Abstract
Arenaviruses have a bisegmented negative-strand RNA genome whose proteomic capability is limited to only four polypeptides, namely, nucleoprotein (NP), surface glycoprotein (GP) that is proteolytically processed into GP1+GP2, polymerase (L), and a small (11-kDa) RING finger protein (Z). The role of Z during the Lymphocytic choriomeningitis virus (LCMV) life cycle is poorly understood. We investigated the function of Z in virus transcription and replication by using a reverse genetic system for the prototypic arenavirus LCMV. This system involves an LCMV minigenome and the minimal viral trans-acting factors (NP and L), expressed from separated cotransfected plasmids. Cotransfection of the Z cDNA strongly inhibited LCMV minigenome expression. The effect required synthesis of Z protein; its magnitude was dose dependent and occurred with levels of Z protein substantially lower than those observed in LCMV-infected cells. Coexpression of Z did not prevent the encapsidation of plasmid supplied minigenome, but it affected both transcription and RNA replication similarly. Mutations in Z that unfolded its RING finger domain eliminated its inhibitory activity, but RING proteins not related to Z did not affect LCMV minigenome expression. Consistent with the minigenome results, cells transiently expressing Z exhibited decreased susceptibility to infection with LCMV.
Lymphocytic choriomeningitis virus (LCMV) is the prototypic member of the family Arenaviridae, which includes important human pathogens such as Lassa fever and Junin viruses (11, 41, 43, 53, 55). In addition, LCMV provides one of the most widely used model systems to study viral persistence and pathogenesis (7, 10, 37, 38, 39, 40, 45).
The LCMV genome consists of two negative-sense single-stranded RNA segments, L and S, with approximate sizes of 7.2 and 3.4 kb, respectively (44, 48, 52, 53). Each RNA segment has an ambisense coding strategy, encoding two proteins in opposite orientations, separated by an intergenic region (IGR) (48, 53). The S RNA directs synthesis of the three major structural proteins: the nucleoprotein, NP (ca. 63 kDa) (44, 48, 53), and the two virion glycoproteins, GP-1 (40 to 46 kDa) and GP-2 (35 kDa), which are derived by posttranslational cleavage of a precursor polypeptide, GP-C (75 kDa) (48, 53, 54, 60). Tetramers of GP-1 and GP-2 make up the spikes on the virion envelope and mediate virus interaction with host cell surface receptor (6, 13). The L RNA segment encodes a high-molecular-mass protein (L, ca. 200 kDa) (52), which has the characteristic motifs conserved in all of the viral RNA-dependent RNA polymerases (46), and a small (11-kDa) RING finger protein (Z) (50). The NP, the most abundant viral protein in virus-infected cells, is associated with the viral RNA to form the nucleocapsid (NC), which is the template for the viral RNA polymerase (23, 24). The L protein is thought to be the main viral component of the arenavirus polymerase (23, 48, 53). The NC associated with the viral polymerase constitutes the viral ribonucleoprotein (RNP), which is active in virus transcription and replication (23, 48, 53). As with other negative-strand RNA viruses, this RNP is the minimum unit of LCMV infectivity.
The investigation of the molecular mechanisms underlying LCMV persistence and its associated disorders has been hampered by the lack of a genetic system to analyze the structure and function of the LCMV genome and its gene products. We recently described a system in which RNA synthesis, both transcription and replication, mediated by LCMV polymerase is reconstituted by intracellular coexpression of an LCMV minigenome and viral proteins from transfected plasmids (33). This allowed us to initiate the analysis of cis- and trans-acting elements required for replication and transcription of the LCMV genome. Using this system we showed that the 5′ and 3′ untranslated regions (UTRs), together with the IGR of the S RNA, are sufficient cis-acting signals to allow RNA synthesis mediated by LCMV RNA polymerase (33), although it remains to be determined which of these elements is strictly required for virus replication and transcription. We also demonstrated that NP and L are the minimal trans-acting viral factors required for replication and transcription of LCMV genome analogues (33).
The Z proteins are highly conserved among the arenaviruses (27). Results from in vitro transcription and immunodepletion studies have implicated Z in both genome replication and mRNA synthesis in Tacaribe virus (25). However, Z was not required for intracellular transcription and replication of the LCMV minigenome (33). Biochemical and immunological studies have suggested that Z might be the arenavirus counterpart of the matrix (M) protein found in other negative-strand RNA viruses (48, 49). Moreover, Z has been shown to interact with several host cell proteins. The association of Z with the eukaryotic initiation factor 4E (eIF-4E) has been implicated in repression of protein synthesis in an eIF-4E-dependent manner (12). Z interacts also with the promyelocytic leukemia (PML) protein, leading to the relocation of PML nuclear bodies to the cytoplasm, a process which has been proposed to be responsible for the noncytolytic nature of LCMV (3, 5). Together, these observations raise intriguing questions about a spectrum of potential functions played by the Z protein in the biology of arenavirus. Here, we present evidence that the Z protein, in a dose-dependent manner, is a potent inhibitor of intracellular transcription and replication of the LCMV minigenome. We also show that the structural integrity of the RING finger domain is required for this inhibitory activity, whereas other RING proteins, not related to Z, do not cause inhibition of RNA synthesis mediated by LCMV polymerase in the minigenome assay. Moreover, transiently transfected cells expressing Z at levels similar to those seen at 24 h postinfection (p.i.) during the natural course of LCMV infection exhibited decreased susceptibility to infection with LCMV.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney cells (BHK-21) were grown in Dulbecco modified minimal essential medium supplemented with heat-inactivated (55°C for 30 min) 10% fetal calf serum, 2 mM l-glutamine, 1× tryptose phosphate broth, 1 mM sodium pyruvate, and 0.5% glucose. BSRT7 cells (9) were grown in minimal essential medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 1× tryptose phosphate broth, 1× minimal essential medium amino acids. Selection of BSRT7 cells was performed using G418 (1 mg/ml). Infections with LCMV strain Armstrong (Arm) were done using a plaque isolated clonal virus population (47).
Plasmids and DNA transfection.
Plasmids pLCMVSCAT2, pCITE-NP, pGEM-L, and pUCIRES-Z have been described (33), as well as plasmids pT7110 (20) and RMX-PML (31). Plasmid pTMI-GFP was generated by cloning the open reading frame (ORF) of the green fluorescent protein (GFP) into the NcoI site of pTMI (34). Plasmid pCITE-rho was generated by inserting a XbaI/BamHI DNA fragment containing the chicken rhomboid full-length cDNA (courtesy of A. Bang) into XbaI/BamHI-digested pCITE2a(+) vector (Novagen). To generate plasmid pGEM-IE86, the IE84 ORF of human cytomegalovirus (huCMVIE84), present in plasmid RSV-IE86 (26), was amplified by PCR using specific primers FIE86#1 (5′-CGGGATCCATGGAGTCCTCTGCCAAGAGAAAG) and BIE86#2 (5′-GCTCTAGAAGTTTTTCACTTACTGAGACTTGTTC) and the following PCR conditions: 5 min at 94°C; 25 cycles of 1 min at 94°C, 1 min at 58°C, and 3 min at 72°C; and finally 10 min at 72°C with Pfu polymerase (Roche). PCR product was purified with the PCR purification kit Concert rapid PCR purification system (Life Technologies). Purified PCR product was digested with XbaI and BamHI, purified, and cloned into XbaI/BamHI sites of pGEM-3Z (Promega). Restriction sites XbaI and BamHI at the 5′ end of the primers are underlined.
To generate plasmid pUCIRESZ-stop, two stop codons and a frameshift insertion were introduced immediately after the start codon of the Z ORF by site-directed mutagenesis. For this, the Z gene was amplified by PCR from pUCIRES-Z with two specific primers: FLCMVZ#1 (5′-CATGCCATGGGTTAATAAACAAGGCAAGTCCAGAGGAGAAAG), carrying the mutations, and the reverse primer BLCMVZ#2 (5′- AAAAGGCCTCCCGGGTTACTCTTCGTAGGGAGGTGGAG). Restriction sites NcoI and StuI at the 5′ end of the primers are underlined. PCR conditions were as follows: 5 min at 94°C; 25 cycles of 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C; and finally 10 min at 72°C, using Taq polymerase (Roche). PCR product was purified on a column with the PCR purification kit (Life Technologies), digested with NcoI and StuI and cloned in the backbone fragment of pUCIRES-Z, which also was digested with NcoI and StuI.
Plasmid pUCIRESZ-F32G35 was generated by overlap extension PCR in two steps. In the first step, plasmid pUCIRES-Z (10 ng) was used as a template for PCR fragment 1 using primers FLCZtag1 (5′-CATGCCATGGGTCAAGGCAAGTCC) and BLCZF32-G35 (5′-TCTGCCAGCCAGATTTGAAGCTTAAAGGGCCAAGATAGG) carrying the point mutations. In a parallel step, plasmid pUCIRES-Z (10 ng) was used as a template for PCR fragment 2 with primers FLCZF32-G35 (5′-CTTCAAATCTGGCTGGCAGAAATTTGACAG), carrying the point mutations, and BLCMVZ#2 (previously described). The two PCR fragments (i.e., from independent PCRs 1 and 2) were generated with Pfu polymerase (Roche) by using the same cycling parameters: 5 min at 94°C; 25 cycles of 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C; and finally 10 min at 72°C. Fragments were gel purified with the QIAquick gel extraction kit (Qiagen). In a second step, PCR fragments 1 and 2 were combined and fused by overlap extension PCR using primers FLCZtag1 and BLCMVZ#2 and the following cycling conditions: 5 min at 94°C; 25 cycles of 1 min at 94°C, 1 min 30 s at 58°C, and 1 min at 72°C; and finally 10 min at 72°C. The final 300-bp product was gel purified, digested with NcoI and StuI, and then ligated into an NcoI/StuI backbone fragment of pUCIRES-Z. The restriction site NcoI in primer FLCZtag1 is underlined.
