Abstract
Placozoa are millimeter-sized, flat, irregularly shaped ciliated animals that crawl on surfaces in warm oceans feeding on biofilms, which they digest externally. They stand out from other animals due to their simple body plans. They lack organs, body cavities, muscles and a nervous system and have only seven broadly defined morphological cell types, each with a unique distribution. Analyses of single cell transcriptomes of four species of placozoans revealed greater diversity of secretory cell types than evident from morphological studies, but the locations of many of these new cell types were unknown and it was unclear which morphological cell types they represent. Furthermore, there were contradictions between the conclusions of previous studies and the single cell RNAseq studies. To address these issues, we used mRNA probes for genes encoding secretory products expressed in different metacells in Trichoplax adhaerens to localize cells in whole mounts and in dissociated cell cultures, where their morphological features could be visualized and identified. The nature and functions of their secretory granules were further investigated with electron microscopic techniques and by imaging secretion in live animals during feeding episodes. We found that two cell types participate in disintegrating prey, one resembling a lytic cell type in mammals and another combining features of zymogen gland cells and enterocytes. We identified secretory epithelial cells expressing glycoproteins or short peptides implicated in defense. We located seven peptidergic cell types and two types of mucocytes. Our findings reveal mechanisms that placozoans use to feed and protect themselves from pathogens and clues about neuropeptidergic signaling. We compare placozoan secretory cell types with cell types in other animal phyla to gain insight about general evolutionary trends in cell type diversification, as well as pathways leading to the emergence of synapomorphies.
Introduction
Placozoans are millimeter-sized, flat, irregularly shaped animals that crawl on surfaces in shallow zones of tropical and subtropical oceans [1–3]. They have no nervous system or muscles, and they lack an axis of symmetry. They feed on microalgae and cyanobacteria, which they digest externally in the space between their lower epithelium and the substrate [4]. They are of interest in an evolutionary context due to their phylogenetic position as sister to the clade that includes Cnidaria and Bilateria[5–7] and their simple body plans and lifestyles. Ctenophora and Porifera are thought to be more ancient phyla[8–12]. Several dozen distinct placozoan haplotypes have been identified based on sequencing the mitochondrial large ribosomal subunits of specimens collected in different parts of the globe. Based on analyses of nuclear gene sequences of 26 haplotypes, Placozoa is proposed to include 2 classes, 4 orders, 5 families, 8 genera and 26 species/haplotypes [13]. Placozoans primarily propagate by binary fission or budding [14], processes that generate genetically identical clones. However, genomic analysis of individual animals collected in a field study and identified as belonging to the same haplotype (H8) provided evidence that sexual reproduction occurs among members of this species [15].
Although placozoans are genetically diverse, they look alike when observed by light microscopy except for variations in size and shape. Electron microscopic studies of representatives of two proposed orders, Trichoplax adhaerens (H1, H2, order Trichoplacea), and Hoilungia hongkongensis (H13, order Hoilungea) showed that they have morphologically identical cell types and body plans [16–20]. Functions of some of the cell types have been inferred based on light and electron microscopic studies. The most prevalent cells in the lower epithelium are columnar ventral epithelial cells (VEC), which have an apical cilium surrounded by a collar of microvilli. The cilia are motile and adhere to the substrate during their effective strokes, thereby allowing the animal to crawl on the substrate and to change shape [4,21,22]. Interspersed among the ciliated VEC are lipophil cells (LC), so called because they contain multiple large lipophilic secretory granules. When T. adhaerens pause to feed on algae, LC secrete a large apical granule whose contents lyses nearby algae [4]. The lower epithelium also contains mucocytes that secrete mucus [17], which the animal requires to crawl on the substrate. The upper epithelium is composed primarily of monociliated cells with broad apical endings that comprise the upper the surface. Their cell bodies are narrower and protrude below. Both dorsal and ventral epithelia contain several morphologically distinct types of gland cells that possess a cilium and may be sensory [17,22,23]. The epithelial cells are interconnected by adherens junctions [24]; occluding junctions and gap junctions are absent and there is no basal lamina. The space between the upper and lower epithelial layers is occupied by a layer of fiber cells, which have long branching processes that contact each other as well as the other cell types. Fiber cells phagocytose bacteria and cellular debris and engage in wound healing [25], functions associated with macrophage-like cells in other animals. Inside the rim of the animal, where dorsal and ventral epithelia meet, are regularly spaced crystal cells that appear to be functional statocysts [26].
Bioinformatic analyses of T. adhaerens genomes, transcriptomes and proteomes [5,6,27–30]reveal a rich repertoire of secretory proteins including digestive enzymes, proteins implicated in innate immunity, and prepropeptide precursors of peptides, some of which elicit changes in the behavior of the animals when added to the ambient seawater [22,23] as well as multiple secretory proteins with no known functional domains. Single cell RNA sequencing (RNA-Seq) analyses of T. adhaerens and three other species of Placozoa [6,31] revealed that many of these secretory products are differentially expressed. Cells expressing several digestive enzymes have been localized by fluorescence in situ hybridization (FISH) [6,32]and some peptidergic cell types have been localized by immunolabeling [17,22,23]. However, the cells that secrete many secretory products remained unidentified.
In the present study, we used FISH probes for genes encoding secretory products that are specifically expressed in T. adhaerens metacells classified as lipophil, gland, or epithelial and a subset of the peptidergic metacells to localize the cells in whole mounts and in dissociated cell cultures, where the shapes and morphological features of the cells could be visualized by differential-interference contrast (DIC) microscopy and by staining them with fluorescent dyes or immunolabels. The compositions of the secretory granules in LC, VEC and dorsal epithelial cells were further investigated with electron microscopic techniques. We used fluorescent dyes and light microscopy to visualize secretory processes in live animals feeding on algae.
Our findings yield a more detailed picture of the placozoan body plan than was apparent from previous morphological studies and provide insight into the mechanisms that the animals use to feed and maintain homeostasis. Comparison of the secretomes of metacells representing different morphological cell types in TaH1 with corresponding metacells in TaH2 and Hoilungia H13 and H23 revealed numerous similarities and several intriguing differences. Some of the secretory products expressed in different cell types in placozoans are expressed in morphologically similar cell types in Cnidaria and/or Bilateria and likely have analogous functions. One placozoan cell type (VEC) combines features that are expressed in distinct cell types in Cnidaria and many Bilateria. These shared cell types may be descendants of cell types that were present in the common ancestor of Placozoa, Cnidaria and Bilateria (Planuloza) and some of them may have been present in the common ancestor of all animals.
Results
Main secretory cell types in the ventral epithelium
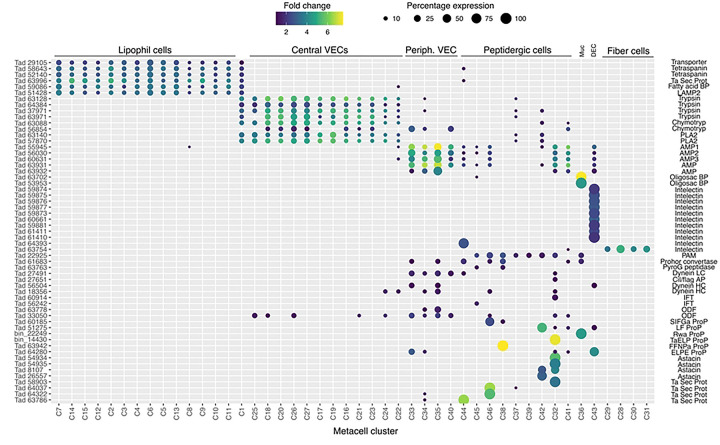
Trichoplax adhaerens H1 lipophil cell (LC) metacell clusters highly express multiple genes that are not expressed or expressed at much lower levels in other metacells (Fig1, S3 Fig) identified by single cell RNA-Seq studies [6,31] We obtained fluorescence in situ hybridization (FISH) probes for three of them: Ta 58643 (Ta Tetraspanin); Ta 29105 (Ta GABA transporter); Ta 63996 (TaH1 secretory protein). We used probes for TaH1orthologs of the precursors of trypsin, chymotrypsin and secretory phospholipase A2 (sPLA2) to label the cell types classified as “digestive gland cells” in the RNA-Seq studies.
Fig 1. Expression of selected genes across T. adhaerens metacells (columns) identified by single cell RNA sequencing.
Data are from [31]. Cell types are identified based on data from the present study and [6,17,25,31]. Dot color represents fold change of gene expression and dot size represents percentage of the total UMI in the given metacell.
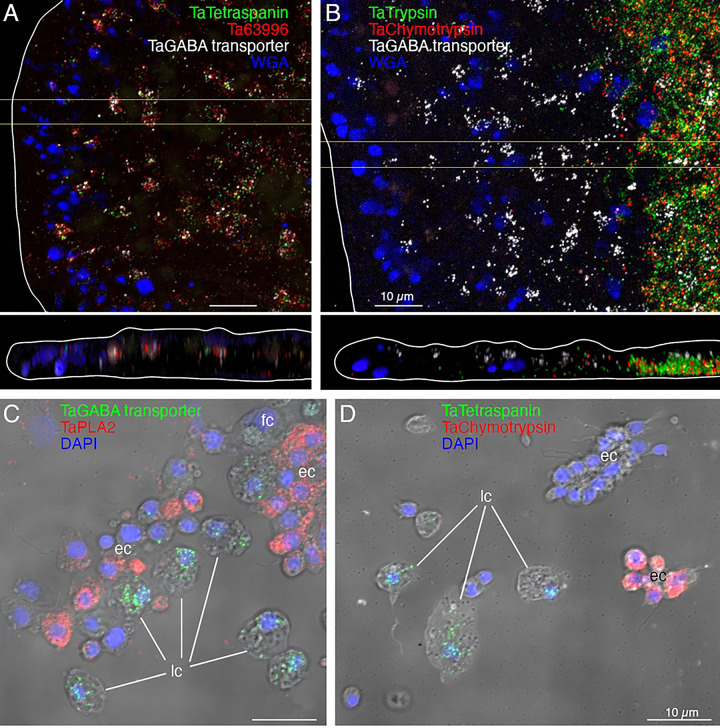
Cells co-labelled with probes for Ta tetraspanin, Ta GABA transporter and Ta 63996 were present throughout the central region of the ventral epithelium but absent in a zone <10 μm from the rim (Fig2A). Cells labeled with probes for digestive enzymes (Ta trypsin, Ta chymotrypsin) were interspersed with LC in the central part of the animal but were absent <60 μm from the rim (Fig 2B). The label in these cells was closer to the ventral surface (Fig 2B, bottom) than the label in LC (Fig 2A, B). Dissociated cells expressing LC specific markers were larger (~8 μm) than cells expressing digestive enzymes (<5 μm; Fig 2C, D) and contained large granules, a hallmark of LC. Unlabeled ventral epithelial cells, identified by their small sizes and cylindrical shapes, also were present (Fig 2D). We previously reported that LC express digestive enzymes based on sequential labeling with vital dyes in live dissociated cultures and FISH probes for precursors of digestive enzymes after fixation of the cells [17]; in this study, we found that the expression level of digestive enzymes by LC was very low, as reported [6].
Fig 2. Fluorescence in situ hybridization (FISH) localization of expression of lipophil and digestive gland cells markers in T. adhaerens H1 wholemounts (A, B) and dissociated cell preparations (C, D).
Images of wholemounts are horizontal (xy) and vertical (xz, from boxed region on xy) maximum intensity projections encompassing ~1/4 of the diameter of the animal (edge of the animal is outlined white). Mucocytes are labeled with WGA. (A) Lipophil cell markers (Ta Tetraspanin, Ta 63996, and Ta GABA transporter) are co-expressed in scattered clusters ~8 μm in diameter throughout the central region of the animal, starting ~ 10 μm from the rim. (B) Digestive cell markers (Ta Trypsin and Ta Chymotrypsin) are highly expressed in a region starting ~60 μm from the rim. Cells expressing a lipophil specific marker (Ta GABA transporter) are interspersed among the digestive gland cells in this region. (C and D) Lipophil cell markers (C, Ta GABA transporter, and D, Ta Tetraspanin) and digestive gland cell markers (C, Ta PLA2, and D, Ta Chymotrypsin) are expressed in different populations of cells. Nuclei are labelled with DAPI (maximum intensity projections merged with DIC). fc – fiber cells; ec – epithelial cells; lc – lipophil cells. Scale bars 10 μm.
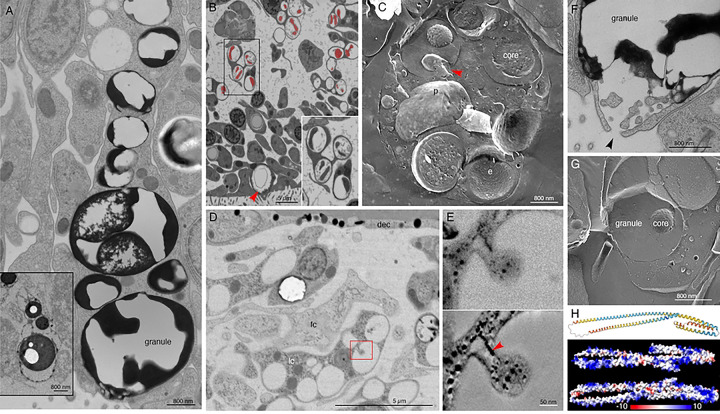
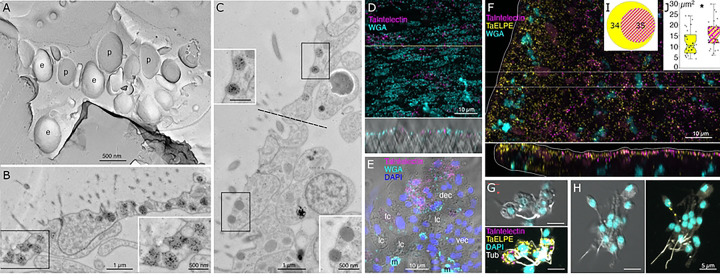
Lipophil cells in thin section from animals prepared by high pressure freezing and freeze substitution contained granules of varying sizes and appearances (Fig 3A). Granules in the basal part of the cell, the location of the Golgi complex where the granules originate [4], ranged in size from a few hundred nm to 2 μm and were electron lucent (the same as background) (Fig 3A,B). Some granules contained membrane bound profiles with content that resembled cytoplasm (Fig 3B). Analysis of serial thin sections (Fig 3B), freeze fracture replicas (Fig 3C), and tomograms (Fig 3D, E) of these granules showed that the profiles represented cytoplasmic protrusions into the granule. Granules closer to the apical pole of the cell were >500 nm in diameter and contained variable amounts of electron dense material as well as areas that were electron lucent (Fig 3A). The largest granule resided close to the apical surface of the cell. These apical granules often had a ring of electron dense material under their membrane (Fig 3A, F), but some were completely devoid of electron dense material. Small (<100 nm) membrane bound inclusions typically were present near the portion of the granule membrane that was closest to the apical surface of the cell (Fig 3A,F). Examination of these small inclusions in serial sections confirmed that they resided in the interior of the granule. We saw a single example of a LC granule with an open fusion pore and the content of the granule, including small membrane bound inclusions, exposed to the exterior (Fig 3F). The continuity of the granule and plasma membranes at the edge of the pore leads us to believe that this profile represents a stage in exocytosis of a LC granule.
Fig 3. Ultrastructural features of lipophil cells and the structure of a highly expressed lipophil cell secretory protein.
(A) Transmission electron microscopy (TEM) image of a thin section from a lipophil cell (LC) with osmiophilic material in its granules. The inset shows a TEM image of a section from an animal fixed with osmium, which better preserved the lipidic content of the granules and their cores. (B) SEM image of a section taken in a backscatter mode shows that granules in basal portions of LC have a dense core (artificially colored red) surrounded by electron lucent content. Inset is an enlarged view of the boxed region. Arrowhead indicates an apical LC granule. (C) Freeze fracture replica showing a basal part of a LC with multiple granules. Note dual component nature of the granules. A fracture through the interior of a core reveals heterogeneous content resembling cytoplasm. Arrowhead marks a protrusion emanating inward from the granule wall. (D) TEM image of 100 nm thick section showing basal region of a LC as inferred from its proximity to the dorsal side of the animal. Red box depicts a region with a protrusion inside a granule, further studied by EM tomography in (E). (E) Upper panel is an EM projection showing direct connection between the protrusion and the inner surface of the granule. Lower panel is a 1.75 nm thick virtual slice through a reconstructed volume of the tomogram. Note that ER (arrowhead) penetrates the protrusion. (F) TEM image of an apical part of an LC showing exocytosis (arrowhead) of a large granule. Note osmiophilic material and membranous particles located near the site of exocytosis. (G) Freeze fracture replica showing an apical part of a LC and its apical granule. (H) A protein present exclusively in LC secretome is largely composed of alpha helices (AlphaFold per-residue confidence score, pLDDT, color-coded: dark blue > 90 very high confidence; light blue 90 > pLDDT > 70 confident; yellow 70 > pLDDT > 50 low confidence; and orange < 50 very low confidence) and is positively charged (electrostatic map created with ChimeraX). dec – dorsal epithelial cell; e – e-face; fc – fiber cell; lc – lipophil cell; p – p-face. Scale bars 5 μm (B, D), 800 nm (A, C, F, G), and 50 nm (E).
