Abstract
Mason-Pfizer monkey virus immature capsids selected from the cytoplasm of baculovirus-infected cells were imaged by scanning transmission electron microscopy. The masses of individual selected Gag particles were measured, and the average mass corresponded to 1,900 to 2,100 Gag polyproteins per particle. A large variation in Gag particle mass was observed within each population measured.
The retroviral assembly process begins within the infected cell as retroviral Gag polyproteins associate with one another to enclose the RNA viral genome within a spherical particle (an immature capsid) with an electron-dense rim and a lucent center (19). Despite extensive study, the fine structure of an immature retroviral particle and the nature, of the molecular interactions that drive particle assembly remain unresolved. While the structures for the matrix and capsid proteins have been determined for several different retroviruses (5, 10, 11, 13, 14, 16, 17, 23), the structure of the complete Gag polyprotein precursor remains unknown. A hexagonal lattice of Gag polyproteins beneath the plasma membrane has been visualized for human immunodeficiency virus (HIV), and a similar arrangement was recently observed for Mason-Pfizer monkey virus (M-PMV) Gag particles assembled in bacteria (M. V. Nermut, P. Bron, D. Thomas, M. Rumlova, T. Ruml, and E. Hunter, submitted for publication), suggesting that the spherical retroviral immature capsid assembles in a manner that involves at least local symmetry (1, 2, 18–20). However, high-resolution cryo-electron microscopy (cryo-EM) images of both murine leukemia virus and HIV type 1 immature particles have failed to identify icosahedral symmetry within immature retroviral particles. While these particles were found to be ordered with radial symmetry and distinct layers, they were heterogeneous in size (9, 32). In addition, measurements of the masses of individual Rous sarcoma virus (RSV) virions by quantitative dark-field scanning transmission electron microscopy (STEM) have demonstrated a significant standard deviation in viral mass (30, 33). Taken together, these views of immature retroviral particles without a well-defined size or mass are inconsistent with a well-ordered icosahedral structure and suggest an alternative assembly mechanism.
In order to define the mass and population diversity of immature capsids of a retrovirus that assembles in the cytoplasm without a requirement for membranes, we analyzed M-PMV immature Gag particles by STEM. M-PMV Gag particles, which do not include viral membranes or surface glycoproteins, can be extracted by detergent cell lysis and purified by a series of sucrose gradients. The mass measurements generated by STEM demonstrate that M-PMV particles are heterogeneous in mass.
Isolated M-PMV Gag particles contain predominantly Gag and Gag-Pro polypeptides.
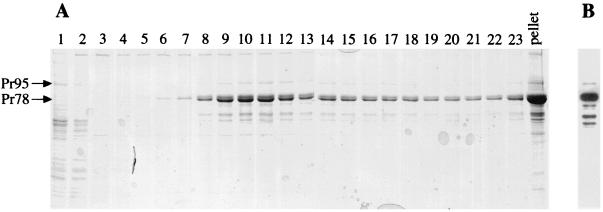
A recombinant baculovirus expressing the M-PMV gag, pro, and pol reading frames, with an A18V point mutation in the matrix domain of the corresponding Gag polyprotein, has been described previously (22). Spodoptera frugiperda (Sf9) cells were grown at 27°C in suspension in Grace's supplemented insect medium in a 1-liter spinner flask to a density of 0.5 × 106 to 1.0 × 106 cells per ml. At 60 h postinfection with recombinant baculovirus, the cells were harvested, concentrated by centrifugation at 100 × g, and resuspended on ice in 20 ml of hypotonic cell lysis buffer, which consisted of 10 mM Tris (pH 7.6), 1 mM EDTA, 1% Triton X-100 (Sigma-Aldrich Corp., St. Louis, Mo.), 1 mM phenylmethylsulfonylfluoride, and 2 μg of leupeptin (Sigma)/ml. After 30 min, the cell lysate was centrifuged at 3,000 × g for 10 min to remove nuclei and cell debris. The cleared cell lysate was loaded onto a 10-ml cushion of 35% (wt/vol) sucrose in 10 mM Tris (pH 7.6) buffer on top of 3 ml of 75% (wt/vol) sucrose in water in an SW28 centrifuge tube (Beckman Instruments, Fullerton, Calif.). The immature Gag particles were separated from the cell lysate by centrifugation at 25,000 rpm in an SW28 rotor (Beckman) for 2 h at 4°C and removed from the 35% sucrose–75% sucrose interface, followed by dialysis against a 10 mM Tris (pH 7.6) buffer at 4°C to remove the sucrose. The dialyzed sample was concentrated by centrifugation at 20,800 × g in a refrigerated microcentrifuge for 30 min, and the pellet was resuspended in 100 μl of cell lysis buffer with 500 mM NaCl. The