Transfection of BHK-21 cells was performed with 10 μl of Lipofectamine (Life Technologies) per 35-mm well and the indicated amount of plasmid DNA. BSRT7/5 cells stably expressing T7 polymerase were transfected with 2 μl of SuperFect (Qiagen) and 1 μg of DNA
Analysis of minigenome expression.
The analysis of LCMV minigenome expression was done essentially as described previously (33). Briefly, BHK-21 cells were infected with vTF7.3 at a multiplicity of infection (MOI) of 3 and transfected with pLCMVSCAT2, pCITE-NP, pGEM-L, pTMI-GFP, and different amounts of plasmid DNA encoding Z-wt, Z-mutant, and other RING finger proteins as indicated in the corresponding figure legends. Plasmid pLCMVSCAT2 directs intracellular T7-mediated synthesis of the LCMVSCAT2 minigenome in which the viral NP and GP ORF encoded by the S RNA have been replaced by an antisense (AS) copy of the chloramphenicol acetyltransferase (CAT) ORF. The CAT AS ORF is flanked by the 5′ UTR-IGR and 3′ UTR of the LCMV S RNA. Plasmid pTMI-GFP was included to assess the efficiency of transfection based on GFP expression. In all cases, the amount of DNA used in the transfections was normalized to 2.7 μg/M6 well by using pTMI plasmid. Transfections were performed by using Lipofectamine. We included 1-β-d-arabinofuranosylcytosine (50 μg/ml) to inhibit the late phase of vaccinia virus expression. Cells were resuspended in 2 ml of phosphate-buffered saline (PBS) and split into two aliquots. After the cells were spun, one aliquot was resuspended in 50 μl of 0.25 M Tris-HCl (pH 7.5), and cell lysates were prepared and assayed for CAT activity. The second aliquot was resuspended in lysis buffer (50 mM Tris-HCl, pH 8; 62.5 mM EDTA; 1% NP-40; 0.4% deoxycholate) and used for analysis of protein expression.
Western blot assay.
Protein expression from pT7-110 (20), RMX-PML (31), pUCIRESZ-stop, and pUCIRESZ-F32G35, as well as pCITE-NP and pGEM-L, was analyzed by Western blot. For this, BHK-21 cells were infected with vTF7.3 at an MOI of 3, followed by transfection using Lipofectamine with the above-mentioned plasmids. Cells were harvested in lysis buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by blotting on a polyvinyidene difluoride Immobilon-P membrane (Millipore). Expression of Vmw110 was detected using a mouse monoclonal antibody to VmW110 (ascites 1106 [20]). PML expression was detected by using a rabbit anti-PML antibody. The LCMV Z wild-type and mutant proteins were detected with a rabbit anti-Z antiserum. Rabbit antisera to PML and LCMV Z were obtained from R. Evans (31) and M. Salvato (49), respectively. Appropriate secondary antibodies (anti-mouse and anti-rabbit) conjugated to peroxidase were used at dilutions recommended by the supplier (Roche). Expression of LCMV NP and L proteins was detected using a guinea pig and a rabbit polyclonal serum to LCMV (54) and to L (33), respectively. Proteins were visualized by enhanced chemiluminescence (ECL; Roche). The amount of protein in the lysates was quantified with a Bio-Rad protein assay kit. The mouse monoclonal antibody to huCMVIE84 did not work in a Western blot. Hence, detection of this protein was done by immunofluorescence.
CAT assays.
Whole-cell extracts were prepared by three freeze-thawing cycles in a dry ice-ethanol bath and at 37°C in a water bath. Cell extracts were clarified by centrifugation: 12,000 × g for 5 min at 4°C. Equal amounts (5 μl) of each sample were incubated 30 min at 37°C in the presence of 0.25 M Tris (pH 7.8), 0.6 mg of acetyl coenzyme A (Roche)/ml, and 0.05 μCi of 14C-labeled chloramphenicol (ICN). The reaction was stopped by adding 1 ml of ethyl acetate, and chloramphenicol was extracted by separating the phases by centrifugation. Then, 900 μl of supernatant was dried, resuspended in 25 μl of ethyl acetate, and analyzed by thin-layer chromatography (TLC). Samples were run for 30 min in CHCl3-methanol (95:5). The TLC plate was dried and exposed to an X-ray film.
Immunofluorescence.
Cells grown onto coverslips and placed on the bottom of the wells of an M24 plate were washed once with PBS and fixed in 100% methanol for 5 min at −20°C. After several washes with PBS and a blocking step with 10% normal goat serum in PBS for 15 min at room temperature, cells were incubated for 1 h at room temperature with the appropriate primary antibodies, i.e., mouse monoclonal antibodies to huCMVIE86, c-myc, or vesicular stomatitis virus (VSV) M; a guinea pig polyclonal antibody to LCMV; and a rabbit polyclonal antibody to Z or to the cytoplasmic tail of VSV G. After several washes with PBS–0.1% Triton X-100, samples were incubated for 45 min at room temperature with appropriate secondary fluorescent antibodies: fluorescein isothiocyanate (FITC)-labeled goat anti-mouse immunoglobulin G (IgG), Texas red (TXR)-labeled goat anti-mouse IgG, FITC-labeled goat anti-guinea pig IgG, and TXR-labeled goat anti-rabbit IgG. After extensive washes with PBS–0.1% Triton X-100, coverslips were mounted using mowiol and analyzed by fluorescence microscopy. Slides were digitized by using Adobe Photoshop and Canvas software.
Analysis of RNA by Northern blot hybridization.
BHK-21 cells infected with LCMV Arm5 at an MOI of 3 or else transfected with plasmids LCMV Z-wt and Z-stop were harvested at the time points indicated and homogenized in TRIreagent (Molecular Research Center, Inc.). RNA was extracted according to the supplier's protocol and resuspended in Formazol (Nuclear Research Center, Inc.). RNA (5 μg) was fractionated by 2.2 M formaldehyde agarose (1.2%) gel electrophoresis. The gel was washed first in warm diethyl pyrocarbonate-H2O and then in warm 10 mM NaPO4. Transfer was performed overnight in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with a Nytran 0.22-μm (pore-size) membrane (Schleicher & Schuell). RNA was cross-linked by exposure to UV and, after a brief wash with H2O, the membrane was prehybridized in QuickHyb (Stratagene) for 30 min at 65°C. Hybridization was done at 65°C for 2 h with an NPα Z-DNA [α-32P]dCTP probe, using 5 ng of probe (specific activity, >109 cpm/μg)/ml and 100 μg of denatured salmon sperm DNA. For detection of CAT mRNA and the antiminigenome RNA, the membrane was hybridized with a [32P]UTP-labeled AS CAT riboprobe. Riboprobe was prepared according to the supplier's protocol (Ambion). Briefly, linearized DNA was incubated for 1 h at 37°C with T7 polymerase, [32P]rUTP, rATP, rCTP, rGTP, and RNase inhibitor; subsequently, the DNA template was digested for 15 min at 37°C with DNase RNase Free (Boehringer Mannheim), and finally the RNA probe was precipitated and resuspended in 0.5 mM EDTA. The membrane was prehybridized for 1 h at 65°C in Ambion Zip hybridization solution (Ambion) and hybridized overnight with the riboprobe (3 × 106 cpm/ml). After hybridization (in both cases), the membranes were washed twice with warm 2× SSC–0.2% SDS, washed twice with warm 0.2× SSC–0.2% SDS, and then exposed to an X-ray film.
Analysis of encapsidated RNA.
Cytosolic extracts were prepared from transfected and LCMV-infected cells by incubating them on ice for 15 min in hypotonic buffer (10 mM Tris, pH 7.5; 10 mM KCl; 1.5 mM MgCl2; 5 mM dithiothreitol [DTT]; 0.5% NP-40). Clarified lysates were incubated 5 min with NaCl to a final concentration of 0.5 M and then loaded onto the top of a discontinuous 5 and 0.5 M CsCl2 gradient in TNE buffer (10 mM Tris, pH 7.8; 150 mM NaCl; 5 mM EDTA). After 3 h of centrifugation, the interphase between the 5 and 0.5 M CsCl2 containing the RNP was removed, diluted 1:1 with TNE buffer, and recentrifuged for 1 h to pellet the RNP. RNP-containing pellets were resuspended in 500 μl of TRIreagent, and encapsidated RNA was isolated and analyzed by reverse transcription-PCR (RT-PCR). RNA isolated from S-segment minigenome RNP was reverse transcribed with Superscript (Life Technologies) and the CAT sense primer BSM1CAT (5′- GATGAATGCTCATCCGGAATTCCG). The resulting cDNA was amplified by PCR using primer BSM1CAT and the CAT AS primer PFLM1CAT (5′-CCCAGGGATTGGCTGAGACAAAAAACATATTC). RNA isolated from bona fide LCMV RNP was reverse transcribed using the NP sense primer NP2750F (5′-GTCTGTGACTGTTTGGCCATAC), and the resulting cDNA was amplified by PCR with primers NP2750F and NP3103R (5′-CCAGACTGTACATTCTCTTGTGG), which produced an NP fragment of 350 bp. PCR conditions were as follows: 5 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C; and finally 10 min at 72°C. PCR was performed using Taq polymerase (Roche). PCR products were resolved by agarose (2%) gel electrophoresis and visualized by ethidium bromide staining.
Preparation of LCMV RNP and transfection of cells with RNP. (i) RNP preparation.
Cells were washed once with ice-cold PBS, collected (5 min, 800 × g, 4°C), and resuspended (107 cells/ml) in ice-cold hypotonic HB buffer (10 mM Tris-HCl, pH 7.5; 10 mM KCl; 1.5 mM MgCl2; 5 mM DTT) containing 0.5% of the nonionic detergent NP-40. After a 15-min incubation in ice, samples were centrifuged (5 min, 1,000 × g, 4°C). The supernatant (cytosolic fraction) was saved on ice, and the nuclear pellet washed with 0.5 volume of HB buffer–0.5% NP-40. Both supernatants were combined (cytosolic fraction) and clarified by centrifugation (10 min, 15,000 × g, 4°C). Clarified cytosolic fraction was adjusted to 1% NP-40, and RNP was collected by ultracentrifugation (60 min, 100,000 × g, 4°C) through a discontinuous 50 to 25% glycerol gradient in buffer I (75 mM NaCl; 10 mM Tris-HCl, pH 7.5; 2 mM DTT; 1% NP-40). The microsome pellet containing RNP was resupended in HB buffer containing 40% glycerol. Samples were stored at −80°C.
