Abstract
One of the most audacious goals of modern neuroscience is unraveling the complex web of causal relations underlying the activity of neuronal populations on a whole-brain scale. This endeavor, which was prohibitive only a couple of decades ago, has recently become within reach owing to the advancements in optical methods and the advent of genetically encoded indicators/actuators. These techniques, applied to the translucent larval zebrafish have enabled recording and manipulation of the activity of extensive neuronal populations spanning the entire vertebrate brain. Here, we present a custom two-photon optical system that couples light-sheet imaging and 3D excitation with acousto-optic deflectors for simultaneous high-speed volumetric recording and optogenetic stimulation. By employing a zebrafish line with pan-neuronal expression of both the calcium reporter GCaMP6s and the red-shifted opsin ReaChR, we implemented a crosstalk-free, noninvasive all-optical approach and applied it to reconstruct the functional and effective connectivity of the left habenula.
Subject terms: Neural circuits, Optogenetics
Two-photon light-sheet imaging and AOD-based optogenetic stimulation enable the detailed reconstruction of the left habenula functional connectivity in translucent larval zebrafish, offering a promising avenue towards brain function unraveling.
Introduction
Understanding the functional connectivity of intricate networks within the brain is a fundamental goal toward unraveling the complexities of neural processes. This longtime focal point in neuroscience requires methodologies to trigger and capture neuronal activity in an intact organism. Critical insights into the complex interplay among large populations of neurons have been provided by electroencephalography and functional magnetic resonance imaging1–4. These gold standard methods, however, do provide a noninvasive means to detect neuronal activity, but with limited spatial (the former) and temporal resolution (the latter), and lack equally noninvasive possibilities to precisely elicit and control it. Therefore, it is evident that deciphering how individual neurons communicate to shape functional neural circuits on a whole-organ scale demands further technological advances. Over the last few decades, with the advent of optogenetics5 and the widespread adoption of genetically encoded fluorescent indicators6, all-optical methods have gained traction for their ability to simultaneously monitor and manipulate the activity of multiple neurons within the intact brain7–10.
In this framework, the ever-increasing use of the tiny and translucent zebrafish larva as a reliable animal model recapitulating manifold features of vertebrate species physiology11, has provided momentum for the development and enhancement of optical technologies aimed at imaging and controlling neuronal activity with light at high spatio-temporal resolution12–14. On the imaging side, previous high-resolution all-optical investigations of zebrafish have made use of two-photon (2P) point scanning methods15–17 or, more rarely, of one-photon (1P) excitation light-sheet fluorescence microscopy (LSFM)18. Compared to point scanning approaches, LSFM19, allowing parallelization of the detection process within each frame, enables concurrent high spatio-temporal resolution and extensive volumetric imaging. However, the use of visible excitation in 1P LSFM represents an undesired source of strong visual stimulation for the larva, often requiring tailored excitation strategies at least to prevent direct illumination of the eyes20.
On the photostimulation side, most advanced all-optical setups typically adopt parallel illumination approaches, making use of spatial light modulators (SLM) or digital micromirror devices to generate multiple simultaneous holographic spots of excitation16,21–23. In computer-generated holography, the input laser power is subdivided among the various spots, resulting in increasing energies released on the specimen as the number of effective targets rises and, consequently, in increasing probability of photodamage24. Conversely, scan-based sequential stimulation allows a fraction of the power needed by parallel approaches to be deposited at any time on the sample, regardless of the number of targets. As a drawback, however, scan-based methods typically employ mechanical moving parts that constrain the stimulation sequence speed and thus the maximum temporal resolution achievable. An exception is represented by acousto-optic deflectors (AODs)25,26 which are not affected by mechanical inertia and thus enable discontinuous three-dimensional trajectories with constant repositioning time. In particular, featuring an ultrashort access time (μs range), AODs represent the scanning technology that gets closest to parallel illumination performance. Indeed, AODs enable quasi-simultaneous three-dimensional targeting of multiple spots, yet keeping low the global energy delivered. However, despite their extensive use for fast 3D imaging27–31, these devices have been rarely employed for photostimulation so far32–36.
In this work, we present an all-optical setup consisting of a light-sheet microscope and a light-targeting system equipped with AODs, both employing nonlinear excitation. The light-sheet microscope enables high spatio-temporal resolution volumetric imaging of the larval zebrafish brain, while the light-targeting system is employed to perform concurrent three-dimensional optogenetic stimulation. Using a double transgenic line pan-neuronally expressing both the green fluorescent calcium indicator GCaMP6s37 and the red-shifted light-gated cation channel ReaChR38, we demonstrate a crosstalk-free experimental approach for all-optical investigation of brain circuitries. Leveraging two-photon excitation and the inertia-free light targeting capabilities of AODs, we validated the system functionality by reconstructing the efferent functional and effective connectivity of the left habenula, a cerebral nucleus mainly composed of excitatory neurons, linking forebrain and midbrain structures.
Results
A crosstalk-free approach for two-photon all-optical investigations in zebrafish larvae
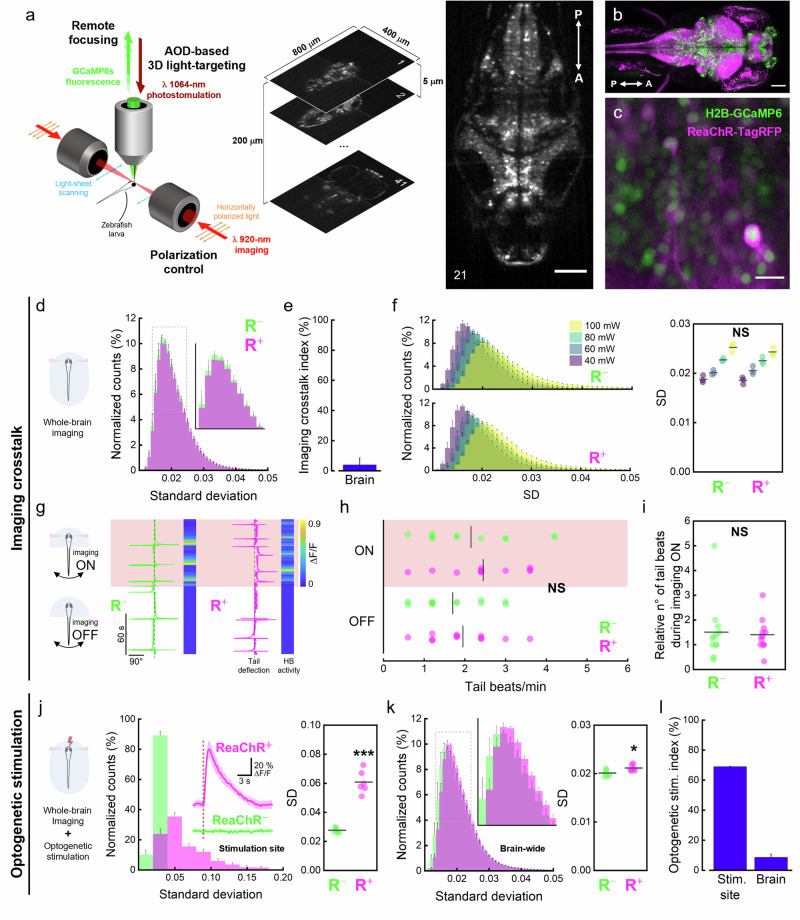
To explore brain functional connectivity in zebrafish larvae, we devised an integrated all-optical 2P system capable of simultaneously recording and stimulating neuronal activity. The setup (Fig. 1a and Supplementary Fig. 1), consists of a light-sheet fluorescence microscope and a light-targeting system, specifically designed for fast whole-brain calcium imaging and 3D optogenetic stimulation, respectively. Both optical paths employ pulsed near-infrared (NIR) laser sources for 2P excitation39. The 2P LSFM module employing digitally scanned mode, double-sided illumination, control of excitation light polarization40 and remote focusing of the detection objective, is capable of recording the entire larval brain (400 × 800 × 200 μm3) at volumetric rates up to 5 Hz41 (Supplementary Movie 1 and Supplementary Fig. 2). On the other hand, the light-targeting system incorporates two couples of acousto-optic deflectors to move the excitation focus to arbitrary locations inside a 100 × 100 × 100 μm3 volume, guaranteeing constant repositioning time (4 μs) independently of the relative distance between sequentially illuminated points, and equal energy delivered independently from the number of targets36.
Fig. 1. A crosstalk-free experimental paradigm for two-photon all-optical electrophysiology of the larval zebrafish brain.
a Left, Schematic representation of the 2P all-optical system employed in the experiments with its main features (see Supplementary Fig. 1 and Optical setup for a full description of the setup components). Center, volumetric sampling of the larval brain. 41 planes (z step, 5 μm) were acquired over a depth of 200 μm at a rate of 2.5 Hz (see also Supplementary Fig. 2 and Supplementary Movie 1). Right, exemplary medial plane (depth: 100 μm from brain surface) extracted from a typical volumetric acquisition. Image is an average projection over 30 s of functional imaging. Scale bar, 100 μm. b Maximum intensity projection of a confocal image stack encompassing the entire head of a 5 dpf double transgenic Tg(elavl3:H2B-GCaMP6s; elavl3:ReaChR-TagRFP) zebrafish larva used for all-optical electrophysiology (see also Supplementary Movie 2). Scale bar, 100 μm. c Higher magnification confocal image of reticulospinal neurons of the same transgenic line displayed in panel b showing the nuclear expression of the calcium reporter GCaMP6s (green) and membrane localization of the fusion protein ReaChR-TagRFP (magenta). Scale bar, 10 μm. d–f Quantification of crosstalk activation of ReaChR channels during 5 min of whole-brain light-sheet imaging. N = 3 ReaChR+ and 3 ReaChR− larvae. d Average normalized distributions of the SD values of voxels composing the larval brain over 5 min of continuous light-sheet imaging at λex 920 nm and 60 mW power for ReaChR+ and ReaChR− larvae. Inset shows a zoom-in of the distributions’ peaks. e Imaging crosstalk index, calculated as the percentage Hellinger distance (see Data analysis for equation) between the average normalized distributions shown in Fig. 1d. f Left, average normalized distributions of the SD values of voxels composing the larval brain over 5 min of continuous light-sheet imaging at increasing laser power (40, 60, 80, and 100 mW) for ReaChR− (R−, upper) and ReaChR+ (R+, lower) larvae. Right, median SD values calculated from individual SD distributions of brain-wide voxels of ReaChR− and ReaChR+ larvae. Statistical comparisons (two-way ANOVA and post-hoc Tukey’s test) highlight no statistical significance (P > 0.05) between SD values of ReaChR− and ReaChR+ larvae exposed to the same imaging power and between SD values of ReaChR+ or ReaChR− larvae exposed to 40 and 60 mW power. g–i Evaluation of the effect of whole-brain light-sheet imaging on behavior. g Tail deflection traces in absence (imaging OFF) and presence (imaging ON) of whole-brain light-sheet imaging (red shaded time window) for individual ReaChR− and ReaChR+ representative larvae (corresponding behavioral recordings are shown in Supplementary Movies 3 and 4, respectively). Dark gray vertical dashed lines indicate fish midline. Each behavioral recording is coupled to the corresponding neuronal activity (heatmaps) extracted from the hindbrain (HB). In the imaging OFF time window, where no volumetric imaging was performed, heatmap values were artificially set to 0. h Tail beats per minute during imaging OFF and ON windows, for both ReaChR− and ReaChR+ larvae. NS P = 0.3330 (factor: zebrafish strain), P = 0.0980 (factor: imaging), two-way ANOVA and post-hoc Tukey’s test. i Relative number of tail beats during imaging ON with respect to the imaging OFF condition. NS P = 0.7903, unpaired t test. For both (h, i), N = 4 ReaChR+ and 4 ReaChR− larvae, n = 3 experimental trials for each larva. j–l Quantification of the effect of the optogenetic stimulation on neuronal activity. j Inset, nuclear GCaMP6s signals induced by 500 ms stimulation at 1064 nm while performing whole-brain light-sheet imaging at 920 nm. Larvae expressing ReaChR and GCaMP6s show stimulation-induced calcium transients (magenta trace, ReaChR+) whereas larvae expressing only GCaMP6s (green trace, ReaChR−) do not. Dark red dashed line indicates the stimulation event. N = 3 ReaChR+, 3 ReaChR− larvae; n = 5 calcium transients per larva. Data are presented as mean ± sd. Left, average normalized distributions of the SD values of voxels inside the stimulation site for ReaChR+ and ReaChR− larvae over 100 s of whole-brain imaging during which five optogenetic stimulation were delivered. N = 6 ReaChR+, 6 ReaChR− larvae. Right, median SD values calculated from individual SD distributions of stimulation site voxels for ReaChR− and ReaChR+ larvae. ***P < 0.0001, unpaired t test. k Left, average normalized distributions of the SD values of brain-wide voxels for ReaChR+ and ReaChR− larvae in the same condition as in (j). Inset shows a zoom-in of the distributions’ peaks. Right, median SD values calculated from individual SD distributions of brain-wide voxels for ReaChR− and ReaChR+ larvae. *P = 0.0218, unpaired t test. l Optogenetic stimulation index, percentage Hellinger distance between the average normalized distributions shown in (j) (stim. site) and (k) (brain), respectively. Error bars of average distributions indicate the sem. Error bars of imaging crosstalk and optogenetic stimulation indices were calculated according to uncertainty propagation theory (see Data analysis for details). Gray horizontal bars in (f, h, i, j and k) represent intragroup mean values.
