Abstract
Metabolism within an organism is regulated by various processes, including post-translational modifications (PTMs). These types of chemical modifications alter the molecular, biochemical, and cellular properties of proteins and allow the organism to respond quickly to different environments, energy states, and stresses. Malate dehydrogenase (MDH) is a metabolic enzyme that is conserved in all domains of life and is extensively modified post-translationally. Due to the central role of MDH, its modification can alter metabolic flux, including the Krebs cycle, glycolysis, and lipid and amino acid metabolism. Despite the importance of both MDH and its extensively post-translationally modified landscape, comprehensive characterization of MDH PTMs, and their effects on MDH structure, function, and metabolic flux remains underexplored. Here, we review three types of MDH PTMs – acetylation, ADP-ribosylation, and methylation – and explore what is known in the literature and how these PTMs potentially affect the 3D structure, enzymatic activity, and interactome of MDH. Finally, we briefly discuss the potential involvement of PTMs in the dynamics of metabolons that include MDH.
General overview of PTMs and malate dehydrogenase
Post-translational modifications (PTMs) are covalent alterations of amino acids in a peptide chain. The dynamics of PTMs rely on either non-enzymatic or enzymatic mechanisms, with a ‘writer’ that adds a specific PTM, and an ‘eraser’ that removes it. These PTMs can be recognized by ‘readers’ which are proteins with specialized domains that recognize and bind to post-translationally modified proteins to mediate specific cellular outcomes. While protein expression levels may not change, protein function and behavior may be altered as a result of PTMs on various protein isoforms in different cellular compartments. More than 400 different PTMs have been identified, such as phosphorylation, acetylation, ADP-ribosylation, and methylation, which differentially modulate the charge, structure, activity, location, and molecular interactions of a protein [1]. Histones are DNA-binding proteins that are components of chromatin which undergo extensive PTM and have been shown to contribute to epigenetic mechanisms involved in diseases like cancers [2]. The extensive study of chromatin regulation by PTMs has led to the recognition of a ‘histone code’ which is ‘read’ by specialized proteins that interact with histones and other proteins to render the chromatin either more or less accessible, for the modulation of DNA replication, gene expression, and the DNA damage response. This elaborate system is difficult to characterize. For example, the choice between gene expression and silencing is dependent on the specific amino acid within the histone protein sequence that is methylated [3]. Moreover, the quantity, duration, and location of the PTM on proteins have importance, and the combination of PTMs directly balances the equilibrium between healthy and disease states [4]. While the precise mechanisms involving writing and reading the histone code are still not perfectly understood, these mechanisms have been extensively studied and can serve as a model for other cellular processes in which enzymes are regulated by PTM. Enzymes involved in metabolic pathways, for example, have been shown to be abundantly post-translationally modified, which provides a mechanism to finely tune their activity to rapidly respond to changes in the surrounding environment. However, the characterization of these PTMs on metabolic enzymes has not, so far, led to the establishment of an analogous ‘metabolic code’. Despite this lack of code, increasing evidence suggests a link between the PTMs on hundreds of metabolic enzymes, metabolic modulation, and diseases [5,6].
Malate dehydrogenase (MDH) is an essential metabolic enzyme, expressed from different genes and localized in various cellular compartments, such as the cytosol, mitochondria, chloroplasts, and peroxisomes (Figure 1) [7,8]. All MDH isoforms catalyze the reversible interconversion of malate and NAD(P)+ to oxaloacetate and NAD(P)H. The roles of MDH vary widely from producing cellular energy via the Krebs cycle to maintaining redox balance, energy flux between cellular compartments, and the cross-talk between metabolic pathways via the malate-aspartate shuttle [7,8]. Physiologically, the directionality of the reaction is regulated by the presence of various metabolites and the energetic needs of the cell [9]. Structurally, MDH is a homodimer or homotetramer and is subjected to extensive allosteric regulation through protein-protein interactions and by small molecule binding such as L-malate, oxaloacetate, citrate, glutamate, and α-ketoglutarate. In addition to fundamental studies of the biochemical, structural, and functional relationships of various MDH isoforms, this protein has been widely analyzed from a physiological perspective for its roles in metabolic disorders, neurodegenerative diseases, and cancer [10,11].
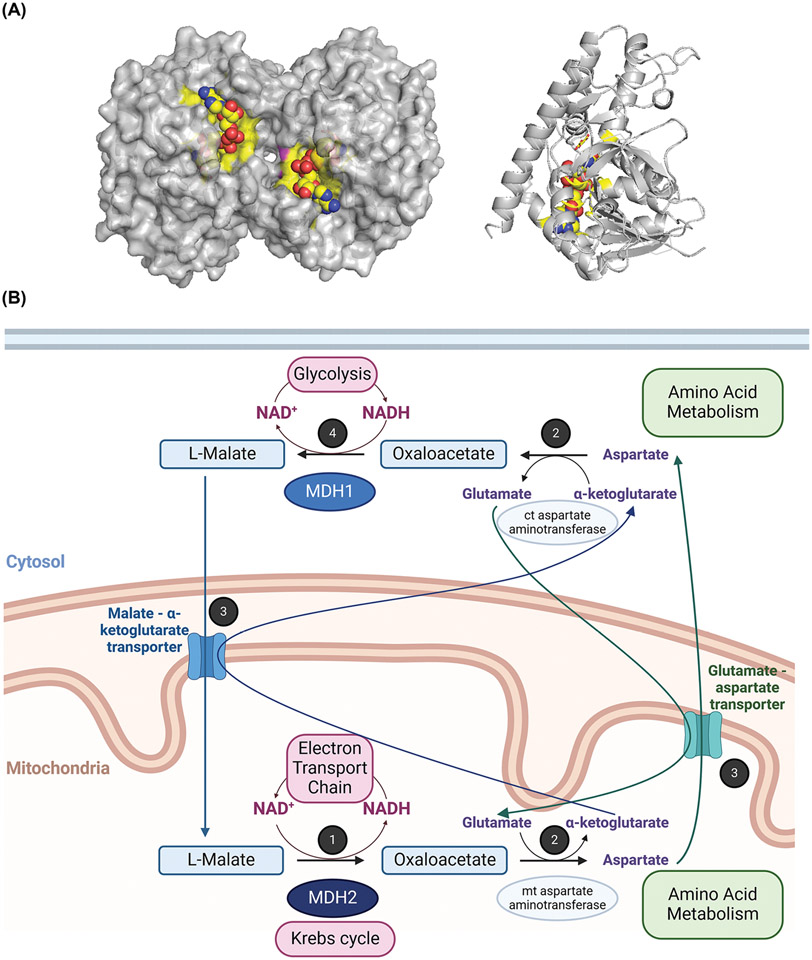
Figure 1. Structure and roles of human MDH in the metabolism.
(A) Three-dimensional structures of porcine cytoplasmic MDH dimer (surface view) and left monomer (ribbon view) are represented (PDB: 5mdh). The active site is highlighted in yellow, and the NAD+ ligand is shown in yellow spheres. Monomeric MDH is presented in the same orientation in Figures 2-4, with the active site facing back compared to the dimeric representation. (B) Overview of the human MDH roles and link to energy production by the electron transport chain, redox balance, and cross-talk between metabolic pathways. 1: Reaction 8 of the Krebs cycle: Oxidation of malate associated with the reduction of NAD+ by mitochondrial MDH2. NAD+ is regenerated via the electron transport chain. 2: A transamination reaction links the malate shuttle to amino acid metabolism. 3: Conversion of oxaloacetate to malate in the malate-aspartate shuttle requires the assistance of the glutamate-aspartate antiporter and the malate-α-ketoglutarate antiporter. 4: Reduction of oxaloacetate with concomitant oxidation of NADH by cytosolic MDH1. NAD+ is used by metabolic pathways like glycolysis. This figure was generated using PyMOL and BioRender.com.
