Abstract
Juvenile myelomonocytic leukemia (JMML) is a disease that occurs in young children and is associated with a high mortality rate. In most patients, JMML has a progressive course leading to death by virtue of infection, bleeding, or progression to acute myeloid leukemia (AML). As it is known that children with neurofibromatosis type 1 syndrome have a markedly increased risk of developing JMML, we have previously developed a mouse model of JMML through reconstitution of lethally irradiated mice with hematopoietic stem cells homozygous for a loss-of-function mutation in the Nf1 gene (D. L. Largaespada, C. I. Brannan, N. A. Jenkins, and N. G. Copeland, Nat. Genet. 12:137–143, 1996). In the course of these experiments, we found that all these genetically identical reconstituted mice developed a JMML-like disorder, but only a subset went on to develop more acute disease. This result strongly suggests that additional genetic lesions are responsible for disease progression to AML. Here, we describe the production of a unique tumor panel, created using the BXH-2 genetic background, for identification of these additional genetic lesions. Using this tumor panel, we have identified a locus, Epi1, which maps 30 to 40 kb downstream of the Myb gene and appears to be the most common site of somatic viral integration in BXH-2 mice. Our findings suggest that proviral integrations at Epi1 cooperate with loss of Nf1 to cause AML.
Juvenile myelomonocytic leukemia (JMML) is a disease characterized by a young age of onset, a tendency to affect boys, prominent enlargement of the liver and spleen, leukocytosis, and the absence of the Philadelphia chromosome. JMML has a poor prognosis, with either progression to acute myeloid leukemia (AML) or death from bleeding or infection (36). It has been estimated that at least 10% of children with JMML also have neurofibromatosis type 1 (NF1) syndrome, an autosomal dominant disorder found in 1/3500 individuals (1, 7, 14, 32). However, the actual frequency of children with NF1 and JMML is likely higher than 10% as the peak incidence of childhood leukemia occurs at an age when NF1 often goes undiagnosed (13, 34). In fact, one study found that 15% of JMML patients had mutations in the NF1 gene even though there was no previous clinical diagnosis of NF1 (33), suggesting that approximately 25% of JMML cases are associated with NF1.
While RAS gene point mutations are commonly found in JMML patients without NF1, they are not found in JMML patients with NF1 (20), providing genetic evidence that NF1 and RAS are involved in the same pathway. This idea is supported by the fact that neurofibromin, the protein product of NF1, contains a region that has extensive homology with the catalytic domain of GTPase activating proteins that are known to accelerate the intrinsic GTPase activity of Ras, thereby negatively regulating Ras GTP levels (15). This suggests that inactivating mutations in NF1 are equivalent to activating mutations in RAS. Consistent with the hypothesis, analysis of bone marrow taken from children with NF1 and JMML revealed that close to half of the samples had somatic loss of heterozygosity (LOH) for markers within and near the NF1 locus (31). In these LOH samples, it was determined that the somatic deletion removed the remaining normal NF1 allele. Together, these results indicated that homozygous NF1 loss predisposes myeloid cells to leukemic transformation in children by activation of the Ras pathway.
To examine the direct consequence of loss of NF1 in the hematopoietic lineage, we previously developed a mouse model for NF1-associated JMML (22). This model relied on reconstitution of lethally irradiated mice with hematopoietic stem cells homozygous for a mutant Nf1 allele, Nf1Fcr, in which an insertion mutation in exon 31 results in the formation of a null allele (4). While we found that all the mice transplanted with homozygous mutant cells developed a myeloproliferative syndrome similar to JMML, only a subset of these genetically identical reconstituted mice went on to develop more acute disease. These results suggested that additional somatic genetic mutations are required for disease progression in mice and humans.
To identify somatic mutations that cooperate with the loss of Nf1 to cause progression to acute leukemia, we have crossed the Nf1Fcr allele onto the BXH-2 mouse genetic background. BXH-2 mice express an ecotropic murine leukemia virus (MuLV) passed from mother to offspring by infection in utero (3). By 1 year of age, nearly 100% of these viremic BXH-2 mice develop an AML that is very similar to AML in humans (28). Previous analysis of BXH-2 tumors has shown that they are nearly all monoclonal in origin and contain one or more somatically acquired MuLV integrations (2). Several investigators have used the BXH-2 system to identify genes that are causally associated with the development of murine myeloid disease by cloning chromosomal sites from tumor DNA into which the MuLV has integrated (6, 16, 23, 25, 26). In many of these cases, specific regions were found to harbor proviral insertions in multiple tumors, each derived from independent mice. Therefore, the identification of a common site of viral integration has served as a good indicator that the region harbors a gene that, when mutated by proviral insertion, contributes to the development of myeloid leukemia. While most of the common sites of viral integration identified in BXH-2 mice are thought to activate the expression of dominant proto-oncogenes (16, 23, 25, 26), one notable exception has been the identification of a common site of viral integration which inactivates the tumor suppressor gene, Nf1. This common site of viral integration, named Evi2 (ectotropic viral integration 2), was identified in 10 to 15% of BXH-2 tumors and found to be within a large intron of the Nf1 gene (6, 8). Using BXH-2 tumors in which biallelic Evi2 integrations had occurred, it was shown that the presence of the virus resulted in premature truncation of the Nf1 transcript, such that no full-length product could be produced (6, 21).
Because loss of Nf1 expression has been shown to be a mechanism of myeloid tumorigenesis in BXH-2 mice, we have exploited this system as a means to identify cooperating events in the progression of JMML. Our strategy has been to backcross C57BL/6J mice containing the Nf1Fcr mutation onto the BXH-2 genetic background for three generations, then age the resulting Nf1Fcr heterozygous N3 mice until they develop acute leukemia, and subsequently collect tumors from each animal. In this manner, we have established a panel of tumors derived from 66 independent N3 BXH-2 Nf1Fcr/+ animals. By cloning sites of viral integration from individual tumors in the panel, we have identified a major common site of viral integration, Epi1, occurring in 44% of tumors in the panel. Interestingly, we also found that Epi1 is affected at nearly the same frequency in BXH-2 tumors without an Nf1 mutation. This indicates Epi1 represents the most common site of somatic viral integration identified to date in BXH-2 mice.
