Abstract
Aberrant caspase recruitment domain family member 14 (CARD14) signaling has been strongly associated with inflammatory skin conditions. CARD14 acts as a scaffold protein, ultimately activating the transcription factor NF-KB. Although primarily studied in the context of inflammation, recent research has suggested its potential implications in tumorigenesis. In this study, we gathered The Cancer Genome Atlas (TCGA) tumor data to gauge the involvement of CARD14 in cancer, including genetic alterations, expression patterns, survival correlations, immune cell infiltration and functional interactions across diverse cancer types. We found heightened CARD14 expression in most tumors and there was a significant correlation between CARD14 expression and the prognosis of patients for certain tumors. For instance, patients with higher CARD14 expression had a better prognosis in sarcoma, lung, cervix and head and neck cancers. Moreover, CARD14 expression positively correlated with neutrophil infiltration in most of the cancer types analyzed. Finally, enrichment analysis showed that epithelial development and differentiation pathways were involved in the functional mechanism of CARD14. Our results show that CARD14 may have the potential to become a prognostic biomarker in several cancers, hence, further prospective studies will be required for its validation.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-74565-4.
Keywords: CARD14, Cancer, Inflammation, TCGA, Prognosis
Subject terms: Cancer, Immunology, Biomarkers, Oncology, Risk factors
Introduction
CARD14 (CARMA2) belongs to the CARD-containing family of scaffolding proteins, which includes CARD9, CARD10 (CARMA3) and CARD11 (CARMA1)1. CARD-containing proteins primarily transmit upstream signals to activate the NF-κB transcription factor, which controls genes associated with immune, inflammatory, cell survival, proliferation and cellular stress responses2. The details of how CARD14 functions within the cell are just beginning to be elucidated; CARD14 requires interaction with BCL10 and the paracaspase MALT1 to form a CBM complex3,4; however, we know more about the role of CARD14 in pathology than its role in physiology.
Gain-of-function (Gof) mutations in CARD14 are linked to inflammatory skin conditions such as psoriasis5 and pityriasis rubra pilaris (PRP)6. Because of its association with psoriasis, there has been a focused interest in describing CARD14 biology with mouse models. Strikingly, the absence of CARD14 is not detrimental to life and protects mice in psoriasis models7,8. On the other hand, different Gof CARD14 mutant mice presented with skin inflammation similar to that of psoriatic lesions8–11. CARD14 Gof mutations disrupt the autoinhibitory conformation of the protein, resulting in spontaneous oligomerization and sustained activation of NF-kB, hence leading to the upregulation of genes and chemokines associated with psoriasis (TNF, CCL20, and IL-8)12. Importantly, in addition to rare mutations described in humans, the common polymorphism CARD14 rs11652075 predisposes people to psoriasis5 and atopic dermatitis13, potentially affecting the health of many individuals.
Psoriasis is a chronic autoimmune skin disorder characterized by the emergence of red and scaly patches on the skin14. Due to persistent inflammation in the skin, psoriasis is considered a systemic disease that can result in tissue damage to other organs. Numerous comorbidities are associated with psoriasis, such as psoriatic arthritis (affecting 20–30% of patients), inflammatory bowel disease and cancer15. Abnormal activation of both adaptive and innate immune cells or dysregulation of signaling pathways in nonimmune cells, such as NF-κB16, chronifies inflammation facilitating cancer onset17.
Notably, increased activity or Gof mutations in the family members CARD9, CARD10 and CARD11 activate NF-KB, promoting cell proliferation in a manner similar to that of CARD14, and have been linked to liver metastasis18, solid tumors19 and lymphoma20. In fact, psoriatic patients have an increased risk of developing lymphomas, keratinocyte, lung and bladder cancer21. It is impossible to predict whether a psoriatic patient will develop a comorbidity or not and which type of comorbidity will develop. Because CARD14 expression is not limited to keratinocytes9 in genetically predisposed individuals, aberrant signaling of CARD14 in tissues other than the skin might be responsible for tumor progression.
In this work, we have investigated the involvement of CARD14 in cancer via several means of large data set analyses. Specifically, through mining TCGA data, we have assessed the mutational status and RNA and protein expression of CARD14 across clinical samples. Strikingly, correlations were identified for the degree of immune cell infiltration and overall patient survival with perturbations in CARD14 sequence and regulation. Moreover, instances of tumor prognosis associated with CARD14 status have been described herein. Collectively, pan-cancer data analysis presents CARD14 as a novel candidate for the further investigation in cancer biology.
Results
Genetic alteration of the CARD14 gene in cancer
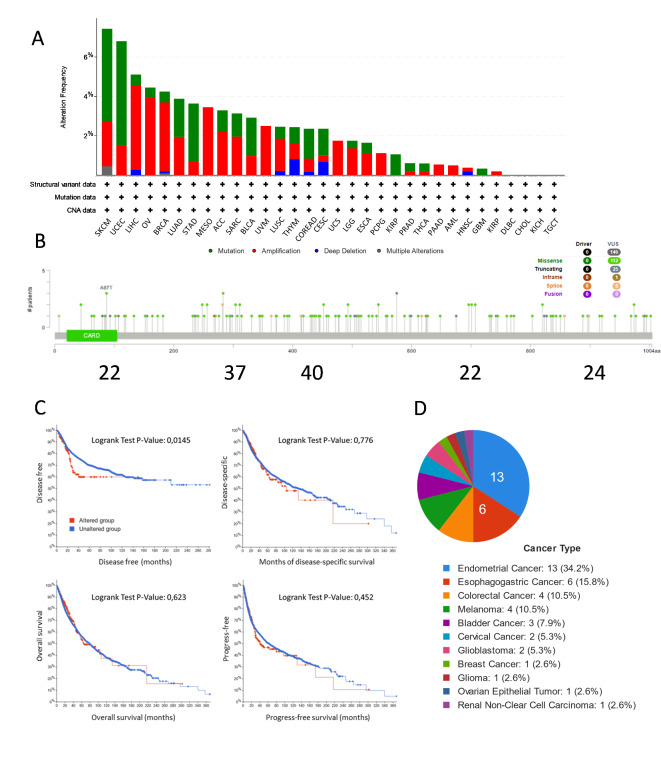
Mutations and gene variations in CARD14 have been found to cause or predispose patients to psoriasis and other inflammatory diseases. Because psoriasis is a common pathology and patients with psoriasis have an increased probability of developing cancer, we wanted to determine whether CARD14 mutations were also present in tumor samples. We used the cBioPortal website to identify CARD14 (NM_024110) genetic alterations, including mutation types, frequencies, copy number alterations (CNAs), and structural variants, across a spectrum of cancer types (Fig. 1A). CARD14 gene was altered in 293 out of 10,950 patients (2,7%), with somatic mutations in approximately 1.2% of patients. The three cancers with the highest alteration frequencies were 7.43% in SKCM (comprising 4.73% mutations and 2.25% amplifications), 6.31% in UCEC (comprising 4.78% mutations and 1.54% amplifications), and 5.11% in LIHC (comprising 0.54% mutations and 4.3% amplifications). Of note, amplifications were more common than deletions, while missense mutations in CARD14 were the predominant mutation type, and a greater frequency of these mutations occurred between aminoacids 200 and 600 of CARD14, where the coiled coil and linker domains are located3 (Fig. 1B).
