Abstract
Patients with germline pathogenic variants of BRCA1/2 genes have a particular predisposition to develop breast cancer. No clinical test has been developed to accurately and quantitatively evaluate their risk of developing breast cancer. We hypothesized that aberrant cell clonal expansion may be initiated in normal breast tissues without manifesting pathologic changes. To assess the prevalence of clonal expansion in the normal breast, we collected normal breast tissue from 24 breast cancer patients who had undergone surgical resection and 5 carriers of pathogenic BRCA1/2 variant who had undergone prophylactic mastectomy. Whole-exome sequencing (WES) was conducted in 97 specimens from 14 individuals, and TOP panel, a gene panel targeting 464 genes, was conducted in 321 specimens from 26 individuals, including 8 individuals with germline pathogenic variants of BRCA1/2 genes. Recurrent oncogenic mutations within PIK3CA, ARHGAP35, HRAS, and NF1 were identified in normal breast tissue at considerable variant allelic frequencies (VAF), suggesting clonal expansion. In addition, 937 normal breast tissues were evaluated using the Breast Cancer Panel (BCP) targeting 25 genes to determine the exact prevalence and distribution of clonal expansion. To assess the clonal expansion, we developed the clonality score, which is the mean value of clonal cell fractions for samples obtained from a given breast. The average clonality score in macroscopically normal breast tissue was 0.95 (0–2.46), with a significant difference between cases with and without a history of breast cancer of stage 2 or more advanced stage (p = 0.01). Additional WES on 42 samples with relatively large clone size (VAF > 3%) confirmed that these cell clones harbored multiple mutations (10.7 mutations/sample), and the number of existing mutations was consistent with the clone size (R = 0.50). The results suggest that clonal changes occur in normal breast tissue of women at high risk for breast cancer even before cancer is detected pathologically and/or radiologically, and the clonality score shows the potential to be a valid method of evaluating clonal expansion for cancer-risk assessment that provides appropriate preventive options for patients at high risk for breast cancer.
Subject terms: Cancer genomics, Cancer prevention, Breast cancer
Introduction
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer-related deaths in women worldwide1. Large cohort studies across various tumor types have identified somatic driver mutations in respective cancers, such as frequent aberrations within PIK3CA, TP53, MAP3K1, CDH1, AKT1, and RB1 for breast cancer2,3. Moreover, recent studies have identified that the expansion of cell clones carrying mutated genes is associated with aging and/or in response to environmental insults and chronic inflammation4 among phenotypically normal or non-cancer tissues such as the skin5, esophagus6,7, colon8,9, bronchus10, liver11, endometrium12, and bladder13. However, extensive analysis of normal breast tissues have not been reported, probably due to the structural complexity of the breast ducts for genomic analyses. Meanwhile, some studies have identified somatic genetic alterations in uninvolved mammary glands (histologically normal glandular tissue distant from the primary tumor site) of patients with breast cancer14–17.
Hereditary breast and ovarian cancer is a syndrome defined by an increased risk of developing breast and/or ovarian cancer that is most commonly caused by germline pathogenic variants of BRCA1/2 genes18. Approximately 5% of patients with breast cancer may carry germline BRCA1 or BRCA2 variants that are either or likely pathogenic19,20. For women at an increased risk for developing breast cancer, effective surveillance strategies and preventive measures, such as risk-reducing surgical options, are important21,22. Prophylactic mastectomy could reduce the risk for breast cancer by >90% among BRCA1/2 variant carriers23.
There are no quantitative assessments to evaluate how “normal” breast tissue may be prone to tumor formation. Since the presence of common cancer driver mutations in normal tissues suggests a strong link to cancer development, we sought to investigate the mutational profile of breast tissues, including those in BRCA1/2 variant carriers, and attempted to create a formula that may predict clonal expansion of precancerous cells.
Results
Patient characteristics
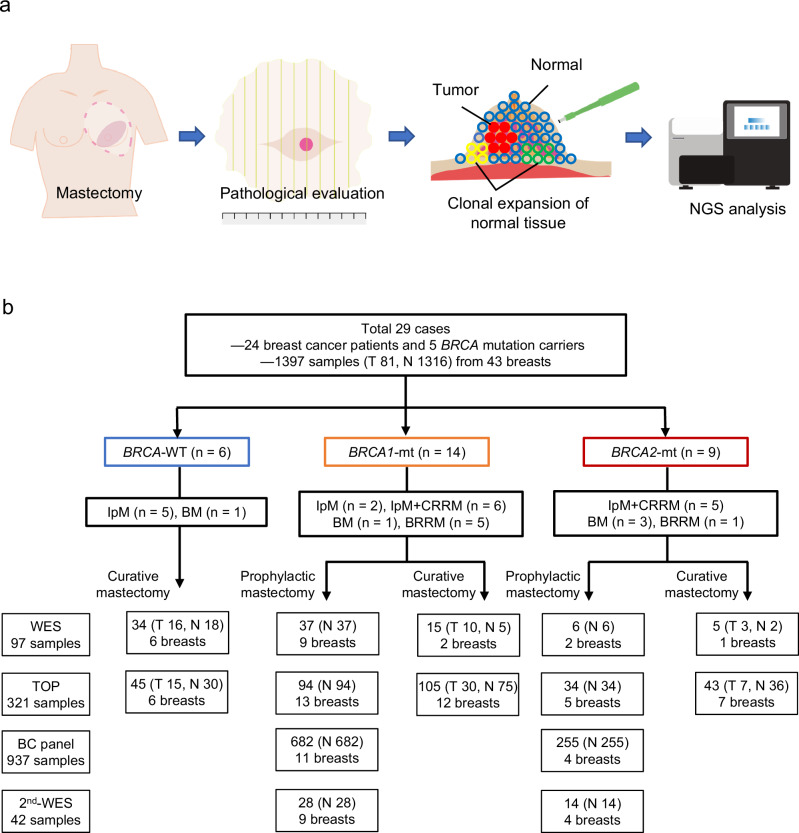
The study cohort comprised of 24 patients with breast cancer who underwent surgical resection and 5 carriers for pathogenic BRCA1/2 variant who underwent prophylactic mastectomy without history of cancer. The clinical data of the patients are summarized in Table 1. The cohort included 14 (48.3%) individuals with germline BRCA1 variants (vBRCA1), 9 (31.0%) with germline BRCA2 variants (vBRCA2), and 6 (20.7%) with wild-type (wt) BRCA1/2. The mean age of the patients in each subgroup was 43.1, 42.9, and 55.2, years old, respectively; individuals with BRCA1/2 variants were significantly younger than those with wt BRCA1/2 (43.0 vs. 55.2, p = 0.01, student’s t test). Treatment performed for the 23 individuals with BRCA1/2 mutations were the following: 6 with curative mastectomy (ipsilateral or bilateral), 11 with ipsilateral curative mastectomy with contralateral risk-reducing mastectomy (2 cases with ipsilateral curative mastectomy previously), and 6 with bilateral risk-reducing mastectomy (1 case with a prior curative partial mastectomy). Meanwhile, all 6 patients with wt BRCA1/2 underwent curative mastectomy. Macroscopical and microscopical assessment in all section was conducted to determine the precise pathological diagnosis. Case #03, #23, #29 had neoadjuvant chemotherapy (NAC) one month before surgery. The pathological staging for the 29 patients who underwent mastectomy in this study was as follows: 1 (3.4%) with stage 0, 11 (37.9%) with stage I, 7 (24.1%) with stage II, and 3 (10.3%) with stage III, 7 (24.1%) with no stage (no residual tumor after surgery or NAC, or no cancer observed in BRCA1/2 variant carriers). Case #12 was the only case which showed the local recurrence after the surgery in the cohort. All tissue samples were subjected to multisampling for whole-exome sequencing (WES) and targeted sequencing of cancer-associated genes to evaluate clonal expansion of normal breast tissues compared with breast cancer tissues (Fig. 1). The frozen whole section of breast was subjected for multiple sampling with Torepan biopsy (cat #BP-L30K, Kaijirushi, Japan). Individual samples were precisely annotated as tumor (T) or normal (N) according to the microscopic assessment of the section facing to the sampling section as well as macroscopic assessment by a medically trained clinical pathologist. A tumor sample is defined as a sample from the tumor lesion by microscopic or macroscopic assessment, or boundary region of macroscopic tumor with pathogenic somatic mutations.
