ABSTRACT
Research on the origin of life investigates the transition from abiotic chemistry to the emergence of biology, with the ‘RNA world hypothesis’ as the leading theory. RNA’s dual role in storage and catalysis suggests its importance in this narrative. The discovery of natural ribozymes emphasizes RNA’s catalytic capabilities in prebiotic environments, supporting the plausibility of an RNA world and prompting exploration of precellular evolution. Collective autocatalytic sets (CASs) mark a crucial milestone in this transition, fostering complexity through autocatalysis. While modern biology emphasizes sequence-specific polymerases, remnants of CASs persist in primary metabolism highlighting their significance. Autocatalysis, driven by CASs, promotes complexity through mutually interdependent catalytic sets. Yet, the transition from ribonucleotides to complex RNA oligomers remains puzzling. Questions persist about the genesis of the first self-replicating RNA molecule, RNA’s stability in prebiotic conditions, and the shift to complex molecular reproduction. This review delves into diverse facets of the RNA world’s emergence, addressing critical bottlenecks and scientific advances. Integrating insights from simulation and in vitro evolution research, we illuminate the multistep biogenesis of catalytic RNA from the abiotic world. Through this exploration, we aim to elucidate the journey from the primordial soup to the dawn of life, emphasizing the interplay between chemistry and biology in understanding life’s origins.
KEYWORDS: RNA-world, autocatalytic-networks, RAF, Eigen’s-threshold, Ribozymes
1. The RNA world hypothesis
We can approximate when life began on Earth, but how life emerged remains unsolved. The RNA world hypothesis, among many theories, promotes RNA as a self-replicating molecule of life and the cornerstone of the transition from abiotic chemistry to biology. The RNA world hypothesis, proposed by Walter Gilbert in 1986 [1], suggests RNA as a key self-replicating molecule and a transition point from abiotic chemistry to biology. It does not assert RNA as the initial replicator but proposes an ‘RNA world’ preceding complex DNA-RNA-protein-based life. Understanding how extraneous factors influenced abiogenesis from chemical soups within such ‘RNA-only’ paradigms in early Earth remains elusive [1–8], given the challenges of recognizing biotic principles in that era.
Unlike DNA and proteins, RNA is capable of both storing information and performing catalytic functions (ribozymes) (Figure 1). Discoveries of naturally occurring ribozymes such as self-splicing introns [2] and ribonuclease P [3] are irrefutable credentials of RNA as a catalyst. Such evidence also hints towards the existence of an RNA world, where critical functions could have been performed solely by RNA. RNA, recognized for its Darwinian behaviour even in the absence of cellular machinery, hence presents a compelling avenue for investigating precellular evolutionary events. In such scenarios, environmental selection could have acted directly on this polymeric material using metals and mineral clusters as active primitive catalysts [1,8]. Nevertheless, the exact process of emergence of such RNA from a purely abiotic chemical environment remains obscure.
Figure 1.
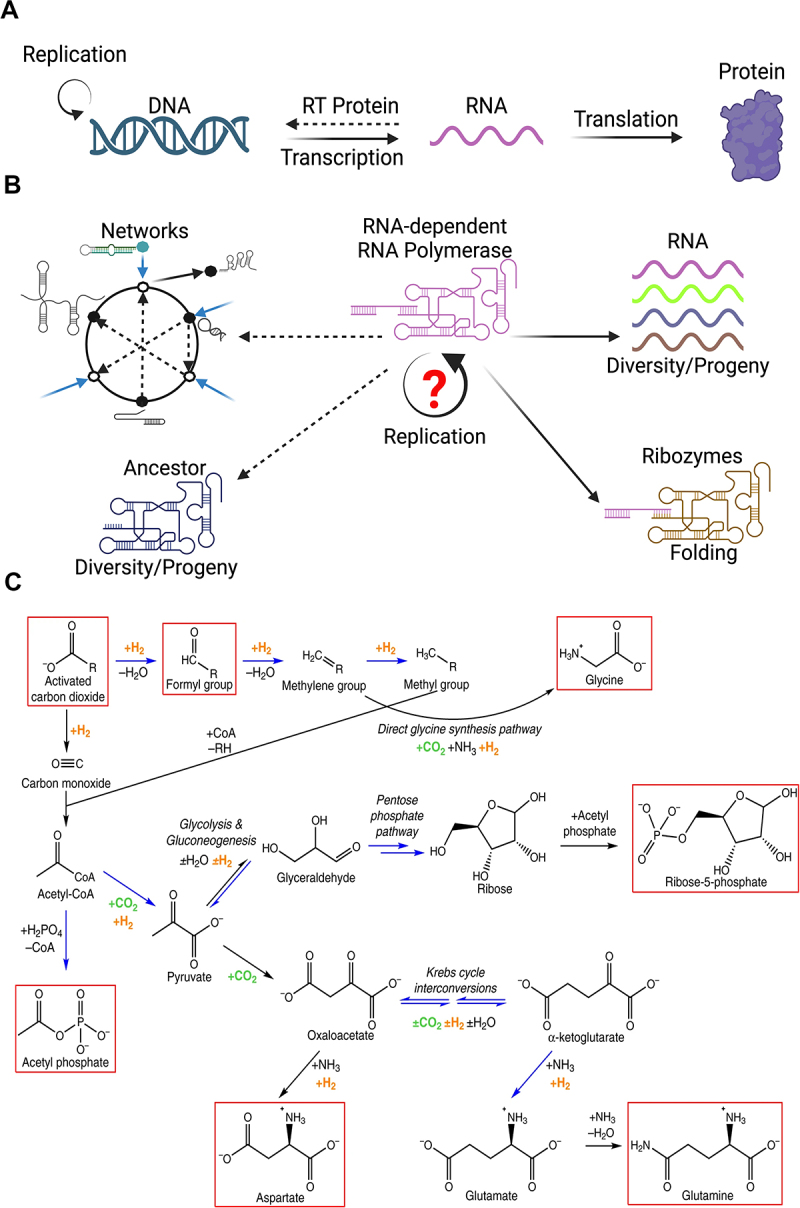
A: central dogma. This schematic depicts the unidirectional flow of genetic information from DNA to RNA to protein. While exceptions to this flow exist, such as reverse transcription in retroviruses, the central dogma remains a foundational principle in molecular biology. B: this schematic represents all the functions that would be necessary for achieving exponential growth within Eigen’s error threshold for a primordial RNA-dependent RNA polymerase ribozyme. Its self-replication is its biggest challenge along with folding and diversification. Meanwhile, each of these folded RNAs would act as a node within a network of short catalytic oligomers. C: example of the earliest reactions that could have dominated the early earth – adapted from “life as a guide to prebiotic nucleotide synthesis” [9]. William Martin and colleagues used many of these as the starter set for formation of an autocatalytic network which show promising networks with emergent properties.
RNA also serves as the critical agent in replication [4], meeting the criteria of a plausible self-replicating entity (Figure 1B). The entire RNA world hypothesis rests on this very dogma [1–8]. To test the limits of this self-replicating capability, Joyce and others conducted groundbreaking research elucidating the evolutionary dynamics of self-duplicating nucleic acids under selection pressure for rapid growth [5]. Their findings indicated that over several generations of molecular replication, the rate of RNA synthesis became faster, and the products turned successively smaller. Mills et al. also provided a tantalizing opportunity to explore the evolution of self-replicating nucleic acid molecules beyond the confines of living cells [6,7].
Basic RNA sequences, featuring elementary secondary configurations like hairpin or stem-loop motifs could have adhered to these mineral surfaces, often originating from meteoric impacts [10], facilitating the assembly of specific structures conducive to molecular interactions [11–14]. In theory, these surfaces could have hosted a complex network of molecular synthesis and degradation [15–17], akin to metabolism, long before the inception of the first cellular life forms (Figure 1C). Simultaneously, RNA-small molecule complexes, primarily through aptamers, facilitate the assembly of complex tertiary structures via non-covalent interactions, enhancing stability and structural diversity, thereby endowing RNA with new functional capabilities such as cofactors for catalysis, modulating folding dynamics, or acting as signalling molecules [4,5,18–20]. These complexes also acted as scaffolds for binding other RNA molecules, forming larger networks that facilitated cooperative interactions and the emergence of rudimentary cellular processes [4,18,20,21]. This integration, driven by aptamers, marked a crucial step in early life evolution through the evolution of the networks, contributing to RNA’s structural and functional diversity and laying the groundwork for complex biological systems [4,5,22,23].
Given the fact that RNA can be easily established as a substantially catalytic macromolecule, which also has the potential to form structured polymers, it stands to reason that minimal networks of rudimentary molecules, with catalytic influences on the other subset of the network (catalytic networks): could have been formed as a consequence [22,24,25]. Such networks, reminiscent of collective autocatalytic sets (CASs) [26–29], represent a critical phase in the transition from a stochastic ensemble of arbitrary molecules to a more complex interdependent pool. Recent advances in the field suggest that CASs played a pivotal role in the early stages of molecular evolution. Autocatalysis, a fortunate by-product of CAS systems, is often thought to have led to the emergence of higher levels of complexity [29–32]. Although relics of CASs persist in primary metabolism, its influence in the genesis of the first self-replicating RNA molecule remains understudied.
2. Challenge of exponential growth against hydrolysis
One major hurdle concerns how RNA overcame hydrolysis to achieve exponential growth during polymerization (Figure 2A). Product inhibition, common in small molecule-dependent propagation like RNA, often leads to parabolic amplification, although an exponential growth pattern is critical for Darwinian behaviour of selection [7]. Additionally, theories suggest that polymers of 30–60 monomers are necessary for a viable genetic system [8,22,33], yet synthesizing such prebiotic polymers in aqueous solutions is hindered by hydrolysis competing with polymerization [18]. Thus, the question arises: how did early RNA molecules reach the critical size range amidst high hydrolysis rates? Furthermore, polymerase activity itself entails a couple of prerequisites not met by purely recombination-based systems. Firstly, it demands that the catalytic function of the polymer is processive involving a cyclic process of binding, catalysing, releasing, and rebinding, which is considerably more intricate than, for example, trans-esterification [18,22,25,26,34]. Additionally, modern polymerases utilize activated substrates, unlike recombination which primarily interconverts bonds of equivalent energy [34,35]. These complexities raise questions about how early RNA achieved exponential growth through simple recombination-based replication.
Figure 2.
