Abstract
Immune evasion represents a crucial milestone in the progression of cancer and serves as the theoretical foundation for tumor immunotherapy. In this study, we reveal a negative association between Human Papillomavirus (HPV)-encoded circular RNA, circE7, and the infiltration of CD8+ T cells in head and neck squamous cell carcinoma (HNSCC). Both in vitro and in vivo experiments demonstrate that circE7 suppresses the function and activity of T cells by downregulating the transcription of LGALS9, which encodes the galectin-9 protein. The molecular mechanism involves circE7 binding to acetyl-CoA carboxylase 1 (ACC1), promoting its dephosphorylation and thereby activating ACC1. Activated ACC1 reduces H3K27 acetylation at the LGALS9 gene promoter, leading to decreased galectin-9 expression. Notably, galectin-9 interacts with immune checkpoint molecules TIM-3 and PD-1, inhibiting the secretion of cytotoxic cytokines by T cells and promoting T cell apoptosis. Here, we demonstrate a mechanism by which HPV promotes immune evasion in HNSCC through a circE7-driven epigenetic modification and propose a potential immunotherapy strategy for HNSCC that involves the combined use of anti-PD-1 and anti-TIM-3 inhibitors.
Subject terms: Cancer epigenetics, Head and neck cancer, Tumour immunology
Immune evasion is a feature of HPV-positive head and neck squamous cell carcinoma (HNSCC). Here the authors report that a circular RNA, circE7, encoded by HPV promotes immune evasion in HNSCC by promoting downregulation of galectin-9.
Introduction
Head and neck squamous cell carcinoma (HNSCC) stands as one of the most prevalent malignant tumors worldwide. In 2020, ~880,000 new cases were confirmed, resulting in over 440,000 fatalities1,2. The conventional treatment options for HNSCC include surgical resection, radiotherapy, and chemotherapy3. Recently, immunotherapy utilizing immune checkpoint inhibitors, such as PD-1/PD-L1 monoclonal antibodies, has emerged as a promising approach for the treatment of refractory recurrent/metastatic HNSCC4. This groundbreaking therapy has significantly enhanced overall patient survival. Nonetheless, it is essential to recognize that more than 80% of patients still exhibit inherent drug resistance and do not benefit from PD-1/PD-L1 immunotherapy alone5–9. These observations underscore the potential of other immune checkpoint molecules in the immune evasion in HNSCC, influencing the efficacy of anti-PD-1/PD-L1 immunotherapy. Therefore, further exploration of the mechanisms underlying immune escape in HNSCC assumes paramount importance. Such research has the potential to reveal targets and provide innovative strategies for HNSCC immunotherapy.
Human Papillomavirus (HPV) infection can instigate the onset and progression of diverse forms of HNSCCs, affecting regions such as the oropharynx, larynx, and oral cavity5,10,11. HPV, a non-enveloped, spherical, double-stranded DNA virus with a genome spanning ~7–8 kb, primarily targets human skin and mucosal epithelial cells. To date, the genomes of almost 500 distinct HPV types have been isolated and sequenced. Among these, high-risk types like HPV16, 18, 31, 33, and 35, along with others, are prone to inducing various human malignancies12,13. The incidence of HNSCC attributed to high-risk HPV infections is steadily increasing at a rate of 5% per year, with HPV16 being the most prevalent high-risk subtype12. HNSCC linked to high-risk HPV infections exhibits distinctive clinical characteristics, including a higher incidence of TP53 mutations14,15. The HPV genome encompasses three functional regions: the early coding region (E), the late coding region (L), and the upstream regulatory region (URR). The early proteins encoded by the E region (E1, E2, E4, E5, E6, and E7, etc.) serve crucial roles in HPV replication, transcription, translation, and cell transformation. Notably, E6 and E7 are the most critical oncogenic proteins, as they facilitate tumorigenesis by suppressing the expression of tumor suppressor factors like p53 and Rb. They also contribute to tumorigenesis by enhancing genomic instability, inhibiting telomere shortening, promoting tumor cell immortalization, and fostering processes such as angiogenesis, invasion, and metastasis16. Furthermore, HPV can influence immune cell populations, immune checkpoints, effector molecules, and both innate and adaptive immune responses within the tumor microenvironment. These alterations aid HPV-related cancers in evading immune surveillance15,17,18. For instance, the HPV-encoded E5 protein can mediate immune tolerance to PD-L1 immune therapy, while proteins E2, E6, and E7 are closely associated with the dysfunction of cytotoxic T lymphocytes19,20.
Circular RNA is generated by the reverse splicing of exons from precursor mRNA, forming a ring structure connected at the head and tail without a 5’ cap and 3’ tail. Circular RNA participates in many biological processes of tumor development through mechanisms such as molecular sponges with miRNA and proteins, influencing transcription, interfering with the normal splicing of precursor mRNA, regulating the translation of mRNA molecules, forming circular RNA-protein complexes, and competitively binding proteins with mRNA21,22. Recently, it has come to light that HPV can also encode circular RNA, with circE7 being the sole identified circular RNA specifically encoded by high-risk HPV16. circE7 spans 472 nucleotides and is formed through the process of reverse splicing involving the 3’-terminal sequence of the HPV gene E6 and E7, and the 5’-terminal sequence of the E1 gene23. Despite this discovery, the biological function of circE7 remains largely unknown, and its involvement in tumor immune evasion and immunotherapy resistance has not been reported.
In this work, we observed that the expression of circE7 was negatively correlated with infiltrating T cells in HPV+ HNSCC clinical tissues. Both in vitro and in vivo experiments further affirmed that circE7 hampered T cell function and activity by downregulating the LGALS9 gene through an unappreciated epigenetic mechanism, consequently promoting immune evasion in HNSCC. Notably, the LGALS9 gene encodes the galectin-9 protein, which can bind to immune checkpoint molecules like TIM-3 and PD-1 on the surface of T cells. This binding activates T cells to release cytotoxic cytokines and inhibits T cell apoptosis. Importantly, combining monoclonal antibodies targeting both PD-1 and TIM-3 significantly enhances the effectiveness of HNSCC immunotherapy. This study uncovers a mechanism by which HPV-encoded circE7 orchestrates tumor immune evasion, thereby advancing the progression of HNSCC. Moreover, it underscores a combinational therapeutic option for enhanced HNSCC immunotherapy.
Results
circE7 is expressed in HPV+ HNSCC and negatively correlated with T cell infiltration and galectin-9 expression
To explore the role of HPV circE7 in head and neck tumors, we assessed the expression of circE7 in 23 pairs of HNSCC tumor samples by PCR experiments. The data showed that circE7 could be detected in 7 tumor tissues of 23 patients, while it was nearly absent in the adjacent normal epithelial tissues (Supplementary Data 1, Supplementary Fig. S1A). circE7 was also detected in HPV-positive SCC090, SCC154, and Caski cell lines, but not in HPV-negative HN30 and CAL27 cell lines (Supplementary Fig. S1A). The specific primers targeting the splicing sites of the circE7 sequence were designed and circE7 was validated to exist in HPV-positive HNSCC SCC090 and SCC154 cells through RT-qPCR, followed by Sanger sequencing (Fig. 1A). To explore the circular nature of circE7, actinomycin D treatment and RNase R digestion experiments were conducted and showed that circE7 was more stable than linear HPV-encoded mRNA E6, aligning with typical circular RNA properties (Supplementary Fig. S1B, C). RNA fluorescence in situ hybridization (FISH) and RNA nucleosome separation experiments showed that the distribution of circE7 was within both the nucleus and cytoplasm (Supplementary Fig. S1D, E).
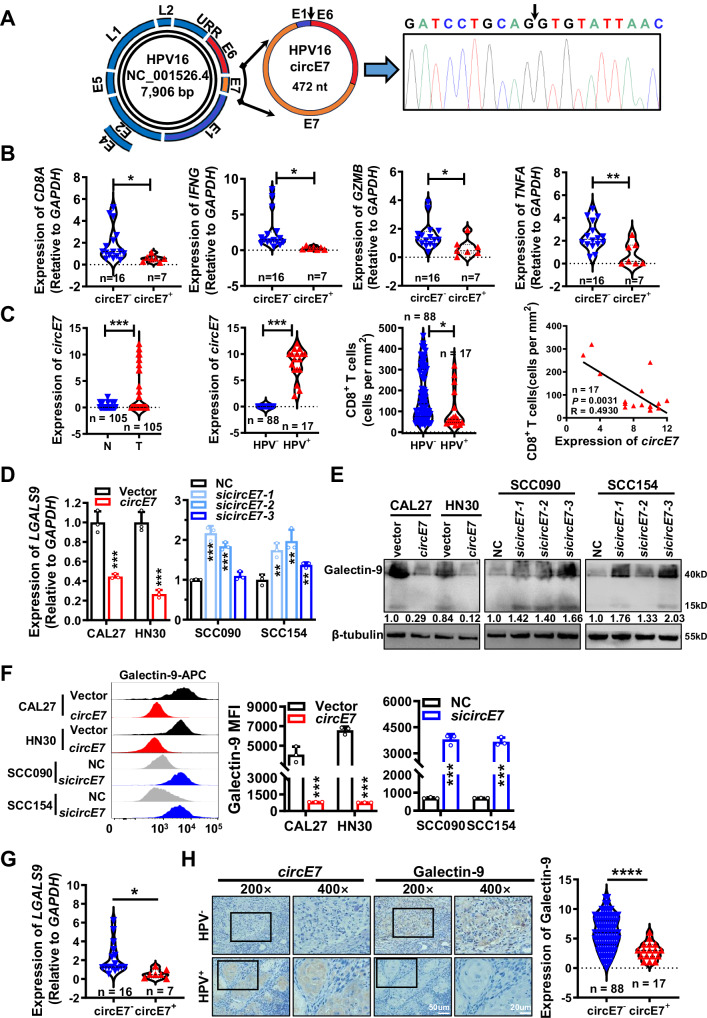
Fig. 1. circE7 is highly expressed in HPV+ HNSCC and negatively correlated with CD8+ T cell function and the expression of immune checkpoint galectin-9.
A By designing primers across the splicing sites and using Sanger sequencing, circE7 was identified as a 472nt circular RNA formed through the reverse splicing of HPV16 E6 tail, E7, and partial E1 sequences. B In the 23 HNSCC tissues used in Supplementary Fig. S1A, RT-qPCR was performed to detect the expression of CD8A, IFNG, GZMB, and TNFA, and their correlation with circE7 expression was analyzed. The expression of CD8A, IFNG, GZMB, and TNFA was significantly lower in circE7+ HNSCC samples compared to that in circE7- samples. (circE7-, n = 16; circE7+, n = 7). C In 105 HNSCC tissues, circE7 expression was detected by in situ hybridization. Expressions of p16, HPV16 E7 protein, and CD8A in infiltrating CD8+ T cells were detected by immunohistochemistry. Among these 105 HNSCC cases, 17 were HPV+ and all of which were HPV16+ with circE7 expression. The expression level of circE7 was inversely correlated with CD8+ T cell infiltration. (HPV-, n = 88; HPV+, n = 17). D–F. circE7 was overexpressed in HPV- HNSCC cell lines CAL27 and HN30, or siRNA targeting circE7 was transfected into HPV16+ HNSCC cell lines SCC090 and SCC154. RT-qPCR was used to detect LGALS9 mRNA (D), western blot (E), and Flow cytometry (F) were used to detect galectin-9 protein expression. F Left panel, flow cytometry analysis; right panel, statistical results. MFI: median fluorescence intensity, mean ± SD of three independent experiments. G RT-qPCR was performed to detect LGALS9 mRNA in the 23 fresh HNSCC tissue samples used in (B). LGALS9 mRNA level was significantly lower in circE7+ HNSCC tissues compared to that in circE7- tissues. (circE7-, n = 16; circE7+, n = 7). H Galectin-9 expression was detected by immunohistochemistry in the 105 HNSCC tissues used in (C), and its correlation with circE7 expression was analyzed. Left panel, representative images; right panel, statistical results. Magnification, ×200, scale bar, 50 μm; Magnification, ×400, scale bar, 20 μm. (circE7-, n = 88; circE7+, n = 17). Correlations were calculated using the parametric two-tailed Pearson correlation test for (C) and unpaired two-tailed Student’s t test for (B, C, D, F, G, H), *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Each western blots were reproduced three times with similar results (E). Data are presented as mean ± SD. Source data are provided as a Source Data File.