To perform simultaneous recording and stimulation of neuronal activity, we employed the pan-neuronal Tg(elavl3:H2B-GCaMP6; elavl3:ReaChR-TagRFP) zebrafish line (Fig. 1b, Supplementary Movie 2). Larvae of this double transgenic line express the green fluorescent calcium indicator GCaMP6s inside neuronal nuclei and the red-shifted light-gated cation channel ReaChR (as a fusion protein with the red fluorescent protein TagRFP) on neuronal membranes (Fig. 1c).
We initially investigated the possible presence of crosstalk activation of ReaChR channels due to the excitation wavelength used for functional imaging. To this end, we employed two complementary approaches. First, light-sheet imaging of both double transgenic larvae (ReaChR+) and GCaMP6s-expressing larvae (ReaChR−, lacking the light-gated channel) was performed for 5 min (volumetric rate: 2.5 Hz, λex: 920 nm, laser power at the sample: 60 mW). To evaluate the level of neuronal activity, we computed the standard deviation (SD) over time for each voxel belonging to the brain (image processing voxel size = 4.4 × 4.4 × 5 μm3, see Data analysis for details). We adopted SD as a metric for neuronal activity since we found it more sensitive in discriminating between different conditions with respect to the number of calcium peaks per minute, and equally sensitive to the average peak amplitude, yet not necessitating the setting of arbitrary thresholds (Supplementary Fig. 3). No major differences could be observed in the average SD distributions computed over a 5-minute exposure to the imaging laser between the two groups (Fig. 1d). Indeed, the resulting imaging crosstalk index (calculated as the Hellinger distance between the two average distributions, see Data analysis for details) was extremely low (3.9% ± 4.5%; Fig. 1e). However, since crosstalk activation of light-gated channels by a spurious wavelength is typically power-dependent42, we then investigated whether higher powers of the laser used for imaging could induce a significant effect on ReaChR+ larvae. Figure 1f shows the average SD distributions obtained from ReaChR+ and ReaChR− larvae at imaging powers ranging from 40 to 100 mW. Despite higher laser powers producing a shift of the distributions towards higher SD values, this shift equally affected the neuronal activity of both ReaChR+ and ReaChR− larvae (see also Supplementary Fig. 4a). The differences between the median values of the SD distributions of the two groups (ReaChR+ and ReaChR−) at the same imaging power were not statistically significant (Fig. 1f, right) and, indeed, the imaging crosstalk index remained essentially constant in the power range tested (Supplementary Fig. 4b).
In addition to this, we investigated the presence of imaging-related crosstalk also from a behavioral standpoint. We performed high-speed tail tracking of head restrained ReaChR− and ReaChR+ larvae in absence (OFF) and in presence (ON) of whole-brain light-sheet imaging (Fig. 1g and Supplementary Movies 3 and 4). As shown in Fig. 1h, compared to the OFF period, during 920 nm laser exposure (ON) both strains showed a slight but not significant increase in the number of tail beats per minute, suggesting that the power applied (60 mW) was quite well tolerated by the animals. Moreover, the relative number of tail beats during imaging ON was not significantly different between ReaChR+ and ReaChR− larvae (Fig. 1i), providing additional proof of the absence of spurious excitation in ReaChR+ larvae by the 920 nm laser used for imaging.
After demonstrating the absence of cross-talk activation of ReaChR channels upon 2P light-sheet scanning, we investigated the ability of our AOD-based photostimulation system to effectively induce optogenetic activation of targeted neurons. For this purpose, we selected a stimulation wavelength (1064 nm) that is red-shifted relative to the opsin’s 2P excitation peak (975 nm). By doing so, we increased the separation between the wavelength used for optogenetic stimulation and the 2P excitation peak of GCaMP6s (920 nm), thus further reducing the potential for stimulation-induced artifacts. We thus stimulated ReaChR+ and ReaChR− larvae at 1064 nm (laser power at the sample: 30 mW, stimulation volume: 30 × 30 × 30 μm3) while simultaneously recording whole-brain neuronal activity via light-sheet imaging. Larvae expressing the opsin showed strong and consistent calcium transients evoked at the stimulation site (Fig. 1j, inset). Conversely, stimulating ReaChR− larvae did not result in any detectable response (Fig. 1j, inset). We quantified the effect of the optogenetic stimulation by computing the distributions of SD values of the voxels inside the stimulation site for ReaChR+ and ReaChR− larvae (Fig. 1j, left). 1064 nm stimulation induced statistically significant optogenetic activation of opsin-expressing neurons in ReaChR+ larvae (ReaChR− = 0.0277 ± 0.0017, ReaChR+ = 0.0608 ± 0.0077, mean ± sem; Fig. 1j, right). Despite the small stimulation volume with respect to the entire brain size, the effect of the photostimulation was also noticeable in the whole-brain SD distribution, where ReaChR+ larvae showed a peak slightly shifted toward higher SD values (Fig. 1k, left), which produced a significantly greater average SD (ReaChR− = 0.0201 ± 0.0007, ReaChR+ = 0.0211 ± 0.0006, mean ± sem; Fig. 1k, right). This appreciable difference was due to the high-amplitude calcium transients evoked by the stimulation and by the activation of the neuronal population synaptically downstream of the stimulation site. Figure 1l shows the optogenetic activation indeces (calculated as the Hellinger distance between the two average distributions, see Data analysis for details) for the stimulation site and the entire brain (stimulation site = 68.9% ± 0.3%, brain = 8.6% ± 2.1%). To rule out any possible spurious activation effect not related to the optogenetic excitation of ReaChR channels (e.g., sensorial perception of the laser stimulus), we also compared the SD distributions of ReaChR− larvae subjected to imaging only (ReaChR−i) or to simultaneous imaging and stimulation (ReaChR−i+s). The analysis highlighted no statistically significant effects of the photostimulation in the absence of opsin expression at either the stimulus site (Supplementary Fig. 5a) or at a brain-wide level (Supplementary Fig. 5b).
Characterization of calcium transients evoked by 3D optogenetic stimulation
After assessing the absence of opsin crosstalk activation upon light-sheet imaging and verifying the ability of our system to consistently induce optogenetic activation of ReaChR+ neurons, we characterized the neuronal responses to identify optimal stimulation parameters. We decided to target the stimulation at an easily recognizable cerebral nucleus mainly composed of excitatory neurons. Neurons having their soma inside the habenulae express vesicular glutamate transporter 2 (VGLUT2, also known as SLC17A6), representing a coherent group of excitatory glutamatergic neurons43,44. We therefore directed the stimulation onto the left habenula, an anatomically segregated nucleus that is part of the dorsal-diencephalic conduction system45. We adopted a stimulation volume of 50 × 50 × 50 μm3, sufficient to cover the entire habenula (Fig. 2a). This volume was populated with 6250 points distributed across 10 z-planes (z step: 5 μm). With a point dwell-time of 20 μs, a complete cycle over all the points in the volume took only 125 ms. We first characterized the calcium transients as a function of the stimulation duration (scan time, Fig. 2b) in the range of 125 to 625 ms (1–5 iterations over the volume). Figure 2c shows the amplitude of the calcium peaks as a function of the scan time. Increasing scan durations produced a progressive increase in peak amplitude until a plateau was reached between 4 and 5 volume cycles (scan time 500-625 ms). From a kinetic point of view, increasing scan durations led to a significant decrease in the rise time of calcium transients (Fig. 2d). Additionally, the decay time of the calcium transients progressively increased with increasing scan time (Fig. 2e). We also characterized the neuronal response as a function of the 1064 nm excitation power (ranging from 10 to 40 mW, Fig. 2f). The amplitude of the calcium transients showed a strong linear dependence on the stimulation power (Fig. 2g, R2: 0.89). While the rise time did not seem to be affected by the laser intensity (Fig. 2h), the decay time showed a strong linear proportionality (Fig. 2i, R2: 0.82). The duration of calcium transients (Supplementary Fig. 6a, b), instead, increased with increasing stimulation power but was not significantly affected by scan time. Given the small variation in rise time, in both cases the overall duration of the calcium transient was largely determined by the decay time trend.
Fig. 2. Characterization of the neuronal activation in response to optogenetic stimulation.
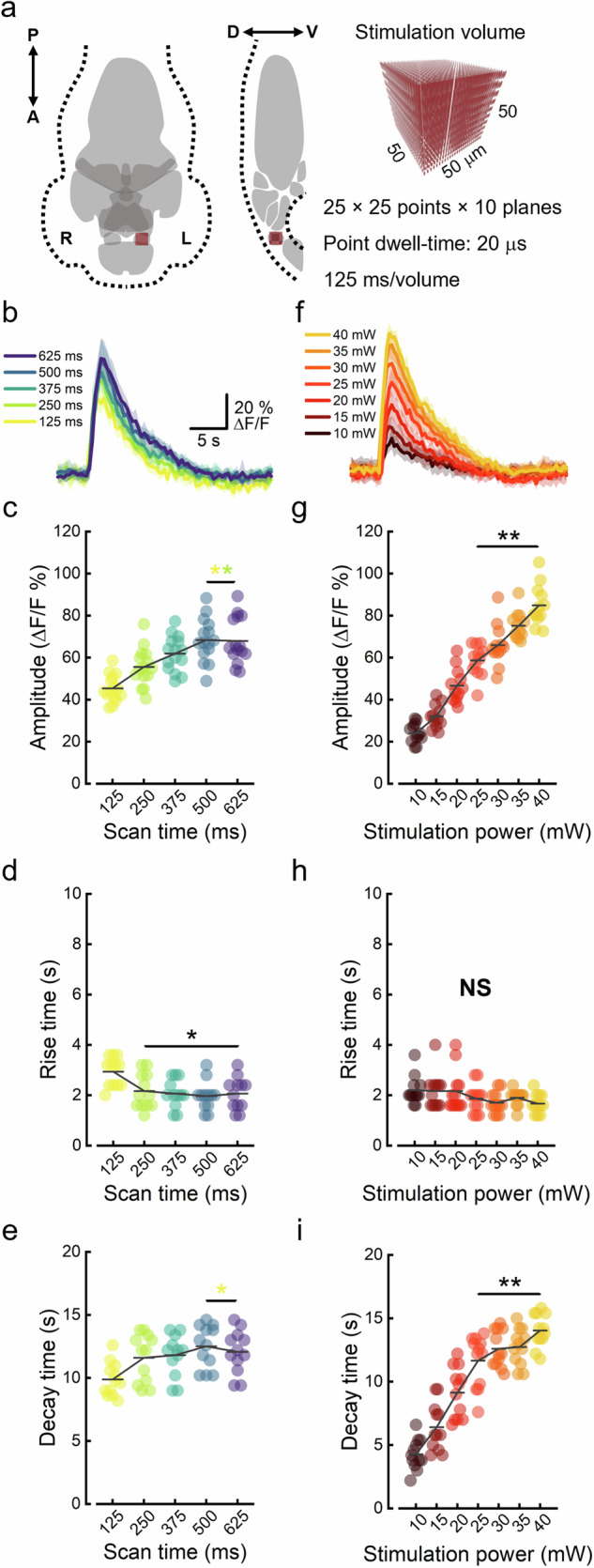
a Schematic representation of the larval brain in dorso-ventral projection (left) and midline sagittal section (right). The brain is segmented into ten gray anatomical districts used for further analysis in Figs. 3 and 4. In the sagittal section, habenulae are depicted with a checkered pattern since they would not be included in the represented midline plane. Dark red squares indicate the 50 × 50 × 50 μm3 stimulation volume to scale with the brain and located over the stimulation site (left habenula). The illumination spot (represented with the excitation PSF dimensions in the rendering on right) was discontinuously scanned to 6250 different locations inside the stimulation volume. With a point every 2 μm, along ten planes displaced every 5 μm, and a dwell-time of 20 μs, a complete iteration took 125 ms. A, anterior; P, posterior; D, dorsal; V, ventral; R, right hemisphere; L, left hemisphere. b Optogenetically evoked calcium transients as a function of scan time (stimulation duration). Amplitude (c), rise (d) and decay time (e) of stimulation-evoked calcium transients as a function of scan time. Color code as in (b). c **P < 0.00304, (d) *P < 0.02575, (e) *P < 0.01798, one way ANOVA and post-hoc Tukey’s test. Colored asterisks indicate with respect to which scan time data are significantly different. f Optogenetically evoked calcium transients as a function of stimulation power. Same scale as in (b). Amplitude (g), rise (h) and decay time (i) of evoked calcium transients as a function of stimulation power. Data are color-coded as in (f). g **P < 0.00431, (h) NS P > 0.2377, (i) **P < 0.00589, one way ANOVA and post-hoc Tukey’s test. Data in panels b and f are presented as mean ± sd. Data in (c, d, e, g–i) represent quantities measured from individual calcium transients. Black horizontal bars underlying asterisks embrace conditions which are statistically significant with respect to: all the other conditions in the plot (black asterisk), the condition(s) indicated by the color(s) of the asterisk(s). Gray horizontal bars in (c, d, e, g–i) represent intragroup mean values. N = 4 ReaChR+ larvae, n = 3 calcium transients per larva.