The MDH enzyme is substantially modified through a variety of PTMs, including phosphorylation, acylation, ADP-ribosylation, and methylation. However, the roles of these PTMs on MDH structure, function, and cellular physiology are complex. One challenge to studying the biochemical and cellular implications of PTMs on metabolic enzymes is that the precise function of a PTM may differ based on the organism or tissue in which it is analyzed. Moreover, the mere presence of a PTM at a site on a protein does not necessarily confer a specific functional or regulatory alteration. In this review, we discuss examples of MDH acylation, ADP-ribosylation, and methylation PTMs and their consequences on metabolism, when known. We conclude by briefly reviewing the potential role of MDH PTMs in the integrity of metabolons. A review of the impacts of MDH phosphorylation is described elsewhere (covered in this issue).
MDH acetylation and acylation
Acetylation is one of the most abundant PTMs identified on proteins [12]. This modification reduces the overall electrostatic positive charge of the amino group, either on the N-α-amino group of the N-terminus or the N-ε-amino group of a lysine side chain (Figure 2A). While N-α-acetylation is more common, here we will focus on the latter modification. N-ε-acetylation (AcK) PTM occurs via both non-enzymatic and enzymatic mechanisms. Non-enzymatic protein acetylation occurs using a reactive acyl donor such as acetyl phosphate (AcP) or acetyl CoA (AcCoA) [13,14]. In contrast, enzymatic acetylation is mediated by N-acetyltransferases that require AcCoA as a substrate [13,14]. The enzymes that acetylate lysine residues specifically are referred to as lysine (K) acetyltransferases (KATs) and typically belong to the Gcn5-related N-acetyltransferase (GNAT), MYST, and p300/CBP protein families [15]. Some acetyl groups can be removed by deacetylase enzymes, such as CobB in Escherichia coli, and SIRT3 in eukaryotic systems [16-18]. Yet, it is currently unclear how these enzymatic and non-enzymatic acetylation/deacetylation mechanisms affect the stoichiometry of protein acetylation and their influence and impact on protein function [19]. Hence, the reversible nature of acetylation makes it an effective strategy for rapidly modulating protein charge, solubility, structure, and interactions with other biomolecules. N-ε-AcK acetylation as a response to a variety of growth conditions and stimuli is a conserved PTM across the domains of life [12,13]. Acetylated proteins from eukaryotic to prokaryotic systems have been observed, but not all acetylation sites on all proteins are conserved across different organisms or isoforms [20]. Furthermore, the functional importance of AcK residues, when known, is not necessarily identical.
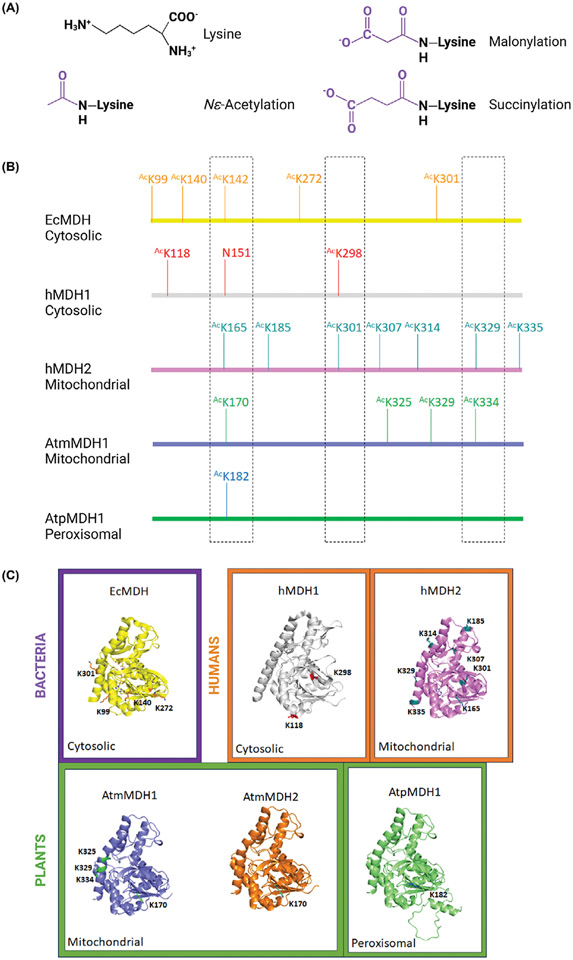
Figure 2. Sequence and structural comparison of acetylated lysine residues identified on proteins from different domains of life.
(A) Chemical structures of known lysine acylations. (B) A cartoon representation of locations of acetylated lysine residues on the linear amino acid sequences based on the structural alignment of multiple MDH isoforms from different organisms. (C) Three-dimensional structure of multiple monomeric MDH isoforms from different organisms. Bacterial proteins are surrounded by a purple box, human proteins with orange boxes, and plants with green boxes. The cellular localization is noted (i.e., cytosolic, mitochondrial, and peroxisomal). The crystal structure of the E. coli MDH enzyme (PDB: 1emd; UniProtID: P61889) is shown in yellow ribbons; citrate and NAD are in yellow sticks. Lysine residues identified as acetylated are in orange sticks. The crystal structure of the human MDH1 enzyme (PDB: 7rm9; UniProtID: P40925) is shown in grey ribbons, with identified AcK residues in red sticks. The crystal structure of the human MDH2 enzyme (PDB: 4wlo; UniProtID: P40926) is shown in pink ribbons; oxaloacetate and NADH are in pink sticks, and identified AcK residues are in teal sticks. AlphaFold models for A. thaliana proteins with identified AcK residues are represented in purple ribbons and green sticks for AtmMDH1 (UniProtID: Q9ZP06), in orange ribbons with cyan sticks for AtmMDH2 (UniProtID: Q9LKA3) and in green ribbons with blue sticks for peroxisomal AtpMDH1 (UniProtID: O82399). The cartoon sequence alignment shows conserved residues in the three-dimensional space with dashed outlined boxes. Not all residues are included. Three-dimensional structures were generated using PyMOL and the figure was constructed using BioRender.com.
MDH acetylation in bacteria
Several studies have investigated E. coli MDH (EcMDH) acetylation in vitro and in vivo. In vivo, AcP and/or AcCoA acetylation of EcMDH has been described on K99, K140, K272, and K301 (Figure 2B,C) [21-23]. These studies indicate that MDH acetylation in this bacterium is strictly non-enzymatic. Functional analysis using site-specific AcK incorporation on AcK99 and AcK140 in EcMDH showed acetylation increases enzyme turnover without changing the apparent affinity for either substrate, malate, or NAD+ [23,24]. Acetylation of K140 on Vibrio cholerae MDH has also been observed and corresponds to the EcMDH K140 residue [25]. Finally, acetylation of EcMDH has been detected in the presence of glucose, but at a lower level in citrate-containing media [22,26]. This observation suggests that in E. coli MDH acetylation is dependent upon the energy state of the cell.
