Abstract
Background
Ejection fraction (EF) estimation informs patient plans in the ICU, and low EF can indicate ventricular systolic dysfunction, which increases the risk of adverse events including heart failure. Automated echocardiography models are an attractive solution for high-variance human EF estimation, and key to this goal are echocardiogram vector embeddings, which are a critical resource for computational researchers.
Objectives
The authors aimed to extract the vector embeddings from each echocardiogram in the EchoNet dataset using a classifier trained to classify EF as healthy (>50%) or unhealthy (<= 50%) to create an embeddings dataset for computational researchers.
Methods
We repurposed an R3D transformer to classify whether patient EF is below or above 50%. Training, validation, and testing were done on the EchoNet dataset of 10,030 echocardiograms, and the resulting model generated embeddings for each of these videos.
Results
We extracted 400-dimensional vector embeddings for each of the 10,030 EchoNet echocardiograms using the trained R3D model, which achieved a test AUC of 0.916 and 87.5% accuracy, approaching the performance of comparable studies.
Conclusions
We present 10,030 vector embeddings learned by this model as a resource to the cardiology research community, as well as the trained model itself. These vectors enable algorithmic improvements and multimodal applications within automated echocardiography, benefitting the research community and those with ventricular systolic dysfunction (https://github.com/Team-Echo-MIT/r3d-v0-embeddings).
Key words: dataset, EchoNet, echocardiography, embeddings, R3D transformer, video
Central Illustration
Background
Heart failure is the chronic impairment of the heart’s ability to pump blood, affecting approximately 6.2 million people in the US and appearing on 350,000 death certificates in 2018.1 Moreover, it accounts for $30.7 billion of annual healthcare expenditures.2 Fortunately, effective interventions prolong life and reduce morbidity when heart failure is detected expediently.3 Left ventricular dysfunction, caused by defectiveness of the left ventricle, raises the risk of heart failure,4 making its diagnosis critical to improving patient care.
Key to heart failure diagnosis is echocardiography, ultrasound of the heart, which measures left ventricular ejection fraction (EF). EF is the fraction of blood that exits the left ventricle during the systolic phase of the cardiac cycle. It is calculated by dividing end-diastolic volume from end-systolic volume (EF = EDV/ESV).
Currently, manual echocardiography suffers from physician bias and lack of standardization. Further, a dearth of trained cardiologists in rural or low resource settings limits its accessibility to patients.2 Automated EF prediction is thus a key goal that would facilitate early ventricular dysfunction detection.5
Related work
Available to researchers, the EchoNet Dynamic dataset was published in 2020, as were several video-based EF classifiers. The best of these models, an architecture with R2+1D spatial + temporal convolutions, achieved an AUC of 0.97 when utilized for the same healthy/unhealthy EF classification task.6 Another group approached the regression problem by predicting continuous EF values with an R-squared of 0.95 on their proprietary database of 50,000 echocardiograms.7
In 2022, video action recognition neural networks achieved an accuracy of 90.17% for binary EF classification using a Gate Shift Network with a BNInception backbone.8 Other echocardiogram models have detected hypertrophic cardiomyopathy, cardiac amyloidosis, and pulmonary arterial hypertension with C statistics of 0.93, 0.87, and 0.85, respectively.9 Automated echocardiography is achievable, with deep learning being its key enabling technology.
Vector embeddings
Key to deep learning research are trained models and vector embeddings. Vector embeddings are lists of numbers, numerical representations of input data learned by a model. Their applications to deep learning and thus to automated echocardiography are vast.
Firstly, vector embeddings enable active learning or the optimal selection of training data that maximizes its representativeness, uncertainty, and diversity. Vector embeddings of the chest X-rays have allowed active selection methods to improve the F1 score of disease classification algorithms by 27%.10 Vector embeddings of echocardiograms could enable active learning to improve cardiac disease classification too.
As lists of numbers, vector embeddings are valuable features for medical classification models. Vector embeddings of Mel Frequency Cepstral Coefficient features from heart sounds were found to achieve a 16% performance improvement on heart sound classification performance.11
Each dimension of an embedding shares a representative meaning. This means that knowing the values of one embedding allows one to find similar embeddings, enabling similarity search. Pretrained embeddings have allowed researchers to retrieve similar CT and MR scan frames from a query image.12 With echocardiogram embeddings, such retrieval systems could be implemented in echocardiography.
Tantalizingly, visual embeddings can combine with embeddings of other data types for multimodal applications. Using a TriNet that incorporates both chest x-ray embeddings and medical report text embeddings, one group developed a model that can generate the text of a medical report given a chest x-ray image.13 This is possible because both data types can be encoded into embeddings which could be compared.
