Abstract
Objectives
This study aims to analyze the results of comprehensive genetic testing in patients presenting to a dedicated multidisciplinary inherited heart disease clinic in India.
Methods
All patients presenting to our clinic from August 2017 to October 2023 with a suspected inherited heart disease and consenting for genetic testing were included. The probands were grouped into familial cardiomyopathies namely hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), arrhythmogenic cardiomyopathy (ACM) and peripartum cardiomyopathy (PPCM), channelopathies namely congenital long QT syndrome (LQTS) and Brugada syndrome (BrS), and heritable connective tissue disorder namely Marfan Syndrome (MFS). Next generation sequencing (NGS) was used, and pre-test and post-test counseling were provided to probands and cascade screening offered to relatives.
Results
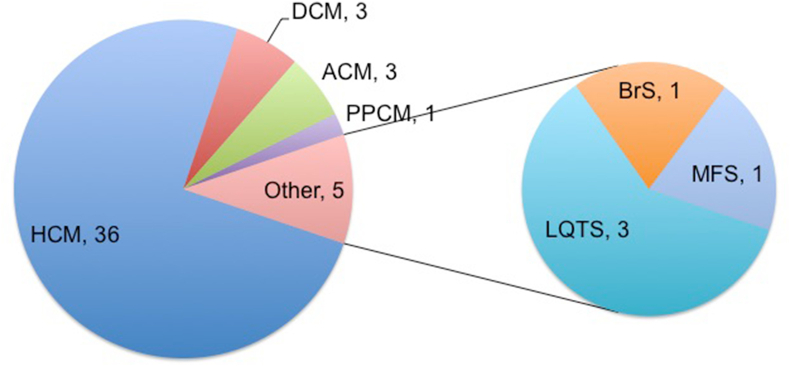
Mean age of the subjects (n = 77; 48 probands, 29 relatives) was 43 ± 18 years, 68 % male and 44 % symptomatic, with 36 HCM, 3 DCM, 3 ACM, 1 PPCM, 3 LQTS, 1 BrS and 1 MFS probands. The diagnostic yield of NGS-based genetic testing was 31 %; variants of uncertain significance (VUS) were identified in 54 %; and 15 % were genotype-negative. Twenty-nine relatives from 18 families with HCM (n = 12), DCM (n = 3), ACM (n = 2) and MFS (n = 1) underwent genetic testing. The genotype positive probands/relatives and VUS carriers with strong disease phenotype and/or high risk variant were advised periodic follow-up; the remaining probands/relatives were discharged from further clinical surveillance.
Conclusions
Genetic testing guides treatment and follow-up of patients with inherited heart diseases and should be carried out in dedicated multidisciplinary clinics with expertise for counseling and cascade screening of family members.
Keywords: Genetic testing, Cardiomyopathies, Channelopathies, Multidisciplinary clinic, Sudden cardiac death prevention
1. Introduction
Inherited heart diseases, the leading cause of unexplained sudden cardiac death (SCD) in young and apparently healthy individuals, are grouped mainly into familial cardiomyopathies or genetic disorders of the myocardium and channelopathies or inherited arrhythmias caused by cardiac ion channel mutations. As individuals with inherited heart diseases are known to present with a wide clinical spectrum ranging from asymptomatic to life-threatening arrhythmias, heart failure, sudden cardiac arrest and SCD, it is imperative to diagnose, risk stratify and treat them in a timely and efficient manner. The recent advances in human genomics and cardiovascular genetics have helped unravel the causative genes for several of these conditions and have also paved the way for the development of a new and exciting field of medicine termed ‘Cardiogenetics’.1, 2, 3
The role of a dedicated multidisciplinary inherited heart disease clinic with expertise in a multimodal diagnostic and therapeutic approach as well as a shared decision making model to optimize patient care, cannot be overemphasized.4 Despite some noteworthy attempts in the right direction, there is a severe paucity of these centers of excellence in India and other developing countries, where the disease load is high but comprehensive care is lacking.5, 6, 7, 8 The dawn of the era of next generation sequencing (NGS) has undoubtedly revolutionized the scope of genetic testing in individuals and families with inherited heart diseases but the awareness and application of this diagnostic tool in the clinical setting is poor. Despite genetic testing becoming more accessible and affordable in the recent past, it is vastly underutilized in our country and this situation needs to change.9,10
The aim of this study is to analyze the results of comprehensive genetic testing in patients presenting to a dedicated inherited heart disease clinic in India.
2. Methods
2.1. Study cohort
All patients presenting to our clinic from August 2017 to October 2023 with a suspected inherited heart disease and consenting for genetic testing, or referred to us for post-test genetic counseling and management, were included in the study. Patients with symptoms and signs suggestive of inherited heart diseases were referred to our clinic for comprehensive care by cardiologists and other physicians from within the tertiary care hospital housing the clinic or from other centers. In the earlier study period (2017–2021), first-degree relatives were included for genetic testing along with the probands; whereas in the latter study period (2022–2023), targeted genetic testing was offered for relatives of probands harboring pathogenic and likely pathogenic (P/LP) mutations only.
Patients referred to us after the clinical evaluation and genetic test were completed, were provided with post-test genetic counseling and a guideline-based management plan and the care was transferred back to the referring physician, or tertiary care was provided at our center, as appropriate, including implantable cardioverter defibrillator (ICD) therapy where indicated.