Table 1.
Clinical information of 29 breast cancer cases
| Case# | Sex | Age | Diagnosis | Treatment | TNM staging | gBRCA status | ClinVar | |||
|---|---|---|---|---|---|---|---|---|---|---|
| T | N | M | Stage | |||||||
| #01 | F | 44 | Past Rt BC, R0 | BRRM | 0 | 0 | 0 | BRCA1 p.Q1281* | Pathogenic:3 | |
| #02 | F | 33 | No cancer history | BRRM | 0 | 0 | 0 | BRCA1 p.L63* | Pathogenic:3 | |
| #03 | F | 39 | Rt BC | NAC + Rt Bt + CRRM (Lt) | 2 | 0 | 0 | IIA | BRCA1 p.L63* | Pathogenic:3 |
| #04 | F | 45 | Lt BC | Lt Bt + CRRM (Rt) | 1c | 3 | 0 | IIIC | BRCA2 p.Ile1065fs | Pathogenic:3 |
| #05 | F | 69 | Lt BC | Lt Bt + CRRM (Rt) | 1c | 0 | 0 | IA | BRCA1 c.5278-1 G > A | Pathogenic/Likely_pathogenic:2 |
| #06 | F | 44 | Past Lt BC, R0 | CRRM (Rt) | 0 | 0 | 0 | BRCA2 p.Ser2835* | Pathogenic:3 | |
| #07 | F | 44 | Lt BC and Rt DCIS* | Lt Bt + CRRM (Rt) | 2 | 0 | 0 | IIA | BRCA1 p.Ser1241fs | Pathogenic:3 |
| #08 | F | 45 | Lt BC | Lt Bt | 3 | 1 | 0 | IIIA | BRCA WT | |
| #09 | M | 66 | Lt BC | Lt Bt | 2 | 0 | 0 | IIA | BRCA WT | |
| #10 | F | 36 | No cancer history | BRRM | 0 | 0 | 0 | BRCA1 p.L63* | Pathogenic:3 | |
| #11 | F | 47 | Rt BC and Lt DCIS | BM | 2 | 0 | 0 | IIA | BRCA WT | |
| #12 | F | 43 | Rt BC | Rt Bt | 1c | 1 mi | 0 | IB | BRCA WT | |
| #13 | F | 49 | Rt BC | Rt Bt | 3 | 1 | 0 | IIIA | BRCA WT | |
| #14 | F | 81 | Rt BC | Rt Bt | 1c | 0 | 0 | IA | BRCA WT | |
| #15 | F | 43 | Past Lt BC, R0 | BRRM | 0 | 0 | 0 | BRCA1 p.Ser1241fs | Pathogenic:3 | |
| #16 | F | 58 | Lt BC and Rt DCIS* | Lt Bt + CRRM (Rt) | 1c | 0 | 0 | IA | BRCA2 p.Tyr1710fs | Pathogenic:3 |
| #17 | F | 39 | Past Lt BC, R0 | CRRM (Rt) | 0 | 0 | 0 | BRCA2 p.Arg2318* | Pathogenic:3 | |
| #18 | F | 43 | Lt BC and Rt DCIS | BM | 1c | 0 | 0 | IA | BRCA2 p.Trp194* | Pathogenic:3 |
| #19 | F | 39 | Bil BC | BM | 2 | 0 | 0 | IIA | BRCA2 p.Asn43fs | Pathogenic:3 |
| #20 | F | 45 | Bil BC | BM | 1a | 0 | 0 | IA | BRCA1 p.Leu63* | Pathogenic:3 |
| #21 | F | 32 | No cancer history | BRRM | 0 | 0 | 0 | BRCA2 p.Arg2494* | Pathogenic:3 | |
| #22 | F | 39 | Lt DCIS | Lt Bt + CRRM (Rt) | is | 0 | 0 | 0 | BRCA2 p.Ala2864fs | Pathogenic:1 |
| #23 | F | 45 | Lt BC | NAC + Lt Bt | 1c | 0 | 0 | IA | BRCA1 c.5278-1 G > A | Pathogenic:2 |
| #24 | F | 46 | Rt BC | Rt Bt + CRRM (Lt) | 1b | 0 | 0 | IA | BRCA1 p.Gln1447fs | Pathogenic:3 |
| #25 | F | 40 | Rt BC and past Lt BC | Rt Bt | 1b | 0 | 0 | IA | BRCA1 p.Leu152fs | Pathogenic:3 |
| #26 | F | 39 | Rt BC | Rt Bt + CRRM (Lt) | 2 | 1 | 0 | IIB | BRCA1 c.4485-2 A > G | Pathogenic/Likely_pathogenic:2 |
| #27 | F | 47 | Rt BC | Rt Bt + CRRM (Lt) | 1b | 0 | 0 | IA | BRCA2 p.Asp1033fs | Pathogenic:3 |
| #28 | F | 40 | Past Lt BC, R0 | BRRM | 0 | 0 | 0 | BRCA1 p.Leu1216* | Pathogenic:3 | |
| #29 | F | 40 | Rt BC | NAC + Rt Bt + CRRM (Lt) | 2 | 0 | 0 | IIA | BRCA1 c.5278-1 G > A | Pathogenic:2 |
gBRCA status indicates the germline variants of BRCA1 and BRCA2. ClinVar column indicates the classification of the germline variant and score of review status. F female, M male, Lt, left, Rt right, BC breast cancer R0 no residual tumor, DCIS ductal carcinoma in situ, DCIS* ductal carcinoma in situ identified by pathological diagnosis after surgery, BRRM bilateral risk-reducing mastectomy, CRRM, contralateral risk-reducing mastectomy, BM bilateral mastectomy, Bt total mastectomy, NAC neoadjuvant chemotherapy, WT wild type.