(ii) RNP transfection.
Procedures for RNP transfection were essentially as described previously (16). Briefly, cell monolayers were washed with Opti-MEM containing 100 μg of autoclaved gelatin (OpM-G)/ml. After treatment for 30 min at room temperature with OpM-G containing DEAE-dextran (5 × 105 Da; 300 μg/ml) and dimethyl sulfoxide (0.5%), cells were washed once with OpM-G. RNP, diluted in OpM-G, and adsorbed (300 μl/35-mm well) for 60 min at 37°C; cells were then washed twice with OpM-G and 2.5 ml of medium containing 1% fetal bovine serum was added per M6 well.
RESULTS
Effect of LCMV Z on the expression of an LCMV S-segment minigenome.
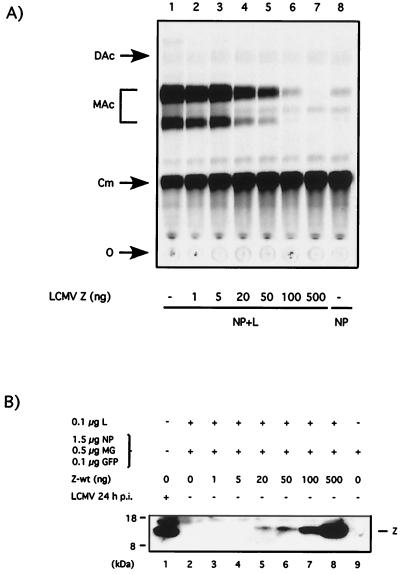
To investigate the role played by Z in the control of LCMV RNA synthesis, we used a reverse genetic system that enabled us to rescue the expression of a plasmid-derived LCMV minigenome. In this system the Z protein is not necessary for minigenome RNA synthesis mediated by the LCMV polymerase (33). Moreover, Z appeared to have a negative impact on the minigenome expression (33). To better understand the mechanism underlying this effect, we evaluated the consequences of incrementally increasing amounts of the Z expression plasmid in the minigenome rescue assay. Z caused inhibition of CAT enzymatic activity in a dose-dependent manner (Fig. 1A). Transfection of 20 ng of Z plasmid was sufficient to significantly decrease CAT activity (Fig. 1A, lane 4), whereas 100 ng of the Z plasmid was able to completely abolish CAT activity specifically mediated by the LCMV polymerase (compare lanes 6 and 8 in Fig. 1A).
FIG. 1.
LCMV Z inhibits CAT expression by the LCMVSCAT2 minigenome in a dose-dependent manner. BHK-21 cells were infected with vTF7.3 (MOI = 3) and subsequently cotransfected with 0.5 μg of pLCMVSCAT2, 1.5 μg of pCITE-NP, 0.1 μg of pGEM-L, 0.1 μg of pTMI-GFP, and increasing amounts of pUCIRES-Z as indicated. Plasmid pGEM-L was not added in lane 8. In all of the samples the total amount of DNA was kept constant (2.7 μg) by adding the appropriate amount of plasmid pTMI. The use of pTMI-GFP allowed us to determine the efficiency of transfection, based on number of GFP-positive cells, prior harvesting of the cells for CAT assays, and protein expression analysis. (A) LCMVSCAT2 minigenome expression. At 24 h after transfection, cell lysates were prepared for measuring CAT activity as described in Material and Methods. (B) Z protein expression. Cell lysates were subjected to SDS-PAGE, and the levels of Z protein expression were determined by Western blot using a rabbit antibody to Z. Lysate applied in lane 1 was prepared from cells infected with LCMV (MOI = 3) at 24 h p.i. The position of the Z protein is indicated on the right, and the molecular size markers are indicated on the left. O, origin; Cm, chloramphenicol; MAc, monoacetylated chloramphenicol; DAc, diacetylated chloramphenicol.
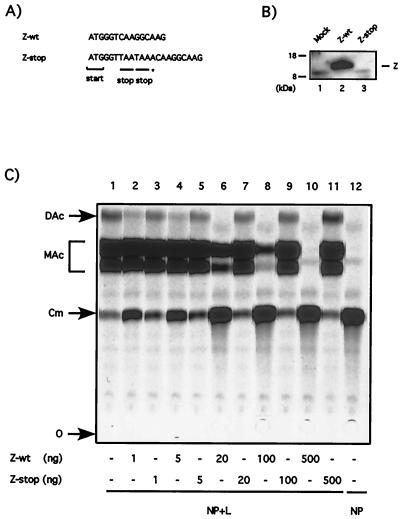
Coexpression of increasing amounts of Z plasmid resulted in concomitant increase in the expression of Z protein (Fig 1B). The amount of Z protein required to completely inhibit minigenome derived CAT activity was slightly lower than that found in LCMV infected cells (MOI = 3) at 24 h p.i. (compare lanes 1, 6, and 7 in Fig. 1B). We next examined whether the inhibitory effect associated with the Z plasmid depended upon its ability to express Z protein or whether this was a consequence of the synthesis of its mRNA. For this, we generated the construct Z-stop by insertion of two stop codons and a frameshift mutation right after the AUG translational start site of Z (Fig. 2A). These mutations would enable transcription of the Z gene but should abolish translation of the protein. As predicted, cells transfected with Z and Z-stop expressed similar levels of Z RNA (data not shown), but Z protein was not detected in cells transfected with Z-stop (Fig. 2B). We then evaluated the effect of Z-stop mutant on the expression of CAT enzyme by the LCMVSCAT2 minigenome complemented by NP and L plasmids. The Z-wt plasmid caused the expected strong inhibition of CAT expression, whereas the Z-stop plasmid had no effect on CAT expression (Fig. 2C).
FIG. 2.
Inhibitory effect of the Z plasmid requires synthesis of the Z protein. (A) Nucleotide sequence differences between Z-wt and the Z-stop mutant. The Z-stop mutant contains two stop codons (underlined) inserted downstream of the ATG start codon. The insertion of an additional A (✽) into the Z-stop construct created a frameshift of the downstream reading frame. (B) The Z-stop mutant is unable to produce the Z protein. BHK-21 cells were infected with vTF7.3 (MOI = 3) and transfected with 0.5 μg of pUCIRES-Z or 0.5 μg of pUCIRESZ-stop. Lysates were prepared 24 h after transfection and subjected to SDS-PAGE. Z protein expression was detected by Western blotting with a rabbit antiserum to Z. The position of the Z protein is indicated on the right, and the molecular size markers are indicated on the left. (C) The Z-stop mutant does not inhibit CAT activity by the LCMVSCAT2 minigenome. BHK-21 cells were infected with vTF7.3 (MOI = 3) and cotransfected with 0.5 μg of pLCMVSCAT2, 1.5 μg of pCITE-NP, 0.1 μg of pGEM-L, 0.1 μg of pTMI-GFP, and increasing amount of pUCIRES-Z or pUCIRESZ-stop as indicated. Plasmid pGEM-L was not added in lane 12. In all of the samples the amount of DNA was kept constant by adding plasmid pTMI to 2.7 μg. At 24 h after transfection, the samples were assayed for CAT activity as described in Materials and Methods. O, origin; Cm, chloramphenicol; MAc, monoacetylated chloramphenicol; DAc, diacetylated chloramphenicol.
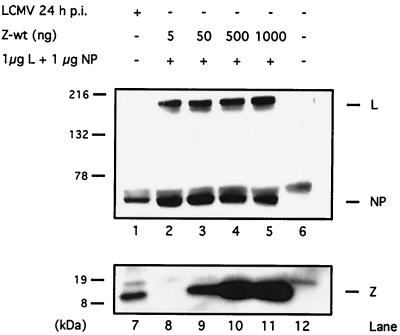
Since the LCMV NP and the L proteins are required to rescue LCMV minigenome expression (33), we assessed whether in our transfection assay coexpression of Z protein could interfere with NP and/or L expression. For this purpose, cells were cotransfected with plasmids expressing NP and L, together with increasing amounts of Z, and cell lysates were analyzed for NP, L, and Z expression by Western blot. Expression levels of NP and L proteins were not affected by coexpression of Z (Fig. 3).
FIG. 3.
Expression of Z protein does not affect plasmid-derived expression of NP or L. Equal amounts of cell lysates were separated either on an SDS–10% PAGE gel for the detection of NP and L protein expression or on an SDS–16% PAGE gel to detect Z protein expression. Lysates in lanes 2 to 5 and lanes 8 to 11 were prepared from cells infected with vTF7.3 (MOI = 3) and transfected with 1 μg of pCITE-NP and 1 μg of pGEM-L. The amount of cotransfected pUCIRES-Z is indicated at the top. Lysate prepared from mock-infected cells was loaded in lanes 6 and 12, whereas lysate from LCMV-infected cells was loaded in lanes 1 and 7. The positions of the L, NP, and Z protein are indicated on the right, and the molecular size markers are indicated on the left. Consistent with previous findings, L protein was not detected by Western blotting in the cell lysates from LCMV-infected cells (33).
Levels of Z associated with infectious LCMV RNP.