2.2. Evaluation protocol
A multidisciplinary cardiogenetic team consisting of a cardiologist, physician coordinator, clinical geneticist, genetic counselor and cardiac nurse was responsible for managing the patients presenting to the clinic. The clinical diagnostic process included a detailed clinical history, a three generational family history, electrocardiogram (ECG) and echocardiogram in all patients and exercise stress test, Holter monitoring and cardiac magnetic resonance imaging (MRI), where applicable. A discussion was held with the referring physician and past medical records reviewed, where applicable. The relevant clinical and genetic reports of affected family members were documented, when available. The probands were grouped into familial cardiomyopathies namely hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), arrhythmogenic cardiomyopathy (ACM) and peripartum cardiomyopathy (PPCM), channelopathies namely congenital long QT syndrome (LQTS), Brugada syndrome (BrS) and heritable connective tissue disorders such as Marfan Syndrome (MFS).
2.3. Genetic counseling and testing
After providing pre-test genetic counseling and obtaining written informed consent, genetic testing was performed using genomic DNA isolated from whole blood samples. Next generation sequencing (NGS) based targeted gene panel (TGP) was used with a backdrop of whole exome sequencing and ACMG criteria were applied for interpretation of results in the proband. The diagnostic yield of genetic testing was calculated by the percentage of pathogenic and likely pathogenic (P/LP) variants in the probands. Individuals with more than one plausible variant were documented as multiple variant carriers. ClinVar, Insilico Prediction tools and population databases were used for comprehensive cataloguing of genetic variants, particularly the variants of uncertain significance (VUS).
Once the genetic test result of the proband became available, an interdisciplinary discussion was held to discuss the findings, post-test counseling was provided to the proband and cascade screening offered to the relatives where applicable.
3. Results
There were 77 subjects who underwent genetic testing. The characteristics of the subjects and the outcomes of the genetic testing are described below.
3.1. Demographic and clinical characteristics
Age of the subjects (n = 77) was 43 ± 18 years (range 5 months–76 years) and 68 % were male (Table 1). Probands (n = 48) presented with cardiovascular symptoms (44 %), family history of young SCD (2 %), syndromic features with cardiovascular manifestation (2 %) or were identified incidentally (52 %). The presenting symptoms in the probands are described in Table 2. Syncope, pre-syncope and dyspnea were the commonest symptoms; the most pathognomonic symptom was considered in individuals with multiple symptoms. Fig. 1 shows the distribution of disease phenotypes in the probands. HCM was the commonest amongst the cardiomyopathies and LQTS amongst the channelopathies. Guideline-based management was provided for all the probands based on the phenotype.
Table 1.
Demographic and clinical characteristics of the study subjects.
| Characteristic | All subjects n = 77 |
Probands n = 48 |
Relatives n = 29 |
|---|---|---|---|
| Age, years (mean ± SD) | 43 ± 18 | 45 ± 17 | 40 ± 19 |
| Male, n (%) | 52 (68) | 36 (75) | 16 (55) |
| Symptomatic, n (%) | 21 (27) | 21 (44) | – |
| Positive family history, n (%) | 8 (10) | 8 (17) | – |
| Known with co-morbidities, n (%) | 15 (19) | 15 (31) | – |
| Cardiomyopathy, n (%) | 69 (90) | 43 (90) | 26 (90) |
| Channelopathy, n (%) | 4 (5) | 4 (8) | 0 |
| Other, n (%) | 4 (5) | 1 (2) | 3 (10) |
Table 2.
Presenting symptoms in the probands.
| Symptom | Symptomatic probands (n = 21) |
|---|---|
| (Pre) Syncope | 6 |
| Dyspnea | 6 |
| Palpitations | 4 |
| Chest pain | 2 |
| Stroke | 1 |
| Sudden cardiac arrest | 1 |
| Fetal bradycardia | 1 |
Fig. 1.
Distribution of diseases in the probands.
3.2. Genetic characteristics
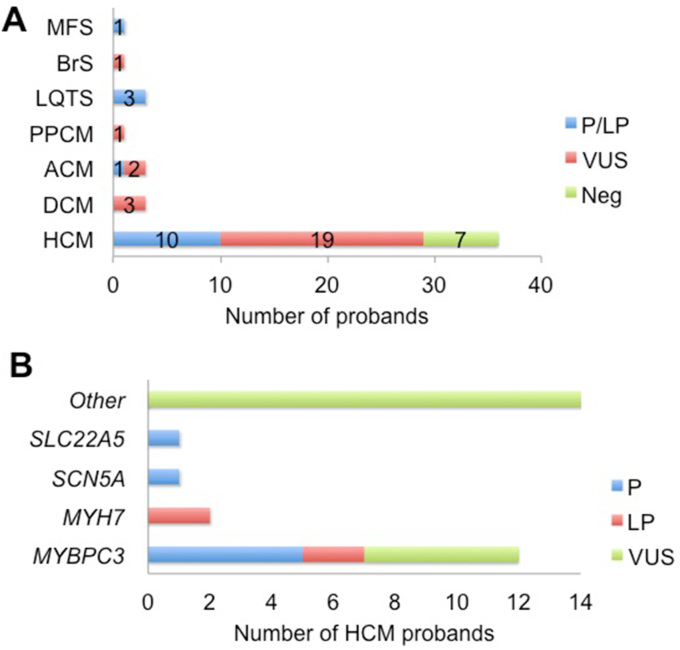
The diagnostic yield (P/LP variants) of genetic testing in the probands was 31 %; VUS were identified in 54 % of probands; and 15 % were genotype-negative. The P/LP variants are described in detail in Table 3. Interestingly, 50 % of all P/LP mutations and 47 % of VUS were novel. Table 4 provides the details of VUS identified in probands along with their clinical details and segregation in family members. Neither a positive family history nor the presence of symptoms was associated with a significantly higher yield. Almost half (49 %) of the probands with P/LP/VUS variants had multiple variants. Fig. 2 Panel A shows the results of genetic testing per disease group in the probands, considering the highest ranked variant in those with multiple variants.
Table 3.