MATERIALS AND METHODS
Mice and harvesting of tumor tissue.
All animal studies were performed according to federal guidelines and University of Florida institutional policies. Mice were aged up to 8 months. When a mouse became moribund it was sacrificed and a necropsy was performed. The necropsy included the following: a physical characterization of the mouse; determination of lymph nodes, spleen, and liver relative size; and liver, spleen, bone marrow, and lymph node fixation in 10% neutral buffered formalin with subsequent embedding in paraffin, sectioning, and staining with hematoxylin and eosin (H&E). Tissue was also frozen in liquid nitrogen to make RNA or DNA. Finally, four lymph nodes were perfused with media, and half of the resulting cells were used to make DNA and half were used to make frozen aliquots of viable cells.
DNA and RNA preparation.
DNA was made from brain and lymph node to use for molecular characterization of the tumors. Tissue (0.4 g) was lysed in homogenizing solution (1× SSC [1× SSC is 150 mM sodium chloride, 15 mM sodium citrate, 0.02 mM citric acid], 1% sodium dodecyl sulfate [SDS], and pronase E [0.25 mg/ml; Sigma]). Samples were vortexed well to mix, and were incubated at 37°C for 1 h and vortexed every 15 min during the incubation. The samples were then extracted once each with phenol (U.S. Biochemical) and phenol-chloroform-isoamyl alcohol (50:49:1; Fisher Scientific). The DNA was precipitated and resuspended in 9 ml of 1× SSC. To this, 0.5 ml of RNase A (2 mg/ml; Sigma) was added and incubated at 37°C for 30 min; this was followed by the addition of 0.5 ml of pronase E (5 mg/ml), and the mixture was then incubated at 37°C for 30 min. The DNA was then extracted again as above and precipitated in ethanol. RNA was made according to the protocol provided with RNAzol B (Tel-Test, Inc.).
Southern and Northern blots.
Five micrograms of genomic DNA was digested with the appropriate restriction enzyme in a total volume of 40 μl at 37°C (or appropriate temperature) for at least 4 h. After 4 h 20 more units of enzyme was added and incubated for 2 more hours at 37°C. The digests were electrophoresed on a 0.8% agarose TPE gel (90 mM Tris-HCl, 26 mM phosphoric acid, 2 mM EDTA). The gel was then soaked in denaturing solution (1.48 M sodium chloride, 0.5 M sodium hydroxide) for 45 min with gentle shaking. The gel was then transferred to neutralizing solution (1 M Tris-HCl, 3 M sodium chloride, 0.2 M hydrochloric acid) and soaked for 1.5 h with gentle shaking. The gel was then blotted onto Hybond (Amersham) membrane in 10× SSC overnight. The membrane was then baked for 2 h at 80°C.
Ten micrograms of total RNA was run on the gel for Northern blots. The 1% agarose gel was made in 1× MOPS [20 mM 3-(N-morpholino)propanesulfonic acid, 5 mM sodium acetate, 1 mM EDTA] and 18% formaldehyde. The RNA samples were mixed with 16 μl of sample buffer (300 μl of formamide, 105 μl of formaldehyde, 60 μl of 10× MOPS, 60 μl of 10% bromophenol blue, and 3 μl of ethidium bromide [10 mg/ml]) and incubated at 65°C for 5 min and then put on ice until loaded. After electrophoresis, the RNA was then transferred to Hybond membrane by blotting overnight in 10× SSC. The membrane was then baked for 2 h at 80°C.
The [α-32P]dCTP-labeled, random-primed probes were made using a random priming kit (Stratagene). The nick-translated probes were labeled using a nick translation kit (Amersham). The Nf1 probe for detecting LOH was hybridized as previously described (4). All other probes were hybridized according to the Church and Gilbert method (9). The membranes were hybridized for 2 h at 65°C, and then 5 ml of fresh hybridization buffer was added. The denatured probes were added, and the membranes were hybridized overnight at 65°C. The next day the membranes were washed at 65°C in 0.2× SSCP and 0.1% SDS (9). The membranes were then subjected to autoradiography.
Cloning flanking DNA.
BamHI-restricted tumor DNA was size fractionated on a sucrose gradient. Then, the fraction with the viral integration was identified and a phage library was made using that fraction. The library was screened to isolate the viral-cellular DNA junction fragment. Specifically, the sucrose gradient was prepared by placing 11.5 ml of 25% sucrose in STE buffer (10 mM Tris, 6 mM EDTA, 10 mM NaCl) in polyallomer tubes (Beckman) and freeze-thawing the solution twice. Fifty micrograms of tumor DNA was digested with BamHI in a volume of 300 μl for about 2 h, and then more enzyme was added and the DNA was digested for another 2 h. The DNA was then extracted twice by the phenol–phenol-chloroform-isoamyl alcohol method and ethanol precipitated. The DNA was washed with 70% ethanol, dried, and then was resuspended in 110 μl of STE buffer. The DNA was layered on the sucrose gradient and spun at 30,000 rpm at 20°C overnight using an SW41Ti rotor (Beckman). Fractions (0.5 ml) were collected from the gradient, and 60 μl was saved to run on a gel as a control. Ethanol (100%) was added to the rest of the fraction and stored at −20°C. The 60-μl aliquots from the fractions were run on a 0.8% TPE gel along with the unfractionated DNA. The gel was blotted and probed with pAKV5 (17) to determine which fraction had the viral integration. BamHI restricted genomic DNA from the positive fraction was then ligated into the Lambda DASH II vector (Stratagene), and the resulting Gigapack III Gold packaged (Stratagene) phage library was screened according to the manufacturer's instructions using the pAKV5 (17) viral probe. Once plaque-pure phage was obtained, phage DNA was prepared and the insert was subcloned into pBluescript KS(+) (Stratagene).
Interspecific backcross mapping.
