Abstract
Mycobacterial species in nature are found in abundance in sphagnum peat bogs where they compete for nutrients with a variety of microorganisms including fungi. We screened a collection of fungi isolated from sphagnum bogs by co-culture with Mycobacterium tuberculosis (Mtb) to look for inducible expression of antitubercular agents and identified five fungi that produced cidal antitubercular agents upon exposure to live Mtb. Whole genome sequencing of these fungi followed by fungal RNAseq after Mtb exposure allowed us to identify biosynthetic gene clusters induced by co-culture. Three of these fungi induced expression of patulin, one induced citrinin expression and one induced the production of nidulalin A. The biosynthetic gene clusters for patulin and citrinin have been previously described but the genes involved in nidulalin A production have not been described before. All three of these potent electrophiles react with thiols and treatment of Mtb cells with these agents followed by Mtb RNAseq showed that these natural products all induce profound thiol stress suggesting a rapid depletion of mycothiol. The induction of thiol-reactive mycotoxins through three different systems in response to exposure to Mtb suggests that fungi have identified this as a highly vulnerable target in a similar microenvironment to that of the caseous human lesion.
Introduction
Tuberculosis remains a leading threat to global health and new chemotherapies to shorten the duration of treatment are urgently needed1. Current strategies to shorten the duration of treatment focus on attempting to understand bacterial metabolism in the microenvironment in which the bacilli persist and have centered on the caseous granuloma as one of the most difficult to sterilize Mtb lesion types2. Within the caseous granuloma nutrients are scarce, oxygen is limiting, and bacterial replication is slow. Pyrazinamide is perhaps the prototypical treatment-shortening drug, and its activity is highly pH dependent which has led to the suggestion that bacteria limiting treatment efficacy are likely within an acidic microenvironment3. There has been considerable effort to identify more “in vivo relevant” screening methods to identify potentially treatment-shortening leads and targets, however, these have largely been used to screen traditional compound libraries4–7.
In considering what other sources of compounds might be used to identify targets that could result in treatment shortening, we considered environmental sources of competitors for slow-growing mycobacteria. In nature Mycobacteria are found in a wide variety of reservoirs, largely, but not exclusively, aquatic in nature8. One important environmental reservoir for slow-growing mycobacteria is in the decomposing “gray-layer” underneath the zone of active growth of sphagnum peat bogs9. Mycobacteria are abundantly found in this layer reaching concentrations of up to 106 cells per gram9. Microbiome sampling of sphagnum-associated bacteria have shown that 8% of the total community is composed of Actinobacteria, including Mycobacteria10,11. There is epidemiological evidence in Norway linking proximity to sphagnum peat bogs and the risk of developing Leprosy, caused by the Mtb-related organism Mycobacterium leprae 12,13. The characteristics of the “gray-layer” of a sphagnum peat bog has many similarities to the microenvironment within the caseous lesion of an Mtb-infected human lung in that it is nutrient-poor, hypoxic, and acidic.
Sphagnum bogs support a rich assemblage of microorganisms that have adapted to the acidic nutrient-poor environment created by these Bryophytes including many unique bacterial and fungal constituents14. We sampled sphagnum bogs throughout the Northeastern United States to identify fungal species occurring within this ecosystem that naturally compete with Mycobacteria to identify antitubercular compounds they may produce and what they have identified as the most vulnerable targets under these conditions.
MATERIAL AND METHODS
Sampling sites, sphagnum collection and fungal isolation
Sampling sites and peat collection:
Sphagnum core samples were obtained from Cranberry Glades Botanical Area (Monongahela National Forest, WV) guided by Rosanna Springston (US Forest Service) on August 10, 2017; Hobart Bog (Moosehorn National Wildlife Refuge, ME, Special Use Permit MSH-18–015) on September 15, 2018; and Sunkhaze Meadows National Wildlife Refuge, ME, Special Use Permit MSH-18–015) on September 16–18, 2018 (Table S7). Soil was collected using a Russian Peat Borer (Aquatic Research Instruments, Hope, ID) from various depths ranging from the top layer to about 50 inches deep under the surface. About 50 g of soil mass was carefully collected in 50 mL falcon tubes following sterile procedures and stored at 4°C until processed.
Fungus Isolation:
5 g of sphagnum core mass was suspended in 30 mL 0.1 M phosphate buffer containing 0.01% Tween-80, pH 4.0 in a 50 mL falcon tube with 4 mm glass beads (about 5 mL). The samples were vortexed vigorously for 2 min and allowed to settle for 30 min. The top layer of the soil suspension was plated at different dilutions on different solid agar media; potato dextrose agar (PDA, pH 5.4), pectin enriched minimal media15 (pH 4.0) and sphagnum enriched media (pH 4.0) with 100 μg/mL chloramphenicol and 50 μg/mL gentamicin sulfate. Plates were incubated for 5–15 days at 25°C and visible fungal colonies were sub-cultured by transferring spores onto fresh plates. Pure spores were finally collected using phosphate buffer with 0.1% Tween-80 (pH 6.0). Spores were counted using an automated cell counter (Nexcelom Bioscience, Lawrence, MA) and the suspensions were stored in 2 mL aliquots in 96-well deep-well plates at −80°C.
Bacterial strains and culture media
Mtb H37Rv was grown in Middlebrook 7H9 medium (Difco Laboratories) supplemented with 0.5% Albumin, 0.2% D-Glucose, 0.2% Glycerol, 0.085% NaCl, 0.05% Tween-80 (ADGNTw) or 7H11 media supplemented with ADGN and 0.006% Oleic acid (OADGN). The reporter strain pOLYG-mScarlet Mtb H37Rv has been previously described16. Cells expressing pPMF62a-3054p (see below) as well as Mrx1-roGFP2 reporter plasmids17 were grown in 7H9+ADGNTw containing 50 μg/mL hygromycin. To prepare aliquots of Mtb for fungal induction 2 L detergent and BSA-free 7H9 media (supplemented with 4 g/L Glucose, 0.81 g/L NaCl and 0.3 g/L Casitone) was inoculated with 200 mL of wild-type Mtb H37Rv (OD600 1.0) in roller bottles and incubated at 37°C for 2 weeks. The cells were harvested by centrifuging at 4000 rpm for 30 min and were washed with 200 mL phosphate buffer saline (PBS). The cells were re-washed as above and finally resuspended in 200 mL PBS to form 10X concentrate (X refers to the density of a fully saturated culture). The cells were homogenized using glass beads (4 mm, 20 mL) by vortexing for 5 min. This 10X cell suspension was then aliquoted into cryovials and frozen at −80°C for further use. Concentrated Mtb stocks were serially diluted in phosphate buffer (pH 6.0) and homogenized by using glass beads (4 mm, 5 mL) followed by vortexing for at least 2 min. Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species were cultured in brain-heart infusion broth (Difco Laboratories). These bacteria were recent clinical isolates obtained from Dr. Adrian Zelazny in the NIH Clinical Microbiology laboratory.
Co-cultivation based fungal hits screening and growth inhibition assays
Preliminary screening of the induced fungal hits was performed in either soft-solid PDA containing 0.75% agar in 96-well trans-well plates (Corning Inc.) and/or potato-dextrose broth (PDB) in 12- or 24-well multi-dishes. Within each well of the insert plate, 100–1000 fungal spores from frozen stocks were added on the surface of soft-solid potato dextrose agar. Each well of the donor plate was immersed in 200 μL of phosphate buffer (pH 6.0) within the receiver plate. The plates were incubated at 28°C for 24 h followed by the addition of different concentrations of Mtb inducer. The plates were further incubated for 9 to 15 days at 28°C – 32°C. Liquid co-cultivation was performed in either 12 or 24 well plates in PDB media (1–1.2 mL/well) containing ~1000 spores grown for 24 h at 28°C before adding Mtb inducer and incubating at 28°C – 32°C for 9 to 15 days with shaking (45 rpm). The contents of the receiver plate and the liquid cultures (mono and co-cultures) were filtered through 0.2 μm 96-well filter plates (MilliporeSigma, Burlington, MA) and two-fold serial dilutions were done in 96 well clear-bottom black plates (Corning Inc.) in 7H9 media supplemented with ADGNTw and hygromycin (50 μg/mL). 104 cells of pOLYG mScarlet Mtb H37Rv were added, and the plates were incubated at 37°C. Growth of the reporter strain was monitored on Day 7 and Day 14 using a Clariostar (BMG Labtech, Cary, NC) plate reader at Em: 610 nm (Ex: 570 nm). The volume of the filtrate at which there was 90–100% growth inhibition was defined as the ‘Minimum Inhibitory Volume’ or ‘MIV’. The cidality assay for the Mtb H37Rv strain was performed in 24 well plates after 7 days of exposure of cells to the induced fungal culture filtrates. Mtb H37Rv culture in 7H9-ADGNTw media was grown to an OD600 of 0.6 and was filtered through a 5 μm filter to remove cell clumps. Cells were diluted to OD600 0.02–0.03 and appropriate dilutions plated on 7H11-OADGN (ADGN supplemented with oleic acid) media to enumerate Day 0 inoculum. The remaining culture was dispensed in 24-well tissue culture plates (0.5 mL/ well) containing concentrations of fungal filtrates corresponding to 2X and 10X MIV and incubated at 37°C, 40 rpm for 7 days. Treated cells were washed and plated on 7H11-OADGN media for CFU enumeration. The assay was performed for two biological replicates. CFU was counted after incubation of the plates at 37°C for at least 3–4 weeks and bactericidal activity was determined in terms of decrease in log10CFU/mL.
Fungal genomic DNA Isolation
Fungal cultures for the positively induced hits were obtained by inoculating about 50,000 spores in 50 mL PDB media (100 μg/mL chloramphenicol and 50 μg/ml gentamicin sulfate) in 250 mL roller bottles (28°C, 5–7 days, 75 rpm). Genomic DNA isolation was performed using the protocol described elsewhere18 with some modifications. Mycelia were harvested and washed with phosphate buffer (pH 6.0) 3 times by centrifuging at 4000 rpm, 10 min, RT followed by vacuum filtering to remove excess media components. The mycelia were then ground to fine powder with liquid N2 by mortar-pestle and stored at −80°C until further use. Cell lysis was performed in tubes with ground mycelia by adding Zr/Si (0.1 mm, 375 μL) beads in 800 μL extraction buffer (0.1M Tris-HCl, pH 8.0; 10 mm EDTA, pH 8.0; 2.5 N NaCl; 3.5 % CTAB) and 20 mg/mL proteinase K (180 μL). The lysate was mixed and incubated at 65°C for 45 min. After centrifuging (13,000 rpm, 15 min, RT), an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added to the supernatant. Samples were mixed by inverting for 15 sec and kept at RT for 1h followed by centrifuging as above. An equal volume of chloroform-isoamyl alcohol was added to the aqueous phase and mixed. After centrifugation, an equal volume of ice-cold isopropanol was added to the aqueous phase. Samples were incubated at −20°C for at least 2 h to precipitate the DNA followed by centrifugation as above. The supernatant was completely decanted, and the DNA pellet was washed with ice-cold 70% ethanol (1 mL, 3 times) by centrifugation (13,000 g, 1 min, RT). The DNA pellets were air-dried overnight and dissolved in 30–40 μL molecular biology grade DNase free water and stored at −80°C. To ensure no contamination of fungal genomic DNA with bacterial DNA, PCR amplification for 16s rRNA was carried out using Q5 high fidelity polymerase and primers 16s-27F and 16s-805R (Table S1).
