Abstract
Background
Recently, various polygenic risk score (PRS)-based methods were developed to improve stroke prediction. However, current PRSs (including cross-ancestry PRS) poorly predict recurrent stroke. Here, we aimed to determine whether the best PRS for Japanese individuals can also predict stroke recurrence in this population by extensively comparing the methods and maximizing the predictive performance for stroke onset.
Methods
We used data from the BioBank Japan (BBJ) 1st cohort (n=179,938) to derive and optimize the PRSs using a 10-fold cross-validation. We integrated the optimized PRSs for multiple traits, such as vascular risk factors and stroke subtypes to generate a single PRS using the meta-scoring approach (metaGRS). We used an independent BBJ 2nd cohort (n=41,929) as a test sample to evaluate the association of the metaGRS with stroke and recurrent stroke.
Results
We analyzed recurrent stroke cases (n=174) and non-recurrent stroke controls (n=1,153) among subjects within the BBJ 2nd cohort. After adjusting for known risk factors, metaGRS was associated with stroke recurrence (adjusted OR per SD 1.18 [95% CI: 1.00–1.39, p=0.044]), although no significant correlation was observed with the published PRSs. We administered three distinct tests to consider the potential index event bias; however, the outcomes derived from these examinations did not provide any significant indication of the influence of index event bias. The high metaGRS group without a history of hypertension had a higher risk of stroke recurrence than that of the low metaGRS group (adjusted OR 2.24 [95% CI: 1.07–4.66, p=0.032]). However, this association was weak in the hypertension group (adjusted OR 1.21 [95% CI: 0.69–2.13, p=0.50]).
Conclusions
The metaGRS developed in a Japanese cohort predicted stroke recurrence in an independent cohort of patients. In particular, it predicted an increased risk of recurrence among stroke patients without hypertension. These findings provide clues for additional genetic risk stratification and help in developing personalized strategies for stroke recurrence prevention.
Keywords: recurrent stroke, stroke, polygenic risk score, LDpred2, risk factor, index event bias, hypertension, metaGRS
Introduction
Stroke is a major cause of mortality in Japan, with 56,000 deaths reported in 2020.1 The conventional risk factors for stroke include hypertension, high waist-to-hip ratio, smoking, cardiac causes, dyslipidemia, and diabetes mellitus.2 In Japan, the stroke recurrence rate is up to 30–50% during 5–10 years of follow-up after the first stroke.3,4 Accordingly, it will be medically beneficial to stratify high-risk groups for recurrent stroke among those who have experienced a stroke to potentially generate more intensive secondary prevention strategies than current recommendations.
Genome-wide association studies (GWAS) have identified many disease-susceptibility variants associated with complex traits.5 A polygenic risk score (PRS) is the weighted summation of the individual genetic effects of these variants. Its weighting strategy varies depending on the construction method; traditionally, only significant variants are used in developing this score. The recently developed PRS methods involve non-significant variants and updated effect weights and consider the linkage disequilibrium (LD) structure. The development of PRS methods has helped stratify high-risk groups for complex traits,6–11 including stroke.12
Polygenic risk scores developed using the 32 genome-wide significant (p< 5.0×10−8) variants or 90 marginally associated (p<1.0×10−5) variants (PRS90) from the MEGASTROKE study12 are associated with stroke onset in subjects of European ancestry.12,13 The meta-scoring PRS approach (metaGRS) includes 3.2 million variants by combining PRSs for stroke subtypes, risk factors, and comorbidities by adjusting the effect weight via elastic-net logistic regression; this approach has an improved predictive performance for stroke compared to that of PRS90.14 MetaGRS can predict stroke incidence independent of environmental factors and could help motivate individuals with high genetic risk to make lifestyle changes for stroke prevention (although not yet implemented in clinical practice outside a research setting).15 The PRS shows reduced transferability between populations. Additionally, a PRS developed using various variants derived from Japanese GWAS successfully predicted stroke onset in the Japanese population.16,17 Most recently, the GIGASTROKE study proposed an integrated PRS approach among PRSs derived from populations of multiple ancestries using the metaGRS framework (iPGS), which showed a better predictive ability than the MEGASTROKE European or East Asian PRS.18 However, the PRS did not successfully predict stroke recurrence; for example, PRS32 and iPGS did not significantly predict stroke recurrence after adjusting for clinical comorbidities, with notably smaller effect sizes than for non-recurrent stroke.12,18 Furthermore, the potential effect of index event (also known as “collider”) bias that may distort the association of PRS was suspected.19,20
The optimal method to improve the predictive accuracy of PRS depends on the population- and trait-specific genetic architecture.21–30 Therefore, we compared different PRS methods and determined whether the best PRS can predict the onset of recurrent stroke in a Japanese population.
