Abstract
The latency-associated nuclear antigen (LANA) encoded by the Kaposi's sarcoma-associated herpesvirus (KSHV) is expressed in the majority of KSHV-infected cells and in cells coinfected with Epstein-Barr virus (EBV). In coinfected body cavity-based lymphomas (BCBLs), EBV latent membrane protein 1 (LMP1), which is essential for B-lymphocyte transformation, is expressed. EBNA2 upregulates the expression of LMP1 and other cellular genes through specific interactions with cellular transcription factors tethering EBNA2 to its responsive promoters. In coinfected BCBL cells, EBNA2 is not detected but LANA, which is constitutively expressed, contains motifs suggestive of potential transcriptional activity. Additionally, recent studies have shown that LANA is capable of activating cellular promoters. Therefore, we investigated whether LANA can affect transcription from two major EBV latent promoters. In this study, we demonstrated that LANA can efficiently transactivate both the LMP1 and C promoters in the human B-cell line BJAB as well as in the human embryonic kidney 293 cell line. Moreover, we demonstrated that specific domains of LANA containing the putative leucine zipper and the glutamic acid-rich region are highly effective in upregulating these viral promoters, while the amino-terminal region (435 amino acids) exhibited little or no transactivation activity in our assays. We also specifically tested truncations of the LMP1 promoter element and showed that the −204 to +40 region had increased levels of activation compared with a larger region, −512 to +40, which contains two recombination signal-binding protein Jκ binding sites. The smaller, −204 to +40 promoter region contains specific binding sites for the Ets family transcription factor PU.1, transcription activating factor/cyclic AMP response element, and Sp1, all of which are known to function as activators of transcription. Our data therefore suggest a potential role for LANA in regulation of the major EBV latent promoters in KSHV- and EBV-coinfected cells. Furthermore, LANA may be able to activate transcription of viral and cellular promoters in the absence of EBNA2, potentially through association with transcription factors bound to their cognate sequences within the −204 to +40 region. This regulation of viral gene expression is critical for persistence of these DNA tumor viruses and most likely involved in mediating the oncogenic process in these coinfected cells.
Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV; also referred to as human herpesvirus type 8) are two closely related human gammaherpesviruses (10, 49, 61). EBV primarily infects nasopharyngeal epithelia and B lymphocytes and is associated with a number of human malignancies, including nasopharyngeal carcinoma, Hodgkin's disease, Burkitt's lymphoma, immunoblastic lymphomas, T-cell lymphomas, gastric carcinoma, breast carcinoma, and body cavity-based lymphomas (BCBLs) (3, 9, 37, 56, 82, 84). EBV typically establishes distinct types of viral latency based on specific latent-gene expression profiles in the infected cells (56). These latent-gene expression profiles include the expression of EBV nuclear antigens (EBNAs) 1, 2, 3A, 3B, 3C, and LP, the EBV early RNAs (EBERs), latent membrane proteins (LMPs) 1, 2A, and 2B, and BARF0 transcripts (56). LMP1 is essential for mediating EBV-induced proliferation of infected B cells through ligand-independent events and signal transduction (37, 48). EBNA1 is required for episomal maintenance and stability of the newly replicated episomes, ensuring that they are not lost during segregation of progeny cells (1, 26, 32, 39, 83). EBNAs LP, 2, 3A, and 3C are critical whereas the EBERs, EBNA-3B, LMP2A, LMP2B, and BARF0 are dispensable for growth transformation of primary B cells (12, 22, 37, 43–45, 59, 60, 70, 72).
Typically, in healthy individuals, the virus remains tightly latent and proliferation of the infected B cell is controlled by the cytotoxic T-cell response (36, 56). However, in the case of an immunocompromised patient, this stringently balanced mechanism is disrupted and the infected cells are dysregulated, leading to proliferation in the infected host (11, 56). Previous studies have shown that BCBLs establish a predominantly type II latency program (5, 24), although one report indicated that a type I latency program was established in one BCBL cell line (71). Type II latency is characterized by the expression of EBNA1, the LMPs, and BARF0 transcripts and is notably lacking expression of EBNA2 (37). This type of infection is seen in all non-B cells infected with EBV, such as in T-cell lymphomas, Hodgkin's disease, and nasopharyngeal as well as other carcinomas (56). The expression of the entire repertoire of latent antigens is considered to be a type III latency program and is typically seen in lymphoblastoid cell lines (LCLs) and in lymphoproliferative disorders occurring in AIDS patients and others with severe immunosuppression (56). Expression of the EBERs and BARF0 transcripts is usually seen in the majority of EBV-associated cancers, with few exceptions (11, 56).
LMP1 of EBV is an integral membrane protein that is essential for growth transformation of B lymphocytes (27, 34, 78). This viral oncogene is involved in triggering many of the morphologic changes associated with EBV infection, including the expression of adhesion molecules and activation markers as well as NF-κB activation (16, 28, 78, 79). The transforming ability of LMP1 is tightly linked to its association with the tumor necrosis factor family receptor-associated factors and may function similarly to the tumor necrosis factor family receptor, aggregating in the plasma membrane, where it is capable of binding tumor necrosis factor family receptor-associated factors (15, 48). Numerous studies of LMP1 regulation have demonstrated that expression of LMP1 is dependent on regulation of its promoters by viral and cellular transcription factors (20, 21, 30, 42, 64, 65, 76).
Previous genetic and biochemical studies have demonstrated that EBNA2 is essential for EBV-mediated growth transformation of B lymphocytes and is a potent activator of transcription in EBV infection (12, 22). EBNA2 lacks the ability to directly bind DNA but targets and activates EBV promoter elements through its association with RBP-Jκ and other cellular transcription factors, including PU.1 and AML1 (21, 23, 30, 42, 64–66, 81). EBNA2 also associates with the basal transcription machinery and other coactivators of transcription and has been shown to transactivate the major EBV latent promoters and other responsive cellular promoters (73–75).
The two major essential latent EBV promoters, the C promoter and the LMP1 promoter, generate the transcripts of the EBNA family and LMP1 during latent infection in vitro and are also active in lesions of patients with lymphoproliferative disease (21, 23, 37, 56, 81, 85, 89). Both of these EBV latent promoters are upregulated through indirect interaction with EBNA2 (29, 76). The interaction between EBNA2 and the LMP1 promoter is dependent on the PU.1 Ets family protein, the RBP-Jκ transcription factor, and other factors which target their cognate sequences within the promoter regulatory region (23, 30, 64). Transactivation is significantly reduced in the absence of the RBP-Jκ consensus sites, and the transcription activating factor/cyclic AMP response element (ATF/CRE) has also been shown to mediate EBNA2-dependent and -independent activation of LMP1p (30, 66).
KSHV is the potential etiological agent of Kaposi's sarcoma and is associated with primary-effusion lymphomas (PELs; also referred to as BCBLs) and multicentric Castleman's disease; more controversially, it has been found in dendritic cells of individuals with multiple myeloma (7, 10, 17, 19, 55, 67). The KSHV genome is detected in endothelial and tumor cells of Kaposi's sarcoma lesions, where the virus expresses a number of critical viral antigens during a persistent latent infection (9, 24, 54, 68, 88). Similar to EBV, KSHV shows distinct patterns of gene expression in different KSHV-associated diseases (33, 50). However, the latency-associated nuclear antigen (LANA) is constitutively expressed in all known KSHV-infected cells.
