Abstract
Coronaviruses rely on the viral-encoded chymotrypsin-like main protease (Mpro or 3CLpro) for replication and assembly. Our previous research on Mpro of SARS-CoV-2 identified cysteine 300 (Cys300) as a potential allosteric site of Mpro inhibition. Here, we identified tixocortol (TX) as a covalent modifier of Cys300 which inhibits Mpro activity in vitro as well as in a cell-based Mpro expression assay. Most importantly TX inhibited SARS-CoV-2 replication in ACE2 expressing HeLa cells. Biochemical analysis and kinetic assays were consistent with TX acting as a non-competitive inhibitor. By contrast, TX was a weaker inhibitor and modifier of C300S Mpro, confirming a role for Cys300 in inhibition of WT Mpro but also providing evidence for an additional Cys target. TX pivalate (TP), a prodrug for TX that was previously marketed as a nasal spray, also inhibited SARS-CoV-2 replication in HeLa–ACE2 cells at low micromolar IC50s. These studies suggest that TX and/or TP could possibly be repurposed for the prevention and/or treatment of SARS-CoV-2 infection.
Tixocortol, the active component in a nasal spray, inhibits SARS-CoV-2 main protease and viral replication. This highlights its potential for use in preventing and/or treating SARS-CoV-2 infections.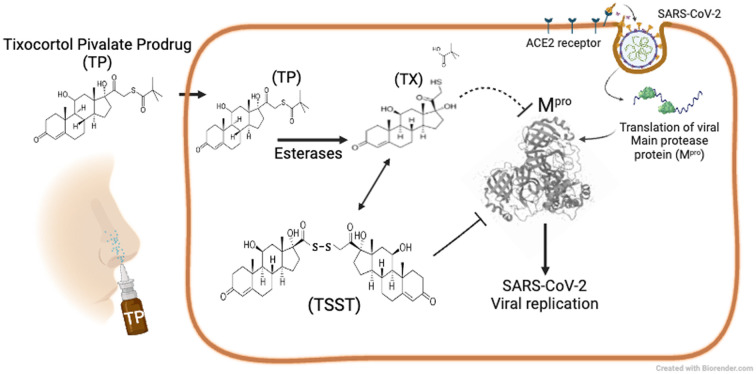
Introduction
Coronaviruses make up a large family of RNA viruses of which SARS-CoV-2 is a member. SARS-CoV-2 is one of seven known coronaviruses that infect humans.1 The COVID-19 pandemic caused by SARS-CoV-2 has caused millions of deaths worldwide, reinforcing the need to find ways to stop the spread and treat infection with SARS-CoV-2 and other highly pathogenic viruses.2 Although recent advances in COVID-19 vaccines have saved millions of lives by preventing severe disease and illness, they only provide partial protection against infection by SARS-CoV-2.3 More recently, drugs have been developed to treat SARS-CoV-2 infection, adding to the arsenal of treatments for SARS-CoV-2 infection. The primary approved anti-SARS-CoV-2 therapy is a fixed combination of two drugs, nirmatrelvir and ritonavir that is sold under the brand name Paxlovid.4 Nirmatrelvir targets the active site cysteine of 3-chymotrypsin-like main protease (Mpro also known as 3CLpro) of SARS-CoV-2 and covalently modifies and inactivates the enzyme, while ritonavir is needed to maintain effective plasma levels of nirmatrelvir.5,6 Mpro is essential for viral replication as it cleaves SARS-CoV-2-encoded polyproteins (PP1a and PP1ab) in at least 11 locations to release mature viral proteins that are required for virus replication.7 While nirmatrelvir/ritonavir can rapidly eliminate SARS-CoV-2 and resolve symptoms of infection, the virus can often rebound, and symptoms can return days after the 5-day treatment and possibly necessitate subsequent therapy.8–10 While these strategies have been greatly successful, COVID-19 remains a substantial public health problem and additional strategies are urgently needed. The importance of Mpro as a target for blocking virus infection was appreciated during the outbreak of SARS (now called SARS-1) in 2003 during which 9% of patients with SARS-CoV-1 died.11 Basic research on SARS-1 led to the development of potential inhibitors of the Mpro.12 However, due to the containment of the SARS-1 outbreak, the need for vaccines and therapeutics on a large scale was no longer thought necessary and therefore research on new drugs against SARS-1 faded. However, with the advent of SARS-CoV-2 outbreak which began in 2019, there was suddenly an urgent interest for preventive and therapeutic strategies against this new worldwide pandemic. Compounds have now been identified from various screening approaches as potential Mpro inhibitors, including FDA-approved drugs that could possibly be repurposed.13 Nirmatrelvir, identified through rational drug design, acts as active site Mpro inhibitor by covalently binding to the active site cysteine 145 to inhibit activity14,15 During replication, SARS-CoV-2 Mpro becomes active by forming a homodimer, which is the active form of Mpro. Our previous research revealed that Mpro could be reversibly inhibited through reversible oxidation of Cys300. Modification of Cys300 with glutathione causes Mpro to form an inactive monomer.16 Activity can be restored with reducing agents or with the addition of the ubiquitous cellular enzyme glutaredoxin, both of which removed the covalently bound glutathione. Interestingly, each monomer of Mpro contains an intact active site. However, the monomer remains inactive unless dimerization occurs, leading to the functional conformation of the active site pocket.17 While it remains uncertain if Cys300 acts to regulate Mpro within cells, it seemed reasonable to hypothesize that this allosteric site could be targeted for inhibition of Mpro. We hypothesized that targeting this site with small molecule drugs could interfere with dimerization, leading to inactivation of Mpro and could provide an alternative way to block SARS-CoV-2 replication.16 Further supporting this idea, another group recently demonstrated allosteric inhibition of the Mpro dimer with colloidal bismuth subcitrate that involves Cys300 and another unidentified cysteine.18 In this report, we describe the identification of tixocortol (TX) as an Mpro inhibitor that acts primarily at Cys300 to inhibit Mpro. Our data suggests that TX is an allosteric covalent inhibitor that, in part, impairs Mpro dimerization. TX, as well as the prodrug form tixocortol pivalate (TP) and TX disulfide (TSST), block SARS-CoV-2 replication in HeLa–ACE2 cells. The fact that TX is part of a previously approved nasal spray suggests it could be repurposed to help prevent SARS-CoV-2 infections or treat SARS-CoV-2 infections and symptoms.
Methods
Chemicals and reagents
ACV tripeptide, zofenoprilat, epicaptropril, bicisate were from BOC Sciences (Shirely, NY), phosphopantetheine, dl-thiorphan, captopril, R-dihydrolipoic acid and hydrocortisone were from Sigma-Aldrich (St Louis, MO), omapatrilat, emerimide were from Cayman Chemicals (Ann Arbor, MI), tixocortol, and tixocortol pivalate were from Medical Isotopes Inc. (Pelham, NH) and bucilliamine was from AK Scientific (Union City, CA) nirmatrelvir (NM) was from Med Chem Express (Monmouth Junction, NJ). All compounds were dissolved in 100% cell culture-grade DMSO from Sigma (St Louis, MO) and stored as 10–20 mM stock solutions at −20 °C until use. TKB-198, 5H, and GRL-0920 were obtained as described previously19–21 and stored as stocks in 100% DMSO. Ammonium hydrogen carbonate for use in SEC/MS chromatography was obtained from Millipore (Oakville, Ontario, CA). Formic acid and trifluoroacetic acid were from Pierce Chemical company (Rockford, IL). Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) solution, dithiothreitol (DTT), and reduced glutathione (GSH) were from Sigma. Sequencing grade chymotrypsin and AccuMAP Low pH Digestion kit was from Promega (Madison, WI). The disulfide of TX was generated by incubation of TX at 1 mM in 50 mM HEPES buffer pH 7.8 and 10% DMSO overnight at 35 °C. The resultant precipitate was dissolved in 100% DMSO and then run on RP-HPLC to purify TSST from remaining TX using a Vydac C18 column and a 2% ACN gradient.
Molecular docking of glutathione and compound discovery procedure
The objective was to form an all-atom detailed molecular model around l-glutathione. Among the SARS-CoV-2 main protease structures examined from the Protein Data Bank, there are marked similarities in their secondary and tertiary features. For the preliminary model, we chose the Mpro X-ray structure PDB: 7AXM, the monomeric main protease that has been co-crystalized with pelitinib. In this initial phase, the preliminary model served as a basis for ligand discovery. For details on the creation of the preliminary model and refinement please see the ESI.† To determine the feasibility of covalent binding of compounds of interest at the identified Cys300 pocket, these compounds were covalently docked by virtual modeling onto the SARS-CoV-2 main protease crystal structure. Maestro 12.7 (Schrödinger, Inc.) was used to prepare the template Mpro crystal structure of PDB: 7AXM and the covalent docking module was used to model the covalent ligand–protein complex using the reactive residue set at Cys300, reaction type (disulfide formation), and docking mode set at virtual screening.