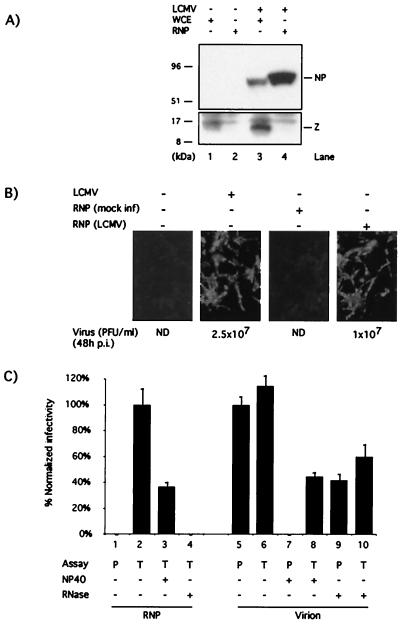
Z was not required for RNA synthesis mediated by the LCMV polymerase in the minigenome assay. This finding led us to examine whether Z was also dispensable for the biosynthetic processes involved in the initiation of LCMV productive infection. For this, we prepared intracellular LCMV RNPs and characterized them both biochemically and biologically. LCMV RNPs prepared following the procedures here described have been shown to be transcriptionally active in vitro (23). These RNPs did not contain detectable levels of Z protein (Fig. 4A). However, they were infectious on the basis of their ability to direct synthesis of LCMV macromolecules and production of infectious virions upon transfection of BHK-1 cells (Fig. 4B). Infectivity associated with LCMV RNP depended on transfection conditions and was resistant to treatment with the nonionic detergent NP-40 but was sensitive to RNase treatment (Fig. 4C).
FIG. 4.
Biochemical and biological characterization of LCMV RNP. (A) Levels of Z associated with LCMV RNP. Whole-cell extracts (lanes 1 and 3) and RNP (lanes 2 and 4) were prepared from mock (lanes 1 and 2) and LCMV-infected (lanes 3 and 4) cells and analyzed by Western blotting using a guinea pig antiserum to LCMV and a rabbit antiserum to Z. The amount of RNP loaded in lane 4 corresponded to a sixfold-higher amount (cell equivalent) of that loaded in lane 3. (B) Infectivity of LCMV RNP. Cells were transfected with RNP prepared from mock- and LCMV-infected cells. As controls, cells were also either LCMV or mock infected. At 48 h p.i. or posttransfection, cells were fixed and analyzed by immunofluorescence by using a guinea pig polyclonal antiserum to LCMV. Levels of infectious LCMV in the corresponding supernatants (48 h p.i.) were determined by plaque assay (18). (C) Characterization of infectivity associated with LCMV RNP. LCMV RNP (lanes 1 to 4) or virions (lanes 5 to 10) were treated with NP-40 (0.05% for 15 min on ice) or pancreatic RNase (25 μg for 30 min at 20°C) or were left untreated. Infectivity associated with treated and untreated virions and RNP was assayed by direct plaque assay (P) or by transfection (T). RNP-associated infectivity was normalized to that obtained by transfection with untreated RNP. Virion-associated infectivity was normalized to that obtained by direct plaque assay with untreated virions. Three independent assays were done. Average values and standard deviations are shown.
Kinetics of Z expression during the natural course of LCMV infection.
The inhibitory effect of Z on LCMV minigenome expression would seem to be counterproductive for virus replication, especially if Z reaches high levels of expression at early stages during the natural life cycle of LCMV. Therefore, we examined the expression levels of Z, both mRNA and protein, during LCMV infection.
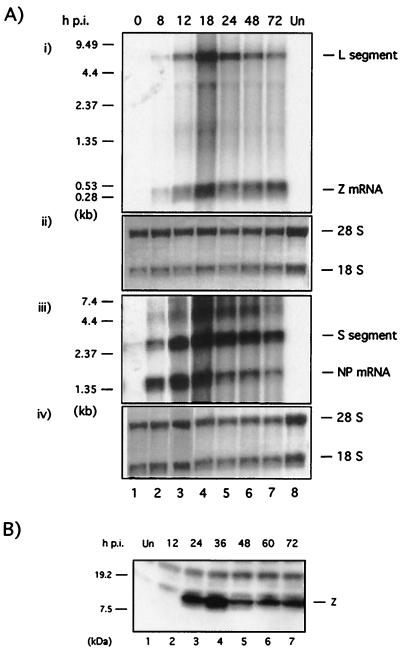
BHK-21 cells were infected with LCMV Arm5 at an MOI of 3 and harvested at the indicated time points (Fig. 5). Total cellular RNA was isolated and analyzed by Northern blot hybridization by using specific probes to detect Z and NP mRNAs, as well as the L and S RNA segments (Fig. 5A). We used Z and NP cDNA probes of similar size and specific activity. S-segment-derived RNA species (NP mRNA and genomic plus antigenomic S RNAs) were readily detected at 8 h p.i., whereas at this time p.i., the levels of L-segment-derived RNA species (Z mRNA and genomic plus antigenomic L RNAs) were barely detectable. Steady-state levels of NP mRNA and S RNA, as well as those of Z mRNA and L RNA, peaked at ca. 18 h p.i. This was followed by a decrease at 24 h p.i. in the steady-state levels of all four RNA species. The levels of NP mRNA and S and L RNA species, but not of Z mRNA, exhibited a slow steady decrease between 24 and 72 h p.i. During the first 12 h of infection, the levels of Z mRNA were significantly lower than those of NP mRNA (Fig. 5A). Consistent with previous findings (24), we also observed a preferential accumulation of genomic S RNA over genomic L for all time points analyzed (Fig. 5A).
FIG. 5.
Kinetic of Z expression during the natural course of LCMV infection. (A) Kinetic of Z mRNA synthesis. BHK-21 cells were infected with LCMV (MOI = 3), and total cell RNA isolated at the indicated time points was analyzed by Northern blot hybridization. For each time point, equal amounts of RNA were loaded in two different gels. Lane 7, in both gels, corresponds to RNA from mock-infected cells. Both gels were identically processed for Northern blot analysis. One membrane was used to detect Z mRNA and L segment by using a [32P]dCTP labeled Z-DNA probe (panel i). The other membrane was used to detect NP mRNA and the S segment using a [32P]dCTP labeled NP DNA probe (panel iii). Molecular markers are indicated on the right. Prior to hybridization, each membrane was stained with methylene blue to assess the total amount of RNA in each sample based on levels of 28S and 18S RNA (panels ii and iv). (B) Kinetics of Z protein expression. BHK-21 cells were infected with LCMV (MOI = 3) and harvested at the indicated time points. Cell lysates were subjected to SDS-PAGE and analyzed for the expression of Z protein by Western blotting with a rabbit antibody to Z. Lane 1 corresponds to mock-infected cells.
Equal amounts of whole lysates for each time point indicated were subjected to Western blot analysis using a specific anti-Z rabbit serum (Fig. 5B). Expression of Z protein was first detected at 24 h p.i. and peaked by 24 to 36 h p.i., followed by a slight decrease of its expression at later times (Fig. 5B).
Effect of Z on the encapsidation of the minigenome RNA.
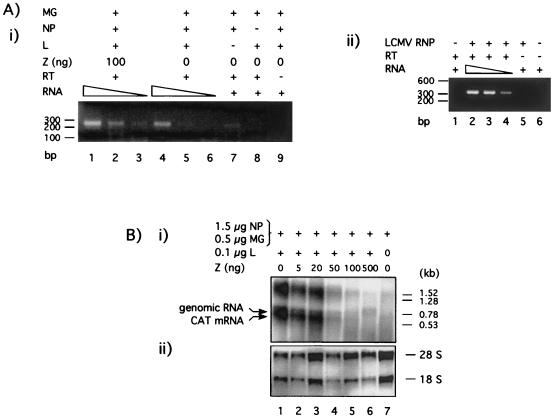
We expected that, as with other negative-strand RNA viruses (35, 36), viral encapsidated RNA would be the template for RNA synthesis mediated by the LCMV polymerase. In this case, encapsidation of plasmid-supplied minigenome RNA would be a first and required step for the synthesis of subgenomic mRNA and full-length antiminigenome RNA mediated by the LCMV polymerase. Therefore, we investigated whether Z could exert its inhibitory activity by preventing encapsidation of the minigenome RNA. For this analysis, we transfected BHK-21 cells with plasmids carrying the LCMVSCAT2 minigenome and the L and NP proteins, together with 100 ng of either a plasmid encoding Z or an empty plasmid vector as a negative control. Cytosolic cell extracts were prepared 24 h after transfection and loaded on a discontinuous CsCl2 gradient to separate the RNPs from the free RNA. In the case of LCMV, the susceptibility of encapsidated RNA to nucleases prevented us from using micrococcal nuclease treatment to assess levels of encapsidated minigenomes. Encapsidated RNA was isolated from purified S-segment minigenome RNPs and subjected to semiquantitative RT-PCR using specific primers to amplify an internal fragment of the minigenome, corresponding to sequences within the CAT ORF. The predicted 250-bp PCR product was obtained by using encapsidated RNA from cells that were transfected only with the NP and L supporting plasmids. The amount of PCR product generated decreased concomitantly with the decrease in the amount of input RNA used for the RT-PCR (Fig. 6A, panel i, lanes 4 to 6). Cotransfection with 100 ng of Z expression plasmid (lanes 1 to 3) did not prevent amplification of the same PCR fragment (Fig. 6A, panel i, lanes 1 to 3). As expected, no minigenome sequence was amplified in the absence of NP (Fig. 6A, panel i, lane 8). The absence of L decreased but did not prevent, the amplification of PCR product (Fig. 6A, panel i, lane 7). Omission of the RT step prevented amplification of minigenome sequences (Fig. 6A, panel i, lane 9). As a control, we used the same procedures to isolate bona fide LCMV RNPs from an amount of infected cells similar to that of transfected cells used to prepare the S-segment minigenome RNPs. RNA present in LCMV RNPs was isolated and characterized by RT-PCR as described in Materials and Methods (Fig. 6A, panel ii).
FIG. 6.
