Abstract
TL-3 is a protease inhibitor developed using the feline immunodeficiency virus protease as a model. It has been shown to efficiently inhibit replication of human, simian, and feline immunodeficiency viruses and therefore has broad-based activity. We now demonstrate that TL-3 efficiently inhibits the replication of 6 of 12 isolates with confirmed resistance mutations to known protease inhibitors. To dissect the spectrum of molecular changes in protease and viral properties associated with resistance to TL-3, a panel of chronological in vitro escape variants was generated. We have virologically and biochemically characterized mutants with one (V82A), three (M46I/F53L/V82A), or six (L24I/M46I/F53L/L63P/V77I/V82A) changes in the protease and structurally modeled the protease mutant containing six changes. Virus containing six changes was found to be 17-fold more resistant to TL-3 in cell culture than was wild-type virus but maintained similar in vitro replication kinetics compared to the wild-type virus. Analyses of enzyme activity of protease variants with one, three, and six changes indicated that these enzymes, compared to wild-type protease, retained 40, 47, and 61% activity, respectively. These results suggest that deficient protease enzymatic activity is sufficient for function, and the observed protease restoration might imply a selective advantage, at least in vitro, for increased protease activity.
Highly active antiretroviral therapy (HAART) with reverse transcriptase inhibitors in combination with protease inhibitors has proven to suppress human immunodeficiency virus type 1 (HIV-1) replication to undetectable levels in patients (11, 12, 17). HIV-1 variants frequently evolve that escape HAART by developing resistance to the inhibitors used (4, 15, 19, 28, 40, 42). Of patients first treated with a single drug regimen and then going onto HAART, as many as 40% have a viral rebound within the first 3 years, and this number is likely to be higher outside of controlled studies (20, 35). Moreover, transmission of drug-resistant HIV has been observed and is likely to increase with more patients on combination therapy (25, 38). Thus, there is a need to fully understand the sequence of molecular changes to HIV concomitant with development of resistance to protease inhibitors. In turn, this knowledge should facilitate the development of new inhibitors with activities against drug-resistant isolates.
The protease structure of feline immunodeficiency virus (FIV) was used as a model for the development of a series of protease inhibitors with broad efficacy (21). This strategy is based on our observation that various potent HIV inhibitors are less-efficient inhibitors of FIV protease by a factor of more than 100 (21). Moreover, although HIV and FIV protease have only 23% amino acid identity, the active-site structures are almost superimposable (39). We therefore reasoned that by designing inhibitors of proteases with a similar alpha-carbon structure and diverse substrate specificities, such as FIV protease, the resulting inhibitors may be effective against drug-resistant HIVs as well. Our first studies documented that one of these protease inhibitors, TL-3, was truly a broad-based inhibitor as shown by its ability to efficiently inhibit in vitro replication of human, simian, and feline immunodeficiency viruses in the micromolar range (21).
We report here on the antiviral activity of TL-3 on clinical isolates from patients failing potent antiretroviral therapy and viruses engineered to be drug resistant. We demonstrate that TL-3 was active against 6 of 12 isolates tested. Common to isolates resistant to TL-3 was a V82A mutation in combination with additional mutations. Generation of TL-3 escape variants in culture documented that an initial V82A mutation was required and was followed by subsequent protease mutations. Biochemical analysis of protease from TL-3-resistant viruses with three and six mutations demonstrated a progressive decrease of affinity (higher Km) for substrate and increased turnover (kcat). The secondary mutations resulted in the maintenance of an acceptable catalytic efficiency, kcat/Km, and therefore, an effective mutant protease. Comparison of wild-type- and TL-3-resistant protease structural models, derived by energy minimization, offer a sterically plausible explanation for the loss of potency to TL-3 in the protease with six mutations. The protease mutations contributed to diminished protease-TL-3 affinity while maintaining a catalytically efficient enzyme possibly due to the mutations in the flap domain. These secondary mutations are necessary for the generation of a fit virus and the broadening of cross-resistance. It is anticipated that the panel of well-defined TL-3-resistant viral isolates may prove useful for the understanding and design of inhibitors less susceptible to resistance development.
MATERIALS AND METHODS
Cells and viruses.
The MT-2 cell line was obtained through the AIDS Research and Reference Program. The R8 virus was a kind gift of D. Trono (University of Geneva, Geneva, Switzerland) (10). Primary HIV isolates were obtained from the peripheral blood mononuclear cells (PBMC) of patients failing antiretroviral therapy with regimens containing the protease inhibitors as identified in Fig. 1. All PBMC samples were obtained from patients treated at the University of California at San Diego Medical Center or at the San Diego VA Health Care Facility. Patient PBMC were activated with phytohemagglutinin (PHA) and interleukin-2 (IL-2) and cocultivated with PBMC from seronegative donors, and HIV stocks were prepared by low-level passage.
