Abstract
Lung cancer remains the leading cause of cancer mortality, despite declining smoking rates. Previous lung cancer GWAS have identified numerous loci, but separating the genetic risks of lung cancer and smoking behavioral susceptibility remains challenging. Here, we perform multi-ancestry GWAS meta-analyses of lung cancer using the Million Veteran Program cohort (approximately 95% male cases) and a previous study of European-ancestry individuals, jointly comprising 42,102 cases and 181,270 controls, followed by replication in an independent cohort of 19,404 cases and 17,378 controls. We then carry out conditional meta-analyses on cigarettes per day and identify two novel, replicated loci, including the 19p13.11 pleiotropic cancer locus in squamous cell lung carcinoma. Overall, we report twelve novel risk loci for overall lung cancer, lung adenocarcinoma, and squamous cell lung carcinoma, nine of which are externally replicated. Finally, we perform PheWAS on polygenic risk scores for lung cancer, with and without conditioning on smoking. The unconditioned lung cancer polygenic risk score is associated with smoking status in controls, illustrating a reduced predictive utility in non-smokers. Additionally, our polygenic risk score demonstrates smoking-independent pleiotropy of lung cancer risk across neoplasms and metabolic traits.
Subject terms: Genome-wide association studies, Lung cancer, Cancer genetics
Lung cancer is the leading cause of cancer mortality, despite declining smoking rates. Gorman et al. report multi-ancestry GWAS meta-analyses of lung cancer providing insights into smoking-independent genetic predisposition to the disease.
Introduction
Lung cancer remains the leading cause of overall cancer mortality, as the most prevalent cancer type in men, and the second highest in women after breast cancer1–3. Despite declines in smoking rates in the US since the 1980s4, tobacco use is currently implicated in upwards of 80% of lung cancer diagnoses1. Even in those who have never smoked, nor had meaningful exposure to environmental carcinogens1,5, there exists a heritable risk component of lung cancer conferred by genetic factors6–8. Differentiating the mutations that directly predispose an individual to lung cancer from those whose effect is mediated through environmental components remains challenging.
Genome-wide association studies (GWAS) have identified lung cancer risk variants associated with oncogenic processes such as immune response7, cell cycle regulation9, and those affecting DNA damage response and genomic stability8. Several lung cancer GWAS have also reported strong effects of genes such as CHRNA nicotine receptor genes which putatively increase the risk of lung cancer through behavioral predisposition towards smoking5. Characteristic molecular markers and genetic risk factors in smokers and never-smokers have been identified10,11, though fewer variants have been found in GWAS performed exclusively in never-smokers12.
Lung cancer has a heterogeneous genetic architecture across ancestral groups13,14. In the two most well-studied ancestries, European (EA) and East Asian (EAS), the majority of genome-wide significant loci are not shared15,16; this is in agreement with molecular studies showing differences in tumor characteristics between EA and EAS17. Smaller African ancestry (AA) cohorts have replicated known loci from EA or EAS8,18, though no AA-specific GWAS loci have been reported.
In this study, we examined lung cancer genetic variation in EA as well as in the largest AA cohort to date. Our discovery analysis is performed in an older cohort of mostly male US veterans in the Department of Veterans Affairs Million Veteran Program (MVP)19. Lung cancer incidence is approximately twice as high in men than in women2, and additionally, MVP contains a large number of cigarette smokers, positioning this biobank as particularly valuable for these analyses. We performed GWAS in overall cases of lung cancer as well as two non-small cell lung cancer (NSCLC) subtypes, adenocarcinoma (LUAD) and squamous cell lung carcinoma (LUSC).
Results
Genome-wide association studies for lung cancer
We performed a GWAS on overall lung cancer within EA participants in MVP (10,398 lung cancer cases and 62,708 controls; Supplementary Data 1), followed by a meta-analysis with the EA International Lung Cancer Consortium OncoArray study (ILCCO)7, for a total of 39,781 cases and 119,158 controls (Supplementary Fig. 1). The EA meta-analysis for overall lung cancer identified 26 conditionally independent SNPs within 17 genome-wide significant loci (P < 5 × 10−8; Supplementary Fig. 2a; Supplementary Data 2). All 12 loci reported by ILCCO7 were confirmed, with consistent direction of effect in all single nucleotide polymorphisms (SNPs) with P < 1 × 10−5, as well as high correlation of effect sizes and allele frequency (Supplementary Fig. 3). Of the 17 genome-wide significant loci for overall lung cancer, four were novel with respect to the broader literature: neuronal growth regulator LSAMP, Wnt signaling regulator NMUR2, DNA damage repair protein XCL2, and hedgehog signaling regulator TULP3, (Table 1; Supplementary Fig. 4a–d).
Table 1.
Novel GWAS loci
| Lung cancer subtype | rsID | Chromosome: Position (hg19) |
Candidate gene | EA/ NEA |
EAF | Discovery P | Replication P | Combined meta-analysis OR (95% CI) | Combined meta-analysis P |
|---|---|---|---|---|---|---|---|---|---|
| Novel loci from the European ancestry GWAS meta-analysis | |||||||||
| Overall | rs77045810a | 1:168,505,017 | XCL2 | A/C | 0.89 | 1.43 × 10−10 | 5.72 × 10−3 | 1.09 (1.07, 1.12) | 3.94 × 10−12 |
| Overall | rs144840030 | 3:117,147,326 | LSAMP | T/G | 0.01 | 1.09 × 10−8 | 0.487 | 1.26 (1.16, 1.37) | 5.01 × 10−8 |
| Overall | rs62400619 | 5:152,343,053 | NMUR2 | T/C | 0.68 | 6.33 × 10−9 | 0.156 | 1.05 (1.03, 1.07) | 1.10 × 10−8 |
| Overall | rs9988980a | 12:3,038,917 | TULP3 | T/C | 0.39 | 5.34 × 10−8 | 2.16 × 10−3 | 1.05 (1.04, 1.07) | 3.72 × 10−10 |
| LUAD | rs67824503a | 8:129,535,264 | MYC | T/C | 0.75 | 1.81 × 10−8 | 5.05 × 10−5 | 1.10 (1.07, 1.14) | 4.09 × 10−12 |
| LUAD | rs11855650a | 15:70,431,773 | TLE3 | T/G | 0.38 | 1.12 × 10−8 | 1.22 × 10−7 | 1.10 (1.07, 1.13) | 1.15 × 10−14 |
| LUSC | rs36229791a | 10:101,991,135 | BLOC1S2 | A/T | 0.04 | 4.04 × 10−8 | 1.49 × 10−4 | 1.26 (1.18, 1.35) | 2.48 × 10−11 |
| Novel loci from the African ancestry GWAS | |||||||||
| Overall | rs78994068 | 12:127,225,803 | LINC00944 | C/A | 0.01 | 1.87 × 10−9 | 0.901 | 1.76 (1.42, 2.17) | 1.81 × 10−7 |
| Novel loci from the multi-ancestry meta-analysis (not genome-wide significant in the European meta-analysis) | |||||||||
| Overall | rs329122a | 5:133,864,599 | JADE2 | A/G | 0.43 | 1.12 × 10−8 | 0.0529 | 0.96 (0.94, 0.97) | 3.69 × 10−9 |
| Overall | rs7300571a | 12:47,857,826 | RPAP3 | T/C | 0.11 | 3.47 × 10−8 | 4.44 × 10−3 | 1.08 (1.06, 1.11) | 6.48 × 10−10 |
| Novel loci after conditioning the European ancestry GWAS meta-analysis on cigarettes per day | |||||||||
| Overall | rs1124241a | 6:97,722,453 | MMS22L | A/G | 0.22 | 1.26 × 10−8 | 6.15 × 10−3 | 1.07 (1.05, 1.10) | 3.39 × 10−10 |
| LUSC | rs61494113a | 19:17,401,859 | ABHD8 | A/G | 0.29 | 4.90 × 10−8 | 3.12 × 10−3 | 1.11 (1.08, 1.15) | 6.39 × 10−10 |
Novel genome-wide significant GWAS index variants for lung cancer risk in European-ancestry meta-analyses from MVP and ILCCO7 cohorts, MVP African ancestry, multi-ancestry meta-analyses, and in European-ancestry meta-analyses after conditioning on cigarettes per day. Replication of these novel variants was conducted in the external OncoArray dataset. Finally, we performed a combined meta-analysis of our discovery results with the OncoArray replication results. Discovery P-values were assessed for genome-wide significance (P < 5 × 10−8), replication using a nominal significance threshold (P < 0.05), and combined meta-analysis using the conservative threshold of P < 4.17 × 10−9 (5 × 10−8/12 total GWAS analyses). Significant alleles meeting this threshold after replication are indicated witha. All meta-analyses in this table refer to fixed-effect inverse variance-weighted meta-analysis.