MDH acetylation in humans
Unlike bacteria, where only one MDH is present, multiple MDH isoforms are found in different cellular compartments in higher organisms. In humans, two isoforms hMDH1 and hMDH2 are located in the cytosol and mitochondria, respectively. Numerous AcK residues have been identified for human MDH isoforms in different cell types (Figure 2B,C) [23,27-32]. For example, hMDH1 and hMDH2 were shown to be acetylated in acute myeloid leukemia MV4-11 cells on K118 and K298 for hMDH1, and K165, K185, K301, K329, and K335 for hMDH2 [27]. In Chang liver cells, acetylation was identified on K185, K301, K307, and K314 of hMDH2. Acetylation was increased in these cells in the presence of a high glucose concentration and the acetylated hMDH2 enzyme showed higher activity [32]. However, only hMDH2 AcK307 was shown to alter enzyme turnover, without changing the apparent affinity for the malate and NAD+, similar to AcK99 and AcK140 in EcMDH [23,24]. While numerous studies have shown that acetylation level in the nucleus has a direct effect on metabolism reprogramming and tumorigenesis, the direct contribution of MDH acetylation in these processes has not been specifically addressed [33].
The physiological roles of acetylated hMDH1 and hMDH2 lysine residues have been examined in the context of adipogenesis and caloric restriction, respectively. For example, hMDH1 is overexpressed and acetylated on K118 during adipogenesis and adipocyte differentiation [30]. Acetylated hMDH1 shows increased activity, generating malate in the cytosol that will be used by the citrate shuttle, a metabolic pathway that produces AcCoA and NADPH required for fatty acid synthesis (Figure 3). In response to caloric restriction, hMDH2 K239 is deacetylated in mouse liver, but is acetylated in the absence of the deacetylase SIRT3 [28]. This residue is located at the dimer interface, and its mutation to glutamine, an uncharged polar amino acid used to crudely mimic acetylated lysine, decreases enzymatic activity, which may suggest a role for acetylation in the stability of the hMDH2 quaternary structure [34]. This same study showed that other enzymes involved in the Krebs cycle, including citrate synthase (CS), aconitase, and succinate dehydrogenase, are acetylated in response to SIRT3 loss, affecting their enzymatic activity and the metabolite profile [28]. Altogether, the alteration of acetylation dynamics is associated with a rewiring of mitochondrial-dependent metabolism [28].
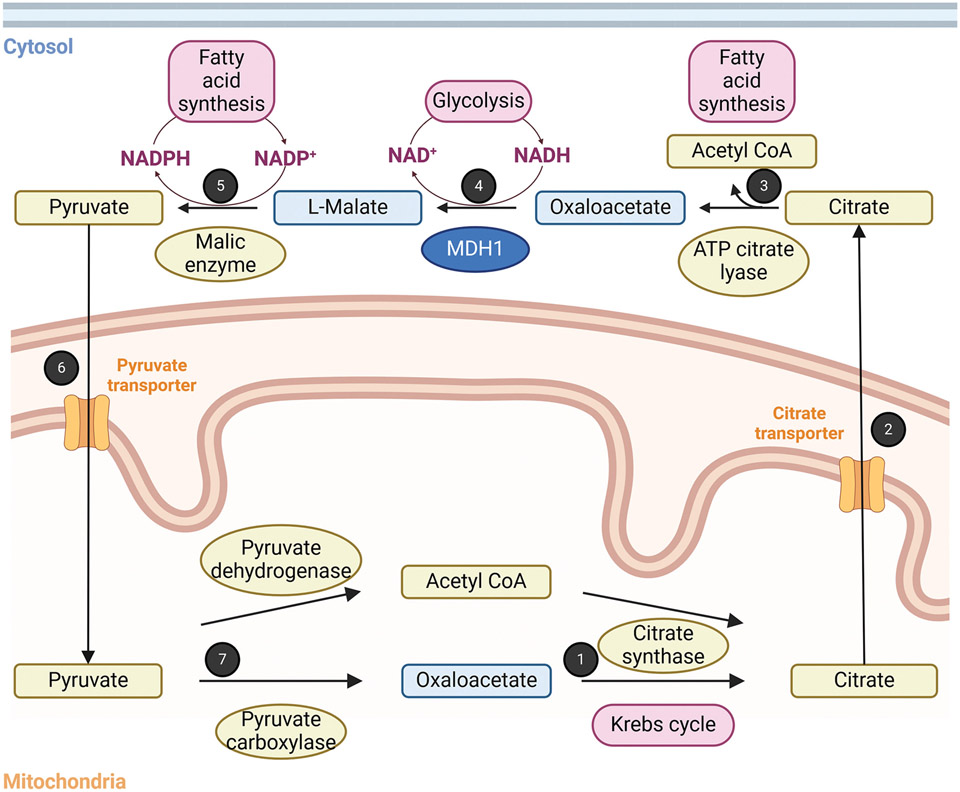
Figure 3. Role of MDH1 in the citrate shuttle and in fatty acid synthesis.
1: Reaction 1 of the Krebs cycle: Condensation of acetyl unit from the acetyl CoA with oxaloacetate by citrate synthase. 2: Transport of citrate from the mitochondria to the cytosol by the citrate transporter. 3. Cleavage of citrate to acetyl CoA and oxaloacetate by ATP citrate lyase. Acetyl CoA can be used as a precursor of fatty acids. 4: Reduction of oxaloacetate with concomitant oxidation of NADH by cytosolic MDH1. NAD+ is used by metabolic pathways like glycolysis. 5: Oxidative decarboxylation of malate by the NADP+-dependent malic enzyme to generate pyruvate. The generated NADPH is used for fatty acid synthesis. 6: Transport of the pyruvate from the cytosol to the mitochondria by the pyruvate transporter. 7: Pyruvate is used to form oxaloacetate via a carboxylation reaction by pyruvate carboxylase, and acetyl CoA by an oxidative decarboxylation reaction by pyruvate dehydrogenase. This figure was generated with BioRender.com.
MDH acetylation in plants
Like humans, plants have multiple MDH isoforms in various cellular compartments. In Arabidopsis thaliana (At), at least eight NAD+-dependent MDH isoforms have been observed in the cytosol, mitochondria, chloroplasts, and peroxisomes [35,36]. However, each isoform displays unique characteristics, including substrate specificity, tissue expression, and PTM profile. For instance, the mitochondrial isoform AtmMDH1 is more widely expressed than AtmMDH2 [37]. Lysine acetylation has been identified on AtmMDH1 (K170, K325, K329, and K334), AtmMDH2 (K170), peroxisomal isoform AtpMDH1 (K182), and cytosolic AtMDH2 (K3) (Figure 2B,C) [29,31,38,39].
The consequences of acetylation have been further studied in AtmMDH1 to determine kinetic parameters in both reaction directions [38]. The site-specific incorporation of AcK325 and AcK329 did not reveal changes in MDH kinetic parameters for oxaloacetate but decreased enzyme turnover was observed for AcK170 and AcK334 in vitro. On the other hand, AcK325, AcK329, and AcK334 were associated with an increased apparent affinity for malate and decreased turnover number, while AcK170 reduced the apparent affinity for malate. Similar effects were observed in another plant model, Physcomitrella patens [38]. Consequently, acetylated lysine residues may play a role in the directionality of the reaction mediated by MDH, impacting metabolic flux. The widespread occurrence of this PTM in plants highlights the relevance of acetylation in modulating the structure and function of MDH (for a review, see [33]).
Other MDH acylation modifications
Acylation involves the transfer of an acyl chain from an acyl CoA donor molecule to a lysine or cysteine residue [40]. The diversity of acyl groups that can decorate protein residues increases the possibility of finely tuning the charge, hydrophobicity, and regulation of a particular protein. Depending on the identity of the acyl group, the charge of a lysine residue can vary from +1 to neutral for an acetyl group or from +1 to −1 for succinyl or malonyl groups under physiological conditions (Figure 2A) [41]. In addition to acetylation, both hMDH1 and hMDH2 have been found to be succinylated and malonylated on lysine residues, including succinylation of hMDH1 K298, malonylation of hMDH2 K185 and K301, and both succinylation and malonylation of hMDH2 K301 and K329 (Table 1) [29,31].