The variable appearance and patchy distribution of the content of LC granules suggested that the content was partially extracted by the procedure used for freeze substitution and fixation. The apical granule in LC in thin sections from animals fixed in glutaraldehyde and osmium simultaneously in aqueous solution at room temperature was a large electron dense sphere with one or two electron lucent inclusions inside (clearer than background plastic; Fig 3A, inset). The electron dense material may represent unsaturated lipids, which are osmiophilic [33] and might be extracted by the organic solvents used for freeze-substitution of frozen specimens. The clear inclusions contain substances that do not bind the uranyl acetate and lead citrate grid stains suggesting that they are not proteinaceous. Analysis of freeze fracture replicas from rapidly frozen animals provided further evidence of the heterogeneous nature of the content of LC granules (Fig 3C, G). A fracture face through the outer content of the granule was smooth in appearance while the fracture face of the granule core was rough.
The uncharacterized protein Ta 63996 that served as one of our markers for LC is the second most highly expressed genes in TaH1 LC metacells (Fig 1, S3 Fig) and the identical gene in TaH2 (006628) is the most highly expressed gene in TaH2 LC metacells (S4 Fig). The gene was annotated as laminin subunit beta, but it lacks a laminin domain and laminin type EGF domains. It has only weak homology with a region located downstream of the critical laminin domains in laminin beta subunit orthologs in other animals and is much shorter (346 aa versus ~1700 aa) than in other species. The gene has a signal peptide and no transmembrane domains. Only LC express this putative secretory protein in TaH1 although some cells classified as “upper-epithelial-like” express it in TaH2. The protein structure predicted by AlphaFold includes extended helical regions with abundant basic amino acids (Fig. 3H). Many lysines are grouped in pairs, composing a KK motif, a well-known cleavage site in neuropeptide prohormones. However, this motif is not conserved beyond Trichoplacidae, and LC do not express any known prohormone convertases or cathepsins. We found no other highly expressed secretory proteins that lack transmembrane domains in Trichoplax LC metacells.
Two LC metacell clusters, LC1 and LC2, were identified in Hoilungia H13 (S5 Fig) and Hoilungia H23 (S6 Fig). The H13 LC2 metacell cluster expressed a gene whose sequence was 34% identical and 52% positive to the uncharacterized secretory protein Ta 63996. In H23, both LC1 and LC2 metacells expressed an ortholog (e−45) of Ta 63996. All LC metacells in TaH1 and H2 and Hoilungia H13 and H23 contained a gene identical to the Tetraspanin (Ta 58643) we used as a second marker for LC. The third marker we used for LC was annotated as a solute carrier (Ta GABA transporter). It was expressed in all metacells classified as LC in TaH1 and TaH2 but only in LC1 metacells in Hoilungia H13 and H23. All LC metacell clusters in H1, H2, H13 and H23 highly expressed several fatty-acid binding proteins, V-type ATPases and solute carriers, and the lysosomal membrane glycoprotein, LAMP2 (S3–6 Figs).
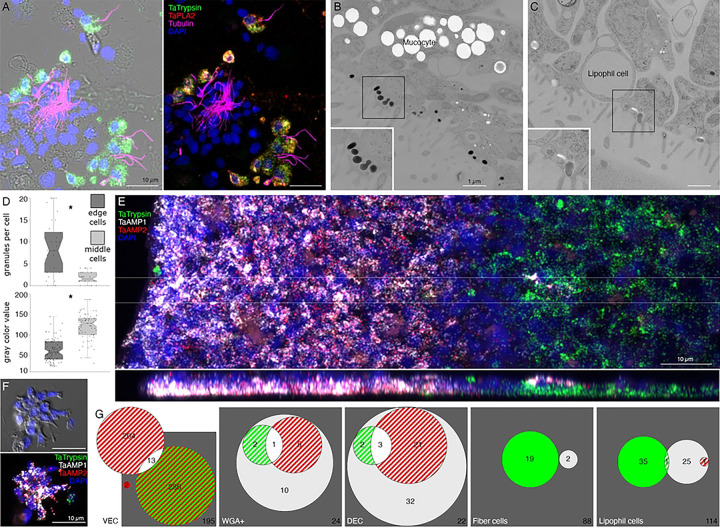
We further investigated the morphology of cells that express digestive enzymes by co-labeling dissociated cell preparations with FISH probes for the precursors of trypsin and sPLA2 and an antibody against acetylated tubulin, a component of cilia. Cells co-expressing Ta trypsin and Ta sPLA2 often occurred in clusters (Fig 4A). The cells were small and cylindrical in shape and many of them possessed a cilium. Labeled cells lacking a cilium may have lost their cilium during dissociation since cilia that were not attached to a cell also were present. Ciliated ventral epithelial cells (VEC) are the most prevalent cell type in the ventral epithelium. Given the high density of cells expressing digestive enzymes in the central part of the ventral epithelium and their possession of a cilium, we identify “digestive gland cells” as a subtype of VEC (Fig 1, S3–6 Figs).
Fig 4. Two monociliated ventral epithelial cell (VEC) types: digestive gland cells and cells expressing putative antimicrobial peptides (AMPs).
(A) Cells secreting digestive enzymes bear a cilium (combination of FISH for Ta Trypsin and Ta PLA2 and immunolabelling for tubulin; left panel shows fluorescence merged with DIC). (B, C) TEM shows that both peripheral (B) and central (C) ciliated epithelial cells have ~500 nm diameter granules located close to the apical surface. Boxed regions are magnified in insets. (D) The granules in peripheral cells are more electron dense and abundant than those in cells in more central regions. * p<0.05. (E) Trypsin is highly expressed in the central region of the ventral epithelium, while AMP1 expression is restricted to the peripheral region. (F) In dissociated cell preparations, two distinct subpopulations of cells expressing either AMP1 or Ta Trypsin are apparent. Most AMP1 expressing cells strongly express AMP2 whereas Ta Trypsin expressing cells show weak AMP2 expression (not visible in E). A dissociated trypsin+ cell (F, green) contains one red AMP2 grain. (G) Venn diagrams based on cell counts show that three quarters of VEC express either AMP1 or Ta Trypsin along with AMP2. Some mucocytes and DEC express AMP1 with or without either AMP2 or Ta Trypsin. Only a few lipophil and fiber cells express AMP1 or Ta Trypsin. Color coding for Venn diagrams is the same as the fluorescence colors on E and F; double co-expression is indicated as strips of respective colors, triple co-expression is white, and the absence of expression is dark gray. Numbers of counted cells are shown. Scale bars: 10 μm (A, E, F) and 1 μm (B, C).
As VEC in the peripheral part of the ventral epithelium (pVEC; within ~60 μm of the rim) do not express digestive enzymes we speculated that their ultrastructure might differ from that of the centrally located VEC (cVEC) that express genes encoding precursors of digestive enzymes. Comparison of pVEC and cVEC within the same thin sections revealed differences in their secretory granules (Fig 4B-D). Peripheral VEC, identified based on their proximity to the rim, had numerous electron dense secretory granules near their apical surfaces (Fig 4B, D). Ventral epithelial cells located in the central region, in the vicinity of LC, had fewer granules (p=1.96E-05) and the granules were less electron dense (p=1.26E-17) (Fig 4C, D).
Metacells identified as “epithelial” in TaH1 [31] or “lower epithelial” [6] expressed multiple genes that were predicted precursors of secretory proteins/peptides (InterPro). We noticed that the sequences of five of them resembled those of arminins, a type of antimicrobial peptide (AMP) expressed in Hydra [34], in that they were ~150– 250 aa in length, had a signal peptide, a highly acidic N-terminal region and alkali C-terminal region that includes aliphatic amino acids and DE cleavage motifs (S2 Fig) common in placozoan and cnidarian prepropeptides [35,36]. The N-terminal domain of one of them, Ta 55945, was annotated “alpha defensin” (Interpro). The signal peptide and acidic N-terminal regions of arminin prepropeptides are removed to produce active arminin peptides. Arminin peptides have a C-terminal glycine and are thought to be amidated. The sequences of the Trichoplax arminin-like peptides have a C-terminal glycine, as is required for amidation by peptidylglycine α-amidating monooxygenase (PAM). An AMP analysis tool (Antimicrobial Peptide Data, Univ. Nebraska Medical Center) identified hydrophobic and basic regions in the C-terminal part of the Ta AMP sequences that might interact with membranes.
We obtained FISH probes for two of the arminin-like genes, Ta 55945 and Ta 56030, and will refer to them as AMP1 and AMP2, respectively. Both probes strongly labeled cells in the peripheral part of the ventral epithelium (Fig 4E). The AMP expressing cells were densely packed and showed little overlap with cells expressing digestive enzymes. Scattered cells in the dorsal epithelium showed moderate expression of AMP1 or both AMP1 and AMP2. Most of the AMP-expressing cells in dissociated cell preparations had morphological features typical of VEC: they were small, cylindrical in shape and possessed a cilium (Fig 4F). They often occurred in clusters with other AMP-expressing cells. Most AMP1 expressing VEC strongly expressed AMP2. Ta Trypsin-expressing VEC weakly expressed AMP2, containing fewer and smaller fluorescent grains than cells co-expressing AMP1 and AMP2. Some WGA-positive mucocytes and DEC expressed AMP1 with or without AMP2. A small fraction of LC and fiber cells contained one or a few AMP1 or Ta trypsin positive grains (Fig 4G) and may represent cells transdifferentiating from cVEC to lipophil or fiber cells, respectively [6].
The genes encoding AMP1, AMP2, and a third arminin-like prepropeptide (Ta 60631) are the three most highly expressed genes in Trichoplax H1 classified as “lower epithelial” metacells [6]. Our findings demonstrate that these metacells correspond to peripheral VEC (pVEC; Fig 1, S3 Fig). The TaH2 metacells classified as “lower epithelial” contain genes that are nearly identical to AMP1, AMP2 and Ta 60631 (S4 Fig). The AMP1 and Ta 60631 genes are the most highly expressed genes these metacells. Hoilungia H13 and H23 metacells classified as “lower epithelial” contain a gene that is more than 55% identical and 70% similar to Ta 60631 and a gene that is more than 40% identical and 60% similar to AMP2 (S5, 6 Figs). No AMP1 ortholog was found in Hoilungia H13 or H23.
Main secretory cell types in the dorsal epithelium
The most prevalent cells in the dorsal epithelium are monociliated dorsal epithelial cells (DEC) [18]. Their broad (~10 μm) polygonal-shaped apices pave the dorsal surface. Their cell bodies are narrower and extend into the interior, where they are surrounded by processes of fiber cells. Freeze fracture replicas of the apices of DEC revealed exoplasmic (e) faces and protoplasmic (p) faces of numerous, large (500 nm) secretory granules (Fig 5A). Electron micrographs of transverse thin sections through the apices of DEC showed dark secretory granules near their dorsal surfaces (Fig 5B). The dark granules bound the lectin wheat-germ agglutinin (WGA) conjugated to nanogold (Fig 5B, inset). WGA also labeled the outer surfaces of DEC. At the rim, cells with morphological characteristics of DEC were adjacent to cells with morphological characteristics of VEC (transition zone). Secretory granules in pVEC were grey or dark like those in DEC but bound much less WGA label (Fig 5C, insets).
Fig 5. Main secretory cells in the dorsal epithelium.
(A) Freeze fracture replica at the apex of a dorsal epithelial cell (DEC) imaged in TEM reveals e- or p-faces of numerous ~500 nm diameter secretory granules. (B, C) TEM of ultrathin sections labelled with nanogold conjugated WGA. Transverse section in the dorsal epithelium (B) shows multiple WGA-stained granules in ciliated DECs; insets show enlarged view of boxed regions. Transverse section at the transition region between dorsal and ventral epithelia (C, the border is demarcated by dotted line) shows that DEC granules bind more WGA than do morphologically similar granules in VEC. (D-H) Confocal images of wholemounts (D, F) and dissociated cell preparations (E, G, H). Many DEC express Ta Intelectin 60661 (D, whole mount; E, dissociated cells) as evident from co-labeling with WGA. Mucocytes (m) label intensely with WGA, but do not express Ta Intelectin 60661. Other cell types (VEC, lipophil (lc) and fiber (fc) cells) are not labeled. (F) Ta ELPE is expressed in nearly all DEC and a few VEC (see xz inset); (G, I) About half of Ta ELPE+ co-expresses Ta Intelectin 60661. (H) Some VEC, identified based on their small sizes and cylindrical shapes express Ta ELPE but not Ta Intelectin. (J) Cells co-expressing Ta ELPE and Ta Intelectin are larger than those expressing only Ta ELPE. e – e face; fc – fiber cells; lc – lipophil cells; m – mucocytes; p – p face.
Many DEC in wholemounts and dissociated cell preparations, identified based on their content of granules stained with WGA conjugated to a fluorescent dye, were labeled by a FISH probe for Ta 60661 (Fig 4D, E), a gene that is highly expressed in TaH1 metacells classified as “epithelial” or “upper epithelial” [6,31]. Labeled cells were present throughout the dorsal epithelium except in a region within ~15 μm of the rim (Fig 4 D, E). Their distribution was patchy, suggesting that only a subset of DEC expressed this gene.
Trichoplax H1 gene Ta 60661 is identical to a gene discovered in a search for orthologs of genes implicated in innate immunity in TaH2 [37]. The gene was identified as belonging to a class of secreted glycoproteins called “intelectins” based on its content of a fibrinogen-related domain (FReD). Intelectins have been implicated in defense against bacteria in both vertebrates and invertebrates [38]. Ten of thirty-one intelectin-like genes identified in TaH2 are uniquely expressed in TaH1 and TaH2 metacells classified as “upper epithelial” (Fig 1, S3, 4 Figs). These metacells also contain a gene annotated “membrane-associated mucin 4-like glycoprotein” based on the presence of von Willebrand factor type D (VWD), Scavenger Receptor Cysteine-Rich (SRCR) and multiple EGF-like domains. Hoilungia 13 metacells classified as “upper epithelial” include genes encoding five of the intelectins and the mucin-4 gene, but not intelectins Ta 60661 and Ta 61411 (S5 Fig). Instead, these genes are expressed in a metacell classified as “gland”. Hoilungia H23 metacells classified as “upper epithelial” express four of the intelectins expressed in upper epithelial metacells in TaH1, TaH2 and Hoilungia H13 (S6 Fig).
The metacells representing DEC also express a prepropeptide identified in TaH1 genomes and transcriptomes that is predicted to produce peptides with C-terminal amino acids Glu-Leu-Pro-Glu, or ELPE [6,23,35]. We will refer to this gene as Ta ELPE. The names, sequences and predicted mature peptides produced by this and other prepropeptide genes investigated in this study are listed in S7 Text.
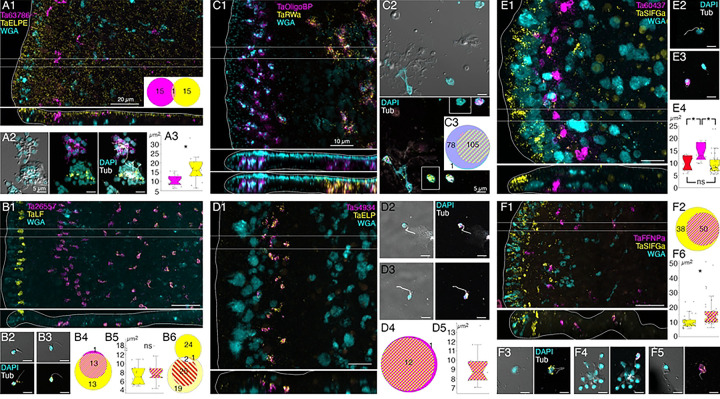
A FISH probe for Ta ELPE prepropeptide labeled cells throughout the dorsal epithelium including those <15 μm of the rim (Figs 5F, 6A1). Approximately half of the Ta ELPE-positive DEC co-expressed Ta 60661 intelectin (Fig 5G-I). A few VEC, identified based on their small sizes and cylindrical shapes, expressed Ta ELPE but not intelectin Ta 60661 (Fig 5F, H). The mean area of cells expressing both Ta 60661 and Ta ELPE was greater (p=0.01) than that of cells expressing Ta 60661 without Ta ELPE (Fig 5G, H, J) because the latter group included the Ta ELPE-positive VEC, which were smaller than DEC.