To identify the function of circE7 on immune evasion of HNSCC, we detected the expression of CD8A in the above 23 samples, as well as the T cell effector molecules TNF-α, GZMB, and IFN-γ within CD8+ T cells using RT-qPCR. Interestingly, the CD8A expression, as well as the T cell effector molecules TNF-α, GZMB, and IFN-γ, within CD8+ T cells in 7 circE7 positive HNSCC tissues (n = 7) was obviously lower than 16 circE7 negative HNSCC tissues (Fig. 1B). In situ hybridization also showed that 17 of 105 HNSCC patients could be detected the existence of circE7 (Supplementary Data 2). Remarkably, the circE7 positive tumor tissues exhibited fewer infiltrating lymphocytes, such as CD8+ T cells, which are pivotal in tumor immune surveillance and immune escape (Fig. 1C and Supplementary Fig. S1F). These findings suggest that HPV16-encoded circE7 may contribute to HNSCC immune evasion by inhibiting T cell infiltration, particularly CD8+ T cells.
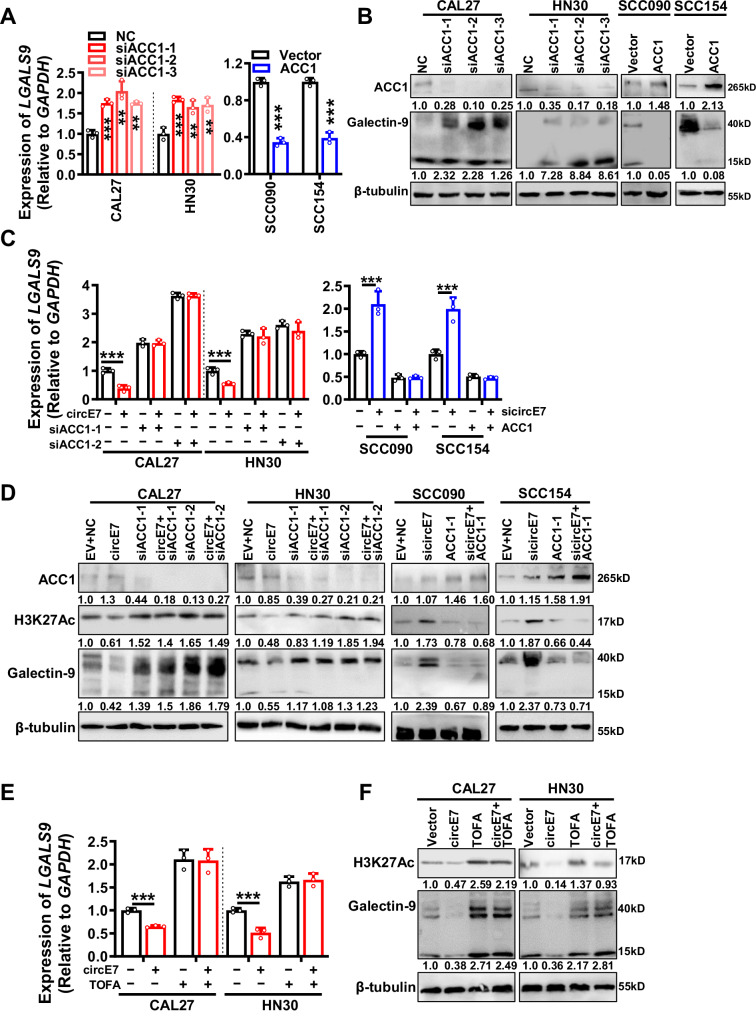
Immune checkpoint molecules play an important role in the immune surveillance of tumor cells. To investigate the function of immune checkpoint molecules in HNSCC, we analyzed the expression of immune checkpoint molecules using the TCGA database. The data showed that a total of 15 immune checkpoint molecules were significantly differential expressed in HNSCC tissues (Supplementary Fig. S2A). Then circE7 was overexpressed in HPV- HNSCC cell lines CAL27 and HN30 or knocked down in HPV16+ HNSCC cell lines SCC090 and SCC154 using three siRNAs specifically targeting circE7 (Supplementary Fig. S2B). Both RT-qPCR and western blotting showed that circE7 significantly only inhibited the expression of galectin-9 encoded by the LGALS9 gene (Fig. 1D, E and Supplementary Fig. S2C). Flow cytometry experiments (Fig. 1F) and immunofluorescence (Supplementary Fig. S2D) also confirmed the inhibition of galectin-9 by circE7. Further, we evaluated the correlation between the expression of LGALS9 and circE7 in HNSCC clinical samples. RT-qPCR analysis showed that LGALS9 expressed lower in 7 circE7 positive samples, compared with 16 circE7 negative samples, and LGALS9 expression negatively correlated with the expression of CD8A, IFNG, GZMB, and TNFA (Fig. 1G and Supplementary Fig. S2E). Immunohistochemistry confirmed the correlation between galectin-9 and circE7 in 105 HNSCC samples (Fig. 1H). Low LGALS9 expression was closely associated with a poor prognosis in the above 105 HNSCC patients (clinical information was lacking in 4 cases, Supplementary Fig. S2F). The TCGA data also showed a strong correlation between low LGALS9 expression and the function of CD8+ T cells and poor prognosis in HNSCC patients (Supplementary Fig. S2G, H). These results collectively suggest that circE7 may impede the function of cytotoxic T cells by downregulating LGALS9.
circE7 promotes HNSCC immune escape by downregulating LGALS9 expression
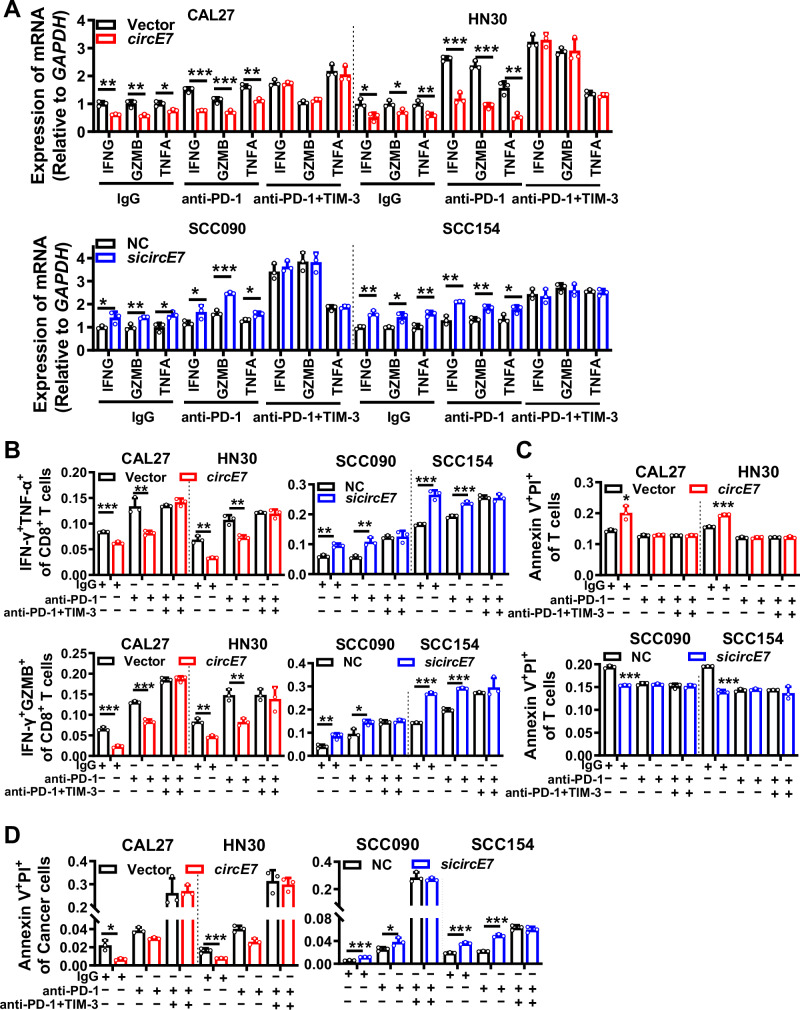
To explore the function of LGALS9 in HNSCC immune evasion, LGALS9 was overexpressed or knocked down in HNSCC cells before co-culture with human primary T cells (Supplementary Fig. S3A, B). RT-qPCR (Fig. 2A) and ELISA assays (Supplementary Fig. S3C) showed that LGALS9 collectively revealed a significant decrease in the levels of effector cytokines IFN-γ, GZMB, and TNF-α in T cells. Flow cytometry analysis further demonstrated a noteworthy rise in the proportion of IFN-γ+ GZMB+ and IFN-γ+ TNF-α+ cells within the CD8+ T cell population upon LGALS9 overexpression, whereas knockdown of LGALS9 had the opposite effect (Supplementary Fig. S3D, E). Additionally, overexpression of LGALS9 in HNSCC cells was associated with a reduction in the apoptosis of co-cultured CD8+ T cells, while knocking down LGALS9 promoted CD8+ T cell apoptosis (Supplementary Fig. S3F, G). After co-culturing with CD8+ T cells, the apoptosis ratio of head and neck tumor cells significantly increased in the LGALS9 overexpressed group, while it significantly decreased in the LGALS9 knockdown group (Supplementary Fig. S3H, I). These compelling findings collectively indicate that LGALS9 enhances the cytotoxic potential of CD8+ T cells against tumor cells in HNSCC.
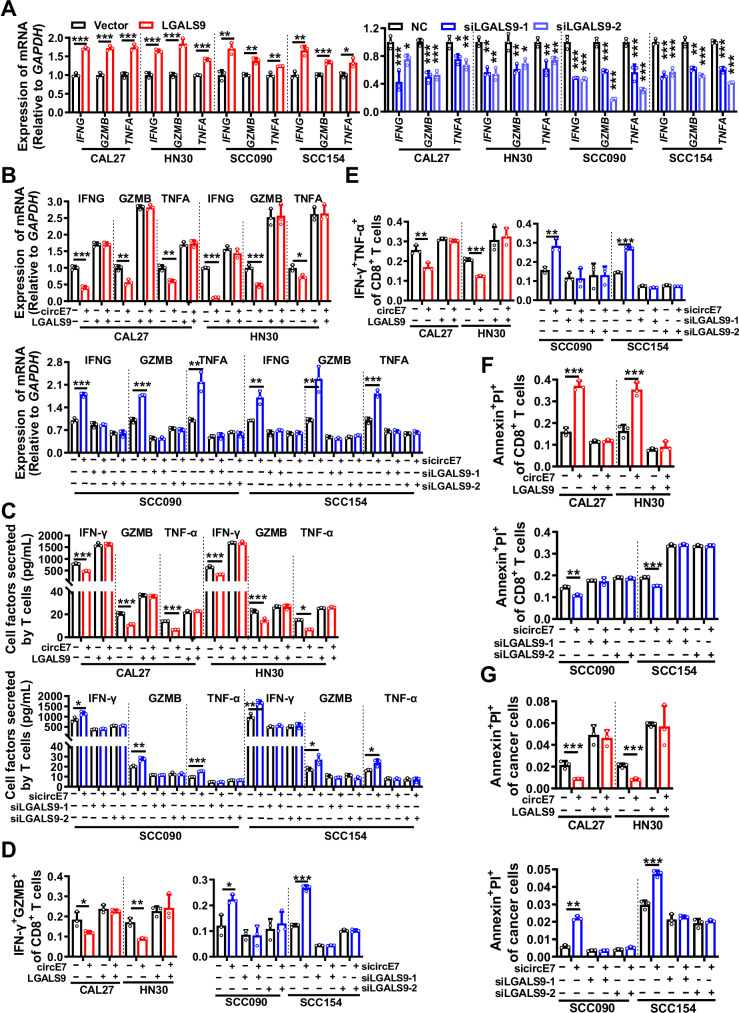
Fig. 2. circE7 downregulates galectin-9 expression in HNSCC cells to inhibit CD8+ T cell function in vitro.