Whole-brain functional circuitry of the left habenular nucleus
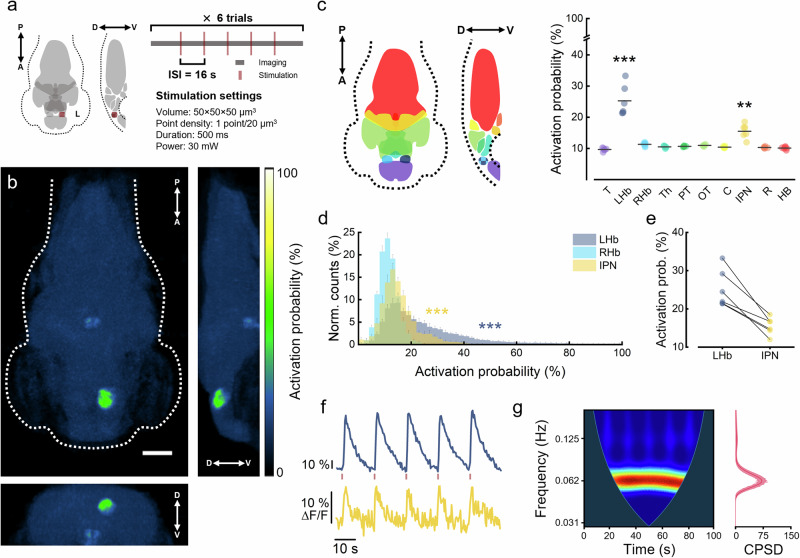
After this initial technical validation, we employed our all-optical setup to identify cerebral regions functionally linked to the left habenular nucleus. To this end, we designed the following stimulation protocol (Fig. 3a). For each zebrafish larva, we performed six trials consisting of 5 optogenetic stimuli (interstimulus interval: 16 s) during simultaneous whole-brain light-sheet imaging. Based on the characterization performed, we adopted a stimulus duration of 500 ms (4 complete consecutive iterations over the 50 × 50 × 50 μm3 volume, point density: 0.5 point/μm) and a laser power of 30 mW to maximize the neuronal response while keeping the laser intensity low (Supplementary Movie 5).
Fig. 3. Brain-wide activation probability in response to optogenetic stimulation of the left habenula.
a Scheme of the stimulation protocol adopted for the investigation of habenular functional connectivity. The protocol consists of six trials, each composed of 100 s of whole-brain imaging (dark gray) during which 5 stimuli (dark red) are delivered with an interstimulus interval (ISI) of 16 s (see also Supplementary Movie 5). The stimulation volume (50 × 50 × 50 μm3) is centered in xyz on the left habenula and discontinuously scanned with 6250 focal points (scan time: 500 ms; 1064 nm power: 30 mW). A, anterior; P, posterior; D, dorsal; V, ventral; L, left hemisphere. b Activation probability maps showing the probability of each brain voxel to exceed a 3-SD threshold during stimulation with respect to pre-stimulus levels. Maps are maximum intensity projections of the average 3D map (Supplementary Movie 6) along the transverse (left), sagittal (right) and coronal (bottom) axis. N = 6 ReaChR+ larvae, n = 30 stimulations per larva. Scale bar, 100 μm. c Activation probability of different regions composing the larval brain. Median probability values are obtained from individual probability maps (Supplementary Movie 7). Regions are color-coded as in the schematic representation on the left. In the sagittal section, habenulae are depicted with a checkered pattern since they would not be included in the represented midline plane. T, telencephalon; LHb, left habenula; RHb, right habenula; Th, thalamus; PT, pretectum; OT, optic tectum; C, cerebellum; IPN, interpeduncular nucleus; R, raphe; HB, hindbrain. Gray horizontal bars represent intragroup mean values. ***P < 0.0001, **P < 0.0059, one way ANOVA and post-hoc Tukey’s test. d Average normalized distributions of activation probability values extracted from voxels inside LHb, IPN and RHb. ***P < 0.0001, KS test with Bonferroni correction (α = 0.01667). N = 6 ReaChR+ larvae. e. Median activation probability extracted from LHb and IPN of individual larvae. Lines connect symbols belonging to the same individual. Color code as in (c). N = 6 ReaChR+ larvae. f Representative average ΔF/F activity traces extracted from the LHb (blue) and IPN (yellow) during an individual trial of the stimulation protocol. At a glance we can observe the perfect match between the response of the two regions (see also Supplementary Movie 5). N = 1 ReaChR+ larva. g Left, cross-wavelet power spectral density (CPSD) of the two time-traces reported in (f). Warmer colors indicate higher power density. Right, time-averaged CPSD. The CPSD peak is centered at 0.0625 Hz, which corresponds to the inverse of the stimulation period (16 s). See Supplementary Fig. 7 for the CPSD obtained from trials of other larvae.
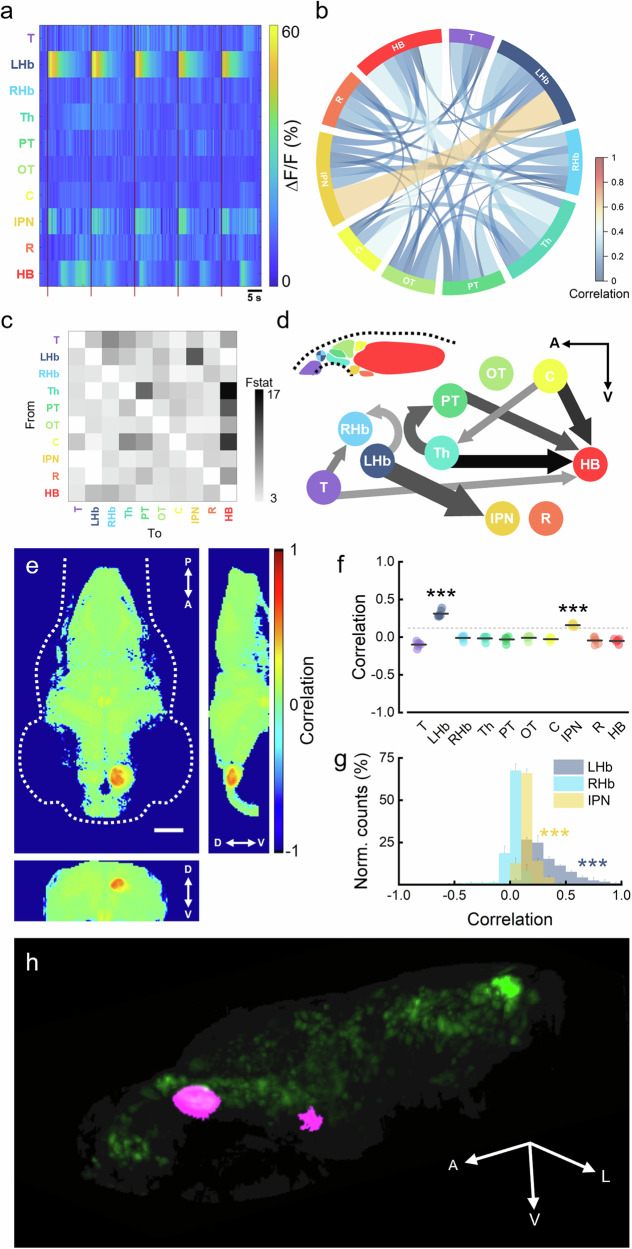
First, we evaluated the brain voxel activation probability in response to the optogenetic stimulation of the left habenula (LHb). Figure 3b shows different projections of the whole-brain average activation probability map (Supplementary Movies 6 and 7). The LHb, the site of stimulation, predictably showed the highest activation probability values. In addition to the LHb, an unpaired nucleus located at the ventral midline of the posterior midbrain showed an increased activation probability with respect to the surrounding tissue. Then, we segmented the entire larval brain into ten different anatomical regions according to structural boundaries (Fig. 3c, left). By extracting the average activation probability from each region (Fig. 3c, right), we found that the deep midbrain district corresponded to the interpeduncular nucleus (IPN). The IPN is a renowned integrative center and relay station within the limbic system that receives habenular afferences traveling through the fasciculus retroflexus46–48. Figure 3d shows the average normalized activation probability distributions for voxels inside the LHb, IPN and right habenula (RHb, the region with the highest activation probability after LHb and IPN). LHb and IPN neurons exhibited activation probabilities as high as 100% and 51%, respectively. Notably, despite LHb presenting higher activation probabilities than IPN across larvae, higher LHb probabilities did not necessarily correspond to higher IPN probabilities (Fig. 3e). Figure 3f shows representative mean ΔF/F signals obtained from the LHb (blue) and IPN (yellow) regions during a stimulation trial. The LHb consistently responded to the photostimulation with high-amplitude calcium transients. The IPN showed lower amplitude activations (~1/10 of the LHb), yet reproducibly following the pace induced by LHb stimulation (as also visible in Supplementary Movie 5, yellow arrowhead). The coherence between these time traces was confirmed by their cross-wavelet power spectrum, showing highest density around the optogenetic trigger rate (1/16 Hz, Fig. 3g; see also Supplementary Fig. 7). As a comparison, Supplementary Fig. 8 shows the cross-wavelet power spectral density of the LHb and RHb activities, where null to low coupling levels emerge.
We then examined whole-brain functional connectivity during optogenetic stimulation. To this end, we first extracted the neuronal activity from previously segmented brain regions. Figure 4a shows, as an example, a heatmap of neuronal activity over time during a single stimulation trial. The LHb and IPN were apparently the only two regions following the photostimulation trigger (dark red vertical bars). This result is confirmed and generalized by the chord diagram presented in Fig. 4b. This chart presents the average all-against-all correlation between the neuronal activity of different brain regions. The LHb and IPN were the two anatomical districts that showed the strongest functional connectivity during stimulation (Pearson’s correlation coefficient = 0.605 ± 0.079, mean ± sem). To explore the causal relationships among observed interactions between brain regions, what is known as effective connectivity49, we analyzed the Granger causality (GC) of their spatially averaged activities50. By examining the added predictability of one time series based on the past values of another, GC analysis allows to draw inferences about directional cause-and-effect relationships between brain activities51. In Fig. 4c, the average strength of the directed interaction among brain regions is depicted using the F statistic. The results from GC analysis showed that the activity recorded in the IPN have a strong causal link only with the activity triggered in the LHb (88.83% ± 8.24% of trials are significant for the LHb→IPN direction while only 2.78% ± 2.78% of them are significant for the opposite direction IPN→LHb, mean ± sem). Furthermore, Fig. 4d illustrates the significant directional causality links between brain regions. Notably, compared to the naturally occurring interacting pairs, the optogenetically revealed LHb-IPN pair showed increased consistency among trials in the significance of their interaction direction (arrow width; percentage of significant trial for the directed interaction: Th-HB, 61.11% ± 9.29%; C-HB, 61.11% ± 10.24%; C-Th, 36.11% ± 5.12%; mean ± sem). On the other hand, the strength of the causal link (represented by the F statistic and graphically depicted by arrow color) for the stimulated LHb-IPN pair was comparable to that of spontaneously occurring pairs (F value: LHb-IPN, 12.16 ± 1.29; PT-HB, 12.10 ± 1.48; Th-PT, 11.40 ± 1.64; PT-HB, 13.98 ± 2.33; mean ± sem). Interestingly, among the causal connections highlighted by GC analysis, causality links (albeit of a lesser extent) emerged also between LHb-RHb and T-RHb pairs.