Effect of Z on encapsidation and RNA synthesis of LCMV S-segment minigenome. (A) Z does not inhibit encapsidation of plasmid supplied LCMVSCAT2 minigenome. (i) BHK-21 cells (106 cells/well, six-well plate) were infected with vTF7.3 (MOI = 3) and cotransfected with 0.5 μg of pLCMVSCAT2, 1.5 μg of pCITE-NP, 0.1 μg of pGEM-L, and 0.1 μg of pTMI-GFP. Plasmids expressing L and NP were omitted in lanes 7 and 8, respectively. Samples from lanes 1 to 3 were also transfected with 100 ng of pUCIRES-Z. In all of the samples the amount of DNA was kept constant (2.7 μg) by adding plasmid pTMI. At 24 h after transfection, cells (three wells/sample) were harvested and RNP was obtained as described in Materials and Methods. Encapsidated RNA was isolated and subjected to semiquantitative RT-PCR by using decreasing amounts of input RNA: 1/4, 1/8, and 1/16 of the total RNA were used in lanes 1 to 3 and lanes 4 to 6. In lanes 7 and 8, 1/8 of the total RNA was amplified. In lane 9, 1/4 of total RNA was analyzed, omitting the RT step as a control for plasmid DNA contamination in the RNP preparation. Molecular size markers are indicated on the left. MG refers to minigenome LCMVSCAT2. (ii) Bona fide LCMV RNPs were prepared from LCMV-infected BHK-21 cells (3 × 106 cells) at 48 h p.i. (MOI = 3). Encapsidated RNA was isolated and analyzed by semiquantitative RT-PCR as described in Materials and Methods. In this case, primers used for PCR amplified a segment (ca. 350 bp) of the S segment corresponding to NP sequences. (B) Effect of Z protein on RNA synthesis mediated by the LCMV polymerase. BHK-21 cells were infected with vTF7.3 (MOI = 3) and cotransfected with 0.5 μg of pLCMVSCAT2, 1.5 μg of pCITE-NP, 0.1 μg of pGEM-L, 0.1 μg of pTMI-GFP, and increasing amounts of pUCIRES-Z as indicated. Plasmid pGEM-L was not added in lane 7. Total cellular RNA was isolated, and equal amounts (5 μg) were analyzed by Northern blot hybridization. (i) Subgenomic CAT mRNA and full-length antigenomic RNA species were detected with a [32P]UTP AS CAT riboprobe. Molecular size markers are indicated on the left. (ii) Methylene blue staining of the membrane was done to determine the positions and levels of the 28S and 18S RNA.
Effect of Z protein on RNA synthesis.
The next event to follow encapsidation of the minigenome RNA is its transcription and replication. Decreased levels of transcription and/or replication can negatively affect minigenome-mediated CAT expression. Hence, we evaluated the effect of Z on these biosynthetic processes. After transfection of BHK-21 cells with increasing amounts of Z, together with plasmids encoding the LCMVSCAT2 minigenome, as well as NP and L proteins, RNA was isolated and analyzed by Northern blot using an AS CAT riboprobe. This probe hybridizes to both CAT mRNA (ca. 700 nucleotides) and the miniantigenomic (1,100 nucleotides) RNA species. The agarose gel electrophoresis conditions used did not separate well the two RNA species, resulting in the appearance of a broad band in the Northern blot (Fig. 6B). LCMV mRNAs are not polyadenylated. Hence, it was not possible to use oligo(dT) chromatography to distinguish between CAT mRNA and antigenomic RNA species.
Coexpression of the LCMV Z protein affected similarly both the synthesis of subgenomic CAT mRNA and the positive-sense antigenome RNA from the LCMVSCAT2 minigenome template RNA (Fig. 6B, lane 4). As predicted above, in the absence of L no synthesis of RNA mediated by the LCMV polymerase was detected (Fig. 6B, lane 7). The CAT AS probe also detected an additional broad-appearing band of ca. 1.5 to 1.7 kb. Synthesis of this RNA species also required L and was prevented by Z in a dose-dependent manner; therefore, it appears to be specifically produced by the virus polymerase. However, the precise nature of this RNA is currently unknown. It is worth noting that a similar finding was observed when RNA from LCMV-infected cells were analyzed by Northern blot using an NP probe. In addition to the expected subgenomic NP mRNA (1.5 kb) and S segment (3.5 kb) RNA species, an RNA of ca. 5 kb also hybridizes specifically to the NP probe (see Fig. 5A). It has been proposed that terminal complementarity may be responsible for the generation of unresolved dimeric RNA species that will migrate with an apparent molecular mass higher than that of the genomic S RNA. This situation could also apply to the S-segment minigenome RNA, which also has terminal complementarity.
The RING finger domain of the LCMV Z protein is required for the Z inhibitory activity.
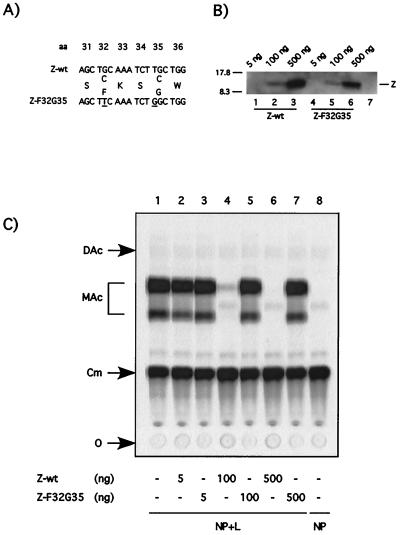
All arenavirus Z proteins thus far characterized contain a RING finger domain of Z (17, 27, 28, 48, 50). The RING finger domain has been identified in a large number of both cellular and viral proteins (51). We sought to determine whether the RING domain was required for the inhibition of RNA synthesis mediated by the LCMV polymerase. For this, we constructed the mutant Z-F32G35, in which C at positions 32 and 35 was changed to F and G, respectively (Fig. 7A). These mutations have been shown to disrupt the Z RING domain (2, 3). We first verified that Z-F32G35 could be expressed to levels similar to those of the parental Z-wt protein, as determined by Western blotting. Subsequently, we tested the mutant Z protein for its ability to inhibit CAT expression by the LCMV minigenome. Transfection with up to 500 ng of plasmid expressing Z-F32G35 did not interfere with CAT expression (Fig. 7C). However, 500 ng of plasmid Z-F32G35 resulted in levels of expression of the mutated Z protein significantly higher than those of Z-wt protein required to cause a strong inhibition of the minigenome expression (compare lanes 2 and 6 in Fig. 7 B).
FIG. 7.
Disruption of the RING finger motif annuls the inhibitory effect of the Z protein. (A) Amino acid changes introduced in the RING domain of mutant Z-F32G35. The RING finger mutant F32G35 contains two nucleotide changes at codon positions 32 and 35 that disrupt the RING finger motif by changing amino acid residues from C to F and from C to G, respectively. (B) The RING finger mutant Z F32G35 is stably expressed. BHK-21 cells were infected with vTF7.3 (MOI = 3) and transfected with the indicated amounts of pUCIRES-Z or pUCIRESZ-F32G35 plasmid DNA. Lane 7 shows results for mock-transfected cells. Lysates were subjected to SDS-PAGE, and immunodetection of Z proteins was performed with a rabbit anti-Z antibody. The position of Z is indicated on the right, and the molecular size markers are indicated on the left. (C) Mutant Z-F32G35 does not inhibit CAT activity mediated by the LCMVSCAT2 minigenome. Monolayers of BHK-21 cells were infected with vTF7.3 (MOI = 3) and cotransfected with 0.5 μg of pLCMVSCAT2, 1.5 μg of pCITE-NP, 0.1 μg of pGEM-L, 0.1 μg of pTMI-GFP, and increasing amounts of pUCIRES-Z and pUCIRESZ-F32G35, as indicated at the bottom. Plasmid pGEM-L was not added in lane 8. In all of the samples, the amount of DNA was kept constant by adding plasmid pTMI to 2.7 μg. Samples were assayed for CAT activity as described in Materials and Methods. O, origin; Cm, chloramphenicol; MAc, monoacetylated chloramphenicol; DAc, diacetylated chloramphenicol.
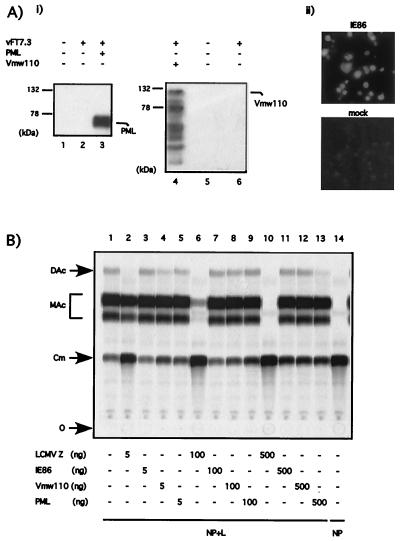
This finding raised the question whether the inhibitory effect could be observed with other RING motif containing proteins not related to Z. To investigate this question, we determined the effect of three other RING finger proteins, unrelated to LCMV Z, on LCMV minigenome expression. These were PML (31), a cellular RING finger protein which has been shown to interact with the LCMV Z protein (3, 4); protein Vmw110 of herpes simplex virus type 1 (HSV 1) (20); and the human cytomegalovirus protein IE86 (26). We first determined that we could express at high levels these three RING proteins (Fig. 8A). Expression of PML and Vmw110 was detected by Western blotting (Fig. 8A, panel i). However, the mouse antibody to IE86 did not work in Western blots; hence, the expression of this RING protein was verified by immunofluorescence (Fig. 8A, panel ii). Different amounts of PML, Vmw110, and IE86 plasmids were transfected in the context of a rescue experiment with the LCMVSCAT2 minigenome. The LCMV Z plasmid exhibited the expected dose response inhibition of CAT activity mediated by the LCMV minigenome. In contrast, PML, Vmw110 and IE86 all failed to inhibit CAT expression even at the highest (500 ng) amount of DNA plasmid tried (Fig. 8B).
FIG. 8.
