Abstract
A direct comparison demonstrates that Rous sarcoma virus is capable of infecting aphidicolin-arrested cells 10-fold more efficiently than murine leukemia virus but less efficiently than human immunodeficiency virus. The efficiency of infection of nondividing cells by the three viruses correlates with the respective ability of each viral DNA to enter the nucleus.
Following entry into the cell cytoplasm, the retroviral genome must gain access to host cell chromosomes within the nucleus to achieve stable integration. During this process the retroviral DNA is present in a large complex with a subset of retroviral proteins known as the preintegration complex (PIC) (2). The physical size of the PIC is thought to exceed the upper limit for passive diffusion through nuclear pores. In nondividing cells, viral entry, uncoating, DNA synthesis, and formation of the murine leukemia virus (MLV) PIC occur at the same rate as in dividing cells, but integration fails to occur (20, 24). During mitosis, however, the nuclear membrane disassembles, rendering the chromosomes accessible to the MLV PIC (24). These findings could explain the conclusion that infection by oncoretroviruses such as MLV and Rous sarcoma virus (RSV) requires cell division (14, 15, 20, 24, 28–30).
The requirement for mitosis during infection is not common to all retroviruses. Indeed, lentiviruses are able to infect certain types of nondividing cells (19, 20). This property is thought to be due to the ability of the lentiviral PIC to be actively transported across the nuclear membrane (6). Although the precise mechanism by which nuclear entry is achieved remains elusive, nuclear localization signals (NLS) have been identified on both the Vpr and matrix proteins of human immunodeficiency virus type 1 (HIV-1) and were initially thought to direct nuclear localization of the PIC (5, 11). However, mutant viruses lacking both Vpr and the matrix NLS were shown to be capable of replicating in nondividing cells (9). Moreover, both Vpr and the matrix protein can be completely removed without affecting infection of nondividing cells (23). Recently, the HIV-1 integrase protein has been shown to be nucleophilic, and this property has been proposed to direct the PIC to the nucleus (10). The identification of an NLS in the integrase of RSV (17, 18) prompted us to reexamine the ability of this virus to infect nondividing cells in direct comparison with HIV and MLV.
To compare the cell cycle dependence of HIV, MLV, and RSV, target cell growth was arrested using aphidicolin. Aphidicolin is a reversible inhibitor of eukaryotic nuclear DNA synthesis, and treatment of cells with this inhibitor arrests the cell cycle in the G1/S phase. Various aphidicolin concentrations and target cell lines were tested, and the human fibrosarcoma cell line HT1080 was chosen for detailed study because the cell cycle could be effectively arrested by aphidicolin without overt cytotoxicity. To efficiently infect this human cell line, and to eliminate any potential artifacts arising from the use of different cell surface receptors, all viruses were pseudotyped with the vesicular stomatitis virus G envelope glycoprotein. In addition, the infectivity of each virus was measured using the same methodology and with vectors capable of only a single round of infection.
HIV-1 and MLV viral stocks were generated by transient transfection of 293T cells. For HIV, the Gag-Pol expression plasmid pΔ8.2 and the vector plasmid pHRlacZ (22) were cotransfected with the vesicular stomatitis virus G envelope glycoprotein expression plasmid pHCMV.G (7). For MLV, the Gag-Pol expression plasmid pHCMV.GagPol (a gift from F.-L. Cosset) was cotransfected with the vector plasmid pLMN1 (a gift from P. D. Bieniasz) and pHCMV.G. RSV vector stocks were produced in DF1 chicken cells transiently transfected with the full-length RSV viral plasmid modified to also express the lacZ gene (kindly provided by Y. Li) (see reference 8) and pHCMV.G. In each case, virus-containing supernatants were harvested 24, 48, and 72 h posttransfection. The supernatants were filtered through a 0.2-μm-pore-size filter, and the virions were purified by ultracentrifugation through a 25% sucrose cushion, resuspended in phosphate-buffered saline, and stored at −70°C until use.
