Abstract
Background
While previous research has shown the potential links between taste perception pathways and brain-related conditions, the area involving Alzheimer’s disease remains incompletely understood. Taste perception involves neurotransmitter signaling, including serotonin, glutamate, and dopamine. Disruptions in these pathways are implicated in neurodegenerative diseases. The integration of olfactory and taste signals in flavor perception may impact brain health, evident in olfactory dysfunction as an early symptom in neurodegenerative conditions. Shared immune response and inflammatory pathways may contribute to the association between altered taste perception and conditions like neurodegeneration, present in Alzheimer’s disease.
Methods
This study consists of an exploration of expression-quantitative trait loci (eQTL), utilizing whole-blood transcriptome profiles, of 28 taste perception genes, from a combined cohort of 475 African American subjects. This comprehensive dataset was subsequently intersected with single-nucleotide polymorphisms (SNPs) identified in Genome-Wide Association Studies (GWAS) of Alzheimer’s Disease (AD). Finally, the investigation delved into assessing the association between eQTLs reported in GWAS of AD and the profiles of 741 proteins from the Olink Neurological Panel.
Results
The eQTL analysis unveiled 3,547 statistically significant SNP-Gene associations, involving 412 distinct SNPs that spanned all 28 taste genes. In 17 GWAS studies encompassing various traits, a total of 14 SNPs associated with 12 genes were identified, with three SNPs consistently linked to Alzheimer’s disease across four GWAS studies. All three SNPs demonstrated significant associations with the down-regulation of TAS2R41, and two of them were additionally associated with the down-regulation of TAS2R60. In the subsequent pQTL analysis, two of the SNPs linked to TAS2R41 and TAS2R60 genes (rs117771145 and rs10228407) were correlated with the upregulation of two proteins, namely EPHB6 and ADGRB3.
Conclusions
Our investigation introduces a new perspective to the understanding of Alzheimer’s disease, emphasizing the significance of bitter taste receptor genes in its pathogenesis. These discoveries set the stage for subsequent research to delve into these receptors as promising avenues for both intervention and diagnosis. Nevertheless, the translation of these genetic insights into clinical practice requires a more profound understanding of the implicated pathways and their pertinence to the disease’s progression across diverse populations.
Keywords: Taste Genes, GWAS, Transcriptome, Proteome, Alzheimer’s Disease, African American
INTRODUCTION
Taste perception is a complex sensory phenomenon that involves the detection and interpretation of various chemical stimuli by specialized receptors located on the tongue and oral cavity 1. The human sense of taste, or gustation, is primarily categorized into five modalities: sweet, salty, sour, bitter, and umami. The intricate interplay between taste perception and the brain constitutes a process, integrating peripheral sensory signals with central neural processing. Neuroimaging studies, such as functional magnetic resonance imaging and positron emission tomography, have provided valuable insights into the neural mechanisms underlying taste perception 2,3. These studies reveal the involvement of various brain regions in different aspects of taste processing, highlighting the distributed nature of the taste neural network.
There is evidence to suggest potential links between taste perception pathways and brain-related conditions 4,5; however, the connections are still not fully understood. For instance, taste perception involves neurotransmitter signaling in the gustatory system, particularly through molecules such as serotonin, glutamate, and dopamine 6. Disruptions in these neurotransmitter systems have been implicated in mood disorders, schizophrenia, and neurodegenerative diseases 7,8. The integration of olfactory and taste signals in flavor perception may have implications for brain health. For example, olfactory dysfunction is a common early symptom in neurodegenerative conditions like Alzheimer’s and Parkinson’s diseases 9. Futhermore, inflammation plays a role in both alterations in taste perception and neuroinflammatory conditions. Shared pathways involving immune response and inflammatory mediators may contribute to the link between altered taste perception and conditions like depression and neurodegeneration 10,11.
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder that significantly impacts cognitive function and daily living activities. As the most common cause of dementia, AD is characterized by the accumulation of amyloid-beta plaques and tau tangles, leading to neuronal loss and a decline in cognitive abilities 12. Alongside these well-documented features, AD is increasingly associated with sensory dysfunctions, including alterations in taste and smell perception 9,13. These sensory changes are not just secondary symptoms but are thought to be integral to the disease process, potentially serving as early indicators of neurodegeneration 9.
Taste alterations in individuals with Alzheimer’s diseases have been documented in various studies 13–15. Individuals with AD often experience changes in food preferences, decreased taste sensitivity or increase taste preference for sweet and salty tastes, and sometimes a complete loss of taste perception. These alterations can lead to nutritional imbalances and affect the quality of life and overall health 16. The underlying mechanisms are believed to involve both central and peripheral pathways, including changes in taste bud integrity, central processing of taste information and taste receptor expression.
Genetic factors influencing taste receptor expression may have broader implications for many diseases including brain conditions 17–20. Polymorphisms in taste receptor genes loci may contribute to variations in gene expression and protein levels that could be associated with neurodenerative conditions such as Alzheimer’s disease (AD) 21. Functional genomics approaches, leveraging transcriptomics and proteomics can serve as valuable supplements to genetic inquiries and offer perspectives into the molecular mechanisms that link polymorphisms associated with taste perception to brain diseases. Such strategies can help decipher the complex biological pathways linked to the initiation and advancement of those diseases.
Notably, AD and associated sensory dysfunctions may vary significantly across different populations, influenced by a complex interplay of genetic, environmental, and social factors. African Americans are disproportionately affected by AD, experiencing higher incidence rates and more severe cognitive deterioration compared to other racial groups22,23. These disparities are not fully understood and are attributed to factors including genetic predisposition, health comorbidities, socioeconomic status, and access to healthcare. Understanding the specific genetic and molecular basis of taste alterations in an African American cohort can provide insights into the unique progression of AD in this population and highlight potential areas for targeted intervention and care. The objectives of this project are twofold: (1) identify expression-quantitative trait loci (eQTL) impacting the expression of taste-related genes that have been reported in genome-wide association studies (GWAS) of brain conditions, and (2) evaluate the connections between these variants and proteins involved in neurological processes relevant to neurodegenerative conditions and specifically AD.
MATERIAL AND METHODS
Phenotype data
The GENomics, Environmental FactORs and the Social DEterminants of Cardiovascular Disease in African-Americans STudy (GENE-FORECAST) is a research platform that establishes a strategic, multi-omics systems biology approach amenable to the deep, multi-dimensional characterization of minority health and disease in AA (African American). GENE-FORECAST is designed to create a cohort based on a community-based sampling frame of U.S.- born, AA men and women (ages 21–65) recruited from the metropolitan Washington D.C. area.