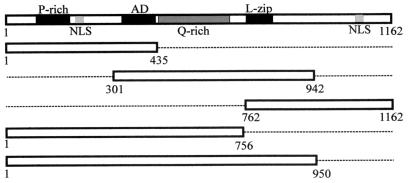
LANA is encoded by KSHV open reading frame 73 and is required for the persistence of the KSHV episome in latently infected cells (35, 52). LANA is a large nuclear phosphoprotein with three distinct domains (Fig. 1): a proline-rich N-terminal region that contains a putative nuclear localization signal (NLS), a repeating internal acidic domain that is rich in glutamic acid, and a carboxy terminus containing a potential leucine zipper motif (61). Both the N and C termini are sufficient to localize LANA to the nucleus of the host cell (63). The efficient segregation of KSHV episomes to progeny cells during mitosis is most likely facilitated by LANA via tethering of KSHV DNA to host chromosomes (2, 13). It was also recently shown that LANA inhibits p53 function, disrupting the cell death pathway and thereby contributing to the persistence of KSHV (18) and associated oncogenes. Additionally, LANA can regulate an E2F-responsive promoter through association with the retinoblastoma protein, suggestive of functions similar to the large T antigen of the polyomaviruses (51).
FIG. 1.
Schematic diagram of LANA showing its structural properties. Note that the clone consisting of amino acids 1 to 435 contains the proline-rich region (P-rich), a nuclear localization sequence (NLS), and part of the acidic region (AD). The LANA construct consisting of amino acids 301 to 942 contains the entire acidic domain, including the glutamine-rich region (Q-rich) and the entire putative leucine zipper (L-zip). The amino acid 762 to 1162 clone contains the putative leucine zipper and a potential nuclear localization sequence. The LANA construct with amino acids 1 to 756 lacks the putative leucine zipper but contains the acidic domains, the proline-rich region, and a nuclear localization sequence. The amino acid 1 to 950 clone is identical to the amino acid 1 to 756 construct except that it includes the putative leucine zipper.
Coinfection of KSHV and EBV has been seen in the majority of AIDS-associated PELs and in a minority of cases not associated with AIDS (6, 19, 24, 62). This unique dual herpesvirus infection provides a useful model to study the interaction between these two human DNA tumor viruses. In the case of KSHV- and EBV-coinfected cells, many of the major essential EBV latent antigens are not detected (5, 24, 71). However, EBNA1 and LMP1 expression was shown to occur, but at significantly lower levels than in cells infected with only EBV (5). In the absence of viral proteins EBNA2, EBNA3A, and EBNA3C, it is likely that KSHV plays a very significant role in regulating EBV gene expression, hence contributing to the pathogenesis of PELs. This body of data suggests that KSHV-encoded proteins or KSHV-induced cellular proteins might regulate EBV latent gene expression. Due to the ubiquitous expression of LANA in KSHV- and EBV-coinfected cells, we chose to investigate this protein in terms of its role in regulating gene expression from two major EBV latent promoters.
MATERIALS AND METHODS
Plasmids.
The pGL reporter plasmids (Promega Inc.) contain the luciferase gene and lack eukaryotic promoter and enhancer regulatory sequences. The LMP1 and C promoter elements, which were cloned into the basic reporter plasmid to assess the ability of LANA to influence expression from these promoters by using luciferase assays, were described previously (41, 80). The pGL2 −512/+72 plasmid has two directly repeated copies of the LMP1 −512/+72 promoter element, and pGL −512/+40 contains a single copy of the −512/+40 promoter element (obtained from Jeffery Lin and Elliott Kieff). pGL −272/+40 and pGL −204/+40 have truncations of the −512/+40 LMP1 promoter region, (consecutive deletions of the two RBP-Jκ binding sites) (30). The reverse primer 5′-TATAGATCTCTCAGGGCAGTGTGTCAGGAG-3′ was used along with the forward primer 5′-TAGGTACCCTGATTGCCGCACTGCCTTTCC-3′ or 5′-TAGGTACCGGGACCCGCTTTTCTAACACAAAC-3′ for pGL2 −272/+40 and pGL2 −204/+40, respectively. The PCR fragments were cloned into KpnI and BglII sites of the pGL3 basic luciferase vector.
The LANA fragments were cloned into pCDNA3 (Invitrogen) or a modified pCDNA3 expression vector, pA3M, containing three myc epitopes for detection of the fusion protein. To amplify regions of the LANA molecule, the sense primer 5′-GAGAATTCTTATGGCGCCCCCGGGAATG-3′ and the antisense primer 5′-GCAGATATCAGCGTTTTGTTTCCATCGCCCCCGTC-3′ were used for amino acids 1 to 435, sense primer 5′-GCAGGTACCATGGAAAATAATCAGGCTGGCGAGGA-3′ and antisense primer 5′-GCAGATATCAGTGACGACCCGTGCAAGATTATGGG-3′ were used for amino acids 301 to 942, and sense primer 5′-GCAGGTACCATGGAGCAGGAGCAGGAGTTAGAG-3′ and antisense primer 5′-GAGATATCCCTGCATTTCCTGTGGAGA-3′ were used for amino acids 762 to 1162. Clones containing amino acids 1 to 756 or 1 to 950 were obtained from Joonho Choe (40).
Cell lines and culture systems.
BJAB is an EBV-negative, human B-cell line obtained from Elliott Kieff (69). The B95-8 cells are singly infected with type I EBV (46, 47). The LCL-1 cell line was created in our lab by infection of primary human B lymphocytes with EBV (14). P3HR-1, an EBV-positive cell line from which portions of the EBNALP and EBNA2 open reading frames have been deleted, expresses low levels of LMP1 (59). BC-1 and BC-2, EBV- and KSHV-coinfected cell lines derived from patients with BCBLs, were purchased from the American Type Culture Collection (8, 24). The BJAB, P3HR-1, B95-8, and LCL-1 cell lines were grown in RPMI 1640 (Gibco-BRL Life Technologies Inc.) supplemented with 10% fetal bovine serum (Gemini Bioproducts Inc.), 2 mM l-glutamine, gentamicin (20 μg/ml), and penicillin-streptomycin (5 U/ml and 5 μg/ml, respectively). BC-1 and BC-2 cells were cultured as described above but in medium supplemented with 20% fetal bovine serum. Human embryonic kidney (HEK) 293 cells were grown in Dulbecco's modified Eagle medium (Gibco-BRL Life Technologies Inc.) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, gentamicin (20 μg/ml), and penicillin-streptomycin (5 U/ml and 5 μg/ml, respectively).
Transfections and luciferase assays.
BJAB, P3HR-1, or 293 cells (107) were transfected with 5-μg quantities of the various promoter constructs or vector and increasing concentrations (5, 10, 15, or 20 μg) of the pCDNA3 LANA clones in 400 μl of RPMI 1640 (BJAB cells) or Dulbecco's modified Eagle medium (293 cells) containing 10% fetal bovine serum, using an electroporator at 210 V and 975 μF. Vector DNA was added to equalize the total amount of DNA used in all transfections, and its levels were normalized for transfection efficiency. After electroporation, cells were resuspended in 10 ml of complete medium, as described above, in 100-mm-diameter plates and incubated at 37°C for 20 h. Cells were harvested and then washed once with phosphate-buffered saline (PBS) and resuspended in the appropriate buffer. For luciferase assays, the cells were resuspended in 200 μl of 1× Reporter Lysis Buffer (Promega Inc.), placed in a dry ice-isopropanol bath for 5 min, thawed, vortexed for 15 s, and then centrifuged at 13,000 rpm for 20 s. Samples (45 μl each) were immediately combined with luciferase substrate in an Optocomp1 luminometer (MGM Instruments, Inc.) and counted for 10 s to determine the relative light units (RLU). The basic pGL vector alone was used in these assays to determine the ability of LANA to activate the vector alone. All assays were done in triplicate and repeated multiple times.
Western blot assays.