Fig. 1. Study overview.
a Schematic diagram of the study. Resected breasts from prophylactic mastectomy of BRCA1/2 carriers and curative mastectomy of patients with breast cancer are subjected to pathological and genomic analysis. Multisampling from breast tissue samples was performed to evaluate the molecular profiling of the tumor and to identify and assess clonal expansion of normal tissue. b Study profile. A total of 24 patients with breast cancer and 5 with BRCA1/2 mutations participated in the study. IpM ipsilateral mastectomy, BM bilateral mastectomy, BRRM bilateral risk-reducing mastectomy, CRRM contralateral risk-reducing mastectomy.
Mutational profile of BRCA-mutated breast cancer with WES and cancer gene panel sequencing
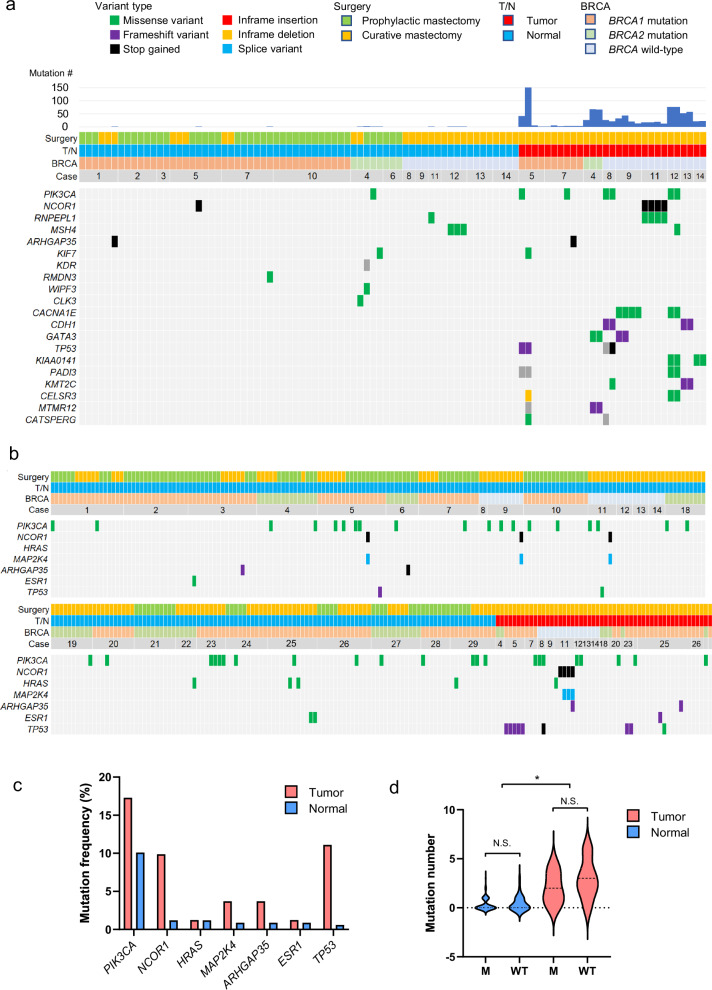
Firstly, the frozen whole section of 20 surgically resected breasts from 12 patients and 2 carriers (6 with vBRCA1, 2 with vBRCA2, and 6 with wt BRCA1/2) was subjected for sampling. A total of 97 specimens (29 tumor tissues and 68 normal tissues) were collected to perform WES (Fig. 2a, Supplementary Figs. 1–14, and Supplementary Data 1). WES of paired-matched blood samples was performed to call somatic mutations subtracting the germline variant. Somatic nonsynonymous mutations were recurrently identified across 14 cases in PIK3CA, NCOR1, ARHGAP35, KIF7, CACNA1E, CDH1, GATA3, TP53, KIAA0141, PADI3, KMT2C, CELSR3, MTMR12, and CATSPERG. Additionally, normal breast tissues carried mutations in PIK3CA, NCOR1, RNPEPL1, MSH4, ARHGAP35, KIF7, KDR, RMDN3, WIPF3, and CLK3. The average tumor mutation burden (TMB) was 0.85/Mbp for cancer specimens, while that was 4.8 × 10−3/Mbp for normal tissues, which were regardless of BRCA1/2 status. Of the 29 tumor samples, copy number alterations (CNA) were observed in 21 samples, whereas the 68 normal breast tissue samples did not show any CNA (Supplementary Figs. 15 and 16).
Fig. 2. Mutational profile of macroscopically normal breast tissues and breast cancer tissues.
a Frequently mutated genes by WES with color coding of their alteration status for each tumor. The case number, BRCA status, sample origin, types of surgery, and total mutation number are shown at the top. b Frequently mutated genes by TOP panel with color coding of their alteration status for each tumor. The case number, BRCA status, sample origin, and types of surgery are shown at the top. The blank case numbers are 19, 22, 24, 27, and 29 in order. c Oncogenic mutations frequently identified in macroscopically normal breast tissues and breast cancer tissues. d The mutation number was compared between tumor and normal samples, and between samples with BRCA1/2 mutations and wild-type BRCA. The dotted lines in the violin plot indicate the average. N.S. not significant, *p < 0.05, student’s t test.
To enhance the sensitivity to detect mutations with a variant allelic frequencies (VAF) of <1%, 321 specimens from 22 patients and 4 carriers were further sequenced at a higher coverage (843×) with a cancer gene panel test (TOP panel24), which evaluates somatic single nucleotide variations and insertions/deletions of 464 genes in the panel as well as TMB and CNA (Supplementary Figs. 1–14). Microscopical assessment was similarly conducted to distinguish T or N. Interestingly, we discovered that normal breast tissues of both the vBRCA1/2 and wt BRCA harbored frequent oncogenic mutations in PIK3CA, NCOR1, HRAS, MAP2K4, ARHGAP35, ESR1, and TP53, albeit at a lower VAF compared to matched tumor specimens (Figs. 2b, c, and Supplementary Data 2). According to the TOP panel analyses, the average TMB was 0.67/Mbp for tumors and 0.1/Mbp for normal tissues. However, the TMB was not different between the vBRCA1/2 and wt BRCA samples (Fig. 2d).
