Abstract
Microglia play important roles in brain development and homeostasis by removing dying neurons through efferocytosis. Morphological changes in microglia are hallmarks of many neurodegenerative conditions, such as Niemann-Pick disease type C. Here, NPC1 loss causes microglia to shift from a branched to an ameboid form, though the cellular basis and functional impact of this change remain unclear. Using zebrafish, we show that NPC1 deficiency causes an efferocytosis-dependent expansion of the microglial gastrosome, a collection point for engulfed material. In vivo and in vitro experiments on microglia and mammalian macrophages demonstrate that NPC1 localizes to the gastrosome, and its absence leads to cholesterol accumulation in this compartment. NPC1 loss and neuronal cell death synergistically affect gastrosome size and cell shape, increasing the sensitivity of NPC1-deficient cells to neuronal cell death. Finally, we demonstrate conservation of cholesterol accumulation and gastrosome expansion in NPC patient-derived fibroblasts, offering an interesting target for further disease investigation.
Subject terms: Cell death and immune response, Organelles, Cell death in the nervous system
In Niemann-Pick type C, microglia display an ameboid shape. Here, the authors show that this is tied to neuronal engulfment and the expansion of the gastrosome, where cholesterol accumulates, making microglia more sensitive to neuronal cell death levels.
Introduction
Efferocytosis, the phagocytosis of dying cells, plays a crucial role in various biological processes, including development, tissue homeostasis and numerous diseases. Microglia, the major efferocytes of the brain, must efficiently digest engulfed neurons, and process the resulting breakdown products, through mechanisms that are poorly understood. Recent data indicate that such internal processes become limiting in contexts where microglia engulf large numbers of dead neurons, such as during brain development and neurodegenerative disease1,2. In these contexts, microglia also adopt a similar gene expression signature that is characterized by the upregulation of genes involved in phagocytosis and lipid metabolism and transport3,4 reviewed in ref. 5). Indeed, the processing of lipids originating from neuronal degradation appears to be particularly challenging for microglia as in many neurodegenerative conditions these cells often contain abnormal lipid inclusions6–9.
One such condition is Niemann-Pick disease type C (NPC), caused by mutations in the NPC intracellular cholesterol transporter 110, resulting in the accumulation in endo-lysosomal compartments of cholesterol, its derivatives and glycolipids11–14. In NPC, these endo-lysosomal defects have been linked with the loss of Purkinje cells15–17 via mechanisms that are compatible with neuronal apoptosis18–20 and necroptosis21,22. Microglia have been shown to also express NPC1 and NPC223–25 and to display impaired lipid processing in NPC25–27. These cells show the acquisition of a DAM-like signature25 and a disease-associated ameboid morphology characterized by swelling and the loss of long extensions, a phenotype that gets stronger as the disease progresses27–30. The cellular mechanisms directing this branched-to-ameboid transition remain elusive. It is unclear whether this transformation is solely prompted by lysosomal defects or if the efferocytic function of microglia also plays a role. Indeed, there is a significant knowledge gap regarding how microglia process lipids, particularly when these cells are under high phagocytic stress.
An intriguing player in efferocytosis in microglia and macrophages is the gastrosome, a single compartment that serves as a point of convergence for all phagosomes containing engulfed apoptotic material2. This compartment is characterized by a set of unique features such as a distinct molecular signature, being LAMP1 positive and Cathepsin D negative, and an electron-lucent lumen containing membrane fragments2. Furthermore, it has been observed to expand in response to increased cell death and because of mutations in SLC37A2, a putative Glucose-6-phosphate transporter responsible for phagosomal shrinkage2.
In this study, in vivo imaging and perturbation in the zebrafish identify the gastrosome as an important link between neuronal cell death, cholesterol transport defects in NPC and microglial morphological changes. We show that NPC1 localizes to the gastrosome, which becomes the major site of cholesterol accumulation in the absence of this transporter. The dynamic expansion of the gastrosome leads to NPC-deficient microglia adopting an ameboid phenotype, which can be further enhanced by increasing neuronal apoptosis, providing a mechanistic explanation for this known hallmark of NPC disease progression. We show that NPC1 loss renders microglia more sensitive to increased neuronal apoptosis, leading to the cell death of microglia themselves, a finding that has important implications for our understanding of NPC disease. Finally, we demonstrate cholesterol accumulation in the gastrosome of fibroblasts derived from NPC patients, which could have significant implications for understanding the cellular mechanisms underlying this and other neurodegenerative diseases.
Results
Loss of NPC1 results in phagocytic microglia adopting an ameboid morphology
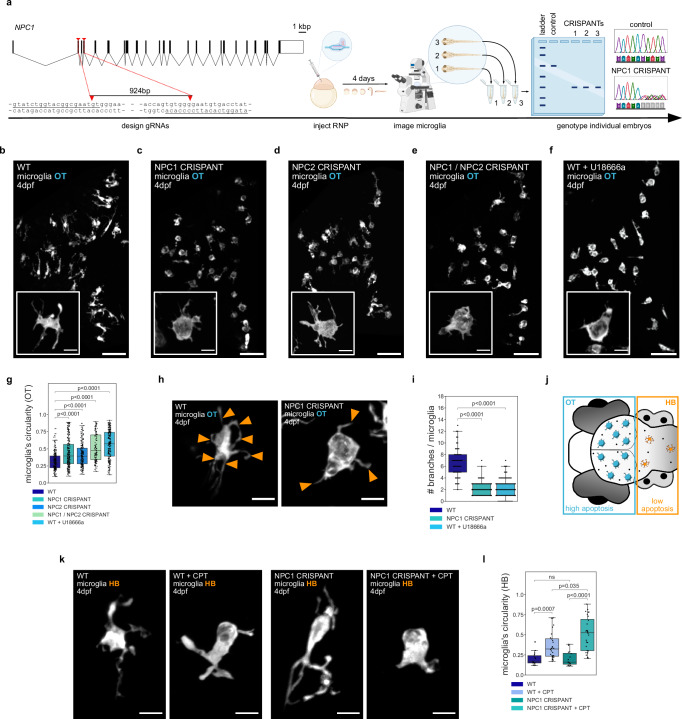
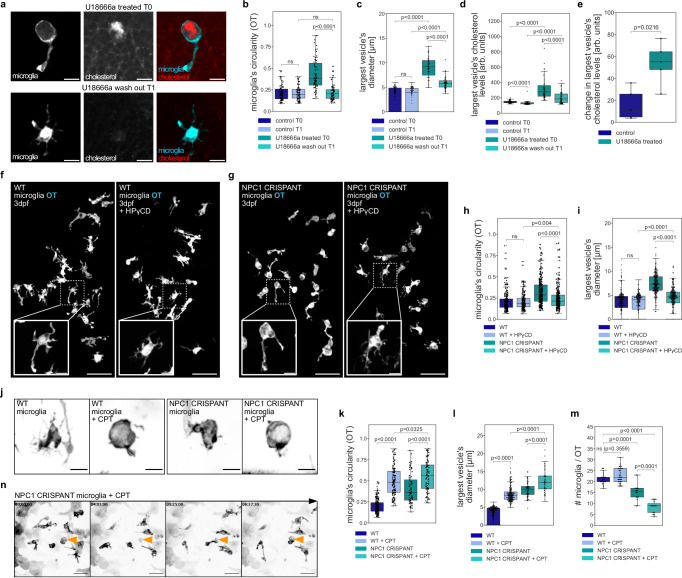
We investigated the role of NPC1 in microglia using a CRISPR/Cas9 strategy31 see schematics in Fig. 1a) to generate F0 somatic NPC1 mutants (referred to as CRISPANTs and NPC1 defective) directly in the background of fluorescently labeled microglia (Tg(mpeg1.1:EGFP-CAAX))2 or (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry))32. Guide RNAs (gRNAs) were designed to create a 924bp-long deletion in the region spanning from exon 2 to exon 6 in NPC1 locus and were injected together with Cas9 into one-cell stage zebrafish eggs. Embryos were examined four days after injection to investigate the role of NPC1 in microglia that populate the optic tectum (OT), as these cells are phagocytically active due to neuronal developmental apoptosis33. Microglia in these larvae showed an ameboid morphology, being more circular with fewer cellular extensions (Fig. 1b, c; Fig. 1g; Fig. 1h, i; Supplementary Movie 1), a phenotype that is characteristic of NPC27–30. Genomic (Supplementary Fig. 1a) and Western Blot analysis (Supplementary Fig. 1b, c; see Material and Methods) confirmed editing of the NPC1 locus and a significant protein loss in NPC1 deficient embryos. We further investigated the role of NPC1 in microglia by using the NPC1 inhibitor U18666a34 and by targeting NPC2, known to work together with NPC1 in cholesterol transport35,36. Deletion in NPC2 locus resulted in ameboid microglia (Fig. 1d, g), and embryos with double NPC1/NPC2 deficiency exhibited a phenotype similar to that of single NPC1 or NPC2 deficient embryo (Fig. 1e, g). In addition, wild-type embryos displayed ameboid microglia 4–6 h after incubation with the NPC1 inhibitor U18666a (Fig. 1f, g, i). We also examined a published NPC1 allele (npc1hg37 37) and found that these embryos were also characterized by the presence of ameboid microglia (Supplementary Fig. 1d). As shown in mice26, fish embryos deficient in NPC1 also exhibited upregulation of pro-inflammatory markers such as Interleukin-1 (IL1) and Nuclear factor NF-kappa-B subunit 2 (NFKB2)38, and to a lesser extent Interleukin-6 (IL6)39,40 (Supplementary Fig. 1g). Interestingly, in the absence of microglia, as observed in irf8(−/−) mutants41 that are also NPC1 deficient, the expression of these markers increased further, indicating a potential anti-inflammatory role for microglia in NPC1 deficient embryos (Supplementary Fig. 1g).