Interspecific backcross progeny were generated by mating (C57BL/6J × Mus spretus)F1 females and C57BL/6J males as described (11). A total of 205 N2 mice were used to map the Epi1 locus (see text for details). DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blot transfer and hybridization were performed essentially as described previously (17a). All blots were prepared with Hybond-N+ nylon membrane (Amersham). The probe, a 1.3-kb BamHI/PstI fragment of mouse genomic DNA, was labeled with [α-32P]dCTP using a random-primed labeling kit (Stratagene); washing was done to a final stringency of 1.0× SSCP and 0.1% SDS at 65°C. A fragment of 10.5 kb was detected in BamHI-digested C57BL/6J DNA, and a fragment of 7.8 kb was detected in BamHI-digested M. spretus DNA. The presence or absence of the 7.8-kb BamHI M. spretus-specific fragment was monitored in backcross mice. A description of the probes and restriction fragment length polymorphisms for the loci linked to Epi1, including Estra, Myb, and Tcf21, has been reported previously (29). Recombination distances were calculated using Map Manager, version 2.6.5. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Immunophenotyping of leukemias.
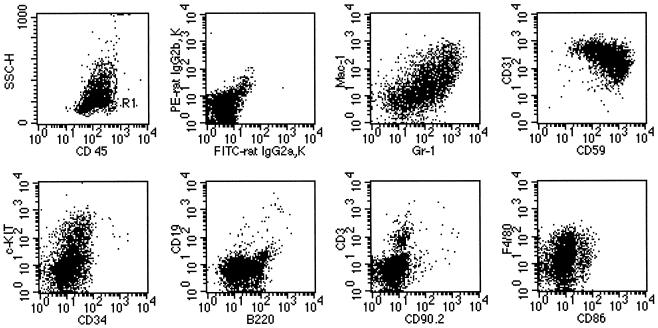
Cryopreserved leukemic cells harvested from enlarged lymph nodes were thawed and washed twice in phosphate-buffered saline with 2% heat-inactivated fetal bovine serum (wash buffer). Single cell suspensions were incubated with antibodies directly conjugated to fluorochromes for 20 to 30 min on ice, washed, and resuspended in wash buffer. Analysis was carried out using a FACScan flow cytometer. A total of 10,000 events were collected on each sample, and results were analyzed using Cell Quest Software (Becton Dickinson). All antibody combinations included anti-CD45-TRI-COLOR. Other antibodies used in combination were anti-Ly-6G(Gr-1)-fluorescein isothiocyanate (FITC) and anti-CD11b(Mac-1)-phycoerythrin (PE), anti-CD59-FITC and anti-CD31-PE, anti-CD34-FITC and anti-CD117(c-kit)-PE, anti-CD45R(B220)-FITC and anti-CD19-PE, anti-CD90.2(Thy1.2)-FITC and anti-CD3-PE, anti-CD41-FITC and anti-CD61-PE, and anti-CD86-FITC and anti-Ly-71(F4/80)-PE. The dominant leukemic cell population was gated using forward scatter, side scatter, and CD45. Expression of antigens on the leukemic cells was assessed relative to isotype controls. Cytospins were also prepared from thawed cells and stained with Wright's Giemsa stain prior to morphological examination. Maturation of cells was assessed by the method of Brecher et al. (5).
RESULTS
Development and characterization of the tumor panel.
We placed the Nf1Fcr mutation on the BXH-2 genetic background by backcrossing for three generations. All N3 mice tested were found to be viremic (data not shown). We aged Nf1Fcr heterozygous N3 mice until they became moribund due to the development of AML and then collected tumor material from each animal. In an initial pilot study, in which we aged both Nf1Fcr heterozygous N3 mice and their wild-type control littermates, we found that the Nf1Fcr heterozygous mice developed AML at a significantly higher rate than controls: 50% of heterozygous mice required sacrifice at 5.5 months versus 8.5 months for controls (P = 0.003 using the rank sum test). This indicated that BXH-2 mice containing one mutant Nf1Fcr allele exhibited a decreased latency for myeloid disease. Based on these data, we initiated a larger aging study using only heterozygous mice and accumulated a panel of tumors isolated from a total of 66 independent heterozygous Nf1Fcr N3 mice. Using Southern blot analysis, we determined that 89% of the tumors in the panel have sustained a second genetic hit to the Nf1 locus, either by LOH or by Evi2 integration. Furthermore, we found that all the tumors from heterozygous Nf1Fcr N3 mice had at least one somatically acquired viral integration. Even tumors which sustained a viral integration at Evi2 contained at least one additional somatic viral integration. This indicates that loss of Nf1 on the BXH-2 background alone is not sufficient to induce acute myeloid disease, but the additional somatic mutations are necessary for tumor progression to AML.
Identification of a common site of viral integration, Epi1
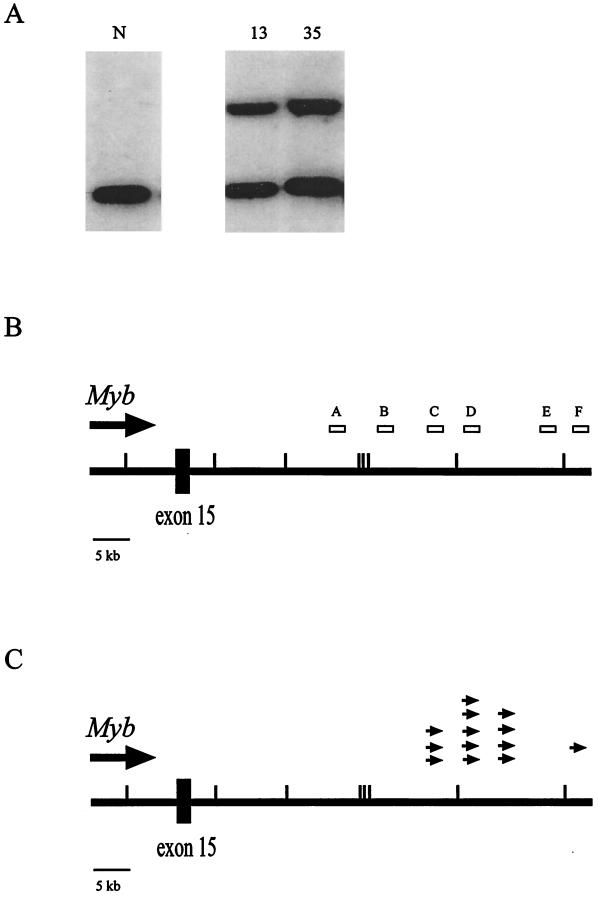
To identify potential cooperating genes for myeloid tumor progression, we cloned genomic DNA flanking sites of somatic viral integration from individual tumors in the panel. Using the ecotropic retroviral probe pAKV5 (17), we isolated a unique BamHI restriction fragment from tumor DNA isolated from animal 355. After isolation of a nonrepetitive probe from the cloned fragment, dubbed probe E, we screened the entire panel of tumors for rearrangements by Southern blot. Not only were we able to confirm a rearrangement in the original tumor (data not shown), but we also detected rearrangements in many additional tumor DNA samples (Fig. 1A). This indicated that we had identified a common site of viral integration, which we designated Epi1 for ecotropic proviral integration site 1.