In CAL27, HN30, SCC090, and SCC154 cells, LGALS9 was overexpressed or knocked down, followed by coculture with primary human T cells at a ratio of 1:10 for 6 h. A RT-qPCR was performed to measure the expression levels of IFNG, GZMB, and TNFA in primary T cells. In CAL27 and HN30 cell lines, both circE7 and LGALS9 were simultaneously overexpressed. In SCC090 and SCC154 cell lines, circE7 and LGALS9 were simultaneously knocked down. These cells were then cocultured with human primary T cells for 6 h. B RT-qPCR was performed to measure the expression of IFNG, GZMB, and TNFA mRNA. C ELISA was conducted to measure the levels of IFN-γ, GZMB, and TNF-α in the coculture media. D–E Flow cytometry was conducted to determine the proportion of IFN-γ+ GZMB+ and IFN-γ+ TNF-α+ T cells among total CD8+ T cells, as shown in Supplementary Fig. S4A. F Annexin V/PI staining followed by flow cytometry was employed to measure the proportion of apoptotic CD8+ T cells, as shown in Supplementary Fig. S4B. G Annexin V/PI staining followed by flow cytometry was used to assess the proportion of apoptotic tumor cells, as shown in Supplementary Fig. S4C. Correlations were calculated using unpaired two-tailed Student’s t test for (A–G), *p < 0.05, **p < 0.01, and ***p < 0.001. These experiments were derived from three independent repetitions. Data are presented as mean ± SD. Source data are provided as a Source Data File.
To identify if circE7 may hinder T cell function and activity by downregulating LGALS9 expression to facilitate immune evasion in HNSCC, circE7 and LGALS9 were simultaneously overexpressed in HPV16- HNSCC cell lines CAL27 and HN30, or both circE7 and LGALS9 were simultaneously knocked down in HPV16+ HNSCC cell lines SCC090 and SCC154 before co-culture with T cells for 6 h. RT-qPCR (Fig. 2B), ELISA (Fig. 2C), and flow cytometry analysis (Fig. 2D, E and Supplementary Fig. S4A) showed that overexpression of circE7 significantly suppressed the expression of effector cytokines IFN-γ, GZMB, and TNF-α in CD8+ T cells. When LGALS9 expression was restored, the inhibition of CD8+ T cell effector cytokine expression caused by circE7 could be counteracted. The opposite results were obtained in HPV16+ HNSCC cell lines SCC090 and SCC154 after circE7 and LGALS9 simultaneous knockdown (Fig. 2B–E, Supplementary Fig. S4A). To assess apoptosis in co-cultured primary T cells and tumor cells, we employed Annexin V/PI staining. The results revealed that overexpression of LGALS9 in HNSCC cells could reverse the apoptosis of CD8+ T cells induced by circE7, while reducing tumor cell apoptosis. Conversely, the simultaneous knockdown of circE7 and LGALS9 in HNSCC cells resulted in the opposite results (Fig. 2F, G and Supplementary Fig. S4B, C). These findings provide convincing evidence that LGALS9 is a critical downstream molecule of circE7 that promotes immune evasion in HNSCC.
Galectin-9 enhances the anti-tumor activity of CD8+ T cells via binding to TIM-3 and PD-1
TIM-3 acts as the receptor for galectin-9 on T cell’s surface. To explore whether galectin-9 augments T cell’s anti-tumor activity via TIM-3, HNSCC cells were transfected with the LGALS9 overexpression vector and incubated with a TIM-3 monoclonal antibody. Flow cytometry analysis disclosed that blocking TIM-3 with a monoclonal antibody impeded galectin-9’s capacity to enhance the secretion of cytotoxic cytokines by co-cultured T cells (Supplementary Fig. S5A). This implies that galectin-9 promotes the release of cytotoxic cytokines by binding to TIM-3 on T cell surface. However, the addition of a TIM-3 monoclonal antibody did not influence galectin-9’s suppressive effect on CD8+ T cell apoptosis (Supplementary Fig. S5B), suggesting that galectin-9 may inhibit T cell apoptosis through alternative pathways.
Furthermore, recent studies have reported that galectin-9 can interact with the PD-1 receptor on CD8+ T cell surface, thereby inhibiting T cell apoptosis. This interaction between galectin-9 and PD-1 can be disrupted by lactose24. When HNSCC cells were transfected with the galectin-9 expression vector and incubated with a PD-1 monoclonal antibody, galectin-9 could continue to regulate the secretion of cytotoxic cytokines by T cells (Supplementary Fig. S5C), but its capacity to inhibit CD8+ T cell apoptosis significantly diminished (Supplementary Fig. S5D). Similarly, blocking galectin-9’s binding to PD-1 with lactose yielded analogous outcomes to the addition of a PD-1 monoclonal antibody (Fig. 3A, B and Supplementary Fig. S5E, F). When HNSCC cells were incubated with both PD-1 and TIM-3 monoclonal antibodies, the regulatory effects of galectin-9 on T cell secretion of cytotoxic cytokines (Fig. 3C and Supplementary Fig. S5G) and apoptosis (Fig. 3D and Supplementary Fig. S5H) were obstructed. These findings suggest that galectin-9 can bind to both TIM-3 and PD-1 on T cell surface, promoting the secretion of cytotoxic cytokines by CD8+ T cells and inhibiting CD8+ T cell apoptosis, thereby synergistically enhancing T cell function and activity.
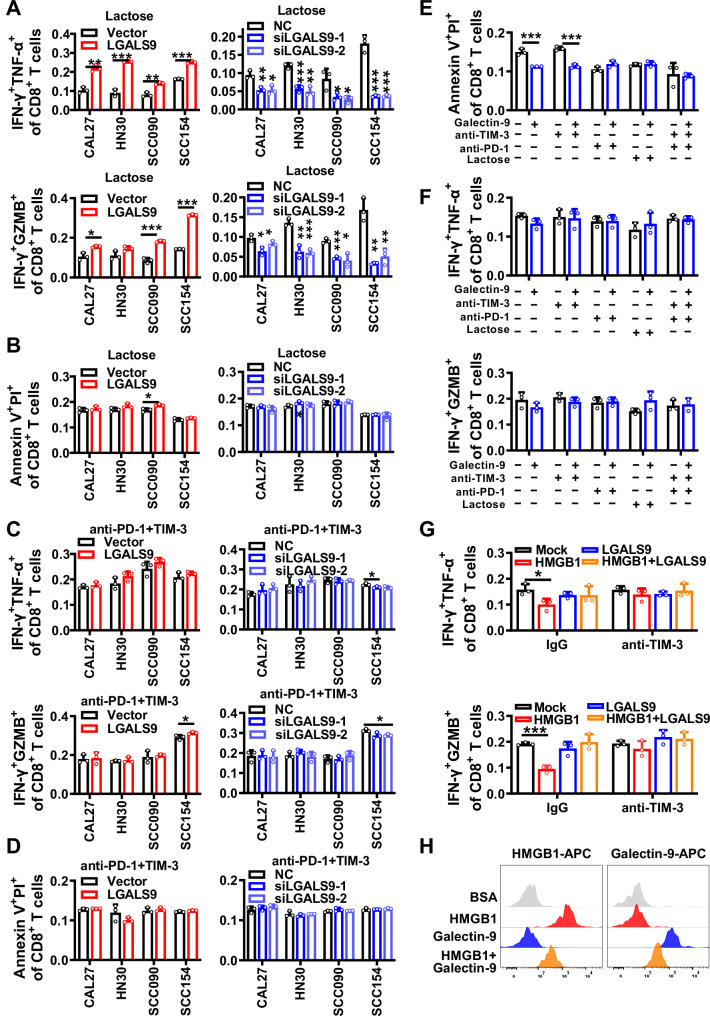
Fig. 3. Galectin-9 enhances the function and activity of T cells via binding with TIM-3 and PD-1.
In CAL27, HN30, SCC090, and SCC154 cells, overexpression or knockdown of galectin-9 was performed, followed by the addition of 10 mg/mL lactose and co-cultured with human primary T cells at 1:10 ratio for 6 h. A Flow cytometry to detect the percentage of IFN-γ+ GZMB+ and IFN-γ+ TNF-α+ cells within the total CD8+ T cell population, as shown in Supplementary Fig. S5E. B Annexin V/PI assay to assess the apoptosis level of CD8+ T cells, as shown in Supplementary Fig. S5F. In CAL27, HN30, SCC090, and SCC154 cells, overexpression or knockdown of galectin-9 was performed, followed by the addition of 5 μg/mL anti-PD-1 and anti-TIM-3 and co-cultured with human primary T cells were co-cultured at 1:10 ratio for 6 h. C Flow cytometry to detect the percentage of IFN-γ+ GZMB+ and IFN-γ+ TNF-α+ cells within the total CD8+ T cell population, as shown in Supplementary Fig. S5G. D Annexin V/PI assay to assess the apoptosis level of CD8+ T cells. Furthermore, human primary T cells were incubated with 1 μg/mL recombinant galectin-9 protein, along with the addition of anti-TIM-3, anti-PD-1, and lactose, as shown in Supplementary Fig. S5H. E Annexin V/PI assay was used to assess the apoptosis level of CD8+ T cells, as shown in Supplementary Fig. S6A. F Flow cytometry to detect the percentage of IFN-γ+ GZMB+ and IFN-γ+ TNF-α+ cells within the total CD8+ T cell population, as shown in Supplementary Fig. S6B. Human primary CD8+ T cells were co-cultured with 1 μg/mL recombinant galectin-9, HMGB1 protein and anti-TIM-3, as shown in Supplementary Fig. S6C. G Flow cytometry to detect the percentage of IFN-γ+ GZMB+ and IFN-γ+ TNF-α+ cells within the total CD8+ T cell population. BSA was used as a negative control, as shown in Supplementary Fig. S6D. H Flow cytometry to detect the level of galectin-9 and HMGB1 proteins attached to the surface of CD8+ T cells. Correlations were calculated using unpaired two-tailed Student’s t test for (A–H), *p < 0.05, **p < 0.01, and ***p < 0.001. These experiments were derived from three independent repetitions. Data are presented as mean ± SD. Source data are provided as a Source Data File.
Interestingly, when primary T cells were incubated with the galectin-9 recombinant protein for 24 h, flow cytometry analysis revealed a significant reduction in CD8+ T cell apoptosis (Fig. 3E and Supplementary Fig. S6A), but no noticeable alteration in the expression of cytotoxic cytokines (Fig. 3F and Supplementary Fig. S6B). This implies that the interaction of galectin-9 with TIM-3 promotes the secretion of cytotoxic cytokines, possibly involving other molecules on the surface of HNSCC cells as when co-cultured with T cells.
There were four known ligands of TIM-3 exist, including galectin-9, high mobility group box-1 protein (HMGB1), carcino-embryonic antigen-related cellular adhesion molecule 1 (CEACAM-1), and phosphatidylserine (PS). The TCGA database showed that CEACAM-1 and PS exhibit low or nearly negligible expression in HNSCC, while HMGB1 is highly expressed. When human primary T cells were incubated with the HMGB1 recombinant protein in vitro, the expression of effector cytokines was inhibited. In contrast, treatment of human primary T cells with excessive galectin-9 and HMGB1 recombinant proteins together partially counteracted the effects of HMGB1 on CD8+ T cell function (Fig. 3G and Supplementary Fig. S6C, D). Flow cytometry analysis also confirmed the competitive binding of galectin-9 and HMGB1 to TIM-3 (Fig. 3H). These results collectively indicate that galectin-9 boosts the secretion of cytotoxic cytokines through competitive binding with HMGB1 to TIM-3.
circE7 binds to and activates ACC1 to suppress acetyl-CoA and inhibit the expression of galectin-9
To explore the molecular mechanism responsible for the reduction of galectin-9 expression by circE7, RNA pull-down in conjunction with high-throughput mass spectrometry identification was utilized and 136 proteins were successfully identified with potential binding capacity to circE7 (Supplementary Fig. S7A and Supplementary Data 3). The top 10 circE7 binding proteins with the highest scores were confirmed by RNA pull-down followed by western blot. The investigation unveiled that circE7 possesses the ability to interact with acetyl-CoA carboxylase 1 (ACC1) (Fig. 4A and Supplementary Fig. S7B). RIP experiments employing the ACC1 antibody (Fig. 4B) also confirmed this. Then the potential binding site between the circE7 sequence and ACC1 was predicted by the catRAPID software25. The truncated mutants for circE7 and ACC1 were constructed according to the UniProt26 software and the RNAfold27 software to predict the structure of ACC1 and circE7, respectively (Supplementary Fig. S7C, D). The implementation of RNA pull-down followed by western blot analysis and RIP experiments confirmed the binding between the 1-500 amino acid region (MT1) in the N-terminal of the ACC1 protein and the 51–76 nucleotides (DEL-1) of circE7 (Fig. 4C–F).