Fig. 1. NPC1 and NPC2 deficiencies lead to ameboid microglia under phagocytic stress.
a Schematic of the CRISPANT workflow created in BioRender. Zareba, J. (2022) BioRender.com/v30d049. b–f Representative images of OT zebrafish microglia (Tg(mpeg1.1:EGFP-CAAX)). Scale bar 50 µm. Boxes with magnifications of single microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)). Scale bar 10 µm. Wild-type (b), NPC1 CRISPANT (c), NPC2 CRISPANT (d), NPC1 / NPC2 CRISPANT (e), wild-type treated with U18666a (f). g Circularity of OT microglia in wild-type (N = 10, n = 207), NPC1 CRISPANT (N = 14, n = 230), NPC2 CRISPANT (N = 10, n = 193), NPC1 / NPC2 CRISPANT (N = 6, n = 115) and wild-type zebrafish treated with U18666a (N = 10, n = 223). h Representative images of OT zebrafish microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)). Branches indicated by orange arrowheads. Scale bar 10 µm. i Number of branches per OT microglia from wild-type (N = 3, n = 54), NPC1 CRISPANT (N = 6, n = 85) and wild-type treated with U18666a (N = 4, n = 95) zebrafish. j Schematic of the zebrafish brain regions. k Representative images of HB zebrafish microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)). Scale bar 10 µm. l Circularity of HB zebrafish microglia from wild-type (N = 8, n = 16), wild-type treated with CPT (N = 15, n = 28), NPC1 CRISPANT (N = 7, n = 16) and NPC1 CRISPANT treated with CPT (N = 12, n = 28). N refers to the number of zebrafish embryos and n to the number of microglia examined. Boxplots represent the median value and interquartile range; the ends of the whiskers correspond to the minimum and maximum values. Statistical tests: Mann-Whitney-Wilcoxon test two-sided with Bonferroni correction. ns without any additional p value on the graph stands for p = 1. OT stands for optic tectum and HB stands for hindbrain. Source data are provided as a Source Data file.
We next took advantage of brain-wide imaging in the zebrafish model to investigate potential regional differences in the microglial phenotype. This revealed that compared to ameboid microglia found in the optic tectum (OT), microglia in the adjacent hindbrain (HB) maintained their typical branched appearance (see schematics in Fig. 1j; Supplementary Fig. 1e, f). First, loss of NPC1 has no impact on the overall rate of neuronal apoptosis and neuronal removal at this stage, as shown by quantifying neuronal cell death in NPC1 deficient brains that also lack microglia (Supplementary Fig. 1h, see Material and Methods41) and the rate of phagosome formation in NPC1 deficient microglia (Supplementary Fig. 1i; see Material and Methods42). However, compared to the HB, the OT is known to display higher levels of developmental neuronal apoptosis33,43, suggesting that NPC1 might exert its influence specifically on phagocytically active microglia. We tested this hypothesis by taking three different approaches. First, we increased apoptosis in the HB using Camptothecin (CPT), an established compound that kills proliferating neurons2,44. Adding CPT significantly increased the rate of neuronal cell death, affecting both wild-type and NPC1 deficient microglia (Fig. 1k, l). However, the NPC1 deficient microglia underwent a more pronounced change (Fig. 1k, l). Second, we used a genetic approach based on nitroreductase (NTR) expression in HB neurons (Tg(NBT:DLexPR::NTR-mCherry), referred to as HB-NTR-mCherry; see schematic in Supplementary Fig. 1j)33,45,46. While NTR-based induction of apoptosis did not have a detectable effect on the morphology of HB microglia in wild-type controls, in the absence of NPC1, HB microglia became more ameboid (Supplementary Fig. 1k and l). Finally, we did the opposite experiment and reduced neuronal efferocytosis by knocking down BAI1, a phagocytic receptor active in microglia46. In NPC1-deficient embryos, this led to a shift in the microglia morphology from ameboid to more ramified (Supplementary Fig. 1m, n).
Thus, our data reveal that NPC1 loss impacts microglia morphology, with these cells adopting an ameboid phenotype. Cells exposed to high neuronal apoptosis show this phenotype, while others adopt it when neuronal apoptosis is increased locally, indicating that the effect is more prevalent in phagocytically active microglia.
NPC1 deficiency leads to the expansion of the gastrosome in phagocytic macrophages and microglia
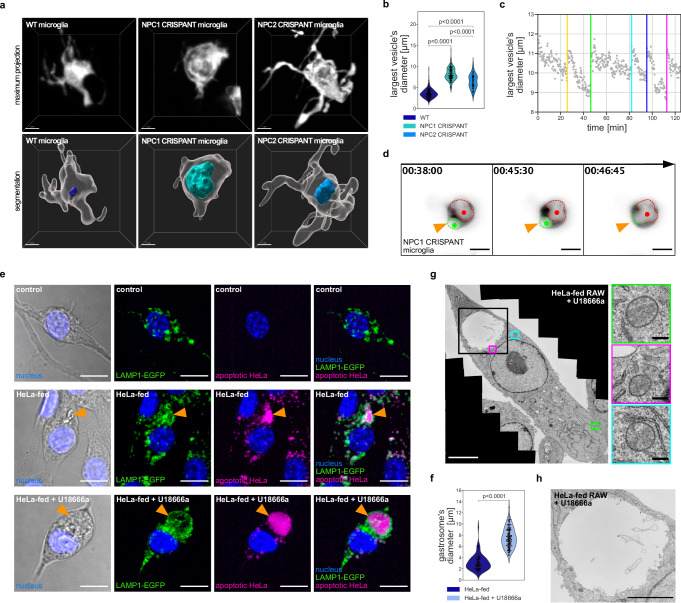
Imaging of NPC1 and NPC2 deficient microglia revealed the presence of a large vesicle that occupied most of their soma (Fig. 2a, b, Supplementary Movie 2), a feature absent in control cells (Fig. 2a, b; Supplementary Movie 2). This vesicle exhibited hallmarks of the gastrosome, as, for example, its size fluctuated over time in response to fusions with all incoming phagosomes (Fig. 2c, d, Supplementary Movie 32). Moreover, the inhibition of efferocytosis through BAI1 knock-down reduced the diameter of the enlarged vesicle observed in NPC1-deficient cells (Supplementary Fig. 2a), linking the size of this vesicle to the phagocytic activity of microglia. In RAW 264.7 mammalian macrophages (referred to as RAW macrophages) the gastrosome can be visualized by feeding these cells with fluorescently tagged apoptotic HeLa cells (referred to as HeLa-fed RAW macrophages)2. Indeed, following feeding, the gastrosome becomes both fluorescent and light-scattering, a characteristic that allows us to differentiate it from other HeLa cell inclusions (Fig. 2e, orange arrowhead2). When we treated HeLa-fed RAW macrophages with U18666a to inhibit NPC1, the labelled compartment expanded significantly (Fig. 2e, f; orange arrowhead), supporting the hypothesis that the large vesicle found in the absence of NPC1 is indeed the gastrosome. Moreover, this compartment displayed the typical molecular and ultrastructural features of the gastrosome. It was LAMP1 positive (Fig. 2e) and Cathepsin D negative (Supplementary Fig. 2b, c) and had an electron-lucent lumen that contained membrane fragments (Fig. 2g, h2). In comparison, lysosomes in U18666a treated HeLa-fed RAW macrophages are smaller and electron-dense (Fig. 2g; the gastrosome and lysosomes are marked by black and colored boxes, respectively). Moreover, an impact of NPC1 knockout on autophagy appears unlikely, as the gastrosome is negative for LC3B, a canonical autophagy marker (Supplementary Fig. 2d, e). Taken together these data indicate that lack of the NPC1 cholesterol transporter in phagocytic macrophages and microglia leads to the expansion of the gastrosome, suggesting a possible role for this compartment in NPC1-mediated cholesterol trafficking.
Fig. 2. NPC1 deficiency causes expansion of the gastrosome in phagocytic cells.
a Representative images of OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)) in the zebrafish displayed as maximum projections and segmented images. The largest vesicle present in each cell is displayed as a segmentation. Scale bar 5 µm. b Diameter of the largest vesicle in wild-type (N = 26, n = 102), NPC1 CRISPANT (N = 29, n = 232) and NPC2 CRISPANT (N = 7, n = 126) OT microglia. N refers to the number of zebrafish embryos and n to the number of microglia examined. c Graph representing the diameter of the largest vesicle found in NPC1 CRISPANT OT microglia over time. Colored vertical lines indicate fusion with incoming phagosomes. d Representative images of fusion events between the large vesicle (marked with red) and incoming phagosome (marked with green) in NPC1 CRISPANT OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)). Scale bar 10 µm. Related to Supplementary Movie 3. e Representative images of RAW LAMP1-EGFP macrophages. Orange arrowhead indicates the gastrosome. Scale bar 10 µm. f Diameter of the gastrosome in HeLa-fed RAW LAMP1-EGFP macrophages, untreated (n = 69) and treated with U18666a (n = 115). n refers to the number of cells examined. g EM image of HeLa-fed RAW treated with U18666a. Lysosomes are framed in colored squares and the gastrosome is framed in the black square (enlargement in Fig. 3h). Scale bar 5 µm. Enlarged regions with lysosomes on the right. Scale bar 0.25 µm. h Electron microscope image of the gastrosome in HeLa-fed RAW treated with U18666a. Scale bar 3 µm. Statistical tests: Mann-Whitney-Wilcoxon test two-sided with Bonferroni correction. OT stands for optic tectum. Source data are provided as a Source Data file.
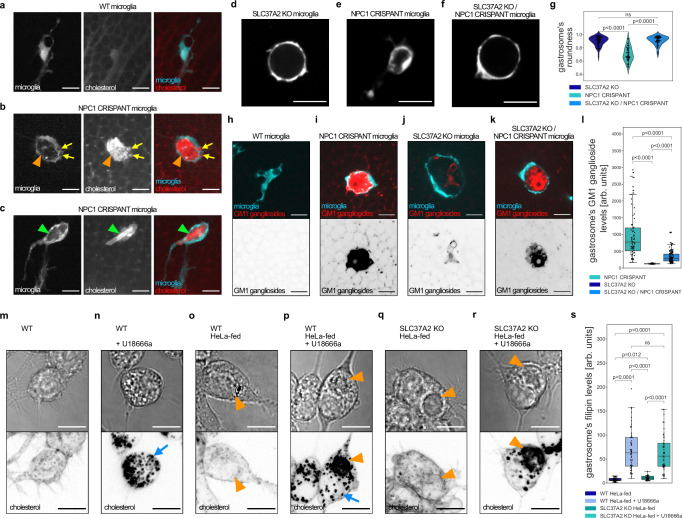
NPC1 loss leads to cholesterol and GM1 ganglioside accumulation in the gastrosome
The absence of NPC1 is known to result in the accumulation of unesterified cholesterol and gangliosides within endo-lysosomal compartments14. Embryos lacking NPC1 exhibited cholesterol accumulation (Supplementary Fig. 3a) as shown by using TopFluor cholesterol and large needle-shaped crystals, a recognized sign of cholesterol overload47, colocalizing with the microglia (Supplementary Fig. 3b). At higher magnification, these crystals were found to be inside the gastrosome (green arrowhead in Fig. 3c) that also contained high TopFluor cholesterol levels (orange arrowhead in Fig. 3b) and was surrounded by TopFluor cholesterol-rich small vesicles (yellow arrows in Fig. 3b), likely to be endo-lysosomal compartments11–14. The accumulation of cholesterol in the gastrosome is linked to efferocytosis. Indeed, knocking down BAI1, which reduces efferocytosis, resulted in a reduction in both the size of the gastrosome (Supplementary Fig. 2a) and its TopFluor cholesterol content (Supplementary Fig. 3c)46.