The Minority Health Genomics and Translational Research Bio-Repository Database (MH-GRID) project is a study of hypertension (HTN) in AA aged 30 to 55 years. The data included in this analysis is from an MH-GRID sub-study of samples from the Morehouse School of Medicine (MSM), in Atlanta (Georgia).
Transcriptome data
The transcriptome data consisted of the mRNA sequencing data of whole blood (buffy coat). Total RNA extraction was carried out using MagMAXTM for Stabilized Blood Tubes RNA Isolation Kit as recommended by vendor (Life Technologies, Carlsbad, CA). For library preparation, total RNA samples are concentration normalized, and ribosomal RNA (rRNA) is removed. Illumina paired end sequencing was performed on HiSeq2000 analyzer (Illumina, USA) with an average sequencing depth of 50 million reads per sample. The mRNA expression was quantified using a bioinformatics pipeline developed by the Broad Institutes and used by the Genotype-Tissue Expression (GTEx). The pipeline is detailed in the GitHub software development platform 24. Transcripts that did not achieve an expression of one read count per million (CPM) in at least three samples were excluded. The expression data was normalized using the Trimmed Mean of M-values (TMM), an optimal method for read count data 25. Principal component analysis was conducted to identify and exclude sample and gene outliers. A set of 17,948 protein coding mRNA passed all quality control (QC) filters including a total of 28 taste perception related genes considered for this analysis. The 28 genes are listed in Supplemental Table T1A with their raw read counts reported the supplemental document named raw_count_data_for_RNA_expression and the distribution of the counts shown graphically in Supplemental Material M1. The genes consist of 22 TAS2R genes related to bitter taste perception, 3 SCNN1 genes related to salty taste perception and 3 TAS1R genes related to to umami (glutamate) perception.
Genotype Data
For all the samples DNA was extracted from blood collected in PAXgene Blood DNA Tube and plated for genotyping on the Illumina Multi-Ethnic Genotyping Array (MEGA) version 2 which includes more than two million loci. The loci targeted by this Illumina product have been specified by the PAGE (Population Architecture in Genetics and Epidemiology) consortium and the ADPC (African Diaspora Power Chip) consortium. The raw intensity data were analyzed using Illumina proprietary software, the Illumina® GenomeStudio Genotyping Module™ Genotyping Module Software v2.0 and clustering for genotype assignment was of high quality (GenTrain Score > 0.9). Pre-analysis QC were carried out according to best practice. Briefly, markers with missing rate > 0.1 were exclude as well as those that failed Hardy-Weinberg Equilibrium test; principal component analysis (PCA) was conducted to identify and exclude sample outliers. Finally, samples with discordant sex information between genotype and phenotype data were also excluded. For the purpose of this work, unrelated samples that have proteome and/or transcriptome data in both cohorts were included in the analyses.
Proteome data
The ethylenediamine tetraacetic acid (EDTA) plasma samples from the GENE-FORECAST and MH-GRID cohorts were sent to the Olink Proteomics Analysis Service in Boston, USA. Proteomic analyses were conducted collectively in a single batch, and the data were delivered in November 2022. The Explore 3072 assay, utilizing eight Explore 384 Olink panels (Cardiometabolic, Cardiometabolic II, Inflammation, Inflammation II, Neurology, Neurology II, Oncology, Oncology II), was run to assess the relative expression of a total of 2,947 proteins. Proximity Extension Assay (PEA) technology was conducted according to the Olink AB manufacturer procedures by the certified laboratory. Briefly, the technique relies on the use of antibodies labelled with unique DNA oligonucleotides that bind to their target protein present in the sample. The DNA oligonucleotides, when in proximity on a target protein, undergo hybridization and act as a template for DNA polymerase-dependent extension, forming a unique double-stranded DNA barcode proportionate to the initial protein concentration. Quantification of resulting DNA amplicons is accomplished through high-throughput DNA sequencing, generating a digital signal reflective of the number of DNA hybridization events corresponding to the protein concentration in the original sample. The measurement of protein levels is based on Normalized Protein eXpression (NPX) values, serving as a relative protein quantification unit. This quantification is normalized to account for systematic noise arising from sample processing and technical variation, leveraging internal controls and sample controls. NPX units are on a log2 scale, where a one NPX unit increase indicates a two-fold rise in the concentration of amplicons representing the target protein compared to the internal control. A total of 2,941 passed QC filtering and a subset of 741 of those from the neurological panel were included in the protein quantitative trait loci (pQTL) analysis with 585 samples (458 from GENE-FORECAST and 127 from MH-GRID) which have genotype and proteome data available.
Statistical Analyses
- eQTL analysis
The analysis included all bi-allelic single nucleotide polymorphisms (SNPs) with a minor allele frequency (MAF) ≥ 0.01 within the combined GENE-FROECAST and MH-GRID dataset (n = 475). Only SNPs within the cis-region (within one megabase) of the 28 taste perception genes in the transcriptome data were considered. The decision to focus on bi-allelic SNPs in the analyses is informed by various considerations, such as the superior accuracy and reliability associated with genotyping techniques for bi-allelic SNPs when compared to multi-allelic SNPs. This enhanced precision contributes to the robustness of the eQTL findings. Moreover, the emphasis on bi-allelic SNPs is strategically guided by their binary allele composition, which facilitates the interpretation of genetic effects and simplifies the identification and characterization of associations between specific alleles and mRNA and protein levels. A SNP was designated as an eQTL if the false-discovery rate (FDR) adjusted p-value of the association with a gene was ≤ 0.05.
- eQTL overlap with variants reported in GWAS studies of AD
The summary statistics of 4 recent large studies of AD outlined in Table 2 were downloaded from the the GWAS Catalogue 26 (version of November 2023). SNPs identified as eQTL in our analysis and reported in the GWAS with a p-value ≤ 0.0001 (GWAS follow up threshold) were identified.
