Abstract
We have characterized the interaction and nuclear localization of the nucleocapsid (N) protein and phosphoprotein (P) of sonchus yellow net nucleorhabdovirus. Expression studies with plant and yeast cells revealed that both N and P are capable of independent nuclear import. Site-specific mutagenesis and deletion analyses demonstrated that N contains a carboxy-terminal bipartite nuclear localization signal (NLS) located between amino acids 465 and 481 and that P contains a karyophillic region between amino acids 40 and 124. The N NLS was fully capable of functioning outside of the context of the N protein and was able to direct the nuclear import of a synthetic protein fusion consisting of green fluorescent protein fused to glutathione S-transferase (GST). Expression and mapping studies suggested that the karyophillic domain in P is located within the N-binding domain. Coexpression of N and P drastically affected their localization patterns relative to those of individually expressed proteins and resulted in a shift of both proteins to a subnuclear region. Yeast two-hybrid and GST pulldown experiments verified the N-P and P-P interactions, and deletion analyses have identified the N and P interacting domains. N NLS mutants were not transported to the nucleus by import-competent P, presumably because N binding masks the P NLS. Taken together, our results support a model for independent entry of N and P into the nucleus followed by associations that mediate subnuclear localization.
Plant-infecting members of the family Rhabdoviridae have been classified into two genera (56). Members of the genus Cytorhabdovirus replicate in the cytoplasm of infected cells, a trait they share with all rhabdoviruses infecting animal hosts. In contrast, members of the genus Nucleorhabdovirus, of which Sonchus yellow net virus (SYNV) is the most extensively characterized species, appear to replicate in the nucleus and undergo morphogenesis at the inner nuclear envelope. SYNV is transmitted by an aphid (Aphis coreopsidis) in nature, is widespread in sowthistle (Sonchus oleraceus L.) and beggar ticks (Bidens pilosa) in the southern United States, and occurs sporadically in lettuce (Lactuca sativa) in Florida (7, 20).
The bacilliform particles of SYNV are composed of a nucleocapsid core surrounded by a phospholipid membrane. The membrane fraction contains a glycoprotein (G) that protrudes from the surface of the virion (11) and an associated protein designated sc4 (46). The membrane fraction also contains an additional protein that is thought to be an analogue of the matrix protein (M) of the animal-infecting prototype rhabdovirus, Vesicular stomatitis virus (VSV) (16), which associates with the G protein and the nucleocapsid core during morphogenesis to stabilize the virus particle. The infectious SYNV core, which can be purified by density gradient centrifugation of nonionic-detergent-treated virions (53, 54), consists of the negative-strand genomic RNA (19) encapsidated by three core polymerase proteins. Structural studies reveal that purified nucleocapsid cores contain the nucleocapsid (N) protein (60), the phosphoprotein (P), formerly designated M2 (14), and the polymerase (L) protein (4). Thus, in many respects, the SYNV proteins correspond to those of the cytoplasmically replicating animal rhabdoviruses typified by VSV and Rabies virus. However, unlike its animal-infecting counterparts, SYNV replicates in virus-induced viroplasms located in the nuclei of infected cells (32). Data consistent with the formation of a nuclear viroplasm include the isolation of an active polymerase complex from the nuclei of infected cells that is capable of transcribing a polyadenylated leader RNA and the six SYNV mRNAs (53, 54). In situ hybridization studies have revealed that the minus-strand genomic RNA of SYNV is restricted to the nucleus while the plus-strand antigenomic RNAs are present in both the nucleus and the cytoplasm (32). Additional evidence consistent with a nuclear site for replication is the accumulation of the N and P proteins in the nuclei of virus-infected cells or in the nuclei of protoplasts infected with a plus-strand RNA virus vector that replicates in the cytoplasm (32). Treatment of SYNV-infected protoplasts with tunicamycin prevents morphogenesis and also results in the accumulation of viral core particles in the nucleus (52). These studies raise a number of questions relevant to the mechanisms involved in formation of viroplasms during nucleorhabdovirus infections. To begin to address these questions, we have analyzed the nuclear import of the N and P proteins in plant cells and have also found that Saccharomyces cerevisiae provides a useful model for refined analysis of nuclear import.
MATERIALS AND METHODS
General.
Healthy and infected Nicotiana benthamiana plants were maintained as described by Martins et al. (32). Standard methods were used to maintain yeast strains. Untransformed strains were maintained on YPD medium, and transformed strains were maintained on appropriate synthetic dropout media (12). Yeast transformations were conducted using a lithium acetate protocol (12, 18). The yeasts used in this study were the protease-deficient BJ2407 strain (MATa/MATα prb1-1122/prb1-1122 prc1-407/prc1-407 pep4-3/pep4-3 leu2/leu2 trp1/trp1 ura3-52/ura3-52) for nuclear localization and the EGY48 (MATa ura3-52 his3 trp1 leu2::LexAop6-LEU2) or PJ694A (MATa trp1-901 leu2–3,112 ura3–52 his3–200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-His3 met2::GAL7-lacZ) strain for two-hybrid analyses.
Restriction enzymes were obtained from New England Biolabs (Beverly, Mass.) or Promega (Madison, Wis.). Chemicals were purchased from Sigma Chemical (St. Louis, Mo.), or Fisher (Springfield, N.J.). Unless otherwise noted, all plasmids were maintained in Escherichia coli strain DH5α or TOP10. Large-scale preparation of plasmid DNA was performed using the polyethylene glycol precipitation method described by Nicoletti and Condorelli (39). DNA fragments were purified from gel slices using the UltraClean 15 kit from Mo Bio Laboratories (Solano Beach, Calif.), and site-specific mutagenesis of the N protein was conducted according to the Kunkel (25) method using the mutagenesis primers described in Table 1. Proteins were quantified using the Bio-Rad colorimetric protein assay II with gamma-globulin as a standard. Sodium-dodecyl sulfate-polyacrylamide gel electrophoresis was performed essentially as described by Laemmli (27) using a Bio-Rad mini-Protean system.
TABLE 1.
Synthetic oligonucleotides used in this study
| Name | Primer sequencea | Description |
|---|---|---|
| NIF | CGCGGATCCTTATGAGCACTACACCAACAATCACTCT | In frame with BamHI of pEG202 |
| N-GFP-F | CGCGGATCCATGAGCACTACACCAACAATCACTCT | In frame with BglII of pRSGFP-C1 |
| NIF-2 | CGCGAATTCATGAGCACTACACCAACAATCACTCT | In frame with EcoRI of pJG4-5 |
| N221F | GGGATCCCAATGACAAATGAGGAGATTGTTACCTTC | In frame with BamHI of pEG202 |
| N971F | GGGATCCTCATGAACACTCTTGATAATGGA | In frame with BamHI of pEG202 |
| N913F | GGAATTCTTATTAGGCATATATATTTAACGGTGAATTATATC | In frame with EcoRI of pEG202 |
| N587F | GGAATTCTTATTAGATGGATGATGATGTACTATAAA | In frame with EcoRI of pEG202 |
| N-KK | ACAACTGAGAAGCCTGCGGCGGCTCTCCCTGCA | N NLS KK→AA mutagenesis primer |
| N-RR | GCCCCATCCAGGAAAGCAGCGAGTGACGCTCTT | N NLS RR→AA mutagenesis primer |
| NR | GGGAATTCTTATTAAAAGTCCGGTATGTTTGGTAGTT | N full-length reverse primer |
| P1F | CGCGGATCCTATGGAAATCGATCCAAATTACGTTA | In frame with BamHI of pEG202 |
| P-GFP-F | CGCGGATCCGCAATGGAAATCGATCCAAATTACGTTA | In frame with BglII of pRSGFP-C1 |
| PR | GGGAATTCTTAGATATAGATGGGTGAACT | P full-length reverse primer |
| P247F | GGGATCCTTCAAGAGAACAGAGCAGGAGATCTCATA | In frame with EcoRI of pEG202 |
| P753F | GGGATCCTGATGAGGCAATCCAAAGAA | In frame with BamHI of pGAD |
| P377R | GGAATTCTTATTTGGTTATTAATTTGAGCTCATGGT | Reverse primer from ntb 877 |
| P377F | GGGATCCCATGAGCTCAAATTAATAACCAAA | In frame with BamHI of pGAD |
| P745R | GGAATTCTTATTTGGATTGCCTCATCACTGTGATAACTG | Reverse primer from nt 745 |
| P814F | GGAATTCATGTGGCAATCCAAAGAA | In frame with BamHI of pGAD |
| P27R | CAAGCTTTTAGTTAACGTAATTTGG | Reverse primer from nt 27 |
| P121R | GAATTCTTATGCTAGTATATCATCCTCTAA | Reverse primer from nt 121 |
| P60R | CAAGCTTTTACATCACTGTGCTCTT | Reverse primer from nt 60 |
| P120R | CAAGCTTTTATTCTCCATCACCTGC | Reverse primer from nt 120 |
| GFP-F | GGGATCCCCATGGGTAAAGGAGAAGAACTTTTC | GFP forward primer |
| GFP-R | GCTCGAGTTACTTGTATAGTTCATCCATGC | GFP reverse primer |
| GST-F | GCCCGGGAATGTCCCCTATACTAGGTTATTGGA | GFP forward primer |
| GFP:GST:KK-R | AAGCTTTCAAGCCTTCTTCGGCTTTCCACGCGGAACCAGATCCGA | Introduces KPKK (aa 458–461 of N) into GFP-GST |
| GFP:GST:-NP-R | AAGCTTTCACATATGAGTGTGCTTGTCGGAGGTACGCGGAACCAGATCCGA | Introduces NP-like NLS (aa 69–76 of N) into GFP-GST |
| GFP:GST:-RR-R | AAGCTTTCACCTTCTTTTCCTGGACGGAGATCCACGCGGAACCAGATCCGA | Introduces PSRKRR (aa 445–450 of N) into GFP-GST |
| GFP:GST:-WT-R | AAGCTTTCAGTCACGATGAATACCCGGAGAACCACGCGGAACCAGATCCGA | Maintains coding sequence of MCS of pGEX-2T |
| GFP:GST:-RR/sp | AAGCTTTCACTCAGTTGTAAGAGCGTCACTCCTTCTTTTCCT | aa 447–457 of N protein. Overlaps GFP-GST-RR-R |
| GFP:GST:-sp/KK | AAGCTTTCAAGCCTTCTTAGGCTTCTCAGTTGTAAG | aa 454–465 of N protein. Overlaps GFP-GST-RR/sp |
| GFP:GST:KK+7 | AAGCTTTTATTTGATTATTGCAGGGAGAGCCTTCTTAGG | aa 459–468 of N protein. Overlaps GFP:GSTsp/KK |
| GFP:GST:+7/end | AAGCTTTTAAAAGTCCGGTATGTTTGGTAGTTTGATTATTGCAGGGAG | aa 463–475 of N protein. Overlaps GFP:GST KK+7 |
| SV40apgpGFP-F | GGGATCCATGCCAAAAAAAAAACGTAAAGTTGCACCAGGCCCAATGGGTAAAGGAGAAGAACTTTTC | SV40 NLS and 4 aa spacer at 5′ end of GFP |
| SV40gnspGFP-F | GGGATCCATGCCAAAAAAAAAACGTAAAGTTGGAAACTCACCAATGGGTAAAGGAGAAGAACTTTTC | SV40 NLS and 4 aa spacer at 5′ end of GFP |
Restriction sites are in boldface.
nt, nucleotide.