Internal transcribed spacer analysis and whole genome sequencing
Preliminary identification of the fungal hits with induced activity was carried by PCR amplifying the Internal Transcribed Spacer (ITS) region using the ITS5 and ITS4 primers19 (Table S1). The ITS PCR products were extracted from a 1% agarose gel and were sequenced using ITS5 primers. NCBI BLASTn was used to identify the closest match using the ITS database.
Whole genome sequencing of the fungal hits was done utilizing Large Nanopore Combo and Hybrid Assembly service from SeqCenter, PA. For short read sequencing, sample libraries were prepared using the Illumina DNA Prep kit and IDT 10 bp UDI indices, and sequenced on an Illumina NextSeq 2000, producing 2×151bp reads. Demultiplexing, quality control and adapter trimming was performed with bcl-convert (v3.9.3). For long read sequencing sample libraries were prepared using Oxford Nanopore Technologies (ONT) Ligation Sequencing Kit (SQK-LSK109) with NEBNext® Companion Module (E7180L) in addition to Native Barcode Kits (EXP-NBD104, EXP-NBD114) to manufacturer’s specifications. All samples were run on Nanopore R9.4.1 flow cells and MinION Mk1B device. Post-sequencing, Guppy (v5.0.16) was used for high accuracy base calling (HAC) and demultiplexing20. Quality control and adapter trimming was performed with bcl-convert 21 and porechop (https://github.com/rrwick/Porechop) for Illumina and ONT sequencing respectively. Long read assembly with ONT reads was performed with flye version 2.822. The long read assembly was polished with pilon23. To reduce erroneous assembly artifacts caused by low quality nanopore reads, long read contigs with an average short read coverage of 15x or less were removed from the assembly. Assembly statistics were recorded with QUAST24. Assembly annotation was performed with funannotate25. BUSCO (Benchmarking Universal Single-Copy Ortholog) version 4.1.3 was used to assess genome completeness while N50 and N75 were calculated to determine genome contiguity. Phylogenetic analysis of the fungal isolates was done utilizing the online version of CVTree 3.0. Genome sequences (.faa) of the closest neighbors based on the BLAST search of Internal Transcribed Spacer (ITS), β-tubulin and calmodulin regions of the induced fungal isolates were used to perform the phylogenetic assessments. The fungal genome assemblies are deposited in the NCBI database under the BioProject PRJNA767328 with accession numbers in Table 1.
Table 1:
Summary of assembly metrics for the fungal genomes identified in the study
| Fungal Id | # Contigs | Total Length (Mb) | GC (%) | N50 (Mb) | N75 (Mb) | BUSCO Score (%) | Nearest Neighbor (ITS BLAST) | Accession |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| F2 | 219 | 32.92 | 49.74 | 0.56 | 0.30 | 80.8 | Penicillium rolfsii | JAYXSC000000000 |
| F50 | 85 | 33.04 | 49.57 | 2.68 | 1.59 | 82.4 | Penicillium rolfsii | JAYXSA000000000 |
| F51 | 78 | 32.98 | 49.60 | 2.76 | 1.89 | 83.9 | Penicillium rolfsii | JAYXSB000000000 |
| C7 | 163 | 32.29 | 47.92 | 2.38 | 0.93 | 80.0 | Penicillium lividum | JAYXSD000000000 |
| F31 | 61 | 34.80 | 45.69 | 2.59 | 1.66 | 74.1 | Talaromyces stollii | JAYXSF000000000 |
Fungal RNAseq and differential gene expression
Fungal biomass from mono- and co-cultures of the induced potential hits was obtained by inoculating about 300,000 spores in 4 mL PDB media in 6 well plates (Corning Inc.). The cultures were grown at 28°C for one day. For co-cultures, 450 μL of 1X (for F2/F50/F51 fungus), 0.1X (for C7 fungus) and 0.001X (for F31 fungus) dilutions of Mtb inducer stock resuspended in phosphate buffer (pH 6.0) were added to fungal cultures on the next day and the cultures were incubated at 32°C at 45 rpm. All culture sets were carried out in at least two biological replicates. For monocultures an equivalent volume of phosphate buffer (pH 6.0) was added. Mycelia were harvested on days 6, 9 and 12 by centrifuging at 4000 rpm for 20 min followed by a brief washing with DEPC-treated water (10 mL, 4000 rpm, 20 min). The harvested fungal biomass was ground to fine powder in liquid nitrogen with a mortar-pestle, which was then transferred to 2–3 mL TRIzol Reagent (Invitrogen, Waltham, MA) to form a slurry and finally thawed. The samples were pipetted several times to ensure complete homogenization and were split into 2–3 RNase-free microcentrifuge tubes (1 mL each) and incubated for 5 min at RT. Chloroform (300 μL) was added to each tube and these were shaken vigorously. The samples were centrifuged at 13,000 x g at 4°C for 15 min. To the upper aqueous layer, an equal volume of 70% ethanol was added to precipitate the RNA, which was then vortexed and loaded onto spin cartridge columns provided by PureLink RNA isolation kit (Invitrogen, Waltham, MA). Further purification steps were done according to the manufacturer’s guidelines with an on-column DNase I treatment (Zymo Research, Irvine, CA). The RNA was eluted in 40 μL DEPC-treated RNase-free water and stored at −80°C. RNA concentration and purity were analyzed using Nanodrop spectrophotometer (ThermoFisher Scientific, Waltham, MA) followed by visual assessments using an Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA) and denaturing formaldehyde agarose gel electrophoresis (1.3% agarose, formaldehyde in 1X MOPS buffer). Total RNA (at least in two biological replicates) from mono- and co-culture conditions were used for mRNA sequencing following ribosomal RNA depletion and cDNA Library prep. Likewise, for expression analysis of citrinin in C7-Mtb and C7+Mtb cultures, total RNA was extracted from both conditions in at least two biological replicates. cDNA preparation and real time quantification for the targeted regions of citrinin gene cluster (C7_116, C7_118 and C7_120; primers listed in Table S1) was performed using One step Luna Universal RT-qPCR kit (New England Biolabs, Ipswich, MA) using an Applied Biosystems QuantStudio 3 (Thermofisher Scientific, Waltham, MA).
Libraries for mRNA sequencing were constructed from total RNA sample using the Zymo-Seq RiboFree Total RNA Library Prep Kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions. Libraries were amplified to incorporate full length adapters. RNAseq libraries were sequenced on an Illumina NovaSeq 6000 to a sequencing depth of at least 30 million read pairs (150 bp paired-end sequencing) per sample. The Zymo Research RNA-Seq pipeline was originally adapted from nf-core/rnaseq pipeline v1.4.2 (https://github.com/nf-core/rnaseq)26. Briefly, adapter and low-quality sequences were trimmed from raw reads using Trim Galore! v0.6.6 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore). Trimmed reads were aligned to the reference genome using STAR v2.6.1d (https://github.com/alexdobin/STAR)27. BAM file filtering and indexing was carried out using SAMtools v1.9 (https://github.com/samtools/samtools)28. RNAseq library quality control was implemented using RSeQC v4.0.0 (http://rseqc.sourceforge.net/)29 and QualiMap v2.2.2-dev (http://qualimap.conesalab.org/)30. Duplicate reads were marked using Picard tools v2.23.9 (http://broadinstitute.github.io/picard/). Reads overlapping with exons were assigned to genes using featureCounts v2.0.1 (http://bioinf.wehi.edu.au/featureCounts/)31. Differential gene expression analysis was completed using DESeq2 v1.28.0. Differentially expressed sequences were assembled using the annotation .gbk files for their respective genomes obtained from SeqCenter. Fungal differential expression was depicted as Magnitude-Amplitude (MA) plot representing log ratio fold change (log2FC) as ‘M’ on y-axis while mean average of gene expression in terms of DESeq2 normalized mean read counts is shown on x-axis using DESeq2 v1.28.0 (https://bioconductor.org/packages/DESeq2/)32. Genes with negative log2FC have higher expression in co-culture conditions, while positive log2FC represents the upregulation in monoculture conditions. Nucleotide and protein sequences for contiguously placed induced genes unique to co-culture conditions were extracted using Geneious Prime33. Protein homology was assessed using BLASTx against experimental clustered Nr database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Protein domain and functional annotation was also determined using InterPro (https://www.ebi.ac.uk/interpro/search/sequence/) and UniProt (https://www.uniprot.org/). Sequence alignments were done by CLUSTALO (https://www.ebi.ac.uk/Tools/msa/clustalo/) and phylogenetic analysis by MEGAX34 and Geneious prime software using the Maximum Likelihood algorithm as well Neighbor-joining methods. BGC annotation and characterization in the fungal genomes was done using antiSMASH fungal version 7.035. Gene expression data was submitted to Gene Expression Omnibus (GEO)-NCBI database with GEO accession GSE271121 (F2); GSE271124 (F50); GSE271125 (F51); GSE271119 (C7); GSE271126 (F31) in mono- and co-culture conditions.