Target HT1080 cells were seeded at 5 × 104 cells/well (24-well plate) and treated with aphidicolin (2 μg/ml) for 24 h prior to infection. The cells were inoculated with serial dilutions of the viral stocks and supplemented with DEAE dextran (10 μg/ml) and aphidicolin (2 μg/ml). After 4 h, the cells were washed three times with phosphate-buffered saline and incubated in the presence or absence of aphidicolin for a further 48 h. Viral titers were determined by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining.
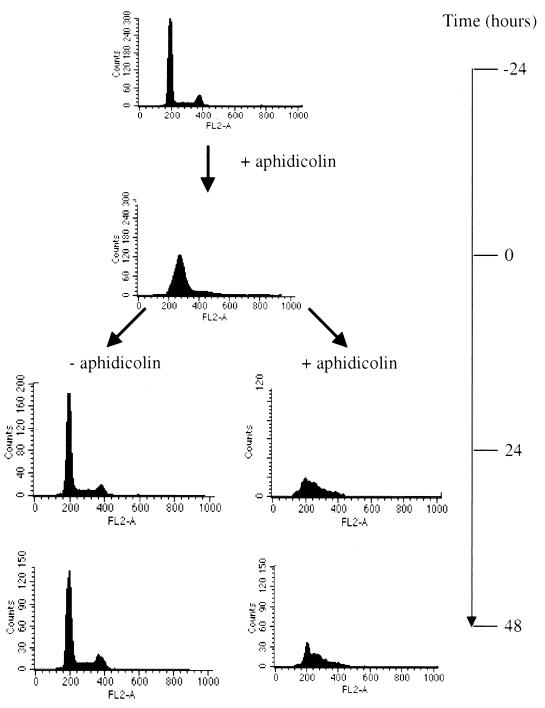
To confirm that aphidicolin blocked target cell division, cells were harvested at several time points during treatment, fixed with cold 80% ethanol, and treated with DNase-free RNase and propidium iodide (Boehringer). The DNA content was subsequently analyzed by a fluorescence-activated cell sorter. As can be seen in Fig. 1, cells treated with aphidicolin for 24 h accumulated in the G1/S phase, demonstrating that the cells were arrested at the time of infection. After the viral supernatant had been removed and the cells had been washed, cells that were maintained in aphidicolin remained arrested in G1/S, whereas cells from which aphidicolin was removed reverted to a normal proliferating profile.
FIG. 1.
Cell cycle analysis of HT1080 cells treated with aphidicolin. Cells were treated with aphidicolin for 24 h, washed, and maintained in the absence (−) or presence (+) of aphidicolin. The histograms show the results of fluorescence-activated cell sorter analyses after the cells had been stained with propidium iodide at the indicated times.
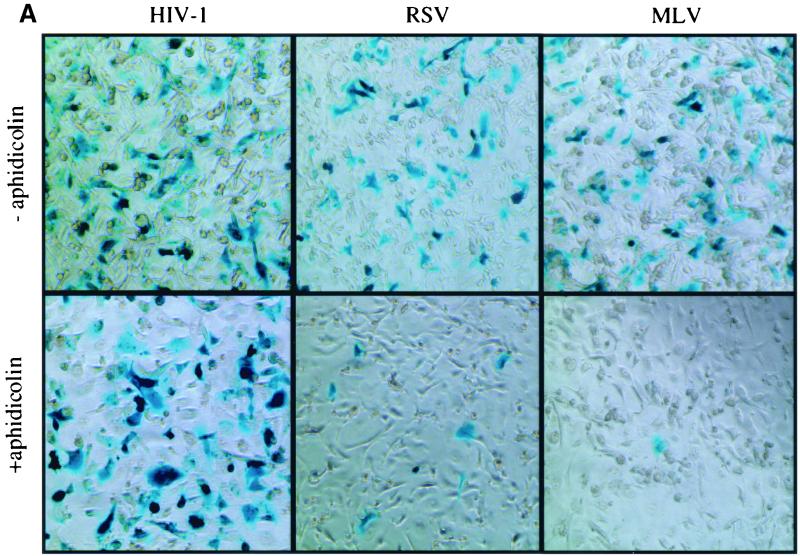
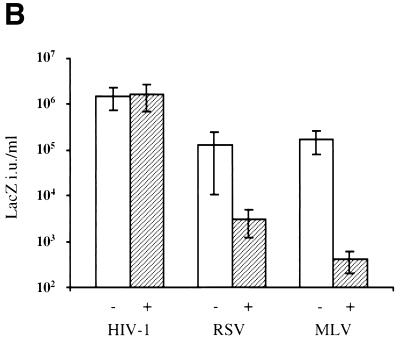
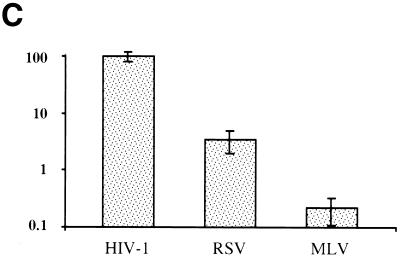
X-Gal staining of HT1080 cells infected with the different viral vectors revealed that the presence of aphidicolin did not affect HIV-1 titers but reduced RSV and, to a greater extent, MLV titers compared to titers in cells released from aphidicolin treatment (Fig. 2A). Quantitative analysis of data obtained from four independent experiments confirmed that the presence or absence of aphidicolin had no measurable effect on HIV-1 infectivity (Fig. 2B). This finding demonstrated that the target cells remained viable and fully competent to support lentiviral infection. In contrast, infectious titers of MLV measured in G1/S-arrested cells were reduced 500-fold more than those in dividing cells (Fig. 2B). These results are similar to those previously reported and further