Novelty and contribution
To the best of our knowledge, none have released a dataset of vector embeddings for echocardiograms. Vector embeddings must be generated by a trained model, which is a time-consuming and computationally expensive process. Releasing pre-trained embeddings would therefore spare researchers’ significant time and computational costs when developing the embedding applications described earlier.
To this end, we analyzed echocardiograms from the EchoNet dataset to predict low EF (below 50%) utilizing a video transformer. We provide both the vector embeddings for all 10,030 echocardiograms learned by our model and the trained model itself for researchers to generate embeddings for their own echocardiograms. These resources are accessible through the GitHub repository link and will democratize access to echocardiogram representations for researchers, thus advancing automated echocardiography for patient care.
Methods
R3D transformer model
Embeddings come from trained models. For videos, an initial neural network (the “backbone”) first extracts visual features from each frame which video transformers then process. An ideal backbone is deep, as architectures with more layers tend to perform better on image data.14 Further, it should be memory efficient and ideally pretrained.
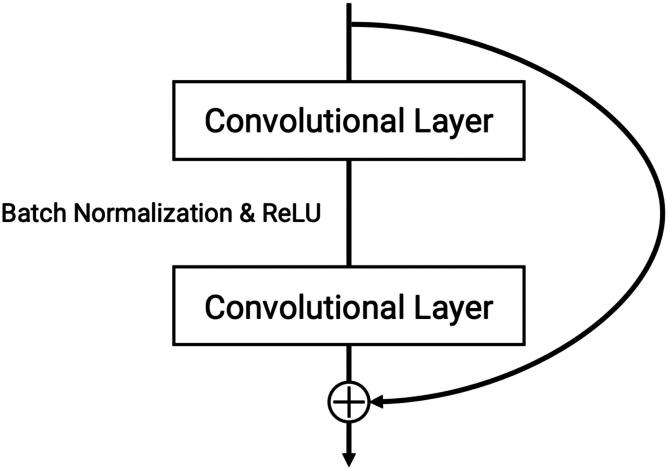
Visual Geometry Group (VGG16), Residual Networks (ResNet18), and InceptionV3 are all deep and pre-trained backbones with 16, 18, and 43 layers respectively. ResNet18, however, has the least parameters (11.2 M) than InceptionV3 (21.8 M) and VGG16 (138.4 M), making it the most memory-efficient option. It also owes its performance to residual blocks with a shortcut connection that allows the direct backpropagation of a gradient to earlier layers. This eases gradient calculations and mitigates overfitting (see Figure 1).
Figure 1.
Example of a Shortcut (“Skip”) Connection Within a Residual Block
ResNet models use residual blocks with shortcut (“skip”) connections across their convolutional layers. This connects layers within a residual block that otherwise have no direct backpropagation between them.
Of ResNet18-based video transformers, 3-Dimensional ResNet (R3D) is among the best-performing approaches to the EF classification problem.5 Designed for video learning,15 R3D is especially apt for videos because it uses 3D convolutional filters that span both spatial and temporal dimensions, allowing it to learn spatiotemporal patterns. R3D uses spatiotemporal kernels of shape 3 × 3 × 3.
Our R3D model comes from PyTorch and was pre-trained on the Kinetics-400 dataset, which consists of clips of human actions.16 With this transfer learning, we initialized our R3D with weights already tuned to human action recognition.
We fine-tuned the R3D as a classifier to predict whether EF was above or below 50%, which is the cutoff for reduced EF used in related papers. In classification, the resulting embeddings represent the binary difference between healthy and unhealthy hearts, which is our current interest. R3Ds can also act as regressors, and the embeddings from these models reflect variation in a continuous way. This approach is future work our group intends to develop.
Cohort selection
The EchoNet dataset contains 10,030 echocardiograms. Each video has frame dimensions 112 × 112 and 173 frames on average (SD = 47). We did not standardize frame lengths in our data, as adaptive pooling layers and other R3D model features support data with inconsistent dimensions.
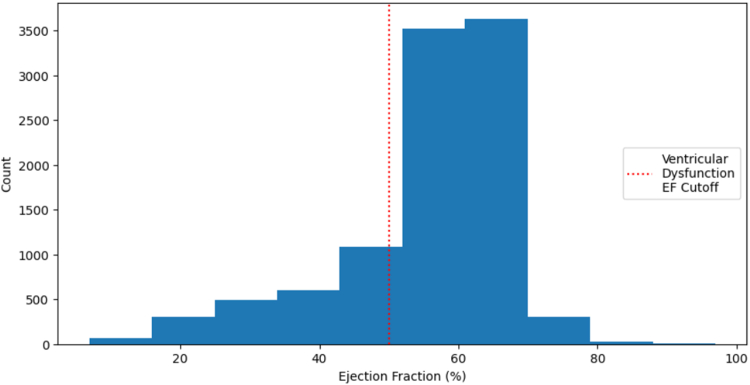
The EF distribution was left-skewed with a mode around 60%. The mean EF is 0.55, held down by a group of EFs between 20 to 50% (see Figure 2). EF almost never exceeded 80% or fell below 10%. The standard deviation is 12.