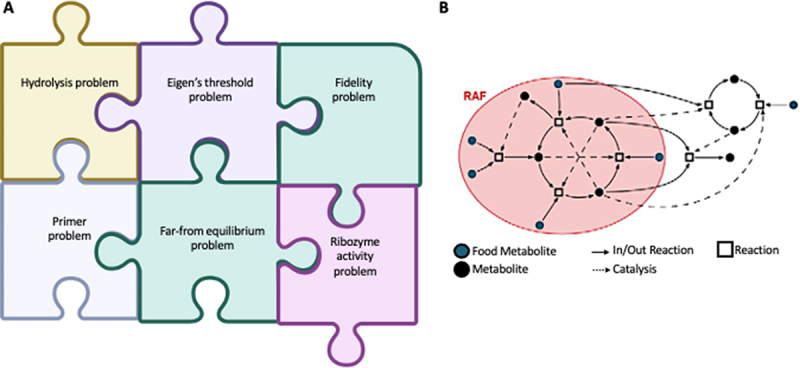
A: challenges of a primordial ribozyme. This cartoon represents a plethora of problems that the primordial ribozyme needed to solve for maintaining exponential growth, without compromising on the Eigen’s error threshold. We discuss each of these and the possible pathways by which the RNA navigated the environment of an early earth. B: this cartoon, adapted from “Autocatalytic chemical networks preceded proteins and RNA in evolution” [32] represents a rudimentary Reflexively Autocatalytic Food-generated network and their interdependence in maintaining the network.
Joyce and co-workers had uncovered a self-replicating ligase ribozyme over 20 years ago that catalyses the assembly of additional copies of itself through an RNA-catalysed RNA ligation reaction [23,36].
Although the initial rate of synthesis seemed conducive towards exponential growth, the two fragments of the evolved ribozyme failed to form functional complexes after few iterations [23]. Networks of RNA can potentially circumvent this problem of exponential growth while maintaining Eigen’s error threshold through several mechanisms such as dynamic equilibrium, competition and selection (Figure 2B). As networks can exist in dynamic equilibrium with their environment, where the rates of replication, degradation, and mutation are balanced to prevent uncontrolled growth while still allowing for adaptation and evolution. Moreover, within a population of molecules comprising the network, competition for resources and selection pressure can act to limit the proliferation of less efficient or deleterious variants, helping to maintain the overall stability and functionality of the network. Autocatalytic networks seem to be the perfect candidate showcasing such attributes.
Autocatalytic reaction networks are defined as a class of chemical or biochemical systems where the products of a reaction within the network act as catalysts for subsequent reactions [14,15,22,29,31,32]. In other words, the reaction network is self-sustaining or self-propagating due to the autocatalytic nature of its components (Figure 2B). In the context of the origin of life, autocatalytic networks can undergo evolution and selection [27,28,37–40]. Variations in the network’s structure or chemistry may lead to more stable or efficient self-propagating systems over time. This is the concept that has been used time and again by Szostak as well as Joyce to initiate and study test tube evolution processes [22,25,41]. Autocatalytic sets and theoretical constructs in the study of the origin of life have been substantiated by numerous experimental examples involving nucleic acids or proteins [27,37,42–44]. These sets range from simple systems of mutually catalytic nucleotide sequences to more complex networks involving multiple peptides or ribozymes [27,42–45]. Experimental studies, such as those examining peptide formation or ribozyme networks, have provided valuable insights into the behaviour of autocatalytic systems, often corroborating theoretical frameworks [38,41,46–50]. By employing these strategies, autocatalytic networks of RNA can potentially achieve a delicate balance between stability and adaptability, allowing them to evolve and persist in complex and fluctuating environments while avoiding the pitfalls of uncontrolled exponential growth.
To tackle this problem in a laboratory setting, Joyce and Inoue described a chemical replication protocol that allows exponential increase in the concentration of RNA oligomers [51]. Their innovative use of surfaces for immobilization and displacement of templates and to separate complementary templates from stable duplexes in solution allowed for a much more robust synthesis [51]. Similar processes may also have played a role in the origin of life on Earth because the earliest replication systems may have proliferated by spreading on mineral surfaces. Similarly, Deck et al. showed that inhibition by used monomers, formed by the hydrolysis of the activated nucleotides, is often the cause for incomplete extension of growing daughter strands in RNA templates [52–55]. By immobilizing strands and incorporating periodic displacement of the solution containing the activated monomers, inhibition of extension can be overcome [23]. This would be akin to a rock surface of an early earth environment (like montmorillonite or hydroxylapatite), which would be frequently washed away by rains and often induce formation of larger polymers [11].
Huang et al. describes the synthesis of a 40-mer oligonucleotide within a day using 1-methyladenine as an activating group when montmorillonite was used as a catalyst. This study substantiated that naturally occurring minerals contain the potential to catalyse synthesis of oligomers that kindled initial lifeforms on early Earth [12]. Innovations such as enhanced ligase ribozyme polymerization through eutectic phases in water-ice mixtures and the discovery of RNA molecules catalysing polymerization highlight ongoing efforts to elucidate the mechanisms underlying the early RNA-driven evolution [13,14]. The aforementioned work and several other researches highlight the mechanisms by which exponential growth can be achieved. The pivotal role played by exponential growth in the emergence of self-replicating entities remains one of the critical challenges of early evolution.
3. Challenges of maintaining Eigen’s error threshold
Another challenge for the establishment of the RNA world came in the form of Eigen’s error threshold (Figures 2B, 3). In 1971, Eigen and co-workers introduced this concept of an error threshold for replicating macromolecules, which sets an upper limit on tolerable copying errors [24]. This model involves a population of replicating polynucleotides relying on a finite supply of activated mononucleotides to create more copies of themselves. The net rate of production depends on the difference between the rate of formation of error-free copies and the rate of decomposition of existing copies. To outcompete others, an advantageous RNA must produce error-free copies more rapidly than other RNAs. Below the error threshold, replication fidelity ensures that mutations are sufficiently rare for the system to retain its genetic information accurately over generations. However, as the mutation rate increases beyond this threshold, the system enters a phase where errors accumulate faster than they can be corrected by selection or other mechanisms. This leads to a collapse of the genetic information, causing the system to lose its ability to replicate faithfully and ultimately resulting in an error catastrophe of the population [24]. The outstanding question is, ‘as the primary replicative molecule leading the transition of abiotic to biotic era, how did RNA manage to stay below Eigen’s error threshold while also maintaining exponential growth (Figures 2B,3)?’
Figure 3.
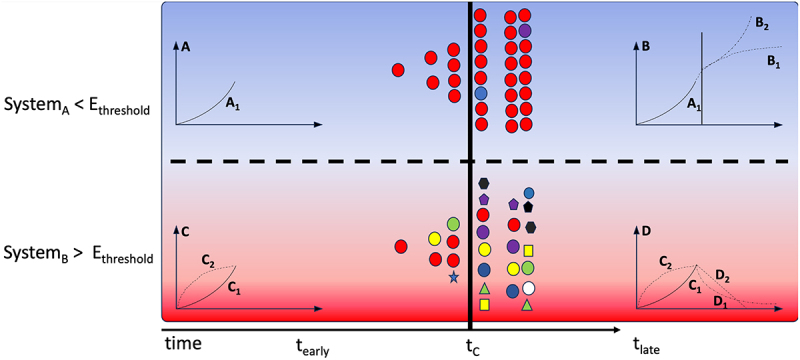
Eigen’s error threshold. This is a pictorial depiction of how RNA evolution takes place in systems maintaining below Eigen’s error threshold (Ethreshold) shown in the top half of the figure. In comparison a different system is shown below that crosses the threshold. RNA molecules (red circles) starting with exponential growth reach criticality (solid black vertical line at time tC). SystemA maintained below Ethreshold reaches exponential growth (A1) in the near term (tearly) followed by stagnation in the form of a parabolic stability (B1) when there is product-dependent inhibition or emergence of another criticality leading to a subsequent exponential growth (B2) within the network of RNA molecules. Whereas, SystemB harboring a faster mutation rate, quickly switches from exponential (C1) to parabolic (C2) nature in tearly followed by exponential (D1) or linear decay (D2).
Over evolutionary time scales, recombination of the constituent nucleotides will eventually yield polymers exhibiting minimal polymerase activity, which can be further refined through selection to enhance replication efficiencies to the extent that they fall below Eigen’s error threshold [22,25,56]. This is the ethos of a plausible RNA world composed of primordial ribozymes and fragments of short-chain nucleotide oligomers. RNA replication in a laboratory setting is supported by three approaches: RNA polymerization by an RNA polymerase ribozyme [22,57–59,59,60], non-enzymatic template-directed ligation [19,20,61–66], and mutually interdependent catalytic RNA networks (Figure 2C) [27,70–73].
Over the last few decades, in vitro evolution experiments have vastly improved ribozyme functionality and ability to self-replicate. More recently, Joyce showed that an RNA polymerase ribozyme can synthesize its own precursor [23,31,36]. The innovation of Joyce and colleagues, was to evolve a polymerase ribozyme that can synthesize the class I ligase into three fragments. While this was innovative and successful in the synthesis step, the activity was often lost within a few generations because of the accumulation of mutations. Thus, the fidelity of RNA polymerization should be considered a major bottleneck to the evolution of an RNA-only network system that can maintain stability in the form of autocatalysis [22]. When pushed to the limits of its activity, the polymerase operates with lower fidelity, which is a critical impediment to maintaining functional information, as would be needed to provide an RNA-based world. For polymerization of abiotic RNA oligomers, phosphodiester exchange reaction can rearrange two sequences to generate a new set of sequences with different lengths (e.g. combining an 8-mer with another 6-mer to produce 10-mer and a 4-mer) [67]. Introduction of stability through secondary structures could drive such energy-neutral reactions providing them the far-from equilibrium impetus in a prebiotic context [18,23,63]. The resulting products could act as templates for reactions, that in turn, catalyse their own production, a process often defined as ‘reproduction’ to distinguish it from the typical ‘replication’ reaction that generates a complementary RNA strand through polymerization chemistry.
Template-directed ligation is also an alternate route for molecular-reproduction [22,23,56,57,62], which involves the concatenation of two sequences. Various RNA molecules, including hc ligase ribozyme and in vitro-evolved ligases, catalyse template-directed polymerization of NTPs, each with distinct characteristics [68]. Evolution experiments starting from the class I ligase led to a ribozyme with robust NTP polymerization activity, capable of incorporating 14 consecutive nucleotides with an average fidelity of ~97% per nucleotide – a remarkable feat [68]. Self-assembling fragments, like the class I ligase and its descendants, form functional units, showing the potential for ribozymes that can polymerize RNA to integrate and develop into mutually interdependent networks of RNA polymers− [69] through cyclic physicochemical processes driving the expansion of RNA complexity.
