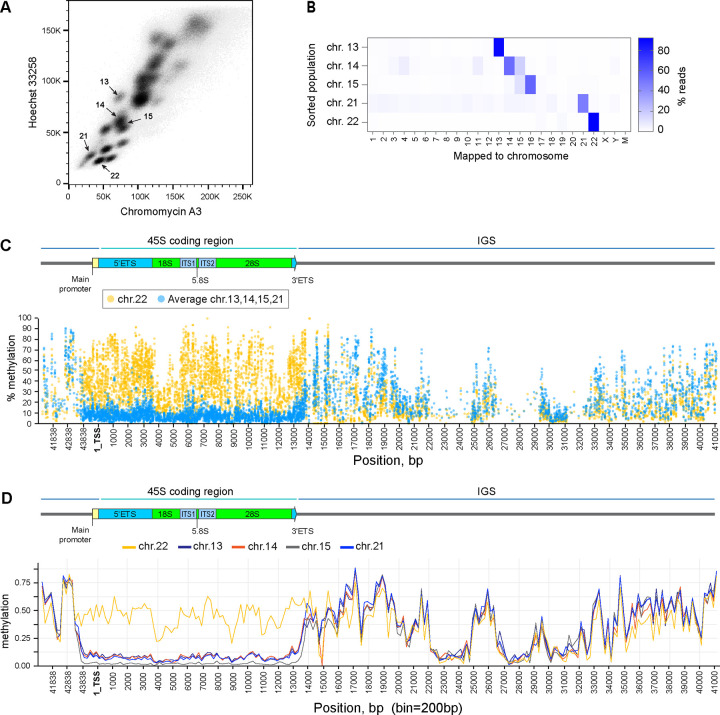
Figure 3. Epigenetic silencing of rDNA by methylation in CHM13 cell line.
A. Acrocentric chromosome sorting by flow cytometry. Chromosomes were isolated from CHM13 cell line as detailed in the Materials and Methods and labeled with Chromomycin A3 and Hoechst 33258. Arrows indicate sorted populations.
B. Illumina short read mapping from flow-sorted chromosomes. The purity of sorted populations was confirmed by the highest fraction of reads mapped to expected chromosomes, indicated by the color scale.
C. Percent methylation of each cytosine base in reads mapped to the rDNA reference sequence across the rRNA gene determined by short-read methyl-sequencing analysis. The promoter and the coding region of transcriptionally inactive chromosome 22 rDNA (yellow circles) are highly methylated compared to averages of all other acrocentric chromosomes with transcriptionally active rDNA (blue circles). Individual plots for each chromosome are shown in Supplementary Figure 5 A–D.
D. Methylation calls from ONT long-read sequencing of rDNA reads mapped to specific acrocentric chromosomes. Reads mapped to chromosome 22 (yellow line) are highly methylated in the promoter and coding region compared to reads mapped to other acrocentric chromosomes. Bin size 200bp.