Cloning of PCR products.
All oligonucleotide primers for PCR (Table 1) were purchased from Life Technologies (Bethesda, Md.) or Operon Technologies (Alameda, Calif.). Primer sequence names indicate the specific gene, N or P, followed by the location of the terminal nucleotide in the N or P sequence. The F or R designation denotes that the primer is a forward (5′) or a reverse (3′) primer. Forward primers contained 5′ BamHI restriction sites, and reverse primers contained 5′ stop codons followed by EcoRI sites, unless otherwise noted. PCRs were performed with the high-fidelity polymerase Vent (New England Biolabs), Pfu (Stratagene, La Jolla, Calif.), or Taq Polymerase-Hi Fidelity (Life Technologies). To take advantage of the efficiency of T/A cloning, PCR products were terminally deoxyadenylated with Taq polymerase by adding 1U of Taq polymerase and 2 μl of 10 mM dATP per 50 μl of PCR mixture and incubating the mixture at 65°C for 15 min. PCR products were cloned directly from this cocktail by topoisomerase cloning (pTOPO; Invitrogen).
Construction of plant and yeast expression cassettes.
Full-length clones of the N and P genes were PCR amplified using Vent polymerase from the PVXM2 and PVXN constructs described by Martins et al. (32). The PCR products were blunt-end ligated to EcoRV-digested pBluescriptII SK(+) vector (Stratagene). Recombinant plasmids were checked for orientation, and clones that could be liberated from the vector by BamHI and HindIII or XhoI double digestion were selected for further use. Three N clones (pSKN1, pSKN2, and pSKN6) and two P clones (pSKP3 and pSKP5) were selected for construction of expression plasmids. Sequence analyses, coupled in vitro transcription-translation using a T3 polymerase-rabbit reticulocyte system (TNT; Promega), and reactivity of N and P polyclonal antibodies to the correct-size polypeptides expressed in yeast confirmed that the five clones selected were in accordance with the published characteristics for the N and P genes (14, 60).
To generate yeast plasmids for the expression of native N protein, N gene BamHI/HindIII fragments were excised from the SK clones described above and ligated to BamHI/HindIII-digested YEp351-GAL1. This vector is a Leu2-selected 2μm yeast episomal shuttle vector whose foreign genes are expressed under the control of a galactose-inducible (GAL1) promoter (15). Native P protein-expressing constructs were generated by ligating the BamHI/XhoI P gene-containing fragment into BamHI/SalI-digested YEp353-GAL1, a Trp1-selected vector derived from YEp352-GAL1 which is identical to YEp351-GAL1 except that it has a Ura3-selectable marker instead of Leu2. Recombinant plasmids were transformed into the protease-deficient yeast strain BJ5460 (MATa ura3-52 trp1 lys2-801 leu2-1 his3-200 pep4::HIS3 prb1-1.6R can1 GAL) or BJ2407. Identical expression and localization results were obtained with either strain; however, the larger size of the diploid strain (BJ2407) facilitated the microscopy studies shown in the micrographs.
Green fluorescent protein (GFP) fusion constructs were generated using red-shifted GFP (pRSGFP-C1) from Clontech (Palo Alto, Calif.). The N- and P-containing SK clones described earlier were initially used for the production of the LexA two-hybrid constructs. These clones were in an inappropriate reading frame for cloning in frame with GFP, so a second set of N and P clones with the correct frame were generated as described above and subjected to translation and sequence analyses to confirm their utility. To generate GFP fusion derivatives, GFP was excised from pRSGFP-C1 as an NheI/BglII fragment, and the N and P genes were excised from the SK clones as BamHI/HindIII fragments. Triple ligations were performed using XbaI/HindIII-digested YEp351-GAL1 or YEp352-GAL1. Correct orientations were determined by restriction analysis, and fusion junctions were verified by sequencing. Since the GFP gene in pRSGFP-C1 does not contain a stop codon at the 3′ end of the gene, we introduced a TAA codon by PCR amplification to generate pTOPO-GFP. BamHI/XhoI double digestion of this plasmid liberated an approximately 700-bp fragment, which was cloned into BamHI/SalI-digested YEp352-GAL1.
In order to characterize the nuclear localization signal (NLS) of the N protein, we constructed a fusion between GFP and glutathione S-transferase (GST). GST was PCR amplified from pGEX-2T (Pharmacia, Piscataway, N.J.), cloned into pTOPO, and subcloned as a BamHI/EcoRI fragment in frame with GFP in BglII/EcoRI-digested pRSGFP-C1. The N NLS (PSRKRRSDALTTEKPKK) was incorporated into the carboxy terminus of the GFP-GST fusion by four rounds of PCR using overlapping 5′ primers that successively incorporated portions of the N NLS (Table 1). GFP-GST or GFP-GST-N NLS clones were subcloned from pTOPO vectors into pBEVY-GU (36) for expression in yeast cells or into pBS316 for transient expression in plant cells.
GST affinity assays.
To facilitate biochemical analyses, yeast vectors were constructed that expressed GST fused to the amino terminus of the N or P gene. The N and P genes were each cloned in frame with the GST coding region in pGEX-2T, and correct fusions were verified by expressing the recombinant plasmids in E. coli strain BL21(DE3)/pLysS (Novagen, Madison, Wis.). GST fusions were purified from induced cells using glutathione-Sepharose (Pharmacia) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Recombinant plasmids that yielded the correct-size fusion product were used as templates for Pfu-mediated PCR (Stratagene). PCR products were cloned into the pTOPO vector, and the GST fusion construct was released from the vector by a SmaI/EcoRI double digestion. The resulting GST fusion fragments were cloned into vectors, pBEVY-GT, pBEVY-GL, and pBEVY-L (36), and transformed into yeast strain BJ2407. The transformants were grown overnight in 5 ml of synthetic dropout (SD)-glucose medium, diluted fourfold in SD-glucose or SD-galactose, depending on the promoter present in the vector, and grown for a further 12 to 18 h at 28°C unless otherwise noted. The cells were harvested by centrifugation and broken by vortexing them in the presence of glass beads in yeast lysis buffer (0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, 10 mM Tris-HCl [pH 7.4], 1 mM phenylmethylsulfonylfluoride, 1 μg of antipain/ml, 1 μg of leupeptin/ml) (37). The cell lysates were clarified by centrifugation at 14,000 × g for 10 min at 4°C. The lysates were used immediately or were adjusted to 20% glycerol and stored at −80°C. GST fusions were purified by glutathione-Sepharose affinity chromatography (Pharmacia) according to the manufacturer's instructions.
Two-hybrid analyses.
Two yeast two-hybrid systems were utilized in this study. The majority of the experiments to identify interacting domains in the N and P proteins were performed using the LexA system developed by Gyuris et al. (13) in the yeast strain EGY48. All binding domain fusions were generated by cloning BamHI/XhoI fragments into pEG202 digested with these two enzymes. Activation domain fusions were generated using the vector pJG4–5. For experiments to test the effects of P on the N-N interaction, the plasmid YEp352GAL1-P or YEp352GAL1-GFP:P was used. Additional two-hybrid experiments were conducted with the GAL4-based system described by James et al. (21) in the yeast strain PJ694A. Binding domain fusions were constructed using the pGBDU Ura3-selected vector, while activation domain fusions were generated using the pGAD vectors.
Deletion mutants of N and P were cotransformed into EGY48 along with pJG4-5 alone or pJG4-5 containing full-length N or P in frame with the activation domain. Transformed colonies were first selected on minimal medium containing glucose as a carbon source, and interactions were tested on minimal medium containing glucose or galactose. Cotransformed yeast colonies capable of growth on galactose medium lacking leucine but not on glucose medium lacking leucine were scored as positive interactions. Unfortunately, the LexA fusion constructs containing the carboxy terminus of the P protein, but not the full-length fusion, activated transcription in the absence of pJG4-5 constructs. To overcome this problem, the yeast strain PJ69-4A was used to provide a more stringent activation system (21). When using this strain, we utilized the Ade2 reporter, which is the most stringent of the three reporter genes in the PJ69-4A system.
Transient-expression assays in plant cells.
To generate constructs for transient expression in plant cells, DNA fragments containing the GFP-N fusion from YEp351-GFP:N and GFP-P from YEp352-GAL1 were isolated as BamHI/EcoRI fragments and ligated to BglII/EcoRI-digested pBS316. This vector, which was derived from pBluescript KS(+), contains in the following (5′-3′) order a T3 promoter, a Cauliflower mosaic virus 35S promoter, a BglII/ClaI/SmaI/XhoI/EcoRI-multiple cloning site, and the nopaline synthase 3′ transcriptional terminator. For GFP expression, the 700-bp BamHI/XhoI GFP-containing fragment from pTOPO-GFP was isolated from agarose gels and ligated to BglII/XhoI-digested pBS316. Native N and P transient-expression plasmids were generated by ligating the BamHI/EcoRI N or P gene fragments from pSKN1, pSKN6, pSKP3, or pSKP5 to BglII/EcoRI-digested pBS316.