show that target cell proliferation was effectively blocked. Simultaneous determination of RSV vector titers revealed that RSV infectivity was also reduced by target cell growth arrest but less dramatically, 50-fold, than that of MLV (Fig. 2B). Compared to dividing cells, the titer in arrested cells was preserved at 100% for HIV, reduced to 3% for RSV, and reduced to 0.2% for MLV (Fig. 2C). These results demonstrate that RSV is capable of infecting nondividing cells less efficiently than HIV but 10-fold more efficiently than MLV. This phenotypic difference between MLV and RSV was not dependent on the cells in which the virus was produced. Specifically, MLV vectors produced in chicken DF1 cells exhibited a sensitivity to aphidicolin identical to that of MLV vectors produced in human 293T cells (data not shown).
FIG. 2.
Infectivity of HIV, RSV, and MLV on dividing and nondividing cells. Infections were carried out in aphidicolin-arrested cells that were released from or maintained in aphidicolin after infection. (A) Representative experiment of HT1080 cells infected with equivalent titers of HIV, RSV, or MLV and stained by X-Gal at 48 h postinfection. (B) Viral titers are expressed as LacZ infectious units per milliliter on HT1080 cells maintained in the absence (−) or presence (+) of aphidicolin. (C) Viral titers on nondividing cells are expressed as a percentage of titers obtained on dividing cells.
To determine which step during the early events of infection differed between the three viruses, the synthesis of viral DNA by each virus was analyzed using PCR. To ensure that the template amplified during the PCR had arisen from viral DNA synthesis during infection and was not contaminating DNA carried over from the transfection during virus preparation, the following steps were taken. Prior to infection, viral supernatants were treated with RNase-free DNase I (1 U/ml; Boehringer) in the presence of MgCl2 (10 mM) for 1 h at 37°C. In addition, as a negative control for de novo viral DNA synthesis, viruses were heat inactivated by incubation at 65°C for 30 min. HT1080 cells were seeded at 2 × 105 cells/plate (60-mm diameter) and treated with aphidicolin (2 μg/ml) for 24 h prior to infection. Cells were then infected at a multiplicity of infection of 1 for 4 h in the presence of aphidicolin, washed, and maintained or released from aphidicolin treatment for a further 24 h. The cells were then lysed and low-molecular-weight DNA was extracted as previously described (12).