Screening assay for Mpro inhibitory activity and determination of Mpro covalent modification
Compounds identified from the docking procedure were screened for activity against Mpro using a peptide based (TSAVLQSGFRKM) RP-HPLC assay as described previously,16 using 100 nM Mpro (final) and 50 μM of each drug in assay buffer (50 mM HEPES buffer, 1 mM EDTA, 50 mM NaCl pH 7.8) (see Table S1†). Briefly, compounds were incubated at 37 °C with Mpro for 1 h, then the assay was started with addition of peptide substrate (2 mM final) and stopped after 10 minutes with TFA. Products were determined by measuring their absorbance at 205 nm. Control was 5% DMSO and all samples contained 5% DMSO in assay. Further assays involving kinetics and further characterization were performed the same except 50 nM Mpro was used in these assays. For assessing covalent modification, compounds dissolved in 100% DMSO were diluted to 50 μM (5% DMSO final) and incubated with Mpro (5 μM) for 1 h in assay buffer and then analyzed by SEC/MS for covalent modification by protein deconvolution as described previously.16 Mpro samples (1–10 μM as indicated in figures) were injected (2–10 μL) and run with an isocratic flow rate of 0.35 mL min−1 and where indicated, cmBSA was used as an internal standard and carrier to help prevent nonspecific losses of protein during the analysis. Mpro eluting from the column was monitored using an Agilent DAD UV detector in series with the Agilent 6230 MS-TOF detector. After different treatments and preincubation times of Mpro with test compounds, the eluting Mpro peaks detected by the ion current from MS analysis were deconvoluted using Agilent's Mass Hunter software to assess the molecular mass of Mpro and to obtain evidence of covalent modification of the eluting Mpro as described previously.16
Mpro kinetic analysis
Wild type and C300S Mpro's were prepared and purified as described previously.16 To perform Michaelis–Menten kinetics the assay was done in 96 well cell culture treated round bottom plates (Corning). The enzyme was made up as a 60 nM stock in HEPES assay buffer. Eight wells received 38 μL of enzyme solution and each well was then treated with 2 μL of DMSO for controls or 2 μL of 20× stock of TX dissolved in DMSO to bring to 1×. The plate was incubated for 1 h at 37 °C and then 5× substrate stocks in HEPES assay buffer were added to give final substrate concentrations of 0.1, 0.2, 0.4, 0.8, 1.6, 3.2 and 6.4 mM for the DMSO controls and inhibitor treatments (50 nM Mpro following substrate addition). The assay was carried out at 37 °C for 8 minutes (confirmed to be within the linear range of the assay to obtain initial velocities) and the assay was stopped with 10 μL of 4% TFA solution in water. Samples were analyzed by RP-HPLC as described above and Lineweaver–Burke (double-reciprocal plots) plots were used to calculate the values of KM, Vmax. and Ki. The data presented represents the average of three independent experiments.
In cellulo Mpro assay
Active Mpro was expressed from the pcDNA3 H2B-mIFP T2A Mpro (WT) plasmid (Addgene 163079). The NanoLuc plasmid containing the Mpro cleavage site (AVLQSGFR) was constructed as reported by Chen et al.22 Briefly, a permuted NanoLuc construct in the pcDNA3.1 vector expressing amino acids 66–171 in the N-terminus was linked using a linker containing the Mpro cleavage site (GGGSGNGSAVLQSGFRSLKACGGG) with amino acids 2–66 of the Nanoluciferase protein in the C-terminus. The NanoLuc construct was codon-optimized for mammalian expression. One hundred thousand early passage healthy HeLa cells were plated in each well of a 12-well plate and transfected 24 h later with FUGENE6 (Promega). A ratio of 1 : 3 : 3 (NanoLuc : Mpro : Bgal) of plasmid was used for transfection along with pcDNA3 as carrier DNA for maintaining transfection efficiency. Briefly, in each well of the 12-well plate, 80 ng of NanoLuc, 240 ng of Mpro, and 240 ng of the beta-gal control vector were transfected. Drugs (TX or NM) were added to the cells 20 h post-transfection and assayed for NanoLuc activity 46 h post-drug treatment.
SARS-Cov-2 antiviral assay
For SARS-CoV-2 replication assays, HeLa–ACE2 cells obtained from Creative Biogene (Shirley, NY) were seeded into 384-well tissue culture treated plates at 6000 per well in 30 μL DMEM (Gibco, Gaithersburg, MD, USA) supplemented with either non-heat inactivated, or heat inactivated (HI) 10% fetal bovine serum (FBS; Sigma, St. Louis, MO, USA) the day prior to infection. One h or 18 h before infection, compounds (dissolved in DMSO) were digitally dispensed directly into cell plates using a Tecan D300e digital dispenser to generate a twelve-point dose curve for each compound, replicated in quadruplicate (n = 4). Under biocontainment conditions, SARS-CoV-2 virus (WA-01) was diluted in DMEM with 10% FBS to a concentration of 3000 plaque forming units (PFU) (60 000 PFU mL−1) (MOI of 2). This inoculum was added to the assay plates for a final assay volume of 50 μL. Plates were fixed in 10% neutral buffered formalin 18 h post-infection; upon removal from biocontainment, cells were stained with a SARS-CoV/SARS-CoV-2 nucleoprotein-specific primary antibody followed by a secondary antibody conjugated to Alexa647 fluorophore. Hoechst dye was added for detection of cell nuclei. Fluorescence readout was quantitated using a PerkinElmer Operetta high-content imaging system. Cytotoxicity on mock-infected plates was determined using the Promega Cell Titer-Glo Luminescent Cell Viability Assay (Catalog #G7571) at the time of fixation of the infected plate. Half-maximal inhibitory concentration (IC50) and 50% cytotoxic concentration (CC50) were calculated using GraphPad Prism Software (La Jolla, CA). Z′ factor scores were assessed as quality-control parameters for each plate.
Metabolism of tixocortol pivalate to tixocortol and related metabolites
To determine conversion of TP to TX and related compounds under various conditions, we incubated TP (50 μM) in water, DMEM, DMEM supplemented with heat inactivated serum or DMEM with 10% serum that was not heat inactivated. Following an 18 h incubation overnight at 37 °C the samples were acidified with 0.2% TFA to stop the reaction and then diluted with DMSO to give a final of 50% DMSO. This was done to ensure solubility of the compounds prior to RP-HPLC/MS analysis. The samples were treated with 10 mM TCEP for 15 min (to reduce any disulfide bound TX or TSST in the samples) and then separated on a Vydac-5205 C18 column and the amount of TP and TX was determined based on the elution time and molecular mass obtained with the standards. To assess the conversion of TP by cells, HeLa cells were plated overnight and then cells were treated with TP (50 μM). Cells were then extracted for metabolites after 1, 4, 24, and 48 h incubation. Cells were washed with PBS and then trypsinized followed by three additional washes with PBS. The pellet was then extracted with 60% methanol solution/0.2% TFA and heated at 95 °C for 5 min. Cellular debris was removed by centrifugation and the methanolic/TFA solution dried by speed vac to dryness. The residue was dissolved in 100% DMSO and analyzed by RP-HPLC/MS. The concentrations of TP, TX, TSSG and the disulfide of TX were determined.
Purification of tixocortol disulfide
To purify tixocortol disulfide for use in Mpro assays, TX was first incubated in HEPES assay buffer pH 7.8 in 5% DMSO. The reaction was left to go overnight at 37 °C. TSST precipitated from the solution and was pelleted and then redissolved in 100% DMSO. The solution was then separated on a Vydac-5205 C18 column and the eluting TSST, as identified by its mass obtained from MS analysis, was collected in eppendorph tubes. The eluates were then dried down in a speed Vac concentrator, incubated with methanol to remove volatiles and dried again. The powder was redissolved in 100% DMSO. The TSST could be converted to TX with 10 mM TCEP and then the concentration determined by comparison to a known concentration of TX standard. The TSST in DMSO was stored at −70 °C until use.
Preparation of TX-Mpro for chymotryptic digestion and RP-HPLC/MS analysis
To determine which cysteines of Mpro were modified by tixocortol treatment, we first prepared TX-modified Mpro (TX-Mpro). Mpro was first pretreated with 10 mM TCEP to ensure full reduction of the 12 cysteines and the correct native mass was verified by SEC/MS analysis as described above. The TCEP was removed by washing through an Amicon 10 kDa cutoff membrane three times using 50 mM HEPES buffer pH 7.8 with 1 mM EDTA, 10% DMSO and 50 mM NaCl. Protease was set at 1 μM and TX was added at a final concertation of 200 μM and incubated at 35 °C for 1 h (total volume 3.5 mL). The preparation was then concentrated using a 10 kDa membrane and washed three times with HEPES buffer pH 7.2 with 1 mM EDTA, 10% DMSO to remove the residual unbound TX. The TX-Mpro preparation was analyzed by SEC/MS to determine the extent of TX modification. To determine which cysteines were modified by TX, we carried out chymotryptic digestions as described previously using the AccuMAP Low pH Digestion kit from Promega.16 Briefly, 20 μg of TX-Mpro was alkylated with NEM in 8 M urea, concentrated by filtration, and put up in final 1 M urea pH 8 in Tris and CaCl2 buffer. The alkylated preparation in 1 M urea was digested with chymotrypsin (1 μg mL−1 in 1 mM HCl) at 28 °C overnight. The solution was stopped with a final of 2% TFA. The samples were then applied to Pierce C18 peptide desalting columns from ThermoFisher Scientific (Waltham, MA) and after washing, the peptides were eluted with acetonitrile. Peptide samples were put up into RP-HPLC running buffer (0.1% FA/0.02% TFA). The peptide samples, treated without or with 10 mM TCEP, as well as the corresponding peptide standards and TX-modified peptide standards were analyzed by RP-HPLC/MS as described previously.16
Results
To further explore the possibility that Cys300 and the surrounding pocket might be a promising target for identifying allosteric inhibitors of Mpro, we developed a model of glutathione anchored to Cys300 through a disulfide bond with Mpro (PDB ID: 7AXM) (Fig. S1A†). A pocket was identified that showed potential binding of glutathione to Mpro through electrostatic interactions with Arg4 and Arg298 of the same monomer and with the –SH group of glutathione juxtaposed to the Cys300 –SH. With the physiologic ligand l-glutathione, a very favourable distance (2.54 angstroms) was achieved once its carboxylate formed a salt bridge with the side chain of Arg 4, enabling disulfide bond formation (Fig. S1A†). By contrast, d-glutathione, upon forming the same salt bridge, produced an inter-thiol distance (5.48 angstroms) that was prohibitive for disulfide bond formation (Fig. S1B†). Therefore, the l-glutathione model could serve as an initial platform for a ligand discovery effort. Next, a database was formed with all thiol-containing compounds present in the DrugBank all drugs structure file, release date 2021-01-03. This database was examined using an automated virtual screening approach at the Cys300 pocket. We initially identified 15 compounds with favorable docking scores and were able to obtain 13 of these compounds from commercially available sources for testing (Table S1†). For screening, each compound was preincubated for 1 h at 50 μM with Mpro and then assayed for activity using a previously described peptide-based (TSAVLQ*SGFRKM) RP-HPLC Mpro method.16 In addition, Mpro was also examined by size exclusion chromatography/mass spectroscopy (SEC/MS) to determine if covalent modification of Mpro was taking place with any of the compounds. While most of the compounds showed no inhibition and even improved Mpro activity to some degree under these conditions, tixocortol (TX) was identified as the only compound of the 13 tested to significantly inhibit Mpro activity in the screen (average 80% inhibition) and to covalently modify Mpro following the 1 h preincubation (Table S1†).