FIG. 1.
The Epi1 locus. (A) Southern blot analysis showing rearrangements of the Epi1 locus in two independent tumors. Normal DNA (N) and tumor DNAs were digested with BglII and hybridized with probe C from the Epi1 locus. (B) Map of the Epi1 region relative to the Myb gene. The large line represents the region 3′ of the Myb gene. The small hash marks represent BamHI sites in the region, the large box is exon 15 of the Myb gene, the large arrow shows the direction of transcription of Myb, and the small boxes above the line are the probes isolated from the region. Probe C is the Ahi1 probe (18). (C) Location and orientation of 13 proviruses integrated in Epi1 relative to the Myb gene. The small arrows above the line show the location and orientation of the viral integrations in the Epi1 locus.
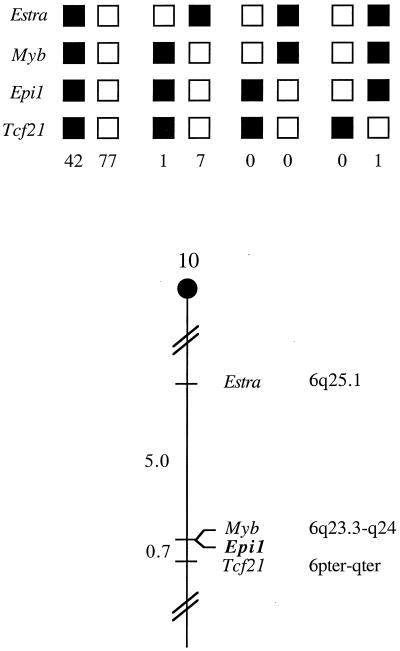
The mouse chromosomal location of Epi1 was determined by interspecific backcross analysis using progeny derived from matings of [(C57BL/6J × M. spretus)F1 × C57BL/6J] mice. This interspecific backcross mapping panel has been typed for over 3,000 loci that are well distributed among all the autosomes as well as the X chromosome (11). C57BL/6J and M. spretus DNAs were digested with several enzymes and analyzed by Southern blot hybridization to obtain informative restriction fragment length polymorphisms using a mouse Epi1 genomic DNA probe. The 7.8-kb BamHI M. spretus restriction fragment length polymorphism (see Materials and Methods) was used to follow the segregation of the Epi1 locus in backcross mice. The mapping results indicated that Epi1 is located in the proximal region of mouse chromosome 10 linked to Estra, Myb, and Tcf21. Although 128 mice were analyzed for every marker and are shown in the segregation analysis (Fig. 2), up to 181 mice were typed for some pairs of markers. Each locus was analyzed in pairwise combinations for recombination frequencies using the additional data. The total number of mice exhibiting recombinant chromosomes and the total number of mice analyzed for each pair of loci are as follows (listed in the most likely gene order): centromere-Estra, 9 of 181; Myb, 0 of 161; Epi1, 1 of 142; Tcf21. The recombination frequencies (expressed as genetic distances ± standard error [in centimorgans]) are as follows: Estra, 5.0 ± 1.6; Myb and Epi1, 0.7 ± 0.7; Tcf21. No recombinants were detected between Myb and Epi1 in 161 animals typed in common, suggesting that the two loci are within 1.9 cM of each other (upper 95% confidence limit).
FIG. 2.
Epi1 maps in the proximal region of mouse chromosome 10. Epi1 was placed on mouse chromosome 10 by interspecific backcross analysis. The segregation patterns of Epi1 and flanking genes in 128 backcross animals that were typed for all loci are shown at the top of the figure. For individual pairs of loci, more than 128 animals were typed (see text). Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M. spretus)F1 parent. The black boxes represent the presence of a C57BL/6J allele, and white boxes represent the presence of a M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 10 linkage map showing the location of Epi1 in relation to linked genes is shown at the bottom of the figure. Recombination distances between loci in centimorgans are shown to the left of the chromosome, and the positions of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from the Genome Data Base, a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore, Md.).
Epi1 maps 3′ of the Myb locus.
Based on the lack of genetic recombination observed between Epi1 and Myb, we sought to determine if these two loci were physically linked. We isolated a bacterial artificial chromosome clone (529J1 BAC; Research Genetics) using the original Epi1 probe (Fig. 1B, probe E) and determined that the Myb gene was also contained on this same BAC clone. We used this BAC clone to generate a restriction map of the Epi1 region and to isolate additional probes (Fig. 1B, probes A, B, D, and F). Using these new probes, as well as the previously isolated Ahi1 probe (18) (referred to here as probe C), we determined that a total of 29 tumors, or 44% of the panel, contained a somatic Epi1 rearrangement (Table 1). While no rearrangements were detected with probe A, the majority of rearrangements were detected by probes B and C (indicating an insertion into the 11-kb BamHI fragment) or by probes D and E (indicating an insertion into the 13.5-kb BamHI fragment). Rearrangements were detected with probe F in four tumors. These data indicate that most of the viral insertions have occurred 30 to 40 kb downstream of the last exon of Myb.
