Abstract
The Kazakh Tobet is an indigenous Kazakh dog breed that has been used to guard livestock since ancient times. To understand the genetic structure and phylogenetic relationship of the Kazakh Tobet breed with other herding and livestock guarding dog breeds, we analysed short tandem repeat data of 107 Kazakh Tobet dogs from different regions of Kazakhstan and Mongolia, as well as whole genome sequencing data from two Kazakh Tobet dogs and 43 dogs from 24 working breeds. Our results indicate a high genetic diversity of the Kazakh Tobet, with the average number of alleles per locus ranging from 6.00 to 10.22 and observed heterozygosity ranging from 76 to 78%. The breed has a complex genetic structure characterised by seven different clusters. The neighbour-joining tree constructed based on 14,668,406 autosomal and the maximum likelihood tree based on mitochondrial D-loop sequences indicate a common genetic heritage between the Kazakh Tobet, the Central Asian Shepherd Dog and the Turkish Akbash. The presence of haplotype A18 in the Kazakh Tobets supports the hypothesis of the ancient origin of the breed, which was previously suggested by archaeological finds and written sources. These results provide an important genetic basis for the ongoing efforts to improve the Kazakh Tobet breed, to ensure its preservation as an independent genetic lineage and to recognise a breed on an international level.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-74061-9.
Subject terms: Genetics, Molecular biology, Zoology
Introduction
Of the historically documented variety of indigenous dog breeds from Southwest Asia and Kazakhstan, only a few have survived to the present day. In addition to the sighthound Turkmen or Kazakh Tazy and the Kyrgyz Taigan, these dog breeds also include various types of livestock guarding and herding shepherd dogs (LGD and HSD, respectively). They are known in Turkmenistan as “gopek” and “gopek-si” (or “alabai”), in Tajikistan as “dahmarda”, in Uzbekistan as “kopek” and “kazakh-it” and in Kazakhstan as “alapar-it”, “arab-it” and “tobet-it”1. The “tobet-it”, also known as the Kazakh Tobet, is an LGD breed in Kazakhstan that has been protecting livestock and guarding nomad camps from night raids since ancient times1. The etymology of the name of the breed is not reliably known. It may be related to the old Turkiс word “töb t”, which can be translated as “male”. Some sources interpret this term as “breed of large dogs” or as a combination of “tobe” (i.e. hill or peak) and “it” (i.e. dog) to form “Tobet”, which could be understood as “dog that lies on the hill” or “supreme guardian“2. The elders in South Kazakhstan believe that the name of this breed is related to the name of the dung heaps pulled out of the “koshara” (roofed sheepfold) and ironically called “tobe”. The Kazakh Tobets lie on them as if on a hill, guarding the flocks, observing the surroundings and digging shelters to keep warm in the cold winter.
t”, which can be translated as “male”. Some sources interpret this term as “breed of large dogs” or as a combination of “tobe” (i.e. hill or peak) and “it” (i.e. dog) to form “Tobet”, which could be understood as “dog that lies on the hill” or “supreme guardian“2. The elders in South Kazakhstan believe that the name of this breed is related to the name of the dung heaps pulled out of the “koshara” (roofed sheepfold) and ironically called “tobe”. The Kazakh Tobets lie on them as if on a hill, guarding the flocks, observing the surroundings and digging shelters to keep warm in the cold winter.
The earliest documented references to this breed can be found in the “Turkish-Arabic Dictionary” from the 13th century, which was published in the Mamluk Sultanate and describes the breed as a “large shepherd dog”3. Evidence of the breed’s even earlier history can be found in petroglyphs in central Kazakhstan, dating from the late third to second millennium BC and attributed to the early Andronovo culture. These ancient images show LGDs with massive bodies and large heads4. Similar depictions have been found on petroglyphs in other regions of Kazakhstan, dating from the Neolithic period (4th to 3rd millennium BC) to the Middle Ages (14th century AD)5–7. The Kazakh Tobet, like other Central Asian LGD breeds, is thought to have descended from dogs bred by nomadic cultures in Central Asia and surrounding areas, selected for their ability to protect livestock from wolves in extreme climates. They played a crucial role in protecting smaller livestock such as sheep and goats, which were more vulnerable to wolf attacks than larger animals. Therefore, the origin of the LGD, including the Kazakh Tobet breed, is closely related to the development of ancient sheep farming, especially transhumance8. Recent studies suggest that the different genetic lineages of sheep were introduced to the Eurasian steppe long before the Late Bronze Age9, confirming the representation of LGDs in early artefacts of the Andronovo culture from Kazakhstan.
Over the centuries, the breeding of these dogs has focused on their working abilities and behavioural traits. Pilshchikov Y.N. described the Kazakh Tobet as a “dog of coarse constitution, of large or medium height, with well-developed muscles, large head and long, thick coat with dense undercoat. The basic colours observed are black, tan, brown and occasionally lighter shades. The dogs appear somewhat sluggish and phlegmatic but are known for their explosive temperament. Within herds, they tend to behave inconspicuously and only become lively when wolves appear”10. When attacked by wolves, the Kazakh Tobets work in a group and initially try to drive the wolves away from the livestock. If the situation forces the Tobets to fight with the wolves to protect the livestock, they inflict traumatic bites on the wolves at high speed. This breed has sometimes been used in hunting, where it co-operates with the Kazakh Tazy, a national sighthound breed. When communicating with people, these dogs are known for their independent and non-aggressive behaviour towards the inhabitants of their village (“aul”), which they can remember perfectly (Fig. 1). At the same time, they can be extremely aggressive when it comes to protecting their owners’ property, themselves and especially children from attack.
Fig. 1.
Photo of the Kazakh Tobet. Materials of the film expedition for the film “Kyz-Zhibek” (1973). Owner K.N. Plakhov. The Kazakhs traditionally cropped the ears of Kazakh Tobet puppies to prevent wolves from grabbing them by the ears. However, the ears of female dogs were sometimes left uncropped.
Despite the traditional recognition of the breed, the survival of the Kazakh Tobet breed has been threatened over the last century by various socio-economic changes such as revolutions, wars, the shift from a nomadic to a sedentary lifestyle and the decline of traditional sheep farming. Biological factors such as the introduction of new breeds, infectious diseases, including rabies, rodent control programmes and the use of poisoned bait have also contributed to the endangerment of the breed1. In uncontrolled breeding, there is a constant risk of crossbreeding with imported breeds, which can lead to the loss of the breed genetic originality.
For the conservation and sustainable management of such endangered valuable native breeds, the study of genetic diversity, which assesses the biological variation within a species, breed, etc., is fundamental. It ensures that individuals have different genetic traits, all of which are characteristic of the breed and some of which may offer advantages under changing environmental conditions. The use of Short Tandem Repeat (STR) loci, also known as microsatellites, is a traditional method for analysing genetic diversity and population structure. These are short nucleotide sequences (typically 1 to 5 bp) that repeat in tandem, are codominant, abundant and multiallelic, making them a suitable tool for molecular population genetics11. They can complement modern whole-genome sequencing (WGS) data by providing additional resolution in detecting genetic variation at a finer scales, particularly in assessing population structure and diversity. While WGS provides valuable insights into the evolutionary history of the breed and the reconstruction of ancestral relatedness12. In addition, WGS data have been recognised as a valuable resource for analysing haplotypes of the mitochondrial genome and could determine haplogroups comparable to targeted sequencing of mitochondrial DNA13,14.
Numerous studies have analysed the genetic structure and ancestry of various dog breeds15–28. However, the genetic background of the indigenous Kazakh Tobet breed is still largely undocumented. Therefore, the STR and WGS data were used in this study to examine the genetic structure of the Kazakh Tobet and the phylogenetic links to LGDs, HSDs and other working breeds. In addition to Kazakh Tobet dogs from Kazakhstan, we also analysed the genetic structure of Kazakh Tobets brought to Mongolia by ethnic Kazakhs. This approach aims to gain a comprehensive understanding of the historical distribution of the breed and the impact of migration on genetic composition. In addition, the haplotype of Kazakh Tobet dogs was determined using WGS data to trace the ancestry of the breed. The results are important for understanding the origin of the Kazakh Tobet and support efforts to preserve and recognise the breed in the international community.