Details of P/LP variants detected in the probands.
| S.No | Age | Sex | Clinical findings | Gene | Variant | Protein | Inheritance/Zygosity | Class | Segregation | Reported in literature | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 16 | M | HCM | MYH7 | c.2221G > T | p.Gly741Trp | AD | P | Present in asymptomatic father | Yes | |
| Heterozygous | |||||||||||
| 2 | 17 | M | HCM | MYH7 | c.2198G > A | p.Gly733Glu | AD | LP | Present in asymptomatic mother and absent in asymptomatic father and sister | Yes | |
| Heterozygous | |||||||||||
| 3 | 33 | M | HOCM | MYBPC3 | c.2905+1G > A | Intronic | AD | P | Not detected in asymptomatic mother | Yes | |
| Heterozygous | |||||||||||
| 4 | 40 | M | Chest pain, | SCN5A | c.5963G > A | p.Arg1898His | AD | P | NA | Yes | |
| Heterozygous | |||||||||||
| FH, HCM | MYBPC3 | c.1224-19G > A | Intronic | AD | LP | ||||||
| Heterozygous | |||||||||||
| 5 | 42 | F | Chest pain | MYBPC3 | c.1021_1028del | p.Gly341Ter | AD | P | Present in asymptomatic sister | This study | |
| FH, HCM | Heterozygous | ||||||||||
| 6 | 45 | M | HCM | MYBPC3 | c.3450_3451del | p.Arg1150SerfsTer18 | AD | P | NA | This study | |
| Heterozygous | |||||||||||
| 7 | 48 | M | Presyncope, | MYBPC3 | c.3372C > A | p.Cys1124Ter | AD | P | Present in 2 asymptomatic children | Yes | |
| HCM | Heterozygous | ||||||||||
| 8 | 48 | M | Chest pain, | MYBPC3 | c.3009_3010del | p.Gln1004GlyfsTer46 | AD | P | NA | This study | |
| HCM | Heterozygous | ||||||||||
| 9 | 52 | M | HCM | MYBPC3 | c.1021_1028del | p.Gly341Ter | AD | P | NA | This study | |
| Heterozygous | |||||||||||
| 10 | 67 | M | HCM | SLC22A5 | c.248G > T | p.Arg83Leu | AR | P | NA | Yes | |
| Heterozygous | |||||||||||
| 11 | 17 | M | Syncope, sports-related SCD | PKP2 | c.235C > T | p.Arg79Ter | AD | P | Present in asymptomatic sister and mother, absent in asymptomatic father | Yes | |
| Heterozygous | |||||||||||
| 12 | 5 months | F | Fetal bradycardia, prolonged QTc, LQTS | KCNQ1 | c.477+1G > A | Intronic | AR | P | NA | Yes | |
| Homozygous | |||||||||||
| 13 | 9 | M | Mildly prolonged QTc, SCD of father | SCN5A | c.5350G > A | p.Glu1784Lys | AD | P | NA | Yes | |
| Heterozygous | |||||||||||
| 14 | 48 | F | Syncope, prolonged QTc, SCD of son | SCN5A | c.3992C > T | p.Pro1331Leu | AD | P | NA | Yes | |
| Heterozygous | |||||||||||
| 15 | 45 | M | Marfanoid features, FH of aortic dissection | FBN1 | c.2113G > T | p.Ala705Ser | AD | LP | LP | This study | |
AD - autosomal dominant, FH – family history, HCM - hypertrophic cardiomyopathy, HOCM – hypertrophic obstructive cardiomyopathy, LP - likely pathogenic, LQTS - congenital long QT syndrome, NA - not available, P - pathogenic, SCD - sudden cardiac death.
Table 4.
Variants of Uncertain Significance (VUS) identified in the probands.
| S. No | Age | Sex | Clinical findings | Gene | Variant | In silico damaging Predictionsa | Percentage allele frequency in gnomADb; TOPMedb | Clinvar/ PMID |
Segregation |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 50 | F | HCM, AF, HTN, T2DM | CRYAB | p.Arg157Cys | SIFT-D; PP2-PrD; LRT-D; FATHMM-D | 0.002; 0.0007 | Clinvar: 41925 (VUS) | NA |
| 2 | 52 | M | HCM, MR and PAH | TNNI3K | p.Val570Phe | SIFT-D; PP2-PrD; LRT-D | 0.0004; Absent | NA | Absent in asymptomatic mother |
| BMPR2 | p.Gly882Asp | PP2-PoD; FATHMM-D; | 0.008; 0.0003 | NA | BMPR2 and OBSCN variants were HET in asymptomatic mother | ||||
| OBSCN | p.Cys4479TrpfsTer38 | NA | 0.037; 0.027 | NA | |||||
| 3 | 30 | F | Hypertrophic obstructive cardiomyopathy | MYBPC3 | c.3628-41_3628-17del; p.? | NA | 0.401; 0.023 | Disease riskc; Clinvar: 177677 (Conflicting classifications of pathogenicity; B, VUS, P) | HET in asymptomatic father and daughter (she is under follow-up) |
| 4 | 31 | M | HCM | RYR2 | p.Val1533Met | PP2-PrD | Absent | NA | HET in the similarly affected brother (he is under follow-up) |
| 5 | 68 | M | HCM, T2DM, HTN, acute pulmonary edema | HCN4 | p.Thr770Met | SIFT_D | 0.002; 0.0007 | NA | NA |
| MYBPC3 | p.Arg1267Pro | SIFT_D | Absent | NA | |||||
| 6 | 66 | F | HCM, CAD, HTN | FLNC | p.Leu1690Val | SIFT-D; LRT-D; FATHMM-D | 0.002; 0.0003 | NA | NA |
| RBM20 | p.Arg513Trp | SIFT-D; FATHMM-D | 0.002; 0.0003 | NA | |||||