Cells were collected by centrifugation and washed in 5 ml of PBS. The pellet was resuspended in an appropriate volume of sodium dodecyl sulfate (SDS) lysis buffer (57); lysates were analyzed by fractionation in SDS–6%, 8%, or 10% polyacrylamide gels for detection of LANA, EBNA2, and LMP1, respectively, and then transferred to 0.45-μm-pore-size nitrocellulose membranes. Membranes were blocked in 5% milk for 1 h at room temperature, rinsed three times with TBST (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20), and then washed three times (5 min each) with TBST. The membranes were then incubated with the specific primary antibody overnight at 4°C before being put through the same washing regimen. The blots were then incubated with the appropriate secondary antibody for 1 h at room temperature and subsequently washed with TBST. Specific signals were detected by enhanced chemiluminescence as per the manufacturer's instructions (Amersham Pharmacia Biotech). LANA was detected with human polyclonal serum (diluted 1:50 in PBS supplemented with 1 mM sodium azide) followed by a protein A-horseradish peroxidase-conjugated secondary antibody (Amersham Pharmacia Biotech; 1:5,000 dilution). Hybridoma cell lines producing monoclonal antibodies PE2 and S12 (diluted 1:2 in hybridoma supernatant) were used to detect EBNA2 and LMP1, respectively, after which was added horseradish peroxidase-conjugated anti-mouse antibody (1:2,500 dilution). The proteins were then detected by enhanced chemiluminescence as suggested by the manufacturer (Amersham Pharmacia Biotech).
Immunofluorescence analysis.
Cells were washed twice in PBS and resuspended at a density of 106 in 20 μl of PBS. A 1-μl aliquot of the cells was placed on a slide and allowed to air dry for 10 min. Slides were then fixed in precooled acetone-methanol (1:1, vol/vol) at −20°C for 10 min and were allowed to air dry. Cells were blocked with 20% goat serum for 30 min at room temperature. To detect LANA, a polyclonal antibody was used at a 1:250 dilution in PBS for 1.5 h at room temperature. Four 5-min washes with PBS were performed before addition of the secondary antibody, a fluorescein isothiocyanate-conjugated goat anti-human antibody used at a dilution of 1:1,000. Cells were incubated with the secondary antibody for 1 h at room temperature. After being washed multiple times, cells were mounted and visualized with an Olympus BX60 fluorescence microscope and captured using the Espirit program, VI.2.
RESULTS
LANA transactivates the EBV major LMP1–512/+72 and C latent promoters in the human BJAB cell line.
In the coinfected BCBL cell lines, LANA is constitutively expressed. Therefore, we examined the effect of LANA on regulation of gene expression from the two major EBV promoters, Cp and LMP1p, in a B-cell background. This analysis in the BJAB cell line would more closely mimic the observed phenomenon in BCBL, which is derived from a B-cell line (4, 8). Cp is the major transcriptional promoter for the EBNA antigens, and LMP1, the viral oncoprotein critical for mediating EBV transformation of primary B lymphocytes, is transcribed from the LMP1 promoter (37, 56). Because LMP1 is expressed in the PEL cell lines, we wanted to compare the effects of LANA on the LMP1 promoter with those on the other major EBV latent promoter, Cp, which also known to be activated by EBNA2 (80).
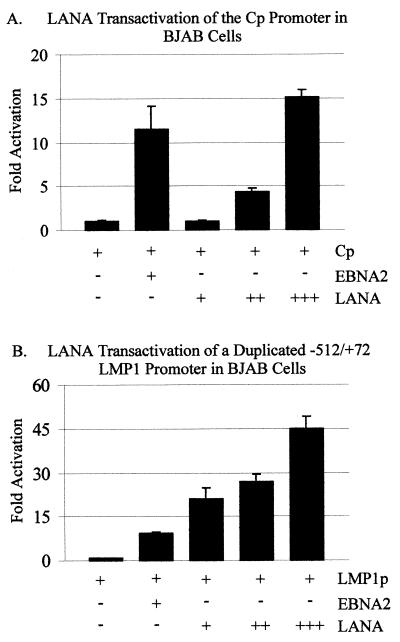
To investigate LANA's ability to act as a transactivator of these promoters, the promoter-luciferase reporter constructs and the LANA expression constructs were transiently transfected, after which luciferase assays were performed. The results of these experiments demonstrate that LANA is a potent activator of the major C promoter, working in a dose-dependent manner (Fig. 2A). LANA upregulated the multimerized EBV Cp 15-fold compared with the promoter-only control. The effects were clearly dose responsive, with increasing amounts of LANA resulting in increased levels of transactivation. In this assay, the EBNA2 transactivator, which is known to activate Cp, activated at levels 10- to 11-fold above those for the promoter alone and therefore was used as a positive control (Fig. 2).
FIG. 2.
LANA transactivation of the major latent EBV promoters, Cp and LMP1, in BJAB cells. The pGL-Cp (A) and pGL-LMP1–512/+72 (B) reporter plasmids (5 μg each) were transfected alone, with 7.5 μg of EBNA2, or with 5, 10, or 15 μg of LANA into 107 BJAB cells as indicated. Equal amounts of cell lysates were assayed for luciferase activity. Luciferase activity is expressed as fold activation relative to that obtained with the pGL promoter alone.
We next determined whether LANA had similar effects on the other major latent EBV promoter, the LMP1 promoter. In similar transient-transfection assays, we demonstrated that LANA also dramatically activated the LMP1 promoter. Moreover, there was a similar dose-response effect with increasing amounts of LANA. In this series of assays, the level of activation was more dramatic than the Cp results provided above. LANA activated the duplicated LMP1 promoter up to 45-fold with 15 μg of LANA (Fig. 2B). Again, and as expected, the positive control EBNA2 activated the promoter to levels similar to those achieved with Cp alone. The basic vector alone had little or no activity in these assays (data not shown). Clearly, these results demonstrate that LANA can activate the EBV major latent C and LMP1 promoters in BJAB cells.
LANA transactivates the EBV major LMP1–512/+72 and C latent promoters in the HEK 293 fibroblast cell line.
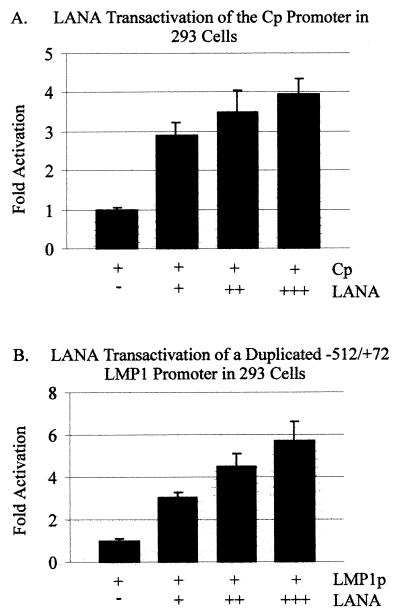
To further elaborate on the significance of the results obtained with BJAB cells, we determined the effects of LANA in another human cell line, HEK 293. Similarly, we showed that LANA could upregulate the major EBV C promoter in HEK 293 fibroblast cells. However, the effects were less striking than those obtained for BJAB cells. LANA consistently activated Cp three- to fourfold in HEK 293 cells compared with the vector-only control (Fig. 3A). These values were approximately threefold lower than those obtained with BJAB cells. In HEK 293 cells, LANA activation of the −512/+72 LMP1 promoter was greater than that with Cp (compare Fig. 3A and B), with the level of activation being three- to sixfold higher than that of the promoter-only control (Fig. 3B). Moreover, the results were dramatically different from those for BJAB cells. A sevenfold reduction in LANA-dependent activation was seen in HEK 293 compared with BJAB cells, suggesting that LANA is a stronger activator of EBV promoters in B cells than in 293 cells.
FIG. 3.
LANA transactivation of the major latent EBV promoters, Cp and LMP1, in HEK 293 cells. The pGL-Cp (A) and pGL-LMP1–512/+72 (B) reporter plasmids (5 μg each) were transfected alone, with 7.5 μg of EBNA2, or with 5, 10, or 15 μg of LANA into 107 293 cells as indicated. Equal amounts of cell lysates were assayed for luciferase activity. Luciferase activity is expressed as fold activation relative to that obtained with pGL-Cp alone.