The third approach of mutually interdependent catalytic RNA networks, akin to a CAS, is a relatively new concept [70–73], often studied using simulations and analytical techniques [30,70–73]. The ability of a set of catalytic reactions to be autocatalytic within its network and be sustained by naturally-occurring nucleotides is observed to be the common requirement for the origin of life [70,71,74]. In a reactionary autocatalytic network of RNA, these nucleotides would be the external supply of substrates that serve as the starting materials for the autocatalytic reactions within the network. These molecules are typically required for the initial steps of the autocatalytic process to occur. As the autocatalytic reactions progress, they generate products that can further catalyse the reactions, leading to self-sustaining cycles of chemical transformations [75]. Without a constant supply of suitable molecules, the autocatalytic reactions would cease, leading to the collapse of the network [76].
A recent breakthrough came from Lehman and co-workers in 2019. In this study, computer simulations were employed to investigate if autocatalytic networks could provide the emergent property of reproduction under prebiotically possible catalytic conditions. It was determined that a small degree of diversity within an RNA population was sufficient to trigger reproduction. In fact, higher diversity tended to reduce the likelihood of reproduction in finite populations. Reproduction at high diversity levels showed some improvement with increased population size [67,77,78]. Conversely, extremely limited variation in RNA sequences also rendered reproduction non-conducive. This intermediate threshold is crucial to initiate reproduction. Within this range, autocatalytic networks were more favourably formed with a high probability. While theoretically, a single RNA could have led to the emergence of the biological world, it is much more feasible to envision a network of RNA that led to the transition. This contrasts with the conventional outlook, which envisions the emergence of a RNA polymerase ribozyme capable of self-copying another long RNA polymer – an unstable concept. Here, Lehman and co-workers introduced the reflexively autocatalytic and food-generated set (RAF): which offers a more stable framework (Figure 2C).
Replication from a truly physio-chemical nature could have occurred from the stacking interactions often seen in short RNA oligomers. Each stacked RNA complex would harbour several diverse 5’ overhangs that can act as intramolecular templates for extension of the paired 3’-terminus from the same or a different RNA oligomer from within the stack [79]. Individually, the RNA oligomers from the stack can also act as a part of the recombinatory collective autocatalytic sets, increasing the likelihood of enhancing polymerase activity [67,80]. Lehman et al. suggests that this process transfers the task of reproduction to a RAF and gradually evolves a polymerase capable of self-replication limited only by its own stacked size as well as complexity [67,80,81]. Both RAFs and constructively autocatalytic food-generated networks (CAFs) are mathematical models that allow the investigations of networks specifically with respect to emergent properties. While RAFs, emerging from ambient food sets, require all necessary catalysts to be produced by the network, modelling the emergence of specificity, speed, and efficiency in autocatalysis, CAFs only require catalysts to be present in the food set.
In a same 2019 study, Lehman and co-workers showed using molecular simulations that for template-directed recombination and ligation, RNA reproduction is unlikely to occur in populations with either low or high sequence diversity, but it exhibits robustness in populations characterized by intermediate sequence diversity. Furthermore, this intermediate range tends to expand towards higher diversity as the population size increases. Interestingly, the emergent property of reproduction seems to favour autocatalytic networks, where each member of the network could facilitate the reproduction of the other, rather than a single RNA ribozyme trying to battle both exponential growth and Eigen’s threshold. Small RNA oligomers, which would lack diversity in the earliest of times, could still rely on network formation for eventual emergence of reproduction from within the reactionary set [67,77–81]. Hordijk et al. deduced an algorithm to determine the feasibility of sustaining such a network within the combination of a group of individual oligomers (RNA or amino acids) while simultaneously maintaining autocatalytic properties. Intriguingly, their findings can be even used to determine an autocatalytic set from within a composite of metabolic pathways. [27,28,82]. Their results indicate that the autocatalytic networks of small oligomeric RNA could circumnavigate the issues of maintaining Eigen’s error threshold, as the mistakes would not be sustainable within the valid frame of the network and as soon as mutations accumulate it would self-correct below the levels of the threshold. Similarly, by regulating the activation threshold for catalysis or replication within the network, autocatalytic reactions can be controlled to prevent runaway growth while still maintaining a sufficient level of activity to sustain the network. Simultaneously, the exponential growth is sustained through other variables creating Le Chatelier’s pull by removal of products as discussed in section 7.
4. Challenges of fidelity in primordial ribozymes
While the emergence of the autocatalytic network could circumnavigate the problems of stability, and competitive rates, it still assumes that the RNA within the network will be enzymatically efficient and processive while also maintaining fidelity. The formation of medium chain RNA fragments relies on efficient catalysis by such primordial ribozymes [83–87]. How these ribozymes maintained secondary structures to induce primitive catalysis while ensuring fidelity remains a question (Figure 2A). For Darwinian-type evolution, RNA polymerase ribozymes must amplify complex RNAs, including themselves, accurately to avoid deleterious mutations [7,22,83]. However, only a few synthetically assembled ribozymes retain catalytic activity as discussed earlier [22,23,36].
Ribozymes must adopt a folded structure to achieve catalysis [3,9,88–94]. Natural and synthetic ribozymes can also be architectured from multiple fragments, which often show higher activity and stability than a single stranded ribozyme [90–99]. A prime example of such a 3-part system is the sunY self-splicing intron [100]. Even with these advances, the feasibility of template-directed ligation under prebiotic conditions remains a well-debated topic. So, what was the source of the RNA primer for polymerization-based propagation of the first templates also remains a mystery.
There have been several attempts to create RNA polymerase ribozymes that can rival the polymerase activity without compromising on fidelity [7,13,57,101–104]. Although existing polymerase ribozymes can synthesize portions of functional RNAs like tRNA and the polymerase’s catalytic subunit, they do so at very low yields and require preformed oligonucleotides [59,104–106]. Zaher et al. stress the importance of selecting an improved RNA polymerase ribozyme with superior extension and fidelity for synthesizing complex functional RNAs effectively [107]. While in vitro evolution of RNA polymerase ribozymes has progressed, achieving a self-replicating RNA-dependent RNA polymerase remains challenging due to the complex structural elements of ribozymes that are difficult to copy [108].
The search for efficient, high-fidelity ribozymes capable of self-replication is not only crucial for understanding the origins of RNA-based life but also for constructing synthetic RNA life in laboratory settings. Pressman et al. propose that prebiotic chemical inventories may have been less complex than previously thought, indicating that ribozyme catalytic capabilities are evolving towards self-replication objectives [109]. Ekland et al. presented a significant advancement in this quest by demonstrating an RNA molecule capable of synthesizing RNA through reactions resembling those catalysed by protein enzymes during RNA polymerization. This ribozyme exhibits remarkable template fidelity and retains ligase functionality [101]. Attwater et al. emphasized the importance of RNA folding in catalytic processes and introduced a breakthrough in RNA-catalysed RNA synthesis on structured templates using triplet substrates. This innovation resolves longstanding challenges in RNA replication and enables ribozyme self-assembly, opening new avenues for understanding the complex interplay between RNA structure and function in the context of early life evolution [13,101–103,103,104].
Attwater et al. describe the evolution of RNA polymerase ribozymes capable of synthesizing RNAs approaching their own size, a crucial step towards RNA self-replication. Through in vitro evolution of water ice, cold-adaptive mutations yield polymerase ribozymes operational at sub-zero temperatures, capable of accurately synthesizing RNA sequences up to 206 nucleotides long. This achievement lays the foundation for RNA self-replication in extreme environments [13,102–104]. Horning et al. further enhances polymerase ribozyme activity and generality via in vitro evolution, enabling synthesis of complex structured RNAs, including aptamers or rudimentary riboswitches, ribozymes, and other forms of catalytic folded RNAs. The improved polymerase can amplify short RNA templates by over 10,000-fold, fulfilling the prerequisites of Darwinian life without protein involvement (Figure 4A) [83]. Additionally, it was demonstrated that ribozyme-catalysed transcription led to the evolution of a polymerase ribozyme synthesizing RNAs up to 95 nucleotides in length. Recombining evolved traits yields a more versatile polymerase ribozyme capable of synthesizing diverse RNA sequences, including enzymatically active ribozymes, echoing a fundamental aspect of RNA-based genetic systems [83,103,103,110]. While origin of the first template still remains a mystery to the curious mind, it goes without saying that proofreading or template-directed replication can help to maintain the fidelity of replication within the network, preventing the accumulation of deleterious mutations beyond Eigen’s error threshold (Figure 4A).
Figure 4.
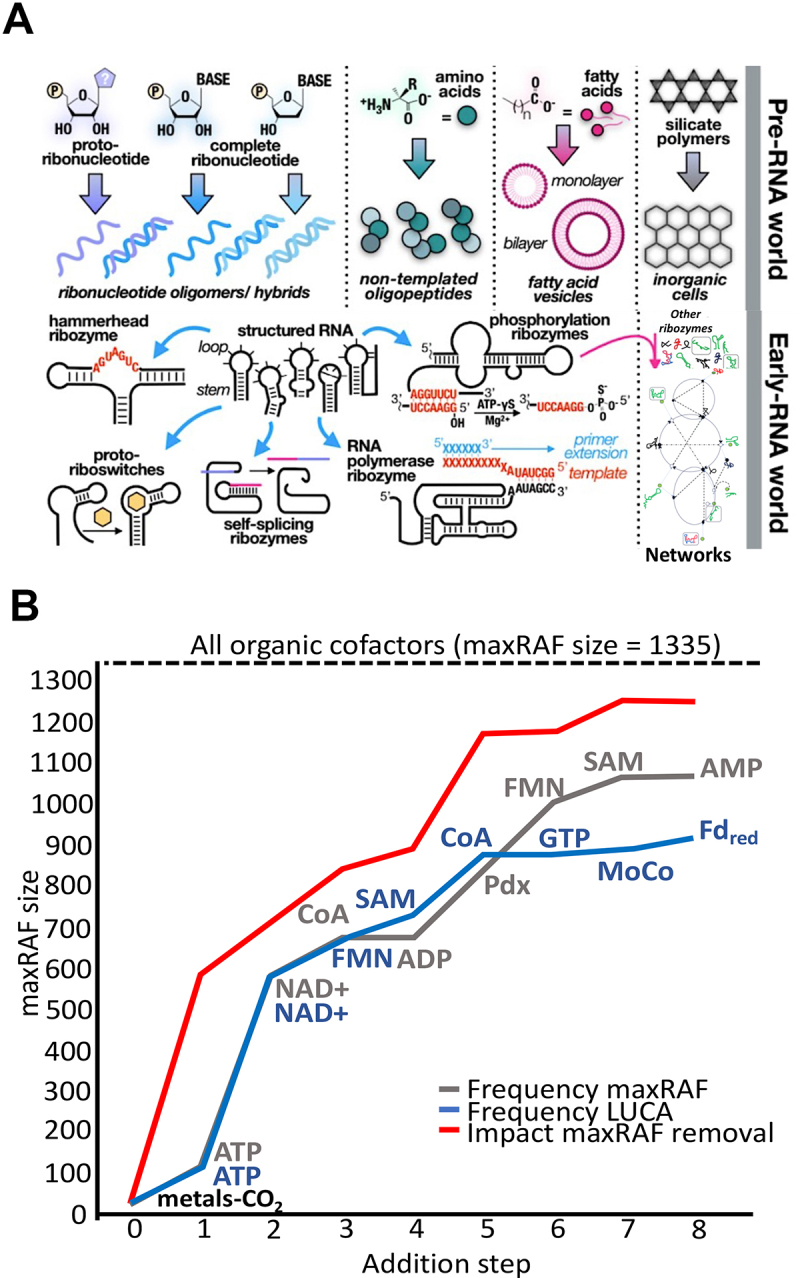
A; non-templated polymerization driving the initial pre-RNA world followed by emergence of ribozymes and eventual autocatalytic network of ribozymes in the Early-RNA world. Specific events are highlighted in cartoons from the non-templated formation of ribonucleotide oligomers to the formation of ribozymes in the later stages and their integration into networks B. This graph, adapted from “autocatalytic chemical networks preceded proteins and RNA in evolution” [32], depicts the influence of individual monomeric molecules and the combinatory size of the conglomeration on the size of the RAF generated.