Fig. 4. Investigation of the efferent connectivity of the left habenular nucleus.
a Heatmap showing the color-coded neuronal activity (ΔF/F) over time of each of the ten brain regions (labels are colored as in Fig. 3c) of one larva during one stimulation trial comprising five photostimulation events (dark red vertical bars). Warmer colors indicate higher activity. b Chord diagram representing the all-against-all neuronal activity correlations of the brain regions (nodes). Link color and width represent the functional connectivity strength. Self-correlations are excluded from the diagram. Data are presented as mean values of N = 6 ReaChR+ larvae. c Granger causality analysis of the larval zebrafish brain (for original causality values see Supplementary Data 1). Each square represents the F statistic which quantifies the statistical strength of the directed interaction from the region localized on the column to the region localized on the row. Only interactions having at least two significant trials are depicted, all the other squares are white (namely, those on the diagonal of the matrix representing self-causality and the Th-LHb, LHb-Hb, OT-T, IPN-T, IPN-LHb, and R-LHb pairs). Average F statistic values range from 3.52 to 16.25 (see Supplementary Data 1). N = 6 ReaChR+ larvae; n = 6 stimulation trials per larva. d Schematic of significant causal interactions among larval brain regions. Arrow color indicates F values (see color bar in c) while arrow width is proportional to the percentage of significant trials over the total. Causality links are shown for percentages of significant trials equal or higher than 33% (see also thresholded causality matrix in Supplementary Fig. 9). Brain regions are presented in the antero-posterior and dorso-ventral directions in their relative anatomical arrangement (see schematic representation of the larval brain in the upper left corner). A, anterior; V, ventral. e Seed-based functional connectivity map showing the Pearson’s correlation coefficient of the activity of each brain voxel with the average activity extracted from the stimulation site (LHb, seed). Maps are maximum intensity projections of the average 3D map (obtained from individual functional connectivity maps of N = 6 ReaChR+ larvae, Supplementary Movie 8) along the transverse (left), sagittal (right) and coronal (bottom) axes. Scale bar, 100 μm. f Average Pearson’s correlation coefficient of different brain regions of the larval brain. Correlation values are obtained from individual average functional connectivity maps. Seed of correlation is represented by the average activity extracted from the LHb (stimulation site). Regions are color-coded as in Fig. 3c. Gray horizontal bars represent intragroup mean values. ***P < 0.0001, *P < 0.0345, one way ANOVA and post-hoc Tukey’s test. Light gray horizontal dashed line represents the correlation coefficient value of 0.12, used as a threshold for Fig. 4h. g Average normalized distributions of Pearson’s correlation coefficient values extracted from voxels inside LHb, IPN and RHb. ***P < 0.0001, KS test with Bonferroni correction (α = 0.01667). h Binarized functional connectivity map (Pearson’s seed-correlation coefficient higher than the 0.12 threshold value, separating regions showing significantly higher correlation) of the left habenula (magenta) overlaid on neurons (green) and larval head structure (gray). A, anterior; V, ventral; L, left. N = 6 ReaChR+ larvae.
After employing GC analysis to assess the direction of the causality links, we employed partial correlation analysis52 to assess the directness of the causal connection observed. Partial correlation analysis represents the remaining correlation between two regions after accounting for the influence of all other regions. Results show with a probability of 88.9 ± 7.0% (mean ± sem) that the LHb-IPN link was direct. In contrast, the LHb-RHb pair produced an opposite result (directness probability 30.4 ± 10.9%, mean ± sem), suggesting an indirect connection, while T-RHb is associated with a more controversial result (directness probability 41.6 ± 17.1%, mean ± sem).
Next, we investigated the seed-based functional connectivity of the left habenular nucleus. To this end, we computed the Pearson’s correlation between the average neuronal activity in the LHb (seed) and the activity in each brain voxel. Figure 4e shows different projections of the average functional connectivity map of the LHb (Supplementary Movie 8). In addition to LHb neurons which exhibited an expected high self-correlation, IPN neurons showed visible higher functional connectivity with respect to other brain regions. This result is confirmed by the analysis of the average correlation coefficient of the different regions (Fig. 4f), where the IPN was the only region presenting a statistically significant functional connectivity with the LHb. Figure 4g shows the average normalized distributions of correlation coefficients computed from voxels inside the LHb, IPN and RHb. With respect to RHb, which had a distribution basically centered at 0 with a short tail towards negative correlation values, neurons in the LHb and IPN showed functional connectivity values as high as 100% and 65%, respectively. In order to visually isolate the neuronal circuit underlying LHb stimulation, we set a threshold on the correlation coefficient. Based on the results shown in Fig. 4f, we chose a threshold of 0.12 as the highest value separating regions showing significantly higher correlation with the seed activity (namely, LHb and IPN). Figure 4h shows the binarized functional connectivity map of the left habenular nucleus in larval zebrafish (Supplementary Movie 9).
Discussion
Dissecting brain functional and effective connectivity requires advanced technology for “reading” and “writing” neuronal activity. Here, we have presented the application of an all-optical 2P system intended for simultaneous imaging and optogenetic control of whole-brain neuronal activity in zebrafish larvae. Our method employs light-sheet microscopy to perform functional imaging, ensuring comprehensive mapping of the entire brain at a significantly improved temporal resolution compared to conventional 2P point-scanning imaging techniques. To elicit precise photoactivation within the larval brain, our light-targeting unit utilizes two pairs of AODs, enabling the displacement of the focal volume to arbitrary locations. Admittedly, the utilization of AODs for optogenetics has been restricted to 1P photostimulation in 2D53–55 owing to the drop in transmission efficiency along the optical axis, which hinders a homogeneous 2P volumetric excitation. However, as we demonstrated in a previous work36, by properly tuning the trains of chirped radio frequency (RF) signals that drive AODs, it is feasible to enhance the uniformity of energy delivery when shifting the focus of the excitation beam. This enhancement has allowed us to proficiently execute optogenetic stimulation of specific targets over a volumetric range of 100 × 100 × 100 μm3. Notably, an intriguing aspect of our approach is that, owing to the use of remote focusing of the detection objective and of AODs for stimulation light defocusing, the localization of the photostimulation volume remains entirely independent of the sequential acquisition of different brain planes, thus affording greater flexibility in our experimental investigations.
As previously mentioned, our setup exploits 2P excitation both for imaging and optogenetic stimulation. On the imaging side, the use of NIR light to produce the sheet of light leads to a significant reduction of common striping artifacts56 that otherwise could severely hinder the interpretation of functional data. Nevertheless, due to the nonlinear nature of its excitation and the need to elongate the axial point spread function (PSF) of the illumination beam to produce the light sheet (thus reducing photon density), 2P LSFM is also typically prone to low signal-to-noise ratio. As a result, despite a voxel size (2.2 × 2.2 × 5 μm3) being 30–35% than the average diameter of a neuronal nucleus (6–7 μm), we did not achieve consistent detection of single neurons throughout the entire brain. On the photostimulation side, the use of nonlinear interaction between light and matter enables precise optical confinement of the stimulation volume, without resorting to narrower genetic control of opsin expression, which is typically required when using 1P excitation57–60. In addition to these aspects, the exclusive use of NIR light as an excitation source, in contrast to visible lasers, dramatically diminishes unwanted and uncontrolled visual stimulation since these wavelengths are scantily perceived by most vertebrate species, including zebrafish61,62. Nevertheless, we observed that 2P light-sheet imaging can elicit a power-dependent increase in the neuronal activity of zebrafish larvae. Despite not significantly affecting zebrafish behavior, this effect, which may be attributed to non-visual sensory perception of the excitation light, remarks once more the significance of maintaining low the overall energy applied to the sample.
To the best of our knowledge, this is the first time that a fully 2P all-optical setup employs light-sheet microscopy for rapid whole-brain imaging and AODs for 3D optogenetic stimulation.
With the aim of establishing an all-optical paradigm for investigating the functional and effective connectivity of the larval zebrafish brain, we considered different pairs of sensor/actuator and eventually opted for the GCaMP6s/ReaChR couple. The green calcium reporter GCaMP6s represents a reliable indicator that has undergone extensive evaluation63–66. On the other hand, the actuator ReaChR, in comparison with other red-shifted opsins, has a slow channel closing mechanism which is particularly suitable for both sequential photostimulation approaches and 2P excitation23. A crucial aspect in all-optical studies lies in the separation between the excitation spectra of proteins used for stimulating and revealing neuronal activity. Previous research has demonstrated that the slow channel closing of ReaChR makes this opsin more susceptible to crosstalk activation when scanning the 920 nm imaging laser at power levels exceeding 60 mW42. However, in our work, we did not observe a significant increase in cross-activation even at power levels as high as 100 mW. This divergence can be attributed to the peculiar excitation features of 2P light-sheet imaging compared to 2P point scanning imaging. In digitally scanned 2P LSFM, the use of low numerical aperture excitation objectives (to obtain a stretched axial illumination PSF, continuously scanned to produce the sheet of light) results in lower intensities (and thus lower photon density) in comparison to point-scanning methods, for equal laser powers. It is worth noting that, despite the negligible crosstalk, 2P light-sheet imaging may still lead to subthreshold activation of ReaChR+ neurons (at 920 nm the opsin retains approximately 25% of the peak action cross-section67), potentially resulting in altered network excitability68.
Previous studies have employed 1030 nm pulsed lasers to stimulate ReaChR42,67. The results of our work demonstrate the feasibility of photostimulating ReaChR at 1064 nm, a wavelength red-shifted by almost 100 nm compared to the ReaChR 2P absorption peak (975 nm67). Furthermore, the use of the 1064 nm wavelength for photostimulation, which is red shifted with respect to the tail of the 2P excitation cross-section of GCaMP6s69, accounts for the absence of fluorescence artifacts potentially caused by the calcium indicator excitation at the wavelength employed for optogenetic intervention.
The characterization of the kinetic features of calcium transients elicited by optogenetic stimulation, which served as a benchmark for identifying the optimal excitation configuration, highlighted two interesting aspects. First, we observed a linear dependence of calcium peak amplitude on the stimulation power applied. This behavior suggests that increasing power produces a proportional increment in the firing rate of ReaChR+ neurons. Secondly, we observed a decrease in the calcium transient rise time in response to longer stimulation durations. This result may be attributed to the fact that ReaChR has a channel off rate (τ-off) of 140 ms38, enabling it to integrate photons beyond the duration of a single volume iteration (125 ms). Supporting this hypothesis is the fact that, after two iterations over the stimulation volume (250 ms and on), the rise time remains constant.
As the system allows accurate identification of groups of neurons functionally connected with the stimulated ones, we exploited the setup to explore the efferent connectivity of the left habenula. The habenulae are bilateral nuclei located in the diencephalon that are highly conserved among vertebrates and connect brain regions involved in diverse emotional states such as fear and aversion, as well as learning and memory70. Like mammals, the habenulae in zebrafish are highly connected hubs receiving afferents from the entopeduncular nucleus71, hypothalamus, and median raphe72, in addition to left-right asymmetric inputs46,73. The habenula can be divided into dorsal (dHb) and ventral (vHb) portions (equivalent to the mammalian medial74 and lateral habenula75, respectively), each exhibiting exclusive efferent connections. Specifically, dHb sends inputs to the IPN while vHb projects to the median raphe74,75. As a consequence, optogenetic stimulation of the entire LHb should lead, in principle, to the activation of both the IPN and the raphe. However, in our experiments, we observed a high probability of activation, strong correlation and causal link only within the IPN population of neurons. This apparent discrepancy can be explained by the fact that, at the larval stage, the vHb represents only a small fraction of the overall habenular volume75. As a result, the limited number of vHb neurons would possess a reduced number of connections with the median raphe, resulting in a weak downstream communication. Furthermore, as described by Amo and colleagues75, although vHb neurons terminate in the median raphe, no direct contact with serotonergic neurons is observed, suggesting the presence of interneurons that may bridge the link, similar to what is observed in mammals76. This inhibitory connection is consistent with the absence of activation of the raphe, which we observed upon left habenular stimulation. Notably, we did not observe any activation in regions downstream of the IPN either. Although adult zebrafish exhibit IPN habenular-recipient neurons projecting to the dorsal tegmental area or griseum centrale77, our results corroborate the structural observations of Ma and colleagues from a functional standpoint. Indeed, using anterograde viral labeling of postsynaptic targets, Ma et al. highlighted that in larval zebrafish habenular-recipient neurons of the IPN do not emanate any efferent axon47.
LHb and the IPN show a high interindividual variability in terms of average activation probability but a lower variability in terms of correlation. This is because larvae may exhibit slightly different opsin expression levels, which result in greater variance in the amplitude of evoked calcium transients and thus a higher activation probability (i.e., the probability of exceeding an arbitrary amplitude threshold). Conversely, the strength of functional connections (i.e., the degree of correlation) appears to not be dependent on the amplitude of evoked neuronal activity. This aspect is also confirmed by the high cross-wavelet power spectral density in a narrow bandwidth centered on the frequency of the triggered optogenetic stimulus, which we observed in the average activity time traces extracted from the LHb and IPN.