TABLE 1.
Tumors derived from N3 BXH-2 Nf1Fcr/+ mice with Epi1 insertionsa
| Tumor no. | Nf1 status | No. of somatic proviruses | Epi1 probe(s) detecting BamHI rearrangement |
|---|---|---|---|
| 10 | Evi2 | 3 | D, E |
| 11 | Nf1Fcr/+ | 1 | D, E |
| 13 | LOH | 3 | D, E |
| 14 | Evi2 | 2 | B, C |
| 33 | LOH | 3 | B, C |
| 35 | LOH | 5 | D, E |
| 46 | Evi2 | 2 | F |
| 56 | LOH | 2 | D, E |
| 84 | LOH | 2 | B, C |
| 103 | LOH | 2 | D, E |
| 355 | LOH | 1 | D, E |
| 356 | Evi2 | 3 | D, E |
| 358 | LOH | 2 | F |
| 368 | LOH | 5 | D, E |
| 371 | LOH | 1 | D, E |
| 419 | LOH | 2 | D, E |
| 468 | LOH | 2 | B, C |
| 522 | LOH | 5 | D, E |
| 570 | LOH | 2 | D, E |
| 603 | Nf1Fcr/+ | 1 | D, E |
| 639 | Evi2 | 2 | F |
| 647 | LOH | 2 | F |
| 660 | Evi2 | 3 | D, E |
| 662 | LOH | 3 | B, C |
| 665 | Evi2 | 3 | B, C |
| 671 | LOH | 2 | B, C |
| 675 | Nf1Fcr/+ | 2 | D, E |
| 725 | Evi2 | 1 | D, E |
| 1858 | Evi2 | 1 | F |
“Tumor no.” refers to the pedigee number of the animal from which the tumor was derived. “Nf1 status” refers to the status of the Nf1 allele in the tumor; all the mice began as heterozygous for the Nf1 mutation (NflFcr/+), but many of the tumors lost the remaining wild-type allele (LOH) or suffered a proviral integration in the Nf1 gene (Evi2). “No. of somatic proviruses” refers to the number of somatically acquired proviral integrations detected in the tumor DNA. “Epi1 probe(s) detecting BamHI rearrangement” indicates the probe(s) from Fig. 1 which detects the somatic rearrangement in the Epi1 locus in the particular tumor.
Based on the location of these somatic insertions, it seemed possible that the integrated proviruses affect Myb. However, all previously reported viral insertions proven to affect Myb occur within the Myb gene. For example, viral integration in the 5′ end has been shown to result in overexpression of a virally promoted chimeric mRNA that lacks the three 5′-most Myb coding exons (36). In addition, integrations in the 3′ end of Myb have been shown to result in the production of a Myb protein that is stabilized due to truncation of the carboxy terminus (36). Therefore, we rescreened our panel using a series of Myb probes, but we found only two tumors that contained insertions within the Myb gene.
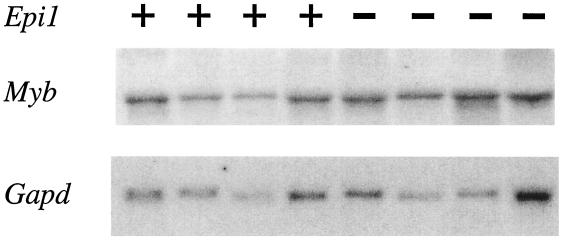
To ascertain if viral integrations at Epi1 affect Myb, we determined the orientation of the integrated provirus in 13 of the tumors in which we were able to confirm the presence of a full-length virus. We found that in all 13 cases, the virus had integrated in the same transcriptional orientation as Myb (Fig. 1C). These data are consistent with a mechanism of viral enhancement in which the viral enhancer located in the 5′ long terminal repeat serves to activate transcription of an upstream gene (19). With this in mind, we assayed the level of Myb expression in the tumors by Northern blotting of total RNA isolated from several tumors with and without a viral integration at Epi1. We found that Myb was expressed in tumors whether or not the tumors harbored Epi1 integrations. Of note, expression levels were not increased in tumors containing an Epi1 insertion relative to expression levels in tumors lacking such a viral insertion (Fig. 3). However, tumors lacking Epi1 insertions may have upregulated Myb by another mechanism. In any case, these data show that the Myb gene is indeed expressed in the Epi1 tumors, but that a viral insertion at Epi1 does not appear to result in marked overexpression of Myb.
FIG. 3.
Northern blot analysis of Myb expression in Epi1 and non-Epi1 tumors. RNA derived from four tumors containing somatic viral insertion at Epi1 (+) and four tumors without this viral insertion (−) were analyzed by Northern blotting. The top panel shows the hybridization with a Myb cDNA probe, and the bottom panel shows the same blot stripped and rehybridized with a control probe for Gapd.
Phenotype of Epi1 tumors.
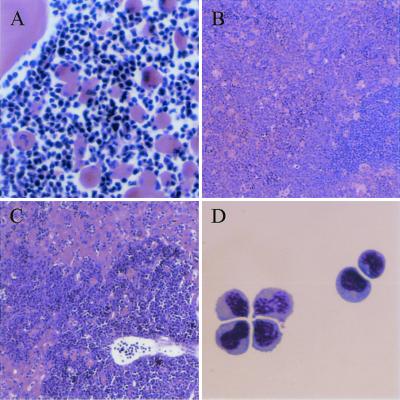
We performed histological analysis of H&E stained tissue sections from a subset of the animals listed in Table 1 (tumors 355, 419, 468, 660, and 671). This analysis revealed that while there was a modest variation in the morphology of the leukemic cells, the overall pattern was similar. Common features included the presence of leukemic cells at the young-intermediate stages of myeloid differentiation accompanied by numerous pseudo-Gaucher cells (indicative of high cell turnover); lymph nodes that were effaced by the leukemia; spleens that exhibited modest areas of residual white pulp but massive expansion of the red pulp predominantly by leukemic cells, but with some admixed megakaryocytes and erythroid precursors; livers that showed leukemic infiltrates with marked accumulations in the periportal areas and with infiltration into the parenchyma and/or sinusoids; and bone marrows filled with leukemic cells (accompanied by pseudo-Gaucher histiocytes) (Fig. 4A to C).
