Abstract
Neonates face unique challenges in the period following birth. The postnatal immune system is in the early stages of development and has a range of functional capabilities that are distinct from the mature adult immune system. Bidirectional immune–microbial interactions regulate the development of mucosal immunity and alter the composition of the microbiota, which contributes to overall host well-being. In the past few years, nutrition has been highlighted as a third element in this interaction that governs host health by modulating microbial composition and the function of the immune system. Dietary changes and imbalances can disturb the immune–microbiota homeostasis, which might alter susceptibility to several autoimmune and metabolic diseases. Major changes in cultural traditions, socioeconomic status and agriculture are affecting the nutritional status of humans worldwide, which is altering core intestinal microbial communities. This phenomenon is especially relevant to the neonatal and paediatric populations, in which the microbiota and immune system are extremely sensitive to dietary influences. In this Review, we discuss the current state of knowledge regarding early-life nutrition, its effects on the microbiota and the consequences of diet-induced perturbation of the structure of the microbial community on mucosal immunity and disease susceptibility.
Introduction
The ‘hygiene hypothesis’ proposed that an increased predisposition to allergies and the rise in the incidence of atopic diseases was linked to a lack of exposure to infectious agents, microorganisms and parasites during childhood that resulted in the development of the immune system being suppressed.1,2 In the past few years, epidemiological studies further showed that children growing up on traditional farms with exposure to livestock and consumption of unprocessed cow’s milk during their early years are resistant to these diseases.3 That errors in the development of the immune system are connected to improved sanitary conditions and the increased use of antibiotics, among other factors, is now evident. The gut microbiota is central to this phenomenon as it responds to changes in the environment and also affects the maturation and function of the immune system. Fluctuations in the composition of this microbiota are also caused by perturbations in diet.4,5 This observation has led to the proposal of the ‘diet hypothesis’ that unifies changes in nutrition with gut microbiota and immune health (Figure 1).6–8
Figure 1 |.
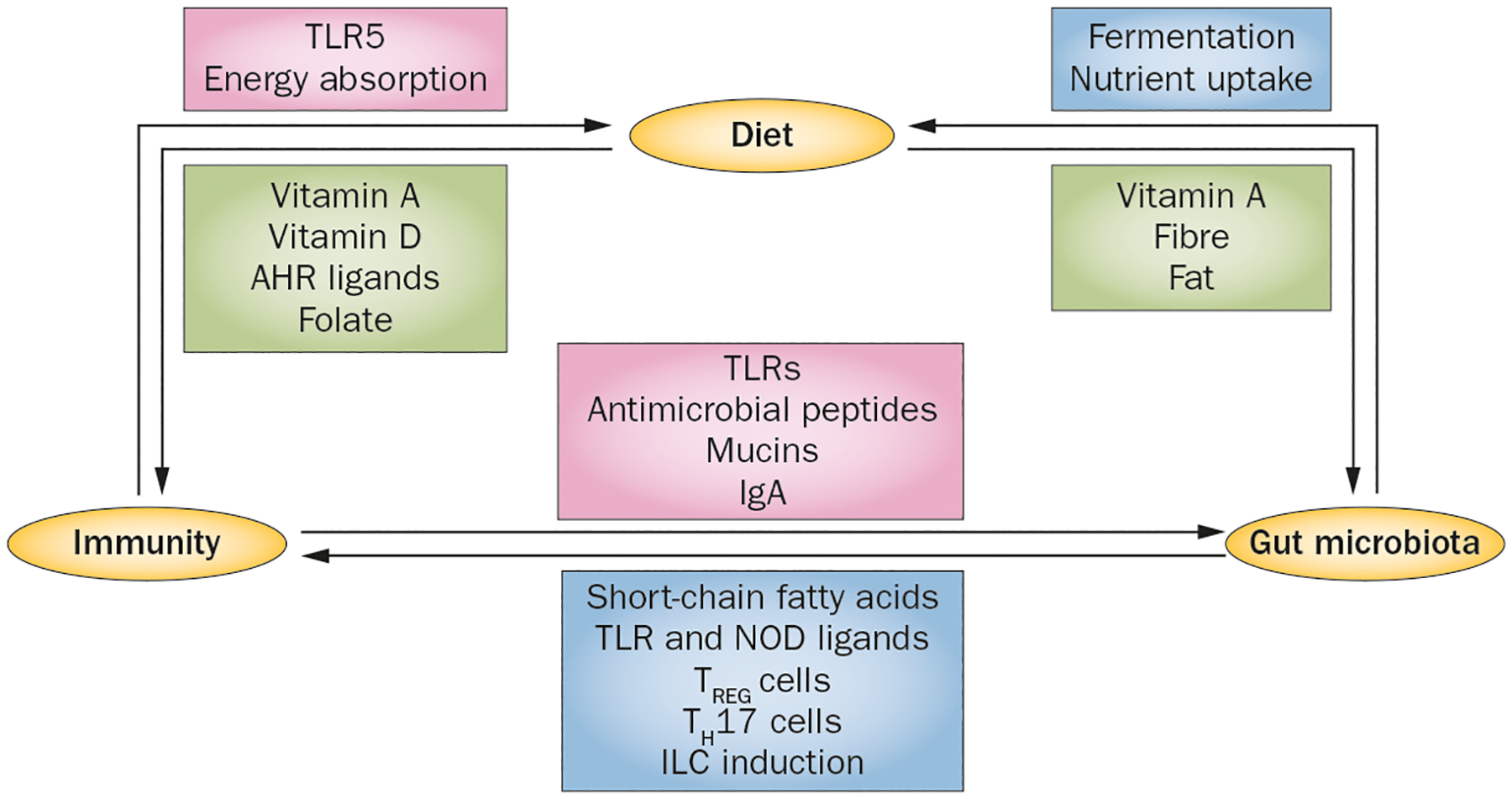
The ‘diet hypothesis’. Diet, gut microbiota and host immunity are intimately connected and their bidirectional communication is central to maintaining intestinal and metabolic homeostasis. The commensal bacteria determine the nutritional value of food by fermenting dietary components to usable energy sources and by affecting nutrient uptake. Specific bacteria and microbial by-products influence the development and function of key components of mucosal immunity. The mucosal immune system shapes the commensal composition and location. Immune–microbial interactions via pattern-recognition receptors (TLRs, NODs) result in secretion of antimicrobial peptides, mucins and IgA, which maintains intestinal homeostasis and barrier function. The mucosal innate immune system also influences dietary energy absorption. Finally, perturbation of microbial community structure leads to dysbiosis, which can precipitate immune-mediated disorders such as IBD and metabolic diseases such as type 2 diabetes. Abbreviations: ILC, innate lymphoid cell; NOD, nucleotide oligomerization domain; TH17 cell, type 17 T helper cell; TLR, Toll-like receptor; TREG cell, regulatory T cell.
Cultural diversity and geographical location contribute to dietary differences that result in distinct patterns of intestinal microbial colonization and disease susceptibility in different populations. The Western diet is generally low in fibre and high in processed foods, which adversely affects the intestinal microbial composition and leads to an obesity-prone metagenome.6,8,9 Conversely, the Japanese diet, which includes rice, beans and fermented foods,10,11 and the diet of Eskimos in Greenland (which is typically high in fish and omega-3 fatty acids) promote resistance to chronic inflammatory diseases and heart diseases.12–14 In mouse experiments, offspring of mice fed a diet rich in omega-3 fatty acids have an altered gut microbiome and have enhanced production of the anti-inflammatory cytokine IL-10 in the colon and spleen, which protects the mice from an allergic challenge.15 The influence of nutrition on the microbiome and disease susceptibility is also specific to age. In newborn babies, the establishment and type of feeding has a considerable effect on the composition of the microbial community.16 In adults, both long-term dietary intake and short-term changes in macronutrients (for example, an animal or plant-product-based diet) influences microbial community structure and microbial gene expression profiles.4 The outcomes of the complex dynamic connections between the microbiota and the immune system are most important during the postnatal period and have consequences on host immunity and on metabolic homeostasis that reach well into adulthood.
In this Review, we discuss the effect of diet on host–microbial interactions in early life and highlight the key aspects of nutritional programming during the postnatal period in influencing the lifelong function of the immune system in health and disease.