Functional connectivity refers to the statistical correlations that signify the synchronous activity between brain regions, without necessarily implying a direct causal interaction. Effective connectivity, on the other hand, takes a step further by seeking to understand the causal influence and the direction of the interaction that one neural population has over another. To delve into the realm of effective connectivity we applied Granger causality analysis. GC results confirmed the presence of a causal link between the LHb and the IPN, with the activity in the latter predicted with high consistency only by the activity triggered in the former. Notably, the magnitude of the causal link strength (F statistic) for the LHb-IPN triggered pair is very similar to that of naturally occurring pairs, underlying the efficacy of our methodology in probing brain connectivity. In addition, partial correlation analysis revealed that the link we observed between LHb and IPN is a direct one, with the interaction between the two not intermediated by any other region, a result which is consistent with the presence of an anatomical connection between LHb and IPN via the fasciculus retroflexus45.
Notably, GC analysis also revealed weaker connections between LHb-RHb and T-RHb. Regarding the former, results from partial correlation analysis highlighted that the link between the two habenulae is most probably indirect. Indeed, no direct connections between the left and right habenulae are known to date, and a crossed feedback circuit passing through the monoaminergic system has been hypothesized78. Concerning T-RHb connection, it is known that in zebrafish a small subset of bilateral pallial neurons sends asymmetric innervations which, passing through the stria medullaris and the habenular commissure, selectively terminate in the RHb79. Despite this direct anatomical connection, partial correlation analysis produced controversial results regarding the directness for this pair of regions. This result is probably due to the limited number of telencephalic cells contacting the RHb45,79, whose activity could have been overshadowed by the averaging of the activity on the entire telencephalon.
In conclusion, we employed optogenetic stimulation to map the whole-brain functional connectivity of the left habenula efferent pathway in zebrafish larvae. This application has showcased the remarkable capabilities of our 2P setup for conducting crosstalk-free all-optical investigations. The use of AODs for precisely addressing the photostimulation is a hot topic in systems neuroscience, as evidenced by recent conference contributions80–83. Owing to their discontinuous scanning and constant access time, these devices indeed enable random-access modality. This feature empowers AODs with the native capability to perform rapid sequential excitation over multiple sparsely distributed cellular targets, a feature recently sought after also by SLM adopters84. Indeed, rapid sequential stimulation enabled by AODs represents an invaluable tool for studies aiming at replicating a physiological neuronal activation pattern.
Future efforts will be devoted to further expanding the volume addressable with AOD scanning while concurrently improving the uniformity of energy delivery. Furthermore, leveraging transgenic strains that express the actuator under more selective promoters (such as vglut2 for glutamatergic and gad1b for GABAergic neurons) will undoubtedly help producing accurate inferences on network structures85,86, thus boosting the quest towards a comprehensive picture of zebrafish brain functional connectivity. On the imaging side, technical implementations will be made, in order to improve image contrast while maintaining a low laser power on the sample. This advancement will enable the use of automated segmentation algorithms for single-neuron detection. Cell-wise analyses will allow to refine the reconstruction of neuronal effective connectivity, capturing the nuanced differences between individual cells. Together, nonlinear light-sheet microscopy and 3D optogenetics with AODs, along with the employment of larval zebrafish, offer a promising avenue for bridging the gap between microscale resolution and macroscale investigations, enabling the mapping of whole-brain functional/effective connectivity at previously unattainable spatio-temporal scales.
Methods
Optical setup
All-optical control and readout of zebrafish neuronal activity is achieved through a custom system that combines a 2P dual-sided illumination LSFM for whole-brain calcium imaging41,87,88 and an AOD-based 2P light-targeting system for 3D optogenetic stimulation36 (Supplementary Fig. 1 and Fig. 1a). The two systems have been slightly modified with respect to the previous published versions to optically couple them. Briefly, the 2P light-sheet imaging path is equipped with a pulsed Ti:Sa laser (Chameleon Ultra II, Coherent), tuned at 920 nm. After a group delay dispersion precompensation step, the near-infrared beam is adjusted in power and routed to an electro-optical modulator (EOM) employed to switch the light polarization orientation between two orthogonal states at a frequency of 100 kHz. A half-wave plate and a quarter-wave plate are used to control the light polarization plane and to pre-compensate for polarization distortions. Then, the beam is routed to a hybrid pair of galvanometric mirrors (GMs). One is a fast resonant mirror (CRS-8 kHz, Cambridge Technology) used to digitally generate the virtual light-sheet scanning (frequency 8 kHz) the larva along the rostro-caudal direction. The second GM is a closed-loop mirror (6215H, Cambridge Technology) used to displace the light-sheet along the dorso-ventral direction. The scanned beam is driven by a scan lens and a tube lens into a polarizing beam splitter, which diverts the light alternatively into either of the two excitation arms, according to the instantaneous polarization state imposed by the EOM. In order to maximize fluorescence collection, after the beam splitter in one of the two arms a half-wave plate is used to rotate the light polarization plane so that light coming from both the excitation paths is polarized parallel to the table surface40. Through a twin relay system, the beams are ultimately routed into the excitation objectives (XLFLUOR4X/340/0,28, Olympus). The excitation light is focused inside a custom fish water-filled imaging chamber, heated to 28.5 °C. The fine positioning of the sample under the detection objective is performed with three motorized stages. The fluorescence emitted by the sample is collected with a water-immersion objective (XLUMPLFLN20XW, Olympus, NA = 1). Finally, a relay system brings the collected signal to an electrically tunable lens (ETL; EL-16-40-TC-VIS-5D-C, Optotune) which performs remote axial scanning of the detection objective focal plane in sync with the light-sheet closed-loop displacement. The signal collected is filtered (FF01-510/84-25, Semrock) to select green emission. The filtered light reaches an air objective (UPLFLN10X2, Olympus, NA = 0.3), which demagnifies the image onto a subarray (512 × 512 pixels) of an sCMOS camera (ORCA-Flash4.0 V3, Hamamatsu) working at 16-bit depth of integer gray levels. The final magnification of the imaging system is 3×, with a resulting pixel size of 2.2 μm.
Below the transparent PMMA bottom of the imaging chamber, a high-speed CMOS camera (Blackfly S USB3, FLIR) equipped with a varifocal objective lens (employed at 50 mm; YV3.3x15SA-2, Fujinon) is positioned to perform behavioral imaging (tail deflections) during light-sheet imaging. Illumination for behavioral imaging is provided by an 850 nm LED (M850L3, Thorlabs) positioned at an angle above the imaging chamber. A bandpass filter (FF01-835/70-25, Semrock) is placed in front of the objective lens for blocking high-intensity light from the 920 nm light-sheet (see Supplementary Fig. 1b). Recordings are performed using a 300 × 300 pixels subarray of the camera chip, covering the entire larval body. This configuration allows to achieve sufficient magnification (pixel size: 15.4 μm) and contrast to enable live tail tracking.
The 3D light-targeting system employs a 1064 nm pulsed laser (FP-1060-5-fs Fianium FemtoPower, NKT Photonics, Birkerød, Denmark) as an excitation source. The output power (max. 5 W) is attenuated and conveyed to a half-wave plate, which is employed to adjust the polarization of the beam, before the first AOD stage (DTSXY-400 AA Opto Electronic, Orsay, France) is reached. The output beam is then coupled with the second AOD stage through two 1:1 relay systems. From the exit of the second stage, by means of a 1:1 relay system, the beam is routed to a pair of galvanometric mirrors (GVS112, Thorlabs). The scanned beam is then optically coupled with a scan lens (AC254-100-B, Thorlabs) and a tube lens (F = 300 mm, in turn formed by two achromatic doublets - AC254-150-C-MLE, F = 150 mm by Thorlabs, so customized to avoid aberrations). The excitation light is finally deflected by a dichroic mirror (DMSP926B, Thorlabs) toward the back pupil of the illumination objective, which is also employed by the imaging system for fluorescence detection.
Optical characterization of the system
The detailed optical characterization of the 2P light-sheet system was described in a previous work of our group41. Summarizing, each of the light sheets coming from the two excitation arms has a transversal full width at half maximum (FWHM) at waist of 6 µm and a longitudinal FWHM of 327 µm. The lateral FWHM of the detection PSF is 5.2 µm. Herein, we describe the optical performance of the AOD-based light-targeting system used for optogenetic stimulation. When using AODs to move the beam away from its native focus, the illumination axial displacement—or defocus—has a linear relation with the chirp parameter α, i.e., the rate of frequency change of the driving radio waves89. We thus measured the axial displacement of the focused beam as a function of α by illuminating a fluorescent solution (Sulforhodamine 101; S7635, Sigma-Aldrich) and localizing the maximum fluorescent peak in the volume as a function of α, which ranged from −1 MHz/µs to 1 MHz/µs (step size 0.1 MHz/µs). For each chirp configuration, the ETL in detection path was used to obtain a 200-µm deep stack (step size: 1 µm) centered at the nominal focal plane of the illumination objective. Supplementary Fig. 10a shows the axial position of the fluorescent intensity peak as a function of the chirp addressed, following an expected linear trend. We evaluated the conversion coefficient from the slope of the linear fit, which was 50.44 ± 3.45 µm/MHz/µs (mean ± sd).
We also measured the amount of energy released on the sample as a function of the chirp parameter or, basically, as a function of the time spent illuminating axially displaced targets. Indeed, the beam would spend slightly different periods lighting spots displaced in different z-planes as the effective frequency ramping time is inversely proportional to the chirp parameter α imposed on the RF signals driving the AODs. As explained in detail in a previous work36, we partially recovered this non-uniformity in the distribution of power deposited along the axial direction by repeatedly triggering equal frequency ramps within the desired dwell time (here, 20 µs each point), using what we called multi-trigger modality. With respect to the conventional single-trigger modality, we effectively multiplied the minimum energy deposited on different focal planes, while keeping a stable dwell time. Supplementary Fig. 10b shows in black the usual light transmission distribution collected as a function of the chirp parameter (single-trigger modality) and in blue the distribution obtained with our multi-trigger approach.
We then measured the point spread function (PSF) of the light-targeting system using subdiffraction-sized fluorescent beads (TetraSpeck microspheres, radius 50 nm; T7279, Invitrogen) embedded in agarose gel (1.5%, w/v) at a final concentration of 0.0025% (vol/vol). The measurements were performed on a field of view of 100 × 100 μm2, performing raster scans of 500 × 500 points. The objective was moved axially covering a 200 μm range (z step: 1 μm) and the emitted signal was conveyed and collected on an auxiliary photomultiplier tube positioned downstream of the fluorescence-collecting objective. The radial and axial intensity profiles of 25 beads were computed using the open-source software ImageJ90 and fitted with Gaussian functions in Origin Pro 2021 (OriginLab Corp.) to estimate FWHM. Supplementary Fig. 10c shows, as an example, the raw fluorescence distributions of 5 beads and the Gaussian fit corresponding to the average FWHM, plotted in red and black for the radial and axial PSF, respectively. We found them to be FWHMr = 0.81 ± 0.06 µm and FWHMa = 3.79 ± 0.66 µm (mean ± sd). This measurement was performed by driving the AODs with stationary RF signals. To evaluate the eventual illumination spatial distortions arising away from the nominal focal plane of the objective, we repeated the same PSF measurement for different chirps or, in other words, for different AOD-controlled axial displacements (80 µm range, step size of 20 µm). The average FWHM obtained for the bead intensity distribution is shown in Supplementary Fig. 10d. The radial PSF of the system remains approximately constant as a function of the chirp parameter. A small change is due to the chromatic dispersion affecting the laser beam interacting with the crystal. The deflection angle induced by the AODs on the incident beam is frequency and wavelength dependent. This means that a broadband laser is straightforwardly spatially dispersed by the crystal and that the frequency variations can slightly affect this distortion. Moreover, the axial PSF tends to become slightly oblong with increasing axial displacement. This effect is attributable to the temporal dispersion affecting a short-pulsed laser beam interacting with the crystal. This temporal broadening reduces the axial 2P excitation efficiency, generating a larger axial PSF. This effect is more evident when a chirp is applied to the RF signals driving the AODs. Under these conditions, the beam reaches the objective back-pupil in a non-collimated state. Future efforts will be devoted to the compensation of chromatic aberration and temporal dispersion, for example employing a highly dispersive prism upstream of AODs91.