FIG. 4.
Leukemia morphology. (A) Bone marrow histology showing immature cells and numerous pseudo-Gaucher histiocytes. Magnification, ×250. (B) Spleen histology showing expanded red pulp with leukemic cells accompanied by large megakaryocytes and few small dark-staining erythroid precursors. Residual white pulp is visible at lower right. Magnification, ×100. (C) Liver histology showing massive infiltration of leukemic cells. Magnification, ×100. H&E stain was used for panels A to C. (D) Cytology (Wright's Giemsa stain) of leukemic cells prepared from lymph node. Magnification, ×500.
Finally, analysis on viable isolates of these same five tumors (tumors 355, 419, 468, 660, and 671; Table 1) revealed that all five were relatively uniform: almost entirely young forms (blasts) by morphology (Fig. 4D) that express an immunophenotype consistent with granulocyte/monocyte precursors [Ly-6G(Gr-1)+, CD11b(Mac-1)variable, CD31+, CD59+, CD117(c-kit)variable, CD34lo, Ly-71(F4/80)lo] (Fig. 5). That the tumor cells had only a weak CD45R (B220) signal and were clearly CD19 negative strongly suggests that the cells are not B-lymphoid cells. In addition, the T-cell markers CD90.2 and CD3 were either negative or weak, indicating the cells are not T lymphocytes. These tumors as well as several others were confirmed to be of myeloid origin as they were found to express the myeloid cell markers c-fms and myeloperoxidase by Northern blot analysis. In further support of this origin, we detected no rearrangements of the T-cell receptor genes or the immunoglobulin heavy chain gene. These data indicate that the mice had developed myeloid leukemias and show that the five tumors examined in detail were relatively homogeneous in character. These five leukemias all contained Epi1 insertions and lacked Nf1. Their common characteristics likely reflect their common genetic pathogenesis: it contrasts with the phenotypic heterogeneity we have observed in unselected, genetically heterogeneous, BXH-2 leukemias (S. Kogan, unpublished observations).
FIG. 5.
Flow cytometric analysis of a representative leukemia. Data in panel CD45/SSC are ungated; for those in the remaining panels, the gate was set at R1.
Analysis of wild-type BXH-2 tumors.
Finally, to determine if viral integrations at Epi1 were restricted to tumors containing mutations at the Nf1 locus, we analyzed a set of BXH-2 tumors that were wild-type at the Nf1 locus, as well as a set of tumors that has suffered one somatic viral integration within Nf1 (Evi2). We found that in both groups of 23 tumors, 10 tumors (43%) exhibited a viral integration in Epi1. Interestingly, however, we found that the distribution of proviral insertions in Epi1 was different between the two groups. Proviral insertions occurred in a discrete region in the tumors with wild-type Nf1, with rearrangements being detected only by probes D and E (Fig. 1B). In contrast, tumors harboring an Evi2 insertion showed a much broader region of proviral insertions in Epi1, with rearrangements being detected by probes B and C, D and E, or F. This difference in distribution of proviral insertions between the two genotypic groups may reflect the differentiation state of the cell at the time of viral infection. For example, myeloid cells with only one intact copy of Nf1 may display a more open chromatin conformation at Epi1 and hence present a wider target area for viral integration than cells with both copies intact. Regardless of the slight difference in the specific region of integration, these data demonstrate that Epi1 represents a major site of viral integration in approximately 43% of all BXH-2 myeloid tumors.
DISCUSSION
We have previously shown that loss of Nf1 in the hematopoietic lineage causes a myeloproliferative syndrome similar to JMML (22). However, we found that only a subset of all of these reconstituted mice went on to develop more acute disease, suggesting that additional somatic genetic mutations are required for disease progression in mice and humans. To identify candidate genes that may cooperate with the loss of the Nf1 gene in progression of myeloid leukemia, we have established a panel of tumors derived from 66 independent N3 BXH-2 Nf1Fcr/+ animals. We have determined that 89% of the tumors in the panel have sustained a second genetic hit to the Nf1 locus, either by LOH or by viral integration within the Nf1 locus (Evi2), demonstrating that the vast majority of the tumors developed in a pathway involving Nf1 gene loss. Therefore, through the introduction of one mutant Nf1 allele, we have obtained a sixfold higher frequency of Nf1-dependent myeloid leukemia compared to the parental BXH-2 strain.
Analysis of the panel revealed that these tumors harbored a mean of three somatically acquired viral insertions. By cloning these sites of viral integration from individual tumors in the panel, we were able to identify a common site of viral integration, Epi1, occurring in 44% of tumors in the panel. The Epi1 locus was found to map 30 to 40 kb downstream of the last exon of the Myb gene. Interestingly, this site has previously been implicated in B-cell lymphoma: Jiang et al. reported that infection of newborn mice with a combination of Abelson MuLV (contains the v-abl oncogene) and Moloney MuLV (the insertional mutagen) resulted in B-cell lymphoma (18). They determined that 16% of these tumors had Moloney MuLV insertions at the Ahi1 locus, located approximately 30 kb 3′ of the last exon of Myb. Again, similar to our findings, they determined that these viral integrations did not result in overexpression of Myb. Based on this result, Jiang et al. concluded that the viral insertions activated an as-yet-unidentified gene. However, one alternate interpretation of these data is that these downstream viral integrations do affect Myb expression, but not by upregulating transcription. Instead, the virus may simply prevent the downregulation of Myb that normally occurs during myeloid differentiation. In fact, it is well known that either constitutive expression of full-length Myb or truncated Myb can block differentiation of leukemic cell lines (10, 12, 24, 30, 35, 37). Furthermore, downregulation of Myb is known to be essential for terminal differentiation of the myeloid and B-cell lineages (27). Therefore, it is possible that constitutive expression of Myb in combination with v-abl induces B-cell lymphoma, whereas constitutive expression of Myb in combination with the loss of Nf1 function leads to AML. Alternatively, it is possible that viral integration into this locus affects a gene other than Myb. In fact, it is possible that viral integration in B-cells affects one gene (the Ahi1 gene), but viral insertion in myeloid cells affects a different gene (the Epi1 gene). In any case, whether viral insertions in Epi1 affect Myb or another gene, we have provided very good evidence that Epi1 does play an important role in BXH-2 myeloid leukemia. The finding that approximately 43% of BXH-2 tumors harbor viral insertion in Epi1 indicates that Epi1 may represent the most common site of viral integration in BXH-2.