LUAD lung adenocarcinoma, LUSC squamous cell lung carcinoma, EA effect allele, NEA non-effect allele, EAF effect allele frequency in the given population, OR (95% CI) odds ratio and 95% confidence interval.
Further association tests stratified by cancer subtypes LUAD and LUSC in MVP EA (Supplementary Fig. 2bc; Supplementary Data 3, 4) replicated associations reported by ILCCO7 (Supplementary Fig. 3) and identified additional loci. Two novel EA meta-analysis loci were identified for LUAD, proto-oncogene MYC, and Wnt signaling inhibitor TLE3 (Table 1; Supplementary Fig. 4e–h). For LUSC, we identified one novel locus at 10q24.31 near NFκB inhibitor CHUK and BLOC1S2. Across all subtypes for EA meta-analysis index variants, the MVP cohort had associations with P < 0.05 in all but one in overall lung cancer, five in LUAD, including approximately nominal significance at rs67824503 (MYC; P = 0.057), and one in LUSC (Supplementary Data 2–4).
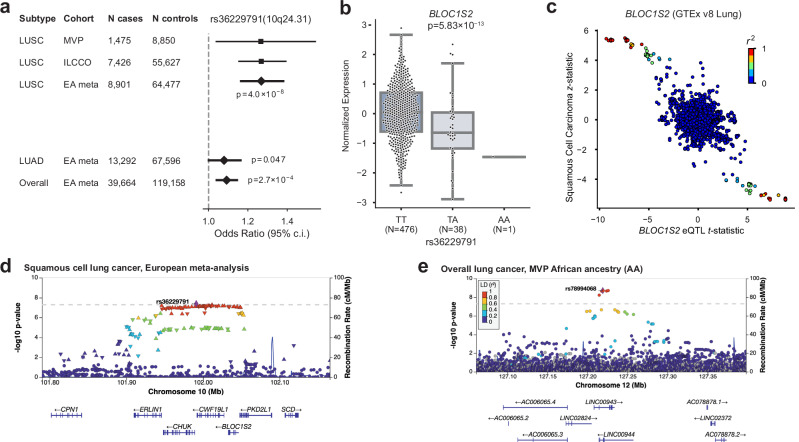
We investigated expression quantitative trait loci (eQTL) relationships between top SNPs from the EA meta-analysis across all lung cancer GWAS in GTEx v8 Lung20 and the Lung eQTL Consortium21 (Supplementary Data 2–4). This analysis showed that the LUSC index SNP rs36229791 on 10q24.31 was associated with the mRNA expression levels of BLOC1S2 (Fig. 1a–d), consistent with previous TWAS22. BLOC1S2 is an oncogene whose gene product is associated with centrosome function; centrosomal abnormalities have previously been observed in vitro in LUSC23,24.
Fig. 1. Highlighted novel GWAS loci.
a–d GWAS of squamous cell lung carcinoma (LUSC) in European ancestry (EA) meta-analysis identifies a novel locus at 10q24.31. a Odds ratios and 95% confidence intervals for rs36229791 in LUSC compared to lung adenocarcinoma (LUAD) and overall lung cancer. Inset p-values were obtained from a fixed-effect inverse variance-weighted meta-analysis. b BLOC1S2 expression varies by genotype at rs36229791 in GTEx v8 Lung tissue samples. t-test was utilized and a two-sided p-value is shown. The center box plot line represents the median (50th percentile). The box edges are the 25th and 75th percentiles (interquartile range; IQR). Whiskers extend to the smallest and largest values within 1.5 times the IQR. Source data are provided as a Source Data file. c BLOC1S2 eQTL t-statistic vs LUSC z-statistic. d Regional association plot showing SNP significance and genes around lead SNP rs36229791. Association p-values were obtained from a fixed-effect inverse variance-weighted meta-analysis. The dashed line marks the threshold for genome-wide significance (P = 5 × 10–8). Source data are provided as a Source Data file. e The African ancestry GWAS highlights a putatively novel locus on chr12 at LINC00944. The risk allele has effectively 0% frequency in EA. Association p-values were obtained from a two-sided test of the z-statistic, calculated using logistic regression. The dashed line marks the threshold for genome-wide significance (P = 5 × 10–8).
We improved our variant selection by fine-mapping and estimating credible sets of candidate causal variants in the EA meta-analysis using sum of single effects (SuSiE)25,26 modeling. For overall lung cancer, LUAD, and LUSC, we identified 23, 23, and 9 high-quality credible sets, respectively, containing 370, 246, and 192 total SNPs (Supplementary Data 5).
GWAS in AA
We analyzed overall lung cancer risk in 2438 cases and 62,112 controls of African ancestry (AA), the largest AA GWAS discovery cohort to date (Supplementary Fig. 5a). Two loci reached genome-wide significance in our discovery scan: 15q25, replicating the association in CHRNA5 for AA populations reported by an earlier GWAS18, and a putative novel locus at 12q23 with index SNP rs78994068 (Table 1; Fig. 1e). We further performed GWAS in AA within LUAD and LUSC subtypes but found no genome-wide significant associations (Supplementary Fig. 5b, c).
The putative AA locus at 12q23 is driven by six SNPs in high linkage disequilibrium (LD; R2 > 0.8) found in long non-coding RNAs LINC00943 and LINC00944 (Fig. 1e). These imputed SNPs all had odds ratios (ORs) close to 2, with 1.3% frequency in AA and 0% in EA, consistent with gnomAD v3. LINC00944 is highly expressed in immune cells and enriched in T cell pathways in lung tissue and cancer27–30. We fine-mapped this locus to define a 95% credible set (Supplementary Data 6) and annotated the functional consequence of the variants using the Variant Effect Predictor (VEP)31. Two variants, rs78994068, and rs115962601, were in a known enhancer regulatory region (ENSR00000974920) and thus may involve regulatory changes. However, this locus was directionally consistent but not significant in our AA replication cohort (discussed below); therefore, larger-scale AA analyses are needed to confirm this finding.
GWAS multi-ancestry meta-analysis
We conducted fixed-effect inverse variance-weighted multi-ancestry meta-analyses, combining the EA meta-analysis and the MVP AA GWAS for overall lung cancer, LUAD, and LUSC (Supplementary Data 7–9; Supplementary Fig. 6a–c). These analyses identified two additional novel genome-wide significant loci in overall lung cancer (Table 1; Supplementary Data 10; Supplementary Fig. 4i, j): ubiquitin ligase JADE2, previously associated with smoking initiation32, and RNA polymerase-associated RPAP3. Neither of these novel multi-ancestry meta-analysis loci were reported in a recent multi-ancestry analysis by Byun et al.8 that included fewer AA and more EAS samples, indicating the value our larger AA sample provided for novel discovery. All genome-wide significant EA meta-analysis associations reached genome-wide significance in the multi-ancestry meta-analyses except rs11855650 (TLE3) in LUAD (P = 6.19 × 10−8). We additionally performed random-effects meta-analyses using the Han-Eskin method (RE2)33 and observed similar P-values to the fixed effect meta-analyses, with all index variants PRE2 < 5 × 10−8 (Supplementary Data 7–9).
