Abstract
Background
The trend of postponing childbearing age is prevalent worldwide. Advanced paternal age (APA) is associated with adverse pregnancy outcomes and offspring health. However, the underlying mechanism by which paternal aging affects the risk of offspring neuropsychiatric disorders is unclear. Our study aims to explore the behavioral phenotypes and the pathologic epigenetic alterations of APA offspring inherited from aging sperm.
Methods
Behavioral tests, ELISA assay, immunofluorescence and western blotting were performed on offspring mice. Methylated RNA immunoprecipitation sequencing (MeRIP-seq) and RNA immunoprecipitation sequencing (RIP-seq) were used to investigate the modified N6-methyladenosine (m6A) profiles of paternal sperm and offspring hippocampus. Intervention of gene expression by lentivirus and adeno-associated virus in both vivo and vitro examined the potential therapeutic targets of intergenerational inherited neuroinflammation.
Results
In our study, APA offspring exhibit cognitive impairment and autism-like behavior. An increase in neuroinflammation in APA offspring is associated with microglial overactivation, which manifests as abnormal morphology and augmented engulfment. MeRIP-seq of F0 sperm and F1 hippocampus reveal that Nr4a2 is hypermethylated with decreased expression in APA offspring involving in synaptic plasticity and microglial function. In addition, Ythdc1, an m6A reader protein, is markedly elevated in aging sperm and remains elevated in adult hippocampus of APA group. Enhanced Ythdc1 recognizes and suppresses the hypermethylated Nr4a2, thereby contributing to the abnormal phenotype in offspring. The overexpression of Ythdc1 triggers microglial activation in vitro and its suppression in the hippocampus of APA progeny alleviates behavioral aberrations and attenuates neuroinflammation.
Conclusion
Our study provides additional evidence of the abnormal behavioral phenotypes of APA offspring and reveals potential epigenetic inheritance signatures and targeted genes for future research.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12974-024-03248-8.
Keywords: Advanced paternal age, Cognitive deficits, Autism, Inheritance, m6A, Microglia
Introduction
The Developmental Origins of Health and Disease (DOHaD) noted that adverse parental exposure and early life traits influence offspring physical and mental health [1]. The underlying biological mechanism of noncommunicable chronic diseases, known as metabolic and psychiatric disorders in adulthood, can be traced back to critical programming periods, including gametogenesis and embryonic development. Early trait investigations and timely interventions could improve the health of the next generation and reduce the economic burden of chronic diseases. Accumulating evidence suggests that maternal stress, immune status and undernutrition influence the behavior and cognitive development of offspring [2–5]. Moreover, the physical fitness and personal lifestyle factors of fathers, such as alcohol consumption, tobacco use, drug addiction and obesity, also lead to alterations in offspring behavior [6–8]. In recent years, the trend toward a postponed childbearing age has received prolonged discussion worldwide [9]. Multiple studies have shown that maternal aging is associated with adverse pregnancy outcomes and various offspring defects [10], but the consequences of advanced paternal age (APA) have not been fully examined. Epidemiological evidence has indicated that APA leads to an increased risk of autism [11, 12] and psychiatric and cognitive morbidities [13–15], including early onset of schizophrenia and impaired neurocognitive outcomes. In addition to clinical evidence, limited research has explicitly studied the behavioral phenotypes and molecular alterations of APA offspring brain. Thus, discussing whether APA influences gametogenic health and how changes are transmitted to the next generation is highly important. Hippocampus serves a critical role in learning, memory and emotional regulation [16]. The neuroinflammatory states in hippocampus contribute to the impaired neurogenesis and synaptic loss which ultimately causing behavior disorders. Therefore, we take the hippocampus as a key brain region for further research.
Aging-associated epigenetic modifications in mammalian sperm, including DNA methylation, small noncoding RNA (ncRNA) alteration and histone modification, have long been discussed [17]. Previous studies have demonstrated that epigenetic alterations in aging sperm contribute to neuropsychiatric disorders of offspring [18–21]. In addition, a study of paternal obesity and offspring metabolic disturbance highlighted the importance of transfer RNA-derived small RNAs (tsRNA) modification through intergenerational transmission [22]. The emerging role of RNA modification in intergenerational inheritance is not negligible. RNA N6-methyladenosine (m6A), stands as the most prevalent endogenous modification of messenger RNAs (mRNAs), has a profound influence on evolutionary adaptation and disease susceptibility [23] as well as gametogenesis and embryo development. Generally, m6A methylation is involved in maternal RNA clearance and chromatin accessibility during the maternal-to-zygote transition [24, 25]. Moreover, m6A modifications in mRNAs modulate murine spermatogenesis [26] and play a critical role in neurodevelopment [27]. Therefore, we conducted the following research with the hypothesis that APA could result in a greater risk of behavioral abnormalities in F1 offspring by affecting the m6A methylation states of functional pathways and key genes.
Materials and methods
Ethics and animal husbandry
All procedures for animal experiments were conducted complied with the Guide for the Care and Use of Laboratory Animals at Shanghai Model Organisms (Shanghai, China) (approval ID for the animal IACUC protocol: 2019-0027-01) and were consistent with ARRIVE guidelines. Male and female C57BL/6J mice (founder mice) were purchased from Shanghai Model Organisms (Shanghai, China). All mice were housed under a light: dark cycle of 12 h:12 h and a temperature of 21 ± 1 °C, with food and water ad libitum. Young fathers (3 months old, YF) and old fathers (21 months old, OF) were mated with virgin 10-week-old female mice at a 1:2 ratio to obtain F1 mice. The female mice with vaginal plugs were singly housed for the completion of gestation and the generation of offspring. The study design is shown in Fig. 1A. F0 mice were sacrificed for sperm collection after successful mating and only those who generated F1 offspring were used for further testing and statistical analysis. One mouse per litter were randomly selected and only male offspring mice were used for consecutive experiments. Every sample of one experiment were come from different litters. The F1 offspring mice were sacrificed and harvested at the age of 14 weeks after the behavioral tests. PLX3397 (pexidartinib, Selleck, Shanghai) was purchased and was added to a mouse chow diet at a concentration of 290 mg/kg (Xietong Pharmaceutical Bio-engineering Co., China). OF-F1 were randomly selected and fed a chow diet or a PLX3397 mixed diet for 4 consecutive weeks. The final dosage of around 20 mg total of drug/mouse over the 28 days of treatment was calculated on an average of 2.5 g of chow consumed by each mouse/day. Three mice per diet group were randomly selected for the examination of drug effectiveness and others were sent to conduct the behavioral tests.
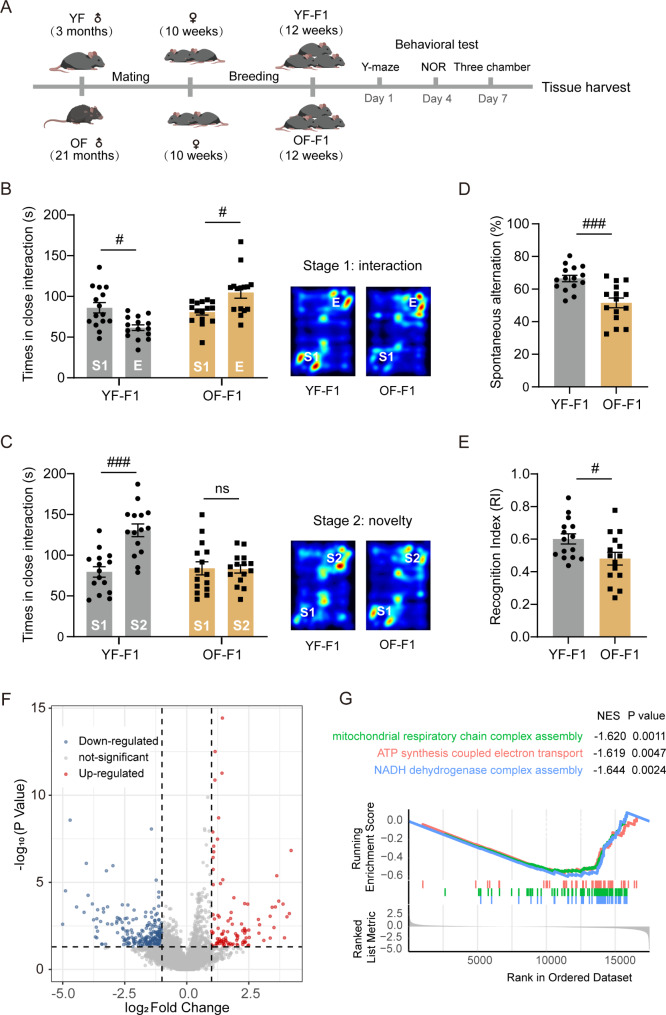
Fig. 1.
Impaired hippocampus-dependent memory and autism-like behavior in advanced paternal age F1 offspring mice. (A) Schematic of the animal model construction. (B, C) The three-chamber test included two stages of social activity recorded by the duration of interaction with each cage including stranger 1 (S1), empty cage (E) and stranger 2 (S2) (YF-F1 vs. OF-F1); two-way ANOVA with Tukey’s multiple comparison test: Stage1: F (1, 28) = 18.03, ηp²=0.2302, P = 0.0002; Stage2: F (1, 28) = 14.58, ηp²=0.1510, P = 0.0007. A heatmap of the two stages is depicted in the right panel. (D) The spontaneous alternations of Y-maze test were measured (YF-F1 vs. OF-F1); unpaired two-tailed Student’s t test: df = 28, R2 = 0.3838, P = 0.0003. (E) The recognition index (RI) of novel object recognition test was recorded (YF-F1 vs. OF-F1); one-sample two-tailed t test for YF-F1: df = 14, R2 = 0.4264, P = 0.0061; unpaired two-tailed Student’s t test: df = 28, R2 = 0.1719, P = 0.0227. (F) The volcano depicted the differentially expressed genes in YF-F1 and OF-F1 hippocampus. Blue dots represent down-regulated transcription and red dots represent up-regulated transcription. (Log2|Fold change| > 1, P < 0.05). (G) GSEA plot of mitochondrial respiratory chain complex assembly, ATP synthesis and NADH dehydrogenase complex assembly pathways in OF-F1. All behavioral tests were performed with 15 mice for each group. #P < 0.05, ##P < 0.01, and ###P < 0.001; ns indicates no significance
Behavioral tests
Behavioral tests of F1 offspring mice started at the age of 12 weeks. A series of tests were conducted with an interval of two days between different tests. All tests were conducted between 12:00 and 18:00 to minimize the effects of the circadian rhythm on performance. All behavior tests were completed at the age of 14 weeks. The recorded videos were further analyzed by EthoVision XT 14.
Novel object recognition
Mice were habituated to an arena (white plastic box, 40 × 40 × 40 cm) for 10 min the day before the test. Then, they were exposed to two unfamiliar but identical objects in the arena for 10 min. After 2 h, the animals were sent back in the arena with one of the objects replaced by a novel object. The time spent exploring each object was recorded. The recognition index (RI) was calculated as the novel object exploration time/ (novel object exploration time + familiar object exploration time). One-sample t-test was conducted on the RI in control group to confirm the proper performance.
Y-maze
Y maze consists of three white arms (40 cm × 3 cm × 12 cm), placed at 120 degrees with respect to each other. Square, circle, triangle shaped patterns were placed along the arms respectively as the internal visual cues. Animals were allowed to freely explore three arms, and the sequence and arm entries were manually recorded for 10 min per mouse. During the trial, all three arms were accessible, and the mice were placed into the same start arm: Spontaneous alternation (%) = (Actual Alternations/Maximum Alternations) *100%.
Three-chamber test
The three-chamber test was performed in a rectangular box with three chambers, each chamber designed as 20 cm (L) × 40 cm (W) × 20 cm (H) [28].The internal walls of chambers have small openings (5 cm × 5 cm) for the free access of test mice. Two empty cages were placed on each side of the chamber. The test mouse was first allowed to habituate to the three chambers for 10 min. At the beginning of the test, a stranger mouse (stranger 1) was placed in one of the empty cages, and the test mouse was subsequently moved into the middle chamber to allow free exploration for 10 min. Then, another mouse (stranger 2) was moved in the other empty cage. Another 10-minute free exploration session of the test mouse was also recorded. Time spent in contact with the cages were analyzed.
Enzyme-linked immunosorbent assay (ELISA)
The hippocampal tissues were washed in phosphate-buffered saline (PBS; Servicebio, Wuhan, China) and were homogenized in lysis buffer (Beyotime, China). After homogenization, the samples were centrifuged at the speed of 12,000 rpm for 10 min at 4 °C. The supernatant was first collected to measure the protein concentration by a bicinchoninic acid assay (Thermo Fisher Scientific, Rochester, NY, USA). The levels of multiple cytokines were measured by ELISA kits (Servicebio, Wuhan, China) according to the manufacturer’s instructions. Each sample was run in triplicate. The final concentration of inflammatory factors was calculated by the specific value of the sample concentration and total protein content.