Methods
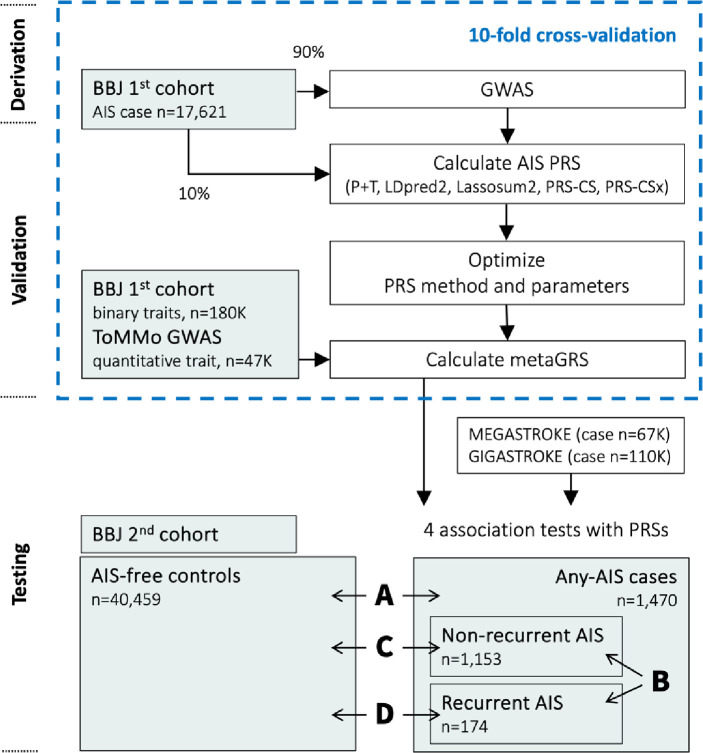
The workflow of this study is shown in Figure 1. This article follows the TRIPOD (Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis) reporting guidelines.
Figure 1. Workflow.
MEGASTROKE AIS summary statistics of European (EUR) studies were only used for PRS-CSx. 1000 Genomes Project super population samples (EAS or EUR) were used for the LD reference panel. Abbreviations: GWAS = genome-wide association study, PRS = polygenic risk score, P+T = pruning and thresholding, OR = odds ratio, AIS = all ischemic stroke, ToMMo = Tohoku Medical Megabank; LD, linkage disequilibrium.
We used a logistic regression model to assess the association of the PRS using the two case-control settings for AIS (A: any-AIS vs. AIS-free controls) and AIS recurrence (B: recurrent AIS vs. non-recurrent AIS). We also applied two other combinations of case-controls (C: recurrent versus AIS-free controls and D: non-recurrent versus AIS-free controls).
Study subjects and quality control
BioBank Japan (BBJ) involves physicians diagnosing all ischemic stroke (AIS) cases at the collaborating hospitals. BBJ was established in 2003 and recruited 267,000 patients from 12 medical institutions (66 hospitals) in two phases.31–33 The recruited patients had at least one of the 51 primarily multifactorial (common) diseases, which accounted for 440,000 cases. We used BBJ 1st cohort (BBJ1) data to derive PRSs and available independent BBJ 2nd cohort (BBJ2) data to evaluate the performance of PRSs in predicting AIS and recurrent AIS. Recurrent AIS information was unavailable for the BBJ1 data. In BBJ2, any AIS cases (n=1,470), AIS-free controls (n=40,459), recurrent AIS cases (n=174), and non-recurrent AIS controls (n=1,153) were available. The mean duration from the first episode of AIS onset to recurrent AIS onset was 4.88 years. Detailed sample characteristics are listed in Supplementary Methods and Table 1.
Table 1.
Characteristic of the testing sample (BBJ2)
| Any-AIS versus AIS-free controls | Recurrent AIS versus non-recurrent AIS | |||||||
|---|---|---|---|---|---|---|---|---|
| N sample (%, SD) | OR | 95% CI | p-value | N sample (%, SD) | OR | 95% CI | p-value | |
| Total participants | 41,929 (100%) | - | - | - | 1,327 (100%) | - | - | - |
| AIS / AIS recurrent | 1,470 (3.5%) | - | - | - | 174 (13.1%) | - | - | - |
| Age (SD)* | 70.0 (SD=12.6) | 1.18 | [1.06–1.31] | 0.002 | 64.1 (SD=11.5) | 0.83 | [0.59–1.16] | 0.256 |
| Female sex | 19,407 (46.3%) | 0.48 | [0.43–0.54] | <0.001 | 383 (28.9%) | 0.63 | [0.42–0.93] | 0.019 |
| Hypertension | 17,710 (42.2%) | 2.27 | [2.04–2.53] | <0.001 | 838 (63.1%) | 0.92 | [0.66–1.3] | 0.673 |
| Hyperlipidemia | 11,604 (27.7%) | 2.17 | [1.95–2.41] | <0.001 | 603 (45.4%) | 0.92 | [0.66–1.29] | 0.625 |
| Diabetes | 3,622 (8.6%) | 1.16 | [0.97–1.39] | 0.089 | 125 (9.4%) | 0.97 | [0.52–1.7] | 1.000 |
| Smoking** | 21,570 (51.9%) | 1.43 | [1.28–1.59] | <0.001 | 795 (60.5%) | 1.28 | [0.90–1.82] | 0.179 |
| Vascular disease | 3,136 (7.5%) | 1.31 | [1.08–1.56] | 0.005 | 122 (9.2%) | 1.08 | [0.59–1.87] | 0.778 |
| Heart failure | 1,329 (3.2%) | 1.27 | [0.95–1.66] | 0.095 | 48 (3.6%) | 1.79 | [0.78–3.75] | 0.124 |
| Atrial fibrillation | 1,004 (2.4%) | 2.22 | [1.71–2.84] | <0.001 | 65 (4.9%) | 1.07 | [0.46–2.23] | 0.850 |
All risk factor characteristics were derived from history and not at the time of registration. Odds ratio (OR), 95% CI (95% confidence intervals, and p-values were calculated using logistic regression for onset and AIS recurrence (unadjusted for other factors).