| 7 | 34 | M | HCM, mild aortic dilatation, dyslipidemia | MYBPC3 | p.Ser478Leu | SIFT-D | 0.0004; 0.0003 | NA | HET in asymptomatic mother; Absent in asymptomatic father |
| 8 | 35 | M | HCM | RBM20 | p.Glu1026Lys | FATHMM-D | 0.003; 0.002 | Clinvar: 238550 (conflicting classifications of pathogenicity, B, VUS) | Both the variants were HET in asymptomatic father and not detected in asymptomatic mother |
| TTN | p.Leu2306Phe | NA | Absent | NA | |||||
| 9 | 65 | F | HCM, HTN, dyslipidemia | MYBPC3 | p.Arg1267Pro | SIFT-D | Absent | NA | NA |
| NEXN | p.Arg306Ser | LRT-D | 0.0004; Absent | NA | |||||
| 10 | 70 | M | HCM | ILK | p.Arg56Trp | SIFT-D; LRT-D | 0.004; 0.0007 | NA | NA |
| JUP | p.Arg84Gln | SIFT-D; PP2-PrD; LRT-D; | 0.002; 0.0003 | NA | |||||
| 11 | 58 | M | HCM, HTN, T2DM | TTN | p.Thr33486Ser | NA | Absent | NA | NA |
| 12 | 55 | M | HCM, T2DM, HTN, dyslipidemia | CBS | p.Gly151Arge | SIFT-D; PP2-PrD; LRT-D; FATHMM-D | 0.001; 0.002 | Clinvar: 863694 (Pathogenic for homocystinuria)d | NA (proband under follow-up) |
| 13 | 66 | M | HCM | VCL | p.Ser434Tyr | SIFT-D; PP2-PrD; LRT-D | 0.001; 0.002 | Clinvar: 964868 (VUS) | |
| 14 | 56 | M | Extreme ventricular hypertrophy, progressive AV conduction delay | MYBPC3 | p.Leu1236Thr | SIFT_D | Absent; 0.0003 | NA | NA |
| 15 | 52 | M | HCM, nsVT | XK | p.Val11Leuf | LRT-D | 0.002; Absent | NA | NA |
| 16 | 38 | F | HCM, family history of young SCD | BMPR2 | p.Asn565dup | NA | Absent | NA | NA |
| 17 | 60 | M | HCM, T2DM | MYPN | p.Pro1102Leu | SIFT-D; PP2-PrD; LRT-D | 0.004; 0.002 | Clinvar: 520383; (VUS) | NA |
| 18 | 60 | M | HCM, occasional tachycardia | FLNC | p.Gly1760Ser | FATHMM-D | 0.011; 0.009 | Clinvar: | NA |
| 472098 (conflicting classifications of pathogenicity, B, VUS); PMID: 30411535 | |||||||||
| 19 | 50 | M | Syncope, Type 1 Brugada ECG, SCD of sister | HCN4 | p.Pro202Thr | SIFT-D | Absent | Clinvar: | NA |
| 2098283 (VUS) | |||||||||
| 20 | 31 | M | DCM, severe biventricular dysfunction | LMNA | p.Arg331Trpe | SIFT-D; LRT-D; FATHMM-D | 0.001; 0.0003 | ClinVar: 245682 (Conflicting classifications of pathogenicity VUS, LP); | Both the variants were HET in asymptomatic father |
| PMID: 28790152 | |||||||||
| TNNT2 | p.Arg154Trp | SIFT-D; PP2-PrD; LRT-D; FATHMM-D | 0.003; 0.0007 | Clinvar: 132943 (Conflicting classifications of pathogenicity VUS, LP); | |||||
| PMID: 24992688 | |||||||||
| 21 | 33 | M | DCM, severe biventricular dysfunction | TNNT2 | p.Glu262Lys | SIFT-D; FATHMM-D | Absent | Clinvar: 178850 (conflicting classifications of pathogenicity, VUS, LP) | NA |
| 22 | 24 | F | DCM, dyspnea, fatigue, pedal edema, palpitations | NEXN | p.Glu421Ter | NA | Absent | NA | This variant has been reclassified as LP during current re-analysis |
| MYH7 | p.Arg1925Cys | SIFT-D; PP2-PrD | Absent | Clinvar: | HET in affected cousin | ||||
| 1037017 (conflicting classifications of pathogenicity, VUS, LP) PMID:27532257 | (proband is lost to follow-up) | ||||||||
| 23 | 59 | F | ACM, HTN, recurrent VT | KCNH2 | p.Thr760Ile | SIFT-D; LRT-D; FATHMM-D | Absent | NA | NA |
| MYLK2 | p.Gln533Arg | SIFT-D | 0.002; Absent | NA | |||||
| 24 | 42 | M | ACM, cardioembolic stroke, heart failure, recurrent VT | DSP | p.Ala1505Thr | SIFT-BN | Absent | Clinvar: | NA |
| 1171054 (VUS) | |||||||||
| DOLK | p.Cys426Tyr | SIFT-D, LRT-D; FATHMM-D | Absent | NA | NA | ||||
| (proband under follow-up) | |||||||||
| 25 | 29 | F | PPCM, dyspnea, heart failure | SCN5A | p.Ser309Asn | FATHMM-D | Absent | Clinvar: | NA |
| 3073777(VUS) |
ACM - arrhythmogenic cardiomyopathy, AF - atrial fibrillation, CAD - coronary artery disease, DCM - dilated cardiomyopathy, ECG – electrocardiogram, HCM - hypertrophic cardiomyopathy, HET - heterozygous (all the detected variants were heterozygous, other than those marked e homozygous, f hemizygous), HTN - hypertension, LP - Likely pathogenic, LVH - left ventricular hypertrophy, MR - mitral regurgitation, NA - not applicable/not available, P - Pathogenic, PAH - pulmonary arterial hypertension, PPCM – peripartum cardiomyopathy, SCD - sudden cardiac death, T2DM - Type 2 diabetes mellitus, VUS - Variants of uncertain significance, (ns) VT - (non sustained) ventricular tachycardia.