Table 1.
List of identified succinylated and/or malonylated lysine residues on human MDH that are known to also be acetylated
| Modification | hMDH1 | hMDH2 |
|---|---|---|
| Succinylation | K103, K107, K110, K214, K298 | K91, K157, K203, K296, K324, K329, K335 |
| Malonylation | K110 | K78, K185, K203, K239, K296, K301, K307, K314, K324 |
Like acetylation, the transfer of succinyl or malonyl groups from succinyl CoA and malonyl CoA, respectively, to the N-ε-amino group of a lysine residue can occur by enzymatic or non-enzymatic mechanisms [42,43]. KAT2A (or hGCN5) has been identified as both a succinyltransferase and malonyltransferase [44-46]. The removal of these PTMs is mediated by Sirtuin proteins, especially SIRT5 and SIRT7 [47,48]. Currently, these PTMs remain less well-understood than acetylation. However, processes that contribute to the synthesis and degradation of these acyl CoA substrates seem to play an important role, such as altering malonylation levels on proteins in the absence of malonyl CoA synthetase or malonyl CoA decarboxylase [49,50]. Furthermore, both succinylation and malonylation have been described on numerous metabolic enzymes and the loss of SIRT5 is associated with hyper-succinylation and hyper-malonylation of mitochondrial proteins [47,51,52]. Current literature suggests that these PTMs contribute to the overall modulation of carbohydrate and fatty acid metabolism, and they are dysregulated in diseases like diabetes and cancer [52-54]. Understanding what stimulus would lead to different types of acylation and the outcomes associated with each PTM remain important challenges for the future.
MDH ADP-ribosylation
ADP-ribosylation is mediated by ADP-ribosyltransferases, which transfer ADP-ribose from NAD+ onto an amino acid side chain (currently nine are known, including glutamate and serine) [55]. ADP-ribosylation can form N-, O-, or S-glycosidic linkages between the ribose of the ADP-ribose and the side chain of the targeted amino acid [56]. In humans, several members of the poly(ADP-ribose) polymerase protein (PARP) family, possess ADP-ribosyltransferase activity [57]. The biological meaning of this PTM is complex and dependent upon the modified amino acids and the number of ADP-ribose moieties that are added (Figure 4A) [55]. For instance, PARP10 transfers one ADP-ribose moiety in a mono-ADP-ribosylation reaction, whereas PARP1 and PARP2 can add multiple ADP-ribose moieties to form a branched polymer in a poly-ADP-ribosylation process. The dynamics of this reaction are mediated by both PARG (poly(ADP-ribose) glycohydrolase) and ARH3 (ADP-ribosylhydrolase), which trim poly(ADP-ribose) chains or remove ADP-ribose moieties from the amino acid without generating NAD+ [58].
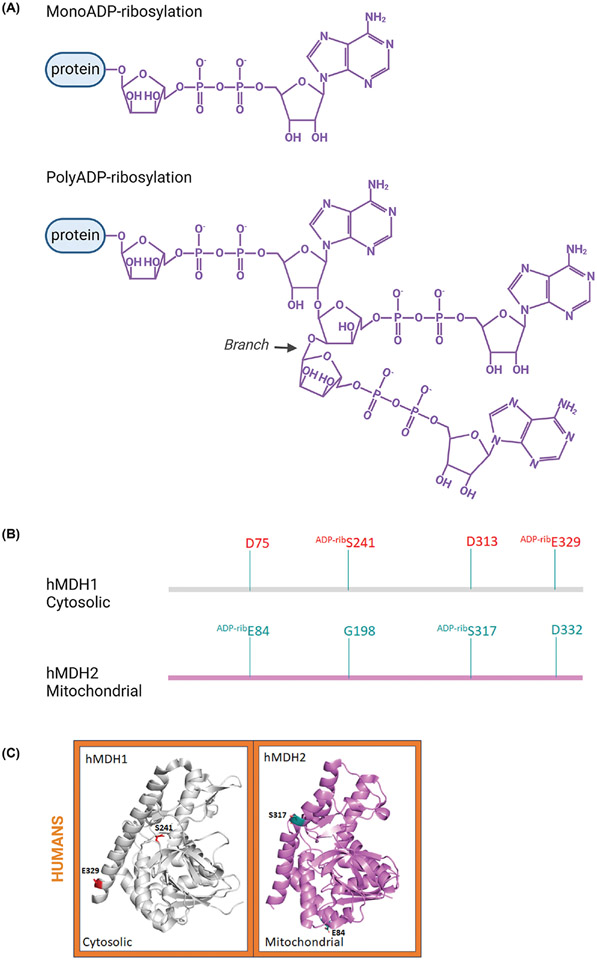
Figure 4. Sequence and structural comparison of ADP-ribosylated (ADP-rib) residues identified on human MDH proteins.
(A) Chemical structures of mono- and poly-ADPribose. (B) A cartoon representation of locations of ADP-ribosylated residues on the linear amino acid sequences based on the structural alignment of human MDH isoforms. (C) The three-dimensional structure of monomeric human MDH1 (PDB: 7rm9; UniProtID: P40925) and MDH2 (PDB: 4wlo; UniProtID: P40926) isoforms with ADP-ribosylated residues. These structures are colored as described in Figure 2. Three-dimensional structures were generated using PyMOL and the figure was constructed using BioRender.com.
The development of mass spectrometry methods to identify ADP-ribosylated proteins has led to the discovery of new PARP targets [59-61]. These targets include numerous proteins involved in metabolism, including hMDH1 and hMDH2 [62-64]. Both isoforms are ADP-ribosylated: S241 and E329 for hMDH1 and E84 and S317 for hMDH2 (Figure 4B,C), but the functional consequences of these modifications are unknown. Yet, their functional meaning may be discerned by considering the location of the ADP-ribosylated residues. The human MDH1 S241 residue is known to be involved in NAD+/NADH binding and is close to the dimerization interface where it interacts with amino acids involved in stabilizing the quaternary structure (e.g. Q238) [65,66]. Thus, S241 ADP-ribosylation may affect enzymatic activity by blocking access to the substrate binding pocket and/or dimerization. Due to its surface exposed location, E329 ADP-ribosylation may have a potential role in protein-protein interactions with molecules containing ADP-ribose binding motifs.
A similar impact of solvent-exposed hMDH2 ADP-ribosylated residues E84 and S317 may contribute to the interaction with proteins specifically recognizing and binding to the ADP-ribose modification. Interestingly, the hMDH1 S241 and hMDH2 S317 residues have also been shown to be phosphorylated, which suggests a potential competition between ADP-ribosylation and phosphorylation [67]. Cross-talk between these two PTMs has already been observed to differentially modulate protein functions [68-73].