Immunofluorescence analysis of brain sections
Offspring mice were anesthetized with isoflurane and perfused transcardially with PBS followed by 4% paraformaldehyde (PFA). The brains were collected and fixed in 4% PFA at 4 °C overnight. After dehydration in 30% sucrose solution in 4% PFA at 4 °C, 30 μm-thick coronal brain sections were cut using a cryostat (CM1950, Leica). Eight to ten sections per mouse were collected. After washing in PBS 3 times (5 min/wash), the brain sections were permeabilized with PBS containing 0.5% Triton X-100 for 30 min, blocked in PBS containing 3% bovine serum albumin (BSA) and 0.2% Triton X-100 for 1 h, and incubated with primary antibodies (Iba1, Cell Signaling Technology, 1:100; PSD95, Invitrogen, 1:150; LAMP1, Santa cruz, 1:100; SYN, Abcam, 1:200) overnight at 4 °C. The sections were washed in PBS 3 times (10 min/wash) and then incubated with secondary antibody (Alexa Fluor 488-conjugated goat anti-rabbit IgG, Abcam, 1:1000; Alexa Fluor 568-conjugated goat anti-mouse IgG, Abcam, 1:1000; Alexa Fluor 647-conjugated goat anti-rat IgG, Abcam, 1:1000) in PBS for 1 h at room temperature. After washing 3 times with PBS (10 min/wash), the sections were stained with the nuclear stain 40,6-diamidino-2-phenylindole (DAPI; Abcam, 1:1000) at room temperature. Confocal microscope was used to detect Immunofluorescence. One randomly chosen slice per animal was selected and two fields of CA1, DG, and CA3 areas per slice were used for measurement. The quantification of microglia abundance was measured by density/mm2. Sholl analysis was used to evaluate microglia complexity as previously described [29, 30]. For microglia phagocytosis analysis by co-staining of Iba1, PSD95 and LAMP1, one slice per animal were acquired and two randomly selected cells per slice were used for analysis.
Western blotting
Tissues and cells were collected and lysed using lysis buffer (Beyotime, China) with protease inhibitor (Yeasen, China). The concentrations of protein were detected by a bicinchoninic acid assay. A total of 15 µg protein per sample was separated via sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (EpiZyme, China) and blotted onto a 0.45 μm polyvinylidene difluoride (PVDF) membrane (Merck Millipore). The PVDF membrane was blocked with 5% milk for 2 h at room temperature. Then, the membrane was incubated with primary antibody (Iba1, Cell Signaling Technology, 1:1000; NeuN, Abcam, 1:1000; GFAP, Invitrogen, 1:1000; Ythdc1, Proteintech, 1:1000; Nr4a2, Proteintech, 1:1000; GAPDH, Proteintech, 1:2000; β-actin, Proteintech, 1:2000; SNAP25, Proteintech, 1:1000; GAP43, Proteintech, 1:1000) at 4 °C overnight. The membrane was incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (Anti-rabbit and anti-mouse, Cell Signaling Technology, 1:5000) after washing three times with Tris-buffered saline containing Tween 20 (TBST). For statistical analysis, relative protein levels were quantified by ImageJ and normalized by the reference protein.
Sperm preparation
Mice were euthanized and the cauda epididymis was dissected and cut into small pieces in 500 µl of BWW solution (Solarbio, Beijing, China) following an incubation at 37 °C for 30 min to release sperm from the epididymis. After being strained with a 40 μm cell strainer, the sperm were centrifuged at 500 × g for 5 min for precipitation. Then, the precipitated sperm were dissolved in lysis buffer (DEPC water containing 0.5% Triton X-100 and 0.1% SDS) for 40 min, further washed with PBS three times and centrifuged at 500 × g for 5 min at room temperature for precipitation. The sperm were obtained and stored at -80 °C.
RNA-seq and methylated RNA immunoprecipitation sequencing (MeRIP-seq)
For RNA-seq, one hippocampi was extracted as one sample. Five samples per group were collected. Total RNA was extracted using RNAisoreagent (Takara) and KAPA RNA HyperPrep Kit with RiboErase (HMR) (KK8561) of Illumina Platforms were used for library construction. For two-cell and blastocyst RNA-seq, 20 embryos from each individual male founder mouse at each stage were pooled as one sample. Three samples per group were collected for future experiments. All the products were preamplified to obtain cDNA following the SMART-Seq2 protocol [31]. cDNA was quantified with a Qubit 3.0 (Thermo Fisher Scientific). 1 ng of cDNA was used for DNA library construction with the TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme) following the manufacturer’s instructions. MeRIP-seq was performed on the hippocampi of F1 mice and the sperm of F0 fathers. The hippocampi of three mice were mixed as one sample. Sperm from five mice were collected and mixed as one sample. Two biological replicates per group were performed on both sperm and hippocampus. Briefly, m6A RNA immunoprecipitation was performed with a GenSeqTM m6A RNA IP Kit (GenSeq, Inc., China) following the manufacturer’s instructions. An NEBNext® Ultra II Directional RNA Library Prep Kit was used to construct RNA sequencing libraries for input RNA samples without immunoprecipitation as well as for IP RNA samples after immunoprecipitation. A library quality check was performed using a Bioanalyzer 2100 analyzer (Agilent). High-throughput sequencing was performed on an Illumina NovaSeq 6000 sequencer using 150 bp paired-end mode. After sequencing, followed by image analysis, base recognition and quality control, the original reads (raw data) were generated. First, Q30 was used for quality control. Then, Cutadapt [32] software (v1.9.3) was used to remove adapters and low-quality reads to obtain high-quality clean read. HISAT2 [33] software (v2.0.4) was used to match the clean reads of all the samples using the mouse genome (assembly GRCm38/mm10). Then, MACS [34] software was used to identify methylated genes in each sample. Differentially methylated sites were identified by diffReps [35]. We screened the peaks located on the exons of the mRNAs and made corresponding annotations. Differentially methylated RNA sites whose |log2(fold change) |> 1, p value < 0.0001 and FDR < 0.01 were filtered to determine the differentially methylated genes. The DEGs identified via RNA-seq analysis were filtered by |log2(fold change) |> 1 and p value < 0.05. Enrichment analysis were performed by “clusterProfiler” (version 3.18.1) R package including Gene Ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) database analysis and gene set enrichment analysis (GSEA).
qPCR
Total embryonic RNA was extracted using the PicoPure RNA Isolation Kit (Thermo Fisher Scientific). Total hippocampus and cell RNA were extracted by FastPure Cell/Tissue Total RNA Isolation Kit (Vazyme, China). RNA was synthesized into cDNA using an RT Reagent Kit and gDNA Eraser (Takara) according to the manufacturer’s instructions. qPCR was conducted on a QuantStudio 7 Flex system (Life Technologies, USA). All samples were run in triplicate. All the data were normalized to the expression level of β-actin to quantify the relative expression of mRNA. The primers used are listed in Table S1.
MeRIP-qPCR and RNA immunoprecipitation (RIP)-qPCR
For MeRIP-qPCR, the MeRIP assay was carried out with a GenSeq® m6A MeRIP Kit (GenSeq, Inc., Shanghai, China) in accordance with the manufacturer’s protocols. The modification of target genes by m6A was determined by qPCR with specific primers. For RIP-qPCR, hippocampal tissues were lysed with lysis buffer (Beyotime, China), and 10% of the lysates were saved as input. The primary antibody, IgG or protein A + G beads (Beyotime, China) were subsequently added to the remaining lysate, which was subsequently incubated on a shaker at 4 °C for 12 h. The beads were subsequently washed with 1 × RIPA buffer supplemented with protein kinase K buffer. After the beads were removed, the supernatant was collected for further RNA isolation. RNA was synthesized into cDNA using an RT Reagent Kit and gDNA Eraser (Takara). The primers used are listed in Table S1.
RIP-seq
Sperm from ten mice were collected and mixed as one sample. Briefly, the RNA immunoprecipitation assay was performed by GenSeq® RIP Kit (GenSeq, Inc.) according to the manufacturer’s instructions. rRNAs were removed by GenSeq® rRNA Removal Kit (GenSeq, Inc.) in both immunoprecipitated RNA and input RNA samples. RNA libraries were constructed by using rRNA-depleted RNAs with the GenSeq® Low Input RNA Library Prep Kit (GenSeq, Inc.). The quality of the libraries was controlled, and the libraries were quantified using a Bioanalyzer 2100 system (Agilent Technologies, Inc.). Library sequencing was performed on Illumina NovaSeq instrument with 150 bp paired-end reads. A similar data analysis method was used, as described in the previous section. The peaks identified with exons of mRNA were identified and chosen for annotation. GO and KEGG analyses were performed on the differentially expressed RIP protein-coding genes.
Culture of BV2 cells and cell transfection
The mouse microglial cell line (BV2) was purchased from the National collection of Authenticated Cell cultures and was seeded at a density of 1–2 × 105 cells/well in a 6-well plate supplemented with 0.1% penicillin-streptomycin (Solarbio, China) and 10% fetal bovine serum (FBS, Sigma) in DMEM (Invitrogen, USA). Cultures were maintained at 37 °C in a 5% CO2 humidified atmosphere for 3 days and were subsequently collected for further experiments. The lentivirus was constructed by GeneChem (Shanghai, China). For transfection, cells were cultured at a density of 40–60%. The negative control or Ythdc1-specific lentivirus was transfected into BV2 cells at an MOI of 50 for 12 h according to the manufacturer’s instructions. The efficiency of lentivirus infection was determined by calculating the percentage of GFP-expressing cells by fluorescence microscopy after transfection for 72 h. Next, 3 mg/ml puromycin was supplemented to the medium to remove the untransfected cells. qPCR and western blotting were used to determine the efficiency of overexpression. The cells were plated on confocal dishes, fixed with 4% paraformaldehyde for 15 min, and washed three times with PBS for 3 min each. Then, the cells were incubated with 5% BSA at 37 °C for 1 h, followed by incubation with primary antibodies (anti-CD68, Proteintech, 1:200) overnight at 4 °C. Then, the sections were washed in PBS 3 times (10 min/wash) and incubated with Alexa Fluor 568-conjugated goat anti-rabbit IgG (1:1000, Abcam) for 60 min at room temperature. After staining with DAPI (Abcam, 1:1000), immunofluorescence was detected using a confocal microscope.
Virus injection
Constructed adeno-associated virus (AAVMG1.2-F4/80p-sh-Ythdc1) and control vector (AAVMG1.2-F4/80p-sh-Ctrl) were purchased from GeneChem (Shanghai, China). OF-F1 mice were randomly subjected to either shYthdc1 or control group and were in situ injected respectively into hippocampus region (1 µl per side) at the rate of 0.10 µl/min bilaterally (coordinates: anteroposterior (AP), -2.0 mm; mediolateral (ML), -1.55 mm; dorsoventral (DV), -1.55 mm). After injection, the needle was maintained in place for additional 5 min. After 4 weeks, the mice underwent behavioral analysis and tissue samples were collected for further study.
Embryo collection and immunofluorescence
Ten-week-old female mice were mated with male mice overnight after human chorionic gonadotropin (hCG) injection. Two-cell embryos were collected 24 h after hCG administration by removing cumulus cells. Morula and blastocyst embryos were collected 66–78 h after hCG administration by flushing the oviduct and the upper part of the uterus. The embryos were washed in M2 medium (Sigma) before further experiments. For the embryo immunofluorescence test, embryos were exposed to Tyrode’s solution (Sigma) to remove the zona pellucida. After washed in M2 medium for three times, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature and permeated with 0.5% Triton X-100 for 30 min. After washing in PBS containing 0.1% BSA solution three times, the zygotes were blocked in 5% BSA solution diluted in PBS for 1 h. The samples were stained overnight at 4 °C with primary antibodies (Ythdc1: Proteintech, 1:200; m6A: Abcam, 1:200). The next day, the samples were incubated with the secondary antibodies Alexa Fluor 488 goat anti-rabbit IgG (1:1000; Abcam) and Alexa Fluor 568 goat anti-mouse IgG (1:1000; Abcam) for 2 h at room temperature. The samples were mounted onto slides with antifade medium containing DAPI (Abcam, 1:1000) and examined under a confocal microscope.
Statistical analysis
All calculations were performed with GraphPad Prism software (GraphPad, La Jolla, CA, USA). Statistical analysis was performed using the unpaired two-tailed Student’s t test for two-group comparisons. One-sample two-tailed t test was performed to examine the efficacy of control groups. One-way ANOVA or two-way ANOVA combined with Tukey’s multiple comparison test to determine differences between more than two groups. Differences were considered significant at P < 0.05. All the data are presented as the means ± SEMs. The significance levels are presented as follows: #P < 0.05, ##P < 0.01, and ###P < 0.001; ns indicates no significance.