Table 2:
Baseline characteristics of GENE-FORECAST and MH-GRID samples included in .
| Characteristics | GENE-FORECAST (n = 342) | MH-GRID (n = 133) | ||
|---|---|---|---|---|
| Mean or Count | SD or Proportion | Mean or Count | SD or Proportion | |
|
| ||||
| Age (years) | 48 | 12 | 45 | 7 |
|
| ||||
| Sex | ||||
| Female | 235 | 69% | 45 | 34% |
| Male | 107 | 31% | 88 | 66% |
|
| ||||
| Systolic Blood Pressure, SBP (mmHg) | 154 | 38 | 120 | 16 |
|
| ||||
| Diastolic Blood Pressure, DBP(mmHg) | 76 | 10 | 77 | 11 |
|
| ||||
| Body Mass Index, BMI (kg/m2) | 32 | 7 | 31 | 9 |
|
| ||||
| Low Density Lipoprotein, LDL (mg/dL) | 112 | 39 | 109 | 32 |
|
| ||||
| FasAng Blood Glucose, FBG (mg/dL) | 103 | 35 | 90 | 9 |
|
| ||||
| Current Smoker | ||||
| No | 399 | 87% | 68 | 51% |
| Yes | 58 | 13% | 62 | 47% |
- pQTL analysis
eQTLs reported in GWAS as associated with AD were tested for association with any of the 741 proteins in the Olink Neurological panel to gain functional insights by understanding the genetic basis of the regulation of proteins involved in the development or progression of AD. A SNP was designated as a pQTL when its association with a protein yielded an FDR-adjusted p-value of ≤ 0.05.
Both eQTL and pQTL analyses were conducted utilizing the MatrixEQTL 27 R library. MatrixEQTL applies a regression model with mRNA/protein levels as the outcome and additive genotypes as independent variables. The regression model was adjusted for covariates, including age, sex, and principal components (PCs) 1 to 6 that explain most of the variance, to account for genetic ancestry admixture.
RESULTS
Data Description
GENE-FORECAST and MH-GRID data were combined in the eQTL and pQTL analyses to maximize the sample sizes. A description of the baseline characteristics of the 342 GENE-FOREAST and 133 MH-GRID samples included in the eQTL analysis are outlined in Table 1.
Table 1:
Four recent large GWAS studies of AD with summary statistics available for download from the GWAS catalogue.
| GWAS Catalogue Study Accession | PubMed ID | Publication Date (Author) | Study Title | Sample Size |
|---|---|---|---|---|
| GCST9002715 | 35379992 | 2022-04-04 (Bellenguez et al.) | New insights into the genetic etiology of Alzheimer’s disease and related dementias | 111,326 cases 677,663 controls |
| GCST013196 | 34493870 | 2021-09-07 (Wightman et al.) | A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease | 90,338 cases 1,036,225 controls |
| GCST007320 | 30617256 | 2019-01-07 (Jansen et al.) | Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk | 71,880 cases 383,378 controls |
| GCST005922 | 29777097 | 2018-05-18 (Marioni et al.) | GWAS on family history of Alzheimer’s disease | 39,918 cases 101,742 controls |
Principal component analysis
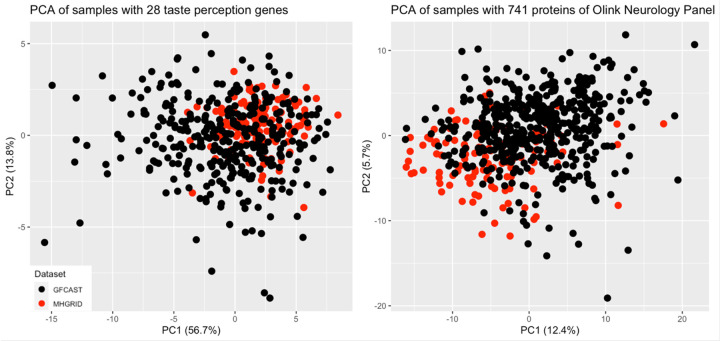
Because the analysis combined two datasets, principal component analysis (PCA) was undertaken to ensure the two datasets are homogeneous across the transcriptome and proteome data analyzed and the ensuing eQTL and pQTL results are not due to batch effects. The plots in Figure 1 indicate that the two datasets cluster together across the 28 taste perception-related mRNAs and the 741 proteins in the Olink’s neurological panel.
Figure 1:
PCA results indicate that the two datasets combined in this analysis cluster well together across PC1 which explains most the variance within the data.
eQTL analysis and overlap with GWAS findings
A comprehensive total of 3,547 SNP-mRNA associations were statiscally significant after adjustment for multiple testing. These associations comprised of 412 distinct SNPs and encompassed all 28 taste genes. Details of all the eQTL associations and the number of eQTL associated with each gene are outlined respectively in Supplemental Tables T1B and T1C.
A set of 11 eQTLs associated with 16 mRNAs, in the eQTL analysis, were reported in 17 independent GWAS studies of a dozen traits. The intersection with GWAS, succinctly presented in Table 3, includes three SNPs (rs11771145, rs11762262, rs10228407) consistently associated with AD across multiple studies (Table 4). The comprehensive information of SNP, mRNA and GWAS is provided in Supplemental Tables T1D.
Table 3:
GWAS studies reporting associations with SNPs revealed as eQTL linked to taste genes in our analysis.
| GWAS Trait/Disease | PubMed IDs | SNPs |
|---|---|---|
| Bitter taste perception (multivariate analysis) | 30223776 | rs10772420, rs10261515 |
| Bitter taste perception | 23966204, 30223776 | rs1031391, rs10772420 |
| Idiopathic intracranial hypertension | 29608535 | rs200288366 |
| Height | 18391951, 25282103, 20881960, 23563607 | rs2187642, rs2856321 |
| Alzheimer’s disease | 35379992, 31473137, 34493870 | rs11771145, rs11762262, rs10228407 |
| Alzheimer’s disease (late onset) | 24162737, 30617256 | rs11771145, rs11762262, rs10228407 |
| Alzheimer’s disease or family history of Alzheimer’s disease | 29777097, 30617256 | rs11771145, rs11762262, rs10228407 |
| Body size at age 10 | 32376654 | rs2187642 |
| Tea with sugar liking | 35585065 | rs10772380 |
| Gamma glutamyl transferase levels | 33462484 | rs11978404 |
| Bitter taste perception (phenylthiocarbamide) in obesity with metabolic syndrome | 31005965 | rs13231650 |
| Type 2 diabetes | 35893037 | rs4920461 |
Table 4:
Effect allele, effect size and p-values reported by the 4 GWAS studies of AD for the 3 SNPs identified as eQTLs associated with TAS2R41 and TAS2R60. Two of the SNPs are not reported by Marioni et al.