Age at AIS case-control was that at recruitment, while age at recurrent AIS case-control was that at first incidence. The numbers indicate the median threshold age.
The total number of missing values of smoking was 389 for all case-control and 12 for recurrent AIS case-control samples.
Abbreviations: BBJ2, BioBank 2nd cohort; AIS, all ischemic stroke; SD = standard deviation
This study was approved by the ethics committee of the Institute of Medical Science, the University of Tokyo, Japan. Quality control, pre-phasing, and genotype imputation were conducted using PLINK(v2.0),34–37 Eagle (v2.4.1), and Minimac4 (v1.0.2), respectively. The detailed processes are presented in the Supplementary Methods.
Constructing PRSs for AIS
Unbiased PRSs were obtained by applying a 10-fold cross-validation to select the model and optimize the parameters.26,38 Briefly, BBJ1 samples were randomly split into ten equal-sized subsamples. We retained one subsample for validation and the others for training. We repeated this process 10 times, with each of the ten subsamples used exactly once for validation. A GWAS was conducted on the training set in each iteration, adjusted for age, sex, and the first 10 principal components (PCs) via Firth logistic regression using PLINK (v.2.0).34
We obtained the weights of variants for PRS from the GWAS summary statistics of the training set using five PRS methods—P+T (PLINK (v.1.9)35 for clumping), LDpred2,39 Lassosum2,40 (LDpred2 and Lassosum2 by bigsnpr package (v.1.7.2) in R (v.3.5.0)), PRS-CS (v.1.0.0),41 and PRS-CSx (v.1.0.0).42 The PRS-CSx integrated BBJ1 with the European stroke GWAS summary statistics (MEGASTROKE; the largest study available at this study design)43 by learning an optimal linear combination. We used combinations of parameters for P+T (1,224 parameters), LDpred2 (126 parameters), Lassosum2 (200 parameters), PRS-CS (9 parameters), and PRS-CSx (9 parameters), as described in the Supplementary Methods. Subsequently, the PRSs for the validation sample were calculated using the weights obtained from the training samples. The accuracy for predicting AIS cases was evaluated from Nagelkerke’s R2 (simply “R2” from this point onwards)29,44 after adjusting for age, sex, and the first 10 PCs. We calculated the mean R2 over 10 cross-validation results for each method with each parameter after a 10-fold cross-validation. We chose the method and parameters that maximized incremental R2 (PRSAIS) among these PRSs.
We further integrated the PRSAIS with PRSs of vascular risk factors, such as stroke subtypes and comorbid diseases presence, using the elastic net framework to construct a metaGRS using the glmnet package (v.4.1.3) in R (v.4.1.0). Nine binary traits and eight quantitative traits of vascular risk factors reported in a previous study14 are described in the Supplementary Methods. Binary traits were determined by conducting GWAS and attempting to obtain unbiased weights using cross-validations. The effect weights from the derivation sample every 17 traits were calculated using PRS-CS-auto since it did not require an independent validation sample set for parameter optimization and performed well for various traits.21,26,39,41,45 Subsequently, we used a validation sample to calculate the weight of AIS and the 17-trait PRSs to predict AIS using elastic-net logistic regression. We conducted a 10-fold cross-validation and used the mean weight for testing.
We used PLINK (v2.0)34 to calculate the individual PRS by aggregating the effect estimates multiplied by each imputed dosage into a single score per person.
Risk factors
The following seven risk factors that were previously utilized as covariates12 were used as covariates for testing: hypertension (systolic blood pressure>140 mmHg, diastolic blood pressure>90 mmHg, or hypertension history), hyperlipidemia, diabetes (all types), smoking (current smoker), vascular disease (myocardial infarction, peripheral artery disease, stable angina pectoris, and unstable angina pectoris), congestive heart failure, and atrial fibrillation (including atrial flutter). A sample was considered to have a risk factor status if it had that status at enrollment or from historical records (Tables 1 and S1).
Assessment of the association of PRS with AIS and AIS recurrence
We used a single selected method with optimized parameters and calculated the metaGRS in independent testing sample sets. We used a logistic regression model to assess the association of the PRS using the two case-control settings for AIS (any-AIS versus AIS-free controls) and AIS recurrence (recurrent AIS versus non-recurrent AIS). We also applied two other combinations of case-controls: recurrent AIS versus AIS-free controls and non-recurrent AIS versus AIS-free controls (Figure 1). Furthermore, we examined additional PRS contributions of the seven risk factors to predictive accuracy and discriminative performance using the values of R2 and area under the curve (AUC), according to our previous studies.14,46
Additionally, we evaluated the performance of the following PRSs derived from other milestone studies for stroke prediction: 32 genome-wide significant variants (for any stroke, ischemic stroke, or ischemic stroke subtypes) from the MEGASTROKE cross-ancestry study of 524,354 individuals (PRS32),43 89 genome-wide significant variants of 1,614,080 multi-population individuals (PRS89), and 6,010,730 variants of the East Asian PRS developed from 9,809 individuals (iPGSEAS),18 both from the GIGASTROKE study. The PRS calculation process is described in the Supplementary Methods.
