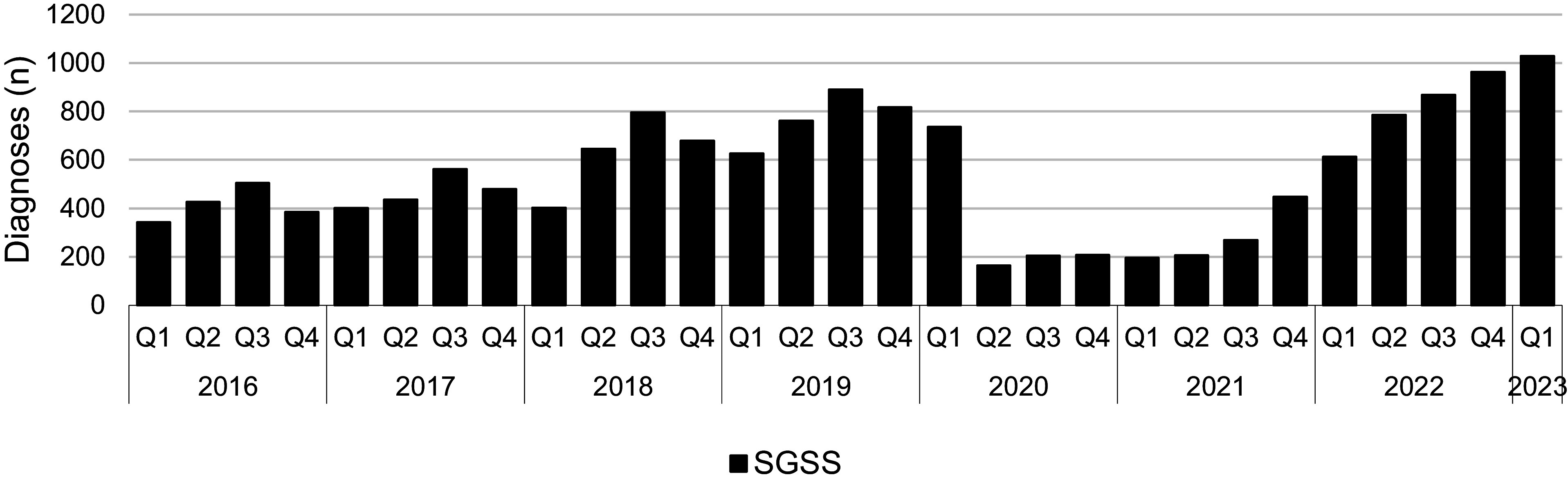
Total number of Shigella spp. diagnoses, England, Q1 2016 to Q1 2023 (data source: SGSS).
Table 1.
Average quarterly number of reported Shigella spp. diagnoses pre COVID-19, during COVID-19 restrictions, and post COVID-19 in SGSS, England
| Pre COVID-19 (Q1 2016 to Q1 2020) | COVID-19 restrictions (Q2 2020 to Q3 2021) | Post COVID-19 restrictions (Q4 2021 to Q1 2023) | % change from pre COVID-19 to post COVID-19 | |
|---|---|---|---|---|
| All persons | 581 | 208 | 785 | 37.2% increase |
| Adults | 465 | 175 | 644 | 38.5% increase |
| Children | 105 | 32 | 138 | 31.4% increase |
| Males | 322 | 151 | 458 | 42.2% increase |
| Females | 246 | 55 | 324 | 31.7% increase |
| Travel | 111 | 8 | 80 | 27.9% decrease |
| Non-travel/unknown | 461 | 200 | 705 | 52.9% increase |
Figure 2.