Mtb RNA isolation and differential gene expression
Mtb (OD600 0.2, 10 mL) cells were exposed to concentrations equivalent to 1X and 10X MIV of induced fungal filtrates (F2+Mtb and F31+Mtb). Cells were harvested after 6 h and 12 h exposure and were washed twice with 7H9-ADGNTw broth by centrifuging (4000 rpm, 4°C, 10 min). Cells were resuspended in 0.9 mL TRIzol and were lysed by bead beating using 0.1 mm Zr/Si beads (6000, 2–3 cycles, 30 sec; 5 mins on ice between pulses). The resulting lysate was centrifuged (13,000 rpm, 1 min, 4°C) to remove the cell debris and 0.3 mL chloroform (Sigma) was added to the supernatant. Tubes were thoroughly shaken, allowed to sit for 5 min and centrifuged (13,000 rpm, 15 min, 4°C) to separate the upper aqueous layer. Total RNA was precipitated using equal volume of 70% ethanol and vortexed for 30 sec. The solution containing RNA was loaded onto spin cartridge columns and the RNA was eluted according to the manufacturer’s guidelines (PureLink, Invitrogen, Waltham, MA). Total RNA was stored in aliquots at −80°C. Total Mtb RNA was used for cDNA library prep and mRNA sequencing in three biological replicates for four different conditions; 6h, 1X; 12h, 1X; 6h, 10X and 12h, 10X. mRNA sequence obtained from untreated Mtb was used as a control for differential gene expression analyses. About 100 ng of Total RNA was used to produce mRNAseq samples using the Illumina TruSeq Stranded Total RNA Gold kit and sequenced as 151 bp paired-end libraries on an Illumina NextSeq 2000 P2. cDNA prep and mRNA sequencing were done at the sequencing facility of Fredrick National Laboratory for Cancer Research (FNLCR, Fredrick, MD, USA). The data was processed using the RNA-seek workflow v1.8.0 (https://doi.org/10.5281/zenodo.5223025) on the NIH HPC Biowulf cluster (http://hpc.nih.gov) with minor adaptations. References were created using the –small-genome build command. Reads were then trimmed using cutadapt v1.18 and aligned to the NC_000962.3 reference genome using STAR v2.7.6a in 2-pass basic mode with the following adaptations to handle the small, non-intronic genome: ALIGNINTRONMAX= 1, ALIGNINTRONMIN= 0, and ALIGNMATESGAPMAX= 500027. Expression levels were quantified with RSEM v1.3.036. For differential analyses, genes with a read count per million (CPM) less than 0.5 in at least 3 samples were filtered before running limma 3.52.437. Genes were identified as significant with an FDR of 0.1 and an absolute fold-change of 2. Venn diagrams were created with ggvenn 0.1.10 (https://www.rdocumentation.org/packages/ggvenn/versions/0.1.10). Log-fold change heatmaps were created using ComplexHeatmap 2.12.1 with circlize 0.4.15 and RcolorBrewer 1.1–3 to control the colors. Cluster networks was performed using STRING db (https://string-db.org/) and functional annotation of the genes was according to Mycobrowser (https://mycobrowser.epfl.ch/) and TB Database (http://tbdb.bu.edu/). Differential expressions at different treatment conditions are represented by MA plot where x-axis represents the log CPM while y-axis is log2FC using Glimma v2.6.038. Gene expression data for Mtb was submitted to GEO-NCBI database with accession number GSE255435.
Reactivity based screening by DTT adduct assay
DTT adduct assay was performed as previously described by treating 20 μL of induced F31+Mtb media filtrate with 20 μL of 1 M dithiothreitol (DTT) in basic conditions (diisopropylethylamine 2.5 μL/mL, final concentration 20 mM) in methanol 39. Control samples were treated similarly but without DTT. Following 16 h of incubation protected from light, metabolite analysis was done by LC-MS/MS by scanning both positive and negative mode ESI from m/z 100 to 1000 for F31+Mtb samples with and without DTT. Chromatograms were compared by examining the −DTT samples for peaks absent in the +DTT samples and vice versa with signal abundance of more than 100,000.
Extraction, fractionation and identification of active metabolite in F31+Mtb and C7+Mtb culture filtrate
Fungal filtrates from a 15-day co-cultivation of F31+Mtb and C7+Mtb in 6 well plates were filtered twice and pooled separately to obtain about 400 mL total filtrate volume from each. Filtrates obtained were mixed with 250 mL ethyl acetate (EtOAc) and were extracted twice. For C7+Mtb, a brine solution was additionally used to facilitate the separation of organic layer from the aqueous layer. The organic layer obtained following ethyl acetate extraction was evaporated to dryness at 30°C in a rotary evaporator. The total EtOAc extract from F31+Mtb and C7+Mtb filtrate was confirmed to have anti-Mtb activity and fractions were collected manually from LC-MS C-18 column detected by UV at either 254/280 nm for F31+Mtb or 334 nm for C7+Mtb between 5 min to 12 min of run. Each fraction collected in multiple replicates were pooled and tested for activity against Mtb H37Rv. F31+Mtb EtOAc extract was taken for further purification of the fraction responsible for anti-Mtb activity using prep-TLC in ethyl-acetate:hexane (1:1) solvent system. Flash column chromatography using silica gel and ethyl-acetate:methanol (10:1) solvent system was used to purify active component in C7+Mtb extract. The purified fraction with anti-Mtb activity was checked for purity on LC-MS and structure identification was done using proton and carbon NMR. The final product was dried evaporated and stored at −80°C.
Mrx1-roGFP2 mediated redox screen
The Mrx1-roGFP2 reporter plasmid was a kind gift from Dr. Amit Singh17. Mtb H37Rv expressing Mrx1-roGFP2 reporter construct was grown in 7H9-ADGNTw media containing hygromycin (50 μg/mL). Cultures of OD600 = 0.2 were treated with patulin, nidulalin A and auranofin each at 0.5X - 10X MIC while diamide at 0.5 to 10 mM concentrations in 96 well black plates with clear bottom at 37°C. Ratiometric fluorescence at emission 510 nm (Ex: 405 nm/ 480 nm) was recorded at different time points from 0h – 48h. Assays were performed in at least two biological replicates.
Quantification of free thiol pools
Mtb H37Rv cultures (0.5 mL) were grown to OD600 0.2 in 7H9-ADGNTw media and were treated with patulin and nidulalin A (0.5X-10X MIC) for 24h (37°C, 45 rpm). Untreated cultures were used as negative control group while auranofin (0.5X-10X MIC) and diamide (0.5 mM-10 mM) were used as additional controls to determine the free thiol levels under the same conditions. Post-treatment, the cells were harvested, washed in phosphate buffer saline, and lysed by bead beating for 2–3 times for 45s at 7000 rpm in a Roche MagnaLyzer and intermittent cooling on ice for 5 min in thiol detection buffer (100 mM potassium phosphate, pH 7.4, 1 mM EDTA). The supernatants collected were used for free intracellular thiol measurements using a thiol detection kit (Cayman Chemical) as reported previously40. Assays were done in triplicates and were representative of two biological replicates. Thiol concentration was calculated in nM using a glutathione standard curve and the effect of all the agents is represented as % free intracellular thiol with respect to untreated control taken as 100%. OD600 was measured before cell harvesting and the relative fluorescence was normalized to measure the thiol levels in equal number of cells.
Mycothiol Adduct Assay
Mtb H37Rv cultures were grown to OD600 = 0.8 to 1.0 in 50 mL 7H9-ADGNTw media. Cells were harvested and washed thrice with phosphate buffer saline by centrifuging at 4000 rpm for 10 min. Cells were resuspended in 10 mL BSA and detergent free 7H9 media supplemented with glucose (4 g/L) and NaCl (0.8 g/L) and aliquoted in 12 well plates where they were treated with either nidulalin A or patulin at different concentrations (0X-20X). At the indicated time points (ranging from 0 – 48 h). the cells were killed with 60% acetonitrile and frozen at −80°C. Supernatants obtained after centrifugation at 13,000 g for 10 min were tested for the released mercapturic acid adduct of nidulalin A/patulin indicative of the intracellular mycothiol adducts in Mtb by LC-MS on C-18 column in positive and negative ion modes.
Rv3054c promoter-reporter construction
The fluorescent reporter construct for the Rv3054c promoter was constructed with an episomal plasmid expressing eGFP under the Psmyc promoter41, conferring hygromycin resistance, and replicating in mycobacteria using the pAL5000 replicon. This has previously been described as pOLYG Psmyc eGFP16. We also used the plasmid pGiles pL* mScarlet16 as the source of DNA encoding the mScarlet fluorescent protein. We PCR amplified pOLYG Psmyc eGFP using primers oPMF373 and oPMF374 (Table S1), obtaining a linearized version of the entire plasmid with flanking PaqCI restriction sites added via 5’ overhangs. The 174 bp region immediately upstream of Rv3054c was PCR amplified using primers oPMF369 and oPMF370, adding flanking PaqCI sites via 5’ overhangs, and amplified DNA encoding mScarlet using primers oPMF371 and oPMF372 utilizing Q5 DNA polymerase (NEB), and a touch-down PCR protocol42. PCR products were gel purified and Golden Gate assembly43 was performed with PaqCI and T4 ligase (NEB). Products of the Golden Gate assembly were transformed into 5-alpha competent E. coli cells (NEB) via heat-shock and plated onto LB agar plates supplemented with 200 μg/mL of hygromycin (Sigma). The sequence of the insert (plasmid pPMF62a) was confirmed by whole-plasmid nanopore sequencing (Plasmidsaurus). Plasmid pPMF62a was subsequently transformed into Mtb via electroporation44,45. Primers used for the construction of pPMF62a are mentioned in Table S1 and the vector map for the reporter plasmid pPMF62a can be seen in Fig. S1. Mtb H37Rv expressing pPMF62a-Rv3054c promoter vector construct was grown in 50 mL ink-well bottles containing 10 mL 7H9-ADGNTw media supplemented with 50 μg/mL hygromycin. Cultures with the final OD600 = 0.2 were treated with different concentrations (0.5X - 10X MIC/MIV) of induced F2+Mtb, F31+Mtb, C7+Mtb, D6+Mtb and D9+Mtb fungal filtrates as well as patulin, nidulalin A and citrinin in 96-well black clear-bottom plates. Untreated/DMSO-treated cells containing reporter plasmid were used as a control group. Diamide and iodoacetamide (0.005 mM-10 mM) were taken as additional controls. The plates were scanned at two different emission wavelengths (Red - Ex.: 488 nm, Em.: 590 nm and Green - Ex.: 488 nm, Em.: 530 nm) at different time points (0h – 24h). The ratio of Red/Green fluorescence was used as a measure to determine the Rv3054c mediated effect of drugs and fungal filtrates on Mtb H37Rv cells. The assay was done with at least two biological replicates each from two individual experiments.
Results
Fungal co-cultivation with Mtb induces antitubercular metabolites.
