Abstract
A Carnation ringspot virus (CRSV) variant (1.26) was identified that accumulates virions but is incapable of forming a systemic infection. The 1.26 capsid protein gene possesses a Ser→Pro mutation at amino acid 282. Conversion of 1.26 amino acid 282 to Ser restored systemic infection, while the reciprocal mutation in wild-type CRSV abolished systemic infection. Similar mutations introduced into the related Red clover necrotic mosaic virus capsid protein gene failed to induce the packaging but nonsystemic movement phenotype. These results provide additional support for the theory that virion formation is necessary but not sufficient for systemic movement with the dianthoviruses.
Carnation ringspot virus (CRSV) is the type species of the Dianthovirus genus of plant viruses (6). The CRSV genome is composed of two nonhomologous, positive-sense single-stranded RNAs of 3.9 and 1.5 kb that are encapsidated into 30- to 32-nm T=3 icosahedral virions by 180 copies of the 37-kDa capsid protein (CP) (6, 7, 11). The larger genomic RNA-1 carries two open reading frames (ORFs). The 5′ proximal ORF, interrupted by a −1 ribosomal frameshifting signal, produces a prereadthrough 27-kDa protein and an 88-kDa polypeptide which is predicted to be the virally encoded replicase (8, 9, 16, 19) (Fig. 1A). The CP is expressed from a subgenomic RNA in vivo (20). RNA-2 is monocistronic and encodes the 35-kDa movement protein (MP) that directs cell-to-cell movement and long-distance transport of the viral infection (7). Like the plant tobamoviruses (1, 3), the dianthoviruses do not require the CP for cell-to-cell movement but do require it for systemic infection in all-systemic hosts (16, 18, 19). It is hypothesized that the CP in the form of a virion interacts with MP and possibly with host factors to transport the infection across the plasmodesmata between phloem parenchyma and the sieve element to facilitate long-distance transport and systemic infection (17).
FIG. 1.
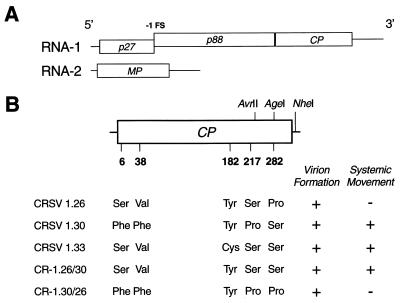
CRSV genome, location of CP amino acid polymorphisms, and corresponding assembly and systemic movement phenotypes. (A) Genome organization. RNA-1 contains ORFs (denoted by open boxes) for p27 and the CP. Occasionally, translation of p27 is extended by a −1 ribosomal frameshift (FS) event to produce p88. RNA-2 codes for the MP. (B) CRSV RNA-1 constructs. Schematic diagram displaying the relative positions of amino acid polymorphisms in the 339-amino acid CP (denoted by numbers below the CP ORF). Locations of restriction sites used for the construction and differentiation of amino acid exchanges are indicated above and to the right of the CP ORF. The AgeI site is unique to CRSV 1.26. The detection of virions in the inoculated leaves and systemic movement to the upper, noninoculated leaves is displayed to the right of the various RNA-1 constructs.
In this study, the movement phenotypes of three independent cDNA constructs of CRSV RNA-1 (CRSV 1.26, 1.30, and 1.33) were examined. All three constructs were wild type for virion formation and symptoms on the inoculated leaves. But CRSV RNA-1 clone 1.26 could not form a systemic infection. A single amino acid sequence polymorphism in CRSV 1.26 determined the localized infection phenotype. Comparable mutations in the red clover necrotic mosaic virus (RCNMV) CP were unable to duplicate the uncoupling of virion formation from systemic movement, suggesting the requirement for a distinct, compatible CP-MP interaction that facilitates the systemic movement of the various dianthoviruses. These results provide additional support that virion formation is necessary but not sufficient for systemic movement.