5. Challenges of far-from-equilibrium thermodynamics
In previous sections, we explored the origins of RNA fragments and how primordial ribozymes catalysed polymerization steps, ultimately leading to replication and the assembly of reactive CASs. However, a few crucial questions remain: How were these improbable events, far from equilibrium, sustained? What effectors of the early-earth paradigm compensated for entropic loss in transitioning from a random pool of RNA to the establishment of the first templates (Figure 2A)? Branscomb et al. stress the necessity of catalysis for life’s emergence on prebiotic Earth, where specific reaction networks require acceleration. Living systems maintain far-from-equilibrium states, comprising numerous subordinate disequilibria, each driven thermodynamically uphill and coupled to dissipation of a greater disequilibrium, transcending mass-action solution chemistry [111].
The potential of RNA-directed recombination processes to establish networks filled with interacting RNA as an answer to the challenge of far-from-equilibrium thermodynamics is becoming clearer [112]. One crucial aspect of autocatalytic networks is their robust network structure, comprising interconnected reactions that facilitate self-propagation. This structure is fundamental for the self-sustaining behaviour observed in autocatalytic systems, making them pertinent to the study of life’s emergence on Earth [27,28,37,40,113–115]. The initial formation of RNA polymers involves recombination, where RNA fragments interact to form recombined versions through transesterification reactions with near-zero Gibbs free energy [4,63]. Primordial ribozymes likely catalysed these energy-neutral reactions, facilitating network diversification and the development of CASs (Figure 4B). Stuart Kauffman and colleagues highlighted recombination’s role in closing autocatalytic sets and transitioning randomness to non-random nucleotide polymers [37,40,115]. Autocatalytic closure occurs as polymers reach a certain length threshold, typically 12–27-mers, depending on the probability of catalysed reactions. Recombination could significantly reduce the required polymer length range, enhancing obtainability through abiotic processes. Polymerase ribozymes with multiple fragments could feasibly replicate, possibly arising from prebiotic RNA synthesis [18,21,24,27,28,116,117]. Autocatalytic networks often exhibit feedback loops, where products generated from reactions act as catalysts, accelerating further product formation. This feedback mechanism, evident in studies by Gerald Joyce and others, contributes to the system’s self-propagation [7,27,28,37,42,118]. Additionally, experiments on the evolution of polymerase ribozymes have revealed the sensitivity of autocatalytic systems to initial conditions, a characteristic shared with chaotic systems and considered significant in the context of the origin of life (Figure 4B) [4,13,23,70].
5.1 Autocatalytic networks solely made of RNA
Given the laboratory success of RNA polymerase and RNA ligase ribozymes, it is not far-fetched to envision a cyclic autocatalytic reaction network, purely composed of RNA fragments. Joyce and co-workers showed that RNA autocatalytic networks can undergo Darwinian evolution [4,7,22]. Variations in the sequence or structure of RNA molecules can lead to changes in their catalytic activity. Natural selection can then act on these variations, favouring those that improve replication efficiency and stability (Figure 4C). This can explain the molecular cascade of exponential rise in the complexity and diversity of the RNA molecules [22,25,38,68,119].
Recently, Lehman et al. discusses a recurrent complication with too much success in the synthesis of a complementary copy [26,29,30,34,77,78,80]. The longer the product, the more tighter it becomes bound to the template and the less likely it is to dissociate, creating another bottleneck for the exponential progression. A better model of prebiotic reproduction involves the existence of an autocatalytic network, instead of binary template-product pairs, where each molecule is multiplied by another, facilitating collective reproduction. This was found to be the case even in scenarios involving template-directed ligation [22,23,26,29,30,34,77,78,80]. Meanwhile, the transition from simple networks to RAFs likely occurred when monomeric networks and their by-products yielded a sufficiently large number of oligomers of lengths conducive to folding and self-catalysis [27,28,40,82,120]. This expansion led to autocatalytic closure and the effective transition from an unbound to a bound set of monomers within the reactionary network (Figure 4C) [35-37, 122–124].
In the context of RAF sets of polymers, a system for the reproduction of stored information emerged when the chemically feasible framework attained stability [15,43–45,122,123]. Unlike the conventional RNA world perspective, which emphasizes polymerization, this model suggests that information reproduction primarily relied on recombination, and it was distributed within the population in an altruistic or cooperative manner [16,17,124]. Recombination is less efficient at exploring diversity for optimal solutions, than mutations, which is sudden and significant [125–129]. Additionally, reproduction through recombination could be overshadowed by template-directed replication when the environment favoured the latter, allowing for inter-self-competition [70,130–133]. Similar kinetic constraints could have acted upon the earliest forms of the RAF in the manner of physical and chemical variables which we discuss later, thereby limiting the rate of replication or catalysis, and thus mitigating the potential for uncontrolled exponential growth.
5.2 Catalysts of the early evolution of RNA networks
Employing the Azoarcus group I intron ribozyme fragments, Lehman and his team demonstrated the self-assembly of these fragments through specific genotype interactions, culminating in the creation of full ribozymes [134]. Their research delved deeply into the dynamics of small networks, emphasizing those with three or four members, and the influence of a tri-member core on the addition of subsequent nodes. Their empirical data not only validated the theory that augmenting connections to a central network could boost its growth rate but also highlighted the consequential role of the direction of these connections. Furthermore, the team formulated a model considering primary interaction effects, which proficiently anticipated the expansion trajectory of more intricate networks when equilibrium was achieved [134]. The relevance of RAFs and CAFs in understanding modern metabolism also allows us to gain insights into the early earth [77,78,80], where many enzymatic reactions involve cofactors and metals that can themselves perform catalysis. It suggests that prebiotic chemical networks maintain vestiges within modern biochemistry [16,27,28,114]. The role of catalysts, including inorganic catalysts (Figure 5) and constructively autocatalytic sets, is crucial in establishing the conditions for life’s emergence on Earth, possibly facilitated by physical entrapment, surface catalysis, and microenvironments like rock crevices and drying ponds (Figure 5) [135–138].
Figure 5.
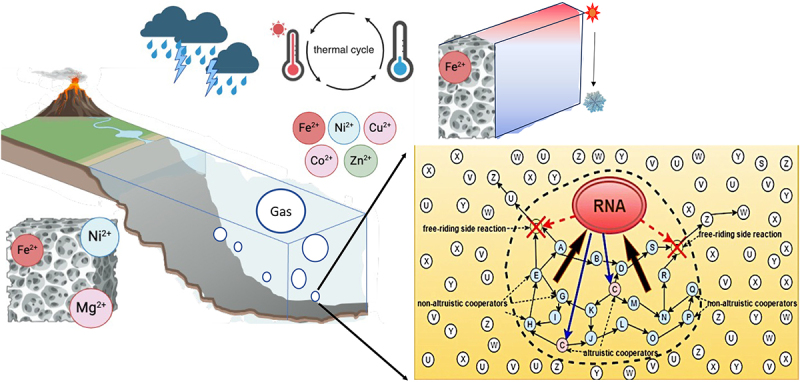
Physical and chemical variables that influence far-from equilibrium evolution with inset of an RNA network that could exclude side reactions potentially forming within rock clusters, adapted from “the origins of life: the Managed-Metabolism Hypothesis”. [121].
The environmental conditions of the early Earth, characterized by higher temperatures and salinity, played a crucial role in ribozyme activity and evolution [7,12,22]. Non-equilibrium environments, like hydrothermal vents, facilitated the assembly and segregation of early life molecules, promoting continuous enrichment of prebiotic molecules [13]. Recent studies demonstrated that thermal gradients drove a complex enrichment process of prebiotic molecules, cycling RNA precursors, monomers, ribozymes, and oligonucleotides between dry and wet states, enhancing enzymatic activity (Figure 5) [14]. Non-equilibrium systems, prevalent on early Earth, particularly through hydrothermal activity, provided conditions essential for biological processes [7]. Gas–water interfaces in thermal gradients played a crucial role in the emergence and evolution of life (Figure 5) [56], facilitating processes like lipid and DNA adsorption and peptide bond formation [10,30,31,139]. Approaches to the origin of life require high concentrations of probiotic molecules, likely facilitated by niche environments such as small pools, that are away from equilibrium states [65,66]. For example, heat currents in porous mineral precipitates drove extreme accumulation of molecules, particularly single nucleotides, across pores (Figure 5), often facilitated by trapped gases [58]. Understanding primitive enzyme evolution in different environmental conditions is a current focus, revealing changes in function and adaptations over time [19,20]. Reconstruction of Precambrian thioredoxin enzymes has shown them to be more stable and active than modern counterparts, offering insights into ancient protein adaptation [20].
6. Advances in the research of RNA catalytic networks
In some RNA autocatalytic networks, ribozymes can also cross-catalyse each other, creating a more robust and interconnected system. This leads to enhanced self-replication and overall network stability [23,140–143]. This is critical for the co-development of the ligase and polymerase functionalities. Moreover, given the possibilities of a metabolism-only protocell, stability of such a system is crucial. In an RNA-dominated world, the challenge faced by singular self-replicating RNAs involved maintaining a sufficiently low mutation rate [144–146]. This is crucial to both preserve genetic integrity and to successfully diminish chances of evolution for ‘virus-like’ molecular entities. The research conducted by the Lehman and Hayden groups introduced a theoretical perspective suggesting that networks of interacting molecules were more likely to show emergent properties including cooperativity, anabolism and catabolism from spontaneously assembled networks [77,78,80]. This underscores the inherent capacity of RNA populations to evolve increased complexity through collaboration [77,78,80,147]. The observed evolvability of networks via in vitro selection further underscores the advantages of cooperative behaviour even during the early molecular stages of life’s emergence.