Zebrafish lines and maintenance
The double Tg(elavl3:H2B-GCaMP6s; elavl3:ReaChR-TagRFP) zebrafish line was obtained from outcrossing the Tg(elavl3:H2B-GCaMP6s)20,92 and the Tg(elavl3:ReaChR-TagRFP)36,63 lines on the slc45a2b4/- heterozygous albino background93, which we previously generated. The double transgenic line expresses the fluorescent calcium reporter GCaMP6s (nucleus) and the red-shifted light-activatable cation channel ReaChR (plasma membrane) in all differentiated neurons. ReaChR is expressed as a fusion peptide with the red fluorescent protein TagRFP to ensure its localization. Zebrafish strains were reared according to standard procedures94, and fed twice a day with dry food and brine shrimp nauplii (Artemia salina), both for nutritional and environmental enrichment. For the experiments, we employed N = 20, 5 dpf Tg(elavl3:H2B-GCaMP6s; elavl3:ReaChR-TagRFP) and N = 13, 5 dpf Tg(elavl3:H2B-GCaMP6s), both of which were on the slc45a2b4/b4 homozygous albino background. Zebrafish larvae used in the experiments were maintained at 28.5 °C in fish water (150 mg/L Instant Ocean, 6.9 mg/L NaH2PO4, 12.5 mg/L Na2HPO4, 1 mg/L methylene blue; conductivity 300 μS/cm, pH 7.2) under a 14/10 light/dark cycle, according to standard protocols94. Experiments involving zebrafish larvae were carried out in compliance with European and Italian laws on animal experimentation (Directive 2010/63/EU and D.L. 4 March 2014, n.26, respectively), under authorization n.606/2020-PR from the Italian Ministry of Health.
Zebrafish larvae preparation
To select calcium reporter/opsin-expressing larvae for use in the experiments, 3 dpf embryos were subjected to fluorescence screening. The embryos were first slightly anesthetized with a bath in tricaine (160 mg/L in fish water; A5040, Sigma-Aldrich) to reduce movement. Using a stereomicroscope (Stemi 508, Carl Zeiss) equipped with LEDs for fluorescence excitation (for GCaMP6s: blue LED, M470L3; for TagRFP: green LED, M565L3, both from Thorlabs) and fluorescence filters to block excitation light (for GCaMP6s: FF01-510/84-25; for TagRFP: FF01-593/LP-25, both from Semrock), embryos were selected according to the presence of brighter green/red fluorescent signals in the central nervous system. Screened embryos were transferred to a Petri dish containing fresh fish water and kept in an incubator at 28.5 °C until 5 dpf. Zebrafish larvae were mounted as previously described95. Briefly, each larva was transferred into a reaction tube containing 1.5% (w/v) low-gelling temperature agarose (A9414, Sigma-Aldrich) in fish water, maintained fluid on a heater set at 38 °C. Using a plastic pipette, larvae were then placed on a microscope slide inside a drop of melted agarose. Before gel polymerization, their position was adjusted using a couple of fine pipette tips for the dorsal portion to face upwards. To avoid movement artifacts during the measurements, larvae were paralyzed by a 10-min treatment with 2 mM d-tubocurarine (93750, Sigma-Aldrich), a neuromuscular blocker. For tail-free preparations, upon gel polymerization, agarose caudal to the swimming bladder was removed using a scalpel. In this case, no paralyzing agent was applied. Mounted larvae were then placed inside the imaging chamber filled with fish water and thermostated at 28.5 °C for the entire duration of the experiment.
Structural imaging to evaluate expression patterns in double transgenic zebrafish larvae
Confocal imaging of a 5 dpf Tg(elavl3:H2B-GCaMP6s; elavl3:ReaChR-TagRFP) larva on an albino background was performed to evaluate the spatial expression of the two proteins. The larva was mounted in agarose as described above and deeply anesthetized with tricaine (300 mg/L in fish water). We employed a commercial confocal microscope (Ti2, Nikon) equipped with two continuous wavelength lasers emitting at 488 and 561 nm for GCaMP6s and TagRFP excitation, respectively. Imaging was performed using a 10× objective, allowing the entire head of the animal to fit into the field of view. Using a piezo-electric motor (PIFOC, Physik Instrumente - PI), the objective was moved at 182 consecutive positions (z step: 2 μm) to acquire the volume of the larval head.
Simultaneous whole-brain and behavioral imaging
Head restrained larvae, capable of performing wide tail deflections, were imaged from below the 2P LSFM imaging chamber using a dedicated high-speed camera (see Optical setup for details). Images were streamed at 300 Hz via a USB3 connection to a workstation running a custom tool for live tail movement tracking, developed using the open-source Python Stytra package96. Larval tail length was divided into 9 segments, and the sum of their relative angles was employed to quantify tail deflection. Tail movements of both ReaChR+ and ReaChR− larvae were tracked for 200 s. During the first half 2P LSFM imaging was off (imaging OFF). During the second half larvae were subjected to whole-brain light-sheet imaging (imaging ON) with the same parameters described in the previous section. Each larva measured underwent 3 consecutive 200-s simultaneous whole-brain/behavioral recordings (inter-measurement interval less than 1 min).
Simultaneous whole-brain imaging and optogenetic stimulation
Whole-brain calcium imaging was performed at 2.5 Hz (a more than optimal volumetric rate considering the typical time constant of the exponential decay for the nuclear localized version of the GCaMP6s sensor τ: 3.5 s97) with 41 stacked z-planes spanning a depth of 200 μm. An interslice spacing of 5 μm was chosen because it coincides with the half width at half maximum of the detection axial PSF. Before each measurement, the scanning amplitude of the resonant galvo mirror was tuned to produce a virtual light-sheet with a length matching the size of the larval brain in the rostro-caudal direction. The laser wavelength was set to 920 nm to optimally excite the GCaMP6s fluorescence. Unless otherwise stated, the power at the sample of the 920 nm laser was set to 60 mW.
Optogenetic stimulation was performed at 1064 nm with a laser power at the sample of 30 mW (unless otherwise specified). Before each experimental session, the 1064 nm stimulation laser was finely aligned to the center of the camera field of view. Then, by means of the galvo mirrors present in the stimulation path, the offset position of the stimulation beam was coarsely displaced in the x-y direction toward the center of the area to be stimulated. During the optogenetics experiment the stimulation volume was covered by discontinuously scanning the beam focus via the two pairs of AODs. A typical volume of 50 × 50 × 50 μm3 was covered with 6250 points (point x-y density: 1 point/0.25 μm2; z step: 5 μm) with a point dwell time of 20 μs (overall time: 125 ms). The medial plane of the stimulation volume (chirp = 0 MHz/μs, null defocus) was adjusted to overlap with the medial plane of the LHb. Unless otherwise stated, each stimulus consisted of four complete cycles of the entire volume, lasting 500 ms. Each stimulation trial consisted of 100 s of whole-brain calcium imaging, during which 5 optogenetic stimuli (interstimulus interval: 16 s, based on the characterization experiments performed, in order to trigger activation events only after the end of the previous calcium transient) were applied at the same volumetric site. Six trials were performed on each larva, with an intertrial interval ranging from 1 to 3 min. Overall, each larva was imaged for 10 min during which it received 30 stimuli.
Data analysis
Preprocessing
Whole-brain calcium imaging data were processed as follows. Images composing the hyperstacks were first 2 × 2 binned (method: average) in the x and y dimensions to obtain a quasi-isotropic voxel size (4.4 × 4.4 × 5 μm3). Then, employing a custom tool written in Python 3, we computed the voxel-wise ΔF/F0 of each volumetric recording, after background subtraction. F0 was calculated using FastChrom’s baseline estimation method98.
Quantification of imaging crosstalk and optogenetic activation extent/specificity
To quantify crosstalk during imaging we first considered different metrics to evaluate neuronal activity levels (Supplementary Fig. 3). We computed the standard deviation (SD) over time, the number of calcium peaks per minute, and the average peak amplitude of each voxel composing the larval brain during 5 min of whole-brain calcium imaging (Supplementary Fig. 3a). For automatic calcium peaks identification, we set the following thresholds: minimum peak prominence 0.05; minimum peak FWHM 2.5 s, minimum peak distance 5 s. We found the SD to have improved sensitivity in discriminating between diverse conditions compared to the number of peaks per minute (Supplementary Fig. 3 b, c). These results reflected those observed by adopting the average amplitude of calcium peaks (Supplementary Fig. 3 d–f) as an activity metric. We thus employed SD over time as a proxy of neuronal activity levels since its results do not depend on predefined thresholds. Therefore, the distribution of SD values calculated for each brain was first normalized with respect to the total number of voxels and then pooled (method: average) according to the larval strain (ReaChR+ and ReaChR−). Similarly, the normalized distributions of SD values for ReaChR+ and ReaChR− larvae subjected to 100 s of whole-brain imaging during which they received 5 photostimulations (1064 nm) were calculated to evaluate the effect of the optogenetic stimulation. Imaging crosstalk and optogenetic stimulation indices were calculated using the Hellinger distance99 as a measure of dissimilarity between two probability distributions P and Q:
The errors in the Hellinger distances were calculated according to error propagation theory as follows:
Finally, normalized distributions of SD values for ReaChR− larvae exposed either to imaging (100 s) only or to imaging and photostimulation (100 s and 5 stimuli at 1064 nm) were calculated to evaluate the specificity of the effect observed.
Quantification of tail movements during whole-brain light-sheet imaging
Tail deflection (i.e., sum of relative tail segments angles) time traces, were processed to detect and count the number of tail beats. In detail, deflection peaks were considered as tail beats if exceeding an absolute threshold of 20°. Consecutive tail deflections that did not come back to resting position for at least 0.5 s were considered part of the same movement. The relative number of tail beats during imaging ON (Fig. 1i) was calculated for each trial of each larva dividing the number of tail movements during the imaging ON period by that quantified during the imaging OFF period.
To combine behavioral and brain activity recordings (Fig. 1g), the average fluorescence time trace of the hindbrain acquired at 2.5 Hz was first interpolated to match the frequency of behavioral recordings (300 Hz). Then, ΔF/F0 was calculated as previously described.
Characterization of stimulation-induced calcium transients
To characterize neuronal activation as a function of stimulation parameters (scan time and laser power), we first extracted the voxel time series averaged over the entire stimulation site (i.e., left habenula) from 4D ΔF/F0 hyperstacks. Time traces were windowed to isolate and align the three stimulation events contained in a single trial. Isolated calcium transients were analyzed using the peak analyzer function in Origin Pro 2021 (OriginLab Corp.) to obtain peak amplitude, rise/decay time (i.e., time from baseline to peak and time from peak to baseline, respectively) and duration values. Pooled peak duration data were obtained by first averaging three events of the same larva (intra-individual) and then averaging data between larvae (inter-individual).
Activation probability and correlation maps
Using a custom Python tool, we calculated the probability of each voxel composing the brain to be active in response to the optogenetic stimulation. For each stimulation event, a voxel was considered active if its change in fluorescence in a 2 s time window after the stimulation exceeded three standard deviations above its baseline level (2 s pre-stimulation). Only events in which the voxels inside the stimulation volume met the activation criterion were considered effective optogenetic stimulations. By iterating this process for all the stimulation events performed (on the same site of the same larva), we calculated the activation probability of each voxel as the number of times the voxel exceeded the threshold divided by the total number of valid stimulations. Employing a second Python tool, we then computed activity correlation maps showing Pearson’s correlation coefficient between each voxel and the activity extracted from the stimulation site (seed). The 3D maps of correlation and activation probability obtained were subsequently aligned. First, the acquired 4D hyperstacks were time averaged. Second, the resulting 3D stack of each larva was registered to a reference brain. Nonrigid image registration was performed using the open source software Computational Morphometry Toolkit (CMTK 3.3.1, https://www.nitrc.org/projects/cmtk/) and the ImageJ user interface100, employing the command string (-awr 01 -X 52 -C 8 -G 80 -R 3 -A “--accuracy 1.6” -W “--accuracy 0.4”). The calculated morphing transformations were ultimately applied to the corresponding 3D maps.
Following the zebrafish brain atlases101,102, the volumetric regions of interest (ROIs) used in the analysis were manually drawn onto the reference brain (employing ImageJ), based on anatomical boundaries. The 10 volumetric ROIs were then adopted to extract from each map the voxel-wise distribution of activation probability/correlation coefficient values used for further analyses.
The binarized functional connectivity map shown in Fig. 4h was obtained after applying a threshold on Pearson’s correlation coefficient to the average correlation map shown in Fig. 4e. The 0.12 value adopted represented the correlation coefficient threshold separating significant from non-significant correlations among brain regions (see Fig. 4f).