Fig 6. Localization and characterization of peptidergic secretory cells.
Trichoplax wholemounts and dissociated cell preparations were labeled with FISH probes for different prepropeptides and other cell-type specific proteins. Mucocytes were labeled with fluorescent-conjugated WGA in wholemounts. Cilia were immunolabeled with an antibody against acetylated tubulin and nuclei were labeled with DAPI in dissociated cell preparations. Circle plots and box plots use the same color coding as fluorescence images; co-expression is indicated as stripes of respective colors. (A). Prepropeptide Ta ELPE (A1) was expressed by DEC throughout the animal and by some VEC near the rim. Interspersed among the Ta ELPE+ DEC were scattered cells that expressed an uncharacterized protein (Ta 63786). Probes for Ta ELPE and Ta 63786 labeled distinct populations of ciliated cells in dissociated cell preparations (A1, circle plot; A2). Ta ELPE+ cells were larger in area than Ta 63786+ cells (A3). (B). Ta LF prepropeptide (B1) was strongly expressed in a row of cells in the dorsal epithelium 5 – 10 μm from the rim and more weakly expressed in scattered cells in the ventral epithelium in a region starting 40 μm from the rim. The Ta LF+ ventral epithelial cells co-expressed an astacin-like metalloendopeptidase (Ta 26557; B1, B2), while dorsal Ta LF+ cells did not (B1, B3, B4). Both Ta LF+/Ta 26557+ cells (B2) and LF+/Ta 26557− cells (B3) were small ciliated cells (B5). Circle plot B6 shows that Ta LF+ cells (bright yellow) were distinct from Ta ELPE+ cells (light yellow) and Ta Intelectin 60661+ cells (red stripes). (C). Mucocytes were labeled with fluorescent WGA and a probe for a secreted oligosaccharide binding protein (Ta OligoBP; C1). The central population of mucocytes co-expressed Ta RWa prepropeptide (C1–3). Both Ta RWa− and Ta RWa+ mucocytes lacked a cilium (C2, tubulin label absent). The rectangular insets show Ta RWa− (top) and Ta RWa+ mucocytes (bottom) without the magenta (Ta OligoBP) fluorescence channel. (D). Ta ELP prepropeptide and an astacin-like metalloendopeptidase (Ta 54934; D1) were co-expressed in a row of cells in the ventral epithelium 15 to 30 μm from the rim. The labeled cells were ciliated (D2, D3), co-expressed both genes (D1–4) and had an area of about 9 mm2 (D5). (E). Probes for Ta SIFGa prepropeptide and for a second uncharacterized secretory protein (Ta 60437) labeled separate populations of cells: Ta SIFGa+ cells were prevalent in the dorsal epithelium of the rim (E1) and more sparsely distributed in the dorsal and ventral epithelium further in the interior; Ta 60437+ cells were in a row 20 to 30 μm from the rim (E1). Both the Ta SIFGa+ and Ta 60437+ cells were ciliated (E2, E3). The Ta 60437+ cells were larger than the Ta SIFGa+ cells (E4,) and larger than Ta ELP+/Ta 54934+ cells (E4, red), which were in the same area (D1). (F). A subset of Ta SIFGa+ cells in the ventral epithelium >10 μm from the rim expressed the Ta FFNP prepropeptide (F1, F2). Both Ta SIFGa+/Ta FFNP− cells (F3) and Ta SIFGa+/Ta FFNP+ cells (F4, F5) bear a cilium. The Ta SIFGa+/Ta FFNP+ cells were larger than the Ta SIFGa+/Ta FFNP− cells (F6). * p<0.05. Scale bars 20 μm (A1, B1, F1), 10 μm (C1, D1, E1) and 5 μm in all dissociated cells images.
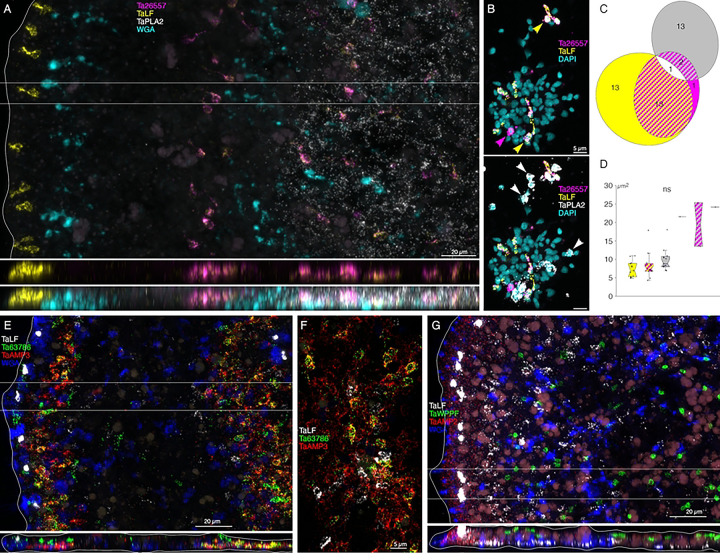
Scattered cells in the dorsal epithelium and a few cells in the ventral epithelium located 10 to 40 μm from the rim were labeled with a probe for a gene encoding a secretory protein with no known functional domains, Ta 63786 (Fig 6A1). The labeled cells did not express Ta ELPE (Fig 6A1 inset, A2). The Ta 63786+ cells in the ventral epithelium, but not those in the dorsal epithelium, co-expressed a gene encoding a secretory peptide that bears the same Hydra arminin-like features as AMP1 and AMP2, including a signal for C-terminal amidation, which we call AMP3 (Fig 7E, F). Dissociated Ta 63786+ cells had a cilium but were smaller and narrower (p=0.004) than Ta ELPE+ and Ta 60661+ DEC cells (Fig 6A2, A3). The metacells that express Ta 63786 and AMP3 were classified as “peptidergic” or “epithelial unknown” [6,31]. These metacells also contain a gene encoding an intelectin (Ta 64393) that is not expressed in any other metacell (Fig 1, S3 Fig). Trichoplax H2 metacells classified as “gland”, or “epithelial lower” [6], express genes identical to Ta 63786 and Ta 64393, and a gene that is nearly identical to AMP3 (S4 Fig). The DEC labeled by the probe for Ta 63786, but not the probe for AMP3 likely correspond to TaH1 metacells C102–107 (S3 Fig) and TaH2 metacells C133 and 134 (S4 Fig), which highly express Ta 63786 but do not express AMP3 or the intelectin. Metacells TaH1 C103 and TaH2 C133 and 134 were reported to co-express Ta 63786 and Ta ELPE [6] but we did not observe cells co-expressing these genes in our microscopy studies. Hoilungia H13 metacells C177–183 express orthologs of Ta 63786 and Ta 64393 intelectin (S5 Fig), but no AMP3 ortholog was found in this species. Hoilungia H23 lacks orthologs of Ta 63786, Ta 64393 intelectin and AMP3. No ortholog of Ta ELPE was found in H13 or H23.
Fig 7. Localization and characterization of peptidergic cells.
(A) Trichoplax wholemount (top panel, horizontal maximum intensity projection; bottom two panels, vertical projection of boxed region). Mucocytes were labeled with fluorescent WGA. Cells co-expressing Ta LF and astacin Ta 26557 are interspersed among mucocytes and Ta PLA2+ cells in the central part of the ventral epithelium while cells that express Ta LF prepropeptide without astacin are most prevalent close to the rim. (B) Dissociated cell preparation. Nuclei were labeled with DAPI. Color separated and merged views show multiple cells that co-express Ta LF and astacin Ta 26557 (yellow arrowheads), a cell that expresses astacin without Ta LF (magenta arrowhead) and cells that express Ta PLA2 (white arrowheads). (C) Many cells co-express Ta LF and astacin Ta 26557, but few cells co-express Ta LF+ or Ta 26557 and Ta PLA2. (D) All labeled cells are similar in size with Ta PLA2+/Ta 26557+ cells slightly larger. (E) Horizontal and vertical maximum intensity projections of a wholemount labeled with probes for Ta LF prepropeptide, secretory protein Ta 63786, putative antimicrobial peptide AMP3 and fluorescent WGA. The images span nearly the entire width of the animal – note the mucocyte and the Ta LF+ cell at the upper right. Cells in a region of the ventral epithelium within 10 to 40 μm of the rim co-express Ta 63786 and AMP3; cells in the peripheral part of the dorsal epithelium express Ta LF or Ta 63786 but do not express AMP3. (F) Enlarged view of partially dispersed cells co-expressing Ta 63786 and AMP3. (G) Horizontal and vertical maximum intensity projections of an animal labeled with probes for Ta LF, Ta WPPF prepropeptides, AMP2, and the lectin WGA. The images encompass ~1/3 the diameter of the animal. Cells expressing Ta WPPF prepropeptide are distributed in the dorsal epithelium in a region starting ~40 μm from the rim and do not express Ta LF or AMP2. Scale bars 20 μm in whole mount images and 5 μm in dissociated cells images.
Morphology and locations of peptidergic secretory cells
A probe for the precursor of Leu-Phe (Ta LF) peptides labeled a row of cells in the dorsal epithelium 5–10 μm from the rim and more weakly labeled scattered cells in the ventral epithelium >40 μm from the rim (Fig 6B1). A probe for a secreted astacin-like metalloendopeptidase, Ta 26557, intensely labeled the Ta LF+ cells in the ventral epithelium but not the Ta LF+ cells in the dorsal epithelium (Figs 6B1–4, 7A). Both Ta LF+/Ta astacin+ and Ta LF+/Ta astacin− cells were cylindrical in shape, possessed a cilium (Figs 6B2, B3, 7B), and were similar in size (Figs 6B5, 7C, D). The positions of Ta LF+/Ta astacin+ cells are like those of cells classified as ventral Type 3 gland cells in ultrastructural studies of Trichoplax [17]. Type 3 gland cells contain small granules with textured content. The Ta LF+/Ta astacin+ cells were intermingled with cells expressing Ta sPLA2, but only a few cells co-expressed Ta astacin and Ta sPLA2 (Fig 7A-D). The Ta LF+/Ta astacin− cells located near the rim of the dorsal epithelium did not express Ta ELPE or the DEC-specific intelectin Ta 60661 (Fig 6B6). Based on the high expression of Ta LF and lack of expression of astacin Ta 26557, these cells represent TaH1 metacell C41 (Fig1) and metacells TaH1 C188, TaH2 C210, and Hoilungia H13 C244 (S3–5 Figs). The cells that co-express Ta LF and astacin Ta 26557 represent TaH1 metacells C42 (Fig 1) and C185 and C186 (S3 Fig), TaH2 C215 (S4 Fig), and Hoilungia H13 C251 and C252 (S5 Fig). No ortholog of LF prepropeptide was found in Hoilungia H23 but metacell C233 contained an ortholog of astacin Ta 26557 (S6 Fig). This metacell contained prepropeptide genes that bore no resemblance to Ta LF prepropeptide.
Mucocytes identified by labeling with WGA conjugated to a fluorescent dye were in the ventral epithelium in a zone 10 and 30 μm from the rim and in the central region >50 μm from the rim (Fig 6C1), as reported [17]. A probe for an oligosaccharide binding protein, Ta 63702, labeled both peripheral and central mucocytes (Fig 6C1). A probe for the Arg-Trp-amide (Ta RWa) prepropeptide labeled the central mucocytes but not the peripheral mucocytes (Fig 6C1–3). Dissociated mucocytes were ovoid cells ~8 μm in length and lacked a cilium, as evident from the absence of staining for acetylated tubulin (Fig 6C2). Based on co-expression of Ta 63702 and the Ta RWa prepropeptide, the central population of mucocytes corresponds to TaH1 metacell C36 (Fig 1), TaH2 metacells C195 and C196 (S4 Fig), Hoilungia 13 metacells C213, C216, and C277 (S5 Fig) and Hoilungia H23 metacell C209 (S6 Fig). These metacells express PAM, the enzyme that converts a glycine at the C terminal of a peptide to an amide, a secreted glycoprotein with VWD, TIL, C8 and PTS domains characteristic of gel-forming mucins[39], and an intelectin, Ta 62229, that is not expressed in other metacells. Trichoplax H2 metacell C196, Hoilungia H13 metacell C218, and Hoilungia H23 metacell C210 contain orthologs of Ta 63702 and the gel-forming mucin but lack the RWa prepropeptide gene, express very little PAM and express only low levels of intelectin Ta 62229. These metacells likely represent the RWa negative mucocytes that are in the peripheral part of ventral epithelium. Expression of Ta RWa prepropeptide was not reported in TaH1 in the more recent scRNAseq study [6]. However, TaH1 metacells C165 and C166 contain orthologs of Ta 63702 and the gel-forming mucin (S3 Fig) suggesting that these metacells represent mucocytes. Metacell C165 contains PAM and intelectin Ta 62229 but C166 does not, suggesting C165 represents central mucocytes and C166 peripheral mucocytes. The metacells representing mucocytes in TaH2, Hoilungia H13 and Hoilungia H23 contain orthologs of a homeobox DBX1-B-like transcription factor that is not present in any other metacells. The GenBank sequence for the TaH1 ortholog of DBX1 is incomplete and consequently the gene was not included in the scRNAseq data for TaH1.
Cells labeled by probes for the Ta-endomorphin-like peptide (TaELP) prepropeptide [22] (S7 Text) and a secreted astacin-like metalloendopeptidase, Ta 54934, were in a narrow zone in the ventral epithelium 10–20 μm from the rim (Fig 6D1). The positions of these cells correspond with those classified as Type 1 gland cells in ultrastructural studies of Trichoplax [17]. Type 1 gland cells contain larger and more electron dense granules than Type 3 gland cells. Dissociated TaELP -expressing cells had a cilium (Fig 6D2, D3), nearly always co-expressed both genes (Fig 6D4), and were relatively small (<5 μm in diameter, Fig 6D5). The TaH1 metacell that expresses TaELP prepropeptide and the astacin Ta 54934 (C32, Fig 1) highly expresses an additional astacin-like gene, Ta 54935, that is not expressed in any other metacells. TaELP expression was not reported in TaH1[6], but a metacell expressing Ta 54934 and Ta 54935 was found: C183 (S3 Fig). A metacell expressing TaELP ortholog was reported in TaH2: C210. However, H2 C210 did not express astacin orthologs Ta 54934 and Ta 54935. Instead, these astacins were expressed in H2 C214 (S4 Fig). Moreover, H2 C210 highly expresses LF prepropeptide, which we found was expressed in distinct sets of cells (Fig 5B1). We suspect that H2 metacell C214 may represent TaELP expressing cells. Hoilungia H13 metacells C248 and C249 expressed orthologs of the TaELP prepropeptide and both astacin-like metalloendopeptidases (S5 Fig). No ortholog of TaELP prepropeptide was found in Hoilungia H23. However, H23 metacell C232 expresses genes identical to astacins Ta 54934 and Ta 54935. Interestingly, metacells that express these astacins in all four species also express one or two G-protein coupled receptors (GPCR) with a C-type lectin domain (CTLD) [37].
A probe for the Ser-Ile-Phe-Gly-amide (Ta SIFGa) prepropeptide labeled cells in the dorsal epithelium near the rim and scatted cells in the dorsal and ventral epithelium further in the interior (Fig 6E1). The metacell expressing high levels of Ta SIFGa prepropeptide (C46; Fig 1) was reported to express a secreted protein with no known functional domains, Ta 60437, that was not expressed in any other metacell. Our probe for Ta 60437 did not label Ta SIFGa-expressing cells but instead labeled a row of cells in the ventral epithelium 20–30 μm from the rim (Fig 6E1). The positions of these cells correspond with those of Type 1 gland cells. Both cell types possessed a cilium (Fig 5E2, E3). Ta 60437+ were larger than cells expressing Ta SIFGa (p = 3.32E-4, Bonferroni corrected p value) and cells co-expressing Ta ELP and astacin-like metalloendopeptidase Ta 54934 (p = 0.02, Bonferroni corrected p value), which were in the same area (Fig 5E4). This fact, along with the location of Ta 60437+ cells, makes it likely that these are Type 1 gland cells, which are larger than other ciliated cells due to their possession of big secretory granules [17]. The Ta 60437 gene is highly expressed in metacells C167 in TaH1 (S3 Fig), C197 in TaH2 (S4 Fig), C197 and C219 -C222 in Hoilungia H13 (S5 Fig) and C111–112 in Hoilungia H23 (S6 Fig).