Because proviral integrations in Epi1 are so common in BXH-2 tumors, one might be tempted to conclude that this event alone is sufficient to cause AML. However, data obtained from our N3 BXH-2 Nf1Fcr/+ tumors suggest that additional cooperating mutations are required for progression to AML: 27 out of 29 of these tumors have another obvious somatic mutation in addition to a proviral insertion in Epi1. Furthermore, we can conclude that at least in these tumors, the cooperating mutation is the loss of the Nf1 gene since 26 out of these 29 tumors (90%) with viral integrations at Epi1 also exhibit loss of Nf1 function. But loss of Nf1 is clearly not the only possible cooperating mutation since Nf1 is mutated in only 10 to 15% of BXH-2 tumors. Therefore, other genes must be able to serve as a cooperating gene for Epi1-associated AML. Since Nf1 is known to affect the Ras pathway, it is tempting to speculate that these other genes might be involved in this same pathway.
Finally, it is unclear whether the loss of Nf1 plus a proviral insertion at Epi1 is sufficient to cause AML. Among our panel of tumors, five appear to have the Epi1 integration as the only other somatic mutation (Table 1, tumors 14, 46, 355, 371, and 639), suggesting that loss of Nf1 plus insertion at Epi1 is acutely leukemogenic. However, it is possible that these five tumors harbor other somatic mutations that we are unable to detect. Further studies of BXH-2 and Nf1 murine models of myeloid neoplasms should allow the combinations of events sufficient for leukemogenesis to be definitively assessed.
ACKNOWLEDGMENTS
We thank Michael Elmore and Deborah B. Householder for excellent technical assistance. We thank Linda Wolff for Myb probes and Paul Jolicoeur for the Ahi1 probe (Probe C, Fig. 1B).
This research was supported by a grant from the Department of Defense, U.S. Army Medical Research and Materiel Command (DAMD17-97-1-7339), to C.I.B.; by grants from the NIH/NCI (CA75986 and CA84221) to S.C.K.; by a Leukemia Research Fund grant to D.A.L.; and, in part, by the National Cancer Institute, DHHS. S.C.K. is a recipient of a Burroughs Wellcome Fund Career Award and is an Edward Mallinckrodt Jr. Foundation Scholar.
REFERENCES
- 1.Bader J L, Miller R W. Neurofibromatosis and childhood leukemia. J Pediatr. 1978;92:925–929. doi: 10.1016/s0022-3476(78)80362-x. [DOI] [PubMed] [Google Scholar]
- 2.Bedigian H G, Johnson D A, Jenkins N A, Copeland N G, Evans R. Spontaneous and induced leukemias of myeloid origin in recombinant inbred BXH mice. J Virol. 1984;51:586–594. doi: 10.1128/jvi.51.3.586-594.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedigian H G, Shepel L A, Hoppe P C. Transplacental transmission of a leukemogenic murine leukemia virus. J Virol. 1993;67:6105–6109. doi: 10.1128/jvi.67.10.6105-6109.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brannan C I, Perkins A S, Vogel K S, Ratner N, Nordlund M L, Reid S W, Buchberg A M, Jenkins N A, Parada L F, Copeland N G. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 5.Brecher G, Endicott K, Gump H, Brawner H P. Effects of X-ray on lymphoid and hemopoietic tissues of albino mice. Blood. 1948;3:1259–1274. [PubMed] [Google Scholar]
- 6.Buchberg A M, Bedigian H G, Jenkins N A, Copeland N G. Evi-2, a common integration site involved in murine myeloid leukemogenesis. Mol Cell Biol. 1990;10:4658–4666. doi: 10.1128/mcb.10.9.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro-Malaspina H, Schaison G, Passe S, Pasquier A, Berger R, Bayle-Weisgerber C, Miller D, Seligman M, Bernard J. Subacute and chronic myelomonocytic leukemia in children (juvenile CML) Cancer. 1984;54:675–686. doi: 10.1002/1097-0142(1984)54:4<675::aid-cncr2820540415>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 8.Cawthon R M, Andersen L B, Buchberg A M, Xu G F, O'Connell P, Viskochil D, Weiss R B, Wallace M R, Marchuk D A, Culver M, et al. cDNA sequence and genomic structure of EV12B, a gene lying within an intron of the neurofibromatosis type 1 gene. Genomics. 1991;9:446–460. doi: 10.1016/0888-7543(91)90410-g. [DOI] [PubMed] [Google Scholar]
- 9.Church G M, Gilbert W. The genomic sequencing technique. Prog Clin Biol Res. 1985;177:17–21. [PubMed] [Google Scholar]
- 10.Clarke M F, Kukowska-Latallo J F, Westin E, Smith M, Prochownik E V. Constitutive expression of a c-myb cDNA blocks Friend murine erythroleukemia cell differentiation. Mol Cell Biol. 1988;8:884–892. doi: 10.1128/mcb.8.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Copeland N G, Jenkins N A. Development and applications of a molecular genetic linkage map of the mouse genome. Trends Genet. 1991;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 12.Cuddihy A E, Brents L A, Aziz N, Bender T P, Kuehl W M. Only the DNA binding and transactivation domains of c-Myb are required to block terminal differentiation of murine erythroleukemia cells. Mol Cell Biol. 1993;13:3505–3513. doi: 10.1128/mcb.13.6.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Bella K, Szudek J, Freidman J M. Use of National Institutes of Health criteria for diagnosis of neurofibromatosis 1 in children. Pediatrics. 2000;105:608–614. doi: 10.1542/peds.105.3.608. [DOI] [PubMed] [Google Scholar]
- 14.Gadner H, Haas O A. Experience in pediatric myelodysplastic syndromes. Vol. 6. Philadelphia. Pa: W. B. Saunders Company; 1992. [PubMed] [Google Scholar]