Considering potential index event bias
An index event bias may be induced when the samples are only selected from cases.47 We evaluated the extent to which the index event bias affected our results since we used case-only samples in this study. The association of PRSs with recurrent AIS was evaluated after adjusting for seven risk factors, in addition to age, sex, and the first 10 PCs. Adjusting for such confounding bias will not be enough to eliminate bias for a recurrence association study.48 Therefore, we sought to mitigate a potential index event bias by applying three distinct methodologies.48,49 First, we utilized linear and logistic regression models to assess the relationships between metaGRS and covariates within the any-AIS case (n=1,327) and AIS-free control groups (n=40,459). Initially, we did not adjust for age, sex, the seven risk factors, or the first 10 PCs. Subsequently, we observed the distributions of covariate values across the metaGRS quintiles. We performed statistical tests to detect heterogeneity in the estimates between the prevalent case and control groups, following the methodology of a prior study.19,50 Second, we refined our analysis by adjusting for associations between metaGRS and AIS recurrence while considering the differential effects of covariates, according to a previous method.19 Finally, we applied the inverse probability weighted (IPW) approach47 to comprehensively account for index event bias. Collectively, these analytical approaches were adopted to enhance the validity of our findings.
Association of metaGRS and AIS recurrence in patients with/without hypertension
Logistic regression was conducted in subgroups with and without hypertension and metaGRS tertiles among recurrent AIS cases (with hypertension n=107, without hypertension n=67) and non-recurrent AIS controls (with hypertension n=731, without hypertension n=422) to assess the relationship between the PRS and the risk of AIS recurrence. The low metaGRS tertile was set as the reference group and adjusted for age, sex, and the first 10 PCs.
Statistical analysis
The mean with standard deviation (SD) or proportion of factors was reported for the baseline characteristics of testing samples. The incremental value (R2 or AUC) was estimated from the differences between patients with and without PRSs of the fitted values of age, sex, first 10 PCs, and seven risk factors48,57,58 and calculated as the 95% confidence interval. The pROC package (v.1.18.0) in R was used to determine the discriminative ability of the AUC. The IPW package (v.1.2) in R was used for the IPW approach. R (v. 3.5.0) was used to perform logistic regression to calculate R2, Pearson’s correlation coefficient, and linear regression. All statistical tests were two-sided. The significance level was set at p = 0.05.
Results
Derivation of effect weight
The imputed genotype data of 17,621 AIS cases and 162,317 controls without an AIS diagnosis were used for 9,622,629 autosomal variants after implementing quality control of the BBJ1 dataset (Tables S2–S4).
We conducted a 10-fold cross-validation to adjust the parameters and select the best PRS associated with AIS. We performed GWAS 10 times on 90% of the randomly selected BBJ1 dataset (training data). We successfully detected previously reported43,51 signals in each dataset (p<5×10−8), including SH3PXD2A, CCDC63 (eight times), CUX2, and LINC02356 (every time) (Figure S1, Table S5).
We confirmed some expected characteristics of each PRS method (such as low accuracy) using only genome-wide significant variants (Tables S6–10 and Supplementary Notes). The mean incremental R2 values of each scoring method with the best-performed parameters were 0.0038 (95% CI: 0.0030–0.0046), 0.00443 (95% CI: 0.0035–0.0054), 0.0039 (95% CI: 0.0030–0.0048), 0.00441 (95% CI: 0.0036–0.0053), and 0.0037 (95% CI: 0.0031–0.0042) for P+T, LDpred2, Lassosum2, PRS-CS, and PRS-CSx, respectively (Table 2). We chose LDpred2 with the parameter set of ρ-value = 0.0056, a heritability-value = 1.0×h2LDSC, where h2LDSC is the heritability estimate from the constrained LD score regression52, and a no-sparse model for subsequent analyses, since it showed the best mean incremental R2 value among the five methods.
Table 2.
Polygenic risk score performance at validation
| Method | Best parameters | Mean number of variants | Best mean incremental Nagelkerke R2 | Standard deviation | 95% confidence interval |
|---|---|---|---|---|---|
| P+T | Clumping R2=0.95, Clumping kb=526, Imputation R2=0.8, p-value threshold=1 | 3,144,737 | 0.0038 | 0.0012 | 0.0030–0.0046 |
| LDpred2 | p value=0.0056, heritability value × 1.0, no sparse | 898,456 | 0.00443 | 0.0013 | 0.0035–0.0054 |
| Lassosum2 | S=0.9, lambda=0.00388 | 282,520 | 0.0039 | 0.0013 | 0.0030–0.0048 |
| PRS-CS | Phi=1.00E-04 | 985,439 | 0.00441 | 0.0012 | 0.0036–0.0053 |
| PRS-CSx | Phi=1.00E-05 | 1,016,745 | 0.0037 | 0.0008 | 0.0031–0.0042 |
We observed an average number of nonzero weights for 8.4 traits after computing the metaGRS via elastic net regularization 10 times (10-fold). The metaGRS weight of AIS was highest (mean=0.123, SD=0.026), followed by diastolic blood pressure (mean=0.039, SD=0.039), atrial fibrillation (mean=0.023, SD=0.024), and myocardial infarction (mean=0.018, SD=0.025) (Figure S2 and Table S11). Only the triglyceride weights were zero at all 10 measurements among the 18 traits included in the metaGRS calculation. The number of variants used for metaGRS was 1,014,026; a total of 1,011,847 variants (99.8%) remained after matching with the BBJ2 dataset.