A probe for the Phe-Phe-Asn-Pro-amide (Ta FFNPa) labeled the Ta SIFGa+ cells in the central part of the ventral epithelium, but not those in the peripheral part of the ventral epithelium or in the dorsal epithelium (Fig 6F1, F2). The positions of the cells that co-express these genes correspond with those classified as ventral Type 3 gland cells [17]. Both FFNPa+ and FFNPa− cells possessed a cilium (Fig 6F3-F5), but the FFNPa+ cells were larger than the FFNPa− cells (p=7.86E-06; Fig5F6). Cells expressing Ta SIFGa and Ta FFNPa correspond to metacell C38 (Fig 1) and C168 (S3 Fig). No expression of SIFGa was reported in the metacell expressing FFNPa in TaH2 (S4 Fig), but Hoilungia H13 metacell C223 and H23 metacell C213 express both SIFGa prepropeptide and very high levels of FFNPa prepropeptide (S5 and S6 Figs). The SIFGa+ cells in the dorsal epithelium and in the rim of the ventral epithelium correspond to TaH1 metacells C46 (Fig 1) and C189 (S3 Fig), TaH2 C217 (S4 Fig), Hoilungia H13 C253–254 (S5 Fig), and H23 C221 (S6 Fig) based on high expression of SIFGa prepropeptide and absence of FFNPa prepropeptide.
A probe for the prepropeptide precursor of the peptide WPPF labeled small (diameter ~5 μm) cells distributed throughout the central part of the dorsal epithelium starting ~40 μm from the rim (Fig 7G). Cells expressing Ta WPPF represent metacell TaH1 C181 (S3 Fig), TaH2 C213 (S4 Fig), Hoilungia H13 C246 (S5 Fig), and Hoilungia H23 C230 (S6 Fig).
Roles of lipophil cells and digestive gland cells in feeding
Both main secretory cell types in the ventral epithelium, LC and ciliated VEC, have been implicated in feeding. Lipophil cells secrete granules whose content lyses algae [4]. VEC constitutively pinocytose extracellular macromolecules such as ferritin [40], HRP, dextran [24] and mucus [17]and, likely, the contents of lysed algae. The expression of high levels of Ta Trypsin, Ta Chymotrypsin and Ta sPLA2 in centrally located VEC led us to investigate whether they secrete digestive enzymes during feeding.
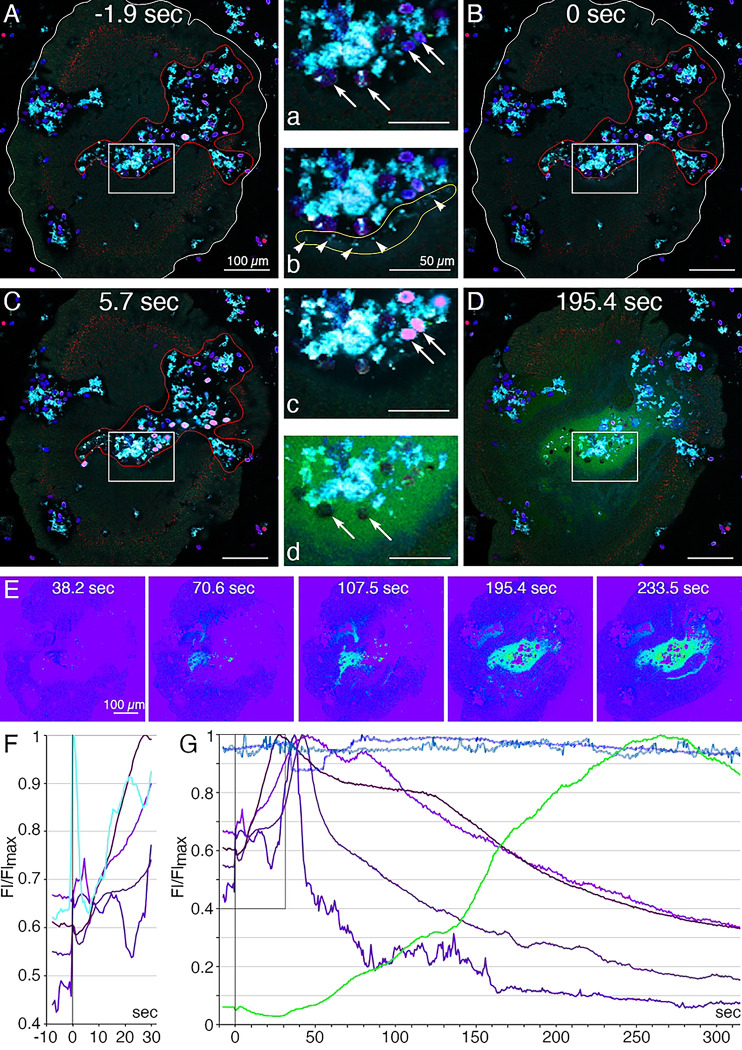
Lipophil granule secretion in living animals can be visualized with the fluorescent dye FM1–43, which stains the contents of secreted LC granules [4]. We used a fluorescent indicator for trypsin activity, BZiPAR [41], to test whether digestive gland cells secrete trypsin. Intracellular LC granules were visualized with a lipophilic dye, LipidTOX. Animals encountering Rhodamonas salina algae on the substrate manifested behaviors typical of Trichoplax during feeding episodes [4]: they ceased gliding and changing shape, and their margins spread and became more closely attached to the substrate (Fig 8A; S2 Fig; S8 and S9 Movies). Some, although not all, LC in the vicinity of algae released their large apical secretory granule, the contents of which became stained with FM1–43 (Fig 8B, F; S2 Fig). The stained content initially was a diffuse cloud but then became more concentrated. When a LC released its granule, the surrounding area of the ventral epithelium moved farther away from the substrate. These movements, together with the slow scan speeds (~1 frame/sec) used for these experiments, made it difficult to visualize secretion of LipidTOX-stained granules, which happens rapidly, so we used the appearance of FM1–43-stained spots to monitor granule secretion. Some animals contracted during lipid granule secretion and their rims detached from the substrate, but then they flattened and reattached (not illustrated). Thereafter, the rim remained close to the substrate, but the central portion of the animal moved further away, forming a “feeding pocket” between the lower surface and the substrate. The secretion of a granule often was followed within <2 sec by lysis of algae in the vicinity (Fig 8C, F). As algae lysed, they became intensely stained with LipidTOX and FM1–43. Following lysis of algae, cells in the central region of the animal began to display “churning” movements, slow swirling movements of large groups of cells and, sometimes, faster oscillations of small groups of cells (S8 and S9 Movies) [4]. Trypsin activity monitored with BZiPAR became detectable ~ 1 minute after LC granule secretion and continued to increase over the course of 3 to 10 minutes (Fig 8D, E, G; S2 Fig). Algae that had been lysed by the content of LC granules gradually disappeared but algae that were not lysed remained intact (Fig 8G). Lysed algae that became trapped under the rim and therefore were protected from enzymes in the feeding pocket did not disappear, implicating digestive enzymes in the disappearance of the lysed algae. The partially digested content of the lysed algae likely was pinocytosed by VEC and transferred to lysosomes for intracellular digestion [42]. When the animal resumed gliding, the remaining BZiPAR fluorescence diffused from the feeding pocket into the surrounding seawater. Some animals pausing over algae did not secrete LC granules and, consequently, no algae were lysed. Although these animals displayed movements typical of animals feeding on algae, no trypsin activity was detected at their lower surfaces (not illustrated).
Fig 8. Secretory behaviors of Trichoplax associated with external digestion of algae.
(A-E) Confocal images of a Trichoplax feeding on R. salina algae; (F, G) normalized fluorescence intensity measured in selected regions. A starved animal was transferred to a cover glass chamber containing algae in seawater with a lipophilic dye (LipidTOX, red) that stains lipophil cell granules; FM1–43 (cyan), a membrane dye used here to visualize the secreted contents of lipophil cell granules; and a fluorescent indicator for trypsin activity (BZiPAR, green). The insets numbered with lowercase letters are enlarged view of rectangular region on a respective image numbered with an uppercase letter. (A, a) At t=−1.9 sec, the animal had ceased moving in a region containing algae (blue, phycoerythrin autofluorescence, arrows) and debris (cyan) representing algae remnants from an earlier feeding episode. The animal body is outlined white, and the feeding pocket is outlined red. (B, b) At t=0 sec, lipophil cell granule secretion was evident in the feeding pocket due to the sudden appearance of small (<5 μm) diffuse clouds and bright particles (arrowheads on b) of FM1–43-stained material. (C, c) At 5.7 sec, some algae near sites of lipophil granule secretion swelled, lysed, and became intensely stained with LipidTOX and FM1–43 (pink in merged, arrows in c). Other algae (blue) remained intact and were not stained with LipidTOX or FM1–43. (D, d) By 195.4 sec, diffuse BZiPAR fluorescence (green) filled the feeding pocket. The lysed algae no longer were visible, but the intact algae remained (arrows in d). (E) Intensity encoded BZiPAR fluorescence images show increasing trypsin activity between 70.6 and 233.5 sec. (F) Details of first events in feeding: Lipophil granules (cyan; the fluorescence profile obtained for the area outlined yellow in b) were secreted approximately synchronously at 0 sec and this was followed by a rapid increase of fluorescence in nearby algae (four fluorescence profiles obtained for four pink algal cells in c; different shades of magenta). (G) Evidence of digestion: algae affected by lipophil granules (different shades of magenta) burst and released their content. Secretion of trypsin, as indicated by BZiPAR fluorescence (green; fluorescence profile obtained for the region outlined red in B), began 40–50 sec after lipophil discharge and was associated with a decline of fluorescence intensity of the lysed algal cells. Those algae not affected by lipophil granules remained constant in intensity throughout the feeding episode (two fluorescence profiles obtained for two individual algal cells; different shades of blue). Scale bars 100 μm.
Discussion
We used FISH probes for genes specifically expressed in metacells identified by analysis of single cell transcriptomes of Trichoplax adhaerens H1 (TaH1) [6,31] to map the distributions of different cell types in TaH1 whole mounts and to identify the cells in dissociated cell cultures where their morphology could be visualized by differential interference contrast microscopy and fluorescent labels used to identify hallmark features. We found that two cell types in the central part of the ventral epithelium, lipophil cells (LC) and a subset of the monociliated ventral epithelial cells (VEC), participate in lysis and digestion of microalgae in animals pausing to feed. The peripheral part of the ventral epithelium and the dorsal epithelium contain secretory cells that express peptides and/or glycoproteins implicated in defense. Electron microscopic studies of LC, dorsal epithelial cells (DEC) and VEC revealed ultrastructural features of their secretory granules that, together with the transcriptomes of these cell types, provide clues about the compositions and biogenesis of their secretory granules. We identified metacells representing mucocytes and found that mucocytes in the central part of the ventral epithelium differ from those in the periphery. We mapped the distributions and identified morphological features of seven peptidergic cell types. Our findings provide a more detailed picture of the placozoan body plan than was apparent from previous microscopic studies and reveal cellular mechanisms placozoans use to obtain nutrients and defend against pathogens. We compare the sequences of the main secreted proteins and expression profiles of single cell transcriptomes of four placozoan species and confirm a close relationship between Trichoplax H1 and H2, partial divergence of Hoilungia H13 and greater divergence of Hoilungia H23. Comparison of cell types in representatives of Planuloza (Placozoa, Cnidaria and Bilateria) with those in Ctenophora and Porifera provides insight into cell types that may have been present in their common ancestor.
A subset of monociliated ventral epithelial cells secrete digestive enzymes
Probes for Ta trypsin, Ta chymotrypsin and Ta sPLA2, which are highly expressed in metacells classified as “digestive gland cells” [6,31] labeled cells in the central part of the ventral epithelium with morphological features typical of ventral epithelial cells (VEC): they possessed an apical cilium and microvilli and had electron dense secretory granules near their apical surface. Although genes associated with cilia were not reported in these metacells, we found that they express genes implicated in cilia structure and function.
We used fluorescent dyes that label the content of lipophil cell granules and a fluorescent indicator for trypsin activity to monitor secretion by lipophil cells (LC) and VEC in Trichoplax while they were consuming algae. After the animal crawled onto a patch of algae and ceased moving, LC in the vicinity of algae secreted a large granule whose content lysed nearby algae within less than 2 seconds[4] Activation of the fluorescent trypsin indicator became detectable after about one minute and the fluorescence increased over the duration of the feeding episode. Detection of trypsin activity provides indirect evidence that central VEC (cVEC) secrete digestive enzymes during feeding episodes. Lysis of algae apparently is required to allow the digestive enzymes to penetrate algae because intact algae were not decomposed even when exposed to digestive enzymes in the feeding pocket.
Orthologs of the digestive enzymes that are co-expressed in Trichoplax cVEC are expressed in different combinations in zymogen gland cells in the digestive cavity of Cnidaria [43–47], the gut of invertebrate Bilateria [48–51] and in pancreatic acinar cells in mammals[52,53]. Zymogen consists of inactive precursors of digestive enzymes and is cleaved to release active enzymes [54]. Zymogen appears homogeneously electron dense or granular when examined by TEM in thin sections of cnidarian and bilaterian zymogen gland cells [43,47,51,55,56]. The secretory granules in Trichoplax cVEC are electron dense, consistent with the idea they contain zymogen. Cells expressing precursors of trypsin and chymotrypsin were found in single cell transcriptomes of the ctenophore Mnemiopsis leidyi [31]and ultrastructural studies of the digestive tracts of ctenophores revealed a type of gland cell that contains vesicles with granular content that may represent zymogen [57]. Searches in the NCBI database for genes encoding trypsin, chymotrypsin and sPLA2 in Porifera yielded only one hit: a trypsin-like serine protease in the homoscleromorphid sponge Corticum candelabrum.
Trichoplax VEC are thought to take up nutrients and digest them intracellularly because markers such as ferritin and fluorescent dextran were observed inside pinocytotic and endosomal vesicles in VEC after addition to the ambient seawater [24,40]. The gastrodermis in many animals with internal digestive systems contains absorptive cells, called enterocytes, that take up nutrients released from partially digested food via pinocytosis or phagocytosis [42]. Enterocytes in Ctenophora, Cnidaria and many Bilateria possess a cilium and microvilli, like Trichoplax VEC [57]. However, enterocytes in different animal lineages also have distinctive characteristics; for example, cnidarian enterocytes are epitheliomuscular cells [55,56]. Enterocytes in insects, nematodes, and some parts of the digestive tracts of vertebrates secrete digestive enzymes [42], as do cVEC in Trichoplax. However, enterocytes in Cnidaria and most Bilateria are not thought to secrete digestive enzymes.
Lipophil cells – a placozoan synapomorphy?
The observation that the content of granules secreted by LC during feeding episodes in Trichoplax rapidly lyses algae led us to search LC metacells for genes that might encode proteins with sequences resembling those of pore-forming proteins [59–62]. We found identical genes in TaH1 and TaH2 LC metacells that encode a protein with a signal peptide and no transmembrane domain; this protein has no orthologs in the NCBI database of non-redundant protein sequences and bears no known domains. The gene is the most highly expressed gene in TaH2 LC metacells, and the second most highly expressed gene in TaH1 LC metacells. The predicted protein is mainly composed of helical regions with positively charged areas that might interact with membranes, like the alpha-helical class of pore-forming proteins [63], nucleoporins [64], or ninjurin [65]. A protein with high sequence similarity to the TaH1/H2 genes is expressed in one of the two metacell clusters classified as LC in Hoilungia (LC2) and an ortholog (e−45) is expressed in both LC1 and LC2 metacells in Hoilungia H23. We found no other highly and uniquely expressed genes encoding secreted peptides or proteins in TaH1 and TaH2 LC metacells. We suggest that this secretory protein is packaged in LC granules and participates in permeabilizing prey organisms. A thin section from a LC revealed an apical granule with an open fusion pore, suggesting that LC granule secretion occurs by conventional exocytosis.