Breast cancer panel analyses for 25 breast cancer-related mutations
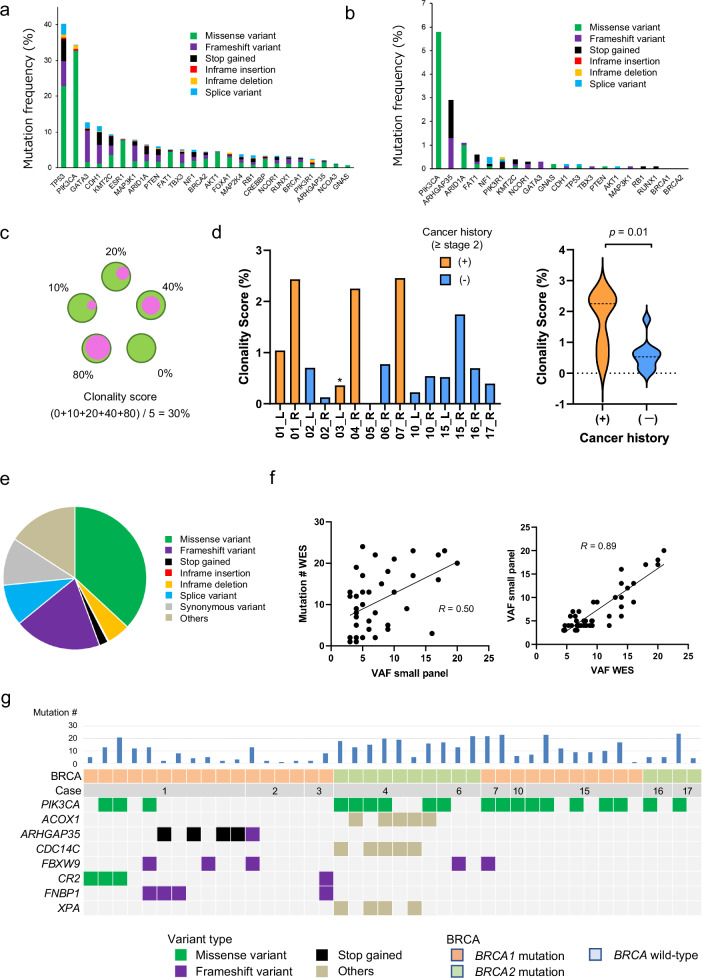
Since the VAF of mutations found in normal breast tissues was low, the number of cells consisting of one clone was expected to be small. To reliably investigate clonal expansion and their distribution in normal breast tissue, we constructed a custom-made breast cancer panel (BCP) to examine 25 genes frequently mutated in breast cancer and performed extra-deep sequencing (average coverage = 2865×) on 937 samples obtained from macroscopically apparent normal breast tissues from 11 patients. Figure 3a summarizes the mutation profile of the 25 genes in breast cancer from the GENIE dataset25. Our BCP analyses of normal tissues identified somatic PIK3CA mutations in 54 samples (5.8%) obtained from 10 (91%) patients, ARHGAP35 mutations in 27 samples (2.9%) from 9 (82%) patients, and ARID1A mutations in 10 samples (1.1%) from 2 (18%) patients. Notably, TP53 mutations, one of the most frequent mutations in breast cancer, were only identified in 2 samples (0.21%), and somatic mutations on BRCA1/2 genes were not observed in normal breast tissues (Fig. 3b and Supplementary Data 3).
Fig. 3. Mutational profile using BCP and its correlation with WES.
a The mutation frequency of the 25 breast cancer-related genes in the GENIE database. The mutation frequencies in the breast cancer cohort in GENIE database are indicated as bar graphs with the variant type categories. b The mutation frequency of the 25 breast cancer-related genes identified in macroscopically normal breast tissues. The mutation frequencies in macroscopically normal breast tissues of this study are indicated as bar graphs with the variant type categories. c The scheme depicts the concept of clonality score, which is defined as the average of the clonal cell fraction in the samples of individual breast. For instance, the clonality score of the breast in which the clonal cell fractions in five samples are 0, 10, 20, 40, and 80% is (0 + 10 + 20 + 40 + 80)/5 = 30%. d The clonality score for 15 individual breasts from 11 patients is shown as bar graphs on the left. *; this breast was resected one month after NAC. The clonality scores were compared between samples from breasts with and without a history of breast cancer, and revealed as violin plots on the right (average; 1.71 vs. 0.57, p = 0.01, student’s t test). e The distribution of the variant types of 451 mutations identified by WES in 42 macroscopically normal breast samples from 10 patients harboring mutations with VAF (>3%). f The correlation between the mutation number identified by WES and highest VAF values identified by BCP is shown on the left. The correlation between the highest VAF identified by WES and that by small panel is shown on the right. g Frequently mutated genes identified by WES in 42 macroscopically normal breast samples with color coding of their variant types. The case number, BRCA status, and total mutation number are shown at the top.
Clonality score to evaluate the extent of clonal expansion in individual breasts
To evaluate the clonal expansion of mutation-bearing cells within individual samples, we developed the clonality score, which is defined as the average of the clonal cell fraction in multiple samples of individual breast. If the clonal cell fraction in five samples are 0, 10, 20, 40, and 80%, the clonality score of the breast is (0 + 10 + 20 + 40 + 80)/5 = 30% (Fig. 3c). The average clonality score in macroscopically normal breast tissue was 0.95 (0–2.46), with a significant difference between cases with and without a history of breast cancer of stage 2 or more advanced stage (average; 1.71 vs. 0.57, p = 0.01, student’s t test) (Fig. 3d), but between breasts with vBRCA1 and vBRCA2 (data not shown). NAC might impact on the clonality score of case #03 as the score is low compared to other cases with past cancer history.