Results
Diversity analysis based on the STR dataset
A total of 18 STR loci were used to genotype 107 Kazakh Tobet dogs from four populations (Table 1), resulting in a total number of 193 alleles. Pop1 had the highest average number of alleles per locus (Na = 10.22 ± 0.50). In comparison, Pop2 and Pop3 had moderate genetic diversity, with average Na values of 7.00 ± 0.34 and 6.11 ± 0.25, respectively. Pop4 had the lowest genetic diversity among the four populations, with an average Na value of 6.00 ± 0.33. The loci with the highest number of alleles were REN162C04, AHT137, AHTh260, FH2054, AHT121 and AHTh171, each of which had between 12 and 15 alleles. The lowest number of alleles was observed for locus AHTk211, which had 6 alleles. The average number of effective alleles for all analysed Kazakh Tobet dogs was 5.47 ± 0.32 and ranged from 4.14 ± 0.28 in Pop4 to 5.42 ± 0.34 in Pop1. Pop2 had the highest observed heterozygosity (Ho = 0.79 ± 0.03), closely followed by Pop1 (Ho = 0.78 ± 0.03) and Pop3 (Ho = 0.76 ± 0.04). Pop3 had the highest expected heterozygosity (uHe = 0.83 ± 0.01), while Pop1 (uHe = 0.81 ± 0.01) and Pop2 (uHe = 0.80 ± 0.01) also showed considerable but slightly lower heterozygosity. In contrast, Pop4 had the lowest heterozygosity measures, with Ho at 0.77 ± 0.05 and uHe at 0.77 ± 0.03. The fixation index F was not significantly negative in Pop2 (uF=-0.02 ± 0.03, 95% CI: -0.07–0.04) and Pop4 (uF=-0.05 ± 0.09, 95% CI: -0.22–0.13), indicating an excess of heterozygotes in these populations. Conversely, non-significant positive F-values were observed in Pop1 (uF = 0.03 ± 0.01, 95% CI: -0.19–0.25) and Pop3 (uF = 0.02 ± 0.07, 95% CI: -0.12–0.16), indicating a slight lack of heterozygotes. A statistically significant absence of inbreeding was demonstrated for all analyzed Kazakh Tobet dogs (uF = 0.03 ± 0.01, 95% CI: 0.02–0.05).
Table 1.
Polymorphism analysis of 18 STR markers in four populations of Kazakh Tobet dogs.
| Pop | Locus | Na | Ne | Ho | uHe | uF |
|---|---|---|---|---|---|---|
| Pop1 | AHTk211 | 6.00 | 3.63 | 0.62 | 0.73 | 0.15 |
| CXX0279 | 10.00 | 5.61 | 0.79 | 0.83 | 0.04 | |
| REN169O18 | 11.00 | 5.72 | 0.81 | 0.83 | 0.02 | |
| INU055 | 9.00 | 4.99 | 0.85 | 0.81 | -0.06 | |
| REN54P11 | 10.00 | 5.71 | 0.88 | 0.83 | -0.06 | |
| INRA21 | 8.00 | 5.56 | 0.92 | 0.83 | -0.12 | |
| AHT137 | 14.00 | 6.34 | 0.90 | 0.85 | -0.07 | |
| REN169D01 | 11.00 | 6.31 | 0.93 | 0.85 | -0.11 | |
| AHTh260 | 12.00 | 4.33 | 0.71 | 0.77 | 0.07 | |
| AHTk253 | 9.00 | 2.79 | 0.47 | 0.65 | 0.28 | |
| INU005 | 10.00 | 3.34 | 0.60 | 0.70 | 0.14 | |
| INU030 | 7.00 | 4.32 | 0.78 | 0.77 | -0.02 | |
| FH2848 | 9.00 | 5.79 | 0.88 | 0.83 | -0.06 | |
| AHT121 | 12.00 | 7.86 | 0.80 | 0.88 | 0.09 | |
| FH2054 | 12.00 | 6.56 | 0.77 | 0.85 | 0.10 | |
| REN162C04 | 13.00 | 6.15 | 0.83 | 0.84 | 0.00 | |
| AHTh171 | 12.00 | 8.17 | 0.88 | 0.88 | 0.00 | |
| REN247M23 | 9.00 | 4.37 | 0.69 | 0.78 | 0.11 | |
| Mean | 10.22 | 5.42 | 0.78 | 0.81 |
0.03 (95% CI: -0.19–0.25) |
|
| SE | 0.50 | 0.34 | 0.03 | 0.01 | 0.01 | |
| Pop2 | AHTk211 | 5.00 | 4.30 | 0.81 | 0.79 | -0.06 |
| CXX0279 | 6.00 | 3.66 | 0.81 | 0.75 | -0.12 | |
| REN169O18 | 6.00 | 4.70 | 0.88 | 0.81 | -0.12 | |
| INU055 | 7.00 | 5.39 | 0.88 | 0.84 | -0.08 | |
| REN54P11 | 7.00 | 4.03 | 0.94 | 0.78 | -0.25 | |
| INRA21 | 6.00 | 3.97 | 0.63 | 0.77 | 0.17 | |
| AHT137 | 9.00 | 7.31 | 0.94 | 0.89 | -0.09 | |
| REN169D01 | 8.00 | 5.07 | 0.88 | 0.83 | -0.09 | |
| AHTh260 | 7.00 | 4.30 | 0.69 | 0.79 | 0.11 | |
| AHTk253 | 7.00 | 3.74 | 0.75 | 0.76 | -0.02 | |
| INU005 | 7.00 | 4.10 | 0.63 | 0.78 | 0.18 | |
| INU030 | 5.00 | 4.20 | 0.63 | 0.79 | 0.19 | |
| FH2848 | 6.00 | 4.45 | 0.81 | 0.80 | -0.05 | |
| AHT121 | 10.00 | 7.01 | 0.94 | 0.89 | -0.10 | |
| FH2054 | 9.00 | 5.33 | 0.81 | 0.84 | 0.00 | |
| REN162C04 | 8.00 | 4.53 | 0.75 | 0.80 | 0.04 | |
| AHTh171 | 8.00 | 3.58 | 0.75 | 0.74 | -0.04 | |
| REN247M23 | 5.00 | 2.93 | 0.63 | 0.68 | 0.05 | |
| Mean | 7.00 | 4.59 | 0.79 | 0.80 |
-0.02 (95% CI: -0.07–0.04) |
|
| SE | 0.34 | 0.26 | 0.03 | 0.01 | 0.03 | |
| Pop3 | AHTk211 | 4.00 | 3.05 | 0.38 | 0.72 | 0.47 |
| CXX0279 | 6.00 | 5.12 | 0.88 | 0.86 | -0.09 | |
| REN169O18 | 5.00 | 3.56 | 0.88 | 0.77 | -0.23 | |
| INU055 | 5.00 | 3.88 | 0.75 | 0.79 | -0.01 | |
| REN54P11 | 6.00 | 4.41 | 0.75 | 0.83 | 0.03 | |
| INRA21 | 6.00 | 4.74 | 1.00 | 0.84 | -0.29 | |
| AHT137 | 6.00 | 4.92 | 1.00 | 0.85 | -0.27 | |
| REN169D01 | 8.00 | 5.82 | 0.88 | 0.88 | -0.06 | |
| AHTh260 | 8.00 | 6.40 | 0.75 | 0.90 | 0.12 | |
| AHTk253 | 8.00 | 6.40 | 0.88 | 0.90 | -0.04 | |
| INU005 | 6.00 | 3.20 | 0.63 | 0.73 | 0.10 | |
| INU030 | 6.00 | 4.41 | 0.88 | 0.83 | -0.14 | |
| FH2848 | 5.00 | 4.74 | 0.75 | 0.84 | 0.05 | |
| AHT121 | 7.00 | 5.33 | 0.75 | 0.87 | 0.08 | |
| FH2054 | 6.00 | 5.12 | 0.88 | 0.86 | -0.09 | |
| REN162C04 | 6.00 | 4.27 | 0.63 | 0.82 | 0.20 | |
| AHTh171 | 6.00 | 4.13 | 0.63 | 0.81 | 0.19 | |
| REN247M23 | 6.00 | 3.77 | 0.50 | 0.78 | 0.34 | |
| Mean | 6.11 | 4.63 | 0.76 | 0.83 |
0.02 (95% CI: -0.12–0.16) |
|
| SE | 0.25 | 0.23 | 0.04 | 0.01 | 0.07 | |
| Pop4 | AHTk211 | 5.00 | 3.70 | 0.90 | 0.77 | -0.25 |
| CXX0279 | 7.00 | 2.94 | 0.90 | 0.69 | -0.38 | |
| REN169O18 | 5.00 | 4.08 | 0.90 | 0.79 | -0.20 | |
| INU055 | 6.00 | 4.00 | 0.60 | 0.79 | 0.21 | |
| REN54P11 | 5.00 | 3.45 | 0.50 | 0.75 | 0.31 | |
| INRA21 | 6.00 | 5.00 | 0.80 | 0.84 | 0.00 | |
| AHT137 | 9.00 | 6.45 | 0.90 | 0.89 | -0.07 | |
| REN169D01 | 8.00 | 5.71 | 0.90 | 0.87 | -0.10 | |
| AHTh260 | 6.00 | 3.70 | 0.90 | 0.77 | -0.25 | |
| AHTk253 | 3.00 | 1.50 | 0.20 | 0.35 | 0.42 | |
| INU005 | 4.00 | 2.67 | 0.70 | 0.66 | -0.13 | |
| INU030 | 5.00 | 4.17 | 0.90 | 0.80 | -0.19 | |
| FH2848 | 7.00 | 4.76 | 0.90 | 0.83 | -0.15 | |
| AHT121 | 7.00 | 5.41 | 0.80 | 0.86 | 0.02 | |
| FH2054 | 7.00 | 4.00 | 0.30 | 0.79 | 0.63 | |
| REN162C04 | 6.00 | 4.35 | 0.90 | 0.81 | -0.18 | |
| AHTh171 | 6.00 | 3.57 | 1.00 | 0.76 | -0.41 | |
| REN247M23 | 6.00 | 5.13 | 0.90 | 0.85 | -0.12 | |
| Mean | 6.00 | 4.14 | 0.77 | 0.77 |
-0.05 (95% CI: -0.22–0.13) |
|
| SE | 0.33 | 0.28 | 0.05 | 0.03 | 0.09 | |
| All | AHTk211 | 6.00 | 3.81 | 0.65 | 0.74 | 0.11 |
| CXX0279 | 11.00 | 5.34 | 0.81 | 0.82 | 0.00 | |
| REN169O18 | 11.00 | 5.63 | 0.83 | 0.83 | -0.01 | |
| INU055 | 10.00 | 5.11 | 0.82 | 0.81 | -0.02 | |
| REN54P11 | 10.00 | 5.52 | 0.84 | 0.82 | -0.03 | |
| INRA21 | 8.00 | 5.41 | 0.87 | 0.82 | -0.07 | |
| AHT137 | 14.00 | 7.20 | 0.92 | 0.87 | -0.06 | |
| REN169D01 | 11.00 | 6.44 | 0.92 | 0.85 | -0.08 | |
| AHTh260 | 13.00 | 4.73 | 0.73 | 0.79 | 0.08 | |
| AHTk253 | 11.00 | 3.09 | 0.51 | 0.68 | 0.24 | |
| INU005 | 10.00 | 3.43 | 0.62 | 0.71 | 0.13 | |
| INU030 | 7.00 | 4.50 | 0.78 | 0.78 | 0.00 | |
| FH2848 | 9.00 | 5.94 | 0.86 | 0.84 | -0.03 | |
| AHT121 | 12.00 | 8.26 | 0.81 | 0.88 | 0.08 | |
| FH2054 | 13.00 | 6.58 | 0.74 | 0.85 | 0.13 | |
| REN162C04 | 15.00 | 6.29 | 0.81 | 0.85 | 0.04 | |
| AHTh171 | 12.00 | 6.90 | 0.85 | 0.86 | 0.01 | |
| REN247M23 | 10.00 | 4.23 | 0.68 | 0.77 | 0.11 | |
| Mean | 10.72 | 5.47 | 0.78 | 0.81 |
0.03 (95% CI: 0.02–0.05) |
|
| SE | 0.55 | 0.32 | 0.03 | 0.01 | 0.01 |
Na Average alleles/locus, Ne Average effective alleles/locus, Ho Observed heterozygosity, uHe Unbiased expected heterozygosity, F Unbiased fixation index.