Cross-wavelet power spectrum analysis
The possible coupling between the delineated brain ROIs and the stimulation site was also characterized in the spectral domain by quantifying and inspecting their cross-wavelet power spectral density (CPSD)103. The wavelet transforms of the average activity signals extracted from each ROI were computed using the Morlet mother wavelet, adopting a central frequency f0 = 1 Hz as time-frequency resolution parameter, and 256 voices per octave for fine frequency discretization. Spurious time-boundary effects were addressed by first applying a zero-padding scheme to the original time series, and then isolating the so-called cone of influence, i.e., the time–frequency region where boundary distortions in the CPSD estimates are negligible104.
Granger causality analysis
The causal link between the activity of different brain regions was explored by analyzing their Granger causality105. GC analysis among ΔF/F0 time series of brain regions was performed in R, with the “lmtest” library106. To select an appropriate lag order, we computed both the Akaike (AIC) and Bayesian (BIC) information criterions of the complete autoregressive model for each comparison (each trial and each possible regions pair) for lag orders from 1 to 8 (0.4–3.2 s). Then, for each comparison we selected the lag order associated with the minimum value of the information criterions. Finally, we computed the mode value of this list and used this unique lag order value for every comparison of the final GC analysis. The mode values based on both AIC and BIC resulted the same: a lag order equal to 2, which corresponds to a 0.8 s lag.
For each larva, trial, pairs of regions’ activity and causality direction we computed the average F statistic value of the tests. Finally, multiplicity correction for the p-values was performed with a false discovery rate approach using the Benjamini–Hochberg method107 (GC analysis results are reported in Supplementary Data 1). The F statistic was presented in Fig. 4c as average values of all pairs having at least two significant trials. The F statistic in the graph of Fig. 4d was presented as arrows color-mapped according to the average F value found between brain regions’ connections. Direction of the arrow indicates direction of the causality interaction, while arrow width represents the proportion of significant trials over the total. Only causal links having at least 33% of significant trials were depicted (see thresholded matrix in Supplementary Fig. 9).
Partial correlation analysis
In order to gain insight into the directness of the interactions between brain regions, we analyzed the partial correlation between pairs of region-wise mean ΔF/F0 time series, aiming to capture their residual coupling after the influence of all other regions was accounted for51. Pairwise partial correlation coefficients were obtained as described by Han and colleagues108. In detail, the partial correlation between a pair of brain regions (i.e., LHb-IPN, LHb-RHb, and T-RHb), A and B, was evaluated as the Pearson’s correlation coefficient between regressed time series ΔF/F0 A,R and ΔF/F0 B,R, suitably corrected for the contribution of each other regions’ mean activity signal. These time series were estimated by multiple regression on the original traces ΔF/F0 A and ΔF/F0 B, through the evaluation of the Moore-Penrose pseudoinverse of the remaining regions’ time series matrix, C:
where C+ is the Moore-Penrose pseudoinverse matrix:
here computed using the Python SciPy library109. The regressed time series were then obtained as:
The directness of the mutual interaction between two brain regions was finally detected from the presence of both statistically significant Pearson’s and partial correlation coefficients. When only the Pearson’s correlation is significant the interaction is defined as indirect. While in case only the partial correlation is significant, we are observing what is defined as pseudo-correlation108. Results of partial correlation analysis can be found in Supplementary Data 2.
Statistics and reproducibility
To guarantee reproducibility of the findings and avoid bias, the larvae employed in the experiments never belonged to a single batch of eggs. No a priori sample size calculation was performed. The sample size employed was justified by the high grade of consistency in the results obtained from different larvae. The expression pattern of GCaMP6s and ReaChR were evaluated in N = 1 ReaChR+ larva by confocal imaging. Crosstalk activation of ReaChR by 920 nm excitation light-sheet imaging was evaluated on N = 3 ReaChR+ and N = 3 ReaChR− larvae, in the brain activity experiment, and N = 4 ReaChR+ and N = 4 ReaChR− larvae in the combined brain/behavioral activities experiment. The effect of optogenetic stimulation was evaluated on N = 6 ReaChR+ and N = 6 ReaChR− larvae. Characterization of optogenetically induced calcium transients as a function of stimulation settings was performed on N = 4 ReaChR+ larvae (n = 3 calcium transients per larva). The activation probability, correlation, and causality were evaluated on N = 6 ReaChR+ (n = 30 stimulations per larva).
OriginPro 2021 (OriginLab Corp.) was used to carry out all the statistical analyses. Unless otherwise stated, results were considered statistically significant if their corresponding p-value was less than 0.05 (*P < 0.05; **P < 0.01; ***P < 0.0001). Both intergroup and intragroup statistical significance of imaging crosstalk (Fig. 1f and Supplementary Fig. 3c, e) were performed using two-way ANOVA (factors: zebrafish strain, imaging power) followed by post-hoc comparisons with Tukey’s method. Two-way ANOVA and Tukey’s post-hoc comparison were employed also for quantifying the statistical significance of tail beats between imaging OFF and ON conditions (Fig. 1h; factors: zebrafish strain, imaging presence). For intergroup statistical evaluations of both activation probability (Fig. 3c) and Pearson’s correlation coefficient (Fig. 4f), we first verified the normality distribution of data using the Shapiro-Wilk test (see Supplementary Fig. 11a,b for test results) and then performed one way ANOVA (factor: brain region), followed by post-hoc comparisons employing Tukey’s method. Statistical comparisons of relative number of tail beats during 920 nm imaging (Fig. 1i) and median SD values to evaluate the effect of optogenetic stimulation (Fig. 1j, k and Supplementary Fig. 5) were performed using unpaired t test.
Statistical comparisons of the average distributions of SD (Fig. 3d) and Pearson’s correlation coefficient (Fig. 4e) values were performed with the two-sample Kolmogorov-Smirnov test (KS test), applying the Bonferroni correction (α = 0.05/3 = 0.01667, in both cases).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Dr. Ludovico Silvestri (University of Florence), Dr. Leonardo Sacconi (INO-CNR), and Domenico Alfieri (Light4Tech) for fruitful discussions regarding AOD implementation. We thank Dorotea Nardini (University of Florence) for the helpful discussion about error propagation theory. We thank Lizzy Griffiths for the larval zebrafish drawing used in Supplementary Fig. 1b. This project was funded by: European Union’s NextGenerationEU Program with the I-PHOQS Infrastructure [IR0000016, ID D2B8D520, CUP B53C22001750006] “Integrated infrastructure initiative in Photonic and Quantum Sciences” and the Italian Ministry for University and Research in the framework of the Advanced Light Microscopy Italian Node of Euro-Bioimaging ERIC, and the Bank Foundation Fondazione Cassa di Risparmio di Firenze with grant “Human Brain Optical Mapping”.
Author contributions
L.T. and F.S.P. conceived the study, L.T. designed the experiments, L.T. performed the experiments, L.T. and P.R. performed the characterization of calcium activity response to optogenetic stimulation, P.R. optically coupled the stimulation and imaging systems, P.R. performed optical characterization of the photostimulation system, M.S. developed the Python and C++ software of the custom RF signal generator driving the AOD-based photostimulation system, M.S. and M.M. programmed the LabVIEW software used for operating the photostimulation system, L.T. conceived data analysis, M.S. developed the Python tools for ΔF/F0 calculation, automatic calcium peak detection, cross-wavelet power spectrum, correlation/partial correlation analyses and live tail tracking, G.d.V. programmed the Python software for activation probability and the R Granger causality computation, L.T. and G.d.V. set up the statistical analysis, L.T., M.S., G.d.V. and P.R. performed data analysis, F.V. supervised zebrafish experiments, F.S.P. acquired funding, L.T. wrote the manuscript. All authors reviewed and agreed to the published version of the manuscript.
Peer review
Peer review information
Communications Biology thanks Mahdi Zarei and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Chao Zhou and Benjamin Bessieres. A peer review file is available.
Data availability
All data source underlying plots are provided as Supplementary Data 3. Raw data are available from the corresponding authors upon reasonable request.
Code availability
Custom-written LabVIEW/Python/C++ codes are available from the corresponding authors upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/16/2025
In the version of the article initially published, refs. 80–83 were incomplete and have now been corrected in the HTML and PDF versions of the article.
Contributor Information
Lapo Turrini, Email: turrini@lens.unifi.it.
Francesco Saverio Pavone, Email: francesco.pavone@unifi.it.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06731-3.
References
- 1.Rizzolatti, G., Fadiga, L., Gallese, V. & Fogassi, L. Premotor cortex and the recognition of motor actions. Brain Res. Cogn. Brain Res.3, 131–141 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Sargolini, F. et al. Conjunctive representation of position, direction, and velocity in entorhinal cortex. Science312, 758–762 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Fabbri-Destro, M. & Rizzolatti, G. Mirror neurons and mirror systems in monkeys and humans. Physiology23, 171–179 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Norman-Haignere, S. V. et al. A neural population selective for song in human auditory cortex. Curr. Biol.32, 1470–1484.e1412 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deisseroth, K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci.18, 1213–1225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin, M. Z. & Schnitzer, M. J. Genetically encoded indicators of neuronal activity. Nat. Neurosci.19, 1142–1153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Packer, A. M. et al. Two-photon optogenetics of dendritic spines and neural circuits. Nat. Methods9, 1202–1205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hochbaum, D. R. et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods11, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou, S. et al. Genetically targeted all-optical electrophysiology with a transgenic cre-dependent optopatch mouse. J. Neurosci.36, 11059–11073 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, L. Z. et al. All-optical electrophysiology reveals the role of lateral inhibition in sensory processing in cortical layer 1. Cell180, 521–535.e518 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, T. Y., Choi, T. I., Lee, Y. R., Choe, S. K. & Kim, C. H. Zebrafish as an animal model for biomedical research. Exp. Mol. Med.53, 310–317 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Power, R. M. & Huisken, J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat. Methods14, 360–373 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Chen, I. W., Papagiakoumou, E. & Emiliani, V. Towards circuit optogenetics. Curr. Opin. Neurobiol.50, 179–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turrini, L., Roschi, L., de Vito, G., Pavone, F. S. & Vanzi, F. Imaging approaches to investigate pathophysiological mechanisms of brain disease in zebrafish. Int. J. Mol. Sci.24, 10.3390/ijms24129833 (2023). [DOI] [PMC free article] [PubMed]
- 15.Rickgauer, J. P., Deisseroth, K. & Tank, D. W. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat. Neurosci.17, 1816–1824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dal Maschio, M., Donovan, J. C., Helmbrecht, T. O. & Baier, H. Linking neurons to network function and behavior by two-photon holographic optogenetics and volumetric imaging. Neuron94, 774–789.e775 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Forster, D., Dal Maschio, M., Laurell, E. & Baier, H. An optogenetic toolbox for unbiased discovery of functionally connected cells in neural circuits. Nat. Commun.8, 116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiao, Z. F. et al. All-optical imaging and manipulation of whole-brain neuronal activities in behaving larval zebrafish. Biomed. Opt. Express9, 6154–6169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E. H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science305, 1007–1009 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Vladimirov, N. et al. Light-sheet functional imaging in fictively behaving zebrafish. Nat. Methods11, 883–884 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Packer, A. M., Russell, L. E., Dalgleish, H. W. & Hausser, M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat. Methods12, 140–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shemesh, O. A. et al. Temporally precise single-cell-resolution optogenetics. Nat. Neurosci.20, 1796–1806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronzitti, E. et al. Recent advances in patterned photostimulation for optogenetics. J. Optics19, 113001 (2017).
- 24.Picot, A. et al. Temperature rise under two-photon optogenetic brain stimulation. Cell Rep.24, 1243–1253.e1245 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Duocastella, M., Surdo, S., Zunino, A., Diaspro, A. & Saggau, P. Acousto-optic systems for advanced microscopy. J. Phys-Photonics3, 012004 (2021).
- 26.Ricci, P. et al. Acousto-optic deflectors in experimental neuroscience: overview of theory and applications. J. Phys-Photonics6, 022001 (2024).