resuspended Gag particles were further purified by centrifugation through a 5 to 20% (wt/vol) sucrose velocity gradient as described previously (22). Gradient fractions (0.5 ml each) were removed from the top of the gradient by a piston gradient fractionator (BioComp Industries, New Brunswick, Canada). A volume consisting of 5 μl (1%) of each gradient fraction and a resuspension of the gradient pellet was analyzed by polyacrylamide gel electrophoresis, and the gel was stained with Coomassie blue as described previously (27) (Fig. 1A). In Fig. 1, bands that correspond to Pr78Gag and Pr95Gag-Pro, as well as two smaller bands, are identified in the gel. Both smaller bands (Fig. 1B) are M-PMV Gag products, since they are found in a Western blot probed with rabbit anti-M-PMV Gag polyclonal serum. Moreover, trypsin digestion of each band after removal from an sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel generated several peptide fragments with a mass equal to that predicted for a tryptic digestion of the middle and the C-terminal regions of the Pr78Gag polyprotein, when measured by matrix-assisted laser desorption ionization–time of flight mass spectroscopy (data not shown). The smaller bands were not distinctly visible when 35S-labeled cell lysates containing assembled Gag particles were run directly on sucrose velocity gradients (without first being pelleted through a sucrose cushion; data not shown) and thus may represent Pr95 and/or Pr78 cleavage by cellular proteases after assembly and during the process of particle preparation for STEM. It is possible, but less likely, that the 50- and 54-kDa Gag proteins are present in the cell and are included with full-length Gag polyproteins during the particle assembly events. If so, an adjustment would be required in the calculation of the number of Gag polyproteins per particle, which is discussed below. As described previously (22), a significant portion of the Gag polyproteins are found in the gradient pellet, due to aggregation and/or to a persistent association with cellular elements. The Gag polyproteins distributed through the middle fractions of the gradient are those particles separated from the cytosol which were used for STEM imaging.
FIG. 1.
SDS-PAGE of velocity gradient fractions during the purification of M-PMV Gag particles. (A) Pr78Gag and Pr95Gag-Pro are shown in a Coomassie blue-stained gel with each fraction labeled from top (fraction 1) to bottom (fraction 23). The results for the resuspension of the pellet at the bottom of the tube is also shown (lane labeled “pellet”). (B) Western blot of an SDS-PAGE gel (using one gradient fraction containing Gag particles) with anti-Gag antiserum demonstrates that all major bands in the gel are Gag-related products.
M-PMV Gag particle mass.
For STEM analysis, each fraction from the velocity gradient was dialyzed and concentrated as described above, with a final resuspension in 100 μl of 10 mM Tris buffer (pH 7.6). For each gradient sample (fractions 8 through 18), a set of STEM digital images were made (30 to 35 images per sample), using methods previously defined at the Brookhaven National Laboratory STEM facility (31). Purified Gag particles were freeze-dried with tobacco mosaic virus (TMV) particles (as an internal control) onto a thin (2 to 3-nm) carbon film and imaged by a 40-keV electron beam on a low-temperature stage (−150°C). In this process, the electron beam is focused to 0.25 nm at a series of points (pixels) within a 512- by 512-nm grid, with each pixel center separated from the next by 1 nm. At each pixel, the scattered electrons at large (40- to 200-milliradian) and at small (15- to 40-milliradian) angles are measured by annular dark-field detectors. The number of electrons scattered at any one point within the sample is proportional to the mass, and the value is multiplied by an established mass coefficient to determine the mass at each point on the grid. The dose used in imaging is less than 500 electrons/nm2, and the loss of mass within the sample during imaging is less than 1%. The mass values are used to generate a digital image, and the summation of the mass values for an entire particle, after subtraction of an averaged background value, yields the mass of each individual Gag particle. TMV particles have a constant known value for mass per unit length, and the experimental measurement of this value within each sample ensures that the mass measurements are accurate.