Figure 2.
Distribution of Ejection Fraction for All Echocardiograms
EF is a percentage between 0 to 100. Echocardiograms in the EchoNet dataset had an EF distribution specified by this figure, with a left skew and a mode around 60%. A class cutoff of EF = 50% results in a class imbalance.
Our dataset split resulted in 7,458 training, 1,284 validation, and 1,277 test videos. Only 22% of the training videos were class 1 (EF <50%), constituting a class imbalance. The precise figures are in Table 1. To mitigate this class imbalance, we experimented with oversampling and undersampling techniques. Using the original data resulted in the lowest binary cross entropy loss, so the final model utilizes it (see Table 2).
Table 1.
Cohort Split Based on Ejection Fraction
| Class | Description | Num Videos | Percentage | Name |
|---|---|---|---|---|
| 1 | EF <50% | 2,246 | 22.4% | Unhealthy |
| 0 | EF ≥50% | 7,784 | 77.6% | Healthy |
This is an imbalanced dataset, as 22% of echocardiograms are of the positive class. We attempted using undersampling and oversampling to mitigate the class imbalance problem.
EF = ejection fraction.
Table 2.
R3D Model Performance Across Class Imbalance Mitigation Strategies
| Mitigation Strategy | Accuracy | AUC | Precision | Recall | Specificity | F1 Score | AUPRC | BCE Loss |
|---|---|---|---|---|---|---|---|---|
| Undersample | 0.774 | 0.897 | 0.496 | 0.842 | 0.754 | 0.624 | 0.776 | 0.465 |
| None | 0.875 | 0.916 | 0.789 | 0.604 | 0.954 | 0.684 | 0.798 | 0.310 |
| Oversample | 0.865 | 0.918 | 0.682 | 0.744 | 0.900 | 0.711 | 0.807 | 0.318 |
To undersample, we sampled from the majority class to reduce it in size to that of the minority class. To oversample, we sampled from the minority class with replacement to inflate in size to that of the majority class. Using the data without these measures resulted in the lowest BCE loss, so we did not use oversampling or undersampling for our final model.
AUC = area under the receiver-operating characteristic curve; AUPRC = area under precision recall curve; BCE = Binary Cross Entropy; F1 = Harmonic Mean of Precision and Recall.
Training
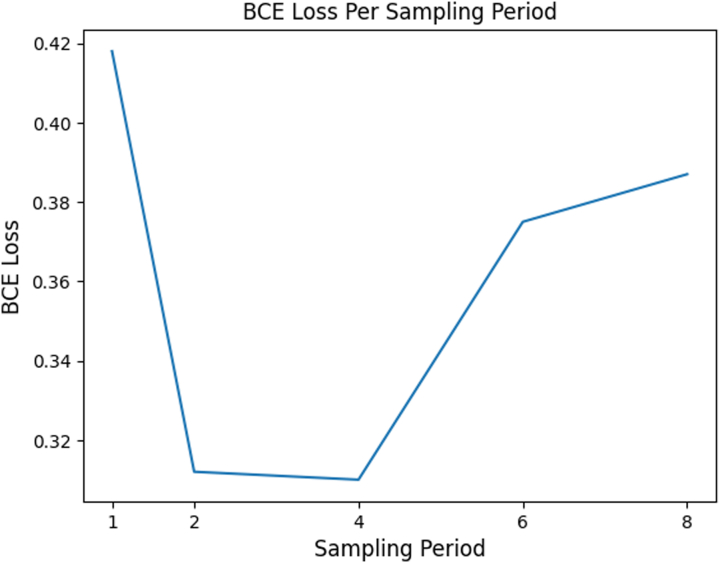
To increase memory efficiency, we sampled 1 of the every 4 frames from each echocardiogram. To ensure this did not exclude end-systolic and end-diastolic frames, which are critical to EF measurement, we repeated training with sampling periods of 8, 6, 4, 2, and 1 (original video) to compare the effects of sampling period on model performance. Because a sampling period of 4 resulted in similar performance to keeping all frames (see Figure 3), we concluded enough end-systolic and end-diastolic frames were retained with this frame sampling strategy to warrant its use.
Figure 3.
R3D Model Performance for Various Sampling Periods
Binary cross entropy (BCE) loss only increases once we sample from every 6 frames. This means 1-in-4 frame sampling likely does not exclude key frames necessary for EF classification.