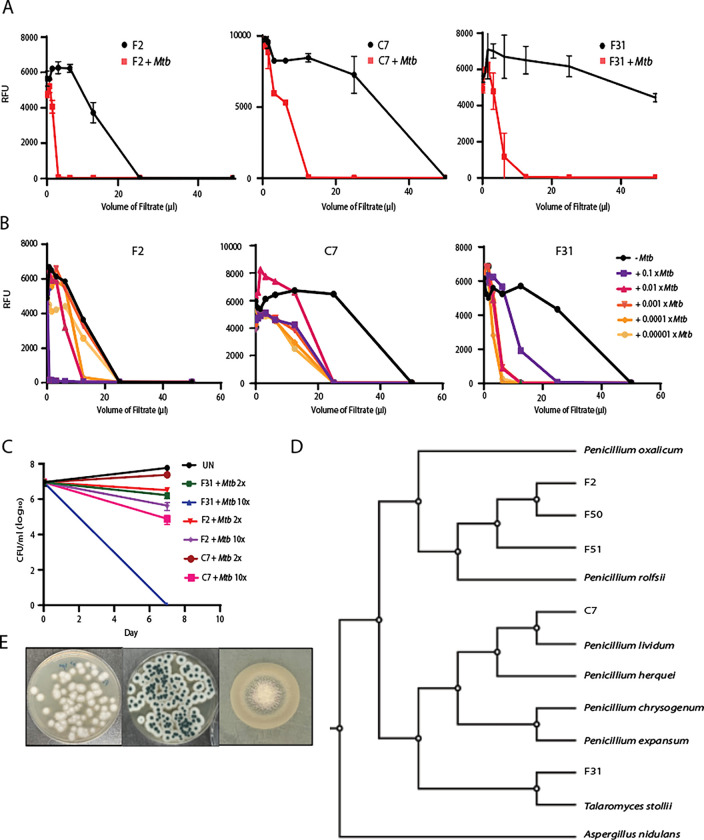
To identify differentially expressed fungal metabolites we screened about 1500 environmental fungal isolates collected from sphagnum peat bogs grown in the presence or absence of live Mtb cells. Culture supernatants were collected and assayed against Mtb constitutively expressing an mScarlet fluorescent protein. Hit fungi were selected based upon significant shifts in the supernatant volume required for growth inhibition when the fungi were co-cultivated with Mtb (Fig. 1A). Varying concentrations of inducer Mtb cells were required for optimal inhibitor production dependent on the individual fungal species (Fig. 1B). In this screen, we obtained five unique fungal isolates displaying induced activity upon co-cultivation with Mtb, three of which were Penicillium sp. with identical ITS sequences (F2, F50 and F51). Another fungi (C7) was an unrelated Penicillium sp. and the final hit (F31) was a Talaromyces sp.
Fig. 1: Activity based screening of Induced Fungal Hits.
A. Growth Inhibition assays using culture filtrates of three hits of mono- and co-culture conditions (n=2–5), B. Dose-dependence of induction with various Mtb inocula (X in this figure refers to the density of a stationary phase culture of H73Rv), growth inhibition assays from the co-cultures were done in at least three biological replicates. C. Bactericidal activity in terms of decline in log10 CFU/mL of co-culture filtrates at 2X and 10X minimum inhibitory volume (MIV) at Day 7. D. Phylogenetic relationship using CVtree46 alignment between F2, F50, F51; C7 and F31 with their nearest neighbors by their ITS/β-tubulin/calmodulin regions and whole genome sequencing (P. rolfsii, P. expansum; P. lividum P. oxalicum, P. herquei, P. chrysogenum and T. stollii). Aspergillus nidulans was used as an outgroup. E. Morphology of F2 (Left), C7 (Right) and F31 (Bottom) on PDA media plates.
To assess the specificity of the induced fungal products for Mtb, filtrates from F2, C7 and F31 co-cultures with Mtb were counter-screened against a panel of ESKAPE pathogens and the non-pathogenic Mycobacterium smegmatis which showed that all three filtrates were inactive against the other bacterial species at the concentrations produced in co-culture supernatant (Table S2). To rule out the production of siderophores competing for available iron, the induced fungal filtrates were screened for growth inhibition of Mtb in the presence of 250 μM Fe+3 which revealed that none of the Mtb active metabolites were likely siderophores (Fig. S3). The antitubercular metabolite produced by C7 was modestly cidal against Mtb reducing CFU by about one log after one week of exposure, F2 had slightly less cidal activity but the metabolite produced by F31 was highly cidal with no viable Mtb detected after 7 days of exposure at this concentration of extract (Fig. 1C).
Determination of fungal reference genomes
Reference genomes for all five hits, including the three fungi that appeared identical by ITS sequencing (F2, F50 and F51), the Penicillium sp. (C7) and the Talaromyces sp. (F31), were obtained by whole genome sequencing followed by hybrid assembly. Induced filtrates of F50 and F51 were equipotent against Mtb compared to F2 (Fig. S2D), although the organisms appeared morphologically unique (Fig. 1E & Fig. 2E) and all three of these fungi were obtained from different geographical locations (Table S7). The average assembled genome size for all fungal hits was about 33 Mb (~32 – 35 Mb) and the number of contigs ranged from 61 to 219. Genome data for all the fungal hits is summarized in Table 1. The ITS, β-tubulin, and calmodulin regions of F2, F50 and F51 displayed the closest identity to Penicillium rolfsii (>99.5%). Phylogenetic analysis using the alignment-free CVTree also indicated P. rolfsii to be the closest neighbor of these three fungi (Fig. 1D). F31 was most closely related to Talaromyces stolli, while C7 appeared more related to Penicillium lividum.
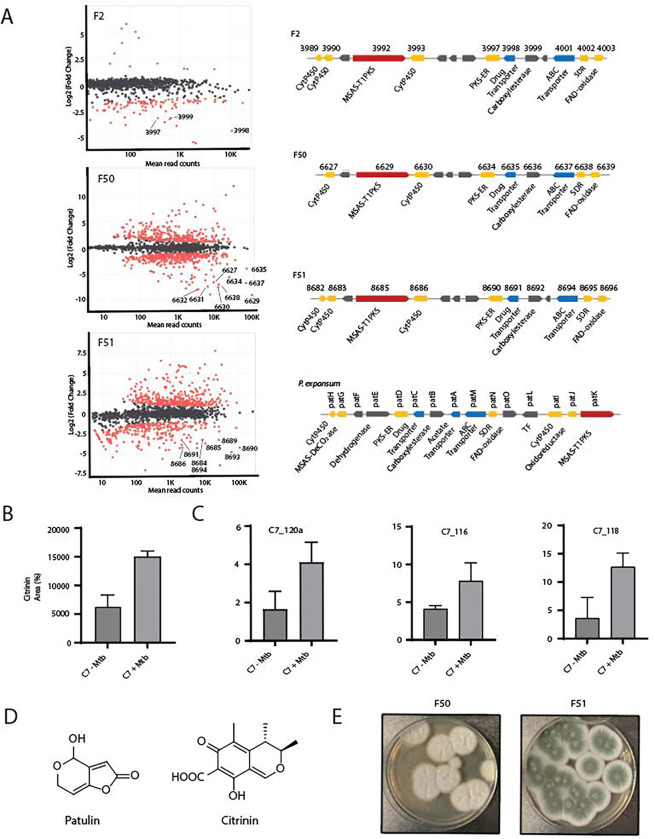
Fig. 2: Four different Penicillium sp. produces patulin and citrinin upon co-cultivation with Mtb.
A. Differential gene expression of three related Penicillium sp. upon co-cultivation with live Mtb. Differentially expressed genes along with read counts are shown in MA plots49. Upregulated genes have a negative value in this analysis and red dots represent differentially expressed while grey/black dots are non-differentially expressed genes. The topmost upregulated BGC for each fungus is shown to the right of each plot and points that are part of this cluster are labeled for each RNAseq experiment. For comparison, the patulin cluster for P. expansum was adapted from elsewhere (bottom right)48. B. Area percentage of citrinin peaks in mono- and co-culture conditions obtained from the Total Ion Chromatogram (TIC) of C7 −/+ Mtb filtrates detected at 334 nm on C-18 liquid chromatography column (Tr=9.2 min). C. RT-qPCR of about a 500 bp region of Type I PKS gene in the citrinin BCG (C7_120a) as well as two other genes involved in biosynthetic pathway (C7_116 and C7_118) amplified from the total RNA isolated from C7-Mtb and C7+Mtb conditions. Y-axis is expression fold relative to expression of the ITS region. D. The chemical structures for patulin and citrinin. E. Morphology of F50 and F51 fungal strains on PDA media.
Three related Penicillium sp. produce patulin upon co-cultivation with Mtb
To identify the biosynthetic gene clusters (BGCs) responsible for anti-Mtb activity produced by these fungi we used RNAseq of co-cultures compared to monocultures for each organism. The three related Penicillium species (F2, F50 and F51) all showed an induction of a single related BGC upon co-culture with Mtb (Fig. 2A). In fungus F2, three genes of this BGC were amongst the most highly upregulated, in F51 9 genes, and in F51 8 genes of this BGC were highly upregulated compared to monocultured cells. In two of three of these upregulated genes included a Type I polyketide synthase. AntiSMASH analysis of this BGC revealed that it shared >78% homology with the patulin producing gene cluster in P. expansum (Fig. 2A right bottom and also see Table S6)47. While the patulin gene clusters in F2, F50 and F51 fungal genomes are comparable, the gene organization was found to be distinct from that in P. expansum48. Table S6 shows the induced genes in F2, F50 and F51 and their amino acid sequence identity with their orthologs in P. expansum.
We confirmed the presence of a peak co-eluting with authentic patulin (Fig. 2D) in co-culture filtrates of F2+Mtb, F50+Mtb and F51+Mtb. In each case, this peak was confirmed to be patulin by MS-MS fragmentation of the parent (m/z 155) into four product ions (109, 99, 81, 71) by LC-MS/MS (Fig. S8). Patulin was only detected in cultures during co-culture with Mtb supporting the notion that synthesis of this metabolite is specifically induced by Mtb. Commercially available patulin was confirmed to inhibit growth of Mtb with an MIC90 of 4.7 μM (Table S5).
For the Pencillium sp. C7, transcriptional analysis of Mtb-induced versus monocultures did not result in strong upregulation of a candidate BGC across biological replicates. Notably C7 had the least increase in activity upon co-culture where it showed only about a two-fold increase in potency (Fig. 1B). Organic extraction of the co-culture supernatant and activity-guided LC-MS/MS analysis revealed an increased production of the polyketide citrinin in induced filtrates (Fig. 2B). The chemical identity of citrinin was confirmed by 2D-NMR (Supplemental Experimental 3.2). Purified citrinin had an MIC90 value of 62 μM against Mtb (Table S5). Despite the apparent lack of differential regulation of the citrinin BGC by RNAseq, qRT-PCR analysis of a portion of a core Type I-PKS gene (C7_120a) as well as two other biosynthetic genes for citrinin production (C7_116 and C7_118) indicated 2–3-fold higher expression of these genes during co-culture with Mtb compared to non-induced cultures supporting the induced biosynthesis of citrinin upon exposure of C7 to Mtb (Fig. 2C).