Particle bombardments (10, 23) were conducted on the adaxial surfaces of fully expanded greenhouse-grown N. benthamiana leaves. Sixty micrograms of 1-μm-diameter tungsten beads (Bio-Rad) were washed in 1 ml of 70% ethanol at room temperature for 15 min. Following a 5-min centrifugation, the supernatant was removed and the pellet was washed three times without resuspending the beads. The beads were resuspended in 1 ml of 50% glycerol and stored at −20°C for up to 3 weeks. To prepare the beads prior to bombardment, 50 μl of the bead-glycerol suspension was added to a 1.5-ml microcentrifuge tube. The following reagents were added in order with vortex mixing prior to the addition of each reagent: 5 μl of plasmid DNA (at 1 μg/μl in water), 50 μl of 2.5 M CaCl2, and 20 μl of freshly prepared 0.1 M spermidine. The resultant suspension was incubated on ice for 15 min with intermittent mixing every 1 to 2 min to keep the beads suspended. The beads were collected by a 5-s pulse centrifugation. After the supernatant was removed, the beads were washed with 140 μl of 70% ethanol followed by the same volume of 100% ethanol without resuspension between washes. The beads were then resuspended in 50 μl of 100% ethanol. Six microliters of this suspension was dried on the center of an aluminum foil rupture disk, and the disk was placed DNA-side down in the acceleration chamber of a helium carrier “gene gun” constructed by the staff in the machine shop of the Department of Molecular and Cell Biology at the University of California (UC)—Berkeley. Leaves to be bombarded were placed approximately 6 cm from the acceleration chamber of the gene gun on 100-mm-diameter petri plates containing Mirashige-Skoog salts agar lacking sucrose. Bombardments were conducted using helium as a carrier gas at 350 kPa in a −20 InHg vacuum.
Epifluorescence and laser scanning confocal microscopy.
Epifluorescence microscopy was conducted using a Zeiss Axiophot microscope. Yeast cells (3 ml) were collected at mid-log growth phase, centrifuged at 3,000 × g for 1 min in an Eppendorf microcentrifuge, and prepared for fluorescence microscopy by freezing the pellet in powdered dry ice for 10 min. The frozen pellets were resuspended in 1 ml of 0.4 M sorbitol in phosphate-buffered saline (PBS) (5 mM Na2HPO4, 5 mM NaH2PO4 [pH 6.8], 0.15 M NaCl) containing 4′-6′-diamidino-2-phenylindole (DAPI) at 1 μg/ml. Following resuspension, the cells were incubated at room temperature for 20 min on a rocker-shaker (Tek-Pro; American Dade, Miami, Fla.). Images were captured using NIH Image as modified by Scion and renamed Color Image 1.60 for CG-7. DAPI staining was visualized by UV epifluorescence through a band pass 450- to 490-nm cutoff filter. GFP fluorescence was visualized by epifluorescence through a long pass 520-nm cutoff filter. Captured images were converted from 72 to 300 dots/in in Adobe Photoshop to generate high-resolution micrographs. All subsequent image manipulation and figure preparation were carried out in Adobe Photoshop 5.0 and Canvas 5.03.
Confocal analyses were conducted with a Molecular Dynamics (Sunnyvale, Calif.) Sarastro 1000 laser scanning confocal microscope. As a UV laser was unavailable on this microscope, propidium iodide was used instead of DAPI to stain plant cell nuclei. Bombarded leaves expressing GFP were submerged 18 h postbombardment in PBS containing 0.5% NP-40 and 0.1 μg of propidium iodide/ml. The submerged leaves were incubated at room temperature for 3 h, rinsed once in PBS, and examined. GFP was visualized using a 530-nm excitation filter long pass, and captured images were manipulated as described above.
RESULTS
N and P GFP fusions localize to the nuclei of plant and yeast cells.
In previously published experiments with N. benthamiana plants, we obtained immunocytological evidence that the N and P proteins accumulate in the nuclei of SYNV-infected plant cells or in cells infected with potato virus X vectors that expressed N or P (32). In addition, fusions of N and P to the carboxy terminus of β-glucuronidase resulted in nuclear staining following inoculation with the potato virus X derivatives (32). Limited mapping studies were conducted with the N fusion derivatives, but these experiments were complicated by an inability to obtain reproducible coinfections with the vectors and by instability of the constructs during systemic invasion. Therefore, alternative strategies of expression in plant and yeast cells were investigated in the present study to more clearly define the N and P NLSs.
In order to provide a visible and nondestructive assay for detection of the localization of N and P in vivo, the N and P proteins were fused to the carboxy terminus of GFP. After biolistic introduction of transient-expression plasmids into N. benthamiana leaf tissue, both the GFP-N and GFP-P proteins exhibited fluorescence in the nuclei (Fig. 1). GFP-N fluoresced in relatively discrete regions of the nuclei, whereas GFP-P fluoresced throughout most of the nucleus, although some cytoplasmic fluorescence was also observed. Examination of plant nuclei under high magnification suggested that the area occupied by GFP-N was smaller than that occupied by GFP-P but was distributed throughout the majority of the nuclear volume (Fig. 1).
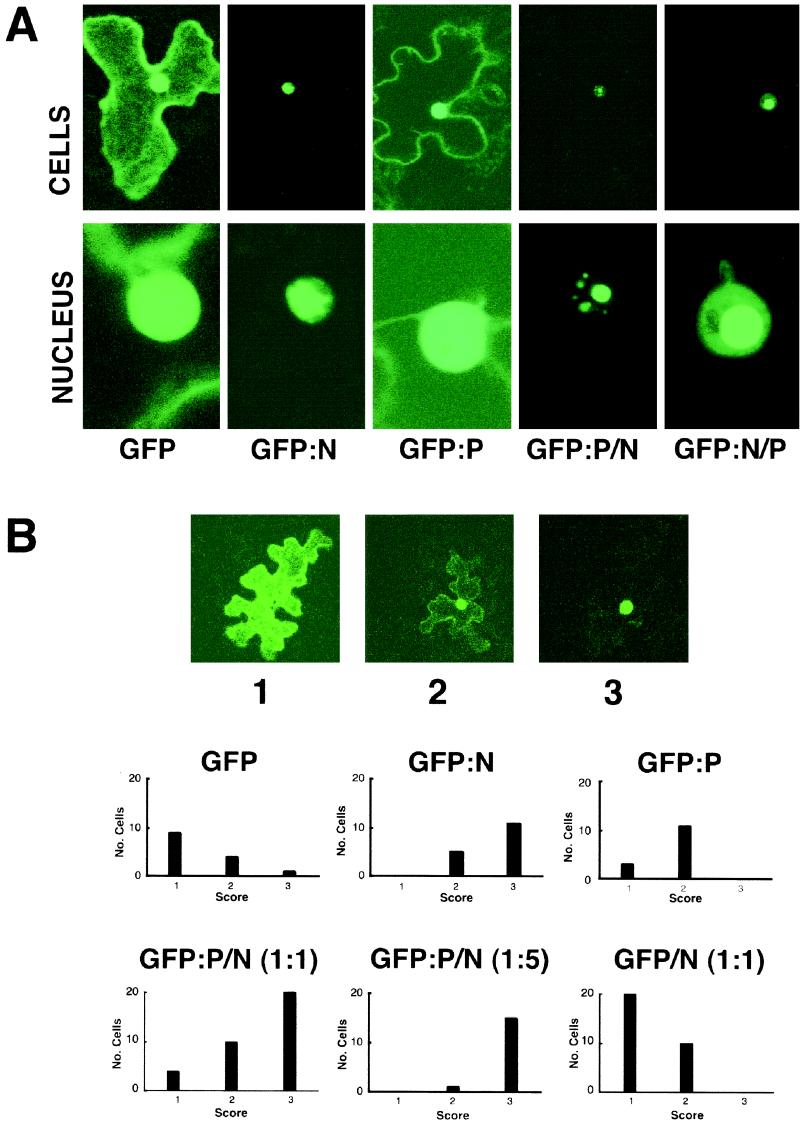
FIG. 1.
Localization of GFP constructs during transient expression in plant cells. (A) GFP fluorescence in epidermal cells of Nicotiana benthamiana 24 h after bombardment of leaves with GFP, GFP-N, and GFP-P plasmids and cobombardment with GFP-P and native N and with GFP-N and native P plasmids. The top row shows single epidermal cells, and the bottom row focuses on the nuclei of those cells. (B) Quantitative examination of bombarded leaves expressing GFP fusions. Micrographs 1, 2, and 3 illustrate categories of expression of single plasmid bombardments resulting from transient expression of GFP, GFP-P, and GFP-N. The three different categories of expression are designated type 1 (mostly cytoplasmic), type 2 (mostly nuclear), and type 3 (completely nuclear). The histograms show the results following cobombardment of GFP-P and native N plasmids at 1:1 and 1:5 ratios and cobombardment of a 1:1 ratio of GFP and native N plasmids. Note that cobombardment of the N and P plasmids at 1:1 and 1:5 ratios resulted in progressive shifts of the proportion of cells showing complete nuclear localization of fluorescence, whereas cobombardment of GFP plasmids and N plasmids at 1:1 ratios failed to result in substantial shifts in the patterns of nuclear localization from those seen with GFP alone.
When plant cells were cobombarded with plasmids expressing GFP-P and N, a different localization pattern emerged (Fig. 1). In these cases, a substantial proportion of the nuclei exhibited highly localized fluorescence of GFP-P in subnuclear sites. Identical results were obtained in reciprocal experiments using GFP-N/P. Since the bombardment system is amenable to titration experiments, the GFP-P plasmids were cobombarded with various amounts of the N plasmid. As the ratio (wt/wt) of the GFP-P/N-expressing plasmids was increased from 1:0 to 1:5, the probability of moving GFP-P completely to the nucleus increased from 9 to 94% (Fig. 1B). This effect was specific for N and P coexpression, because control experiments indicated that there was no effect on GFP localization during coexpression with N or on GFP-P localization by coexpression of the VirD2 protein of Agrobacterium tumefaciens, which is capable of independent nuclear import (data not shown). The GFP fusion results gave higher resolution than the β-glucuronidase fusion experiments conducted by Martins et al. (32), and they provide additional evidence that coexpression of N and P results in localization of a complex to discrete regions of the nuclei.
In addition to experiments conducted with plants, we also expressed the N and P proteins in yeast to determine whether this system might reflect the localization observed in plant cells and thus provide a system suitable for molecular genetic analysis of nuclear localization. Despite differences in the sizes of yeast nuclei (≈1- to 2-μm diameter) in comparison to those of plants (≈10-μm diameter), yeasts have the distinct advantage that coexpression of proteins can be obtained in all cells and the uniformity of fluorescence of cells expressing GFP-P or GFP-N greatly increases the efficiency and reproducibility of the assays.
Extensive observations indicated that the nuclear localization patterns of the N and P proteins in yeast cells were very similar to those observed in plant cells when GFP-P and GFP-N were expressed individually (Fig. 2A and B) or coexpressed with the N or P protein, respectively (Fig. 2C). Although cytoplasm fluorescence was not evident at 30°C from N protein expressed alone from episomal plasmids and N antigen could not be detected in Western blots of yeast cell extracts, high levels of fluorescence appeared throughout the nucleus at 22°C (Fig. 2A). GFP-P, when expressed alone, also exhibited bright fluorescence that occupied the entire nucleus and, as was the case with bombarded plant cells, fluorescence was also detected around the peripheries of the cells (Fig. 2B). As was the case with the plant cell titration experiments, coexpression of N or P with their heterologous GFP fusion proteins resulted in a striking relocalization of fluorescence to subnuclear foci at both 22 and 30°C. Thus, the expression patterns in yeast appeared to be very similar to those observed in plant cells, and coexpression of P appeared to have a major effect on the stability of N at 30°C.