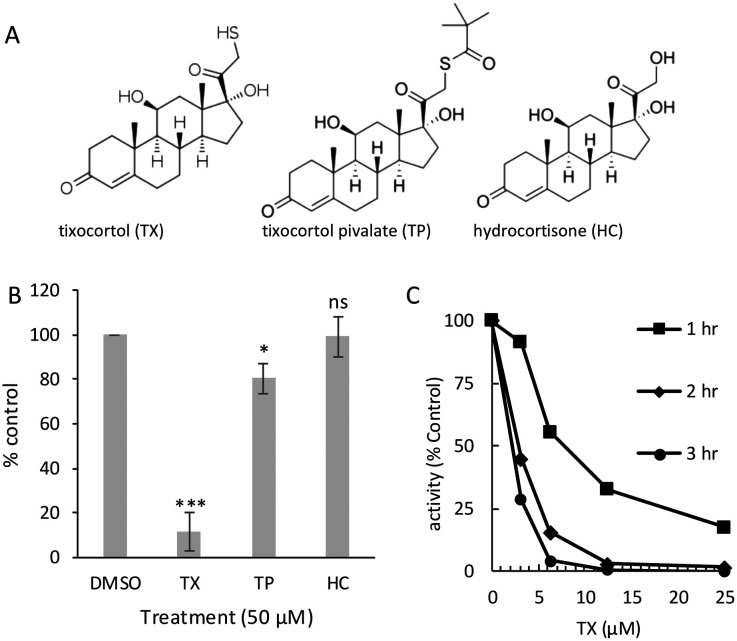
Based on these preliminary results, we explored in more detail the effects of TX on inhibition of Mpro and SARS-CoV-2 replication. TX is a 21-thiol derivative of hydrocortisone (HC) that was developed as a prodrug in the form of a tixocortol pivalate (TP) suspension for use as a nasal spray for rhinitis (brand name Pivalone) (Fig. 1A).23 In the presence of serum or cellular esterase, TP releases the pivalate group to produce TX, the active form of the steroid.24 The 21-sulfur in TX participates in a thiol ester bond with pivalate to form TP (Fig. 1A). To better understand the mechanism of the inhibitory activity of TX, we assayed the effects of TX, TP, and HC on Mpro activity following a 1 h preincubation with each compound. TX inhibited Mpro activity an average of 90%, while TP was weakly active, while HC showed no inhibitory activity (Fig. 1B). A dose response and time course study using TX as an inhibitor of Mpro revealed that inhibition was time-dependent with longer pre-incubation times leading to improved IC50's (IC50 of 8 μM at 1 h and IC50 of <2 μM at 3 h) (Fig. 1C). These data suggest that the 21-thiol group is needed for good inhibitory activity of TX and suggests that a time-dependent covalent modification of Mprovia the 21-thiol group is likely required for TX inhibition. Michaelis–Menten (MM) kinetics were performed and a Lineweaver–Burk plot of the MM data (Fig. S2A†) indicated that TX had little effect on Km but decreased the Vmax 5-fold consistent with non-competitive inhibition (Fig. S2B†). From the MM data, the Ki for TX was calculated to be approximately 2.5 μM.
Fig. 1. Structures of tixocortol (TX), tixocortol pivalate (TP) and hydrocortisone (HC) and characterization of TX inhibition of SARS-CoV-2 Mpro. (A) Structure for tixocortol (TX), tixocortol pivalate (TP) and hydrocortisone (HC). (B) Comparison of the inhibition of SARS-CoV-2 Mpro enzyme activity using 50 μM of each compound after 1 h preincubation compared to DMSO control. Shown is the mean and standard deviation from three separate experiments, * p < 0.05, *** p < 0.005, ns = not significant. (C) Dose response curves for tixocortol at three different preincubation times (1 h, 2 h, and 3 h) prior to substrate addition. Mpro (60 nM,) was incubated with TX in assay buffer for the indicated times and then assayed (final Mpro concentration of 50 nM) for activity (10 min).
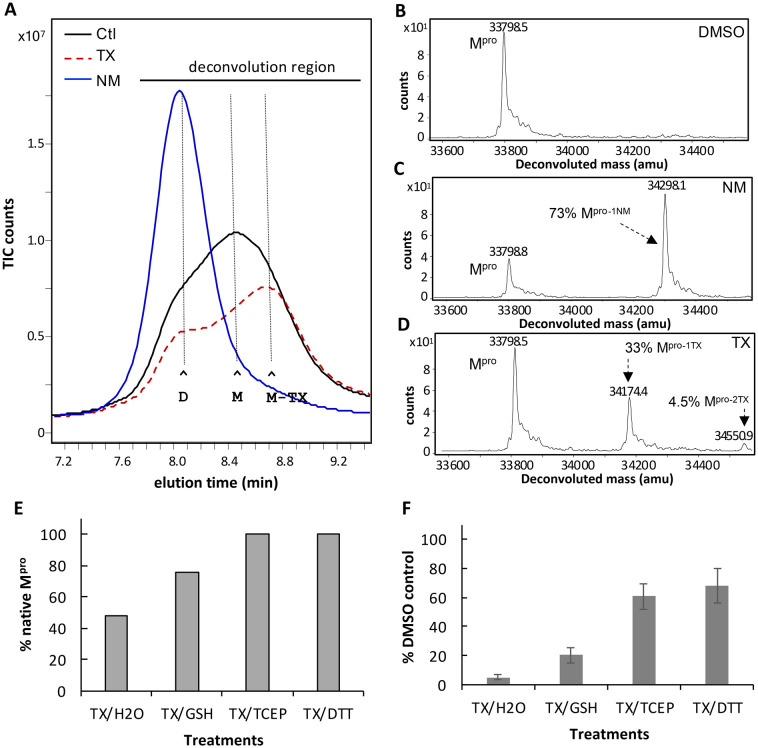
Mpro inhibitors that bind to and covalently modify the active site of Mpro are known to promote dimerization of Mpro resulting in an inactive dimer.19,25 We therefore compared the effect of TX on Mpro dimerization to that of nirmtrelvir and several other active site inhibitors using SEC/MS analysis. Mpro (10 μM) was treated with each compound (200 μM) (20:1 ratio drug to Mpro) for 1 h in HEPES assay buffer, and then analyzed by SEC/MS. The Mpro control (5% DMSO) eluted as a mix of dimer (D) and monomer (M) (Fig. 2A, black tracing). Treating Mpro with NM clearly induced Mpro dimerization, consistent with that described previously (Fig. 2A blue tracing).25 We also observed this shift to dimer when Mpro was treated with active site inhibitors TKB-198,19 GRL-0920,26 and 5 h,21 (Fig. S3A–E†). By contrast, TX treatment did not induce dimer but instead increased the proportion of monomer compared to dimer and shifted the peak for the monomer to a later elution time (denoted as Mpro-TX) (Fig. 2A, red dashed tracing). TX also led to a decrease in the amount of Mpro recovered over time (Fig. 2A, red dashed tracing). These results suggest that TX may covalently modify and destabilize the dimer leading to dissociation of dimers and may subsequently lead to protein aggregation. This is consistent with what was previously reported for colloidal bismuth subcitrate where Mpro dissociation ultimately led to protein aggregation.18 The fact that TX did not shift Mpro to dimer suggests that it may not act as an active site inhibitor at Cys145 but instead forms covalent adducts with a different cysteine or cysteines of Mpro and alters dimerization. Protein deconvolution following MS of the eluting Mpro revealed only native Mpro in the control (Fig. 2B). As expected, protein deconvolution of Mpro treated with NM revealed that approximately 79% of Mpro was covalently modified with NM based on the mass increase of +499.3 (Fig. 2C). Similarly, treatment of Mpro with TX led to approximately 33% of Mpro covalently modified with TX based on the observed mass increase of +375.9 (Fig. 2D). A smaller percentage of Mpro (4.5%) showing a mass increase of +752.4 (Fig. 2D) indicated that an additional cysteine was also modified by TX but to a lesser extent (Fig. 2D). Since the inhibition of Mpro by TX was time dependent, we carried out an experiment to follow the SEC eluting behavior and covalent modification of Mpro over time. In these experiments, a ratio of TX (200 μM) to Mpro (1 μM) of 200 : 1 was used and BSA was included to protect from protein loss and as an internal standard. By 60 minutes, approximately 50% of Mpro was modified with TX (40% with 1 TX and 10% with 2 TX) (Fig. S4A†) and over time the Mpro-1TX monomer elution shifted to the right of unmodified Mpro, indicating a later elution time (Fig. S4B†). To distinguish the elution profiles for the 3 different forms of Mpro that were detected by MS (native Mpro, Mpro–1TX, and Mpro-2TX) we extracted the specific ions for the native, 1TX and 2TX Mpro species (list of ions shown in Fig. S4C†) from the total ion current (TIC) chromatogram for the eluting Mpro's after the 60 min incubation with TX (Fig. S4D†). When this was done, while Mpro-1TX eluted in both the dimer and monomer regions (compare blue tracing for Mpro-TX vs. black hashed tracing for native Mpro) the monomer form eluted later than native monomer Mpro (Fig. S4D†). Also, Mpro-2TX eluted almost exclusively as a later eluting monomer (Fig. S4D†), (compare red tracing with black dashed tracing) providing evidence that TX modification disrupts the ability of the Mpro to form dimers. To determine if TX-modification and inhibition of Mpro was reversible, we tested three different reducing agents (GSH, TCEP, DTT). Mpro (1 μM) was treated with TX (50 μM) for 60 min and then either exposed to buffer (control) or reducing agents in buffer and incubated for an additional 15 minutes. Samples were then analyzed by SEC/MS to assess the extent of TX modification. The TX treated Mpro showed 52% Mpro modified with TX and 48% native Mpro (Fig. 2E). Treatment with GSH (20 mM) increased the amount of native Mpro from 48% to 76% while 10 mM TCEP or 10 mM DTT treatment restored all the Mpro to its native form (Fig. 2E). Protein deconvolutions following the treatment of TX-Mpro with the different reducing agents are provided in Fig. S5.† To determine if covalent modification of Mpro was required for TX inhibition of Mpro activity, we first preincubated Mpro (60 nM) with TX (50 μM) for 60 minutes in assay buffer and then added the different reducing agents (glutathione, TCEP, and DTT) at 10 mM for 15 minutes prior to assaying for activity. GSH restored about 20% of the DMSO/GSH Mpro control activity, while TCEP and DTT restored over 60% of their respective control activities indicating that covalent modification with TX plays a role in the inhibition of Mpro by TX (Fig. 2F).