The Hardy-Weinberg equilibrium (HWE) test, which was performed separately for each locus for all dogs, showed no significant deviation from the expected frequencies (P > 0.05), with the exception of the loci INRA21, AHT137, AHTh260, AHTk253, FH2054, REN162C04 and AHTh171, which each showed a significant deviation with P < 0.001 (Table 2).
Table 2.
HWE for 18 STR markers.
| Locus | ChiSq | Prob | Signif |
|---|---|---|---|
| AHTk211 | 12.86 | 0.61 | ns |
| CXX0279 | 45.88 | 0.81 | ns |
| REN169O18 | 45.07 | 0.83 | ns |
| INU055 | 31.95 | 0.93 | ns |
| REN54P11 | 44.17 | 0.51 | ns |
| INRA21 | 65.96 | 0.00 | * |
| AHT137 | 153.32 | 0.00 | * |
| REN169D01 | 41.03 | 0.92 | ns |
| AHTh260 | 308.44 | 0.00 | * |
| AHTk253 | 169.84 | 0.00 | * |
| INU005 | 54.37 | 0.16 | ns |
| INU030 | 18.17 | 0.64 | ns |
| FH2848 | 24.93 | 0.92 | ns |
| AHT121 | 72.84 | 0.26 | ns |
| FH2054 | 255.68 | 0.00 | * |
| REN162C04 | 159.49 | 0.00 | * |
| AHTh171 | 176.66 | 0.00 | * |
| REN247M23 | 46.81 | 0.40 | ns |
Ns not significant.
*P < 0.001.
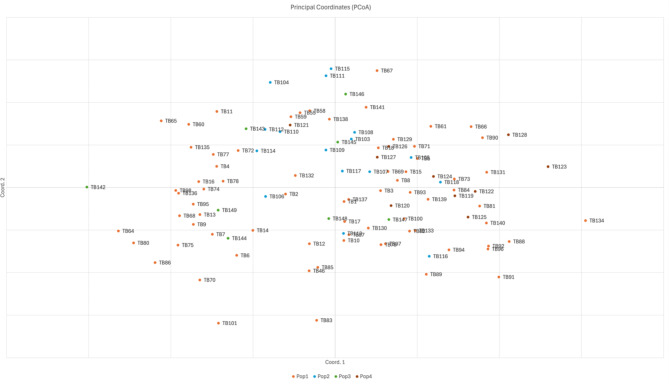
The PCoA of the STR data for the four populations revealed three significant axes of genetic variation: axis one accounted for 4.68%, axis two for 9.06% and axis three for 13.10% of the total variation (Fig. 2). The analysis showed that all four populations were admixed.
Fig. 2.
PCoA plot of 107 Kazakh Tobet dogs from four populations based on STR data.
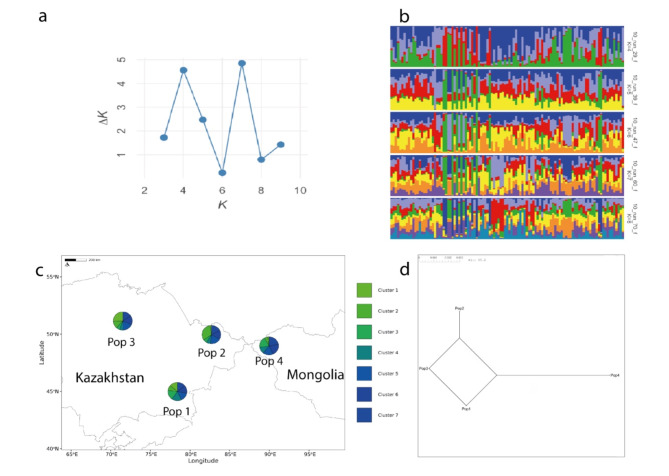
To further elucidate the genetic variation within the Kazakh Tobet breed, a STRUCTURE analysis was performed (Fig. 3). The ΔK method revealed that K = 7 is the optimal number of genetic clusters representing the most genetically similar groups (Fig. 3a), suggesting that seven distinct gene pools form the genetic architecture of Kazakh Tobet dogs (Fig. 3b). In addition, the second highest ΔK value at K = 4 was much larger than the other values, indicating another significant clustering pattern. Remarkably, at K = 7 all genetic clusters were present in all four populations (Fig. 3c).
Fig. 3.
Genetic structure of the Kazakh Tobet dogs. Bayesian clustering on the STR dataset of 107 dogs performed with STRUCTURE v2.3.4 after correction by Evanno et al.29. (CLUMPAK): (a) results of the ΔK method; (b) bar plots where each dog is represented by a single vertical line and this line has coloured segments representing the relative percentage of membership of the cluster; (c) the admixture structure of four populations in geographic space for K = 7; (d) neighbour-net tree for four populations based on the pairwise Fst values.
The analysis of pairwise Fst values showed different degrees of genetic similarity and divergence (Table 3; Fig. 3d). Pop1 and Pop3 showed no genetic differentiation (Fst = 0.000), while Pop2 was minimally different from Pop3 (Fst = 0.003) and moderately different from Pop1 (Fst = 0.017). Pop4 showed the highest genetic differentiation from Pop3 (Fst = 0.023) and moderate differentiation from Pop1 (Fst = 0.015) and Pop2 (Fst = 0.020), making it the most genetically differentiated population in this analysis.
Table 3.
Pairwise fst matrix for four populations of Kazakh Tobet dogs.
| Pop1 | Pop2 | Pop3 | Pop4 | |
|---|---|---|---|---|
| 0,000 | Pop1 | |||
| 0,017 | 0,000 | Pop2 | ||
| 0,000 | 0,003 | 0,000 | Pop3 | |
| 0,015 | 0,020 | 0,023 | 0,000 | Pop4 |
Preparing whole-genome sequencing data
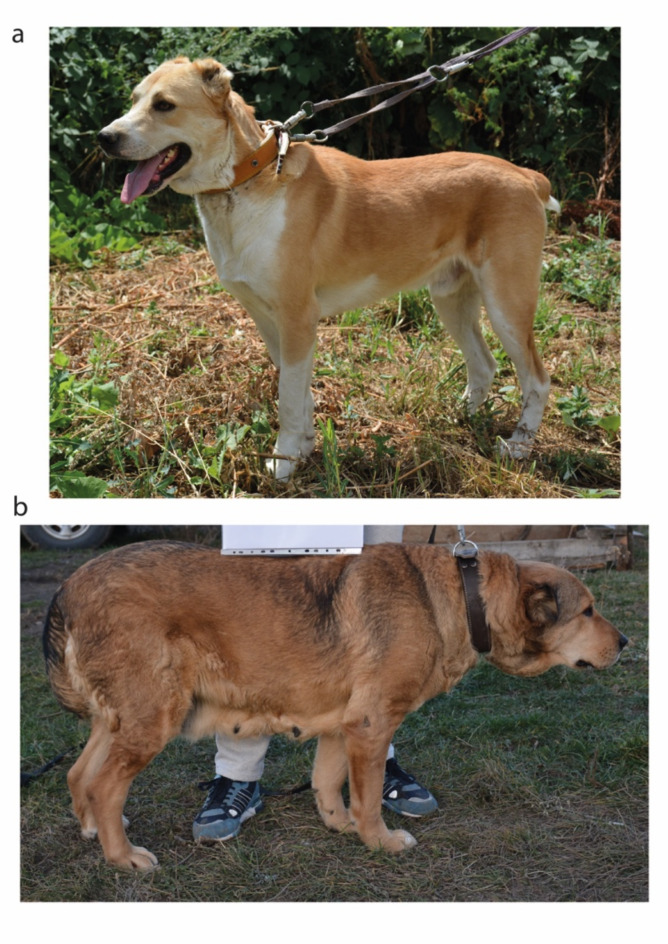
We performed WGS on two dogs of the indigenous Kazakh Tobet breed (BioProject ID PRJNA1144634): TB1 (male) and TB63 (female) (Fig. 4). The selection of TB1 was supported by its position close to the intersection of the axes in the PCoA plot, suggesting that the genetic composition of TB1 may be representative for the overall genetic structure of all analysed Kazakh Tobet dogs, while TB63 received the highest expert scores.