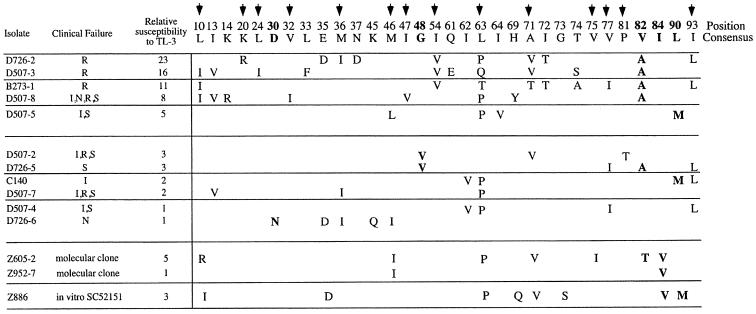
FIG. 1.
TL-3 cross-resistance to and amino acid sequence of protease inhibitor-resistant HIV-1 isolates and isolates obtained from patients failing antiretroviral therapy. The numbered protease consensus amino acid sequence is shown. Bold arrows indicate amino acids associated with resistance when present with other mutations, and amino acids in boldface are active-site mutations. Viral isolates, in the first column, starting with B, C, or D, were obtained from patients experiencing a viral rebound during treatment with the protease inhibitor(s) indicated in the second column. Inhibitors: I, indinavir; N, nelfinavir; S, saquinavir; R, ritonavir. The Z886 isolate was resistant to SC52151. Relative susceptibility to TL-3 is expressed as the relative IC50 compared to the control virus, R8, and values shown are the means from two independent experiments. Significant resistance was defined as a >4-fold-higher IC50 compared to R8.
Resistance to TL-3.
To determine TL-3 susceptibility of the isolates, 106 PHA-stimulated PBMC were exposed to 5,000 to 50,000 50% tissue culture infective dose(s) (TCID50) for 3 h, washed, and plated in duplicate at 20,000 cells per well in 96-well culture plates in the presence of various concentrations of TL-3 in complete RPMI 1640, and 20 U of IL-2/ml (29). Three-day supernatants were used to determine p24 antigen production (30). The drug concentration resulting in 50% inhibition (IC50) was determined by comparison to the drug-free cultures. The relative resistance was determined by comparing the IC50 value to that of the wild-type R8 virus.
Generation of TL-3-resistant HIV mutants.
Viral mutants resistant to TL-3 were generated by cultivating R8 wild-type virus at an initial concentration of 35 nM TL-3 in MT-2 cells grown in complete medium (29). When viral growth had resulted in abundant syncytium formation, usually days 4 to 10, one-half of the viral supernatant was passaged to uninfected MT-2 cells containing a higher TL-3 concentration. Culture medium was exchanged with fresh medium containing TL-3 at the desired concentration every 3 days to maintain selective pressure. The remaining supernatant was stored in aliquots at −80°C for further analysis or as a backup if virus did not grow at the higher TL-3 concentration.
Sequence analysis of protease from selected passages.
The protease gene was amplified from proviral DNA present in infected cells by using primers PR1s (5′-GCCGATAGACAAGGAACTGT) and PR2AS (5′-TTCCTGGCTTTAATTTTACTG). The PCR products were cloned into pCR2.1 (Invitrogen) or gel purified for direct sequencing.
Cloning and purification of HIV proteases.
The entire protease gene of wild-type and TL-3 resistant viruses were cloned for protein expression and the protein was expressed, purified, and verified, as previously described (21).
Protease assay.
Proteolytic activity and cleavage efficiencies (percent cleavage) and the Km and Vmax values were determined as previously described (21, 24, 36). TL-3 IC50 was as reported previously (24).
Molecular modeling of protease structure.
Modeling of TL-3 with the wild-type and TL-3-resistant 6x mutant HIV-1 protease was performed using the AMBER force field as implemented in the Discover 3 module of InsightII version 98.0 (MSI). All atoms were allowed to move, and energy minimization was performed for 500 iterations of conjugate gradients with both wild-type and the 6x mutant viruses. The starting structure of the HIV-1 protease inhibitor, TL-3, complexed with the wild-type form of HIV-1 protease was taken from the crystal structure (Protein Data Bank accession code 3TLH) (23). The presumed structure of the 6x mutant of HIV-1 protease was constructed by using homology modeling, based on wild-type HIV-1 protease complexed with TL-3 (Protein Data Bank accession code 3TLH). The side chains in both chains were mutated as follows: L24I, M46I, F53L, L63P, V77I, and V82A. Simulations began with the same backbone conformations of the crystal structure 3TLH. Since only one monomer of HIV-1 protease was seen in the asymmetric unit cell, the dimer was constructed by applying the appropriate transformation. This asymmetric unit cell also contained only one half of the TL-3 inhibitor, which is in itself a symmetric molecule. The resulting two halves of the TL-3 inhibitor were connected by a single bond. One of the two catalytic aspartic acid side chains was protonated, while the other was treated as charged. In both cases, water-301 and the four water molecules that bind adjacent to positions R8 and R8′ were also included.