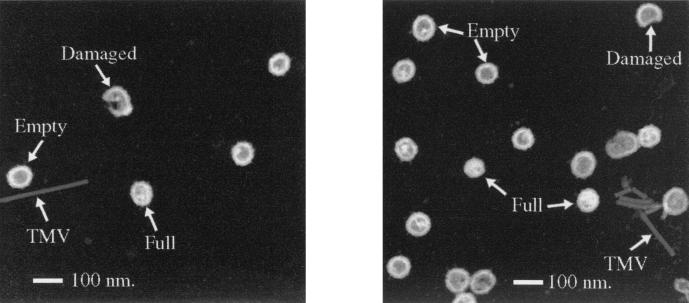
An automated selection routine was used to compare the particles in the digital images with a theoretical model of a Gag particle: a hollow sphere with internal and external diameters of 640 and 900 Å, respectively. Particles that fitted the model and also satisfied requirements for spherical symmetry and low backgrounds relative to particle mass were selected for mass measurements. Two dark-field images containing M-PMV Gag particles are shown in Fig. 2. By conventional thin-section electron microscopy, intracellular M-PMV Gag particles appear as a dense ring with a relatively hollow core. While most of the particles selected for mass measurement have similar appearances (“empty” particles), some have additional mass within the center, with a higher average mass value (“full” particles). The full particles were found to be a minor component (16% of all particles) in the middle fractions of the velocity gradient (fractions 8 through 12), where most of the Gag particles are found, while in the fractions associated with a higher S value (fractions 13 through 18), up to 50% of selected particles had a significant central density (data not shown). The full particles are inconsistent with viral particles imaged by thin-section electron microscopy in other expression systems (15, 24, 26) and, because they segregate into the lower fractions of the velocity gradient that are associated with a higher S value, may represent the introduction of additional material into the center of the particle during assembly rather than artifacts of particle preparation for imaging. Thus, particles taken from the lower fractions (fractions 13 through 18) of the velocity gradients were not used for mass analysis.
FIG. 2.
STEM images. Two representative dark-field STEM images that depict the complete Gag particles (full and empty) and damaged or incomplete particles found during image analysis are shown. TMV is also shown as an internal control for mass measurements.
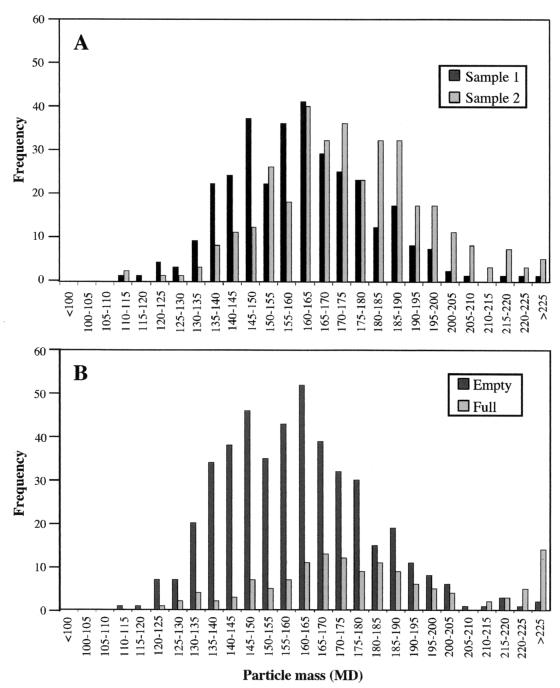
In two independent preparations with clean backgrounds and a large number of particles available for analysis (388 Gag particles in the first sample and 348 in the second sample), the average mass for all particles selected for mass measurement ranged from 164 to 174 MDa, with a standard deviation in mass of 20 to 21 MDa. A histogram of particle mass measurements from each of these samples demonstrates the considerable diversity in particle preparation (Fig. 3A). The difference in mass between empty and full particles taken from all fractions of the velocity gradient is also shown (Fig. 3B). Mass-per-unit-length measurements for the TMV standards (13.4 ± 0.8 and 13.6 ± 0.9 kDa/Å) were within 5% of the predicted value (13.1 kDa/Å) for both sets of samples.