LANA is expressed and localizes to the nucleus when transiently transfected in the human cell lines BJAB and HEK 293.
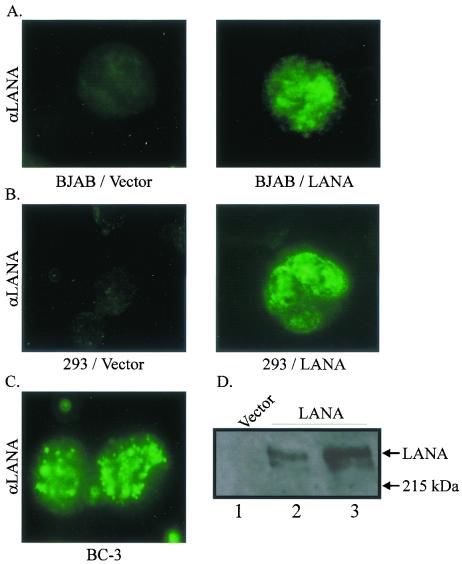
To determine if transiently transfected LANA was expressed and localized to the nucleus in a fashion similar to that seen in KSHV-infected B cells, we performed Western blot and immunofluorescence assays. Cells harvested 20 h after transient transfection and tested by Western blot and immunofluorescence analyses indicated that LANA was expressed. In addition, BJAB cells transfected with LANA had the characteristic punctate pattern, similar to the staining pattern seen in the nucleus of the KSHV-positive cell line BC-3 (Fig. 4). Similarly, in 293 cells, LANA staining in the nucleus was punctate compared with that seen in BJAB cells. The BC-3 KSHV-positive cell line was used as a control for LANA signal in the nucleus (Fig. 4). The level of detection in the transfected cells was similar to that seen in BC-3 (compare Fig. 4A, B, and C). However, the LANA signal was slightly less punctate, although the nuclear staining in specific regions was clearly seen. This may have been an effect of overexpression and localization when a larger amount of LANA was introduced transiently in cells compared with the amount used in the KSHV-infected cell line BC-3. No specific signals were seen for LANA in the cells transfected with vector alone (Fig. 4).
FIG. 4.
LANA is expressed when transiently transfected into BJAB and HEK 293 cells. BJAB (A) or HEK 293 (B) cells were transfected with vector alone or with LANA, and slides were prepared and analyzed by immunofluorescence microscopy; a KSHV-infected positive-control cell line for LANA, BC-3, is also shown (C). (D) HEK 293 cells transfected with either vector alone or LANA were electrophoresed on a 6% polyacrylamide–SDS gel and analyzed by Western blotting. LANA-specific signal is seen slightly above the 215-kDa molecular marker. αLANA was obtained from polyclonal rabbit serum made against the carboxy-terminal 300 amino acids of the LANA protein.
To detect LANA expression by an alternative method, Western blotting for LANA was performed with protein lysates prepared from transfected HEK 293 cells, using a specific polyclonal serum produced against LANA. A specific signal was seen when 10 or 15 μg of LANA was transfected (Fig. 4D, lanes 2 and 3, respectively). Moreover, the signature doublet (5) migrating at over 200 kDa was seen. As expected, the vector-only lane showed no specific signal representative of LANA. These results clearly show that LANA transiently transfected from the expression constructs was expressed in BJAB as well as in HEK 293 cell lines and predominantly localized to the nucleus in all transfected cells analyzed.
The truncated −204/+40 LMP1 promoter element is highly upregulated by LANA in both BJAB and HEK 293 human cell lines.
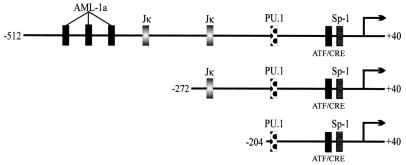
To determine the specific regions of LMP1p involved in LANA-mediated transactivation of this promoter, two LMP1p reporter truncations were constructed; one had a deletion of the AML1 binding site and the leftmost of the RBP-Jκ binding sites (−272/+40 construct), and the second construct had deletions of the AML1 binding site and both RBP-Jκ sites (Fig. 5). The smaller construct, −204/+40, contains PU.1, Sp1, and ATF/CRE-responsive target sequences (30, 66). These sites were determined using a motif search program (TFSEARCH, version 1.3). These constructs were cotransfected in BJAB cells, and LANA and luciferase assays were performed.
FIG. 5.
Schematic diagram of the truncated LMP1–512/+40 promoter elements. The −272/+40 construct does not contain the AML1a consensus sites or one Jκ site that is present in the −512/+40 promoter. The −204/+40 promoter is missing these elements as well as an additional Jκ consensus site. All of the promoter truncations contain the PU.1, ATF/CRE, and Sp1 sites.
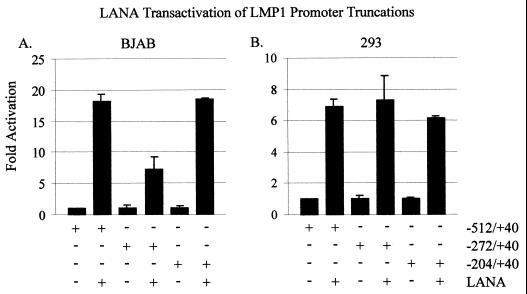
In these assays, we demonstrated that LANA activated each promoter 5- to 20-fold (−512/+40, −272/+40, and −204/+40 constructs) compared with the promoter alone in 293 and BJAB cells (Fig. 6). Surprisingly, −272/+40, containing a single RBP-Jκ binding site, activated to lower levels than did −512/+40 and −204/+40 in BJAB cells. It is possible that the repressive effects of RBP-Jκ tempered the increased activation mediated through the other downstream factors.
FIG. 6.
The −204/+40 LMP1 promoter element is upregulated by LANA in both BJAB and HEK 293 cells. pGL-LMP1–512/+40, −272/+40, or −204/+40 reporter plasmid (5 μg each) was transfected alone or with 20 μg of LANA into 107 BJAB (A) or HEK 293 (B) cells. Luciferase activity is shown as fold activation relative to that obtained with each of the promoters alone. Equal amounts of cell lysates were assayed for luciferase activity.
We therefore compared the fold activity of each promoter construct when LANA was expressed in BJAB to the fold activity of that promoter construct in the HEK 293 cell line. The results from these assays showed that there were no dramatic differences in fold activity versus the promoter constructs alone when LANA was expressed in HEK 293 cells compared with the results seen for the −272/+40 levels in BJAB cells (Fig. 6). These results suggest that although LANA activates the different truncated promoter elements to similar levels, the overall transcriptional activity of each promoter element can differ in specific cell lines.
The glutamic acid-rich central portion and carboxy terminus of LANA activate transcription of the LMP1 promoter.
Based on the secondary structure of LANA, specific motifs suggest that the central glutamic acid-rich region, which is similar to that found in known transcription factors of the c-Fos/c-Jun family and other viral transcription factors, including EBNA3A and EBNA3C, can function as an activator of transcription (58, 77, 86). Additionally, the carboxy-terminal 400 amino acids of LANA contains a leucine zipper motif that is potentially involved in homo- or heterodimerization with other transcription factors (35, 52).