LCMVSCAT2 minigenome expression is not inhibited by RING finger proteins unrelated to Z. (A) Expression of viral (Vmw110 and IE86) and cellular (PML) RING finger proteins in BHK cells. BHK-21 cells were infected with vTF7.3 (MOI = 3) and transfected with 0.5 μg of RMX-PML (panel i, lane 3), 0.5 μg of pT7-110 (panel ii, lane 4), or 0.5 μg of pGEM-IE86 (panel ii). Protein expression was assayed by Western blotting with either a rabbit anti-PML antibody (panel i, lanes 1 to 3) or a mouse anti-Vmw110 antibody (panel i, lanes 4 to 6) or by immunofluorescence with a mouse anti-IE86 antibody (panel ii). The positions of PML and Vmw110 are indicated on the right, and the molecular size markers are indicated on the left. (B) Inhibition of CAT expression by the LCMVSCAT2 minigenome is not observed with RING finger proteins unrelated to Z. BHK-21 cells were cotransfected with 0.5 μg of pLCMVSCAT2, 1.5 μg of pCITE-NP, 0.1 μg of pGEM-L, 0.1 μg of pTMI-GFP, and increasing amount of pUCIRES-Z, pGEM-IE86, pT7-110, or RMX-PML as indicated at the bottom of the figure. Plasmid pGEM-L was omitted in lane 14. In all of the samples the amount of DNA was kept constant by adding vector pTMI to 2.7 μg. Samples were assayed for CAT activity as described in Materials and Methods. O, origin; Cm, chloramphenicol; MAc, monoacetylated chloramphenicol; DAc, diacetylated chloramphenicol.
Effect of early expression of Z on LCMV multiplication in vivo.
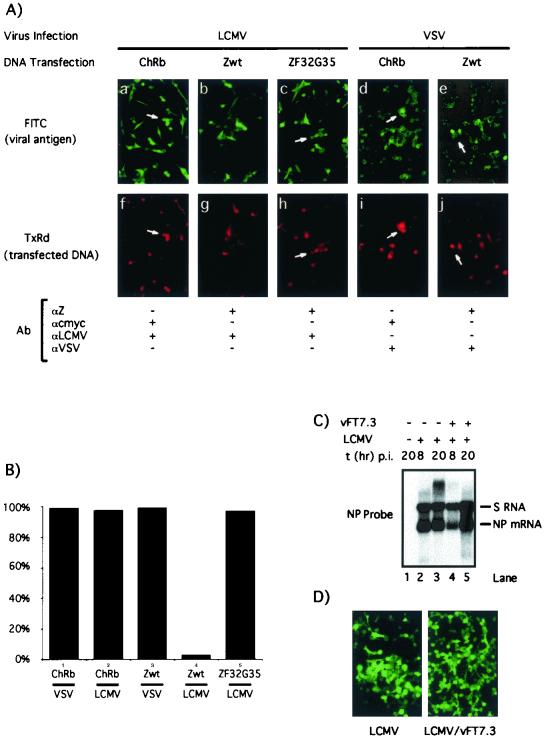
The strong inhibitory effect of Z on the expression of the LCMV minigenome led us to test whether virus multiplication could be also affected by increased expression of Z at early times during the natural course of LCMV infection. For this, we used vTF7.3 to mediate expression of plasmid-derived Z in the context of a LCMV infection. We first examined whether vTF7.3 infection had a significant negative impact on LCMV replication that would preclude the use of this approach. Cells infected with LCMV alone and with LCMV plus vTF7.3 exhibited similar levels of LCMV S RNA and NP mRNA (Fig. 9C). In addition, vTF7.3 infection did not affect significantly the number of LCMV NP-positive cells detected by immunofluorescence at 24 h p.i. (Fig. 9D). As controls, vTF7.3-infected cells were transfected also with ZF32G35 and with a plasmid encoding chicken rhomboid (ChRb), a nonviral protein unrelated to Z. To facilitate its detection, ChRb was tagged at its C terminus with a c-myc epitope. Five hours after transfection, cells were infected either with LCMV or with VSV, both at an MOI of 3. At 12 and 24 h after infection with VSV and LCMV, respectively, cells were fixed and analyzed by immunofluorescence with the following antibodies: (i) a guinea pig polyclonal antibody to LCMV that recognizes the NP and GP, but not Z, viral proteins; (ii) a rabbit serum to Z; (iii) a mouse monoclonal antibody to the c-myc epitope; and (iv) a rabbit polyclonal antibody to the cytoplasmic tail of VSV G. It should be mentioned that levels of Z protein present in LCMV infected cells at early times (<24 h p.i.) could not be detected by immunofluorescence with the rabbit antibody to Z used in these studies. This allowed identification of cells expressing plasmid derived Z protein and virus NP antigen. Cells expressing ChRb were readily infectible by LCMV and VSV, as determined by the colocalization of transfected ChRb and viral antigens (Fig. 9A, panels a, f, d, and i, and B). Most cells expressing the transfected Z did not express levels of LCMV NP antigen that could be detected by IF (Fig. 9A, panels b and g, and B). In contrast, Z-transfected cells exhibited a susceptibility to VSV similar to that observed in ChRb-transfected cells (Fig. 9A, panels d, i, e, and j, and B). This Z-mediated decreased susceptibility to LCMV was statistically significant (chi-squared test, P < 0.0001). Consistent with the CAT results, cells expressing the mutant ZF32G35 did not exhibit decreased susceptibility to LCMV infection (Fig. 9A, panels c and h, and B).
FIG. 9.
Effect of vTF7.3-mediated expression of Z on LCMV infection. (A) Effect of vTF7.3-mediated Z and ChRb expression on LCMV and VSV infections. BHK-21 cells were infected with vTF7.3 (MOI = 3) and subsequently transfected with ChRb (a, f, d, and i), Zwt (b, g, e, and j), or ZF32G35 (c and h) and infected with either LCMV (MOI = 3) (a to c and f to h) or VSV (MOI = 3) (d, e, i, and j). Cells were fixed at 12 and 24 h p.i. with VSV and LCMV, respectively, and analyzed by immunofluorescence by using the indicated primary antibodies, together with the appropriate secondary antibodies coupled to either FITC or TXR. (B) Quantitation of immunofluorescence results shown in panel A. In each case a minimum of 100 positive transfected cells were counted, and the percentage of cells also expressing the corresponding viral antigen was determined. (C) Effect of vTF7.3 infection on LCMV RNA synthesis. Cells were infected with LCMV alone (lanes 2 and 3) or with both LCMV and vTF7.3 (lanes 4 and 5). At the indicated times p.i., RNA was prepared and analyzed by Northern blotting using an LCMV NP probe. (D) Effect of vTF7.3 infection on LCMV antigen expression. Cells were infected with LCMV or coinfected with LCMV plus vTF7.3. After 24 h the cells were fixed and examined by immunofluorescence using a guinea pig polyclonal antiserum to LCMV.
DISCUSSION
In this study, we present evidence that LCMV Z has a strong inhibitory effect on the expression of an LCMV S-segment minigenome. Moreover, cells expressing Z via plasmid transfection appeared to have decreased susceptibility to LCMV infection but not to VSV infection. The inhibitory effect requires an intact Z ORF, and the magnitude of inhibition correlates directly with the amount of Z protein produced (Fig. 2). In addition, plasmid-mediated expression of LCMV trans-acting factors NP and L was unaffected by the coexpression of Z (Fig. 3). Together, these results support the conclusion that the inhibitory effect observed is mediated by the Z protein rather than the consequence of an artifact of transfection of the Z plasmid or intracellular synthesis of Z mRNA. Moreover, 20 ng of input Z plasmid caused an approximately 50% decrease in CAT activity by the LCMV minigenome (Fig. 1A), with the corresponding levels of plasmid supplied Z protein being below those naturally occurring in LCMV-infected cells (Fig. 1B). This suggests that the Z inhibitory effect could very well operate during the natural course of LCMV infection. Because our studies were only done with an S-segment minigenome, we cannot rule out that the observed effects of Z are unique to the LCMV S segment. However, it seems reasonable to assume, though it is as yet unproved, that similar mechanisms would be responsible for transcription and RNA replication of both S and L segments. If we assume that this hypothesis as correct, then Z would be expected to exert a similar inhibitory effect on the transcription and RNA replication of both S and L segments.
Early studies using in vitro transcription combined with immunodepletion experiments of the Z protein from Tacaribe virus (TACV) indicated that Z is required for both genome replication and mRNA synthesis (25). The apparent discrepancies between those published findings and our results may be related to differences between TACV and LCMV with respect to Z functions. It is also possible that the antibody to TACV Z protein used in the immunodepletion studies may have also affected cellular factors required for virus RNA synthesis. It is well documented that cellular components, especially proteins associated with the cytoskeleton, can frequently play important roles in virus replication and transcription (14, 32). This interpretation, however, cannot explain how in vitro synthesis of TACV RNA could be restored by adding an extract of cells infected with vTF7-3 and transfected with a T7-Z plasmid but not by one of cells infected with vTF7-3 alone. However, LCMV RNP preparations with activity in in vitro synthesis of RNA (23) and the ability to resume infection after transfection into cells (Fig. 4B and C) were devoid of detectable levels of Z protein (Fig. 4A). Recent evidence has shown that the bunyamwera virus nonstructural (NS) proteins also inhibit viral RNA synthesis in a minireplicon system (59). Interestingly, the NS proteins of RVFV, another bunyavirus, did not exhibit such inhibitory effect on minigenome RNA synthesis. This suggests that NS proteins of viruses in different Bunyaviridae genera may share only limited functional similarities with respect to the viral replication cycle. Likewise, the apparent discrepancies between previously published findings with TACV and our results with LCMV may be related to differences between these two arenaviruses with respect to their Z functions. In this regard, it is worth noting that TACV and LCMV belong to the two different arenavirus groups or complexes: the TACV complex, which includes all of the known New World arenaviruses, and the LCMV-Lassa complex, which includes the Old World arenaviruses (8).