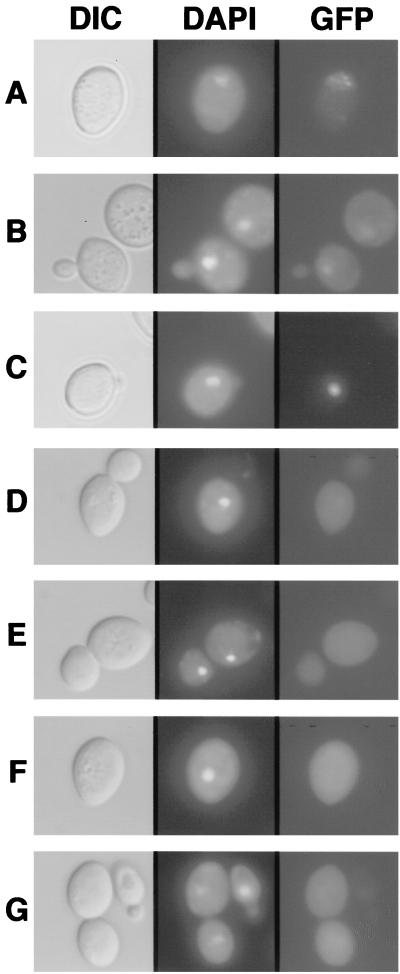
FIG. 2.
Localization patterns of the N and P proteins in yeast cells. The left column shows differential interference contrast (DIC) micrographs, the central column illustrates UV epifluorescence micrographs of DAPI-stained cells (DAPI), and the right column shows GFP fluorescence of yeast cells expressing GFP-N (A), GFP-P (B), GFP-P and N (C), GFP-GST (D), N and GFP-GST (E), P and GFP-GST (F), and N, P, and GFP-GST (G). Note that the yeast expression patterns corresponded to those obtained with plant cells bombarded with transient-expression plasmids. GFP-N was predominately nuclear, and GFP-P exhibited both nuclear and cytoplasmic staining, whereas coexpression of GFP-P and N resulted in a pronounced subnuclear fluorescence. In contrast, patterns of GFP-GST expression were unaffected and remained cytoplasmic in the presence of N and P. The positions of the nuclei were determined by counterstaining with the fluorescent DNA selective dye DAPI. The yeasts were grown overnight in SD-glucose medium and harvested by centrifugation. The pellets were resuspended in SD-galactose medium to an A600 of 0.2 and grown for an additional 12 to 18 h in appropriate selection medium. Expression of the proteins was verified by Western blotting (data not shown).
Control experiments were conducted to determine whether N or P expression might have some aberrant effects on unfused-GFP localization. These results suggest that neither of these proteins when expressed alone or together affected the localization of unfused GFP, which occurs throughout the cell (Fig. 2D to G). Control experiments also revealed no discernible difference in the localization of GFP-P, GFP-P/N, GFP-N/P, or GFP-P/GFP-N expressed at 22 or 30°C (data not shown). To determine whether relocalization of N and P after coexpression might be due to interference with nuclear import per se, we investigated the effects of N and P expression on the nuclear import of other proteins. For these experiments, N or P was coexpressed with a nucleoplasmin-GFP fusion protein that localizes completely to the nuclei in yeast cells (28). Expression of N or P singly or together did not affect the import of nucleoplasmin into the nucleus, nor was the compartmentalization of nucleoplasmin altered (not shown). These results, in conjunction with the GFP controls described above, support our contention that relocalization to a subnuclear compartment requires N-P interactions.
N protein contains a C-terminal bipartite NLS.
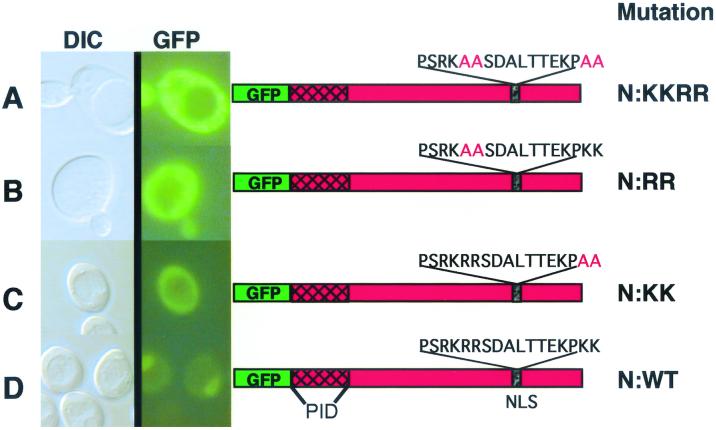
Given that both N and P are capable of independent nuclear import, a computer analysis was conducted to determine karyophillic regions in these proteins. In the previous study (32), we postulated that N must contain NLSs and noted a bipartite signal near the carboxy terminus. However, the exact requirement for this motif was not clearly resolved. In the present study, we first investigated the NLSs in the N protein by fusing portions (ca. 100 amino acids [aa]) of the protein to GFP and expressing these fusions in yeast and plant cells. However, these constructs were highly unstable and could not be detected immunologically or by fluorescence. Therefore, to further address the functionality of the putative NLSs in the N protein, mutations were introduced into the protein by site-specific mutagenesis. For this purpose, we concentrated on the carboxy-terminal signal between aa 465 and 481 predicted to contain the bipartite motif. This motif contains arginine-rich PSRKRR (aa 465 to 470) and lysine-rich KPKK (aa 478 to 481) regions separated by seven residues. Site-specific mutagenesis was used to introduce alanines in both portions of the putative bipartite signal. The sequence PSRKRR was changed to PSRKAA (henceforth called the N-RR mutant), and KPKK was changed to KPAA (the N-KK mutant). The mutant N proteins were then expressed in yeast as GFP-N fusions or as untagged proteins coexpressed with GFP-P. As shown in Fig. 3, mutations in either of the two C-terminal signals resulted in GFP fusions that failed to exhibit nuclear fluorescence. Therefore, basic residues within each component of the motif are required for nuclear import of the N protein.
FIG. 3.
Mutational analyses of the carboxy-terminal NLS in the N protein. Shown are differential interference contrast microscopy (DIC) and UV epifluorescence microscopy of yeast cells transformed with plasmids expressing GFP fusions to mutant (A to C) or wild-type (D) N proteins. Note that the GFP–wild-type N fusion (N:WT) localized almost completely to the nucleus, whereas the N-RRKK, N-RR, and N-KK amino acid substitutions of the italicized residues of the bipartite nuclear localization signal (PSRKRRSDALTTEKPKK) resulted in pronounced cytoplasmic fluorescence. The location of the P-interaction domain (PID) of N as defined by two-hybrid experiments is shown. The yeasts were grown and visualized as outlined in the legend to Fig. 2.
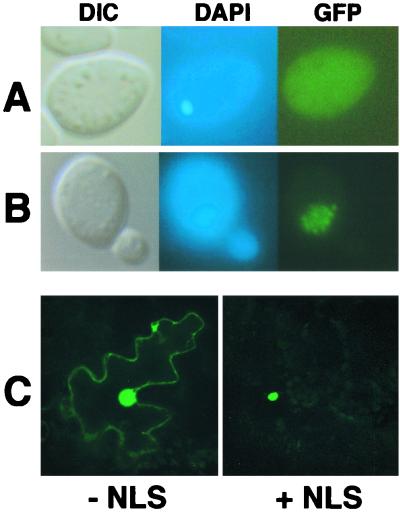
In order to determine whether the bipartite N NLS is sufficient for nuclear localization, a GFP-GST fusion was constructed to serve as a fluorescent probe for NLS function. GST remains primarily cytoplasmic when introduced into mammalian cells (41), and GFP-GST fusions have recently provided valuable markers for nuclear transport experiments (3, 45). As was the case with mammalian cells, a general cytoplasmic fluorescence was observed during expression of GFP-GST in yeast (Fig. 4A). Expression of GFP-GST-N NLS resulted in fluorescence only within the nucleus (Fig. 4B). Identical results were obtained when the simian virus 40 (SV40) large-T-antigen NLS (PKKKRVK) was incorporated into GFP-GST in place of the N NLS (data not shown). Although substantial nuclear fluorescence was noted in plant cells bombarded with the GFP-GST plasmids, fluorescence was restricted entirely to the nucleus in plant cells bombarded with the GFP-GST-N NLS fusion (Fig. 4C). In contrast, a second putative N protein NLS (TSDKHTHM), which resides between residues 69 and 76 and resembles the influenza NP NLS (35, 57), failed to alter the fluorescence patterns when fused to GFP-GST (data not shown). These results demonstrate that the carboxy-terminal N protein NLS is capable of functioning outside of its native environment and that the N protein influenza NP-like motif does not provide an independent nuclear localization function. Therefore, the N protein NLS signal appears to encompass the carboxy-terminal bipartite motif.
FIG. 4.
Fusion of the N NLS to a heterologous protein. A synthetic GFP-GST fusion was used as a fluorescent probe to determine if the N NLS was capable of functioning outside of the context of the N protein in both yeast and plant cells. (A and B) Images of yeast cells examined by differential interference contrast (DIC) and epifluorescence of DAPI-stained (DAPI) cells and GFP fluorescence images (GFP). (A) GFP-GST expression showing nonspecific accumulation of the fluorescent probe. (B) GFP-GST-N NLS expression showing a shift in accumulation to the nucleus. The yeasts were grown and visualized as outlined in the legend to Fig. 2. (C) Laser-scanning confocal micrographs of N. benthamiana leaf epidermal cells expressing GFP-GST without (− NLS) or with (+ NLS) an incorporated N NLS. GFP fluorescence was found exclusively in the nucleus in those cells expressing GFP-GST-N NLS
Carboxy-terminal fusions of each of the basic components of the bipartite motif to GFP-GST were also constructed to determine whether either of these constituents could function as a nuclear import signal. Neither the PSRKRR (aa 465 to 470) nor the KPKK (aa 478 to 481) element was able to mediate nuclear import of GFP-GST (not shown). These results suggest that the complete bipartite NLS (aa 465 to 481) is required for N protein recognition by the nuclear import apparatus, and the site-specific mutations introduced into the intact N protein indicate that arginine and lysine residues within each component of the bipartite motif provide critical import recognition determinants.