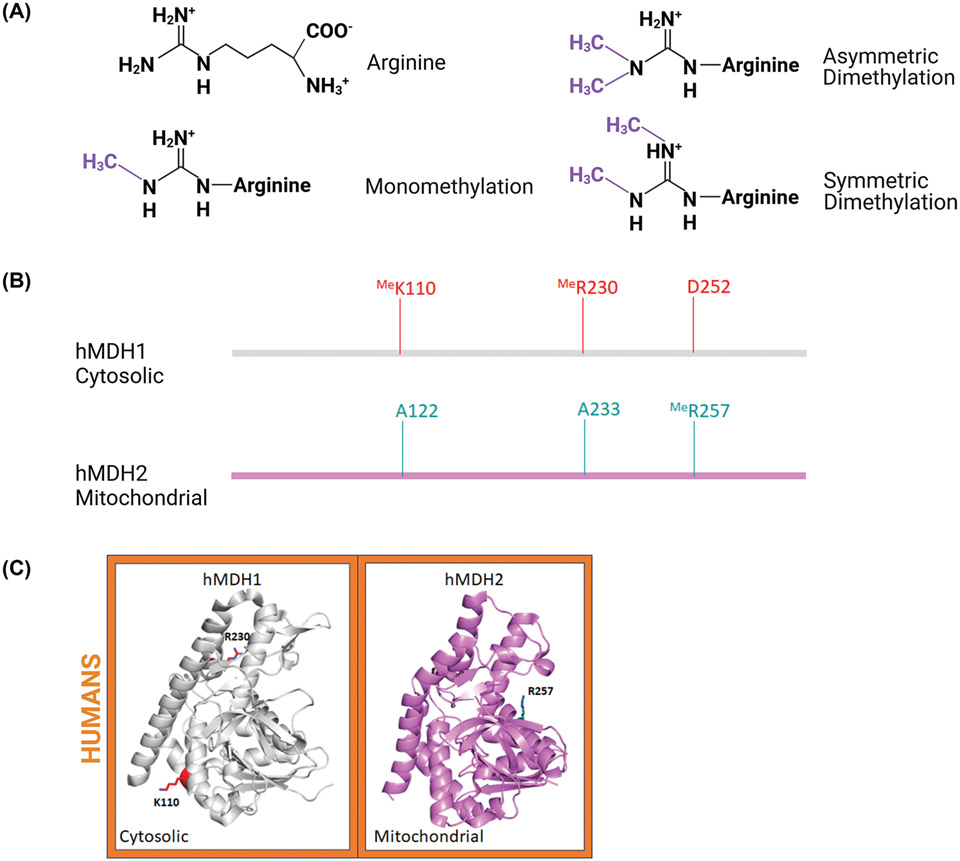
MDH methylation
Once thought to be a modification exclusive to histones, it is now well known that lysine and arginine residues of numerous proteins are substrates for S-adenosylmethionine (AdoMet)-dependent methyltransferases, such as AdoMet-protein arginine methyltransferases (PRMTs) [74]. Unlike lysine, arginine methylation maintains the side chain positive charge (Figure 5A). Additionally, while lysine residues can only be modified by a single acyl group, arginine residues can be mono- or di-methylated. Di-methylated arginine residues can be asymmetrical (two methyl groups on a single nitrogen) or symmetrical (two nitrogen atoms each with a single methyl group). A wide variety of methods have been developed to identify methylated proteins with an array of antibodies specific for mono-, symmetrical di-, and asymmetrical dimethylated arginine [75,76]. In addition to immunological methods, ion exchange techniques can be used to identify methylated arginine residues; these techniques can be used together for enhanced identification.
Figure 5. Sequence and structural comparison of methylated (Me) residues identified on human MDH proteins.
(A) Chemical structures of known arginine methylations. (B) Methylated residues represented on the linear sequence of human MDH isoforms. (C) Crystal structure of monomeric human MDH1 (PDB: 7rm9; UniProtID: P40925) and MDH2 (PDB: 4wlo; UniProtID: P40926) isoforms with methylated residues. These structures are colored as described in Figure 2. Three-dimensional structures were generated using PyMOL and the figure was constructed using BioRender.com.
To date, no methylation sites have been identified on EcMDH or AtMDH. In contrast, both hMDH1 and hMDH2 isoforms are methylated (Figure 5B,C). Cytosolic MDH1 is asymmetrically di-methylated on R230 located at the dimer interface [77,78]. The protein methyltransferase CARM1/PRMT4 catalyzes methylation of this arginine, which inhibits MDH1 activity by disrupting the ability of the enzyme to dimerize. By altering the quaternary structure, this CARM1-dependent methylation diminishes flux through the malate-aspartate shuttle to reduce the level of mitochondrial respiration. This modification also seems associated with the cellular stress level, as CARM1 activity is reduced in response to oxidative stress. Human MDH1 has also been found monomethylated on K110 by SMYD2, though the functional significance of this modification is unknown [79]. It is noteworthy that CARM1 and SMYD2 have been shown to be overexpressed in cancers [79]. To date, methylation of mitochondrial hMDH2 R257 has been shown in two methyl-proteomics studies [80,81]. However, the functional relevance of this particular modification has not been investigated. While PRMT inhibitors have been used in the proteomic study, the identity of the specific methyltransferase(s) responsible for this MDH methylation remains elusive [81].
Potential PTM role in the organization of metabolon
Substantial evidence supports the existence of an in vivo metabolon including Krebs cycle proteins MDH2 and CS, as well as aconitase, succinyl CoA synthetase (SCS), fumarase, and isocitrate dehydrogenase (IDH), in eukaryotes and bacteria [82-84]. Co-localization of multiple enzymes involved in a metabolic pathway offers certain advantages such as channeling of metabolic intermediates directly from one active site to another and provides an opportunity for regulation [85-87]. Recent studies show that MDH2 and CS share a common interface [84,85]. Interestingly, that interface includes hMDH2 E84, which, as noted above, can be ADP-ribosylated. Work defining the MDH2-CS interface also identified K301 of MDH2 through a chemical cross-link which may indicate that this acetylatable residue is involved in the interaction with CS. As in the case of hMDH1 R230me2a which inactivates the enzyme by disrupting subunit-subunit interactions, other PTMs may offer a similar regulatory function by affecting metabolon assembly.
A multiprotein complex in the fungus Cunninghamella bainieri involved in lipid biosynthesis, has been identified and is composed of MDH1, malic enzyme (ME), fatty acid synthase (FAS), ATP:citrate lyase (ACL), acetyl CoA carboxylase (ACC), pyruvate carboxylase (PC), and MDH [88]. Further characterizations are needed to understand how this metabolon is organized and regulated and whether it is conserved across species. Thus, unveiling the roles of PTMs on MDH and other metabolic enzymes like CS will provide a better understanding of the dynamics of metabolons in response to environmental conditions.
Future perspectives
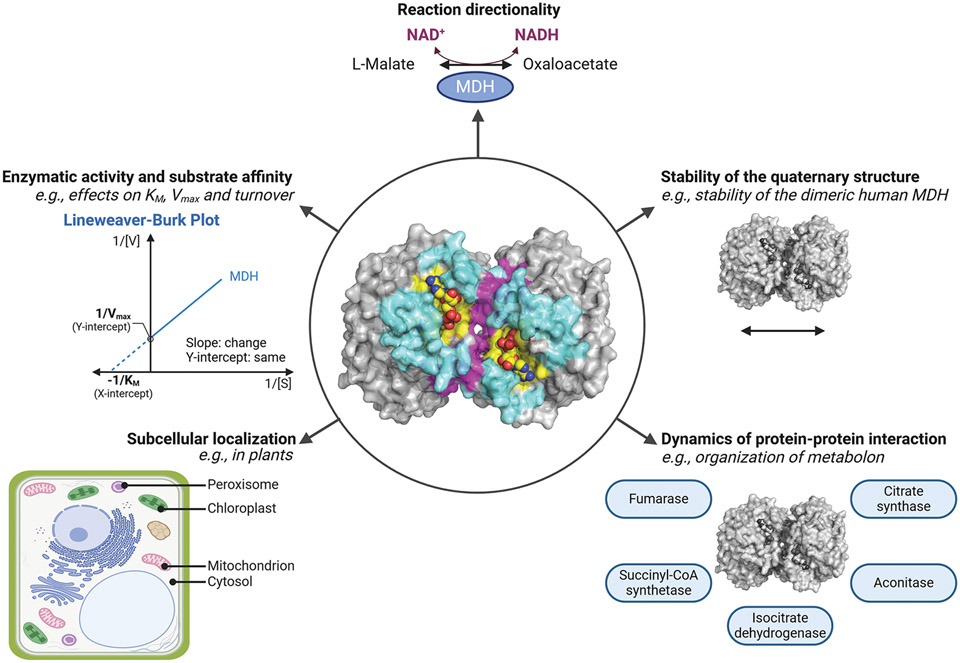
The development of suitable tools, such as mass-spectrometry methods, has unveiled a large repertoire of protein PTMs, including acylation, ADP-ribosylation, and methylation [40,60,75]. Yet, identifying their functional implications, if any, is still challenging. For each PTM, the environmental conditions and the enzymes involved in the PTM dynamics need to be determined to understand the impacts on the enzymatic activity, three-dimensional structure, subcellular localization, and interactome (Figure 6). By cross-listing PTMs on MDH, a comprehensive picture of how MDH modulates metabolism and contributes to diseases will be established.