Training utilized an L4 GPU from Google Cloud Platform, an Adam optimizer, binary cross entropy loss, a learning rate of 1e-5, and a weight decay of 5e-4. We also had gradients accumulate over successive mini-batches of size 20 before they updated the model. Model performance peaked at 4 epochs of training, which convergence likely accelerated.
Classification metrics
Accuracy is a necessary metric to compare the results of our model with those from literature. It can become misleading, however, when the label distribution is imbalanced. Precision describes how well a classifier identifies relevant (positive) data. Recall describes how well a classifier identifies any relevant data to begin with. F1 score is the harmonic mean of both. Specificity measures how well a classifier identifies negative data.
In addition to AUC, we recorded AUPRC, the area under the precision-recall curve, since it tends to remain stable for imbalanced data like our dataset. This is because a small number of correct or incorrect predictions can result in large changes in the receiver operating characteristic curve but not the precision-recall curve for imbalanced data.
Embeddings extraction
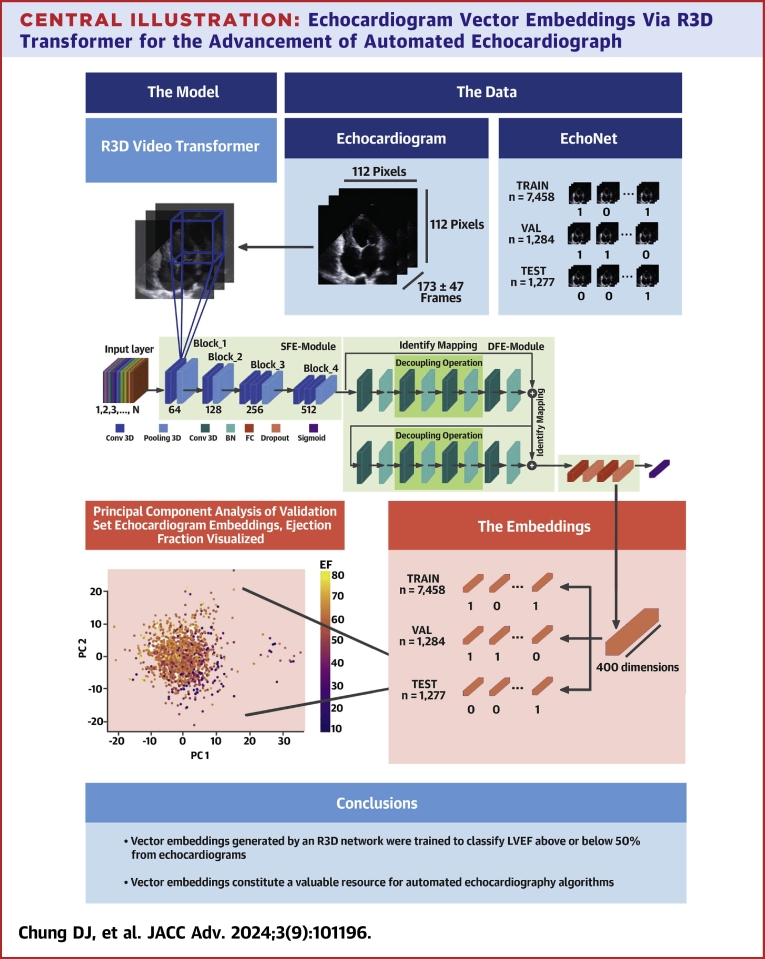
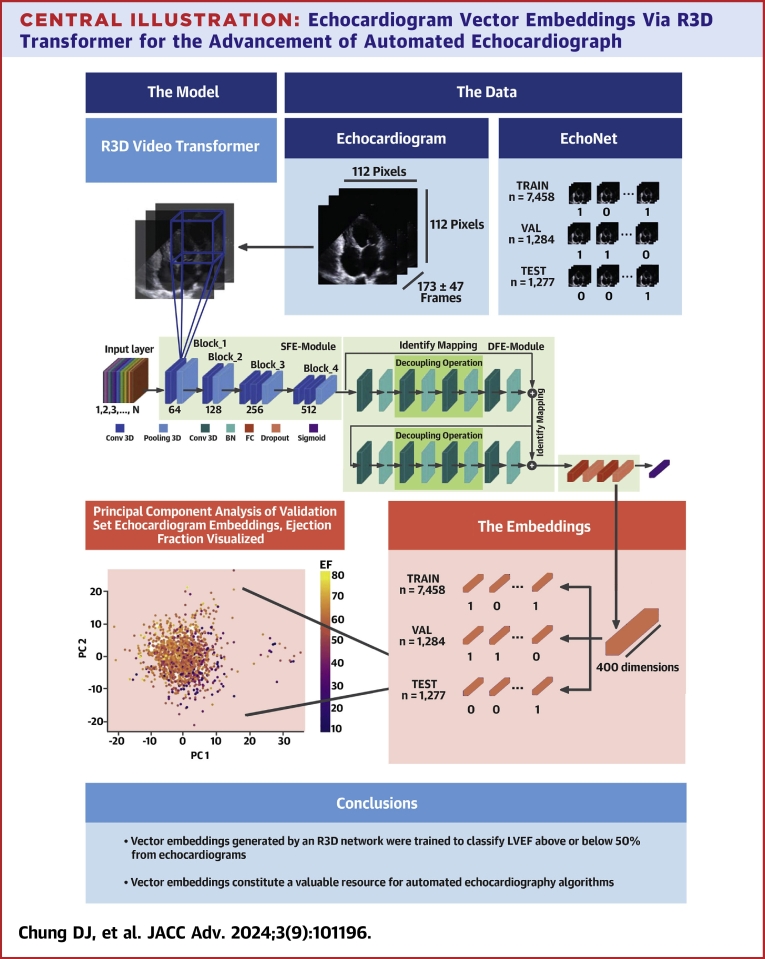
Vector embeddings for each echocardiogram are the values of the final, 400-dimensional hidden layer of the trained R3D transformer given that echocardiogram as the input. To extract EchoNet embeddings, we fed them through the trained R3D and extracted the final hidden layer each time (see Central Illustration).
Central Illustration.
Echocardiogram Vector Embeddings Via R3D Transformer for the Advancement of Automated Echocardiograph
Echocardiograms, represented as black frame sequences in the blue-shaded boxes, are fed into the R3D model, where 3D convolutions pick up spatiotemporal patterns and eventually reduce the video into a 400-dimensional fully connected layer before the sigmoid head. We extract the last fully-connected layer as the vector embeddings of an echocardiogram, and the principle component analysis (PCA) of these vector embeddings shows that EF patterns are preserved among them. Black arrows = show equivalence; black lines = highlight dimensions; blue lines = represent a 3D convolution; purple shapes = the vector embeddings; blue and green shapes = convolutional and pooling layers; orange shapes = fully connected and dropout layers; purple shape = sigmoid layer.