We noted that coexpression of Z did not prevent encapsidation of the plasmid-supplied LCMV minigenome (Fig. 6A, panel i) but affected both transcription and replication of the minigenome similarly (Fig. 6B). Based on results shown in Fig. 6A, panel i, it appears as though the presence of Z causes a modest increase (two- to fourfold) on the levels of encapsidated minigenome RNA (compare lanes 1, 3, 4, and 6 in Fig. 6A, panel i). Nevertheless, the semiquantitative nature of the assay should caution about the significance of these apparent differences. The results shown in Fig. 6A, panel i, however, provide strong evidence against the possibility that Z has a significant negative effect on the encapsidation of the minigenome RNA. Amplification of minigenome sequences by RT-PCR in the encapsidation assay could have been due to contaminant plasmid DNA. We rule out this possibility based on the lack of amplified sequences when the RT step was omitted (Fig. 6A, panel i, lane 9). These findings suggest that Z is acting at a very early stage after encapsidation. Z could act on the encapsidated template, the polymerase complex, or both. The amount of Z plasmid that caused ca. 50% inhibition was 15-fold lower and 4-fold higher than the molar concentrations of the NP and L plasmids, respectively, suggesting that it is more likely that Z is acting on L.
Biochemical and immunological studies have shown that Z is a structural component of the virion, where it is closely associated with NP. Treatment of LCMV with nonionic detergents showed that Z partitions into the hydrophobic phase rather than remaining associated with the viral nucleocapsid. Moreover, infectious and transcriptionally active LCMV RNP did not contain detectable levels of Z. These findings suggest that Z might be the arenavirus counterpart of the matrix (M) proteins found in other negative-strand RNA viruses (48, 49). For several negative-strand RNA viruses, the association of the M protein with the RNP core appears to shut down virus transcription (15, 42, 56, 58). Whether LCMV Z plays an equivalent role in LCMV infections remains to be determined. LCMV Z has also been shown to interact with several host cell proteins (3, 4, 12). The association of Z with the eukaryotic initiation factor 4E (eIF-4E) has been implicated in the repression of protein synthesis in an eIF-4E-dependent manner (12). Therefore, it is possible that low levels of Z protein could be sufficient to affect the expression of a host cell factor required to initiate synthesis of RNA by the LCMV polymerase.
All arenavirus Z genes sequenced thus far contain a conserved RING finger domain of the type RING HC (C1-X2-C2-X9-C3-X2-H1-X2-C4-X2-C5-X10-C6-X2-C7) (22). Double-point mutations of the zinc-binding residues in the RING finger domain have been shown to unfold the RING and to affect the function of these proteins (2, 3). Using site-directed mutagenesis, we generated an LCMV Arm Z protein wherein the C1 and C2 positions within its RING finger were changed to F and G, respectively (Z-F32G35). This mutated Z-F32G35 protein was stable and could be expressed at levels similar to that of Z-wt, but the mutant Z lost the inhibitory activity (Fig. 7). This result indicated a key role of the RING domain in Z-mediated inhibition of the LCMV minigenome. However, other viral and nonviral RING proteins not related to Z did not have any effect on the LCMV minigenome expression (Fig. 8), suggesting that the presence of a RING-finger domain per se is not responsible for the Z inhibitory effect. The RING domain is thought to mediate protein-protein interaction rather than binding to nucleic acids (12, 19, 21, 30, 57). Hence, the inhibitory activity of Z may involve its interaction with L and/or specific host cell factors through its RING finger domain. Recent findings from a number of laboratories have led to the realization that many RING finger proteins can play a key role in the control of intracellular protein degradation by acting as ubiquitin (Ub)-protein ligases (or E3s) (29, 30). E3s provide specificity to Ub conjugation by facilitating the transfer of Ub from a Ub-conjugating enzyme (Ubc or E2) to ubiquitination targets. It is appealing to speculate that the Z inhibitory effect might be related to its potential E3 activity. This E3 activity would provide Z with the capability to specifically target for degradation proteins required at early stages of RNA synthesis mediated by LCMV polymerase. Nevertheless, our attempts to demonstrate an E3 activity associated with Z yielded negative results. These assays were done using E2 Ubc4 and Z itself as a target for ubiquitination. Therefore, we cannot rule out that Z has an E3 activity that requires interaction with a different E2 or that Z itself is not a target for ubiquitination. In the future, it would be of interest to analyze whether Z has E3 activity when examined in the context of different E2s, as well as using a variety of potential ubiquitination targets.
The NS1 protein of respiratory syncytial virus (RSV) has been shown to have an inhibitory activity (1) similar to that described here for Z. These findings raise the obvious question of why RSV and LCMV encode a protein with an activity that would seem to be counterproductive for viral replication. It may not be accidental that, like RSV, many arenaviruses are characterized by their relative modest growth properties compared to other negative-strand RNA viruses. Negative regulatory proteins such as NS1 and Z could contribute to restricted virus replication and gene expression. Z likely plays different roles during the life cycle of LCMV. Its expression at low levels early during the life cycle of LCMV (Fig. 5) could have a negative regulatory effect on RNA synthesis that might contribute to the restricted replication and noncytopathic properties of many arenaviruses. Increased levels of Z protein at later times of the LCMV life cycle could be required for virus maturation and budding. Levels of Z mRNA increased abruptly by between 12 and 18 h p.i., and we could not detect Z protein prior to 24 h p.i, which postdated the onset of synthesis of L RNA. Thus, the expression of Z during the progression from early to late phases of the LCMV life cycle may be highly regulated. Expression of high levels of Z protein earlier than needed during the virus life cycle could have a very strong negative impact on virus multiplication. This possibility is likely to be recreated in our minigenome rescue assay with use of increasing amounts of Z plasmid. Consistent with this hypothesis, cells transfected with Z, but not with ZF32G35, exhibited decreased susceptibility to LCMV (Fig. 9). This effect of Z appears to be specific since cell susceptibility to VSV infection was not altered in cells transfected with Z. In addition, cells expressing a Z not related to the transfected gene and untransfected control cells showed similar susceptibilities to LCMV infection. These findings suggest that Z could play previously unsuspected roles in the control of arenavirus RNA synthesis. A detailed understanding of the mechanisms by which LCMV Z exerts its inhibitory activity could provide valuable information for the design of new antiviral strategies against highly pathogenic human arenaviruses such as Lassa fever virus.
ACKNOWLEDGMENTS
We thank L. Whitton and M. B. A. Oldstone for valuable discussions, A. Bang for the Ch-Rb cDNA, R. Evans for the PML cDNA, R. D. Everett for the Vmw110 cDNA and the monoclonal antibody to Vmw110, P. Ghazal for the IE86 cDNA and the monoclonal antibody to IE86, M. Salvato for the polyclonal antibody to Z, and M. Whitt for the rabbit polyclonal serum to the C-tail of VSV G and the mouse monoclonal antibody to VSV M.
This work was supported by NIH grant AG04342 (J.C.D.L.T.) and AI09484 (T.I.C.).
Footnotes
Publication 13869-NP from The Scripps Research Institute.
REFERENCES
- 1.Atreya P L, Peeples M E, Collins P L. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J Virol. 1998;72:1452–1461. doi: 10.1128/jvi.72.2.1452-1461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borden K L, Boddy M N, Lally J, O'Reilly N J, Martin S, Howe K, Solomon E, Freemont P S. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borden K L, Campbell-Dwyer E J, Salvato M S. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J Virol. 1998;72:758–766. doi: 10.1128/jvi.72.1.758-766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden K L, Campbell-Dwyer E J, Carlile G W, Djavani M, Salvato M S. Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J Virol. 1998;72:3819–3826. doi: 10.1128/jvi.72.5.3819-3826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borden K L, Campbell-Dwyer E J, Salvato M S. The promyelocytic leukemia protein PML has a pro-apoptotic activity mediated through its RING domain. FEBS Lett. 1997;418:30–34. doi: 10.1016/s0014-5793(97)01344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrow P, Oldstone M B. Mechanism of lymphocytic choriomeningitis virus entry into cells. Virology. 1994;198:1–9. doi: 10.1006/viro.1994.1001. [DOI] [PubMed] [Google Scholar]
- 7.Borrow P, Oldstone M B A. Lymphocitic choriomeningitis virus. In: Nathanson N, editor. Viral pathogenesis. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 593–627. [Google Scholar]
- 8.Bowen M D, Peters C J, Nichol S T. Phylogenetic analysis of the Arenaviridae: patterns of virus evolution and evidence for cospeciation between arenaviruses and their rodent hosts. Mol Phylogenet Evol. 1997;8:301–316. doi: 10.1006/mpev.1997.0436. [DOI] [PubMed] [Google Scholar]
- 9.Buchholz U J, Finke S, Conzelmann K K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchmeier M J, Zajac A J. Lymphocytic choriomeningitis virus. In: Ahmed R, Chen J, editors. Persistent viral infections. Vol. 1. London, England: John Wiley & Sons, Ltd.; 1999. pp. 575–605. [Google Scholar]
- 11.Buckley S M, Casals J, Downs W G. Isolation and antigenic characterization of Lassa virus. Nature. 1970;227:174. doi: 10.1038/227174a0. [DOI] [PubMed] [Google Scholar]