RESULTS AND DISCUSSION
Some protease inhibitor-resistant patient isolates are susceptible to TL-3.
We have shown that the protease inhibitor TL-3 was active against wild-type HIV and FIV and drug-resistant proteases containing G48V and V82F mutations (22). To further define the inhibitory activity of TL-3 on drug-resistant viral isolates, we tested a panel of 11 primary viral isolates obtained from individuals that failed treatment with regimens containing the protease inhibitors indicated in Fig. 1. No cross-resistant assessment of viral isolates to currently approved protease inhibitors was performed. In Fig. 1, the viral isolates, information on the patient's drug regime, and the protease gene sequence are shown. Protease sequence determination allowed the verification of mutations associated with inhibitor resistance. Two molecular clones with known mutations and well-documented cross-resistance to most inhibitors (clone Z605-2 [L10R, M46I, L63P, V82T, and I84V] and clone Z952-7 [M46I and I84V], respectively) (3, 27) and one isolate, Z886, selected in vitro for resistance to SC-52151, were also tested (9). Nine of the eleven patient isolates have protease mutations at D30, G48, V82, I84, and/or L90, all of which are mutations associated with clinical resistance to protease inhibitors (4, 32, 34). Two isolates, D507-4 (V77I and I93L) and D507-7 (M36I), were found to contain only mutations generally considered as secondary, rather than primary active-site mutations. Thus, these isolates were not considered resistant to approved protease inhibitors.
IC50 was determined for each isolate by measuring viral p24 levels on day 3 over a concentration range of 10 to 1,250 nM TL-3 in PHA- and IL-2-stimulated and infected PBMC. Figure 1 shows the relative IC50 values of all isolates compared to the control virus, R8. Six of the isolates were found to have a >4-fold relative resistance, and five of these TL-3-resistant isolates shared a V82A/T mutation (Fig. 1). Nonetheless, the V82A change in itself does not lead to significant resistance, as evidenced by the D726-5 isolate with only the G48V and V82A active-site mutations and threefold relative resistance. The V82A/T mutation is commonly found in isolates from patients who failed ritonavir or indinavir therapy (4, 28, 34, 37). Besides the mutation at position 82, an I54V mutation was observed in the three isolates with the highest TL-3 resistance, a combination found in 27% of highly cross-resistant isolates (13). Of the three laboratory-derived resistant isolates, only one was found to be >4-fold resistant. In general, the predominant mutation associated with resistance to TL-3 in five of six isolates was a change at V82 to either an A or a T in combination with additional mutations. A V82A change in itself did not lead to high-level resistance to TL-3, since the D726-5 isolate had a threefold increase in resistance only. However, the sample size is too small to determine which mutation initially confers resistance and which combination of mutations is involved in increasing the resistance to TL-3.
TL-3-resistant virus.
To determine chronological changes in the protease gene conferring TL-3 resistance, R8 virus was passaged in MT-2 cells in the presence of increasing concentrations of the drug. R8 was initially grown in the presence of 35 nM TL-3, which is approximately the IC50, and the concentration was gradually increased to 46 μM during 56 passages over a 5-month period. At concentrations greater than 46 μM, TL-3 begins to precipitate out of the solution, therefore defining the TL-3 concentration limit. Susceptibility to TL-3 was determined every five to seven passages during the 5-month selection period. Figure 2 documents the relative resistance of isolates from passages 21, 31, 37, 44, and 56 compared to the wild-type R8 virus. The relative resistance increased by passage 56 to 17-fold.
FIG. 2.
Amino acid sequences of proteases from TL-3 escape mutants resistant to TL-3 at passages (P) 21, 31, 37, 44, and 56. The fraction of clones containing non-wild-type R8 protease sequences is indicated in the rightmost column. The sequence of the R8 virus is shown in the top row. Identity with R8 at individual amino acid positions is indicated by dashes. Superscripts: a, not applicable (NA) since the PCR product was sequenced directly; b, position 53 was ambiguous at passage 56, but clonal analysis demonstrated two independent mutations to L (codons CTT and TTG).
The proviral protease genes from passages 21, 31, 37, 44, and 56 were cloned from each viral stock and sequenced. For the later passages 44 and 56, the amplified protease region was directly sequenced to determine the predominant mutations in the virus stock. Wild-type sequence was seen at passage 21 in the majority of clones, although two clones were found to have an S-to-N mutation at the −P2 position of the p6*-protease cleavage site, and one clone demonstrated an additional mutation at position 35 (Fig. 2). By passage 31, resistance increased twofold over that of the wild-type virus. All of the six clones sequenced had a V82A substitution, a position associated with the S1 and S3 pocket in the active site of the enzyme and often found to be mutated in drug-resistant viral isolates (8, 30). At passage 37, two additional mutations, M46I and F53L, were observed in five of six clones isolated. These additional mutations were associated with a tripling of resistance to TL-3 relative to resistance of the single V82A mutant observed at P31. By passage 44, a V77I mutation was apparent in all clones, but TL-3 resistance had increased very little. Interestingly, this mutation was already present in one of six clones at P37. At passage 56 two more mutations, L24I and L63P, were observed and were associated with a doubling of resistance to TL-3 compared to the 4x mutant from passage 44. Thus, prolonged cultivation of HIV-1 in TL-3 results in a sequential increase of drug resistance, which is paralleled by acquisition of mutations in the protease gene.