Population databases.
Insilico Prediction tools: SIFT (Sorting Intolerant From Tolerant); PolyPhen-2 (Polymorphism Phenotyping v2); FATHMM (Functional Analysis through Hidden Markov Models) and LRT (the likelihood ratio test).
Leaky deletions have been reported i.e both residual normally spliced and deleted transcripts. MYBPC3Δ25 has also been seen (in co-occurrence) with pathogenic variants in HCM genes. This allele may contribute (modifying effect) to the secondary risk factors and/or may enhance the phenotypic mutation and has been reported in ClinVar with conflicting interpretation.
Homocystinuria has also been associated with premature cardiovascular complications induced by accelerated atherosclerosis and/or thromboembolism leading to arterial ischemic events such as acute myocardial infarction, common carotid wall hypertrophy, peripheral vascular disease, stroke and heart failure [PMID: 9826314, 28445283]. The CBS (Gly151Arg) is pathogenic variant reported for homocystinuria, however its relevance in cardiac abnormalities is not well established.
Fig. 2.
Panel A. Results of genetic testing in the probands
Fig. 2 Panel B. Genes harboring mutations in the HCM probands.
4. Cardiomyopathies
4.1. HCM
The age of the HCM probands (n = 36) was 49 ± 15 years and 31 % were symptomatic. P/LP variants were identified in 10 probands resulting in a yield of 28 % and 60 % of them harbored multiple variants. Probands with P/LP variants were significantly younger than those with VUS and negative genotyping (41 ± 16 vs. 53 ± 14, p = 0.03). Genes harboring variants in the HCM probands are shown in Fig. 2 Panel B.
A 40-year old male (No. 4 in Table 3) with history of exertional angina, presyncope, QTc of 500 ms, interventricular septal thickness of 27 mm, left ventricular ejection fraction (LVEF) of 57 % and SCD of an apparently healthy cousin at 39 years of age, was identified with a pathogenic SCN5A mutation (c.5693G > A) and a likely pathogenic MYBPC3 mutation. He has been managed with beta-blockers and surgical myomectomy and counseled about the coexistence of HCM and LQTS type 3 (LQT3). The medications to be avoided and exercise precautions have been explained to the patient.
An asymptomatic 67-year old male (No. 10 in Table 3) was incidentally diagnosed with HCM and found to be a carrier of a single copy of an autosomal recessively inherited variant, implicated in a metabolic condition called carnitine deficiency that can affect the heart and skeletal muscles. His serum carnitine assay was normal; he is under regular cardiology follow-up.
A 30-year old female (No. 3 in Table 4), under follow-up since adolescence for biventricular HCM, presented with worsening of symptoms and progression to severe heart failure. Her genetic test reports from two different labs dated 2016 and 2017 were different; however, the revised report from 2022 as per current criteria concluded the MYBPC3 intronic variant (c.3628-41_3628-17del) she harbored to be a VUS; the variant was also detected in her father and asymptomatic 9-year old daughter. She underwent successful heart transplantation at the age of 31 years but succumbed to refractory sepsis and multi-organ failure subsequently.
4.2. DCM
The age of the DCM probands (n = 3, 2 males, Nos. 20–22 in Table 4) was 29 ± 5 years and all were symptomatic. Five VUS were identified in LMNA, MYH7, TNNT2, and NEXN genes with two of them carrying two variants. Guideline-based therapy was provided to all patients and periodic reviews with the cardiogenetic team advised.
An asymptomatic sibling of a proband (whose reports were unavailable for review) with unexplained heart failure and a likely pathogenic TNNT2 LP variant (c.523A > C) presented to us for post-test counseling as he was also identified to harbor the same variant. His cardiovascular evaluation was normal and he was advised yearly cardiology review.
4.3. ACM
The age of the ACM probands (n = 3, 2 males) was 17, 42 and 59 (female) years, respectively. The 17-year old male (No. 11 in Table 3) presented with sports-related SCD with one episode of sports-related syncope in the past that was not evaluated. He harbored a pathogenic PKP2 mutation (c.235C > T), which was also detected in his asymptomatic mother and sister for whom periodic reviews have been advised.
The 42-year old male (No. 24 in Table 4) presented with cardioembolic stroke and later developed recurrent VT, was identified with ACM morphology, and harbored 2 VUS in the DSP and DOLK genes; he was managed with beta-blockers, oral anticoagulation and ICD therapy.
The 59-year old female (No. 23 in Table 4) presented with recurrent ventricular tachycardia (VT), had ACM morphology and was found to harbor 2 VUS in the KCNH2 and MYLK2 genes; she had an ICD inserted and is under continuous surveillance.
4.4. PPCM
A 29-year old female (No. 25 in Table 4) presented in her second pregnancy with history of PPCM after the birth of her first child, when she was admitted to the hospital with severe heart failure (LVEF 20 %) and discharged after return of LVEF to normal. She was found to harbor a novel missense variant in the SCN5A gene (c.926G > A) classified as a VUS. Her second pregnancy and the peripartum period were uneventful.
5. Channelopathies
5.1. LQTS
All 3 (100 %) LQTS probands were found to harbor pathogenic variants. A female infant (No.12 in Table 3) born of a consanguineous marriage, presented with clinical symptoms of Jervell and Lange-Nielsen syndrome, namely fetal bradycardia, severe QTc prolongation in the neonatal ECG and severe bilateral hearing loss. Her genetic test showed two mutations: a pathogenic autosomal recessive homozygous mutation in the KCNQ1 gene (c.477+1G > A) and a VUS in the KCND3 gene (c.146G > A). Pharmacotherapy was initiated and the family counseled on the role of ICD and medications to be avoided.