Fig. 3. Gastrosome in NPC1 deficient cells accumulates cholesterol and GM1 gangliosides.
a–c Representative images of OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)) with TopFluor cholesterol. Scale bar 10 µm. b Orange arrowhead indicates gastrosome and yellow arrows point to smaller cholesterol-rich vesicles. c Green arrowhead points to a visible cholesterol crystal. d–f Representative images of OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)). Scale bar 20 µm. g Roundness of the gastrosome in SLC37A2 KO (N = 3, n = 69), NPC1 CRISPANT (N = 4, n = 59) and NPC1 CRISPANT in SLC37A2 KO background (N = 4, n = 61) in OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)). N refers to the number of zebrafish embryos and n to the number of microglia examined. h–k, Representative images of OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)) fixed and stained for GM1 gangliosides. Scale bar 10 µm. l GM1 ganglioside levels in the gastrosome from NPC1 CRISPANT (N = 9, n = 68), SLC37A2 KO (N = 7, n = 69) and NPC1 CRISPANT in SLC37A2 KO background (N = 9, n = 120) fixed OT microglia. N refers to the number of zebrafish embryos and n to the number of microglia examined. m–r Representative images of RAW macrophages stained with filipin. Scale bar 10 µm. Blue arrows point to cholesterol-rich small vesicles while the orange arrowheads indicate the gastrosome. s Filipin levels in the gastrosome of RAW macrophages: WT HeLa-fed (n = 40), WT HeLa-fed and U18666a treated (n = 54), SLC37A2 KO HeLa-fed (n = 30), SLC37A2 KO HeLa-fed and U18666a treated (n = 50). n refers to the number of cells examined. Boxplots represent the median value and interquartile range; the ends of the whiskers correspond to the minimum and maximum values. Statistical tests: Mann-Whitney-Wilcoxon test two-sided with Bonferroni correction. ns without any additional p-value on the graph stands for p = 1. OT stands for optic tectum. Source data are provided as a Source Data file.
Next, we turned to RAW macrophages and treated these cells with U18666a to inhibit NPC1. As expected, 24 hours after treatment, filipin staining48 showed unesterified cholesterol accumulation in a puncta-like pattern, consistent with previous research (Fig. 3n, p, blue arrow)11–14. Interestingly, feeding U18666a-treated RAW macrophages with apoptotic HeLa cells led to a dramatic change in unesterified cholesterol distribution characterized by an additional buildup in the expanded gastrosome (Fig. 3p, indicated by an orange arrowhead). Lipidomic analysis further supported these data, showing that free (or unesterified) cholesterol and esterified cholesterol accumulated upon NPC1 inhibition in both RAW macrophages and HeLa-fed RAW macrophages (Supplementary Fig. 3d, Supplementary Table 1 and 2).
To test the involvement of the gastrosome in lipid handling we exploited the previous finding that the gastrosome expands when the Glucose-6-phosphate transporter SLC37A2 is absent2. In SLC37A2 mutants, the expanded gastrosome was characterized by a distinct bubble-like morphology (Fig. 3d, g2) and contained low GM1 ganglioside levels (Fig. 3j, l). In contrast, it appeared looser (Fig. 3e, g) with elevated GM1 ganglioside levels in the absence of NPC1 (Fig. 3i, l). Interestingly, in double NPC1 and SLC37A2 embryos, microglia were characterized by one expanded gastrosome with features of both single knockouts, namely a bubble-like shape (Fig. 3f, g) and accumulation of GM1 gangliosides (Fig. 3k, l), indicating that both SLC37A2 and NPC1 play distinct yet complementary roles in regulating gastrosome morphology and handling of sugars and lipids. Similarly, HeLa-fed RAW macrophages mutant for SLC37A2 showed increased cholesterol levels in the gastrosome after inhibition of NPC1 (Fig. 3q, r, s) highlighting a potential role for this compartment in NPC1-mediated cholesterol handling.
Interestingly, real-time imaging revealed normal phagosomal behavior and shrinkage in NPC1 deficient microglia (Supplementary Fig. 4a, b, c, Supplementary Movie 4), supporting the hypothesis of NPC1 might exert its function downstream of phagosomal shrinkage at the level of the gastrosome.
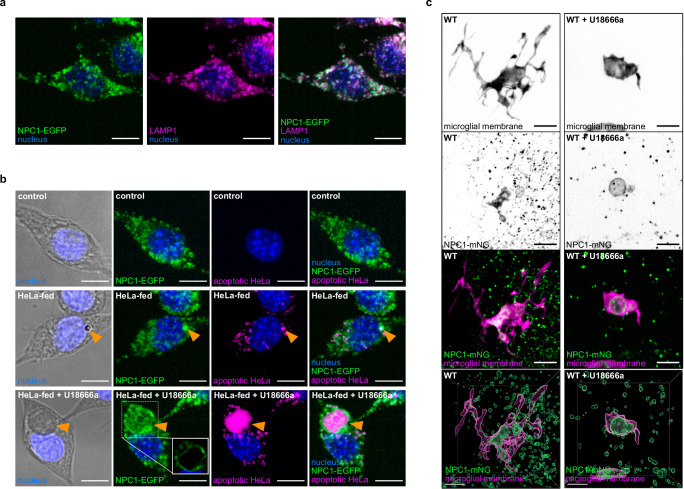
Next, we utilized a CRISPR-mediated approach to tag the endogenous NPC1 protein in both zebrafish and RAW macrophages to investigate its expression and localization on the gastrosome. As expected NPC1 is expressed in RAW macrophages and localized on small vesicles that are also LAMP1 positive (Fig. 4a49). Feeding these RAW macrophages with apoptotic HeLa cells revealed localization of NPC1 also on the gastrosome (Fig. 4b, orange arrowhead) which expanded upon NPC1 inhibition using U18666a (Fig. 4b, orange arrowhead). In line with this, the endogenous tagging of NPC1 in zebrafish revealed expression in microglia and localization of this transporter on the gastrosome, which expanded when embryos were treated with U18666a (Fig. 4c). Additionally, NPC1 was observed on smaller vesicles likely of endo-lysosomal nature (Fig. 4c and Supplementary Fig. 4d).
Fig. 4. NPC1 localizes on the gastrosome that accumulates apoptotic material.
a Representative image of RAW NPC1-EGFP macrophages stained for LAMP1 (N = 2, more than 4 cells examined per condition in each repetition). Scale bar 10 µm. b Representative images of RAW NPC1-EGFP macrophages from in vitro feeding experiment. Gastrosome indicated by orange arrowhead. For HeLa-fed U18666a treated example, single plane view shown in a box for NPC1-EGFP (N = 2, more than 4 cells examined per condition in each repetition). Scale bar 10 µm. c, Representative images of OT (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry; npc1:mNeonGreen)) zebrafish microglia showing localization of NPC1 displayed as maximum projection and segmentation (N = 4). Scale bar 10 µm. N refers to the number of experimental replicates. OT stands for optic tectum.
NPC1 impact on the gastrosome is conserved in Niemann-Pick type C human patient-derived cells
We next asked if the mechanism linking NPC1 to changes in gastrosomal morphology was conserved in human Niemann-Pick type C syndrome. We began by first establishing the impact of NPC1 inhibition on the gastrosome in human microglia, using the cell line HMC350. Feeding these cells fluorescent apoptotic HeLa cells resulted in the accumulation of the labeled material in a compartment that had defining hallmarks of the gastrosome (Supplementary Fig. 5a, c). In addition, this compartment contained high cholesterol levels (Supplementary Fig. 5d) and expanded upon U18666a treatment (Supplementary Fig. 5b), pointing to the high conservation of cholesterol sorting mechanisms in human microglia.
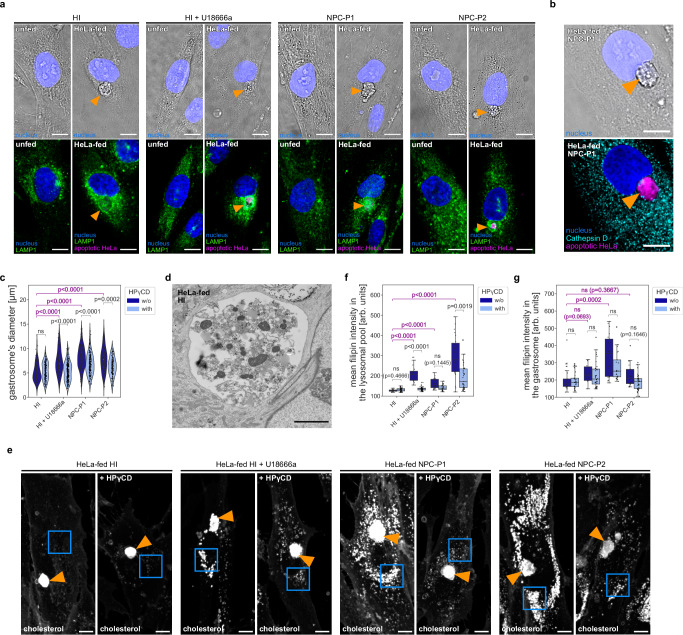
Next, we obtained two NPC human patient-derived fibroblast lines with different mutations in NPC1 (GM18393, referred to as NPC-P1 and GM17919, referred to as NPC-P2) and one healthy individual control (GM03652, referred to as HI). As the size of the gastrosome is coupled to efferocytosis, we first determined the ability of fibroblasts to engulf apoptotic cells. Twenty-four hours after exposure to fluorescently labeled apoptotic HeLa cells, we observed the accumulation of fluorescence in the gastrosome, as defined using standard criteria (Fig. 5a–d). A conspicuous ultrastructural difference between the gastrosome in immune cells and fibroblasts was the elevated content within the lumen of the fibroblast gastrosome, suggesting that fibroblasts exhibit a lower efficiency in digesting apoptotic material compared to professional phagocytes such as microglia and macrophages (Fig. 5d compare with Fig. 2g, h). Nevertheless, the size of the gastrosome was increased in HeLa-fed NPC-P1 and NPC-P2 cells when compared to HI control cells (Fig. 5a, c, dark blue violin plot), as was the accumulation of cholesterol in the gastrosome (Fig. 5e, indicated by an orange arrowhead and Fig. 5g, dark blue box plot) and in the endo-lysosomal pool (Fig. 5e, marked by blue box and Fig. 5f, dark blue box plot). Both features of NPC patient derived cells could be recapitulated in HI fibroblasts treated with U18666a, supporting the conclusion that these gastrosomal changes in patient-derived cells are due to reduced NPC1 activity (Fig. 5a, c, e, f, g, dark blue plots).