Results
Animal model construction and abnormal behavior test performance
An APA mouse model was established (Fig. 1A). Three-month-old young fathers (YFs) and 21-month-old older fathers (OFs) were mated with ten-week-old female mice, and the offspring were generated as YF offspring (YF-F1) and OF offspring (OF-F1), respectively. At 12 weeks of age, mice underwent a series of behavioral tests. The three-chamber test was performed as a classical method for detecting autism spectrum disorder [36]. YF-F1 mice preferred to interact with stranger 1, while OF-F1 mice spent less time interacting with stranger 1 (Fig. 1B). In the novelty section (stage 2), YF-F1 mice spent more time interacting with stranger 2 than with stranger 1, while OF-F1 mice did not (Fig. 1C). The NOR test and Y-maze test were used to evaluate short-term object memory [37] and spatial working memory [38], individually. In the Y-maze test, the OF-F1 mice exhibited fewer spontaneous alternations (Fig. 1D). In the NOR test, the OF-F1 mice spent less time exploring the novel object and had a lower recognition index (RI) (Fig. 1E). These findings demonstrated the occurrence of behavioral disorders in OF offsprings.
Advanced paternal age mice exhibited neuroinflammation and microglial overactivation
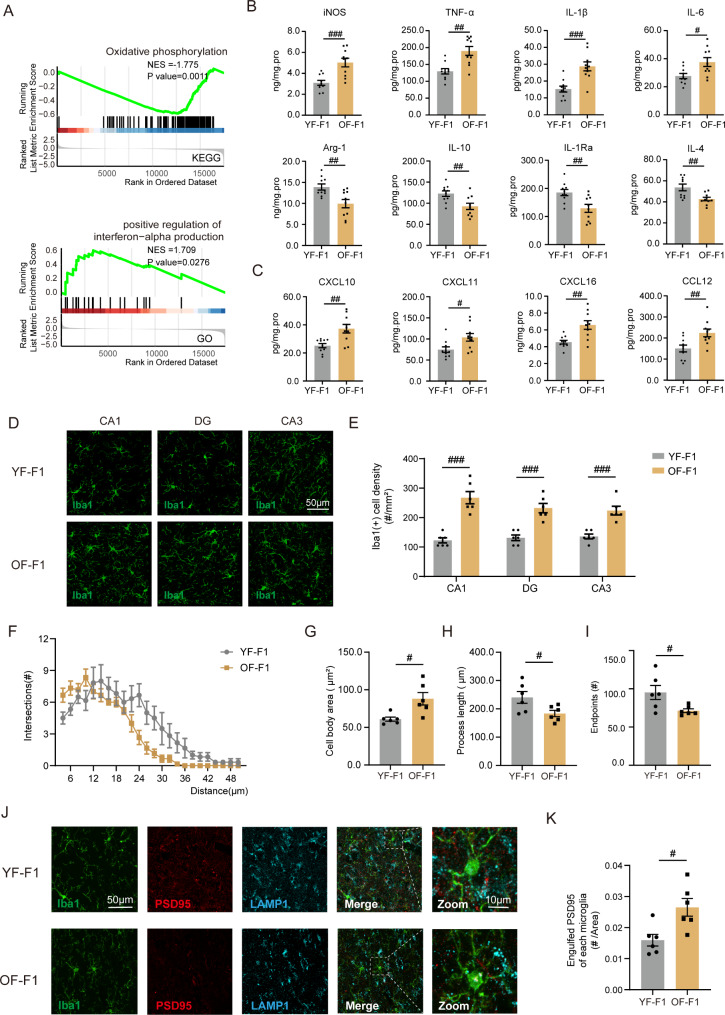
To determine the possible molecular mechanism involved in the changes in the OF offspring, we investigated differences in the transcriptome of YF-F1 and OF-F1. The hippocampal RNA-seq results revealed multiple differentially expressed genes (DEGs) between the two groups (Fig. 1F). Gene set enrichment analysis (GSEA) revealed the downregulation of mitochondrial respiratory chain complex assembly, ATP synthesis and NADH dehydrogenase complex assembly [39] (Fig. 1G). Besides, inflammation-related pathways, including the oxidative phosphorylation pathway and the positive regulation of interferon (IFN)-alpha production, were significantly altered (Fig. 2A). Hyperactive IFN signaling may contribute to neuroinflammatory states in the OF-F1 hippocampus. Thus, inflammatory cytokines levels were measured in hippocampal lysates from the two groups using ELISA. Significant increases were observed in proinflammatory cytokines, including inducible nitric oxide synthase (iNOS), tumor necrosis factor α (TNF-α), interleukin-1β (IL-1β) and interleukin-6 (IL-6), which are normally secreted by activated microglia and repress the expression of anti-inflammatory cytokines, including arginase 1 (Arg1), interleukin-10 (IL-10), interleukin-1 receptor antagonist (IL1-Ra), and interleukin-4 (IL-4), in OF-F1 hippocampal lysates (Fig. 2B). We also measured several representative factors associated with classic microglial activation in the hippocampus, including C-X-C motif ligand 16 (CXCL10), C-X-C motif ligand 11 (CXCL11), C-X-C motif ligand 16 (CXCL16) and chemokine (C-C motif) ligand 12 (CCL12) [40–42] (Fig. 2C). The levels of all of these chemokines were elevated in OF-F1. Next, we examined the protein levels of neuronal and glial cell types by performing western blotting for a neuronal marker (NeuN), an astrocyte marker (GFAP) and a microglial marker (Iba1). We found that the expression of Iba1 was significantly increased in OF-F1 mice (Fig. S1). We performed immunostaining in three regions of hippocampus including cornus ammonis 1 (CA1), cornusammonis 3 (CA3) and dentate gyrus (DG). We found that the number of microglia (defined as Iba1 + cells) increased consistently in these three areas, especially in the CA1 region (Fig. 2D, E). Sholl analysis revealed a reduction in the number of intersections in the hippocampus of OF-F1 mice (Fig. 2F). In addition, an enlargement of the microglial cell body area (Fig. 2G) was accompanied by a reduction in process length (Fig. 2H) and fewer endpoints (Fig. 2I), suggesting significant changes in microglial morphological characteristics and dendritic complexity in the OF-F1 hippocampus. PSD95 is a marker of postsynaptic components and LAMP1 is a maker of lysosome [43]. By immunostaining of Iba1, PSD95 and LAMP1 (Fig. 2J), the colocalization of LAMP1 and PSD95 in Iba1 + cells were defined as engulfed PSD95. We detected increased engulfed PSD95 in microglia in OF-F1, suggesting the activation of microglial phagocytosis. In addition, by immunostaining of PSD95 and SYN, a presynaptic marker, we found that the colocalization of PSD95 + and SYN + puncta were reduced in OF-F1, indicating the synaptic loss in paternal aging offsprings (Fig. S2). Further, we detected a decrease in presynaptic protein including SNAP25 and GAP43 (Fig. S3). These results collectively support the conclusion that OF offspring exhibit neuroinflammation and microglial overactivation, thereby these molecular changes may contribute to the behavioral deficits of social interaction and cognition.
Fig. 2.
Neuroinflammatory state and microglial overactivation in APA offspring mice. (A) GSEA of RNA-seq data showing alterations in pathways between the YF-F1 and OF-F1 hippocampi. (B) The differential expression levels of proinflammatory factors (iNOS, TNF-α, IL-1β, and IL-6) and anti-inflammatory factors (Arg1, IL-10, IL-1Ra, and IL-4) between the two groups were determined via ELISA (n = 10/group); unpaired two-tailed Student’s t test: iNOS: df = 18, R2 = 0.4763, P < 0.001; TNF-α: df = 18, R2 = 0.4617, P = 0.0010; IL-1β: df = 18, R2 = 0.5055, P < 0.001; IL-6: df = 18, R2 = 0.2918, P = 0.0139; Arg1: df = 18, R2 = 0.3643, P = 0.0048; IL-10: df = 18, R2 = 0.3411, P = 0.0069; IL-1Ra: df = 18, R2 = 0.3515, P = 0.0059; IL-4: df = 18, R2 = 0.3449, P = 0.0065. (C) Microglial activation-associated cytokine expression in YF-F1 and OF-F1 hippocampus (n = 10/group); unpaired two-tailed Student’s t test: CXCL10: df = 18, R2 = 0.4142, P = 0.0022; CXCL11: df = 18, R2 = 0.2737, P = 0.0179; CXCL16: df = 18, R2 = 0.4296, P = 0.0017; CCL12: df = 18, R2 = 0.3395, P = 0.0070. (D) Confocal images of Iba1 (green)-stained CA1, DG and CA3 regions from YF-F1 and OF-F1 mice (n = 6/group). (E) Quantification of microglial numbers in the CA1, DG, and CA3 regions (YF-F1 vs. OF-F1); unpaired two-tailed Student’s t test: CA1: df = 10, R2 = 0.8037, P < 0.0001; DG: df = 10, R2 = 0.7482, P = 0.0003; CA3: df = 10, R2 = 0.7299, P = 0.0004. (F) Sholl analysis of microglial cells in YF-F1 and OF-F1 (n = 6/group). Data are displayed as the number of intersections at each distance from the cell bodies of each cell; two-way ANOVA with Tukey’s multiple comparison test: F (23, 230) = 2.775, ηp²=0.05688, P < 0.0001. (G) Quantification of microglial cell-body area in YF-F1 and OF-F1 (n = 6/group); unpaired two-tailed Student’s t test: df = 10, R2 = 0.4945, P = 0.0107. (H) Quantification of process length per microglial cell in YF-F1 and OF-F1 (n = 6/group); unpaired two-tailed Student’s t test: df = 10, R2 = 0.3692, P = 0.0361. (I) Quantification of endpoints per microglial cell (n = 6/group); unpaired two-tailed Student’s t test: df = 10, R2 = 0.3846, P = 0.0314. (J) Representative images of Iba1 (green)-, PSD95 (red)- and LAMP1 (blue)- stained CA1 region from YF-F1 and OF-F1 mice. An enlarged image is used to show the engulfment of PSD95. (K) The number of engulfed PSD95 in each microglia of YF-F1 and OF-F1(n = 6/group); unpaired two-tailed Student’s t test: df = 10, R2 = 0.4899, P = 0.0113. #P < 0.05, ##P < 0.01, and ###P < 0.001; ns indicates no significance
Depletion of microglia rescues cognitive impairment and autism-like behavior in OF offspring
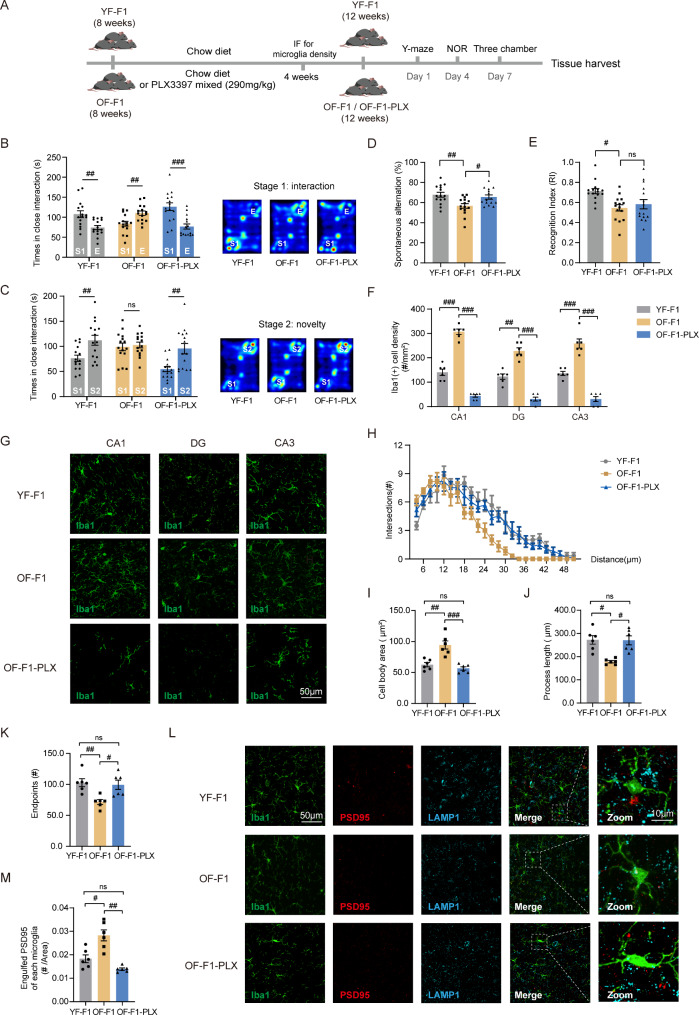
To directly address whether overactivated microglia play a role in the decreased performance of OF-F1 mice in cognitive tests and three-chamber tests, we constructed a mouse model supplied with PLX3397. PLX3397 has been proven to decrease the microglial population without causing significant cognitive impairment in mice [44]. Thus, we divided the OF-F1 mice randomly into two groups at the age of 8 weeks; one group had PLX3397 in their food supply (290 mg/kg of food available ad libitum) for 4 weeks, namely, the OF-F1-PLX group, and the other group (the remaining OF-F1 mice) continued to consume a chow diet (Fig. 3A). We then evaluated their cognitive performance and social interaction at the age of 12 weeks. Remarkably, microglia ablation by PLX3397 successfully rescued their cognitive deficits and autism-like behavior. PLX3397-treated OF-F1 mice spent much more time interacting with stranger 1 mice than an empty cage (Fig. 3B) and had closer interactions with stranger 2 mice in the novelty section (Fig. 3C). In the Y-maze, OF-F1 mice exhibited decreased spontaneous alternations, and OF-F1-PLX mice exhibited better performance (Fig. 3D). In the NOR test, OF-F1-PLX mice did not exhibit greater interest in exploring novel objects (Fig. 3E). We checked the number and morphology of microglia in three groups. The number of microglia was decreased by 90% in three regions of the hippocampus after drug supplementation (Fig. 3F, G). Sholl analysis revealed an increase in the number of intersections in the hippocampus of OF-F1-PLX mice (Fig. 3H). In addition, the cell body area (Fig. 3I) decreased with increasing process length (Fig. 3J) and number of endpoints (Fig. 3K) in the OF-F1-PLX hippocampus. Similarly, the engulfed PSD95 decreased in OF-F1-PLX mice (Fig. 3L, M). Besides, PLX3397 treatment increased the expression of synapse in OF-F1 hippocampus (Fig. S4). The morphological changes and the engulfment of synapse in OF-F1-PLX mice did not appear statistically significant when comparing to YF-F1 group indicating that the treatment of PLX3397 brings OF-F1 microglia back to homeostasis without significant overshooting. Thus, microglial ablation attenuated the learning and memory defects and autism-like behaviors of OF offspring.