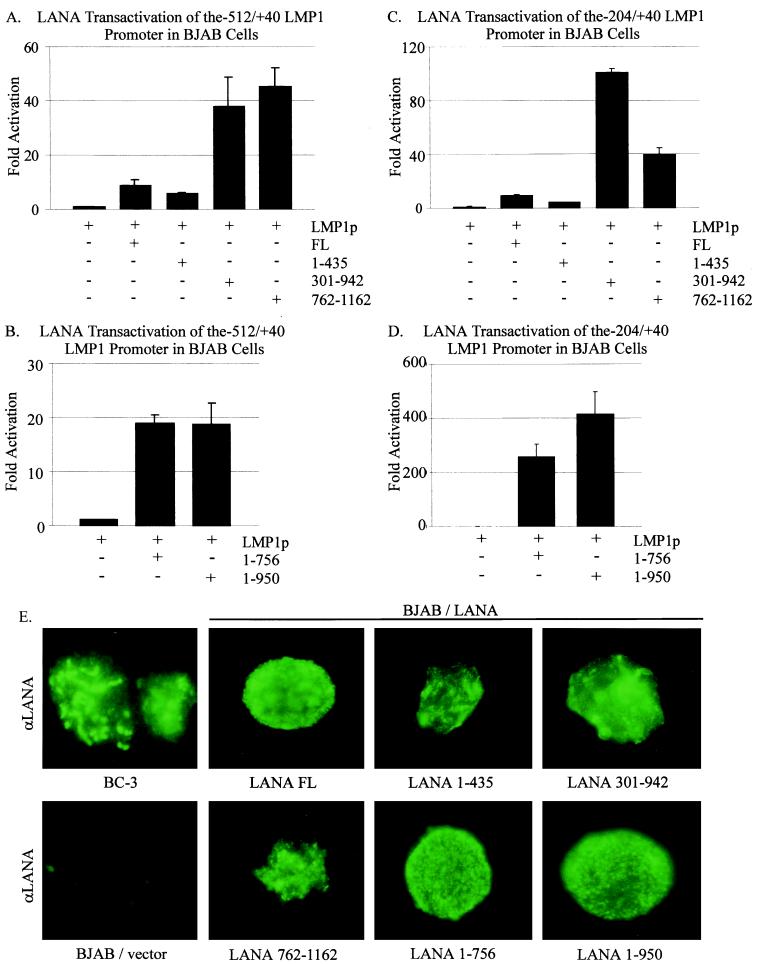
Specific domains of LANA (Fig. 1) containing the amino terminus (amino acids 1 to 435), the central glutamic acid-rich domain (amino acids 301 to 942), or the carboxy terminus (amino acids 762 to 1162) were cloned into expression vectors and tested in these transient reporter assays. As expected, LANA activated the −512/+40 LMP1 promoter 10-fold over the promoter alone control. The level of activation obtained with the amino-terminal LANA truncation mutant (amino acids 1 to 435) was always consistently about 50% (or less) of that seen with full-length LANA. Surprisingly, the central LANA domain (amino acids 301 to 942) and the carboxy-terminal LANA (amino acids 762 to 1162) constructs resulted in activation levels 40-fold higher than that attained with the promoter alone control and 4-fold higher than that observed for full-length LANA (Fig. 7A). The fact that the central-domain and the carboxy-terminus constructs had similar levels of activation suggests that these two regions may have a common motif, since they overlap at amino acids 762 to 942 (Fig. 1). These results suggested that a major activator of transcription might lie within amino acids 762 to 942 of LANA and that the amino terminus, although not completely devoid of activation functions, was clearly not the major contributor to LANA's activity on the LMP1 promoter.
FIG. 7.
The central glutamine-rich region as well as the carboxy terminus of LANA is involved in transactivation of the LMP1 promoter. (A) pGL-LMP1–512/+40 (5 μg) was transfected alone or with 20 μg of one of the following LANA clones: full-length LANA (FL), LANA amino acids 1 to 435 (1–435), LANA amino acids 301 to 942 (301–942), or LANA amino acids 762 to 1162 (762–1162). (B) pGL-LMP1–512/+40 (5 μg) was transfected alone or with 20 μg of LANA 1–756 or LANA 1–950. Luciferase activity is shown as fold activation above that attained with the promoter alone. Equal amounts of cell lysates were assayed for luciferase activity. (C) pGL-LMP1–204/+40 (5 μg) was transfected alone or with 20 μg of full-length LANA, LANA 1–435, LANA 301–942, or LANA 762–1162. (D) pGL-LMP1–204/+40 (5 μg) was transfected alone or with 20 μg of LANA 1–756 or LANA 1–950. (E) The LANA constructs are expressed when transiently transfected into BJAB cells. Slides were prepared and analyzed by immunofluorescence microscopy, as indicated, 20 h after 20 μg of LANA construct was transfected into BJAB cells. A KSHV-positive control cell line, BC-3, is also shown, as is BJAB transfected with promoter alone as a negative control.
The putative leucine zipper of LANA is not solely responsible for mediating the transactivation functions of LANA.
The results of the domain analysis indicated that the LANA region between amino acids 762 and 942 contains a leucine zipper motif which may be the predominantly responsible for mediating activation of transcription (35, 52). Therefore, we wanted to determine whether constructs lacking the leucine zipper and the carboxy terminus and constructs lacking but retaining the leucine zipper differed in their ability to activate the LMP1 promoter. Both LANA truncation constructs resulted in transcriptional activation of the −512/+40 LMP1 promoter. Surprisingly, the LANA construct containing amino acids 1 to 756, which lacks the putative leucine zipper, resulted in 20-fold activation, a level similar to that seen with the LANA construct consisting of amino acids 1 to 950, which contains the leucine zipper (Fig. 7B). These results suggest that the glutamine-rich domain may contribute levels of activation similar to those of the leucine zipper domain and that the domain downstream of the leucine zipper, amino acids 950 to 1162, may also contribute to the overall activation activity encoded by LANA when tested on the −512/+40 LMP1 reporter construct.
Since the −204/+40 construct had the highest activity, we wanted to determine whether the effects on LANA domains would be more pronounced, and therefore more clearly delineated, when tested on this smaller construct. As described above, the LANA constructs consisting of amino acids 1 to 435, 301 to 942, or 762 to 1162 were tested on the −204/+40 LMP1 reporter construct. In this assay, the construct which expresses the glutamic acid-rich central domain and the leucine zipper had the highest level of activity (Fig. 7C), about twofold greater than that seen for the carboxy terminus lacking the glutamic acid-rich central domain but containing the leucine zipper. When the constructs consisting of amino acids 1 to 756 or 1 to 950 of LANA, containing the glutamic acid-rich region without and with the leucine zipper, respectively, were tested on the −204/+40 reporter construct, the results showed that the portion of LANA that contains the glutamic acid-rich region and the leucine zipper had an approximately twofold higher level of activity than the construct with the glutamic acid-rich region when compared with the activity of the promoter alone (Fig. 7D). These results suggest that although the glutamic acid-rich region makes the greatest contribution to the activity of the LMP1 promoter, the leucine zipper and the carboxy terminus of LANA are important and contribute to the overall transcriptional activation of the LMP1 promoter seen with LANA. Immunofluorescence analysis indicated that these LANA-truncated constructs all localized to the nucleus in BJAB cells (Fig. 7E).
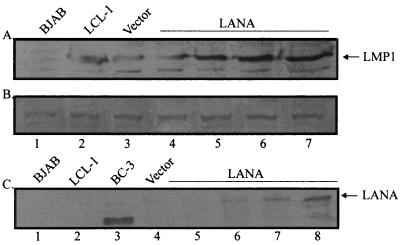
Western blot analysis shows that LMP1 is expressed in the coinfected BC-1 and BC-2 cell lines expressing LANA.
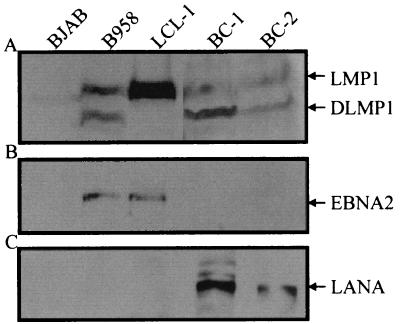
As previously shown, LANA is expressed in the coinfected BCBLs BC-1 and BC-2. In the same cell lines, LMP1 is also expressed (5). We wanted to show that the expression of LMP1 was maintained in these cell lines in the absence of EBNA2 expression even after prolonged passage of these cultures. BC-1 and BC-2 cell lines coinfected with KSHV and EBV were passaged in culture over a 24-month period. From these long-term-passage cultures, lysates were obtained and analyzed by Western blotting for latent antigen expression. LANA was clearly expressed in both cell lines (Fig. 8C); however, EBNA2 was not detected (Fig. 8B). Again, LMP1 was expressed, as evidenced in lanes 4 and 5 of Fig. 8A. LMP1 was also expressed in the positive-control cell lines B95-8 and LCL-1. Interestingly, the D1LMP1 signal seen in the B95-8 cell line was also seen in the coinfected cell lines. These results corroborate the previous data suggesting that LANA may contribute to activation of the EBV major latent LMP1 promoter.