Polygenic risk scoring
To gain an understanding of the penetrance and pleiotropy of lung cancer risk, we constructed polygenic risk scores (PRSs) based on the ILCCO summary statistics7 for every EA subject in MVP. As expected, the PRS was highly associated with both lung cancer risk as well as smoking behavior (Supplementary Fig. 7a, b). Even after removing individuals with any history of lung cancer risk to prevent the enrichment of risk factors and comorbidities, the association with smoking behavior remained, suggesting that the PRS is partially capturing genetic smoking behavioral risk factors (Supplementary Fig. 7c). In all groups, individuals at the top decile of the PRS were at significantly higher risk of lung cancer than those in the lowest decile.
Multi-trait conditional analysis for smoking status
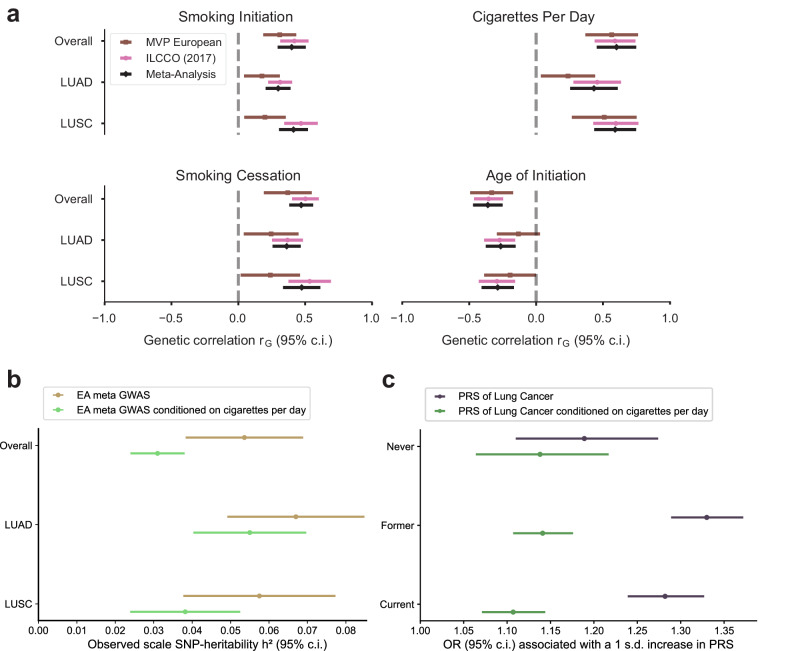
Despite adjusting for smoking status, both in MVP EA and ILCCO7, a significant genetic correlation was observed between all subsets of lung cancer GWAS and a recently published GWAS of smoking behaviors34 (Fig. 2a, Supplementary Data 11). In order to remove all residual effects of smoking on lung cancer susceptibility, we conducted a multi-trait-based conditional and joint analysis (mtCOJO)35,36, conditioning on a GWAS for cigarettes per day34, which was the smoking trait most strongly correlated with overall lung cancer and subtype GWAS from the EA meta-analysis. Because lung cancer case selection also preferentially selects smokers, conventional adjustment for smoking may inadvertently cause selection bias, which functions as a collider to induce biased genetic effects37. mtCOJO is considered more robust to potential collider bias than conventional covariate adjustment35,36. The total observed-scale SNP-heritability38 of lung cancer risk decreased substantially after conditioning on cigarettes per day, from 5.4% to 3.1% in overall LC, from 6.7% to 5.5% in LUAD, and from 5.8% to 3.8% in LUSC (Fig. 2b; Supplementary Data 12).
Fig. 2. Association of lung cancer GWAS with smoking behaviors.
a Genetic correlations between the lung cancer GWAS and smoking behaviors, including smoking initiation, cigarettes per day, smoking cessation, and age of initiation. Error bands represent the 95% confidence interval of the genetic correlation point estimates. b SNP heritability for the meta-analysis and conditional meta-analysis. The heritability decreases in the conditional analysis for overall lung cancer as well as both subtypes, suggesting that some portion of the heritability of lung cancer is due to smoking behavior. Error bands represent the 95% confidence interval of the SNP-heritability point estimates. c Polygenic risk scores (PRS) based on standard lung cancer GWAS (blue) perform worse in never-smokers than former or current smokers, while conditioning on smoking behavior (orange) results in similar performance. Error bands represent the 95% confidence interval of the odds ratio (OR) associated with a one standard deviation increase in PRS.
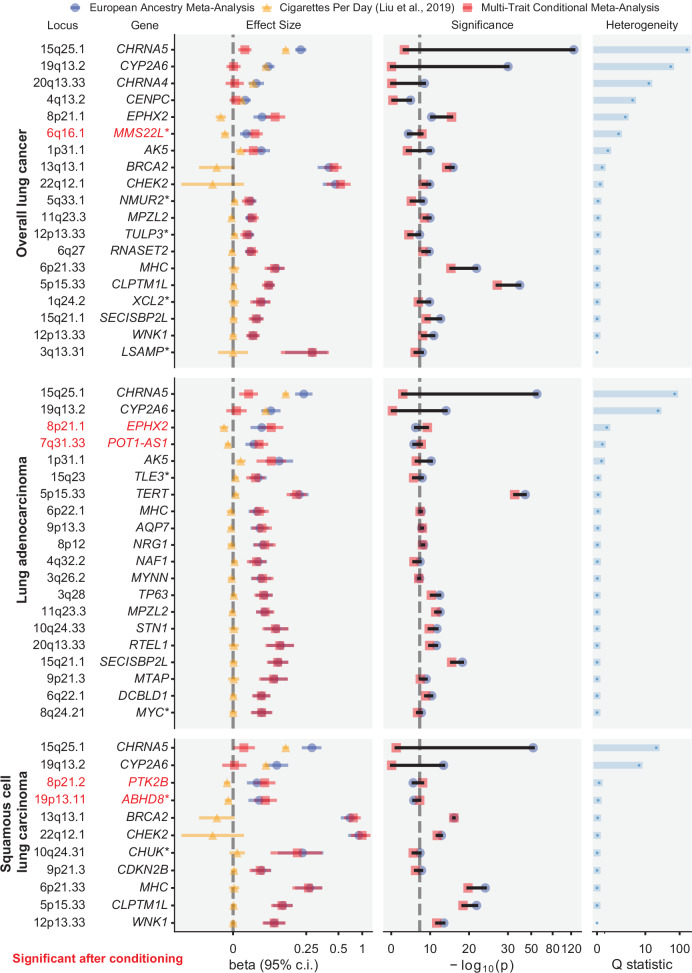
Significant loci from the conditional analyses are shown in Supplementary Figs. 8 and 9 and Supplementary Data 13–15. As expected, the statistical significance of loci harboring smoking-related genes (e.g., CHRNA5, CYP2A6, CHRNA4) dropped to below genome-wide significance after conditioning (Fig. 3). Conversely, five signals (four loci) became significant only after conditioning, including novel signals at MMS22L in overall lung cancer and 19p13.1 (ABHD8) in LUSC. MMS22L is a novel GWAS signal but was previously identified as overexpressed in lung cancer in a genome-wide gene expression scan39. These may represent biological lung cancer signals partially masked by countervailing genetic effects on smoking behavior. We performed fine-mapping to identify candidate causal variants in the conditioned EA meta-analysis summary statistics, and for overall lung cancer, LUAD, and LUSC, we identified 11, 15, and 6 high-quality credible sets, respectively, containing a total of 243, 277, and 78 SNPs (Supplementary Data 5).
Fig. 3. Forest plot of genome-wide significant associations.
Within each cancer subtype, changes in effect size and significance are shown before and after conditioning on cigarettes per day in the European meta-analysis cohort. Novel loci are indicated by an asterisk after the gene name (*). Loci that became significant after conditioning (P < 5 × 10−8) are in red. Ncases = 39,664; Ncontrols = 119,158. Association p-values were obtained from fixed-effect inverse variance-weighted meta-analysis of MVP European and ILCCO7 cohorts. The dashed line in −log10(p) marks the threshold for genome-wide significance (P = 5 × 10–8).
We constructed PRS based on mtCOJO-conditioned ILCCO summary statistics7 to directly compare the predictive performance of PRS derived from the conditioned and non-conditioned GWAS in MVP EA. While the PRS based on the non-conditioned overall lung cancer GWAS exhibited reduced performance in never-smokers compared to ever-smokers, the PRS based on the conditional analysis resulted in similar performance across smoking status (Fig. 2c; Supplementary Data 16).