6.1 Cooperativity as the basis of early evolution of RNA networks
Vaidya et al. demonstrate that mixtures of self-assembling RNA fragments generate cooperative catalytic cycles and networks with an essential emergent property being cooperativity. Their work indicates the intrinsic ability of RNA populations to evolve complexity through cooperation [15,70]. Through in vitro selection, they observe the evolvability of these networks, emphasizing the advantages of cooperative behaviour even at the molecular stages of nascent life [72]. Review from Higgs et al. underscores key concepts in RNA world research, particularly the mechanisms of RNA replication and the role of cooperation in molecular evolution. Autocatalytic feedback mechanisms are highlighted as potential drivers of RNA replication. Cooperation among molecules is identified as crucial during the origins of life, with three types of molecular cooperation proposed. The review also discusses the possibility of chemical alternatives to RNA and the importance of network establishment in organizing the living state, from small molecules to cell-like structures (Figure 5- inset) [28]. Sievers et al. reported on the replication of nucleic acids using complementary strands and demonstrated self-replication of complementary templates, providing kinetic evidence for selective stimulation of synthesis [148]. This study also offered insights into the mechanisms underlying the replication of nucleic acids and the potential implications for early Earth conditions [148].
The 2018 study made a giant leap by simulating a prebiotic environment and fuelled the Azoarcus ribozyme system solely with modified RNA substrates that cannot directly form autocatalytic ribozymes [149]. Surprisingly, unmodified fragments are reconstituted, resulting in the production of wild-type catalysts. Detailed analyses reveal that this transformation of raw materials is catalysed by the reaction products through a multi-step reaction pathway, involving specific but unexpected binding interactions between RNA substrates and the ribozyme. This combination of catabolic and anabolic steps facilitated collective autocatalysis [147,149]. The study also demonstrated that CASs composed of multiple species are maintained under similar conditions. This discovery underscores the presence of rudimentary catabolism, where catalysts convert available resources into building blocks that drive their own formation [149]. Nghe’s subsequent breakthrough demonstrates how anabolic and catabolic processes may cooperatively lead to the formation of autocatalytic networks based on RNA. These CASs utilize available environmental substrates, showcasing their potential as a mechanism for life’s origin, especially in environments with limited resources [147,150]. The 2018 study focusing on Azoarcus ribozymes also revealed how catabolic processes within CASs can broaden the range of usable substrates in diverse prebiotic environments. It illustrates the role of cooperative networks fuelled by modified substrates in producing catalysts efficiently, highlighting the importance of coupled catabolism and anabolism even in the early RNA world [147,149]. Additionally, a 2019 study by Lehman et al. demonstrates that RNA reproduction exhibits robustness in populations characterized by intermediate sequence diversity, facilitating autocatalytic network formation and increasing information capacity [50,67,149].
6.2 Influence of catabolic reactions in the development of the network
In the same work of 2019, Phillippe Nghe’s group showed how anabolic and catabolic processes might have cooperatively led to the formation of autocatalytic networks purely based on RNA [150]. Collective autocatalytic sets, where RNA molecules serve both as carriers of genetic information and as catalysts, remain a potential mechanism for the origin of life. A crucial feature of CASs is their ability to self-sustain using available environmental substrates, often referred to as the ‘food set’. In contemporary biological systems, this property has evolved into highly complex and diverse metabolic processes. In the context of the origin of life and the RNA world, where readily usable substrates were limited, it would have been advantageous for self-replicating systems to thrive on a wide range of resources by processing them before utilization [150]. The 2018 study centred on RNAs derived from the group I intron of the Azoarcus bacterium, which are 200-nucleotide RNA recombinases capable of self-reproduction and recycling of RNA materials through recombination reactions. These ribozymes can catalyse their self-assembly from fragments [149]. Previous studies on this system primarily relied on artificially designed substrates obtained through ribozyme fragmentation. In a more realistic prebiotic setting, the initial reaction mixtures would likely contain a broader spectrum of molecules, potentially inhibiting or disrupting the self-reproduction of CASs [149]. This could occur for various reasons, such as the inability of available molecules to serve as catalyst substrates or the production of futile products that lack catalytic abilities.
These findings illuminated how catabolic processes within collectively autocatalytic sets can broaden the range of usable substrates (Figures 4B, 5), a crucial advantage in a diverse prebiotic environment. The assimilation of modified substrates is enabled by the recombination activity of Azoarcus ribozymes, featuring a multi-step reaction pathway distinct from other experimental autocatalytic systems [149,150]. Moreover, cooperative networks, comprising several species and fuelled by modified substrates, prove to be almost as efficient at producing catalysts as those with unmodified substrates while maintaining the relative distribution of species. Although the RNA fragments used in this study were relatively long, Azoarcus ribozymes can be formed from much shorter RNA fragments, making them relevant to prebiotic scenarios [149,150]. The study also introduced an inherent form of molecular-level selection, arising from the differential protection of functional and parasitic folds from recombination. This mechanism aligns with concepts from dynamical combinatorial chemistry, offering insights into preventing the extinction of early metabolic cycles [149,150]. The coexistence of coupled catabolic and anabolic processes, although different from contemporary living systems, reflects a universal aspect of biology, where complex molecules are broken down into simpler building blocks and utilized to synthesize new biomolecules. This observation suggests that even in the early RNA world, coupled catabolism and anabolism could have played a pivotal role, despite the system’s lower complexity at that time.
7. Physical and chemical variables affecting Eigen’s threshold and far-from-equilibrium dynamics
7.1 Physical variables
It is important to discuss the physical and chemical variables that would have influenced the coupled stabilization of the far-from-equilibrium thermodynamics. Here we will discuss some of these recent discoveries. These are just highlights of some of the variables at play and in no way representing an exhaustive list of all variables. The RNA world likely originated in aqueous environments, such as hydrothermal vents or warm little ponds, facilitated by meteoritic delivery of nucleobases (Figure 5). A numerical model developed by the authors integrates Earth’s early evolution with prebiotic chemistry, suggesting rapid RNA polymer appearance after meteorite deposition, predominantly from meteoritic nucleobases [10]. Wet-dry cycles in warm little ponds promote RNA polymerization, contrasting with hydrothermal vent conditions that yield shorter RNA chains. Another theory suggests the geochemical origin for chemiosmotic coupling and Na+/H+ transporters, proposing proton gradients across FeS walls in alkaline vents could drive carbon assimilation and protocell emergence. Previous studies demonstrate RNA-like polymer production in hydrothermal processes, enhanced by amphiphilic phospholipids [151]. These findings illuminate the plausibility of RNA polymerization in hydrothermal environments, enriching our understanding of early Earth chemistry and life’s origins [151].
Hydration-dehydration cycles with monovalent salts significantly enhance polymer yields, as evidenced by ethidium bromide-stained products showing base pairing. LiCl and other monovalent cationic chlorides (Figure 5) yield synthesis of A and U oligomers (10–300 in size) [151]. Bouossau et al. (2013) propose a novel approach to explore the ecology of the last universal common ancestor (LUCA). Their analysis of resurrected proteins suggests a thermophilic bacterial ancestor, followed by adaptation to lower temperatures, supported independently by ribosomal RNAs and protein sequences. This approach reconciles conflicting results and offers insights into the evolution of ecological traits across life’s tree [77,78,80] highlights constraints on RNA availability on the early Earth, including biases in abiotically synthesized RNAs, a bias towards shorter RNA molecules during prebiotic polymerization and limitations imposed by the need for compartments for sustainable genetic information propagation [67].
7.2 Chemical variables
Originally proposed by Orgel in 1980, mineral surfaces were identified to have the potential to selectively adsorb longer RNAs, primarily on hydroxyapatite, suggesting a chromatographic effect [5,20]. Despite its initial presentation, this concept has not been extensively explored. The selective accumulation of longer RNA molecules on mineral surfaces carries significant implications for the RNA world hypothesis (Figure 5). Firstly, it can counteract the bias towards shorter RNAs typically observed in abiotic or ribozyme-mediated RNA synthesis [152,153]. Secondly, mineral surfaces can protect longer RNAs from loss in self-replicating systems, where shorter sequences tend to replicate more rapidly [65,154]. Thirdly, the specific accumulation of long informational RNAs may facilitate cooperative interactions among RNA molecules, promoting complex reactions and collective reproduction [155,156]. This underscores the importance of investigating the collaborative effects of minerals and ribozymes in promoting cooperative phenomena. Further exploration is needed to fully understand these dynamics (Orgel).
Lehman’s group also presented some interesting research on the general enrichment of longer RNAs on mineral surfaces involving various types of mineral grains and environmental conditions [80]. They observed the preferential accumulation of longer RNAs when they were incubated with these minerals, with the extent of enrichment influenced by factors such as temperature. Additionally, they explored the integration of this size-selection capability with the catalytic activity of a ribozyme, ultimately demonstrating that mineral surfaces can enhance genetic complexity, especially when combined with ribozymes. Lehman and Baum observed from theoretical calculations that empirically several prebiotic minerals have the capability to preferentially accumulate longer RNA molecules, and this selectivity is notably heightened at elevated temperatures [80,157]. Additionally, they illustrated the ability to combine surfaces with a catalytic RNA to produce extended RNA polymers, indicating the potential of minerals in the development of genetic information during the early stages of Earth’s history [80,157].
Minerals on early Earth likely played a crucial role in various processes relevant to the origins of life, offering a diverse range of catalytic possibilities due to the presence of co-precipitating ions and multiple cationic species within the same mineral [8,33,155]. Numerous experiments exploring the origins of life have utilized minerals to produce biomolecules and mimic biological pathways. Life is thought to have originated on Earth around 4.4 ~ 3.5 billion years ago, possibly in the presence of minerals, where organic molecules evolved into systems capable of self-organization and reproduction. Mineral surfaces provide opportunities for catalytic reactivity and concentration, potentially shaping the complexification of the molecules of the early earth that led to the emergence of life [158–160]. Recent studies have delved into prebiotic mineral-organic interfacial processes, shedding light on macromolecular interactions between geophysical scaffolds and activated nucleotides, and offering insights into the origin of life (Figure 5). In particular, the catalysed formation of 50-mer oligonucleotides by minerals holds significant potential for the RNA world scenario, highlighting the role of minerals in the emergence of life based on RNA [8,33,155].