CRSV cDNA clones were created by reverse transcribing CRSV virion RNA followed by amplification with Taq DNA polymerase and ligation of the amplification products into the pGEM-T Vector (Promega Corp., Madison, Wis.) (T. L. Sit and S. A. Lommel, unpublished data). Several independent full-length cDNA clones of CRSV RNA-1 (CRSV 1.26, 1.30, and 1.33) and RNA-2 (CRSV 2.4) were isolated and linearized with HindIII prior to in vitro transcription with T7 RNA polymerase (10). Five microliters of each infectious RNA-1 and RNA-2 transcript was combined with 100 μl of 10 mM sodium phosphate buffer, pH 7.2, and then rub inoculated onto carborundum-dusted Nicotiana benthamiana leaves. The inoculated plants were placed in a glasshouse at 20 to 25°C under ambient light conditions. Plants were monitored for symptom development up to 20 days postinoculation (dpi). Each inoculation was repeated at least three times.
N. benthamiana plants inoculated with CRSV RNA-2 and either CRSV 1.30 or CRSV 1.33 displayed typical necrotic ringspot and wilting symptoms on the inoculated leaves 4 to 5 dpi. Systemic symptoms were observed 7 to 8 dpi and consisted of mild chlorosis and ringspots. Virions and virion RNA were isolated from inoculated and noninoculated leaves 4 to 20 dpi and were visualized by transmission electron microscopy (magnification, ×50,000) after negative staining with 2% uranyl acetate (16). Virus particles with wild-type morphology were abundant in both inoculated and systemically infected leaves (Fig. 2). Inoculation with CRSV 1.26 plus CRSV RNA-2 also yielded typical symptoms and virions (Fig. 2) on the inoculated leaves. However, no symptoms of infection were ever observed on the noninoculated upper leaves of these plants up to 20 dpi. Furthermore, virions could not be detected from the upper leaves (Fig. 2).
FIG. 2.
Detection of CRSV particles. Representative electron micrographs are shown for negatively stained virion preparations from the various CRSV inoculations listed to the left. Images in the left column represent preparations from inoculated tissue, while those on the right are from upper, noninoculated tissue. Scale bar, 50 nm.
Passaging of sap from the upper noninoculated leaves of plants infected with CRSV 1.30 plus CRSV RNA-2 by rub inoculation with an autoclaved sponge in a 1:1 dilution of potassium phosphate buffer, pH 7.2, to carborundum-dusted uninfected plants led to local and systemic symptom development on N. benthamiana plants. However, the same passaging experiment with N. benthamiana plants inoculated with CRSV 1.26 plus CRSV RNA-2 did not lead to symptom development (Table 1).
TABLE 1.
Serial passage of CRSV virion preparations to examine defects in systemic movement and disassembly
| Inoculum | Leaf tissuea | Treatment prior to passagingb | Symptom
|
|
|---|---|---|---|---|
| Inoculated | Noninoculated | |||
| Mock inoculated | NI | — | − | − |
| CRSV 1.26 plus RNA-2 | NI | — | − | − |
| CRSV 1.30 plus RNA-2 | NI | — | + | + |
| Mock inoculated | I | — | − | − |
| Mock inoculated | I | RNase A | − | − |
| CRSV 1.26 plus RNA-2 | I | — | + | − |
| CRSV 1.26 plus RNA-2 | I | RNase A | + | − |
| CRSV 1.30 plus RNA-2 | I | — | + | + |
| CRSV 1.30 plus RNA-2 | I | RNase A | + | + |
Leaf tissue source of virion preparation. NI, upper, noninoculated tissue; I, inoculated tissue.
Treatment of virion preparations performed to prevent infection by unencapsidated viral RNA. —, no treatment; RNase A, 1 μg of RNase A for 10 min at 25°C.