Nutritional programming
Nutritional deficiencies are a major cause of death in children in developing countries. Globally, undernutrition contributes to ~45% of the deaths of the ~6.6 million children <5 years old who die each year.17,18 The effects of childhood malnutrition are long-term and can even affect subsequent generations.19 Nutrition is an important driver of the convergence in gut microbial composition across mammalian phylogeny and within humans.20,21 The diet shapes the composition of the gut microbiota and the expression of microbial genes. In turn, the structure and activity of the gut microbiota determines the nutritional value the host will extract from the food they consume. Faecal microbiota profiles in adults with dietary habits that are specific to their culture, region and socioeconomic status are fairly stable over time.21 By contrast, the composition of the microbiota undergoes considerable changes during the period from birth to weaning.16 Microarray studies based on small subunit ribosomal DNA have identified intrapersonal and interpersonal variation in the structure of faecal bacterial communities during the first year of life.22 The sterile fetal intestine receives its first inoculum in the hours after birth and the colonization events that follow shape the structure of the microbial community and influence health outcomes as the child grows up (Figure 2).16
Figure 2 |.
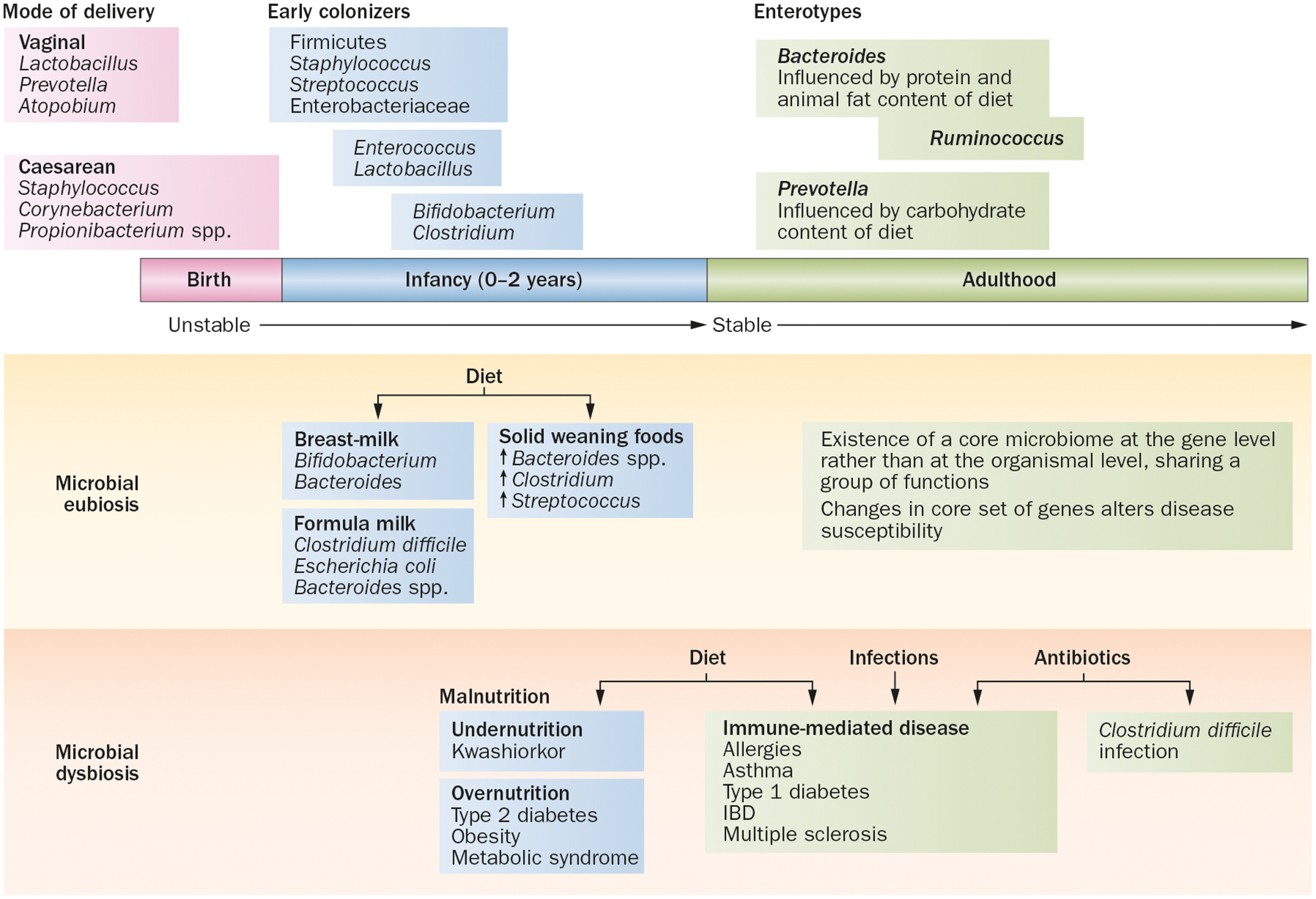
Diet, gut microbiota and dysbiosis. Several features regulate the establishment and composition of the microbiota and their effect on the health and immune function of the host. Eubiosis or a normal microflora structure that protects against infections educates the immune system and contributes to nutrient digestion. Energy harvest is established by early intestinal colonization with specific microbes immediately after birth. An ordered process of subsequent colonization and expansion shaped by diet results in the establishment of distinct ‘enterotypes’, or clusters of microbial communities, that remains fairly stable in adults. Perturbations in the microbial community structure or dysbiosis are induced by factors such as diet, use of antibiotics or infection, which can alter susceptibility to several diseases.
Early intestinal colonization
Infant nutrition, either breast-milk or formula milk, has a defining role in establishing early colonization patterns of the gut microbiota. In breast-fed infants, the gut microbiota is dominated by species from the Bifidobacterium family, whereas formula-fed babies harbour a more diverse microbiome containing Escherichia coli, Clostridium difficile, Bacteroides spp. and Lactobacillus spp.23 In general, a step-wise progression towards adult-like enterotypes has been documented, with initial colonization primarily by Firmicutes and aerobic and facultative anaerobic bacteria such as Staphylococcus, Streptococcus and Enterobacteriaceae.24,25 These species are followed by colonization by Enterococcus and Lactobacillus-like species, which generate an anaerobic intestinal environment that provides favourable conditions for subsequent anaerobic colonizers.26 After 1 week of life, Bifidobacterium, Bacteroides and Clostridium are detected in the faeces; the dominance of Bifidobacterium species in breast-fed infants is usually accompanied by a decrease in the amount of Enterobacteriaceae.16,27
Breast-milk contains functional nutrients such as carbohydrates, nondigestible carbohydrates, fatty acids and lactoferrin that provide the microenvironment for gut protection and maturation.28,29 The oligosaccharides found in human breast-milk are nondigestible carbohydrates that are fermented in the colon and stimulate the growth of specific bacteria, including Bifidobacterium and Bacteroides, that contribute to the development of the immune system.27,30 Compared with that of adults, the infant metabolome is rich in acetate but deficient in propionate and butyrate,31 which correlates with the established microbial ecosystem as Bifidobacterium spp. and lactic acid bacteria do not produce butyrate. The infant intestinal environment is adapted to this butyrate deficit and enterocytes probably utilize alternative energy sources (these sources have yet to be identified).32 In addition to encoding genes whose products are involved in carbohydrate metabolism, bifidobacteria also express genes involved in folate biosynthesis,33 which contribute to the unique nutritional requirement for the development of infants’ intestine and immune system. Human milk is also a source of live bacteria such as staphylococci, streptococci, bifidobacteria and lactic acid bacteria.34–36
The introduction of solid foods containing insoluble nondigestible carbohydrates, and weaning from breast-milk or formula milk at age 4–6 months introduces another shift in the composition of the intestinal microbiome with increased counts of Bacteroides, Clostridium and anaerobic streptococci.37–39 This stage in the development of infants also marks the point when adult-like microbes that express genes involved in the degradation of xenobiotic compounds and vitamin biosynthesis are detectable. The gut microbiota begins to reach adult-like composition by the time an infant is 1 year old,40 and is stable and firmly established by 2–3 years of age.16,41 However, in a paper published in 2012, it was suggested that the population of adult-type anaerobes might reach densities similar to those seen in adults within 1 week of birth and that the switch from facultative to strict anaerobes in healthy breast-fed neonates might occur earlier than previously assumed.42
Bacterial enterotypes
The adult human gut microbiota can be clustered into three enterotypes of co-occurring microbial species defined by the levels of bacterial genera—Bacteroides, Prevotella and Ruminococcus.43 The separation between the Ruminococcus cluster and the Bacteroides-led enterotype does not seem to be very discrete; however, the two groups remain distinct from the Prevotella-driven group.44 These enterotypes are strongly associated with long-term diets; high protein and animal fat content of the diet increases levels of Bacteroides whereas a high carbohydrate content leads to dominance of Prevotella.9,44 Metagenomic studies comparing the gut microbiomes of members of agrarian Malawian41 and Bangladeshi45 societies to the gut microbiomes of residents of the USA also found a similar inverse relationship between Bacteroides and Prevotella genera, underscoring a link between diet and structure of the gut microbiota. Whether these enterotypes can be used to predict disease incidence and outcome and whether long-term dietary interventions can stably switch individuals to a disease-resistant microbiome will be important to determine. Furthermore, the origin of these enterotypes has not yet been characterized. A longitudinal study of the faecal microbial composition of Danish infants found that the establishment of the enterotype occurred between 9 and 36 months of age and correlated with the time of cessation of breast-feeding.46 Patterns of initial intestinal colonization that are influenced by early childhood nutrition might be involved in the establishment of adult enterotypes.