Various studies have explored the chemical synthesis of the earliest molecules of earth and replication systems, leading to the RNA world proposal [63,103,103,104]. From a geological perspective, thermal gradients are prevalent in dissipative systems on early Earth, surrounded by porous rock clusters which can accumulate single nucleotides [144,161–164]. The accumulation process is highly resistant to changes in shape and sizes, thereby allowing for various molecular compositions, including short and long RNA oligomers to be concentrated [58,164]. Submarine hydrothermal environments have been considered as potential sites for life’s emergence due to their contemporary habitability and supply of essential materials [59,60]. Combining the multiplexing of hydrothermal environments with a concentration mechanism in porous rocks strengthens the case for an environment conducive to the origin of life. Considering multiple variables together, rather than in isolation, is crucial for accurately recapitulating early Earth environments. It is evident that exponential growth can help surpass the threshold for the emergence of complexity by allowing for the accumulation and interaction of molecules beyond a critical mass. This threshold is essential for the transition from simple chemical systems to more organized and functionally integrated biological systems and could not have been driven by an oversimplified mechanism of single-variable.
8. Concluding remarks
In the progression of primitive life on Earth, the transition from polymers to polymerases and the emergence of complex life remain one of the major events which we have thoroughly discussed here with respect to Eigen’s error threshold requirement. Initially, the focus lies on the establishment of polymerase activity, enabling efficient exploration of phenotypic phase space and overcoming challenges like the error threshold and hydrolytic elements. The subsequent integration of genetic information encoding into the system follows, laying the foundation for further developments. Metabolic autocatalytic sets, potentially including amino acids and nucleotides, played a crucial role in this process through wet-dry cycles. If metabolic RAFs could produce essential nucleotides, then RNA RAFs would naturally select for their production. This led to the emergence of polymer RAF, likely dominated by RNA. Polymers acted as catalysts facilitating the exploration of molecular diversity space and enabling intermolecular recombination. These recombination reactions, demonstrated experimentally, allowed for rapid traversal of genotype and phenotype space, contributing to the formation of extensive networks of RNA-driven RNA recombinations [26,29,67,77,78,80,157].
Molecular evolution is a complex entity that has been tested mostly in experimental isolation. Although necessary to experiment with one variable at a time, it is critical to consider the integration of multiple factors from chemical and physical environments. As most lab research occurs in ‘test tube’ settings, new designs of experimental advances allow for the investigation of complex variables that can mimic particular real-life situations. For example, bubbling experiments can provide gas–liquid phase separation in a thermal gradient similar to geothermal vent systems that existed on early Earth. By testing the RNA world hypothesis and ribozyme experiments in such settings, we can better recapitulate the native environment of where these cycles of molecular evolution were presumed to take place in contrast to previous experiments that take place in isolated settings. Understanding how catalytic RNA is involved in the propagation and establishment of this self-replicatory cycle will afford an understanding of molecular evolution on Earth and guide the establishment for life on other planets. As humans continue to venture into space and aim to eventually colonize other planets, an inclusive knowledge about molecular evolution is vital to ensure a successful colonization and establishment of sustainable life in these new ‘Earths’.
Supplementary Material
Acknowledgments
We thank Dr. Deepto Mozumdar (UCSF) for the development of sections of the figure 4 and Dr. Nidhi Walia (Purdue University) for help with the corrections and critical reading of the manuscript.
Funding Statement
The author(s) reported that there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2024.2405757
References
- [1].Gilbert W. The RNA world. Nature. 1986;319(6055):618–618. [Google Scholar]
- [2].Cech TR, Bass BL. Biological catalysis by RNA. In: Gesteland RF, and Atkins JF, editors. The RNA world. Long Island, NY: Cold Spring Harbor Laboratory Press; 1982. p. 133–165. [Google Scholar]
- [3].Guerrier-Takada C, Gardiner K, Marsh T, et al. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35(3 Pt 2):849–857. [DOI] [PubMed] [Google Scholar]
- [4].Joyce GF, Orgel LE. Prospects for understanding the origin of the RNA world. In: The RNA world. Long Island, NY: Cold Spring Harbor Laboratory Press; 1999. p. 49–77. [Google Scholar]
- [5].Joyce GF, Schwartz AW, Miller SL, et al. “The case for an ancestral genetic system involving simple analogues of the nucleotides”. In: Proceedings of the National Academy of Sciences; 1987. 84(14): 4398–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mills DR, Peterson RL, Spiegelman S. ”An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule”. In: Proceedings of the National Academy of Sciences of the United States of America (PNAS); 1967. 58. p. 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Joyce GF. RNA evolution and the origins of life. Nature. 1989;338(6212):217–224. [DOI] [PubMed] [Google Scholar]
- [8].Ferris JP, Ertem G. Oligomerization of ribonucleotides on montmorillonite: reaction of the 5’-phosphorimidazolide of adenosine. Science. 1992;257(5070):1387–1389. [DOI] [PubMed] [Google Scholar]
- [9].Harrison SA, Lane N. Life as a guide to prebiotic nucleotide synthesis. Nat Commun. 2018;9, article number 5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sleep NH, Zahnle K, Kasting JF, et al. Annihilation of ecosystems by large asteroid impacts on the early earth. Nature. 1989;342(6246):139–142. doi: 10.1038/342139a0 [DOI] [PubMed] [Google Scholar]
- [11].Ferris JP, Hill AR Jr, Liu R, et al. Synthesis of long prebiotic oligomers on mineral surfaces. Nature. [1996 May 2];381(6577):59–61. [DOI] [PubMed] [Google Scholar]
- [12].Huang W, Ferris JP. One-step, regioselective syntheses of RNA oligomers. J Am Chem Soc. 2006;128(27):8914–8919. [DOI] [PubMed] [Google Scholar]
- [13].Attwater J, Holliger P. In-ice evolution of RNA polymerase ribozyme activity. Nat Chem. 2014;6(10):774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vlassov AV, Kazakov SA, Johnston BH, et al. The RNA world on ice: a new scenario for the emergence of RNA information. J Mol Evol. 2005;61(2):264–273. [DOI] [PubMed] [Google Scholar]
- [15].Vaidya N, Walker SI. A biological evolution driven by natural selection on molecule-like matter. Nat Rev Chem. 2018;2(10):10–14. [Google Scholar]
- [16].Vasas V, Szathmáry E, Santos M, et al. Evolution before genes. Biol Direct. 2010;5(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fernando C, Rowe JE, Szathmáry E. Evolution of mutualistic cooperation from reciprocal altruism without individual recognition. Sci Rep. 2017;7(1):1–12. doi: 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Orgel LE. Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol. 2004;39(2):99–123. [DOI] [PubMed] [Google Scholar]
- [19].Lancet D, Zidovetzki R. Template-directed synthesis of oligoguanylates in a model prebiotic reaction. J Mol Evol. 1991;33(4):321–332.1774787 [Google Scholar]
- [20].Lohrmann R, Orgel LE, Robertson MP. Template-directed synthesis of adenine peptides. J Am Chem Soc. 1990;112(6):2435–2436. [Google Scholar]
- [21].Orgel LE. The origin of life: a review of facts and speculations. Trends Biochem Sci. 2004;29(7):429–435. [DOI] [PubMed] [Google Scholar]
- [22].Joyce GF. The antiquity of RNA-based evolution. Nature. 2002;418(6894):214–221. [DOI] [PubMed] [Google Scholar]
- [23].Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009;323(5918):1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58(10):465–523. [DOI] [PubMed] [Google Scholar]
- [25].Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409(6818):387–390. [DOI] [PubMed] [Google Scholar]
- [26].Lehman N. The emergence of life on earth: a historical and scientific overview. In: Gargaud M, Martin H, López-García P, and Montmerle T, editors. Lectures in astrobiology. Vol. 1. New Brunswick, NJ: Springer; 2003. p. 491–540. [Google Scholar]
- [27].Hordijk W, Steel M. Detecting autocatalytic, self-sustaining sets in chemical reaction systems. J Theor Biol. 2004;227(4):451–461. [DOI] [PubMed] [Google Scholar]
- [28].Hordijk W, Steel M. Autocatalytic sets and the origin of life. Entropy. 2008;10(2):196–214. [Google Scholar]
- [29].Lehman N. A recombination-based model for the origin and early evolution of genetic information. Chem Biodivers. 2015;12(12):1840–1848. [DOI] [PubMed] [Google Scholar]
- [30].Lehman NE, Kauffman SA. Constraint closure drove major transitions in the origins of life. Entropy (Basel). 2021;23(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Joyce GF. Directed evolution of nucleic acid enzymes. Annu Rev Biochem. 2004. July;73(1):791–836. doi: 10.1146/annurev.biochem.73.011303.073717 [DOI] [PubMed] [Google Scholar]
- [32].Xavier JC, Hordijk W, Kauffman S, et al. ”Autocatalytic chemical networks at the origin of metabolism”. In: Proceedings of the Royal Society B: Biological Sciences; 1923. 287. p. 20192377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ferris JP, Ertem G. Oligomerization of ribonucleotides on montmorillonite: reaction of the 5’-phosphorimidazolide of adenosine. Science. 1993;261(5126):1564–1566. [DOI] [PubMed] [Google Scholar]
- [34].Lehman N. The origins of life on earth. Scientia. 2003;1(1):65–79. [Google Scholar]
- [35].Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69(1):497–529. [DOI] [PubMed] [Google Scholar]
- [36].Paul N, Joyce GF. ”A self-replicating ligase ribozyme”. In: Proceedings of the National Academy of Sciences; 2002. 99(20): 12733–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kauffman S. The origins of order: self-organization and selection in evolution. Oxford, UK: Oxford University Press; 1993. [Google Scholar]
- [38].Eigen M, Schuster P. The hypercycle: a principle of natural self-organization. New York, NY: Springer; 1977. [DOI] [PubMed] [Google Scholar]