Additional WES of normal samples with somatic mutations
Overall, 42 macroscopically normal samples (4.6%) from 10 patients harbored somatic mutations with a relatively high VAF ( > 3%). Those specimens were further subjected to WES at a relatively high coverage (442×), leading to the detection of 451 mutations (10.7 mutations/sample) (Fig. 3e and Supplementary Data 4). The mutation number detected was concordant with the highest VAF values identified with the BCP (Fig. 3f). Additionally, the highest VAF values calculated from the WES dataset was also concordant with those measured with the BCP (Fig. 3f), suggesting that the clonality score calculated using the representative mutations detected by BCP may reliably evaluate the clonality expansion in the samples. Notably, WES detected recurrent mutations in PIK3CA, ARHGAP35, FBXW9, and FNBP1 (Fig. 3g)
Phylogenetic tree of clonal evolution
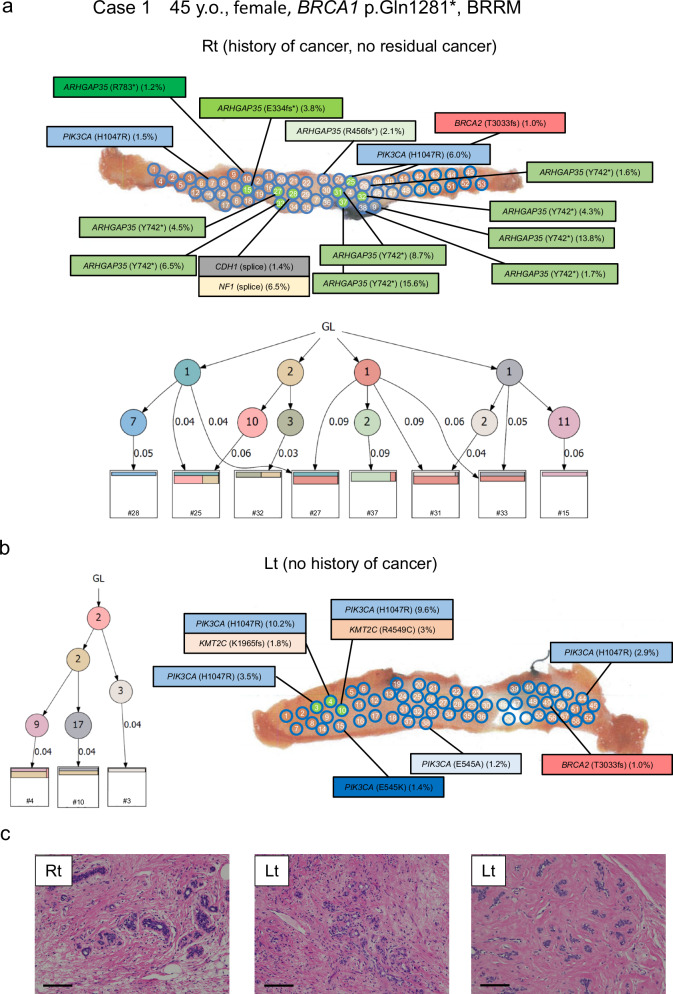
Case 1 is a 44-year-old woman with a germline BRCA1 p.Q1281* variant who underwent bilateral risk-reducing mastectomy. At 42 years old, she was diagnosed with right breast cancer and underwent partial mastectomy followed by adjuvant chemoradiotherapy. In this analysis, 53 samples obtained from the remaining right breast tissue were subjected to BCP examination. Surprisingly, 14 specimens revealed mutations in PIK3CA, ARHGAP35, CDH1, and NF1; the clonality score was 2.43. Likewise, 6 of 58 samples obtained from the left breast were analyzed with the BCP, detecting mutations in PIK3CA with a clonality score of 1.04 (Figs. 3c and 4a, b). Based on the WES data from 8 samples, a phylogenetic tree was constructed and revealed that some mutations are shared with multiple samples, suggesting spatially wide-range clonal expansion. Notably, there were no residual microscopic tumors in the right breast. Tissues from both breasts did not show pathological changes except for focal sclerosing adenosis (Fig. 4c).
Fig. 4. Mutational mapping and phylogenetic tree analysis following bilateral risk-reducing mastectomy.
a Case 1 is an analysis of bilateral risk-reducing mastectomy in a 45-year-old woman with BRCA1 p.Q1281*. She has a history of right breast cancer and underwent partial mastectomy followed by adjuvant chemoradiotherapy. In the right breast, 14 specimens from 53 samples had mutations in PIK3CA, ARHGAP35, CDH1, and NF1, and the clonality score was 2.42. Phylogenetic tree analysis of 8 samples (green highlighted) using WES data are shown. b Overall 6 of 58 samples in the left breast had mutations in PIK3CA, and the clonality score was 0.99. Phylogenetic tree analysis of 3 samples (green highlighted) using WES data are shown. c The representative microscopic images of breast tissue with hematoxylin and eosin (HE) staining. The left picture shows sclerosing adenosis. Magnification, ×100. Scale bars represent 200 μm. Lt left, Rt right, GL germline.
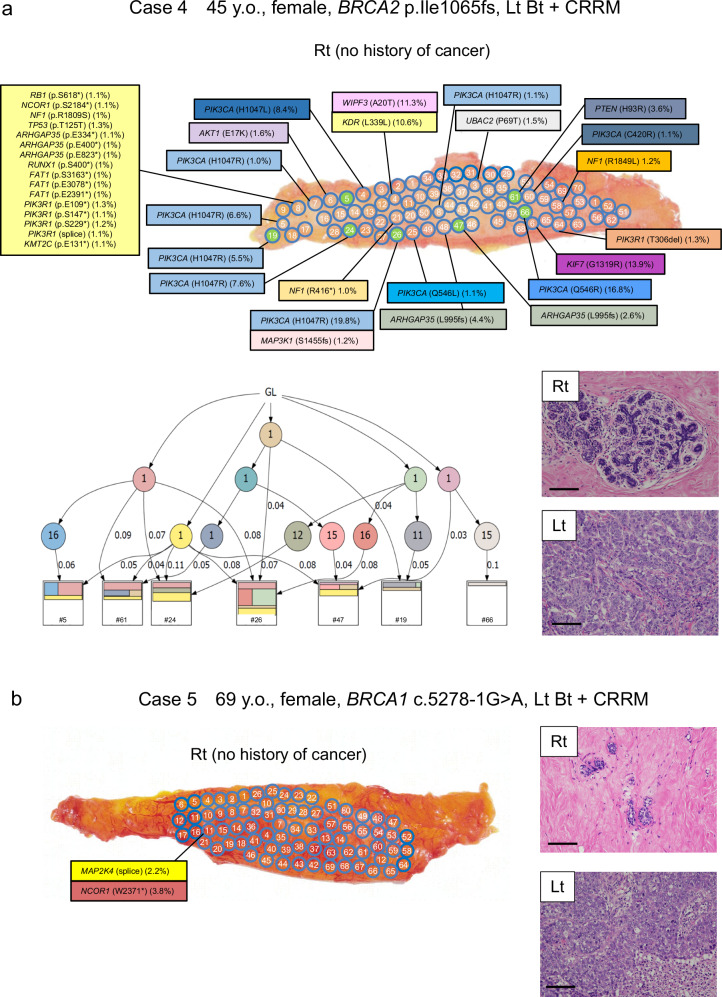
Both cases 4 and 5 were patients who had left breast cancer who had undergone total mastectomy of the left breast and had received risk-reducing mastectomy of the right breast. Case 4 was a 45-year-old woman with a germline BRCA2 p.I1065fs variant, and case 5 was a 69-year-old woman with a germline BRCA1 c.5278-1 G > A variant; both variants were reported as pathogenic. In case 4, although cancer was not pathologically identified in the right breast, 16 specimens from 70 right breast tissue samples had mutations in PIK3CA, ARHGAP35, and PIK3R1, with a clonality score of 2.25 (Figs. 3c and 5a). Similar to case 1, phylogenetic tree analysis for case 4 showed distant clonal expansion within normal breast tissues. Interestingly, sample #8 harbored multiple mutations, including an oncogenic TP53 mutation. In contrast, all 66 samples in the normal breast of case 5 had no mutations (Fig. 5b). Extensive hyalinization and ductal and lobular atrophy were observed in the right breast of case 5.