Fig. 2. SEC elution behavior and covalent modification of Mpro following treatment with NM or TX. (A) SEC/MS analysis of Mpro following treatment with DMSO (black tracing -), NM (blue tracings) or TX (red dashed tracing). Mpro (10 μM) was incubated at 37 °C in 50 mM HEPES buffer pH 7.8 with 1 mM EDTA and treated with DMSO control or 200 μM of NM or TX (20 : 1 ratio drug to Mpro) for 1 h and then analyzed by SEC–MS. The peak elution time for Mpro dimer (D), monomer (M) and TX-shifted monomer (M-TX) are indicated by the horizontal lines. (B–D) Protein deconvolution of eluting Mpro showing the masses obtained for (B) control, (C) 200 μm NM treated Mpro and (D) TX-treated Mpro. (E) Reversibility of TX modification and (F) TX inhibition of Mpro with different reducing agents. (E) Restoration of native Mpro following treatment with reducing agents. Mpro (1 μM) was first modified with 200 μM TX for 1 h in assay buffer then reducing agents were added for 15 minutes. Mpro was then analyzed by SEC/MS followed by protein deconvolution (deconvolution region indicated by black horizontal line)) to determine the % native Mpro restored. (F) Activity of TX-treated Mpro after treatment with reducing agents. Mpro (60 nM) was incubated with 5% DMSO (control) or 50 μM TX in assay buffer for 1 h then reducing agents were added (diluted 1 : 20 as 20× stocks) to give a final concentration of 10 mM and incubated for 15 minutes. Samples were then assayed (Mpro 50 nM final) for activity (10 min). Shown is the mean and standard deviation from three separate experiments.
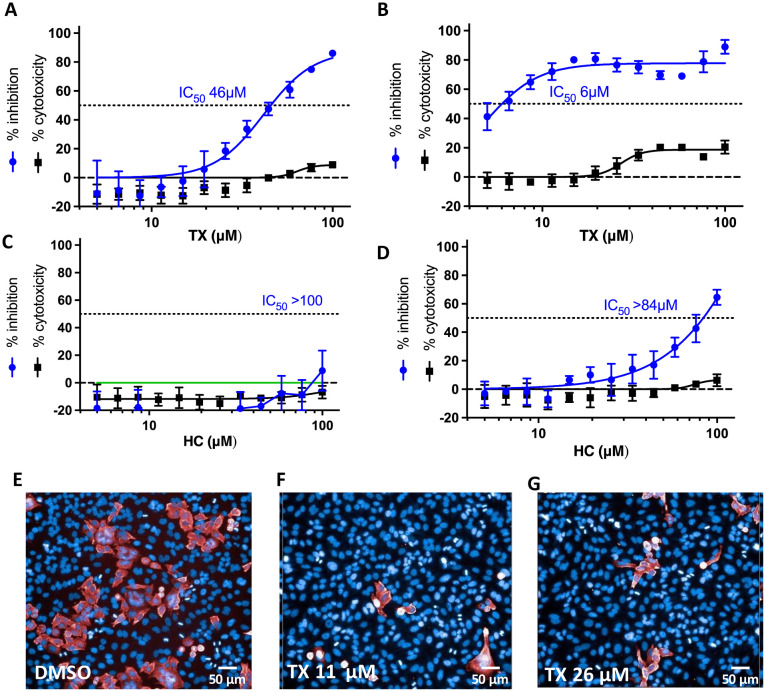
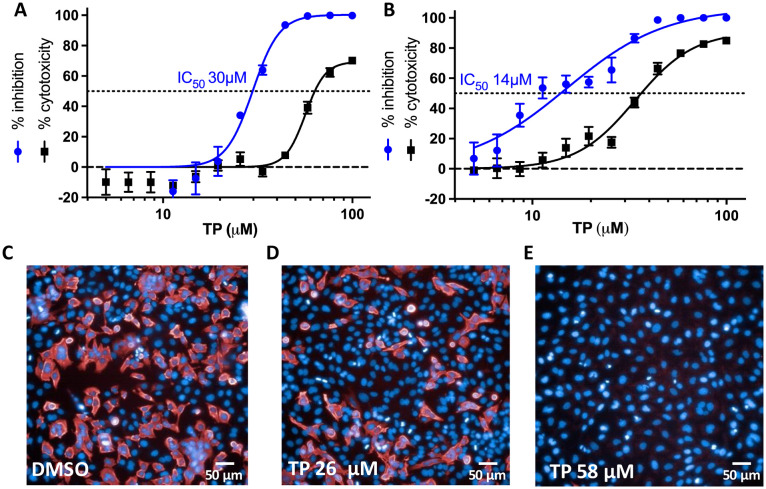
Since reducing agents did not completely reverse TX inhibition of Mpro, we decided to assess the ability of TX and/or TP to inhibit SARS-CoV-2 replication in cells by targeting Mpro. In addition, even though cells usually maintain a reducing environment, it's been reported that SARS-CoV-2 infection leads to dramatically lower cellular thiol levels in SARS-CoV-2 infected cells within the first 24 h of infection resulting in a pro-oxidant environment.27 This could therefore favor TX covalent modification and inhibition of Mpro in infected cells as well as other cysteine modifying compounds. To assess TX activity against SARS-CoV-2 infected cells, we tested the ability of TX to inhibit SARS-CoV-2 replication in a HeLa–ACE2 cellular system from Creative Biogene (Shirley, NY). These epithelial cells express the ACE-2 receptor, allowing for SARS-CoV-2 infection. We tested the effect of pretreating cells with TX for 1 h or 18 h before virus infection. HC, which was inactive in the Mpro assay, was used as a control. Pretreatment of cells with TX for 1 h resulted in an IC50 for viral inhibition of 45 μM and was associated with little or no toxicity at the concentrations used (up to 100 μM) (Fig. 3A). Interestingly, if cells were pre-treated with TX for 18 h prior to virus infection the IC50 for TX substantially improved (6 μM) with <20% toxicity observed (Fig. 3B). Yet, for reasons that are not clear, TX inhibitory activity plateaued at 80–85% inhibition for the 18 h pretreatment group (Fig. 3B). By contrast, HC was inactive if cells were pretreated for 1 h prior to infection and showed relatively weak activity in the overnight pretreatment with HC (IC50 84 μM) (Fig. 3C and D). Representative images of the SARS-CoV-2 nucleoprotein (in red) and Hoechst staining for cell nuclei are shown for the Fig. 3B 18 h pretreatment control (Fig. 3E), 11 μM TX (Fig. 3F), and 26 μM TX (Fig. 3G). We next investigated the ability of TP to inhibit SARS-CoV-2 replication since this is the form previously marketed for use as a nasal spray for rhinitis. TP dose dependently inhibited viral replication (IC50 of 30 μM) although TP treatment (unlike TX) also affected cell viability at higher doses with a TC50 of about 60 μM (Fig. 4A). However, TP at 44 μM inhibited more than 90% of the replication with less than 10% toxicity seen (Fig. 4A). If cells were pretreated for 18 h with TP, then like TX, the IC50 improved (14 μM) but this was also associated with greater toxicity (TC50 of approximately 35 μM) (Fig. 4B). Representative images of the SARS-CoV-2 nucleoprotein (in red) and the Hoescht staining for cell nuclei are shown for the 1 h pretreatment control (Fig. 4C), 26 μM TP, (Fig. 4D), and 58 μM TP (Fig. 4E). The SARS-CoV-2 replication data shown in Fig. 3 and 4 were acquired from cells grown in serum that had not been heat inactivated (nonHI) to better mimic natural serum which could provide active serum esterases to convert TP to TX. However, we also tested these compounds in cells grown in standard HI serum. The results with HI-serum mostly paralleled that seen with non-HI serum although TP toxicity was greater in HI serum and there was a somewhat higher calculated IC50 for virus inhibition in the 18 h pretreatment in HI serum (compare IC50 of 14 μM in non-HI serum vs. 21 μM in HI serum) (Fig. S6A–D†). With HI-serum HC did not inhibit viral replication nor was it toxic to the cells (Fig. S6E and F†). Taken together, these data indicate that the 21-thiol group is required for inhibition of Mpro activity as well as inhibition of SARS-CoV-2 replication.
Fig. 3. Inhibition of SARS-CoV-2 replication in HeLa–ACE2 cells following a 1 h or 18 h pretreatment with TX or HC. Cells were pretreated with TX or HC (as a control) for 1 h (A and C) or 18 h (B, and D) prior to exposure to SARS-CoV-2. Results are percentage inhibition of virus replication (blue values) and cytotoxicity (black values) relative to untreated controls. Error bars represent standard deviations (SDs) from tests run for each concentration with 4 replicates. IC50, half-maximal inhibitory concentration is indicated for each drug on the dotted line. (E–G)) representative images from Fig. 3B 18 h pretreatment. (E) Infected control, (F) TX at 11 μM and (G) TX at 26 μM. Red staining represents SARS-CoV-2 nucleoprotein with percentage infectivity measured as the number of SARS-CoV-2–positive cells relative to total cells measured by Hoechst 33342 nuclear staining (blue).