Fig. 3.
Rescuing overactivated microglia could attenuate adverse behavioral performance in advanced paternal age mice. (A) Experimental design for PLX3397 supplementation. (B, C) The three-chamber test included two stages of social activity recorded by the duration of interaction with each cage including stranger 1 (S1), empty cage (E) and stranger 2 (S2) (YF-F1 vs. OF-F1 and OF-F1-PLX); two-way ANOVA with Tukey’s multiple comparison test: Stage1: F (2, 42) = 12.32, ηp²=0.246, P < 0.0001; Stage2: F (2, 42) = 4.046; ηp²=0.0568, P = 0.0247. Heatmaps of the two stages is depicted in the right panel. (D) The Y-maze test was used to detect spontaneous alternations in YF-F1, OF-F1 and OF-F1-PLX; two-way ANOVA with Tukey’s multiple comparison test: F (2, 24) = 7.465, ηp²=0.255, P = 0.0030. (E) The novel object recognition test was performed to measure the time spent exploring the new object in YF-F1, OF-F1 and OF-F1-PLX mice; one-sample two-tailed t test for YF-F1: df = 14, R2 = 0.8349, P < 0.0001; two-way ANOVA with Tukey’s multiple comparison test: F (2, 22) = 4.392, ηp²=0.155, P = 0.0248. (F) Quantification of microglial numbers in the CA1, DG, and CA3 regions (YF-F1 vs. OF-F1 and OF-F1-PLX); two-way ANOVA with Tukey’s multiple comparison test: CA1: F (2, 10) = 141.0, ηp²=0.9564, P < 0.0001; DG: F (2, 10) = 120.7, ηp²=0.9160, P < 0.0001; CA3: F (2, 10) = 59.76, ηp²=0.9094, P < 0.0001. (G) Confocal images of Iba1 (green)-stained CA1, DG and CA3 regions from YF-F1, OF-F1 and OF-F1-PLX mice (n = 6/group). (H) Sholl analysis of microglial cells in YF-F1, OF-F1 and OF-F1-PLX (n = 6/group). Data are displayed as the number of intersections at each distance from the cell bodies of each cell; two-way ANOVA with Tukey’s multiple comparison test: F (46, 345) = 2.298, ηp²=0.0490, P < 0.0001. (I) Quantification of microglial cell-body area in YF-F1, OF-F1 and OF-F1-PLX (n = 6/group); two-way ANOVA with Tukey’s multiple comparison test: F (2, 10) = 18.87, ηp²=0.7223, P = 0.0004. (J) Quantification of process length per microglial cell in YF-F1, OF-F1 and OF-F1-PLX (n = 6/group); two-way ANOVA with Tukey’s multiple comparison test: F (2, 10) = 8.550, ηp²=0.6083, P = 0.0068. (K) Quantification of endpoints per microglial cell in YF-F1, OF-F1 and OF-F1-PLX (n = 6/group); two-way ANOVA with Tukey’s multiple comparison test: F (2, 10) = 8.464, ηp²=0.5104, P = 0.0071. (L) Representative images of Iba1 (green)-, PSD95 (red)- and LAMP1 (blue)- stained CA1 from YF-F1, OF-F1 and OF-F1-PLX mice. An enlarged image is shown to show engulfment of PSD95. (M) The number of engulfed PSD95 in each microglia of YF-F1, OF-F1 and OF-F1-PLX groups (n = 6/group); two-way ANOVA with Tukey’s multiple comparison test: F (2, 10) = 21.00, ηp²=0.7199, P = 0.0003. All behavioral tests were performed with 15 mice for each group. #P < 0.05, ##P < 0.01, and ###P < 0.001; ns indicates no significance
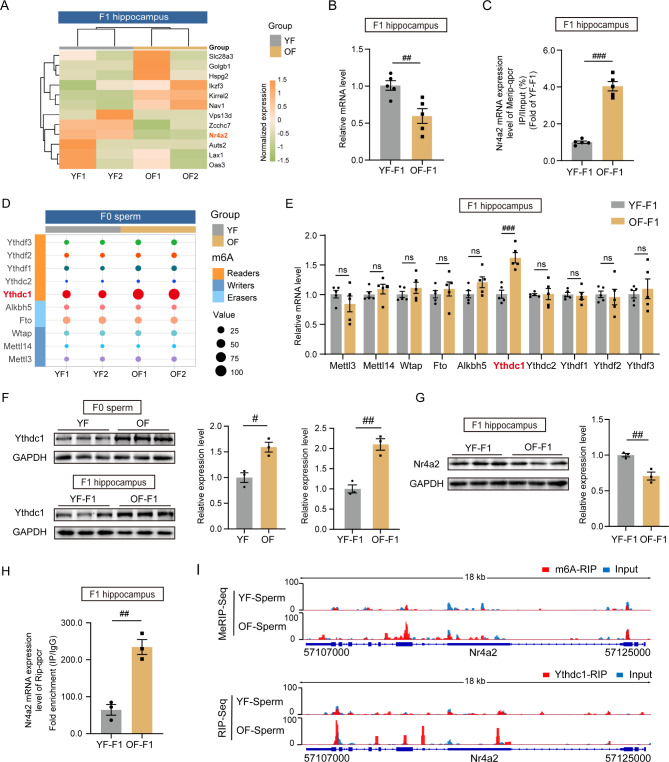
mRNA m6A modifications in sperm and F1 hippocampus
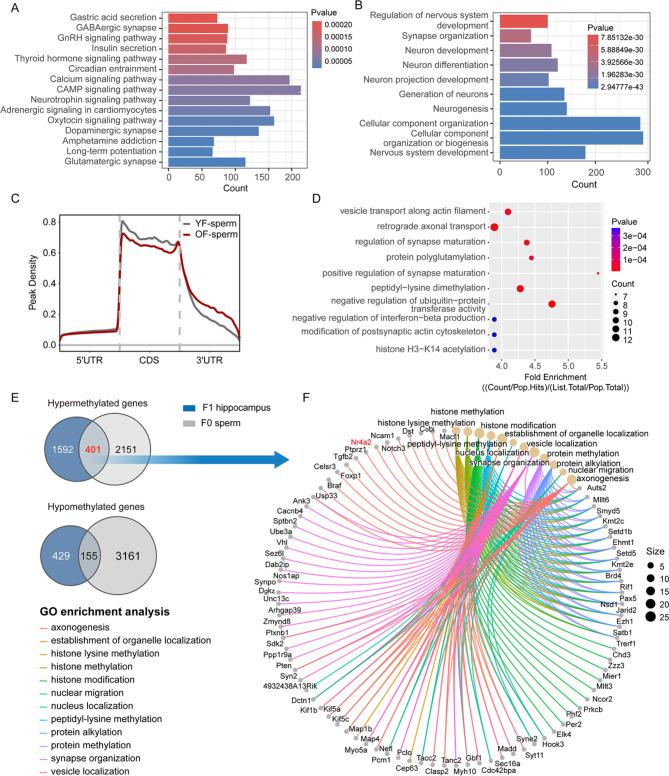
Various epigenetic modifications may be involved in intergenerational inheritance. The emerging candidate RNA N6-methyladenosine (m6A) has been proven to be an essential epigenetic modifying factor [45]. After RNA extraction, library construction and high-throughput sequencing, MeRIP-seq of the hippocampi from the offspring was performed, and the results revealed differences in the distribution of peak density between the two groups (Fig. S5A). According to the KEGG enrichment analysis of the differentially methylated genes, the m6A level was elevated in multiple processes, including GABAergic synapses, dopaminergic synapses, glutamatergic synapses and long-term potentiation (Fig. 4A). The top 10 hypomethylated pathways in the hippocampus, as determined by GO analysis, also revealed pathways related to neuron development, synapse organization and neurogenesis (Fig. 4B). GO enrichment revealed hypermethylated genes related to synaptic transmission, postsynaptic structure and axodendritic protein transport (Fig. S5B). We then performed MeRIP-seq on paternal sperm to determine the possible changes in the associated pathways. In OF sperm, the peak density of m6A modification was also distinctly distributed, with a decreased preference for the coding sequence (CDS) region but a greater peak in the 3’ untranslated region (3’UTR) (Fig. 4C). The top 10 upregulated pathways in aging sperm included synaptic maturation and the regulation of interferon production (Fig. 4D). The genes whose methylation level was downregulated according to KEGG enrichment analysis are mapped in Fig. S5C. Interestingly, when the differentially methylated m6A genes in the hippocampi of the offspring and paternal sperm were overlapped, many genes were the same, with 401 upregulated genes and 155 downregulated genes (Fig. 4E). We selected common genes with increased m6A methylation to construct a gene-GO term network and found that the axonogenesis and synapse organization terms were enriched (Fig. 4F). Importantly, the joint analysis of MeRIP-seq data for paternal sperm and offspring hippocampus indicate potential links between m6A modifications in paternal sperm and offspring hippocampus, suggesting further investigation into the mechanisms of epigenetic inheritance.
Fig. 4.
m6A modifications in the sperm and F1 hippocampus were altered in OF group. (A) Differentially hypermethylated genes in the hippocampus of the offspring were analyzed via KEGG enrichment (YF-F1 vs. OF-F1, 14-weeks). Fold change > 2.0, P < 0.0001. (B) Differentially hypomethylated genes in the hippocampi of the offspring were analyzed by GO analysis (YF-F1 vs. OF-F1, 14-weeks). Fold change > 2.0, P < 0.0001. (C) The peak density of YF (3-month) and OF (21-month) sperm was analyzed by metagene using MetaPlotR [74]. (D) Differentially hypomethylated genes were analyzed by GO enrichment analysis in sperm (YF vs. OF). Fold change > 2.0, P < 0.0001. (E) Overlapping genes of hypermethylated and hypomethylated genes in APA sperm and the hippocampi of offspring. (F) Network plot of common hypermethylated genes enriched by GO analysis
Hypermethylation and downregulation of Nr4a2 in the OF-F1 hippocampus, was modulated by the reader Ythdc1
To determine the possible gene candidates associated with behavioral defects and microglial overactivation, we overlapped three gene sets including hypermethylated genes in OF sperm and OF-F1 hippocampus and the differentially expressed mRNA in OF-F1 hippocampus (Fig. S6). Then, we generated a heatmap of normalized expression to visualize 12 genes with hypermethylated m6A sites and differentially mRNA expression in Fig. 5A. Among them, Nr4a2 was identified as the targeted gene with high abundance and biological significance in OF-F1 hippocampus. Next, we confirmed the relative mRNA expression level (Fig. 5B) and methylation status in the F1 hippocampus (Fig. 5C). Furthermore, we aimed to identify the candidate regulator of Nr4a2 in paternal gametes. Among the key epigenetic modifiers of m6A (writers, erasers and readers) expressed in F0 sperm, Ythdc1 exhibited the highest abundance and the most significant increase in transcription in the OF group (Fig. 5D). We also measured the mRNA expression levels of these regulators in the F1 hippocampus (Fig. 5E), and Ythdc1 had the most significant effect. We examined the protein level of Ythdc1 in both YF and OF sperm and in the offspring’s hippocampus by Western blotting, and the expression of both was greater in the OF group (Fig. 5F). Similarly, the protein level of Nr4a2 decreased in agreement with the decrease in the mRNA level in the hippocampus of OF-F1 (P = 0.0042) (Fig. 5G). We subsequently detected elevated Nr4a2 expression in Ythdc1 pulled-down mRNAs in F1 hippocampus (Fig. 5H). Then, Ythdc1-RIP-seq was performed on F0 sperm to identify Ythdc1-targeted genes (Fig. S7A). The Integrative Genomics Viewer (IGV) tracks clearly depicted the Nr4a2 expression pattern in the YF and OF sperm according to the MeRIP-seq and Ythdc1-RIP-seq data (Fig. 5I). Thus, we hypothesized that Ythdc1 modulates Nr4a2 expression via m6A recognition.