An asymptomatic 9-year old male (No. 13 in Table 3) with borderline prolonged QTc and normal cardiac evaluation was screened for channelopathies. His father, with history of syncope and unexplained seizures in the past that were not evaluated systematically, had a SCD at the age of 35 years. The child's genetic test revealed a pathogenic gain-of-function SCN5A mutation (c.5350G > A), was initiated on beta-blocker therapy and the family counseled accordingly. His female sibling was advised targeted genetic testing.
A 48-year old female (No. 14 in Table 3) presented with syncope at rest, history of SCD of her 4 year-old son and of a paternal aunt in infancy. She had been taking beta-blockers for over a decade for incidental observation of prolonged QTc on ECG. She was identified to harbor a pathogenic gain-of-function mutation in the SCN5A gene (c.3992C > T) and counseled accordingly. She had an apparently healthy 17-year old son and was advised targeted genetic testing.
5.2. BrS
A 50-year old male (No. 19 in Table 4) presented with syncope, SCD of sister at 40 years of age and type 1 Brugada ECG pattern; he was found to harbor a VUS in the HCN4 gene (c.604C > A). As the history of syncope was suggestive of vasovagal etiology, the patient is under close follow-up.
6. Connective tissue disorders
6.1. MFS
A 45-year old male (No. 15 in Table 3) presented with Marfanoid features and history of fatal aortic dissection in his male sibling. He harbored a LP heterozygous variant in the FBN1 gene (c.2113G > T) and a VUS each in the DSP and the HCN4 genes; while his asymptomatic daughter was found to be genotype negative, two of his affected second-degree relatives were found to be carriers of the pathogenic LP variant. The family is under close follow-up.
6.2. Family screening and follow-up
Family members of 18 (44 %) probands with P/LP (n = 11) and VUS (n = 7) variants were screened for disease. Twenty-nine relatives were evaluated clinically and genetically for HCM (n = 19), DCM (n = 3), ACM (n = 4) and MFS (n = 3). The genotype positive probands/relatives and VUS carriers with strong disease phenotype and/or high-risk variant were advised periodic follow-up; the remaining probands/relatives were discharged from further clinical surveillance.
7. Discussion
India is leading the way in medical advancements in general and in cutting edge cardiovascular therapies including heart transplantation in particular, in the South Asian region. However, there is a lack of focus on cardiogenetics whereby systematic efforts are taken to evaluate, treat and prevent young sudden cardiac arrest and death. The absence of specialized multidisciplinary clinics and the paucity of scientific literature pertaining to the population at hand are a major concern as well as a cause for knowledge being confined to small pockets, respectively. To the best of our knowledge, this is the first detailed report of the results of comprehensive genetic testing in an Indian cohort with clinical suspicion of inherited heart diseases.
With NGS revolutionizing the genetics and genomics space in the recent years, genetic aberrations underlying cardiovascular diseases and SCD are getting unveiled at a rapid pace and the awareness of genotype-phenotype correlations is paving the way for personalized care for affected individuals and their families.11 Despite NGS being available in India for nearly a decade, there is a poor uptake of the technology even amongst tertiary care centres mainly due to the inability of clinicians to find genetic links to complex conditions, genetic data interpretation issues and lack of actionable genetic insights.12 The authors of this study have a solid expertise in cardiogenetics and a clear vision of collaborating with healthcare professionals across the country and beyond geographic borders to further the science and the care of families with inherited heart diseases in the country and the region.
The yield of genetic testing (31 %) in the present study was comparable with the global yield as reported by Stafford et al (37 %) and Hofman et al (31 %).13,14 However, the detection rate for VUS (54 %) was higher than previously reported, attributable mainly to the lack of a curated ethnicity specific database for genetic disorders in the region and the dearth of functional genomics tools. Though a TGP was used to evaluate the cohort, the backdrop of phenotype based WES ensured that no significant genotype related to the phenotype was missed based on current available literature evidence. A re-analysis in future or new evidence may re-classify these VUS. The probands were counseled that irreversible decisions cannot be taken based on VUS, about the need for follow-up with the genetic counselor every 6–12 months for potential changes in the variant classification and about the benefits of clinical screening of family members followed by genetic screening of individuals with the disease phenotype.