The outer part of the LC apical granule binds osmium, as evident by electron microscopy in thin sections from samples fixed with osmium, indicating the presence of unsaturated lipids [33]. In thin sections of LC from frozen and freeze substituted samples, the apical granules contained variable amounts of electron dense material as well as small vesicles whose content resembled cytoplasm. Granules deeper in the cell bodies of LC contained progressively less electron dense material and larger membrane enclosed vesicles as well as finger-shaped protrusions from the membrane surrounding the granule. The ultrastructure of LC apical granules resembles that of Type 1 granules in rat natural killer cells (NKC), which have an electron dense core and small vesicles near the granule membrane, while the deeper granules resemble Type 2 granules, which contain smaller volumes of electron dense material and numerous small vesicles [66–68]. Antisera against cytolysin (perforin) and serine proteases labeled the electron dense material whereas the antisera against lysosomal enzymes or a lysosomal associated membrane protein labeled the small vesicles [67]. A histochemical marker for acidic compartments showed that the content of both Type 1 and Type 2 granules was comparable in acidity to lysosomes and more acidic than nuclei or cytoplasm. Natural killer cell granules are considered secretory lysosomes or lysosome related organelle (LRO) because they possess characteristics of both secretory granules and lysosomes. The affinity of acidophilic Lysotracker dyes for LC apical granules demonstrates that their content is acidic [4,18]. Moreover, LC metacells highly express lysosomal proteins including lysosomal membrane associated protein 2 (LAMP2), hydrolases and multiple H-transporting V-type ATPases, leading us to conclude that LC granules may likewise represent a type of LRO.
The presence of lipids in LC granules is intriguing and not a feature shared with NKC granules. However, LRO in other types of cells do contain lipids. For example, mammalian epidermal cells contain LRO with lipid lamellae in their interiors that they secrete to create a moisture-retaining barrier on the surface of the skin [69]. Intestinal cells in C. elegans package lipids into LRO called “fat droplets” that are stored intracellularly and used for nutrition [70]. The precursors of epidermal lamellar bodies and intestinal fat droplets are produced in the Golgi complex and the granules acquire their lipidic content during transport to their destinations via interactions with the trans Golgi complex (lamellar bodies) or ER (fat droplets). The higher osmiophilic content of apical LC granules compared to granules closer to the Golgi complex suggests that LC granules in placozoans likewise acquire their lipid content during transport. Lipophil metacells highly express several fatty-acid binding proteins [6] that might serve as chaperones to deliver lipids to their granules. Trichoplax can lyse and consume cyanobacteria in addition to microalgae [4], indicating that the content of LC granules can penetrate both bacterial and eukaryotic membranes. The identities of the lytic components and their molecular targets in microalgae and cyanobacteria are important questions that remain to be addressed.
The morphology and secretome of LC appear unlike those of cells in the digestive systems of other animals. However, many animal lineages have evolved unique types of secretory cells that they use to capture, incapacitate, or digest prey and to defend against predators. Well documented examples include cnidarian cnidocytes [55,71–73], ctenophore colloblasts [57,72,74], and venom secreting cells in arthropods, for example [75,76].
Cells in the peripheral part of the ventral epithelium express putative antimicrobial peptides
Metacells classified as “epithelial” or “lower epithelial” in scRNAseq studies of TaH1 [6,31] based on expression of genes associated with the structure and function of cilia highly express genes with structural similarities to precursors of arminins, antimicrobial peptides found in Hydra magnipapillata endodermal cells [34,77]. Probes for the mRNA encoding two of these arminin-like genes (referred to here as AMP1 and AMP2) labeled monociliated cells in the peripheral part of ventral epithelium (pVEC). These cells had ultrastructural features like digestive cVEC but possessed darker and more numerous secretory granules. All TaH1 metacells classified as “lower epithelial” express both AMP1 and AMP2 and a third arminin-like gene, Ta 60631. Metacells classified as “lower epithelial” in TaH2 express genes that are nearly identical to Ta AMP1 and Ta 60631 and a subset of them express an ortholog of AMP2, but Hoilungia H13 and H23 “lower epithelial” metacells express only an ortholog of AMP2.
A probe for a different arminin-like Trichoplax gene, AMP3, labeled cells in a narrow zone in the ventral epithelium ~15 to 30 μm from the rim. Their positions correspond to those of cells classified as Type 1 gland cells in ultrastructural studies of Trichoplax [17]. The metacells that contain AMP3 also express genes encoding an intelectin and an uncharacterized secretory protein, Ta 63786 that were present only in this metacell cluster.
The genes encoding AMP1, AMP2, and a third arminin-like prepropeptide (Ta 60631) are the three most highly expressed genes in Trichoplax H1 “lower epithelial” metacells and the TaH2 orthologs of AMP1 and Ta 60631 are the most highly expressed genes in TaH2 “lower epithelial” metacells. Similarly, arminins are among the most highly expressed genes in most Hydra species that have been studied [77,78]. Labeling with mRNA probes for nine arminin paralogs in Hydra wholemounts showed that eight were expressed in the gastrodermis (endoderm) and one in the epidermis. Hydra vulgaris metacells containing arminins were classified as “endodermal epithelial” [79] and likely correspond to absorptive gastrodermal cells (enterocytes). Different species of Hydra express different arminins and their epithelia are inhabited by different species of bacteria [78]. Genetic knockdown of arminin expression impairs the ability of H. magnipapillata to reestablish a microbiome resembling its native microbiome following destruction of its microbiome by antibiotic treatment. Although arminins have been found only in Hydra, antimicrobial peptides have been identified in the gastrodermis of many cnidarians and bilaterians [80–82].
Two types of mucocytes arrayed in distinct patterns in the ventral epithelium
We identified metacells representing mucocytes in whole mounts and dissociated cell cultures of TaH1 based on labeling with fluorescent WGA and a probe for an oligosaccharide binding protein, Ta 63702, that was highly expressed in a single TaH1 metacell (C36) [31]. Mucocytes were abundant in the peripheral part of the ventral epithelium and more sparsely distributed further in the interior, as reported [17]. Mucocytes in the central region of the ventral epithelium, but not those in the periphery, co-expressed a gene for prepropeptide that is predicted to produce RWamide peptides. We identified metacells in TaH2 and Hoilungia H13 and H23 that express a gene identical to Ta 63702 and a subset of these metacells contain a gene encoding the RWamide prepropeptide. Although no Ta RWa gene was found in the second RNAseq analysis of TaH1 [6], we identified metacells that likely represent central and peripheral mucocytes in this dataset. All metacells representing mucocytes in the more recent RNAseq datasets contain a gene encoding an intelectin (Ta 62229) and a protein with VWD, TIL, C8 and PTS domains characteristic of gel-forming mucins [39]. The RWamide peptides and intelectin likely are packaged and secreted along with mucus since mucocytes contain only one type of secretory granule [17]. Animal mucosa often contain lectins and antimicrobial peptides [83]. However, the predicted products of the Ta RWa do not have electrostatic properties typical of antimicrobial peptides. It is possible that the peptides are used for intercellular signaling, although synthetic RWa peptides had no apparent effect on the behavior of Trichoplax when added to their culture dishes [23]
Genes encoding gel-forming mucins with VWD, TIL, C8 and PTS domains were found in single cell transcriptomes of the cnidarian Hydra vectensis and the ctenophore M. leidyi [31,84]. These metacells did not contain orthologs of the transcription factors found in metacells representing placozoan mucocytes [6]. So far, no ortholog of a gel-forming mucin has been identified in Porifera [39], although genomic sequences are available for only 13 of the 9500 known species of Porifera [85]. Mucus-like substances have been observed on epithelia of adult demosponges [86] and cells resembling mucocytes were found by electron microscopy in thin sections of metamorphosing larvae of several classes of sponges at a stage when the larvae secrete substances that mediate attachment [87–89] Moreover, genes encoding VWD, C8 and PTS (but not TIL) domains have been found in two species of choanoflagellates (Salpingoeca rosetta and M. brevis), the eukaryotes that are the closest sister to Metazoa [90], although whether the products of these genes are mucins is uncertain [91]. The distribution of mucussecreting cells across animal phyla and presence of mucus-like genes in Choanoflagellates suggests that the common ancestor of Metazoa may have possessed cells that secreted a mucus-like substance.
Dorsal epithelial cells secrete lectins implicated in defense
Cells with morphological characteristics of DEC were labeled with a probe for mRNA encoding the ELPE prepropeptide and many of them were labeled with a probe for an intelectin, Ta 60661. The TaH1 metacells expressing these genes were classified as “epithelial” or “upper epithelial” [6,31] and include genes encoding ten intelectins that are not expressed in any other metacells. Trichoplax H2 “upper epithelial” metacells contain identical intelectin genes. Hoilungia 13 “upper epithelial” metacells contain nearly identical orthologs of five of them and H23 “upper epithelial” metacells contain four of them. These intelectins likely are packed inside the electron dense granules in DEC, like intelectins in goblet cells in mammalian intestine [92]. These metacells also express a gene encoding a membrane associated mucin 4-like glycoprotein that may be a component of the glycocalyx observed on the apical surfaces of DEC by electron microscopy.
Genes encoding intelectins have been found in members of all metazoan phyla except Ctenophora [38,93]. However, the distributions of intelectins in different lineages are patchy. Among vertebrates, zebrafish, humans and mice have them but birds and dogs do not [93]. The pattern of expression across tissues varies between species. Expression of intelectins is most observed in the intestine of vertebrates, although expression also has been found in other tissues such as heart and liver. The first discovered intelectin-like gene was found in oocytes of Xenopus laevis and named “oocyte lectin” [94]. In Amphioxus, intelectins are expressed in the intestine and epidermis [93]. Intelectins in chordates have been implicated in multiple processes including prevention of polyspermy, activation of immune responses, and defense. Intelectins are thought to protect epithelia from invasion by bacteria by binding bacterial glycans and aggregating bacterial cells. Less is known about the prevalence and functions of intelectins in non-chordates, although the available evidence suggests that the primary function of FReD-containing proteins in invertebrates is defense [38,95]. Twenty-eight genes containing FReD domains were identified in the cnidarian H. vectensis and more than ninety were identified in the poriferan Amphimedon queenslandica [38]. We found genes with FReD in N. vectensis scRNAseq metacells classified as “neuro/gland” and “cnidocytes” and in A. queenslandica metacells classified as “pinacocytes” and “archeocytes” [31]. A FReD containing protein in the demosponge Suberites domuncula was detected in the pinacoderm in animals that were incubated with a polysaccharide that is present in the cell walls of fungi and bacteria, but not in animals that were not exposed to the polysaccharide[96], suggesting intelectins may function in defense in S. domuncula. Although many animals, including Trichoplax, have intracellular endosymbionts [16,97], bacteria are rarely observed in the interstitial space in morphological studies of Ctenophora, Placozoa, Cnidaria and Bilateria indicating that they have effective mechanisms for deterring microbial invaders. By contrast, the extracellular matrices in the interiors of many species of Porifera harbor large numbers of bacteria[98,99].
Peptidergic cell types
We mapped the positions and studied the morphology of cells expressing seven of the fourteen prepropeptides identified by scRNAseq in TaH1 [6]. Peptides that are the predicted/possible products of the six of these prepropeptides (WPPF, ELPE, SIFGa, FFNPa, LF, TaELP) elicited changes in the behavior of Trichoplax when added to their culture dishes[22,23]. We anticipated that the positions of the peptidergic cells and the responses elicited by peptides they secrete might provide clues about their functions.
Cells that expressed the Ta SIFGa prepropeptide were most prevalent in a narrow zone near the rim of the dorsal and ventral epithelium but were also present in more central regions of the epithelia. The Ta SIFG+ cells in the central part of the ventral epithelium co-expressed the Ta FFNPa peptide precursor. Addition of low concentrations (<500 nM) of SIFGa to dishes containing Trichoplax rapidly (<1 min) elicited contraction of the entire animal. Higher concentrations of the peptide caused the animal to fold up and detach from the substrate[22,23] The peptide FFNPamide elicited an expansion of the area of the animal and an increase in the frequency of spontaneous pauses in movement. The latencies of the responses to FFNPa were longer (> 7 minutes) than those of the contractions elicited by SIFGamide, suggesting that simultaneous secretion of SIFGa and FFNPa by cells that co-express these peptides might elicit contraction followed by relaxation. Detachment from the substrate elicited by secretion of endogenous SIFGa may allow the animal to move between the substrate and air/water interface, as animals maintained in culture often do. The location of the cells that co-expressed Ta SIFGa and Ta FFNPa suggests that they may participate in orchestrating behaviors that occur during feeding.
Cells expressing the gene for LF prepropeptide were in a narrow zone near the rim of the dorsal epithelium and distributed throughout the central part of the ventral epithelium. The Ta LF+ cells in the ventral epithelium co-expressed a secreted astacin-like metalloendopeptidase. Applying LF to Trichoplax elicited a large expansion in the animals’ area. In addition, animals that were translocating stopped and rotated in place [23], indicating a change in the collective behavior of the monociliated VEC that propel crawling. Secretion of endogenous LF by cells located at the rim could promote adhesion of the rim to the substrate by relaxing the contractile cytoskeletons of cells in the rim. The location of the cells that co-expressed Ta LF and astacin Ta 26557 suggests that they may have roles in feeding.
Cells co-expressing the TaELP prepropeptide and an astacin-like metalloendopeptidase were in the ventral epithelium near the rim. Their locations did not correspond to those of cells labeled with antiserum against TaELP prepropeptide or against the peptide YPFFamide (human endomorphin 2), which instead labeled mucocytes[17,22]. We previously reported that adding YPFFamide or QDYPFFamide to a dish containing moving Trichoplax consistently caused them to stop moving and arrested ciliary beating, but the predicted products of the TaELP prepropeptide (QDYPFFGN or pQDYPFFGN) arrested movement of fewer than half of the animals tested [22]. The observation that antiserum against YPFFamide did not label the cells that express TaELP prepropeptide suggests that these cells do not produce the peptide QDYPFFamide and casts doubt on the idea that the function of these cells is to detect food and arrest movement during feeding episodes[22]
The Ta ELPE prepropeptide was expressed in a large fraction of DEC and by cells near the rim of the ventral epithelium. Cells expressing the Ta WPPF prepropeptide were interspersed among DEC in the central part of the dorsal epithelium. Bath application of ELPE or WPPF were reported to elicit behaviors reminiscent of behaviors observed in animals feeding on biofilms: periodic cessation of gliding accompanied by churning movements of cells in central regions of the ventral epithelium[23]. The ELPE peptide elicited a small expansion in the animals’ area, comparable to that observed in animals pausing to feed, whereas the WPPF peptide elicited a much larger expansion. The positions of the cells that express Ta ELPE or Ta WPPF make it unlikely that their primary function is to initiate feeding behaviors.
The cells that expressed TaELP prepropeptide and the central population of Ta LF prepropeptide expressing cells co-express different astacin-like metalloendopeptidases. Astacins are widespread among animals and have been implicated in a wide variety of physiological processes, including digestion [100,101] and development [102,103], as well as breakdown of mucus, which supports adhesion to the substrate in aquatic animals [104,105]. Astacin secreting cells in placozoans could be functionally antagonistic to mucocytes. The mucocytes in the rim of the ventral epithelium are close to the cells that co-express an astacin and TaELP prepropeptide while mucocytes in the central part of the ventral epithelium are interspersed among cells that co-express an astacin and Ta LF prepropeptide.
Multiple secretory peptides have been identified by bioinformatics and mass spectroscopy in representatives of all metazoan phyla except Porifera [6,29,35,36,106–108]. Sequences encoding the precursors of phoenixin and nesfatin-like peptides were reported to be present in representatives of all metazoan phyla except Placozoa as well as in the choanoflagellate S. rosetta [109]. However, no phoenixin and nesfatin-like peptides were detected by mass spectroscopy in the poriferan Ephydatia fluvitilis or the ctenophore Bolinopsis mikado [106]. Although Porifera have genes implicated in processing precursors of secretory peptides (prohormone convertase, PAM, carboxypeptidases) [111], peptidergic signaling has not yet been definitively demonstrated in this phylum.
Conclusions
Placozoa provide an opportunity to study the biology of animals whose body plans and lifestyles may resemble those of the ancestors of the complex animals that now inhabit the earth [112–117]. The availability of single cell transcriptomes of four placozoan species [6,31] provides a rich resource for research aimed at understanding the functions and evolutionary histories of placozoan cell types. We built upon this foundation by associating placozoan single cell transcriptomes with previously described morphological cell types and identifying functions of secretory products they express. Placozoan secretory cell types share morphological features with cell types found in representatives of other metazoan phyla and some of them express orthologs or analogs of functionally characterized secretory proteins/peptides. Lipophil cell granules, which secrete lytic substances, morphologically resemble granules in mammalian natural killer cells. Placozoan VEC resemble enterocytes in cnidarians and many bilaterians in that they have an apical cilium and multiple microvilli and take up nutrients by endocytosis. The VEC in the central part of the animal express digestive enzymes, as do zymogen gland cells in cnidarians and bilaterians, while VEC in the periphery express putative AMPs, as do gastrodermal cells in many animals. Placozoan DEC bear morphological resemblances to poriferan pinacocytes and cnidarian epidermal cells and express glycoproteins implicated in defense against pathogens in poriferans, cnidarians, invertebrate bilaterians and chordates. Placozoan mucocytes contain electron lucent secretory granules like mucocytes in ctenophores, cnidarians and bilaterians, and express orthologs of a gel-forming mucin, a major component of mucus. Placozoans have multiple peptidergic cell types. Comparison of the gene expression profiles of placozoan peptidergic cells with those of cnidarian and bilaterian neurons revealed global transcriptional similarities and gene gains that accompanied evolution of nervous systems in Eumetazoa[6]. Similar analyses across metazoan phyla of transcriptional profiles of other functionally analogous cell types can be expected to provide further insights into the evolution of metazoan cell type programs.