FIG. 8.
Expression of LANA, EBNA2, and LMP1 in cells singly infected with EBV and in coinfected PEL cell lines. BJAB is an EBV-negative cell line, B958 and LCL-1 are infected with EBV only, and BC-1 and BC-2 are PEL cell lines coinfected with KSHV and EBV. LANA was fractionated on a 6% polyacrylamide–SDS gel. Polyclonal human serum specific for the LANA protein was used as the primary antibody. EBNA2 and LMP1 were fractionated on an 8% polyacrylamide–SDS gel and detected with EBNA2- and LMP1-specific monoclonal antisera (PE2 and S12, respectively). Protein extracts from BJAB, B958, and LCL-1 cells (5 × 105 each) were fractionated, while 106 BC-1 and BC-2 cells were electrophoresed in lanes 4 and 5, respectively.
Exogenously expressed LANA upregulates LMP1 in transiently transfected P3HR-1 cells.
To determine if the levels of LMP1 change when LANA is expressed from a heterologous promoter, we transfected an EBV-positive cell line exhibiting low levels of LMP1 expression to determine whether increased expression would occur when the promoter was transactivated by LANA. P3HR-1 cells (107) were transfected with increasing amounts of LANA, and the transfected cells were harvested and analyzed for LMP1 by Western blotting with a monoclonal antibody (S12) specific for LMP1. The results demonstrated that LMP1 levels increased with increasing amounts of LANA transfected (Fig. 9A). Vector-alone controls with no LANA exhibited relatively low levels of LMP1 in P3HR-1, as was expected (Fig. 9A, lane 4). It should be noted that no change in levels of EBNA1 was seen by Western blot analysis. Moreover, EBNA3C was not detected, suggesting that any effects on Cp in the background of an endogenous genome were not detectable (data not shown). This is consistent with the lower levels of Cp activation observed in the reporter assays. The weak background bands evident in the EBV-negative BJAB lane are nonspecific. Figure 9B shows a cross-reactive band with equivalent intensities in all lanes, indicating that similar levels of protein were loaded in all lanes. Figure 9C is a Western blot showing that the levels of LANA increased with increasing amounts of LANA transfected, as expected. The results from this assay suggest that LANA expressed from a heterologous promoter can upregulate LMP1 levels in the context of the EBV genomic, native promoter elements.
FIG. 9.
LMP1 is upregulated in P3HR-1 EBV-positive cells transiently transfected with LANA. P3HR-1 cells contain an EBV genome from which EBNA2 has been deleted. BJAB is an EBV-negative cell line used as a negative control. LCL-1 is an EBV-positive, EBNA2-positive cell line. (A) A Western blot for LMP1 prepared using the S12 monoclonal antibody. Lanes 4 to 7 contain lysates from P3HR-1 cells transfected with increasing amounts of LANA. (B) Verification of loading of equivalent levels of protein by comparing band intensities with that of a nonspecific band. (C) A Western blot for LANA prepared using polyclonal human antiserum which is specific for LANA. Lanes 5 to 8 contain lysates from P3HR-1 cells transfected with increasing amounts of LANA and correspond to lanes 4 to 7 in panel A. The positive-control cell line BC-3 exhibits a slightly weaker LANA signal. The transfected LANA construct was cloned from the BC-1 KSHV-positive cell line. P3HR-1 cells without LANA were transfected with vector alone in lane 3 (A and B) or lane 4 (C).
DISCUSSION
LANA is consistently detected in all KSHV-infected cells as well as in cells coinfected with both KSHV and EBV (7, 10). Since LMP1 is expressed in these cells in the absence of EBNA2, LANA may be capable of performing functions typically associated with the EBNA family of proteins encoded by EBV because of its localization in the nucleus, its restriction to the latent phase of infection of KSHV, and the apparent structural similarities among these proteins (5, 24, 35, 50, 71). LANA has already been shown to function, like EBNA1, by tethering the KSHV genome to chromosomal DNA, thereby facilitating viral episome persistence (2, 13). Also, LANA was shown to function as a transcriptional regulator in a number of studies, regulating KSHV and cellular promoters (38, 40, 51, 53, 63). Additionally, LANA has structural features typical of transcriptional activators, namely the central region containing repetitive blocks of acidic amino acids as well as a putative leucine zipper (35, 52). In fact, LANA has recently been shown to act as a transcriptional repressor and activator (38, 40, 63). These findings, as well as our inability to detect EBNA2 and other important EBV proteins in coinfected cells, led us to investigate LANA's potential role as a transcriptional activator of the EBV latent promoters in the case of KSHV-EBV coinfection.
By performing transient reporter assays, we have shown that LANA is an efficient transactivator of the two major latent promoters of EBV. Our experiments demonstrate that LANA can function similarly to EBNA2 in upregulating these promoters (24, 38). A recent study demonstrated that when LANA was cotransfected with EBNA2 and a Cp-chloramphenicol acetyltransferase reporter, LANA modulated the ability of EBNA2 to transactivate this promoter and LANA alone had no affect on Cp-chloramphenicol acetyltransferase activity (38). This is in contrast to our results, which demonstrate that LANA in fact can activate this promoter in two human cell lines. There are many experimental differences that could account for this discrepancy, including the cell lines utilized in the assay, the choice of reporters, and the amount of LANA expressed in these assays. Additionally, the effect on Cp by LANA in coinfected cells may be masked by expression of other viral or cellular proteins involved in viral gene regulation.
LANA's ability to transactivate the EBV LMP1 promoter triggers important questions about the cross talk between KSHV and EBV in the case of coinfected BCBL cells. Although EBNA2 has not been detected in any EBV- and KSHV-coinfected BCBL, LMP1 is consistently present in these cells, as documented in two separate reports (5, 24). Also, it should be noted that in another study LMP1 expression was not detected as is expected for a type I latency program (71). Since EBNA2 is the primary transactivator of the LMP1 promoter in EBV latency, it seems likely that another viral or cellular transactivator is involved in LMP1 upregulation. Therefore, like EBNA2, LANA can function as a transactivator of LMP1p in the case of KSHV-EBV coinfection, albeit through potentially separate mechanisms associated with only a portion of the overall role of LANA in these cell lines.
The fact that the basal activity of the truncated LMP1 promoters increased with the deletion of upstream elements that include two CBF1/RBP-Jκ consensus sites corresponds with the fact that RBP-Jκ is a known repressor of transcription (21, 23, 25, 89). EBNA2 interacts with RBP-Jκ as well as other cellular proteins that tether to the LMP1 promoter. The other significant factors involved in this mechanism of activation are the PU.1 factor (a member of the Ets family), an ATF/CRE, and the Sp1 transcription factor (31, 66).