FIG. 3.
Distribution of particle mass. (A) Particle frequency is plotted versus particle mass (in megadaltons) for all particles used for mass measurements from gradient fractions 8 through 12 from two independent samples. These data were used in the calculation of the average mass for M-PMV Gag particles. (B) Particle frequency is plotted as a function of particle mass for both empty and full particles found within velocity gradient fractions 8 through 18 from sample 1.
Protein and RNA content of M-PMV Gag particles.
In order to estimate the number of Gag and Gag-Pro polyprotein molecules that participate in a particle assembly event, the protein and RNA contents were measured in each of the velocity gradient fractions used for STEM imaging. The RNA content for each sample of Gag particles was extracted from particle lysates with RNA columns, using a QIAamp viral RNA kit (Qiagen, Valencia, Calif.). The viral RNA was quantified by UV absorption at 260 nm, with a conversion factor of 40 μg per ml per optical density unit, using a Beckman DU 530 spectrophotometer. The protein content of the same samples was estimated by a modified Lowry assay using a DC protein assay kit (Bio-Rad Laboratories, Hercules, Calif.) after establishing an approximate absorbance-to-concentration curve with purified bovine serum albumin. The average Gag/RNA mass ratio was 30.1:1, which yielded a calculated average genomic RNA content of 3.2%, ranging from 2.1 to 4.2% (data not shown).
In order to calculate the relative amounts of Pr78Gag and Pr95Gag-Pro that were assembled into Gag particles in insect cells, [35S]methionine-labeled immature capsids were separated from cell lysates directly by velocity gradient sedimentation and the ratio was determined by quantitative phosphorimaging of dried SDS-PAGE gels (data not shown). The intensity of each Pr78 and Pr95 band in several velocity gradient fractions from independent preparations was measured and adjusted for a difference in the number of methionine residues between Pr78 and Pr95. The protein content averaged 92% Pr78 and 8% Pr95, with a small amount (<1%) of Pr180Gag-Pro-Pol. Although there are no other significant protein bands seen in the samples prepared for STEM, the contents of a sample of purified [35S]methionine-labeled Gag particles were separated by SDS-PAGE and quantitatively analyzed by phosphorimaging. The bands corresponding to Gag polyproteins represented 91% of the activity across an entire lane (data not shown), and this value was used as the percentage of Gag relative to the amount of all proteins within a particle in order to calculate the number of Gag polyproteins per particle.
Using M-PMV particles purified and concentrated for STEM, the mass for Pr78Gag and Pr95Gag-Pro was determined by matrix-assisted laser desorption ionization–time of flight mass spectroscopy. Purified M-PMV Gag particles were disrupted by mixing them with 6 M guanidine at a 1:1 ratio and then boiling for 5 min. The sample was diluted 1:50 with a matrix (sinapinic acid [Sigma] dissolved in acetonitrile and mixed at a 1/1 ratio with trifluoroacetic acid), and 1 μl was pipetted onto a smooth plate. Samples were analyzed in the positive mode on a Voyager Elite mass spectrometer with delayed-extraction technology (PerSeptive Biosystems, Framingham, Mass.). The acceleration voltage was set at 25 kV, and 50 to 100 laser shots were summed. The mass spectrometer was calibrated with bovine serum albumin. The mass values for Pr78 and Pr95 were 73.0 and 92.5 kDa, respectively. The smaller Gag-related proteins discussed above were measured at 50.1 and 54.4 kDa. After 3% was subtracted from each averaged particle mass measurement (above) to account for RNA content, and after the remaining mass was multiplied by 0.91 to exclude protein content unrelated to Gag, the values for the masses of Pr78Gag and Pr95Gag-Pro and their relative amounts in Gag particles were used to calculate the number of Gag and/or Gag-Pro molecules included in a particle assembly event. These values ranged from 1,900 for an average particle mass of 164 MDa to 2,100 for an average particle mass of 174 MDa, with a standard deviation of 270 or 280 molecules per particle, respectively.