- [39].Smith J, Morowitz HJ. The origin and nature of life on earth: the emergence of the fourth geosphere. Cambridge, UK: Cambridge University Press; 2016. [Google Scholar]
- [40].Kauffman SA. Autocatalytic sets of proteins. J Theor Biol. 1986;119(1):1–24. [DOI] [PubMed] [Google Scholar]
- [41].Szostak JW, Ellington AD. In vitro selection of functional RNA sequences. Methods Enzymol. 2009;468:419–439. [Google Scholar]
- [42].Segre D, Ben-Eli D, Lancet D. Composing life. EMBO Rep. 2000;1(3):217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Szathmáry E, Gladkih I. Sub-exponential growth and coexistence of non-enzymatically replicating templates. J Theor Biol. 1989;138(1):55–58. [DOI] [PubMed] [Google Scholar]
- [44].Pross A. On the emergence of biological complexity: life as a kinetic state of matter. Origins Life Evol Biospheres. 2005;35(2):151–166. [DOI] [PubMed] [Google Scholar]
- [45].Pascal R, Pross A, Sutherland JD. Toward an evolutionary theory of the origin of life based on kinetics and thermodynamics. Open Biol. 2013;3(11):130156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lee DH, Granja JR. An autocatalytic peptide replicator. Nature. 2007;412(6844):564–567. [Google Scholar]
- [47].Adamala K, Szostak JW. Nonenzymatic template-directed RNA synthesis inside model protocells. Science. 2013;342(6162):1098–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Swinney HL, Wolynes PG. Theory of microphase separation in block copolymers. J Chem Phys. 2016;121(22):10814–10831. [Google Scholar]
- [49].Lehn JM. Supramolecular chemistry: concepts and perspectives. Weinheim, Germany: Wiley-VCH; 2002. [Google Scholar]
- [50].Monnard PA, Deamer DW. Membrane self-assembly processes: steps toward the first cellular life. Anatomical Rec. 2002;268(3):196–207. [DOI] [PubMed] [Google Scholar]
- [51].Joyce GF, Inoue T. A novel method for the rapid polymerization of RNA. Nucleic Acids Res. 1989;17(21):711–722. [PMC free article] [PubMed] [Google Scholar]
- [52].Kruger K, Grabowski PJ, Zaug AJ, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31(1):147–157. [DOI] [PubMed] [Google Scholar]
- [53].Milligan JF, Groebe DR, Witherell GW, et al. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15(21):8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Eckstein F, Gish G. Inhibition of the RNA polymerase-catalyzed elongation of RNA chains by oligonucleotide-directed RNA hydrolysis. Nucleic Acids Res. 1989;17(22):8309–8320. [Google Scholar]
- [55].Deck C, Jauker M, Richert C. Efficient enzyme-free copying of all four nucleobases templated by immobilized RNA. Nat Chem. 2011;3(8):603–608. [DOI] [PubMed] [Google Scholar]
- [56].Orgel LE. ”Self-organizing biochemical cycles”. In: Proceedings of the National Academy of Sciences; 2000. 97(23): 12503–12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261(5127):1411–1418. [DOI] [PubMed] [Google Scholar]
- [58].Ekland EH, Szostak JW, Bartel DP. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995;269(5224):364–370. [DOI] [PubMed] [Google Scholar]
- [59].Johnston WK, Unrau PJ, Lawrence MS, et al. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science. 2001;292(5520):1319–1325. [DOI] [PubMed] [Google Scholar]
- [60].Sievers D, von Kiedrowski G. Self-replication of complementary nucleotide-based oligomers. Nature. 1994;369:221–224. [DOI] [PubMed] [Google Scholar]
- [61].Szostak JW, Bartel DP. Directed evolution of RNA with catalytic activity. Trends Genet. 1993;9(2):69–75.8488562 [Google Scholar]
- [62].Walde P, Ichihashi N. Template-directed synthesis in a model protocell. Acc Chem Res. 2001;34(11):828–837. [Google Scholar]
- [63].Sutherland JD. The origin of life—out of the blue. Angew Chem Int Ed. 2016;55(4):104–121. [DOI] [PubMed] [Google Scholar]
- [64].Zhou C, Avins JL, Klauser PC, et al. Nonenzymatic template-directed RNA synthesis inside model protocells. Science. 2012;336(6079):341–344.22517858 [Google Scholar]
- [65].Breslow R, Cheng ZL. Evidence for an evolutionary path leading to the catalytic activity of ribonuclease. J Am Chem Soc. 2009;131(23):7492–7499.19435346 [Google Scholar]
- [66].Höcker B. Template-directed formation of peptide bonds by non-enzymatic reactions. Hel (Rome) Acta. 2006;89(3):474–484. [Google Scholar]
- [67].Mizuuchi R, Lehman N. Limited sequence diversity within a population supports prebiotic RNA reproduction. Life (Basel). 2019;9(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Jaeger L, Wright MC, Joyce GF. A complex ligase ribozyme evolved in vitro from a group I ribozyme domain. In: Proceedings of the National Academy of Sciences of the United States of America (PNAS); 1999. 96(26):14712–14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mutschler H, Wochner A, Holliger P. Freeze-thaw cycles as drivers of complex ribozyme assembly. Nat Chem. 2015;7(6):502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Vaidya N, Manapat ML, Chen IA, et al. Spontaneous network formation among cooperative RNA replicators. Nature. 2012;491(7422):72–77. [DOI] [PubMed] [Google Scholar]
- [71].Saha R, Yomo T. Construction of mutual catalytic network replicating ribosome ribozyme complex for high yield of RNA replication. ACS Synth Biol. 2017;6(8):1539–1548. [Google Scholar]
- [72].Bares JM, Belcher AM, Epstein IR. Complex networks in chemical kinetics. PNAS. 1997;94(22):11709–11715. [Google Scholar]
- [73].Wakamoto Y, Maniwa F, Shingaki R, et al. Single-cell-cooperative regulation of mutually exclusive cell fates in Bacillus subtilis colonies. J Bacteriol. 2009;191(13):4441–4444.19376854 [Google Scholar]
- [74].Kaneko K. Prebiotic self-organization processes as dynamic phase transitions of molecular systems. Top Curr Chem. 2006;259:71–111. [Google Scholar]
- [75].Ameta S, Kumar M, Chakraborty N, et al. Multispecies autocatalytic RNA reaction networks in coacervates. Commun Chem. 2023;6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Lu H, Blokhuis A, Turk-MacLeod R, et al. Small-molecule autocatalysis drives compartment growth, competition and reproduction. Nat Chem. 2024;16(1):70–78. [DOI] [PubMed] [Google Scholar]
- [77].Smail BA, Clifton BE, Mizuuchi R, et al. Spontaneous advent of genetic diversity in RNA populations through multiple recombination mechanisms. RNA. 2019;25(4):453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mathis C, Ramprasad SN, Walker SI, et al. Prebiotic RNA network formation: a taxonomy of molecular cooperation. Life (Basel). 2017;7(4):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mutschler H, Holliger P. Non-canonical 3′-5′ extension of RNA with prebiotically plausible ribonucleoside 2′,3′-cyclic phosphates. J Am Chem Soc (JACS). 2014;136(14):5193–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Mizuuchi R, Blokhuis A, Vincent L, et al. Mineral surfaces select for longer RNA molecules. Chem Commun (ChemComm). 2019;55(14):2090–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yeates JAM, Nghe P, Lehman N. Topological and thermodynamic factors that influence the evolution of small networks of catalytic RNA species. RNA. 2017;23(7):1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hordijk W, Steel M. A formal model of autocatalytic sets emerging in an RNA replicator system. J Syst Chem. 2012;4(1):4. Previously 120. doi: 10.1186/1759-2208-4-3 [DOI] [Google Scholar]
- [83].Horning DP, Joyce GF. ”Amplification of RNA by an RNA polymerase ribozyme”. In: Proceedings of the National Academy of Sciences; 2016. 113(34): 9786–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Higgs PG. The origin of short RNA template-directed synthesis of RNA. IUBMB Life. 2009;61(2):99–109.19117371 [Google Scholar]
- [85].Ma W, Yu C. Autocatalytic sets in the RNA world. J Mol Evol. 2018;86(11–12):725–737. [Google Scholar]
- [86].Plasson R, Bersini H. An evolutionary transition towards primitive multicellularity could have increased the maximal length of genes. J Theor Biol. 2015;382:370–379. [Google Scholar]
- [87].Czárán T, Szathmáry E. Coexistence of replicators in prebiotic evolution: a network model. Artif Life. 2002;8(1):25–37.12020420 [Google Scholar]
- [88].Scott WG, Finch JT, Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995;81(7):991–1002. [DOI] [PubMed] [Google Scholar]
- [89].Cech TR, Zaug AJ. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1986;44(2):213–223. [DOI] [PubMed] [Google Scholar]
- [90].Noller HF. RNA structure: reading the ribosome. Science. 2005;309(5740):1508–1514. [DOI] [PubMed] [Google Scholar]
- [91].Steitz TA, Moore PB. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem Sci. 2003;28(8):411–418. [DOI] [PubMed] [Google Scholar]
- [92].Cech TR. The RNA worlds in context. Cold Spring Harb Perspect Biol. 2012;4(7):a006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7(4):499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Staley JP, Woolford JL Jr. Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol. 2009;21(1):109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Beaudry AA, Joyce GF. Directed evolution of an RNA enzyme. Science. 1992;257(5075):635–641. [DOI] [PubMed] [Google Scholar]
- [96].Green AA, Silver PA, Collins JJ, et al. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159(4):925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Robertson MP, Ellington AD. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat Biotechnol. 1999;17(1):62–66. [DOI] [PubMed] [Google Scholar]
- [98].Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. [DOI] [PubMed] [Google Scholar]
- [99].Win MN, Smolke CD. ”A modular and extensible RNA-based gene-regulatory platform for engineering cellular function”. In: Proceedings of the National Academy of Sciences; 2007. 104(36): 14283–14288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Doudna JA, Couture S, Szostak JW. A multisubunit ribozyme that is a catalyst of and template for complementary strand RNA synthesis. Science. 1991;251(5001):1605–1608. [DOI] [PubMed] [Google Scholar]
- [101].Ekland EH, Bartel DP. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature. 1996;382(6592):373–376. [DOI] [PubMed] [Google Scholar]
- [102].Attwater J, Wochner A, Pinheiro VB, et al. Ice as a protocellular medium for RNA replication. Nat Commun. 2013;4:2627. [DOI] [PubMed] [Google Scholar]
- [103].Attwater J, Raguram A, Morgunov AS, et al. Ribozyme-catalysed RNA synthesis using triplet building blocks. Elife. 2018;7:e35255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Attwater J, Wochner A, Pinheiro VB, et al. Ice as a protocellular medium for RNA replication. Nat Commun. 2010;1(1):1–6. [DOI] [PubMed] [Google Scholar]