P cannot facilitate nuclear import of the N protein.
Because coexpression of N and P results in subnuclear localization of these proteins, we investigated whether P could facilitate the nuclear import of cytoplasmically restricted N mutants lacking functional NLSs. The ability of one protein to assist in the nuclear import of another has been demonstrated in a number of instances. For example, the VirD2 protein of A. tumefaciens can mediate nuclear import of cytoplasmically localized VirD1 (43). To determine whether P is capable of providing import assistance for N, the P protein was coexpressed with GFP-N NLS mutants or GFP-P was coexpressed with NLS-mutagenized N protein. In both cases, a highly localized pattern of GFP fluorescence resulted that was superficially similar to the compact subnuclear fluorescence observed with the wild-type fusion proteins. However, DAPI staining revealed that this complex was restricted to cytoplasmic aggregates rather than localized to the nuclei (Fig. 5). This highly discrete area of GFP fluorescence appeared to be very similar to the “false nuclei” observed when nuclear import of GFP-nucleoplasmin is blocked in yeast cells (22). In toto, the results of these experiments revealed that P could not facilitate the nuclear localization of N NLS mutants. The experiments also suggested that the import-compromised mutant N protein retained the ability to interact with P in the cytoplasm and that this interaction interfered with the normal nuclear import of the P protein.
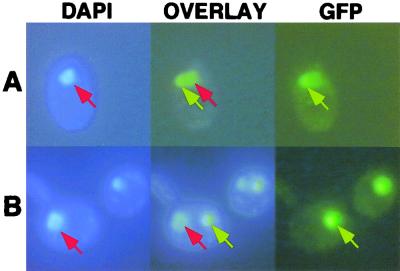
FIG. 5.
Inability of P to facilitate nuclear localization of N NLS mutants. Wild-type GFP-N (A) or GFP-N NLS (B) mutant coexpressed with P. DAPI-stained cells are shown on the left. GFP epifluorescence micrographs are shown on the right, and overlays of these images are shown in the middle. Wild-type proteins accumulated in a subnuclear site (top row), and the NLS mutant combination accumulated in cytoplasmic aggregates (bottom row). The yeasts were grown and visualized as for Fig. 2. The red arrows indicate sites stained with DAPI, while GFP fluorescence is indicated by yellow arrows.
The P protein contains an amino-terminal karyophillic region.
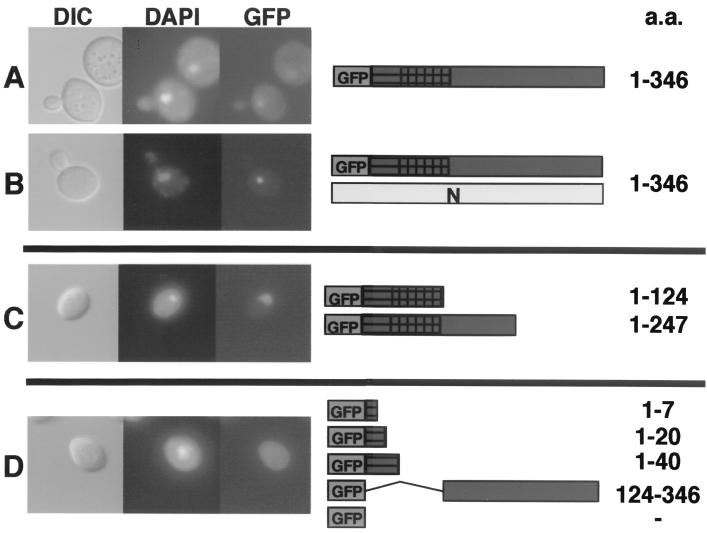
The ability of N NLS mutants to retard the nuclear import of the unmutagenized GFP-P protein prompted us to determine the location of the P NLS. The two-hybrid mapping experiments described below suggested that the amino terminus (aa 1 to 124) of P contains the N-interacting domain. When this domain (P-124) was fused to GFP, it was capable of independent localization to the nucleus (Fig. 6C). Further deletion analyses revealed that only those GFP-P deletions containing an 84-aa fragment (aa 40 to 124) were capable of directing GFP to the nucleus when expressed in yeast cells (Fig. 6C). Thus, amino acids residing between positions 40 and 124 appear to contain the P karyophyllic domain, because GFP-P fusions lacking this domain failed to exhibit nuclear fluorescence (Fig. 6D). Moreover, when the GFP-P deletions were coexpressed with N, relocalization of the GFP signal to a subnuclear locale was not observed.
FIG. 6.
Deletion analysis of the P protein karyophillic domain. Full-length copies or portions of the P protein were cloned in frame with GFP and expressed in the presence of N or without the N protein. (A) Wild-type GFP-P had nuclear and cytoplasmic accumulation when expressed alone. (B) GFP-P coexpressed with N resulted in a complete shift of fluorescence to the nucleus. (C) GFP-P lacking the C terminus of P exhibited a shift from the cytoplasm to the nucleus. (D) GFP-P deletions lacking the N terminus of P, aa 40 to 124, accumulated in the cytoplasm. Coexpression of the N protein did not affect the localization of the GFP-P deletions (data not shown). The yeast cells were grown and visualized as described in the legend to Fig. 2.
The N and P proteins interact physically.
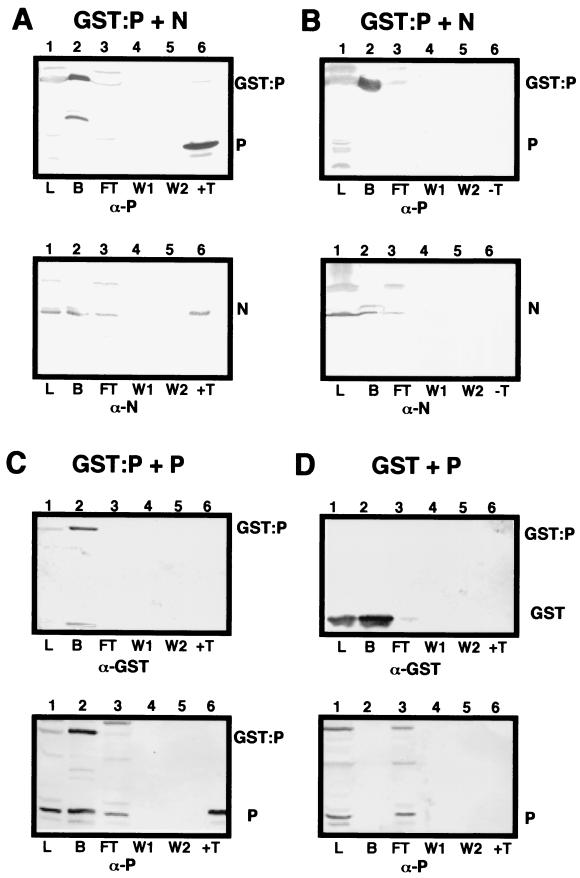
Our previous studies (54) have revealed that N, P, and L interact with the genomic RNA to form a rapidly sedimenting nucleocapsid polymerase core complex. More slowly sedimenting fractions also contain complexes of N-P and P-P (54). To confirm the N and P interactions, supporting results were obtained by GST-N and GST-P affinity chromatography. In yeast cells, coexpression of native N protein with GST-P resulted in the formation of N-P complexes that could be recovered by glutathione affinity chromatography following purification from cell lysates and thrombin digestion (Fig. 7). Similar results were obtained during chromatography of GST-N and native P, although much lower levels of GST-N than of GST-P were recovered from cell lysates (data not shown). The data in Fig. 7 show that N and GST-P complexes are retained on Sepharose columns (Fig. 7A, lane 2) and were not removed by extensive washing (Fig. 7A, lanes 4 and 5). However, thrombin digestion liberated both N and P (Fig. 7A, lane 6). Overnight incubation in wash buffer lacking thrombin did not result in leaching of either GST-P or N from the column, suggesting that these proteins are tightly bound to each other (Fig. 7B, lane 6). Further controls indicated that neither the native N nor P proteins bound to GST (Fig. 7C and D). Therefore, these results clearly indicate that N-P interactions similar to those observed in plants also occur in yeast cells. Homologous P-P interactions were also revealed by incubating lysates containing GST-P and P with glutathione-Sepharose. After extensive washing, both proteins remained bound to the column (Fig. 7D, lane 2), and additional washing did not result in further leaching of these proteins from the column (Fig. 7C, lanes 4 and 5). These results verify that both N-P and P-P interactions occur during expression in yeast cells.
FIG. 7.
Glutathione affinity chromatography of N and P interactions with GST-P. Lanes 1, total yeast lysate (L) containing GST-P or GST coexpressed with either the N or P protein; lanes 2, fraction bound (B) to glutathione-Sepharose columns; lanes 3, flow-through (FT) fraction; lanes 4, first 5-ml wash (W1); lanes 5, second 5-ml wash (W2); lanes 6, thrombin (T) digestion to release the P protein from GST. In control experiments, the columns were incubated overnight in wash buffer lacking thrombin (−T), and the resulting eluate was checked for the presence of GST-P, N, and P. (A) Yeast lysate containing GST-P coexpressed with N incubated with glutathione-Sepharose beads. On top is shown a Western blot probed with P-specific antibodies (α-P), and below is shown a replica blot probed with N-specific antibodies (α-N). GST-P and N were detected in lanes 1 (L), 2 (B), and 3 (FT). P, N, and the GST fusion were not detected in the wash fractions (lanes 4 and 5). After thrombin digestion, P was released from the column along with the N protein (lanes 6, top and bottom). (B) Control experiment identical to that shown in panel A except that the column was incubated overnight in wash buffer lacking thrombin. Note that neither GST-P nor the N protein was released from the column (lanes 6), despite being present in the bound fraction (lanes 2, top and bottom). ( C) Expression of GST-P plus P. Native P bound to GST-P (lanes 2) and was retained on the column during both washes (lanes 4 and 5). Thrombin treatment liberated P, but GST was retained on the column. (D) Control showing coexpression of GST and P. Note that P did not bind GST or the glutathione affinity matrix (lanes 2) and was present only in the lysate and flow-through fractions (lanes 1 and 3). As expected, GST bound strongly to the column and was not released in either wash (lanes 4 and 5) or by thrombin digestion (lanes 6).