Fig. 4.
Kazakh Tobet dogs TB1 (a) and TB63 (b).
A description of the sequence data can be found in Table 4.
Table 4.
Basic statistics of WGS data for two Kazakh Tobet dogs.
| Sample code | Clean reads | Clean base | Read length | Q20(%) | Q30(%) | GC(%) |
|---|---|---|---|---|---|---|
| TB1 | 288,467,303 | 86,540,190,900 | PE150 | 98.36 | 94.32 | 41.54 |
| TB63 | 288,180,253 | 86,454,075,900 | PE150 | 98.29 | 94.10 | 41.41 |
In addition, genome sequences of 43 dogs from 24 breeds traditionally used for guarding, herding or serving livestock and other work were downloaded from public databases. A total of 21,852,067 autosomal variants were called for all 45 dogs. After applying the GATK criteria for variant filtering, 15,995,420 SNVs were selected for subsequent analyses. The SNV set was further filtered using Plink 1.9, resulting in 14,668,406 SNVs, that were used as the input dataset for the construction of the phylogenetic tree.
Determining mitochondrial haplogroup
The haplotype A18 (C15814T) was identified for both Kazakh Tobet dogs. The haplotypes for the other dogs can be found in Supplementary Table 1. New haplotypes were identified in five dogs.
Phylogenetic tree
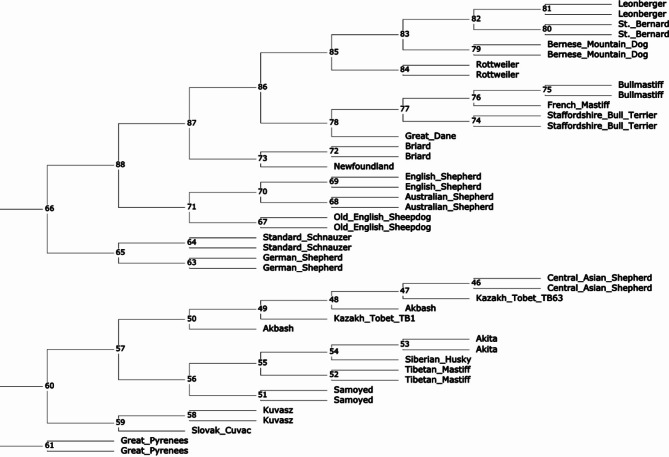
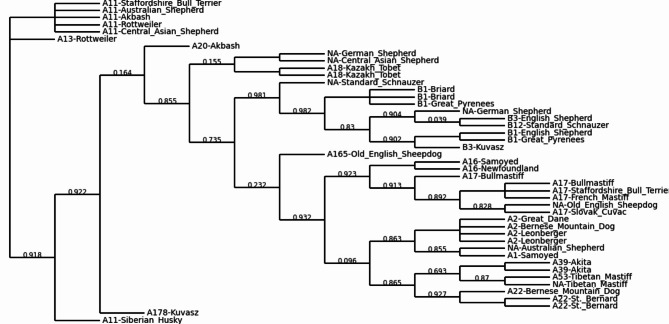
We constructed a neighbor-joining phylogenetic tree based on WGS data for 45 dogs from 25 breeds that have been used in the past as LGD, HSD and for other work, including two Kazakh Tobet dogs (Fig. 5). The Kazakh Tobets (TB1 and TB63) and the Central Asian Shepherd Dogs showed a close genetic relationship and were clustered with the Akbash. However, the Kazakh Tobet dogs were not grouped as a separate breed. This group of three breeds had the closest common node with a group of four breeds that included Samoyeds, Tibetan Mastiffs, Huskies and Akita dogs, with the Samoyeds forming a more distinct cluster. The Great Pyrenees also showed a separate genetic lineage. The Old English Sheepdog was part of a large group that also included Australian Shepherds and English Shepherds. Breeds such as the Great Dane, the Staffordshire Bull Terrier, the French Mastiff and the Bullmastiff, as well as the Bernese Mountain Dog, the St. Bernard, the Leonberger and the Rottweiler, formed two further large groups of clades. The Newfoundland and the Briard, the German Shepherd and the Standard Schnauzer, as well as the Slovak Cuvac and the Kuvasz, formed their own narrow groups.
Fig. 5.
Neighbour-joining phylogenetic tree constructed for working breeds, including Kazakh Tobet dogs (the internal vertices are labelled).
In addition, a maximum likelihood tree was constructed based on the mitochondrial D-loop sequences of 45 dogs (Fig. 6). The breeds were grouped mainly according to their mitochondrial haplotypes in clades. The two Kazakh Tobet dogs (haplotype A18) were close to each other with a branch length of zero and formed a large central cluster alongside Briard and Great Pyrenees (haplotype B1), English Shepherds (haplotype B3) and the Standard Schnauzer (haplotype B12). Central Asian Shepherds were found in haplogroup A11 as well as in a new haplotype. Similarly, the Akbash appeared in haplotypes A11 and A20. From a broader perspective, the Kazakh Tobet (A18), the Akbash (A20) and the Central Asian Shepherd were part of a larger group, although this grouping had a relatively low bootstrap value of 0.155. This result supports the topology and placement of these breeds in the WGS tree. For the other breeds, the clustering of dogs of the same breed to a single clade in the mtDNA D-loop tree was less well resolved compared to the WGS tree, as mitochondrial DNA only captures maternal lineage. Therefore, several breeds in the tree were divided into different clades (e.g. Bernese Mountain Dogs, Standard Schnauser, Samoed, etc.).
Fig. 6.
Maximum likelihood tree for working breeds, including Kazakh Tobet dogs, based on mitochondrial D-loop sequences (new haplogroups are labelled as NA).
Discussion.
To improve our understanding of the genetic structure and phylogenetic relationship of the Kazakh Tobet, especially with other breeds traditionally used for guarding and herding livestock, in this study we analysed STR data from 107 Kazakh Tobet dogs from the south, east and north regions of Kazakhstan and from the Bayan-Ulgii district of Mongolia, as well as WGS data from two Kazakh Tobet dogs and 43 dogs from 24 different breeds.
Genetic diversity and structure of the Kazakh Tobet dogs
The Kazakh Tobet dogs showed high genetic variability and diversity, which is reflected in the average number of alleles per locus (Na) and the observed heterozygosity (Ho). The mean Na value in the four different populations ranged from 6.00 to 10.22. This level of genetic diversity is comparable to that observed in our earlier study in a smaller group of Kazakh Tobet from the southern region of Kazakhstan (Na = 7.11)30 as well as in other breeds within the molossoid group. The Tibetan Mastiff, for example, has an average Na value of 7.70, based on a panel of 10 STR loci31. Similarly, the English Bulldog has an average Na value of 6.46, derived from 33 STR loci32. In contrast, the genetic diversity of the Kazakh Tobet exceeds that of the French bulldog, which has an Na value of 5.1033. The observed heterozygosity was over 78% in all Kazakh Tobet dogs, with a range of 76.4–78.5% between the four populations, which was higher than the Ho values observed in other molossoid breeds such as Boxer, Staffordshire Bull Terrier and Rottweiler (Ho = 0.51, 0.63 and 0.47, respectively) when analysing a panel of 15 STRs23, and Tibetan Mastiff and French Bulldog (Ho = 0.69–0.76 and 0.61, respectively) when analysing a panel of 10 STRs31,33,34. In comparison, non-molossoid breeds, such as the Korean Donggyeongi dog, Italian Pointer, Podenco, Jack Russell Terrier and Yorkshire Terrier, showed similar levels of observed heterozygosity, with Ho values of 0.73, 0.72, 0.71–0.72, 0.76 and 0.73, respectively, when analysed with panels of 10–19 STR loci23,35,36. Previous studies on the Tazy, another national Kazakh breed belonging to the sighthound group, also reported high Ho values (Ho = 0.75)37. Heterozygosity is often used to assess the degree of mixing with another breed. Low observed heterozygosity usually indicates purebred dogs, while high levels of observed heterozygosity are associated with mixed breeds or village dogs. Village dogs, for example, generally have Ho values between 0.73 and 0.8038. The high Ho values observed across all four Kazakh Tobet populations indicate significant crossbreeding.