It has been reported that some mutations in the protease gene result in a reduced viral replicative ability in vitro (5, 14, 27). The analysis of TL-3-resistant viruses with one, three, and six protease gene mutations (i.e., 1x, 3x, and 6x, respectively) were tested using MT-2 cells for alterations in viral replication. MT-2 cells were infected at a multiplicity of infection of 0.001, and viral replication was monitored for 7 days by determining the p24 content. Although viral isolates varied in their protease gene mutations, their replication rates, as judged by p24 increase over time, were indistinguishable (data not shown). Impairment of viral fitness is hypothesized to be a result of defective processing of the structural proteins or the cassette encoded by pol (6, 7, 26, 41). It has been reported that HIV carrying the V82A mutation in combination with M46I causes impaired growth kinetics (42). However, in some studies with viruses with mutant proteases the growth kinetics were found to be similar to those of wild-type viruses (16). Although the V82A mutation impaired purified protease activity, it is estimated that protease activity must fall below 10% of that of wide-type protease activity to disrupt the production of infectious viral particles (18, 27, 31, 33). Therefore, TL-3 mutant protease activity may not be rate-limiting for viral replication at a level of 40% activity of the wild-type in the case of the V82A mutation. Alternately, the simple comparisons of in vitro replication, rather than a competitive analysis of viral fitness of TL-3-resistant mutants versus wild-type virus, may have underestimated the true extent of loss of viral fitness in TL-3-resistant isolates (27). Although gag/pol cleavage site mutations that could allow more efficient cleavage by the mutated protease were not observed (data not shown), we cannot rule out the possibility that additional mutations outside of the protease gene may be present in TL-3-resistant viruses that compensate for defective protease function (1, 42).
The catalytic activity of protease from TL-3-resistant virus is reduced.
To quantify biochemical changes in the protease activity of TL-3 selected mutants, purified wild-type and 1x (passage 31), 3x (passage 37), and 6x (passage 56) mutant proteases were tested for relative enzymatic activity using a fluorogenic substrate (21) and a 16-amino-acid peptide representing the matrix-capsid (MA-CA) cleavage site. The results for the affinity (Km), the turnover rate (kcat), and the catalytic efficiency (i.e., kcat/Km) for the fluorogenic substrate and the percent cleavage of the MA-CA peptide are presented in Table 1. The Km for the fluorogenic substrate increased from 45 to 73 μM with the V82A position change in the protease, whereas the kcat dropped from 2.73 to 1.74 s−1. Thus, the resulting kcat/Km for the V82A mutation was only 40% of the wild-type catalytic efficiency (0.060 to 0.024 s−1 μM−1). With subsequent mutations in the protease, the Km increased even further but was compensated for by an increase in the catalytic activity to levels higher than that of wild-type protease. This results in a net increase of the catalytic efficiency of the 3x and 6x mutants to 47 and 62%, respectively, of the wild-type levels. These findings are paralleled by results obtained from analyzing the cleavage of the MA-CA peptide (Table 1). After 30 min of incubation of the MA-CA peptide with a constant amount of V82A mutant protease, only 18% of the peptide was cleaved. In contrast, the wild-type enzyme cleaved 31% of the peptide under identical assay conditions. With increasing mutations, the protease completely regained enzyme activity for this substrate, with 32% of the peptide being cleaved by the 6x mutant in the equivalent time interval. Thus, evaluation of enzymatic activity using two different substrates demonstrated that the initial V82A mutation decreased enzymatic activity, but this activity was restored with subsequent mutations.
TABLE 1.
Comparison of the enzyme kinetic constants for wild-type and mutant TL-3-resistant HIV proteases
| HIV proteasea | Fluorogenic substrateb
|
MA-CA peptidec (% cleavage ± SD) | ||
|---|---|---|---|---|
| Mean Km ± SD (μM) | kcat (s−1) | kcat/Km (s−1 μM−1)d | ||
| Wild type | 45.4 ± 3.9 | 2.73 | 0.060 | 31.1 ± 3.3 |
| V82A (1x) | 73.5 ± 9.1 | 1.74 | 0.024 (40) | 18.2 ± 1.3 |
| M46I/F53L/V82A (3x) | 194.2 ± 19.5 | 5.45 | 0.028 (47) | 26.4 ± 3.8 |
| L24I/M46I/F53L/L63P/V77I/V82A (6x) | 187.2 ± 31.9 | 6.86 | 0.037 (60) | 32.2 ± 2.7 |
Indicated are the viral isolates from which the cloned protease was obtained. The term within the parentheses reflects the number of mutations present.