Fig. 4. Inhibition of SARS-CoV-2 replication in HeLa–ACE2 cells following a 1 h or 18 h pretreatment with TP. Cells were pretreated with TP for 1 h (A) or 18 h (B) prior to exposure to SARS-CoV-2. Results are percentage inhibition of virus replication (blue values) and cytotoxicity (black values) relative to untreated controls. Error bars represent standard deviations (SDs) from tests run for each concentration with 4 replicates. IC50, half-maximal inhibitory concentration is indicated for each drug on the dotted line. (C–E) Representative images from results in Fig. 4A. (C) Infected control, (D) TP at 26 μM and (E) TP at 58 μM. Red staining represents SARS-CoV-2 nucleoprotein with percentage infectivity measured as the number of SARS-CoV-2–positive cells relative to total cells measured by Hoechst 33342 nuclear staining (blue).
To further explore the need for TP to convert to TX to inhibit Mpro during viral replication, we also investigated the ability of serum (heat inactivated and non-heat inactivated) and cells to convert TP to TX since this conversion would be needed to inhibit Mpro activity if the free thiol group is required to inhibit Mpro in SARS-CoV-2 infected cells. TP was incubated for 18 h with H2O, DMEM, DMEM with 10% HI-serum or DMEM with 10% non-heat inactivated serum and then analyzed for TP and TX by RP-HPLC/MS analysis. Recovery of TP from the samples was similar for H2O, DMEM and DMEM with HI-serum (15–17 μM recovered from 25 μM initially) (Fig. S7A†). However, only about 7 μM of TP was recovered from the DMEM with non-HI serum (Fig. S7A†). This corresponded with more TX detected by RP-HPLC/MS analysis from the non-HI serum sample (3.1 μM) than the other samples (0.6–1.3 μM) (Fig. S7A†). The detection of TX in the absence of serum is likely due to solvolysis of TP over time in the overnight incubation. To determine if TP was converted to TX when added to cells, HeLa cells were treated with TP (50 μM) for 1, 4, 24 and 48 h and then trypsinized, washed and extracted with 60% methanol/TFA to determine cellular levels of TP and its metabolites. The major metabolite detected from TP-treated cells was TX which peaked at 24 h incubation (Fig. S7B†). Also detected were low levels of TP and TX conjugated to glutathione but at levels only 10–20% of that of TX (Fig. S7B†). These data confirmed that when TP is added to cells, it rapidly undergoes conversion (presumably via cellular esterases) to TX.
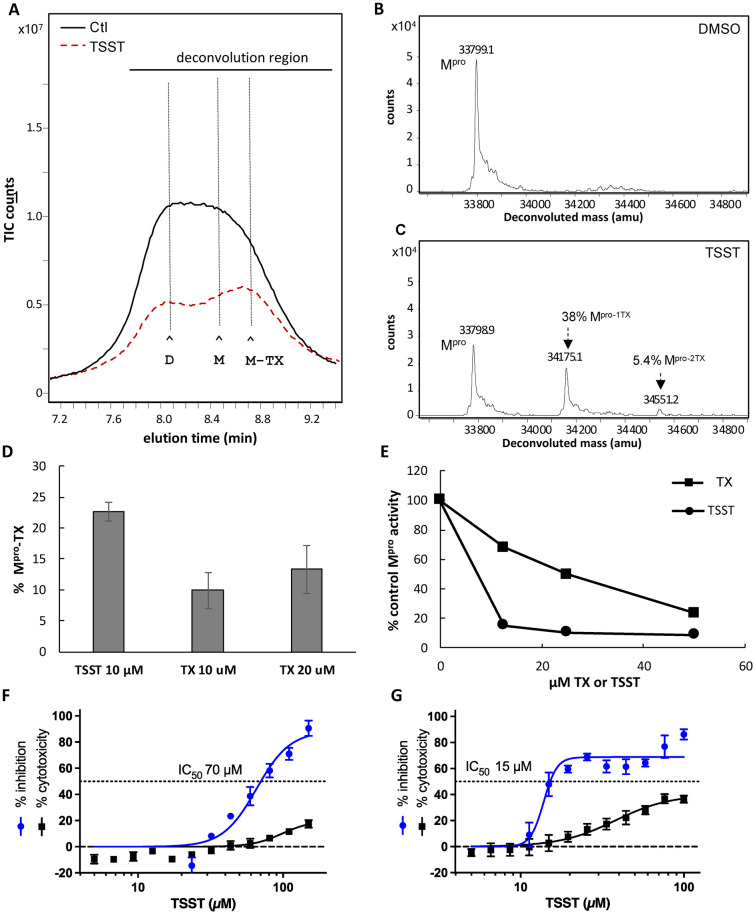
When TX was tested in the Mpro enzyme assay, we also learned that TX would undergo oxidation over time to its disulfide (TSST) (Fig. S8A–C†). We considered the possibility that TSST may be the form of TX that covalently modifies Mpro through disulfide exchange with cysteines of Mpro. To examine this, TSST was separated and purified as described in supplemental information (Fig. S8C†) and its ability to covalently modify Mpro and inhibit Mpro activity was determined. The effect of TSST on Mpro dimerization was like that seen for TX, causing a shift to monomer, a later elution time for the eluting monomer, and partial loss of total recoverable Mpro (Fig. 5A). TSST covalently modified about 43% of Mpro (Fig. 5B and C). Although TSST did modify Mpro to a similar degree as TX in a 1 h preincubation with Mpro, when tested at lower concentrations and shorter incubation times, TSST more rapidly modified Mpro compared to TX (Fig. 5D). For example, TSST at 10 μM modified more than 20% of Mpro in 15 minutes as compared to an average of 10% and 13% modification with 10 μM or 20 μM TX, respectively (Fig. 5D). The increased modification by TSST also correlated to greater inhibition of Mpro. In a typical 1 h preincubation assay with 50 nm Mpro, TSST at 12.5 μM decreased Mpro activity to about 10% of the control as compared to a decrease in activity to about 70% of the control with 12.5 μM TX (Fig. 5E). Yet, at the higher concentration of 50 μM inhibition of Mpro by TX and TSST was comparable (Fig. 5E). These data demonstrate that TSST is superior to TX at covalently modifying and inhibiting Mpro and suggests that TX may first oxidize to TSST before covalently modifying Mpro. However, from this data we cannot rule out direct modification of Mpro by the reduced form of TX. TSST was also tested in the SARS-CoV-2 viral replication assay. Pretreatment of cells with TSST for 1 h prior to infection led to an IC50 of 70 μM with minimal toxicity (<20%) (Fig. 5F). Pretreatment of cells with TSST for 18 h led to an improved IC50 of 15 μM although this was also associated with some toxicity and inhibitory activity plateaued at around 80% like that seen with TX (Fig. 5G). The lower activity of the TSST in the cellular replication assays as compared to the in vitro assays with Mpro may be due to the lower solubility of TSST as compared to TX when used in cell culture due to the limitation on DMSO concentrations when used on cells.
Fig. 5. SEC elution behavior and covalent modification of Mpro following treatment with DMSO or TSST. (A) SEC/MS analysis of Mpro following treatment with DMSO (black tracing -) or TSST (red dash tracing). Mpro (10 μM) was incubated at 37 °C in 50 mM HEPES buffer pH 7.8 with 1 mM EDTA and treated with DMSO (control) or 200 μM of TSST (20 : 1 ratio drug to Mpro) for 1 h and then analyzed by SEC–MS. The peak elution time for Mpro dimer and monomer (D and M) as well as TX-shifted monomer (M-TX) are indicated by the horizontal lines. (B and C) Protein deconvolution of eluting Mpro showing the masses obtained for (B) control, (C) TSST-treated Mpro. (D) TX modification of Mpro using TSST (10 μM) or TX (at both 10 μM and 20 μM) for 15 min. Values are the average of 3 separate experiments. (E) Effect of TSST on SARS-CoV-2 replication. Cells were pretreated with TSST for (F) 1 h or (G) 18 h prior to exposure to SARS-CoV-2. Results are percentage inhibition of virus replication (blue values) and cytotoxicity (black values) relative to untreated controls. Error bars represent standard deviations (SDs) from tests run for each concentration with 4 replicates. IC50, half-maximal inhibitory concentration is indicated for TSST on the dotted line.
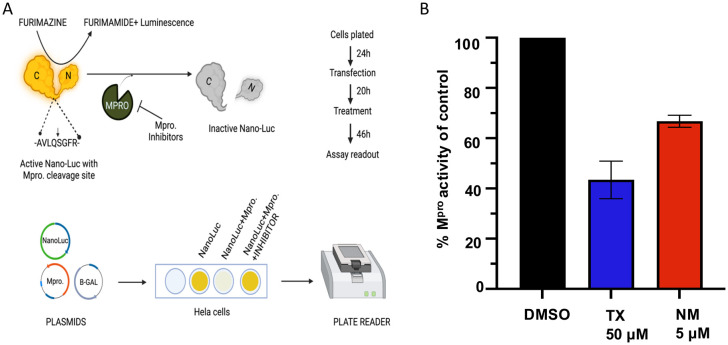
Although TX and TP have activity against SARS-CoV-2 replication it remains possible that inhibition of replication is due to blocking different aspects of infection rather than inhibiting Mpro. To assess whether TX could inhibit Mpro activity expressed within cells absent the virus, we used a previously described cell-based luciferase assay developed by Chen et al.22 The assay uses a modified nano-luciferase which contains an Mpro cleavage site. Upon cleavage by Mpro, the nano-luciferase activity is eliminated while in the presence of inhibitors the activity is protected (see schematic in Fig. 6A). Treatment of HeLa–ACE2 cells with 50 μM TX decreased Mpro activity by an average of 55% providing further evidence that TX can inhibit Mpro activity within cells and may at least be one of its targets leading to SARS-CoV-2 inhibition in infected cells (Fig. 6B). NM was tested at 5 μM as a positive control for Mpro inhibition and inhibited Mpro activity by an average 35% (Fig. 6B).
Fig. 6. TX inhibition of Mpro activity expressed in cells. (A) Schematic depiction of the nano-luciferase-based Mpro cellular assay. Inhibition of Mpro leads to increased luciferase activity by preventing substrate cleavage. (B) Inhibition of Mpro activity in the cellular assay as compared to the DMSO treated control. Cells were seeded (24 h) then transfected with Mpro expression plasmid and 18 h later treated with DMSO, TX (50 μM), or NM (5 μM) and then luciferase activity was measured 24 h later. NM was only used as a positive control at 5 μM to verify the assay.