The average expected heterozygosity for all analysed Kazakh Tobet samples was 0.81, which is higher than the observed heterozygosity of 0.78. When these parameters are equal, this usually indicates that crossing within the population occurs almost randomly. In cases where the observed heterozygosity is lower than the expected heterozygosity, the population is considered inbred, and conversely, if the observed heterozygosity exceeds the expected values, the population is considered outbred. In the Kazakh Tobet dogs, the slightly higher value of expected heterozygosity compared to observed heterozygosity indicates that random mating rather than inbreeding occurs in this cohort, which is also supported by the significant value of the inbreeding coefficient of almost zero (F = 0.03), indicating minimal inbreeding overall.
The analysis of the genetic structure of the analysed sample using PCoA confirms the high genetic diversity of the Kazakh Tobet dogs. As is well known, PCoA measures the genetic relatedness of individuals within a population. In the PCoA graph, the Kazakh Tobet dogs form a group with considerable diversity, as shown by the diffuse distribution along the Y-axis and the genetic outliers. Furthermore, based on the average values of the logarithm of the likelihood function and the dispersion of the estimates obtained in ten runs of STRUCTURE with a selected set of appropriate parameters, the optimal number of clusters in the analysed sample was seven. And all genetic clusters were present in all four populations, as shown by the admixture structure of the four populations in geographic space.
Although the Kazakh Tobet breed has considerable genetic diversity, there are notable differences between the four populations. The population from South Kazakhstan shows the highest genetic diversity, with the highest average number of alleles per locus (Na = 10.22) and number of effective alleles (Ne = 5.42), but a positive F-value (F = 0.03) indicates a slight deficit of heterozygotes and suggests some degree of inbreeding or the influence of population structure effects. The populations of East Kazakhstan and North Kazakhstan show moderate genetic diversity. The East Kazakhstan population has an average Na of 7.00 and the highest observed heterozygosity (Ho = 0.79). The fixation index for the population (F=-0.02) is slightly negative, indicating a slight excess of heterozygotes. The population from North Kazakhstan also shows considerable diversity, with an average Na of 6.11 and Ho of 0.76, but with a slight heterozygote deficiency (F = 0.02). Meanwhile, the population from Mongolia is characterized by the lowest genetic diversity, with an average Na of 6.00 and the lowest Ne (4.14), and has a negative F-value (F=-0.04), reflecting a possible trend towards crossbreeding. However, the interpretation of the obtained F values should be treated with caution, as the small sample sizes in the populations make these values statistically insignificant.
The analysis of the genetic distance between the four Kazakh Tobet populations shows different levels of genetic divergence. The population from South Kazakhstan shows the closest genetic relationships to the populations from East and North Kazakhstan. It is possible that the frequent gene flow has led to a low degree of genetic differentiation between these populations. In contrast, the population from Mongolia is the most genetically differentiated population, especially compared to the population from the northern region with the highest Fst values of 0.023. This considerable genetic differentiation becomes more understandable when one considers that migration to Mongolia began as early as the 19th century, when Kazakhs living in the Chinese province of Xinjiang began to leave their homeland because they were oppressed by the Dungan and Uyghurs. It can also be assumed that a high genetic diversity was already characteristic of the Kazakh Tobet at that time, as all seven genetic clusters of the Kazakh Tobet from Kazakhstan can also be found among the Kazakh Tobet dogs in Mongolia.
Phylogenetic relationships and ancestry of the Kazakh Tobet dogs
Possibly due to the high genetic diversity, the Kazakh Tobet dogs, unlike most other breeds we have observed, did not form a distinct cluster in the phylogenetic tree constructed based on the WGS data. Nevertheless, this breed clearly showed its common genetic origin with the Central Asian Shepherd Dog and the Turkish Akbash breed. It is known that the Central Asian Shepherd Dog is an established breed that originated from several indigenous populations in Central Asia in the 20th century39. Our phylogenetic analysis suggests that the Kazakh Tobet may have been one of these ancestral forms, as well as the Akbash, a Turkish shepherd dog common in the western regions of Turkey. Previous mitochondrial analyses have already shown the close relationship between the Turkish breeds Akbash and Kangal and the Central Asian Shepherd Dog39. The tree we constructed based on the mitochondrial D-loop sequences also confirms the genetic relationship and a possible recent divergence between the Kazakh Tobet, Akbash and the Central Asian Shepherd. The Kazakh Tobet and Akbash may be descended from ancient guard dogs that spread throughout the region thousands of years ago, before the modern state borders were established. Kazakhstan’s strategic location in Central Asia made it an important link in the Silk Road network, facilitating interaction and exchange between East and West. Routes ran through the Ili Valley in Kazakhstan, connecting the region with various parts of Eurasia, including Turkey40. The ancient guard dogs may have been exchanged or bred along these routes, resulting in a common genetic heritage between the Kazakh Tobet and the Akbash and providing for the extremely low genetic differentiation in the Asian LGDs, previously demonstrated in the Caucasian Shepherd Dog, North Caucasian Volkodav, Central Asian Shepherd Dog and Turkish Akbash and Kangal based on analyses of mitochondrial DNA39 and also observed in the Kazakh Tobet in this study. Interestingly, our haplotype analysis based on mitochondrial read extraction from the WGS showed that both samples of Kazakh Tobets had haplogroup A (haplotype A18), further supporting the ancient origin of the breed. Previous research has shown that haplogroups A, B and C together account for approximately 97.40% of the global dog population, with haplogroup A alone accounting for approximately 72.34% of dogs41, suggesting that haplogroup A has played a crucial role in the evolution of different dog breeds. Recent extensive analyses of haplotype networks have confirmed that haplogroup A was introduced into dog populations during the early stages of domestication42. It is noteworthy that 11 haplotypes of haplogroup A, including haplotype A18, had significantly high betweenness values and were clearly recognisable in this network. Haplotype A18 ranks sixth after A3, A9, A15, A29 and A11 and can rightly be described as ancient. It has already been identified in various regions of the world, including village dogs from Southeast Asia and the Middle East, Vietnamese dogs, European and Middle Eastern breeds43–45. The widespread distribution of haplotype A18 in breeds such as the Serra da Estrela Mountain Dog, Central Asian Shepherd Dog, North Caucasian Volkodav, Caucasian Shepherd Dog, Turkish Akbash and Kangal and Tibetan Mastiff highlights its importance in the historical development of LGDs39,43,46.
However, the hypothesis about the close phylogenetic relationship between the Kazakh Tobets and the Tibetan Mastiffs, which is widespread among dog breeders in Kazakhstan, is refuted by our phylogenetic analysis. According to the genetic distances determined, the Kazakh Tobet has as long an evolutionary history as the Tibetan Mastiff. As far as we know, this is the first phylogenetic study of the Kazakh Tobet from Kazakhstan. In a recent study by Yang et al., a phylogenetic analysis of 15 indigenous Chinese dog breeds, including the Kazakhstan Shepherd Dog, was conducted using genotyping data from 170,000 SNP chips47. This Shepherd Dog may belong to the Kazakh Tobets, which were brought to the Xinjiang Uygur Autonomous Region of China by Kazakhs. Yang et al. also showed that the Kazakhstan Shepherd Dog is grouped into clades that are distinct from the large Chinese clade including the Tibetan Mastiff and do not show close genetic relationship with western breeds such as the Bernese Mountain Dog, German Shepherd, Newfoundland and Rottweiler.
The WGS included only two dogs of the Kazakh Tobet breed, which is a significant limitation of this study given their high genetic diversity. Due to this diversity, the selection of a suitable dog for the WGS is not a trivial endeavour. It is expected that increasing the sample size for phylogenetic analysis will more accurately confirm or refute the current results. However, the relevance of the phylogenetic relationships of the Kazakh Tobet that have been revealed remains substantiated, as our results regarding the other breeds are consistent with previous studies27,48. In the cladogram of 161 domestic dog breeds based on the genotyping data of 170,000 SNP chips, Samoyeds, Tibetan Mastiffs, Huskies and Akitas were also grouped together. The Great Dane clade was clearly separated from the clades of breeds such as the Staffordshire Bull Terrier, the French Mastiff and the Bullmastiff. The Old English Shepherd was genetically related to the Australian Shepherd and the Bernese Mountain Dog was related to the St Bernard, the Leonberger and the Rottweiler27. In addition, a genetic relationship between the German Shepherds and the Standard Schnauzers has already been established48. It seems that in this study, where we aimed to evaluate the genetic distances between dog breeds with a limited sample, the neighbour-joining tree was well suited to highlight the clear breed-specific divergence. Nevertheless, in future work with larger datasets, the neighbour-net tree would allow deeper insights into non-tree evolutionary processes between breeds and within breeds, such as hybridisation, recombination or gene flow49. In addition, not only SNVs but also indels, which were excluded from our analysis, may improve the accuracy of phylogenetic reconstruction in our future work. Studies have shown that while SNVs are more frequent, stable and more easily aligned across sequences, making them ideal for measuring genetic divergence and evolutionary relationships, indels can also be reliable for phylogenetic analyses50,51. However, there is disagreement about the best method for defining homologous character states and coding strategies50,51. Another drawback of our research is that while microsatellite markers indicate similar biological processes and patterns as SNPs, it is essential to verify these results with genome-wide commercial SNPs. Moving forward, we plan to conduct a comprehensive genome-wide SNP analysis of Kazakh Tobet dogs to confirm their high genetic diversity, identify selective breeding signatures and assess the genetic distinctiveness of the breed compared to related breeds. Both SNP and STR markers will provide stronger support for the robustness of our results. However, given the evolving understanding of genetic diversity theories, our results may still require careful interpretation, especially when assessed solely through the lens of neutral theory, which has recently come under heavy criticism. Neutral theory, which assumed that most of the genome is variable and neutral and formed the basis for understanding genetic variation52,53, has been challenged by recent empirical studies54 showing that it does not fully explain genetic diversity. STRs, traditionally considered neutral, have been shown to have functional roles, such as binding transcription factors55, thus challenging previous assumptions. As an alternative, the theory of maximum genetic diversity has emerged56, which assumes that genetic diversity reaches a maximum saturation point. This could influence the interpretation of our results, so that they will have to be re-evaluated in the future.