Our workflow is 2-fold: we train the R3D transformer to discriminate between high and low EF and use the trained R3D to generate vector embeddings for each echocardiogram in the EchoNet dataset.
Ethics statement
Our study utilized data collected in an ethical manner by the Stanford School of Medicine. Medical data such as echocardiograms were obtained by controlled access and were not distributed outside our research team. Our work received proper ethical oversight and does not require IRB approval.
Results
The AUPRC and AUC of a random classifier is ∼0.5, and for a perfect classifier, it is 1.0. Our R3D test AUC was 0.916. (see Table 3, Figure 4). Test AUPRC was also high at 0.798. Given how AUC can be misleading in contexts of class imbalance, this high AUPRC value suggests it is genuinely the strength of the R3D classifier itself that explains the high AUC.
Table 3.
R3D Transformer Results
| Model Metric |
Data Split |
||
|---|---|---|---|
| Training | Validation | Test | |
| Accuracy | 0.951 | 0.889 | 0.875 |
| AUC | 0.979 | 0.912 | 0.916 |
| Precision | 0.924 | 0.809 | 0.789 |
| Recall | 0.85 | 0.661 | 0.604 |
| Specificity | 0.98 | 0.955 | 0.954 |
| F1 Score | 0.885 | 0.728 | 0.684 |
| AUPRC | 0.949 | 0.811 | 0.798 |
| BCE Loss | 0.151 | 0.306 | 0.31 |
Our R3D model performance is demonstrated for each data split. Metrics like AUC and F1 score gauge the model’s holistic discriminative ability while others like AUPRC verify that these strong results are not the result of imbalanced data. Metrics like recall prioritize different objectives like minimizing false negatives. Across all metrics, model results are strong (>0.8).
AUC = area under the receiver-operating characteristic curve; AUPRC = area under precision recall curve; BCE = binary cross entropy; F1 = harmonic mean of precision and recall; R3D = 3-Dimensional ResNet.