Dysbiosis and disease
The composition of the microbial community is dynamic in early life but can also shift in adulthood under the influence of diet, antibiotic use or infection with invasive pathogens (Figure 2). These changes in the gut microbiota, referred to as dysbiosis, can disturb the homeostasis of microorganisms in the intestine and favour the outgrowth of potentially pathogenic constituents. Ultimately, dysbiosis can perturb the regulatory circuits of the immune system that restrain intestinal inflammation, leading to immune-mediated diseases directed against antigens of the gut microbiota.47 As discussed earlier, the postnatal microbiota is not stable and is profoundly influenced by diet. However, the role of the evolving and fluctuating neonatal microbiota in influencing the development and function of the immune system is still unclear. Furthermore, the degree of involvement of the neonatal immune system in shaping the composition of the gut microbiota is also unknown.
Basis for and effect of dysbiosis
Dysbiosis is characterized by a reduction in taxonomic diversity and species representation of the gut microbiota,48 and can be accompanied by a decrease in bacterial biomass, depending on the nature of the infection and inflammatory insult.49,50 Pathogenic bacteria exploit conditions of dysbiosis, including decreased and altered metabolic capacity of the intestine,51 and gain access to intestinal nutrients and niches, displacing commensal bacteria and leaving the host increasingly susceptible to both pathogens and pathobionts. The developing microbiota in early postnatal life is fragile and extremely susceptible to external influences that affect overall host health. For instance, a correlation exists between the use of antibiotics during infancy and childhood and increased incidences of asthma, atopic dermatitis, multiple sclerosis and IBD.52
The effects of dysbiosis on host physiology have been studied extensively in gnotobiotic mice (Box 1).53,54 Faecal microbial communities and the abundance of bacterial genes in experimentally colonized germ-free mice are highly sensitive to different foods.5,55 Furthermore, dysbiosis induced by antibiotics in newborn mice leads to alterations in the development of the immune system that are similar to those observed in germ-free mice,56–58 with reduced numbers of intestinal regulatory T cells (TREG cells),59 increased colonic infiltration of invariant natural killer T cells60,61 and increased susceptibility to allergen-induced airway hyper-reactivity and colitis.60 Thus, host–microbe interactions at the postnatal stage are critical for the establishment of protective immunity. Disruption of this process by changes in diet, the use of antibiotics or by infectious inflammation might alter the susceptibility to immune-mediated diseases in adulthood.
Box 1 |. Gnotobiotic mice for investigating host–microbiota relationships.
Germ-free animals are reared in a sterile environment (special positive-pressure isolators that maintain a ‘germ-free’ environment) and can be used to establish simplified microbial ecosystems to study the effect of eubiosis and dysbiosis on host immune function and physiology. The absence of a microbiota affects several developmental and physiological aspects in these mice as outlined below.53
Developmental defects
Intestine
IECs with altered patterns of microvilli formation and decreased rates of turnover
Reduced capillary network formation
Fewer and less cellular Peyer’s patches
Thinner and less cellular lamina propria
Fewer plasma cells in germinal centres
Smaller and less cellular isolated lymphoid follicles Secondary lymphoid organs
Spleen and lymph nodes lack structure and have poorly formed B-zones and T-zones
Abnormal high-endothelial venule morphology
Mesenteric lymph nodes with smaller germinal centres that are less cellular with fewer plasma cells
Immune defects
Increased susceptibility to infection by certain bacteria, viruses and parasites (Shigella flexneri, Listeria monocytogenes, Salmonella enterica subsp. enterica serova Typhimurium)
Display atopy with a characteristic TH2-cell gene signature, airway eosinophilia and increased IgE production upon antigen challenge
Fewer CD8+ intraepithelial lymphocytes
Fewer CD4+ lamina propria lymphocytes; decreased levels of TH17 cells in small intestines
Fewer FOXP3+ T regulatory cells in mesenteric lymph nodes with decreased suppressive capacity
Reduced IgA production by B cells
Reduced expression of REG3γ and angiogenin-4 (both antimicrobial peptides) by Paneth cells
Altered gene expression profiles of IECs
Reduced expression of major histocompatibility complex class II, Toll-like receptor 9 and IL-25 by IECs
Metabolic defects
Decreased levels of ATP in intestines
Altered glycosylation pattern of lumenally exposed surface proteins on IECs
Reduced energy absorption capacity
Abbreviations: FOXP3, forkhead box P3; IEC, intestinal epithelial cell; REG3, regenerating islet-derived protein 3; TH, T helper.
Diet, dysbiosis and disease
Malnutrition (which includes overnutrition and undernutrition) is a global health problem and a major contributor to childhood morbidity and mortality. Food insecurity has been linked to this epidemic,62 but it is becoming increasingly evident that the gut microbiota is closely connected to the problem of malnutrition. In developing countries, inadequate nutrition and micronutrient deficiencies delay the growth of children and result in stunting that affects a staggering 27% of children <5 years old.63 Interestingly, discordance for moderate to severe malnutrition between twins (monozygotic and dizygotic) within the same household and fed similar diets is only ~40%, which suggests that additional factors such as individual gut community configurations might influence the severity of undernutrition.64 By contrast, in developed countries, modern societal practices such as improved hygiene, reduced family size, an increased incidence of Caesarean deliveries, reduced rates of breast-feeding and widespread antibiotic use in children affects the transmission and maintenance of the indigenous gut microbiota.65 The consequent effect on the development and function of the immune system has contributed to the increase in chronic inflammatory and autoimmune disorders.66 Thus, in addition to a genetic predisposition and biological mechanisms, a ‘shift’ in the resident microbiota from a ‘healthy’ to a ‘diseased’ state underlies disease development and progression.
In a study published in 2013, the gut microbiota was implicated in the propagation of kwashiorkor, a form of severe acute malnutrition.64 Longitudinal studies in Malawian twin pairs showed that the overall gene content of the faecal microbiota in children with kwashiorkor did not develop with increasing age.64 In addition, the disease phenotype, including weight loss, was transmissible to germ-free mice that received faecal transplants from sick children but not from healthy ones.64 Importantly, treatment with ready-to-use therapeutic food did not change the microbial community profiles of affected children in the long term, supporting the notion that multiple determinants shape the fitness landscape of the gut; this landscape can be used to predict disease severity and progression.