Fig. 5.
Deficient Nr4a2 in OF group was modulated by paternal aging induced of Ythdc1 overexpression. (A) Heatmap of normalized mRNA expression level among overlapped m6A-modified genes in YF-F1 and OF-F1 hippocampus. (B) qPCR results showing the Nr4a2 mRNA expression level in the YF-F1 and OF-F1 hippocampi (replicates of five mice per group); unpaired two-tailed Student’s t test: df = 8, R2 = 0.6024, P = 0.0083. (C) The m6A-targeted Nr4a2 expression level in the F1 hippocampus was analyzed by qPCR (replicates of five mice per group); unpaired two-tailed Student’s t test: df = 8, R2 = 0.9445, P < 0.0001. (D) The mRNA expression levels of key epigenetic modifiers of m6A in F0 sperm. (E) qPCR results showing the m6A modifier mRNA expression levels in the YF-F1 and OF-F1 hippocampi (replicates of five mice per group); Ythdc1: unpaired two-tailed Student’s t test: df = 8, R2 = 0.796, P = 0. 0005. (F) Western blotting of Ythdc1 expression in YF and OF sperm as well as in the YF-F1 and OF-F1 hippocampi (replicates of three mice per group); unpaired two-tailed Student’s t test: sperm, df = 4, R2 = 0.8289, P = 0.0117; hippocampus, df = 4, R2 = 0.9101, P = 0.0031. (G) Western blotting of Nr4a2 expression in the YF-F1 and OF-F1 hippocampi (replicates of three mice per group); unpaired two-tailed Student’s t test: sperm, df = 4, R2 = 0.8546, P = 0.0083. (H) Ythdc1-mediated inhibition of Nr4a2 expression in the F1 hippocampus was analyzed via qPCR (replicates of three mice per group); unpaired two-tailed Student’s t test: df = 4, R2 = 0.9216, P = 0.0024. (I) The IGV plot of Nr4a2 in the MeRIP-seq and Ythdc1-targeted RIP-seq in sperm. #P < 0.05, ##P < 0.01, and ###P < 0.001; ns indicates no significance
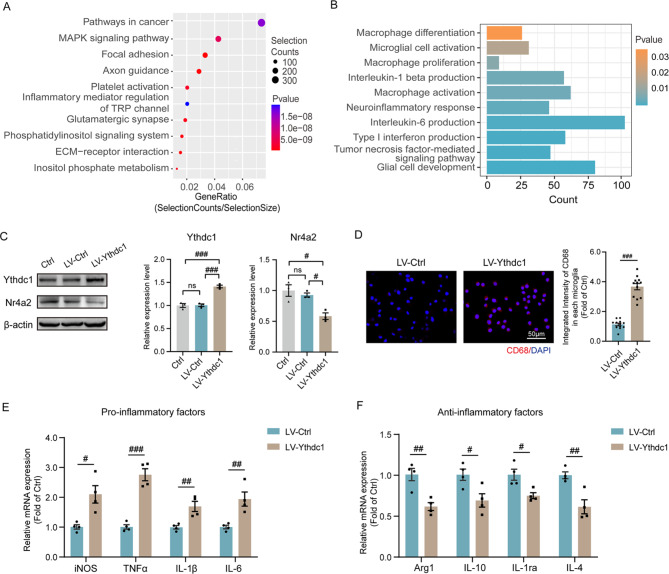
Elevated Ythdc1 contributes to microglial overactivation
When the Ythdc1 antibody pull-down mRNAs were compared between the two groups, KEGG analysis of the upregulated genes revealed enrichment of axon guidance-, glutamatergic synapse- and inflammatory mediator-related pathways in APA sperm (Fig. 6A). Inflammation-associated pathways, including glial cell development, microglial activation, interleukin-1β production, the TNF signaling pathway, type-I IFN production and the neuroinflammatory response, were also upregulated in the APA group (Fig. 6B). The most significantly altered genes identified by GO analysis are also listed in Fig. S7B, C. The hypo-binding genes enriched in the modification of synaptic structure and the hyper-binding genes enriched in synaptic transmission, indicating an association between Ythdc1 and neural transcriptome.
Fig. 6.
Elevation of Ythdc1 was associated with microglial activation. (A) KEGG analysis of genes upregulated in Ythdc1-binding between OF and YF sperm. (B) GO analysis of upregulated genes related to inflammation and microglia between OF and YF sperm. (C) Western blotting showing Ythdc1 and Nr4a2 expression in the Ctrl, LV-Ctrl and LV- Ythdc1 groups; one-way ANOVA with Tukey’s multiple comparison test: Ythdc1, F = 48.56, ηp²=0.9418, P = 0.0002; Nr4a2, F = 11.63, ηp²=0.7949, P = 0.0086. (D) Confocal images of CD68 (red)-stained Ctrl, LV-Ctrl and LV-Ythdc1 cells (three replicates per group). The integrated intensity of CD68 in LV-Ctrl and LV-Ythdc1 groups was examined by immunofluorescence (three cells per image, four replicates per group); unpaired two-tailed Student’s t test: sperm, df = 22, R2 = 0.8506, P < 0.0001. (E) qPCR results of pro-inflammatory mRNA expression level in LV-Ctrl and LV-Ythdc1 (four replicates per group); unpaired two-tailed Student’s t test: iNOS, df = 6, R2 = 0.6863, P = 0.0111; TNFα, df = 6, R2 = 0.9164, P = 0.0002; IL-1β, df = 6, R2 = 0.7193, P = 0.0078; IL-6, df = 6, R2 = 0.710, P = 0.0086. (F) qPCR results of anti-inflammatory mRNA expression level in LV-Ctrl and LV-Ythdc1(four replicates per group); unpaired two-tailed Student’s t test: Arg1: df = 6, R2 = 0.7627, P = 0.0046; IL-10: df = 6, R2 = 0.5904, P = 0.0259; IL-1Ra: df = 6, R2 = 0.6507, P = 0.0155; IL-4: df = 6, R2 = 0.7327, P = 0.0067.#P < 0.05, ##P < 0.01, and ###P < 0.001; ns indicates no significance
To evaluate the influence of Ythdc1 on microglia, we transfected a lentivirus into BV2 cells to overexpress Ythdc1 and evaluated its effect on microglial activation. The LV-Ythdc1 group presented a significant increase in Ythdc1 protein (Fig. 6C). Similarly, the Nr4a2 expression level decreased in the LV-Ythdc1 group. Immunofluorescence analysis revealed that microglia in the LV-Ythdc1 group were activated, as indicated by elevated CD68 expression (Fig. 6D). Then, we used qPCR to measure the mRNA expression levels of cytokines in LV-Ctrl and LV-Ythdc1 groups after transfection. Compared with those in the LV-Ctrl group, we found an increase in the levels of proinflammatory factors (iNOS, TNF-α, IL-1β and IL-6) and a decrease in the levels of anti-inflammatory factors (Arg1, IL-10, IL-1Ra and IL-4) (Fig. 6E, F). Taken together, these findings indicate that the upregulation of Ythdc1 leads to downregulation of Nr4a2 which is associated with microglial overactivation.
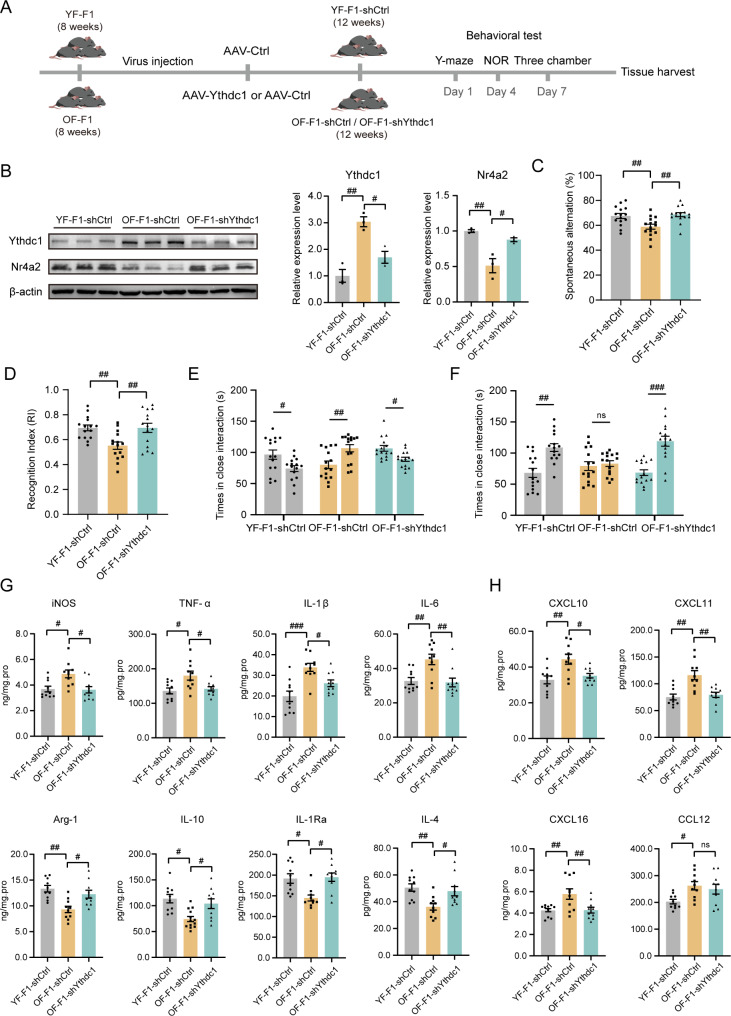
Microglia-specific knockdown of Ythdc1 in OF-F1 mice improved their behavioral performance
Ythdc1 elevation was verified in OF-F1 hippocampus and was associated with an abnormal phenotype and microglial activation in OF offspring. However, the causal relationship between microglia-specific Ythdc1 elevation and abnormal behaviors of offspring of fathers with APA is still unclear. Thus, we performed AAV-Ctrl and AAV-shYthdc1 virus intracranial injection, as illustrated in Fig. 7A, and examined the expression of Ythdc1 4 weeks after injection (Fig. 7B). The specific diagram of injection was displayed in Fig. S8. The OF-F1-shYthdc1 group presented a significant decrease in Ythdc1 protein. Similarly, the Nr4a2 expression level increased in the OF-F1-shYthdc1 group Furthermore, we investigated whether the abnormal phenotype in OF-F1 cells could be alleviated by virus injection. In the Y-maze test, the OF-F1-shYthdc1 mice exhibited greater spontaneous alternations than the OF-F1-shCtrl mice (Fig. 7C). In the NOR test, the OF-F1-shYthdc1 mice spent more time exploring the novel object and had a greater RI than did the OF-F1-shCtrl mice (Fig. 7D). In the three-chamber test, the OF-F1-shYthdc1 mice preferred to interact with stranger 1 and spent more time interacting with stranger 2 in the novelty section than the OF-F1 shCtrl mice (Fig. 7E, F). Therefore, we found that autism-like behavior and cognitive defects could be attenuated by Ythdc1 suppression. To evaluate neuroinflammation after Ythdc1 suppression, we measured cytokine expression in the hippocampus of YF-F1-shCtrl, OF-F1-shCtrl and OF-F1-shYthdc1 mice. We detected a decrease in the levels of proinflammatory factors (iNOS, TNF-α, IL-1β and IL-6) and an increase in the levels of anti-inflammatory factors (Arg1, IL-10, IL-1Ra and IL-4) in the OF-F1-shYthdc1 group compared to the OF-F1-shCtrl group (Fig. 7G). The levels of chemokines associated with microglial activation, including CXCL10, CXCL11, CXCL16 and CCL12, were also decreased in the OF-F1-shYthdc1 group, indicating that neuroinflammation was alleviated (Fig. 7H). In summary, suppressing Ythdc1 expression in the hippocampus of OF offspring reversed behavioral defects and neuroinflammation.
Fig. 7.