Fig. 1.
Analysis of CARD14 gene mutations in cBioPortal (TCGA PanCancer Atlas Studies). (A) Types of mutations and their frequencies in each cancer type. (B) Location of cancer mutations in the CARD14 gene sequence. (C) Survival analysis comparing DFS, DSS, OS and PFS in cancer patients with and without CARD14 gene alterations. The log-rank test was used to test the null hypothesis, we considered significance when, p < = 0.05. (D) Percentage of samples with mutations described in ClinVar. ACC: Adrenocortical carcinoma. BLCA: Bladder Urothelial Carcinoma. BRCA: Breast invasive carcinoma. CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma. CHOL: Cholangio carcinoma. COAD: Colon adenocarcinoma. DLBC: Lymphoid Neoplasm Diffuse Large B-cell Lymphoma. ESCA: Esophageal carcinoma. GBM: Glioblastoma multiforme. HNSC: Head and Neck squamous cell carcinoma. KICH: Kidney Chromophobe. KIRC: Kidney renal clear cell carcinoma. KIRP: Kidney renal papillary cell carcinoma. LAML: Acute Myeloid Leukemia. LGG: Brain Lower Grade Glioma. LIHC: Liver hepatocellular carcinoma. LUAD: Lung adenocarcinoma. LUSC: Lung squamous cell carcinoma. MESO: Mesothelioma. OV: Ovarian serous cystadenocarcinoma. PAAD: Pancreatic adenocarcinoma. PCPG: Pheochromocytoma and Paraganglioma. PRAD: Prostate adenocarcinoma. READ: Rectum adenocarcinoma. SARC: Sarcoma. SKCM: Skin Cutaneous Melanoma. STAD: Stomach adenocarcinoma. TGCT: Testicular Germ Cell Tumors. THCA: Thyroid carcinoma. THYM: Thymoma. UCEC: Uterine Corpus Endometrial Carcinoma. UCS: Uterine Carcinosarcoma. UVM: Uveal Melanoma.
Subsequently, we investigated whether CARD14 mutations had an impact on the outcome of patients (prognosis). We found that individuals with altered CARD14 exhibited a notably less favorable prognosis in terms of disease-free survival (DFS), with a statistically significant log-rank test P value of 0.0145. However, this group did not show statistical significance in terms of disease-specific survival (DSS), overall survival (OS), or progression-free survival (PFS) (Fig. 1C). We were interested in knowing whether the CARD14 mutations found in TCGA had been previously described in individuals suffering from CARD14-induced skin inflammatory conditions. Notably, of the 146 TCGA CARD14 mutations, 44 (30.5%) were described in ClinVar to be associated with autoinflammatory syndrome, psoriasis, PRP or inborn genetic disease. The top five cancers harboring ClinVar mutations were endometrial cancer (34.2%), esophagogastric cancer (15.8%), colorectal cancer (10.5%), melanoma (10.5%) and bladder cancer (7.9%) (Fig. 1D). Hence, CARD14 gene mutations are present in cancer and seem to have an impact on patient outcome.
CARD14 gene expression is dysregulated in a broad variety of cancers
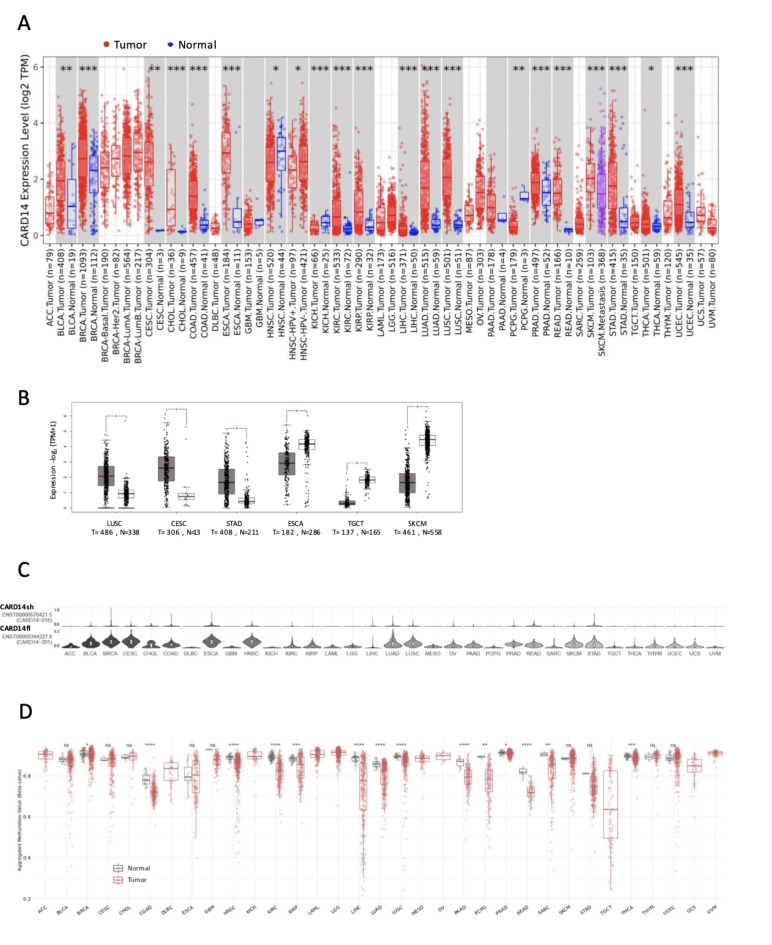
Gene mutations are not the only possible cause of cancer, as dysregulated expression of genes might also promote tumorigenesis22. Hence, we analyzed RNA-seq datasets to examine the expression of CARD14 in both normal and cancer tissues. Strikingly, CARD14 upregulation was observed in several tumor types (Fig. 2A), including BLCA, BRCA, CESC, CHOL, COAD,
Fig. 2.
CARD14 gene expression across cancers. (A) TIMER2.0 was used to plot CARD14 RNA levels in TCGA samples, statistical significance was calculated with the Wilcoxon test (*p < 0.05; **p < 0.01; ***p < 0.001). (B) Expression levels of the CARD14 gene in tumor and normal tissues from the TCGA and GTEx datasets, respectively, were analyzed with GEPIA2. The log2FC cutoff was 1, and the p value cutoff was 0.01. (C) Analysis of CARD14 RNA isoform expression in different cancer types with GEPIA2. (D) Methylation status of the CpG islands in the CARD14 promoter across various cancers analyzed with the SMART app. One-way ANOVA: ns, p > 0.05; *, p < = 0.05; **, p < = 0.01; ***, p < = 0.001; ****, p < = 0.0001.
ESCA, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD, READ, SKCM, STAD, THCA and UCEC, which is surprising due to the limited expression of CARD14 in healthy tissues (Supplementary Fig. 1A), particularly in epithelial cells from the lining epithelium (Supplementary Fig. 1B). In contrast, it was downregulated in HNSC, KICH, and PCPG. CARD14 expression remained unaltered in other cancer types, such as GBM and PAAD. Due to the reduced number of normal samples in the TCGA for some tumors, we used GEPIA2 to compare tumor expression versus the GTEx dataset, which contains expression data for a large number of normal tissues. CARD14 was upregulated in LUSC, CESC, and STAD but was significantly downregulated in ESCA, TGCT, and SKCM (Fig. 2B).