Methods
Animals
Trichoplax adhaerens (Schultze, 1883) of the Grell (1971) strain, a gift from Leo Buss (Yale University), were kept in Petri dishes with artificial seawater (ASW; Instant Ocean, Blacksburg, VA, USA) supplemented with 1% Micro Algae Grow (Florida Aqua Farms, Dade City, FL, USA) and red algae (Rhodomonas salina, Provasoli-Guillard National Center for Culture of Marine Plankton, East Boothbay, ME, USA), as described previously [18]. Water was partially changed once a week.
Cell dissociation
To prepare dissociated cells, a group of animals was rinsed in calcium and magnesium free ASW (calcium-free ASW; CSP Prot) and then incubated in 0.25% trypsin in calcium-free ASW for 2 h. The animals were transferred to normal ASW and triturated with a glass Pasteur pipette until the suspension was homogeneous.
Fluorescence in situ hybridization
In situ hybridization was performed with probes and reagents from Advanced Cell Diagnostics, Inc. (ACD, Hayward, CA, USA) using protocols developed to optimize staining in Trichoplax. The RNA sequences were retrieved from PubMed or from a T. adhaerens transcriptome database, access to which we were granted by Adriano Senatore (University of Toronto). RNAscope probes for multiplex fluorescence in situ hybridization were designed by ACD. Catalog names and numbers, gene names, and annotations, and mRNA accession numbers are listed in Table 1. Further details are available in the ACD product catalog.
Table 1.
RNAscope probes for multiplex fluorescence in situ hybridization.
| RNAscope probe | Catalog # | Gene | Annotation | Accession number |
|---|---|---|---|---|
|
| ||||
| Ta-57870-C2 | 561141-C2 | Ta-57870 | PLA2 | XM_0021139 |
| Ta-63128 | 561031 | Ta-63128 | Trypsin | XM_0021155 |
| Ta-Chymotrypsin-C2 | 572831-C2 | Ta-63088 | Chymotrypsin | XM_0021092 |
| Ta-PhospholipaseA2 | 572841 | Ta-63140 | PLA2 | XM_0021162 |
| Ta-57870-C2 | 561141-C2 | Ta-57870 | PLA2 | XM_0021139 |
| Ta-56030-C2 | 56030-C2 | Ta-56030 | AMP2 | XM_0021116 |
| Ta-55945-C3 | 55945-C3 | Ta-55945 | AMP1 | XM_0021116 |
| Ta-64402-C2 | 1313051-C2 | Ta- 64402 | AMP3 | XM_0021180 |
| Ta-58643-C1 | 1138211-C1 | Ta-58643 | Tetraspanin | XM_0021147 |
| Ta-63786 | 1209181-C1 | Ta-63786 | Ta Sec Prot | XM_0021105 |
| Ta-64037-C2 | 1222681-C2 | Ta-64037 | Ta Sec Prot | XM_0021134 |
| Ta-63996-C2 | 1138221-C2 | Ta-63996 | Ta Sec Prot | XM_0021135 |
| Ta-29105-C3 | 1138231-C3 | Ta-29105 | Tansporter | XM_0021153 |
| Ta-63702-C3 | 823661-C3 | Ta-63702 | Oligosac BP | XM_0021101 |
| Ta-60185-C2 | 487491-C2 | Ta-60185 | SIFGamide Pre | XM_0021161 |
| None | 856541-C2 | EST | RWamide Pre | GR951923.1 |
| Ta-63942-C2 | 823731-C2 | Ta-63942 | FFNPamide pre | XM_0021127 |
| Ta-51275 | 561121 | Ta-51275 | LF Pre | XM_0021083 |
| Ta-51275-C3 | 561121-C3 | Ta-51275 | LF Pre | XM_0021083 |
| Ta-YPFFamide-O1-C2 | 1228931-C2 | AQX36197.1 | TaELP Pre | KY675296.1 |
| Ta-56359 | 561051 | Ta- 56359 | WPPF Pre | XM_0021123 |
| Ta-60185 | 487491 | Ta-60185 | SIFGa Pre | XM_0021161 |
| Ta-64280-C3 | 8236681-C3 | Ta-64280 | ELPE Pre | XM_0021163 Tricho evg15 |
| Ta-26557-C3 | 561111-C3 | Ta-26557 | Astacin | 92 |
| Ta-54934 | 560991 | Ta-54934 | Astacin | XM_0021106 |
| Ta-60661-C3 | 1241961-C3 | Ta-60661 | Intelectin | XM_0021166 |
| Ta-60661-C2 | 1217761-C2 | Ta-60661 | Intelectin | XM_0021166 |
| 3-Plex Neg. Control | 320871 | |||
Abbreviations: Ta – Triaddraft-; PLA2 – PhopholipaseA2; AMP– Putative antimicrobial peptide; Ta Sec Prot – Predicted T. adhaerens secretory protein; Oligosac BP – Oligosaccharide binding protein; Pre – Prepropeptide
Animals and dissociated cells were transferred to silanized cover slips or Superfrost Plus Gold glass slides (Thermo Fisher Scientific, Pittsburgh, PA, USA) with a drop of ASW mixed 1:1 with 0.97 M mannitol (in water). Samples were left on cover slips/slides to adhere for about 2 h. Then the liquid was blotted, and the cover slips/slides were plunged into prechilled tetrahydrofuran on dry ice and kept overnight. Wholemount samples were transferred to 3% acetic acid in methanol at − 20 °C for 30 min followed by a mixture of formalin and methanol (1:10), initially at − 20 °C and then at room temperature (RT) for 30 min. The wholemount samples were rinsed twice in methanol, dried for 5 min and then treated with Protease IV for 30 min. The dissociated cell samples were transferred directly to a mixture of formalin and methanol (1:10), initially at − 20°C and then at RT for 30 min. The cell samples were then rinsed once with methanol, twice with ethanol followed by descending concentrations of ethanol in PBS (70% and 50%) and PBS. The cell samples were treated with Protease III diluted 1:15 in PBS for 15 min. Hybridization was performed with RNAscope Fluorescent Multiplex Reagent Kit (# 320850) according to supplier’s instructions. The negative control 3-Plex Negative Control Probe gave no labelling. Samples were counterstained with 1:200 wheat germ agglutinin (WGA) conjugated to Alexa 647 (# W32466, Thermo Fisher Scientific, Waltham, MA) or CF405M (# 29028, Biotium, Hayward, CA, USA) and/or DAPI, and mounted in ProLong™ Gold antifade reagent (# P36934, Invitrogen, Eugene, OR, USA). Dissociated cell preparations were also subjected to immunolabelling for tubulin after RNAscope steps to visualize cilia. For that, samples rinsed with wash buffer after the last (AMP4) hybridization step were further washed twice with PBS (pH7.4) and once with blocking buffer (BB, 3% goat serum, 2% horse serum, 1% BSA in PBS). Then, primary anti-acetylated tubulin mouse antibody (#T7451; Sigma) diluted 1:500 in BB was applied. After overnight incubation with the primary antibody, samples were washed with PBS and incubated in secondary Atto-655 goat anti-mouse antibody (#50283; Sigma) or Alexa 555 goat anti-mouse (#A21422; ThermoFisher) diluted 1:200 in blocking buffer. Finally, samples were rinsed from secondary antibody with PBS, briefly incubated in DAPI, and mounted in ProLong™ Gold antifade reagent (ThermoFisher). The samples were examined with a Plan-Apochromat 40X NA 1.3 or 63X NA 1.4 Plan-Apochromat objective on a LSM 800 or LSM 880 AIry laser scanning confocal microscope with DIC and fluorescence optics (Carl Zeiss Microscopy, LLC). The areas of dissociated cells visualized with DIC optics were estimated by measuring the length of the long axis of the cell body and the perpendicular axis and calculated using the equation for an ellipse. Cells were considered labeled if they possessed at least two fluorescent grains. To demonstrate expression patterns in wholemounts, we created xy maximum intensity projections in Zen software (Carl Zeiss Microscopy, LLC). In order to show the distribution of a signal across the animal thickness, we made xz projections for selected strip regions, indicated on the xy images with a narrow box. Labelling with each probe was done at least twice; the results of independently repeated experiments were similar.
Time-Lapse Microscopy of Living Animals
To monitor secretion of lipophil cell granules and trypsin during feeding episodes, animals first were transferred to a 35 mm petri dish containing ASW and kept for one to three hours and then incubated for 10 minutes in ASW containing FM1–43 (N-(3-Triethylammoniumpropyl)-4-(4-(Dibutylamino) Styryl) Pyridinium Dibromide; Thermo Fisher; #T3163; 1μg/ml) and LipidTOX (HCS LipidTOX Deep Red neutral lipid stain; Thermo Fisher; #34477; 3 μl/ml). The animals were transferred to a RC-40LP chamber containing ASW with FM1–43, LipidTOX, BZiPAR (Rhodamine 110, bis-(N-CBZ-L-isoleucyl-L-prolyl-L-arginine amide), dihydrochloride; Biotium; #10208; 8 μM) and Rhodamonas salina algae. The behavior of the animals was monitored with a 32-channel spectral detector and a 10X NA 0.45 Plan-Apochromat objective on a LSM880 confocal microscope with 488 nm, 561 nm and 647 nm illumination. Images (512 X 512 bits) were captured at ~1.7 frames/second. Reference spectra used for linear unmixing were collected from the following samples: (1) FM1–43, ventral epithelium of an animal labeled with FM1–43; (2) LipidTOX, lipophil cell granule in an animal labeled with LipidTOX; (3) algae; autofluorescence; (4) BZiPAR; seawater containing BZiPAR and 0.25% trypsin. Bicubic interpolation was used to enlarge the images and reduce pixilation. Still images used for illustration in Fig 8 were created by averaging two successive frames to improve signal. Fiji software was used to build fluorescence profiles shown in Fig 8.
Electron microscopy
For transmission electron microscopy (TEM), scanning electron microscopy (SEM), and electron microscope tomography animals were high pressure frozen, freeze-substituted and embedded as described previously [18]. WGA-nanogold (EY Laboratories, San Mateo, CA, USA) labelled thin sections were prepared as described[17]
For transmission electron microscopy, the ultrathin sections were observed in a JEOL 200-CX (JEOL, Japan) at 120–200 kV and were imaged with an AMT camera mounted below the microscope column. Image processing and color intensity measurement was done with Fiji.
The protocol for serial sectioning followed by SEM imaging in backscatter mode was published elsewhere [17]. Series of these images were used to study the nature of lipophil granules protrusions and to count granules in the pVEC and cVEC.
For EM tomography, we used ~100 nm sections and a transmission electron microscope, JEOL 1400 (JEOL, Japan) at an accelerating voltage of 120kV. Images were taken at ~1° tilt intervals from −69° to 69° at a single tilt axis at the magnification x5000. The tilt images were aligned using an electron tomography software package, EM3D (em3d.org), upgraded by Jae Hoon Jung and Eun Jin Choi in Dr. Reese’s laboratory, and reconstructed to generate a reconstructed volume or tomogram by Simultaneous Iterative Reconstruction Technique (SIRT) with 10 iterations using the algorithm implemented in a tomography program, called VEM (Volume Electron Microscopy), developed by Jae Hoon Jung and Eun Jin Choi. The examination of the virtual slices was carried out using both EM3D and VEM. The virtual slices through the tomogram were one-voxel thick, which is 1.8 nm.
To prepare freeze fractured samples, T. adhaerens were placed on the surface of a gelatin cushion and directly frozen by contact with a sapphire disk glued to the surface of a copper block cooled to −186 °C using a LifeCell CF-100 slam-freezing machine. Samples were transferred to a Balzer freeze-fracture apparatus. Replicas were prepared by freeze-fracturing the specimen at −110 °C and at a vacuum of 10−7 Torr, rotary shadowing with platinum and carbon, and backing the replica with carbon. The replicas were cleaned with sodium hypochlorite, mounted on grids, and examined using a JEOL 200-CX at 120 kV.
Protein/peptide predictions
Deep TMHMM [118] was used to predict signal peptides and transmembrane alpha helices. InterPro online tool [119]was used to predict protein topology and conserved domains. AlphaFold protein database [120] was used to predict secondary structures of the Trichoplax proteins. ChimeraX-1.5 [121,122]was used to render a secondary structure of the proteins and color protein surface by electrostatic potential.
Regions of prepropeptides were annotated and classified into signal peptide region using SignalP6.0 [https://pubmed.ncbi.nlm.nih.gov/34980915/], prohormone convertase cleavage sites (dibasic residues), mature peptide (based on characteristics of cnidarian neuropeptide processing [123]. C-terminal glycine residues were predicted to be processed into amide-groups, and N-terminal glutamates were predicted to be processed into pyroglutamates.
Quantitative image processing and Statistics
We used Paleontological Statistics (PaSt), freely available software [124], to do statistical comparisons and build boxplots. Mann-Whitney test was applied, since many variables did not follow the Gaussian distribution. For multiple samples, we used an ANOVA test followed by Mann-Whitney post hoc pairwise test with the Bonferroni correction. In boxplots, the box represents 25–75 quartiles, and the median is shown with a horizontal line inside the box. “Whiskers” show the minimal and maximal values excluding outliers. Each value is plotted as a dot on top of a boxplot. Accurate area-proportional Venn diagrams were drawn using eulerAPE program [125] or PaSt.
Gene expression profiles
The data for Fig 1 was downloaded from [31]. The fold change and percentage expression were extracted for the selected genes across all the metacells and visualized using the tidyverse package in R. Gene expression heatmaps were generated from [https://sebelab.crg.eu/placozoa_cell_atlas/] using orthologous genes from H1, H2, H13, and H23. The genes and meta cells were manually annotated using InkScape.
Acknowledgements
We thank the Electron Microscopy Facility of the National Institute of Neurological Disorders and Stroke, the Central Microscopy Facility of the Marine Biological Laboratory, Christine A. Winters (NINDS) and Katherine Hammar (MBL) for sample preparations and technical support. We thank Professor Adriano Senatore for valuable comments on the manuscript. This work was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, United States of America.
Footnotes
S1 Fig. Secondary structure predictions and electrostatic potential maps for putative Trichoplax AMPs (AMP1 (A), AMP2 (B), and AMP3 (C) and Hydra vulgaris arminin (D). Secondary structures are predicted with Alpha Fold and peptide surfaces are colored by electrostatic potential with ChimeraX.
S2 Fig. Secretory behaviors of Trichoplax feeding on R. salina algae. Lipophil cell granules were labeled with LipidTOX (red). The fluorescent membrane dye FM1–43 (cyan) was added to the seawater to label cell membranes and the contents of LC granules. BZiPAR, a fluorescent indicator of trypsin activity (green), was added to detect secretion of trypsin. Algae are visible by autofluorescence and FM1–43 staining (pink in merged images). At the beginning of the sequence, the peripheral part of the animal was closely attached to the substrate, while the central part was invaginated, forming a feeding pocket enclosing algae. At t=0 sec, lipophil granule secretion was evident due to the appearance of small FM1–43-stained spots (cyan/white) in the feeding pocket. At t=12 sec, several algae (pink) in the feeding pocket were lysed and released material stained by FM1–43 (cyan). By 150 – 287 sec, trypsin activity was evident in the feeding pocket and the lysed algae were decomposing. Bottom panels show intensity-coded images of BZiPAR trypsin activity indicator at sequential timepoints for the boxed region.
S3–6 Fig. Normalized expression of selected genes across metacells for TaH1, TaH2, Hoilungia H13, and Hoilungia H23. Cell types are identified based on data from the present study and [6,25,31,126]. Expression data from: https://sebelab.crg.eu/placozoa_cell_atlas/
S7 Text. Placozoan prepropeptides, predicted cleavage sites and processing motifs. Underlined regions represent predicted signal peptides. Highlighted yellow represents the mature peptide region, green represents prohormone convertase cleavage sites, cyan represents pyroglutamate, and magenta represents C-terminal amide.