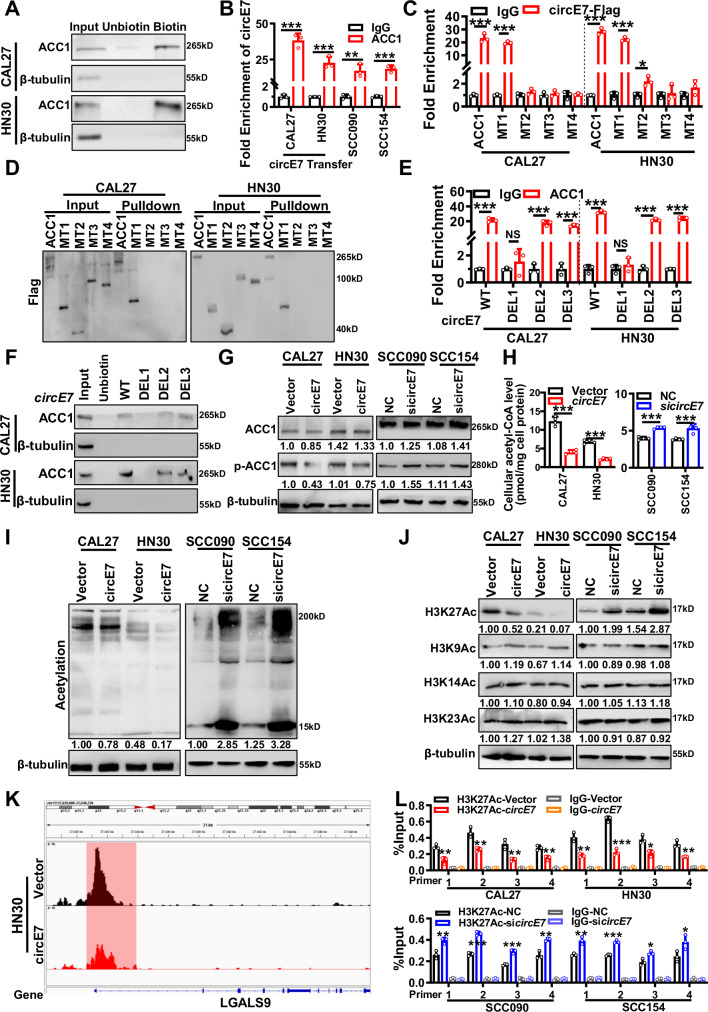
Fig. 4. circE7 epigenetically downregulates galectin-9 expression mediated by ACC1.
A RNA pulldown analysis was performed using biotin-labeled circE7 probe to detect the binding between circE7 and ACC1 protein in CAL27 and HN30 cells. Non-biotin-labeled circE7 probe was used as a control. B. Binding of ACC1 protein with circE7 was evaluated by RIP assay using anti-ACC1 antibody in CAL27 and HN30 cells overexpressing circE7, or in HPV-positive SCC090 and SCC154 cells, followed by quantification using RT-qPCR. C RNA immunoprecipitation using anti-Flag antibody to evaluate the binding between ACC1 protein and its truncation mutants with circE7, followed by RT-qPCR analysis of circE7. D RNA pulldown using biotin-labeled circE7 probe and western blot using anti-Flag antibody to detect the binding between circE7 and ACC1 and its truncation mutants. E RNA immunoprecipitation using anti-ACC1 antibody to evaluate the direct binding between ACC1 protein and circE7 and its truncation mutants, followed by RT-qPCR analysis of circE7 and its truncation mutants. F RNA pulldown using biotin-labeled circE7 probe to investigate the binding between circE7 and its truncation mutants with ACC1 protein. circE7 were overexpressed in CAL27 and HN30 cells, or si-circE7 was transfected into SCC090 and SCC154 cells. G Western blot to detect the expression levels of total ACC1 protein and p-ACC1 protein. H Acetyl-CoA content in cell lysate was measured using the Acetyl-CoA detection kit. I Western blot to assess the expression levels of global acetylation. J Western blot to assess the expression levels of H3K27 Ac, H3K9 Ac, H3K14 Ac, and H3K23 Ac. as shown in Supplementary Fig. S7L. K Overexpression of circE7 in the HN30 cells, ChIP-seq peaks show the levels of H3K27ac at the LGALS9 promoter region. The horizontal axis represents genomic coordinates, and the vertical axis represents ChIP-seq signal intensity. L ChIP-qPCR to investigate the changes in H3K27 acetylation level in the LGALS9 promoter region in CAL27 and HN30 cells overexpressing circE7 or SCC090 and SCC154 cells with circE7 knocked down. Correlations were calculated using unpaired two-tailed Student’s t test for (B, C, E, H, L), *p < 0.05, **p < 0.01, and ***p < 0.001. These experiments were derived from three independent repetitions. Each western blots were reproduced three times with similar results (A, D, F, G, I, J). Data are presented as mean ± SD. Source data are provided as a Source Data File.
circE7 is formed by reverse splicing the 3’ end sequence of HPV gene E6, E7 gene, and the 5’ end sequence of E1 gene; it has the potential to complete the E7 sequence and encode the E7 protein. To clarify whether the binding of circE7 to ACC1 is dependent on its ability to encode the E7 protein, we constructed a circE7 mutant (circE7-MT) by mutating the E7 promoter sequence. RNA pulldown experiments found that circE7-MT also binds to ACC1 (Supplementary Fig. S7E, F). Further, when E7 was overexpressed in CAL27 and HN30 cells (Supplementary Fig. S7G, H), it could not regulate the phosphorylation of ACC1 (Supplementary Fig. S7I). The RIP experiments showed that there was no direct binding between the ACC1 protein and E7 mRNA using the ACC1 antibody (Supplementary Fig. S7J). Since the direct binding site of circE7 and ACC1 (circE7-DEL1 51-76 nt) is in the E6 mRNA region, RIP assays with ACC1 antibody were also performed in SCC090 and SCC154, and we found that ACC1 and E6 mRNA also did not bind with each other (Supplementary Fig. S7K). These results indicate that the binding of circE7 to ACC1 mainly depends on the molecule itself, and does not depend on its partially homologous linear molecules, such as E6 and E7.
The ACC1 enzyme plays a pivotal role in the regulation of cellular lipid metabolism by converting acetyl-CoA into malonyl-CoA. Previous research has pointed to ACC1 activation being linked to the dephosphorylation of its N-terminal serine 80 (Ser80) site, situated within the 1–500 amino acid region (MT1)28. The activation of ACC1 is affirmed through western blotting, revealing that circE7 enhances the dephosphorylation of the ACC1 Ser80 (Fig. 4G) through binding to ACC1. The intracellular acetyl-CoA content was also quantified in cells after circE7 overexpression, and the data showed that circE7 overexpression led to a reduction in intracellular acetyl-CoA, while knockdown of circE7 increased intracellular acetyl-CoA (Fig. 4H).
Acetyl-CoA serves as the substrate for cellular protein acetylation, and the intracellular acetyl-CoA concentration exerts a notable influence on protein acetylation, particularly histone acetylation. This epigenetic regulatory mechanism assumes a pivotal role in shaping downstream gene expression. In HNSCC cells, we observed that overexpression of circE7 led to a reduction in intracellular protein acetylation levels (Fig. 4I). Importantly, circE7 displayed the most pronounced impact on the acetylation modification of H3K27 (Fig. 4J and Supplementary Fig. S7L). By performing ChIP-seq experiments after overexpressing circE7 in HPV16- HN30 cell lines (Supplementary Fig. S7M), we found that overexpression of circE7 significantly reduced the H3K27ac levels in the promoter regions of LGALS9 and other genes (Fig. 4K and Supplementary Fig. S7N). Furthermore, the acetylation level of the LGALS9 promoter region decreased in HPV16- HNSCC cells after overexpression of circE7 using the ChIP-qPCR method (Fig. 4L and Supplementary Fig. S7O).
To identify the regulation of ACC1 on LGALS9, ACC1 was knocked down in HPV16- cells or overexpressed in HPV16+ cells. The data showed that there was a significant change in LGALS9 transcription (mRNA) and a concomitant downstream modulation of galectin-9 protein levels by ACC1 (Fig. 5A, B). The inhibitory effect of circE7 on galectin-9 expression was partially reversed upon ACC1 knockdown in HPV16- cells. Likewise, galectin-9 induced by circE7 knockdown was counteracted by ACC1 in HPV16+ cells (Fig. 5C, D). Furthermore, ACC1 inhibitor TOFA could revert the galectin-9 expression level to normal in HPV16- cells after overexpression of circE7 (Fig. 5E, F). These findings strongly suggest that ACC1 is a pivotal downstream molecule of circE7 to inhibit LGALS9 transcription. circE7, by binding to ACC1, promotes ACC1 dephosphorylation, which subsequently leads to a reduction in intracellular acetyl-CoA content and inhibition of histone acetylation in the vicinity of the LGALS9 promoter. Ultimately, this epigenetic modification suppresses galectin-9 expression.
Fig. 5. ACC1 is a key downstream target regulated by circE7 in the epigenetic modulation of LGALS9 transcription.
Three ACC1 siRNAs were transfected into CAL27 and HN30 cells, or ACC1 was overexpressed in SCC090 and SCC154 cells. A RT-qPCR was used to detect the expression level of LGALS9 mRNA. B Western blot was used to detect the expression levels of ACC1 and galectin-9 proteins. Overexpression of circE7 in CAL27 and HN30 cells was accompanied by knockdown of ACC1, or knockdown of circE7 in SCC090 and SCC154 was accompanied by overexpression of ACC1. C RT-qPCR was used to detect the expression level of LGALS9 mRNA. D Western blot was used to detect the protein levels of ACC1, H3K27 Ac and galectin-9. Overexpression of circE7 in CAL27 and HN30 cells was accompanied by the addition of a small molecule inhibitor of ACC1, TOFA. E RT-qPCR was used to detect the expression level of LGALS9 mRNA. F Western blot was used to detect the protein levels of H3K27 Ac and galectin-9. Correlations were calculated using unpaired two-tailed Student’s t test for (A, C, E), *p < 0.05, **p < 0.01, and ***p < 0.001. These experiments were derived from three independent repetitions. Each western blots were reproduced three times with similar results (B, D, F). Data are presented as mean ± SD. Source data are provided as a Source Data File.
Co-application of TIM-3 and PD-1 monoclonal antibodies substantially improves the efficacy of HNSCC immunotherapy
Currently, immune checkpoint inhibitor therapy utilizing PD-1/PD-L1 monoclonal antibodies in HNSCC clinical trials yields an overall response rate of less than 30%29. This suggests that apart from the PD-1/PD-L1 immune checkpoint, other immune checkpoints also hold significance in HNSCC immune evasion and may serve as targets for HNSCC immunotherapy. The GEO datasets GSE159067 (HNSCC) and GSE91061 (melanoma) revealed a positive correlation between galectin-9 expression (Supplementary Fig. S8A), CD8+ T cell infiltration, and T cell function (Supplementary Fig. S8B) with a favorable prognosis in anti-PD-1 therapy, suggesting that simultaneous inhibition of galectin-9 receptors TIM-3 and PD-1 may enhance the efficacy of HNSCC immunotherapy. So, the PD-1 and TIM-3 monoclonal antibodies were simultaneously used to treat HNSCC tumor cells coculturing with T cells. RT-qPCR and ELISA experiments demonstrated that overexpression of circE7 in CAL27 and HN30 cell lines reduced the expression of CD8+ T cell cytotoxic cytokines promoted by anti-PD-1 alone. However, the combination of PD-1 and TIM-3 monoclonal antibodies significantly enhanced the expression of CD8+ T cell cytotoxic cytokines. Knockdown of circE7 in SCC090 and SCC154 cell lines increased the expression of CD8+ T cell cytotoxic cytokines promoted by anti-PD-1 alone, and the addition of TIM-3 monoclonal antibody further boosted it (Fig. 6A and Supplementary Fig. S9A). Flow cytometry results yielded similar conclusions (Fig. 6B and Supplementary Fig. S9B). Additionally, flow cytometry was utilized to assess the apoptosis of CD8+ T cells and tumor cells under various conditions. The results revealed that the combination of PD-1 and TIM-3 monoclonal antibodies significantly preserved CD8+ T cell and promoted tumor cell apoptosis (Fig. 6C, D and Supplementary Fig. S9C, D). These findings suggest that co-application of TIM-3 monoclonal antibody can enhance the efficacy of PD-1 monoclonal antibody in HNSCC immunotherapy, particularly for HNSCC with high circE7 expression.