Conclusion
The Kazakh Tobet dogs exhibit considerable genetic diversity and a complex genetic structure characterised by seven distinct clusters found in all populations studied from three regions of Kazakhstan and Mongolia. This intricate genetic landscape is likely the result of a historical nomadic lifestyle, regional selective breeding practises and extensive crossbreeding, which together have shaped the breed’s complex and diluted genetic profile. Phylogenetic analysis revealed the proximity of the Kazakh Tobet to the Akbash and Central Asian Shepherd Dogs, which, combined with morphological similarities, suggests a common genetic heritage or historical crossbreeding that was likely favoured by migratory events in early Asian history. The detection of haplotype A18 in both Kazakh Tobet dogs supports the hypothesis of the ancient origin of the breed. Our study provides an important clue for understanding the ancestry of LGDs in Asia and the first scientific basis for the improvement and preservation of the Kazakh Tobet breed in Kazakhstan.
Methods
Sample collection and DNA extraction
All applicable international, national and institutional guidelines for the care and use of animals were strictly followed. All procedures were approved by the Bioethics Committee of the Institute of Molecular Biology and Biochemistry named after M.A. Aitkhozhin, Almaty, Kazakhstan (# 1, 18 August 2023). The study is reported in adherence with the ARRIVE guidelines (https://arriveguidelines.org).
DNA samples from Kazakh Tobet dogs were collected using cheek swabs and/or blood samples at various dog shows, special events and through mail-in contributions. An expert from the national breed-affiliated organisation “Kansonar” evaluated the samples for compliance with the breed standard. The owners gave their informed consent for the samples and images of their dogs to be used for the research. A total of 107 samples were collected from three regions in Kazakhstan: 73 from South Kazakhstan (Pop1), 16 from East Kazakhstan (Pop2) and 8 from North Kazakhstan (Pop3) (Supplementary Fig. 1). In addition, DNA samples were collected from Kazakh Tobet dogs in Bayan-Ulgei, Mongolia, a district inhabited by ethnic Kazakhs (Pop4, n = 10). The DNA samples were transported in a portable cooler and stored at -20 °C before DNA extraction. Genomic DNA was extracted using the QIAamp DNA kit (Qiagen, MD, USA) according to the protocol specified by the manufacturer.
STR genotyping
A total of 19 highly polymorphic STR markers with a wide range of allele variations (AHTk211, CXX279, REN169O18, INU055, REN54P11, INRA21, AHT137, REN169D01, AHTh260, AHTk253, INU005, INU030, FH2848, AHT121, FH2054, REN162C04, AHTh171, REN247M23 and amelogenin for sex determination), as recommended by the International Society for Animal Genetics (ISAG), were amplified for 107 DNA samples using the Canine ISAG STR Parentage Kit (Thermo Fisher Scientific, CA, USA). Genotyping was performed using the SeqStudio™ Genetic Analyser (Thermo Fisher Scientific, CA, USA). Alleles at each microsatellite locus were then processed and manually confirmed using GeneMapper™ Software 6 (Thermo Fisher Scientific, CA, USA). Сommon thresholds were used to ensure accurate allele determination. Peak height thresholds generally varied but started at 50–150 RFUs (relative fluorescence units) to distinguish between true alleles and noise. Stutter peaks, which are common artefacts in microsatellite amplification, were filtered out with a stutter ratio threshold of approximately 15–20% of the main peak.
Analysis of the STR data
The allele frequencies of 18 STR loci were used to calculate key genetic diversity parameters, including average alleles per locus (Na), average effective alleles per locus (Ne), Shannon information index (I) and observed heterozygosity (Ho). To avoid bias due to the different population sizes, the unbiased expected heterozygosity and the unbiased inbreeding coefficient were estimated instead of the standard estimates. All calculations were performed with GenAIEX 6.557, which was also used to analyse pairwise Fst values and for principal coordinate analysis (PCoA). Amelogenin was excluded from the analysis as it does not show allelic variation. Bayesian clustering was performed using STRUCTURE v2.3.429,58. The following analysis parameters were defined: Admixture model algorithm, correlation with allele frequency, 10,000 burn-in iterations and 100,000 MCMC repeats. Ten independent analyses were performed for each K value in the range between 2 and 10 to ensure sufficient coverage of possible population structures, from minimal differentiation (K = 2) to more complex scenarios (K = 10). The R package Pophelper59 was used to calculate the most probable number of clusters according to the method of Evanno et al.29 and to create a bar plot for the best K. The R package mapmixture was used to visualise the admixture structure in geographic space60.
WGS and variant calling
The construction of DNA libraries and WGS of two DNA samples using the paired-end 150 bp (PE150) sequencing strategy was performed by BGI, Shenzhen, China on the DNBSEQ platform (average sequencing coverage 200×, with minimal variation across samples). In addition, the WGS data of 43 dogs from 24 breeds were downloaded from the NCBI SRA database using the Sratoolkit v 3.1.0 (Supplementary Table 2). Raw reads were checked for quality using FastQC v0.12.1. Metrics such as GC content, per-base sequence quality and sequence length distribution were monitored to ensure that most bases achieved a quality score of Q30 for further analysis. Adapter sequences and low-quality bases were removed with Trim-Galore v0.6.1061, using the default settings: reads with a Phred score below 20 were trimmed and sequences shorter than 20 base pairs were discarded. The resulting reads were then aligned to the dog reference genome Dog10K_Boxer_Tasha (canFam6, NCBI RefSeq Assembly GCF_000002285.5) with BWA mem v0.7.1262. The mapped reads were sorted with Samtools v1.2063. PCR duplicates were marked and read groups were added with Picard v2.27.564. Base qualities were recalibrated with BaseRecalibrator and ApplyBQSR of Genome analysis toolkit (GATK) v4.2.6.065. The genomic variants per sample were called with GATK HaplotypeCaller. A combined variant set for all samples per autosome was created with GATK GenomicsDBImport. Subsequently, GATK GenotypeGVCFs was then used to generate a set of variants for each autosome. The variants from different autosomes were consolidated with bcftools v1.20. Any potential batch effects were minimised by uniform processing of all samples, from consistent read trimming to variant calling steps. The single nucleotide variants (SNVs) and indels were then separated using vcftools v0.1.16 and the SNVs were filtered using the GATK VariantFiltration tool according to the recommended criteria for hard filtering: QD < 2.0, FS > 60.0, MQ < 40.0, MQRankSum < 12.5, ReadPosRankSum < -8.0 and SOR > 3.066.
mtDNA analysis
Reconstruction of the mitochondrial genome from WGS data is challenging due to the presence of nuclear mitochondrial DNA segments (NUMTs), which are very similar to mitochondrial DNA and can mimic true heteroplasmy13. In this study, a bioinformatic pipeline was developed to prevent the mapping of NUMT reads. The raw WGS reads were aligned to the mitochondrial dog reference genome (Canis lupus familiaris mitochondrion complete genome, NCBI RefSeq Assembly NC_002008.4) using BWA mem v0.7.1262. The resulting alignments were converted to BAM format and sorted using Samtools v1.2063. The D-loop region (positions 15458–16727) was then extracted from the BAM file and converted to FASTA using Samtools v1.2063. To exclude nuclear mitochondrial DNA sequences (NUMTs), the resulting mitochondrial sequences were then compared to the dog nuclear genome using blastn from BLAST v2.16.0. The dog nuclear genome was generated with makeblastdb from BLAST v2.16.0 based on the dog reference genome Dog10K_Boxer_Tasha (canFam6, NCBI RefSeq Assembly GCF_000002285.5). A Python script (available at https://github.com/Anastassiya2024/KazakhTobet2024/blob/main/Script.py) was then applied to filter the results and exclude NUMs with a percentage identity greater than 95%. The filtered FASTA files were aligned with auto from MAFFT v7.47567 and compared to the reference database with blastn from BLAST v2.16.0. The haplogroup was determined using the Canis mtDNA HV1 database (http://chd.vnbiology.com)68.