Fig. 5. Patient-derived fibroblasts accumulate apoptotic material in the gastrosome that is larger in NPC deficient cases.
a Representative images of human patients’ fibroblasts stained for LAMP1. The gastrosome is indicated with an orange arrowhead. Scale bar 10 µm. b Representative image of HeLa-fed human patients’ fibroblasts stained for Cathepsin D. The gastrosome is indicated with an orange arrowhead. Scale bar 10 µm. c Diameter of the gastrosome in HeLa-fed human patients’ fibroblasts: HI (n = 114), HI treated with HPγCD (n = 113), HI treated with U18666a (n = 126), HI treated with U18666a and HPγCD (n = 176), NPC-P1 (n = 156), NPC-P1 treated with HPγCD (n = 146), NPC-P2 (n = 113), NPC-P2 treated with HPγCD (n = 94). d Representative EM image of the gastrosome in HeLa-fed human patients’ fibroblast. Scale bar 3 µm. e Representative images of HeLa-fed human patients’ fibroblast stained with filipin. The gastrosome is indicated with an orange arrowhead and lysosomal pools are framed in blue squares. Scale bar 10 µm. f, g Mean intensities of the filipin staining from HeLa-fed human patients’ fibroblasts: HI (n = 25), HI treated with HPγCD (n = 30), HI treated with U18666a (n = 16), HI treated with U18666a and HPγCD (n = 32), NPC-P1 (n = 21), NPC-P1 treated with HPγCD (n = 21), NPC-P2 (n = 23), NPC-P2 treated with HPγCD (n = 33) in the lysosomal pool (framed in blue squares in Fig. 5e) (f), in the gastrosome (indicated with an orange arrowhead in Fig. 5e) (g). n refers to the number of cells examined. Boxplots represent the median value and interquartile range; the ends of the whiskers correspond to the minimum and maximum values. Statistical tests: Mann-Whitney-Wilcoxon test two-sided with Bonferroni correction. ns without any additional p-value on the graph stands for p = 1. HI stands for healthy individual. NPC1-P1 corresponds to GM18393 NPC patient and NPC1-P2 corresponds to GM17919 NPC patient. Source data are provided as a Source Data file.
To determine if the gastrosomal expansion observed in HeLa-fed patient fibroblasts is due to cholesterol trafficking defects, we conducted experiments using 2-hydroxypropyl-γ-cyclodextrin (HPγCD)51,52. This drug has shown efficacy in reducing cholesterol accumulation in fibroblasts from NPC patients by activating the transcription factor EB (TFEB), boosting cholesterol transport to the endoplasmic reticulum and autophagy53. Incubation of HeLa-fed HI fibroblasts with HPγCD did not affect gastrosomal size (Fig. 5c, light blue violin plot) nor cholesterol levels (Fig. 5e, f, g, light blue box plots). In contrast, NPC1 deficient fibroblast lines exhibited a gastrosomal size reduction (Fig. 5c, light blue violin plot), decreased cholesterol levels in the endo-lysosomal pool (Fig. 5e, marked by blue box and Fig. 5f, light blue box plot), and a noticeable downward trend of cholesterol levels in the gastrosome (Fig. 5e, indicated by an orange arrowhead and Fig. 5g, light blue box plot). While we cannot rule out the possibility that the impact of HPγCD on the gastrosome is indirect and potentially mediated by its influence on upstream components of the phagocytic pathway, such as lysosomes, the modifications observed in gastrosome size and content resulting from HPγCD strongly suggest a substantial involvement of this vesicle in lipid metabolism. These observations could offer mechanistic explanations for the poorly understood late-onset phenotypes observed in NPC and other lysosomal storage disorders, where cells have been shown to expand and form vacuoles (for review see ref. 54).
Changes in the gastrosome and increased cholesterol accumulation underlie NPC1’s impact on microglial morphology
Next, we investigated the impact of NPC1 activity on the gastrosome and its influence on microglial morphology and function. The transition from branched to ameboid morphology in microglia caused by NPC1 inhibition via U18666a (Fig. 1f, g, i) could be reversed by changing the medium and rinsing off the NPC1 inhibitor. This led to deflation of the gastrosome and microglia to regain their branched morphology within a few hours (Supplementary Fig. 6a, b–d and Supplementary Movie 5 and 6). This suggests a direct link between NPC1 activity and the physical characteristics of the gastrosome as well as microglial shape. Quantifying gastrosome size, cholesterol content, and microglial morphology before (T0) and after (T1) washing off the drug further validated this relationship (Fig. 6a–e). Resumed NPC1 activity led to reduced cholesterol levels in the gastrosome, decreased vesicle size, and the recovery of the microglial morphology, indicating a causal connection between these features. As upon NPC1 inhibition, the gastrosome becomes a significant site of cholesterol accumulation with a dramatic effect on microglial morphology, we wondered if boosting cholesterol trafficking in NPC1-deficient microglia using HPγCD would lead to improved microglial phenotypes. First, treating wild-type embryos with HPγCD had no effect on cell morphology (Fig. 6f, h), however, HPγCD treatment of NPC1 deficient embryos reduced the size of the gastrosome (Fig. 6g, i) and the severity of the ameboid microglia phenotype (Fig. 6g, h), supporting our hypothesis that the ameboid microglial phenotype observed in phagocytic NPC deficient cells is linked to cholesterol buildup and expansion of the gastrosome.
Fig. 6. NPC deficient microglia are sensitive to elevated neuronal cell death.
a Representative images from U18666a treated (T0) OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)) and from wash out (T1) with TopFluor cholesterol. Scale bar 10 µm. b–e Measurements from control T0 (N = 5, n = 76), control T1 (N = 5, n = 76), U18666a treated T0 (N = 5, n = 71) and U18666a wash out T0 (N = 5, n = 71) zebrafish with TopFluor cholesterol of OT microglia circularity (b), quantification of the largest vesicle’s diameter (c), quantification of the largest’s vesicle cholesterol levels (d) and its changes per individual zebrafish brains (e). f, g Representative images of zebrafish OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)) from HPγCD experiment. Scale bar 50 µm. Examples of single microglia in boxes. Scale bar 10 µm. h, i Measurements from wild-type (N = 10, n = 201), wild-type treated with HPγCD (N = 10, n = 191), NPC1 CRISPANT (N = 18, n = 291) and NPC1 CRISPANT treated with HPγCD (N = 18, n = 291) zebrafish of OT microglia circularity (h) and quantification of the largest vesicle’s diameter (i). j Representative images of OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)) from Camptothecin (+ CPT) experiment. Scale bar 10 µm. k Circularity of OT microglia from wild-type (N = 10, n = 198), wild-type treated with CPT (N = 10, n = 123), NPC1 CRISPANT (N = 8, n = 84) and NPC1 CRISPANT treated with CPT (N = 15, n = 95) zebrafish. l Quantification of the largest vesicle’s diameter in wild-type (N = 10, n = 218), wild-type treated with CPT (N = 10, n = 221), NPC1 CRISPANT (N = 8, n = 80) and NPC1 CRISPANT treated with CPT (N = 9, n = 66) zebrafish OT microglia. m Number of OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)) in wild-type (N = 23), wild-type treated with CPT (N = 31), NPC1 CRISPANT (N = 16), NPC1 CRISPANT treated with CPT (N = 15) zebrafish. n Representative images of OT microglia (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)) treated with CPT. Left side of Supplementary Movie 7. Dying cell and its subsequent engulfment indicated by orange arrowhead. Scale bar 40 µm. N refers to the number of zebrafish embryos and n to the number of microglia examined. Boxplots represent the median value and interquartile range; the ends of the whiskers correspond to the minimum and maximum values. Statistical tests: Mann-Whitney-Wilcoxon test two-sided with Bonferroni correction. ns without any additional p-value on the graph stands for p = 1. OT stands for optic tectum and CPT stands for Camptothecin. Source data are provided as a Source Data file.
Next, we tested how NPC1 deficient microglia respond to challenges, particularly heightened neuronal death, a characteristic of later stages of NPC disease27–30. To increase neuronal cell death, we used Camptothecin (CPT), an established inducer of neuronal apoptosis44,2. Wild-type microglia responded by adopting a more ameboid morphology (Fig. 6j, k and Supplementary Fig. 6e). Notably, CPT administration to NPC1-deficient microglia intensified the ameboid morphology (Fig. 6j, k and Supplementary Fig. 6e), further strengthening the link between NPC1 loss and the rate of neuronal apoptosis on microglial morphology. The experiments also revealed a significant increase in the size of the gastrosome (Fig. 6l), especially notable in NPC1-deficient microglia, indicating a synergistic effect between NPC1 loss and increased phagocytosis on gastrosomal expansion. We next leveraged our ability to monitor microglial dynamics in intact living zebrafish brains to investigate the subsequent impact of high neuronal cell death and NPC1 deficiency on microglia. While CPT treatment alone does not affect microglial viability in wild-type, in NPC1 deficient embryos, CPT-induced neuronal apoptosis led to a further reduction in the already low number of microglia compared to wild-type (Fig. 6m). Live imaging of CPT-treated NPC1 deficient embryos revealed that microglial reductions can be explained by the death of these cells, which are subsequently cannibalized via efferocytosis by surrounding microglia (Fig. 6n, Supplementary Fig. 6h, Supplementary Movie 7). We tested whether the death of NPC1-deficient microglia depends on efferocytosis by reducing neuronal uptake via BAI1 knockdown, which led to increased microglial survival (Supplementary Fig. 6g). Similarly, improving cholesterol trafficking using HPγCD showed a trend toward fewer microglia dying (Supplementary Fig. 6f). Thus, the loss of NPC1 renders microglia highly sensitive to neuronal cell death. In response to high neuronal apoptotic levels, phagocytic microglia die and are eliminated by other microglia. This finding can have implications for understanding microglial responses in NPC and other neurodegenerative diseases.