Replication of novel variants and combined meta-analysis
We queried the OncoArray Consortium Lung Study (OncoArray)8,40 as an external non-overlapping replication dataset for our significant GWAS signals (Supplementary Data 1, 17, and 18). For GWAS in the EA meta-analyses for overall lung cancer, LUAD, and LUSC, we replicated five of seven novel loci (P < 0.01) in an OncoArray European ancestry cohort: XCL2 and TLE3 in overall lung cancer, MYC and TLE3 in LUAD, and BLOC1S2 in LUSC. The novel African ancestry association for overall lung cancer at LINC00944 was not replicated. We meta-analyzed OncoArray European and African ancestry participants to replicate our multi-ancestry meta-analysis signals (Supplementary Fig. 10, Supplementary Data 18) for overall lung cancer at RPAP3 (P = 0.0044) and JADE2 which bordered on nominal significance (rs329122; P = 0.053). For the two novel loci that were identified in the EA meta-analysis conditioned on cigarettes per day, we included smoking as a covariate for association analysis in the OncoArray European ancestry cohort. These association signals were replicated for overall lung cancer at MMS22L (P = 0.006) and LUSC at ABHD8 (P = 0.003). In a variant-level replication of 137 conditionally independent discovery associations that fell within ≤1 Mb of a previously reported lung cancer GWAS signal, 134 had P < 0.05 in OncoArray, and 42 had P < 5 × 10−8 (Supplementary Data 19).
We then performed a combined meta-analysis of our discovery results with OncoArray replication results (Supplementary Data 19). We considered a conservative threshold of P < 4.17 × 10−9 (P < 5 × 10−8/12 total GWAS analyses) to be significant, which was met by 9 of the 12 loci. Because rs329122 in JADE2 achieved the more conservative significance threshold (P = 3.69 × 10−9), and has also been associated with smoking behavior32 and identified as a splicing-related variant associated with lung cancer41, we considered this locus to be replicated. In the combined meta-analysis we observed similar P-values in fixed effects and random effects (RE2) models.
Next, for all previously reported lung cancer and subtype loci in this study, we identified lung cancer associations from the GWAS Catalog which fell within the same loci as our index variants (Supplementary Data 20). We confirmed two loci that previously had been reported only in a recent genome-wide association by proxy (GWAx) of lung cancer42: CENPC (rs75675343) in overall lung cancer in the EA meta-analysis (P = 2.40 × 10−8) and the multi-ancestry meta-analysis, and TP53BP1 in overall lung cancer in the multi-ancestry meta-analysis (rs9920763; P = 1.63 × 10−8). Our multi-ancestry meta-analysis for overall lung cancer also confirmed a recently reported locus at 4q32.2 (NAF1)15 in EAS.
Multi-trait analysis with breast cancer
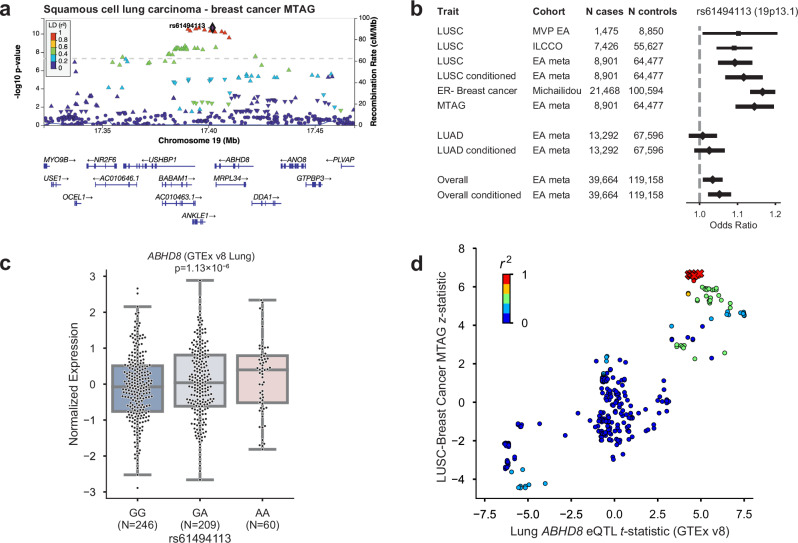
At 19p13.1, a known pleiotropic cancer locus43,44, the index SNP of LUSC conditioned on smoking (rs61494113) sits in a gene-rich region where a recent fine-mapping effort of breast cancer risk loci45 proposed two independent associations, one affecting the regulation of ABHD8 and MRPL34, and another causing a coding mutation in ANKLE1. Here, we used the increased power provided by a multi-trait analysis of GWAS (MTAG)46 of LUSC and estrogen receptor negative (ER−) breast cancer47 to disentangle the complex relationships between cancer risk and the genes in this locus (Fig. 4a). Overexpression of ABHD8 has been shown to significantly reduce cell migration43,44. Similar odds ratios at rs61494113 were observed across LUSC and breast cancer, and MTAG enhanced the GWAS signal at this locus (Fig. 4b).
Fig. 4. Significant locus after conditioning on smoking behavior, 19p13.11, has pleiotropic associations with ER-negative breast cancer.
a Regional association plot of the 19p13.11 multi-trait analysis of GWAS (MTAG) locus. P-values are derived from the two-tailed MTAG test. The dashed line marks the threshold for genome-wide significance (P = 5 × 10–8). b Odds ratios and 95% confidence intervals for lead SNP rs61494113 in squamous cell lung carcinoma (LUSC), before and after conditioning on cigarettes per day, compared to lung adenocarcinoma, overall lung cancer, and MTAG analysis with estrogen receptor-negative (ER−) breast cancer. c ABHD8 expression varies by genotype at rs61494113 in GTEx v8 Lung tissue samples. t-test was utilized and a two-sided p-value is shown. The center box plot line represents the median (50th percentile). The box edges are the 25th and 75th percentiles (interquartile range; IQR). Whiskers extend to the smallest and largest values within 1.5 times the IQR. Source data are provided as a Source Data file. d ABHD8 eQTL t-statistic vs LUSC z-statistic; red X’s indicate the 95% credible set. Source data are provided as a Source Data file.
We used the coloc-SuSiE method48 to assess colocalized associations between pairs of credible sets in this locus underlying the risk of LUSC and ER− breast cancer, allowing for multiple causal signals. We found evidence for a shared causal signal between credible sets in the LUSC conditional meta-analysis and ER− breast cancer (97.7% posterior probability; Supplementary Data 21). The index SNPs for the credible sets of LUSC conditioned on smoking and ER− breast cancer (rs61494113 and rs56069439, respectively) have r2 = 0.99.
The eQTL effect of ABHD8 was replicated in multiple tissues of GTEx v8, including Lung (Fig. 4c). Interestingly, the group of SNPs in the LUSC-BC credible set did not have the most significant eQTL effect, suggesting a complex relationship between the multiple causal variants at the locus and gene expression (Fig. 4d). For instance, a recent splice variant analysis49 implicated splicing of BABAM1 (a BRCA1-interacting protein) as a culprit of the associations observed in 19p13.1. Consistent with previous reports43,44, the cancer risk-increasing haplotype was correlated with increased expression of ABHD8 and alternative splicing of BABAM1. However, there was no overlap between the 95% eQTL credible sets of ABHD8 and BABAM1, and neither of the credible sets included rs61494113.