A number of assumptions that could have a small effect on the calculations have been made in arriving at these figures. When M-PMV Gag particles are prepared for STEM imaging, the dominant portion of the protein content is composed of Pr78Gag and Pr95Gag-Pro polyproteins. For reasons described above, we have assumed that the smaller Gag-related proteins (50 and 54 KDa) seen in the velocity gradient fractions are a consequence of Pr78Gag and/or Pr95Gag-Pro cleavage after particle assembly. However, if these smaller Gag products arise before or during particle assembly and are incorporated into Gag particles as truncated Gag polyproteins, the calculated number of Gag polyproteins per particle increases by a small amount (40 molecules per particle, or 2% of the total). We have also presumed that the salt and Triton X-100 detergent used during particle purification is removed by extensive washing of the samples in salt-free Tris buffer prior to STEM imaging. It is possible, however, that salts and/or detergent remained associated with the Gag particles and thus increased the measured particle mass. The genomic RNA content of each sample of M-PMV Gag particles was measured after column extraction. The columns used are designed for the extraction of genomic RNA and may not efficiently bind smaller RNAs (less than 200 bp), such as tRNAs, that may represent up to one-third of the RNA found in the viral core (3, 4, 6, 7, 28, 29). Thus, the viral RNA content may be moderately underestimated and the protein (Gag) content may be slightly overestimated in this study. If one-third of the RNA content of the M-PMV Gag particles has been lost and an adjusted RNA content of 4.5% is used in place of the measured value of 3%, the number of Gag molecules per particle is reduced by ∼30. In all studies to date, RNA is a relatively minor component of the virion, and we feel that the uncertainty in the mass of small RNAs in the Gag particle represents a small error in our calculations.
In a previous STEM analysis of RSV virions and RSV Gag particles (30, 33), the calculated number of Gag polyproteins per virion was significantly less than that we report here (1,500 and 1,200, respectively, versus ∼2,000). In a recent cryo-EM study of in vitro-assembled RSV particles, the radii of the immature particles were significantly smaller than those observed for HIV, murine leukemia virus, or M-PMV (33 versus ∼55 nm) (9, 33; S. D. Parker et al., unpublished data), suggesting that there may be significant differences among the different genera of retroviruses in the number of protein subunits used to assemble an immature Gag particle.
In both studies, the distribution of particle mass is remarkable in its breadth. M-PMV Gag particles and enveloped HIV type 1 virus-like particles, both expressed from baculovirus-insect cell expression systems, have been studied and compared with similar products from mammalian expression systems by conventional electron microscopy (12, 20, 25; Parker et al., unpublished). Although no dramatic differences have been observed between the products of different expression systems, the baculovirus system may produce Gag particles which are more heterogeneous than those found in naturally infected cells. We have assumed that any variations in particle mass are due to differences in the numbers of Gag polyproteins within the particle and that the RNA-to-protein ratios, as well as the ratio of Pr78 to Pr95, remain constant among all particles measured. These assumptions are very difficult to address experimentally, and some of the variation in particle mass may be due to differences in RNA content and/or Pr78/Pr95 ratios. Nevertheless, high-resolution cryo-EM imaging of a variety of different retroviruses, including M-PMV (9, 32, 33; Parker et al., unpublished), demonstrates a similarly considerable diversity in particle size, suggesting that a variable number of Gag polyproteins are included in the assembly process. Several high-resolution microscopic studies have demonstrated a well-ordered arrangement of Gag domains, with a threefold axis of symmetry near the surfaces of both immature Gag particles and mature virions similar to that seen with icosahedral viruses (1, 2, 18–20, Nermut et al., submitted). However, it is difficult to define an assembly model with a high degree of order in two dimensions on a local level but with large variations in particle mass and size.