We later interrogated whether differential transcription occur for CARD14 in cancers, as three protein-coding transcripts of CARD14 have been reported to exist in human cells: full-length (fl.) (ENST00000344227, consensus sequence), short (sh) (ENST00000570421) and cardless (cl) (ENST00000573754.5) CARD14. We investigated the cancer isoform expression of CARD14 with GEPIA2. Surprisingly, CARD14fl was the most abundantly expressed isoform across cancer types, and virtually no expression of CARD14sh was found, despite previous reports of this isoform as the most abundant isoform in different tissues12,23 (Fig. 2C and Supplementary Fig. 2A). We checked the GTEX dataset to determine whether the increased expression of CARD14fl could be a cancer-specific feature and found that in normal tissues, CARD14fl was the most highly expressed isoform, whereas the expression of CARD14sh was very low (Supplementary Fig. 2B).
Promoter methylation controls gene transcription and plays a pivotal role in neoplasia24. Consequently, we examined CARD14 promoter methylation levels in cancerous and normal tissues. We observed a significant decrease in CARD14 promoter methylation in numerous malignancies, including BRCA, COAD, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, PAAD, PCPG, PRAD, READ, SARC, and THCA. However, promoter methylation levels remained unaltered in BLCA, CESC, CHOL, ESCA, GBM, SKCM, STAD, THYM, and UCEC (Fig. 2D), all of which exhibited higher CARD14 gene expression in tumor samples than in controls. These findings suggest that CARD14 promoter demethylation is involved in the increased CARD14 expression observed in most tumors; however, other mechanisms, such as transcription factor activity, or histone post-translational modifications could regulate the expression of CARD14 without affecting promoter methylation.
In conclusion, these results show that important dysregulations of CARD14 levels occur in cancer. In this manner, the considerable big amount of significant differences found in Fig. 1 indicates that CARD14 may have a potential role in several cancer types, either as diagnostic/prognostic marker or as therapeutic target.
CARD14 protein levels in cancer are positively correlated with RNA levels
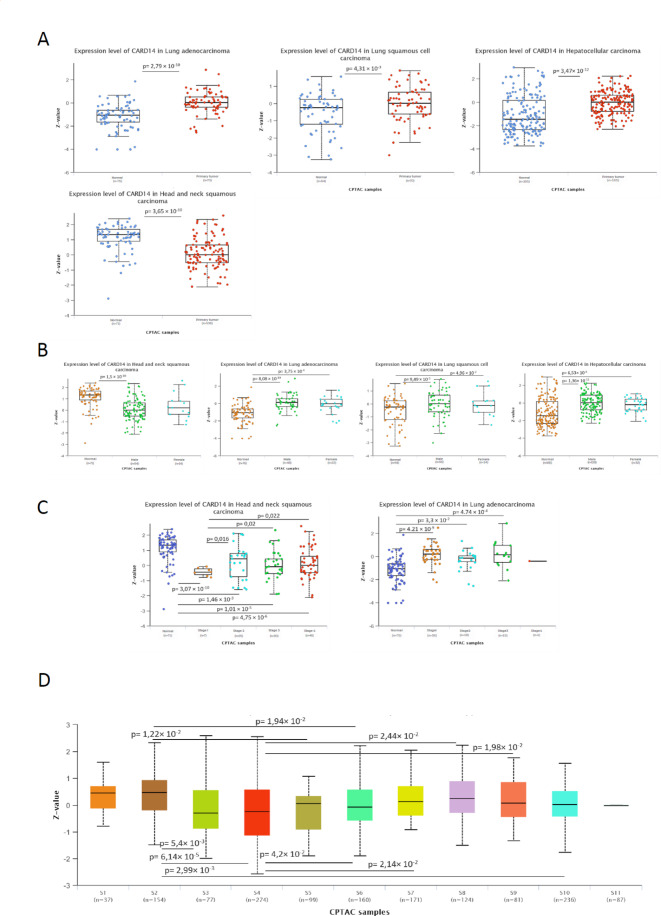
We wanted to determine whether the changes in CARD14 RNA in cancer were maintained at the protein level, since discrepancies between RNA and protein levels are commonly observed with potential implications for diagnosis and therapy25. We used UALCAN26,27 to compare tumor and normal tissues within the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) database. CARD14 protein levels were significantly elevated in LUAD, LUSC, and LIHC, with p values of 2.79 × 10− 10, 4.3 × 10− 3, and 3.4 × 10− 12, respectively. In contrast, HNSC protein levels were lower than controls, with a p value 3.65 × 10− 10, (Fig. 3A).
Fig. 3.
CARD14 protein expression in cancer. UALCAN CARD14 protein expression from the CPTAC database in (A) LUAD, LUSC, LIHC and HNSC. (B) CARD14 protein expression in female and male cancer patients. (C) CARD14 expression levels according to cancer stage in HNSC and LUAD patients. (D) CARD14 expression levels in cancer subtypes. Protein expression values downloaded from the CPTAC were log2 transformed. Then a Z-value for each sample for each protein was calculated as standard deviations from the median across samples. Welch’s T-test estimated the significance of differences in expression levels between normal and primary tumors or tumor subgroups. Anova was used for multiple comparisons. P value < 0.05 was considered to be statistically different.
We next assessed whether clinical parameters were associated with CARD14 protein levels but found no statistically significant differences between sexes in HNSCC, LUAD, LUSC or HCC patients (Fig. 3B), indicating that sex hormones may not play a role in the regulation of CARD14. As cancers progress, they can be classified in different stages depending on the extent to which they have grown and spread. We analyzed the levels of CARD14 protein through tumor stages in HNSC and LUAD (the only cancer types with available data). There was a significant decrease in the CARD14 protein level in stage 1, with subsequent stages exhibiting greater expression than stage 1 but less expression than normal tissue in HNSC (Fig. 3C). Moreover, a significantly greater amount of the CARD14 protein was detected in all LUAD stages than in normal tissues. Recently, tumor subtypes have been proposed based on their proteomes28. Subtypes S1 (associated with the proteasome complex and ubiquitin) and S2 (associated with T cells and the immune response) had the highest CARD14 protein levels, whereas subtypes S3 (associated with basal-like breast cancer) and S4 (associated with the epithelium and oxidative phosphorylation and TCA cycle pathways) had the lowest levels (Fig. 3D). These data indicate that, in the assessed tumors, the changes in RNA levels are followed by the same protein changes.