S8 Movie. Trichoplax feeding on R. salina algae (same animal as illustrated in Fig 8. Lipophil cell granules were labeled with LipidTOX (red). The fluorescent membrane dye FM1–43 (cyan) was added to the seawater to label cell membranes and the contents of LC granules. BZiPAR, a fluorescent indicator of trypsin activity, was added to detect secretion of trypsin. Algae are visible by autofluorescence (blue). Time stamp is at upper left. Lipophil cell granule secretion begins at ~50 sec as evident from the appearance of small FM1–43-stained puncta in the vicinity of algae. Green fluorescence generated by proteolysis of BZiPAR becomes visible at ~110 sec and reaches a peak at ~270 sec. Content of lysed algae is stained by FM1–43, but staining declines as the algae decompose. The rim of the animal remains closely attached to the substrate forming a feeding pocket that confines the contents of the lysed algae and the BZiPAR fluorescence.
S9 Movie. Trichoplax feeding on R. salina algae (same animal as illustrated in S2 Fig). Staining is the same as in S8 Movie. Lipophil granule secretion begins at ~14 sec as evident from the appearance of small clouds of FM1–43-stained material in the vicinity of algae. The clouds coalesce into puncta within seconds. Algae begin to lyse at ~20 sec. Trypsin activity becomes visible at ~45 sec.
References
- 1.Grell KG, Ruthmann A. Placozoa. In: Harrison FW, Westfall JA, editors. Microscopic Anatomy of Invertebrates. New York: Wiley-Liss; 1991. pp. 13–27. Available: http://download.springer.com/static/pdf/813/art%3A10.1007%2Fs00427-004-0390-8.pdf?auth66=1363711205_d5c9623bde6301d7ab57239eaefe9a93&ext=.pdf [Google Scholar]
- 2.Pearse VB, Voigt O. Field biology of placozoans (Trichoplax): distribution, diversity, biotic interactions. Integr Comp Biol. 2007;47: 677–92. doi: 10.1093/icb/icm015 [DOI] [PubMed] [Google Scholar]
- 3.Eitel M, Schierwater B. The phylogeography of the Placozoa suggests a taxon-rich phylum in tropical and subtropical waters. Mol Ecol. 2010;19: 2315–27. doi: 10.1111/j.1365-294X.2010.04617.x [DOI] [PubMed] [Google Scholar]
- 4.Smith CL, Pivovarova N, Reese TS. Coordinated feeding behavior in trichoplax, an animal without synapses. Steele RE, editor. PLoS One. 2015;10: e0136098. doi: 10.1371/journal.pone.0136098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454: 955–60. doi: 10.1038/nature07191 [DOI] [PubMed] [Google Scholar]
- 6.Najle SR, Grau-Bové X, Elek A, Navarrete C, Cianferoni D, Chiva C, et al. Stepwise emergence of the neuronal gene expression program in early animal evolution. Cell. 2023;186: 4676–4693.e29. doi: 10.1016/j.cell.2023.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schierwater B, DeSalle R. Placozoa. Current Biology. 2018;28: R97–R98. doi: 10.1016/J.CUB.2017.11.042 [DOI] [PubMed] [Google Scholar]
- 8.Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510: 109–114. doi: 10.1038/nature13400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pisani D, Pett W, Dohrmann M, Feuda R, Rota-Stabelli O, Philippe H, et al. Genomic data do not support comb jellies as the sister group to all other animals. Proceedings of the National Academy of Sciences. 2015;112: 201518127. doi: 10.1073/pnas.1518127112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan JF, Pang K, Schnitzler CE, Nguyen A-D, Moreland RT, Simmons DK, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342: 1242592. doi: 10.1126/science.1242592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz DT, Haddock SHD, Bredeson J V., Green RE, Simakov O, Rokhsar DS. Ancient gene linkages support ctenophores as sister to other animals. Nature. 2023;618: 110–117. doi: 10.1038/s41586-023-05936-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simion P, Philippe H, Baurain D, Jager M, Richter DJ, Di Franco A, et al. A Large and Consistent Phylogenomic Dataset Supports Sponges as the Sister Group to All Other Animals. Current Biology. 2017;27: 958–967. doi: 10.1016/j.cub.2017.02.031 [DOI] [PubMed] [Google Scholar]
- 13.Tessler M, Neumann JS, Kamm K, Osigus HJ, Eshel G, Narechania A, et al. Phylogenomics and the first higher taxonomy of Placozoa, an ancient and enigmatic animal phylum. Front Ecol Evol. 2022;10: 1016357. doi: 10.3389/FEVO.2022.1016357/BIBTEX [DOI] [Google Scholar]
- 14.Thiemann M, Ruthmann A. Alternative modes of asexual reproduction in Trichoplax adhaerens (Placozoa). Zoomorphology. 1991;110: 165–174. doi: 10.1007/BF01632872 [DOI] [Google Scholar]
- 15.Signorovitch AY, Dellaporta SL, Buss LW. Molecular signatures for sex in the Placozoa. Proc Natl Acad Sci U S A. 2005;102: 15518–22. doi: 10.1073/pnas.0504031102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruber-Vodicka HR, Leisch N, Kleiner M, Hinzke T, Liebeke M, McFall-Ngai M, et al. Two intracellular and cell type-specific bacterial symbionts in the placozoan Trichoplax H2. Nature Microbiology 2019 4:9. 2019;4: 1465–1474. doi: 10.1038/s41564-019-0475-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayorova TD, Hammar K, Winters CA, Reese TS, Smith CL. The ventral epithelium of trichoplax adhaerens deploys in distinct patterns cells that secrete digestive enzymes, mucus or diverse neuropeptides. Biol Open. 2019;8. doi: 10.1242/bio.045674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith CL, Varoqueaux F, Kittelmann M, Azzam RN, Cooper B, Winters CA, et al. Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Current Biology. 2014;24: 1565–1572. doi: 10.1016/j.cub.2014.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romanova DY, Varoqueaux F, Daraspe J, Nikitin MA, Eitel M, Fasshauer D, et al. Hidden cell diversity in Placozoa: ultrastructural insights from Hoilungia hongkongensis. Cell Tissue Res. 2021;385: 623–637. doi: 10.1007/S00441-021-03459-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eitel M, Francis WR, Varoqueaux F, Daraspe J, Osigus H-J, Krebs S, et al. Comparative genomics and the nature of placozoan species. Tyler-Smith C, editor. PLoS Biol. 2018;16: e2005359. doi: 10.1371/journal.pbio.2005359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bull MS, Prakash VN, Prakash M. Ciliary flocking and emergent instabilities enable collective agility in a non-neuromuscular animal. 2021. [cited 20 May 2024]. Available: https://arxiv.org/abs/2107.02934v1
- 22.Senatore A, Reese TS, Smith CL. Neuropeptidergic integration of behavior in Trichoplax adhaerens, an animal without synapses. Journal of Experimental Biology. 2017;220: 3381–3390. doi: 10.1242/JEB.162396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varoqueaux F, Williams EA, Grandemange S, Schierwater B, Jeke, Fasshauer Correspondence D. High Cell Diversity and Complex Peptidergic Signaling Underlie Placozoan Behavior. Current Biology. 2018;28: 3495–3501. doi: 10.1016/j.cub.2018.08.067 [DOI] [PubMed] [Google Scholar]
- 24.Smith CL, Reese TS. Adherens junctions modulate diffusion between epithelial cells in Trichoplax adhaerens. Biological Bulletin. 2016;231. doi: 10.1086/691069 [DOI] [PubMed] [Google Scholar]
- 25.Mayorova TD, Hammar K, Jung JH, Aronova MA, Zhang G, Winters CA, et al. Placozoan fiber cells: mediators of innate immunity and participants in wound healing. Sci Rep. 2021;11: 23343. doi: 10.1038/S41598-021-02735-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayorova TD, Smith CL, Hammar K, Winters CA, Pivovarova NB, Aronova MA, et al. Cells containing aragonite crystals mediate responses to gravity in Trichoplax adhaerens (Placozoa), an animal lacking neurons and synapses. Steele RE, editor. PLoS One. 2018;13: e0190905. doi: 10.1371/journal.pone.0190905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong YY, Le P, Elkhatib W, Piekut T, Senatore A. Transcriptome profiling of Trichoplax adhaerens highlights its digestive epithelium and a rich set of genes for fast electrogenic and slow neuromodulatory cellular signaling. 2019. doi: 10.21203/RS.2.14504/V1 [DOI] [Google Scholar]
- 28.Ringrose JH, van den Toorn HWPP, Eitel M, Post H, Neerincx P, Schierwater B, et al. Deep proteome profiling of Trichoplax adhaerens reveals remarkable features at the origin of metazoan multicellularity. Nat Commun. 2013;4: 1408. doi: 10.1038/ncomms2424 [DOI] [PubMed] [Google Scholar]
- 29.Jékely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci U S A. 2013;110: 8702–7. doi: 10.1073/pnas.1221833110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikitin M. Bioinformatic prediction of Trichoplax adhaerens regulatory peptides. Gen Comp Endocrinol. 2014;212: 145–155. doi: 10.1016/j.ygcen.2014.03.049 [DOI] [PubMed] [Google Scholar]
- 31.Sebé-Pedrós A, Chomsky E, Pang K, Lara-Astiaso D, Gaiti F, Mukamel Z, et al. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat Ecol Evol. 2018;2: 1176–1188. doi: 10.1038/s41559-018-0575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayorova TD, Hammar K, Winters CA, Reese TS, Smith CL. The ventral epithelium of trichoplax adhaerens deploys in distinct patterns cells that secrete digestive enzymes, mucus or diverse neuropeptides. Biol Open. 2019;8. doi: 10.1242/bio.045674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belazi D, Solé-Domènech S, Johansson B, Schalling M, Sjövall P. Chemical analysis of osmium tetroxide staining in adipose tissue using imaging ToF-SIMS. Histochem Cell Biol. 2009;132: 105–115. doi: 10.1007/S00418-009-0587-Z/FIGURES/8 [DOI] [PubMed] [Google Scholar]
- 34.Augustin R, Anton-Erxleben F, Jungnickel S, Hemmrich G, Spudy B, Podschun R, et al. Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob Agents Chemother. 2009;53: 5245–5250. doi: 10.1128/AAC.00826-09/ASSET/DDE78552-AE28-472D-8CBE-1EE103DCACEB/ASSETS/GRAPHIC/ZAC0120985750004.JPEG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch TL. Molecular Evolution of Peptide Signaling in Placozoa and Cnidaria. Ph. D., University of Copenhagen. 2020. [Google Scholar]
- 36.Hauser F, Koch TL, Grimmelikhuijzen CJP. Review: The evolution of peptidergic signaling in Cnidaria and Placozoa, including a comparison with Bilateria. Front Endocrinol (Lausanne). 2022;13: 973862. doi: 10.3389/FENDO.2022.973862/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamm K, Schierwater B, DeSalle R. Innate immunity in the simplest animals - placozoans. BMC Genomics. 2019;20: 5. doi: 10.1186/s12864-018-5377-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doolittle RF. Structural and Functional Diversity of Fibrinogen-Related Domains. The Evolution of the Immune System: Conservation and Diversification. 2016; 275–294. doi: 10.1016/B978-0-12-801975-7.00011-6 [DOI] [Google Scholar]
- 39.Lang T, Hansson GC, Samuelsson T. Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci U S A. 2007;104: 16209–14. doi: 10.1073/pnas.0705984104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruthmann A, Behrendt G, Wahl R. The ventral epithelium of Trichoplax adhaerens (Placozoa): Cytoskeletal structures, cell contacts and endocytosis. Zoomorphology. 1986;106: 115–122. doi: 10.1007/BF00312113 [DOI] [Google Scholar]
- 41.Leytus SP, Melhado LL, Mangel WF. Rhodamine-based compounds as fluorogenic substrates for serine proteinases. Biochemical Journal. 1983;209: 299–307. doi: 10.1042/BJ2090299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartenstein V, Martinez P. Phagocytosis in cellular defense and nutrition: a food-centered approach to the evolution of macrophages. Cell and Tissue Research 2019 377:3. 2019;377: 527–547. doi: 10.1007/S00441-019-03096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babonis LS, Ryan JF, Enjolras C, Martindale MQ. Genomic analysis of the tryptome reveals molecular mechanisms of gland cell evolution. Evodevo. 2019;10. doi: 10.1186/S13227-019-0138-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chari T, Weissbourd B, Gehring J, Ferraioli A, Leclère L, Herl M, et al. Whole-animal multiplexed single-cell RNA-seq reveals transcriptional shifts across Clytia medusa cell types. Sci Adv. 2021;7: 1683. doi: 10.1126/SCIADV.ABH1683/SUPPL_FILE/SCIADV.ABH1683_TABLES_S3_AND_S5.ZIP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siebert S, Farrell JA, Cazet JF, Abeykoon Y, Primack AS, Schnitzler CE, et al. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science. 2019;365. doi: 10.1126/science.aav9314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinmetz PRH, Aman A, Kraus JEM, Technau U. Gut-like ectodermal tissue in a sea anemone challenges germ layer homology. Nat Ecol Evol. 2017;1: 1535–1542. doi: 10.1038/s41559-017-0285-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takashima S, Gold D, Hartenstein V. Stem cells and lineages of the intestine: a developmental and evolutionary perspective. Dev Genes Evol. 2013;223: 85–102. doi: 10.1007/s00427-012-0422-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piovani L, Leite DJ, Guerra LAY, Simpson F, Musser JM, Salvador-Martínez I, et al. Single-cell atlases of two lophotrochozoan larvae highlight their complex evolutionary histories. Sci Adv. 2023;9. doi: 10.1126/SCIADV.ADG6034/SUPPL_FILE/SCIADV.ADG6034_SM.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Annunziata R, Perillo M, Andrikou C, Cole AG, Martinez P, Arnone MI. Pattern and process during sea urchin gut morphogenesis: The regulatory landscape. genesis. 2014;52: 251–268. doi: 10.1002/DVG.22738 [DOI] [PubMed] [Google Scholar]
- 50.Burke RD. Structure of the digestive tract of the pluteus larva of Dendraster excentricus (Echinodermata: Echinoida). Zoomorphology. 1981;98: 209–225. doi: 10.1007/BF00312050/METRICS [DOI] [Google Scholar]
- 51.Robertson HE, Sebé-Pedrós A, Saudemont B, Loe-Mie Y, Zakrzewski AC, Grau-Bové X, et al. Single cell atlas of Xenoturbella bocki highlights limited cell-type complexity. Nat Commun. 2024;15. doi: 10.1038/S41467-024-45956-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leung PS. Physiology of the pancreas. Adv Exp Med Biol. 2010;690: 13–27. doi: 10.1007/978-90-481-9060-7_2/FIGURES/4 [DOI] [PubMed] [Google Scholar]
- 53.Pandol SJ. The Exocrine Pancreas. The Exocrine Pancreas. Morgan & Claypool Life Sciences; 2010. doi: 10.4199/C00026ED1V01Y201102ISP014 [DOI] [PubMed] [Google Scholar]
- 54.Whitcomb DC, Lowe ME. Human Pancreatic Digestive Enzymes. Dig Dis Sci. 2007;52: 1–17. doi: 10.1007/s10620-006-9589-z [DOI] [PubMed] [Google Scholar]
- 55.Fautin DG, Mariscal RN. Cnidaria: anthozoa. In: Harrison FW, Ruppert EE, editors. Microscopic Anatomy of Invertebrates Volume 2: Placozoa, Porifera, Cnidaria, and Ctenophora. Siley-Lis; 1991. pp. 267–358. [Google Scholar]
- 56.Thomas MB, Edwards N. Microscopic Anatomy of Invertebrates. Volume 2. Placozoa, Porifera, Cnidaria, and Ctenophora. Chapter 4. Cnidaria: Hydrozoa. Harrison FW, Westfall JA, editors. Wiley-Liss, Inc. NY; 1991. [Google Scholar]
- 57.Hernandez-Nicaise M-L. Ctenophora. In: Harrison F, Westfall JA, editors. Microscopic Anatomy of Invertebrates Volume 2. New York: Wiley-Liss; 1991. pp. 359–418. [Google Scholar]
- 58.Hartenstein V, Martinez & P. Phagocytosis in cellular defense and nutrition: a food-centered approach to the evolution of macrophages. [cited 21 May 2024]. doi: 10.1007/s00441-019-03096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parker MW, Feil SC. Pore-forming protein toxins: from structure to function. Prog Biophys Mol Biol. 2005;88: 91–142. doi: 10.1016/J.PBIOMOLBIO.2004.01.009 [DOI] [PubMed] [Google Scholar]