Discussion
In the absence of the cholesterol transporter NPC1, microglia are known to undergo a branched-to-ameboid transition. This investigation unveils the role of the gastrosome in linking neuronal cell death and cholesterol transport defects with morphological changes in microglia in NPC disease. While not excluding the potential roles of NPC1 in other endo-lysosomal system compartments, this work uncovers a function for NPC1 in regulating cholesterol trafficking at the level of the gastrosome during efferocytosis. The transmembrane transporter NPC1 localizes to the gastrosome, acting as a player in preventing cholesterol accumulation within this compartment downstream of neuronal engulfment. Furthermore, it shows the exquisite sensitivity of the gastrosomal shape and size to alterations in lipid transport. The expansion of the gastrosome in the absence of NPC1’s activity coincides with microglia becoming ameboid, but when these cells regain NPC1 function in U18666a wash-out experiments, the gastrosome undergoes a reduction in size and the microglia re-establish their complex, branched morphology. This suggests the existence of a link between the gastrosome and microglial morphology. As cholesterol serves as a fundamental component of the plasma membrane, crucial for maintaining its structural integrity and fluidity (for review see ref. 55), NPC1-mediated cholesterol transport at the level of the gastrosome might enable microglia to form branches and sustain a high rate of neuronal engulfment. Therefore, understanding the signaling pathways and molecular interactions downstream of NPC1 activity becomes very important. Further research into these pathways could provide valuable insights into how cholesterol affects microglia and their functions, elucidating the broader implications of cholesterol dysregulation in neurodegenerative diseases and other conditions associated with defective neuronal clearance.
Interestingly, we show that in response to elevated neuronal apoptosis, phagocytic NPC1 deficient microglia undergo cell death and are subsequently cleared by other microglia. This finding has implications for understanding microglial responses in NPC as the death of these cells could effectively reduce the brain’s ability to clear dead neurons, leading to the accumulation of neuronal corpses and increased inflammation. Several mechanisms could be responsible for the death of microglia in this context. One is the presence of large cholesterol crystals in these cells (Fig. 3c and Supplementary Fig. 3b), which have been shown to damage cells by physically disrupting their integrity56,57. Alternatively, microglia could undergo pyroptotic death, as suggested by high IL1 expression observed in NPC1 deficient embryos58.
The discovery of the gastrosome and its central role in lipid handling post-engulfment brings to light a critical aspect of efferocytosis, namely the collection of apoptotic material into a single compartment. This suggests a coordinated strategy in dealing with engulfed cellular material, potentially specialized for certain molecules like lipids. On the other hand, this compartment might also serve a more general function handling a wider range of different products resulting from the breakdown of apoptotic material. Indeed, while we have demonstrated that the gastrosome contains lipids whose transport depends on NPC1, this compartment may also contain additional important molecules. In this context, it is crucial to understand the role of the gastrosome in relation to other key components of the phagocytic pathway, such as, lysosomes and LAMP1-labeled non-degradative compartments that lack lysosomal hydrolases and have been identified in neurons59. It is well-established that the vesicular components of the phagocytic pathway undergo constant fusion-fission events and share common molecular markers and structural features. However, the gastrosome stands out because it is a single compartment characterized by electron-lucent properties while lysosomes, telolysosomes, and other non-degradative vesicles are typically numerous, small, and electron-dense60. This distinction adds depth to our understanding of intracellular processing and lysosomal storage disorders. The lack of Cathepsin D observed in the gastrosome strongly supports a scenario in which this compartment is non degradative fulfilling a storage function, which will be further explored.
Efferocytosis-dependent collection of cholesterol into one compartment is also found in human non-myeloid cells, such as fibroblasts, demonstrating the remarkable conservation of this compartment and its function across different cell types in vertebrates. This collecting function of the gastrosome is also reminiscent of similar processes occurring in single-celled organisms. For example, in malaria plasmodium, where small hemoglobin digestive compartments fuse, creating a single food vacuole with known storage and transport functions (for review see ref. 61). Hence, the gastrosome might be compared to a food vacuole seen in single-celled organisms, emerging as an optimal hub for lipid sensing and trafficking within phagocytic cells.
Finally, this study highlights the importance of NPC1 in phagocytic microglia, as interfering with its function under phagocytic stress expands the gastrosome affecting microglial morphology and viability. In NPC disease, microglia are exposed to increasingly high levels of neuronal cell death, suggesting that NPC1’s localization on the gastrosome could be relevant for understanding this disease and developing treatments.
Methods
Animal handling
Zebrafish (Danio rerio) were raised, maintained, and bred according to standard procedures as described in “Zebrafish – A practical approach” (Nüsslein-Volhard, 2012). All experiments were performed on embryos younger than 5dpf, in accordance with the European Union Directive 2010/62/EU and local authorities (Kantonales Veterinäramt; Fishroom licence TVHa Nr. 178). Live embryos were kept in E3 buffer at 28–30 °C until the desired developmental stage. Pigmentation was prevented during experiments by treating the embryos with 0.003% N-phenylthiourea (PTU) (Sigma-Aldrich, St. Louis, US-MO, #P7629) from 1dpf onwards. Embryos were anaesthetized during mounting procedures and experiments using 0.01% tricaine (Sigma-Aldrich, #A5040).
Fish lines
The following transgenic animals were used in this study: (Tg(mpeg1.1:EGFP-CAAX))2, (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry))32, (Tg(NBT:DLexPR::NTR-mCherry)46.
The following mutant lines were used in this study: slc37a2NY007 2, irf8st95 41, npc1hg37 37.
Cell lines
RAW 264.7 mouse macrophages (ATCC) and HeLa Kyoto H2B-mCherry cells (Mitocheck consortium, EMBL) were cultured as previously described2, in DMEM (Gibco) plus 10% FBS (Gibco) and in DMEM plus 1% Penicillin/Streptomycin (Gibco) and 1% GlutMAX (Gibco) respectively. Human microglia cell line 3 (HMC3) was obtained from ATCC and cultured in Minimum Essential Medium (MEM) (Gibco) supplemented with 1% sodium pyruvate (Gibco) and 10% FBS (Gibco). Human patients’ fibroblasts (GM03652, GM18393, GM17919) were obtained from the NHGRI Sample Repository for Human Genetic Research at the Coriell Institute for Medical Research (informed consent obtained by the provider) and cultured in Minimum Essential Medium (MEM) with NEAA (Gibco) supplemented with 1% GlutMAX (Gibco) and 15% FBS (Gibco).
CRISPR Knock-out in zebrafish
gRNAs (NPC1: 5’-GTATCTGGTACGGCGAATGT-3’, 5’-ATAGGTCACATTCCCCACAC-3’; NPC2: 5’-GGACTACAGAGTGCTCGGCG-3’, 5’-GGTAGACGGAAAAGTAGTTC-3’) were designed using the CRISPR/Cas9 target online predictor CHOPCHOP and ordered from IDT as crRNA. crRNA was mixed in a 1:1 ratio with tracrRNA (IDT) and incubated at 95 °C for 5 min to obtain a 10 µM gRNA solution. gRNAs for either NPC1 or NPC2 were mixed in a 1:1 ratio and incubated at 37 °C for 20 min to form a RNP complex with in-house Cas9 protein (Protein Expression and Purification Facility, EMBL, Heidelberg, Germany) and injected into one cell stage zebrafish eggs. Embryos were raised to 4 days post fertilization and experiments were performed. Afterwards, genomic DNA was extracted with QuickExtract™ DNA Extraction Solution (Lucigen) to confirm genomic deletions by PCR (primers NPC1: 5’-TGGCTGCCAATGGTTACAGG-3’, 5’-ACCATCCAAACTAAGCGTATAGCA-3’; primers NPC2: 5’-AGTGTACGCATATTCGCTGT-3’, 5’-TAAAGCAACTCACCACTGCT-3’).
CRISPR Knock-out in RAW 264.7 mouse macrophages
gRNAs targeting the fifth and seventh exon of SLC37A2 were designed using the CRISPR/Cas9 target online predictor CHOPCHOP. The template for the sgRNAs (5’-TTACTACCTCTCGGCTGGAA-3’, 5’-TTCAGGCGGGGATTCATCAT-3’) were produced by designing primers containing the T7 promotor. Amplicons were transcribed using the MEGAshortscript T7 transcription kit (Ambion). RAW 264.7 mouse macrophages were transfected using Viromer CRISPR kit (Lipocalyx) with gRNAs and in-house Cas9 protein (Protein Expression and Purification Facility, EMBL, Heidelberg, Germany) in a 1:1 ratio. Cells were diluted and seeded as single cells to obtain monoclonal lines. Successful SLC37A2 targeting was confirmed by PCR and scoring for the expected phenotype by imaging cells fed with apoptotic material. CRISPR/Cas9-based editing generated a trans-heterozygous line characterized by several deletions in exons 5 (5 bp) and 7 (3 and 117 bp).
CRISPR Knock-in in zebrafish
A sgRNA (5’-ACGCTAAAATTGCTGCATCT-3’) was designed using the CRISPR/Cas9 target online predictor tool CRISPR scan (https://www.crisprscan.org) and ordered from IDT as crRNA. crRNA was mixed in a 1:1 ratio with tracrRNA (IDT) and incubated at 95 °C for 5 min to obtain 10 μM gRNA solution. NPC1-mNG PCR fragment was used as the homology recombination template. gRNA was mixed with PCR fragment and in-house Cas9 protein (Protein Expression and Purification Facility, EMBL, Heidelberg, Germany). The mix was incubated at 37 °C for 20 min to form a RNP complex that was injected into zebrafish eggs at the one cell stage. Embryos were raised to 3dpf and screened on a fluorescence microscope. Experiments were performed at 4dpf.