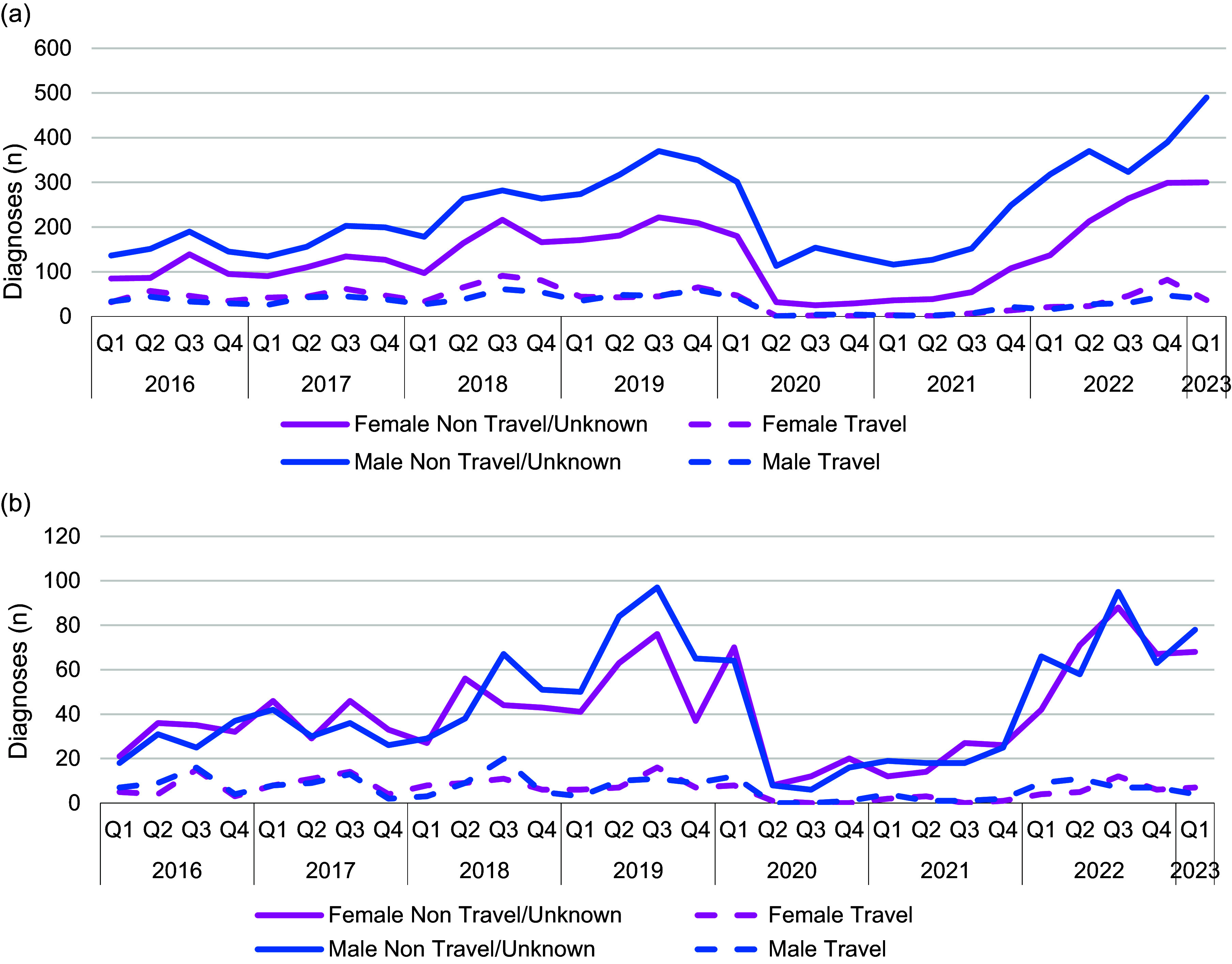
Number of Shigella spp. diagnoses associated with sex and travel, England, Q1 2016 to Q1 2023 in adults (a) and children (b) (data source: SGSS).