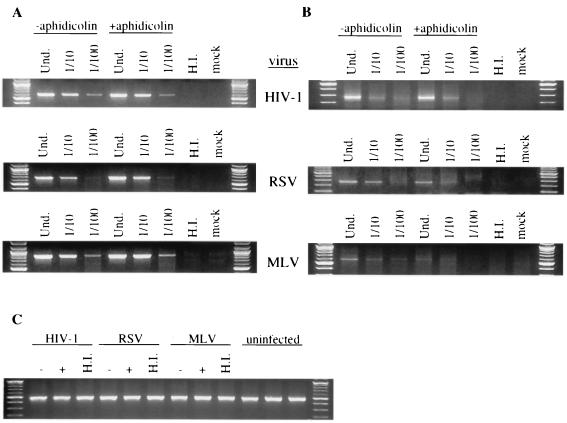
The presence of aphidicolin had no effect on the synthesis of viral DNA. Specific primers that hybridize to the R/U5 and Gag regions of each virus were used to detect elongated plus-strand DNA, a late product of reverse transcription. The primers used for HIV have been described elsewhere (3). The following primers were used to detect viral DNA: 5′-GCCATTTGACCATTCACCACATTGG-3′ (forward primer) and 5′-CTAATCCCAACCAAAACTTTGCTTG-3′ (reverse primer) for RSV and 5′-GCGCCAGTCCTCCGATTGACT-3′ (forward primer) and 5′-GACCTTGATCTTAACC TGGG-3′ (reverse primer) for MLV. The standard program for amplification was 30 to 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. The number of amplification cycles was chosen so that the assay was below saturation; i.e., the amount of product was related to the amount of DNA used. Similar levels of each viral DNA product were detected in both arrested and nonarrested cells for all viruses (Fig. 3A). No viral DNA products were detected in uninfected cells or in cells incubated with the heat-inactivated viruses (Fig. 3A). To verify that equivalent amounts of total cellular DNA were present in each sample, human mitochondrial DNA was amplified under the conditions described above using forward primer 5′-GAATGTCTGCACAGCCACTTTCCAC-3′ and reverse primer 5′-GATCGTGGTGATTTAGAGGGTGAAC-3′ (Fig. 3C). These data demonstrate that HIV, RSV, and MLV entry, uncoating, and reverse transcription proceed in nondividing cells as efficiently as in dividing cells.
FIG. 3.
PCR analysis of Hirt extracts from infected and uninfected cells. Aphidicolin-arrested HT1080 cells were infected with the indicated virus and released from or maintained in aphidicolin after infection. Hirt DNA was harvested 24 h postinfection and subjected to PCR analysis (see the text for details). (A) PCR amplification of elongated plus-strand viral DNA. Undiluted (Und.) Hirt DNA or Hirt DNA diluted 10-fold (1/10) or 100-fold (1/100) was used as a template for the PCR. (B) The same sample dilutions were used for PCR amplification of 2-LTR circular DNA. (C) PCR amplification of mitochondrial DNA from undiluted Hirt extracts from uninfected or infected cells that were maintained in the absence (−) or presence (+) of aphidicolin. H.I., undiluted Hirt DNA from cells incubated with heat-inactivated virus; mock, undiluted Hirt DNA from uninfected cells.
Once reverse transcription is completed and the linear viral DNA enters the nucleus, circular DNA forms containing one or two copies of the long terminal repeat (LTR) are generated, probably by the action of nuclear host enzymes (2, 4). Although these DNA forms are not involved in productive viral integration, they can be used as a hallmark of viral DNA entry into the nucleus. The levels of the 2-LTR circular DNA in infected cells were determined using PCR primers designed to amplify the LTR-LTR junction of each virus. The primers and conditions used for each virus are listed below. For HIV, forward primer 5′-GGTACTAGCTTGAAGCACCATCC-3′ and reverse primer 5′-GCCTCAATAAGCTTGCCTTGAGTG-3′ were used for 45 cycles of 94°C for 1 min, 64.5°C for 1 min, and 72°C for 1 min. For RSV, forward primer 5′-GGACCGTTGATTCCCTGAC-3′ and reverse primer 5′-CACTTAAATACAATATCT C-3′ were used for 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min. For MLV, primers MR5784 and MR4091 have been previously described (27) and were used for 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min.