Phylogenetic and genetic network analysis
SplitsTree v6.0.069 was used to construct the Neighbour-Net tree based on the matrix of pairwise Fst values calculated from the STR data.
To construct phylogenetic tree based on WGS data SNVs of autosomal chromosomes were additionally filtered using Plink 1.970. Variants with a missing genotype rate more than 0.05 and a minor allele frequency (MAF) of less than 0.01 were excluded from the analysis. Genomic distance (1-IBS) was calculated in PLINK v1.9 using this dataset and converted to PHYLIP format using Phylogeny.fr71. A neighbour-joining tree was created in PHYLIP v3.69672 and visualised using ggplot2 v.3.5.073 and APE v.5.874.
To construct a phylogenetic tree of mitochondrial D-loop sequences, the aligned FASTA files for all samples were merged into a single file. Positions with more than 50% missing data (the default threshold) were filtered out using Biopython. A maximum likelihood phylogenetic tree was then generated using FastTree v2.1.1175 in Newick format and visualised using the Phylogenetic Tree (Newick) Viewer (http://etetoolkit.org/treeview/).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the dog breeders and owners who provided us with samples and information about this unique breed. We would like to thank K. T. Plakhov for providing a photo from his personal archive for this article.
Author contributions
A.P. and K.B. designed and supervised the study; A.P. and Y.P. wrote the manuscript; K.P., B.B., G.Zh. and L.Dj. edited the manuscript; K.B. visualisation; A.A., O.V., I.N., T.Dz. and A.Kh. performed the sample collection; M.B. performed formal analysis; A.Z. and A.Y. purified the DNA; Ye.K. and A.T. performed the STR analysis; A.P. and R.M. performed the bioinformatic analysis. All authors have read and approved the final version of the manuscript.
Data availability
The genomes of two Kazakh Tobet dogs sequenced for this work are available via the Short Read Archive (ncbi.nlm.nih.gov/sra; BioProject ID PRJNA1144634).
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Plakhov, K. N. & Plakhova, A. S. Kazakh Tobet - myth, reality or necessity. (2003). https://ptic-gol.forum2x2.ru/t57-topic (in Russian).
- 2.Shcherbak, A. M. Names of Domestic and wild Animals in Turkic Languages in Historical Development of the Vocabulary of Turkic Languages82–172 (Publishing House of the USSR Academy of Sciences, Institute of Linguistics, 1961). (in Russian).
- 3.Kuryshzhanov, A. Research on the vocabulary of the Old Kypchak written monument of the 13th century in Turkic-Arabic Dictionary (Alma-Ata: Nauka, (1970). (in Russian).
- 4.Novozhenov, V. Petroglyphs of Sary-Arka (Almaty, 2002). (in Russian).
- 5.Marikovskiy, P. In the Tien Shan Mountains (Alma-Ata: Kazakhstan,, 1981). (in Russian).
- 6.Sala, R. & Deom, J-M. Rock Art of Southern Kazakhstan (Laboratory of Geoarchaeology, 2005). (in Russian).
- 7.Medoev, A. G. Engravings on the Rocks (Alma-Ata: Zhalyn, 1979). (in Russian).
- 8.Gorelov, Y. K. On the Origin and Evolution of Central Asian Shepherd Dogs and Other Molossoids. ASKA Magazine (Aboriginal Dogs of the Caucasus and Asia).https://yoltay-allan.jimdofree.com/%D1%81%D1%82%D0%B0%D1%82%D1%8C%D0%B8/%D1%8E-%D0%B3%D0%BE%D1%80%D0%B5%D0%BB%D0%BE%D0%B2/%D0%BE%D1%82%D0%BA%D1%83%D0%B4%D0%B0-%D0%B2%D0%B7%D1%8F%D0%BB%D0%B8%D1%81%D1%8C-%D0%BE%D0%B2%D1%86%D1%8B-%D0%B8-%D0%BF%D1%80%D0%B0%D0%BC%D0%BE%D0%BB%D0%BE%D1%81%D1%81%D1%8B/. (2004).
- 9.Tarlykov, P. et al. Mitochondrial DNA analysis of ancient sheep from Kazakhstan: evidence for early sheep introduction. Heliyon7, e08011. 10.1016/j.heliyon.2021.e08011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pil’shchikov Yu. N. Shepherd dog breeding in Kazakhstan. Inform. Works Kazakh Res. Inst. Anim. Husb.2 (1965).
- 11.Marwal, A., Sahu, A. K. & Gaur, R. K. Molecular markers: tool for genetic analysis. Anim. Biotechnology: Models Discovery Translation. 289–305. 10.1016/B978-0-12-416002-6.00016-X (2014).
- 12.Delsuc, F., Brinkmann, H. & Philippe, H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet.6, 361–375. 10.1038/nrg1603 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Sturk-andreaggi, K. et al. The value of whole‐genome sequencing for mitochondrial DNA Population studies: strategies and Criteria for Extracting High‐Quality Mitogenome Haplotypes. Int. J. Mol. Sci.. 2310.3390/ijms23042244 (2022). [DOI] [PMC free article] [PubMed]
- 14.Chen, R. et al. Comparison of whole genome sequencing and targeted sequencing for mitochondrial DNA. Mitochondrion 58. 10.1016/j.mito.2021.01.006 (2022). [DOI] [PMC free article] [PubMed]
- 15.Parker, H. G. Genomic analyses of modern dog breeds. Mamm. Genome23, 19–27. 10.1007/s00335-011-9387-6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigi, D., Marelli, S. P., Randi, E. & Polli, M. Genetic characterization of four native Italian shepherd dog breeds and analysis of their relationship to cosmopolitan dog breeds using microsatellite markers. Animal9. 10.1017/S1751731115001561 (2015). [DOI] [PubMed]
- 17.Yang, Z. et al. Genetic characterization of four dog breeds with Illumina CanineHD BeadChip. Forensic Sci. Res.10.1080/20961790.2019.1614292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciampolini, R., Cecchi, F., Bramante, A., Casetti, F. & Presciuttini, S. Genetic variability of the Bracco Italiano dog breed based on microsatellite polimorphism. Italian J. Anim. Sci.10, 267–270. 10.4081/IJAS.2011.E59 (2016). [Google Scholar]
- 19.Wiener, P. et al. Genomic data illuminates demography, genetic structure and selection of a popular dog breed. BMC Genom.18. 10.1186/s12864-017-3933-x (2017). [DOI] [PMC free article] [PubMed]
- 20.Boccardo, A. et al. The German shorthair pointer dog breed (Canis lupus familiaris): genomic inbreeding and variability. Animals10. 10.3390/ani10030498 (2020). [DOI] [PMC free article] [PubMed]
- 21.Gajaweera, C. et al. Genetic diversity and population structure of the Sapsaree, a native Korean dog breed. BMC Genet.20. 10.1186/s12863-019-0757-5 (2019). [DOI] [PMC free article] [PubMed]
- 22.Bigi, D. et al. Investigating the population structure and genetic differentiation of livestock guard dog breeds. Animal12. 10.1017/S1751731117003573 (2018). [DOI] [PubMed]
- 23.Mellanby, R. J. et al. Population structure and genetic heterogeneity in popular dog breeds in the UK. Veterinary Journal 196. 10.1016/j.tvjl.2012.08.009 (2013). [DOI] [PubMed]
- 24.Ali, M. B. et al. Genetic analysis of the modern Australian labradoodle dog breed reveals an excess of the poodle genome. PLoS Genet.16. 10.1371/journal.pgen.1008956 (2020). [DOI] [PMC free article] [PubMed]
- 25.Pfahler, S. & Distl, O. Effective population size, extended linkage disequilibrium and signatures of selection in the rare dog breed lundehund. PLoS One10. 10.1371/journal.pone.0122680 (2015). [DOI] [PMC free article] [PubMed]
- 26.Mastrangelo, S. et al. Genome-wide diversity and runs of homozygosity in the ‘Braque Français, type Pyrénées’ dog breed. BMC Res. Notes 11. 10.1186/s13104-017-3112-9 (2018). [DOI] [PMC free article] [PubMed]
- 27.Parker, H. G. et al. Genomic Analyses Reveal the Influence of Geographic Origin, Migration, and hybridization on Modern Dog Breed Development. Cell. Rep.19. 10.1016/j.celrep.2017.03.079 (2017). [DOI] [PMC free article] [PubMed]
- 28.Ahn, B. et al. Origin and population structure of native dog breeds in the Korean peninsula and East Asia. iScience26. 10.1016/j.isci.2023.106982 (2023). [DOI] [PMC free article] [PubMed]
- 29.Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol.14, 2611–2620. 10.1111/j.1365-294X.2005.02553.x (2005). [DOI] [PubMed] [Google Scholar]
- 30.Perfilyeva, A. et al. Assessment of the genetic diversity of the Kazakh national dog breed Tobet in the southern region of Kazakhstan. 3i: Intellect. idea Innov. - интеллект идея инновация. 1, 58–67. 10.52269/22266070_2024_1_58 (2024). [Google Scholar]
- 31.Ye, J. H. et al. Microsateliite-based genetic diversity and evolutionary relationships of six dog breeds. Asian-Australasian J. Anim. Sci.22. 10.5713/ajas.2009.80493 (2009).