Total RNA was extracted from the upper, noninoculated leaves of plants coinoculated with CRSV RNA-2 and either CRSV 1.26 or 1.30. Virion RNA was detected by reverse transcription (RT)-PCR by priming cDNA synthesis with either CRSV RNA-1 primer (5′-ATAAGCTTaggggcccgggggttatac-3′) or CRSV RNA-2 primer (5′ ATTCtaGAttgtaccaacctcacccgttg-3′) and PCR amplification with the same initial primers in combination with a second upstream primer (RNA-1, 5′-gccaacaaagttgcaagagc-3′; RNA-2, 5′-TACCatggcagttcatgtagataacg-3′) using standard protocols (12). Upper-case letters in the primer sequences indicate nucleotides differing from wild-type viral sequences. Both RNA-1 and RNA-2 could be detected by RT-PCR in the upper leaves of N. benthamiana plants coinoculated with CRSV 1.30 plus CRSV RNA-2 (Fig. 2). In the upper leaves of the CRSV 1.26 plus CRSV RNA-2 coinoculations, no viral RNA could be detected. Consequently, neither virus particles nor viral RNA were detectable in the upper noninoculated leaves of plants inoculated with CRSV 1.26 plus CRSV RNA-2. Together with the electron microscopy and bioassay observations, these findings show that CRSV 1.26 is incapable of systemic movement either as free RNA or as virions.
To eliminate a disassembly defect as the cause for the lack of symptoms in the upper noninoculated leaves of plants infected with CRSV 1.26 plus CRSV RNA-2, virions purified from the initially inoculated leaves were used to inoculate N. benthamiana plants. The addition of 1 μl of RNase A (1 mg/ml) ensured that any unencapsidated viral RNA would not affect infectivity. CRSV 1.26 plus CRSV RNA-2 inoculations led to symptom development only on the inoculated leaves (Table 1). Total RNA was harvested from inoculated tissue 8 dpi and was subjected to RT-PCR analysis. Again, viral RNA was detected from tissue inoculated with virions of CRSV 1.26 plus CRSV RNA-2, indicating that disassembly and replication does occur for this construct (data not shown). Furthermore, the CRSV 1.26 movement phenotype was maintained after both passaging experiments, suggesting that progeny virions retained their variant sequence.
The CP sequences of the three CRSV RNA-1 clones were determined by the University of Georgia Molecular Genetics Facility (Athens, Ga.). Four deduced amino acid polymorphisms were detected between CRSV 1.30 and 1.26 (Fig. 1B) at residue number 6 (Phe versus Ser), 38 (Phe versus Val), 217 (Pro versus Ser), and 282 (Ser versus Pro). Ser 282 is conserved between CRSV 1.30 and 1.33. A comparison of the CRSV 1.33 and 1.26 CP sequences revealed only two amino acid polymorphisms: Cys versus Tyr at amino acid (aa) 182 and Ser versus Pro at aa 282. Because aa 282 is the only amino acid change unique to CRSV 1.26 among the three CRSV CP genes examined, it was further investigated. To determine whether the Pro 282 in CRSV 1.26 CP is responsible for the arrest of systemic movement, a sequence fragment was exchanged between CRSV 1.26 and 1.30 containing the codon for aa 282. CRSV 1.26 and 1.30 were digested at unique AvrII and NheI restriction sites, resulting in a 362-nucleotide fragment. Both fragments and the resultant CRSV 1.26 and CRSV 1.30 vector backbones were gel purified (Qiagen Inc., Valencia, Calif.) and exchanged between CRSV 1.30 and CRSV 1.26 in reciprocal ligations. A unique Agel site found only in the CRSV 1.26 fragment was used to differentiate the resultant clones. Clones CR-1.26/30 1 and 3 (CRSV 1.26 backbone with the CRSV 1.30 fragment) and clones CR-1.30/26 3 and 5 (CRSV 1.30 backbone with the CRSV 1.26 fragment) were isolated and inoculated to plants.
At 4 dpi, typical local symptoms appeared on plants coinoculated with CRSV RNA-2 and all recombinant or parental (CRSV 1.26 or 1.30) RNA-1 constructs. At 7 dpi, systemic symptoms on the upper leaves could be observed on plants inoculated with CRSV 1.30 plus CRSV RNA-2. No symptoms were observed on upper leaves of N. benthamiana plants inoculated with CRSV 1.26 plus CRSV RNA-2. Virion purifications performed on both inoculated and upper noninoculated leaves followed by electron microscopy and RT-PCR analysis (Fig. 2 and 3) confirmed the presence of virions from both locations for plants inoculated with CRSV 1.30 plus CRSV RNA-2 but only in the inoculated leaves of plants inoculated with CRSV 1.26 plus CRSV RNA-2.