Analyses of data on GDW from isolates referred to the reference laboratory
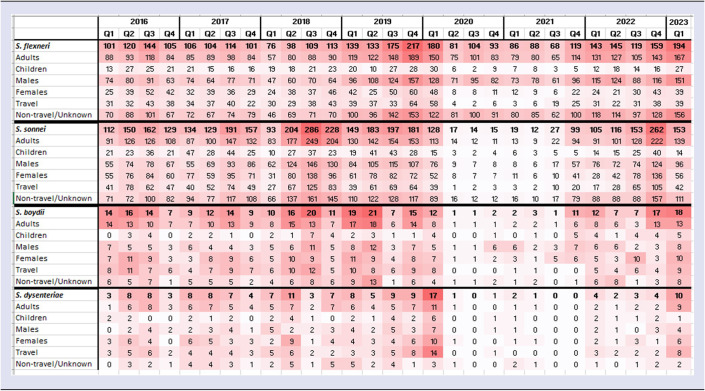
Shigella speciation data for isolates referred to GBRU between Q1 2016 and Q1 2023 showed the majority were S. sonnei or S. flexneri (94%, n = 7 339/7 782). From Q1 2016 to Q3 2019, the highest number of diagnoses were S. sonnei (Figure 3). However, notifications of S. flexneri had been increasing since 2018 and just prior to the pandemic lockdown, S. flexneri became the dominant species in Q4 2019 (Figure 3). In Q2 2020, notifications of both S. sonnei and S. flexneri declined steeply, but the decrease was more striking for S. sonnei (from 128 in Q1 2020 to 17 in Q2 2020, 87% decrease). Following the removal of lockdown restrictions in Q3 2021, case numbers increased for both S. sonnei and S. flexneri, with S. sonnei exhibiting the largest increase, peaking at 262 cases in Q4 2022 before decreasing in Q1 2023 (Figure 3). There was an increase in notifications of S. flexneri during the latter half of 2022, and S. flexneri was the most frequently reported species in Q1 2023.
Figure 3.

Number of all Shigella spp. diagnoses by species, England, Q1 2016 to Q1 2023 (all persons) (data source: GDW).
The trends of S. flexneri and S. sonnei among adults and children, males and females, individuals reporting travel, and those with no or unknown travel history, revealed that S. sonnei diagnoses declined universally during the COVID-19 pandemic (Table 2). Conversely, S. flexneri diagnoses continued to be reported among adult males and those reporting no or unknown travel status, yet declined among children, females and people reporting travel (Table 2). A more detailed analysis of the serotypes influencing the trends in S. flexneri notifications showed that prior to the implementation of lockdown restrictions, serotype 2a was dominant in England. However, the increase in S. flexneri in Q3 2019 was driven by a combination of serotypes 1b, 2a, and 3a, and all three serotypes continued to circulate during the pandemic (Figure 4), mainly amongst adult males. After lockdown restrictions were lifted, diagnoses of S. flexneri serotypes 1b and 2a were most common, with reports of S. flexneri 2a rising sharply in the first quarter of 2023.
Table 2.
Quarterly number of Shigella diagnoses by species and population group, England, Q1 2016 to Q1 2023
Figure 4.
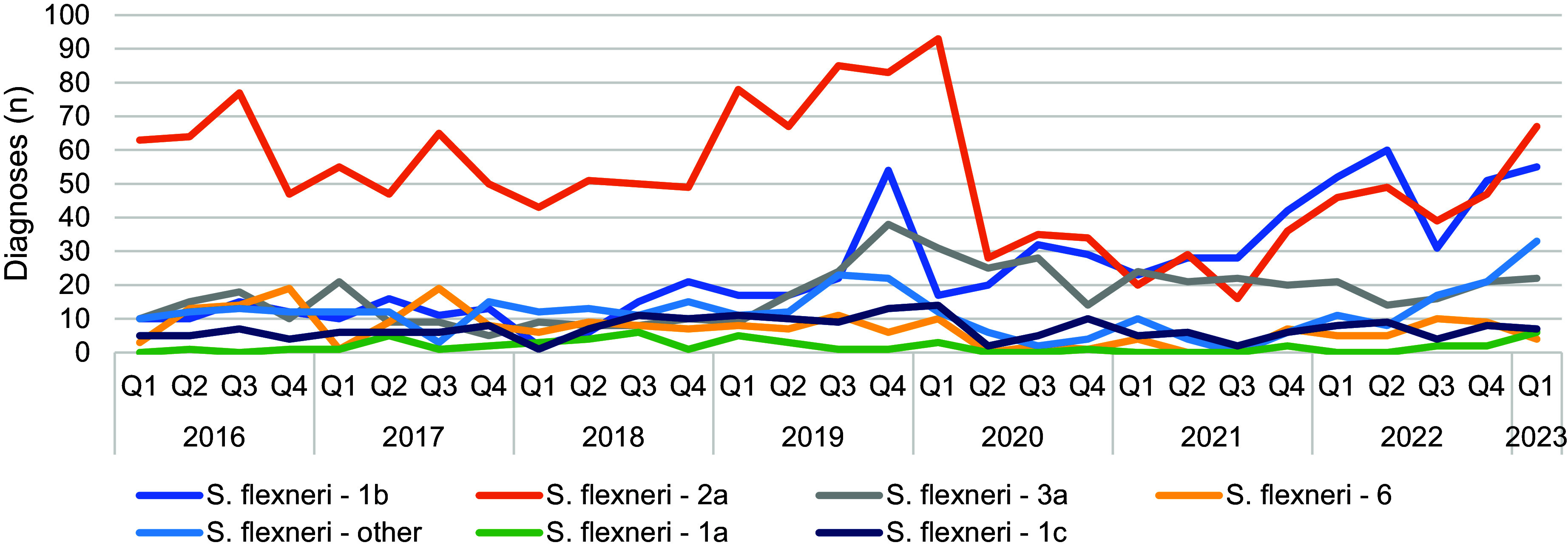
Number of all S. flexneri diagnoses by serotype, England, Q1 2016 to Q1 2023 (all persons) (data source: GDW).
Analysis of antimicrobial resistance data
Overall, the number and proportion of extensively drug resistant (XDR) isolates increased over the period between Q1 2016 and Q1 2023. The proportion of XDR isolates was higher for S. sonnei compared to S. flexneri, such that in 2022, 34% (219/636) of S. sonnei isolates and 6% (34/537) S. flexneri isolates were defined as XDR (Figure 5a,b).