- 32.Pedersen, N. C., Pooch, A. S. & Liu, H. A genetic assessment of the English bulldog. Canine Genet. Epidemiol.3. 10.1186/s40575-016-0036-y (2016). [DOI] [PMC free article] [PubMed]
- 33.Radko, A. & Podbielska, A. Microsatellite dna analysis of genetic diversity and parentage testing in the popular dog breeds in Poland. Genes (Basel) 12. 10.3390/genes12040485 (2021). [DOI] [PMC free article] [PubMed]
- 34.Ren, D. R. et al. Strong heterozygote deficit in tibetan Mastiff of China based on microsatellite loci. Animal3, 1213–1215. 10.1017/S1751731109004704 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Lee, E. W., Choi, S. K. & Cho, G. J. Molecular genetic diversity of the Gyeongju Donggyeong dog in Korea. J. Vet. Med. Sci.76. 10.1292/jvms.14-0189 (2014). [DOI] [PMC free article] [PubMed]
- 36.S García, L. et al. (ed, A.) Genetic structure of the ca Rater Mallorquí Dog Breed inferred by microsatellite markers. Animals12. 10.3390/ani12202733 (2022). [DOI] [PMC free article] [PubMed]
- 37.Perfilyeva, A. et al. Kazakh national dog breed tazy: what do we know? PLoS One18. 10.1371/journal.pone.0282041 (2023). [DOI] [PMC free article] [PubMed]
- 38.Pedersen, N., Liu, H., Theilen, G. & Sacks, B. The effects of dog breed development on genetic diversity and the relative influences of performance and conformation breeding. J. Anim. Breed. Genet.130. 10.1111/jbg.12017 (2013). [DOI] [PubMed]
- 39.Riabinina, O. M. Mitochondrial DNA variation in Asian guardian dogs. Genetika42 (2006). [PubMed]
- 40.Kazakhstan – Silk Road Research. https://silkroadresearch.blog/silk-road-countries/kazakhstan/ (2018).
- 41.Pang, J. F. et al. MtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol. Biol. Evol.26. 10.1093/molbev/msp195 (2009). [DOI] [PMC free article] [PubMed]
- 42.Thai, Q. K., Nguyen, T. T. & Pham, H. T. mtDNA haplotype network analysis: exploring genetic relationships and diversity in dog haplogroups. GSC Biol. Pharm. Sci.24. 10.30574/gscbps.2023.24.1.0284 (2023).
- 43.van Asch, B. et al. MtDNA diversity among four Portuguese autochthonous dog breeds: a fine-scale characterisation. BMC Genet.6. 10.1186/1471-2156-6-37 (2005). [DOI] [PMC free article] [PubMed]
- 44.Thai, Q. K. et al. HV1 mtDNA reveals the high genetic diversity and the ancient origin of Vietnamese dogs. Animals13. 10.3390/ani13061036 (2023). [DOI] [PMC free article] [PubMed]
- 45.Brown, S. K. et al. Phylogenetic distinctiveness of Middle Eastern and Southeast Asian village dog Y chromosomes illuminates dog origins. PLoS One6. 10.1371/journal.pone.0028496 (2011). [DOI] [PMC free article] [PubMed]
- 46.Li, Y. & Zhang, Y. P. High genetic diversity of tibetan mastiffs revealed by mtDNA sequences. Sci. Bull.10.1007/s11434-012-4995-4 (2012). [Google Scholar]
- 47.Yang, Q. et al. Genetic diversity and signatures of selection in 15 Chinese indigenous dog breeds revealed. By genome-wide SNPs. Front. Genet.10. 10.3389/fgene.2019.01174 (2019). [DOI] [PMC free article] [PubMed]
- 48.Vonholdt, B. M. et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature464. 10.1038/nature08837 (2010). [DOI] [PMC free article] [PubMed]
- 49.Woolley, S. M., Posada, D. & Crandall, K. A. A comparison of phylogenetic network methods using computer simulation. PLoS One3(4), 10.1371/journal.pone.0001913 (2008). [DOI] [PMC free article] [PubMed]
- 50.Young, N. D. & Healy, J. GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinform.4. 10.1186/1471-2105-4-6 (2003). [DOI] [PMC free article] [PubMed]
- 51.Donath, A. & Stadler, P. F. Split-inducing indels in phylogenomic analysis. Algorithms Mol. Biol. 13. 10.1186/s13015-018-0130-7 (2018). [DOI] [PMC free article] [PubMed]
- 52.Nei, M., Suzuki, Y. & Nozawa, M. The neutral theory of molecular evolution in the genomic era. Annu. Rev. Genom Hum. Genet.11, 265–289. 10.1146/annurev-genom-082908-150129 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Nei, M. Selectionism and neutralism in molecular evolution. Mol. Biol. Evol.22. 10.1093/molbev/msi242 (2005). [DOI] [PMC free article] [PubMed]
- 54.Lynch, M., Wei, W., Ye, Z. & Pfrender, M. The genome-wide signature of short-term temporal selection. Proc. Natl. Acad. Sci. U. S. A.121, 720–731. 10.1073/pnas.2307107121 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horton, C. A. et al. Short tandem repeats bind transcription factors to tune eukaryotic gene expression. Science381, 6664. 10.1126/science.add1250 (2023). [DOI] [PubMed] [Google Scholar]
- 56.Huang, S. The overlap feature of the genetic equidistance result—a fundamental biological phenomenon overlooked for nearly half of a century. Biol. Theory5, 1–9. 10.1162/BIOT_a_00021 (2010). [Google Scholar]
- 57.Peakall, R. & Smouse, P. E. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes6. 10.1111/j.1471-8286.2005.01155.x (2006). [DOI] [PMC free article] [PubMed]
- 58.Falush, D., Stephens, M. & Pritchard, J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics164. 10.1093/genetics/164.4.1567 (2003). [DOI] [PMC free article] [PubMed]
- 59.Francis, R. M. Pophelper: an R package and web app to analyse and visualize population structure. Mol. Ecol. Resour.10.1111/1755-0998.12509 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Jenkins, T. L. & mapmixture An R package and web app for spatial visualisation of admixture and population structure. Mol. Ecol. Resour. 24. 10.1111/1755-0998.13943 (2024). [DOI] [PubMed]
- 61.Krueger, F. Trim Galore! Babraham Bioinformatics (2019).
- 62.Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics26. 10.1093/bioinformatics/btp698 (2010). [DOI] [PMC free article] [PubMed]
- 63.Li, H. et al. The sequence alignment / map (SAM) format and SAMtools 1000 Genome Project Data Processing Subgroup. Bioinformatics25 (2009). [DOI] [PMC free article] [PubMed]
- 64.Broad Institute. Picard toolkit. (2019).
- 65.Van der Auwera, G. A. et al. From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protocols Bioinf. SUPL4310.1002/0471250953.bi1110s43 (2013). [DOI] [PMC free article] [PubMed]
- 66.Jónás, D., Sándor, S., Tátrai, K., Egyed, B. & Kubinyi, E. A preliminary study to investigate the genetic background of Longevity based on Whole-Genome Sequence Data of Two Methuselah Dogs. Front. Genet.11. 10.3389/fgene.2020.00315 (2020). [DOI] [PMC free article] [PubMed]
- 67.Katoh, K., Rozewicki, J. & Yamada, K. D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform.20, 1160–1165. 10.1093/bib/bbx108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thai, Q. K., Chung, D. A. & Tran, H. D. Canis mtDNA HV1 database: a web-based tool for collecting and surveying Canis mtDNA HV1 haplotype in public database. BMC Genet. 18. 10.1186/s12863-017-0528-0 (2017). [DOI] [PMC free article] [PubMed]
- 69.Bryant, D. & Huson, D. H. NeighborNet: improved algorithms and implementation. Front. Bioinform. 3. 10.3389/fbinf.2023.1178600 (2023). [DOI] [PMC free article] [PubMed]
- 70.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4. 10.1186/s13742-015-0047-8 (2015). [DOI] [PMC free article] [PubMed]
- 71.Dereeper, A. et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res.36. 10.1093/nar/gkn180 (2008). [DOI] [PMC free article] [PubMed]
- 72.Baum, B. R. P. H. Y. L. I. P. Phylogeny inference Package. Version 3.2. Joel Felsenstein. Q. Rev. Biol.64. 10.1086/416571 (1989).
- 73.Wilkinson, L. ggplot2: Elegant Graphics for Data Analysis by WICKHAM, H. Biometrics 67. 10.1111/j.1541-0420.2011.01616.x (2011).
- 74.Paradis, E., Claude, J., Strimmer, K. & APE Analyses of phylogenetics and evolution in R language. Bioinformatics10.1093/bioinformatics/btg412 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol.26, 1641–1650. 10.1093/molbev/msp077 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomes of two Kazakh Tobet dogs sequenced for this work are available via the Short Read Archive (ncbi.nlm.nih.gov/sra; BioProject ID PRJNA1144634).