Despite the ability of LANA to directly bind DNA to perform functions associated with episomal maintenance (2, 13), it is possible that LANA also utilizes its ability to associate with cellular proteins to transactivate EBV LMP1p as well as other viral and cellular promoters (38, 40). Whether LANA is directly binding to the LMP1 promoter or interacting with cellular proteins, the region of the promoter downstream of −204, shown to have enhanced activity, is likely to contain sequences targeted by known potent activators of transcription, such as ATF/CRE and Sp1 (31, 66, 87). Three binding sites known to be important for EBNA2-mediated transactivation of LMP1—namely, those for PU.1, ATF/CRE, and Sp1—are all located within this region (31, 66). Further truncations and mutagenesis of LMP1p will be performed and similar experiments will be done to determine the sequences required for LANA to efficiently transactivate the LMP1 promoter. Performing electrophoretic mobility shift assays with LANA and these cellular factors having target sequences within this region would provide clues to whether LANA can directly complex with these factors bound to their cognate sequences. Immunoprecipitation experiments will demonstrate their associations in the KSHV- and EBV-coinfected cells.
To better characterize the region(s) of LANA responsible for mediating transactivation of the EBV LMP1 promoter, we tested a number of LANA constructs lacking specific domains of LANA in the transient reporter assays. The amino terminus of LANA has minimal activity on EBV LMP1p, while the constructs containing the central domain or the carboxy terminus were potent activators of transcription. In comparing constructs consisting of LANA amino acids 1 to 756 or 1 to 950, we demonstrated that the putative leucine zipper domain contributes only partially to the upregulation of the LMP1 promoter. There was little difference between the LANA amino acid 301 to 942 and amino acid 762 to 1162 constructs, or between the constructs containing amino acids 1 to 756 or 1 to 950, with regard to −512/+40 promoter activation. The constructs lacking the amino-terminal 301 amino acids consistently had levels of activation higher than those lacking the region downstream of the leucine zipper domain. This might imply that the amino terminus contains repressive elements which may function in balancing the regulatory activities of LANA in these coinfected cell lines. Different LANA constructs will be made to further elucidate the specific regions of the protein responsible for transactivation of the LMP1 promoter. It should be noted that the central glutamic acid-rich domain had a significantly higher level of activation when tested on the −204/+40 reporter construct. This suggests that the amino acid 301 to 942 domain complexes with activators of transcription and is not regulated or modulated by other cellular factors which associate with the domain downstream of amino acid 950.
It is clear that LANA can upregulate LMP1p in the context of the virus. Our data demonstrate that LMP1 is consistently upregulated with increased levels of LANA exogenously expressed from a heterologous promoter. Therefore, LANA is capable of activating LMP1 at the level of its native promoter. This suggests a true transcription-regulatory function for LANA in affecting an EBV major essential latent promoter in coinfected PELs.
The accumulated data are beginning to suggest that proteins encoded by KSHV and EBV have the ability to regulate gene expression and have downstream oncogenic effects on each other in coinfected PEL cell lines. It appears that LANA is able to partially facilitate the role of EBNA2 as a transactivator of EBV latent promoters in EBV- and KSHV-coinfected lymphomas.
ACKNOWLEDGMENTS
We thank Elliott Kieff for the BJAB cell line and Jon Aster and Jeffrey Sklar for the HEK 293 cell line. Joonho Choe kindly provided the amino acid 1 to 756 and amino acid 1 to 950 LANA constructs, and Elliott Kieff and Jeff Lin provided the pGL-LMP1–512/+40 promoter construct. We would thank Vojo Deretic and Jens Poschet for providing assistance and for use of the BX60 fluorescence microscope.
This work was supported by grants from the National Institutes of Health (NCI CA72150-01 to E.S.R.) and the Lymphoma and Leukemia Society of America. E.S.R. is a Scholar of the Lymphoma and Leukemia Society. M.A.C. is a fellow of the Lady Tata Memorial Trust and is supported by Medical Scientist Training Program grant T32 GM07863 to the University of Michigan.
REFERENCES
- 1.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet M, Guinebretiere J M, Kremmer E, Grunewald V, Benhamou E, Contesso G, Joab I. Detection of Epstein-Barr virus in invasive breast cancers. J Natl Cancer Inst. 1999;91:1376–1381. doi: 10.1093/jnci/91.16.1376. [DOI] [PubMed] [Google Scholar]
- 4.Burger R, Neipel F, Fleckenstein B, Savino R, Ciliberto G, Kalden J R, Gramatzki M. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human myeloma cells. Blood. 1998;91:1858–1863. [PubMed] [Google Scholar]
- 5.Callahan J, Pai S, Cotter M, Robertson E S. Distinct patterns of viral antigen expression in Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus coinfected body-cavity-based lymphoma cell lines: potential switches in latent gene expression due to coinfection. Virology. 1999;262:18–30. doi: 10.1006/viro.1999.9876. [DOI] [PubMed] [Google Scholar]
- 6.Carbone A, Gloghini A, Vaccher E, Zagonel V, Pastore C, Dalla Palma P, Branz F, Saglio G, Volpe R, Tirelli U, Gaidano G. Kaposi's sarcoma-associated herpesvirus DNA sequences in AIDS-related and AIDS-unrelated lymphomatous effusions. Br J Haematol. 1996;94:533–543. doi: 10.1046/j.1365-2141.1996.d01-1826.x. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 9.Cesarman E, Nador R G, Aozasa K, Delsol G, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus in non-AIDS-related lymphomas occurring in body cavities. Am J Pathol. 1996;149:53–57. [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 11.Cohen J I. Epstein-Barr virus infection. N Engl J Med. 2000;343:481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 12.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotter M A, II, Robertson E S. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 14.Cotter M A, II, Robertson E S. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol Cell Biol. 2000;20:5722–5735. doi: 10.1128/mcb.20.15.5722-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devergne O, McFarland E C, Mosialos G, Izumi K M, Ware C F, Kieff E. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J Virol. 1998;72:7900–7908. doi: 10.1128/jvi.72.10.7900-7908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flowers C C, Flowers S P, Nabel G J. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor confers resistance to the antiproliferative effect of interferon-alpha. Mol Med. 1998;4:402–412. [PMC free article] [PubMed] [Google Scholar]
- 18.Friborg J, Jr, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 19.Ganem D. KSHV and Kaposi's sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh D, Kieff E. cis-acting regulatory elements near the Epstein-Barr virus latent-infection membrane protein transcriptional start site. J Virol. 1990;64:1855–1858. doi: 10.1128/jvi.64.4.1855-1858.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 23.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 24.Horenstein M G, Nador R G, Chadburn A, Hyjek E M, Inghirami G, Knowles D M, Cesarman E. Epstein-Barr virus latent gene expression in primary effusion lymphomas containing Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8. Blood. 1997;90:1186–1191. [PubMed] [Google Scholar]
- 25.Hsieh J J-D, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJκ-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung S C, Kang M S, Kieff E. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc Natl Acad Sci USA. 2001;98:1865–1870. doi: 10.1073/pnas.031584698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumi K M, Kaye K M, Kieff E D. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izumi K M, Kieff E D. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc Natl Acad Sci USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin X W, Speck S H. Identification of critical cis elements involved in mediating Epstein-Barr virus nuclear antigen 2-dependent activity of an enhancer located upstream of the viral BamHI C promoter. J Virol. 1992;66:2846–2852. doi: 10.1128/jvi.66.5.2846-2852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johannsen E, Miller C L, Grossman S R, Kieff E. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJκ in Epstein-Barr virus-transformed B lymphocytes. J Virol. 1996;70:4179–4183. doi: 10.1128/jvi.70.6.4179-4183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapoor P, Shire K, Frappier L. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 2001;20:222–230. doi: 10.1093/emboj/20.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katano H, Sato Y, Kurata T, Mori S, Sata T. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi's sarcoma, and multicentric Castleman's disease. Virology. 2000;269:335–344. doi: 10.1006/viro.2000.0196. [DOI] [PubMed] [Google Scholar]