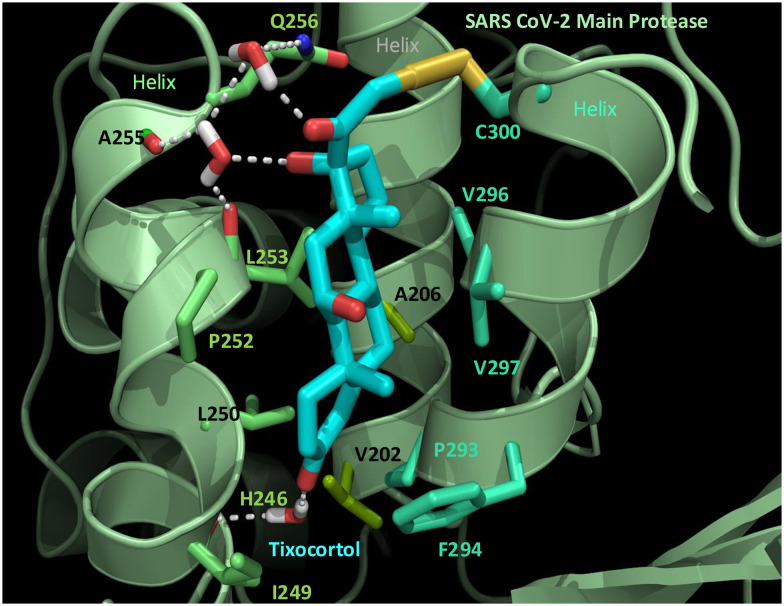
To identify which cysteines of Mpro were becoming modified by TX, we made a preparation of TX-modified Mpro (TX-Mpro), and then removed the excess TX using molecular weight cut-off filtration membranes. The final prep contained 49% of Mpro modified with one TX and 11% with two TX's (Fig. S9A and B†). As seen before, by extracting the ions associated with for each Mpro species, we could see that the single TX-modified Mpro eluted more as monomer than native Mpro while the 2TX-Mpro eluted >90% as monomer (Fig. S9C†). We next carried out chymotrypsin digestions of Mpro as described previously16 to determine which of the 12 cysteines of Mpro were becoming modified. Based on RP-HPLC/MS analysis we identified two tixocortol labeled peptides: 295DVVRQCTXSGVTF305 and 295DVVRQCTXSGVTFQ306 each harboring Cys300 (Table 1).16 Treatment of the peptide digest with the reducing agent TCEP revealed the masses of the native forms of these same peptides (Table 1 and Fig. S10A–F†). Unfortunately, we were not able to identify additional Cys residues modified with TX based on our analysis. However, it should be noted that alkylated peptides for Cys38, Cys44 and Cys160 were not detected (see Fig. S11† for list of peptides detected in the chymotryptic analysis) so these cysteines can't be ruled out as secondary targets for TX. To assess the extent to which TX modification of Cys300 was playing a role in the inhibition of Mpro activity, we tested the effect of TX on C300S Mpro, prepared as described previously.16 A time course study was carried out to assess the elution profile and modification rate C300S Mpro with TX (Fig. S12A†). Several differences from WT Mpro were noted. First, the monomer and dimer peaks for C300S treated with TX had retained the same retention times as the control over the 90 minute period examined (Fig. S12A†) which is unlike what we saw for WT Mpro where the monomer peak shifted to later elution times (see Fig. 2A and 5A). Second, protein deconvolution of the eluting C300S Mpro region (S12B and C†) revealed that only 23% of the total C300S Mpro was modified with a single TX even after 90 minutes of incubation (Fig. S12C†) as compared to 65% modification seen for WT under the same conditions (Fig. S12D†). Also, even after 90 minutes of incubation with TX, TX appeared to modify only a one cysteine in C300S (Fig. S12C†) consistent with Cys300 being one of the two cysteines modified in WT Mpro. In addition to less modification of C300S with TX, TX was also a weaker inhibitor of C300S Mpro compared to WT, but some inhibition was still observed (Fig. S12E†). TX at 12.5 μM inhibited 62% of WT Mpro activity as compared to approximately 27% inhibition of C300S Mpro (Fig. S12E†). To determine if the active site Cys145 might be the target in C300S modified with TX, we first occupied Cys145 by covalent modification with GRL-0920 since our studies showed >90% modification of Cys145 with this drug (see Fig. S3D†). When this was done, there was only a small decrease in the extent of TX modification (23.5% TX modification without GRL-0920 vs. 19.8% TX modification with GRL-0920) (Fig. S13A–C†) suggesting Cys145 is unlikely to be a primary target for TX in C300S Mpro. Together, these data support a primary role for Cys300 in the inhibition of Mpro activity with an additional role for another unidentified cysteine. Since Cys300 was identified as one of the primary targets for TX, we generated a biochemically feasible covalent binding model for the TX-Cys300 modification of Mpro (Fig. 7). A scheme for TX in inhibition of SARS-CoV-2 replication is proposed based on our data (Fig. S14†).
RP/HPLC/MALDI-TOF MS identification of TX modified Cys300 peptides from chymotryptic digestion.
| Peptide founda | TCEP | M r (calc) | M r (expt) | % TX or Alk |
|---|---|---|---|---|
| 295DVVRQCTXSGVTF305 | — | 1585.77 | 1584.80 | 25 |
| 295DVVRQCSGVTF305 | + | 1209.57 | 1209.60 | 75 |
| 295DVVRQCTXSGVTFQ306 | — | 1713.83 | 1713.00 | 37 |
| 295DVVRQCSGVTFQ306 | + | 1209.57 | 1209.60 | 63 |
The Cys300 295–306 peptide is a result of incomplete cleavage at the 305:306 predicted chymotryptic cleavage site. Peptide extracts were treated with or without TCEP to reveal the native reduced form of the TX modified peptides. Molecular masses for these peptides were confirmed with the use of synthetic peptides that were run on RP-HPLC/MALDI-TOF as native, alkylated or tixocortol modified peptides. Shown are the calculated native masses [Mr (calc)], the experimental masses [Mr (expt)]. The % modified shown in the last column was determined based on the area obtained for the native peptide after TCEP treatment to the total area for that peptide (native + alkylated). RTs for the two TX-peptides were 26.6 and 25.2 respectively, and 16.2 and 15.3 for the native forms.
Fig. 7. Molecular model of the interaction of TX with SARS-CoV-2 Mpro. TX (carbons in cyan) binds within a pocket formed by 3 helices (pale green cartoon). Displayed Mpro residues consist of the first neighbor contacts between Mpro and TX. These contacts comprise 10 hydrophobic residues and three polar residues (Cys 300, Gln 256, and his 246). In addition to the hydrophobic interactions, a hydrogen-bonded network (dashed white lines) bridges TX through three water molecules (rendered in stick) to the polar contacts in Mpro. Side chain carbon atoms are color-coordinated by helix (from left to right: Lime green, olive green and blue green). The TX-Cys300 disulfide bond (2.54 angstroms) is shown in yellow, nitrogen (blue), oxygen (red), and water hydrogens are displayed as white sticks. The proposed mechanism is the formation of a disulfide bond between TX and Cys300 of SARS-CoV-2 Mpro.
Discussion
We provide evidence that TX acts as an allosteric covalent inhibitor of Mpro, in part, by covalently modifying Cys300 in the C-terminal helical domain. TX also inhibits Mpro activity in cells and shows inhibitory activity in SARS-CoV-2 replication assays at low micromolar levels when preincubated with ACE2–HeLa cells. TP, the prodrug form of TX previously marketed as a nasal spray, undergoes conversion to TX in cells and showed good inhibitory activity against SARS-CoV-2 replication, although TP is associated with some cellular toxicity in this system. The disulfide of TX, TSST, more rapidly covalently modified and inhibited Mpro suggesting that the covalent attachment of TX to Mpro may first go through a TSST intermediate. Since longer preincubation times of HeLa–ACE2 cells with TP or TX may generate TSST within infected cells, this may explain the improved IC50's obtained with longer preincubation times. Based on our results, we propose the following scheme whereby cellular esterases remove the pivalate moiety leading to the production of TX. TX can then react with Mpro or undergo further conversion to TSST in cells under a pro-oxidant state, and this will more readily react with Mpro and form covalent disulfides with Cys300 of Mpro and another unidentified Cys residue(s) leading to disruption of Mpro dimerization, aggregation and loss of activity (Fig. S14†).
Cys300 is the most solvent-exposed cysteine residue in Mpro18,28 and we've previously demonstrated that modification of Cys300 with glutathione inhibits Mpro dimerization and activity.16 Tao et al. have previously demonstrated disruption of Mpro dimerization and activity by colloidal bismuth subcitrate through interaction with Cys300.18 It is not surprising that certain sulfhydryl reactive compounds have the potential to inhibit Mpro, given that Mpro contains 12 cysteines all in their reduced state. It has been suggested that the usefulness and specificity of various sulfhydryl reactive compounds for Mpro, for example ebselen or carmofur, would be better assessed with enzyme assays carried out under reducing conditions, by either including DTT, or perhaps more relevantly, the cellular thiol, glutathione.29,30 However, our studies here suggest that the reversibility of inhibition of Mpro by reducing agents may not mean that the compound would not be useful in cellular assays. Several active site inhibitors that covalently modify Cys145 have been developed, yet few compounds have been reported to inhibit Mpro by modifying cysteines other than the active site.18 Unlike active site inhibitors such as nirmatrelvir, which covalently modify the active site Cys145 and stabilize the dimeric state,25 TX appears to disrupt protease dimerization resulting in inhibition of activity. Although TX inhibition was partly reversible with GSH, TX was still able to inhibit Mpro expressed in cells and inhibit viral replication suggesting the reducing environment was not sufficient to prevent TX inhibition of Mpro. Due to several potential advantages with the use of nasal sprays for the treatment of SARS-CoV-2 infection, several groups have pursued their development for use in SARS-CoV-2 infections.31–34 Through intranasal delivery or inhalation, nasal sprays can be concentrated at the primary site of SARS-CoV-2 infection without causing systemic toxicity. They may also have use as prophylactic agents to prevent infection when an individual is exposed.35 TP is the prodrug form of TX; it was approved as a 1% nasal spray suspension for treating rhinitis in the mid 1980’s and was found to have an excellent safety profile.36,37 There is essentially no literature available on why TX was developed as a pivalate prodrug but it was perhaps done to prevent TX oxidation to TSST. However, TP is presumably hydrolysed to TX by cellular and other esterases to form the active steroid24 and we demonstrated that in this study as well. It should be noted that TP has glucocorticoid effects, which may also be of clinical benefit in SARS-CoV-2 infection.38 Although TX is less potent against Mpro (μM range vs. nM range) than other drugs, such as NM, the ability to use it as a nasal spray allows for application directly to the site of infection. TP is administered as a 21 mM suspension which greatly exceeds the micromolar concentrations of TP/TX required for inhibition of SARS-CoV-2. Our studies on HeLa–ACE2 cells demonstrate that preincubation of TX, TP or TSST improved the ability to block SARS-CoV-2 infection up to 7-fold, suggesting that TP may be useful as a prophylactic during potential exposure during crowded events or to prevent SARS-CoV-2 infection in individuals following potential exposure. To our knowledge there are no approved products for this purpose. Finally, although GSH appears to impair TX activity somewhat against Mpro, this may be less of an issue in SARS-CoV-2 infected cells as virus infection can lead to a prooxidant state with dramatically lower cellular thiol and GSH levels in SARS-CoV-2 infected Vero cells within the first 24 h of infection.27 These types of changes in redox parameters also appear to be involved in the pathogenesis of SARS-CoV-2-mediated disease.39–41 Interestingly, lowered cellular thiols in infected cells could promote the formation of TSST which was found in our hands to be the most active form of TX for inhibiting Mpro activity.