- 15.Gutmann D H, Collins F S. The neurofibromatosis type 1 gene and its protein product, neurofibromin. Neuron. 1993;10:335–343. doi: 10.1016/0896-6273(93)90324-k. [DOI] [PubMed] [Google Scholar]
- 16.Hansen G M, Skapura D, Justice M J. Genetic profile of insertion mutations in mouse leukemias and lymphomas. Genome Res. 2000;10:237–243. doi: 10.1101/gr.10.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herr W, Gilbert W. Somatically acquired recombinant murine leukemia proviruses in thymic leukemias of AKR/J mice. J Virol. 1983;46:70–82. doi: 10.1128/jvi.46.1.70-82.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Jenkins N A, Copeland N G, Lee B K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang X, Villeneuve L, Turmel C, Kozack C A, Jolicoeur P. The Myb and Ahi-1 genes are physically very closely linked on mouse chromosome 10. Mamm Genome. 1994;5:142–148. doi: 10.1007/BF00352344. [DOI] [PubMed] [Google Scholar]
- 19.Jonkers J, Berns A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim Biophys Acta. 1996;1287:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- 20.Kalra R, Paderanga D C, Olson K, Shannon K M. Genetic analysis is consistent with the hypothesis that NF1 limits myeloid cell growth through p21ras. Blood. 1994;84:3435–3459. [PubMed] [Google Scholar]
- 21.Largaespada D A, Shaughnessy J D, Jenkins N A, Copeland N G. Retroviral insertion at the Evi-2 locus in BXH-2 myeloid leukemia cell lines disrupts Nf1 expression without changes in steady-state Ras-GTP levels. J Virol. 1995;69:5095–5102. doi: 10.1128/jvi.69.8.5095-5102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Largaespada D L, Brannan C I, Jenkins N A, Copeland N G. Nf1 deficiency causes Ras-mediated granulocyte/macrophage stimulating factor hypersensitivity and chronic myeloid leukemia. Nat Genet. 1996;12:137–143. doi: 10.1038/ng0296-137. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Shen H, Himmel K L, Dupuy A J, Largaespada D L, Nakamura T, Shaughnessy J D J, Jenkins N A, Copeland N G. Leukemia disease genes: large-scale cloning and pathway predictions. Nat Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- 24.McClinton D, Stafford J, Brents L, Bender T P, Kuehl W M. Differentiation of mouse erythroleukemia cells is blocked by late up-regulation of a c-myb transgene. Mol Cell Biol. 1990;10:705–710. doi: 10.1128/mcb.10.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moscow J J, Bullrich F, Huebner K, Daar I O, Buchberg A M. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura T, Largaespada D A, Shaughnessy J D, Jenkins N A, Copeland N G. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 27.Oh I-H, Reddy E P. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 28.Perkins A S. The pathology of murine myelogenous leukemias. Curr Top Microbiol Immunol. 1989;149:3–21. doi: 10.1007/978-3-642-74623-9_1. [DOI] [PubMed] [Google Scholar]
- 29.Robb L, Mifsud L, Hartley L, Biben C, Copeland N G, Gilbert D J, Jenkins N A, Harvey R P. Epicardin: a novel basic helix-loop-helix transcription factor gene expressed in epicardium, branchial arch myoblasts, and mesenchyme of developing lung, gut, kidney, and gonads. Dev Dyn. 1998;213:105–113. doi: 10.1002/(SICI)1097-0177(199809)213:1<105::AID-AJA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 30.Selvakumaran M, Liebermann D A, Hoffman-Liebermann B. Deregulated c-myb disrupts interleukin-6- or leukemia inhibitory factor-induced myeloid differentiation prior to c-myc: role in leukemogenesis. Mol Cell Biol. 1992;12:2493–2500. doi: 10.1128/mcb.12.6.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon K M, O'Connell P, Martin G A, Paderanga D, Olson K, Dinndorf P, McCormick F. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N Engl J Med. 1994;330:597–601. doi: 10.1056/NEJM199403033300903. [DOI] [PubMed] [Google Scholar]
- 32.Shannon K M, Watterson J, Johnson P, O'Connell P, Lange B, Shah N, Steinherz P, Kan Y W, Priest J R. Monosomy 7 myeloproliferative disease in children with neurofibromatosis, type 1: epidemiology and molecular analysis. Blood. 1992;79:1311–1318. [PubMed] [Google Scholar]
- 33.Side L E, Emanuel P D, Taylor B, Franklin J, Thompson P, Castleberry R P, Shannon K M. Mutations of the NF1 gene in children with juvenile myelomonocytic leukemia without clinical evidence of neurofibromatosis, type 1. Blood. 1998;92:267–272. [PubMed] [Google Scholar]
- 34.Stiller C A, Chessells J M, Fitchett M. Neurofibromatosis and childhood leukaemia/lymphoma: a population-based UKCCSG study. Br J Cancer. 1994;70:969–972. doi: 10.1038/bjc.1994.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todokoro K, Watson R J, Higo H, Amanuma H, Kuramochi S, Yanagisawa H, Ikawa Y. Down-regulation of c-myb gene expression is a prerequisite for erythropoietin-induced erythroid differentiation. Proc Natl Acad Sci USA. 1988;85:8900–8904. doi: 10.1073/pnas.85.23.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolff L. Myb-induced transformation. Crit Rev Oncog. 1996;7:245–260. doi: 10.1615/critrevoncog.v7.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 37.Yanagisawa H, Nagasawa T, Kuramochi S, Abe T, Ikawa Y, Todokoro K. Constitutive expression of exogenous c-myb gene causes maturation block in monocyte-macrophage differentiation. Biochim Biophys Acta. 1991;1088:380–384. doi: 10.1016/0167-4781(91)90128-9. [DOI] [PubMed] [Google Scholar]