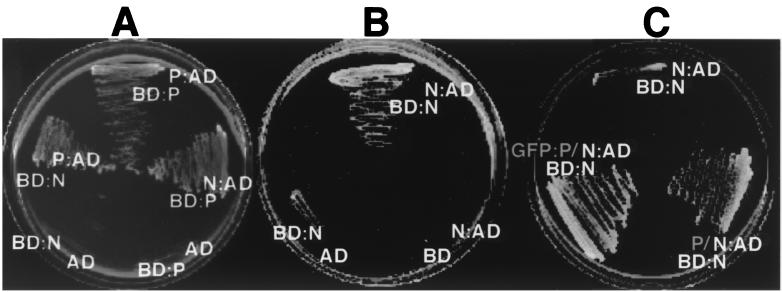
In order to examine the N and P interactions in more detail, yeast two-hybrid analyses were conducted. Interaction tests with the yeast two-hybrid system revealed the presence of homologous N-N and P-P complexes and heterologous complexes between N and P (Fig. 8A). The N-P and P-P complexes were easily detected by rapid growth in 3 days, but growth was inhibited substantially in cells expressing both activation and binding domain fusions to the N protein (Fig. 8B). This growth inhibition was pronounced at 30°C and, as was the case with the GFP fusions noted above, colonies failed to grow by 9 days after streaking. However, distinct colony growth was evident after 9 days at 22°C, but very little colony growth could be detected at 3 days at 22°C. Growth inhibition was released by coexpressing either native P or GFP-P from a 2μm plasmid. In these cases, extensive growth occurred within 3 days at 30°C (Fig. 8C). This growth was strictly dependent on N and P interactions, because cells lacking either the activation or binding domain fusion failed to grow.
FIG. 8.
Homologous and heterologous interactions of N and P. Yeast two-hybrid analyses were used to determine the specificity of the N-N, N-P, and P-P interactions. (A) Interactions of P fusions to the Gal4 activation domain with N or P fusions to the Gal4 DNA-binding domain. Neither N or P was able to activate transcription when coexpressed as a binding domain fusion with unfused activation domain vectors. Identical results were obtained in a reciprocal experiment in which P was expressed as a binding domain fusion and N was expressed as an activation domain fusion. The plates were incubated at 30°C for 4 days. (B) N and N interaction. Note that the growth of yeast coexpressing the N-activation (N:AD) and N-binding domain (BD:N) fusions required incubation at 21°C for 9 days to achieve appreciable growth. Growth failed to occur when the plates were incubated at 30°C. (C) N-N interactions in the presence of P. The plates were incubated at 30°C for 4 days. Enhanced growth of yeast transformants was achieved when either P or GFP-P was coexpressed with the N-N binding and activation domain fusions.
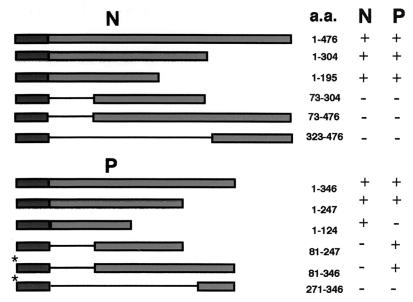
To determine the domains that mediate the interactions of the N and P proteins, portions of N and P were fused to the LexA DNA-binding domain and coexpressed with full-length N and P activation domain fusions. A reciprocal strategy had to be conducted to test interactions of the carboxy terminus of the P protein, because all tested P protein binding domain fragments lacking the first 100 amino-terminal amino acids activated transcription in the absence of the activation domain fusions. Thus, this activity prohibited the use of these constructs in interaction studies because bona fide interactions could not be distinguished from spurious activation by the binding domain fusions. To circumvent this problem, all carboxy-terminal fragments were expressed as activation domain fusions and tested against full-length proteins expressed as binding domain fusions. Interestingly, the full-length P protein did not activate transcription when expressed as a binding domain fusion in the absence of activation domain fusions. These results suggest that the P protein has a carboxy-terminal transcription activation domain that is revealed when the amino terminus is deleted. Although we have not explored the molecular basis of the effect of the amino terminus of P on transcriptional activation, portions of P that activate transcription when bound to a DNA-binding domain also lack the N-interacting domain (Fig. 9). This observation suggests that conformational changes, possibly in association with the N protein, may lead to similar viral transcriptional activation in vivo. This raises some interesting questions about regulation of the multiple activities of the P protein that can be addressed in future studies.
FIG. 9.
Yeast two-hybrid analysis of N- and P-interacting domains. Full-length copies, or portions thereof, of the N and P genes were cloned in frame with the LexA DNA-binding domain of plasmid pEG202 and transformed into yeast strain EGY48. Full-length and deletion mutants were tested against full-length copies of N and P cloned in frame with the B42 activation domain of plasmid pJG4-5. Cotransformants with the ability to grow on galactose medium lacking leucine were scored as a positive (+) interaction. Cotransformants that failed to grow on such media were scored as a negative (−) interaction. (Note that portions of the P gene lacking the amino terminus were able to activate transcription when fused to the LexA DNA-binding domain. To circumvent this problem, these fragments [*] were tested for interactions against full-length N and P in a Gal4-based system using the Ade2 reporter in yeast strain PJ694-A.)
A summary of the two-hybrid assay results demonstrates that the amino-terminal portion of N (aa 1 to 73) is involved in both N-N and N-P interactions (Fig. 9). This observation suggests that P may prevent N-N aggregation by blocking the site for N-N interactions. Similarly, the amino terminus of P (aa 1 to 81) is involved in P-N interactions, but P-P interactions are mediated by a centrally located domain between positions 40 and 124. Additional experiments under way to more precisely resolve the complexities of these interactions will be described in a subsequent communication.
DISCUSSION
The SYNV N and P proteins are representative of two classes of essential nucleocapsid core components that are present in all monopartite negative-strand RNA viruses. The N protein encapsidates both the full-length genomic and antigenomic viral RNAs as the nascent molecules are transcribed. The N, P, and L proteins also interact to form ribonucleoprotein complexes with the viral genomic RNAs, which then serve as templates for secondary rounds of transcription and for replication. Naked RNAs are not infectious, and the encapsidated genomic and antigenomic RNAs serve as the minimal infectious units of the virus (48). N also encapsidates nascent leader RNAs, and in the case of the prototypical rhabdovirus, VSV, this interaction has been proposed to play a role in the switch from transcription to replication (55). The P proteins are multifunctional proteins believed to serve as chaperones for the typically insoluble N protein and unstable polymerase (L) protein (33, 34, 49, 50). In addition, phosphorylated and unphosphorylated complexes of P have also been implicated in regulation of transcription (9) and replication (6, 17), respectively. However, in contrast to those of all known cytoplasmically replicating vertebrate rhabdoviruses, the SYNV N, P, and L transcription complexes are present in the nuclei of infected cells (53, 54), where they appear to form replicating viroplasms (32).
Our present studies provide further insight into the interaction and localization of the SYNV N and P proteins and also demonstrate the utility of yeast as a model to facilitate studies of these proteins. In both plant and yeast cells, the N protein has distinct patterns of localization in comparison to those of the P protein (Fig. 1 and 2). These patterns of localization may reflect the mechanisms by which these proteins are imported into the nucleus. The mutagenesis and localization experiments demonstrate that the N protein contains only a single functional NLS, since full-length proteins containing mutations in the carboxy-terminal NLS failed to enter the nucleus. Several negative-strand viruses, notably influenza viruses and Borna disease virus, are known to replicate in the nuclei of their hosts. However, there appears to be little conservation in the location or numbers of the NLSs in the nucleocapsid proteins of these viruses. This disparity is exemplified by the influenza A (57) and Thogoto (58) virus NP proteins, which, in contrast to the SYNV N protein, have bona fide bipartite NLSs at both their amino and carboxy termini. The carboxy-terminal SYNV N protein NLS also differs from that of the analogous borna disease virus p39 protein, which contains an amino-terminal NLS (42). Among the plant nucleorhabdoviruses that have been characterized, our comparisons reveal that the N protein of Rice yellow stunt virus (RYSV) has a single putative bipartite NLS located near the carboxy terminus. Thus, the positions of NLSs in the N proteins of the two sequenced nucleorhabdoviruses appear to be similar, and differences in their sequences probably reflect particular evolutionary selections mediated by individual relationship constraints imposed by replication in their diverse plant hosts and insect vectors.
The SYNV N NLS contains two basic regions (RKRR and KPKK) separated by 7 aa. In addition, the N NLS contains two prolines, an alpha-helix-breaking amino acid shown to be commonly associated with NLSs (2). The N NLS is sufficient to direct the nuclear import of proteins outside of the context of the N protein, because a GFP-GST-N NLS fusion was quantitatively directed to the nucleus in both yeast and plant cells. This bipartite NLS is characteristic of proteins imported into the nucleus by the importin-α/importin-β pathway (44), as is also the case with the influenza A virus, Thogoto virus, and nucleoplasmin NLSs. The suggestion that the N protein utilizes the importin-α/importin-β pathway is supported by our observations that recombinant N, expressed in either E. coli or yeast, binds to a GST fusion of yeast importin-α (M. M. Goodin and A. O. Jackson, unpublished data). We have not yet determined whether the N NLS resides within the RNA-binding domain of the N protein. However, this may be a distinct possibility, because NLSs often reside within nucleic acid-binding domains (26).
Our results also clearly demonstrate that the P protein is capable of independent nuclear localization. However, in contrast to the N protein, the SYNV P protein does not contain a canonical arginine-lysine-rich NLS and it does not bind importin-α in vitro (Goodin and Jackson, unpublished). This suggests that P utilizes an alternative import pathway to that used by the N protein. This hypothesis is supported by our finding that P possesses a karyophillic region within an 84-aa sequence located in the N-interacting domain. In addition, the P protein fails to bind to a GST fusion of yeast importin-α (Goodin and Jackson, unpublished). Moreover, some unexpected nuclear import complexity exists in the P protein interactions, because deletion mutants lacking the carboxy-terminal amino acid residues at positions 247 to 346 have much more pronounced nuclear accumulation than that of the wild-type GFP-P fusion or the native P protein (32). These results suggest that P may have the ability to shuttle between the nucleus and cytoplasm. Experiments are in progress to address these putative nuclear-shuttle functions.
In addition to characterizing the nuclear localization patterns and identification of the N and P NLS signals, our results show that coexpression of N and P results in a dramatic shift of both proteins to a distinct subnuclear location in both plant and yeast cells. The specificity of this shift is supported by control experiments during which GFP (pI 6.0) or GFP-nucleoplasmin (pI 4.5) were coexpressed with the N (pI 8.8) and/or P (pI 5.3) protein. Neither the localization of GFP nor that of GFP-nucleoplasmin was affected by N or P, suggesting that the N-P localization is not an adventitious consequence of electrostatic interactions. In addition, no effect on GFP-P/N localization was noted when these proteins were coexpressed with either the SYNV M or sc4 protein (Goodin and Jackson, unpublished). These results, and our previous findings with proteins recovered from isolated nuclei and protoplasts, indicate that the shift in accumulation results from direct interactions of N and P. In this regard, the yeast two-hybrid and GST pulldown experiments demonstrate that specific physical interactions occur between N and P, and our mutagenesis experiments demonstrate that these interactions are essential for subnuclear localization.