Fig. 6. Combination treatment with exogenous TIM-3 and PD-1 monoclonal antibodies enhances CD8+ T cell cytotoxic function.
CAL27 and HN30 cells were overexpressed with circE7, or SCC090 and SCC154 cells were knocked down with circE7, and then co-cultured with primary human T cells with the addition of 5 μg/mL anti-PD-1 or anti-PD-1/TIM-3 for 6 h. A. RT-qPCR was used to measure the expression levels of IFNG, GZMB, and TNFA mRNA in CD8+ T cells. B. Flow cytometry was used to detect the proportion of IFN-γ+ GZMB+ and IFN-γ+ TNF-α+ T cells among all CD8+ T cells. Left panel shows representative images; right panel shows the statistical results, as shown in Supplementary Fig. S9B. C, D Annexin V/PI staining was used to assess the apoptosis levels of CD8+ T cells (C) and tumor cells (D), as shown in Supplementary Fig. S9C, D. Correlations were calculated using unpaired two-tailed Student’s t test for (A, B, C, D), *p < 0.05, **p < 0.01, and ***p < 0.001. These experiments were derived from three independent repetitions. Data are presented as mean ± SD. Source data are provided as a Source Data File.
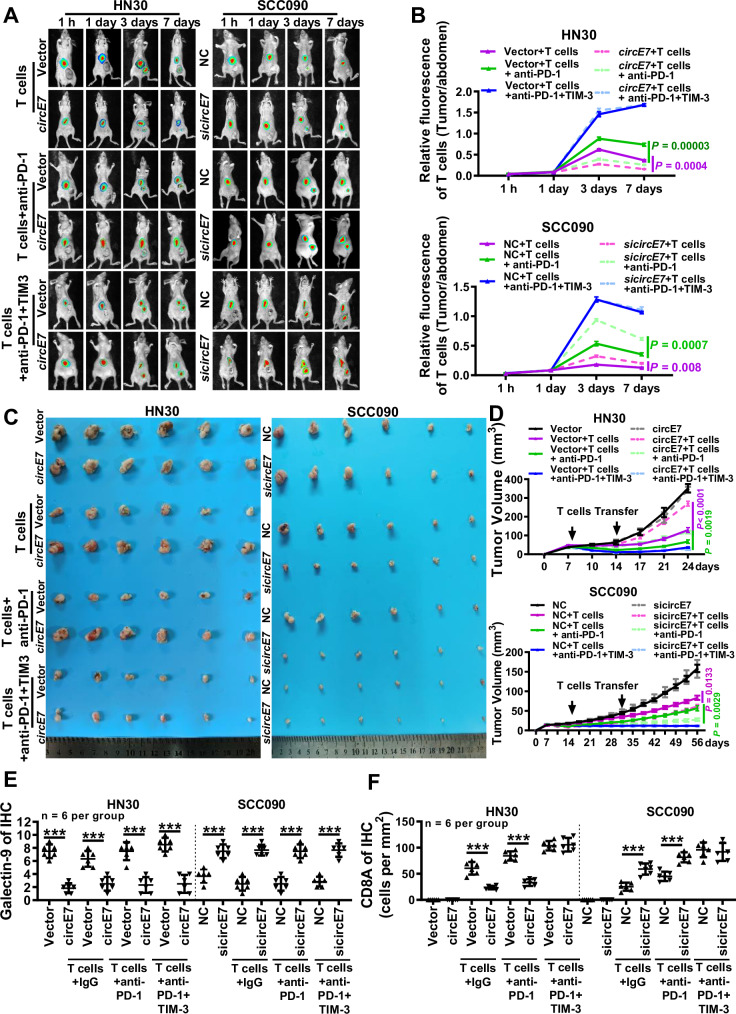
To further validate the immune therapeutic potential of the combination of TIM-3 and PD-1 monoclonal antibodies, the in vivo experiments were performed. HN30 with circE7 overexpression or SCC090 with circE7 knockdown were subcutaneously injected into nude mice through the tail vein for 7 days, following staining with the live fluorescent dye DIR. Next, co-cultured human primary T cells with dendritic cells (DCs) stimulated by lysates of tumor cells and sufficient PD-1 and TIM-3 monoclonal antibodies were injected into mice (Supplementary Fig. S10A). After 7 days of T cell injection, the transplanted tumor tissues formed by overexpressing circE7 in HN30 cells exhibited the lowest T cell distribution, while the group treated with PD-1 monoclonal antibody displayed an increase in T cell infiltration within the transplanted tumor. Notably, the combination of PD-1 and TIM-3 monoclonal antibodies resulted in even higher T cell accumulation within the transplanted tumor. Conversely, when circE7 was knocked down, T cell accumulation at the transplanted tumor site significantly increased. Subsequent treatments with PD-1 and TIM-3 monoclonal antibodies further promoted T cell infiltration within the transplanted tumor (Fig. 7A, B).
Fig. 7. Combination therapy with TIM-3 and PD-1 monoclonal antibodies significantly improves the efficacy of immunotherapy in HNSCC.
Nude mice were injected with 1 × 106 HN30 cells overexpressing circE7 or vector control in the right hind limb, or with SCC090 cells knocking down circE7 or NC control to form xenograft tumors. After 7 and 14 days, human primary T cells and 20 μg/each of Anti-PD-1 or Anti-PD-1/TIM-3 were injected for immunotherapy. A, B T cells were labeled with DeepRed and in vivo imaging was performed to detect the survival and distribution of human primary T cells in tumor-bearing mice (n = 3 for each group). A representative images; B statistical results of the ratio of the DiR fluorescence signal from the tumor site (at the base of the right thigh) to T cell fluorescence intensity, collected 1, 3, 5, and 7 days after injection. The color scale represents the DiR fluorescence intensity of T cells in mice. C Representative tumor images. left, 24 days after HN30 cell injection (n = 6 for each group); right, 56 days after SCC090 cell injection (n = 6 for each group). D Real-time measurement of tumor volume in mice injected with HN30 or SCC090 cells once a week (n = 6 for each group). E Immunohistochemistry was performed to detect the expression of galectin-9 in paraffin-embedded HN30 and SCC090 xenograft tumors, and the results were statistically analyzed (n = 6 for each group). F Immunohistochemistry was performed to detect the expression of CD8A in HN30 and SCC090 xenograft tumors, and the results were statistically analyzed (n = 6 for each group). Correlations were calculated using unpaired two-tailed Student’s t test for (B, D), *p < 0.05, **p < 0.01, and ***p < 0.001. Data are presented as mean ± SD. Source data are provided as a Source Data File.
Also, the volume and weight of the transplanted tumors were assessed after injection of T cells (24 days for HN30 cells and 56 days for SCC090 cells). For the HN30 transplanted tumor, the group without T cell treatment exhibited no significant impact on tumor volume and weight. However, the T cell treatment group with circE7 overexpression resulted in an increased tumor volume, indicating that circE7 primarily promotes HNSCC tumor progression by driving tumor immune escape. Within the T cell treatment group, overexpression of circE7 diminished the therapeutic effect of anti-PD-1. The group receiving a combination of PD-1 and TIM-3 monoclonal antibodies exhibited the smallest tumor volume and weight, demonstrating the most favorable therapeutic effect and significant suppression of immune resistance mediated by circE7. Correspondingly, SCC090 cells transplanted tumor without T cell treatment showed no significant effect on tumor volume and weight after knockdown of circE7. In the T cell therapy group, cells with circE7 knockdown exhibited smaller tumor volume and weight. This indicates that the knockdown of circE7 can inhibit HNSCC cells’ immune evasion and enhance the therapeutic efficacy of the PD-1 monoclonal antibody. Collectively, the combination of PD-1 and TIM-3 monoclonal antibodies displayed a more pronounced inhibitory effect on tumor progression (Fig. 7C, D and Supplementary Fig. S10B).
ELISA and RT-qPCR were also used to measure the peripheral serum cytotoxic cytokine secretion and expression in T cells (Supplementary Fig. S10C, D). In the HN30 xenograft tumor model, overexpression of circE7 led to a decrease in T cell cytotoxic cytokines. In contrast, the combination treatment with PD-1 and TIM-3 antibodies significantly increased the secretion of cytotoxic cytokines. In the SCC090 xenograft tumor model, the knockdown of circE7 significantly enhanced T cell cytotoxic activity. The combination treatment with PD-1 and TIM-3 further increased T cell cytotoxic activity.
Flow cytometry showed that overexpression of circE7 led to a decrease in CD8+ T cells and an increase in T cell apoptosis in the HN30 transplant tumor model. Conversely, the combination treatment with PD-1 and TIM-3 antibodies significantly increased the number of CD8+ T cells and reduced T cell apoptosis. In the SCC090 transplant tumor model, knockdown of circE7 resulted in an increase in CD8+ T cells and a decrease in apoptosis. The combination treatment of PD-1 and TIM-3 further inhibited T cell apoptosis and significantly increased CD8+ T cells (Supplementary Fig. S10E–G). Then, the ISH method verified the efficiency of circE7 overexpression and knockdown in nude mice tumors and immunohistochemistry (IHC) revealed that overexpression of circE7 led to a downregulation of galectin-9 expression in tumor tissues and a reduction in infiltrating CD8+ T cells, whereas knockdown of circE7 exhibited the opposite effect (Fig. 7E, F and Supplementary Fig. S10H, I).