2-LTR circles were readily detectable in both dividing and nondividing cells infected with HIV (Fig. 3B). In contrast, circular MLV DNA was detected in dividing cells but was barely detectable in the aphidicolin-treated cells (Fig. 3B). The detection of MLV 2-LTR circles was difficult even in dividing cells, and infection at a multiplicity of infection of 100 was necessary to obtain the results shown. RSV-infected cells showed an intermediate phenotype in that the levels of circular DNA were detectable in both dividing and arrested cells but were clearly present at reduced levels in the latter (Fig. 3B). Although the relationship between DNA input and the quantity of amplified product is not linear, it can be estimated that RSV and MLV 2-LTR circles were reduced approximately 10- and 100-fold, respectively, as a result of target cell arrest. These results correlate with the infectivity data (Fig. 2) and suggest that the ability of each virus to infect nondividing cells is a consequence of the ability of the viral DNA to enter the nondividing target cell nucleus.
These data clearly demonstrate that RSV can infect aphidicolin-arrested cells with moderate efficiency. It has been previously reported that aphidicolin treatment blocked the formation of 2-LTR circles in quail embryonic fibroblasts (13). However, in that study, 2-LTR circle formation was detected using Southern blot analysis that is not as sensitive as the PCR used in this study, which could explain the discrepancy in the results. It is unlikely that the difference in results arises from the use of different target cell lines since we have obtained infectivity results similar to those presented here in a variety of different cell lines, including chicken fibroblasts (DF1) (data not shown). Additionally, these findings were not restricted to cells arrested at the G1/S border, because analogous results were obtained using mitomycin C, a drug that arrests cells in the G2 phase of the cell cycle. Although the arrest induced by mitomycin C treatment in HT1080 cells was not as potent as that induced by aphidicolin, preliminary data showed that virus titers in treated cells were reduced to 1% (MLV), 5% (RSV), and 67% (HIV) of the titers obtained in untreated cells.
Although the efficiency of RSV infection in arrested cells is lower than that of HIV-1 infection, it is significantly higher than that of MLV infection. The ability of RSV to generate 2-LTR circles in arrested cells suggests that the differences in infectivity reflect the different abilities of these viruses to enter the nucleus of nondividing cells. The mechanism used by lentiviruses to gain access to the nucleus of nondividing cells has not been definitely elucidated; however, the identification of several NLS in different HIV-1 proteins raises the possibility that these signals mediate nuclear entry. While no NLS have been identified in MLV, it has been shown that the RSV integrase contains a potent NLS (17, 18). It is reasonable to speculate that the presence of this NLS is responsible for the ability of RSV to infect nondividing cells more efficiently than MLV. The difference in efficiencies of arrested-cell infection between RSV and HIV-1 could be due to the fact that multiple NLS have been found in HIV-1 PIC components compared to only one (thus far) in RSV. Potentially, the HIV-1 NLS could act cooperatively to result in more efficient nuclear entry. Alternatively, it is possible that the RSV NLS is partly occluded in the context of a PIC. Ultimately, further blocks to RSV replication, following nuclear entry of the viral DNA, could exist.
Other retroviruses may ultimately prove to have the ability to infect nondividing cells with intermediate efficiency, as shown here for RSV. It has been demonstrated that foamy virus Gag and Pol proteins, including the integrase, contain NLS (16), and the viral DNA was detected in the nucleus of cells arrested in G1/S (26). However, the ability of foamy viruses to infect nondividing cells remains controversial. Certain studies have demonstrated that foamy viruses cannot infect aphidicolin-arrested cells (1, 26), while others suggest that they can infect nondividing cells to some extent (21, 25). It is likely that the nucleophilic properties of these viruses allow infection of nondividing cells, but the efficiency remains significantly lower than that of lentiviruses.
Finally, the ability of lentiviruses to infect nondividing cells has been exploited in the development of vectors for human gene therapy, but their application is limited by safety concerns. Since RSV can infect nondividing cells with low efficiency, future efforts could focus on improving this ability, in order to generate vectors useful for gene therapy strategies in which the infection of nondividing cells is required.
Acknowledgments
We thank F.-L. Cosset, Y. Li, G. Gao, D. Lim, and D. Braaten for the gifts of plasmids and primers. We are also grateful to P. D. Bieniasz for critical discussions and for the gift of pLMN1 plasmid.
T.H. is an associate and S.P.G. is an investigator of the HHMI.