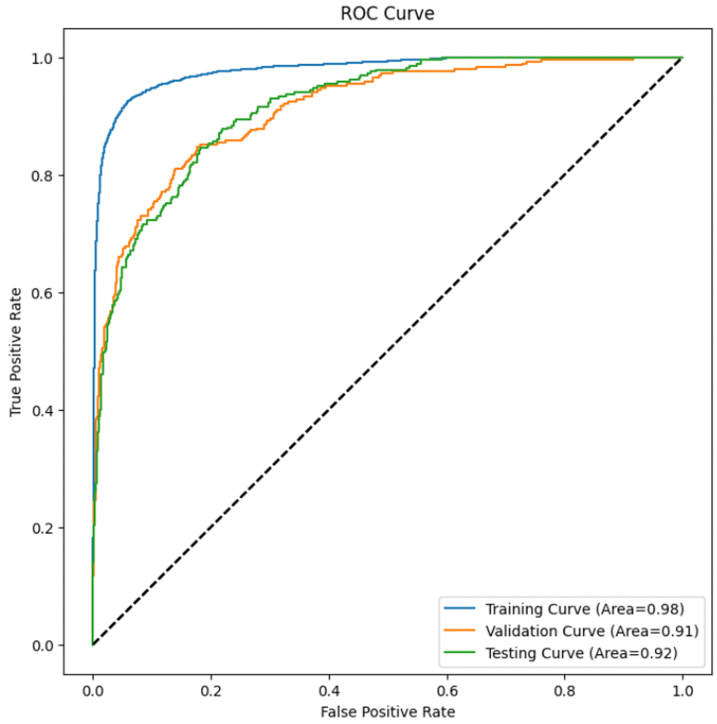
Figure 4.
AUC Curve for Training, Validation, and Test Sets
AUC (area under the curve) measures holistic discriminative ability and is 1.0 for a perfect binary classifier. Our test AUC of 0.92 thus demonstrates a strong learned representation by our model.
Test specificity was 0.954, meaning the classifier makes negative predictions correctly almost every time. Test recall was lower at 0.604 but means that the model still predicts the majority of ventricular dysfunction cases correctly. Test precision, finally, was also high at 0.789, meaning that most positive predictions were truly ventricular dysfunction. Together, these metrics demonstrate the strong discriminative ability of this EF classifier. The vector embeddings of this model, therefore, reflect an effective learned representation of echocardiogram data.
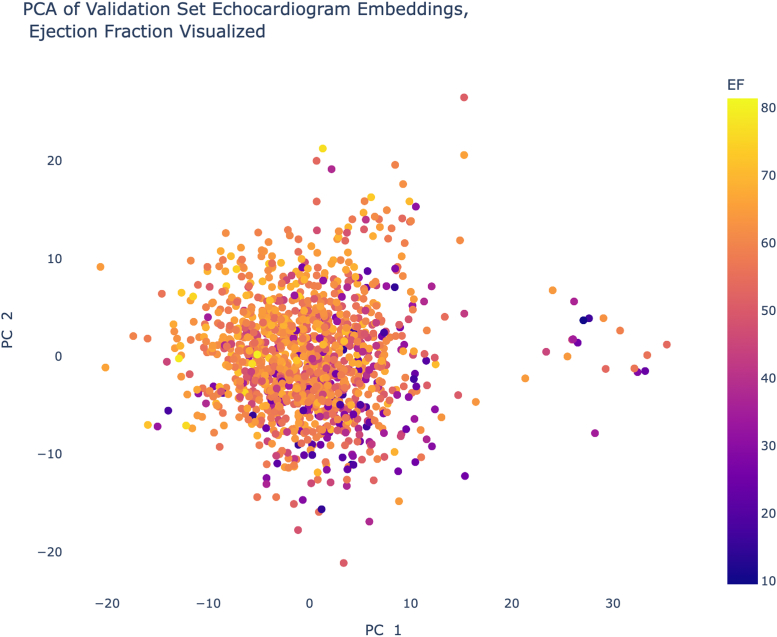
Using principal component analysis, a form of dimensionality reduction, we visualized the embeddings in 2 dimensions to verify that they capture EF patterns, which are visible in Figure 5.
Figure 5.
A 2-Dimensional View of the EchoNet Embeddings
Condensing the embeddings from 400 dimensions to 2 principle components (PCs) results in the above scatterplot, where visible trends in EF show that our embeddings successfully capture EF variation.
Discussion
Embedding quality checks
Between 2019 and 2023, 4 studies also developed EF classification models, the results of which are visible in Table 4. Our R3D achieves an accuracy (0.875) higher than that of the most recent paper (0.87), which uses a Mobile U-Net,17 and it comes within 5 points of the best classification accuracy achieved back in 2019 (0.92).7 Our AUC (0.916) is also higher than that of an Inception-based classifier, the latest model with a published AUC (0.847).8 Finally, it comes within 6 points of the best AUC (0.97) among related literature, achieved by a 3D CNN with atrous convolutions.6 These comparisons bode well for our embeddings because they show that the R3D model that trained them has comparable performance to those in the current literature.
Table 4.