While the activity of TP and TX is somewhat less than systemic agents, such as nirmatrelvir, the ability to deliver it locally at the site of infection may compensate for this lower activity. Also, there is substantial clinical experience with TP use in humans as a nasal spray and it has an excellent safety profile. Given this, the results here suggest that it is worth exploring as a potential treatment or preventive agent for SARS-CoV-2. In addition, the results suggest that it may be worthwhile to identify or develop other compounds that target Cys300 pocket in Mpro.
Data availability
The data that support the findings of this study are in the main text and ESI.† Additional data that support the findings of this study are available from the author [D. A. D.], upon reasonable request and/or will be deposited in the Dryad repository.
Author contributions
D. A. D. and R. Y. conceived the project and designed the experiments. A. N. and P. S. designed and carried out the Mpro cellular assays, D. A. D., Y. A. and E. T. carried out the in vitro Mpro proteolytic assays, D. A. D. carried out the RP/HPLC/MS SEC assays. B. P., R. G. and T. N. carried out the compound identification and Mpro modelling assays for glutathione and TX. H. B. carried out Mpro protein purification. B. E., E. P., M. M., H. M, and S.-I. H. supervised and carried out the virus inhibition experiments.
Conflicts of interest
D. A. D, Y. A., B. P, R. G., T. N., R. Y. and B. E. are co-inventors on US provisional Patent, U.S. Patent application No. 63/599446 entitled “Tixocortol for use in the Prevention and/or Treatment of Coronavirus Infections”. This invention was made as full-time employees of the US government under 45 Code of Federal Regulations Part 7. All rights, title, and interest to these patents have been or should by law be assigned to the U.S. Department of Health and Human Services. The government conveys a portion of the royalties it receives to its employee inventors under the Federal Technology Transfer Act of 1986 (P. L. 99-502).
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute (grant number ZIA BC011945). We also thank Dr John Mieyal (CWRU). Dr Rodney Levine (NIH), and Douglas J. Underhill for their encouragement, helpful comments, and suggestions throughout this work.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4md00454j
References
- Harrison C. M. Doster J. M. Landwehr E. H. Kumar N. P. White E. J. Beachboard D. C. Stobart C. C. Evaluating the Virology and Evolution of Seasonal Human Coronaviruses Associated with the Common Cold in the COVID-19 Era. Microorganisms. 2023;11:445. doi: 10.3390/microorganisms11020445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M. Liang C. Human coronaviruses: The emergence of SARS-CoV-2 and management of COVID-19. Virus Res. 2022;319:198882. doi: 10.1016/j.virusres.2022.198882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboob T. Ismail A. A. Shah M. R. Rahmatullah M. Paul A. K. Pereira M. L. Wiart C. Wilairatana P. Rajagopal M. Dolma K. G. Nissapatorn V. Development of SARS-CoV-2 Vaccine: Challenges and Prospects. Diseases. 2023;11:64. doi: 10.3390/diseases11020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews H. S. Herman J. D. Gandhi R. T. Treatments for COVID-19. Annu. Rev. Med. 2024;75:145. doi: 10.1146/annurev-med-052422-020316. [DOI] [PubMed] [Google Scholar]
- Brewitz L. Dumjahn L. Zhao Y. Owen C. D. Laidlaw S. M. Malla T. R. Nguyen D. Lukacik P. Salah E. Crawshaw A. D. Warren A. J. Trincao J. Strain-Damerell C. Carroll M. W. Walsh M. A. Schofield C. J. Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine. J. Med. Chem. 2023;66:2663–2680. doi: 10.1021/acs.jmedchem.2c01627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemian S. M. R. Sheida A. Taghizadieh M. Memar M. Y. Hamblin M. R. Bannazadeh Baghi H. Sadri Nahand J. Asemi Z. Mirzaei H. Paxlovid (Nirmatrelvir/Ritonavir): A new approach to Covid-19 therapy? Biomed. Pharmacother. 2023;162:114367. doi: 10.1016/j.biopha.2023.114367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S. A. Banerjee S. Ghosh K. Gayen S. Jha T. Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg. Med. Chem. 2021;29:115860. doi: 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amani B. Amani B. Efficacy and safety of nirmatrelvir/ritonavir (Paxlovid) for COVID-19: A rapid review and meta-analysis. J. Med. Virol. 2023;95:e28441. doi: 10.1002/jmv.28441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D. W. Ju M. J. Wang H. Jia Y. C. Wang X. D. Fang H. Fan J. Proxalutamide for the treatment of COVID-19 rebound following Paxlovid treatment: Report of four cases and review of the literature. J. Clin. Lab. Anal. 2023;37:e24880. doi: 10.1002/jcla.24880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Jeffcoat S. E. Biddle J. E. Talbot H. K. Morrisey K. G. Stockwell M. S. Maldonado Y. McLean H. Q. Ellingson K. D. Bowman N. M. Asturias E. Mellis A. M. Johnson S. Kirking H. L. Rolfes M. A. R. Olivo V. Merrill L. Battan-Wraith S. Sano E. McLaren S. H. Vargas C. Y. Goodman S. Sarnquist C. C. Govindaranjan P. Petrie J. G. Belongia E. A. Ledezma K. Pryor K. Lutrick K. Bullock A. Yang A. Haehnel Q. Rao S. Zhu Y. Schmitz J. Hart K. Grijalva C. G. Salvatore P. P. Symptoms, viral loads, and rebound among COVID-19 outpatients treated with nirmatrelvir/ritonavir compared to propensity score matched untreated individuals. Clin. Infect. Dis. 2023;78:1175. doi: 10.1093/cid/ciad696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Song Y. Structure and function of SARS-CoV and SARS-CoV-2 main proteases and their inhibition: A comprehensive review. Eur. J. Med. Chem. 2023;260:115772. doi: 10.1016/j.ejmech.2023.115772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. Bartlam M. Rao Z. Drug design targeting the main protease, the Achilles' heel of coronaviruses. Curr. Pharm. Des. 2006;12:4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- de Souza A. S. de Souza R. F. Guzzo C. R. Quantitative structure-activity relationships, molecular docking and molecular dynamics simulations reveal drug repurposing candidates as potent SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2022;40:11339–11356. doi: 10.1080/07391102.2021.1958700. [DOI] [PubMed] [Google Scholar]
- Owen D. R. Allerton C. M. N. Anderson A. S. Aschenbrenner L. Avery M. Berritt S. Boras B. Cardin R. D. Carlo A. Coffman K. J. Dantonio A. Di L. Eng H. Ferre R. Gajiwala K. S. Gibson S. A. Greasley S. E. Hurst B. L. Kadar E. P. Kalgutkar A. S. Lee J. C. Lee J. Liu W. Mason S. W. Noell S. Novak J. J. Obach R. S. Ogilvie K. Patel N. C. Pettersson M. Rai D. K. Reese M. R. Sammons M. F. Sathish J. G. Singh R. S. P. Steppan C. M. Stewart A. E. Tuttle J. B. Updyke L. Verhoest P. R. Wei L. Yang Q. Zhu Y. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Joyce R. P. Hu V. W. Wang J. The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): an orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations. Med. Chem. Res. 2022;31:1637–1646. doi: 10.1007/s00044-022-02951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D. A. Bulut H. Shrestha P. Yaparla A. Jaeger H. K. Hattori S. I. Wingfield P. T. Mieyal J. J. Mitsuya H. Yarchoan R. Regulation of the Dimerization and Activity of SARS-CoV-2 Main Protease through Reversible Glutathionylation of Cysteine 300. MBio. 2021;12:e0209421. doi: 10.1128/mBio.02094-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrelle F. X. Boll E. Brier L. Moschidi D. Belouzard S. Landry V. Leroux F. Dewitte F. Landrieu I. Dubuisson J. Deprez B. Charton J. Hanoulle X. NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment. Angew. Chem., Int. Ed. 2021;60:25428–25435. doi: 10.1002/anie.202109965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X. Zhang L. Du L. Liao R. Cai H. Lu K. Zhao Z. Xie Y. Wang P. H. Pan J. A. Zhang Y. Li G. Dai J. Mao Z. W. Xia W. Allosteric inhibition of SARS-CoV-2 3CL protease by colloidal bismuth subcitrate. Chem. Sci. 2021;12:14098–14102. doi: 10.1039/D1SC03526F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi-Kuwata N. Tsuji K. Hayashi H. Bulut H. Kiso M. Imai M. Ogata-Aoki H. Ishii T. Kobayakawa T. Nakano K. Takamune N. Kishimoto N. Hattori S. I. Das D. Uemura Y. Shimizu Y. Aoki M. Hasegawa K. Suzuki S. Nishiyama A. Saruwatari J. Shimizu Y. Sukenaga Y. Takamatsu Y. Tsuchiya K. Maeda K. Yoshimura K. Iida S. Ozono S. Suzuki T. Okamura T. Misumi S. Kawaoka Y. Tamamura H. Mitsuya H. Identification of SARS-CoV-2 M(pro) inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2. Nat. Commun. 