| GWAS Study | GWAS stats/info | rsll762262 | rs11771145 | rs10228407 |
|---|---|---|---|---|
|
| ||||
| Wightman et al. PMID 34493870 | log Odds | −0.07 | −0.05 | 0.06 |
| P-Value | 9.80e-08 | 3.81e-07 | 2.71e-08 | |
| Mapped Gene (location) | EPHA1-AS1 (intron) | EPHA1-AS1 (intron) | EPHA1-AS1 (intron) | |
| Effect allele reported | T | A | T | |
|
| ||||
| Marioni et al. PMID 29777097 | log Odds | Not reported | Not reported | 0.05 |
| P-Value | Not reported | Not reported | 1.28e-09 | |
| Mapped Gene (location) | Not reported | Not reported | EPHA1-AS1 (intron) | |
| Effect allele reported | Not reported | Not reported | T | |
|
| ||||
| Bellenguez et al. PMID 35379992 | Odds-Ratio | 0.94 | 0.94 | 1.05 |
| P-Value | 6.35e-10 | 1.29e-12 | 1.24e-10 | |
| Mapped Gene (location) | EPHA1-AS1 (intron) | EPHA1-AS1 (intron) | EPHA1-AS1 (intron) | |
| Effect allele reported | T | A | T | |
|
| ||||
| Jansen et al. PMID 30617256 | log Odds | −0.016 | −0.011 | −0.01 |
| P-Value | 5.22e-09 | 8.84e-07 | 9.33e-06 | |
| Mapped Gene (location) | EPHA1-AS1 (intron) | EPHA1-AS1 (intron) | EPHA1-AS1 (intron) | |
| Effect allele reported | T | A | G | |
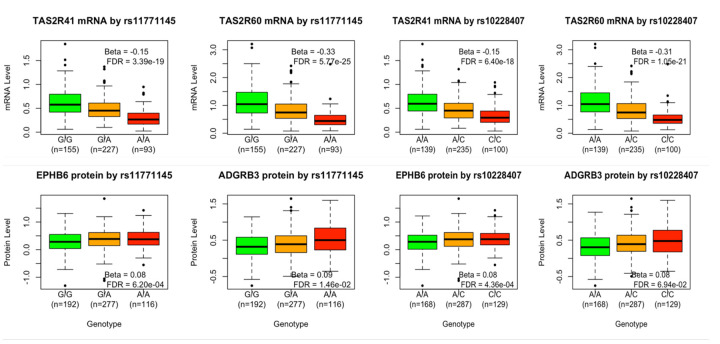
In our eQTL analysis all three SNPs exhibit significant association with a down-regulation of TAS2R41 and two of them (rs11771145, rs10228407) are additionally associated with the down-regulation of TAS2R60, reported in Table 5 and Figures 2. The three SNPs have a common frequency in the analysis datasets; they are all upstream of TAS2R41 and TAS2R60 and are not in significant linkage disequilibrium (LD), in the analysis dataset. The LD values measured as r2 are respectively 0.58 (between rs11771145 and rs10228407), 0.06 (between and rs11771145 and rs10228407) and 0.10 (between and rs11771145 and rs11762262).
Table 5:
eQTL analysis results for the 3 SNPs associated with AD in múltiple GWAS studies.
| SNP/eQTL | Chr. | Position (GRCh38) | Location | mRNA | Beta | P-Value (raw) | P-Value (adjusted) | Allele tested (Minor Allele) | MAF |
|---|---|---|---|---|---|---|---|---|---|
| rs11762262 | 7 | 143410783 | 67.1 kb upstream | TAS2R41 | −0.07 | 9.45e-04 | 1.99e-02 | A | 0.21 |
| rs11771145 | 7 | 143413669 | 64.2 kb upstream | TAS2R41 | −0.15 | 2.58e-21 | 3.39e-19 | G | 0.43 |
| rs11771145 | 7 | 143413669 | 29.8 kb upstream | TAS2R60 | −0.33 | 3.26e-27 | 5.77e-25 | G | 0.43 |
| rs10228407 | 7 | 143430678 | 12.8 kb upstream | TAS2R60 | −0.31 | 7.40e-24 | 1.05e-21 | A | 0.46 |
| rs10228407 | 7 | 143430678 | 47.2 kb upstream | TAS2R41 | −0.15 | 5.11e-20 | 6.40E-18 | A | 0.46 |
Figure 2:
mRNA expression and protein level by rs11771145 and rs10228407 genotypes. The beta and FDR adjusted p-value of each association is provided in the legend.
pQTL analysis of the 3 SNPs reported in GWAS of AD
The association between all 741 proteins in the Olink’s neurological panel and the 3 SNPs was evaluated. A set of three proteins (EPHB6, ARHGEF5, KEL) are encoded by genes in the vicinity (cis) of the three SNPs and the remaining 738 proteins are encoded by genes further away or in another chromosome (trans). The pQTL analysis revealed that rs11771145 was associated with an upregulation of one cis protein (EPHB6, beta=0.8, p-value=0.0002, FDR adjusted p-value=0.0006) and one trans protein (ADGRB3, 0.09, p-value=0.00002, FDR adjusted p-value=0.014) whilst rs10228407 was associated with an upregulation of cis EPHB6 (beta=0.08 and p-value=0.0001, FDR adjusted p-value=0.0004) and associated with trans ADGRB3 (beta=0.08 and p-value=0.00009, FDR adjusted p-value=0.069). These results are depicted graphically in Figure 2.
DISCUSSION
The detection of bitter taste receptors such as TAS2R41 and TAS2R60 beyond the oral cavity has substantially broadened the scope of research revealing their multifaceted biological functions 28. Originally evolved to discern noxious compounds, these receptors are now recognized as integral contributors to immunological defense mechanisms 29. Notably, their localization within the cerebral cortex and choroid plexus represents a seminal expansion of their previously ascribed functional roles, implying a plausible engagement in metabolic and immune processes within the brain 30.
Neurological Implications
Recent investigations have unveiled the prospect that bitter taste receptors may exert their influence on neurological pathways implicated in neurodegenerative conditions, such as AD 31. Traditionally acknowledged as chemical sentinels and more recently recognized as immune modulators 32, these receptors offer novel insights into the mechanisms governing the pathophysiology of neurodegenerative diseases 33. Their involvement in the immune and metabolic regulation within the brain not only broadens our understanding but also prompts the consideration of these receptors as potential therapeutic targets or early-stage biomarkers for neurodegenerative disorders 34.
Genetic Associations Reported
Our research has brought to light noteworthy genetic associations linked to the TAS2R41 and TAS2R60 genes-associations extending beyond taste perception to intersect with AD. Specifically, the SNPs rs11771145 and rs10228407 located upstream of these genes, emerge as potential critical influencers of their expression. While the primary function of TAS2R41 and TAS2R60 is rooted in gustatory detection 35, the association between these SNPs and AD is consistently reported in large GWAS investigations which beckons further explorations into the broader implications of genetic variants. Studies suggest that TAS2Rs might have broader implications for health and disease, including their expression in the brain and potential roles in neuronal signaling and disease pathology 36. The association of TAS2Rs SNPs and their potential modulation during AD could shed light on the mechanisms of taste alterations and offer new perspectives on how sensory changes are linked to neurodegenerative processes.