There are three possible explanations for the variable number of Gag polyproteins found within M-PMV Gag particles. The first possibility is that the otherwise icosahedral Gag particles have random defects or holes as an artifact of assembly or the purification process. Indeed, small defects are found around the perimeters of some M-PMV Gag particles by STEM imaging. However, while particle defects may have a small effect on the variations seen in particle mass, the selection criteria by which M-PMV Gag particles are chosen for STEM imaging make it unlikely that large defects, in the absence of other factors, are present to the degree necessary to explain the large variation in particle mass. The second possible explanation is that Gag particles have strict icosahedral symmetry with variable triangulation numbers and that discrete classes in particle mass are obscured in the measurements by defects in the Gag shell or variable amounts of mass around a particle that is unrelated to Gag (detergent or salt). Particles assembled by a truncated form of the Ty-1 capsid protein in Saccharomyces cerevisiae have been shown by cryo-EM to have icosahedral symmetry but variable triangulation numbers (T=3 and T=4), and strict icosahedral symmetry is lost as the length of the capsid protein is increased (21). However, if the triangulation numbers for M-PMV particles are restricted to those allowed in the model described by Forster et al. (8), the large difference in predicted mass between the classes of particles allowed in the model should not have been obscured by an experimental artifact or relatively small particle defects. The third possible explanation is that the Gag molecules are well ordered in two dimensions into hexagonal or fullerene-like arrays but are not well ordered in the way that Gag sheets are curved or otherwise organized in three dimensions to form a sphere (8, 9). This last explanation may be the most likely and would explain both the negative stain and freeze-fracture electron microscopy studies of HIV and M-PMV particles that have shown the Gag polyproteins to be well organized on a local level in two dimensions (19, 20) as well as the variable size and mass observed for Gag particles by cryo-EM and STEM (9, 30, 33).
Acknowledgments
This investigation was supported by U.S. Public Health Service research grant CA-27834 from the National Institutes of Health. The BNL STEM is an NIH-supported Resource Center, NIH P41-PR01777, with additional support provided by DOE, OBER.
REFERENCES
- 1.Barklis E, McDermott J, Wilkens S, Fuller S, Thompson D. Organization of HIV-1 capsid proteins on a lipid monolayer. J Biol Chem. 1998;273:7177–7180. doi: 10.1074/jbc.273.13.7177. [DOI] [PubMed] [Google Scholar]
- 2.Barklis E, McDermott J, Wilkens S, Schabtach E, Schmid M F, Fuller S, Karanjia S, Love Z, Jones R, Rui Y, Zhao X, Thompson D. Structural analysis of membrane-bound retrovirus capsid proteins. EMBO J. 1997;16:1199–1213. doi: 10.1093/emboj/16.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Bishop J M, Levinson W E, Quintrell N, Sullivan D, Fanshier L, Jackson J. The low molecular weight RNAs of Rous sarcoma virus. I. The 4S RNA. Virology. 1970;42:182–195. doi: 10.1016/0042-6822(70)90251-5. [DOI] [PubMed] [Google Scholar]
- 5.Conte M R, Klikova M, Hunter E, Ruml T, Matthews S. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus, and implications for the morphology of retroviral assembly. EMBO J. 1997;16:5819–5826. doi: 10.1093/emboj/16.19.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erikson R L. Studies on the RNA from avian myeloblastosis virus. Virology. 1969;37:124–131. doi: 10.1016/0042-6822(69)90313-4. [DOI] [PubMed] [Google Scholar]
- 7.Faras A J, Garapin A C, Levinson W E, Bishop J M, Goodman H M. Characterization of the low-molecular-weight RNAs associated with the 70S RNA of Rous sarcoma-virus. J Virol. 1973;12:334–342. doi: 10.1128/jvi.12.2.334-342.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forster M J, Mulloy B, Nermut M V. Molecular modelling study of HIV p17gag (MA) protein shell utilising data from electron microscopy and X-ray crystallography. J Mol Biol. 2000;298:841–857. doi: 10.1006/jmbi.2000.3715. [DOI] [PubMed] [Google Scholar]