Figure 6. Potential roles of PTMs on MDH.
At the center, the three-dimensional structure of the porcine cytoplasmic MDH dimer is represented (PDB: 5mdh). The active site is highlighted in yellow, the dimer interface in magenta, and the citrate synthase surface for the metabolon complex in cyan. The NAD+ ligand is shown in yellow spheres. Properties that can be modulated by PTMs, including defining the directionality of the reaction, altering enzymatic characteristics (e.g. Km and Vmax), defining the subcellular localization, regulating the stability of the quaternary structure and protein–protein interactions protrude around the circle. This figure was generated using PyMOL and BioRender.com.
Summary.
MDH is extensively modified by acylation, ADP-ribosylation, and methylation.
The role of these PTMs remains mostly uncharacterized, but growing evidence indicates their contribution in modulating MDH structure, catalytic activity, and interactome.
Further study of the metabolon will provide an understanding of the importance of PTMs in the stability and regulation of this multiprotein complex.
Funding
This work was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health [grant number R35GM133506 (to M.L.K.)].
Abbreviations
- ACC
acetyl CoA carboxylase
- AcCoA
acetyl CoA
- AcK
acetylated Nε-amine of a lysine
- ACL
ATP:citrate lyase
- AcP
acetyl phosphate
- AdoMet
S-adenosylmethionine
- ARH
ADP-ribosylhydrolase
- CS
citrate synthase
- FAS
fatty acid synthase
- GNAT
Gcn5-related N-acetyltransferase
- IDH
isocitrate dehydrogenase
- KAT
lysine (K) acetyltransferase
- MDH
malate dehydrogenase
- ME
malic enzyme
- NAD
nicotinamide adenine dinucleotide
- NADP
nicotinamide adenine dinucleotide phosphate
- PARG
poly(ADP-ribose)glycohydrolase
- PARP
poly(ADP-ribose)polymerase
- PC
pyruvate carboxylase
- PRMT
AdoMet-protein arginine methyltransferase
- PTM
post-translational modification
- SCS
succinyl CoA synthetase
- SIRT
sirtuin
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Ramazi S and Zahiri J (2021) Posttranslational modifications in proteins: resources, tools and prediction methods. Database (Oxford) 2021, 10.1093/database/baab012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Sun Z, Jia J, Du T, Zhang N, Tang Y et al. (2021) Overview of histone modification. Adv. Exp. Med. Biol 1283, 1–16 [DOI] [PubMed] [Google Scholar]
- 3.Jambhekar A, Dhall A and Shi Y (2019) Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol 20, 625–641, 10.1038/s41580-019-0151-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basavarajappa BS and Subbanna S (2021) Histone methylation regulation in neurodegenerative disorders. Int. J. Mol. Sci 22, 10.3390/ijms22094654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang YH, Wen R, Yang N, Zhang TN and Liu CF (2023) Roles of protein post-translational modifications in glucose and lipid metabolism: mechanisms and perspectives. Mol. Med 29, 93, 10.1186/s10020-023-00684-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong Q, Xiao X, Qiu Y, Xu Z, Chen C, Chong B et al. (2023) Protein posttranslational modifications in health and diseases: Functions, regulatory mechanisms, and therapeutic implications. MedComm (2020) 4, e261, 10.1002/mco2.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borst P. (2020) The malate-aspartate shuttle (Borst cycle): How it started and developed into a major metabolic pathway. IUBMB Life 72, 2241–2259, 10.1002/iub.2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selinski J and Scheibe R (2019) Malate valves: old shuttles with new perspectives. Plant Biol. (Stuttg) 21, 21–30, 10.1111/plb.12869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minarik P, Tomaskova N, Kollarova M and Antalik M (2002) Malate dehydrogenases–structure and function. Gen. Physiol. Biophys 21, 257–265 [PubMed] [Google Scholar]
- 10.Broeks MH, van Karnebeek CDM, Wanders RJA, Jans JJM and Verhoeven-Duif NM (2021) Inborn disorders of the malate aspartate shuttle. J. Inherit. Metab. Dis 44, 792–808, 10.1002/jimd.12402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jitrapakdee S, Wutthisathapornchai A, Wallace JC and MacDonald MJ (2010) Regulation of insulin secretion: role of mitochondrial signalling. Diabetologia 53, 1019–1032, 10.1007/s00125-010-1685-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ali I, Conrad RJ, Verdin E and Ott M (2018) Lysine acetylation goes global: from epigenetics to metabolism and therapeutics. Chem. Rev 118, 1216–1252, 10.1021/acs.chemrev.7b00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen DG, Baumgartner JT, Xie X, Jew KM, Basisty N, Schilling B et al. (2019) Mechanisms, detection, and relevance of protein acetylation in prokaryotes. mBio 10, 10.1128/mBio.02708-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menzies KJ, Zhang H, Katsyuba E and Auwerx J (2016) Protein acetylation in metabolism - metabolites and cofactors. Nat. Rev. Endocrinol 12, 43–60, 10.1038/nrendo.2015.181 [DOI] [PubMed] [Google Scholar]
- 15.Srivastava S, Kumar S, Bhatt R, Ramachandran R, Trivedi AK and Kundu TK (2023) Lysine acetyltransferases (KATs) in disguise: diseases implications. J. Biochem 173, 417–433, 10.1093/jb/mvad022 [DOI] [PubMed] [Google Scholar]
- 16.Li G, Tian Y and Zhu WG (2020) The roles of histone deacetylases and their inhibitors in cancer therapy. Front Cell Dev. Biol 8, 576946, 10.3389/fcell.2020.576946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinert BT, lesmantavicius V, Wagner SA, Scholz C, Gummesson B, Beli P et al. (2013) Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 51, 265–272, 10.1016/j.molcel.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 18.Weinert BT, Moustafa T, Iesmantavicius V, Zechner R and Choudhary C (2015) Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 34, 2620–2632, 10.15252/embj.201591271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen BK, Gupta R, Baldus L, Lyon D, Narita T, Lammers M et al. (2019) Analysis of human acetylation stoichiometry defines mechanistic constraints on protein regulation. Nat. Commun 10, 1055, 10.1038/s41467-019-09024-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jew KM, Le VTB, Amaral K, Ta A, Nguyen May NM, Law M et al. (2021) Investigation of the importance of protein 3D structure for assessing conservation of lysine acetylation sites in protein homologs. Front Microbiol. 12, 805181, 10.3389/fmicb.2021.805181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen DG, Meyer JG, Baumgartner JT, D’Souza AK, Nelson WC, Payne SH et al. (2018) Identification of novel protein lysine acetyltransferases in Escherichia coli. mBio 9, 10.1128/mBio.01905-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn ML, Zemaitaitis B, Hu LI, Sahu A, Sorensen D, Minasov G et al. (2014) Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PloS ONE 9, e94816, 10.1371/journal.pone.0094816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkat S, Gregory C, Sturges J, Gan Q and Fan C (2017) Studying the lysine acetylation of malate dehydrogenase. J. Mol. Biol 429, 1396–1405, 10.1016/j.jmb.2017.