- 60.Bischofberger M, Iacovache I, Gisou Van Der Goot F. Pathogenic Pore-Forming Proteins: Function and Host Response. Cell Host Microbe. 2012;12: 266–275. doi: 10.1016/J.CHOM.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 61.Iacovache I, Bischofberger M, van der Goot FG. Structure and assembly of pore-forming proteins. Curr Opin Struct Biol. 2010;20: 241–246. doi: 10.1016/J.SBI.2010.01.013 [DOI] [PubMed] [Google Scholar]
- 62.Liu X, Lieberman J. Knocking ‘em Dead: Pore-Forming Proteins in Immune Defense. Annu Rev Immunol. 2020;38: 455–485. doi: 10.1146/ANNUREV-IMMUNOL-111319-023800/CITE/REFWORKS [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cosentino K, Ros U, García-Sáez AJ. Assembling the puzzle: Oligomerization of α-pore forming proteins in membranes. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2016;1858: 457–466. doi: 10.1016/J.BBAMEM.2015.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeudy S, Schwartz TU. Crystal Structure of Nucleoporin Nic96 Reveals a Novel, Intricate Helical Domain Architecture. Journal of Biological Chemistry. 2007;282: 34904–34912. doi: 10.1074/JBC.M705479200 [DOI] [PubMed] [Google Scholar]
- 65.David L, Borges JP, Hollingsworth LR, Volchuk A, Jansen I, Garlick E, et al. NINJ1 mediates plasma membrane rupture by cutting and releasing membrane disks. Cell. 2024;187: 2224–2235.e16. doi: 10.1016/J.CELL.2024.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blott EJ, Griffiths GM. Secretory lysosomes. Nat Rev Mol Cell Biol. 2002;3: 122–31. doi: 10.1038/nrm732 [DOI] [PubMed] [Google Scholar]
- 67.Griffiths GM, Argon Y. Structure and Biogenesis of Lytic Granules. Curr Top Microbiol Immunol. 1995;198: 39–58. doi: 10.1007/978-3-642-79414-8_3 [DOI] [PubMed] [Google Scholar]
- 68.Neighbour PA, Huberman HS, Kress Y. Human large granular lymphocytes and natural killing: Ultrastructural studies of strontium-induced degranulation. Eur J Immunol. 1982;12: 588–595. doi: 10.1002/EJI.1830120711 [DOI] [PubMed] [Google Scholar]
- 69.Weaver TE, Na CL, Stahlman M. Biogenesis of lamellar bodies, lysosome-related organelles involved in storage and secretion of pulmonary surfactant. Semin Cell Dev Biol. 2002;13: 263–270. doi: 10.1016/S1084952102000551 [DOI] [PubMed] [Google Scholar]
- 70.Schroeder LK, Kremer S, Kramer MJ, Currie E, Kwan E, Watts JL, et al. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol Biol Cell. 2007;18: 995–1008. doi: 10.1091/MBC.E06-08-0685/ASSET/IMAGES/LARGE/ZMK0030779830009.JPEG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steger J, Cole AG, Denner A, Lebedeva T, Genikhovich G, Ries A, et al. Single-cell transcriptomics identifies conserved regulators of neuroglandular lineages. Cell Rep. 2022;40. doi: 10.1016/J.CELREP.2022.111370 [DOI] [PubMed] [Google Scholar]
- 72.Babonis LS, Martindale MQ. Old Cell, New Trick? Cnidocytes as a Model for the Evolution of Novelty. Integr Comp Biol. 2014;54: 714–722. doi: 10.1093/ICB/ICU027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fautin DG. Structural diversity, systematics, and evolution of cnidae. Toxicon. 2009;54: 1054–1064. doi: 10.1016/J.TOXICON.2009.02.024 [DOI] [PubMed] [Google Scholar]
- 74.Babonis LS, DeBiasse MB, Francis WR, Christianson LM, Moss AG, Haddock SHD, et al. Integrating Embryonic Development and Evolutionary History to Characterize Tentacle-Specific Cell Types in a Ctenophore. Mol Biol Evol. 2018;35: 2940–2956. doi: 10.1093/MOLBEV/MSY171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schendel V, Müller CHG, Kenning M, Maxwell M, Jenner RA, Undheim EAB, et al. The venom and telopodal defence systems of the centipede Lithobius forficatus are functionally convergent serial homologues. BMC Biology 2024 22:1. 2024;22: 1–22. doi: 10.1186/S12915-024-01925-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von Reumont BM, Undheim EAB, Jauss RT, Jenner RA. Venomics of Remipede Crustaceans Reveals Novel Peptide Diversity and Illuminates the Venom’s Biological Role. Toxins 2017, Vol 9, Page 234. 2017;9: 234. doi: 10.3390/TOXINS9080234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klimovich A, Bosch TCG. Novel technologies uncover novel ‘anti’-microbial peptides in Hydra shaping the species-specific microbiome. Philos Trans R Soc Lond B Biol Sci. 2024;379. doi: 10.1098/RSTB.2023.0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TCG, et al. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci U S A. 2013;110: E3730–E3738. doi: 10.1073/PNAS.1304960110/SUPPL_FILE/PNAS.201304960SI.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Siebert S, Farrell JA, Cazet JF, Abeykoon Y, Primack AS, Schnitzler CE, et al. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science (1979). 2019;365. doi: 10.1126/SCIENCE.AAV9314/SUPPL_FILE/AAV9314_SIEBERT_SM.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hancock REW, Haney EF, Gill EE. The immunology of host defence peptides: beyond antimicrobial activity. Nature Reviews Immunology 2016 16:5. 2016;16: 321–334. doi: 10.1038/nri.2016.29 [DOI] [PubMed] [Google Scholar]
- 81.Mansour SC, Pena OM, Hancock REW. Host defence peptides: front-line immunomodulators. Trends Immunol. 2014;35: 443–450. doi: 10.1016/j.it.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 82.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature 2002 415:6870. 2002;415: 389–395. doi: 10.1038/415389a [DOI] [PubMed] [Google Scholar]
- 83.Bakshani CR, Morales-Garcia AL, Althaus M, Wilcox MD, Pearson JP, Bythell JC, et al. Evolutionary conservation of the antimicrobial function of mucus: a first defence against infection. NPJ Biofilms Microbiomes. 2018;4: 14. doi: 10.1038/s41522-018-0057-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sebé-Pedrós A, Saudemont B, Chomsky E, Plessier F, Mailhé M-P, Renno J, et al. Cnidarian Cell Type Diversity and Regulation Revealed by Whole-Organism Single-Cell RNA-Seq. Cell. 2018;173: 1520–1534.e20. doi: 10.1016/J.CELL.2018.05.019 [DOI] [PubMed] [Google Scholar]
- 85.Steffen K, Proux-Wéra E, Soler L, Churcher A, Sundh J, Cárdenas P. Whole genome sequence of the deep-sea sponge Geodia barretti (Metazoa, Porifera, Demospongiae). G3 (Bethesda). 2023;13. doi: 10.1093/G3JOURNAL/JKAD192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kornder NA, Esser Y, Stoupin D, Leys SP, Mueller B, Vermeij MJA, et al. Sponges sneeze mucus to shed particulate waste from their seawater inlet pores. Current Biology. 2022;32: 3855–3861.e3. doi: 10.1016/J.CUB.2022.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leys SP, Degnan BM. Cytological basis of photoresponsive behavior in a sponge larva. Biological Bulletin. 2001;201: 323–338. doi: 10.2307/1543611/ASSET/IMAGES/LARGE/1N0610531011.JPEG [DOI] [PubMed] [Google Scholar]
- 88.Nakanishi N, Stoupin D, Degnan SM, Degnan BM. Sensory Flask Cells in Sponge Larvae Regulate Metamorphosis via Calcium Signaling. Integr Comp Biol. 2015;55: 1018–1027. doi: 10.1093/icb/icv014 [DOI] [PubMed] [Google Scholar]
- 89.Ereskovsky A V., Tokina DB, Bézac C, Boury-Esnault N. Metamorphosis of cinctoblastula larvae (Homoscleromorpha, porifera). J Morphol. 2007;268: 518–528. doi: 10.1002/JMOR.10506 [DOI] [PubMed] [Google Scholar]
- 90.Brunet T, King N. The Origin of Animal Multicellularity and Cell Differentiation. Developmental Cell. Elsevier; 2017. pp. 124–140. doi: 10.1016/j.devcel.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lang T, Klasson S, Larsson E, Johansson MEV, Hansson GC, Samuelsson T. Searching the Evolutionary Origin of Epithelial Mucus Protein Components - Mucins and FCGBP. Mol Biol Evol. 2016;33: 1921–1936. doi: 10.1093/molbev/msw066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wrackmeyer U, Hansen GH, Seya T, Danielsen EM. Intelectin: A novel lipid raft-associated protein in the enterocyte brush border. Biochemistry. 2006;45: 9188–9197. doi: 10.1021/BI060570X/ASSET/IMAGES/LARGE/BI060570XF00009.JPEG [DOI] [PubMed] [Google Scholar]
- 93.Chen L, Li J, Yang Guiwen, Chen CL, Key SP. A comparative review of intelectins. Scand J Immunol. 2020;92: e12882. doi: 10.1111/SJI.12882 [DOI] [PubMed] [Google Scholar]
- 94.Nishihara T, Wyrick RE, Working PK, Chen YH, Hedrick JL. Isolation and Characterization of a Lectin from the Cortical Granules of Xenopus laevis Eggs. Biochemistry. 1986;25: 6013–6020. doi: 10.1021/BI00368A027/ASSET/BI00368A027.FP.PNG_V03 [DOI] [PubMed] [Google Scholar]
- 95.Hanington PC, Zhang SM. The Primary Role of Fibrinogen-Related Proteins in Invertebrates Is Defense, Not Coagulation. J Innate Immun. 2010;3: 17–27. doi: 10.1159/000321882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Perović-Ottstadt S, Adell T, Proksch P, Wiens M, Korzhev M, Gamulin V, et al. A (1-->3)-beta-D-glucan recognition protein from the sponge Suberites domuncula. Mediated activation of fibrinogen-like protein and epidermal growth factor gene expression. Eur J Biochem. 2004;271: 1924–1937. doi: 10.1111/J.1432-1033.2004.04102.X [DOI] [PubMed] [Google Scholar]
- 97.Driscoll T, Gillespie JJ, Nordberg EK, Azad AF, Sobral BW. Bacterial DNA sifted from the Trichoplax adhaerens (Animalia: Placozoa) genome project reveals a putative rickettsial endosymbiont. Genome Biol Evol. 2013;5: 621–45. doi: 10.1093/gbe/evt036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harrison F, Westfall JA. Placozoa, Porifera, Cnidaria, and Ctenophora. In: Harrison F, Westfall JA, editors. Microscopic Anatomy of Invertebrates Volume 2. New York: Wiley-Liss; 1991. [Google Scholar]
- 99.Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, Hacker J, et al. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol. 2002;68: 4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002/ASSET/D970C1FF-9582-4A5D-BDFD-FEDEDD366328/ASSETS/GRAPHIC/AM0920452007.JPEG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Titani K, Torff HJ, Hormel S, Kumar S, Walsh KA, Rodl J, et al. Amino acid sequence of a unique protease from the crayfish Astacus fluviatilis. Biochemistry. 1987;26: 222–226. doi: 10.1021/BI00375A029 [DOI] [PubMed] [Google Scholar]
- 101.Kawaguchi M, Okazawa Y, Imafuku A, Nakano Y, Shimizu R, Ishizuka R, et al. Pactacin is a novel digestive enzyme in teleosts. Scientific Reports 2021 11:1. 2021;11: 1–13. doi: 10.1038/s41598-021-86565-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park JO, Pan J, Möhrlen F, Schupp MO, Johnsen R, Baillie DL, et al. Characterization of the astacin family of metalloproteases in C. elegans. BMC Dev Biol. 2010;10. doi: 10.1186/1471-213X-10-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242: 1528–1534. doi: 10.1126/SCIENCE.3201241 [DOI] [PubMed] [Google Scholar]
- 104.Algrain M, Hennebert E, Bertemes P, Wattiez R, Flammang P, Lengerer B. In the footsteps of sea stars: deciphering the catalogue of proteins involved in underwater temporary adhesion. Open Biol. 2022;12. doi: 10.1098/RSOB.220103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gomis-Rüth FX, Stöcker W. Structural and evolutionary insights into astacin metallopeptidases. Front Mol Biosci. 2023;9: 1080836. doi: 10.3389/FMOLB.2022.1080836/FULL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hayakawa E, Watanabe H, Menschaert G, Holstein TW, Baggerman G, Schoofs L. A combined strategy of neuropeptide prediction and tandem mass spectrometry identifies evolutionarily conserved ancient neuropeptides in the sea anemone Nematostella vectensis. PLoS One. 2019;14: e0215185. doi: 10.1371/JOURNAL.PONE.0215185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koch TL, Grimmelikhuijzen CJP. Global Neuropeptide Annotations From the Genomes and Transcriptomes of Cubozoa, Scyphozoa, Staurozoa (Cnidaria: Medusozoa), and Octocorallia (Cnidaria: Anthozoa). Front Endocrinol (Lausanne). 2019;10. doi: 10.3389/FENDO.2019.00831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koch TL, Grimmelikhuijzen CJP. A comparative genomics study of neuropeptide genes in the cnidarian subclasses Hexacorallia and Ceriantharia. BMC Genomics. 2020;21: 1–21. doi: 10.1186/S12864-020-06945-9/FIGURES/4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yañez-Guerra LA, Thiel D, Jékely G. Premetazoan Origin of Neuropeptide Signaling. Mol Biol Evol. 2022;39. doi: 10.1093/MOLBEV/MSAC051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jekely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proceedings of the National Academy of Sciences. 2013;110: 8702–8707. doi: 10.1073/pnas.1221833110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Colgren J, Burkhardt P. The premetazoan ancestry of the synaptic toolkit and appearance of first neurons. Essays Biochem. 2022;66: 781–795. doi: 10.1042/EBC20220042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arendt D, Benito-Gutierrez E, Brunet T, Marlow H. Gastric pouches and the mucociliary sole: setting the stage for nervous system evolution. Phil Trans R Soc Lond B. 2015;370: 20150286. doi: 10.1098/rstb.2015.0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brunet T, King N. The origin of animal multicellularity and cell differentiation. Dev Cell. 2017;43: 124–140. doi: 10.1016/j.devcel.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cavalier-Smith T. Origin of animal multicellularity: precursors, causes, consequences-the choanoflagellate/sponge transition, neurogenesis and the Cambrian explosion. Phil Trans R Soc Lond B. 2017;372: 20150476. doi: 10.1098/rstb.2015.0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nielsen C. Six major steps in animal evolution: are we derived sponge larvae? Evol Dev. 2008;10: 241–257. doi: 10.1111/j.1525-142x.2008.00231.x [DOI] [PubMed] [Google Scholar]
- 116.Smith CL, Mayorova TD. Insights into the evolution of digestive systems from studies of Trichoplax adhaerens. Cell Tissue Res. 2019;377: 353–367. doi: 10.1007/s00441-019-03057-z [DOI] [PubMed] [Google Scholar]
- 117.Syed T, Schierwater B. Trichoplax adhaerens: Discovered as a missing link, forgotten as a hydrozoan, re-discovered as a key to Metozoan evolution. Vie et Milieu. 2002;52: 177–187. [Google Scholar]
- 118.Hallgren J, Tsirigos KD, Damgaard Pedersen M, Juan J, Armenteros A, Marcatili P, et al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv. 2022; 2022.04.08.487609. doi: 10.1101/2022.04.08.487609 [DOI] [Google Scholar]
- 119.Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, et al. InterPro in 2022. Nucleic Acids Res. 2023;51: D418–D427. doi: 10.1093/NAR/GKAC993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50: D439–D444. doi: 10.1093/NAR/GKAB1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27: 14–25. doi: 10.1002/PRO.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30: 70–82. doi: 10.1002/PRO.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grimmelikhuijzen CJP, Leviev I, Carstensen K. Peptides in the Nervous Systems of Cnidarians: Structure, Function, and Biosynthesis. Int Rev Cytol. 1996;167: 37–89. doi: 10.1016/S0074-7696(08)61345-5 [DOI] [PubMed] [Google Scholar]
- 124.Hammer DAT, Ryan PD, Hammer Ø, Harper DAT. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica. 2001;4: 178. Available: http://palaeo-electronica.orghttp://palaeo-electronica.org/2001_1/past/issue1_01.htm. [Google Scholar]
- 125.Micallef L, Rodgers P. eulerAPE: Drawing Area-Proportional 3-Venn Diagrams Using Ellipses. PLoS One. 2014;9: 101717. doi: 10.1371/journal.pone [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mayorova TD, Smith CL, Reese TS. Secretory cells in the digestive epithelium of Trichoplax adhaerens, an animal that feeds by external digestione. Society for Neuroscience Abstract. 2018;282.15. [Google Scholar]