- 27.Katona, G. et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat. Methods9, 201–208 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Szalay, G. et al. Fast 3D imaging of spine, dendritic, and neuronal assemblies in behaving animals. Neuron92, 723–738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadella, K. M. et al. Random-access scanning microscopy for 3D imaging in awake behaving animals. Nat. Methods13, 1001–1004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villette, V. et al. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell179, 1590–1608.e1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akemann, W. et al. Fast optical recording of neuronal activity by three-dimensional custom-access serial holography. Nat. Methods19, 100–110 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Wang, K. et al. Precise spatiotemporal control of optogenetic activation using an acousto-optic device. PLoS One6, e28468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, K. et al. Parallel pathways convey olfactory information with opposite polarities in Drosophila. Proc. Natl Acad. Sci. USA111, 3164–3169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez, O., Pietrajtis, K., Mathieu, B. & Dieudonne, S. Optogenetic stimulation of complex spatio-temporal activity patterns by acousto-optic light steering probes cerebellar granular layer integrative properties. Sci. Rep.8, 13768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conti, E. et al. Combining optogenetic stimulation and motor training improves functional recovery and perilesional cortical activity. Neurorehabil. Neural Repair36, 107–118 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Ricci, P. et al. Power-effective scanning with AODs for 3D optogenetic applications. J. Biophoton.15, e202100256 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, J. Y., Knutsen, P. M., Muller, A., Kleinfeld, D. & Tsien, R. Y. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci.16, 1499–1508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zipfel, W. R., Williams, R. M. & Webb, W. W. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol.21, 1369–1377 (2003). [DOI] [PubMed] [Google Scholar]
- 40.de Vito, G. et al. Effects of excitation light polarization on fluorescence emission in two-photon light-sheet microscopy. Biomed. Opt. Express11, 4651–4665 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Vito, G. et al. Fast whole-brain imaging of seizures in zebrafish larvae by two-photon light-sheet microscopy. Biomed. Opt. Express13, 1516–1536 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen, I. W. et al. In vivo submillisecond two-photon optogenetics with temporally focused patterned light.J. Neurosci.39, 3484–3497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.deCarvalho, T. N. et al. Neurotransmitter map of the asymmetric dorsal habenular nuclei of zebrafish. Genesis52, 636–655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fore, S. et al. Functional properties of habenular neurons are determined by developmental stage and sequential neurogenesis. Sci. Adv.6, 10.1126/sciadv.aaz3173 (2020). [DOI] [PMC free article] [PubMed]
- 45.Bianco, I. H. & Wilson, S. W. The habenular nuclei: a conserved asymmetric relay station in the vertebrate brain. Philos. Trans. R. Soc. Lond. B Biol. Sci.364, 1005–1020 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreosti, E., Vendrell Llopis, N., Carl, M., Yaksi, E. & Wilson, S. W. Left-right asymmetry is required for the habenulae to respond to both visual and olfactory stimuli. Curr. Biol.24, 440–445 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma, M., Kler, S. & Pan, Y. A. Structural neural connectivity analysis in zebrafish with restricted anterograde transneuronal viral labeling and quantitative brain mapping. Front. Neural Circuits13, 85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buhler, A. & Carl, M. Zebrafish tools for deciphering habenular network-linked mental disorders. Biomolecules11, 10.3390/biom11020324 (2021). [DOI] [PMC free article] [PubMed]
- 49.Stephan, K. E. & Friston, K. J. Analyzing effective connectivity with functional magnetic resonance imaging. Wiley Interdiscip. Rev. Cogn. Sci.1, 446–459 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seth, A. K., Barrett, A. B. & Barnett, L. Granger causality analysis in neuroscience and neuroimaging. J. Neurosci.35, 3293–3297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reid, A. T. et al. Advancing functional connectivity research from association to causation. Nat. Neurosci.22, 1751–1760 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marrelec, G., Kim, J., Doyon, J. & Horwitz, B. Large-scale neural model validation of partial correlation analysis for effective connectivity investigation in functional MRI. Hum. Brain Mapp.30, 941–950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang, L. et al. Task learning promotes plasticity of interneuron connectivity maps in the olfactory bulb. J. Neurosci.36, 8856–8871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allegra Mascaro, A. L. et al. Combined rehabilitation promotes the recovery of structural and functional features of healthy neuronal networks after stroke. Cell Rep.28, 3474–3485.e3476 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Resta, F. et al. Large-scale all-optical dissection of motor cortex connectivity shows a segregated organization of mouse forelimb representations. Cell Rep.41, 111627 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ricci, P. et al. Removing striping artifacts in light-sheet fluorescence microscopy: a review. Prog. Biophys. Mol. Biol.168, 52–65 (2022). [DOI] [PubMed] [Google Scholar]
- 57.Zhang, B. B., Yao, Y. Y., Zhang, H. F., Kawakami, K. & Du, J. L. Left habenula mediates light-preference behavior in zebrafish via an asymmetrical visual pathway. Neuron93, 914–928.e914 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Helmbrecht, T. O., Dal Maschio, M., Donovan, J. C., Koutsouli, S. & Baier, H. Topography of a visuomotor transformation. Neuron100, 1429–1445.e1424 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Andalman, A. S. et al. Neuronal dynamics regulating brain and behavioral state transitions. Cell177, 970–985.e920 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antinucci, P. et al. A calibrated optogenetic toolbox of stable zebrafish opsin lines. Elife9, 10.7554/eLife.54937 (2020). [DOI] [PMC free article] [PubMed]
- 61.Lewis, P. R. A theoretical interpretation of spectral sensitivity curves at long wavelengths. J. Physiol.130, 45–52 (1955). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobs, G. H. The evolution of vertebrate color vision. Adv. Exp. Med. Biol.739, 156–172 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Dunn, T. W. et al. Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion. Elife5, e12741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turrini, L. et al. Optical mapping of neuronal activity during seizures in zebrafish. Sci. Rep.7, 3025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feierstein, C. E. et al. Dimensionality reduction reveals separate translation and rotation populations in the zebrafish hindbrain. Curr. Biol.33, 3911–3925.e3916 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shainer, I. et al. A single-cell resolution gene expression atlas of the larval zebrafish brain. Sci. Adv.9, eade9909 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaigneau, E. et al. Two-photon holographic stimulation of ReaChR. Front. Cell Neurosci.10, 234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ronzitti, E. et al. Submillisecond optogenetic control of neuronal firing with two-photon holographic photoactivation of chronos. J. Neurosci.37, 10679–10689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dana, H. et al. Sensitive red protein calcium indicators for imaging neural activity. Elife5, 10.7554/eLife.12727 (2016). [DOI] [PMC free article] [PubMed]
- 70.Namboodiri, V. M., Rodriguez-Romaguera, J. & Stuber, G. D. The habenula. Curr. Biol.26, R873–R877 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Amo, R. et al. The habenulo-raphe serotonergic circuit encodes an aversive expectation value essential for adaptive active avoidance of danger. Neuron84, 1034–1048 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Turner, K. J. et al. Afferent connectivity of the zebrafish Habenulae. Front. Neural Circuits10, 30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bianco, I. H., Carl, M., Russell, C., Clarke, J. D. & Wilson, S. W. Brain asymmetry is encoded at the level of axon terminal morphology. Neural Dev.3, 9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agetsuma, M. et al. The habenula is crucial for experience-dependent modification of fear responses in zebrafish. Nat. Neurosci.13, 1354–1356 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Amo, R. et al. Identification of the zebrafish ventral habenula as a homolog of the mammalian lateral habenula. J. Neurosci.30, 1566–1574 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, R. Y. & Aghajanian, G. K. Physiological evidence for habenula as major link between forebrain and midbrain raphe. Science197, 89–91 (1977). [DOI] [PubMed] [Google Scholar]
- 77.Chou, M. Y. et al. Social conflict resolution regulated by two dorsal habenular subregions in zebrafish. Science352, 87–90 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Ichijo, H., Nakamura, T., Kawaguchi, M. & Takeuchi, Y. An evolutionary hypothesis of binary opposition in functional incompatibility about habenular asymmetry in vertebrates. Front. Neurosci.10, 595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hendricks, M. & Jesuthasan, S. Asymmetric innervation of the habenula in zebrafish. J. Comp. Neurol.502, 611–619 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Turrini, L. et al. Fast multi-photon brain-wide volumetric imaging and photostimulation. Multiphoton Microscopy in the Biomedical Sciences XXIII,10.1117/12.2649824 (2023).
- 81.Hubert, A. et al. Random-access two-photon holographic optogenetic stimulation combined with brain-wide functional light-sheet imaging in larval zebrafish. Advances in Microscopic Imaging IV, 10.1117/12.2671030 (2023).
- 82.Turrini, L., Sorelli, M., de Vito, G., Vanzi, F. & Pavone, F. Dissecting zebrafish brain functional connectivity with two-photon all-optical electrophysiology. Optica Biophotonics Congress: Biomedical Optics10.1364/TRANSLATIONAL.2024.JS4A.39 (2024).
- 83.Pisoni, M. et al. Towards two-photon all-optical electrophysiology with acousto-optic scanning. Optica Biophotonics Congress: Biomedical Optics10.1364/BRAIN.2024.BS3C.6 (2024).
- 84.Faini, G. et al. Ultrafast light targeting for high-throughput precise control of neuronal networks. Nat. Commun.14, 1888 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Betzel, R. F. Organizing principles of whole-brain functional connectivity in zebrafish larvae. Netw. Neurosci.4, 234–256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chicchi, L. et al. Reconstruction scheme for excitatory and inhibitory dynamics with quenched disorder: application to zebrafish imaging. J. Comput. Neurosci.49, 159–174 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Vito, G. et al. Two-photon high-speed light-sheet volumetric imaging of brain activity during sleep in zebrafish larvae. Neural Imaging Sens.11226, 10.1117/12.2542285 (2020).
- 88.de Vito, G. et al. Two-photon light-sheet microscopy for high-speed whole-brain functional imaging of zebrafish physiology and pathology. Neurophotonics11360, 10.1117/12.2560341 (2020).
- 89.Reddy, G. D. & Saggau, P. Fast three-dimensional laser scanning scheme using acousto-optic deflectors. J. Biomed. Opt.10, 064038 (2005). [DOI] [PubMed] [Google Scholar]
- 90.Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bi, K. et al. Position of the prism in a dispersion-compensated acousto-optic deflector for multiphoton imaging. Appl. Opt.45, 8560–8565 (2006). [DOI] [PubMed] [Google Scholar]
- 92.Mullenbroich, M. C. et al. Bessel beam illumination reduces random and systematic errors in quantitative functional studies using light-sheet microscopy. Front. Cell Neurosci.12, 315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.ZFIN Feature: b4, https://zfin.org/ZDB-ALT-980203-365.
- 94.Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). 4th edn, (University of Oregon Press, 2000).
- 95.Turrini, L. et al. Multimodal characterization of seizures in zebrafish larvae. Biomedicines10, 951 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stih, V., Petrucco, L., Kist, A. M. & Portugues, R. Stytra: an open-source, integrated system for stimulation, tracking and closed-loop behavioral experiments. PLoS Comput. Biol.15, e1006699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Migault, G. et al. Whole-brain calcium imaging during physiological vestibular stimulation in larval zebrafish. Curr. Biol.28, 3723–3735.e3726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Johnsen, L. G., Skov, T., Houlberg, U. & Bro, R. An automated method for baseline correction, peak finding and peak grouping in chromatographic data. Analyst138, 3502–3511 (2013). [DOI] [PubMed] [Google Scholar]
- 99.Hellinger, E. Neue Begründung der Theorie quadratischer Formen von unendlichvielen Veränderlichen. J. Reine Angew. Mathematik, 10.1515/crll.1909.136.210 (1909).
- 100.Ostrovsky, A., Cachero, S. & Jefferis, G. Clonal analysis of olfaction in Drosophila: image registration. Cold Spring Harb. Protoc.2013, 347–349 (2013). [DOI] [PubMed] [Google Scholar]
- 101.Randlett, O. et al. Whole-brain activity mapping onto a zebrafish brain atlas. Nat. Methods12, 1039–1046 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kunst, M. et al. A cellular-resolution atlas of the larval zebrafish brain. Neuron103, 21–38.e25 (2019). [DOI] [PubMed] [Google Scholar]
- 103.Sorelli, M., Hutson, T. N., Iasemidis, L. & Bocchi, L. Linear and nonlinear directed connectivity analysis of the cardio-respiratory system in type 1 diabetes. Front Netw. Physiol.2, 840829 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Iatsenko, D., McClintock, P. V. E. & Stefanovska, A. Linear and synchrosqueezed time-frequency representations revisited: overview, standards of use, resolution, reconstruction, concentration, and algorithms. Digit. Signal Process42, 1–26 (2015). [Google Scholar]
- 105.Granger, C. W. J. Investigating causal relations by econometric models and cross-spectral methods. Econometrica37, 424–438 (1969). [Google Scholar]
- 106.Zeileis, A. & Hothorn, T. Diagnostic cheking in regression relationships. R N.2, 7–10 (2002). [Google Scholar]
- 107.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc.57, 289–300 (1995). [Google Scholar]
- 108.Han, X., Cramer, S. R. & Zhang, N. Deriving causal relationships in resting-state functional connectivity using SSFO-based optogenetic fMRI. J. Neural Eng.19, 10.1088/1741-2552/ac9d66 (2022). [DOI] [PMC free article] [PubMed]
- 109.Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data source underlying plots are provided as Supplementary Data 3. Raw data are available from the corresponding authors upon reasonable request.
Custom-written LabVIEW/Python/C++ codes are available from the corresponding authors upon reasonable request.