Physical interactions occurred between all combinations of the N and P proteins, namely, N-N, N-P, and P-P. However, there was a marked effect of P protein expression on N-N protein interactions that affected yeast growth during two-hybrid analyses and the accumulation of N or GFP-N in yeast expression experiments. These enhanced effects of P expression probably reflect the ability of P interactions to increase the solubility and decrease the turnover rate of the N protein and to facilitate subnuclear localization of the N-P complex. Observations consistent with the notion that P stabilizes N were that the N protein was much more abundant in Western blots when cell lysates containing both N and P or GFP-N and P were compared to those containing N or GFP-N alone and that the fluorescence of the GFP-N fusions was more intense in yeast cells coexpressing P.
As is the case with the N and P proteins of other negative-strand RNA viruses, the terminal regions of the proteins are necessary for N-P interactions (1, 24, 29, 31, 38, 40, 47, 50). The finding that the amino terminus of N is involved in both N-N and N-P interactions suggests that P binding may prevent N-N aggregation by blocking some of the sites necessary for N-N interactions. The N- and P-interacting domains identified by our two-hybrid analyses are supported by computer-assisted analyses of N and P using the COILS algorithm, which predicts the presence of coiled-coil regions in proteins (30). Coiled-coil regions often mediate interactions between proteins, and the presence of these coiled coils has been useful for predicting interacting regions (5). When fused to a heterologous protein, the predicted coiled-coil region of the mumps virus phosphoprotein was demonstrated to be sufficient to mediate protein oligomerization (5). In the case of the SYNV P protein, COILS predicts three possible coiled-coil domains. These domains are located between amino acids 39 and 53 (domain 1), 126 and 139 (domain 2), and 200 and 214 (domain 3). Interestingly, domain 1 resides within the P-N interaction region identified by yeast two-hybrid analyses. Similarly, domains 2 and 3 reside in the region that is required for P-P interactions. The fact that domains 2 and 3 reside in the central portion of the P protein in a locale similar to the coiled-coil regions of the P proteins of Sendai and mumps viruses suggests that functional constraints may have resulted in structural conservation among these proteins. Furthermore, the location of the karyophillic region (aa 40 to 124) in P overlaps that of domain 1. This finding is consistent with our results showing that an N protein rendered incapable of nuclear import has the ability to block nuclear import of the wild-type P protein. This cytoplasmic retention of the N-P protein complex may be explained if the N protein NLS mutation inactivates interactions with importin-α to result in increased amounts of cytoplasmic N that are able to bind to the P protein and interfere with nuclear localization by masking the P NLS. Fine-structure analyses using site-specific mutagenesis are under way to further define the relationship between the P-N interacting domain and the P NLS.
In an attempt to evaluate relationships among plant rhabdovirus P proteins, we compared the results of COILS analyses for SYNV to the putative P proteins of the nucleorhabdovirus RYSV (8; GenBank accession no. AB011257) and the cytorhabdoviruses Lettuce necrotic yellow virus (LNYV) (59; GenBank accession no. AF209035) and Northern cereal mosaic virus (NCMV) (51; GenBank accession no. AB030277). These comparisons revealed that each protein contains central and terminal coiled-coil domains. In contrast to the SYNV P protein, the putative LNYV P protein homologue, 4a, and NCMV P protein homologues have a high degree of predicted coiled-coil character at central domains and carboxy termini but not at their amino termini. In keeping with their assignment to the cytorhabdovirus group, the LNYV 4a protein has no readily predictable NLSs, nor does the NCMV P protein. Interestingly, NS, the P protein homologue of the nucleorhabdovirus RYSV, has a putative bipartite carboxy-terminal NLS sequence (RKDSHHYRTVVSRIEKK) starting at aa 265 that overlaps a coiled-coil domain, so the nuclear import signals of RYSV might rely on the importin-α pathway and hence differ from those of the SYNV P protein. However, the RYSV NS protein had predicted coiled coils in the amino-terminal, central, and carboxy-terminal regions and was similar to the SYNV P protein in this regard.
A similar analysis of the SYNV N protein using COILS failed to reveal clearly defined regions capable of forming coiled coils. However, a small carboxy-terminal region has a minor predicted coiled coil that overlaps the N protein NLS. As is the case with the SYNV N protein, the RYSV N protein has only a minimal predicted coiled-coil structure, aside from a carboxy-terminal domain that is in close proximity to the putative NLS region. Although the predicted RYSV NLS signals (PKRMKAL at aa 358 and KKLGPPRANAHSRRKEP at aa 405) have little direct relatedness to the SYNV N protein NLS sequence, the common carboxy-terminal location and predicted bipartite nature of these motifs provide preliminary evidence that members of the genus Nucleorhabdovirus may utilize similar N protein nuclear import mechanisms. These predictions now provide the basis for biochemical experiments to provide more extensive comparisons of the cellular targeting and protein-protein interactions of the N and P proteins of members of this genus.
In summary, our results demonstrate that the nuclear localization of the N and P proteins in plant and yeast cells is dramatically affected by their coexpression. Although both proteins are capable of independent nuclear localization (32), coexpression results in a shift in accumulation to a subnuclear site. Several lines of evidence indicate that N and P are capable of physical interactions and that subnuclear accumulation is dependent on the formation of N and P complexes. Taken together, these data suggest a model whereby N and P are independently imported into the nucleus and subsequently form protein associations that result in their relocalization to a subnuclear site. According to this model, the inability of native P to mediate the nuclear import of a null NLS mutant N protein argues against a role for the P protein in facilitating import of N from the cytoplasm to the nucleus. Since the karyophillic region of P resides within the N protein-interacting domain of P, the molecular basis for the cytoplasmic retardation of native P by the NLS mutant N protein would appear to be blockage of the P NLS by the N-P association. During normal nuclear import of wild-type N protein, importin-α probably blocks the P-interacting domain to prevent cytoplasmic interactions of N and P. However, once within the nucleus, this domain would become available for N-P interactions as the respective nuclear import proteins release the N protein cargo and recycle back to the cytoplasm. P would then serve as a chaperone to facilitate N solubility and nonspecific RNA binding of the N protein (33, 34) and possibly to mediate viroplasm formation within a subnuclear location. We are presently conducting experiments to test this model and to discern functions that are regulated by P protein phosphorylation and relative levels of free N and P proteins.
ACKNOWLEDGMENTS
We thank the Thorner and Rine laboratories at UC Berkeley for providing plasmids, reagents, and advice. In particular, we thank Tim Durfee for the YEp351 and -352 vectors and BJ5460 yeast strain and Jorg Kulda for the pBS316 vector. We thank Erica Golemis (Fox Chase Cancer Institute) and Roger Brent (Massachusetts General Hospital) for making the complete LexA two-hybrid vectors available to us. Philip James and Christopher Beh supplied all reagents and yeast strains for the GAL4 two-hybrid system used in this study. Steve Ruzin and Denise Schichnes at the Center for Biological Imaging at UC Berkeley were instrumental in obtaining high-quality photomicrographs. The nucleoplasmin-GFP construct used in the study was a kind gift from Kenji Kohno (Nara Institute of Science and Technology). The construction of this vector was described by Lim et al. (28). We thank Jennifer Bragg, Ralf Dietzgen, Angelika Fath, Jennifer Johnson, Diane Lawrence, Robin MacDiarmid, Teresa Rubio, and anonymous reviewers for critical review of the manuscript and helpful comments.
This research was supported by NSF competitive grant MCB-990-4810 awarded to A.O.J.