Co-cultivation mediated identification of induced biosynthetic gene clusters in F31 genome
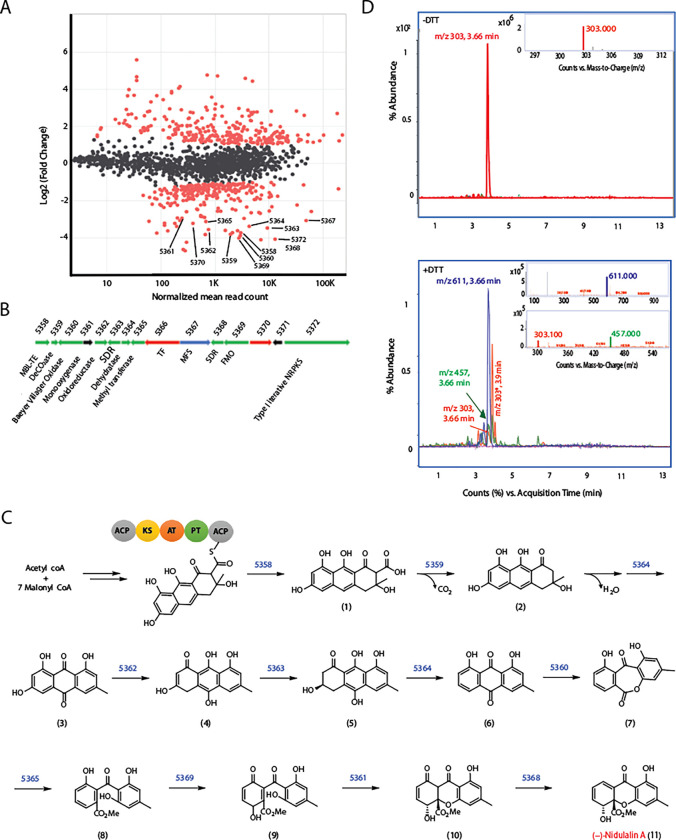
Differential gene expression of the Talaromyces sp. (F31) was also performed on monocultures and co-cultures with Mtb (Fig. 3A). Alignment of the mRNA sequences with the F31 reference genome (> 97% read alignment) revealed a potential BGC of fifteen clustered genes that were highly overexpressed in co-cultured cells. The genetic organization and proposed gene functions for the 15 genes are shown in Fig. 3B and Table S3 (also see Dataset 1). BLAST and Interpro Scan analysis of the protein sequences revealed the core enzyme of this BGC as a Type I non-reducing polyketide synthase (Type I NR-PKS; F31_005372). F31_005372 has about 58% sequence identity with the neosartorin biosynthesis enzyme, NsrB48 containing the characteristic ketoacyl synthase (KS), acyltransferase (AT) and acyl carrier protein (ACP) domains (Fig. S4B). Besides these major domains, other functional domains include starter unit ACP transacylase (SAT), product template (PT), phosphopantatheine (PP-ACP) and a dehydrogenase (DH). The PT domain in the NR-PKSs is responsible for catalyzing the specific aldol cyclization and aromatization of mature precursor polyketides. We compared F31_005372 with 55 fungal NR-PKS full length sequences, which have been previously categorized into eight groups I-VIII 50. This revealed that F31_005372 is an ortholog of Group V members including XP_657754 and XP_001217072, which are known to produce polyketide synthase derived secondary metabolites like emodin and atrochrysone derivatives (Fig. S4A). The PT domain of F31_005372 would therefore most likely result in a C6-C11 cyclization pattern. Domain prediction by InterPro Scan analyses (Fig. S4B) indicated the absence of a thioesterase (TE) domain in the protein, which is another characteristic feature of Group V NR-PKS proteins50. The protein sequence of F31_005358 shares 74% identity with NsrC that functions as a metallo-β-lactamase thioesterase (MβL-TE) likely responsible for releasing the polyketide precursor (1) from the enzyme (Table S3)51. Therefore, based on the sequence identity and homology between most of the proteins encoded by the F31 cluster and their homologues in neosartorin and agnestins pathways52,53, we propose the following biosynthetic scheme (Fig. 3C). Claisen condensation of 7 units of malonyl CoA and 1 acetyl CoA by the F31_005372 would lead to the formation of the cyclized precursor polyketide (1), which is released by the MβL-TE (F31_005358) and further decarboxylated by F31_005359 and dehydrated by F31_005364 (which is 58% identical to AgnL8) to generate emodin (3). This further undergoes a series of oxidation and reduction reactions by the enzymes that share high sequence identity with those involved in agnestin biosynthesis52 including an oxidoreductase (F31_005362: 59% identical to AgnL4), short-chain dehydrogenase (F31_005363: 68% identical to AgnL6) and dehydratase (F31_005364: 54% identical to AgnL8) to form a dihydroxyanthraquinone (6). The Baeyer-Villiger oxidase encoded by F31_005360 shares 60% amino acid identity with NsrF would further oxidize (6) to (7), which can undergo methylation by a methyltransferase (F31_005365: 51% identical to NsrG) as well as opening the ring to form (8). Oxidation of (8) catalyzed by F31_005369 (60% identical to NsrK) encoding an FAD-dependent monooxygenase (FMO) would result in the formation of (9). F31_005361 encodes a monooxygenase that shares a weak sequence identity with NsrQ, probably producing (10). The only orphan that doesn’t share close homology with any protein in the either the agnestin52 or neosartorin53 biosynthetic pathways is a short chain dehydrogenase encoded by F31_005368.
Fig. 3: Proposed biosynthetic pathway for Nidulalin A production.
A) MA plot to represent the log2FC of genes induced with respect to normalized read counts in co-culture vs monoculture conditions with genes in the nidulalin A cluster B) Gene organization of nidulalin A biosynthetic gene cluster in F31 C) Putative biosynthetic pathway for nidulalin A production. Numbers above the arrows in the biosynthetic scheme represents the corresponding F31 genes for the indicated enzymatic reaction D) Top, chromatogram showing the 303 (m/z) ion detected in −DTT conditions as seen at 3.66 min. Inset, MS plot showing 303 m/z. Bottom, Chromatogram representing 457 and 611 m/z for the addition of one or two DTT. Inset: MS plot for 611 m/z ion (top) and 457 m/z ion (bottom). 303* (m/z) is an unrelated ion at Tr 3.9 min.
Knowing that we were likely looking for a lipophilic aromatic polyketide we performed ethyl acetate extraction from F31+Mtb culture filtrates followed by fractionation of the extracts on a C-18 liquid chromatography column which revealed that antitubercular activity eluted in a fraction containing a m/z of 303. The isolated peak had growth inhibitory activity against Mtb cells with an MIC90 of 1.5 μM (Table S5). Because we suspected this would be a thiol-reactive electrophile we also examined the reactivity of this metabolite with DTT39. Reaction with DTT resulted in the loss of the 303 m/z peak along with the appearance of the 457 Da and 611 Da ions corresponding to addition of one (+154 Da) or two (+308 Da) DTT molecules (Fig. 3D). 1H and 13C NMR spectroscopy (Supplemental Experimental 3.1) revealed that the product was a polyketide metabolite previously isolated from Emericella nidulans named nidulalin A54. As expected, there was minimal detection of nidulalin A in filtrates obtained from monocultures as compared to those from co-cultures as detected by LC-MS/MS, confirming the specific induction of nidulalin A upon co-cultivation with Mtb (Fig. S5A).
Fungal metabolites result in a thiol stress response in Mtb
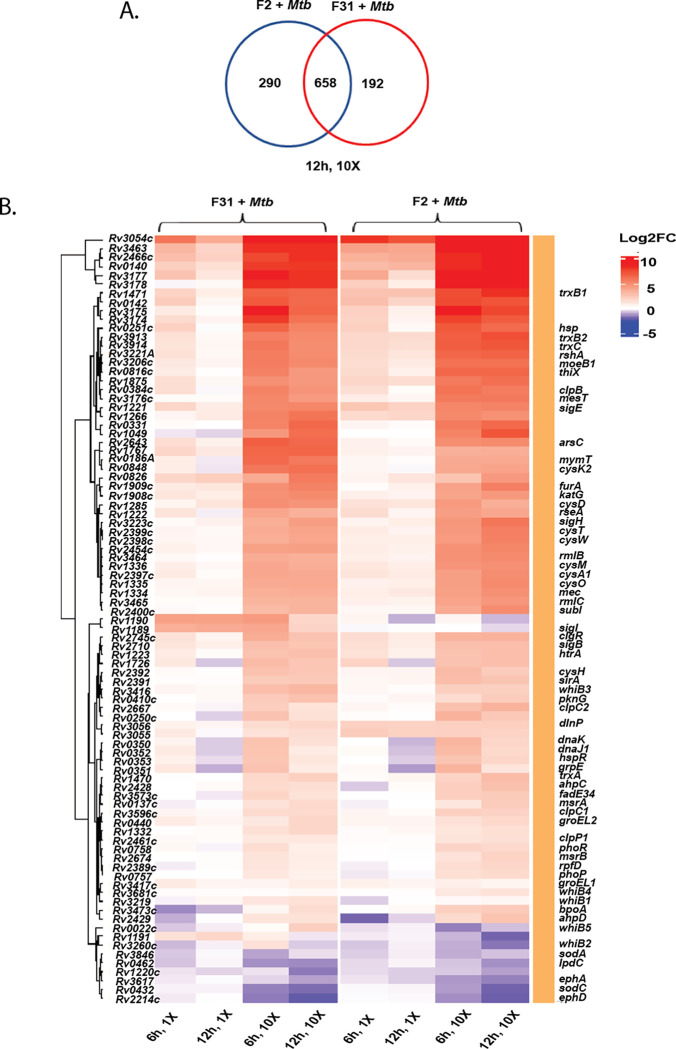
Attempts to raise Mtb mutants resistant to either patulin or nidulalin A consistently failed to generate high-level resistant mutants. We therefore performed global transcriptional profiling of Mtb during exposure to patulin and nidulalin A containing filtrates to identify differentially expressed genes. We excluded citrinin because of its modest activity against Mtb. Comparison of the patulin and nidulalin A exposed cells to control Mtb cultures revealed a concentration and time-dependent upregulation of a substantial set of response genes (see supplementary Table S4 and Dataset 2). Surprisingly, there was a significant overlap between the Mtb genes overexpressed in response to either patulin or nidulalin A at similar concentrations relative to their MIC (Fig. 4A, S7B). The transcriptional profile exhibited by Mtb exposed to the active metabolites from both of these filtrates indicated upregulation of redox stress pathways, in particular thiol reactive oxidative stress (Fig. 4B, S7A)55,56. This signature includes a significant dose-dependent expression of genes encoding sigma factors like sigB, sigE, sigH, and their associated regulons which play a role in thiol and redox homeostasis in Mtb in response to reactive oxygen and nitrogen species57. SigH regulates the expression of SigE, which together control the inducible expression of SigB58. The SigH regulon plays a central role in adjusting the thiol-oxidative stress during Mtb pathogenesis55,58–61. Genes encoding thioredoxin/thioredoxin reductases (trxC, trxB1, trxB2, thiX), methyltransferase (moeZ), cysteine synthesis/sulfur metabolism/transport (cysK2, cysA1, cysH, cysM, cysN, cysO, cysT, cysW, sirA, subI) and lipid synthesis genes (papA4) were all found to be over-expressed. There was also an upregulation of a recently discovered mycothiol dependent reductase mycoredoxin 2 (Rv2466c), several peroxidases, dehydrogenases (katG, Rv3177, Rv3178, ahpC, ahpD), ferric uptake transcriptional regulator (furA), several probable oxidoreductases (Rv1726, Rv2454, Rv3463). In fact, a recent study reported the coordinated expression of SigH and Mrx-2 under oxidative/nitrosative stress is important for the intracellular activation of nitronaphthofuran prodrugs62. Moreover, the lack of superoxide dismutase (sodA, sodC) and membrane associated oxidoreductases (SseA, DoxX family) upregulation in our study indicates that the response to the fungal metabolites was not that of a general oxidative stress response63. Higher concentrations of the co-culture filtrates similarly resulted in the upregulation of the thiol stress response with more extensive upregulation of additional stress responsive genes including several chaperones (Rv0251, clpB, clgR, grpE, htpx, Rv0991c), PhoP/R, protein kinase G (pknG), phosphoenolpyruvate carboxykinase (pckA), several cell envelope lipids or polyketide biosynthesis genes and many members of PE/PPE/PE-PGRS proteins (Fig. 4B and S9). The transcriptional profiling data suggests fungal metabolites such as patulin and nidulalin A result in depletion of free intracellular thiols like low molecular weight mycothiol/ergothioneine and reduced cysteines.