Not surprisingly, the microbiota and excessive caloric intake have also been linked to obesity, the metabolic syndrome and type 2 diabetes. For example, the intestinal microbial profile of obese mice is considerably different to that of normal-weight mice with over-representation of Firmicutes instead of Bacteroidetes that results in increased harvesting of energy from the diet of the host.67 This disease trait is transmissible, as colonization of germ-free mice with an ‘obese microbiota’ results in increased body fat compared with colonization with ‘lean microbiota’.67 In addition, germ-free mice are resistant to diet-induced obesity, but show increased adiposity when colonized with gut flora from leptin-deficient ob/ob mice.68 Similarly, studies of twin cohorts in the USA revealed a distinct bacterial phylogenetic composition and representation of microbial genes involved in nutrient metabolism in lean versus obese twin pairs.69 Interestingly, the immune system might itself regulate obesity as mice deficient in Toll-like receptor (TLR) 5 have an increased incidence of the metabolic syndrome, which has been linked to their unique gut microbiota and outgrowth of E. coli.70,71 Other studies have, however, challenged the magnitude of the effect TLR signalling has on changes to the microbial composition72 and have raised awareness of effects of animal husbandry conditions and lineage or legacy effects in microbiome studies in rodents.73
Substantial changes to the commensal gut microbiota have been observed in patients with type 2 diabetes. These patients had decreased levels of bacteria from the phylum Firmicutes and strains of Clostridia that produce short-chain fatty acids as well as increased levels of the Bacteroidetes and Prevotella groups that correlated with high plasma concentrations of glucose.74–76 In addition, a low-grade systemic and chronic inflammation that results from reduced transepithelial integrity and increased metabolic endotoxaemia that is linked to altered gut microbial profiles has been associated with the development of type 2 diabetes.77 Increased serum levels of lipopolysaccharide and translocation of commensal E. coli have also been observed in mouse models of type 2 diabetes induced by a high-fat diet.78,79
Changes in diet and the composition of the microbiota also contribute to susceptibility to autoimmune diseases such as type 1 diabetes.66,80 For example, low gut microbial diversity has been linked with the development of type 1 diabetes in children.81–83 The presence of segmented filamentous bacteria has a protective effect on the incidence of type 1 diabetes in mice in disease-susceptible female nonobese diabetic mice.84 A meta-analysis of 43 studies found that breast-feeding might offer some limited protection against the development of type 1 diabetes,85 whereas exposure to cow’s milk protein through infant formula might increase the risk of developing β-cell autoimmunity.86 These are as yet unsettled issues, as other studies found no effects of breast-feeding on the susceptibility to developing type 1 diabetes.80 After weaning, the introduction of solid foods, including excessive intake of cow’s milk and consumption of gluten-containing cereals has also been associated with the development of type 1 diabetes.87,88
Thus, dietary patterns and microbiota shape host physiology and effect disease susceptibility at mucosal barrier and systemic sites. The components of the host barrier and immune system that are affected by diet and the microbiota are discussed in the next section.
Mucosal barrier function
The intestinal tract is the largest barrier in the human body. It is made up of a single layer of epithelial cells (intestinal epithelial cells [IECs]) that confines the gut microbiota and other potentially harmful substances to the lumen whilst regulating the flow of solutes, nutrients and ions into the underlying mucosa.89–91 Highly specialized barrier defences have evolved that include multiple layers of resistance, incorporating a stratified mucous layer, a fairly impenetrable but responsive epithelium and a lamina propria that contains innate and adaptive immune cells that maintain intestinal homeostasis (Figure 3). In the event of a barrier breach, all these components are poised to mount antimicrobial clearance responses and to rapidly reseal and repair tissue after injury to restore barrier function. The development and maturation of the intestinal mucosa and its gut-associated lymphoid tissue, including Peyer’s patches, isolated lymphoid follicles and mesenteric lymph nodes, is initiated by and dependent upon intestinal microbial colonization.90
Figure 3 |.
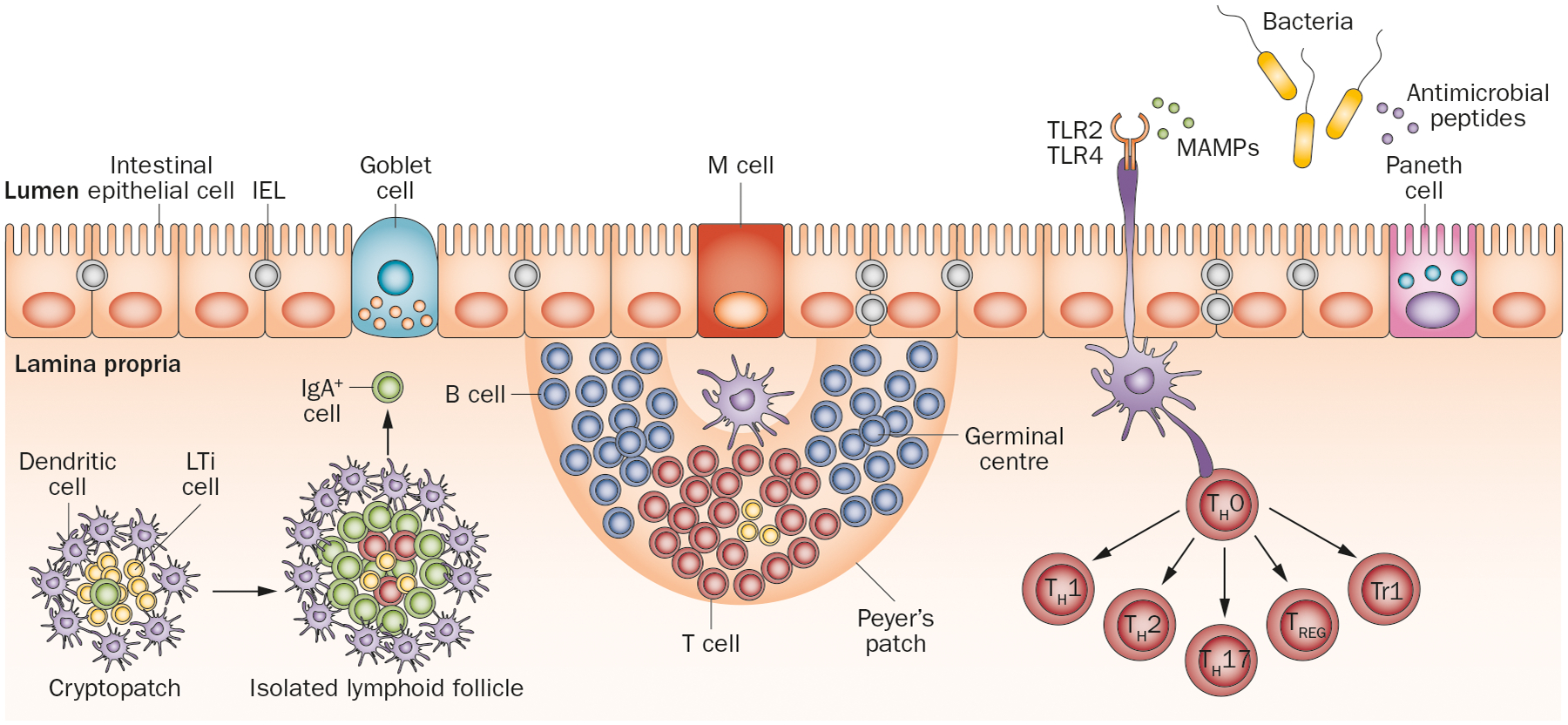
The intestinal barrier. The intestinal barrier is made up of a single layer of epithelium consisting of intestinal epithelial cells and specialized Goblet cells, M cells and Paneth cells (present only in small intestine). Peyer’s patches (found specifically in the ileum) and mesenteric lymph nodes develop prenatally when LTi are recruited to sites of the developing intestines called cryptopatches. Cryptopatches mature into isolated lymphoid follicles when pattern-recognition receptors (TLRs) are triggered by MAMPs, which then release IgA-producing plasma cells into the lamina propria. Dendritic cells in the Peyer’s patches access microbes through M cells or directly from the lumen by extending dendrites through intestinal epithelial cells. These antigen-loaded dendritic cells can induce T-cell differentiation or T-cell-dependent B-cell maturation into germinal centres. Naive T cells (TH0 cells) can differentiate into effector TH1, TH2 or TH17 cells or into regulatory FOXP3+ TREG cells or Tr1 cells. Microbial sensing by intestinal epithelial cells also marks the release of antimicrobial peptides and stimulation of intestinal epithelial cell proliferation in crypts. Abbreviations: IEL, intraepithelial lymphocyte; LTi, lymphoid tissue inducer cell; MAMP, microbe-associated molecular pattern; M cell, microfold cell; TH, T helper cell; TLR, Toll-like receptor; Tr1, type 1 regulatory T cell; TREG, regulatory T cell.
The intestinal epithelium contains five main types of cells, each of which is responsive to and conditioned by the gut microbiota (Box 2). The intestinal epithelium of a newborn baby is immature and therefore ingested foreign particles and bacteria readily gain access to the lymphatic system and bloodstream during this period.92 Maturational changes are rapidly triggered by microbial colonization,93 as well as by dietary substances such as breast-milk or formula milk,94 which prepares the infant intestine to accommodate dense bacterial colonization (Figure 4). This process might take several weeks, and is usually prolonged in infants born prematurely.95 In premature neonates, immaturity of the intestinal barrier can result in devastating diseases such as necrotizing enterocolitis,96 toxigenic diarrhoea and intestinal allergies.97
Box 2 |. Composition of the intestinal epithelium.
Enterocytes: Absorptive cells that secrete hydrolases and absorb nutrients, ions and fluids
Goblet cells: Produce mucin that makes up the mucous layer
Paneth cells: Found primarily in small intestines and secrete antimicrobial peptides such as cryptidins, defensins and enzymes such as lysozyme
Microfold (M) cells: Specialized cells found in epithelium of Peyer’s patches that transport organisms and particles from the gut lumen to immune cells deep in the epithelial barrier
Enteroendocrine cells: Secrete hormones such as serotonin, substance P and secretin
Figure 4 |.