Figure 5.

Number and proportion of extensively drug resistant isolates, England, Q1 2016 to Q1 2023 (all persons) (a) S. flexneri and (b) S.sonnei.
Analyses of the presence of AMR across different demographic groups revealed that among individuals reporting travel, adult females, and children, the proportions of cases infected with S. flexneri isolates that had ermB and/or mphA (travellers, 8.7%; adult female non-travellers, 8.7%; children, 5.7%), gyrA, parC and/or qnr (travellers, 61.7%; adult female non-travellers, 53.8%; children, 66.9%), and bla CTX-M-27 (travellers, 1.2%; adult female non-travellers, 0.3%; children, 0.2%), were similar (Table 3). In comparison, adult male non-travellers were associated with a higher proportion of isolates that had ermB and/or mphA (adult male non-travellers, 45.6%) or bla CTX-M-27 (adult male non-travellers, 2.9%), but a lower proportion of isolates that had gyrA, parC and/or qnr (adult male non-travellers, 45.6%) and bla CTX-M-15 than the other demographic groups (travellers, 7.5%; adult female non-travellers, 9.5%, adult male non-travellers, 1.9%; children, 16.1%) (Table 3).
Table 3.
Presence of AMR determinants encoding resistance to azithromycin, ciprofloxacin and third-generation cephalosporins among S. flexneri isolates, England, Q1 2016 to Q1 2023
| n (%) | ||||
|---|---|---|---|---|
| AMR determinants | All travellers | Adult female non-travellers | Adult male non-travellers | Children |
| Azithromycin (ermB and/or mphA) | 73 (8.7) | 31 (8.7) | 906 (45.6) | 27 (5.7) |
| Ciprofloxacin (gyrA, parC and/or qnr) | 515 (61.7) | 192 (53.8) | 906 (45.6) | 319 (66.9) |
| Third generation cephalosporins | ||||
| bla CTX-M-15 | 63 (7.5) | 34 (9.5) | 37 (1.9) | 77 (16.1) |
| bla CTX-M-27 | 10 (1.2) | 1 (0.3) | 58 (2.9) | 1 (0.2) |
For S. sonnei, the proportions of AMR isolates were generally higher than for S. flexneri, although a similar pattern among the different population groups was observed (Table 4). With the exception of those in the adult male non-travellers group, the proportions of cases with S. flexneri isolates that had ermB and/or mphA (travellers, 15.5%; adult female non-travellers, 14.6%; children, 17.8%), gyrA, parC and/or qnr (travellers, 61.7%; adult female non-travellers, 57.5%; children, 56.3%), and bla CTX-M-27 (travellers, 1.9%; adult female non-travellers, 2.4%; children, 0.5%), were similar (Table 4). In comparison, adult male non-travellers were associated with a higher proportion of isolates that had ermB and/or mphA (adult male non-travellers, 59.5%), gyrA, parC and/or qnr (adult male non-travellers, 79.4%), or bla CTX-M-27 (adult male non-travellers, 15.7%), but a lower proportion of isolates that had bla CTX-M-15 than the other three groups (travellers, 23.8%; adult female non-travellers, 23.0%, adult male non-travellers, 9.8%; children, 33.8%) (Table 4).