The kinetics of the proteases were assayed with the fluorogenic substrate Abz-T-I-Nle∗ (p-NO2)F-Q-R. The protease assay was performed in 0.1 M morpholine ethanesulfonate buffer (pH 5.6), 0.2 M NaCl, 1 mM dithiothreitol, and 5% glycerol.
For the HIV-1 MA-CA junction peptide SSQVSQNY*PIVQNLQG, the percent cleavage was assayed by HPLC and is expressed as the percent cleaved in the 30-min assay. The protease and substrate concentrations were 2 and 100 mM, respectively.
The values within the parentheses indicate the catalytic efficiency as a percentage of wild-type virus (R8).
TL-3-resistant mutants are cross-resistant.
The mutant protease from the 3x mutant (passage 37) was found to be 11-fold and that of the 6x mutant (passage 56) was found to be 30-fold resistant to TL-3 compared to wild-type protease (Table 2). The 3x mutant protease was cross-resistant to saquinavir (5-fold), ritonavir (7-fold), and nelfinavir (4-fold) and by passage 56 the resulting 6x mutant protease demonstrated increased resistance to saquinavir (9-fold), ritonavir (14-fold), and nelfinavir (8-fold) compared to the wild-type protease. Given the primary V82A active site mutation seen after TL-3 passage, a mutation commonly found in the development of resistance to ritonavir, it is not surprising that some cross-resistance to ritonavir was evident. Our earlier studies suggest that a mutation at V82F impart a low level of resistance to TL-3 (22). Although the V82A active-site mutation is not a mutation typically observed during resistance development to saquinavir and nelfinavir, the generation of cross-resistance with the acquisition of secondary mutations is a characteristic seen in some, but not all, viruses isolated from patients treated with single protease inhibitors (32). Our results with TL-3 were not predictable from our earlier studies with single mutations at G48V and V82F (22) and imply that the contributions of secondary mutations in the flap and/or interior of the protease (see below) are critical for enhanced resistance to TL-3 and cross-resistance.
TABLE 2.
IC50 values of the wild-type and mutant proteases to TL-3 and cross-resistance to saquinavir (SQV), ritonavir (RTV), and nelfinavir (NFV)
| HIV proteasea | IC50 (nM) to protease inhibitor (fold increase)b
|
|||
|---|---|---|---|---|
| TL-3 | SQV | RTV | NFV | |
| Wild type | 6 | 5 | 4 | 6 |
| V82A (1x) | 22 (4) | 12 (2) | 7 (2) | 6 (1) |
| M46I/F53L/V82A (3x) | 64 (11) | 25 (5) | 31 (8) | 24 (4) |
| L24I/M46I/F53L/L63P/V77I/V82A (6x) | 183 (30) | 47 (9) | 55 (14) | 46 (8) |
The 1x mutant is from passage 31, the 3x mutant is from passage 37, and the 6x mutant is from passage 56.
The fold increase in IC50 for a mutant protease compared to the wild-type is shown in parentheses.
Computer modeling of TL-3 complexed with wild-type and 6x mutant protease.
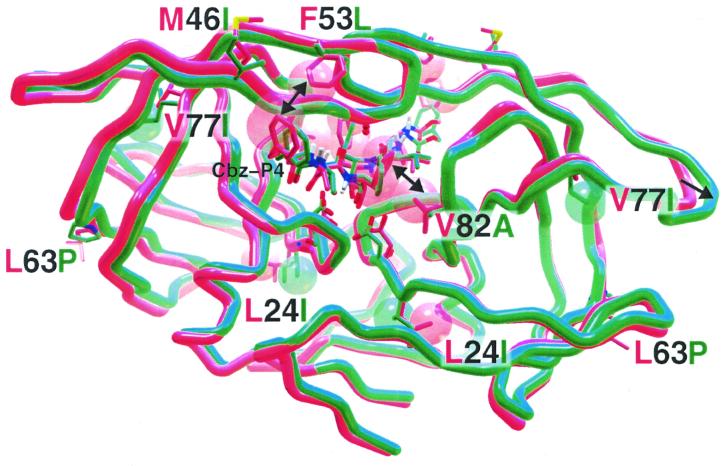
It was possible to postulate structural reasons, after molecular mechanics energy minimization calculations and structural overlays, for the loss in potency of TL-3 when confronted by the changes in the 6x mutant protease (green) compared to the wild-type protease (red) (Fig. 3). Inspection of the energy-minimized protease–TL-3 complexes revealed alterations in the packing of residue side chains of the 6x mutant throughout the structure. The initial V82A mutation, with loss of the larger V side chain (red) when replaced by the smaller A (green), removed van der Waals stabilization of the phenyl side chains present in P1 and P1′ of TL-3 (double-headed arrow; the spheres represent the van der Waals radii). When the F53 side chain was mutated to the smaller, nonaromatic L, favorable pi-pi contacts with the carboxy-benzyl (Cbz) groups at P4 and P4′ of TL-3 were lost (double-headed arrow). The L63P mutation results in the loss of a backbone amido proton. This appears to have a “domino effect” on the neighboring hydrogen bonding network. In combination with the M46I mutation, both presumably affect the flap structure. Overall, the flaps moved outward (arrow) and the backbone of the flaps assumed a more narrow conformation. In addition, the mutations at L24I and V77I occur in the interior of the dimeric protease's subunits, and the introduction of these beta-branched amino acids caused fairly significant local repacking of the neighboring atoms.