- [105].Robertson MP, Ellington AD. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat Biotechnol. 2000;17(1):62–66. [DOI] [PubMed] [Google Scholar]
- [106].Lorsch JR, Szostak JW. In vitro evolution of new ribozymes with polynucleotide kinase activity. Nature. 1994;371(6495):31–36. [DOI] [PubMed] [Google Scholar]
- [107].Zaher HS, Unrau PJ. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13(7):1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Tjhung KF, Shokhirev MN, Horning DP, et al. ”An RNA polymerase ribozyme that synthesizes its own ancestor”. In: Proceedings of the National Academy of Sciences of the United States of America (PNAS); 2020. 117(6):2906–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Pressman A, Blanco C, Chen IA. The RNA world as a model system to study the origin of life. Curr Biol. 2015;25(19):R953–R963. [DOI] [PubMed] [Google Scholar]
- [110].Wochner A, Attwater J, Coulson A, et al. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011;332(6026):209–212. [DOI] [PubMed] [Google Scholar]
- [111].Branscomb E, Russell MJ. Turnstiles and bifurcators: the disequilibrium converting engines that put metabolism on the road. Biochim et Biophys Acta (BBA) - Bioenerg. 2013;1827(2):62–78. [DOI] [PubMed] [Google Scholar]
- [112].Hayden EJ, Lehman N. Self-assembly of a group I intron from inactive oligonucleotide fragments. Chem Biol. 2006;13(9):909–918. doi: 10.1016/j.chembiol.2006.06.014 [DOI] [PubMed] [Google Scholar]
- [113].Ashkenasy G, Jagasia R, Yadav M. Design of molecular and supramolecular systems: challenges, advances, and opportunities. Chem Rev. 2017;117(3):13608–13941. [Google Scholar]
- [114].Ganti T. The principles of life. Oxford, UK: Oxford University Press; 2003. [Google Scholar]
- [115].Bagley RJ, Farmer JD, Kauffman SA, et al. Adaptation, coevolution, and the emergence of complex form. Artif Life. 1989;1(2):129–159. [Google Scholar]
- [116].Shapiro R. Origins: a skeptic’s guide to the creation of life on earth. Manila, Philippines: Summit Books; 1986. [Google Scholar]
- [117].Szostak JW. The eightfold path to non-enzymatic RNA replication. J Syst Chem. 2012;3(1):2. doi: 10.1186/1759-2208-3-2 [DOI] [Google Scholar]
- [118].Luisi PL. The emergence of life: from chemical origins to synthetic biology. Cambridge, UK: Cambridge University Press; 2006. [Google Scholar]
- [119].Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418(6894):222–228. doi: 10.1038/418222a [DOI] [PubMed] [Google Scholar]
- [120].Vasas V, Fernando C, Santos M, et al. Evolution before genes. Biol Direct. 2012;7(1):1–24. doi: 10.1186/1745-6150-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Stewart JE. The origins of life: the managed-metabolism hypothesis. Found Sci. 2019;24:171–195. [Google Scholar]
- [122].Deamer D. The role of lipid membranes in life’s origin. Life. 2017;7(4):52. doi: 10.3390/life7010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Pascal R, Pross A. Stability and its manifestation in the chemical and biological worlds. Nat Rev Chem. 2015;1(1):1–9. [DOI] [PubMed] [Google Scholar]
- [124].Takeuchi N, Hogeweg P, Kaneko K. The origin of cells: from individual to collective entities. In: Proceedings of the National Academy of Sciences; 2011. 108(21): 8693–8698. [Google Scholar]
- [125].Wagner A. Robustness and evolvability: a paradox resolved. In: Proceedings of the Royal Society B: Biological Sciences; 2008. 275(1630): 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Poelwijk FJ, Kiviet DJ, Weinreich DM, et al. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445(7126):383–386. doi: 10.1038/nature05451 [DOI] [PubMed] [Google Scholar]
- [127].Khan AI, Dinh DM, Schneider D, et al. Exploring fitness landscapes by directed evolution. Physica A: Stat Mech Appl. 2011;390(21–22):4382–4403. [Google Scholar]
- [128].Lunzer M, Golding GB, Dean AM, et al. How fast is protein evolution? Science. 2005;275(5308):1326–1330. [Google Scholar]
- [129].Weinreich DM, Delaney NF, Depristo MA, et al. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312(5770):111–114. doi: 10.1126/science.1123539 [DOI] [PubMed] [Google Scholar]
- [130].Vetsigian K, Woese C, Goldenfeld N. Collective evolution and the genetic code. In: Proceedings of the National Academy of Sciences; 2006. 103(28): 10696–10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Takeuchi N, Hogeweg P, Kaneko K. Recombinational repair dominates the repair of random damage in the DNA sequences of genes encoding highly conserved proteins. J Mol Evol. 2005;60(5):673–682. [Google Scholar]
- [132].Blokhuis A, Lacoste D, Nghe P. Mutualistic polymerase ribozes. J R Soc Interface. 2016;13(116):20160554.27733700 [Google Scholar]
- [133].Mansy SS, Szostak JW. Reconstructing the emergence of cellular life through the synthesis of model protocells. Cold Spring Harb Symp Quant Biol. 2008;74:47–54. doi: 10.1101/sqb.2009.74.014 [DOI] [PubMed] [Google Scholar]
- [134].Leontis NB, Lehman N, editors. RNA 3D structure analysis and prediction. Vol. 27. Berlin, Germany: Springer Science & Business Media; 2010. [Google Scholar]
- [135].Hordijk W, Steel M. Autocatalytic sets and the origin of life. Entropy. 2017;19(5):259. [Google Scholar]
- [136].Rasmussen S, Chen L, Deamer D, et al. Transitions from nonliving to living matter. Science. 2004;303(5660):963–965. doi: 10.1126/science.1093669 [DOI] [PubMed] [Google Scholar]
- [137].Pohorille A, Deamer D, editors. Prebiotic chemistry: from simple amphiphiles to protocell models. Boca Raton, FL: CRC Press; 2009. [Google Scholar]
- [138].Ruiz-Mirazo K, Briones C, de la Escosura A. Prebiotic systems chemistry: new perspectives for the origins of life. Chem Rev. 2014;114(1):285–366. doi: 10.1021/cr2004844 [DOI] [PubMed] [Google Scholar]
- [139].Crick FH. Central dogma of molecular biology. Nature. 1970;227(5258):561–563. [DOI] [PubMed] [Google Scholar]
- [140].Ichihashi N, Usui K, Kazuta Y, et al. Darwinian evolution in a translation-coupled RNA replication system within a cell-like compartment. Nat Commun. 2013;4(1):2494. doi: 10.1038/ncomms3494 [DOI] [PubMed] [Google Scholar]
- [141].Poudyal RR, Hud NV, Moroz OV. Short peptides self-assemble to produce catalytic amyloids. Nat Chem. 2014;6(4):303–309. doi: 10.1038/nchem.1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Murray JB, Seyhan AA, Walter NG, et al. Scottie: a database of semantically annotated ribozymes. Nucleic Acids Res. 1998;26(1):347–349. [Google Scholar]
- [143].Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Cold Spring Harb Perspect Biol. 2010;2(7):a005348. [Google Scholar]
- [144].Martin W, Russell MJ, Allen JF, et al. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Phil Trans R Soc B: Biol Sci. 2007;362(1486):59–85. doi: 10.1098/rstb.2002.1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Woese CR. The genetic code: the molecular basis for genetic expression. New york, NY: Harper & Row; 1967. [Google Scholar]
- [146].Baum DA, Smith SD. Tree thinking: an introduction to phylogenetic biology. Greenwood Village, CO: Roberts and Company Publishers; 2013. [Google Scholar]
- [147].Arsène S, Ameta S, Lehman N, et al. Coupled catabolism and anabolism in autocatalytic RNA sets. Nucleic Acids Res. 2018;18:9660–9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Sievers D, von Kiedrowski G. Self-replication of complementary nucleotide-based oligomers. Nature. 1994;369(6477):221–224. [DOI] [PubMed] [Google Scholar]
- [149].Lehman N, Hayden EJ, Um JW. A synthetic azoarcus ribozyme autaposes RNA substrates without protein cofactors. RNA. 2018;24(1):36–43. [Google Scholar]
- [150].Lu H, Blokhuis A, Turk-MacLeod R, et al. Small-molecule autocatalysis drives compartment growth, competition and reproduction. Nat Chem. 2023;16(1):70–78. [DOI] [PubMed] [Google Scholar]
- [151].Rajamani S, Ichida JK, Antal T, et al. Effect of stalling after mismatches on the error catastrophe in nonenzymatic nucleic acid replication. J Am Chem Soc. 2008;130(7):2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Monnard PA, Szostak JW. Metal-ion catalyzed polymerization in the eutectic phase in water–ice: a possible approach to template-directed RNA polymerization. J Inorg Biochem. 2008;102(5–6):1104–1111. doi: 10.1016/j.jinorgbio.2008.01.026 [DOI] [PubMed] [Google Scholar]
- [153].Ricardo A, Carrigan MA, Olcott AN, et al. Borate minerals stabilize ribose. Science. 2004;303(5655):196. doi: 10.1126/science.1092464 [DOI] [PubMed] [Google Scholar]
- [154].Joyce GF, Szostak JW. Protocells and RNA self-replication. Cold Spring Harb Perspect Biol. 2018;10(7):a034801. doi: 10.1101/cshperspect.a034801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Hazen RM, Sverjensky DA. Mineral surfaces, geochemical complexities, and the origins of life. Cold Spring Harb Perspect Biol. 2010;2(5):a002162. doi: 10.1101/cshperspect.a002162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Lambert JF, Brack A. Molecular evolution and the rise of complexity. Naturwissenschaften. 2017;104(3–4):89.28993880 [Google Scholar]
- [157].Woo H-J, Satya RV, Reifman J. Thermodynamic basis for the emergence of genomes during prebiotic evolution. PLoS Comput Biol. 2012;81:e1002534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Cody GD, Boctor NZ, Filley TR, et al. Primordial carbonylated iron-sulfur compounds and the synthesis of pyruvate. Science. 2000;289(5483):1337–1340. doi: 10.1126/science.289.5483.1337 [DOI] [PubMed] [Google Scholar]
- [159].Ferris JP. Caralysis and prebiotic RNA synthesis. Orig Life Evol Biosph. 1993;23(5–6):307–315. [DOI] [PubMed] [Google Scholar]
- [160].Brack A, Borruat FX. Mineral-induced formation of RNA oligomers: the “chimney” hypothesis. Origins Life Evol Biosphere. 1996;26(1–3):59–69. [Google Scholar]
- [161].Yarus M, Widmann JJ, Beintema JJ. Papaya (carica papaya) lysozyme is a member of the family 19 (basic, class II) chitinases. J Mol Evol. 1999;49(6):819–828. doi: 10.1007/pl00000075 [DOI] [PubMed] [Google Scholar]
- [162].Powner MW, Gerland B, Sutherland JD. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature. 2009;459(7244):239–242. doi: 10.1038/nature08013 [DOI] [PubMed] [Google Scholar]
- [163].Hanczyc MM, Fujikawa SM, Szostak JW. Experimental models of primitive cellular compartments: encapsulation, growth, and division. Science. 2003;302(5645):618–622. doi: 10.1126/science.1089904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Ranjan S, Sasselov DD. Influence of the UV environment on the synthesis of prebiotic molecules. Astrobiology. 2016;16(1):68–88. doi: 10.1089/ast.2015.1359 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