Table 4.
Presence of AMR determinants encoding resistance to azithromycin, ciprofloxacin and third-generation cephalosporins among S. sonnei isolates, England, Q1 2016 to Q1 2023
| n (%) | ||||
|---|---|---|---|---|
| AMR determinants | All travellers | Adult female non-travellers | Adult male non-travellers | Children |
| Azithromycin (ermB and/or mphA) | 204 (15.5) | 102 (14.6) | 792 (59.5) | 111 (17.8) |
| Ciprofloxacin (gyrA, parC and/or qnr) | 811 (61.7) | 403 (57.5) | 1 057 (79.4) | 351 (56.3) |
| Third generation cephalosporins | ||||
| bla CTX-M-15 | 313 (23.8) | 161 (23.0) | 131 (9.8) | 211 (33.8) |
| bla CTX-M-27 | 25 (1.9) | 17 (2.4) | 206 (15.5) | 3 (0.5) |
Discussion
Overall, we observed a steady increase in notifications of shigellosis from 2016 to 2023 in children and in adults of both sexes not reporting travel abroad. Although we cannot accurately determine the proportion of the cases that had travelled abroad due to the low completion of travel history information, the numbers of cases reporting travel abroad remained constant during this time. This suggests that domestic transmission was increasing both within GBMSM sexual transmission networks and amongst men, women and children in the wider community. It also suggests that sexual transmission may not be the only factor driving the reproductive rate of shigellosis.
Despite the perceived increase in domestic transmission, previous studies have shown that shigellosis is travel-related [6, 7, 16, 17]. The COVID-19 pandemic and associated lockdown restrictions highlighted the impact of travel and social distancing restrictions on the transmission of shigellosis, as notifications dramatically declined and then gradually increased as restrictions were relaxed [23]. S. sonnei diagnoses were more affected by lockdown restrictions than S. flexneri, indicating that imported, travel-associated infection may be a contributing factor to S. sonnei notifications and that returning travellers may be a source of subsequent ongoing domestic transmission of S. sonnei in England.
Species-specific pathogen characteristics and host attributes that may enhance person to person transmission, and specifically sexual transmission, of S. flexneri over S. sonnei, were considered. Historically, S. flexneri has been associated with more severe clinical outcomes than S. sonnei [24], and it is possible that only the most severe cases were captured by UKHSA surveillance systems, due to the difficulties in accessing health care during the pandemic [25]. We also considered the possibility that chronic infection and long-term carriage may be more prevalent following infection with S. flexneri than with S. sonnei [4]. Another consideration was that S. flexneri and S. sonnei were associated with different host transmission networks: S. flexneri continued to interact and/or travel during the lockdown restrictions, whereas S. sonnei did not.
Genomic epidemiology of these species outside of the COVID-19 pandemic in both South Africa and globally suggests that S. flexneri may have a different ecology that supports local geographical persistence relative to S. sonnei, possibly through greater stability of the S. flexneri virulence plasmid [26–30]. Furthermore, we noted that notifications of S. flexneri were on the rise prior to lock-down whereas S. sonnei notifications were in decline, and this may have facilitated persistence the in community despite the reduced person to person contact [31]. Although AMR is known to drive transmission among GBMSM, it is unlikely that AMR is a factor in this context as the S. sonnei and S. flexneri GBMSM clades exhibit similar AMR profiles [3, 31]. Moreover, we noted that the S. sonnei isolates that persisted during 2020 and 2021 were XDR, although the S. flexneri isolates circulating during the pandemic period, were not [13, 14].