Fig. 5. Mutational mapping and phylogenetic tree analysis following contralateral risk-reducing mastectomy.
a Case 4 was a 45-year-old woman with BRCA2 p.I1065fs who had left breast cancer, underwent total mastectomy of left breast, and underwent risk-reducing mastectomy of the right breast. and case 5 was a 69-year-old woman with BRCA1 c.5278-1 G > A. Although no cancer was identified in the right breast, 14 specimens from 68 samples had mutations in PIK3CA, ARHGAP35, and PIK3R1, and the clonality score was 2.29. Phylogenetic tree analysis of 7 samples (green highlighted) using WES data are shown. The representative microscopic images of breast tissue with HE staining. No pathological finding was observed in the right breast, while the tumor in the left breast was invasive ductal carcinoma. Magnification, ×100. Scale bars represent 200 μm. b Case 5 was a 69-year-old woman with BRCA1 c.5278-1 G > A who had left breast cancer, underwent total mastectomy of left breast, and underwent risk-reducing mastectomy of the right breast. In contrast to case 4, all 68 samples from case 5 had no mutation in the 25 genes. The representative microscopic images of breast tissues with HE staining are shown in the right. Extensive hyalinization and ductal and lobular atrophy were observed in the right breast. The tumor in the left breast was invasive ductal carcinoma. Magnification, ×100. Scale bars represent 200 μm. Lt left, Rt right, GL germline.
Both cases 10 and 15 were patients who had no history of breast cancer that had undergone bilateral risk-reducing mastectomy. Case 10 was a 36-year-old woman with a germline BRCA1 p.L63* variant, and case 15 was a 43-year-old woman with a germline BRCA1 p.S1241fs variant. In both patients, pathological analysis of breast tissues following bilateral risk-reducing mastectomy showed no apparent cancer. In case 10, however, PIK3CA mutations were identified in 2 of 42 samples of the right breast, and ARHGAP35 mutations were identified in 2 of 56 samples of the left breast (Supplementary Fig. 17). In case 15, mutations in PIK3CA, PIK3R1, and FAT1 were identified in 5 of 70 samples of the right breast, and mutations in PIK3CA and PIK3R1 were identified in 8 of 73 samples of the left breast (Supplementary Fig. 17). The clonality scores of the right and left breasts were 0.54 and 0.22 for case 10, and 1.75 and 0.52 for case 15, respectively. The photographs of the breast sections of the other cases subjected to BCP (case 2, 3, 6, 7, 16, and 17) are shown in Supplementary Figs. 18–20.
Discussion
This is the first study to extensively analyze the mutational status of normal breast tissues, including specimens obtained following risk-reducing mastectomy, in women with no history of breast cancer, and to propose a method to potentially assess the breast cancer risk for individuals with germline pathogenic BRCA1/2 variants. Our findings suggest that clonal changes are present in normal breast tissues of high-risk women even before cancer is detected pathologically and/or radiologically. Our study supports a recent study in the literature that have shown similar results regarding genomic/genetic changes in normal or adjacent normal breast tissue from patients with cancer and/or at high risk26. Together with other studies that have identified somatic mutations in normal tissues, our study further corroborates the notion that cancer is a stepwise accumulation of genetic/epigenetic changes. Quantitatively measuring clonality may help evaluate the cancer risk and provide proper prophylactic options for high-risk patients. Considering that TP53 mutations and loss-of-heterozygosity in BRCA1/2 were hardly observed in normal breast tissues, disruption of the TP53 or BRCA pathway may trigger a cell clone to transform into a malignancy.
Importantly, seemingly normal breast tissues harbored several somatic mutations, which may be shared within the cells from the same branch of the breast duct. These alterations occurred in normal tissue without pathological hyperplasia or nuclear atypia. Considering that the quantity of DNA extracted from one sample was around 6 μg, which is roughly equivalent to 1.0 × 106 cells, expanding clones with a VAF of >3% (clone fraction >6%) may consist of as many as 6 × 104 cells. Since the sensitivity of clone analyses is severely affected by the quantity/quality of input DNA and the depth of sequencing, the size of the samples to be analyzed should be optimized.
The clonal score was developed as an index to survey clonal expansion in the breasts. For the clinical application of the clonality score, less invasive testing methods need to be considered. Fine needle aspiration from several sites is an option because it can be used routinely and in clinical trials for tumor molecular profiling. The method also has the advantages of being minimally invasive and can preserve tissue materials for diagnostic, prognostic, or therapeutic purposes27. Given the degree of heterogeneity in clonality scores within each patient, sampling from different sites is needed. Core needle biopsy from several sites may be needed to analyze more than 10 spots to obtain a valid score.
This method of cancer-risk assessment may be applied to patients at high risk for breast cancer who carry cancer-predisposing genes, including BRCA1/2, or those who have a history of breast cancer. However, regarding the additional burden that multiple biopsies places on patients, the potential benefit of a substantially increased biopsy schedule may not outweigh the risks. Therefore, liquid biopsy using cell-free DNA in plasma or urine may be considered for the potential next step to develop a less invasive test. It is challenging to evaluate cfDNA from premalignant region because there are few papers demonstrated the utility of liquid biopsy for the detection of early stage breast cancer so far. The assays need to be very sensitive as reliably detecting mutations in as low as 1% of breast epithelial cells is required. To maximize its sensitivity, it may be better to focus on several frequently mutated genes, such as PIK3CA and ARHGAP35, than conducting comprehensive gene panel testing. Furthermore, not only mutational analysis but multimodal assays such as methylomics, fragmentomics, copy number, and end motifs analysis may contribute to develop a highly sensitive assay28–30.