CRISPR Knock-in in RAW 264.7 mouse macrophages
sgRNAs (LAMP1: 5’-CATGTTGATCCCCATTGCTG-3’; NPC1: 5’-CCACTTACGAGCGCTACAGA-3’) were designed using the CRISPR/Cas9 target online predictor CHOPCHOP and ordered from IDT as crRNA. crRNA was mixed in a 1:1 ratio with tracrRNA (IDT) and incubated at 95 °C for 5 min to obtain a 100 μM gRNA solution. LAMP1-EGFP or NPC1-EGFP PCR fragments were used as homology recombination templates. RAW 264.7 mouse macrophages were transfected using Cell Line Nucleofector™ Kit V (Lonza) with gRNA, PCR fragment and HiFi Cas9 protein (IDT). Cells were sorted as single cells to obtain EGFP positive monoclonal lines. Successful endogenous tagging was confirmed by flow cytometry, light microscopy, PCRs and sequencing.
Morpholino knockdown
To reduce phagocytosis of the optic tectum microglia BAI1 morpholino (CTAGAACTCTAACACACTTACTCAT) was injected into the yolk of zebrafish eggs at the one cell stage as previously described46. Embryos were raised to 4dpf, mounted and imaged.
in vitro phagocytosis assay
HeLa Kyoto H2B-mCherry were seeded on a 60 mm dish and left until adhered. Cells were exposed to UV light (150mJ/cm2) using Stratalinker 1800 and left for at least 8 h in the incubator (5% CO2, 37 °C). Apoptotic cells were collected, centrifuged, 4.2 ml of the supernatant was removed and cells were resuspended in the remaining 800 μl of the supernatant. Concentrated apoptotic cells were added on separately seeded phagocytic cells (either RAW 264.7 mouse macrophages, human microglia HMC3 or human patients’ fibroblasts) and phagocytosis was allowed for the next 24 h before fixation. Such samples are referred to as HeLa-fed. Consistent with previous work on the gastrosome2, HeLa cells were used because they stably express fluorescent proteins that allow for tracking apoptotic cargo when these cells undergo apoptosis and are engulfed by phagocytes.
Chemical perturbations
U18666a
4dpf zebrafish embryos were incubated in 200 μM U18666a (Sigma-Aldrich, #U3633) solution for 6 h, or 3dpf zebrafish embryos were incubated in 100 μM U18666a solution overnight at RT. Embryos were mounted and imaged in the drug. For wash out experiments, the drug was removed from the medium before imaging.
RAW 264.7 mouse macrophages and human microglia HMC3 or human patients’ fibroblasts were incubated with 7 μM and 50 μM U18666a, respectively, in the growth medium for 24 h before fixation.
Camptothecin
To induce higher levels of apoptosis in zebrafish embryos, Camptothecin (CPT) (Sigma-Aldrich, #C9911) was used. Embryos were placed in 1 μM CPT solution in E3 medium with 1% (v/v) DMSO for 7-8 h before mounting and imaging.
Metronidazole
To induce higher levels of apoptosis specifically in the hindbrain, Tg(NBT:DLexPR::NTR-mCherry) zebrafish embryos were incubated with 10 mM MTZ (Sigma, #M3761) solution in E3 medium with 0.2% (v/v) DMSO for 24 h. Embryos were washed extensively with E3 before mounting and imaging.
2-hydroxypropyl-γ-cyclodextrin (HPγCD)
To prevent the NPC1 deficient microglia phenotype, 2-hydroxypropyl-γ-cyclodextrin (HPγCD) (Sigma-Aldrich, #H125) was used. 2.5dpf zebrafish embryos were placed in 1 M HPγCD in E3 medium for 24 h before mounting and imaging.
Human patients’ fibroblasts were incubated with 100 mM HPγCD in the growth medium for 24 h before fixation.
TopFluor cholesterol injections in zebrafish
To label cholesterol, TopFluor cholesterol (Avanti Polar Lipids, #810255 P) was injected into the yolk of zebrafish eggs at the one cell stage as previously described in ref. 37. Embryos were raised to 4dpf, mounted and imaged.
Acridine orange staining in zebrafish
To visualize apoptotic cells in vivo, embryos were stained with acridine orange (Sigma-Aldrich, #A6014). The larvae were incubated in the dark for 1 h at 28 °C in 10 μg/ml acridine orange solution in E3 and washed extensively before imaging.
GM1 ganglioside staining in zebrafish
To visualize GM1 gangliosides, a group of glycophospholipids, Cholera Toxin subunit B (CtxB recombinant) Alexa Fluor 488 conjugate (Invitrogen, #C34775) staining was performed. 4dpf zebrafish embryos were fixed in 4% PFA for 1 h at RT, permeabilized in acetone for 20 min at −20 °C, washed in 0.1% Triton-PBS and incubated in blocking buffer (1% BSA / 5% normal goat serum / 1% Triton-X-100 / PBS) for 1 hour at RT. Afterwards, larvae were placed in 10 μg/ml CtxB solution in a blocking buffer overnight at 4 °C. Embryos were washed extensively in 0.1% Triton-PBS before mounting and imaging.
Filipin staining
Cells were fixed for 10 min with 4% PFA, washed three times with 1xPBS and stained with filipin (Sigma-Aldrich, #SAE0088) for 2 h at 0.05 mg/ml final concentration in the dark. Afterwards cells were washed three times with PBS and imaged directly.
Immunohistochemistry in cell culture
Cells were fixed for 10 min in 4% PFA solution, washed with PBS and permeabilized for 7 min with 0.2% Triton-X-100/PBS solution. After 1 h of blocking solution (3% BSA / 0.2% Triton-X-100 / PBS), cells were incubated with primary antibody in blocking solution overnight at 4 °C: LAMP1 (Abcam, 1:100, ab25245, anti-mouse), LAMP1 (Abcam, 1:1000, ab24170, anti-human), Cathepsin D (Abcam, 1:100, ab75852, anti-mouse), Cathepsin D (Abcam, 1:200, ab72915, anti-human), LC3B (Novus Biologicals, 1:200, nb100-2220, anti-mouse). Cells were washed three times with 1xPBS and incubated for 1 h with fluorescently labeled secondary antibody in blocking solution at RT (1:500 or 1:1000, Thermo Fisher Scientific, Abcam). Cells were mounted using a Vectashield Kit (Vector Laboratories, Inc., Burlingame, CA) with DAPI or stained with 300 nM DAPI for 5 min and directly imaged in PBS.
Immunohistochemistry in zebrafish
Embryos at 4dpf were fixed in 4% PFA and processed for whole-mount antibody staining as previously described in ref. 62. The primary mouse 4C4 monoclonal antibody (gift from Valérie Wittamer laboratory, 1:500) and secondary Alexa Fluor 647-conjugated anti-mouse (Abcam, 1:500, ab150107) antibodies were used.
Western Blot and protein extraction in zebrafish
50 4dpf wild-type and 50 4dpf NPC1 CRISPANT zebrafish embryos were anesthetized in tricaine and placed in 170 μl of RIPA buffer supplemented with protease inhibitor cocktail cOmplete EDTA-free tablets (Roche, #11836170001). After rotating for 30 min at 4 °C, samples were sonicated (3 x 5s) with Bandelin Electronic Sonoplus HD 2070 Homogenisator (Bandelin) and lysates were cleared by centrifugation. Samples were mixed with 4x NuPAGE Lithium Dodecyl Sulphate (LDS) buffer and electrophoresed on a 4–12% Bis-Tris NuPAGE gel (Invitrogen). Proteins were transferred to nitrocellulose membrane (Invitrogen) using XCell II Blot module (Invitrogen). After blocking with 5% milk saturation buffer, blots were incubated overnight at 4 °C with anti-NPC1 primary antibody (Abcam, 1:1000, ab134113) and for 1 h at RT with anti-rabbit secondary antibody HRP-conjugated (Promega, 1:2500, W4018). Proteins were revealed using Immobilion Western Detection System Kit (Millipore) and Amersham Imager 600 (GE Healthcare Life Sciences). Afterwards blots were stripped (Thermo Scientific, Restore™ PLUS Western Blot Stripping Buffer), re-saturated, stained again using anti-γTubulin primary antibody (Sigma-Aldrich, 1:2000, T6557) and anti-mouse secondary antibody HRP-conjugated (Promega, 1:2500, W4028).
qPCR and RNA extraction in zebrafish
For each condition 50 4.5dpf zebrafish embryos were anesthetized with tricaine, heads were cut off and placed in 300 μl of Trizol (Invitrogen, #15596026). Samples were sonicated (3x5s) with Bandelin Electronic Sonoplus HD 2070 Homogenisator (Bandelin) and lysates were cleared by centrifugation. After the addition of 60 μl of chloroform, samples were vortexed and centrifuged at 13,000 x g for 15 min in 4 °C. Supernatants were transferred to new tubes, and RNA extraction was continued using PureLink RNA mini Kit (Invitrogen, #12183018 A) with on-column PureLink DNase treatment (Invitrogen, #12185-010). The RNA template from three independent experiments for each condition was used for cDNA synthesis using the SuperScript IV Reverse Transcriptase (Invitrogen, #18090010). Gene expression levels were analyzed using the CFX96 Real-Time System (Bio-Rad) and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, #1725270). The data were analyzed based on ref. 63 using three reference genes (β-Actin (ACTB)64, Ribosomal Protein L13a (RPL13A)65, TATA-Box Binding Protein (TBP)). For inflammation analysis three markers were used: Interleukin-6 (IL6)40, Nuclear factor NF-kappa-B subunit 2 (NFΚB2) and Interleukin-1 (IL1)38. Table 1 contains primers used in the analysis.
Table 1.