We have previously shown that AMR is a potential driver of epidemic and endemic transmission of shigellosis and here we reviewed the AMR profiles of the isolates in the UKHSA archive, specifically resistance to azithromycin, fluroquinolones, and third-generation cephalosporins [3, 11, 20]. As expected, the proportion of isolates of S. flexneri and S. sonnei that had genes known to confer resistance to azithromycin was higher in the adult males not reporting travel group compared to other population groups. Azithromycin resistance has been driving epidemics of sexually transmissible S. flexneri and S. sonnei for the last decade, and this study indicates that this continues to be a feature [3, 11, 20]. For S. sonnei, we also continue to detect resistance to fluroquinolones in isolates from adult male cases not reporting travel at a higher frequency than those from travel-associated cases but this was not the case for S. flexneri. This is despite the evidence of increasing fluroquinolone resistance worldwide, and particularly in southeast Asia [32].
There was a difference in the distribution of the bla CTX-M variant genes conferring resistance to third-generation cephalosporins across the different population groups, with adult male non-travellers being the only group to exhibit a higher proportion of isolates harbouring bla CTX-M-27 compared to bla CTX-M-15. A high proportion of individuals in the adult male non-travel group identified as GBMSM and during this study period, XDR Shigella species associated with sexual transmission among men almost exclusively had bla CTX-M-27 [13, 15, 33]. In contrast, during this study period, the majority of XDR Shigella species from travellers had bla CTX-M-15. The similar proportion of cases with bla CTX-M-15 in (i) travellers, (ii) females not reporting travel, and (iii) children, may be confounded by the inclusion of travel-associated cases that have been misclassified due to missing travel histories in the latter two groups. Alternatively, this may be evidence of ongoing domestic transmission of imported strains across the wider community, seeded by returning travellers.
Between 2009 and 2019, we recorded a series of epidemics of azithromycin resistance S. flexneri 3a, S. flexneri 2a and S. sonnei in England [3, 12, 34]. Despite fluctuations in notifications, S. sonnei clade 5 carrying resistance to azithromycin and ciprofloxacin, remained the dominant strain until 2019 when we observed a resurgence of S. flexneri, partially but not entirely reduced by the COVID-19 pandemic [3, 31]. Post-pandemic, S. sonnei rapidly re-emerged having acquired a plasmid encoding bla CTX-M-27 conferring the XDR profile [13]. In the first quarter of 2023, we observed the landscape changed once more, with a sharp rise in S. flexneri 2a, a proportion of these carrying bla CTX-M-27, and a corresponding decrease in S. sonnei [14]. Although the drivers of the rise and fall of the different clades of Shigella are likely to be multifactorial, there is accumulating evidence that AMR has a role, and modelling and seroepidemiological work are needed to study this, and other factors, further.
The pandemic presented a unique opportunity to study the microbiological and epidemiological drivers of persistence of shigellosis within GBMSM sexual networks and the wider population. However, it is unclear whether the sustained transmission of S. flexneri during the pandemic was due to a fitness advantage of the strain, differences in access to care or care-seeking behaviour, or clinical manifestation or severity. It could be that travel is a more significant driver of sexual transmission of S. sonnei than previously thought. Travel restrictions during the COVID-19 lockdown were the most likely cause of reduced notifications of both S. flexneri and S. sonnei among children and adult females.
Over the last decade, we have focused on the transmission of MDR and XDR shigellosis within sexual networks. Previous studies have highlighted the global transmission of MDR and XDR Shigella species driven by travel, and this study provides further evidence with nearly a quarter of travel-associated S. sonnei isolates harboured bla CTX-M-15 [6, 7, 16, 17, 26–30]. Currently, in England, it is not mandatory for isolates of S. sonnei to be referred to UKHSA for further characterization by genome sequencing. Given the high incidence of XDR S. sonnei, we recommend that enhanced surveillance is carried out on all isolates of this clinically significant pathogen. We also recommend that accurate and more complete case travel histories should be sought to better understand the impact of travel, and the epidemiological exposures associated with travel, on the acquisition of AMR and domestic transmission.