Phenome-wide association study
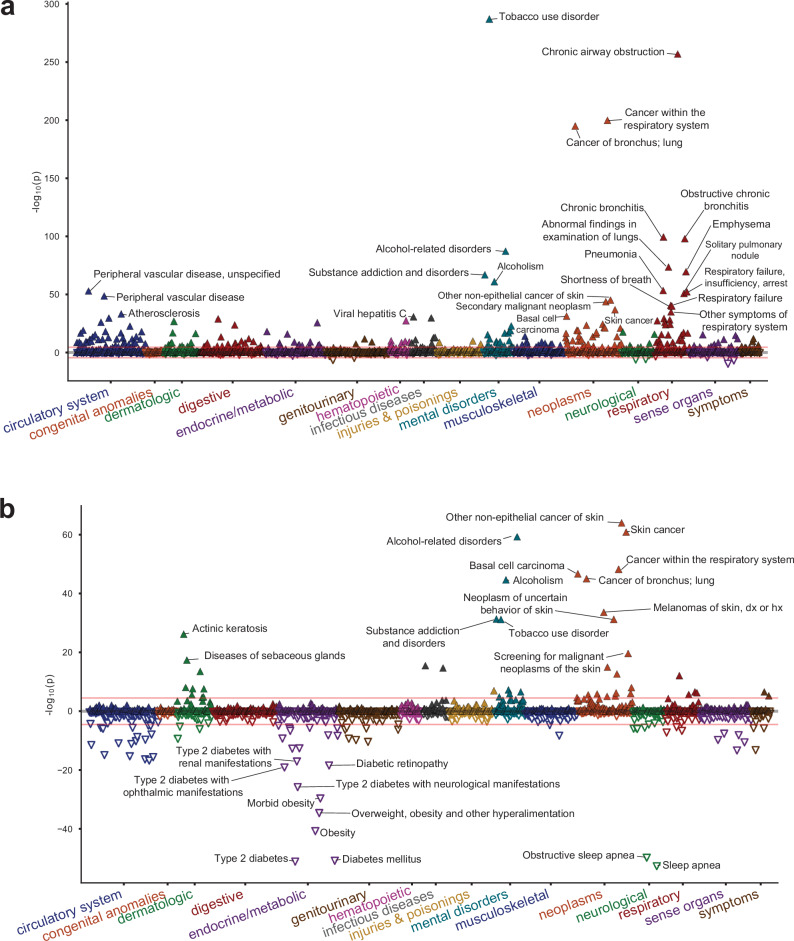
Finally, to investigate the pleiotropy of lung cancer genetic risk in the absence of the overwhelming effect of smoking behavior, we performed phenome-wide association studies (PheWAS) in MVP using the PRS scores constructed from the ILCCO summary statistics7 for overall lung cancer, both based on the standard GWAS (“unconditioned PRS”; Fig. 5a; Supplementary Data 22) and the GWAS conditioned on cigarettes per day using mtCOJO (“conditioned PRS”; Fig. 5b; Supplementary Data 23). Each PRS was tested for association with 1772 phecode-based phenotypes. Overall, 240 phenotypes were associated with the unconditioned PRS and 112 were associated with the conditioned PRS at a Bonferroni-corrected significance threshold (P < 0.05/1772). Although lung cancer remained a top association with the conditioned PRS, the association with tobacco use disorder was greatly reduced, from an OR associated with a standard deviation increase in the PRS of 1.151 [1.142–1.160] (P = 2.32 × 10−237) in the unconditioned PRS to OR = 1.046 [1.038–1.053] (P = 1.05 × 10−32) in the conditioned PRS. However, the effect on alcohol use disorder was only modestly attenuated between the unconditioned (OR = 1.098 [1.089–1.108]; P = 1.05 × 10−87) and conditioned LC (OR = 1.078 [1.069–1.088], P = 4.41 × 10−60) PRSs. Whether a role for alcohol in lung cancer exists independently of smoking is controversial50,51; this analysis suggests that may be the case. Other putatively smoking-related associations, such as chronic obstructive pulmonary disease, pneumonia, and peripheral vascular disease were greatly diminished with the conditioned PRS. Mood disorders, depression, and post-traumatic stress disorder were also significantly associated with the unconditioned PRS but no longer significantly associated with the conditioned PRS, reflecting neuropsychiatric correlates of smoking behavior.
Fig. 5. Phenome-wide association study (PheWAS) of polygenic risk scores (PRS) of lung cancer and lung cancer conditioned on cigarettes per day.
a PheWAS of PRS on lung cancer is mostly confounded with smoking associations. b PheWAS of the conditional meta-analysis PRS shows associations with skin cancer and metabolic traits. Association p-values were obtained from a two-sided test of the z-statistic, calculated using logistic regression. The red line in each plot indicates Bonferroni-corrected significance (P = 2.8 × 10–5).
Intriguingly, a category of metabolic traits that were not associated with the unconditioned PRS was highly associated with the conditioned PRS and in a negative effect direction. We observed protective associations of the conditioned PRS with metabolic traits such as type 2 diabetes (OR = 0.945 [0.938–0.952], P = 9.46 × 10−52) and obesity (OR = 0.952 [0.945–0.959], P = 2.48 × 10−41). Neither were associated with the unconditioned PRS (OR = 1.006 [0.999–1.014]; P = 0.092, and OR = 1.005 [0.998–1.012]; P = 0.183, respectively). Other traits in this category included sleep apnea and hyperlipidemia. These findings are consistent with prior observational findings of an inverse relationship between BMI and lung cancer52 and illustrate the extent to which smoking may be a major confounder of this relationship.
Finally, we observed strong associations of the lung cancer PRS with skin cancer and related traits, such as actinic keratitis. In basal cell carcinoma, the OR increased from 1.087 [1.072–1.102] (P = 6.06 × 10−32) with the unconditioned PRS to 1.105 [1.090–1.120] (P = 1.82 × 10−47) with the conditioned PRS. As a sensitivity analysis, we tested the strength of this association after removing the TERT locus, which is prominently associated with both traits. Doing so only modestly reduced the effect of the conditioned PRS to OR = 1.092 [1.077–1.107] (P = 4.08 × 10−36). Thus, our results are consistent with a genome-wide genetic correlation between lung cancer and basal cell carcinoma that is strengthened when the effect of smoking is removed. Overall, our results suggest that the biology underlying lung cancer risk may be partially masked by the residual genetic load of smoking.
Discussion
We identified novel lung cancer-associated loci in a new cohort of EA and AA participants, including the largest AA cohort analyzed to date. We also show that, despite studies on the genetic basis of lung cancer risk taking smoking status into account, the effects of smoking continue to obfuscate our understanding of lung cancer genetics. In particular, we report two novel loci, at MMS22L (overall) and ABHD8 (LUSC), which may be partially masked by countervailing genetic effects on smoking. Our replication analysis which adjusted for smoking pack-years confirmed these loci. Additionally, our analyses demonstrated that PRSs for lung cancer contain large uncorrected genetic loading for smoking behavioral factors. Our results indicate that controlling for these factors can improve risk assessment models, potentially improving lung cancer screening even for non-smokers. Finally, our phenomic scans comparing PRSs derived from GWAS with and without genomic conditioning on smoking showed divergent associations across numerous traits, especially metabolic phenotypes.
The increased sample size in this study enabled the interpretation of multiple causal variants underlying the gene-rich ADHL8-BABAM1 region, synthesizing prior observations into a clearer understanding of this locus. Our other novel loci strengthen established lung cancer mechanisms. We identify for the first time a susceptibility locus at MYC, a well-known oncogene and master immune regulator. XCL2 is involved in cellular response to inflammatory cytokines53. LSAMP is a tumor suppressor gene in osteosarcoma54, and 3q13.31 homozygous deletions have been implicated in tumorigenesis55. TLE3 is a transcriptional corepressor involved in tumorigenesis and immune function56. The transcription factor TULP3 has been implicated in pancreatic ductal adenocarcinoma and colorectal cancer57. XCL2, NMUR2, and TULP3 may also be related to cancer progression via G-protein-coupled receptor (GPCR) signaling pathways58. JADE2 expression has been experimentally linked to NSCLC59 and has been identified in GWAS of smoking behavior34. Finally, DNA damage repair genes are implicated, including RPAP3, an RNA polymerase that may be involved in DNA damage repair regulation60, and MMS22L which repairs double-strand breaks61.
Although smoking is the major risk factor for lung cancer, it is important to clearly disentangle the effect of smoking to fully understand the complex genetic and environmental causes of the disease. Our approach enables the development of new polygenic scores, which can improve precision medicine applications for lung cancer in both smokers and nonsmokers.