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan C, Xiong H, Reynolds NM and Soll D (2015) Rationally evolving tRNAPyl for efficient incorporation of noncanonical amino acids. Nucleic Acids Res. 43, e156, 10.1093/nar/gkv800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jers C, Ravikumar V, Lezyk M, Sultan A, Sjoling A, Wai SN et al. (2017) The global acetylome of the human pathogen Vibrio cholerae V52 reveals lysine acetylation of major transcriptional regulators. Front Cell Infect Microbiol. 7, 537, 10.3389/fcimb.2017.00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J et al. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007, 10.1126/science.1179687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC et al. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840, 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- 28.Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD et al. (2013) Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol. Cell 49, 186–199, 10.1016/j.molcel.2012.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang KY, Lee TY, Kao HJ, Ma CT, Lee CC, Lin TH et al. (2019) dbPTM in 2019: exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res. 47, D298–D308, 10.1093/nar/gky1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim EY, Kim WK, Kang HJ, Kim JH, Chung SJ, Seo YS et al. (2012) Acetylation of malate dehydrogenase 1 promotes adipogenic differentiation via activating its enzymatic activity. J. Lipid Res 53, 1864–1876, 10.1194/jlr.M026567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Li S, Luo M, Jhong JH, Li W, Yao L et al. (2022) dbPTM in 2022: an updated database for exploring regulatory networks and functional associations of protein post-translational modifications. Nucleic Acids Res. 50, D471–D479, 10.1093/nar/gkab1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T et al. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004, 10.1126/science.1179689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma R, Wu Y, Li S and Yu X (2021) Interplay between glucose metabolism and chromatin modifications in cancer. Front Cell Dev. Biol 9, 654337, 10.3389/fcell.2021.654337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graf LG, Vogt R, Blasl AT, Qin C, Schulze S, Zuhlke D et al. (2021) Assays to study enzymatic and non-enzymatic protein lysine acetylation in vitro. Curr. Protoc 1, e277, 10.1002/cpz1.277 [DOI] [PubMed] [Google Scholar]
- 35.Gietl C. (1992) Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim. Biophys. Acta 1100, 217–234, 10.1016/0167-4838(92)90476-T [DOI] [PubMed] [Google Scholar]
- 36.Liszka A, Schimpf R, Cartuche Zaruma KI, Buhr A, Seidel T, Walter S et al. (2020) Three cytosolic NAD-malate dehydrogenase isoforms of Arabidopsis thaliana: on the crossroad between energy fluxes and redox signaling. Biochem. J 477, 3673–3693, 10.1042/BCJ20200240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee CP, Eubel H, O’Toole N and Millar AH (2008) Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol. Cell. Proteomics 7, 1297–1316, 10.1074/mcp.M700535-MCP200 [DOI] [PubMed] [Google Scholar]
- 38.Balparda M, Elsasser M, Badia MB, Giese J, Bovdilova A, Hudig M et al. (2022) Acetylation of conserved lysines fine-tunes mitochondrial malate dehydrogenase activity in land plants. Plant J. 109, 92–111, 10.1111/tpj.15556 [DOI] [PubMed] [Google Scholar]
- 39.Konig AC, Hartl M, Boersema PJ, Mann M and Finkemeier I (2014) The mitochondrial lysine acetylome of Arabidopsis. Mitochondrion 19, 252–260, 10.1016/j.mito.2014.03.004 [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, Shi Z and Bao L (2022) An expanding repertoire of protein acylations. Mol. Cell. Proteomics 21, 100193, 10.1016/j.mcpro.2022.100193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirschey MD and Zhao Y (2015) Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol. Cell. Proteomics 14, 2308–2315, 10.1074/mcp.R114.046664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saggerson D. (2008) Malonyl-CoA, a key signaling molecule in mammalian cells. Annu. Rev. Nutr 28, 253–272, 10.1146/annurev.nutr.28.061807.155434 [DOI] [PubMed] [Google Scholar]
- 43.Sreedhar A, Wiese EK and Hitosugi T (2020) Enzymatic and metabolic regulation of lysine succinylation. Genes Dis. 7, 166–171, 10.1016/j.gendis.2019.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Guo YR, Liu K, Yin Z, Liu R, Xia Y et al. (2017) KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277, 10.1038/nature25003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Guo YR, Xing D, Tao YJ and Lu Z (2018) Supramolecular assembly of KAT2A with succinyl-CoA for histone succinylation. Cell Discov. 4, 47, 10.1038/s41421-018-0048-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang R, Bons J, Scheidemantle G, Liu X, Bielska O, Carrico C et al. (2023) Histone malonylation is regulated by SIRT5 and KAT2A. iScience 26, 106193, 10.1016/j.isci.2023.106193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H et al. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809, 10.1126/science.1207861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L, Shi L, Yang S, Yan R, Zhang D, Yang J et al. (2016) SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat. Commun 7, 12235, 10.1038/ncomms12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowman CE, Rodriguez S, Selen Alpergin ES, Acoba MG, Zhao L, Hartung T et al. (2017) The mammalian malonyl-CoA synthetase ACSF3 is required for mitochondrial protein malonylation and metabolic efficiency. Cell Chem. Biol 24, 673e674–684e674, 10.1016/j.chembiol.2017.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colak G, Pougovkina O, Dai L, Tan M, Te Brinke H, Huang H et al. (2015) Proteomic and biochemical studies of lysine malonylation suggest its malonic aciduria-associated regulatory role in mitochondrial function and fatty acid oxidation. Mol. Cell. Proteomics 14, 3056–3071, 10.1074/mcp.M115.048850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P et al. (2015) SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell 59, 321–332, 10.1016/j.molcel.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L et al. (2013) SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930, 10.1016/j.molcel.2013.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai X, Zhou Y, Han F and Li J (2022) Succinylation and redox status in cancer cells. Front Oncol. 12, 1081712, 10.3389/fonc.2022.1081712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du Y, Hu H, Qu S, Wang J, Hua C, Zhang J et al. (2018) SIRT5 deacylates metabolism-related proteins and attenuates hepatic steatosis in ob/ob mice. EBioMedicine 36, 347–357, 10.1016/j.ebiom.2018.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suskiewicz MJ, Palazzo L, Hughes R and Ahel I (2021) Progress and outlook in studying the substrate specificities of PARPs and related enzymes. FEBSJ. 288, 2131–2142, 10.1111/febs.15518 [DOI] [PubMed] [Google Scholar]