Supporting information
Charles et al. supplementary material
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268824001006.
click here to view supplementary material
Author contributions
Writing – review & editing: I.S., K.S., K.B.; Conceptualization: G.G., C.J.; Formal analysis: H.C., C.J.; Methodology: H.C., C.J.; Writing – original draft: H.C., C.J.
References
- [1].Charles H, et al. (2023) Spotlight on drug-resistant Shigella: Raising awareness within general practice. British Journal General Practice 73, 187–188. 10.3399/bjgp23X732537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Terry LM, et al. (2018) Antimicrobial resistance profiles of Shigella dysenteriae isolated from travellers returning to the UK, 2004–2017. Journal Medical Microbiology 67, 1022–1030. 10.1099/jmm.0.000779. [DOI] [PubMed] [Google Scholar]
- [3].Bardsley M, et al. (2020) Persistent transmission of shigellosis in England is associated with a recently emerged multidrug-resistant strain of Shigella sonnei. Journal Clinical Microbiology 58, e01692–e01619. 10.1128/JCM.01692-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Allen H, et al. (2021) Evidence for re-infection and persistent carriage of Shigella species in adult males reporting domestically acquired infection in England. Clinical Microbiology Infection 27, e7, e13–e126. 10.1016/j.cmi.2020.03. [DOI] [PubMed] [Google Scholar]
- [5].Bentley CA, et al. (1996) Phage typing and drug resistance of Shigella sonnei isolated in England and Wales. Epidemiology & Infection 116, 295–302. 10.1017/s0950268800052602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dallman TJ, et al. (2016) Use of whole-genome sequencing for the public health surveillance of Shigella sonnei in England and Wales, 2015. Journal Clinical Microbiology 65, 882–884. 10.1099/jmm.0.000296. [DOI] [PubMed] [Google Scholar]
- [7].Chattaway MA, et al. (2017) Whole-genome sequencing for National Surveillance of Shigella flexneri. Frontiers in Microbiology 8, 1700. 10.3389/fmicb.2017.01700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].McDonnell J, et al. (2013) Retrospective analysis of whole genome sequencing compared to prospective typing data in further informing the epidemiological investigation of an outbreak of Shigella sonnei in the UK. Epidemiology & Infection 141, 2568–2575. 10.1017/S0950268813000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Maguire HC, et al. (1998) Shigella outbreak in a school associated with eating canteen food and person to person spread. Communicable Disease Public Health 1, 279–280. [PubMed] [Google Scholar]
- [10].Jenkins C, et al. (2023) Foodborne outbreak of extended Spectrum Beta-lactamase producing Shigella sonnei associated with contaminated spring onions in the United Kingdom. Journal Food Protection 86, 100074. 10.1016/j.jfp.2023.100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Baker KS, et al. (2015) Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: A cross-sectional study. Lancet Infectious Disease 15, 913–921. 10.1016/S1473-3099(15)00002-X. [DOI] [PubMed] [Google Scholar]
- [12].Simms I, et al. (2015) Intensified shigellosis epidemic associated with sexual transmission in men who have sex with men - Shigella flexneri and S. sonnei in England, 2004 to end of February 2015. EuroSurveillance 20, 21097. 10.2807/1560-7917.es2015.20.15.21097. [DOI] [PubMed] [Google Scholar]
- [13].Charles H, et al. (2022) Outbreak of sexually transmitted, extensively drug-resistant Shigella sonnei in the UK, 2021-22: A descriptive epidemiological study. Lancet Infectious Disease 22, 1503–1510. 10.1016/S1473-3099(22)00370-X. [DOI] [PubMed] [Google Scholar]
- [14].Thorley K, et al. (2023) Emergence of extensively drug-resistant and multidrug-resistant Shigella flexneri serotype 2a associated with sexual transmission among gay, bisexual, and other men who have sex with men, in England: A descriptive epidemiological study. Lancet Infectious Disease 23, 732–739. 10.1016/S1473-3099(22)00807-6. [DOI] [PubMed] [Google Scholar]
- [15].Mook P, et al. (2016) ESBL-producing and macrolide-resistant Shigella sonnei infections among men who have sex with men, England, 2015. Emerging Infectious Disease 22, 1948–1952. 10.3201/eid2211.160653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kotloff KL, et al. 20180 shigellosis. Lancet 391, 801–812. 10.1016/S0140-6736(17)33296-8. [DOI] [PubMed] [Google Scholar]