2023;14:1076. doi: 10.1038/s41467-023-36729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K. Ishii T. Kobayakawa T. Higashi-Kuwata N. Azuma C. Nakayama M. Onishi T. Nakano H. Wada N. Hori M. Shinohara K. Miura Y. Kawada T. Hayashi H. Hattori S. I. Bulut H. Das D. Takamune N. Kishimoto N. Saruwatari J. Okamura T. Nakano K. Misumi S. Mitsuya H. Tamamura H. Potent and biostable inhibitors of the main protease of SARS-CoV-2. iScience. 2022;25:105365. doi: 10.1016/j.isci.2022.105365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori S. I. Higashi-Kuwata N. Hayashi H. Allu S. R. Raghavaiah J. Bulut H. Das D. Anson B. J. Lendy E. K. Takamatsu Y. Takamune N. Kishimoto N. Murayama K. Hasegawa K. Li M. Davis D. A. Kodama E. N. Yarchoan R. Wlodawer A. Misumi S. Mesecar A. D. Ghosh A. K. Mitsuya H. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021;12:668. doi: 10.1038/s41467-021-20900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. Y. Krischuns T. Varga L. O. Harigua-Souiai E. Paisant S. Zettor A. Chiaravalli J. Delpal A. Courtney D. O'Brien A. Baker S. C. Decroly E. Isel C. Agou F. Jacob Y. Blondel A. Naffakh N. A highly sensitive cell-based luciferase assay for high-throughput automated screening of SARS-CoV-2 nsp5/3CLpro inhibitors. Antiviral Res. 2022;201:105272. doi: 10.1016/j.antiviral.2022.105272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle P. Du Souich P. Bolte E. Lelorier J. Goyer R. Tixocortol pivalate, a corticosteroid with no systemic glucocorticoid effect after oral, intrarectal, and intranasal application. Clin. Pharmacol. Ther. 1983;33:343–350. doi: 10.1038/clpt.1983.43. [DOI] [PubMed] [Google Scholar]
- Lelievre V. Junien J. L. Goyer R. Russo-Marie F. Affinity of tixocortol pivalate (JO 1016), tixocortol, cortisol acetate and cortisol for dexamethasone receptors of mouse thymus cells and rat renomedullary interstitial cells in culture. Correlation with their biological activities. J. Steroid Biochem. 1984;20:363–366. doi: 10.1016/0022-4731(84)90234-6. [DOI] [PubMed] [Google Scholar]
- Paciaroni A. Libera V. Ripanti F. Orecchini A. Petrillo C. Francisci D. Schiaroli E. Sabbatini S. Gidari A. Bianconi E. Macchiarulo A. Hussain R. Silvestrini L. Moretti P. Belhaj N. Vercelli M. Roque Y. Mariani P. Comez L. Spinozzi F. Stabilization of the Dimeric State of SARS-CoV-2 Main Protease by GC376 and Nirmatrelvir. Int. J. Mol. Sci. 2023;24:6062. doi: 10.3390/ijms24076062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori S. I. Higshi-Kuwata N. Raghavaiah J. Das D. Bulut H. Davis D. A. Takamatsu Y. Matsuda K. Takamune N. Kishimoto N. Okamura T. Misumi S. Yarchoan R. Maeda K. Ghosh A. K. Mitsuya H. GRL-0920, an Indole Chloropyridinyl Ester, Completely Blocks SARS-CoV-2 Infection. MBio. 2020;11:e01833-20. doi: 10.1128/mBio.01833-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini D. Stabile A. M. Bastianelli S. Giustarini D. Pierucci S. Busti C. Vacca C. Gidari A. Francisci D. Castronari R. Mencacci A. Di Cristina M. Focaia R. Sabbatini S. Rende M. Gioiello A. Cruciani G. Rossi R. Galli F. SARS-CoV2 infection impairs the metabolism and redox function of cellular glutathione. Redox Biol. 2021;45:102041. doi: 10.1016/j.redox.2021.102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk L. M. Poschmann G. Rabe von Pappenheim F. Chari A. Stegmann K. M. Dickmanns A. Wensien M. Eulig N. Paknia E. Heyne G. Penka E. Pearson A. R. Berndt C. Fritz T. Bazzi S. Uranga J. Mata R. A. Dobbelstein M. Hilgenfeld R. Curth U. Tittmann K. Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design. Nat. Commun. 2024;15:411. doi: 10.1038/s41467-023-44621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C. Tan H. Choza J. Wang Y. Wang J. Validation and invalidation of SARS-CoV-2 main protease inhibitors using the Flip-GFP and Protease-Glo luciferase assays. Acta Pharm. Sin. B. 2022;12:1636–1651. doi: 10.1016/j.apsb.2021.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C. Hu Y. Townsend J. A. Lagarias P. I. Marty M. T. Kolocouris A. Wang J. Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors. ACS Pharmacol. Transl. Sci. 2020;3:1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colado Simao A. N. Perugini Stadtlober N. Stinghen Garcia Lonni A. A. Venancio L. M. Lerner Trigo G. de Souza Cassela P. L. C. Mastellini Sanches Silva T. De Fatima Oliveira Hirth Ruiz M. Batisti Lozovoy M. A. Tano Z. N. da Fonseca Orcina B. Vieira Vilhena F. da Silva Santos P. S. Effect of phthalocyanine oral and nasal antiseptic solutions on the infectivity of SARS-CoV-2 in patients with COVID-19: a randomized controlled trial. Ger. Med. Sci. 2023;21:Doc07. doi: 10.3205/000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klussmann J. P. Grosheva M. Meiser P. Lehmann C. Nagy E. Szijarto V. Nagy G. Konrat R. Flegel M. Holzer F. Gross D. Steinmetz C. Scherer B. Gruell H. Schlotz M. Klein F. de Aragao P. A. Morr H. Al Saleh H. Bilstein A. Russo B. Muller-Scholtz S. Acikel C. Sahin H. Werkhauser N. Allekotte S. Mosges R. Early intervention with azelastine nasal spray may reduce viral load in SARS-CoV-2 infected patients. Sci. Rep. 2023;13:6839. doi: 10.1038/s41598-023-32546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nittayananta W. Lerdsamran H. Chutiwitoonchai N. Promsong A. Srichana T. Netsomboon K. Prasertsopon J. Kerdto J. A novel film spray containing curcumin inhibits SARS-CoV-2 and influenza virus infection and enhances mucosal immunity. Virol. J. 2024;21:26. doi: 10.1186/s12985-023-02282-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talotta R. Roberston E. S. Perspectives: potential therapeutic approach with inhalation of ACE2-derived peptides for SARS-CoV-2 infection. Am. J. Clin. Exp. Immunol. 2020;9:73–80. [PMC free article] [PubMed] [Google Scholar]
- Chavda V. P. Baviskar K. P. Vaghela D. A. Raut S. S. Bedse A. P. Nasal sprays for treating COVID-19: a scientific note. Pharmacol. Rep. 2023;75:249–265. doi: 10.1007/s43440-023-00463-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumowski P. de Gournay C. Girard J. P. Tixocortol pivalate and beclomethasone dipropionate in seasonal allergic rhinitis: a double-blind study. J. Int. Med. Res. 1985;13:328–331. doi: 10.1177/030006058501300605. [DOI] [PubMed] [Google Scholar]
- Chanoine F. Grenot C. Sellier N. Barrett W. E. Thompson R. M. Fentiman A. F. Nixon J. R. Goyer R. Junien J. L. Isolation and identification of major metabolites of tixocortol pivalate in human urine. Drug Metab. Dispos. 1987;15:868–876. [PubMed] [Google Scholar]
- Fernandez-Cruz A. Ruiz-Antoran B. Munoz-Gomez A. Sancho-Lopez A. Mills-Sanchez P. Centeno-Soto G. A. Blanco-Alonso S. Javaloyes-Garachana L. Galan-Gomez A. Valencia-Alijo A. Gomez-Irusta J. Payares-Herrera C. Morras-Torre I. Sanchez-Chica E. Delgado-Tellez-de-Cepeda L. Callejas-Diaz A. Ramos-Martinez A. Munez-Rubio E. Avendano-Sola C. A Retrospective Controlled Cohort Study of the Impact of Glucocorticoid Treatment in SARS-CoV-2 Infection Mortality. Antimicrob. Agents Chemother. 2020;64(9) doi: 10.1128/AAC.01168-20. https://dx.doi.org/10.1128/aac.01168-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone C. Bizkarguenaga M. Gil-Redondo R. Diercks T. Arana E. Garcia de Vicuna A. Seco M. Bosch A. Palazon A. San Juan I. Lain A. Gil-Martinez J. Bernardo-Seisdedos G. Fernandez-Ramos D. Lopitz-Otsoa F. Embade N. Lu S. Mato J. M. Millet O. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. iScience. 2020;23:101645. doi: 10.1016/j.isci.2020.101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarrere C. A. Kassab G. S. Glutathione deficiency in the pathogenesis of SARS-CoV-2 infection and its effects upon the host immune response in severe COVID-19 disease. Front. Microbiol. 2022;13:979719. doi: 10.3389/fmicb.2022.979719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhail S. Zajac J. Fossum C. Lowater H. McCracken C. Severson N. Laatsch B. Narkiewicz-Jodko A. Johnson B. Liebau J. Bhattacharyya S. Hati S. Role of Oxidative Stress on SARS-CoV (SARS) and SARS-CoV-2 (COVID-19) Infection: A Review. Protein J. 2020;39:644–656. doi: 10.1007/s10930-020-09935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are in the main text and ESI.† Additional data that support the findings of this study are available from the author [D. A. D.], upon reasonable request and/or will be deposited in the Dryad repository.