- 34.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khanna R, Burrows S R, Kurilla M G, Jacob C A, Misko I S, Sculley T B, Kieff E, Moss D J. Localization of Epstein-Barr virus cytotoxic T cell epitopes using recombinant vaccinia: implications for vaccine development. J Exp Med. 1992;176:169–176. doi: 10.1084/jem.176.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kieff E. Epstein-Barr virus and its replication. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 38.Krithivas A, Young D B, Liao G, Greene D, Hayward S D. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J Virol. 2000;74:9637–9645. doi: 10.1128/jvi.74.20.9637-9645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leight E R, Sugden B. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev Med Virol. 2000;10:83–100. doi: 10.1002/(sici)1099-1654(200003/04)10:2<83::aid-rmv262>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 40.Lim C, Sohn H, Gwack Y, Choe J. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J Gen Virol. 2000;81:2645–2652. doi: 10.1099/0022-1317-81-11-2645. [DOI] [PubMed] [Google Scholar]
- 41.Ling P D, Hsieh J J-D, Ruf I K, Rawlins D R, Hayward S D. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J Virol. 1994;68:5375–5383. doi: 10.1128/jvi.68.9.5375-5383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Longnecker R, Miller C L, Miao X-Q, Tomkinson B, Kieff E. The last seven transmembrane and carboxy-terminal cytoplasmic domains of Epstein-Barr virus latent membrane protein 2 (LMP2) are dispensable for lymphocyte infection and growth transformation in vitro. J Virol. 1993;67:2006–2013. doi: 10.1128/jvi.67.4.2006-2013.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longnecker R, Miller C L, Tomkinson B, Miao X-Q, Kieff E. Deletion of DNA encoding the first five transmembrane domains of Epstein-Barr virus latent membrane proteins 2A and 2B. J Virol. 1993;67:5068–5074. doi: 10.1128/jvi.67.8.5068-5074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mannick J B, Cohen J I, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller G, Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci USA. 1973;70:190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller G, Shope T, Lisco H, Stitt D, Lipman M. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci USA. 1972;69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 49.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parravicini C, Chandran B, Corbellino M, Berti E, Paulli M, Moore P S, Chang Y. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am J Pathol. 2000;156:743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radkov S A, Kellam P, Boshoff C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene hras transforms primary rat cells. Nat Med. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 52.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Renne R, Barry C, Dittmer D, Compitello N, Brown P O, Ganem D. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Virol. 2001;75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rettig M B, Ma H J, Vescio R A, Pold M, Schiller G, Belson D, Savage A, Nishikubo C, Wu C, Fraser J, Said J W, Berenson J R. Kaposi's sarcoma-associated herpesvirus infection of bone marrow dendritic cells from multiple myeloma patients. Science. 1997;276:1851–1854. doi: 10.1126/science.276.5320.1851. [DOI] [PubMed] [Google Scholar]
- 56.Rickinson A, Kieff E. Epstein-Barr virus. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 57.Robertson E, Kieff E. Reducing the complexity of the transforming Epstein-Barr virus genome to 64 kilobase pairs. J Virol. 1995;69:983–993. doi: 10.1128/jvi.69.2.983-993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson E S. The Epstein-Barr virus EBNA3 protein family as regulators of transcription. Epstein-Barr Virus Rep. 1997;4:143–150. [Google Scholar]
- 59.Robertson E S, Kieff E. Genetic analysis of Epstein-Barr virus in B lymphocytes. Epstein-Barr Virus Rep. 1995;2:73–80. [Google Scholar]
- 60.Robertson E S, Tomkinson B, Kieff E. An Epstein-Barr virus with a 58-kilobase-pair deletion that includes BARF0 transforms B lymphocytes in vitro. J Virol. 1994;68:1449–1458. doi: 10.1128/jvi.68.3.1449-1458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Said J W, Tasaka T, Takeuchi S, Asou H, de Vos S, Cesarman E, Knowles D M, Koeffler H P. Primary effusion lymphoma in women: report of two cases of Kaposi's sarcoma herpes virus-associated effusion-based lymphoma in human immunodeficiency virus-negative women. Blood. 1996;88:3124–3128. [PubMed] [Google Scholar]
- 63.Schwam D R, Luciano R L, Mahajan S S, Wong L, Wilson A C. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J Virol. 2000;74:8532–8540. doi: 10.1128/jvi.74.18.8532-8540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sjöblom A, Jansson A, Yang W, Lain S, Nilsson T, Rymo L. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J Gen Virol. 1995;76:2679–2692. doi: 10.1099/0022-1317-76-11-2679. [DOI] [PubMed] [Google Scholar]
- 65.Sjöblom A, Nerstedt A, Jansson A, Rymo L. Domains of the Epstein-Barr virus nuclear antigen 2 (EBNA2) involved in the transactivation of the latent membrane protein 1 and the EBNA Cp promoters. J Gen Virol. 1995;76:2669–2678. doi: 10.1099/0022-1317-76-11-2669. [DOI] [PubMed] [Google Scholar]
- 66.Sjöblom A, Yang W, Palmqvist L, Jansson A, Rymo L. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J Virol. 1998;72:1365–1376. doi: 10.1128/jvi.72.2.1365-1376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 68.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steinitz M, Klein G. Comparison between growth characteristics of an Epstein-Barr virus (EBV)-genome-negative lymphoma line and its EBV-converted subline in vitro. Proc Natl Acad Sci USA. 1975;72:3518–3520. doi: 10.1073/pnas.72.9.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szekely L, Chen F, Teramoto N, Ehlin-Henriksson B, Pokrovskaja K, Szeles A, Manneborg-Sandlund A, Lowbeer M, Lennette E T, Klein G. Restricted expression of Epstein-Barr virus (EBV)-encoded, growth transformation-associated antigens in an EBV- and human herpesvirus type 8-carrying body cavity lymphoma line. J Gen Virol. 1998;79:1445–1452. doi: 10.1099/0022-1317-79-6-1445. . (Erratum, 79:2875, 1998.) [DOI] [PubMed] [Google Scholar]
- 72.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tong X, Drapkin R, Reinberg D, Kieff E. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1995;92:3259–3263. doi: 10.1073/pnas.92.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tong X, Wang F, Thut C J, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsang S-F, Wang F, Izumi K M, Kieff E. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J Virol. 1991;65:6765–6771. doi: 10.1128/jvi.65.12.6765-6771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vinson C R, Sigler P B, McKnight S L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 78.Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 79.Wang D, Liebowitz D, Wang F, Gregory C, Rickinson A, Larson R, Springer T, Kieff E. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J Virol. 1988;62:4173–4184. doi: 10.1128/jvi.62.11.4173-4184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang F, Tsang S-F, Kurilla M G, Cohen J I, Kieff E. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J Virol. 1990;64:3407–3416. doi: 10.1128/jvi.64.7.3407-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yalamanchili R, Tong X, Grossman S, Johannsen E, Mosialos G, Kieff E. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204:634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]
- 82.Yanai H, Takada K, Shimizu N, Mizugaki Y, Tada M, Okita K. Epstein-Barr virus infection in non-carcinomatous gastric epithelium. J Pathol. 1997;183:293–298. doi: 10.1002/(SICI)1096-9896(199711)183:3<293::AID-PATH937>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 83.Yates J L, Camiolo S M, Bashaw J M. The minimal replicator of Epstein-Barr virus oriP. J Virol. 2000;74:4512–4522. doi: 10.1128/jvi.74.10.4512-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoshiyama H, Imai S, Shimizu N, Takada K. Epstein-Barr virus infection of human gastric carcinoma cells: implication of the existence of a new virus receptor different from CD21. J Virol. 1997;71:5688–5691. doi: 10.1128/jvi.71.7.5688-5691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Young L, Alfieri C, Hennessy K, Evans H, O'Hara C, Anderson K C, Ritz J, Shapiro R S, Rickinson A, Kieff E, et al. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med. 1989;321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 86.Zhao B, Marshall D R, Sample C E. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao B, Sample C E. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi-1/Spi-B binding site. J Virol. 2000;74:5151–5160. doi: 10.1128/jvi.74.11.5151-5160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]