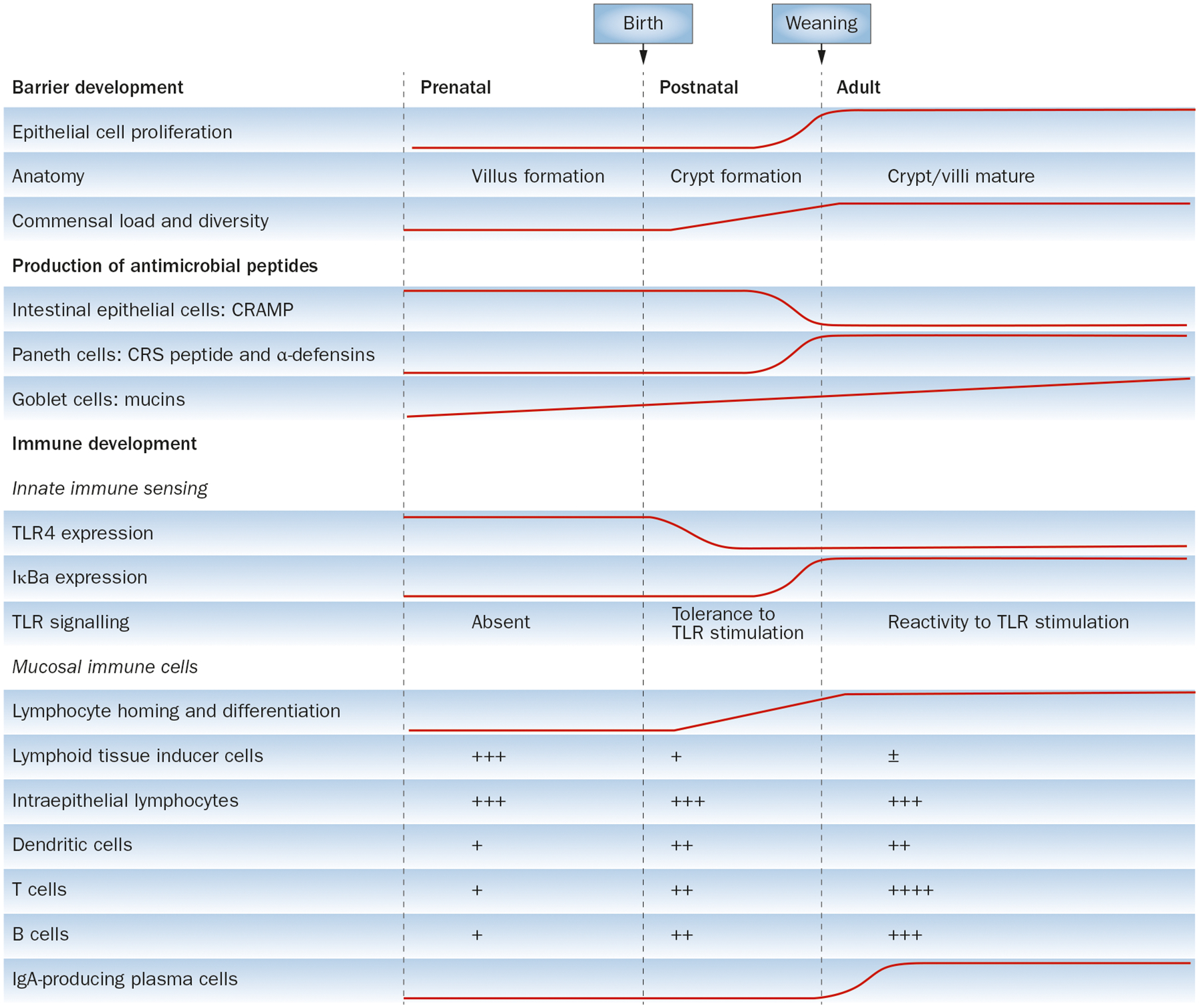
Development and maturation of the intestinal mucosal barrier and mucosal immune system. Developmental changes in the prenatal, postnatal and adult intestine are summarized. Intestinal microbial colonization, as well as dietary components, induces maturation of the intestinal epithelium and initiates development of the mucosal immune system. Complex bidirectional interactions between gut microbiota, diet and the immune system itself regulate the establishment and maintenance of intestinal homeostasis and barrier function. Abbreviations: CRAMP, cathelin-related antimicrobial peptide; CRS, cryptidin-related sequence; TLR, Toll-like receptor; ±, cells might or might not be present; +, ++, +++, ++++, relative numbers of indicated cells.
Toll-like receptors and barrier function
The IECs express a wide array of receptors such as TLRs and C-type lectin receptors that reside in the endosome or on the cell surface, and cytosolic nucleotide oligomerization domain-like receptors that sense microbe-associated molecular patterns (MAMPs) expressed by commensal and pathogenic microbiota.98 Recognition of MAMPs by neonatal IECs is necessary for stimulating the development of isolated lymphoid follicles and the formation of lymphoid structures that are capable of supporting maturation of B cells and production of sIgA.99–103 A singular example of the sensitivity and responsiveness of the infant gut to microbial colonization is regulation of the expression of TLR4 (the receptor for the Gram-negative bacterial product lipopolysaccharide). In mouse IECs, expression of TLR4 is high before intestinal colonization at birth.104 Within hours of exposure to the gut microbiota, the expression of TLR4 in IECs is rapidly downregulated with a concomitant attenuation of components of the TLR4 signalling pathway such as inhibition of IRAK-1 translation by the microRNA miR-146a.105 This desensitization of a microbial sensor is an adaptation to mounting microbial loads, and failure to temper TLR4 signals has been linked to the development of necrotizing enterocolitis, particularly in preterm infants.106
Neonatal intestinal maturation and barrier function is modified by breast-feeding. Intestinal immunomodulation that is mediated by breast-milk might result in subclinical infections that gradually stimulate immunological memory to pathogens while reducing inflammation.107 Breast-milk modulates the expression of TLRs, as well as the signalling pathways they are involved in. For example, soluble TLR2 that is found in human milk can competitively inhibit signalling through membrane TLR2 and helps restrict innate immune stimulation in the neonatal gut.108 Furthermore, reduced TLR2 activity at birth could facilitate the normal establishment of Bifidobacterium in the intestine.108 Immune cells such as macrophages that are present in colostrum and mature breast-milk persist in the neonatal intestine and also translocate to systemic sites where they form a first line of defence against invading pathogens.109 Intestinal growth, maturation and function are also promoted by various maternal factors such as epidermal growth factor and transforming growth factor β, which are found at high levels in the colostrum and mature breast-milk.110
Tight junctions
The barrier function of the intestinal epithelium is maintained by the apical junction complex, which is composed of tight junctions and adherence junctions.111 Adherence junctions and desmosomes provide the strong adhesive bonds between IECs and participate in intercellular communication. Tight junctions, which encircle the apical ends of the lateral membranes of IECs, mediate the selective permeability and paracellular transport function of the epithelial barrier. The permeability of tight junctions is regulated by several factors, including local cytokine milieu,112 age,113 the composition of the commensal bacteria and infection.114 Treatment of epithelial cell monolayers with cytokines such as IFN-γ and TNF increases the permeability of the tight junctions, which can be reversed by commensal bacteria such as Bacteroides thetaiotaomicron, as well as by probiotic Streptococcus thermophilus and Lactobacillus acidophilus.115 The mechanisms by which commensal microbiota protect barrier function include modulation of the expression of the occludin and caludin proteins and utilization of epithelial cell signalling machinery such as Rho GTPases, PKC and MAPK pathways to enhance barrier integrity.114
Enteric pathogens such as Vibrio cholerae116 and enteropathogenic E. coli117 disrupt the tight junction barrier, which leads to abnormal electrolyte and fluid transport and tissue inflammation. Impaired function of the tight junctions has been observed in several autoimmune diseases, including coeliac disease and type 1 diabetes.118 In preterm infants, failure of tight junctions and barrier function has been implicated in the development of necrotizing enterocolitis.119 Finally, tight junctions are also influenced by dietary components, but more work needs to be done to firmly establish the cause–effect relationship.114 Glutamines, polyphenols and probiotics enhance and protect tight junction barrier integrity, whereas alcohol and its metabolites impair tight junction barrier function.114
Immune system development
The mucosal tissues of the gastrointestinal tract harbour more cells from the immune system than all the secondary lymphatic tissues combined. Over the past few years, it has become apparent that the complex microbiota (including commensals, symbiotic bacteria and pathogens) influences immune reactivity in the intestine as well as at extraintestinal sites such as the pancreas84 and the brain.120,121 The infant mucosal immune system matures over several months after birth and the process is closely tied with the development and establishment of the gut microbiota and the quality of dietary nutrients and commensal-derived metabolites (Figure 3).122,123
Organized lymphoid tissues
The development of the mucosal immune system begins at the fetal stage. At this stage in mice, the actions of a subset of innate lymphoid cells (ILCs) called lymphoid tissue inducer cells initiates prenatal organization of secondary lymphoid tissues including Peyer’s patches and mesenteric lymph nodes.101,124 Additional tertiary lymphoid tissue structures such as the isolated lymphoid follicles, cryptopatches in the small intestine and colonic patches in the large intestine develop after birth under the influence of lymphoid tissue inducer cells.124 Cryptopatches consist of a cluster of lymphoid tissue inducer cells that express the transcription factor nuclear receptor ROR-γt, are surrounded by dendritic cells that express CD11c (also known as integrin-αX) and are positioned below the single layer of intestinal epithelium. Colonization by gut microbiota is required before cryptopatches develop into isolated lymphoid follicles.90 Aryl hydrocarbon receptor ligands (which are derived from cruciferous vegetables such as broccoli) provide important signals for the development and maturation of the immune system.125,126 The aryl hydrocarbon receptor is expressed in ILCs positive for nuclear receptor ROR-γt and has an essential role in their function and maintenance.126–128 Ahr-deficient mice lack postnatally imprinted cryptopatches and isolated lymphoid follicles but not Peyer’s patches, as these develop in the embryo.127 Furthermore, these mice are highly susceptible to infection with the intestinal pathogen Citrobacter rodentium, which emphasizes the role of dietary components in controlling mucosal immunity.