FIG. 3.
Energy-minimized model of R8 wild-type (red) protease overlaid with the TL-3 resistant 6x mutant (green) protease. TL-3 is bound to R8 and mutant proteases. Depicted are the protein backbones with mutated residues at numbered positions. The double-headed arrows indicate loss of both the TL-3 P1 interaction (P1′ not shown) with V82 upon mutation to the smaller A and the P4-Cbz interaction with the F53 upon mutation to the smaller L. The single-headed arrow points to distended flaps. The van der Waals radii of the L24I and V77I mutant side chains, resulting in rearrangement of local packing, are also shown as red (wild-type) or green (mutant) spheres, respectively.
Although changes in enzymatic activity based on changes in the flaps cannot be concluded from energy minimization modeling alone, the biochemical analysis (Table 1) demonstrates an increase in kcat consistent with this structural interpretation. TL-3 selection resulted in protease mutations that first affected substrate affinity and drastic changes in enzyme activity were mitigated by increased efficiency of cleavage through subsequent mutations. Therefore, secondary mutations allowed for maintenance of an acceptable kcat/Km, resulting in an effective mutant protease. Taken together, enzymatic results and proposed structural changes offer a sterically plausible explanation for the loss of potency to TL-3 and gain of biochemical function in the 6x mutant.
The demonstration that TL-3 was effective against some, although not all, drug-resistant viral isolates underscores the potential value of using FIV protease as a model for HIV protease inhibitor development. Our chronological panel of well-defined TL-3-resistant HIV isolates should be useful in helping to understand protease variability and substrate and inhibitor recognition. Substrates that are very effectively cleaved by HIV protease have been shown to be useful as models for designing tight binding inhibitors (2). Recently, a phage library was used to display potential substrates to compare FIV and HIV protease substrate specificities (Z. Q. Beck, Y.-C. Lin, and J. H. Elder, unpublished data). This method should prove useful for investigating the substrate specificity and cleavage efficacy of our panel of TL-3-resistant proteases and allow a rapid selection of substrates that can then be employed as templates to generate efficacious inhibitors for evaluation. It is anticipated that that our findings will help to define structural parameters to aid in the development of broad-based inhibitors less susceptible to evasion by mutations within protease.
ACKNOWLEDGMENTS
We gratefully acknowledge the laboratory assistance of Kent A. Smith, Giano P. Panzarella, Hugh B. Perkin, and Danica L. Lerner.
This study was supported by NIH grants DK49886 (B.E.T.), P01GM48870 (J.H.E., A.J.O., and C.-H.W.), and AI27670, AI38585, AI43638, and AI29164 (D.D.R.) and by the Research Center for AIDS and HIV Infection of the San Diego Veterans Affairs Healthcare System. TL-3 was synthesized and supplied under a contract operated by the Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Publication 13925-MEM from The Scripps Research Institute.