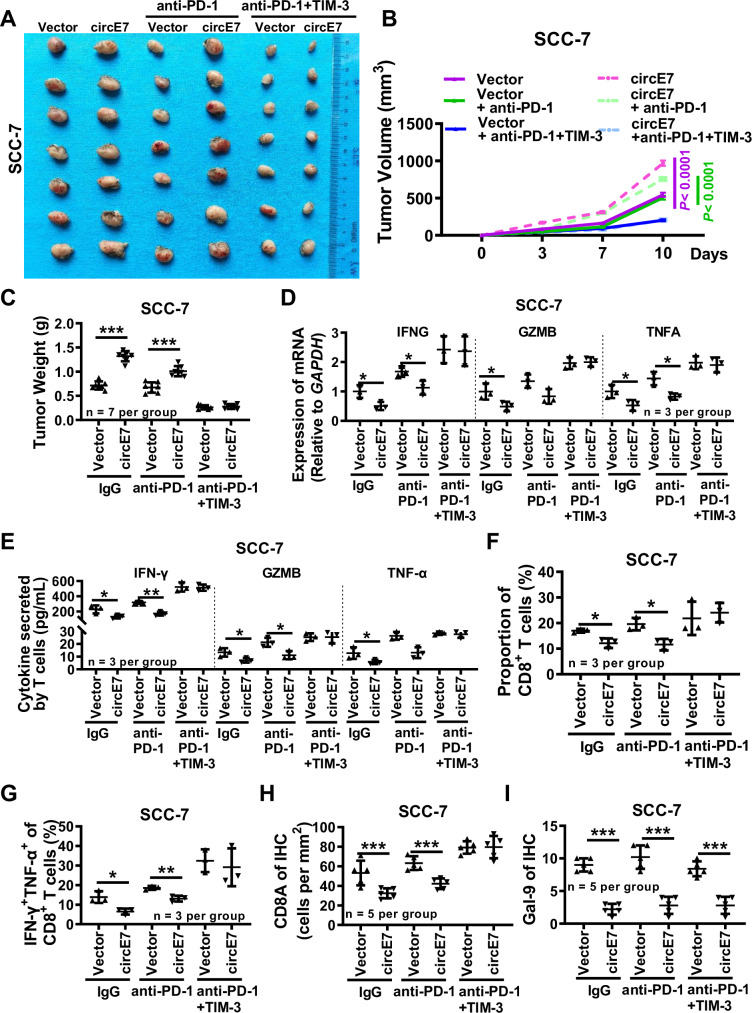
Further, to further verify the immunotherapeutic effect of the combined application of the TIM-3 and PD-1 monoclonal antibodies, the mouse squamous cell carcinoma cell line SCC-7 was injected into the cheeks of C3H mice to establish a homologous in situ tumor model. Because ACC1 is highly conserved between humans and mice, according to the NCBI website, we performed RNA pulldown experiments and found that circE7 could also bind to Acc1 of mice in SCC-7 after overexpression of circE7 (Supplementary Fig. S11A, B). RT-qPCR and western blot showed that circE7 downregulated the expression of Lgals9 in mice (Supplementary Fig. S11C, D). Overexpression of circE7 resulted in increased tumor size in SCC-7 cell transplants. When adequate PD-1 and TIM-3 monoclonal antibodies were injected into C3H mice, the size and weight of tumors got smaller and markedly inhibited the tumor enlargement mediated by circE7 overexpression, suggesting that circE7 significantly reduced the therapeutic effect of anti-PD-1 (Fig. 8A–C and Supplementary Fig. S11E). Both RT-qPCR and ELISA experiments revealed that overexpression of circE7 inhibited the expression of cytotoxic cytokines in tumor tissues. In contrast, the combination therapy with PD-1 and TIM-3 antibodies could reverse the downregulation of cytotoxic cytokine expression mediated by circE7 (Fig. 8D, E). Flow cytometry revealed that overexpression of circE7 led to a decrease in the proportion of CD8+ T cells within the T cell population and a reduction in the proportion of INF-γ+ TNF-α+ cells within the CD8+ T cell population, while combined treatment with PD-1 and TIM-3 could significantly increase the number and killing function of CD8+ T cells (Fig. 8F, G and Supplementary Fig. S11F–H). The ISH and IHC tests also validated the overexpression efficiency of circE7 and the expression of galectin-9 and CD8A expression (Fig. 8H, I and Supplementary Fig. S11I, J). These results underscore that circE7 promotes HNSCC cell immune evasion in vivo and attenuates the therapeutic efficacy of PD-1 monoclonal antibody. However, the co-application of TIM-3 and PD-1 monoclonal antibodies reverses the immune evasion mediated by circE7 (Supplementary Fig. S12).
Fig. 8. The Mouse Orthotopic Transplant Model showed that the combination of PD-1 and TIM-3 monoclonal antibodies enhances the efficacy of immunotherapy in HNSCC.
C3H mice were injected with 2 × 105 SCC-7 cells overexpressing circE7 or vector control in the right cheek to form xenograft tumors. After 3 days, 20 μg/each of anti-PD-1 or anti-PD-1/TIM-3 were injected for immunotherapy. A Representative tumor images. (n = 7 for each group). B Tumor volume measured 3 days, 7 days and 10 days after SCC-7 cell injection (n = 7 for each group). C Tumor weight measured 10 days after SCC-7 cell injection (n = 7 for each group). D Prepare single-cell suspension from mouse tumor tissue, RT-qPCR detection of IFNG, GZMB, and TNFA mRNA expression (n = 3 per group). E ELISA was used to detect the content of IFN-γ, GZMB, and TNF-α (n = 3 per group). Flow cytometry was performed to detect the proportion of CD8+ T cells (F) and the percentage of IFN-γ+ TNF-α+ cells (G) within the total CD8+ T cell population (n = 3 for each group). H Immunohistochemistry was performed to detect the expression of Cd8a in SCC-7 xenograft tumors, and the results were statistically analyzed (n = 5 for each group). I Immunohistochemistry was performed to detect the expression of galectin-9 in paraffin-embedded SCC-7 xenograft tumors, and the results were statistically analyzed (n = 5 for each group). Correlations were calculated using unpaired two-tailed Student’s t test for (B, C, D, E, F, G, H, I), *p < 0.05, **p < 0.01, and ***p < 0.001. Data are presented as mean ± SD. Source data are provided as a Source Data File.
Discussion
Tumorigenic viruses such as Epstein-Barr virus (EBV), Kaposi’s sarcoma herpesvirus (KSHV), Merkel Cell polyomavirus (MCPyV), and Human T-cell lymphotropic virus (HTLV-1) can encode circular RNAs. These circRNAs play a role in regulating the proliferation, invasion, metastasis, and stemness of tumor cells30–36. However, research on the relationship between tumorigenic virus-encoded circular RNA and tumor immune evasion is limited37,38. So far, only circBART2.2 encoded by EBV has been found to promote immune evasion in nasopharyngeal carcinoma by upregulating PD-L1 through its interaction with RIG-I38. In this study, we have identified that circE7, encoded by HPV16, also assumes a critical role in HNSCC immune evasion. circE7 is currently exclusive to high-risk HPV16 strains and has not been identified in other HPV subtypes23. The HPV16 genome can be categorized into three regions: the upstream regulatory region, housing cis-acting elements responsible for transcription and replication control; the early region, encompassing E1, E2, E4, E5, E6, and E7 protein-coding sequences; and the late region, encoding the L1 and L2 capsid proteins39. circE7, encoded by HPV16, originates from the reverse splicing of the HPV16 E6 gene’s tail and the E1 gene’s head, incorporating the complete E7 gene sequence (Figure S1A)23. We have observed circE7 expression in the HPV16+ cervical cancer cell line Caski and the head and neck cancer cell lines SCC090 and SCC154 but not in another common high-risk HPV18+ cell line, Hela, which aligns with prior findings.
Although circE7 carries the full coding sequence of the E7 protein and predominantly localizes in the cytoplasm23, whether it actually encodes a functional E7 protein and its specific biological functions remain unclear. Our research has unveiled that circE7 primarily exerts its biological functions by directly binding to the pivotal enzyme ACC1 in the metabolic pathway. This interaction prevents ACC1 phosphorylation and, through epigenetic regulation, curtails the expression of the LGALS9 gene, thus facilitating HNSCC immune evasion40. ACC1 stands as a rate-limiting enzyme in fatty acid synthesis, primarily residing in the cytoplasm and catalyzing the conversion of acetyl-CoA into malonyl-CoA41. Recent reports have substantiated the close association of ACC1 with the development and progression of various cancers42,43. Our study has further revealed that circE7 promotes ACC1 dephosphorylation by directly binding to ACC1, leading to activated ACC1 catalyzing the conversion of acetyl-CoA into malonyl-CoA, consequently reducing acetyl-CoA levels in tumor cells. Acetyl-CoA serves as the principal substrate for protein acetylation modifications within cells, and its depletion leads to reduced protein acetylation within cells44. Moreover, we have determined that ACC1 activation predominantly instigates a reduction in histone H3K27Ac, ultimately suppressing the transcription of the immune checkpoint gene LGALS9 through epigenetic regulation.
The galectin-9 protein, encoded by the LGALS9 gene, belongs to the galectin family, a group of lectins within the broader lectin superfamily, with widespread distribution in various animals45. Recent research has revealed that galectin-9 serves as one of the ligands for the immune checkpoint TIM-3, participating in intricate biological functions across various immune cell types. In NK cells, galectin-9 binds to TIM-3, thereby enhancing IFN-γ production23. In NKT cells, galectin-9 indirectly contributes to their expansion46–49. Within Th1 cells, the interaction between TIM-3 and galectin-9 results in reduced IFN-γ production and immune suppression50. Our study has demonstrated that galectin-9 on the surface of HNSCC cells can interact with TIM-3 and PD-1 on the surface of CD8+ T cells, each exerting distinct biological effects. Specifically, galectin-9 binds to TIM-3, stimulating T cell secretion of cytotoxic cytokines, while its binding to PD-1 inhibits T cell apoptosis. This dual interaction coordinates and sustains T cell function and activity. However, when galectin-9 recombinant protein was incubated alone in T cell culture medium, its impact on T cell secretion of cytotoxic cytokines in the absence of other factors was not significant.
Further analysis and verification uncovered that HNSCC cells express both galectin-9 and HMGB1 on their surfaces. Galectin-9 competes with HMGB1 for binding to TIM-3 on the surface of T cells, effectively neutralizing the immune suppression caused by HMGB1’s binding to TIM-3. Consequently, this upregulates the expression of cytotoxic cytokines in T cells, mirroring the effect of TIM-3 monoclonal antibodies. In this context, our study revealed that HPV16-derived expression of circE7 in HNSCC binds to and promotes ACC1 dephosphorylation, ultimately activating ACC1. This activation results in a reduction of acetyl-CoA content in HNSCC cells, particularly leading to a decrease in intracellular protein acetylation, particularly histone H3K27Ac levels. This epigenetic alteration downregulates the LGALS9 transcription and subsequently reduces galectin-9 protein levels. Consequently, insufficient galectin-9 binding with PD-1 leads to increased T cell apoptosis, while its competitive binding with TIM-3 inhibits T cell function, thus fostering HNSCC immune evasion. Patients with high galectin-9 expression in HNSCC exhibit a better prognosis, implying that galectin-9 plays a crucial anti-tumor role in HNSCC.
Presently, clinical immunotherapy for HNSCC relies on PD-1/PD-L1 monoclonal antibodies and has shown some degree of effectiveness9,51. Previous clinical investigations have unveiled that immune checkpoint blockade therapy employing PD-1/PD-L1 monoclonal antibodies is marginally more successful in HPV+ HNSCC patients compared to HPV- patients, Nivolumab induced pathologic regression in both HPV-positive (23.5%) and HPV-negative (5.9%) tumors. Nevertheless, the overall effectiveness remains limited52,53. This underscores the impact of HPV on HNSCC immune evasion and the response to immunotherapy. Additionally, while PD-1/PD-L1 may be pivotal immune checkpoint molecules in HNSCC, other immune checkpoint molecules also exert a notable influence on HNSCC immune evasion. This is evidenced by the modest overall efficacy of PD-1/PD-L1-based immune checkpoint inhibitors in HNSCC immunotherapy. The identification of critical immune checkpoint molecules and their regulatory mechanisms holds great potential for enhancing the effectiveness of HNSCC immunotherapy. In this context, this study reveals that circE7, expressed by HPV16, downregulates the co-activator ligand galectin-9 of PD-1 and TIM-3, consequently facilitating HNSCC immune escape. Furthermore, both in vitro and in vivo experiments have validated that the combination of TIM-3 monoclonal antibody and PD-1 significantly enhances the efficacy of HNSCC immunotherapy, particularly in the case of HPV16+ HNSCC. circE7 and galectin-9 hold the promise of serving as potential targets and markers for predicting efficacy and prognosis, ultimately improving the outcomes of immunotherapy for HPV16+ HNSCC patients. This research also proposes a more effective combination regimen for HPV16+ HNSCC patients. The latest single-cell sequencing results indicate that there is a significant individual variability in CD8+ T cells infiltration among patients with HPV- and HPV+ HNSCC18,54. However, our study found a reduced expression of CD8+ T cells in HNSCC patients with HPV infection. This may be due to our samples primarily coming from the Hunan region of China, and all these patients are infected with the HPV16 subtype. Current research has found that only the HPV16 subtype expresses circE7. CircE7 inhibits LGALS9 expression through epigenetic regulation, thereby limiting HPV infection’s ability to activate and recruit CD8+ T cells. It is interesting to question whether the varying degrees of T cell infiltration among patients with different HPV subtypes contribute to this effect, and we plan to explore this further in future studies.
In summary, our work unveils a employed by HPV16 to induce metabolic reprogramming in HNSCC. This mechanism hinges on the expression of circular RNA circE7, which drives epigenetic modifications and promotes immune evasion. Moreover, this study identifies promising therapeutic targets and strategies for improving the effectiveness of HNSCC immunotherapy.