Myeloid cells
Myeloid cells, such as dendritic cells positive for integrin-αE, dendritic cells negative for integrin-αE and positive for CX3CR1 as well as macrophages, are strategically positioned below the intestinal epithelium and act as cellular mediators that translate microbial signals into intestinal homeostasis. Dendritic cells loaded with microbial antigens interact with lymphocytes to induce T-cell differentiation and T-cell-dependent maturation of B cells in the germinal centres.90 CX3CR1+ dendritic cells make direct contact with luminal antigens by extending processes through the tight junction of IECs in a TLR-dependent manner (Figure 3).129 This process is important for tolerance to orally ingested antigens.130 Microbiota-induced production of IL-23 by dendritic cells also recruits ILCs to the sites of mucosal immune responses during infections.131
In newborn mice, delayed developmental maturation of dendritic cells and a reduced number of antigen-presenting cells results in compromised immunity in the first week of life.132 However, the function and homeostasis of myeloid cells in the infant intestine is less clear. An important role for dietary micronutrients in regulating mucosal myeloid cell homeostasis is emerging. In mice, migratory dendritic cells that are positive for integrin-αE have high basal expression of Aldh1a2.133 This gene encodes the enzyme retinal dehydrogenase 2 that synthesizes retinoic acid (a vitamin A metabolite), which imprints gut homing potential on T cells and B cells by inducing expression of migration receptors α4β7 and CCR9.134,135 Thus, these myeloid cells are central to regulation of mucosal immunity mediated by vitamin A. Vitamin A deficiency is a common micronutrient deficiency in children136 and its effect on intestinal myeloid cell homeostasis and function in the young remains to be determined.
Lymphoid cells
Several subsets of lymphocytes reside throughout the intestinal epithelium (intraepithelial lymphocytes [IELs]) and the lamina propria (lamina propria lymphocytes). IELs are heterogeneous populations of antigen-experienced T cells that express either the αβ or γδ T-cell receptor (TCR).137 As IELs have direct contact with enterocytes and close proximity to antigens in the gut lumen, they are geared to provide immediate and heightened immune protection to halt the initial entry and spread of pathogens. At the same time, IELs are under tight regulatory control, as an overzealous inflammatory response could jeopardize barrier integrity. As with other cell types in the gut, the development and function of IELs are also influenced by dietary components such as aryl hydrocarbon receptor ligands125 and vitamin D.138 Mice deficient in vitamin D receptor have reduced numbers of CD8αα+ IELs, which was associated with low levels of IL-10 in the small intestine and increased inflammation in wild-type animals.139
Lamina propria lymphocytes are enriched in differentiated TCRαβ+ CD4+ T cells, including the proinflammatory type 17 T helper (TH17) cell subset and suppressor TREG cells. Segmented filamentous bacteria are key to the differentiation and accumulation of TH17 cells in the terminal ileum of mice,140,141 where they are critical for host defence against infections with extracellular bacteria and fungi.142 TH17 cells have also been associated with numerous autoimmune and chronic inflammatory diseases. For example, increased numbers of TH17 cells have been found in patients with IBD143,144 and polymorphisms in the IL23R gene (which is important for the maintenance and pathogenicity of TH17 cells) are linked to susceptibility to IBD.145 In addition, numerous studies in mouse models of IBD have demonstrated the involvement of TH17 cells in the pathogenesis of IBD.146,147 As a result of the tremendous pathogenic potential of TH17 cells, several mechanisms have evolved to restrain and limit their function and prevent chronic inflammatory episodes. TREG cells expressing the transcription factor FOXP3 and type 1 TREG cells that produce IL-10 are key to this activity and suppress intestinal inflammation.148,149 Genome-wide association studies have linked IL10 polymorphisms (IL-10 produced by TREG cells is critical for suppressive effects by TREG cells150) to susceptibility to IBD.151 In addition, mutations in IL10, IL10RA or IL10RB as well as IL-10R signalling components STAT3, TYK2 and JAK2 lead to the development of early-onset IBD.152,153
Vitamin A is a fat-soluble essential micronutrient whose metabolite, retinoic acid, has several crucial roles in intestinal immunity, including regulating the balance between intestinal TH17 and TREG cells. Retinoic acid promotes the extrathymic generation of TREG cells and induction of mucosal and oral tolerance.154,155 Interestingly, mice fed a diet deficient in vitamin A also have diminished numbers of TH17 cells in the small intestine at steady state, and differentiation of TH17 cells in the presence of low physiological concentrations of retinoic acid can promote their trafficking to the gut.156,157 Products of bacterial fermentation such as short-chain fatty acids regulate the balance of inflammatory and regulatory responses in the colon of mice.158–160 A reduced intake of plant dietary fibres and use of antibiotics negatively affect levels of short-chain fatty acids, which alters intestinal immune regulation by reducing the number of colonic TREG cells.161
ILCs include a variety of specialized cells that secrete a suite of effector cytokines and chemokines to combat infection and repair tissue at mucosal barriers.162 The ILC subsets have considerable developmental and functional plasticity, which might be instructed by the needs of their local microenvironment. Furthermore, ILCs are also influenced by dietary factors and act as sensors of dietary stress. Vitamin A deficiency negatively affects the development of group 3 ILCs, which results in compromised immunity to acute bacterial infection.163 However, this phenomenon was compensated for by the dramatic expansion of group 2 ILCs (which produce IL-13), resulting in resistance to nematode infection in mice.163 In addition, maternal levels of dietary retinoids control the size of secondary lymphoid organs and the efficiency of immune responses in their adult offspring by regulating the differentiation of group 3 ILCs (lymphoid tissue inducer cells) in utero.164
Conclusions
The relationship between diet, microbiota and host immunity is being rapidly unravelled using a combination of epidemiological, immunological, metagenomic and metabolomic approaches. These studies are most pertinent at the postnatal period when dietary intake is closely tied to the development of both the gut microbiota and the immune system. In a study published in 2014, a prenatal placental microbiome was described that could be a source of the infants’ first bacterial inoculum via intrauterine seeding.165 Whether and how this low abundance yet metabolically rich microbiome directs the development of the immune system and the microbial community structure during gestation, as well as the effect of maternal nutrition on these processes, remains to be determined. A systems approach involving both animal studies and analysis of human cohorts are needed to unravel the complexities of microbiota–host crosstalk in early life. Animal models are invaluable and have provided a plethora of information and insights into the interplay between the immune system and host microbiota. However, it is important to caution that a direct correlation from animal studies to humans is not possible, particularly when interrogating immune developmental events in early life. For instance, in humans, αβ TCR+ T cells are seen in peripheral tissue at 10–12 gestational weeks; however, in mice, peripheral T cells are undetected in the fetus and their numbers only increase after birth,166 which is suggestive of distinct developmental cues and immune requirements in human versus mouse neonates.