REFERENCES
- 1.Bally F, Martinez R, Peters S, Sudre P, Telenti A. Polymorphism of HIV type 1 gag p7/p1 and p1/p6 cleavage sites: clinical significance and implications for resistance to protease inhibitors. AIDS Res Hum Retrovir. 2000;16:1209–1213. doi: 10.1089/08892220050116970. [DOI] [PubMed] [Google Scholar]
- 2.Beck Z Q, Hervio L, Dawson P E, Elder J H, Madison E L. Identification of efficiently cleaved substrates for HIV-1 protease using a phage display library and use in inhibitor development. Virology. 2000;274:391–401. doi: 10.1006/viro.2000.0420. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, Li Y, Schock H B, Hall D, Chen E, Kuo L C. Three-dimensional structure of a mutant HIV-1 protease displaying cross-resistance to all protease inhibitors in clinical trials. J Biol Chem. 1995;270:21433–21436. doi: 10.1074/jbc.270.37.21433. [DOI] [PubMed] [Google Scholar]
- 4.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsh P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 5.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Carriere L C, Paulous S, Clavel F, Mammano F. Effects of human immunodeficiency virus type 1 resistance to protease inhibitors on reverse transcriptase processing, activity, and drug sensitivity. J Virol. 1999;73:3455–3459. doi: 10.1128/jvi.73.4.3455-3459.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eastman P S, Mittler J, Kelso R, Gee C, Boyer E, Kolberg J, Urdea M, Leonard J M, Norbeck D W, Mo H, Markowitz M. Genotypic changes in human immunodeficiency virus type 1 associated with loss of suppression of plasma viral RNA levels in subjects treated with ritonavir (Norvir) monotherapy. J Virol. 1998;72:5154–5164. doi: 10.1128/jvi.72.6.5154-5164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischl M A, Richman D D, Flexner C, Para M F, Haubrich R, Karim A, Yeramian P, Holden-Wiltse J, Meehan P M. Phase I/II study of the toxicity, pharmacokinetics, and activity of the HIV protease inhibitor SC-52151. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:28–34. doi: 10.1097/00042560-199705010-00005. [DOI] [PubMed] [Google Scholar]
- 10.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 12.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Jr, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 13.Hertogs K, Bloor S, Kemp S D, Van den Eynde C, Alcorn T M, Pauwels R, Van Houtte M, Staszewski S, Miller V, Larder B A. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS. 2000;14:1203–1210. doi: 10.1097/00002030-200006160-00018. [DOI] [PubMed] [Google Scholar]
- 14.Ho D D, Toyoshima T, Mo H, Kempf D J, Norbeck D, Chen C M, Wideburg N E, Burt S K, Erickson J W, Singh M K. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobsen H, Hanggi M, Ott M, Duncan I B, Owen S, Andreoni M, Vella S, Mous J. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, and frequencies. J Infect Dis. 1996;173:1379–1387. doi: 10.1093/infdis/173.6.1379. [DOI] [PubMed] [Google Scholar]
- 16.Jacobsen H, Yasargil K, Winslow D L, Craig J C, Krohn A, Duncan I B, Mous J. Characterization of human immunodeficiency virus type 1 mutants with decreased sensitivity to proteinase inhibitor Ro 31-8959. Virology. 1995;206:527–534. doi: 10.1016/s0042-6822(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 17.Kirk O, Katzenstein T L, Gerstoft J, Mathiesen L, Nielsen H, Pedersen C, Lundgren J D. Combination therapy containing ritonavir plus saquinavir has superior short-term antiretroviral efficacy: a randomized trial. AIDS. 1999;13:F9–F16. doi: 10.1097/00002030-199901140-00002. [DOI] [PubMed] [Google Scholar]
- 18.Konvalinka J, Litterst M A, Welker R, Kottler H, Rippmann F, Heuser A M, Krausslich H G. An active-site mutation in the human immunodeficiency virus type 1 proteinase (PR) causes reduced PR activity and loss of PR-mediated cytotoxicity without apparent effect on virus maturation and infectivity. J Virol. 1995;69:7180–7186. doi: 10.1128/jvi.69.11.7180-7186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence J, Schapiro J, Winters M, Montoya J, Zolopa A, Pesano R, Efron B, Winslow D, Merigan T C. Clinical resistance patterns and responses to two sequential protease inhibitor regimens in saquinavir and reverse transcriptase inhibitor-experienced persons. J Infect Dis. 1999;179:1356–1364. doi: 10.1086/314751. [DOI] [PubMed] [Google Scholar]
- 20.Ledergerber B, Egger M, Opravil M, Telenti A, Hirschel B, Battegay M, Vernazza P, Sudre P, Flepp M, Furrer H, Francioli P, Weber R. Clinical progression and virological failure on highly active antiretroviral therapy in HIV-1 patients: a prospective cohort study. Swiss HIV Cohort Study. Lancet. 1999;353:863–868. doi: 10.1016/s0140-6736(99)01122-8. [DOI] [PubMed] [Google Scholar]
- 21.Lee T, Laco G S, Torbett B E, Fox H S, Lerner D L, Elder J H, Wong C H. Analysis of the S3 and S3′ subsite specificities of feline immunodeficiency virus (FIV) protease: development of a broad-based protease inhibitor efficacious against FIV, SIV, and HIV in vitro and ex vivo. Proc Natl Acad Sci USA. 1998;95:939–944. doi: 10.1073/pnas.95.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee T, Le V-D, Lim D, Lin Y-C, Morris G M, Wong A L, Olson A J, Elder J H, Wong C H. Development of a new type of protease inhibitors, efficacious against FIV and HIV variants. J Am Chem Soc. 1999;121:1145–1155. [Google Scholar]