REFERENCES
- 1.Bieniasz P D, Weiss R A, McClure M O. Cell cycle dependence of foamy retrovirus infection. J Virol. 1995;69:7295–7299. doi: 10.1128/jvi.69.11.7295-7299.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowerman B, Brown P O, Bishop J M, Varmus H E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 3.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukrinsky M, Sharova N, Stevenson M. Human immunodeficiency virus type 1 2-LTR circles reside in a nucleoprotein complex which is different from the preintegration complex. J Virol. 1993;67:6863–6865. doi: 10.1128/jvi.67.11.6863-6865.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinsky M I, Sharova N, Dempsey M P, Stanwick T L, Bukrinskaya A G, Haggerty S, Stevenson M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Federspiel M J, Hughes S H. Retroviral gene delivery. Methods Cell Biol. 1997;52:179–214. [PubMed] [Google Scholar]
- 9.Freed E O, Englund G, Martin M A. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 13.Hsu T W, Taylor J M. Effect of aphidicolin on avian sarcoma virus replication. J Virol. 1982;44:493–498. doi: 10.1128/jvi.44.2.493-498.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphries E H, Temin H M. Cell cycle-dependent activation of Rous sarcoma virus-infected stationary chicken cells: avian leukosis virus group-specific antigens and ribonucleic acid. J Virol. 1972;10:82–87. doi: 10.1128/jvi.10.1.82-87.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphries E H, Temin H M. Requirement for cell division for initiation of transcription of Rous sarcoma virus RNA. J Virol. 1974;14:531–546. doi: 10.1128/jvi.14.3.531-546.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imrich H, Heinkelein M, Herchenroder O, Rethwilm A. Primate foamy virus Pol proteins are imported into the nucleus. J Gen Virol. 2000;81:2941–2947. doi: 10.1099/0022-1317-81-12-2941. [DOI] [PubMed] [Google Scholar]
- 17.Kukolj G, Jones K S, Skalka A M. Subcellular localization of avian sarcoma virus and human immunodeficiency virus type 1 integrases. J Virol. 1997;71:843–847. doi: 10.1128/jvi.71.1.843-847.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kukolj G, Katz R A, Skalka A M. Characterization of the nuclear localization signal in the avian sarcoma virus integrase. Gene. 1998;223:157–163. doi: 10.1016/s0378-1119(98)00169-3. [DOI] [PubMed] [Google Scholar]
- 19.Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis P F, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mergia A, Chari S, Kolson D L, Goodenow M M, Ciccarone T. The efficiency of simian foamy virus vector type-1 (SFV-1) in nondividing cells and in human PBLs. Virology. 2001;280:243–252. doi: 10.1006/viro.2000.0773. [DOI] [PubMed] [Google Scholar]
- 22.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 23.Reil H, Bukovsky A A, Gelderblom H R, Gottlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell D W, Miller A D. Foamy virus vectors. J Virol. 1996;70:217–222. doi: 10.1128/jvi.70.1.217-222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saïb A, Puvion-Dutilleul F, Schmid M, Périès J, de Thé H. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J Virol. 1997;71:1155–1161. doi: 10.1128/jvi.71.2.1155-1161.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith C M, Potts W B, Smith J S, Roth M J. RNase H cleavage of tRNAPro mediated by M-MuLV and HIV-1 reverse transcriptases. Virology. 1997;229:437–446. doi: 10.1006/viro.1997.8454. [DOI] [PubMed] [Google Scholar]
- 28.Temin H M. Studies on carcinogenesis by avian sarcoma viruses: requirement for new DNA synthesis and for cell division. J Cell Physiol. 1967;69:53–63. doi: 10.1002/jcp.1040690314. [DOI] [PubMed] [Google Scholar]
- 29.Varmus H, Swanstrom R. Replication of retroviruses. In: Weiss R A, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. pp. 467–512. [Google Scholar]
- 30.Weiss R A. Studies on the loss of growth inhibition in cells infected with Rous sarcoma virus. Int J Cancer. 1970;6:333–345. doi: 10.1002/ijc.2910060303. [DOI] [PubMed] [Google Scholar]