Methods
Ethical statement
This research complies with all relevant ethical regulations. All animal experiments were approved by the Institutional Animal Care and Use Committee at the Hunan Cancer Hospital.
Patient samples
All clinical samples were collected from the Second Xiangya Hospital, Central South University. A total of 23 paired tumors and adjacent normal tissues were used for quantitative real-time PCR (qRT-PCR) and were obtained from patients diagnosed with HNSCC who underwent initial surgery (Supplementary Data 1). Paraffin-embedded sections (n = 105) from patients with HNSCC were collected between November 2017 and December 2021, along with complete follow-up data for immunohistochemistry and in situ hybridization (Supplementary Data 2). All samples are collected with the patients’ informed consent and written consent forms signed, and the experiments were approved by the Ethics Committee of Second Xiangya Hospital, Central South University, in accordance with the ethical standards established in the Declaration of Helsinki.
Cell lines
In this study, the HPV- HNSCC cell lines CAL27 and HN30, the HPV16+ cell lines SCC090 and SCC154, and the mouse HNSCC cell line SCC-7 were used. The CAL27(ZQ0606) and SCC090 (ZQ0937) cell lines were purchased from Shanghai Zhongqiao Xinzhou Co (Shanghai, China), HN30(CTCC-400-0142) and SCC154 cell line (CTCC-001-0182) was purchased from Meisen CTCC (Hangzhou, China). SCC-7 cell line was kindly provided by Professor Zhengjun Shang from Wuhan University55. Prior to use, all cell lines underwent cellular STR identification. All cell lines were regularly tested for Mycoplasma using Mycoplasma Detection Kit. Cells were incubated in the following culture conditions: 10% FBS (Gibco, 10270106, USA), 1% penicillin-streptomycin combination (BI, 03-03101B, Israel), and DMEM medium (Gibco, C11995500BT, USA), in a humidified atmosphere with 5% CO2 at 37 °C. Cells were cryopreserved in a −80 °C freezer using serum-free cell cryopreservation solution (NCM Biotech, C40100, Suzhou, China).
RNA extraction, qRT-PCR, and regular PCR
Total RNA was extracted using the Trizol reagent (Invitrogen, 15596018, USA) and reverse transcribed into cDNA using the HiScript cDNA Synthesis Kit (Vazyme, R323-01, China). Quantitative real-time PCR was performed using the universal SYBR qPCR Master Mix (Vazyme, Q511-02, China) and the CFX Connect Real-Time PCR Detection System (Bio-Rad, USA). GAPDH was used as the internal control. The relative expressions of genes were calculated using the 2-ΔΔCT method. PCR amplification of cDNA was performed using the Golden Star T6 Super PCR Mix (Tsingke, TSE101, China) and the Mastercycler® nexus (Eppendorf, Germany). The 2% agarose gels were prepared using UltraPureTM Agarose (ThermoFisher, 16500100, USA), SolarRed (Solarbio, G5560, China), and 1 × TAE Buffer. Images of the target strips were captured after electrophoresis using the Bio-Rad Universal Hood II Gel Doc XR System (Bio-Rad, USA). The primers used are shown in Supplementary Data 4.
Vectors, siRNA sequences, and cell transfection
The circRNA expression plasmid pCirc was generously provided by Professor Li Yong from Baylor College of Medicine. To overexpress circE7, cDNA reverse transcribed from RNA of SCC090 cells was used as the PCR template, and the full length of circE7 was amplified. The siRNA targeting circE7 and the control scramble siRNA were purchased from Genepharma (Shanghai, China). The overexpression vector or control empty plasmid was transfected into HNSCC cells using Neofect DNA transfection reagent (Neofect biotech, Beijing, China). siRNA transfection was performed using Hiperfect (Qiagen, Hilden, Germany). The circE7 sequence and the primer sequences used for constructing the overexpression vector are provided in Supplementary Data 4.
RNA fluorescence in situ hybridization (FISH)
Digoxigenin-labeled oligonucleotide probes were designed for the circE7 splice site and synthesized by Tsingke (China). Cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. After pre-hybridization, the probe was added to slides for overnight hybridization at 37 °C. Cellular nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI) (Invitrogen, D1306, USA). Fluorescent confocal images were captured under STELLARIS 5 and STELLARIS 8 (Leica, Germany).
Immunofluorescence
After plating onto the coverslips, the cells were allowed to grow to a certain density and fixed in 4% paraformaldehyde. Following permeabilization with 0.5% Triton X-100, the cells were incubated overnight at 4 °C with the primary antibody. Subsequently, the cells were incubated with a secondary fluorescent antibody at 37 °C, and the nuclei were stained with 4’, 6-diamidino-2-phenylindole (DAPI) (Invitrogen, D1306, USA). The fluorescent confocal images were captured using STELLARIS 5 and STELLARIS 8 (Leica, Germany). The antibodies used are listed in Supplementary Data 5.
Immunohistochemistry and in situ hybridization
Immunohistochemistry was performed using the ElivisionTM Plus Polymer HRP (Mouse/Rabbit) IHC Kit (Maxim, KIT-9902, China), following the provided protocol. In situ hybridization was done using the Enhanced Sensitive ISH Detection Kit I (POD) (BOSTER, MK1030, China), following the provided manual as well. The digoxigenin-labeled circE7 probe was purchased from Tsingke (China). The score was calculated by multiplying the intensity of stained cells with the positivity score. Staining intensity scores were assigned as follows: 0 = negative, 1 = weak, 2 = moderate, and 3 = strong. Positive rate scores were assigned as follows: 0 = negative, 1 = (1–25%), 2 = (26–50%), 3 = (51–75%), and 4 = (76–100%). All scores were determined by two senior pathologists. Representative images were captured using the Olympus BX51 fluorescence microscope (Olympus, Japan). The antibodies used are listed in Supplementary Data 5.
RNA pulldown and western blotting
Biotin-labeled circE7 probes were transfected into CAL27 or HN30 cells for 48 h. Cells were subsequently lysed with 0.1% Triton-X at 4 °C. Streptavidin affinity magnetic beads (Thermo Scientific, 20164, USA) were added to the cellular lysate and incubated overnight at 4 °C. After washing the magnetic beads 3 times, the supernatant was collected and boiled for western blotting analysis. Proteins were extracted by lysing cells with a RIPA lysis solution (Epizyme Biomedical, PC101) and separated by 10% SDS-PAGE (Servicebio, G2177, China). The proteins were then transferred onto polyvinylidene difluoride membranes (Millipore, IPVH00010, USA). Primary antibodies were incubated overnight at 4 °C, and the secondary antibodies were incubated for 1–2 h at room temperature. Images of the target bands were captured using the Chemiluminescent Imaging and Analysis System (Servicebio, G2020, China). The antibodies used are listed in Supplementary Data 5.
RNA-IP
After washing with pre-cooled PBS, cells were lysed by adding lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 10 mM NaF, 1 mM EDTA, 100 U/ml RNase OUT (Invitrogen, 10777019, USA). Cellular lysates were then incubated with anti-ACC1 or rabbit IgG-coupled magnetic beads. After washing magnetic beads twice, RNA in the immunoprecipitates was isolated with the TRIzol reagent (Invitrogen, Carlsbad, USA) and analyzed by reverse transcription with HiScript cDNA synthesis kit (Vazyme, R323-01, China), followed by RT-qPCR.
Analysis of Acetyl-Coenzyme A
The analysis of Acetyl-Coenzyme A was conducted following the steps outlined in the instruction manual of the Acetyl-Coenzyme A Assay Kit (Sigma-Aldrich, MAK039, USA). The fluorescence intensity data were collected using the SpectraMax M Multifunctional Enzyme Labeler (Molecular Devices, China).
ChIP-qPCR
Cells were fixed with 1% paraformaldehyde and 0.125 M glycine. Subsequently, DNA was extracted following the steps outlined by the Pierce™ Magnetic ChIP Kit (Thermo Scientific, 26157, USA). The LGALS9 promoter region fragment was then enriched using the H3K27 antibody (Supplementary Data 5). The quantity of the enriched fragment was analyzed using RT-qPCR and PCR gel electrophoresis. The primers for PCR analysis of DNA precipitation are provided in Supplementary Data 4. ChIP-Seq was sequenced at E-GENE Co., Ltd.
Preparation of dendritic cells (DCs) and primary T cells
Following the manufacturer’s instructions, peripheral blood mononuclear cells (PBMCs) were isolated from healthy donors’ peripheral blood using the Ficoll method (Cytiva, USA).
To prepare DCs, the isolated PBMCs were added to a mixture containing 50 ng/ml GM-CSF (Sino Biological, 51048-MNAH, China), 20 ng/ml IL-4 (Sino Biological, GMP-11846-HNAE, China) in 10% FBS-1640 medium and cultured for 5 days. Cell differentiation was monitored under light microscopy. DC cells were obtained by adding 25 ng/ml IFN-γ (Sino Biological, 11725-HNAE, China) and continuing the culture for 1 day, followed by co-culture with tumor cell lysates for another day.
For T cells, they were expanded in vitro by adding CD3/CD28 amplified magnetic beads (Miltenyi, 130-091-441, Germany) and 15 ng/ml IL-2, 5 ng/ml IL-7, 10 ng/ml IL-15 (Sino Biological, 10360-HNCE, China) for 8 days. Tumor-specific CTL was obtained by co-culturing prepared DCs with amplified T cells at a ratio of 1:5 in a medium supplemented with IL-2, IL-7, and IL-15. Fresh medium and cytokines were replaced every 2 days during the experimental period. When recruiting volunteers, we selected volunteer T cells with the same HLA-A type as the cell lines for co-culture experiments.
To achieve antigen presentation in tumor cells and generate specific cellular immunity against the cell line, we co-cultured the tumor cell lysate with dendritic cells (DCs) for 24 h. Then, these antigen-presented DCs were co-cultured with T cells for another 24 h to generate T cells with specific cellular immunity against tumor cells. Finally, these antigen-presented T cells were injected subcutaneously into mice via the tail vein.
Enzyme-linked immunosorbent assay (ELISA)
The ELISA assay was performed using the ELISA kits for TNF-α (ZCIBIO, ZC-M6765, China), GZMB (ZCIBIO, ZC-33834, China), and IFN-γ (ZCIBIO, ZC-32271, China). Data was acquired using a SpectraMax M multifunctional enzyme marker from Molecular Devices, China.
Flow cytometry
Flow cytometry was done as previously reported38,56. In brief, apoptosis was detected by flow cytometry analysis using the Annexin V-FITC Apoptosis Detection Kit (BMS500FI, Invitrogen). Cytokine release from cells was blocked using BFA, and intracellular cytokines were then detected by incubating with the corresponding cytokine antibodies after pretreatment with a membrane breaker. All flow cytometry was performed using a DxP AthenaTM flow cytometer (Cytek Biosciences, USA). The antibodies used are listed in Supplementary Data 5.
Animal experiments
All animal experiments were approved by the Institutional Animal Care and Use Committee at the Hunan Cancer Hospital. BALB/c-nu mice (n = 160) and C3H mice (n = 60) were purchased from Hunan Silaikejingda Experimental Animal Co., Ltd. and Shanghai Model Organisms Center, Inc., respectively. To ensure data consistency, we selected a single-gender mouse model for the animal experiments to reduce experimental variables. Since female mice are generally quieter and more docile, we used only female mice in the study. Female mice aged at least 6 weeks were used for all experiments. Mice were housed with ad libitum access to food and water on a 12 h light-dark cycle in cages maintained at 20–22 °C and 22–30% relative humidity. Tumor growth was measured by caliper every 3–4 days. Mice were euthanized once the primary tumor measured a maximal size of 2 cm in at least one dimension. Maximal tumor size was not exceeded in this study.