Association between CARD14 expression levels and patient survival
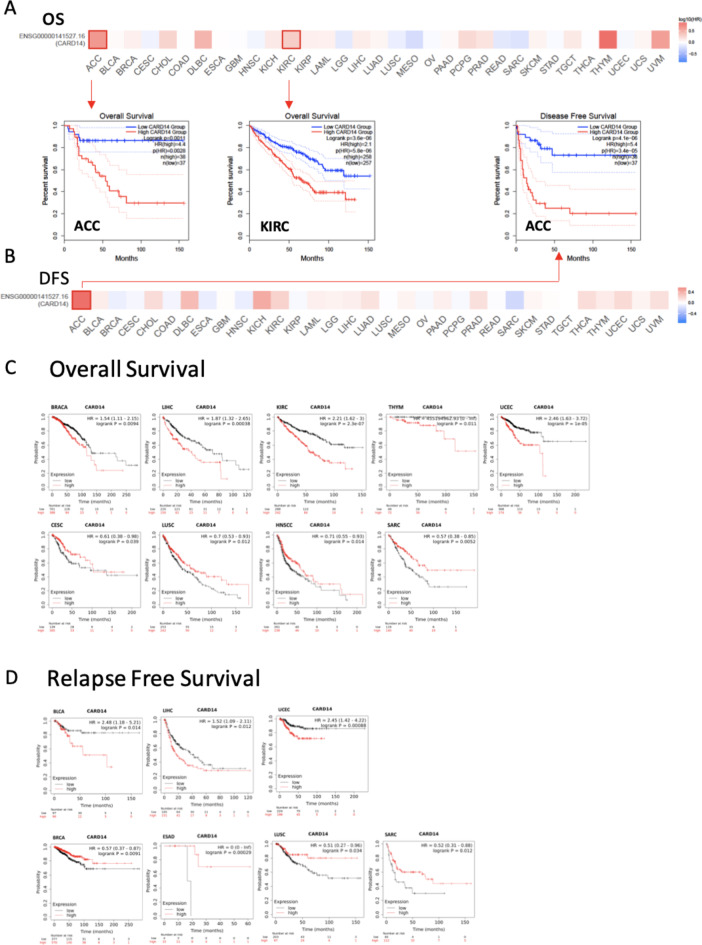
To determine whether the differential expression of RNA has an impact on disease, we analyzed the correlation between CARD14 expression and the prognosis of cancer patients. We used GEPIA 2.0 for the analysis of overall survival (OS) and disease-free survival (DFS). Notably, elevated CARD14 expression was associated with shorter OS (Fig. 4A) and DFS (Fig. 4B) in patients with ACC, with p values of 0.0011 for OS and 4.1e-6 for DFS. Similar trends were noted in KIRC, where the high-CARD14 cohort exhibited significantly poorer OS (Fig. 4A), with a p value of 3.6e − 06, than did the low-CARD14 cohort.
Fig. 4.
Survival analysis of patients stratified by the expression of CARD14 across cancers. Correlation between CARD14 expression and the survival outcomes of patients with diverse tumors, including both (A) overall survival (OS) and (B) disease-free survival (DFS), using the GEPIA2 website. The analysis employed the log-rank test with a significance level of P < 0.05 and a p value adjustment by FDR, and the cutoff was a median of 50%. Outlined squares indicate significance. (C) Kaplan‒Meier plots obtained from Kaplan‒Meier plotter comparing overall survival (OS) and (D) relapse-free survival (RFS) across different types of cancer. The p values were obtained from a log-rank test between two groups, a log rank p value < 0.05 was considered to be statistically significant.
We further investigated patient prognosis associated with CARD14 gene expression with Kaplan‒Meier Plotter. This tool allows rapid assessment of all possible cutoff values, choosing the best performing threshold for the analysis. Patients with high CARD14 expression had shorter OS in the BRACA (p = 0.0094), LIHC (p = 0.00038), KIRC (p = 2.3e − 07), THYM (p = 0.011), and UCEC (p = 1e − 05) cohorts (Fig. 4C). Conversely, the opposite trend was observed for other cancer types, indicating that high CARD14 expression is associated with a better prognosis in CESC (p = 0.039), LUSC (p = 0.012), HNSCC (p = 0.014), and SARC (p = 0.0052). No significant differences in OS were found in BLCA, ESCA, ESCC, KIRP, LUAD, PAAD, TGCT, PCPG, READ, STAD, THCA, or OC (Supplementary Fig. 3A). On the other hand, RFS showed that high CARD14 expression was linked to poor prognosis in BLCA (p = 0.014), LIHC (p = 0.012), and UCEC (p = 0.00088), while it was associated with better prognosis in BRCA (p = 0.0091), ESAD (p = 0.00029), LUSC (p = 0.034), and SARC (p = 0.012) (Fig. 4D). No differences were detected between patients with CARD14 overexpression and those with low CARD14 expression in KIRP, PCPG, READ, TGCT, THCA, CESC, ESCA, HNSC, KIRC, LUAD, OC, PAAD, or STAD (Supplementary Fig. 3A).
In addition, Cox analysis was carried out to determine the associations between CARD14 expression levels and overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) (Supplementary Fig. 4). CARD14 was a high-risk gene for KIRC, UVM, ACC, LIHC, BRCA and LUAD and a low-risk gene for OS in patients with LGG. For DFI, CARD14 was a high-risk gene in ACC and a low-risk gene in BRCA. In terms of the PFI, CARD14 was linked to a worse prognosis in ACC, KIRC, UVM, PRAD and KICH. Finally, CARD14 was a high-risk gene for DSS in KIRC, UVM and ACC patients.
Our observations underscore an association between CARD14 levels and disease outcomes across specific cancer types. For instance, elevated expression of CARD14 associates with survival decline in certain cancer types, hence, we can hypothesize that targeting CARD14 may represent an effective therapeutic option in those cancer types.
Correlation between CARD14 expression and immune cell infiltration
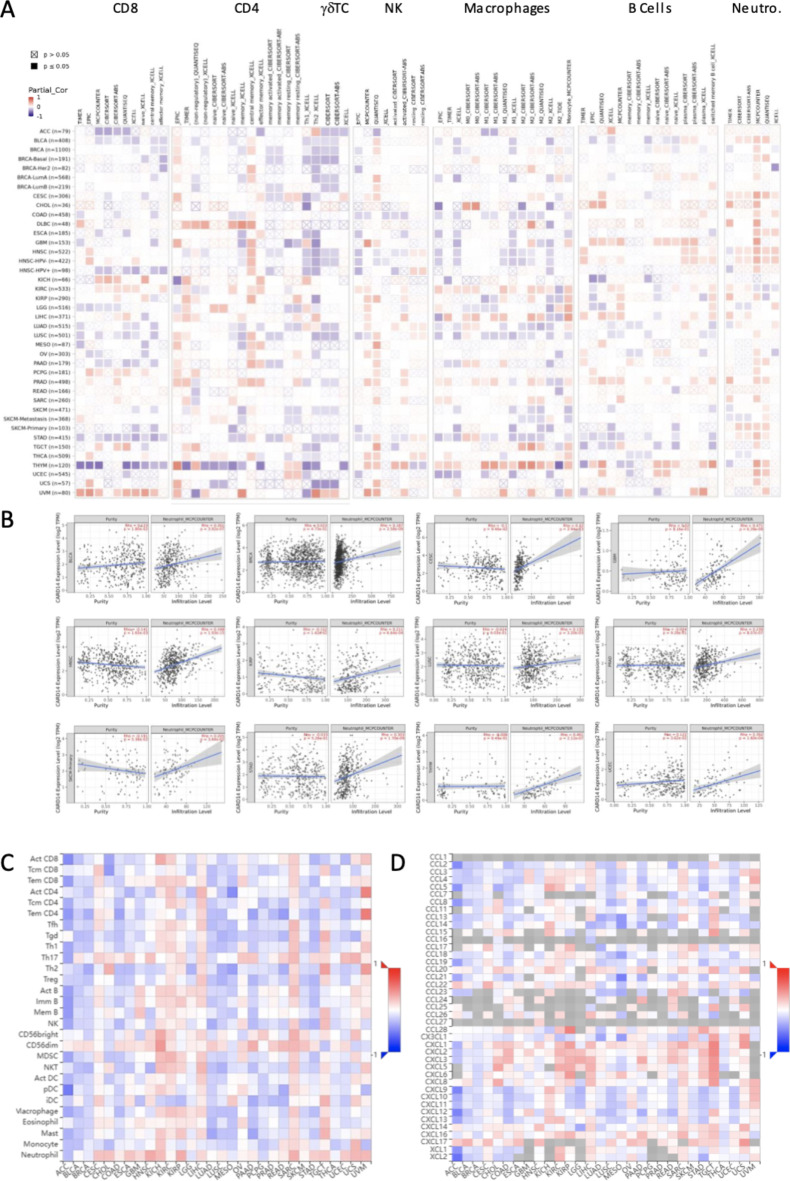
The immune system is an active player in the fight against cancer29, and one-way CARD14 might influence patient prognosis could be by regulating immune cell infiltration. To understand the mechanism by which CARD14 levels associated with survival improvement or decline we examined the relationship between immune cell recruitment and the RNA levels of CARD14 in tumors within the TCGA database using TIMER 2.0 (Fig. 5A). Our findings revealed a negative correlation between CARD14 expression and CD8 + T-cell infiltration in ACC, BLCA, HNSC-HPV+, LGG, LUAD, LUSC, OV, PAAD, STAD, and THYM. Conversely, a positive correlation with infiltrating CD8 + T cells was observed in the KICH, KIRC, PCPG, TGCT, and UVM cohorts. Furthermore, we observed a negative correlation between CARD14 and CD4 + T cells in THYM and a positive correlation between CARD14 and CD4 + T cells in DLBC patients. In most tumors, there was a significant negative correlation between γδ T cells and CARD14 expression. The number of NK cells was negatively correlated with the levels of CARD14 in COAD and HNSC and positively correlated with KIRC, KIRP, PRAD and THCA. For macrophages, positive correlations were detected in KIRC, LIHC, TGCT, and THYM, while negative correlations were detected in ACC, BLCA, BRCA-LumA, COAD, ESCA, GBM, HNSC, HNSC-HPV-, LUSC, SKCM, STAD and UCEC. We detected a positive correlation between B cells and GBM, HNSC, HNSC-HPV-, LIHC, and PRAD and a negative correlation between B cells and KICH, SKCM-Primary, STAD, TGCT, and THCA.