ARHGAP35 encodes Rho GTPase-activating proteins, including p190A, and is frequently mutated in up to 15% of endometrial cancers and 2% of all tumors in a studied cohort31,32. ARHGAP35 is essential for mammary gland branching morphogenesis and mammary epithelial cell differentiation33. Although ARHGAP35 mutations are not common in breast cancer, this study showed an unexpectedly high frequency of ARHGAP35 mutations in normal breast tissues. It is known that some driver mutations are unique to normal tissues or have a mutation frequency that is much higher in normal tissue than in cancer tissue, indicating that the respective clones may not necessarily be destined for evolution to cancer but even negatively selected for carcinogenesis depending on the mutated gene4. Regardless, ARHGAP35 mutations may promote clonal expansion by preventing terminal differentiation, which is required in recycling the epithelium.
There are some limitations of the study. First is the small sample size of the cohort. Especially, this study did not investigate the mammary tissue from people with wild-type BRCA and no cancer history. Therefore, the prevalence of clonal expansion in low-risk people is unknown, which make it difficult to evaluate the performance of the clonality score. Second, it remains to be identified how the clonality score might change overtime and how often a patient would need to be assessed, as a single timepoint cannot not rule out increased risk in the future. Therefore, prospective studies of independent cohorts should be conducted to validate this proof of concept and to determine how we could apply this assessment in clinical practice. Third, the mean age of patients with BRCA mutations and those with wild-type BRCA is different, which may be a confounding factor in the comparative evaluation of the two groups. Forth is the poor resolution of CNA analysis such as detection of loss of heterozygosity in BRCA, which is known to arise in the process of breast cancer initiation in BRCA1/2 mutation carriers. However, no apparent CNA was observed in macroscopically normal breast tissues. It is because the sensitivity of CNA analysis is not as high as that of SNV analysis, and probably because the clonal expansion of the cells with CNA happens in the late phase of tumorigenesis. Single cell analysis is needed to elucidate the prevalence of CNA in normal breast tissue.
In conclusion, our discoveries may have the potential for application in a clinical setting. The clonality score may directly predict the risk of tumor occurrence in macroscopically normal breasts. Our genomic analyses highlight the importance of precise premalignant tissue profiling to provide proper prophylactic options for patients at high risk for breast cancer.
Methods
Study design and patient specimens
The study cohort consisted of 29 patients with breast cancer who underwent surgical resection between October 2005 and September 2017 at hospitals in St. Luke’s International Hospital. A board-certified pathologist (NK) specializing in breast cancer reviewed the histological features based on the World Health Organization classification. Fresh frozen specimens or formalin-fixed paraffin-embedded (FFPE) surgically resected tumors were obtained from all patients. This study was conducted in accordance with the Declaration of Helsinki and the protocol was approved by the Ethics Committee of St. Luke’s International Hospital (18-R106) and National Cancer Center (No. 2019-271). All subjects provided written informed consent, except for those who could not be reached because of loss of follow-up or death at registration. In these cases, the Institutional Review Board at each participating institution granted permission for the existing tissue samples to be used for research purposes. No samples from the patients who had opted out of participation were used in this study.
Sample preparation
The frozen whole section of breast was subjected for sampling with Torepan biopsy (cat #BP-L30K, Kaijirushi, Japan). The approximate size of the specimen was 21 mm3 (column diameter = 3 mm, height = 3 mm). Genomic DNA was extracted using QIAamp Fast DNA Tissue Kit for fresh frozen and GeneRead DNA FFPE Kit for FFPE (Qiagen, Hilden, Germany).
WES, including mutation calls and copy number analysis
A total of 500 ng from fresh frozen samples were subjected to target fragment enrichment using Twist Library Preparation EF Kit (Twist Bioscience, South San Francisco, CA, USA). Massively parallel sequencing of the isolated fragments was performed using the paired-end option on a NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Paired-end WES reads that had nucleotides masked with a quality score <20 were aligned to the human reference genome (hg38) using BWA-MEM. Somatic mutations were called using Genome Analysis Toolkit (https://gatk.broadinstitute.org/hc/en-us) MuTect2, VarScan2 (http://varscan.sourceforge.net) and our in-house somatic caller. The concept of our caller is as follows: in general, it is expected that the distribution of the difference in VAFs between tumor and normal tissues follows a normal distribution. For each position , a difference of VAFs between tumor and normal is defined by , where Σ is the set of nucleotides . Let and be the mean and the standard deviation of the set for the estimated position . Assume that . Then the somatic mutation for is defined by , and , where . We used K = 75, which is half the read length. Mutations were discarded if any of the following criteria were met: read depth was <20, variant allele frequency was <0.05, mutation occurred in only one strand of the genome, mutant read number in the germline control samples was >2, or the variant was present in normal human genomes in either the 1000 Genomes Project dataset (https://www.internationalgenome.org/) or our in-house database. Gene mutations were annotated by SnpEff (http://snpeff.sourceforge.net). Copy number status was analyzed using our in-house pipeline, which determines the logR ratio () as follows: (i) we selected SNP positions in the 1000 Genomes Project database that were homozygous (VAF, ≤0.05 or ≥0.95) or heterozygous (VAF, 0.4–0.6) in the genomes of the white blood cells of the individual cases; (ii) normal and tumor read depths at the selected position were adjusted based on the GC percentage of a 100-bp window flanking the position; (iii) we calculated the , where and are the normal and tumor-adjusted depths at position , respectively; and (iv) each representative LRR was determined by the median of a moving window (4 Mb) centered at position . The values of the copy number of both alleles, major allele, or minor allele were determined for every region of the genome.
DNA sequencing with the TOP cancer gene panel and BCP
Fresh frozen or FFPE specimens were analyzed with TOP panel version 3 or BCP, both of which were hybridization-based assay. Top panel evaluates nucleotide variants and insertions/deletions for 464 genes to calculate the TMB and to infer the copy number variation24. BCP evaluates nucleotide variants and insertions/deletions for 25 genes. A total of 500 ng from fresh frozen samples or FFPE were subjected to target fragment enrichment using Twist Library Preparation EF Kit (Twist Bioscience). Massively parallel sequencing of isolated fragments was conducted with a NovaSeq 6000 (Illumina) using a paired-end option. The analysis algorithms were similar to WES for both panels; however, the mutation detection criteria were different. TOP and BCP discarded mutations that met any of the following criteria: read depth was <100 or VAF < 0.01 (for TOP), and <500 or <0.01 (for BCP), mutation occurred in only one strand of the genome, mutant read number in the germline control samples was >2 for TOP, non-pathogenic mutations shared by across different cases, or the variant was present in normal human genomes in either the 1000 Genomes Project dataset or our in-house database. MicroSEC34 was used to eliminate false-positive mutations related to FFPE derived DNA. The correctness of detecting mutations are confirmed by manually checking of each igv image.