Methods
Ethics/study approval
The VA Central Institutional Review Board (IRB) approved the MVP000 study protocol. Informed consent was obtained from all participants, and all studies were performed with approval from the IRBs at participating centers, in accordance with the Declaration of Helsinki. Only previously generated data were analyzed in this study.
Cohort definition
Patients were identified from MVP participants19 utilizing clinical information available through the United States Department of Veterans Affairs (VA) Corporate Data Warehouse (CDW) with ICD codes for primary lung cancer. Occurrences of the ICD-9 codes 162.3, 162.4, 162.5, 162.8, and 162.9 or the ICD-10 codes C34.10, C34.11, C34.12, C34.2, C34.30, C34.31, C34.32, C34.80, C34.81, C34.82, C34.90, C34.91, and C34.92 were used in case identification. Patients with secondary lung cancer were excluded from the cohort using ICD-9/10 codes 197.x, C78.00, C78.01, and C78.02. Additional patients were identified in the VA Cancer Registry using the ICD-O site, including lung/bronchus, other respiratory system or intrathoracic organs, or trachea. The Cancer Registry was also used to determine the lung cancer subtypes LUAD and LUSC among cases.
Preliminary totals of 18,633 and 10,845 patients with MVP participation were identified from the VA CDW and Cancer Registry, respectively. A combined cohort of 20,631 unique patients was generated for further analysis. The cohort was predominantly male (~95%) with a median age of 64–68 for sub-cohorts, depending on ancestry assignments and cancer subtypes. The cohort was curated further to remove any participant with missing data. The final cohorts are described in Supplementary Data 1.
Once patients were identified from VA’s CDW and Cancer Registry, cases were used to gather records related to age, sex, smoking status, and ancestry. Smoking status included former, current, and never, based on the MVP survey at the time of enrollment and on electronic medical records. Ancestry was defined using a machine learning algorithm that harmonizes self-reported ethnicity and genetic ancestry (HARE)62. All analyses described here were performed on patients of EA or AA ancestry in ancestry-stratified cohorts. Additionally, the cohorts were further stratified by lung cancer subtypes for analysis. Matched controls were selected based on age, gender, smoking status, and HARE assignments. Age was binned into 5-year intervals for this purpose.
Genotyping and principal component analysis
Genotyping and quality control were conducted as described previously63. Briefly, we removed all samples with excess heterozygosity (F statistic < −0.1), excess relatedness (kinship coefficient ≥ 0.1 with 7 or more MVP samples), and samples with call rates <98.5%. Additional samples with a mismatch between self-reported sex and genetic sex were removed.
Principal component (PC) analysis was conducted as described previously63. Briefly, PCs were generated with PLINK 2.064 (v2.00a3LM) using a pruned set of SNPs (window size 1 Mb, step size 80, r2 < 0.1, minor allele frequency (MAF) < 0.01, Hardy–Weinberg equilibrium P < 1 × 10−10, missingness rate < 10%) within unrelated European ancestry (EA) and African ancestry (AA) individuals. (Unrelated individuals were defined as greater than third-degree relatives.) PCs were then projected onto related individuals.
Imputation
Prior to imputation, a within-cohort pre-phasing procedure was applied across the whole cohort by chromosome using Eagle265. Imputation was then conducted on pre-phased genotypes using Minimac466 and the 1000 Genomes Phase 3 (v5) reference panel67 in 20 Mb chunks and 3 Mb flanking regions. The quality of imputation was then re-computed in EA and AA separately to be used as filters for respective GWAS (Minimac Rsq or INFO > 0.3). An MAF cutoff of >0.001 was applied for all analyses. Imputed loci reaching genome-wide significance were tested for deviation from Hardy–Weinberg equilibrium (HWE) in 61,538 EA controls (Supplementary Data 24). Of the 93 conditionally independent SNPs across the GWAS analyses, 6 SNPs had a significant (P < 1 × 10−6) HWE signal; unsurprisingly, the strongest HWE signal was from SNPs in the Major Histocompatibility Complex region. However, none of the 12 novel loci reported in Table 1 significantly deviated from HWE.
Association analyses
For the EA lung cancer overall and subtype GWAS, we performed standard logistic regression using PLINK 2.0 (v2.00a2LM)64 with a matched control design. EA GWAS was performed in unrelated individuals, defined as greater than third-degree relatives. For the AA lung cancer overall and subtype analyses, because the case numbers were smaller, we performed a mixed-model logistic regression using REGENIE (v1.0.6.7)68; REGENIE applies a whole-genome regression model to control for relatedness and population structure and includes a Firth correction to control for bias in rare SNPs as well as case-control imbalance. GWAS covariates for each ancestry included age, age-squared, sex, smoking status as a categorical variable (current, former, never), and the first ten principal components. Participants with missing smoking status (n = 786) were removed. Pearson’s r was calculated for effect size concordance between MVP EA and ILCCO7 cohorts.
EA meta-analysis
We performed inverse-variance weighted meta-analyses of MVP-EA summary statistics and summary statistics previously reported by ILCCO7 using METAL (v20100505)69 with scheme STDERR. Significant inflation across GWAS and meta-analyses was not observed (all genomic control values (λ) for GWAS in this study ≤1.15). Only variants present in both studies were meta-analyzed. We further performed a sensitivity analysis using the Han-Eskin random effects model (RE2) in METASOFT v2.0.133.
Lung eQTL consortium
The lung tissues used for eQTL analyses were from human subjects who underwent lung surgery at three academic sites: Laval University, the University of British Columbia (UBC), and the University of Groningen. Genotyping was carried out using the Illumina Human1M-Duo BeadChip. Expression profiling was performed using an Affymetrix custom array (see GEO platform GPL10379). Only samples that passed genotyping and gene expression quality controls were considered for eQTL analysis, leaving sample sizes of 409 for Laval, 287 for UBC, and 342 for Groningen. Within each set, genotypes were imputed in each cohort with the Michigan Imputation Server66 using the Haplotype Reference Consortium70 version 1 (HRC.r1-1) data as a reference set, and gene expression values were adjusted for age, sex, and smoking status. Normalized gene expression values from each set were then combined with ComBat71. eQTLs were calculated using a linear regression model and additive genotype effects as implemented in the Matrix eQTL package in R72. Cis-eQTLs were defined by a 2 Mb window, i.e., 1 Mb distance on either side of lung cancer-associated SNPs. Pre-computed lung eQTLs were also obtained from the Genotype-Tissue Expression (GTEx) Portal20. Lung eQTLs in GTEx (version 8) are based on 515 individuals and calculated using FastQTL73.
Fine-mapping
We performed Bayesian fine-mapping of the genome-wide significant loci from EA meta-analysis and AA using the FinnGen fine-mapping pipeline74 (https://github.com/FINNGEN/finemapping-pipeline) and the SuSiE R package (v0.9.1.0)25,26. Pairwise SNP correlations were calculated directly from imputed dosages on European-ancestry MVP samples from this analysis using LDSTORE 2.074. The maximum number of allowed causal SNPs at each locus was set to 10. Fine-mapping regions which overlapped the major histocompatibility complex (MHC; chr6:25,000,000–34,000,000) were excluded. High-quality credible sets were defined as those with minimum r2 < 0.5 between variants. The functional consequences of the AA credible set variants were annotated using the Variant Effect Predictor (VEP)31.
Replication analysis
External replication was performed for all genome-wide significant associations in overall lung cancer, LUAD, and LUSC in OncoArray Consortium Lung Study (OncoArray)8,40. Replication for genome-wide significant multi-ancestry associations was performed in a fixed-effects meta-analysis of OncoArray CEU Europeans for significant EA meta-analysis associations, and in a YRI AA meta-analysis composed of 5 studies8 for significant MVP AA associations. Meta-analysis associations from this study were replicated against a meta-analysis of these OncoArray groups; Pearson’s r was calculated for effect size concordance between these groups. To replicate significant variants from EA analysis conditioned on smoking, pack-years was additionally included as a covariate in replication cohorts. There was no participant overlap between the replication cohorts and the ILCCO study7 used in the discovery scan. Covariates included the first five genetic principal components and participant study sites. Proxy SNPs were used to replicate known associations at rs75675343 (rs2318539/4:67831628:C:A; R2EUR = 1) and rs4586884 (rs4435699/4:164019500:C:G; R2EUR = 0.999).