Fig. 5.
Correlation between CARD14 expression and immune cell infiltration. (A) TIMER2 correlation between the RNA expression of the CARD14 gene and immune cell type infiltration in each cancer type. (B) Correlation between CARD14 and neutrophils determined via deconvolution using the MCPCOUNTER algorithm. (C) TISIDB was used to analyze the correlation between CARD14 gene expression and immune cell infiltration or (D) chemokine expression in different cancers. P-values and partial correlation were obtained by Spearman’s rank correlation test after adjusting by purity. And all data were displayed as a heatmap and scatter plot. ns, p > 0.05; *, p < = 0.05; **, p < = 0.01; ***, p < = 0.001; ****, p < = 0.0001.
Neutrophils (Fig. 5A and B) showed a positive correlation in most tumors with little discrepancies between algorithms. A positive correlation between neutrophils and CARD14 expression was also found in TISIDB (Fig. 5C), which uses a different approach to infer immune cell infiltration from bulk RNA-seq data. Notably, we also found that in TISIDB, the expression of CARD14 was positively correlated with the expression of the chemokines CXCL1, CXCL2, and CXCL3 (Fig. 5D), which are known to act as chemoattractants for neutrophils30, suggesting that CARD14 could play a role in the recruitment of this cell type.
CARD14-related genes and their contribution to cancer
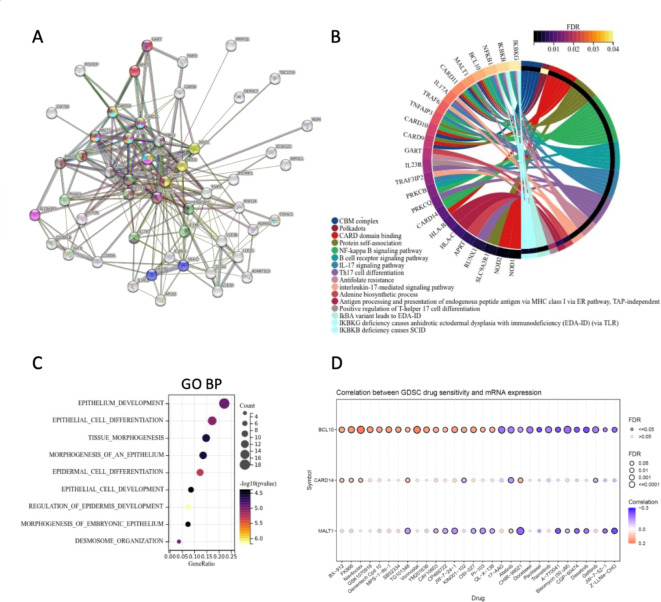
To further determine the molecular function of CARD14 in cancer, we gathered the genes with similar expression patterns and CARD14-interacting proteins for subsequent pathway enrichment analyses. We explored the functional interactions of CARD14 using the known and predicted protein–protein interaction (PPI) database in the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (Fig. 6A). From the fifty top predicted functional partners, we obtained a PPI enrichment p value of < 1.0e-16. The FDRs of the most enriched pathways containing those genes can be found in Supplementary Table 1. Twelve of the genes were from the KEGG31 category NF-kappa B signaling pathway, and other common pathways included CARD domain binding, the B-cell receptor signaling pathway, and the IL-17 signaling pathway. To better visualize the interaction between proteins, we drew circle diagrams of the enrichment results from only the genes contained in the most significant pathways (Fig. 6B and Supplementary Table 1).
Fig. 6.
Function of CARD14 and its associated proteins. (A) The interaction network of CARD14 and its partners was generated with STRING. Proteins are represented as nodes, and lines indicate associations based on known functional interactions in humans. The network was significantly enriched in interactions (PPI enrichment p value: 1.07e–16, FDR < 0.05). (B) Circle diagram of genes and their pathways; only enriched pathways from the String database with FDRs below 0.04 were used. (C) Functional enrichment analysis of Gene Ontology (GO) biological process (GO BP) terms with a false discovery rate (FDR) < 0.1 and a p value < 0.05. (D) Drug sensitivity of CARD14, MALT1 and BCL10 according to the GSCA Lite database. The bubble plot shows the correlation between gene expression and drugs. A positive Spearman correlation coefficient indicates that upregulated gene expression is associated with drug resistance whereas negative correlation indicates a higher gene expression may make drug sensitive.
To explore the pathways in which CARD14 might influence carcinogenesis, we used GEPIA2 to download the 100 genes most commonly expressed with CARD14 in TCGA tumors. Subsequently, we removed pseudogenes and lncRNAs (FAM83H-AS12, CTD-2015H6.3, RP11-429J17.7, RP11-747H7.3, RP11-473M20.5, and BZW1P2) for downstream analysis. With this gene set (Supplementary Table 2), we performed functional analysis with GO biological process (Fig. 6C) and found that epithelial development, differentiation and morphogenesis were the most enriched biological processes.