Clonality score
Clonality score is defined as the average of the clonal cell fraction in multiple samples of individual breast. If the clonal cell fraction in five samples are 0, 10, 20, 40, and 80%, the clonality score of the breast is (0 + 10 + 20 + 40 + 80)/5 = 30%. The clonal cell fraction in individual sample is calculated as 2× (highest VAF of somatic mutation in the sample), assuming that the somatic mutation is heterozygous in a copy number-neutral allele.
Supplementary information
Acknowledgements
The authors would like to thank A. Maruyama-Shiino for technical assistance. This study was supported by the grants from the Practical Research for Innovative Cancer Control (grant no. JP22kk0305018), Program for Promoting Platform of Genomics based Drug Discovery (grant no. JP23kk0305018), and Moonshot Research and Development Program (grant no. JP22zf0127009) from the Japan Agency for Medical Research and Development (AMED). This work was also supported by the JSPS Grants-in-Aid for Scientific Research (C) (grant no. 21K12117).
Author contributions
M.I., H.M., and S.K. conceived the project and designed the study. M.I. and S.K. developed the methodology. T.H. and M.I. performed the experiments. T.H., T.U. and S.K. analyzed and interpreted the data. K.K., L.W., S.T., N.K., and H.Y. provided administrative, technical, or material support. T.H., M.I., K.K., H.Y., H.M., and S.K. wrote and edited the manuscript with feedback from all authors.
Data availability
We deposited the raw sequencing data under accession number JGAS000368 in the Japanese Genotype-Phenotype Archive, which is hosted by the DNA Data Bank of Japan.
Code availability
In-house pipelines for somatic calling and copy number analysis are described in Materials and Methods. The references of all source codes are included within Materials and Methods.
Competing interests
The authors declare no competing interests.
Consent for publication
All authors have approved the manuscript and agree with the submission.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41523-024-00693-9.
References
- 1.Heer, E. et al. Global burden and trends in premenopausal and postmenopausal breast cancer: a population-based study. Lancet Glob. Health8, e1027–e1037 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature490, 61–70 (2012). [DOI] [PMC free article] [PubMed]
- 3.Ciriello, G. et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell163, 506–519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kakiuchi, N. & Ogawa, S. Clonal expansion in non-cancer tissues. Nat. Rev. Cancer21, 239–256 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Martincorena, I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science348, 880–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama, A. et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature565, 312–317 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Martincorena, I. et al. Somatic mutant clones colonize the human esophagus with age. Science362, 911–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kakiuchi, N. et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature577, 260–265 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Nanki, K. et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature577, 254–259 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Yoshida, K. et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature578, 266–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunner, S. F. et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature574, 538–542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore, L. et al. The mutational landscape of normal human endometrial epithelium. Nature580, 640–646 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Lawson, A. R. J. et al. Extensive heterogeneity in somatic mutation and selection in the human bladder. Science370, 75–82 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Kostecka, A. et al. High prevalence of somatic PIK3CA and TP53 pathogenic variants in the normal mammary gland tissue of sporadic breast cancer patients revealed by duplex sequencing. NPJ Breast Cancer8, 76 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronowicz, A. et al. Concurrent DNA copy-number alterations and mutations in genes related to maintenance of genome stability in uninvolved mammary glandular tissue from breast cancer patients. Hum. Mutat.36, 1088–1099 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Forsberg, L. A. et al. Signatures of post-zygotic structural genetic aberrations in the cells of histologically normal breast tissue that can predispose to sporadic breast cancer. Genome Res.25, 1521–1535 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danforth, D. N. Jr Genomic changes in normal breast tissue in women at normal risk or at high risk for breast cancer. Breast Cancer (Auckl.)10, 109–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samadder, N. J., Giridhar, K. V., Baffy, N., Riegert-Johnson, D. & Couch, F. J. Hereditary cancer syndromes-a primer on diagnosis and management: part 1: breast-ovarian cancer syndromes. Mayo Clin. Proc.94, 1084–1098 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Breast Cancer Association, C. et al. Breast cancer risk genes - association analysis in more than 113,000 women. N. Engl. J. Med.384, 428–439 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, C. et al. A population-based study of genes previously implicated in breast cancer. N. Engl. J. Med.384, 440–451 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campeau, P. M., Foulkes, W. D. & Tischkowitz, M. D. Hereditary breast cancer: new genetic developments, new therapeutic avenues. Hum. Genet124, 31–42 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Pashayan, N. et al. Personalized early detection and prevention of breast cancer: ENVISION consensus statement. Nat. Rev. Clin. Oncol.17, 687–705 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Sprundel, T. C. et al. Risk reduction of contralateral breast cancer and survival after contralateral prophylactic mastectomy in BRCA1 or BRCA2 mutation carriers. Br. J. Cancer93, 287–292 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohsaka, S. et al. Comprehensive assay for the molecular profiling of cancer by target enrichment from formalin-fixed paraffin-embedded specimens. Cancer Sci.110, 1464–1479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium, A. P. G. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov.7, 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishimura, T. et al. Evolutionary histories of breast cancer and related clones. Nature620, 607–614 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupain, C. et al. Fine-needle aspiration as an alternative to core needle biopsy for tumour molecular profiling in precision oncology: prospective comparative study of next-generation sequencing in cancer patients included in the SHIVA02 trial. Mol. Oncol.15, 104–115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanley, K. E. et al. Cell type signatures in cell-free DNA fragmentation profiles reveal disease biology. Nat. Commun.15, 2220 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, J. et al. Genome-wide cell-free DNA methylation analyses improve accuracy of non-invasive diagnostic imaging for early-stage breast cancer. Mol. Cancer20, 36 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen, V. T. C. et al. Multimodal analysis of methylomics and fragmentomics in plasma cell-free DNA for multi-cancer early detection and localization. Elife12, RP89083 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature502, 333–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heckman-Stoddard, B. M. et al. P190A RhoGAP is required for mammary gland development. Dev. Biol.360, 1–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikegami, M. et al. MicroSEC filters sequence errors for formalin-fixed and paraffin-embedded samples. Commun. Biol.4, 1396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We deposited the raw sequencing data under accession number JGAS000368 in the Japanese Genotype-Phenotype Archive, which is hosted by the DNA Data Bank of Japan.
In-house pipelines for somatic calling and copy number analysis are described in Materials and Methods. The references of all source codes are included within Materials and Methods.