Comparison of Test Accuracy and AUC With State-of-the-Art EF Classifiers
| Model Metric |
Classification Approach |
||||
|---|---|---|---|---|---|
| Our Approacha | Asch et al7b | Ouyang et alc | Almadani et al8d | Muldoon and Khan17e | |
| Accuracy | 0.875 | 0.92 | N/A | 0.902 | 0.87 |
| AUC | 0.916 | N/A | 0.97 | 0.847 | N/A |
| EF classification cutoff | 50% | 35% | 50% | 50% | 50% |
Our model performance approaches current SOTA (state-of-the-art) classifiers developed for the same EF classification problem, underscoring its quality. Our accuracy is higher than that of the latest model, for instance, while coming within 5 points of the best SOTA accuracy. Our AUC is also higher than that of the latest model and comes within 6 points of the best SOTA AUC.
AUC = area under the receiver-operating characteristic curve; EF = ejection fraction; GSM = gate shift module.
R3D transformer, ResNet18 backbone.
Undisclosed algorithm.
3D convolutional neural network with atrous convolutions.
GSM, inception backbone, 32-frame echocardiograms.
Mobile U-Net.
Applications of vector embeddings
The vector embeddings obtained from our R3D model encapsulate information about echocardiograms in a latent space, enabling diverse cardiac care applications.
Echocardiogram-based clustering
Clustering analyses using echocardiogram vector embeddings could reveal subtypes within cardiac data that represent diverse presentations of ventricular dysfunction. Models trained on echocardiogram embeddings could thus help identify what medical subtype a cardiac patient belongs to.
Disease classification
Vector embeddings can train classifiers to automatically identify specific cardiac conditions beyond EF. This is because a lower EF can be caused by other conditions like coronary artery disease or systolic heart failure.18 This extends the model’s measurement utility to a broader spectrum of cardiac pathologies.
Patient risk stratification
By incorporating vector embeddings, new risk prediction models could assist in stratifying patients based on their risk of developing ventricular dysfunction or related complications, enabling targeted interventions and personalized treatment plans.
Echocardiographic report generators
When algorithms can access both video and text embeddings, it is possible to produce accurate text from medical videos.13 The release of echocardiogram embeddings therefore makes possible multimodal systems that generate medical reports given an echocardiogram input.
Limitations and future work
In binary classification, the nature of a single cutoff value is limiting. Patients with an EF above 50% would classify as “healthy” by our framing, but they could still exhibit myocardial disease and other clinical changes. Future work will therefore develop a regression model to predict continuous EF and use it to extract updated echocardiogram embeddings. We also hope to predict global longitudinal strain as a more dependable indicator of EF.
Due to limited computational resources, this study did not utilize data augmentation to improve R3D performance, which has been demonstrated to successful effect in previous comparable studies.6 Convention is to use 5 augmentations, namely color jitter, grayscaling, grayscaling and color jitter, increased brightness, and increased sharpness. This multiplies the dataset size by a factor of 6.
Further, this study did not assess R3D performance in conditions such as atrial fibrillation or in regards to beat-to-beat variability, as this information was not available.
Finally, we did not validate the trained R3D model on external echocardiogram datasets, which would have shed insight into its broader applicability. Echocardiograms are protected patient data, and the EchoNet dataset is to our knowledge the only one available to researchers, meaning we could not access an external dataset to validate on.
Conclusions
In this paper, we provide vector embeddings generated by an R3D network that we trained to classify the healthiness of a heart based on estimated EF from echocardiograms. This model achieves a high AUC and accuracy, on par with current best-practice models, supporting the quality of the embeddings. These vector embeddings describe each of the 10,030 echocardiograms in the EchoNet dataset in 400-dimensional latent space, constituting a valuable resource that enables the improvement of automated echocardiography algorithms, search tools, and multimodal generative tools. Researchers need not repeat our arduous training and extraction process to develop applications that advance cardiac patient care, as this resource is available to all to improve outcomes for all.
PERSPECTIVES.
COMPETENCY IN SYSTEMS-BASED PRACTICE: Residents are called to participate in systems solutions and advocate for optimal care systems. Crucial to this goal is the improvement of EF estimators that help medical professionals measure EF, as this allows them to identify patients at highest risk of heart failure. Essential to improving these models, finally, are embeddings like the ones presented here.
TRANSLATIONAL OUTLOOK: Vector embeddings themselves are not medical interventions—rather, they enable their development. Barriers to clinical translation therefore include the unfinished work of developing automated echocardiography models from the raw embeddings and echocardiograms available to the research community. Future research directions follow the development of the applications described in the introduction, using our embeddings for active and deep learning algorithms to improve automated echocardiography, for echocardiogram similarity search systems and for automated echocardiography report generation.