Finally, we used Gene Set Cancer Analysis (GSCALite), which contains data from 869 cell lines related to cell death sensitivity after challenge with more than 250 small molecules32. Because CARD14 must form a CBM complex to activate NF-κB3, Pearson correlation analysis of CARD14, MALT1 and BCL10 mRNA expression and drug IC50 values was performed (top 30 correlations) (Fig. 6D). Our search of the Genomics of Drug Sensitivity in Cancer (GDSC) database revealed that increased expression of CARD14 and BCL10 rendered cells more sensitive to afatinib and gefitinib, whereas increased expression of these genes correlated with resistance to BX-912, FK866, Navitoclax and TG101348.
Taken together these results show that CARD14 acts in synergy with other genes involved in epithelial homeostasis and drugs disrupting epithelium proliferation signals through EGF, might be a good way to treat cancers with high CARD14 levels.
Discussion
CARD14 is most known for its association with psoriasis5. Its aberrant signaling due to mutations promotes inflammation through NF-κB activation. Because sustained inflammation is one of the hallmarks of cancer33 and right so, psoriatic patients have an increased risk for cancer, CARD14 might be involved in tumorigenesis. Here, we analyzed the features of CARD14 in cancer, from its expression to its relevance for patient survival, to achieve a better understanding of CARD14 and its implications for human neoplasia.
We found that TCGA CARD14 mutations tend to spread evenly, with a moderate increased frequency between aminoacid 200 and 600 of CARD14 sequence, not resembling the location pattern of psoriasis Gof mutations, which concentrate between aminoacid 100 and 20034. Hence, CARD14 missense mutations might equally predispose to cancer, regardless of their contribution to psoriasis. Interestingly, some of the TCGA CARD14 mutations, such M119T and G117S, have also been described to be associated with psoriasis. Although psoriasis Gof mutations might be highly penetrant and dominant12, they are rare, and the fact that they are also present in some cancer patients might indicate that psoriasis patients with CARD14 variations could have an enhanced susceptibility to a particular type of cancer. However, the number of patients with a given CARD14 mutation was too low to draw meaningful conclusions in this regard. On the other hand, the contribution of common CARD14 variations, while predisposing patients to psoriasis, might have a weaker effect on tumorigenesis35.
Despite the rather limited expression of CARD14 in normal squamous epithelia, prostate, salivary and mammary glands (particularly in epithelial cells), we observed widespread dysregulation of CARD14 expression in tumor tissue, which is overexpressed in many cancers. Notably, among those with higher CARD14 levels, CARD14 is the fourth most differentially expressed gene in gastric cancer, and patients can be stratified according to their CARD14 levels, with reduced survival, among those with higher CARD14 levels36. In agreement with our results, CARD14 was overexpressed and associated with tumor proliferation in breast cancer, since its knockdown induces apoptosis and reduces cell migration37. Moreover, in prostate cancer, overexpression of CARD14 has been associated with poor prognosis and increased invasiveness38. Aside from CARD14 Gof mutations that cause spontaneous CBM assembly, elevated levels of CARD14 alone also trigger the proteolytic function of MALT1, which is essential for CARD14-induced inflammatory responses4; therefore, high CARD14 levels might strongly impact the activation of signaling cascades in cancer cells.
Interrogating datasets generated with RNAseq, we were able to describe the actual abundance of CARD14 isoforms. CARD14sh lacks a small portion of the C-terminus of CARD14fl, whereas CARD14cl contains only a part of the coiled-coil domain and a stretch of sequence thereafter23. To date, no difference in the function of CARD14fl and CARD14sh has been reported, but CARD14cl is known to be unable to activate NF-KB in the same manner12. CARD14sh is thought to be the predominant isoform, and thus, most of the studies on CARD14 function have used CARD14sh12,23,39. We found that CARD14fl is the dominant isoform in both cancers and normal tissues; hence, it should be used in studies aiming to investigate the role of CARD14.
Survival analysis revealed that CARD14 might be a valuable biomarker for patient prognosis in some cancer types. However, in some cancers, high levels of CARD14 are linked to a better prognosis, whereas in other cancers, high levels of CARD14 are linked to a worse prognosis. It is likely that the specific cell types in each cancer might activate different pathophysiologic mechanisms depending on CARD14 expression, with different implications for cell and tumor fate. In this regard, the tumor microenvironment (TME) plays an important role in the progression of cancer; for instance, cancer-associated fibroblasts have been linked to poor prognosis40. Adaptive and innate immune cells are also present in the tumor environment and can have both suppressive and promoting functions in tumor development, potentially allowing their use as biomarkers for patient prognosis29. CARD14 expression, although modestly, tends to positively correlate with neutrophil infiltration in most tumor types. Notably, extensive neutrophil infiltration is a hallmark of psoriasis41, in CARD14-mutant mouse models10 9 11 and in psoriatic patients with Gof CARD14 mutations12. Of note, ten different infiltrating neutrophil states have been described recently with opposing outcomes for cancer patients42; hence, depending on the neutrophil subset attracted by CARD14, patients might see their prognosis worsen or improve. Furthermore, CARD14 may have both pro-tumorigenic and anti-tumorigenic roles depending on the tissue context because of its function activating NF-KB. The transcription factor NF-KB has very diverse roles in most biological processes with both cell-intrinsic and cell-extrinsic mechanisms that control inflammation, survival, proliferation and cell death43. The number and types of genes regulated by NF-KB in one cell might change due to re-stimulation or modulating signals and “noise” from the environment44. Nonetheless, different cell types might have different active promoters available for NF-KB dependent transcription45. For all these, it is no surprise that CARD14 expression level could influence patient prognosis for good and for bad depending on the cancer type. Additional studies focusing on the role of CARD14 in a particular cancer are needed to disentangle the complex role of CARD14 in specific tumors.
CARD14 is functionally associated to proteins involved in inflammation and co-expressed with a set of genes strongly related to epithelia homeostasis processes, such development and differentiation. EGFR (epidermal growth factor receptor) tyrosine kinase is expressed in epithelial tissues, has a pivotal role in epithelial development46 and is frequently mutated in cancers47. In connection with that, we found that cells with higher expression of CARD14 were more susceptible to Afatinib and Gefitinib, which are both EGFR inhibitors48. These indicates that patients with elevated expression of CARD14 in their tumors might particularly benefit from this type of drugs, not only they would reduce the unwanted effects of mutated EGFR but could also indirectly counteract CARD14 activity through their anti-inflammatory effects49.
To summarize, this comprehensive assessment provides insights into the intricate role of CARD14 in cancer, shedding light on its potential as a prognostic marker and therapeutic target across diverse malignancies. Our analysis will serve as a starting point for further studies aiming to elucidate the role of CARD14 in the different cancer types. Understanding the molecular mechanisms underlying CARD14 dysregulation in each cancer could pave the way for novel diagnostic and therapeutic strategies in oncology.