Knockdown of Ythdc1 in OF-F1 improved the behavior performance and alleviated the neuroinflammation state. (A) Experimental design for virus injection. (B) Western blotting showing Ythdc1 and Nr4a2 expression in YF-F1, OF-F1-shCtrl and OF-F1-shYthdc1 mice; two-way ANOVA with Tukey’s multiple comparison test: Ythdc1, F (2, 4) = 20.80, ηp²= 0.8829, P = 0.0016; Nr4a2, F (2, 4) = 22.49, ηp²=0.8528, P = 0.0067. (C) The Y-maze test was used to detect spontaneous alternations in YF-F1, OF-F1-shCtrl and OF-F1-shYthdc1; two-way ANOVA with Tukey’s multiple comparison test: F (2, 25) = 8.471, ηp²= 0.1869, P = 0.0016. (D) The novel object recognition test was performed to measure the time spent exploring the new object in YF-F1, OF-F1-shCtrl and OF-F1-shYthdc1 mice; one-sample two-tailed t test for YF-F1: df = 14, R2 = 0.8313, P < 0.0001; two-way ANOVA with Tukey’s multiple comparison test: F (2, 26) = 8.162, ηp²=0.2502, P = 0.0018. (E, F) The three-chamber test included two stages of social activity recorded by the duration of interaction with each cage (YF-F1 vs. OF-F1-shCtrl and OF-F1-shYthdc1); two-way ANOVA with Tukey’s multiple comparison test: Stage1: F (2, 42) = 11.82, ηp²=0.2123, P < 0.0001; Stage2: F (2, 42) = 5.496, ηp²=0.261, P = 0.0076. (G) The differential expression levels of proinflammatory factors (iNOS, TNF-α, IL-1β, and IL-6) and anti-inflammatory factors (Arg1, IL-10, IL-1Ra, and IL-4) between YF-F1, OF-F1-shCtrl and OF-F1-shYthdc1 were determined via ELISA (n = 10/group); two-way ANOVA with Tukey’s multiple comparison test: iNOS, F (2, 18) = 6.193, ηp²=0.3488, P = 0.0090; TNFα, F (2, 18) = 5.740, ηp²=0.3177, P = 0.0118; IL-1β, F (2, 17) = 10.24, ηp²=0.4114, P = 0.0012; IL-6, F (2, 18) = 9.707, ηp²=0.3855, P = 0.0014; Arg1, F (2, 13) = 9.330, ηp²= 0.4165, P = 0.0031; IL-10, F (2, 14) = 5.720, ηp²=0.3035, P = 0.0153; IL-1Ra, F (2, 18) = 6.835, ηp²=0.3789, P = 0.0062; IL-4, F (2, 17) = 6.992, ηp²=0.2700, P = 0.0061. (H) Microglial activation-associated cytokine expression in YF-F1 and OF-F1 hippocampus (n = 10/group); two-way ANOVA with Tukey’s multiple comparison test: CXCL10, F (2, 18) = 7.240, ηp²=0.3604, P = 0.0049; CXCL11, F (2, 18) = 9.890, ηp²= 0.4781, P = 0.0013; CXCL16, F (2, 18) = 7.614, ηp²=0.3301, P = 0.0040; CCL12, F (2, 18) = 4.461, ηp²=0.2503, P = 0.0267. All behavioral tests were performed with 15 mice for each group. #P < 0.05, ##P < 0.01, and ###P < 0.001; ns indicates no significance
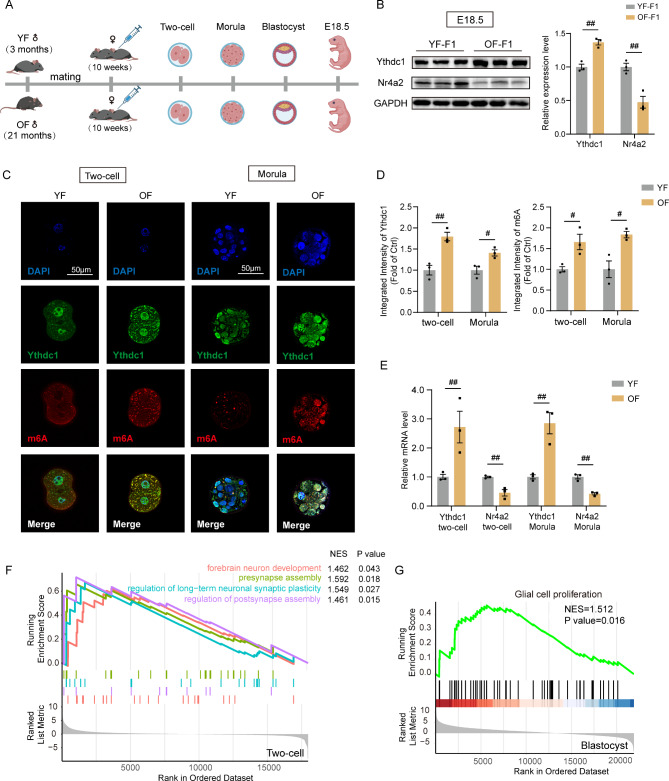
The changes in embryonic and fetal development in the OF group were similar to those in adult offspring
It is widely acknowledged that APA might perturb the epigenetic reprogramming of gametes and cause these changes to be transmitted to offspring. To explore the intergenerational inheritance of paternal gametes, we collected embryos at different stages in vivo and fetal mouse hippocampi at E18.5 from the YF and OF groups (Fig. 8A). We detected increased Ythdc1 protein levels and decreased Nr4a2 levels in the hippocampi of E18.5 fetal mice (Fig. 8B). Early embryos were collected to examine m6A and Ythdc1 expression levels at the two-cell and morula stages (Fig. 8C). Ythdc1 intensity was increased in the OF group (Fig. 8D). The intensity of m6A in the OF group was greater than that in the control group. qPCR further confirmed an increase in Ythdc1 mRNA expression in the two-cell and morula stages and a decrease in the expression of Nr4a2 in the OF group (Fig. 8E). RNA-seq at the two-cell and blastocyst stages was also performed. Pathways associated with embryo brain development were significantly upregulated in the OF two-cell embryo (Fig. 8F) and blastocyst (Fig. S9D) stages. GSEA revealed that the glial cell proliferation pathway was significantly upregulated in OF blastocysts (Fig. 8G). The top 10 up- and downregulated KEGG pathways associated with the two cell stages are depicted in Fig. S9A, C. The volcano plot shows the many DEGs in blastocysts in the two groups (Fig. S9B). The most significant enriched GO terms of blastocyst stage between the two groups in the cellular component (CC), biological process (BP) and molecular function (MF) categories are shown. Terms related to the negative regulation of neurogenesis and multiple synapse-related pathways were significantly enriched in OF blastocysts (Fig. S9E). In conclusion, the modifications in aged sperm were consistent during embryonic and fetal development and were inherited by adult offspring.
Fig. 8.
The common inherited alterations in embryonic and fetal development in the OF group. (A) Illustration of embryo and fetal hippocampus collection. (B) Western blotting of Ythdc1 and Nr4a2 expression in the YF-F1 and OF-F1 hippocampi (replicates of three mice per group); unpaired two-tailed Student’s t test: Ythdc1, df = 4, R2 = 0.9055, P = 0.0035; Nr4a2, df = 4, R2 = 0.8712, P = 0.0073. (C) Confocal images of Ythdc1 (green)- and m6A (red)-stained YF and OF embryos in two-cell and morula states (replicates of three mice per group). (D) Integrated intensity of Ythdc1 and m6A in embryos examined by immunofluorescence (three replicates per group); unpaired two-tailed Student’s t test: Ythdc1: two-cell, df = 4, R2 = 0.8732, P = 0.0063; morula, df = 4, R2 = 0.7447, P = 0.0269; m6A: two-cell, df = 4, R2 = 0.7345, P = 0.0292; morula, df = 4, R2 = 0.7893, P = 0.0180. (E) qPCR results showing Ythdc1 and Nr4a2 expression levels in YF and OF two-cell and morula embryos; unpaired two-tailed Student’s t test: two-cell: Ythdc1: df = 4, R2 = 0.7074, P = 0.0359; Nr4a2: df = 4, R2 = 0.8536, P = 0.0086; morula, Ythdc1: df = 4, R2 = 0.8624, P = 0.0075; Nr4a2: df = 4, R2 = 0.9283, P = 0.0020. (F) GSEA of differentially expressed neuron development and synapse assembly pathways between the OF and YF groups enriched by GO. (G) GSEA of the glial cell proliferation pathway in blastocysts between the OF and YF groups enriched by GO. #P < 0.05, ##P < 0.01, and ###P < 0.001; ns indicates no significance
Discussion
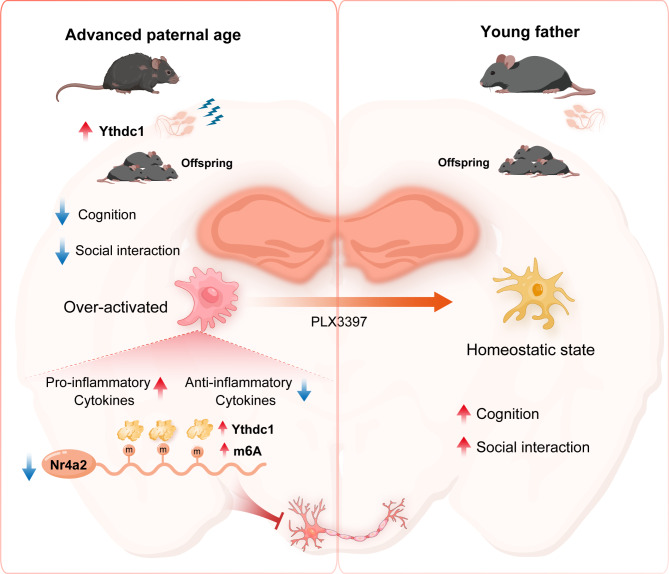
Paternal adverse exposure and unhealthy lifestyles influence the wellbeing of the next generation. Epidemiological evidence has indicated that APA is associated with multiple neurodevelopmental diseases. As a growing number of couples tend to postpone childbearing, the hidden risks and possible preventive measures must be elucidated. However, few studies have evaluated the brain phenotypes of APA offspring and the underlying mechanism of intergenerational inheritance. In this study, we constructed an APA mouse model and detected defective social interaction ability and cognitive function in APA offspring. APA offspring mice exhibited neuroinflammation and microglial overactivation. Microglia ablation attenuated autism-like behavior and cognitive defects in APA offspring. We conducted MeRIP-seq on both paternal sperm and offspring hippocampi and found that the hypermethylated nuclear receptor Nr4a2, along with increased m6A modification and reduced protein expression, is highly important for synaptic plasticity and microglial function. In addition, Ythdc1 was markedly elevated in OF sperm and exhibited a higher binding to Nr4a2 in the OF-F1 hippocampus. Overexpression of Ythdc1 in microglia induced activation and promote inflammation. Knockdown of Ythdc1 in hippocampus of OF-F1 mice attenuated the behavioral defects as well as neuroinflammatory states. After verifying these changes during embryonic and fetal development, we concluded that paternal aging exacerbates offspring neuroinflammation in offspring via m6A modification-mediated intergenerational inheritance.
The offspring of OF are predisposed to an elevated risk of neurodevelopmental abnormalities, with autism-like behaviors being particularly prevalent. Our study introduces, for the first time, cognitive deficits in a mouse model (OF-F1) derived from APA, with significant hippocampal memory impairments observed. Y-maze was used to evaluate working memory and spatial recognition, both of which are associated with hippocampal learning [46]. NOR test measures non-spatial object memory, a function in which hippocampus plays a compelling role [47, 48]. Reduced spontaneous alternations and a preference for familiar objects in OF-F1 indicated hippocampus involvement. RNA sequencing from hippocampal tissues revealed alterations in oxidative phosphorylation and INF production, alongside a neuroinflammatory profile characterized by fluctuating pro- and anti-inflammatory cytokines. The observed increase in INF, decrease in mitochondrial respiratory complex assembly and the shifted metabolism from oxidative phosphorylation to glycolysis suggest microglial activation [49]. Microglia, a resident immune cell type in the CNS, is known to impact neuronal structure and function. Recent studies have highlighted the direct role of microglia in synaptic pruning [50], potentially impairing synaptic plasticity [51] and contributing to memory loss and synapse elimination [43]. In OF-F1 mice, microglial enlargement reflected enhanced phagocytic activity, leading to increased synaptic protein colocalization and reduced synaptic protein expression. Targeted suppression of hyperactive microglia with PLX3397 mitigated learning and memory deficits [29], as well as autism-like behaviors, in these mice. The PLX3397 treatment rescues the levels to those observed in the control group and ensures that the restoration is physiologically relevant. These findings implicate that APA increases risks of cognitive impairment and autism-like behavior in offspring by triggering microglial hyperactivation and neuroinflammation in the hippocampus. However, there are also limitations. We observed that PLX3397 calms overactive microglia and improves mice behavioral performance. Previous studies shows that PLX3397 enhances astrocyte response by increasing inflammatory cytokines release in brain injury models [52]. And microglia-astrocyte interactions are crucial in acute injury recovery [53]. Yet, another study found no significant increase in astrocyte numbers in an Alzheimer’s disease model treated with PLX3397 [54]. As a colony stimulating factor 1 receptor (CSF1R) inhibitor, PLX3397 affects not only microglia [55], but also other cell types, including macrophages and hematopoietic cells. Those are confounding variables involving the interpretation of our findings [56, 57]. Moreover, the increased synapse engulfment impairs synaptic plasticity, underscoring the need for further experiments to fully understand synaptic or electrophysiological changes associated with the conditions studied. Addressing these questions in future research is both challenging and promising.