Within the HCM cohort, MYBPC3 was the most implicated sarcomeric gene, followed by MYH7, and nearly three fourths of all the variants were novel as noted in earlier reports.15,16 Interestingly, the South Asian MYBPC3Δ25 intronic deletion variant, identified as a risk allele for heart failure and recently classified as a VUS was present in a young female patient with a severe HCM phenotype and her asymptomatic daughter.17,18 In the absence of tools such as humanized mouse models, human induced pluripotent stem cells-derived cardiomyocytes and organoids, it is challenging to tease out the actual role of this variant in our population. Given that HCM phenocopies may pose a diagnostic challenge, the role of a thorough three generational family history, systematic phenotyping using the available investigatory armamentarium and pre-test counseling by trained genetic counselors cannot be overemphasized.19
The yield among ACM probands was higher than that among DCM probands but it is well recognized that DCM genes are the most elusive due to its complex genetic architecture. Four of the genes harboring VUS in the DCM probands have been classified as having definitive evidence in the causation and one classified as moderate evidence.20 Of note, two of the probands had multiple variants in genes with moderate to definitive evidence. The study by Bhatt et al reporting on the genetic underpinnings of idiopathic pediatric cardiomyopathies in North India has identified MYBPC3 as the most commonly involved gene.21 PPCM, recognized as an entity resembling and related to DCM, is known to have a genetic explanation in around 15 % cases.22 The proband in this study was identified with a novel missense variant in the SCN5A gene, one of the few genes well implicated in the DCM-PPCM spectrum, highlighting the role for genetic testing in young women within this disease group.23
Among the ACM probands, the dramatic presentation of a young athletic male with sport-related SCD and of a middle-aged man with a cerebrovascular accident leading to the diagnosis of ACM have created increased awareness about the varied presentation of this condition within the healthcare community. Similar presentations have been reported in a 17-year old male with sport-related aborted sudden cardiac arrest and a middle-aged male with stroke.24,25 The largest study on ACM in the Indian population included 34 patients subjected to polymerase chain reaction-based analysis in the pre-NGS era revealing novel RYR2 and PKP2 mutations.26
All three probands with LQTS had a strong phenotype and a high Schwartz score, leading to a 100 % yield from genotyping. The uptake of cascade screening was however poor, mainly attributable to the fact that all the probands were from smaller towns, referred to our centre by their primary care physician/cardiologist and were not directly approachable. All probands were free of symptoms on medical management and were under follow-up. Counseling of affected patients and family members involved lifestyle advice, competitive sports participation guidelines, medications to be avoided and discussion on advanced therapies including ICD and left cardiac sympathetic denervation. The mutation spectrum of LQTS reported by Vyas et al in Asian Indian probands included 17 in KCNQ1, 1 in KCNH2 and 2 in SCN5A genes.27
While SCN5A is the only validated gene in the causation of BrS, HCN4 and a few other genes have been implicated in the past and their role is currently disputed.4 Apart from a couple of review articles describing the prevalence of BrS, there are only a few case reports of BrS in the Indian population and they too do not report on the genetics of the disease.28,29
Nayak et al have reported the clinical and genetic spectrum of the largest series of heritable connective tissue disorders in Indian patients, identifying FBN1 variants in 36 index patients.30 The family reported in the current study had a novel FBN1gene variant that segregated in the affected family members and has paved the way for preemptive medical care.
The relatively small cohort and the lack of complete clinical information are the main limitations of this study. Nevertheless, with the establishment of a multidisciplinary cardiogenetic clinic the authors are confident that the database will expand and that the documentation of the clinical characteristics will be accurate henceforth, thereby overcoming both limitations in the future. With more awareness and accessibility, genetic testing in affected individuals as well as molecular autopsy in deceased individuals should become routine practice in India to completely understand the genotype-phenotype characteristics of these inheritable cardiac conditions in our population.
8. Conclusions
Inherited heart diseases are a major cause of death and disability in the young. Genetic testing guides treatment and follow-up of affected patients and should be carried out in dedicated multidisciplinary clinics with expertise for counseling and cascade screening of family members. The yield of genetic testing can be improved by accurate phenotyping prior to submission to genotyping and by employing systematic variant resolution tools.
Sources of funding
None.
9. Key messages
9.1. What is Already known?
Genetic testing is an integral aspect of patient management and sudden cardiac death prevention in individuals and families with inherited heart diseases such as cardiomyopathies, channelopathies and heritable connective tissue disorders affecting the heart. Unraveling the genetic underpinnings and establishing genotype-phenotype correlations in the population at hand have paved the way for a personalized approach to patient care in this potentially lethal group of diseases.
9.2. What this study Adds?
This is the first detailed report of the results of comprehensive genetic testing in an Indian cohort with a strong clinical suspicion of inherited heart diseases. This study has shown that genetic testing is available, affordable and reliable in India and guides management of affected patients. It should be carried out in dedicated multidisciplinary clinics with expertise for counseling and cascade screening of family members.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Medgenome lab for the research funding and Medgenome, Neuberg and LifeCell labs for the genetic testing.
References