Materials and methods
Genetic alteration analysis of CARD14 in cancer
The cBioPortal database version 5.4.5 (http://www.cbioportal.org/) was used to investigate genetic alterations in the CARD14 gene (NM_024110) across cancer types described in the TCGA. “CARD14” term was searched in the “Quick Search” to obtain details about mutation types, frequencies, copy number alterations (CNAs), and structural variants in the “Cancer Types Summary.” Information about mutation sites in the CARD14 protein was obtained from the “Mutations” module. Additionally, the “comparison/survival” methods were used to assess survival curves, such as disease-free survival (DFS), disease-specific survival (DSS), overall survival (OS), and progression-free survival (PFS), for patients with altered and unaltered CARD14 expression via the log-rank test. ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) was used in this study to link CARD14 mutations with human diseases. ClinVar follows the HGVS standard, accurately representing human variations as sequence alterations in mRNA, genomic and protein reference sequences elucidating their significance for human health.
Differential gene expression analysis of CARD14 in cancer
We used the Timer2.0 database (http://timer.cistrome.org/) to investigate the differences in CARD14 gene expression between tumor and normal tissues via the “Diff Exp” module. For the analysis of differential gene expression between TCGA tumor and GTEX normal tissue data, we used GEPIA2 (http://gepia2.cancer-pku.cn/#analysis). We generated box plots to visualize the differences in the expression of CARD14, and the Wilcoxon test was used to assess differences between groups. Gepia2 was also used to plot CARD14 isoform expression across cancers. Additionally, UALCAN (https://ualcan.path.uab.edu/) was used to acquire data on the differential protein expression of CARD14 in tumor and normal tissues from the CPTAC database. For the normal tissue distribution of CARD14, we used GTEX (https://www.gtexportal.org/).
For CARD14 promoter methylation analysis, we used the SMART (Shiny Methylation Analysis Resource Tool) app (http://www.bioinfo-zs.com/smartapp/). Only promoter CpGs were considered for the analysis (cg14893129, cg20623503, cg06707993, cg11271526, cg05022061, cg25565082, cg22082329, cg06209035, cg03290213, cg08699655, cg26619987, cg13855862, cg18595957, cg26257145, cg25838977 and cg12848405).
Association between CARD14 expression levels and patient survival
The survival map function of Gepia2 was used to stratify cancer patients into high- and low-expression groups based on the median (50%) expression value of CARD14, employing the log-rank test with a significance level of P < 0.05. Survival analysis was also conducted with Kaplan–Meier Plotter (https://kmplot.com/analysis/), which generated Kaplan–Meier survival curves for overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI). These results are presented as Kaplan–Meier curves.
Furthermore, forest plots with Cox proportional hazards analysis were generated with the Sanger-Box tool (http://www.sangerbox.com/home.html), which is based on the R package survival, utilizing the log-rank test and P < 0.05 significance threshold.
Immune cell infiltration analysis
The associations between CARD14 expression and immune cells, including CD8 + T cells, CD4 + T cells, γδ T cells, NK cells, neutrophils, macrophages, and B cells, were examined using TIMER 2.0 (http://timer.cistrome.org/). CARD14 was analyzed through the “Immune” module on the TIMER2 website, and various immune cells of interest were selected. The investigation employed the XCELL, TIDE, EPIC, TIMER, CIBERSORT-ABS, CIBERSORT, MCPCOUNTER, and QUANTISEQ algorithms to explore the relationship between immune inflammatory cells and CARD14 expression in the TCGA database. The correlation between CARD14 expression and immune cells was assessed using Spearman’s correlation coefficient, with statistical significance defined as p < 0.05. Positive correlations are highlighted in red, indicating p > 0. Negative correlations are highlighted in blue, indicating p < 0. Additionally, the relationships between the abundance of tumor-infiltrating lymphocytes (TILs) and chemokines and the expression of CARD14 were analyzed with TISIDB (http://cis.hku.hk/TISIDB/index.php), which uses immune-related signatures of 28 immune cell types from Charoentong’s study50.
CARD14-related genes and enrichment analysis
The top 100 genes with the most comparable expression patterns to CARD14 were extracted from TCGA using the “Similar Gene Detection” module of GEPIA2, and their correlation was assessed through the “correlation analysis” module with Pearson correlation coefficients.
The STRING database has been used to explore protein‒protein interactions (PPIs) and construct a CARD14 organizational network by selecting a physical subnetwork (the edges indicate that the proteins are part of a physical complex, although they may not directly interact) with a maximum of 50 interactors and a minimum required interaction score of medium confidence of 0.4.
For GO (Gene Ontology) enrichment analysis with CARD14-related genes sourced from GEPIA2, we used the R software package clusterProfiler. The gene sets needed to contain at least 5 genes and no more than 5000 genes. P values < 0.05 (adjustable) and a false discovery rate (FDR) < 0.25 (adjustable) were considered to indicate statistically significant results.
CBM drug sensitivity
Data from the TCGA and Drug Sensitivity in Cancer (GDSC) databases were analyzed via Gene Set Cancer Analysis (GSCALite) (http://bioinfo.life.hust.edu.cn/web/GSCALite/).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Natalia Artigas for manuscript reading and discussion. This research was funded by the Spanish Ministry of Science and Innovation PID2021-127724OA-100 to MIS, PID2021-126249OA-I00 to J.M and PID2020-114477RB-I00 to C.S. from the MCIN/AEI/10.13039/501100011033) and by the Catalan Government (2021-SGR-00275, JJR). IF acknowledges the financial support of the Generalitat de Catalunya for the PhD scholarship FI-SDUR (2021 FISDUR 00331).
Author contributions
DB and DP contributed equally to this work.DB, DP, SVG, JF, DLR, IF, LW, and PAO were involved in methodology; formal analysis; data curation; investigation; resources; software; visualization; writing original draft. SCL, SVF and JJR were involved in data analysis and/or interpretation. MIS, CS and JM were involved in supervision; validation; funding acquisition.
Data availability
The data that support the findings of our study is readily available from, CBIOPORTAL, GEPIA2, Kaplan-Meier Plotter, TIMER, UALCAN, The SMART app, TISIDB, GSCALite, GTEX and TCGA database at http://www.cbioportal.org/, http://gepia.cancer-pku.cn/index.html, www.kmplot.com, https://cistrome.shinyapps.io/timer/, https://ualcan.path.uab.edu/, http://www.bioinfo-zs.com/smartapp/, http://cis.hku.hk/TISIDB/index.php, http://bioinfo.life.hust.edu.cn/web/GSCALite, https://www.gtexportal.org/, https://tcga-data.nci.nih.gov/tcga/.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Daniil Bespalov and Dayana Pino.