The interdependence of diet, immune and microbiota interactions and communications between the elements of this triad dictate intestinal mucosal homeostasis as well as metabolic well-being. The mechanisms by which these dialogues occur are only now being elaborated on and major gaps remain in our understanding of how specific nutrients and microbial metabolites regulate microbial composition, host metabolism and immunity. The use of specific dietary components in modulating the gut microbiota and subsequent immune function offers an attractive approach to deliver health benefits to a vulnerable population, such as paediatric and geriatric populations. As such, probiotics and prebiotics are being increasingly used to prevent and treat a variety of gastrointestinal and systemic diseases in infants.167,168 Discoveries aimed at establishing specific features of the immune–microbiota crosstalk will provide useful insights for the development of preventive and therapeutic agents of multiple infectious, autoimmune and metabolic disorders.
Key points.
Infant nutrition, including breast-milk, formula milk and solid weaning foods, is a key determinant of early microbial community structure that influences development of protective immunity and seems to affect health throughout life
Diet-induced dysbiosis changes the species composition of the gut microbiota and leads to immune-mediated inflammatory and metabolic diseases
Diet influences the postnatal development of innate and adaptive defences at the mucosal barrier surface and affects intestinal barrier function
A triad of diet, the microbiota and the immune system regulates postnatal intestinal homeostasis and host physiology, which has consequences through to adulthood
Acknowledgements
N.J. would like to acknowledge the support of the Charles King Trust/Charles Hood Foundation Postdoctoral Fellowship Award. W.A.W. would like to acknowledge the support of grants P30 DK040561, P01 DK33506, R01 HD012447 and R01 HD059126.
Footnotes
Competing interests
The authors declare no competing interests.
Contributor Information
Nitya Jain, Center for Computational and Integrative Biology, Massachusetts General Hospital, 185 Cambridge Street, Boston, MA 02114, USA..
W. Allan Walker, Mucosal Immunology and Biology Research Centre, Massachusetts General Hospital, Building 114, 16th Street, Charlestown, MA 02129-4404, USA..
References
- 1.Strachan DP Hay fever, hygiene, and household size. BMJ 299, 1259–1260 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bach JF The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med 347, 911–920 (2002). [DOI] [PubMed] [Google Scholar]
- 3.von Mutius E & Vercelli D Farm living: effects on childhood asthma and allergy. Nat. Rev. Immunol 10, 861–868 (2010). [DOI] [PubMed] [Google Scholar]
- 4.David LA et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med 1, 6ra14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maslowski KM & Mackay CR Diet, gut microbiota and immune responses. Nat. Immunol 12, 5–9 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Noverr MC & Huffnagle GB Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 12, 562–568 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Devereux G The increase in the prevalence of asthma and allergy: food for thought. Nat. Rev. Immunol 6, 869–874 (2006). [DOI] [PubMed] [Google Scholar]
- 9.De Filippo C et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Acad. Sci. USA 107, 14691–14696 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shoda R, Matsueda K, Yamato S & Umeda N Epidemiologic analysis of Crohn disease in Japan: increased dietary intake of n-6 polyunsaturated fatty acids and animal protein relates to the increased incidence of Crohn disease in Japan. Am. J. Clin. Nutr 63, 741–745 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Iso H Lifestyle and cardiovascular disease in Japan. J. Atheroscler. Thromb 18, 83–88 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Bang HO, Dyerberg J & Nielsen AB Plasma lipid and lipoprotein pattern in Greenlandic West-coast Eskimos. Lancet 1, 1143–1145 (1971). [DOI] [PubMed] [Google Scholar]
- 13.Harris WS et al. Omega-6 fatty acids and risk for cardiovascular disease: a science advisory from the American Heart Association Nutrition Subcommittee of the Council on Nutrition, Physical Activity, and Metabolism; Council on Cardiovascular Nursing; and Council on Epidemiology and Prevention. Circulation 119, 902–907 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Jump DB, Depner CM & Tripathy S Omega-3 fatty acid supplementation and cardiovascular disease. J. Lipid Res 53, 2525–2545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myles IA, Pincus NB, Fontecilla NM & Datta SK Effects of parental omega-3 fatty acid intake on offspring microbiome and immunity. PLoS ONE 9, e87181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholtens PA, Oozeer R, Martin R, Amor KB & Knol J The early settlers: intestinal microbiology in early life. Annu. Rev. Food Sci. Technol 3, 425–447 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Bhutta ZA & Black RE Global maternal, newborn, and child health—so near and yet so far. N. Engl. J. Med 369, 2226–2235 (2013). [DOI] [PubMed] [Google Scholar]
- 18.UN Inter-agency Group for Child Mortality Estimation (IGME). Levels and trends in child mortality [online], http://www.unicef.org/media/files/2013_IGME_child_mortality_Report.pdf (2013).
- 19.Barker DJ Adult consequences of fetal growth restriction. Clin. Obstet. Gynecol 49, 270–283 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Muegge BD et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kau AL, Ahern PP, Griffin NW, Goodman AL & Gordon JI Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer C, Bik EM, DiGiulio DB, Relman DA & Brown PO Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Power SE, O’Toole PW, Stanton C, Ross RP & Fitzgerald GF Intestinal microbiota, diet and health. Br. J. Nutr 111, 387–402 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Fanaro S, Chierici R, Guerrini P & Vigi V Intestinal microflora in early infancy: composition and development. Acta Paediatr. Suppl 91, 48–55 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Mackie RI, Sghir A & Gaskins HR Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr 69, 1035S–1045S (1999). [DOI] [PubMed] [Google Scholar]
- 26.Orrhage K & Nord CE Factors controlling the bacterial colonization of the intestine in breastfed infants. Acta Paediatr. Suppl 88, 47–57 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Sela DA & Mills DA Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18, 298–307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldman AS & Smith CW Host resistance factors in human milk. J. Pediatr 82, 1082–1090 (1973). [DOI] [PubMed] [Google Scholar]
- 29.Walker A Breast milk as the gold standard for protective nutrients. J. Pediatr 156, S3–S7 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Marcobal A et al. Bacteroides in the infant gut consume milk oligosaccharides via mucusutilization pathways. Cell Host Microbe 10, 507–514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knol J et al. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J. Pediatr. Gastroenterol. Nutr 40, 36–42 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Parrett AM, Edwards CA & Lokerse E Colonic fermentation capacity in vitro: development during weaning in breast-fed infants is slower for complex carbohydrates than for sugars. Am. J. Clin. Nutr 65, 927–933 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Klaassens ES et al. Mixed-species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast- and formula-fed infants. Appl. Environ. Microbiol 75, 2668–2676 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collado MC, Delgado S, Maldonado A & Rodriguez JM Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett. Appl. Microbiol 48, 523–528 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Martin R et al. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr 143, 754–758 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Perez PF et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 119, e724–e732 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Stark PL & Lee A The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J. Med. Microbiol 15, 189–203 (1982). [DOI] [PubMed] [Google Scholar]
- 38.Amarri S et al. Changes of gut microbiota and immune markers during the complementary feeding period in healthy breast-fed infants. J. Pediatr. Gastroenterol. Nutr 42, 488–495 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Roger LC, Costabile A, Holland DT, Hoyles L & McCartney AL Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156, 3329–3341 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Koenig JE et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4578–4585 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yatsunenko T et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jost T, Lacroix C, Braegger CP & Chassard C New insights in gut microbiota establishment in healthy breast fed neonates. PLoS ONE 7, e44595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arumugam M et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu GD et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin A et al. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS ONE 8, e53838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergstrom A et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol 80, 2889–2900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pham TA & Lawley TD Emerging insights on intestinal dysbiosis during bacterial infections. Curr. Opin. Microbiol 17, 67–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lupp C et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 204 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Willing BP, Russell SL & Finlay BB Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol 9, 233–243 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Cho I et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeissig S & Blumberg RS Life at the beginning: perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat. Immunol 15, 307–310 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Round JL & Mazmanian SK The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol 9, 313–323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hooper LV & Macpherson AJ Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol 10, 159–169 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Goodman AL et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl Acad. Sci. USA 108, 6252–6257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oyama N, Sudo N, Sogawa H & Kubo C Antibiotic use during infancy promotes a shift in the T(H)1/T(H)2 balance toward T(H)2-dominant immunity in mice. J. Allergy Clin. Immunol 107, 153–159 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Russell SL et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN & Huffnagle GB Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect. Immun 73, 30–38 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atarashi K et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olszak T et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.An D et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Food and Agriculture Organization of the United Nations. The multiple dimensions of food security [online], http://www.fao.org/publications/sofi/en/ (2013).