- 1.Stiles M.K., Wilde A.A.M., Abrams D.J., et al. 2020 APHRS/HRS expert consensus statement on the investigation of decedents with sudden unexplained death and patients with sudden cardiac arrest, and of their families. Heart Rhythm. 2021;18:e1–e50. doi: 10.1016/j.hrthm.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo L., Torii S., Fernandez R., et al. Genetic variants associated with unexplained sudden cardiac death in Adult white and African American individuals. JAMA Cardiol. 2021;6:1013–1022. doi: 10.1001/jamacardio.2021.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilde A.A.M., Nannenberg E. van der Werf C. Cardiogenetics, 25 years a growing subspecialism. Neth Heart J. 2020;28:39–43. doi: 10.1007/s12471-020-01444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilde A.A.M., Semsarian C., Márquez M.F., et al. Expert Consensus Statement on the state of genetic testing for cardiac diseases. EP Europace. August 2022;24(Issue 8):1307–1367. doi: 10.1093/europace/euac030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maron B.J., Kalra A. Hypertrophic cardiomyopathy in the developing world: focus on India. Eur Heart J. 2014;35:2492–2495. doi: 10.1093/eurheartj/ehu280. [DOI] [PubMed] [Google Scholar]
- 6.Chockalingam P., Wilde A.A. Inherited arrhythmia syndromes leading to sudden cardiac death in the young: a global update and an Indian perspective. Indian Heart J. 2014;66:S49–S57. doi: 10.1016/j.ihj.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajan S., Chockalingam P., Koneti N.R., Geetha T.S., Mishra S., Narasimhan C. Initial experience and results of a cardiogenetic clinic in a tertiary cardiac care center in India. Ann Pediatr Cardiol. 2021;14:443–448. doi: 10.4103/apc.apc_123_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahamed H., Balegadde A.V., Menon S., et al. Phenotypic expression and clinical outcomes in a South Asian PRKAG2 cardiomyopathy cohort. Sci Rep. 2020;10 doi: 10.1038/s41598-020-77124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaidya V., Dhiman R.S., Mittal A., Khullar M., Sharma M., Bahl A. Genotyping Indian patients with primary cardiomyopathies-analysis of database. Indian Heart J. 2023;75:43–46. doi: 10.1016/j.ihj.2022.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bajaj A., Senthivel V., Bhoyar R., et al. 1029 genomes of self-declared healthy individuals from India reveal prevalent and clinically relevant cardiac ion channelopathy variants. Hum Genomics. 2022;16:30. doi: 10.1186/s40246-022-00402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musunuru K., Hershberger R.E., Day S.M., et al. Genetic testing for inherited cardiovascular diseases: a scientific statement from the American Heart Association. Circ Genom Precis Med. 2020;13 doi: 10.1161/HCG.0000000000000067. [DOI] [PubMed] [Google Scholar]
- 12.Pemmasani S.K., Raman R., Mohapatra R., Vidyasagar M., Acharya A. A review on the challenges in Indian genomics research for variant Identification and interpretation. Front Genet. 2020;11 doi: 10.3389/fgene.2020.00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stafford F., Krishnan N., Richardson E., et al. The role of genetic testing in diagnosis and care of inherited cardiac conditions in a specialised multidisciplinary clinic. Genome Med. 2022;28(14):145. doi: 10.1186/s13073-022-01149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofman N., Tan H.L., Alders M., et al. Yield of molecular and clinical testing for arrhythmia syndromes: report of 15 years' experience. Circulation. 2013;128:1513–1521. doi: 10.1161/CIRCULATIONAHA.112.000091. [DOI] [PubMed] [Google Scholar]
- 15.Borrelli F., Losi M.A., Canciello G., et al. Sarcomeric versus non-sarcomeric HCM. Cardiogenetics. 2023;13:92–105. [Google Scholar]
- 16.Rani D.S., Kasala A., Dhandapany P.S., et al. Novel MYBPC3 mutations in Indian population with cardiomyopathies. Pharmgenomics Pers Med. 2023;16:883–893. doi: 10.2147/PGPM.S407179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harper A.R., Bowman M., Hayesmoore J.B.G., et al. A Re-evaluation of the South Asian MYBPC3Δ25 intronic deletion in hypertrophic cardiomyopathy. Circ Genom Precis Med. 2020;13 doi: 10.1161/CIRCGEN.119.002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadayappan S., Puckelwartz M.J., McNally E.M. South Asian-specific MYBPC3Δ25bp intronic deletion and its role in cardiomyopathies and heart failure. Circ Genom Precis Med. 2020;13 doi: 10.1161/CIRCGEN.120.002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butters A., Bagnali R.D., Ingles J. Revisiting the diagnostic yield of hypertrophic cardiomyopathy genetic testing. Circ Genom Precis Med. 2020;13 doi: 10.1161/CIRCGEN.120.002930. [DOI] [PubMed] [Google Scholar]
- 20.Jordan E., Peterson L., Ai T., et al. Evidence-based Assessment of genes in dilated cardiomyopathy. Circulation. 2021;144:7–19. doi: 10.1161/CIRCULATIONAHA.120.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatt D.D., Mathews S., Ahuja V., et al. Genetic testing and family screening in idiopathic pediatric cardiomyopathy: a prospective observational study from a tertiary care center in North India. Egypt J Med Hum Genet. 2023;24:35. [Google Scholar]
- 22.Davis M.B., Arany Z., McNamara D.M., Goland S., Elkayam U. Peripartum cardiomyopathy: JACC state-of-the-Art review. J Am Coll Cardiol. 2020;75:207–221. doi: 10.1016/j.jacc.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Arany Z. It is Time to offer genetic testing to women with peripartum cardiomyopathy. Circulation. 2022;146:4–5. doi: 10.1161/CIRCULATIONAHA.122.059177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jan M., Shillingford M.S., Turbendian H.K., Ferns S.J. Cardiac arrest secondary to arrhythmogenic right ventricular cardiomyopathy in an adolescent male. Indian Pacing Electrophysiol J. 2022;22:241–244. doi: 10.1016/j.ipej.2022.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parikh J.M., Ganeshwala G., Mathew N., Venkitachalam A., Natarajan K.U. Young stroke: an Unusual presentation of arrhythmogenic right ventricular Dysplasia. Neurol India. 2019;67:1528–1531. doi: 10.4103/0028-3886.273639. [DOI] [PubMed] [Google Scholar]
- 26.Vutthikraivit W., Rattanawong P., Putthapiban P., et al. Worldwide prevalence of Brugada syndrome: a systematic review and Meta-analysis. Acta Cardiol Sin. 2018;34:267–277. doi: 10.6515/ACS.201805_34(3).20180302B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khawaja M., Qadeer Y.K., Siddiqui R., et al. Brugada syndrome within Asian populations: state-of-the-Art review. Cardiogenetics. 2023;13:61–74. [Google Scholar]
- 28.Pamuru P.R., Maithili D.V.N., Mohiuddin K., Calambur N., Nallari P. Novel mutations in arrhythmogenic right ventricular cardiomyopathy from Indian population. Indian J Hum Genet. 2011;17:70–76. doi: 10.4103/0971-6866.86182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vyas B., Puri R.D., Namboodiri N., et al. Phenotype guided characterization and molecular analysis of Indian patients with long QT syndromes. Indian Pacing Electrophysiol J. 2016;16:8–18. doi: 10.1016/j.ipej.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nayak S.S., Schneeberger P.E., Patil S.J., et al. Clinically relevant variants in a large cohort of Indian patients with Marfan syndrome and related disorders identified by next-generation sequencing. Sci Rep. 2021;11:764. doi: 10.1038/s41598-020-80755-7. [DOI] [PMC free article] [PubMed] [Google Scholar]