List of primers used for gene expression analysis
| gene | Forward primer (5’−3’) | Reverse primer (5’−3’) |
|---|---|---|
| ACTB | CACTGAGGCTCCCCTGAATCCC | CGTACAGAGAGAGCACAGCCTGG |
| RPL13A | TCTGGAGGACTGTAAGAGGTATGC | AGACGCACAATCTTGAGAGCAG |
| TBP | ACAACAGCCTACCTCCTTTCG | CGTCCCATACGGCATCATAGG |
| IL6 | TCAACTTCTCCAGCGTGATG | TCTTTCCCTCTTTTCCTCCTG |
| NFKΒ2 | ACATCTCTGCTCCATGCT | GCAGTGAACTTGCTGAACCA |
| IL1 | TGGACTTCGCAGCACAAAAT | GTTCACTTCACGCTCTTGGATG |
Lipidomic analysis
Lipid extraction, lipidomics, and data analysis were adapted from ref. 66 and performed at the Functional Genomic Center Zurich, ETH Zurich and University of Zurich.
Briefly, each sample (control RAW, NPC1 inhibitor treated RAW, HeLa-fed RAW, HeLa-fed and NPC1 inhibitor treated RAW) was re-suspended in 100 μl of isopropanol / methanol (1:1) by shaking 20 min, 20 °C, 100 x g. Samples were centrifuged (16,000 x g, 10 min, 20 °C) and 70 μL of the supernatant was transferred into glass vial for LC-MS. The QCpool consisted of 10 μL of each sample. For the LC method a Waters Premier BEH C18 column (50 mm × 2.1 mm) was employed at 60 °C, paired with a Thermo Vanquish Horizon Binary Pump. A 2 µL sample was used, with a flow rate of 1 mL for 7.5 min. The gradient transitioned from 15% Buffer B to 99% Buffer B (Buffer A: 60% acetonitrile, 40% H2O, 5 mm NH4acetate; Buffer B: 90% isopropanol, 10% acetonitrile, 5 mm NH4acetate). For the MS method a Thermo Q Exactive HF DDA top5 with two different resolutions (MS1 resolution 60‘000; MS2 resolution 15‘000) was used. The stepped normalized collision energy was 10, 20, and 30. Data analysis was conducted using the Thermo Compound Discoverer software 3.3. Lipid identification was achieved by matching to lipidblast library, followed by visual inspection of the mirror plots (library vs data) to confirm the match and the presence of diagnostic fragments according to the lipids standard initiative guidelines. Only lipids that matched the expected fragmentation spectra were included in the analysis. For identification of cholesterol and its derivatives see Supplementary Table 1.
Light microscopy of fixed cells
Confocal analysis of fixed cells was performed with an Andor Dragonfly 200 Sona spinning-disc microscope with a Nikon 40x/NA 1.25 silicon oil objective and 40 μm spinning disc to capture 15–20 μm thick stacks with a z-step of 0.2 μm.
HMC3 and human patients’ fibroblasts in vitro feeding screens were imaged on a GE IN Cell Analyzer 2500HS microscope with Nikon 40x/NA 0.95 air objective to capture 300 randomized areas per sample.
All images were analyzed in Fiji67.
Light microscopy of live and fixed zebrafish
Zebrafish embryos were anaesthetized with tricaine and pre-selected based on the expression of the desired fluorophore, using a Nikon ZMZ18 fluorescent stereoscope. Embryos were embedded in 1–1.5% low-melting (LM) agarose (PeqGOLD Low Melt Agarose, PeqLab Biotechnologie GmbH), dissolved in E3 medium with tricaine. Embryos were mounted on glass-bottom dishes (Greiner Bio-One, #627871) for confocal microscopy or pulled together with agar, into glass capillaries (Brand, #701904) with a rod (Brand, #701932), and then pushed halfway out, into the microscopy chamber for light-sheet microscopy. Both the imaging dishes and the microscopy chamber were filled with E3 medium containing PTU and tricaine during the entire imaging period. For confocal microscopy, an Andor Dragonfly 200 Sona spinning-disc microscope with a Nikon 20x/NA 0.95 WI objective or Nikon 40x/NA 1.25 silicon oil objective and 40 μm spinning disc were used to capture 70–90 μm thick stacks with a z-step of 0.5 μm. The same conditions were used for fixed zebrafish embryos. For light-sheet microscopy, a Zeiss Z.1 microscope with a W Plan-APO 20x/NA 1.0 WI imaging objective and 10x/NA 0.2 air illumination objectives were used to capture 40–75 μm thick stacks with a z-step of 0.4–0.5 μm, every 15–30 s.
Electron microscopy
Transmission Electron Microscopy on RAW 264.7 mammalian macrophages fed with apoptotic HeLa and treated with U18666a, an NPC1 inhibitor, was performed at the Center for Microscopy and Image Analysis, University of Zurich. Cells were grown on 12 mm cover glasses treated with L-polylysine and containing gold reference markers (20 nm thickness), which were sputter coated onto the discs using an appropriate mask (Leica Microsystems, Vienna, Austria). Cells were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.35, pre-warmed to 37 °C) for 1 h, followed by 1% OsO4 for 1 h in 0.1 M cacodylate buffer, and 1% aqueous uranyl acetate for 1 hour both at RT. Samples then were dehydrated in an ethanol series and embedded in Epon/Araldite (Sigma-Aldrich). To target specific cells from in vitro feeding assays that presented the clear phenotype of the enlarged gastrosome, we imaged the resin block with a fluorescent microscope (Thunder, Leica Microsystems, Vienna, Austria) and identified the cells of interest based on the gold marks and the weak fluorescence of the sample compared with the resin. Ultrathin (70 nm) sections were post-stained with lead citrate and examined with a Talos 120 transmission electron microscope at an acceleration voltage of 120KV using a Ceta digital camera and the MAPS software package (Thermo Fisher Scientific, Eindhoven, The Netherlands).
Light-sheet image pre-processing
For 3D analysis of light-sheet microscopy images, 3D volumes as czi files were converted to H5 files using Fiji67 and BigDataProcessor68. Cells were tracked and individual frames were saved using BigDataTools and BigDataTracker for further analysis. For 2D analysis, maximum projections of hyperstacks were used.
Image analysis
Microglial morphology
Maximum projections of individually cropped cells were used to create a Gaussian blurred image for further thresholding and mask generation in Fiji67. Shape descriptors, including circularity, were measured using the created mask.
Vesicle and phagosome detection
Vesicles were detected as empty spaces in microglia labelled with cytoplasmic fluorescent marker (TgBAC(csf1ra:GAL4-VP16), Tg(UAS-E1B:NTR-mCherry)). Phagosomes were defined as vesicles that form at the plasma membrane upon the closure of a phagocytic cup.
Microglia and vesicle segmentation
3D microglia segmentations were generated in Imaris using surface detection function. The largest vesicle was segmented manually using the same software.
Microglial phagocytosis
Quantification of phagocytosis rate was performed as previously described in ref. 42. Maximum projections of individual cells were created to count successful phagocytic events in Fiji67. The total number of counts from one cell was divided by the duration of the time-lapse to estimate the number of events per hour ratio.
Uncollected apoptotic cells quantification
3D volumes from acridine orange staining experiments were processed in Imaris using the spot detection function. Automated selection was corrected manually to consider only spots in the optic tectum of zebrafish and to estimate the number of uncollected apoptotic neurons in each sample.
Diameter of the vesicle
The area of vesicles was measured in Fiji67 using a polygonal tool on the chosen frame from hyperstacks representing the middle of the vesicle, or in the case of data acquired with widefield microscopy, on the only frame available. Diameter was calculated based on the following formula:
Vesicle tracking
Volumes of single cell crops were used to track individual vesicles using the MTrackJ plug-in69 in Fiji67. For each time point a tracker was placed in the center of the vesicle in the chosen frame representing the middle of the vesicle. The center z-plane was used for measuring the diameter of the vesicle in the subsequent time points.
Statistical testing
N refers to the number of biological replicates (e.g., fish embryos) and n to the number of items (e.g., cells) analyzed, unless otherwise specified. For statistical analysis, Python 3.8 and statannot packages were used. Conditions were compared using an unpaired, two-tailed, nonparametric Mann-Whitney U test with Bonferroni correction.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We are grateful to Cornelia Henkel for helping with fish work, Max Brambach for his assistance with data analysis and Darren Gilmour for providing feedback on the manuscript. We would like to thank Nicolas Rieser for helping with qPCR experiments. The authors acknowledge the support of the Center for Microscopy and Image Analysis, University of Zurich for light and electron microscopy experiments. We are thankful to Forbes D. Porter for sharing the npc1hg37 mutant fishline and to Valérie Wittamer for sharing the 4C4 antibody. This project was supported by Swiss National Science Foundation Grants 310030_212794 and 31003A_182733 to F.P., J.Z. and A.V.
Author contributions
J.Z. and F.P. designed the study and wrote the manuscript. J.Z. conducted the majority of experiments and analyzed the data. E.F.C. participated in the initial characterization of NPC1-deficient embryos. A.V. contributed to EM experiments and cholesterol detection in BAI1 knockdowns. A.O. and S.S. performed lipidomic analysis.
Peer review
Peer review information
Nature Communications thanks Ryuta Koyama, Cody Smith and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data and materials supporting the findings of this study are available from the corresponding author upon request. Source data are provided with this paper. The imaging data generated in this study have been deposited in the EBI BioStudies database under accession number S-BSST1537. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52874-6.