FIG. 3.
Detection of CRSV virion RNA. Representative agarose gel images are shown for RT-PCRs specific for either CRSV RNA-1 (A) or CRSV RNA-2 (B) from purified virion RNA. Each inoculation involved virion purification and subsequent RNA extraction from inoculated (I) and upper, noninoculated (N) leaves. An amplification control reaction (lane denoted DNA) was also performed on a control viral cDNA template of either CRSV RNA-1 or RNA-2. MW denotes the 100-bp DNA ladder (New England Biolabs, Beverly, Mass.), with the 500-bp fragment marked with an arrow to the right of the image.
On plants coinoculated with CR-1.30/26 plus CRSV RNA-2, no systemic symptoms could be observed, and virion purification from inoculated and upper tissue revealed the presence of virions from only the initially inoculated leaves (Fig. 2). These data strongly suggest that the movement phenotype of CRSV 1.26 is due to the single Ser-to-Pro amino acid substitution at aa 282.
N. benthamiana plants coinoculated with CR-1.26/30 plus CRSV RNA-2 exhibited systemic symptoms at 7 dpi, much like those observed for CRSV 1.30 plus CRSV RNA-2. Virion purification and RT-PCR analysis from both inoculated and upper noninoculated leaves of plants infected with CR-1.26/30 plus CRSV RNA-2 revealed the presence of virions from both locations (Fig. 2 and 3). Therefore, restoration of systemic movement in CRSV 1.26 was accomplished by the reversion of aa 282 from Pro to Ser.
To determine if the apparent uncoupling of virion assembly from subsequent systemic movement observed in CRSV 1.26 could be mimicked by similar mutations in the related RCNMV CP gene, site-directed mutagenesis of the RCNMV CP was performed based on the results of a ClustalW alignment (15) between the CRSV and RCNMV CP sequences. The alignment indicated that the CRSV CP Ser 282 is present at a similar position in the RCNMV CP (Fig. 4). However, because a neighboring Thr 280 residue in the RCNMV CP aligns with CRSV Ser 282, and because Thr has structural properties similar to those of Ser, RCNMV Thr 280 was also considered a residue potentially corresponding to CRSV Ser 282. Thus, mutations in the RCNMV CP gene were introduced, resulting in single amino acid substitutions of Ser 279 to Pro (pCPS2P) and Thr 280 to Pro (pCPT2P). Additionally, a double mutant was constructed with Pro at both locations (pST2PP).
FIG. 4.
Schematic of RCNMV CP mutants based on the CRSV 1.26 CP sequence. The local alignment of the CRSV CP with that from RCNMV is depicted. Identical residues are indicated by a double dot, while similar residues are designated with a single dot. The positions of the residues within their respective CPs are indicated above and below the sequences. The altered residues of the RCNMV CP mutants are indicated below the RCNMV sequence, while the altered CRSV 1.26 residue is shown above the CRSV 1.30 sequence. The detection of virions in the inoculated leaves and systemic movement to the upper, noninoculated leaves is displayed to the right of the mutants.
The mutations were introduced into the RCNMV CP ORF by long-inverse PCR mutagenesis (2) of an RCNMV RNA-1 infectious cDNA clone (pRC169) (13). RCNMV Ser 279 was mutated to a proline codon by using the forward primer RCPS2PF (5′-tCcaactctggttggcgacagtagg-3′) to make pRCPS2P. After sequence confirmation of the mutated region, the BglII/MluI fragment from pRCPS2P was reinserted into pRC169 to generate pCPS2P. Similarly, the threonine codon at aa 280 was changed to proline with the forward primer RCPT2PF (5′-ttcaCcCTtAgttggcgacagtagggcg-3′) to make pRCPT2P. The sequence-verified region containing the amino acid substitution was transferred back into pRC169 to make pCPT2P. The forward primer ST2PPF (5′-tCcaCcCctggttggcgacagtagggcg-3′) was used to change both Ser 279 and Thr 280 to proline codons in clone pST2PP, which was used as a template for transcription reactions after sequence verification of the mutated region. In all cases, the reverse primer in the amplification reaction was RCPsubPR (5′-tttcctcctactgtgacgctaccag-3′). Upper-case letters in the primer sequences indicate nucleotides differing from wild-type viral sequences.