- [17].Sadouki Z, et al. (2017) Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Shigella sonnei isolated from cases of diarrhoeal disease in England and Wales, 2015. Journal Antimicrobial Chemotherapy 72, 2496–2502. 10.1093/jac/dkx170. [DOI] [PubMed] [Google Scholar]
- [18].Mitchell HD, et al. (2019) Use of whole-genome sequencing to identify clusters of Shigella flexneri associated with sexual transmission in men who have sex with men in England: A validation study using linked behavioural data. Microbial Genomics 5, e000311. 10.1099/mgen.0.000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chattaway MA, et al. (2017) Identification of Escherichia coli and Shigella species from whole-genome sequences. Journal Clinical Microbiology 55, 616–623. 10.1128/JCM.01790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Baker KS, et al. (2016) Travel- and community-based transmission of multidrug-resistant Shigella sonnei lineage among international orthodox Jewish communities. Emerging Infectious Diseases 22, 1545–1553. 10.3201/eid2209.151953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Baker KS, et al. (2018) Genomic epidemiology of Shigella in the United Kingdom shows transmission of pathogen sublineages and determinants of antimicrobial resistance. Scientific Reports 8, 7389. 10.1038/s41598-018-25764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Baker KS, et al. (2018) Horizontal antimicrobial resistance transfer drives epidemics of multiple Shigella species. Nature Communications 9, 1462. 10.1038/s41467-018-03949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Love NK, et al. (2023) Understanding the impact of the COVID-19 pandemic response on GI infection surveillance trends in England, January 2020–April 2022. Epidemiology & Infection 151, e147. 10.1017/S095026882300136X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hale TL, Keusch GT. (1996) Chapter 22: Shigella. In: Baron S (ed.), Medical Microbiology, 4th Edn. Galveston, TX: University of Texas Medical Branch at Galveston. Available at https://www.ncbi.nlm.nih.gov/books/NBK8038/. [PubMed] [Google Scholar]
- [25].Goyal DK, et al. (2021) Restricted access to the NHS during the COVID-19 pandemic: Is it time to move away from the rationed clinical response? Lancet Regional Health - Europe 8, 100201. 10.1016/j.lanepe.2021.100201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Holt KE, et al. (2012) Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nature Genetics 44, 1056–1059. 10.1038/ng.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Connor TR, et al. (2015) Species-wide whole genome sequencing reveals historical global spread and recent local persistence in Shigella flexneri. eLife 4, e07335. 10.7554/eLife.07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].McVicker G and Tang CM (2016) Deletion of toxin-antitoxin systems in the evolution of Shigella sonnei as a host-adapted pathogen. Nature Microbiology 2, 16204. 10.1038/nmicrobiol.2016.204. [DOI] [PubMed] [Google Scholar]
- [29].McVicker G, et al. (2019) Maintenance of the virulence plasmid in Shigella flexneri is influenced by Lon and two functional partitioning systems. Molecular Microbiology 111, 1355–1366. 10.1111/mmi.14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Stenhouse GE, et al. (2023) The genomic epidemiology of shigellosis in South Africa. Nature Communications 14, 7715. 10.1038/s41467-023-43345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dallman TJ, et al. (2021) Emergence of novel strains of Shigella flexneri associated with sexual transmission in adult men in England, 2019–2020. Journal Medical Microbiology 70, 001437. 10.1099/jmm.0.001437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chung The H and Baker S (2018) Out of Asia: The independent rise and global spread of fluoroquinolone-resistant Shigella. Microbial Genomics 4, e000171. 10.1099/mgen.0.000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mason LCE, et al. (2023) The evolution and international spread of extensively drug resistant Shigella sonnei. Nature Communications 14, 1983. 10.1038/s41467-023-37672-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Borg ML, et al. (2012) Ongoing outbreak of Shigella flexneri serotype 3a in men who have sex with men in England and Wales, data from 2009–2011. EuroSurveillance 17, 20137. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Charles et al. supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268824001006.
click here to view supplementary material