- 12.Campbell-Dwyer E J, Lai H, MacDonald R C, Salvato M S, Borden K L. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J Virol. 2000;74:3293–3300. doi: 10.1128/jvi.74.7.3293-3300.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao W, Henry M D, Borrow P, Yamada H, Elder J H, Ravkov E V, Nichol S T, Compans R W, Campbell K P, Oldstone M B. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 14.Ciampor F. The role of cytoskeleton and nuclear matrix in virus replication. Acta Virol. 1988;32:168–189. [PubMed] [Google Scholar]
- 15.Clinton G M, Little S P, Hagen F S, Huang A S. The matrix (M) protein of vesicular stomatitis virus regulates transcription. Cell. 1978;15:1455–1462. doi: 10.1016/0092-8674(78)90069-7. [DOI] [PubMed] [Google Scholar]
- 16.Cubitt B, de la Torre J C. Borna disease virus (BDV), a nonsegmented RNA virus, replicates in the nuclei of infected cells where infectious BDV ribonucleoproteins are present. J Virol. 1994;68:1371–1381. doi: 10.1128/jvi.68.3.1371-1381.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djavani M, Lukashevich I S, Sanchez A, Nichol S T, Salvato M S. Completion of the Lassa fever virus sequence and identification of a RING finger open reading frame at the L RNA 5′ end. Virology. 1997;235:414–418. doi: 10.1006/viro.1997.8722. [DOI] [PubMed] [Google Scholar]
- 18.Doyle M V, Oldstone M B. Interactions between viruses and lymphocytes. I. In vivo replication of lymphocytic choriomeningitis virus in mononuclear cells during both chronic and acute viral infections. J Immunol. 1978;121:1262–1269. [PubMed] [Google Scholar]
- 19.Duan H, Wang Y, Aviram M, Swaroop M, Loo J A, Bian J, Tian Y, Mueller T, Bisgaier C L, Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett R D. Construction and characterization of herpes simplex type 1 viruses without introns in immediate early gene 1. J Gen Virol. 1991;72:651–659. doi: 10.1099/0022-1317-72-3-651. [DOI] [PubMed] [Google Scholar]
- 21.Fang S, Jensen J P, Ludwig R L, Vousden K H, Weissman A M. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 22.Freemont P S. RING for destruction? Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 23.Fuller-Pace F V, Southern P J. Detection of virus-specific RNA-dependent RNA polymerase activity in extracts from cells infected with lymphocytic choriomeningitis virus: in vitro synthesis of full-length viral RNA species. J Virol. 1989;63:1938–1944. doi: 10.1128/jvi.63.5.1938-1944.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuller-Pace F V, Southern P J. Temporal analysis of transcription and replication during acute infection with lymphocytic choriomeningitis virus. Virology. 1988;162:260–263. doi: 10.1016/0042-6822(88)90419-9. [DOI] [PubMed] [Google Scholar]
- 25.Garcin D, Rochat S, Kolakofsky D. The Tacaribe arenavirus small zinc finger protein is required for both mRNA synthesis and genome replication. J Virol. 1993;67:807–812. doi: 10.1128/jvi.67.2.807-812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghazal P, Young J, Giulietti E, DeMattei C, Garcia J, Gaynor R, Stenberg R M, Nelson J A. A discrete cis element in the human immunodeficiency virus long terminal repeat mediates synergistic trans activation by cytomegalovirus immediate-early proteins. J Virol. 1991;65:6735–6742. doi: 10.1128/jvi.65.12.6735-6742.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibadulinova A, Zelnik V, Reiserova L, Zavodska E, Zatovicova M, Ciampor F, Pastorekova S, Pastorek J. Sequence and characterisation of the Z gene encoding ring finger protein of the lymphocytic choriomeningitis virus MX strain. Acta Virol. 1998;42:369–374. [PubMed] [Google Scholar]
- 28.Iapalucci S, Lopez N, Rey O, Zakin M M, Cohen G N, Franze-Fernandez M T. The 5′ region of Tacaribe virus L RNA encodes a protein with a potential metal binding domain. Virology. 1989;173:357–361. doi: 10.1016/0042-6822(89)90257-2. [DOI] [PubMed] [Google Scholar]
- 29.Joazeiro C A, Weissman A M. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 30.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 31.Kakizuka A, Miller W H, Jr, Umesono K, Warrell R P, Jr, Frankel S R, Murty V V, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 32.Lai M M. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 33.Lee K J, Novella I S, Teng M N, Oldstone M B, de La Torre J C. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol. 2000;74:3470–3477. doi: 10.1128/jvi.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 35.Moyer S A, Smallwood-Kentro S, Haddad A, Prevec L. Assembly and transcription of synthetic vesicular stomatitis virus nucleocapsids. J Virol. 1991;65:2170–2178. doi: 10.1128/jvi.65.5.2170-2178.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers T M, Moyer S A. An amino-terminal domain of the Sendai virus nucleocapsid protein is required for template function in viral RNA synthesis. J Virol. 1997;71:918–924. doi: 10.1128/jvi.71.2.918-924.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oldstone M B. Molecular anatomy of viral persistence. J Virol. 1991;65:6381–6386. doi: 10.1128/jvi.65.12.6381-6386.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oldstone M B. Viral persistence. Cell. 1989;56:517–520. doi: 10.1016/0092-8674(89)90573-4. [DOI] [PubMed] [Google Scholar]
- 39.Oldstone M B. Viral persistence: mechanisms and consequences. Curr Opin Microbiol. 1998;1:436–441. doi: 10.1016/s1369-5274(98)80062-3. [DOI] [PubMed] [Google Scholar]
- 40.Oldstone M B, Ahmed R, Byrne J, Buchmeier M J, Riviere Y, Southern P. Virus and immune responses: lymphocytic choriomeningitis virus as a prototype model of viral pathogenesis. Br Med Bull. 1985;41:70–74. doi: 10.1093/oxfordjournals.bmb.a072029. [DOI] [PubMed] [Google Scholar]
- 41.Parodi A S, Coto C E, Boxaca M, Lajmanovich S, Gonzalez S. Characteristics of Junin virus. Etiological agent of Argentine hemorrhagic fever. Arch Gesamte Virusforsch. 1966;19:393–402. doi: 10.1007/BF01250608. [DOI] [PubMed] [Google Scholar]
- 42.Perez D R, Donis R O. The matrix 1 protein of influenza A virus inhibits the transcriptase activity of a model influenza reporter genome in vivo. Virology. 1998;249:52–61. doi: 10.1006/viro.1998.9318. [DOI] [PubMed] [Google Scholar]
- 43.Peters C J, Buchmeier M, Rollin P E, Ksiazek T G. Arenaviruses. In: Knipe D M, Fields B N, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1521–1551. [Google Scholar]
- 44.Riviere Y, Ahmed R, Southern P J, Buchmeier M J, Dutko F J, Oldstone M B. The S RNA segment of lymphocytic choriomeningitis virus codes for the nucleoprotein and glycoproteins 1 and 2. J Virol. 1985;53:966–968. doi: 10.1128/jvi.53.3.966-968.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salvato M, Borrow P, Shimomaye E, Oldstone M B. Molecular basis of viral persistence: a single amino acid change in the glycoprotein of lymphocytic choriomeningitis virus is associated with suppression of the antiviral cytotoxic T-lymphocyte response and establishment of persistence. J Virol. 1991;65:1863–1869. doi: 10.1128/jvi.65.4.1863-1869.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salvato M, Shimomaye E, Oldstone M B. The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology. 1989;169:377–384. doi: 10.1016/0042-6822(89)90163-3. [DOI] [PubMed] [Google Scholar]
- 47.Salvato M, Shimomaye E, Southern P, Oldstone M B. Virus-lymphocyte interactions. IV. Molecular characterization of LCMV Armstrong (CTL+) small genomic segment and that of its variant, Clone 13 (CTL−) Virology. 1988;164:517–522. doi: 10.1016/0042-6822(88)90566-1. [DOI] [PubMed] [Google Scholar]
- 48.Salvato M S. The Arenaviridae. Vol. 1. New York, N.Y: Plenum Press; 1993. [Google Scholar]
- 49.Salvato M S, Schweighofer K J, Burns J, Shimomaye E M. Biochemical and immunological evidence that the 11 kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Res. 1992;22:185–198. doi: 10.1016/0168-1702(92)90050-j. [DOI] [PubMed] [Google Scholar]
- 50.Salvato M S, Shimomaye E M. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology. 1989;173:1–10. doi: 10.1016/0042-6822(89)90216-x. [DOI] [PubMed] [Google Scholar]
- 51.Saurin A J, Borden K L, Boddy M N, Freemont P S. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 52.Singh M K, Fuller-Pace F V, Buchmeier M J, Southern P J. Analysis of the genomic L RNA segment from lymphocytic choriomeningitis virus. Virology. 1987;161:448–456. doi: 10.1016/0042-6822(87)90138-3. [DOI] [PubMed] [Google Scholar]
- 53.Southern P J. Arenaviridae: the viruses and their replication. In: Knipe D M, Fields B N, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1505–1551. [Google Scholar]
- 54.Southern P J, Singh M K, Riviere Y, Jacoby D R, Buchmeier M J, Oldstone M B. Molecular characterization of the genomic S RNA segment from lymphocytic choriomeningitis virus. Virology. 1987;157:145–155. doi: 10.1016/0042-6822(87)90323-0. [DOI] [PubMed] [Google Scholar]
- 55.Speir R W, Wood O, Liebhaber H, Buckley S M. Lassa fever, a new virus disease of man from West Africa. IV. Electron microscopy of Vero cell cultures infected with Lassa virus. Am J Trop Med Hyg. 1970;19:692–694. doi: 10.4269/ajtmh.1970.19.692. [DOI] [PubMed] [Google Scholar]
- 56.Suryanarayana K, Baczko K, ter Meulen V, Wagner R R. Transcription inhibition and other properties of matrix proteins expressed by M genes cloned from measles viruses and diseased human brain tissue. J Virol. 1994;68:1532–1543. doi: 10.1128/jvi.68.3.1532-1543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi R, Deveraux Q, Tamm I, Welsh K, Assa-Munt N, Salvesen G S, Reed J C. A single BIR domain of XIAP sufficient for inhibiting caspases. J Biol Chem. 1998;273:7787–7790. doi: 10.1074/jbc.273.14.7787. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe K, Handa H, Mizumoto K, Nagata K. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J Virol. 1996;70:241–247. doi: 10.1128/jvi.70.1.241-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weber F, Dunn E F, Bridgen A, Elliott R M. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology. 2001;281:67–74. doi: 10.1006/viro.2000.0774. [DOI] [PubMed] [Google Scholar]
- 60.Wright K E, Spiro R C, Burns J W, Buchmeier M J. Post-translational processing of the glycoproteins of lymphocytic choriomeningitis virus. Virology. 1990;177:175–183. doi: 10.1016/0042-6822(90)90471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]