Fig. 4: Thiol stress homeostasis pathways triggered in Mtb upon exposure to nidulalin A (F31) and patulin (F2).
A) Venn diagram representing the overlap of upregulated genes between F2+Mtb and F31+Mtb exposed cells at 10X MIV concentration after 12h treatment (also see Fig. S7B for other treatment conditions). B) Heat map showing the differential expression of genes involved in redox/thiol stress homeostasis pathways in Mtb upon treatment with 1X and 10X MIV of F31+Mtb and F2+Mtb culture filtrates for 6h and 12h. Differentially expressed genes selected for the heat plot are statistically significant (FDR<0.1 and log2FC>1.0) in at least one treatment condition.
Nidulalin A and patulin deplete the free thiol pool within Mtb
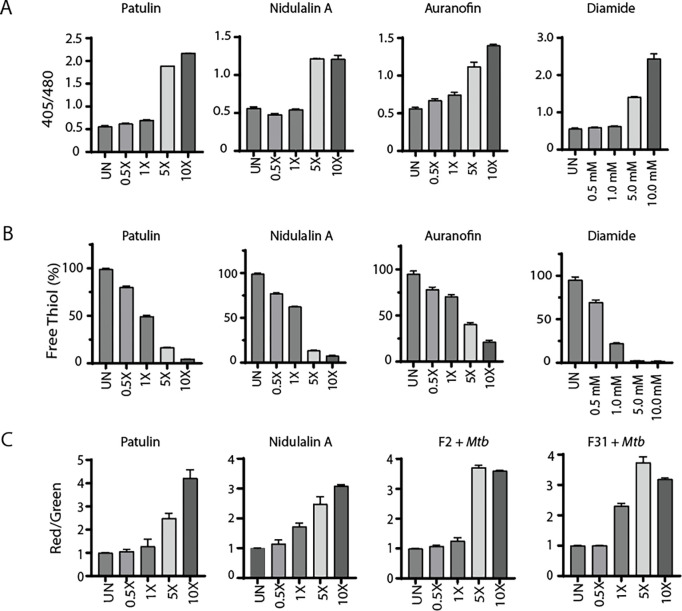
Based on the critical role of mycothiol in protecting against oxidative stress within mycobacterial cells, we used the previously reported64 Mrx1-roGFP2 redox biosensor to measure mycothiol-dependent redox potential during treatment with either patulin or nidulalin A. Cultures were treated with different concentrations of patulin and nidulalin A (0.5X-10X MIC90) for 0–48 h and responses compared to untreated cells or cells treated with auranofin (Aur; 0.5X-10X MIC) and diamide (0.5 mM-10 mM). Auranofin disrupts thiol-redox balance by inhibiting bacterial thioredoxin reductase65,66 whereas diamide causes reversible oxidation of intracellular thiols55,67,68. Both patulin and nidulalin A treatment led to a 2–4 fold increase in Mrx1-roGFP2 fluorescence ratio at 5X and 10X concentrations at 48h, consistent with increased mycothiol oxidation (Fig. 5A) with increased oxidative poise already evident at 12h to 24h (data not shown). As expected, auranofin and diamide similarly resulted in an oxidative shift (~3–5 fold) (Fig. 5A).
Fig. 5: Patulin and nidulalin A induce a thiol reactive oxidative shift within Mtb.
A. Mtb H37Rv cells expressing Mrx1-roGFP2 grown to OD600 0.1 were treated with patulin and nidulalin A at different concentrations for 48 h. Diamide, auranofin were used as positive controls for inducing thiol stress. Ratiometric sensor response (Ex:405 nm/488 nm; Em:510 nm). B. Quantitation of free thiol pool was evaluated in Mtb H37Rv cells treated with patulin and nidulalin A at different concentrations for 24h. Free thiol levels were detected by measuring the relative fluorescence at Em: 510 nm (Ex: 380 nm) using a fluorescent thiol detection assay. Relative fluorescence was normalized for the OD600 for all the samples tested with respect to the control group. A glutathione (GSH) standard curve was used to estimate free thiol levels. Auranofin and diamide were used as thiol stress controls. The assays results presented here were performed in at least two biological replicates. C. Rv3054c promoter driven expression of mScarlet normalized to constitutive eGFP in the same cell. UN represents the untreated control cultures.
We also measured free thiol pools after exposure of Mtb cells to either patulin or nidulalin A at 0.5–1X MIC and observed a 25–50% depletion with complete depletion at 5X - 10X MICs compared to the untreated control. Free thiol depletion during patulin or nidulalin A treatment was higher than that observed for auranofin but comparable to those obtained during diamide treatment (Fig. 5B). Citrinin is also predicted to be a Michael acceptor which was supported by the observation that this metabolite and the corresponding C7+Mtb fungal filtrate also resulted in depletion of cellular thiols (Fig. S6A). Subsequently two other co-culture derived fungal cell-free supernatants (D6 and D9) identified while this work was in progress were also found to result in depletion of intracellular thiols in Mtb (Fig. S6B). We confirmed that nidulalin A formed mycothiol adducts in vivo within Mtb as measured by the release of a nidulalin A-mercapturic acid covalent conjugate in the extracellular milieu 69,70 (Fig. S5B).
The most highly upregulated gene in response to thiol stress was Rv3054c. This gene has previously been reported to be upregulated by diamide and regulated by SigH56,71. We therefore created a reporter plasmid where the Rv3054c promoter drives expression of the mScarlet fluorescent protein with expression of the eGFP fluorescent protein under a constitutive promoter. Mtb carrying this reporter construct demonstrated an increased red to green fluorescence ratio in response to a concentration range of nidulalin A, patulin, their corresponding induced culture filtrates (F2+Mtb, F31+Mtb), diamide and iodoacetamide (Fig. 5C and S6C). Higher concentrations of iodoacetamide resulted in decreased mScarlet fluorescence likely due to a corresponding decline in Mtb viability (Fig. S6C ii). To demonstrate the global relevance of thiol stress as an anti-tubercular mechanism, the Rv3054c promoter-mediated increase in fluorescence was also confirmed for other induced fungal filtrates (C7+Mtb, D6+Mtb, D9+Mtb) (Fig. S6D) identified in our initial screens.
DISCUSSION
Co-cultivation is increasingly being used to unlock cryptic natural products that are not normally expressed under standard in vitro growth conditions72. Mycobacteria, like most members of the Mycobacteriales order, have unique and complex cell surfaces that offer potential for recognition by competitors. In fact, mycolic acid producing bacteria have been shown to induce the expression of silent secondary metabolites in other actinomycetes73. Our results support the use of mycobacteria as inducers of secondary metabolites in fungi.
Both fungi and mycobacteria are found in the gray decomposition layer of the sphagnum peat bog where they compete for limited nutrients as the cell walls of sphagnum moss are slowly decomposed. In this microenvironment, which appears to support the highest concentration of mycobacteria found anywhere in nature, secondary metabolite production by fungi to enhance their ability to compete makes sense. In our initial screen of the ~1500 fungi isolated from sphagnum samples at or near the gray layer cores, we identified a total of five fungi that showed robust induction of antimycobacterial activity upon exposure to live cells of Mtb. Three of these fungi were closely related with identical ITS sequences but were isolated from different geographical areas and upon whole genome sequencing were found to be distinct yet closely related. These three fungi induced expression of patulin, a known mycotoxin, in response to the presence of Mtb. Patulin synthesis in P. expansum is controlled by the transcription factor PatL in response to a wide variety of poorly characterized environmental features including carbon catabolism, light, and pH74. The fourth Penicillium sp. we identified by co-culture weakly induced another complex mycotoxin, citrinin. Like patulin, regulation of citrinin production in fungi is complex and multifactorial.
The fifth fungus induced the expression of a dihydroxyxanthone metabolite that has been previously described from a Penicillium sp. from a Japanese soil sample called nidulalin A75. Initially identified as having antitumour activity nidulalin A and derivatives have been explored as Topoisomerase II inhibitors and have been associated with a range of bioactivities76–78. Several total syntheses of nidulalin A have been reported79–81 but no further details of its biosynthesis have been reported previously. Our results determined the biosynthetic origins of nidulalin A. Nidulalin A biosynthesis shares many steps with the biosynthetic pathways to agnestin52 and neosartorin53, distinguished by a unique short-chain dehydrogenase that catalyzes the penultimate step.
Remarkably, the three metabolites produced by these five unique fungi all target the same physiological process in Mtb cells: thiol homeostasis. This suggests that fungi have selected for the most vulnerable target in mycobacteria under the specific microenvironmental conditions of the gray-layer of a sphagnum bog and identified thiol stress as being the most effective target. These results contrast with the results of a recent genome-wide CRISPR interference screen assessment of target vulnerability82, but this is not surprising as that screen was done under conditions of aerobically replicating cells. Although mycotoxins, per se, are not promising starting points for drug discovery, they do point to a rich target area that could be exploited and there are many other strategies for drug discovery that could lead to enhanced thiol oxidative stress. Recent screening that has been done under acidic conditions, which amplifies the sensitivity towards imbalances in redox poise, has identified active compounds that perturb thiol homeostasis and give a transcriptional response very similar to that reported here83,84. Inhibitors of IspQ, a putative redox sensor, gave rise to compounds that induced thiol stress85. Screening of thiol-deficient strains to look for differential activity of hits has led to the identification of compounds that can modulate thiol and oxidative stress levels86.
Overall, our results suggest that to eradicate the most difficult to kill Mtb that exist within hypoxic, caseous lesions and cavity walls, compounds that target thiol homeostasis have the potential to have significant impact. Improving the clearance of these bacilli has a high likelihood of reducing the overall treatment duration for TB patients.