Association of metaGRS with AIS cases and recurrent AIS
We used the imputed genotype data of 1,470 AIS cases and 40,459 controls without a diagnosis of AIS for 59,387,070 variants from the BBJ2 dataset to test the association of metaGRS with AIS and AIS recurrence. The AIS case-only sample of the BBJ2 was used to analyze AIS recurrence. Table 1 presents the characteristics of the test samples.
MetaGRS was associated with AIS diagnosis after adjusting for age, sex, first 10 PCs, and seven risk factors (adjusted OR, 1.21 [95% CI: 1.15–1.27, p=2.89×10−12]), as previously reported.14,18 MetaGRS was also associated with AIS recurrence compared with recurrence-free AIS (adjusted OR 1.18 [95% CI: 1.00–1.39, p=0.044]; Table 3 and Figure S3). MetaGRS showed stronger association when comparing recurrent AIS with AIS-free controls (adjusted OR 1.37 [95% CI: 1.18–1.59, p=5.35×10−5]; Table S12).
Table 3.
Polygenic risk score performance at testing
| PRS Method | Association tests | OR per SD [95% CI] | p-value | Incremental AUC | Incremental Nagelkerke R2 |
|---|---|---|---|---|---|
| MetaGRS | AIS | 1.21[1.15–1.27] | 2.89E-12 | 0.0087 | 0.0044 |
| Recurrence | 1.18[1.00–1.39] | 0.044 | 0.0123 | 0.0057 | |
| MEGASTROKE 27 SNVs | AIS | 1.11[1.06–1.17] | 4.23E-05 | 0.0033 | 0.0015 |
| Recurrence | 1.07[0.91–1.25] | 0.41 | 0.0032 | 0.0009 | |
| GIGASTROKE 84 SNVs | AIS | 1.08[1.03–1.14] | 2.96E-03 | 0.0022 | 0.0008 |
| Recurrence | 1.17[1.00–1.38] | 0.054 | 0.0125 | 0.0052 | |
| GIGASTROKE iPGS | AIS | 1.26[1.20–1.33] | 1.24E-17 | 0.0130 | 0.0066 |
| Recurrence | 1.08[0.91–1.27] | 0.37 | 0.0044 | 0.0011 |
Polygenic risk score performance was evaluated using an independent testing set for AIS and recurrent AIS. We showed two main association tests; AIS (any-AIS cases vs. AIS-free controls) and AIS recurrence (recurrent AIS vs. non-recurrent AIS). Incremental AUC and R2 are the differences in the values when fitting with/without PRS, along with age, sex, the first 10 PCs, and seven risk factors. Abbreviations: AIS, all ischemic strokes; OR, odds ratio; AUC, area under the curve; PC, principal components; PRS, polygenic risk score; SD = standard deviation
The contribution of the metaGRS and traditional risk factors showed an AIS prediction accuracy with an R2 value of 0.06 and an AUC of 0.689 after constructing the baseline model using age, sex, the first 10 PCs, and seven risk factors. The incremental AUCs were 0.0087 and 0.0123 for AIS and AIS recurrence, respectively when metaGRS was added to the baseline model (Table 3). In our dataset, clinical risk factors (including hypertension) were related to AIS diagnosis but were insignificantly associated with AIS recurrence (Table S1). We assessed the prediction performance of previously developed PRSs for AIS and AIS recurrence in our dataset. After matching with the BBJ2 dataset (Supplementary Methods), 27, 84, and 5,756,652 variants remained in PRS32, PRS89, and iPGS, respectively. We confirmed their association with AIS diagnosis; adjusted ORs were 1.11 [95% CI: 1.06–1.17, p=4.23×10−5], 1.08 [95% CI: 1.03–1.14, p=2.96×10−3], and 1.26 [95% CI: 1.20–1.33, p=1.24×10−17] for PRS32, PRS89, and iPGS, respectively (Table 3, Figure S3); however, a significant association was not observed between PRSs and AIS recurrence (p-values of 0.41, 0.054, and 0.37, respectively; Table 3 and Figure S3). Our Japanese optimized metaGRS was the only PRS significantly associated with AIS recurrence in this study.
Analyzing for potential index event bias
We observed the values of covariates at the AIS-free control group and any-AIS case group in each quintile. We did not find any significant heterogeneous relationships between the covariates and the metaGRS in terms of regression estimates in the prevalent case and control samples (p>0.05, Table S13).
We used three different variable models—i) age and sex; ii) age, sex, and seven risk factors; and iii) age, sex, the first 10 PCs, and seven risk factors—to determine the association between metaGRS and recurrent AIS; none of these confounders significantly influenced our results (Figure S4).
We compared the association results of IPW adjusted (accounting for index event bias) with those of non-adjusted IPW (accounting for confounding bias). The results remained almost unchanged, but the 95% confidence intervals overlapped (Figure S5). A comparison of the three distinct models did not indicate an effect of index event bias.