The resultant wild-type and mutant RCNMV plasmids were linearized with SmaI prior to T7 transcription and inoculation. Typical ringspot symptoms were observed on inoculated leaves 2 to 4 dpi for wild-type RCNMV and all three CP mutants on cowpea, Nicotiana clevelandii, and N. benthamiana. Symptoms on noninoculated leaves appeared 2 days afterwards on N. benthamiana and N. clevelandii plants inoculated with wild-type RCNMV but did not appear on plants inoculated with any of the CP mutants. Virus particles were not detectable by transmission electron microscopy of virion preparations from inoculated leaves 7 dpi (Fig. 4). RT-PCR analysis of RCNMV RNA-1 from these virion preparations, using a primer complementary to the extreme 3′ end for first-strand synthesis and nested amplification primers (forward primer, 5′-gctcccaagaaatccaaacaacggtccc-3′; reverse primer, 5′-CGGGTACCttaaccaagtatgaaagtgttccc-3′), did not detect the presence of viral RNA that may have been packaged into virus particles (data not shown). Thus, all three-substitution mutations blocked the systemic movement of RCNMV in a manner similar to that observed with CRSV 1.26. However, unlike CRSV 1.26, none of the three RCNMV CP mutants tested were able to form virions.
The ability of viruses to move systemically is necessary for the spread of the infection within a susceptible host plant. We have shown that CP, likely in the form of a virion, and possibly MP and host factors are required for the long-distance transport of RCNMV (16, 17). Here we demonstrate with CRSV that virion formation alone does not confer the ability for systemic movement. These findings reaffirm that virion formation in dianthoviruses is necessary for systemic infection of a susceptible host, but it is not sufficient on its own to facilitate this type of movement. The lack of systemic infection may be caused by an altered interaction between the CP and other proteins, specifically the viral MP and/or host factors, which would normally allow virion binding to plasmodesmatal targets at the phloem parenchyma and companion cell-sieve element complex. Previously, RCNMV MP mutants were isolated which allowed cell-to-cell movement but inhibited long-distance transport, suggesting that MP may interact directly with virions to facilitate the crossing of this cellular barrier (4). Conversely, the stability of CRSV 1.26 virions may be reduced compared to that of wild-type virions such that they can survive treatment with RNase A (see Table 1) but may not withstand translocation into and/or long-distance transport within the plant vasculature.
Our refined model for the systemic movement of dianthoviruses involves the binding of MP and/or host factors to the surface of the virus particle, presumably on the P domain, involving aa 282. This interaction may determine a virion's ability to reach a vascular plasmodesmatal channel or, alternatively, to traverse through it. The requirement for a cognate CP-MP interaction to facilitate long-distance movement has been demonstrated for cucumoviruses and tobamoviruses, among others (5, 14).
Acknowledgments
We thank Valerie Knowlton for electron microscope imaging.
This research was supported by USDA NRI competitive grant 98-02298 and National Science Foundation competitive grant MCB-0077964.