Acknowledgements
This work was funded by the Intramural Research Program of NIAID, NIH (CEB3). We thank Dr. Amir Sayed Mousavi and Dr. Pavel Khil (Department of Laboratory Medicine, Clinical Center, NIH) for help with fungal identification. We would also like to thank Dr. Adrian Zelazny (Department of Laboratory Medicine, Clinical Center, NIH) for the ESKAPE pathogens and Dr. Amit Singh (International Centre for Genetic Engineering and Biotechnology, New Delhi, India) for the Mrx-roGFP2 construct. We would like to thank Dr. Robert O’Conner (Laboratory of Bioorganic Chemistry, NIDDK/NIH) for assistance with the 2D NMR spectra. We would also like to thank Rosanna Springston and Gauley District Ranger Richard Raione (US Forest Service) for guiding sampling at Cranberry Glades.
Abbreviations
- RT
Room Temperature
- RPM
Rotation Per Minute
- BGC
Biosynthetic gene cluster
- PKS
Polyketide Synthase
- MA
Magnitude-Amplitude plot
- SD
Standard Deviation
- Log2FC
Log2Fold Change
- MS
Mass Spectrometer
- LC-MS
Liquid Chromatography-Mass Spectrometry
- LC-MS/MS
Liquid Chromatography-tandem Mass Spectrometry
- NaCl
Sodium Chloride
- Mtb
Mycobacterium tuberculosis
- BUSCO
Benchmarking Universal Single-Copy Ortholog
- SDR
Short Chain Dehydrogenase/Reductase
- FMN
Flavin Adenine Mononucleotide
- FAD
Flavin Adenine Dinucleotide
- ER
Enoyl Reductase
- Pat
Patulin
- Tx
Tyloxapol
- Tw
Tween-80
- ADC
Albumin-Dextrose-Catalase
- ADGNTw
Albumin-Dextrose-Glycerol-NaCl-Tween
- OADGN
Oleic acid-Albumin-Dextrose-Glycerol-NaCl
- RFU
Relative Fluorescence Unit
- CPM
Read Count Per Million
- EtOAc
Ethyl Acetate
- ITS
Internal Transcribed Spacer region
- Agn
Agnestin
- Nsr
Neosartorin
- PDA
Potato Dextrose Agar
- PDB
Potato Dextrose Broth
- YEM
Yeast Extract Mannitol
- NP
Natural Product
- TF
Transcription Factor
- MFS
Multi-Facilitator Substrate
- ACP
Acyl Carrier Protein domain
- PKS
Polyketide synthase
- PP
Phosphopatathiene domain
- PT
Product Template domain
- DH
Dehydrogenase domain
- NR-PKS
Non-Reducing Polyketide Synthase
- NMR
Nuclear Magnetic Resonance
- eGFP
Enhanced Green Fluorescent protein
- INH
Isoniazide
- Aur
Auranofin
- DTT
Dithiothrietol
- Padj
Adjusted p-value
- MIC
Minimum Inhibitory Concentration
- MIV
Minimum Inhibitory Volume
- PE
Proline-Glutamic acid
- PPE
Proline-Proline-Glutamic acid
- PGRS
Polymorphic GC-rich Repetitive Sequence
- FDR
False Discovery Rate
- MRM
Multiple Reaction Monitoring
- gDNA
Genomic Deoxyribose Nucleic Acid
- RNA
Ribose Nucleic Acid
- DEPC
Diethyl pyrocarbonate
- TLC
Thin Layer Chromatography
- Mrx
Mycoredoxin
- DMSO
Dimethyl Sulfoxide
- CFU
Colony Forming Unit
- SNP
Single Nucleotide Polymorphism
- BSA
Bovine Serum Albumin
- WGS
Whole Genome Sequencing
- NCBI
National Center for Biotechnology information
- BLAST
Basic Local Alignment Search Tool
- GEO
Gene Expression Omnibus
Footnotes
Supplementary Data
Supplemental Information (.txt) – Tables and Figures supporting our findings.
Dataset 1 (.xml) – Fungal RNAseq data
Dataset 2 (.xml) – Mtb RNAseq data
Data Availability
Data supporting the findings of this study are included in the main manuscript and Supplementary Data. Genome assemblies as well as their Sequence Read Archive (SRA) data files have been submitted to NCBI and are available in the BioProject PRJNA767328. Gene expression data for fungal isolates and Mtb obtained in the study were submitted to GEO-NCBI database and are available online with accession numbers mentioned above. Source data for all the results can be provided upon request.
References
- 1.Singh V. Tuberculosis treatment-shortening. Drug Discov Today 29, 103955 (2024). 10.1016/j.drudis.2024.103955 [DOI] [PubMed] [Google Scholar]
- 2.Sarathy J. P. & Dartois V. Caseum: a Niche for Mycobacterium tuberculosis Drug-Tolerant Persisters. Clin Microbiol Rev 33 (2020). 10.1128/CMR.00159-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Permar S. & Sun Z. Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J Med Microbiol 51, 42–49 (2002). 10.1099/0022-1317-51-1-42 [DOI] [PubMed] [Google Scholar]
- 4.Yuan T. & Sampson N. S. Hit Generation in TB Drug Discovery: From Genome to Granuloma. Chem Rev 118, 1887–1916 (2018). 10.1021/acs.chemrev.7b00602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarathy J. P. et al. A Novel Tool to Identify Bactericidal Compounds against Vulnerable Targets in Drug-Tolerant M. tuberculosis found in Caseum. mBio 14, e0059823 (2023). 10.1128/mbio.00598-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nuermberger E. L. et al. GSK2556286 Is a Novel Antitubercular Drug Candidate Effective In Vivo with the Potential To Shorten Tuberculosis Treatment. Antimicrob Agents Chemother 66, e0013222 (2022). 10.1128/aac.00132-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parish T. In vitro drug discovery models for Mycobacterium tuberculosis relevant for host infection. Expert Opin Drug Discov 15, 349–358 (2020). 10.1080/17460441.2020.1707801 [DOI] [PubMed] [Google Scholar]
- 8.Thorel M. F., Falkinham J. O. 3rd & Moreau R. G. Environmental mycobacteria from alpine and subalpine habitats. FEMS Microbiol Ecol 49, 343–347 (2004). 10.1016/j.femsec.2004.04.016 [DOI] [PubMed] [Google Scholar]
- 9.Kazda J. The Ecology of Mycobacteria. (Kluwer Academic Publishers, 2000). [Google Scholar]
- 10.Bragina A. et al. The Sphagnum microbiome supports bog ecosystem functioning under extreme conditions. Mol Ecol 23, 4498–4510 (2014). 10.1111/mec.12885 [DOI] [PubMed] [Google Scholar]
- 11.Liu L., Wang Z., Ma D., Zhang M. & Fu L. Diversity and Distribution Characteristics of Soil Microbes across Forest-Peatland Ecotones in the Permafrost Regions. Int J Environ Res Public Health 19 (2022). 10.3390/ijerph192214782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irgens L. M. Leprosy in Norway. An epidemiological study based on a national patient registry. Lepr Rev 51 Suppl 1, i-xi, 1–130 (1980). [PubMed] [Google Scholar]
- 13.Kazda J., Irgens L. M. & Muller K. Isolation of non-cultivable acid-fast bacilli in sphagnum and moss vegetation by foot pad technique in mice. Int J Lepr Other Mycobact Dis 48, 1–6 (1980). [PubMed] [Google Scholar]
- 14.Kostka J. E. et al. The Sphagnum microbiome: new insights from an ancient plant lineage. New Phytol 211, 57–64 (2016). 10.1111/nph.13993 [DOI] [PubMed] [Google Scholar]
- 15.Pankratov T. A., Serkebaeva Y. M., Kulichevskaya I. S., Liesack W. & Dedysh S. N. Substrate-induced growth and isolation of Acidobacteria from acidic Sphagnum peat. ISME J 2, 551–560 (2008). 10.1038/ismej.2008.7 [DOI] [PubMed] [Google Scholar]
- 16.Kolbe K. et al. Development and optimization of chromosomally-integrated fluorescent Mycobacterium tuberculosis reporter constructs. Frontiers in microbiology 11, 591866 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhaskar A. et al. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS pathogens 10, e1003902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gontia-Mishra I., Tripathi N. & Tiwari S. A simple and rapid DNA extraction protocol for filamentous fungi efficient for molecular studies. (2014). [Google Scholar]
- 19.Bellemain E. et al. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC microbiology 10, 1–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magi A., Semeraro R., Mingrino A., Giusti B. & D’aurizio R. Nanopore sequencing data analysis: state of the art, applications and challenges. Briefings in bioinformatics 19, 1256–1272 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Illumina. BCL Convert: a proprietary Illumina software for the conversion of BCL files to basecalls. (2021). [Google Scholar]
- 22.Lin Y. et al. Assembly of long error-prone reads using de Bruijn graphs. Proceedings of the National Academy of Sciences 113, E8396–E8405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker Bruce J. et al. An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gurevich A. & Saveliev V. Vyahhi and, N., Tesler G.(2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love J. et al. Nextgenusfs/Funannotate: Funannotate v1. 5.1. Zenodo. doi 10 (2018). [Google Scholar]
- 26.Ewels P. A. et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 38, 276–278 (2020). 10.1038/s41587-020-0439-x [DOI] [PubMed] [Google Scholar]
- 27.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danecek P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10 (2021). 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Wang S. & Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- 30.Okonechnikov K., Conesa A. & Garcia-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32, 292–294 (2016). 10.1093/bioinformatics/btv566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Y., Smyth G. K. & Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 32.Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hidalgo-Cantabrana C., Goh Y. J., Pan M., Sanozky-Dawes R. & Barrangou R. Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proceedings of the National Academy of Sciences 116, 15774–15783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S., Stecher G., Li M., Knyaz C. & Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular biology and evolution 35, 1547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blin K. et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic acids research, gkad344 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B. & Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics 12, 1–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 43, e47–e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kariyawasam H., Su S., Voogd O., Ritchie M. E. & Law C. W. Dashboard-style interactive plots for RNA-seq analysis are R Markdown ready with Glimma 2.0. NAR Genom Bioinform 3, lqab116 (2021). 10.1093/nargab/lqab116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris L. A. & Mitchell D. A. in Methods in enzymology Vol. 665 177–208 (Elsevier, 2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coulson G. B. et al. Targeting Mycobacterium tuberculosis sensitivity to thiol stress at acidic pH kills the bacterium and potentiates antibiotics. Cell chemical biology 24, 993–1004. e1004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaps I. et al. Energy transfer between fluorescent proteins using a co-expression system in Mycobacterium smegmatis. Gene 278, 115–124 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Don R., Cox P., Wainwright B., Baker K. & Mattick J. ‘Touchdown’PCR to circumvent spurious priming during gene amplification. Nucleic acids research 19, 4008 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engler C., Kandzia R. & Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PloS one 3, e3647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parish T. Electroporation of mycobacteria. Mycobacteria Protocols, 273–284 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Parish T. & Stoker N. G. in Mycobacteria Protocols (eds Parish Tanya & Stoker Neil G) 129–144 (Humana Press, 1998). [Google Scholar]