Then, we explored the link between offspring deficits and paternal signatures. Previous studies highlighted the role of RNA modifications in the intergenerational inheritance through male germline [58]. m6A is considered a novel epigenetic regulator, and its high abundance in mRNAs has attracted increased amounts of attention. A pioneer study using a zebrafish model to elucidate the role of m6A in maternal RNA clearance and zygote activation demonstrated its crucial role in embryo development [45]. Several studies have shown that exposure to harmful substances and conditions during pregnancy can induce alterations in the m6A profile in offspring [59–61]. To date, few studies have focused on m6A modification in aged sperm and its role in intergenerational inheritance. Therefore, we aimed to perform a joint analysis of m6A alterations in fathers and offspring to investigate a new epigenetic mechanism involved in aging-related inheritance. For the first time, we performed MeRIP-seq to map the most striking changes in aging sperm. Our KEGG and GO enrichment results showed that multiple pathways related to synaptic transmission, postsynaptic structure and axodendritic protein transport were hypermethylated. These results indicate that a remarkable number of neurodevelopmental genes in the F1 hippocampus were differentially methylated due to APA.
In terms of the F0 sperm MeRIP-seq results, we found that genes in pathways involved in synapse maturation and interferon production were hypermethylated. Interestingly, a substantial number of differentially methylated genes overlapped between F0 sperm and the F1 hippocampus. GO enrichment of common hypermethylated genes also revealed synapse organization and axonogenesis. Notably, previous studies have demonstrated that neurons and sperm share many biological characteristics, surpassing those of other somatic tissues, including similar exocytotic processes and signaling pathways [62, 63]. Epigenetic modifications in APA sperm are potentially transmitted to the offspring, impacting the brain’s epigenetic landscape. Recently, an article demonstrated that sperm-modified m6A traits caused by environmental exposure could induce neuronal senescence in offspring [64], which added evidence of intergenerational m6A inheritance. By overlapping the common differentially methylated genes in sperm and the brain, we identified a competitive candidate gene called Nr4a2, which is responsible for neuroinflammation and cognitive defects. With our model, we examined the association between a higher m6A methylation rate and decreased mRNA expression in the hippocampi of APA offspring by qPCR and also detected a reduction in protein by western blotting. Traditionally, Nr4a2 has been shown to act as an orphan nuclear receptor critical for dopaminergic neuron survival and maintenance [65]. Nr4a2 deficiency could increase the risk of Parkinson’s disease and cause cognitive impairment in patients with Alzheimer’s disease [66, 67]. Although Nr4a2 is expressed mainly in neuronal cells, approximately 10% of which are glial cells, researchers recently found that Nr4a2 serves as a protective factor in chronic neuroinflammation by reversing the overactivation of microglia to prevent dendritic spine loss [68]. LPS treatment led to deficient Nr4a2 expression, while its overexpression alleviated abnormal behaviors and microglial overactivation, which was strongly related to the pathological traits observed in our model.
Furthermore, we aimed to identify the key regulator modulating the Nr4a2 expression level inherited from APA sperm. m6A modification is regulated by methyltransferases, namely, writers, such as Mettl3, Mettl14 and Wtap; demethylases, namely, erasers, including Fto and Alkbh5. After modification, the protein is recognized by m6A-binding proteins, including Ythdf1, Ythdf2, Ythdf3, Ythdc1 and Ythdc2, which are also called readers [69]. By analyzing the transcriptome data of sperm, we found that Ythdc1 was highly expressed in aged sperm. Ythdc1 is the only nuclear reader protein associated with alternative splicing and mRNA export [70]. Moreover, researchers recently confirmed its diverse roles in regulating chromatin states and regulating gene expression. Its extensive functions in both germ cells and the brain have been proven [71, 72]. We considered Ythdc1 to be a key modulator in aged sperm. We examined Ythdc1 protein levels in both F0 sperm and the F1 hippocampus, and both were increased in the OF group. The Ythdc1 protein also had a greater interaction with Nr4a2 in the OF group, indicating reader-dependent gene silencing [73]. In addition, the upregulated Ythdc1 pulled-down mRNAs were enriched in inflammatory regulation and glutamatergic synapse pathways in aging sperm. GO term analysis revealed upregulated genes associated with glial cell proliferation, macrophage activation and the production of multiple cytokines. The increase in microglial activation and neuroinflammatory responses indicated that Ythdc1 might also directly impact microglial status. Thus, we used lentivirus to overexpress Ythdc1 in microglia and evaluated the activation status and expression of inflammatory factors. Interestingly, compared to those in the LV-Ctrl group, the microglia in the LV-Ythdc1 group were activated, the levels of proinflammatory factors were increased, and the levels of anti-inflammatory factors were decreased. Furthermore, suppression of Ythdc1 significantly alleviated neuroinflammation and abnormal behavior, which strongly supports the function of Ythdc1 in pathological processes. Consistent with these findings, we observed changes in Nr4a2 expression in response to Ythdc1 treatment both in vivo and vitro. Thus, the multifaceted functions of Ythdc1 recognition might ultimately contribute to alterations in mRNA expression and translation. Though we mainly focused on the microglia, we did not conduct a cell-type specific sequencing. Single cell transcriptome sequencing in future studies is warranted to have a comprehensive understanding of the molecular changes in offspring hippocampi.
Intergenerational inheritance involves complicated mechanisms, so testing whether paternal modifications are consistent during embryonic and fetal development is crucial. We collected embryos from various stages and E18.5 fetal hippocampi from the two groups to verify whether the expression patterns of Ythdc1 and Nr4a2 persistently changed during embryo and fetal development. Importantly, these two findings were consistent with the findings in APA sperm and offspring hippocampi. Notably, GSEA of the embryo RNA-seq data also revealed changes in brain development and glial proliferation. Thus, our study explored a novel pathogenic mechanism of neuroinflammation via epigenetic reprogramming in parental gamete. Meanwhile, the expression changes in embryo indicated that observed molecular changes may have a broader impact on offspring triggering the importance of studying other brain regions across different development stages.
Conclusion
Stepwise, we revealed the m6A signature in aging paternal gametes and its inheritance traits during embryonic development, which causes defective neuroinflammation in offspring mice (Fig. 9). Raising the public awareness of hidden epigenetic RNA modifications in aged sperm and the tendency to develop memory and learning defects as well as autism-like behaviors in their offspring is an urgent task. We aimed to decipher gamete and embryo-originated disease and to determine the underlying pathways involved, which might be useful in clinical practice.
Fig. 9.
Graphical abstract of this study
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82088102 to H.F.H, 82171688 and 82192873 to J.X.P), CAMS Innovation Fund for Medical Sciences (2019-I2M-5-064 to H.F.H), Collaborative Innovation Program of Shanghai Municipal Health Commission (2020CXJQ01 to H.F.H.), Shanghai Clinical Research Center for Gynecological Diseases, and Shanghai Urogenital System Diseases Research Center.
Abbreviations
- APA
Advanced paternal age
- m6A
N6-methyladenosine
- DOHaD
Developmental origins of health and disease
- mRNA
Messenger RNA
- NOR
Novel object recognition
- RI
Recognition index
- ELISA
Enzyme-linked immunosorbent assay
- MeRIP-seq
Methylated RNA immunoprecipitation sequencing
- DEGs
Differentially expressed genes
- RIP-seq
RNA immunoprecipitation sequencing
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GSEA
Gene set enrichment analysis
- COCs
Cumulus-oocyte complexes
- hCG
Human chorionic gonadotropin
- YF
Young father
- OF
Old father
Author contributions
YTM and HFH conceptualized the study and ensured the validity of the experiments, YTM, YCM, KXZ, NXQ, JY, YYW, YC, WHS, CLZ and HXC performed the research and analyzed data. YTM and JXP drafted and edited the manuscript. YTM and PJC performed the experiment and analyzed the data during revision. HFH, JXP, XML and JZS supervised this study. All authors read and approved the manuscript prior to submission.
Data availability
The original data was available deposited in the NCBI (National Center for Biotechnology Information) and the BioProject ID was PRJNA1053303.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All procedures for animal experiments were conducted complied with the Guide for the Care and Use of Laboratory Animals at Shanghai Model Organisms (Shanghai, China) (approval ID for the animal IACUC protocol: 2019-0027-01) and were consistent with ARRIVE guidelines.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yiting Mao and Yicong Meng contributed equally to this work.
Contributor Information
Xinmei Liu, Email: liuxinmei@fudan.edu.cn.
Jiexue Pan, Email: panjiexue@fudan.edu.cn.
Hefeng Huang, Email: huanghefg@fudan.edu.cn.
References
- 1.Fleming TP, Watkins AJ, Velazquez MA, Mathers JC, Prentice AM, Stephenson J, et al. Origins of lifetime health around the time of conception: causes and consequences. Lancet (London England). 2018;391(10132):1842–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van den Bergh BRH, van den Heuvel MI, Lahti M, Braeken M, de Rooij SR, Entringer S, et al. Prenatal developmental origins of behavior and mental health: the influence of maternal stress in pregnancy. Neurosci Biobehav Rev. 2020;117:26–64. [DOI] [PubMed] [Google Scholar]
- 3.Matteoli M, Pozzi D, Fossati M, Menna E. Immune synaptopathies: how maternal immune activation impacts synaptic function during development. EMBO J. 2023;42(13):e113796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Marroun H, Schmidt MN, Franken IH, Jaddoe VW, Hofman A, van der Lugt A, et al. Prenatal tobacco exposure and brain morphology: a prospective study in young children. Neuropsychopharmacology: Official Publication Am Coll Neuropsychopharmacol. 2014;39(4):792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortés-Albornoz MC, García-Guáqueta DP, Velez-van-Meerbeke A, Talero-Gutiérrez C. Maternal Nutrition and Neurodevelopment: a scoping review. Nutrients. 2021;13(10). [DOI] [PMC free article] [PubMed]
- 6.Ceccanti M, Coccurello R, Carito V, Ciafrè S, Ferraguti G, Giacovazzo G, et al. Paternal alcohol exposure in mice alters brain NGF and BDNF and increases ethanol-elicited preference in male offspring. Addict Biol. 2016;21(4):776–87. [DOI] [PubMed] [Google Scholar]
- 7.Toussaint AB, Ellis AS, Bongiovanni AR, Peterson DR, Bavley CC, Karbalaei R et al. Paternal morphine exposure enhances morphine self-administration and induces region-specific neural adaptations in reward-related brain regions of male offspring. bioRxiv: the preprint server for biology. 2023.