Multi-ancestry meta-analysis
A multi-ancestry meta-analysis of MVP EA and AA cohorts with summary statistics previously reported by ILCCO7 was conducted in METAL69 using an inverse variance-weighted fixed effects scheme. Only variants present in two or more cohorts were meta-analyzed. Index variants were defined using the two-stage “clumping” procedure implemented in the Functional Mapping and Annotation (FUMA) platform75. In this process, genome-wide significant variants are collapsed into LD blocks (r2 > 0.6) and subsequently re-clumped to yield approximately independent (r2 < 0.1) signals; adjacent signals separated by <250 kb are ligated to form independent loci. Novel variants are defined as meta-analysis index variants located >1 Mb from previously reported lung cancer associations. We additionally performed a sensitivity analysis using the random effects model (RE2) in METASOFT v2.0.133.
Polygenic risk score (PRS) calculation
We constructed PRSs based on the ILCCO summary statistics7 and the conditional meta-analysis of ILCCO adjusted for cigarettes per day34 for every EA subject in MVP. We used PRS-CS76 to generate effect size estimates under a Bayesian shrinkage framework and then used PLINK 2.0 (v2.00a3LM)64 to linearly combine weights into a risk score using a global shrinkage prior of 1 × 10−4, which is recommended for less polygenic traits. Finally, scores were normalized to a mean of 0 and a standard deviation of 1.
Multi-trait analyses
In order to remove all residual effects of smoking on lung cancer susceptibility, we conducted a multi-trait meta-analysis35 conditioned on cigarettes per day, which was shown to be most significantly correlated with all lung cancer GWAS34. The meta-analysis was performed on the EA meta-analysis summary statistics using mtCOJO, part of the GCTA software package77. An LD reference was constructed from 50,000 MVP EA samples.
Multi-trait analysis of GWAS (MTAG)46 (v0.9.0) was applied using genome-wide LUSC summary statistics after conditioning on cigarettes per day, and estrogen receptor negative (ER−) breast cancer summary statistics47 (21,468 ER− cases and 100,594 controls) which were munged using LDSC (v1.01)38. Colocalization between LUSC conditioned on cigarettes per day and ER− breast cancer allowing for multiple causal signals was performed using the coloc-SuSiE method48 of coloc (R; v5.2.1)78 for variants at ABHD8 (chr19: 17,350,000 to 17,475,000). A posterior probability >0.9 for Hypothesis 4 was used as the criteria for colocalization.
Heritability and genetic correlations
Linkage Disequilibrium score regression (LDSC) v1.0.1 was used to calculate observed-scale SNP-heritability38 using lung cancer and subtypes summary statistics, before and after conditioning on cigarettes per day. Pairwise genetic correlations were estimated between lung cancer and subtypes from MVP, ILCCO7, and EA meta-analysis, and four smoking traits (smoking initiation, cigarettes per day, smoking cessation, and age of initiation)34.
Conditional and joint SNP analysis
To find independently associated genome-wide significant SNPs at each locus in a stepwise fashion, we used GCTA-COJO77 using the --cojo-slct option. An LD reference was constructed from 50,000 MVP EA samples. Variants with MAF < 0.01 in the COJO reference panel were not included in the identification of independent signals. LDTrait79 was queried to identify previously published significant GWAS variants within 1 Mb of our index variants in all populations. Novel loci were defined as those at which the index variant was located >1 Mb from previously reported genome-wide significant lead SNPs for lung cancer or its subtypes in any ancestry.
Phenome-wide association study (PheWAS)
We conducted a PheWAS of electronic health record-derived phenotypes and lab results in EA subjects using either the normalized PRS as the predictor or independently associated genome-wide significant SNPs. Comparison of unconditioned PRS PheWAS and conditioned PRS PheWAS were based on ILCCO summary statistics7 and used MVP EA as the out-of-sample test set. Associations were tested using the R PheWAS package80 (v0.1) with QC procedures described previously81. Control and sex-based exclusion criteria were applied.
Statistics and reproducibility
Samples sizes for the case-control status of overall lung cancer, LUAD, and LUSC in MVP participants are provided in Supplementary Data 1. EA GWAS meta-analysis was performed across lung cancer and its subtypes using MVP and an external cohort, ILCCO7, comprised of up to 39,664 cases and 119,158 controls (Supplementary Data 1). GWAS replication and combined meta-analysis were performed using an external OncoArray8,40 cohort made up of 19,404 cases and 17,378 controls (Supplementary Data 1); these participants had no overlap with ILCCO7. All statistical tests were two-tailed linear or logistic regressions unless otherwise noted. Nominal significance was defined as P < 0.05. In hypothesis-free scans, we applied strict significance thresholds to account for multiple hypothesis testing. For GWAS analyses, the standard genome-wide significance threshold (P < 5 × 10−8) was used. In PheWAS analyses, we applied Bonferroni-corrected significance thresholds. All P-values are presented without adjustment for multiple hypotheses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Source data
Acknowledgements
This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by award #MVP000. This publication does not represent the views of the Department of Veterans Affairs or the United States Government. R.J.H., J.D.M., J.B., Y.H., X.X., and C.I.A. were supported by the National Institutes of Health (NIH) for Integrative Analysis of Lung Cancer Etiology and Risk (U19CA203654). C.I.A. was supported by Sequencing Familial Lung Cancer (R01CA243483). Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article, and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization. Full consortium acknowledgements for MVP and ILCCO7 are provided in Supplementary Information.
Author contributions
Drafted the manuscript: B.R.G., M.F., S.-G.J., A.K.S., E.P., A.K.D., and S.P. Acquired the data: B.R.G., S.-G. J., A.K.S., Y.S., P.D., U.S., J.B., Y.H., X.X., D.D.S., W.T., J.M., S.M., R.R., R.J.H., J.D.M., Y.B., C.I.A., VA MVP, and S.P. Analyzed the data: B.R.G., S.-G.J., M.F., A.K.S., Y.S., P.D., X.X., U.S., Y.B., and R.S. Critically revised the manuscript for important intellectual content: B.R.G., M.F., S.-G.J., A.K.S., Y.S., P.D., U.S., E.P., A.K.D., J.B., Y.H., X.X., D.D.S., W.T., J.M., S.M., R.R., R.J.H., J.D.M., Y.B., R.S., C.I.A., and S.P.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The full summary level association data from the individual population analyses in MVP are available via the dbGaP study accession number phs001672. ILCCO7 summary statistics can be found in GWAS Catalog accession numbers GCST004748, GCST004744, and GCST004750. OncoArray Consortium8,40 summary statistics used for replication can be found in dbGaP study accession number phs001273. GTEx v8 lung eQTL summary data were accessed on the GTEx portal [https://gtexportal.org]; full data are available via the dbGaP study accession number phs000424.v8.p2. Source data are provided with this paper.
Code availability
This study did not use any custom computer code or algorithms to generate results. All software tools used in this analysis were open source.
Competing interests
S.-G.J. is an employee and shareholder of BridgeBio Pharma, unrelated to the present work. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
A full list of members and their affiliations appears in the Supplementary Information.
Contributor Information
Saiju Pyarajan, Email: saiju.pyarajan@va.gov.
VA Million Veteran Program:
Jennifer Moser, Sumitra Muralidhar, Rachel Ramoni, and Saiju Pyarajan
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52129-4.