Immunomodulation, Neuroinflammation and Alzheimer’s disease
Previous investigations have highlighted the role of the family of TAS2R genes in modulating immune cell activity, specifically in the regulation of antimicrobial peptides and inflammatory responses 37. If these receptors extend a similar influence within the central nervous system, they could potentially modify microglial activity, thereby impacting neuroinflammatory pathways. The crucial function of bitter taste receptors in attenuating neuroinflammatory responses under normal physiological conditions is now well-established 38. Conversely, a reduction in the expression of pivotal components within the taste receptor signaling pathway may escalate oxidative stress and activate the inflammasome, ultimately leading to neuroinflammation 39.
Neuroinflammation, a shared characteristic among various neurodegenerative diseases, including AD, is now acknowledged as central to their pathophysiology 40. The involvement of TAS2R bitter taste receptors in inflammatory responses underscores their significance beyond the realm of taste perception 41. Pioneering work by Du et al. in 2018 unveiled that the loss of α-gustducin, a G-protein subunit integral to taste signal transduction, resulted in inflammatory responses and tissue damage 42. This underscores the pro-inflammatory potential associated with disruptions in taste receptor signaling, specifically through the NF-κB signaling pathway and the NLRP3 inflammasome 43.
Genetic Variations and Alzheimer’s Pathology
The identification of significant SNP-Gene associations across all 28 taste genes, particularly those leading to the down-regulation of TAS2R41 and TAS2R60, presents a notable link to Alzheimer’s disease within the African American cohort studied. The location of rs117771145 and rs10228407 within the regulatory regions of TAS2R41 and TAS2R60, coupled with their association with AD, in GWAS, fortifies the proposition that bitter taste receptors may be involved in the pathological processes of AD. Variations in expression mediated by these SNPs have the potential to modulate receptor activation thresholds, thereby influencing cellular processes pivotal to neuroprotection, management of neuroinflammatory responses, and clearance of amyloid-beta 44. Consequently, such variations may exacerbate neuronal susceptibility to the characteristic pathologies of AD.
Proteomic Insights
To gain further functional insights, we conducted a comprehensive pQTL analysis targeting three SNPs, revealed as eQTL associated with TAS2R45 and TAS2R60 in our analysis, and previously identified in GWAS of AD 45. This exploration unveiled compelling associations, elucidating proteins subject to modulation by these specific SNPs. Particularly noteworthy was the observation that rs11771145 exhibited a pronounced upregulation of EPHB6 (Ephrin type-B receptor 6), a protein encoded by a gene within the cis-region of the SNP involved in many developmental processes including neuronal development, angiogenesis, and cell migration 46. This observation posits a compelling inference of direct regulatory influence. In the context of AD, EPHB6 assumes prominence owing to its extensive relavance in synaptic plasticity 47, neuroprotection, and neuroinflammation 48,49. Its regulatory role in fundamental processes positions EphB receptors as potential modulators of AD pathophysiology 50.
Moreover, rs11771145 manifested an intriguing association with a protein, encoded by a gene, ADGRB3 (Adhesion G Protein-Coupled Receptor B3, also known as BAI3), located on a another chromosome. This finding implies the potential for long-range interactions that exert influence over protein expression levels. These associations collectively suggest that the implicated SNPs not only correlate with alterations in gene expression but also wield downstream effects on protein levels. ADGRB3 has been implicated in synaptic regulation and may influence neural circuit formation and plasticity. Alterations in ADGRB3 expression or function have been linked to various neurological conditions, suggesting that it plays a role in maintaining normal cognitive and neural functions 51. The brain-specific angiogenesis inhibitor 1 (BAI1), also known as Adhesion G protein-coupled receptor B1 (ADGRB1), emerges as a pivotal regulator of synaptic plasticity 52, particularly in the hippocampus. Its involvement in learning and memory processes underscores its significance 53. Furthermore, ADGRB1 has been implicated in neuroprotection, mitigating toxin-induced neuronal cell death, and has known associations with dopaminergic neuronal loss in Parkinson’s disease 54,55. Concurrently, ADGRB3, enriched in post-synaptic density and cerebellar Purkinje cells 56, orchestrates synaptic connections, particularly within the cerebellum 57. Genetic variations in ADGRB3, encompassing SNPs and gene amplifications, have been linked to familial schizophrenia and other psychiatric conditions, including bipolar disorder 58,59.
The association of the identified SNPs with upregulation of EPHB6 and ADGRB3 proteins suggests potential pathways through which genetic variations may contribute to the unique progression of AD in African Americans. As noted EPHB6 and ADGRB3 are involved in neuronal function and development, and their dysregulation could have significant implications for neurodegeneration. In both cases, the specific mechanisms by which TAS2R41 and TAS2R60 influence the expression or function of EPHB6 and ADGRB3 in the context of Alzheimer’s disease remain to be fully elucidated. It is possible that changes in TAS2R expression alter signaling cascades or cellular environments in ways that impact these proteins, which in turn could affect neuronal health and function. Further research is needed to understand these relationships and their implications for AD and sensory health.
Integrated Perspective and Potential Therapeutic Implications
This integrative perspective, elucidating the interplay between SNPs, taste receptor genes and AD through pQTL analysis, offers profound insights into the intricacies of AD’s etiology. Beyond merely illuminating potential mechanistic pathways, it underscores the multifaceted nature of neurodegenerative diseases. This holistic approach encourages nuanced research strategies for the refinement of therapeutic interventions and the development of preventive strategies.
Furthermore, the neuroprotective and anti-inflammatory attributes associated with bitter compounds, known to interact with TAS2Rs, substantiate the hypothesis that these receptors could potentially modulate the pathophysiological conditions of AD 60. Compounds like flavonoids and polyphenols, which engage with TAS2Rs, have exhibited promise in enhancing cognitive function and mitigating markers of neurodegeneration in disease models 61. The exploration of TAS2Rs in orchestrating the therapeutic effects of these compounds represents an intriguing avenue for further investigation 62.
Limitations
Our study, elucidating novel insights into the genetic foundations of taste perception genes and their correlation with AD in an African American cohort, is constrained by several limitations. AD was not assessed in this cohort and it was hence not possible to directly link the mRNA and protein changes to AD.