- 23.Li M, Morris G M, Lee T, Laco G S, Wong C H, Olson A J, Elder J H, Wlodawer A, Gustchina A. Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease. Proteins. 2000;38:29–40. doi: 10.1002/(sici)1097-0134(20000101)38:1<29::aid-prot4>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 24.Lin Y C, Beck Z, Lee T, Le V D, Morris G M, Olson A J, Wong C H, Elder J H. Alteration of substrate and inhibitor specificity of feline immunodeficiency virus protease. J Virol. 2000;74:4710–4720. doi: 10.1128/jvi.74.10.4710-4720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Little S J, Daar E S, D'Aquila R T, Keiser P H, Connick E, Whitcomb J M, Hellmann N S, Petropoulos C J, Sutton L, Pitt J A, Rosenberg E S, Koup R A, Walker B D, Richman D D. Reduced antiretroviral drug susceptibility among patients with primary HIV infection. JAMA. 1999;282:1142–1149. doi: 10.1001/jama.282.12.1142. [DOI] [PubMed] [Google Scholar]
- 26.Mammano F, Petit C, Clavel F. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J Virol. 1998;72:7632–7637. doi: 10.1128/jvi.72.9.7632-7637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Picado J, Savara A V, Sutton L, D'Aquila R T. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol. 1999;73:3744–3752. doi: 10.1128/jvi.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 29.Mosier D E, Gulizia R J, MacIsaac P D, Torbett B E, Levy J A. Rapid loss of CD4+ T cells in human-PBL-SCID mice by noncytopathic HIV isolates. Science. 1993;260:689–692. doi: 10.1126/science.8097595. [DOI] [PubMed] [Google Scholar]
- 30.Patick A K, Rose R, Greytok J, Bechtold C M, Hermsmeier M A, Chen P T, Barrish J C, Zahler R, Colonno R J, Lin P F. Characterization of a human immunodeficiency virus type 1 variant with reduced sensitivity to an aminodiol protease inhibitor. J Virol. 1995;69:2148–2152. doi: 10.1128/jvi.69.4.2148-2152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose J R, Babe L M, Craik C S. Defining the level of human immunodeficiency virus type 1 (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J Virol. 1995;69:2751–2758. doi: 10.1128/jvi.69.5.2751-2758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schinazi R F, Larder B A, Mellors J W. Mutations in the retroviral genes associated with drug resistance. Antiviral News. 1997;5:129–142. [Google Scholar]
- 33.Schock H B, Garsky V M, Kuo L C. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. Compensatory modulations of binding and activity. J Biol Chem. 1996;271:31957–31963. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 34.Shafer R W, Hsu P, Patick A K, Craig C, Brendel V. Identification of biased amino acid substitution patterns in human immunodeficiency virus type 1 isolates from patients treated with protease inhibitors. J Virol. 1999;73:6197–6202. doi: 10.1128/jvi.73.7.6197-6202.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staszewski S, Miller V, Sabin C, Carlebach A, Berger A M, Weidmann E, Helm E B, Hill A, Phillips A. Virological response to protease inhibitor therapy in an HIV clinic cohort. AIDS. 1999;13:367–373. doi: 10.1097/00002030-199902250-00009. [DOI] [PubMed] [Google Scholar]
- 36.Toth M V, Marshall G R. A simple, continuous fluorometric assay for HIV protease. Int J Peptide Protein Res. 1990;36:544–550. doi: 10.1111/j.1399-3011.1990.tb00994.x. [DOI] [PubMed] [Google Scholar]
- 37.Turriziani O, Antonelli G, Jacobsen H, Mous J, Riva E, Pistello M, Dianzani F. Identification of an amino acid substitution involved in the reduction of sensitivity of HIV-1 to an inhibitor of viral proteinase. Acta Virol. 1994;38:297–298. [PubMed] [Google Scholar]
- 38.Wegner S A, Brodine S K, Mascola J R, Tasker S A, Shaffer R A, Starkey M J, Barile A, Martin G J, Aronson N, Emmons W W, Stephan K, Bloor S, Vingerhoets J, Hertogs K, Larder B. Prevalence of genotypic and phenotypic resistance to anti-retroviral drugs in a cohort of therapy-naive HIV-1 infected US military personnel. AIDS. 2000;14:1009–1015. doi: 10.1097/00002030-200005260-00013. [DOI] [PubMed] [Google Scholar]
- 39.Wlodawer A, Gustchina A, Reshetnikova L, Lubkowski J, Zdanov A, Hui K Y, Angleton E L, Farmerie W G, Goodenow M M, Bhatt D, et al. Structure of an inhibitor complex of the proteinase from feline immunodeficiency virus. Nat Struct Biol. 1995;2:480–488. doi: 10.1038/nsb0695-480. [DOI] [PubMed] [Google Scholar]
- 40.Young B, Johnson S, Bahktiari M, Shugarts D, Young R K, Allen M, Ramey II R R, Kuritzkes D R. Resistance mutations in protease and reverse transcriptase genes of human immunodeficiency virus type 1 isolates from patients with combination antiretroviral therapy failure. J Infect Dis. 1998;178:1497–1501. doi: 10.1086/314437. [DOI] [PubMed] [Google Scholar]
- 41.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]