REFERENCES
- 1.Bankamp B, Horikami S M, Thompson P D, Huber M, Billeter M, Moyer S A. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology. 1996;216:272–277. doi: 10.1006/viro.1996.0060. [DOI] [PubMed] [Google Scholar]
- 2.Boulikas T. Putative nuclear localization signals (NLS) in protein transcription factors. J Cell Biochem. 1994;55:32–58. doi: 10.1002/jcb.240550106. [DOI] [PubMed] [Google Scholar]
- 3.Bullido R, Gómez-Puertas P, Albo C, Portela A. Several protein regions contribute to determine the nuclear and cytoplasmic localization of the influenza A virus nucleoprotein. J Gen Virol. 2000;81:135–142. doi: 10.1099/0022-1317-81-1-135. [DOI] [PubMed] [Google Scholar]
- 4.Choi T-J, Kuwata S, Koonin E V, Heaton L A, Jackson A O. Structure of the L (polymerase) protein gene of sonchus yellow net virus. Virology. 1992;189:31–39. doi: 10.1016/0042-6822(92)90678-i. [DOI] [PubMed] [Google Scholar]
- 5.Curran J, Boeck R, Lin-Marq N, Lupas A, Kolakofsky D. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology. 1995;214:139–149. doi: 10.1006/viro.1995.9946. [DOI] [PubMed] [Google Scholar]
- 6.Das T, Pattnaik A K, Takacs A M, Li T, Hwang L N, Banerjee A K. Basic amino acid residues at the carboxy-terminal eleven amino acid region of the phosphoprotein (P) are required for transcription but not for replication of vesicular stomatitis virus genome RNA. Virology. 1997;238:103–114. doi: 10.1006/viro.1997.8823. [DOI] [PubMed] [Google Scholar]
- 7.Falk B W, Purcifull D E, Christie S R. Natural occurrence of sonchus yellow net virus in Florida [USA] lettuce (Lactuca sativa) Plant Dis. 1986;70:591–593. [Google Scholar]
- 8.Fang R-X, Wang Q, Xu B-Y, Pang Z, Zhu H-T, Mang K-Q, Gao D-M, Qin W-S, Chua N-H. Structure of the nucleocapsid protein gene of rice yellow stunt rhabdovirus. Virology. 1994;204:367–375. doi: 10.1006/viro.1994.1541. [DOI] [PubMed] [Google Scholar]
- 9.Gao Y, Greenfield N J, Cleverley D Z, Lenard J. The transcriptional form of the phosphoprotein of vesicular stomatitis virus is a trimer: structure and stability. Biochemistry. 1996;35:14569–14573. doi: 10.1021/bi9613133. [DOI] [PubMed] [Google Scholar]
- 10.Godon C, Caboche M, Daniel-Vedele F. Transient plant gene expression: a simple and reproducible method based on flowing particle gun. Biochimie. 1993;75:591–595. doi: 10.1016/0300-9084(93)90065-z. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg K-B, Modrell B, Hillman B I, Heaton L A, Choi T-J, Jackson A O. Structure of the glycoprotein gene of Sonchus yellow net virus, a plant rhabdovirus. Virology. 1991;185:32–38. doi: 10.1016/0042-6822(91)90750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Methods in enzymology vol. 194. New York, N.Y: Academic Press; 1991. [PubMed] [Google Scholar]
- 13.Gyuris J, Golemis E, Chertokov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1995;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 14.Heaton L A, Zuidema D, Jackson A O. Structure of the M2 protein gene of sonchus yellow net virus. Virology. 1987;161:234–241. doi: 10.1016/0042-6822(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 15.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 16.Hillman B I, Heaton L A, Hunter B G, Modrell B, Jackson A O. Structure of the gene encoding the M1 protein of sonchus yellow net virus. Virology. 1990;179:201–207. doi: 10.1016/0042-6822(90)90289-4. [DOI] [PubMed] [Google Scholar]
- 17.Hwang L N, Englund N, Das T, Banerjee A K, Pattnaik A K. Optimal replication activity of vesicular stomatitis virus RNA polymerase requires phosphorylation of a residue(s) at carboxy-terminal domain II of its accessory subunit, phosphoprotein P. J Virol. 1999;73:5613–5620. doi: 10.1128/jvi.73.7.5613-5620.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson A O, Christie S R. Purification and some physiochemical properties of sonchus yellow net virus. Virology. 1977;77:344–355. doi: 10.1016/0042-6822(77)90431-7. [DOI] [PubMed] [Google Scholar]
- 20.Jackson A O, Francki R I B, Zuidema D. Biology, structure and replication of plant rhabdoviruses. In: Wagner RR, editor. The rhabdoviruses. New York, N.Y: Plenum Press; 1987. pp. 427–508. [Google Scholar]
- 21.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Hirsch J P. A nucleolar protein that affects mating efficiency in Saccharomyces cerevisiae by altering the morphological response to pheromone. Genetics. 1998;149:795–805. doi: 10.1093/genetics/149.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein T M, Wolf E E, Wu R, Sanford J C. High-velocity microprojectiles for delivering nucleic acids into living cells. Nature. 1987;327:70–73. [PubMed] [Google Scholar]
- 24.Krishnamurthy S, Samal S K. Identification of regions of bovine respiratory syncytial virus N protein required for binding to P protein and self-assembly. J Gen Virol. 1998;79:1399–1403. doi: 10.1099/0022-1317-79-6-1399. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaCasse E C, Lefebvre Y A. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lim C R, Kimata Y, Oka M, Nomaguchi K, Kohno K. Thermosensitivity of green fluorescent protein fluorescence utilized to reveal novel nuclear-like compartments in a mutant nucleoporin NSP1. J Biochem. 1995;118:13–17. doi: 10.1093/oxfordjournals.jbchem.a124868. [DOI] [PubMed] [Google Scholar]
- 29.Liston P, Batal R, Diflumeri C, Briedis D J. Protein interaction domains of the measles virus nucleocapsid protein (NP) Arch Virol. 1997;142:305–321. doi: 10.1007/s007050050078. [DOI] [PubMed] [Google Scholar]
- 30.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 31.Mallipeddi S K, Lupiani B, Samal S K. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P interaction using a two-hybrid system. J Gen Virol. 1996;77:1019–1023. doi: 10.1099/0022-1317-77-5-1019. [DOI] [PubMed] [Google Scholar]
- 32.Martins C R F, Johnson J A, Lawrence D M, Choi T-J, Pisi A, Tobin S L, Lapidus D, Wagner J D O, Ruzin S, McDonald K, Jackson A O. Sonchus yellow net rhabdovirus nuclear viroplasms contain polymerase-associated proteins. J Virol. 1998;72:5669–5679. doi: 10.1128/jvi.72.7.5669-5679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masters P S, Banerjee A K. Resolution of multiple complexes of phosphoprotein NS with nucleocapsid protein N of vesicular stomatitis virus. J Virol. 1988;62:2651–2657. doi: 10.1128/jvi.62.8.2651-2657.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masters P S, Banerjee A K. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J Virol. 1988;62:2658–2664. doi: 10.1128/jvi.62.8.2658-2664.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGeoch D J. On the predictive recognition of signal peptide sequences. Virus Res. 1985;3:271–286. doi: 10.1016/0168-1702(85)90051-6. [DOI] [PubMed] [Google Scholar]
- 36.Miller C A, III, Martinat M A, Hyman L E. Assessment of aryl hydrocarbon receptor complex interactions using pBEVY plasmids: expression vectors with bi-directional promoters for use in Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26:3577–3583. doi: 10.1093/nar/26.15.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell D A, Marshall T K, Deschenes R J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 38.Myers T M, Pieters A, Moyer S A. A highly conserved region of the Sendai virus nucleocapsid protein contributes to the NP-NP binding domain. Virology. 1997;229:322–335. doi: 10.1006/viro.1996.8429. [DOI] [PubMed] [Google Scholar]
- 39.Nicoletti V G, Condorelli D F. Optimized PEG method for rapid plasmid DNA purification: high yield from “midi-prep.”. BioTechniques. 1993;14:532–536. [PubMed] [Google Scholar]
- 40.Nishio M, Tsurudome M, Kawano M, Watanabe N, Ohgimoto S, Ito M, Komoda H, Ito Y. Interaction between the nucleocapsid (NP) and phosphoprotein (P) of human parainfluenza virus type 2: one of the two NP binding sites on P is essential for granule formation. J Gen Virol. 1996;77:2457–2463. doi: 10.1099/0022-1317-77-10-2457. [DOI] [PubMed] [Google Scholar]
- 41.O'Neil R E, Talon J, Palese J T. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pyper J M, Gartner A E. Molecular basis for the differential subcellular localization of the 38- and 39-kilodalton structural proteins of borna disease virus. J Virol. 1997;71:5133–5139. doi: 10.1128/jvi.71.7.5133-5139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Relic B, Andjelkovic M, Rossi L, Nagamine Y, Hohn B. Interaction of the DNA modifying VirD1 and VirD2 of Agrobacterium tumefaciens: analysis by subcellular localization in mammalian cells. Proc Natl Acad Sci USA. 1998;95:9105–9110. doi: 10.1073/pnas.95.16.9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 45.Rosorius O, Heger P, Stelz G, Hirschmann N, Hauber J, Stauber R H. Direct observation of nucleocytoplasmic transport by microinjection of GFP-tagged proteins in living cells. BioTechniques. 1999;27:350–355. doi: 10.2144/99272rr02. [DOI] [PubMed] [Google Scholar]
- 46.Scholthof G K-B, Hillman B I, Modrell B, Heaton L A, Jackson A O. Characterization and detection of sc4: a sixth gene encoded by sonchus yellow net virus. Virology. 1994;204:279–288. doi: 10.1006/viro.1994.1532. [DOI] [PubMed] [Google Scholar]
- 47.Slack M S, Easton A J. Characterization of the interaction of the human respiratory syncytial virus phosphoprotein and nucleocapsid protein using the two-hybrid system. Virus Res. 1998;55:167–176. doi: 10.1016/s0168-1702(98)00042-2. [DOI] [PubMed] [Google Scholar]
- 48.Szilagyi J F, Uryvayev L. Isolation of an infectious ribonucleoprotein from vesicular stomatitis virus containing an RNA transcriptase. J Virol. 1973;11:279. doi: 10.1128/jvi.11.2.279-286.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takacs A M, Banerjee A K. Efficient interaction of the vesicular stomatitis P protein with the L or N protein in cells expressing recombinant proteins. Virology. 1995;208:821–826. doi: 10.1006/viro.1995.1219. [DOI] [PubMed] [Google Scholar]
- 50.Takacs A M, Das T, Banerjee A K. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc Natl Acad Sci USA. 1993;90:10375–10379. doi: 10.1073/pnas.90.21.10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanno F, Nakatsu A, Toriyama S, Kojima M. Complete nucleotide sequence of Northern cereal mosaic virus and its genome organization. Arch Virol. 2000;145:1373–1384. doi: 10.1007/s007050070096. [DOI] [PubMed] [Google Scholar]
- 52.Van Beek N A M, Derksen A C G, Dijkstra J. Polyethylene glycol-mediated infection of cowpea protoplasts with sonchus yellow net virus. J Gen Virol. 1985;66:551–557. [Google Scholar]
- 53.Wagner J D O, Choi T-J, Jackson A O. Extraction of nuclei from Sonchus yellow net rhabdovirus-infected plants yields a polymerase that synthesized viral mRNAs and polyadenylated plus-strand leader RNA. J Virol. 1996;70:468–477. doi: 10.1128/jvi.70.1.468-477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner J D O, Jackson A O. Characterization of the components and activity of sonchus yellow net rhabdovirus polymerase. J Virol. 1997;71:2371–2382. doi: 10.1128/jvi.71.3.2371-2382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner R R. Rhabdovirus biology and infection. In: Wagner R R, editor. The rhabdoviruses. New York, N.Y: Plenum Press; 1987. pp. 9–74. [Google Scholar]
- 56.Walker P J, Benmansour A, Dietzgen R, Fang R-X, Jackson A O, Kurath G, Leong J C, Nadin-Davies S, Tesh R B, Tordo N. Family Rhabdoviridae. In: van Regenmortel M H V, Fauquet C M, Bishop D H L, Carstens E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. San Diego, Calif: Academic Press; 2000. [Google Scholar]
- 57.Wang P, Palese P, O'Neill R E. The NPI-1/NIP-3 (karyopherin alpha) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol. 1997;71:1850–1856. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weber F, Kochs G, Gruber S, Haller O. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology. 1998;250:9–18. doi: 10.1006/viro.1998.9329. [DOI] [PubMed] [Google Scholar]
- 59.Wetzel T, Dietzgen R G, Geering A D W, Dale J L. Analysis of the nucleocapsid gene of lettuce necrotic yellow rhabdovirus. Virology. 1994;202:1054–1057. doi: 10.1006/viro.1994.1438. [DOI] [PubMed] [Google Scholar]
- 60.Zuidema D, Heaton L A, Jackson A O. Structure of the nucleocapsid protein gene of sonchus yellow net virus. Virology. 1987;159:373–380. doi: 10.1016/0042-6822(87)90476-4. [DOI] [PubMed] [Google Scholar]