- 9.Fuller S D, Wilk T, Gowen B E, Krausslich H G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 10.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 11.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 12.Gheyson D, Jacobs E, da Foresta F, Thiriart C, Francotte M, Thines D, de Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 13.Khorasanizadeh S, Campos-Olivas R, Summers M F. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J Mol Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- 14.Kingston R L, Fitzon-Ostendorp T, Eisenmesser E Z, Schatz G W, Vogt V M, Post C B, Rossmann M G. Structure and self-association of the Rous sarcoma virus capsid protein. Structure Fold Des. 2000;8:617–628. doi: 10.1016/s0969-2126(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 15.Klikova M, Rhee S S, Hunter E, Ruml T. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J Virol. 1995;69:1093–1098. doi: 10.1128/jvi.69.2.1093-1098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthews S, Barlow P, Clark N, Kingsman S, Kingsman A, Campbell I. Refined solution structure of p17, the HIV matrix protein. Biochem Soc Trans. 1995;23:725–729. doi: 10.1042/bst0230725. [DOI] [PubMed] [Google Scholar]
- 17.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 18.Nermut M V, Hockley D J. Comparative morphology and structural classification of retroviruses. Curr Top Microbiol Immunol. 1996;214:1–24. doi: 10.1007/978-3-642-80145-7_1. [DOI] [PubMed] [Google Scholar]
- 19.Nermut M V, Hockley D J, Bron P, Thomas D, Zhang W H, Jones I M. Further evidence for hexagonal organization of HIV gag protein in prebudding assemblies and immature virus-like particles. J Struct Biol. 1998;123:143–149. doi: 10.1006/jsbi.1998.4024. [DOI] [PubMed] [Google Scholar]
- 20.Nermut M V, Hockley D J, Jowett J B, Jones I M, Garreau M, Thomas D. Fullerene-like organization of HIV gag-protein shell in virus-like particles produced by recombinant baculovirus. Virology. 1994;198:288–296. doi: 10.1006/viro.1994.1032. [DOI] [PubMed] [Google Scholar]
- 21.Palmer K J, Tichelaar W, Myers N, Burns N R, Butcher S J, Kingsman A J, Fuller S D, Saibil H R. Cryo-electron microscopy structure of yeast Ty retrotransposon virus-like particles. J Virol. 1997;71:6863–6868. doi: 10.1128/jvi.71.9.6863-6868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker S D, Hunter E. A cell-line-specific defect in the intracellular transport and release of assembled retroviral capsids. J Virol. 2000;74:784–795. doi: 10.1128/jvi.74.2.784-795.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao Z, Belyaev A S, Fry E, Roy P, Jones I M, Stuart D I. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 24.Rhee S, Hunter E. Amino acid substitutions within the matrix protein of type D retroviruses affect assembly, transport and membrane association of a capsid. EMBO J. 1991;10:535–546. doi: 10.1002/j.1460-2075.1991.tb07980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Royer M, Cerutti M, Gay B, Hong S-S, Devauchelle G, Boulanger P. Functional domains of HIV-1 Gag-polyprotein expressed in baculovirus-infected cells. Virology. 1991;184:417–422. doi: 10.1016/0042-6822(91)90861-5. [DOI] [PubMed] [Google Scholar]
- 26.Sakalian M, Parker S D, Weldon R A, Jr, Hunter E. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Manatis T. Molecular cloning, a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Sawyer R C, Dahlberg J E. Small RNAs of Rous sarcoma virus: characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J Virol. 1973;12:1226–1237. doi: 10.1128/jvi.12.6.1226-1237.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stromberg K, Hurley N E, Davis N L, Rueckert R R, Fleissner E. Structural studies of avian myeloblastosis virus: comparison of polypeptides in virion and core component by dodecyl sulfate-polyacrylamide gel electrophoresis. J Virol. 1974;13:513–528. doi: 10.1128/jvi.13.2.513-528.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt V M, Simon M N. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wall J S, Hainfeld J F, Simon M N. Scanning transmission electron microscopy of nuclear structures. Methods Cell Biol. 1998;53:139–164. doi: 10.1016/s0091-679x(08)60878-x. [DOI] [PubMed] [Google Scholar]
- 32.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu F, Joshi S M, Ma Y M, Kingston R L, Simon M N, Vogt V M. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J Virol. 2001;75:2753–2764. doi: 10.1128/JVI.75.6.2753-2764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]