Whole-blood transcriptome may imperfectly reflect brain gene expression, the primary site of AD pathology.
An additional limitation is our reliance on GWAS data predominantly derived from European populations. This underrepresentation of African American and other non-European populations in genetic research can limit the generalizability of our findings. The genetic architecture of AD may vary across different ancestries, and findings from European cohorts may not fully capture the genetic risk factors pertinent to populations of African ancestry. However, the main variants we reported are all common, suggesting they are not ancestry-specific and that our findings might be transferable across populations. Future studies should prioritize the inclusion of diverse populations to enhance the applicability of genetic research across different ethnic groups and to address health disparities in the understanding and treatment of AD.
The cross-sectional design hampers capturing the dynamic nature of gene expression changes over the disease course. Unaccounted environmental and lifestyle factors may confound genetic associations. Lack of direct correlation between genetic variations and clinical manifestations of AD, absence of detailed pathway analysis for SNP-gene and SNP-protein associations, and omission of microRNA exploration further limit our study. Notably, functional consequences of identified expression and protein QTLs lack experimental validation. Future research necessitates in vitro studies to discern genetic variant impact on gene expression and protein levels and function. Subsequent in vivo studies are vital for establishing their role in disease processes, fostering a comprehensive understanding of these associations in AD context.
Conclusions
Our investigation has unveiled novel insights into the genetic determinants implicated in AD, underscoring the potential important role of bitter taste receptor genes in its pathophysiological mechanisms. The identification of SNPs exhibiting robust associations with both taste perception genes and AD provides evidence for the plausible involvement of these genes in the intricate pathways of neurodegenerative diseases 63. These findings posit genetic variations influencing taste receptor expression as potential contributors to brain health, positioning these receptors as plausible biomarkers for AD 64.
The implications of our study for unraveling the genetic architecture of Alzheimer’s disease have valuable significance, offering prospects for the development of targeted interventions aimed at modulating the pathways identified. Our findings also raise new questions and avenues for future research. Subsequent investigations should prioritize the validation of these associations within broader and more genetically diverse cohorts to ascertain the generalizability of our observations. Furthermore, longitudinal studies are warranted to elucidate the causal relationships between these genetic variations, gene expression regulation, and the progression of AD.
Our study highlights a significant step towards understanding the complex interplay of genetics, sensory function, and neurodegeneration in Alzheimer’s disease within an African American cohort. Critical to the translation of genetic insights into therapeutic strategies, future research endeavors should delve into the direct impact of these SNPs on the neural circuitry and cognitive functions affected by AD. Such investigations will be instrumental in advancing our understanding and, consequently, facilitating the development of precise therapeutic interventions.
ACKNOWLEDGEMENTS
PVJ is supported by National Institute of Alcohol Abuse and Alcoholism under award number, Z01AA000135, the National Institute of Nursing Research and the Rockefeller University Heilbrunn Nurse Scholar Award. PVJ is supported by the Office of Workforce Diversity, and the Office of Workforce Diversity, National Institutes of Health Distinguished Scholar Program.
We are grateful to Dr Gary H. Gibbons, the initial PI of the GENE-FORECAST study.
FUNDING
This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
ABBREVIATIONS
- eQTL
Expression-Quantitative Trait Loci
- pQTL
Protein-Quantitative Trait Loci
- GWAS
Genome-Wide Association Study
- SNP
Single-Nucleotide Polymorphisms
- AD
Alzheimer’s Disease
- GENE-FORECAST
GENomics, Environmental FactORs and the Social DEterminants of Cardiovascular Disease in African-Americans Study
- MH-GRID
Minority Health Genomics and Translational Research Bio-Repository Database
- MSM
Morehouse School of Medicine
- PCA
Principal Component Analysis
- LD
Linkage Disequilibrium
- SBP
Systolic Blood Pressure
- DBP
Diastolic Blood Pressure
Footnotes
COMPETING INTERESTS
The authors have no financial and/or personal relationships with other people or organizations that could inappropriately influence (bias) this work.
ETHICS APPROVAL AND CONSENT TO PARTICPATE
This study was approved by National Institutes of Health Institutional Review Board (IRB). The study was conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIAL
Additional information can be found in the Supplementary Material of this article. The datasets presented in this article cannot be publicly shared due to privacy restrictions. Requests to access the datasets should be directed to the corresponding author.
REFERENCES
- 1.Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. Nov 16 2006;444(7117):288–94. doi: 10.1038/nature05401 [DOI] [PubMed] [Google Scholar]
- 2.Small DM, Prescott J. Odor/taste integration and the perception of flavor. Exp Brain Res. Oct 2005;166(3–4):345–57. doi: 10.1007/s00221-005-2376-9 [DOI] [PubMed] [Google Scholar]
- 3.Kure Liu C, Joseph PV, Feldman DE, et al. Brain Imaging of Taste Perception in Obesity: a Review. Curr Nutr Rep. Jun 2019;8(2):108–119. doi: 10.1007/s13668-019-0269-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heckmann JG, Lang CJG. Neurological causes of taste disorders. Adv Otorhinolaryngol. 2006;63:255–264. doi: 10.1159/000093764 [DOI] [PubMed] [Google Scholar]
- 5.Ram S, Wada T, Sahai-Srivastava S. Neurosensory Disturbances Including Smell and Taste. In: Farah CS, Balasubramaniam R, McCullough MJ, eds. Contemporary Oral Medicine: A Comprehensive Approach to Clinical Practice. Springer International Publishing; 2018:1–27. [Google Scholar]