References
- 1.Sierra, A. et al. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell7, 483–495 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villani, A. et al. Clearance by Microglia Depends on Packaging of Phagosomes into a Unique Cellular Compartment. Dev. Cell49, 77–88 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Keren-Shaul, H. et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell169, 1276–1290 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Li, Q. et al. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron101, 207–223.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zareba, J. & Peri, F. Microglial ‘fat shaming’ in development and disease. Curr. Opin. Cell Biol.73, 105–109 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Cantuti-Castelvetri, L. et al. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science359, 684–688 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Brekk, O. R., Honey, J. R., Lee, S., Hallett, P. J. & Isacson, O. Cell type-specific lipid storage changes in Parkinson’s disease patient brains are recapitulated by experimental glycolipid disturbance. Proc. Natl Acad. Sci. USA117, 27646–27654 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marschallinger, J. et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci.23, 194–208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arbaizar‐Rovirosa, M. et al. Aged lipid‐laden microglia display impaired responses to stroke. EMBO Mol. Med.15, 1–21 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polese-Bonatto, M. et al. Niemann-Pick Disease Type C: Mutation Spectrum and Novel Sequence Variations in the Human NPC1 Gene. Mol. Neurobiol.56, 6426–6435 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Kwon, H. J. et al. Structure of N-Terminal Domain of NPC1 Reveals Distinct Subdomains for Binding and Transfer of Cholesterol. Cell137, 1213–1224 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, X., Saha, P., Lib, J., Blobel, G. & Pfeffer, S. R. Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proc. Natl. Acad. Sci. USA113, 10079–10084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trinh, M. N., Brown, M. S., Seemann, J., Goldstein, J. L. & Lu, F. Lysosomal cholesterol export reconstituted from fragments of Niemann-Pick C1. ELife7, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, N. L. et al. The Presence of Vacuolated Kupffer Cells Raises a Clinical Suspicion of Niemann-Pick Disease Type C in Neonatal Cholestasis. Front. Genet.13, 1–5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appelqvist, H. et al. Attenuation of the lysosomal death pathway by lysosomal cholesterol accumulation. Am. J. Pathol.178, 629–639 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Appelqvist, H. et al. Sensitivity to Lysosome-Dependent Cell Death Is Directly Regulated by Lysosomal Cholesterol Content. PLoS ONE7, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung, C., Puthanveetil, P., Ory, D. S. & Lieberman, A. P. Genetic and pharmacological evidence implicates cathepsins in Niemann-Pick C cerebellar degeneration. Hum. Mol. Genet.25, 1434–1446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higashi, Y., Murayama, S., Pentchev, P. G. & Suzuki, K. Cerebellar degeneration in the Niemann-Pick type C mouse. Acta Neuropathologica85, 175–184 (1993). [DOI] [PubMed] [Google Scholar]
- 19.Erickson, R. P. & Bernard, O. Studies on neuronal death in the mouse model of Niemann-Pick C disease. J. Neurosci. Res.68, 738–744 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Wu, Y. P. et al. Apoptosis accompanied by up-regulation of TNF-α death pathway genes in the brain of Niemann-Pick type C disease. Mol. Genet. Metab.84, 9–17 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Cougnoux, A., et al. Necroptosis in Niemann–Pick disease, type C1: a potential therapeutic target. Cell Death Disease710.1038/cddis.2016.16 (2016). [DOI] [PMC free article] [PubMed]
- 22.Funakoshi, T., Aki, T., Tajiri, M., Unuma, K. & Uemura, K. Necroptosis-like neuronal cell death caused by cellular cholesterol accumulation. J. Biol. Chem.291, 25050–25065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasad, A., Fischer, W. A., Maue, R. A. & Henderson, L. P. Regional and developmental expression of the Npc1 mRNA in the mouse brain. J. Neurochem.75, 1250–1257 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci.34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colombo, A. et al. Loss of NPC1 enhances phagocytic uptake and impairs lipid trafficking in microglia. Nat. Commun.12, 1158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cologna, S. M. et al. Human and mouse neuroinflammation markers in Niemann-Pick disease, type C1. J. Inherit. Metab. Dis.37, 83–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cougnoux, A. et al. Microglia activation in Niemann-Pick disease, type C1 is amendable to therapeutic intervention. Hum. Mol. Genet.27, 2076–2089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.German, D. C. et al. Neurodegeneration in the Niemann-Pick C mouse: Glial involvement. Neuroscience109, 437–450 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Baudry, M., Yao, Y., Simmons, D., Liu, J. & Bi, X. Postnatal development of inflammation in a murine model of Niemann-Pick type C disease: Immunohistochemical observations of microglia and astroglia. Exp. Neurol.184, 887–903 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Kavetsky, L. et al. Increased interactions and engulfment of dendrites by microglia precede Purkinje cell degeneration in a mouse model of Niemann Pick Type-C. Sci. Rep.9, 14722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroll, F. et al. A simple and effective f0 knockout method for rapid screening of behaviour and other complex phenotypes. eLife10, e59683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray, C. et al. Simultaneous intravital imaging of macrophage and neutrophil behaviour during inflammation using a novel transgenic zebrafish. Thrombosis Haemost.105, 811–819 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Casano, A. M., Albert, M. & Peri, F. Developmental Apoptosis Mediates Entry and Positioning of Microglia in the Zebrafish Brain. Cell Rep.16, 897–906 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Lu, F. et al. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife4, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naureckiene, S. et al. Identification of HE1 as the second gene of Niemann-Pick C disease. Science290, 2298–2301 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Subramanian, K. & Balch, W. E. NPC1/NPC2 function as a tag team duo to mobilize cholesterol. Proc. Natl Acad. Sci. USA105, 15223–15224 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng, W. C. et al. Modeling Niemann-Pick disease type C1 in zebrafish: A robust platform for in vivo screening of candidate therapeutic compounds. DMM Dis. Models Mechanisms11, dmm034165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiweger, M., Majewski, L., Adamek-Urbanska, D., Wasilewska, I. & Kuznicki, J. npc2-Deficient Zebrafish Reproduce Neurological and Inflammatory Symptoms of Niemann-Pick Type C Disease. Front. Cell. Neurosci.15, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, M. et al. Endosomal accumulation of Toll-like receptor 4 causes constitutive secretion of cytokines and activation of signal transducers and activators of transcription in Niemann-Pick disease type C (NPC) fibroblasts: A potential basis for glial cell activation in the NPC brain. J. Neurosci.27, 1879–1891 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varela, M., Dios, S., Novoa, B. & Figueras, A. Characterisation, expression and ontogeny of interleukin-6 and its receptors in zebrafish (Danio rerio). Dev. Comp. Immunol.37, 97–106 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Shiau, C. E., Kaufman, Z., Meireles, A. M. & Talbot, W. S. Differential Requirement for irf8 in Formation of Embryonic and Adult Macrophages in Zebrafish. PLoS ONE10, e0117513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Möller, K. et al. A role for the centrosome in regulating the rate of neuronal efferocytosis by microglia in vivo. eLife11, e82094 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva, N. J., Dorman, L. C., Vainchtein, I. D., Horneck, N. C. & Molofsky, A. V. In situ and transcriptomic identification of microglia in synapse-rich regions of the developing zebrafish brain. Nat. Commun.12, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pommier, Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer6, 789–802 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Curado, S. et al. Conditional targeted cell ablation in zebrafish: A new tool for regeneration studies. Dev. Dyn.236, 1025–1035 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Mazaheri, F. et al. Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat. Commun.5, 4046 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Varsano, N. et al. Two polymorphic cholesterol monohydrate crystal structures form in macrophage culture models of atherosclerosis. Proc. Natl Acad. Sci. USA115, 7662–7669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maxfield, F. R. & Wüstner, D. Analysis of Cholesterol Trafficking with Fluorescent Probes. Methods Cell Biol.108, 367–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen, J. W., Murphy, T. L., Wlllingham, M. C., Pastan, I. & August, J. T. Identification of two lysosomal membrane glycoproteins. J. Cell Biol.101, 85–95 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janabi, N., Peudenier, S., Heron, B., Ng, K. H. & Tardieu, M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci. Lett.195, 105–108 (1995). [DOI] [PubMed] [Google Scholar]
- 51.Soga, M. et al. Erratum: HPGCD Outperforms HPBCD as a Potential Treatment for Niemann-Pick Disease Type C during Disease Modeling with iPS Cells. Stem Cells33, 2885–2886 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Davidson, C. D. et al. Efficacy and ototoxicity of different cyclodextrins in Niemann-Pick C disease. Ann. Clin. Transl. Neurol.3, 366–380 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singhal, A., Krystofiak, E. S., Jerome, W. G. & Song, B. 2-Hydroxypropyl-gamma-cyclodextrin overcomes NPC1 deficiency by enhancing lysosome-ER association and autophagy. Sci. Rep.10, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferreira, C. R. & Gahl, W. A. Lysosomal storage disease. Transl. Sci. Rare Dis.2, 1–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, S. T., Kreutzberger, A. J. B., Lee, J., Kiessling, V. & Tamm, L. K. The role of cholesterol in membrane fusion. Chem. Phys. Lipids199, 136–143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kellner-Weibel, G. et al. Crystallization of free cholesterol in model macrophage foam cells. Arteriosclerosis, Thrombosis, Vasc. Biol.19, 1891–1898 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Shu, F. et al. Cholesterol crystal-mediated inflammation is driven by plasma membrane destabilization. Front. Immunol.9, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He, W. T. et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res.25, 1285–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng, X. T. et al. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J. Cell Biol.217, 3127–3139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hartenstein, V. & Martinez, P. Phagocytosis in cellular defense and nutrition: a food-centered approach to the evolution of macrophages. Cell Tissue Res.377, 527–547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matz, J. M. Plasmodium’s bottomless pit: properties and functions of the malaria parasite’s digestive vacuole. Trends Parasitol.38, 525–543 (2022). [DOI] [PubMed] [Google Scholar]
- 62.Rovira, M. et al. Zebrafish Galectin 3 binding protein is the target antigen of the microglial 4C4 monoclonal antibody. Dev. Dyn.252, 400–414 (2022). [DOI] [PubMed] [Google Scholar]
- 63.Taylor, S. C. et al. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol.37, 761–774 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Tsarouchas, T. M. et al. Dynamic control of proinflammatory cytokines Il-1β and Tnf-α by macrophages in zebrafish spinal cord regeneration. Nat. Commun.9, 4670 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin, C., Spikings, E., Zhang, T. & Rawson, D. Housekeeping genes for cryopreservation studies on zebrafish embryos and blastomeres. Theriogenology71, 1147–1155 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Ganguin, A. A., Skorup, I., Streb, S., Othman, A. & Luciani, P. Formation and Investigation of Cell-Derived Nanovesicles as Potential Therapeutics against Chronic Liver Disease. Adv. Healthc. Mater.12, e2300811 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tischer, C. et al. BigDataProcessor2: a free and open-source Fiji plugin for inspection and processing of TB sized image data. Bioinformatics37, 3079–3081 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meijering, E., Dzyubachyk, O., & Smal, I. Methods for cell and particle tracking. In Methods in Enzymology (1st ed., Vol. 504) (2012). 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data and materials supporting the findings of this study are available from the corresponding author upon request. Source data are provided with this paper. The imaging data generated in this study have been deposited in the EBI BioStudies database under accession number S-BSST1537. Source data are provided with this paper.