References
- 1.Lu, H. Y. et al. Germline CBM-opathies: from immunodeficiency to atopy. J. Allergy Clin. Immunol.143, 1661–1673 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Liu, T., Zhang, L., Joo, D. & Sun, S. C. NF-κB signaling in inflammation. Signal. Transduct. Target. Ther.2, 17023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howes, A. et al. Psoriasis mutations disrupt CARD14 autoinhibition promoting BCL10-MALT1-dependent NF-κB activation. Biochem. J.473, 1759–1768 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afonina, I. S. et al. The paracaspase MALT 1 mediates ARD 14-induced signaling in keratinocytes.EMBO Rep.17, 914–927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan, C. T. et al. Rare and common variants in CARD14, encoding an epidermal Regulator of NF-kappaB, in Psoriasis. Am. J. Hum. Genet.90, 796–808 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs-Telem, D. et al. Familial pityriasis rubra pilaris is caused by mutations in CARD14. Am. J. Hum. Genet.91, 163–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka, M. et al. Essential role of CARD14 in murine experimental psoriasis. J. Immunol.200, 71–81 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Wang, M. et al. Gain-of-function mutation of Card14 leads to spontaneous psoriasis-like skin inflammation through enhanced keratinocyte response to IL-17A. Immunity. 49, 66–79e5 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Manils, J. et al. CARD14E138A signalling in keratinocytes induces TNF-dependent skin and systemic inflammation. Elife. 9, (2020). [DOI] [PMC free article] [PubMed]
- 10.Mellett, M. et al. CARD14 gain-of-function mutation alone is sufficient to drive IL-23/IL-17–Mediated psoriasiform skin inflammation in vivo. J. Invest. Dermatol.138, 2010–2023 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Van Nuffel, E. et al. MALT 1 targeting suppresses < scp > CARD 14-induced psoriatic dermatitis in mice. EMBO Rep.21, (2020). [DOI] [PMC free article] [PubMed]
- 12.Jordan, C. T. et al. PSORS2 is due to mutations in CARD14. Am. J. Hum. Genet.90, 784–795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeVore, S. B. et al. Novel role for caspase recruitment domain family member 14 and its genetic variant rs11652075 in skin filaggrin homeostasis. J. Allergy Clin. Immunol.149, 708–717 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong, A. W. & Read, C. Pathophysiology, clinical presentation, and treatment of psoriasis. JAMA. 323, 1945 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Daugaard, C., Iversen, L. & Hjuler, K. F. Comorbidity in adult psoriasis: considerations for the clinician. Psoriasis: Targets Therapy. 12, 139–150 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi, K. & Karin, M. NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol.18, 309–324 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Zhao, H. et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal. Transduct. Target. Ther.6, 263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang, M. et al. Tumor cell-activated CARD9 signaling contributes to metastasis-associated macrophage polarization. Cell. Death Differ.21, 1290–1302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McAuley, J. R., Freeman, T. J., Ekambaram, P. & Lucas, P. C. & McAllister-Lucas, L. M. CARMA3 is a critical mediator of G protein-coupled receptor and receptor tyrosine kinase-driven solid tumor pathogenesis. Front. Immunol.9, (2018). [DOI] [PMC free article] [PubMed]
- 20.Lenz, G. et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Sci. (1979). 319, 1676–1679 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Vaengebjerg, S., Skov, L., Egeberg, A. & Loft, N. D. Prevalence, incidence, and risk of cancer in patients with psoriasis and psoriatic arthritis. JAMA Dermatol.156, 421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, Y. et al. A pan-cancer compendium of genes deregulated by somatic genomic rearrangement across more than 1,400 cases. Cell. Rep.24, 515–527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scudiero, I. et al. Alternative splicing of CARMA2/CARD14 transcripts generates protein variants with differential effect on NF-κB activation and endoplasmic reticulum stress‐induced cell death. J. Cell. Physiol.226, 3121–3131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esteller, M. Epigenetics in cancer. N. Engl. J. Med.358, 1148–1159 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Buccitelli, C. & Selbach, M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet.21, 630–644 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Chandrashekar, D. S. et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 19, 649–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandrashekar, D. S. et al. An update to the integrated cancer data analysis platform. Neoplasia 25. UALCAN, 18–27 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, Y., Chen, F., Chandrashekar, D. S., Varambally, S. & Creighton, C. J. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat. Commun.13, 2669 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petralia, F. et al. Pan-cancer proteogenomics characterization of tumor immunity. Cell. 187, 1255–1277e27 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffith, J. W., Sokol, C. L. & Luster, A. D. Chemokines and chemokine receptors: positioning cells for Host Defense and immunity. Annu. Rev. Immunol.32, 659–702 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Kanehisa, M. K. E. G. G. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, C. J. et al. GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief. Bioinform.24, (2023). [DOI] [PubMed]
- 33.Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Israel, L. & Mellett, M. Clinical and genetic heterogeneity of CARD14 mutations in psoriatic skin disease. Front. Immunol.9, (2018). [DOI] [PMC free article] [PubMed]
- 35.Jiang, S. H., Stanley, M. & Vinuesa, C. G. Rare genetic variants in systemic autoimmunity. Immunol. Cell. Biol.98, 490–499 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Carino, A. et al. Analysis of gastric cancer transcriptome allows the identification of histotype specific molecular signatures with prognostic potential. Front. Oncol.11, (2021). [DOI] [PMC free article] [PubMed]
- 37.LIM, J. Y., KIM, S. W., KIM, B. & Park, S. J. Knockdown of CARD14 inhibits cell proliferation and migration in breast cancer cells. Anticancer Res.40, 1953–1962 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Vanneste, D. et al. CARD14 signalling ensures cell survival and cancer associated gene expression in prostate cancer cells. Biomedicines. 10, 2008 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berki, D. M. et al. Activating CARD14 mutations are associated with generalized pustular psoriasis but rarely account for familial recurrence in psoriasis vulgaris. J. Invest. Dermatol.. 135, 2964–2970 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Hanley, C. J. et al. Single-cell analysis reveals prognostic fibroblast subpopulations linked to molecular and immunological subtypes of lung cancer. Nat. Commun.14, 387 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hawkes, J. E., Chan, T. C. & Krueger, J. G. Psoriasis pathogenesis and the development of novel targeted immune therapies. J. Allergy Clin. Immunol.140, 645–653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, Y. et al. Neutrophil profiling illuminates anti-tumor antigen-presenting potency. Cell. 187, 1422–1439.e24 (2024). [DOI] [PubMed] [Google Scholar]
- 43.Zinatizadeh, M. R. et al. The nuclear factor kappa B (NF-kB) signaling in cancer development and immune diseases. Genes Dis.8, 287–297 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, A. G., Son, M., Kenna, E., Thom, N. & Tay, S. NF-κB memory coordinates transcriptional responses to dynamic inflammatory stimuli. Cell. Rep.40, 111159 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ngo, K. A. et al. Dissecting the regulatory strategies of NF-κB RelA target genes in the inflammatory response reveals differential transactivation logics. Cell. Rep.30, 2758–2775.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen, J. et al. Expression and function of the epidermal growth factor receptor in physiology and disease. Physiol. Rev.96, 1025–1069 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Sigismund, S., Avanzato, D. & Lanzetti, L. Emerging functions of the GFR in cancer. Mol. Oncol.12, 3–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park, K. et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol.17, 577–589 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Chen, Y. J. et al. Anti-inflammatory effect of afatinib (an EGFR-TKI) on OGD-induced neuroinflammation. Sci. Rep.9, 2516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charoentong, P. et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell. Rep.18, 248–262 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of our study is readily available from, CBIOPORTAL, GEPIA2, Kaplan-Meier Plotter, TIMER, UALCAN, The SMART app, TISIDB, GSCALite, GTEX and TCGA database at http://www.cbioportal.org/, http://gepia.cancer-pku.cn/index.html, www.kmplot.com, https://cistrome.shinyapps.io/timer/, https://ualcan.path.uab.edu/, http://www.bioinfo-zs.com/smartapp/, http://cis.hku.hk/TISIDB/index.php, http://bioinfo.life.hust.edu.cn/web/GSCALite, https://www.gtexportal.org/, https://tcga-data.nci.nih.gov/tcga/.