- 56.Liu Q, Florea BI and Filippov DV (2017) ADP-Ribosylation goes normal: serine as the major site of the modification. Cell Chem. Biol 24, 431–432, 10.1016/j.chembiol.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 57.Gupte R, Liu Z and Kraus WL (2017) PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 31, 101–126, 10.1101/gad.291518.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rack JGM, Palazzo L and Ahel I (2020) (ADP-ribosyl)hydrolases: structure, function, and biology. Genes Dev. 34, 263–284, 10.1101/gad.334631.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ayyappan V, Wat R, Barber C, Vivelo CA, Gauch K, Visanpattanasin P et al. (2021) ADPriboDB 2.0: an updated database of ADP-ribosylated proteins. Nucleic Acids Res. 49, D261–D265, 10.1093/nar/gkaa941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bashyal A and Brodbelt JS (2024) Uncommon posttranslational modifications in proteomics: ADP-ribosylation, tyrosine nitration, and tyrosine sulfation. Mass Spectrom. Rev 43, 289–326, 10.1002/mas.21811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Challa S, Stokes MS and Kraus WL (2021) MARTs and MARylation in the cytosol: biological functions, mechanisms of action, and therapeutic potential. Cells 10, 10.3390/cells10020313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akram M. (2014) Citric acid cycle and role of its intermediates in metabolism. Cell Biochem. Biophys 68, 475–478, 10.1007/s12013-013-9750-1 [DOI] [PubMed] [Google Scholar]
- 63.Grimaldi G, Catara G, Valente C and Corda D (2018) In vitro techniques for ADP-ribosylated substrate identification. Methods Mol. Biol 1813, 25–40 [DOI] [PubMed] [Google Scholar]
- 64.Hopp AK, Teloni F, Bisceglie L, Gondrand C, Raith F, Nowak K et al. (2021) Mitochondrial NAD(+) controls nuclear ARTD1-induced ADP-ribosylation. Mol. Cell 81, 340e345–354e345, 10.1016/j.molcel.2020.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birktoft JJ, Rhodes G and Banaszak LJ (1989) Refined crystal structure of cytoplasmic malate dehydrogenase at 2.5-A resolution. Biochemistry 28, 6065–6081, 10.1021/bi00440a051 [DOI] [PubMed] [Google Scholar]
- 66.McCue WM and Finzel BC (2022) Structural characterization of the human cytosolic malate dehydrogenase I. ACS Omega 7, 207–214, 10.1021/acsomega.1c04385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ochoa D, Jarnuczak AF, Vieitez C, Gehre M, Soucheray M, Mateus A et al. (2020) The functional landscape of the human phosphoproteome. Nat. Biotechnol 38, 365–373, 10.1038/s41587-019-0344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brustel J, Muramoto T, Fumimoto K, Ellins J, Pears CJ and Lakin ND (2022) Linking DNA repair and cell cycle progression through serine ADP-ribosylation of histones. Nat. Commun 13, 185, 10.1038/s41467-021-27867-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Q, Bian C, Wang X, Liu X, Ahmad Kassab M, Yu Y et al. (2021) ADP-ribosylation of histone variant H2AX promotes base excision repair. EMBO J. 40, e104542, 10.15252/embj.2020104542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daniels CM, Kaplan PR, Bishof I, Bradfield C, Tucholski T, Nuccio AG et al. (2020) Dynamic ADP-ribosylome, phosphoproteome, and interactome in LPS-activated macrophages. J. Proteome Res 19, 3716–3731, 10.1021/acs.jproteome.0c00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daniels CM, Nuccio A, Kaplan PR and Nita-Lazar A (2020) Simultaneous, quantitative characterization of protein ADP-ribosylation and protein phosphorylation in macrophages. Methods Mol. Biol 2184, 145–160 [DOI] [PubMed] [Google Scholar]
- 72.Huang D, Camacho CV, Setlem R, Ryu KW, Parameswaran B, Gupta RK et al. (2020) Functional interplay between histone H2B ADP-ribosylation and phosphorylation controls adipogenesis. Mol. Cell 79, 934e914–949e914, 10.1016/j.molcel.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin J, Yuan Y, Xian W, Tang Z, Fu J and Liu X (2023) The ever-increasing necessity of mass spectrometry in dissecting protein post-translational modifications catalyzed by bacterial effectors. Mol. Microbiol 119, 677–686, 10.1111/mmi.15071 [DOI] [PubMed] [Google Scholar]
- 74.Di Blasi R, Blyuss O, Timms JF, Conole D, Ceroni F and Whitwell HJ (2021) Non-histone protein methylation: biological significance and bioengineering potential. ACS Chem. Biol 16, 238–250, 10.1021/acschembio.0c00771 [DOI] [PubMed] [Google Scholar]
- 75.Brown T, Nguyen T, Zhou B and Zheng YG (2023) Chemical probes and methods for the study of protein arginine methylation. RSC Chem. Biol 4, 647–669, 10.1039/D3CB00018D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carlson SM and Gozani O (2014) Emerging technologies to map the protein methylome. J. Mol. Biol 426, 3350–3362, 10.1016/j.jmb.2014.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA et al. (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 13, 372–387, 10.1074/mcp.0113.027870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang YP, Zhou W, Wang J, Huang X, Zuo Y, Wang TS et al. (2016) Arginine methylation of MDH1 by CARM1 inhibits glutamine metabolism and suppresses pancreatic cancer. Mol. Cell 64, 673–687, 10.1016/j.molcel.2016.09.028 [DOI] [PubMed] [Google Scholar]
- 79.Olsen JB, Cao XJ, Han B, Chen LH, Horvath A, Richardson TI et al. (2016) Quantitative profiling of the activity of protein lysine methyltransferase SMYD2 using SILAC-based proteomics. Mol. Cell. Proteomics 15, 892–905, 10.1074/mcp.M115.053280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Larsen SC, Sylvestersen KB, Mund A, Lyon D, Mullari M, Madsen MV et al. (2016) Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal 9, rs9, 10.1126/scisignal.aaf7329 [DOI] [PubMed] [Google Scholar]
- 81.Maron MI, Lehman SM, Gayatri S, DeAngelo JD, Hegde S, Lorton BM et al. (2021) Independent transcriptomic and proteomic regulation by type I and II protein arginine methyltransferases. iScience 24, 102971, 10.1016/j.isci.2021.102971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bartholomae M, Meyer FM, Commichau FM, Burkovski A, Hillen W and Seidel G (2014) Complex formation between malate dehydrogenase and isocitrate dehydrogenase from Bacillus subtilis is regulated by tricarboxylic acid cycle metabolites. FEBS J. 281, 1132–1143, 10.1111/febs.12679 [DOI] [PubMed] [Google Scholar]
- 83.Mitchell CG (1996) Identification of a multienzyme complex of the tricarboxylic acid cycle enzymes containing citrate synthase isoenzymes from Pseudomonas aeruginosa. Biochem. J 313, 769–774, 10.1042/bj3130769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu F and Minteer S (2015) Krebs cycle metabolon: structural evidence of substrate channeling revealed by cross-linking and mass spectrometry. Angew. Chem. Int. Ed. Engl 54, 1851–1854, 10.1002/anie.201409336 [DOI] [PubMed] [Google Scholar]
- 85.Bulutoglu B, Garcia KE, Wu F, Minteer SD and Banta S (2016) Direct evidence for metabolon formation and substrate channeling in recombinant TCA cycle enzymes. ACS Chem. Biol 11, 2847–2853, 10.1021/acschembio.6b00523 [DOI] [PubMed] [Google Scholar]
- 86.Huang YM, Huber GA, Wang N, Minteer SD and McCammon JA (2018) Brownian dynamic study of an enzyme metabolon in the TCA cycle: Substrate kinetics and channeling. Protein Sci. 27, 463–471, 10.1002/pro.3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Omini J, Wojciechowska I, Skirycz A, Moriyama H and Obata T (2021) Association of the malate dehydrogenase-citrate synthase metabolon is modulated by intermediates of the Krebs tricarboxylic acid cycle. Sci. Rep 11, 18770, 10.1038/s41598-021-98314-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shuib S, Ibrahim I, Mackeen MM, Ratledge C and Hamid AA (2018) First evidence for a multienzyme complex of lipid biosynthesis pathway enzymes in Cunninghamella bainieri. Sci. Rep 8, 3077, 10.1038/s41598-018-21452-4 [DOI] [PMC free article] [PubMed] [Google Scholar]