- 8.Eyolfson E, Bhatt D, Wang M, Lohman AW, Mychasiuk R. Paternal exposure to exercise and/or caffeine and alcohol modify offspring behavioral and pathophysiological recovery from repetitive mild traumatic brain injury in adolescence. Genes, brain, and behavior. 2021:egbb12736. [DOI] [PubMed]
- 9.Lawson G, Fletcher R. Delayed fatherhood. J Family Plann Reproductive Health care. 2014;40(4):283–8. [DOI] [PubMed] [Google Scholar]
- 10.Frederiksen LE, Ernst A, Brix N, Braskhøj Lauridsen LL, Roos L, Ramlau-Hansen CH, et al. Risk Adverse Pregnancy Outcomes Adv Maternal Age. 2018;131(3):457–63. [DOI] [PubMed] [Google Scholar]
- 11.Wu S, Wu F, Ding Y, Hou J, Bi J, Zhang Z. Advanced parental age and autism risk in children: a systematic review and meta-analysis. Acta Psychiatrica Scandinavica. 2017;135(1):29–41. [DOI] [PubMed] [Google Scholar]
- 12.Fico G, Oliva V, De Prisco M, Giménez-Palomo A, Sagué-Vilavella M, Gomes-da-Costa S, et al. The U-shaped relationship between parental age and the risk of bipolar disorder in the offspring: a systematic review and meta-analysis. Eur Neuropsychopharmacology: J Eur Coll Neuropsychopharmacol. 2022;60:55–75. [DOI] [PubMed] [Google Scholar]
- 13.Saha S, Barnett AG, Foldi C, Burne TH, Eyles DW, Buka SL, et al. Advanced paternal age is associated with impaired neurocognitive outcomes during infancy and childhood. PLoS Med. 2009;6(3):e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Onofrio BM, Rickert ME, Frans E, Kuja-Halkola R, Almqvist C, Sjölander A, et al. Paternal age at childbearing and offspring psychiatric and academic morbidity. JAMA Psychiatry. 2014;71(4):432–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang SH, Hsiao PC, Yeh LL, Liu CM, Liu CC, Hwang TJ, et al. Advanced Paternal Age and early onset of Schizophrenia in sporadic cases: not confounded by parental polygenic risk for Schizophrenia. Biol Psychiatry. 2019;86(1):56–64. [DOI] [PubMed] [Google Scholar]
- 16.Lisman J, Buzsáki G, Eichenbaum H, Nadel L, Ranganath C, Redish AD. Viewpoints: how the hippocampus contributes to memory, navigation and cognition. Nat Neurosci. 2017;20(11):1434–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashapkin V, Suvorov A, Pilsner JR, Krawetz SA, Sergeyev O. Age-associated epigenetic changes in mammalian sperm: implications for offspring health and development. Hum Reprod Update. 2023;29(1):24–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denomme MM, Haywood ME, Parks JC, Schoolcraft WB, Katz-Jaffe MG. The inherited methylome landscape is directly altered with paternal aging and associated with offspring neurodevelopmental disorders. Aging Cell. 2020;19(8):e13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang K, Yao L, Wang S, Zheng L, Qian Z, Ge Y, et al. miR-125a-5p increases cellular DNA damage of aging males and perturbs stage-specific embryo development via Rbm38-p53 signaling. Aging Cell. 2021;20(12):e13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, Bai D, Liu W, Liu Y, Zhang Y, Kou X, et al. Altered sperm tsRNAs in aged male contribute to anxiety-like behavior in offspring. Aging Cell. 2021;20(9):e13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Chen ZP, Hu H, Lei J, Zhou Z, Yao B et al. Sperm microRNAs confer depression susceptibility to offspring. Sci Adv. 2021;7(7). [DOI] [PMC free article] [PubMed]
- 22.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Volume 351. New York, NY: Science; 2016. pp. 397–400. 6271. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, et al. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020;48(11):6251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei J, Yu X, Yang L, Liu X, Gao B, Huang B, et al. FTO mediates LINE1 m(6)a demethylation and chromatin regulation in mESCs and mouse development. Sci (New York NY). 2022;376(6596):968–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Xu X, Qi M, Chen C, Li M, Yan R, et al. N(6)-methyladenosine regulates maternal RNA maintenance in oocytes and timely RNA decay during mouse maternal-to-zygotic transition. Nat Cell Biol. 2022;24(6):917–27. [DOI] [PubMed] [Google Scholar]
- 26.Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q, et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 2017;27(10):1216–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafik AM, Zhang F, Guo Z, Dai Q, Pajdzik K, Li Y, et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021;22(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng J, Han L, Zheng N, Wang T, Xu H, Jiang Y, et al. Microglial Tmem59 Deficiency impairs phagocytosis of synapse and leads to Autism-Like behaviors in mice. J Neuroscience: Official J Soc Neurosci. 2022;42(25):4958–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinto B, Morelli G, Rastogi M, Savardi A, Fumagalli A, Petretto A, et al. Rescuing over-activated Microglia restores cognitive performance in Juvenile animals of the Dp(16) mouse model of Down Syndrome. Neuron. 2020;108(5):887–e90412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada J, Jinno S. Novel objective classification of reactive microglia following hypoglossal axotomy using hierarchical cluster analysis. J Comp Neurol. 2013;521(5):1184–201. [DOI] [PubMed] [Google Scholar]
- 31.Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171–81. [DOI] [PubMed] [Google Scholar]
- 32.Kechin A, Boyarskikh U, Kel A, Filipenko M, cutPrimers:. A New Tool for Accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biology: J Comput Mol cell Biology. 2017;24(11):1138–43. [DOI] [PubMed] [Google Scholar]
- 33.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE. 2013;8(6):e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaidanovich-Beilin O, Lipina T, Vukobradovic I, Roder J, Woodgett JR. Assessment of social interaction behaviors. J Visualized Experiments: JoVE. 2011(48). [DOI] [PMC free article] [PubMed]
- 37.Antunes M, Biala G. The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn Process. 2012;13(2):93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prieur EAK, Jadavji NM. Assessing spatial Working Memory using the spontaneous alternation Y-maze test in aged male mice. Bio-protocol. 2019;9(3):e3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Xia X, Wang Y, Zheng JC. Mitochondrial dysfunction in microglia: a novel perspective for pathogenesis of Alzheimer’s disease. J Neuroinflamm. 2022;19(1):248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bsibsi M, Peferoen LA, Holtman IR, Nacken PJ, Gerritsen WH, Witte ME, et al. Demyelination during multiple sclerosis is associated with combined activation of microglia/macrophages by IFN-γ and alpha B-crystallin. Acta Neuropathol. 2014;128(2):215–29. [DOI] [PubMed] [Google Scholar]
- 41.Lepore F, D’Alessandro G, Antonangeli F, Santoro A, Esposito V, Limatola C, et al. CXCL16/CXCR6 Axis Drives Microglia/Macrophages Phenotype in physiological conditions and plays a crucial role in Glioma. Front Immunol. 2018;9:2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo T, Liu C, Yang C, Wu J, Su P, Chen J. Immunoproteasome subunit PSMB8 regulates microglia-mediated neuroinflammation upon manganese exposure by PERK signaling. Food Chem Toxicology: Int J Published Br Industrial Biol Res Association. 2022;163:112951. [DOI] [PubMed] [Google Scholar]
- 43.Wang C, Yue H, Hu Z, Shen Y, Ma J, Li J, et al. Microglia mediate forgetting via complement-dependent synaptic elimination. Sci (New York NY). 2020;367(6478):688–94. [DOI] [PubMed] [Google Scholar]
- 44.Elmore MR, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82(2):380–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, et al. M(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542(7642):475–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian J, Geng F, Gao F, Chen YH, Liu JH, Wu JL, et al. Down-regulation of Neuregulin1/ErbB4 signaling in the Hippocampus is critical for learning and memory. Mol Neurobiol. 2017;54(6):3976–87. [DOI] [PubMed] [Google Scholar]
- 47.Cohen SJ, Munchow AH, Rios LM, Zhang G, Asgeirsdóttir HN, Stackman RW. Jr. The rodent hippocampus is essential for nonspatial object memory. Curr Biology: CB. 2013;23(17):1685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen SJ, Stackman RW. Jr. Assessing rodent hippocampal involvement in the novel object recognition task. A review. Behav Brain Res. 2015;285:105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu Y, Mai W, Chen L, Cao K, Zhang B, Zhang Z, et al. mTOR-mediated metabolic reprogramming shapes distinct microglia functions in response to lipopolysaccharide and ATP. Glia. 2020;68(5):1031–45. [DOI] [PubMed] [Google Scholar]
- 50.Hong S, Dissing-Olesen L, Stevens B. New insights on the role of microglia in synaptic pruning in health and disease. Curr Opin Neurobiol. 2016;36:128–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salter MW, Stevens B. Microglia emerge as central players in brain disease. Nat Med. 2017;23(9):1018–27. [DOI] [PubMed] [Google Scholar]
- 52.Jin WN, Shi SX, Li Z, Li M, Wood K, Gonzales RJ, et al. Depletion of microglia exacerbates postischemic inflammation and brain injury. J Cereb Blood flow Metabolism: Official J Int Soc Cereb Blood Flow Metabolism. 2017;37(6):2224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng J, Lu J, Mei S, Wu H, Sun Z, Fang Y, et al. Ceria nanoparticles ameliorate white matter injury after intracerebral hemorrhage: microglia-astrocyte involvement in remyelination. J Neuroinflamm. 2021;18(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett RE, Bryant A, Hu M, Robbins AB, Hopp SC, Hyman BT. Partial reduction of microglia does not affect tau pathology in aged mice. J Neuroinflamm. 2018;15(1):311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patwardhan PP, Surriga O, Beckman MJ, de Stanchina E, Dematteo RP, Tap WD, et al. Sustained inhibition of receptor tyrosine kinases and macrophage depletion by PLX3397 and rapamycin as a potential new approach for the treatment of MPNSTs. Clin cancer Research: Official J Am Association Cancer Res. 2014;20(12):3146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Wernersbach I, Strehle J, Li S, Appel D, Klein M, et al. Early posttraumatic CSF1R inhibition via PLX3397 leads to time- and sex-dependent effects on inflammation and neuronal maintenance after traumatic brain injury in mice. Brain Behav Immun. 2022;106:49–66. [DOI] [PubMed] [Google Scholar]
- 57.Shaikh SN, Willis EF, Dierich M, Xu Y, Stuart SJS, Gobe GC, et al. CSF-1R inhibitor PLX3397 attenuates peripheral and brain chronic GVHD and improves functional outcomes in mice. J Neuroinflamm. 2023;20(1):300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu D, Zhang K, Guan K, Khan FA, Pandupuspitasari NS, Negara W et al. Future in the past: paternal reprogramming of offspring phenotype and the epigenetic mechanisms. Arch Toxicol. 2024. [DOI] [PubMed]
- 59.Zhang S, Meng P, Cheng S, Jiang X, Zhang J, Qin X, et al. Pregnancy exposure to carbon black nanoparticles induced neurobehavioral deficits that are associated with altered m(6)a modification in offspring. Neurotoxicology. 2020;81:40–50. [DOI] [PubMed] [Google Scholar]
- 60.Kaspi A, Khurana I, Ziemann M, Connor T, Spolding B, Zimmet P, et al. Diet during pregnancy is implicated in the regulation of hypothalamic RNA methylation and risk of obesity in offspring. Mol Nutr Food Res. 2018;62(14):e1800134. [DOI] [PubMed] [Google Scholar]
- 61.Fang J, Wu X, He J, Zhang H, Chen X, Zhang H, et al. RBM15 suppresses hepatic insulin sensitivity of offspring of gestational diabetes mellitus mice via m6A-mediated regulation of CLDN4. Molecular medicine (Cambridge. Mass). 2023;29(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim JM, Kim HG, Son CG. Tissue-specific profiling of oxidative stress-Associated Transcriptome in a healthy mouse model. Int J Mol Sci. 2018;19(10). [DOI] [PMC free article] [PubMed]
- 63.Matos B, Publicover SJ, Castro LFC, Esteves PJ, Fardilha M. Brain and testis: more alike than previously thought? Open Biology. 2021;11(6):200322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, Xiong YW, Tan LL, Zheng XM, Zhang YF, Ling Q, et al. Sperm rhoa m6A modification mediates intergenerational transmission of paternally acquired hippocampal neuronal senescence and cognitive deficits after combined exposure to environmental cadmium and high-fat diet in mice. J Hazard Mater. 2023;458:131891. [DOI] [PubMed] [Google Scholar]
- 65.Jakaria M, Haque ME, Cho DY, Azam S, Kim IS, Choi DK. Molecular insights into NR4A2(Nurr1): an emerging target for neuroprotective therapy against neuroinflammation and neuronal cell death. Mol Neurobiol. 2019;56(8):5799–814. [DOI] [PubMed] [Google Scholar]
- 66.Moon M, Jung ES, Jeon SG, Cha MY, Jang Y, Kim W, et al. Nurr1 (NR4A2) regulates Alzheimer’s disease-related pathogenesis and cognitive function in the 5XFAD mouse model. Aging Cell. 2019;18(1):e12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al-Nusaif M, Yang Y, Li S, Cheng C, Le W. The role of NURR1 in metabolic abnormalities of Parkinson’s disease. Mol Neurodegeneration. 2022;17(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He Y, Wang Y, Yu H, Tian Y, Chen X, Chen C, et al. Protective effect of Nr4a2 (Nurr1) against LPS-induced depressive-like behaviors via regulating activity of microglia and CamkII neurons in anterior cingulate cortex. Pharmacol Res. 2023;191:106717. [DOI] [PubMed] [Google Scholar]
- 69.Chen J, Fang Y, Xu Y, Sun H. Role of m6A modification in female infertility and reproductive system diseases. Int J Biol Sci. 2022;18(9):3592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Widagdo J, Anggono V, Wong JJ. The multifaceted effects of YTHDC1-mediated nuclear m(6)a recognition. Trends Genet. 2022;38(4):325–32. [DOI] [PubMed] [Google Scholar]
- 71.Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14(5):e1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mathoux J, Henshall DC, Brennan GP. Regulatory mechanisms of the RNA modification m(6)A and significance in brain function in Health and Disease. Front Cell Neurosci. 2021;15:671932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, et al. N6-methyladenosine of chromosome-associated Regul RNA Regulates Chromatin State Transcription. 2020;367(6477):580–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olarerin-George AO, Jaffrey SR. MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinf (Oxford England). 2017;33(10):1563–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data was available deposited in the NCBI (National Center for Biotechnology Information) and the BioProject ID was PRJNA1053303.