- 6.Yarmolinsky DA, Zuker CS, Ryba NJ. Common sense about taste: from mammals to insects. Cell. Oct 16 2009;139(2):234–44. doi: 10.1016/j.cell.2009.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howes OD, Kambeitz J, Kim E, et al. The nature of dopamine dysfunction in schizophrenia and what this means for treatment. Arch Gen Psychiatry. Aug 2012;69(8):776–86. doi: 10.1001/archgenpsychiatry.2012.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nutt DJ, Lingford-Hughes A, Erritzoe D, Stokes PR. The dopamine theory of addiction: 40 years of highs and lows. Nat Rev Neurosci. May 2015;16(5):305–12. doi: 10.1038/nrn3939 [DOI] [PubMed] [Google Scholar]
- 9.Doty RL. Olfactory dysfunction in neurodegenerative diseases: is there a common pathological substrate? Lancet Neurol. Jun 2017;16(6):478–488. doi: 10.1016/S1474-4422(17)30123-0 [DOI] [PubMed] [Google Scholar]
- 10.Steiner JE. Human facial expressions in response to taste and smell stimulation. Adv Child Dev Behav. 1979;13:257–95. doi: 10.1016/s0065-2407(08)60349-3 [DOI] [PubMed] [Google Scholar]
- 11.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. Jan 2008;9(1):46–56. doi: 10.1038/nrn2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medeiros R, Baglietto-Vargas D, LaFerla FM. The role of tau in Alzheimer’s disease and related disorders. CNS Neurosci Ther. Oct 2011;17(5):514–24. doi: 10.1111/j.1755-5949.2010.00177.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai M, Kazui H, Shigenobu K, Komori K, Ikeda M, Nishikawa T. Gustatory Dysfunction as an Early Symptom of Semantic Dementia. Dement Geriatr Cogn Dis Extra. Sep-Dec 2017;7(3):395–405. doi: 10.1159/000481854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakai M, Ikeda M, Kazui H, Shigenobu K, Nishikawa T. Decline of gustatory sensitivity with the progression of Alzheimer’s disease. Int Psychogeriatr. Mar 2016;28(3):511–7. doi: 10.1017/s1041610215001337 [DOI] [PubMed] [Google Scholar]
- 15.Schiffman SS, Graham BG, Sattely-Miller EA, Zervakis J, Welsh-Bohmer K. Taste, smell and neuropsychological performance of individuals at familial risk for Alzheimer’s disease. Neurobiology of Aging. 2002/May/01/ 2002;23(3):397–404. doi: 10.1016/S0197-4580(01)00337-2 [DOI] [PubMed] [Google Scholar]
- 16.Steinbach S, Hundt W, Vaitl A, et al. Taste in mild cognitive impairment and Alzheimer’s disease. J Neurol. Feb 2010;257(2):238–46. doi: 10.1007/s00415-009-5300-6 [DOI] [PubMed] [Google Scholar]
- 17.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. Dec 2013;45(12):1452–8. doi: 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marioni RE, Harris SE, Zhang Q, et al. GWAS on family history of Alzheimer’s disease. Transl Psychiatry. May 18 2018;8(1):99. doi: 10.1038/s41398-018-0150-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno-Grau S, de Rojas I, Hernandez I, et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer’s disease and three causality networks: The GR@ACE project. Alzheimers Dement. Oct 2019;15(10):1333–1347. doi: 10.1016/j.jalz.2019.06.4950 [DOI] [PubMed] [Google Scholar]
- 20.Bellenguez C, Kucukali F, Jansen IE, et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet. Apr 2022;54(4):412–436. doi: 10.1038/s41588-022-01024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. Mar 2019;51(3):404–413. doi: 10.1038/s41588-018-0311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lennon JC, Aita SL, Bene VAD, et al. Black and White individuals differ in dementia prevalence, risk factors, and symptomatic presentation. Alzheimers Dement. Aug 2022;18(8):1461–1471. doi: 10.1002/alz.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Association As. Black Americans and alzheimer’s. Accessed 02/05, 2024. https://www.alz.org/help-support/resources/black-americans-and-alzheimers
- 24.Institute B. broadinstitute/gtex-pipeline. https://github.com/broadinstitute/gtex-pipeline
- 25.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sollis E, Mosaku A, Abid A, et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. Jan 6 2023;51(D1):D977–D985. doi: 10.1093/nar/gkac1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. May 15 2012;28(10):1353–8. doi: 10.1093/bioinformatics/bts163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avau B, Depoortere I. The bitter truth about bitter taste receptors: beyond sensing bitter in the oral cavity. Acta Physiol (Oxf). Apr 2016;216(4):407–20. doi: 10.1111/apha.12621 [DOI] [PubMed] [Google Scholar]
- 29.Carey RM, Lee RJ. Taste Receptors in Upper Airway Innate Immunity. Nutrients. Aug 28 2019;11(9)doi: 10.3390/nu11092017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duarte AC, Santos J, Costa AR, et al. Bitter taste receptors profiling in the human blood-cerebrospinal fluid-barrier. Biochem Pharmacol. Jul 2020;177:113954. doi: 10.1016/j.bcp.2020.113954 [DOI] [PubMed] [Google Scholar]
- 31.Duarte AC, Costa AR, Goncalves I, Quintela T, Preissner R, Santos CRA. The druggability of bitter taste receptors for the treatment of neurodegenerative disorders. Biochem Pharmacol. Mar 2022;197:114915. doi: 10.1016/j.bcp.2022.114915 [DOI] [PubMed] [Google Scholar]
- 32.Lu P, Zhang CH, Lifshitz LM, ZhuGe R. Extraoral bitter taste receptors in health and disease. J Gen Physiol. Feb 2017;149(2):181–197. doi: 10.1085/jgp.201611637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Jesus VC, Mittermuller BA, Hu P, Schroth RJ, Chelikani P. Association between Downstream Taste Signaling Genes, Oral Microbiome, and Severe Early Childhood Caries. Int J Mol Sci. Dec 21 2022;24(1)doi: 10.3390/ijms24010081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harmon CP, Deng D, Breslin PAS. Bitter Taste Receptors (T2Rs) are Sentinels that Coordinate Metabolic and Immunological Defense Responses. Curr Opin Physiol. Apr 2021;20:70–76. doi: 10.1016/j.cophys.2021.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Silva EC, de Jager N, Burgos-Paz W, Reverter A, Perez-Enciso M, Roura E. Characterization of the porcine nutrient and taste receptor gene repertoire in domestic and wild populations across the globe. BMC Genomics. Dec 3 2014;15(1):1057. doi: 10.1186/1471-2164-15-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Behrens M, Meyerhof W. Gustatory and extragustatory functions of mammalian taste receptors. Physiol Behav. Nov 30 2011;105(1):4–13. doi: 10.1016/j.physbeh.2011.02.010 [DOI] [PubMed] [Google Scholar]
- 37.Xi R, Zheng X, Tizzano M. Role of Taste Receptors in Innate Immunity and Oral Health. J Dent Res. Jul 2022;101(7):759–768. doi: 10.1177/00220345221077989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welcome MO, Dogo D, Nikos EM. Cellular mechanisms and molecular pathways linking bitter taste receptor signalling to cardiac inflammation, oxidative stress, arrhythmia and contractile dysfunction in heart diseases. Inflammopharmacology. Feb 2023;31(1):89–117. doi: 10.1007/s10787-022-01086-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welcome MO, Mastorakis NE. The taste of neuroinflammation: Molecular mechanisms linking taste sensing to neuroinflammatory responses. Pharmacol Res. May 2021;167:105557. doi: 10.1016/j.phrs.2021.105557 [DOI] [PubMed] [Google Scholar]