Funding support and author disclosures
The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
We thank our mentors, Prabhu Sasankhan, Po-Chih Kuo, George Tang, Brigitte Kazzi, Leo A Celi, and Jacques Kpodonu, for their clinical and computational guidance on this work.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
References
- 1.Virani S.S., Alonso A., Benjamin E.J., et al. Heart disease and stroke statistics-2020 update: a report from the American heart association. Circulation. 2020;141(9):e139–e596. doi: 10.1161/cir.0000000000000757. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin E.J., Muntner P., Alonso A., et al. Heart disease and stroke statistics-2019 update: a report from the American heart association. Circulation. 2019;139(10):e56–e528. doi: 10.1161/cir.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 3.Hunt S.A., Abraham W.T., Chin M.H., et al. 2009 focused update incorporated into the ACC/AHA 2005 guidelines for the diagnosis and management of heart failure in adults: a report of the American college of cardiology foundation/American heart association task force on practice guidelines: developed in collaboration with the international society for heart and lung transplantation. J Am Coll Cardiol. 2009;53:e1–e90. doi: 10.1161/circulationaha.109.192065. [DOI] [PubMed] [Google Scholar]
- 4.Cleland J.G., Torabi A., Khan N.K. Epidemiology and management of heart failure and left ventricular systolic dysfunction in the aftermath of a myocardial infarction. Heart. 2005;91(Suppl 2):ii7–ii13. doi: 10.1136/hrt.2005.062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ouyang D., He B., Ghorbani A., et al. 33rd conference on Neural Information Processing Systems (NeurIPS 2019) 2019. EchoNet-dynamic: a large new cardiac motion video data resource for medical machine learning.https://api.semanticscholar.org/CorpusID:225101287 [Google Scholar]
- 6.Ouyang D., He B., Ghorbani A., et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature. 2020;580(7802):252–256. doi: 10.1038/s41586-020-2145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asch F.M., Polivert N., Abraham T., et al. Automated echocardiographic quantification of left ventricular ejection fraction without volume measurements using a machine learning algorithm mimicking a human expert. Circ Cardiovasc Imaging. 2019;12(9):e009303. doi: 10.1161/circimaging.119.009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almadani A., Shivdeo A., Agu E., Kpodonu J. 2022 IEEE International Conference on Big Data (Big Data); Osaka, Japan: 2022. Deep Video Action Recognition Models for Assessing Cardiac Function from Echocardiograms; pp. 5189–5199. [DOI] [Google Scholar]
- 9.Zhang J., Gajjala S., Agrawal P., et al. Fully automated echocardiogram interpretation in clinical practice. Circulation. 2018;138(16):1623–1635. doi: 10.1161/circulationaha.118.034338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J., Ding W., Zhang J., et al. Variational deep embedding-based active learning for the diagnosis of pneumonia. Front Neurorobot. 2022;16 doi: 10.3389/fnbot.2022.1059739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adiban M., BabaAli B., Shehnepoor S. Statistical feature embedding for heart sound classification. J Electr Eng. 2019;70(4):259–272. doi: 10.2478/jee-2019-0056. [DOI] [Google Scholar]
- 12.Jush F.K., Truong T., Vogler S., Lenga M. Medical image retrieval using pretrained embeddings. arXiv. 2023;2311 doi: 10.48550/arXiv.2311.13547. [DOI] [Google Scholar]
- 13.Yang Y., Yu J., Zhang J., Han W., Jiang H., Huang Q.A. Joint embedding of deep visual and semantic features for medical image report generation. IEEE Trans Multimed. 2023;25:167–178. doi: 10.1109/TMM.2023.243868822. [DOI] [Google Scholar]
- 14.Ososkov G., Goncharov P. Two-stage approach to image classification by deep neural networks. EPJ Web Conf. 2018;173 doi: 10.1051/epjconf/201817301009. [DOI] [Google Scholar]
- 15.Tran D., Wang H., Torresani L., Ray J., LeCun Y., Paluri M. 2018 IEEE/CVF Conference on computer vision and pattern recognition (CVPR) 2018. A closer look at spatiotemporal convolutions for action recognition; pp. 6450–6459. [DOI] [Google Scholar]
- 16.Kay W., Carreira J., Simonyan K., et al. The Kinetics human action video dataset. arXiv. 2017 doi: 10.48550/arXiv.1705.06950. [DOI] [Google Scholar]
- 17.Muldoon M., Khan N. Lightweight and interpretable left ventricular ejection fraction estimation using Mobile U-Net. arXiv. 2023 doi: 10.32920/22734314. [DOI] [Google Scholar]
- 18.Vicent L., Álvarez-García J., Vazquez-Garcia R., et al. Coronary artery disease and prognosis of heart failure with reduced ejection fraction. J Clin Med. 2023;12(8):3028. doi: 10.3390/jcm12083028. [DOI] [PMC free article] [PubMed] [Google Scholar]