Association of metaGRS and AIS recurrence in patients with/without hypertension
We divided the test sample into subgroups according to the presence or absence of a history of hypertension and evaluated the risk effect of the metaGRS tertile. The high metaGRS group without a history of hypertension showed a higher risk effect for AIS recurrence compared to the low metaGRS group (OR of the high metaGRS group compared to that of the low metaGRS group was 2.24 [95% CI: 1.07–4.66, p=0.032], Figure 2 and Table S14). However, no significant association was observed between the metaGRS and AIS recurrence in the group with a history of hypertension (the OR of the high metaGRS group compared to that of the low metaGRS group was 1.21 [95% CI: 0.69–2.13, p=0.50] (Figure 2 and Table S14).
Figure 2. Odds ratio of metaGRS tertiles with/without a history of hypertension.
Association of AIS recurrence and meta-GRS tertiles with or without a history of hypertension (HT) in the testing sample, with reference to the low metaGRS tertiles.
Discussion
We successfully examined the association between recurrent AIS and our best model PRS (metaGRS using LDpred2); the adjusted OR was 1.18 for each unit of SD increase in PRS. Our metaGRS showed stronger (adjusted OR per SD=1.37) association when comparing recurrent AIS with AIS-free controls. Furthermore, a high PRS was associated with AIS recurrence particularly in groups without a history of hypertension (OR of the top vs. bottom metaGRS tertile=2.24). These results are consistent with the result of a previous study wherein the stroke prediction accuracy of the PRS was high in the group with low CHA2DS2-VASc scores.12 These results indicate the utility of the PRS in developing more precise strategies to prevent AIS recurrence in individuals with a high PRS who do not have high profiles based on clinical risk factors.
We attempted to mitigate potential index event bias since our purpose was to specifically determine the efficacy of PRS among AIS patients. It is difficult to predict and provide an accurate assessment of recurrent AIS based on genetic predisposition owing to the possible effect of index event bias leading to a distorted association in studies on recurrent stroke.19,20,53 This study found no evidence of heterogeneous associations between covariates and the metaGRS; we did not find any evidence of a solid collider bias of known variables. By applying IPW, we confirmed that our results support the association between metaGRS and recurrent AIS.
There are three putative reasons our metaGRS could predict AIS recurrence. First, the metaGRS algorithm combines the genetic profiles of related traits and slightly improves the performance, reaching the level of significance. Second, the performances of PRS-CS (incremental R2=0.00441) and LDpred2 (incremental R2=0.00443) in our validation analysis were better than those of other traditional PRS methods, such as P+T (incremental R2=0.0038). This demonstrated the importance of using shrinkage estimation methods that consider LD to predict AIS and AIS recurrence. Third, we restricted to use only single matched ancestry throughout.
Nevertheless, our study had several limitations. First, the sample size for recurrent AIS needs to be increased (n=174 at testing), even in the largest hospital-based biobank in Japan. Compared to our metaGRS, iPGS constructed in GIGASTROKE showed a stronger association for AIS and weaker association for AIS recurrence. Although potential discrepancies exist, both PRSs (metaGRS and iPGS) exhibit the same direction of effects and have overlapping confidence intervals (Table 3, Figure S3). Second, despite using as many covariates (age, sex, the first 10 PCs, and seven risk factors (hypertension, hyperlipidemia, diabetes mellitus, smoking, vascular disease, congestive heart failure, and atrial fibrillation)) as possible based on a previous study,12 other confounders might have affected our results. Finally, there may have been an index event bias that was not fully detected by each method that we implemented; however this risk was minimized using multiple approaches. Further studies using different sample sets (including other ancestry groups) are warranted to confirm the prediction of recurrent stroke using the PRS.
In conclusion, our study indicated that PRS can be applied to predict AIS recurrence in addition to traditional clinical risk factors. This shows the potential utility of PRS in population-based screening and in the clinical setting. Overall, our results indicate that stratifying high-risk groups for recurrent stroke among those who have experienced a stroke could be medically beneficial and help in developing personalized strategies for recurrence prevention. Our results suggest that it might be particularly useful in patients with AIS without hypertension, although this requires confirmation in independent datasets.
Supplementary Material
Acknowledgments
We want to acknowledge all the participants and investigators of BioBank Japan. Supercomputing resources were provided by the Human Genome Center, Institute of Medical Science, and the University of Tokyo (http://sc.hgc.jp/shirokane.html). The ToMMo summary statistics were derived from jMorp (https://jmorp.megabank.tohoku.ac.jp/gwas-studies/TGA000007). The MEGASTROKE project received funding from sources specified at https://www.megastroke.org/acknowledgements.html
Source of Funding
This research was supported by the Ministry of Education, Culture, Sports, Sciences and Technology (MEXT) of the Japanese government and the Japan Agency for Medical Research and Development (AMED) under grant nos. JP18km0605001/JP23tm0624002 (the BioBank Japan project), JP223fa627011 (Y.K.), and JP23tm0524003 (Y.K.).
Non-standard abbreviations and acronyms
- PRS
polygenic risk score
- P+T
pruning and thresholding
- BBJ
BioBank Japan
- BBJ1
BBJ 1st cohort
- BBJ2
BBJ 2nd cohort
- ToMMo
Tohoku Medical Megabank
- AIS
all ischemic stroke
- IPW
inverse probability weighting
- LD
linkage disequilibrium
- GWAS
genome-wide association study
- PC
principal component
- IPW
inverse probability weight
- AUC
area under the curve
- LAS
large artery stroke
- SVS
small vessel stroke
- CES
cardioembolic stroke
- TIS
transient ischemic attack
- HWE
Hardy-Weinberg equilibrium
- WGS
whole genome sequencing
- MI
myocardial infarction
- SAP
stable angina pectoris
- AP
unstable angina pectoris
- AF
atrial fibrillation
- DM
diabetes mellitus
- SM
smoking
- BMI
body mass index
- HE
height
- SBP
systolic blood pressure
- DBP
diastolic blood pressure
- TC
total cholesterol
- TG
triglyceride
- HDL
high-density lipoprotein
- LDL
low-density lipoprotein
Footnotes
Disclosures
Y.K. holds stock of StaGen Co, Ltd.