Transfected cells were injected subcutaneously into the right thigh of each mouse. Once visible tumor formation was observed, primary T cells that had undergone antigenic presentation of tumor cell lysates with anti-PD-1 monoclonal antibody and anti-TIM-3 monoclonal antibody were infused into each cell-derived xenograft (CDX) mouse through the tail vein. The distribution of human primary T cells, labeled with a deep red live cell fluorescent dye (DiR, ThermoFisher), was assessed using a small animal in vivo imaging system (Bruker, USA). After 14 days of secondary T cell injection, peripheral blood was extracted from the mice for RT-qPCR, ELISA, or flow cytometry analysis. The body weight and tumor size of the mice were measured periodically. Once sacrificed, the tumor tissues were paraffin-embedded, and sections were prepared. H&E staining, in situ hybridization, and immunohistochemistry were performed on the sections. Transfect circE7 into SCC-7 cells and inject them into the cheeks of C3H mice. Once visible tumor formation was observed, intraperitoneal injections of anti-PD-1 (Selleck, A2002, China) and TIM-3 (Selleck, A2037, China) were administered every 3 days for treatment. The tumor volume and size in mice were measured regularly. After 10 days, the mice were euthanized, and the mouse tumor tissues were collected for RT-qPCR, ELISA, and flow cytometry analysis. Post-euthanasia, tumor tissues were paraffin-embedded and sectioned. Sections were stained with H&E, and subjected to in situ hybridization, and immunohistochemistry.
Statistical analysis
All continuous variables were presented as mean ± standard deviation (SD), while categorical data were presented as percentages. Statistical analyses were conducted using GraphPad Prism 9.4.1 software. To determine significant differences between the two groups, a Student t-test was employed. Survival analysis was carried out using the Kaplan-Meier curve and log-rank test. For all tests, p < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Source data
Acknowledgements
This work was supported in part by grants from the National Natural Science Foundation of China (U20A20367 to Z.Y.Z, 82372811 to Z.J.G), the Natural Science Foundation of Hunan Province (2023ZK1122 to Z.Y.Z, 2023DK2001 to Z.Y.Z), the Natural Science Foundation of Changsha City (kh2301025 to W.X, kq2208329 to L.S). Diagrams were generated using BioRender (www.biorender.com) under Academic License terms (NT276JXPUQ, SC276JXN82).
Author contributions
J.S.G., Y.M., and J.Y.G. designed the project and completed most experiments. P.C., J.W., D.W., H.K.Q., and P.W. performed some of the experiments. Z.J.G., P.C., L.S., C.M.F., S.S.Z., B.X., F.Y.W., Q.J.L., M.Z., and M.T. collected tissue samples. J.S.G. analyzed the ChIP-Seq data. J.S.G., Y.M., and J.Y.G. analyzed the data and wrote the manuscript. W.X. and Z.Y.Z. revised the manuscript. W.X., Z.Y.Z., and Z.J.G. are responsible for research supervision and funding acquisition. All authors read and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw ChIP-Seq data from generated in this study have been deposited in the Sequence Read Archive (SRA) database under accession code PRJNA1149346. The raw proteomic data are stored in the ProteomeXchange consortium repository under accession code PXD055003. The publicly released data used in this study are available in the GEO database under accession code GSE9106157, GSE15906758, and GSE18553159. The TCGA publicly available data used in this study are available in the Genomic Data Commons database under accession code TCGA-HNSC60. Part of the H3K27 acetylation level analysis is sourced from the ENCODE database61. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Junshang Ge, Yi Meng, Jiayue Guo.
Contributor Information
Zhaojian Gong, Email: gongzhaojian4458@csu.edu.cn.
Wei Xiong, Email: xiongwei@csu.edu.cn.
Zhaoyang Zeng, Email: zengzhaoyang@csu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52981-4.
References
- 1.Johnson, D. E. et al. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim.6, 92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung, H. et al. Global cancer statistics 2020: GLOBOCAN Estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Galot, R. et al. Personalized biomarker-based treatment strategy for patients with squamous cell carcinoma of the head and neck: EORTC position and approach. Ann. Oncol.29, 2313–2327 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Oliva, M. et al. Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann. Oncol.30, 57–67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruffin, A. T. et al. Improving head and neck cancer therapies by immunomodulation of the tumour microenvironment. Nat. Rev. Cancer23, 173–188 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, E. E. W. et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet393, 156–167 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Saba, N. F. et al. Pembrolizumab and cabozantinib in recurrent metastatic head and neck squamous cell carcinoma: a phase 2 trial. Nat. Med.29, 880–887 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burtness, B. et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet394, 1915–1928 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Carlisle, J. W., Steuer, C. E., Owonikoko, T. K. & Saba, N. F. An update on the immune landscape in lung and head and neck cancers. CA Cancer J. Clin.70, 505–517 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Taberna, M. et al. Human papillomavirus-related oropharyngeal cancer. Ann. Oncol.28, 2386–2398 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Kang, J. J. et al. Consensuses, controversies, and future directions in treatment deintensification for human papillomavirus-associated oropharyngeal cancer. CA Cancer J. Clin.73, 164–197 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride, A. A. Human papillomaviruses: diversity, infection and host interactions. Nat. Rev. Microbiol.20, 95–108 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Bao, H. L. et al. Prevalence of cervicovaginal human papillomavirus infection and genotypes in the pre-vaccine era in China: a nationwide population-based study. J. Infect.82, 75–83 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Gillison, M. L., Chaturvedi, A. K., Anderson, W. F. & Fakhry, C. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J. Clin. Oncol.33, 3235–3242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lechner, M., Liu, J., Masterson, L. & Fenton, T. R. HPV-associated oropharyngeal cancer: epidemiology, molecular biology and clinical management. Nat. Rev. Clin. Oncol.19, 306–327 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shamseddine, A. A., Burman, B., Lee, N. Y., Zamarin, D. & Riaz, N. Tumor immunity and immunotherapy for HPV-related cancers. Cancer Discov.11, 1896–1912 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giannella, L. et al. HPV-negative adenocarcinomas of the uterine cervix: from molecular characterization to clinical implications. Int J. Mol. Sci.23, 15022 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cillo, A. R. et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity52, 183–199.e189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyauchi, S. et al. HPV16 E5 mediates resistance to PD-L1 blockade and can be targeted with rimantadine in head and neck cancer. Cancer Res.80, 732–746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishna, S. et al. Human papilloma virus specific immunogenicity and dysfunction of CD8(+) T cells in head and neck cancer. Cancer Res.78, 6159–6170 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Liu, C. X. & Chen, L. L. Circular RNAs: characterization, cellular roles, and applications. Cell185, 2016–2034 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Chen, L. L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol.21, 475–490 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Zhao, J. et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun.10, 2300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang, R. et al. Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat. Commun.12, 832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belk, J. A., Daniel, B. & Satpathy, A. T. Epigenetic regulation of T cell exhaustion. Nat. Immunol.23, 848–860 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic Acids Res46, 2699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenz, R. et al. ViennaRNA package 2.0. Algorithms Mol. Biol.6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunkeler, M. et al. Structural basis for regulation of human acetyl-CoA carboxylase. Nature558, 470–474 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Galon, J. & Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov.18, 197–218 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Müller-Coan, B. G., Caetano, B. F. R., Pagano, J. S. & Elgui de Oliveira, D. Cancer progression goes viral: the role of oncoviruses in aggressiveness of malignancies. Trends Cancer4, 485–498 (2018). [DOI] [PubMed] [Google Scholar]
- 31.McBride A. A. Oncogenic human papillomaviruses. Philos. Trans. R. Soc. Lond. B Biol. Sci.372, 20160273 (2017). [DOI] [PMC free article] [PubMed]
- 32.Lange, P. & Damania, B. Kaposi sarcoma-associated herpesvirus (KSHV). Trends Microbiol28, 236–237 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Hatano, Y. et al. Virus-driven carcinogenesis. Cancers13, 20160273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferris, R. L. & Westra, W. Oropharyngeal carcinoma with a special focus on HPV-related squamous cell carcinoma. Annu. Rev. Pathol.18, 515–535 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrell, P. J. Epstein-Barr virus and cancer. Annu. Rev. Pathol.14, 29–53 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Chang, Y. & Moore, P. S. Merkel cell carcinoma: a virus-induced human cancer. Annu. Rev. Pathol.7, 123–144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan, K. E. & Lim, Y. Y. Viruses join the circular RNA world. FEBS J.288, 4488–4502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge, J. et al. Epstein-Barr virus-encoded circular RNA CircBART2.2 promotes immune escape of nasopharyngeal carcinoma by regulating PD-L1. Cancer Res.81, 5074–5088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crosbie, E. J., Einstein, M. H., Franceschi, S. & Kitchener, H. C. Human papillomavirus and cervical cancer. Lancet382, 889–899 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Yu, L., Lobanov, A. & Zheng, Z. M. Reply to Wang et al., “Assessment of the abundance and potential function of human papillomavirus type 16 circular E7 RNA”. mBio13, e0075822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Batchuluun, B., Pinkosky, S. L. & Steinberg, G. R. Lipogenesis inhibitors: therapeutic opportunities and challenges. Nat. Rev. Drug Discov.21, 283–305 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lally, J. S. V. et al. Inhibition of acetyl-CoA carboxylase by phosphorylation or the inhibitor ND-654 suppresses lipogenesis and hepatocellular carcinoma. Cell Metab.29, 174–182.e175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morelli, E. et al. A MIR17HG-derived long noncoding RNA provides an essential chromatin scaffold for protein interaction and myeloma growth. Blood141, 391–405 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shvedunova, M. & Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol.23, 329–349 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Chou, F. C., Chen, H. Y., Kuo, C. C. & Sytwu, H. K. Role of galectins in tumors and in clinical immunotherapy. Int J. Mol. Sci.19, 430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golden-Mason, L. & Rosen, H. R. Galectin-9: diverse roles in hepatic immune homeostasis and inflammation. Hepatology66, 271–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang, Z. H. et al. Tim-3/galectin-9 regulate the homeostasis of hepatic NKT cells in a murine model of nonalcoholic fatty liver disease. J. Immunol.190, 1788–1796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, Y. et al. Recipient T cell TIM-3 and hepatocyte galectin-9 signalling protects mouse liver transplants against ischemia-reperfusion injury. J. Hepatol.62, 563–572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirao, H. et al. The protective function of galectin-9 in liver ischemia and reperfusion injury in mice. Liver Transpl.21, 969–981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, C. et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat. Immunol.6, 1245–1252 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Leidner, R. et al. Neoadjuvant immunoradiotherapy results in high rate of complete pathological response and clinical to pathological downstaging in locally advanced head and neck squamous cell carcinoma. J. Immunother. Cancer9, e002485 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferris, R. L. et al. Neoadjuvant nivolumab for patients with resectable HPV-positive and HPV-negative squamous cell carcinomas of the head and neck in the CheckMate 358 trial. J. Immunother. Cancer9, e002568 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferris, R. L. et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med.375, 1856–1867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kürten, C. H. L. et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat. Commun.12, 7338 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo, X. et al. Melatonin inhibits EMT and PD-L1 expression through the ERK1/2/FOSL1 pathway and regulates anti-tumor immunity in HNSCC. Cancer Sci.113, 2232–2245 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, J. et al. EBV miRNAs BART11 and BART17-3p promote immune escape through the enhancer-mediated transcription of PD-L1. Nat. Commun.13, 866 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riaz, N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell171, 934–949.e916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foy, J. P. et al. Immunologically active phenotype by gene expression profiling is associated with clinical benefit from PD-1/PD-L1 inhibitors in real-world head and neck and lung cancer patients. Eur. J. Cancer174, 287–298 (2022). [DOI] [PubMed] [Google Scholar]
- 59.Callahan, S. C. et al. High enhancer activity is an epigenetic feature of HPV negative atypical head and neck squamous cell carcinoma. Front Cell Dev. Biol.10, 936168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature517, 576–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
The raw ChIP-Seq data from generated in this study have been deposited in the Sequence Read Archive (SRA) database under accession code PRJNA1149346. The raw proteomic data are stored in the ProteomeXchange consortium repository under accession code PXD055003. The publicly released data used in this study are available in the GEO database under accession code GSE9106157, GSE15906758, and GSE18553159. The TCGA publicly available data used in this study are available in the Genomic Data Commons database under accession code TCGA-HNSC60. Part of the H3K27 acetylation level analysis is sourced from the ENCODE database61. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.