- 40.Guzman-Martinez L, Maccioni RB, Andrade V, Navarrete LP, Pastor MG, Ramos-Escobar N. Neuroinflammation as a Common Feature of Neurodegenerative Disorders. Front Pharmacol. 2019;10:1008. doi: 10.3389/fphar.2019.01008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuzim K, Korolczuk A. An update on extra-oral bitter taste receptors. J Transl Med. Oct 21 2021;19(1):440. doi: 10.1186/s12967-021-03067-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong G, Boothe K, He L, Shi Y, McCluskey LP. Altered peripheral taste function in a mouse model of inflammatory bowel disease. Res Sq. Sep 6 2023;doi: 10.21203/rs.3.rs-3304297/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu T, Zhang L, Joo D, Sun SC. NF-kappaB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023-. doi: 10.1038/sigtrans.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinney JW, Bemiller SM, Murtishaw AS, Leisgang AM, Salazar AM, Lamb BT. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement (N Y). 2018;4:575–590. doi: 10.1016/j.trci.2018.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wooding SP, Ramirez VA. Global population genetics and diversity in the TAS2R bitter taste receptor family. Front Genet. 2022;13:952299. doi: 10.3389/fgene.2022.952299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kania A, Klein R. Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat Rev Mol Cell Biol. Apr 2016;17(4):240–56. doi: 10.1038/nrm.2015.16 [DOI] [PubMed] [Google Scholar]
- 47.Sloniowski S, Ethell IM. Looking forward to EphB signaling in synapses. Semin Cell Dev Biol. Feb 2012;23(1):75–82. doi: 10.1016/j.semcdb.2011.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He CH, Song NN, Xie PX, et al. Overexpression of EphB6 and EphrinB2 controls soma spacing of cortical neurons in a mutual inhibitory way. Cell Death Dis. May 6 2023;14(5):309. doi: 10.1038/s41419-023-05825-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darling TK, Lamb TJ. Emerging Roles for Eph Receptors and Ephrin Ligands in Immunity. Front Immunol. 2019;10:1473. doi: 10.3389/fimmu.2019.01473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dal Pra I, Armato U, Chiarini A. Family C G-Protein-Coupled Receptors in Alzheimer’s Disease and Therapeutic Implications. Front Pharmacol. 2019;10:1282. doi: 10.3389/fphar.2019.01282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanoue V, Usardi A, Sigoillot SM, et al. The adhesion-GPCR BAI3, a gene linked to psychiatric disorders, regulates dendrite morphogenesis in neurons. Molecular Psychiatry. 2013/August/01 2013;18(8):943–950. doi: 10.1038/mp.2013.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stephenson JR, Paavola KJ, Schaefer SA, Kaur B, Van Meir EG, Hall RA. Brain-specific angiogenesis inhibitor-1 signaling, regulation, and enrichment in the postsynaptic density. J Biol Chem. Aug 2 2013;288(31):22248–56. doi: 10.1074/jbc.M113.489757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiu FH, Wong JC, Yamamoto T, et al. Mice lacking full length Adgrb1 (Bai1) exhibit social deficits, increased seizure susceptibility, and altered brain development. Exp Neurol. May 2022;351:113994. doi: 10.1016/j.expneurol.2022.113994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi JS, Bae WY, Nam S, Jeong JW. New Targets for Parkinson’s Disease: Adhesion G Protein-Coupled Receptor B1 is Downregulated by AMP-Activated Protein Kinase Activation. OMICS. Jul 2018;22(7):493–501. doi: 10.1089/omi.2018.0047 [DOI] [PubMed] [Google Scholar]
- 55.Choi JS, Park C, Jeong JW. AMP-activated protein kinase is activated in Parkinson’s disease models mediated by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Biochem Biophys Res Commun. Jan 1 2010;391(1):147–51. doi: 10.1016/j.bbrc.2009.11.022 [DOI] [PubMed] [Google Scholar]
- 56.Purcell RH, Hall RA. Adhesion G Protein-Coupled Receptors as Drug Targets. Annu Rev Pharmacol Toxicol. Jan 6 2018;58:429–449. doi: 10.1146/annurev-pharmtox-010617-052933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lala T, Hall RA. Adhesion G protein-coupled receptors: structure, signaling, physiology, and pathophysiology. Physiol Rev. Oct 1 2022;102(4):1587–1624. doi: 10.1152/physrev.00027.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scuderi C, Saccuzzo L, Vinci M, et al. Biallelic intragenic duplication in ADGRB3 (BAI3) gene associated with intellectual disability, cerebellar atrophy, and behavioral disorder. Eur J Hum Genet. Apr 2019;27(4):594–602. doi: 10.1038/s41431-018-0321-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bipolar D, Schizophrenia Working Group of the Psychiatric Genomics Consortium. Electronic address drve, Bipolar D, Schizophrenia Working Group of the Psychiatric Genomics C. Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell. Jun 14 2018;173(7):1705–1715 e16. doi: 10.1016/j.cell.2018.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zehentner S, Reiner AT, Grimm C, Somoza V. The Role of Bitter Taste Receptors in Cancer: A Systematic Review. Cancers (Basel). Nov 23 2021;13(23)doi: 10.3390/cancers13235891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minocha T, Birla H, Obaid AA, et al. Flavonoids as Promising Neuroprotectants and Their Therapeutic Potential against Alzheimer’s Disease. Oxid Med Cell Longev. 2022;2022:6038996. doi: 10.1155/2022/6038996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nayak AP, Villalba D, Deshpande DA. Bitter Taste Receptors: an Answer to Comprehensive Asthma Control? Curr Allergy Asthma Rep. Sep 5 2019;19(10):48. doi: 10.1007/s11882-019-0876-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oatman SR, Reddy JS, Quicksall Z, et al. Genome-wide association study of brain biochemical phenotypes reveals distinct genetic architecture of Alzheimer’s disease related proteins. Mol Neurodegener. Jan 7 2023;18(1):2. doi: 10.1186/s13024-022-00592-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Romano RR 3rd, Carter MA, Monroe TB. Narrative Review of Sensory Changes as a Biomarker for Alzheimer’s Disease. Biol Res Nurs. Apr 2021;23(2):223–230. doi: 10.1177/1099800420947176 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Additional information can be found in the Supplementary Material of this article. The datasets presented in this article cannot be publicly shared due to privacy restrictions. Requests to access the datasets should be directed to the corresponding author.