Data availability
The weights of metaGRS derived in this study will be publicly available after acceptance. Genotype datasets were deposited in the National Bioscience Database Center Human Database (BBJ1, Research ID: hum0014; BBJ2, Research ID: hum0311).
References
- 1.Ministry of Health L. and Vital W. Statistics of Japan, https://www.mhlw.go.jp/toukei/saikin/hw/jinkou/kakutei20/index.html. (2020).
- 2.O’Donnell M. J. et al. Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. The Lancet 388, 761–775 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Hata J. et al. Ten year recurrence after first ever stroke in a Japanese community: The Hisayama study. J Neurol Neurosurg Psychiatry 76, 368–372 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takashima N. et al. Long-term survival after stroke in 1.4 million japanese population: Shiga stroke and heart attack registry. J Stroke 22, 336–344 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacArthur J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45, D896–D901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torkamani A., Wineinger N. E. & Topol E. J. The personal and clinical utility of polygenic risk scores. Nat Rev Genet 19, 581–590 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Khera A. v. et al. Whole-Genome Sequencing to Characterize Monogenic and Polygenic Contributions in Patients Hospitalized With Early-Onset Myocardial Infarction. Circulation 139, 1593–1602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konuma T. & Okada Y. Statistical genetics and polygenic risk score for precision medicine. Inflamm Regen 41, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin M. G. & Rader D. J. Polygenic Risk Scores and Coronary Artery Disease. Circulation 637–640 (2020) doi: 10.1161/CIRCULATIONAHA.119.044770. [DOI] [PubMed] [Google Scholar]
- 10.Plagnol V. Polygenic score development in the era of large-scale biobanks. Cell Genomics 2, 100088 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khera A. v. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nature Genetics vol. 50 1219–1224 Preprint at 10.1038/s41588-018-0183-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marston NA et al. Clinical Application of A Novel Genetic Risk Score Predicts Ischemic Stroke in Patients With Cardiometabolic Disease. Circulation. 2021 Feb 2;143(5):470–478. doi: 10.1161/CIRCULATIONAHA.120.051927. Epub 2020 Nov 13. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutten-Jacobs L. C. A. et al. Genetic risk, incident stroke, and the benefits of adhering to a healthy lifestyle: Cohort study of 306 473 UK Biobank participants. BMJ (Online) 363, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abraham G. et al. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat Commun 10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas E. A. et al. Polygenic Risk, Midlife Life’s Simple 7, and Lifetime Risk of Stroke. J Am Heart Assoc 11, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hachiya T. et al. Genetic Predisposition to Ischemic Stroke: A Polygenic Risk Score. Stroke 48, 253–258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hachiya T. et al. Genome-wide polygenic score and the risk of ischemic stroke in a prospective cohort: The Hisayama study. Stroke 759–765 (2020) doi: 10.1161/STROKEAHA.119.027520. [DOI] [PubMed] [Google Scholar]
- 18.Mishra A. et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature (2022) doi: 10.1038/s41586-022-05165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe L. J. et al. Polygenic risk scores for coronary artery disease and subsequent event risk amongst established cases. Hum Mol Genet 29, 1388–1395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahabreh I. J. & Kent D. M. Index Event Bias as an Explanation for the Paradoxes of Recurrence Risk Research. https://jamanetwork.com/. [DOI] [PMC free article] [PubMed]
- 21.Ni G. et al. A Comparison of Ten Polygenic Score Methods for Psychiatric Disorders Applied Across Multiple Cohorts. Biol Psychiatry 90, 611–620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn G. et al. A Smoothed Version of the Lassosum Penalty for Fitting Integrated Risk Models Using Summary Statistics or Individual-Level Data. Genes (Basel) 13, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osterman M. D., Kinzy T. G. & Bailey J. N. C. Polygenic Risk Scores. Curr Protoc 1, (2021). [DOI] [PubMed] [Google Scholar]
- 24.Zhao Z., Song J., Wang T. & Lu Q. Polygenic risk scores: effect estimation and model optimization. Quantitative Biology 0, 0 (2021). [Google Scholar]
- 25.Ma Y. & Zhou X. Genetic prediction of complex traits with polygenic scores: a statistical review. Trends in Genetics 37, 995–1011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pain O. et al. Evaluation of polygenic prediction methodology within a reference-standardized framework. PLoS Genet 17, 1–22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q., Privé F., Vilhjálmsson B. & Speed D. Improved genetic prediction of complex traits from individual-level data or summary statistics. Nat Commun 12, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S. & Zhou X. PGS-server: accuracy, robustness and transferability of polygenic score methods for biobank scale studies. Brief Bioinform 23, 1–19 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Tsuo K., Kanai M., Neale B. M. & Martin A. R. Challenges and opportunities for developing more generalizable polygenic risk scores. 293–320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulm S., Marderstein A., Mezey J. & Elemento O. A systematic framework for assessing the clinical impact of polygenic risk scores. medRxiv preprint doi: 10.1101/2020.04.06.20055574 (2021) doi: 10.1101/2020.04.06.20055574. [DOI] [Google Scholar]