REFERENCES
- 1.Arce-Johnson P, Reimann-Philipp U, Padgett H S, Rivera-Bustamante R, Beachy R N. Requirement of the movement protein for long distance spread of tobacco mosaic virus in grafted plants. Mol Plant-Microbe Interact. 1997;10:691–699. [Google Scholar]
- 2.Callaway A S. A genetic approach to study host factors of Arabidopsis (Arabidopsis thaliana) that influence susceptibility to cauliflower mosaic virus. Ph.D. dissertation. Ithaca, N.Y: Cornell University; 1998. [Google Scholar]
- 3.Citovsky V. Tobacco mosaic virus: a pioneer of cell-to-cell movement. Philos Trans R Soc Lond. 1999;B354:637–643. doi: 10.1098/rstb.1999.0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giesman-Cookmeyer D, Lommel S A. Alanine scanning mutagenesis of a plant virus movement protein identifies three functional domains. Plant Cell. 1993;5:973–982. doi: 10.1105/tpc.5.8.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilf M E, Dawson W O. The tobamovirus capsid protein functions as a host-specific determinant of long-distance movement. Virology. 1993;193:106–114. doi: 10.1006/viro.1993.1107. [DOI] [PubMed] [Google Scholar]
- 6.Hiruki C. The dianthoviruses: a distinct group of isometric plant viruses with a bipartite genome. Adv Virus Res. 1987;33:257–300. doi: 10.1016/s0065-3527(08)60320-6. [DOI] [PubMed] [Google Scholar]
- 7.Kendall T L, Lommel S A. Nucleotide sequence of carnation ringspot virus RNA-2. J Gen Virol. 1992;73:2479–2482. doi: 10.1099/0022-1317-73-9-2479. [DOI] [PubMed] [Google Scholar]
- 8.Kim K H, Lommel S A. Identification and analysis of the site of -1 ribosomal frameshifting in red clover necrotic mosaic virus. Virology. 1994;200:574–582. doi: 10.1006/viro.1994.1220. [DOI] [PubMed] [Google Scholar]
- 9.Kim K H, Lommel S A. Sequence element required for efficient -1 ribosomal frameshifting in red clover necrotic mosaic dianthovirus. Virology. 1998;250:50–59. doi: 10.1006/viro.1998.9358. [DOI] [PubMed] [Google Scholar]
- 10.Pokrovskaya I D, Gurevich V V. In vitro transcription: preparative RNA yields in analytical scale reactions. Anal Biochem. 1994;220:420–423. doi: 10.1006/abio.1994.1360. [DOI] [PubMed] [Google Scholar]
- 11.Ryabov E V, Generozov E V, Kendall T L, Lommel S A, Zavriev S K. Nucleotide sequence of carnation ringspot dianthovirus RNA-1. J Gen Virol. 1994;75:243–247. doi: 10.1099/0022-1317-75-1-243. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 13.Sit T L, Vaewhongs A A, Lommel S A. RNA-mediated trans-activation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- 14.Taliansky M E, Garcia-Arenal F. Role of cucumovirus capsid protein in long-distance movement within the infected plant. J Virol. 1995;69:916–922. doi: 10.1128/jvi.69.2.916-922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaewhongs A A, Lommel S A. Virion formation is required for the long-distance movement of red clover necrotic mosaic virus in movement protein transgenic plants. Virology. 1995;212:607–613. doi: 10.1006/viro.1995.1518. [DOI] [PubMed] [Google Scholar]
- 17.Wang H-L, Wang Y, Giesman-Cookmeyer D, Lommel S A, Lucas W J. Mutations in viral movement protein alter systemic infection and identify an intercellular barrier to entry into the phloem long-distance transport system. Virology. 1998;245:75–89. doi: 10.1006/viro.1998.9154. [DOI] [PubMed] [Google Scholar]
- 18.Xiong Z, Kim K H, Giesman-Cookmeyer D, Lommel S A. The roles of the red clover necrotic mosaic virus capsid and cell-to-cell movement proteins in systemic infection. Virology. 1993;192:27–32. doi: 10.1006/viro.1993.1004. [DOI] [PubMed] [Google Scholar]
- 19.Xiong Z, Kim K H, Kendall T L, Lommel S A. Synthesis of the putative RCNMV RNA polymerase by ribosomal frameshifting in vitro. Virology. 1993;193:213–221. doi: 10.1006/viro.1993.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zavriev S K, Hickey C M, Lommel S A. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology. 1996;216:407–410. doi: 10.1006/viro.1996.0076. [DOI] [PubMed] [Google Scholar]