References
- 1.Schabath, M. B. & Cote, M. L. Cancer progress and priorities: lung cancer. Cancer Epidemiol. Biomark. Prev.28, 1563–1579 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leiter, A., Veluswamy, R. R. & Wisnivesky, J. P. The global burden of lung cancer: current status and future trends. Nat. Rev. Clin. Oncol.20, 624–639 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin.72, 7–33 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Bossé, Y. & Amos, C. I. A decade of GWAS results in lung cancer. Cancer Epidemiol. Biomark. Prev.27, 363–379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timofeeva, M. N. et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum. Mol. Genet.21, 4980–4995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKay, J. D. et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet.49, 1126–1132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byun, J. et al. Cross-ancestry genome-wide meta-analysis of 61,047 cases and 947,237 controls identifies new susceptibility loci contributing to lung cancer. Nat. Genet.54, 1167–1177 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, Y. et al. SNP rs17079281 decreases lung cancer risk through creating an YY1-binding site to suppress DCBLD1 expression. Oncogene39, 4092–4102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, T. et al. Genomic and evolutionary classification of lung cancer in never smokers. Nat. Genet.53, 1348–1359 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Govindan, R. et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell150, 1121–1134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, Z. et al. Meta-analysis of genome-wide association studies identifies multiple lung cancer susceptibility loci in never-smoking Asian women. Hum. Mol. Genet.25, 620–629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schabath, M. B., Cress, D. & Munoz-Antonia, T. Racial and ethnic differences in the epidemiology and genomics of lung cancer. Cancer Control23, 338–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long, E., Patel, H., Byun, J., Amos, C. I. & Choi, J. Functional studies of lung cancer GWAS beyond association. Hum. Mol. Genet.31, R22–R36 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi, J. et al. Genome-wide association study of lung adenocarcinoma in East Asia and comparison with a European population. Nat. Commun.14, 3043 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai, J. et al. Identification of risk loci and a polygenic risk score for lung cancer: a large-scale prospective cohort study in Chinese populations. Lancet Respir. Med.7, 881–891 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nahar, R. et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat. Commun.9, 216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zanetti, K. A. et al. Genome-wide association study confirms lung cancer susceptibility loci on chromosomes 5p15 and 15q25 in an African-American population. Lung Cancer98, 33–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaziano, J. M. et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol.70, 214–223 (2016). [DOI] [PubMed] [Google Scholar]
- 20.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao, K. et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet.8, e1003029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bossé, Y. et al. Transcriptome-wide association study reveals candidate causal genes for lung cancer. Int. J. Cancer146, 1862–1878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koutsami, M. K. et al. Centrosome abnormalities are frequently observed in non-small-cell lung cancer and are associated with aneuploidy and cyclin E overexpression. J. Pathol.209, 512–521 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Chan, J. Y. A clinical overview of centrosome amplification in human cancers. Int. J. Biol. Sci.7, 1122–1144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, G., Sarkar, A., Carbonetto, P. & Stephens, M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. Ser. B Stat. Methodol.82, 1273–1300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou, Y., Carbonetto, P., Wang, G. & Stephens, M. Fine-mapping from summary data with the ‘sum of single effects’ model. PLoS Genet.18, e1010299 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Goede, O. M. et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell184, 2633–2648.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y. et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat. Commun.11, 1000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Santiago, P. R. et al. Immune-related IncRNA LINC00944 responds to variations in ADAR1 levels and it is associated with breast cancer prognosis. Life Sci.268, 118956 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Chen, D. et al. Genome-wide analysis of long noncoding RNA (lncRNA) expression in colorectal cancer tissues from patients with liver metastasis. Cancer Med.5, 1629–1639 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaren, W. et al. The ensembl variant effect predictor. Genome Biol.17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saunders, G. R. B. et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature612, 720–724 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han, B. & Eskin, E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet.88, 586–598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, M. et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet.51, 237–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun.9, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue, A. et al. Genome-wide analyses of behavioural traits are subject to bias by misreports and longitudinal changes. Nat. Commun.12, 20211 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munafò, M. R., Tilling, K., Taylor, A. E., Evans, D. M. & Davey Smith, G. Collider scope: when selection bias can substantially influence observed associations. Int. J. Epidemiol.47, 226–235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet.47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen, M.-H., Ueda, K., Nakamura, Y. & Daigo, Y. Identification of a novel oncogene, MMS22L, involved in lung and esophageal carcinogenesis. Int. J. Oncol.41, 1285–1296 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Amos, C. I. et al. The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol. Biomark. Prev.26, 126–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, W. et al. Deciphering associations between three RNA splicing-related genetic variants and lung cancer risk. NPJ Precis. Oncol.6, 48 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabriel, A. A. G. et al. Genetic analysis of lung cancer and the germline impact on somatic mutation burden. J. Natl Cancer Inst.114, 1159–1166 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawrenson, K. et al. Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast-ovarian cancer susceptibility locus. Nat. Commun.7, 12675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lesseur, C. et al. Genome-wide association meta-analysis identifies pleiotropic risk loci for aerodigestive squamous cell cancers. PLoS Genet.17, e1009254 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fachal, L. et al. Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat. Genet.52, 56–73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turley, P. et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet.50, 229–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michailidou, K. et al. Association analysis identifies 65 new breast cancer risk loci. Nature551, 92–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallace, C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet.17, e1009440 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gusev, A. et al. A transcriptome-wide association study of high-grade serous epithelial ovarian cancer identifies new susceptibility genes and splice variants. Nat. Genet.51, 815–823 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brenner, D. R. et al. Alcohol consumption and lung cancer risk: a pooled analysis from the International Lung Cancer Consortium and the SYNERGY study. Cancer Epidemiol.58, 25–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larsson, S. C. et al. Smoking, alcohol consumption, and cancer: a mendelian randomisation study in UK Biobank and international genetic consortia participants. PLoS Med.17, e1003178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrelli, F. et al. Association of obesity with survival outcomes in patients with cancer: a systematic review and meta-analysis. JAMA Netw. Open4, e213520 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lan, T., Chen, L. & Wei, X. Inflammatory cytokines in cancer: comprehensive understanding and clinical progress in gene therapy. Cells10, 100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kresse, S. H. et al. LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer48, 679–693 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Xie, J. et al. Copy number analysis identifies tumor suppressive lncRNAs in human osteosarcoma. Int. J. Oncol.50, 863–872 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Yu, G. et al. Roles of transducin-like enhancer of split (TLE) family proteins in tumorigenesis and immune regulation. Front. Cell Dev. Biol.10, 1010639 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sartor, I. T. S., Recamonde-Mendoza, M. & Ashton-Prolla, P. TULP3: a potential biomarker in colorectal cancer? PLoS ONE14, e0210762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chaudhary, P. K. & Kim, S. An insight into GPCR and G-proteins as cancer drivers. Cells10, 3288 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy, C. et al. An analysis of JADE2 in non-small cell lung cancer (NSCLC). Biomedicines11, 2576 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ni, L. et al. RPAP3 interacts with Reptin to regulate UV-induced phosphorylation of H2AX and DNA damage. J. Cell. Biochem.106, 920–928 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Saredi, G. et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex. Nature534, 714–718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang, H. et al. Harmonizing genetic ancestry and self-identified race/ethnicity in genome-wide association studies. Am. J. Hum. Genet.105, 763–772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunter-Zinck, H. et al. Genotyping array design and data quality control in the million veteran program. Am. J. Hum. Genet.106, 535–548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loh, P.-R. et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet.48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet.48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet.53, 1097–1103 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet.48, 1279–1283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Shabalin, A. A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics28, 1353–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ongen, H., Buil, A., Brown, A. A., Dermitzakis, E. T. & Delaneau, O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics32, 1479–1485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benner, C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics32, 1493–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun.8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ge, T., Chen, C.-Y., Ni, Y., Feng, Y.-C. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun.10, 1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet.88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet.10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Machiela, M. J. & Chanock, S. J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics31, 3555–3557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carroll, R. J., Bastarache, L. & Denny, J. C. R. PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics30, 2375–2376 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klarin, D. et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet.50, 1514–1523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
The full summary level association data from the individual population analyses in MVP are available via the dbGaP study accession number phs001672. ILCCO7 summary statistics can be found in GWAS Catalog accession numbers GCST004748, GCST004744, and GCST004750. OncoArray Consortium8,40 summary statistics used for replication can be found in dbGaP study accession number phs001273. GTEx v8 lung eQTL summary data were accessed on the GTEx portal [https://gtexportal.org]; full data are available via the dbGaP study accession number phs000424.v8.p2. Source data are provided with this paper.
This study did not use any custom computer code or algorithms to generate results. All software tools used in this analysis were open source.