- 31.Nagai A. et al. Overview of the BioBank Japan Project: Study design and profile. J Epidemiol 27, S2–S8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirata M. et al. Cross-sectional analysis of BioBank Japan clinical data: A large cohort of 200,000 patients with 47 common diseases. J Epidemiol 27, S9–S21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirata M. et al. Overview of BioBank Japan follow-up data in 32 diseases. J Epidemiol 27, S22–S28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang C. C. et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marees A. T. et al. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int J Methods Psychiatr Res 27, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi S. W., Mak T. S. H. & O’Reilly P. F. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc 15, 2759–2772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koyama S. et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat Genet 52, 1169–1177 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Privé F., Arbel J. & Vilhjálmsson B. J. LDpred2: Better, faster, stronger. Bioinformatics 36, 5424–5431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Privé F., Arbel J., Aschard H. & Vilhjálmsson B. J. Identifying and correcting for misspecificationsin GWAS summary statistics and polygenic scores. Human Genetics and Genomics Advances 100136 (2022) doi: 10.1016/j.xhgg.2022.100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ge T., Chen C. Y., Ni Y., Feng Y. C. A. & Smoller J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun 10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruan Y. et al. Improving polygenic prediction in ancestrally diverse populations. Nat Genet (2022) doi: 10.1038/s41588-022-01054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malik R. et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 50, 524–537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagelkerke N. J. D. A Note on a General Definition of the Coefficient of Determination. Biometrika 78, 691–692 (1991). [Google Scholar]
- 45.Wang Y. et al. Global Biobank analyses provide lessons for developing polygenic risk scores across diverse cohorts. Cell Genomics 3, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hindy G. et al. Genome-wide polygenic score, clinical risk factors, and long-term trajectories of coronary artery disease. Arterioscler Thromb Vasc Biol 2738–2746 (2020) doi: 10.1161/ATVBAHA.120.314856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell R. E. et al. Strategies to investigate and mitigate collider bias in genetic and Mendelian randomisation studies of disease progression. PLoS Genetics vol. 19 Preprint at 10.1371/journal.pgen.1010596 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sep S. J., Van Kuijk S. M. & Smits L. J. Index event bias: Problems with eliminating the paradox. Journal of Stroke and Cerebrovascular Diseases vol. 23 2464 Preprint at 10.1016/j.jstrokecerebrovasdis.2014.06.025 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Levine D. A. et al. Smoking and mortality in stroke survivors: Can we eliminate the paradox? Journal of Stroke and Cerebrovascular Diseases 23, 1282–1290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altman D. G. & Bland J. M. Interaction revisited: The difference between two estimates. BMJ 326, 219 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishigaki K. et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet 52, 669–679 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bulik-Sullivan B. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho S. M. J. et al. Genetic, sociodemographic, lifestyle, and clinical risk factors of recurrent coronary artery disease events: a population-based cohort study. Eur Heart J (2023) doi: 10.1093/eurheartj/ehad380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weale E. M. Quality control for genome-wide association studies. Methods Mol Biol (2010) doi: 10.1007/978-1-60327-367-1_19. [DOI] [PubMed] [Google Scholar]
- 55.Akiyama M. et al. Characterizing rare and low-frequency height-associated variants in the Japanese population. Nat Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Privé F., Vilhjálmsson B. J., Aschard H. & Blum M. G. B. Making the Most of Clumping and Thresholding for Polygenic Scores. Am J Hum Genet 105, 1213–1221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chagnon M., O’Loughlin J., Engert J. C., Karp I. & Sylvestre M. P. Missing single nucleotide polymorphisms in Genetic Risk Scores: A simulation study. PLoS One 13, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldstein B. A., Yang L., Salfati E. & Assimes T. L. Contemporary Considerations for Constructing a Genetic Risk Score: An Empirical Approach. Genet Epidemiol 39, 439–445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y. et al. Global Biobank analyses provide lessons for developing polygenic risk scores across diverse cohorts. Cell Genomics 3, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tadaka S. et al. jMorp updates in 2020: Large enhancement of multi-omics data resources on the general Japanese population. Nucleic Acids Res 49, D536–D544 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Restuadi R. et al. Polygenic risk score analysis for amyotrophic lateral sclerosis leveraging cognitive performance, educational attainment and schizophrenia. European Journal of Human Genetics (2021) doi: 10.1038/s41431-021-00885-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turley P. et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet 50, 229–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maier R. M. et al. Improving genetic prediction by leveraging genetic correlations among human diseases and traits. Nat Commun 9, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho W. K. et al. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nat Commun 11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amariuta T. et al. Improving the trans-ancestry portability of polygenic risk scores by prioritizing variants in predicted cell-type-specific regulatory elements. Nat Genet 52, 1346–1354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The weights of metaGRS derived in this study will be publicly available after acceptance. Genotype datasets were deposited in the National Bioscience Database Center Human Database (BBJ1, Research ID: hum0014; BBJ2, Research ID: hum0311).