- 46.Zuo G. CVTree: A Parallel Alignment-free Phylogeny and Taxonomy Tool Based on Composition Vectors of Genomes. Genomics Proteomics Bioinformatics 19, 662–667 (2021). 10.1016/j.gpb.2021.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blin K. et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res 51, W46–W50 (2023). 10.1093/nar/gkad344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tannous J. et al. Sequencing, physical organization and kinetic expression of the patulin biosynthetic gene cluster from Penicillium expansum. Int J Food Microbiol 189, 51–60 (2014). 10.1016/j.ijfoodmicro.2014.07.028 [DOI] [PubMed] [Google Scholar]
- 49.Ewels P., Magnusson M., Lundin S. & Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016). 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ballester A. R. et al. Genome, Transcriptome, and Functional Analyses of Penicillium expansum Provide New Insights Into Secondary Metabolism and Pathogenicity. Mol Plant Microbe In 28, 232–248 (2015). 10.1094/MPMI-09-14-0261-FI [DOI] [PubMed] [Google Scholar]
- 51.Liu L., Zhang Z., Shao C. L. & Wang C. Y. Analysis of the Sequences, Structures, and Functions of Product-Releasing Enzyme Domains in Fungal Polyketide Synthases. Front Microbiol 8, 1685 (2017). 10.3389/fmicb.2017.01685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szwalbe A. J. et al. Characterisation of the biosynthetic pathway to agnestins A and B reveals the reductive route to chrysophanol in fungi. Chem Sci 10, 233–238 (2019). 10.1039/c8sc03778g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsuda Y., Gotfredsen C. H. & Larsen T. O. Genetic Characterization of Neosartorin Biosynthesis Provides Insight into Heterodimeric Natural Product Generation. Organic Letters 20, 7197–7200 (2018). 10.1021/acs.orglett.8b03123 [DOI] [PubMed] [Google Scholar]
- 54.Kawahara N., Sekita S., Satake M., Udagawa S. & Kawai K. Structures of a New Dihydroxanthone Derivative, Nidulalin-a, and a New Benzophenone Derivative, Nidulalin-B, from Emericella Nidulans. Chem Pharm Bull 42, 1720–1723 (1994). [Google Scholar]
- 55.Coulson G. B. et al. Targeting Mycobacterium tuberculosis Sensitivity to Thiol Stress at Acidic pH Kills the Bacterium and Potentiates Antibiotics. Cell Chemical Biology 24, 993-+ (2017). 10.1016/j.chembiol.2017.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choudhary E., Sharma R., Pal P. & Agarwal N. Deciphering the Proteomic Landscape of Mycobacterium tuberculosis in Response to Acid and Oxidative Stresses. ACS Omega 7, 26749–26766 (2022). 10.1021/acsomega.2c03092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sachdeva P., Misra R., Tyagi A. K. & Singh Y. The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. FEBS J 277, 605–626 (2010). 10.1111/j.1742-4658.2009.07479.x [DOI] [PubMed] [Google Scholar]
- 58.Raman S. et al. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J Bacteriol 183, 6119–6125 (2001). 10.1128/JB.183.20.6119-6125.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voskuil M. I., Bartek I. L., Visconti K. & Schoolnik G. K. The response of mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front Microbiol 2, 105 (2011). 10.3389/fmicb.2011.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manganelli R. et al. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol Microbiol 45, 365–374 (2002). 10.1046/j.1365-2958.2002.03005.x [DOI] [PubMed] [Google Scholar]
- 61.Dechow S. J., Coulson G. B., Wilson M. W., Larsen S. D. & Abramovitch R. B. AC2P20 selectively kills at acidic pH by depleting free thiols. Rsc Advances 11, 20089–20100 (2021). 10.1039/d1ra03181c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cioetto-Mazzabo L. et al. SigH stress response mediates killing of Mycobacterium tuberculosis by activating nitronaphthofuran prodrugs via induction of Mrx2 expression. Nucleic Acids Res 51, 144–165 (2023). 10.1093/nar/gkac1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nambi S. et al. The Oxidative Stress Network of Mycobacterium tuberculosis Reveals Coordination between Radical Detoxification Systems. Cell Host Microbe 17, 829–837 (2015). 10.1016/j.chom.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhaskar A. et al. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog 10, e1003902 (2014). 10.1371/journal.ppat.1003902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harbut M. B. et al. Auranofin exerts broad-spectrum bactericidal activities by targeting thiol-redox homeostasis. Proc Natl Acad Sci U S A 112, 4453–4458 (2015). 10.1073/pnas.1504022112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruth M. M. et al. Auranofin Activity Exposes Thioredoxin Reductase as a Viable Drug Target in. Antimicrobial Agents and Chemotherapy 63, 10.1128/aac. 00449–00419 (2019). https://doi.org/ARTNe00449-19, 10.1128/AAC.00449-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kosower N. S. & Kosower E. M. [11] Diamide: An oxidant probe for thiols. Methods in enzymology 251, 123–133 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Dosanjh N. S., Rawat M., Chung J. H. & Av-Gay Y. Thiol specific oxidative stress response in Mycobacteria. FEMS Microbiol Lett 249, 87–94 (2005). 10.1016/j.femsle.2005.06.004 [DOI] [PubMed] [Google Scholar]
- 69.Jothivasan V. K. & Hamilton C. J. Mycothiol: synthesis, biosynthesis and biological functions of the major low molecular weight thiol in actinomycetes. Nat Prod Rep 25, 1091–1117 (2008). 10.1039/b616489g [DOI] [PubMed] [Google Scholar]
- 70.Newton G. L., Av-Gay Y. & Fahey R. C. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry 39, 10739–10746 (2000). 10.1021/bi000356n [DOI] [PubMed] [Google Scholar]
- 71.Sharp J. D. et al. Comprehensive Definition of the SigH Regulon of Mycobacterium tuberculosis Reveals Transcriptional Control of Diverse Stress Responses. PLoS One 11, e0152145 (2016). 10.1371/journal.pone.0152145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cueto M. et al. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J Nat Prod 64, 1444–1446 (2001). 10.1021/np0102713 [DOI] [PubMed] [Google Scholar]
- 73.Hoshino S., Onaka H. & Abe I. Activation of silent biosynthetic pathways and discovery of novel secondary metabolites in actinomycetes by co-culture with mycolic acid-containing bacteria. J Ind Microbiol Biotechnol 46, 363–374 (2019). 10.1007/s10295-018-2100-y [DOI] [PubMed] [Google Scholar]
- 74.Wang W., Liang X., Li Y., Wang P. & Keller N. P. Genetic Regulation of Mycotoxin Biosynthesis. J Fungi (Basel) 9 (2022). 10.3390/jof9010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sato S. et al. F390B and C, new antitumor dihydroxanthone derivatives isolated from Penicillium sp. J Antibiot (Tokyo) 50, 614–616 (1997). 10.7164/antibiotics.50.614 [DOI] [PubMed] [Google Scholar]
- 76.Sato S. et al. Syntheses of novel antitumor dihydroxanthone derivatives with inhibitory activity against DNA topoisomerase II. Bioorg Med Chem Lett 9, 2653–2656 (1999). 10.1016/s0960-894x(99)00440-0 [DOI] [PubMed] [Google Scholar]
- 77.Sato S. et al. Inhibition of DNA topoisomerases by nidulalin A derivatives. Biol Pharm Bull 23, 511–512 (2000). 10.1248/bpb.23.511 [DOI] [PubMed] [Google Scholar]
- 78.Fujimoto H., Asai T., Kim Y. P. & Ishibashi M. Nine constituents including six xanthone-related compounds isolated from two ascomycetes, Gelasinospora santi-florii and Emericella quadrilineata, found in a screening study focused on immunomodulatory activity. Chem Pharm Bull (Tokyo) 54, 550–553 (2006). 10.1248/cpb.54.550 [DOI] [PubMed] [Google Scholar]
- 79.Tatsuta K., Yoshihara S., Hattori N., Yoshida S. & Hosokawa S. The first total synthesis of nidulalin A, a dihydroxanthone possessing multiple bioactivities. J Antibiot (Tokyo) 62, 469–470 (2009). 10.1038/ja.2009.52 [DOI] [PubMed] [Google Scholar]
- 80.Ji K., Johnson R. P., McNeely J., Al Faruk M. & Porco J. A. Jr. Asymmetric Synthesis of Nidulalin A and Nidulaxanthone A: Selective Carbonyl Desaturation Using an Oxoammonium Salt. J Am Chem Soc 146, 4892–4902 (2024). 10.1021/jacs.3c13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ji K., Parthiban J., Jockusch S., Sivaguru J. & Porco J. A. Jr. Triple-Dearomative Photocycloaddition: A Strategy to Construct Caged Molecular Frameworks. J Am Chem Soc 146, 13445–13454 (2024). 10.1021/jacs.4c02674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bosch B. et al. Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis. Cell 184, 4579–4592 e4524 (2021). 10.1016/j.cell.2021.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dechow S. J., Coulson G. B., Wilson M. W., Larsen S. D. & Abramovitch R. B. AC2P20 selectively kills Mycobacterium tuberculosis at acidic pH by depleting free thiols. RSC Adv 11, 20089–20100 (2021). 10.1039/d1ra03181c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coulson G. B. et al. Targeting Mycobacterium tuberculosis Sensitivity to Thiol Stress at Acidic pH Kills the Bacterium and Potentiates Antibiotics. Cell Chem Biol 24, 993–1004 e1004 (2017). 10.1016/j.chembiol.2017.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Szekely R. et al. 6,11-Dioxobenzo[f]pyrido[1,2-a]indoles Kill Mycobacterium tuberculosis by Targeting Iron-Sulfur Protein Rv0338c (IspQ), A Putative Redox Sensor. ACS Infect Dis 6, 3015–3025 (2020). 10.1021/acsinfecdis.0c00531 [DOI] [PubMed] [Google Scholar]
- 86.Sao Emani C., Williams M. J., Wiid I. J., Baker B. & Carolis C. Compounds with Potential Activity against Mycobacterium tuberculosis. Antimicrob Agents Chemother 62 (2018). 10.1128/AAC.02236-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the findings of this study are included in the main manuscript and Supplementary Data. Genome assemblies as well as their Sequence Read Archive (SRA) data files have been submitted to NCBI and are available in the BioProject PRJNA767328. Gene expression data for fungal isolates and Mtb obtained in the study were submitted to GEO-NCBI database and are available online with accession numbers mentioned above. Source data for all the results can be provided upon request.