- 63.Lutter CK et al. Undernutrition, poor feeding practices, and low coverage of key nutrition interventions. Pediatrics 128, e1418–e1427 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Smith MI et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blaser MJ Who are we? Indigenous microbes and the ecology of human diseases. EMBO Rep. 7, 956–960 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Belkaid Y & Hand TW Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turnbaugh PJ et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Turnbaugh PJ, Backhed F, Fulton L & Gordon JI Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turnbaugh PJ et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vijay-Kumar M et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carvalho FA et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 12, 139–152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ubeda C et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med 209, 1445–1456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kostic AD, Howitt MR & Garrett WS Exploring host-microbiota interactions in animal models and humans. Genes Dev. 27, 701–718 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karlsson FH et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Qin J et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Larsen N et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 5, e9085 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lam YY et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS ONE 7, e34233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amar J et al. Involvement of tissue bacteria in the onset of diabetes in humans: evidence for a concept. Diabetologia 54, 3055–3061 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Jayashree B et al. Increased circulatory levels of lipopolysaccharide (LPS) and zonulin signify novel biomarkers of proinflammation in patients with type 2 diabetes. Mol. Cell Biochem 388, 203–210 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Nielsen DS, Krych L, Buschard K, Hansen CH & Hansen AK Beyond genetics. Influence of dietary factors and gut microbiota on type 1 diabetes. FEBS Lett. 10.1016/j.febslet.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 81.de Goffau MC et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes 62, 1238–1244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giongo A et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 5, 82–91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murri M et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 11, 46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kriegel MA et al. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl Acad. Sci. USA 108, 11548–11553 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cardwell CR et al. Breast-feeding and childhood-onset type 1 diabetes: a pooled analysis of individual participant data from 43 observational studies. Diabetes Care 35, 2215–2225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rosenbauer J, Herzig P & Giani G Early infant feeding and risk of type 1 diabetes mellitus—a nationwide population-based case-control study in pre-school children. Diabetes Metab. Res. Rev 24, 211–222 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Norris JM et al. Timing of initial cereal exposure in infancy and risk of islet autoimmunity. JAMA 290, 1713–1720 (2003). [DOI] [PubMed] [Google Scholar]
- 88.Ziegler AG, Schmid S, Huber D, Hummel M & Bonifacio E Early infant feeding and risk of developing type 1 diabetes-associated autoantibodies. JAMA 290, 1721–1728 (2003). [DOI] [PubMed] [Google Scholar]
- 89.Artis D Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol 8, 411–420 (2008). [DOI] [PubMed] [Google Scholar]
- 90.Maynard CL, Elson CO, Hatton RD & Weaver CT Reciprocal interactions of the intestinal microbiota and immune system. Nature 489, 231–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Turner JR Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol 9, 799–809 (2009). [DOI] [PubMed] [Google Scholar]
- 92.Stockinger S, Hornef MW & Chassin C Establishment of intestinal homeostasis during the neonatal period. Cell. Mol. Life Sci 68, 3699–3712 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levy O Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat. Rev. Immunol 7, 379–390 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Schwartz S et al. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol. 13, r32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wagner CL, Taylor SN & Johnson D Host factors in amniotic fluid and breast milk that contribute to gut maturation. Clin. Rev. Allergy Immunol 34, 191–204 (2008). [DOI] [PubMed] [Google Scholar]
- 96.Neu J & Walker WA Necrotizing enterocolitis. N. Engl. J. Med 364, 255–264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huffnagle GB The microbiota and allergies/asthma. PLoS Pathog. 6, e1000549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peterson LW & Artis D Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol 14, 141–153 (2014). [DOI] [PubMed] [Google Scholar]
- 99.Lorenz RG, Chaplin DD, McDonald KG, McDonough JS & Newberry RD Isolated lymphoid follicle formation is inducible and dependent upon lymphotoxin-sufficient B lymphocytes, lymphotoxin beta receptor, and TNF receptor I function. J. Immunol 170, 5475–5482 (2003). [DOI] [PubMed] [Google Scholar]
- 100.Pabst O et al. Cryptopatches and isolated lymphoid follicles: dynamic lymphoid tissues dispensable for the generation of intraepithelial lymphocytes. Eur. J. Immunol 35, 98–107 (2005). [DOI] [PubMed] [Google Scholar]
- 101.Cherrier M & Eberl G The development of LTi cells. Curr. Opin. Immunol 24, 178–183 (2012). [DOI] [PubMed] [Google Scholar]
- 102.Bouskra D et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 (2008). [DOI] [PubMed] [Google Scholar]
- 103.Tsuji M et al. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity 29, 261–271 (2008). [DOI] [PubMed] [Google Scholar]
- 104.Lotz M et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med 203, 973–984 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chassin C et al. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe 8, 358–368 (2010). [DOI] [PubMed] [Google Scholar]
- 106.Afrazi A et al. New insights into the pathogenesis and treatment of necrotizing enterocolitis: Toll-like receptors and beyond. Pediatr. Res 69, 183–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Newburg DS & Walker WA Protection of the neonate by the innate immune system of developing gut and of human milk. Pediatr. Res 61, 2–8 (2007). [DOI] [PubMed] [Google Scholar]
- 108.LeBouder E et al. Modulation of neonatal microbial recognition: TLR-mediated innate immune responses are specifically and differentially modulated by human milk. J. Immunol 176, 3742–3752 (2006). [DOI] [PubMed] [Google Scholar]
- 109.Robinson G, Volovitz B & Passwell JH Identification of a secretory IgA receptor on breast-milk macrophages: evidence for specific activation via these receptors. Pediatr. Res 29, 429–434 (1991). [DOI] [PubMed] [Google Scholar]
- 110.Cummins AG & Thompson FM Postnatal changes in mucosal immune response: a physiological perspective of breast feeding and weaning. Immunol. Cell Biol 75, 419–429 (1997). [DOI] [PubMed] [Google Scholar]
- 111.Suzuki T Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci 70, 631–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Capaldo CT & Nusrat A Cytokine regulation of tight junctions. Biochim. Biophys. Acta 1788, 864–871 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mullin JM, Valenzano MC, Verrecchio JJ & Kothari R Age- and diet-related increase in transepithelial colon permeability of Fischer 344 rats. Dig. Dis. Sci 47, 2262–2270 (2002). [DOI] [PubMed] [Google Scholar]
- 114.Ulluwishewa D et al. Regulation of tight junction permeability by intestinal bacteria and dietary components. J. Nutr 141, 769–776 (2011). [DOI] [PubMed] [Google Scholar]
- 115.Resta-Lenert S & Barrett KE Probiotics and commensals reverse TNF-α- and IFN-γ-induced dysfunction in human intestinal epithelial cells. Gastroenterology 130, 731–746 (2006). [DOI] [PubMed] [Google Scholar]
- 116.Fasano A et al. Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc. Natl Acad. Sci. USA 88, 5242–5246 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Muza-Moons MM, Schneeberger EE & Hecht GA Enteropathogenic Escherichia coli infection leads to appearance of aberrant tight junctions strands in the lateral membrane of intestinal epithelial cells. Cell. Microbiol 6, 783–793 (2004). [DOI] [PubMed] [Google Scholar]
- 118.Visser J, Rozing J, Sapone A, Lammers K & Fasano A Tight junctions, intestinal permeability, and autoimmunity: celiac disease and type 1 diabetes paradigms. Ann. NY Acad. Sci 1165, 195–205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chokshi NK et al. The role of nitric oxide in intestinal epithelial injury and restitution in neonatal necrotizing enterocolitis. Semin. Perinatol 32, 92–99 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rhee SH, Pothoulakis C & Mayer EA Principles and clinical implications of the brain–gut–enteric microbiota axis. Nat. Rev. Gastroenterol. Hepatol 6, 306–314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.El Aidy S, Dinan TG & Cryan JF Immune modulation of the brain–gut–microbe axis. Front. Microbiol 5, 146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Veldhoen M & Brucklacher-Waldert V Dietary influences on intestinal immunity. Nat. Rev. Immunol 12, 696–708 (2012). [DOI] [PubMed] [Google Scholar]
- 123.Renz H, Brandtzaeg P & Hornef M The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol 12, 9–23 (2012). [DOI] [PubMed] [Google Scholar]
- 124.van de Pavert SA & Mebius RE New insights into the development of lymphoid tissues. Nat. Rev. Immunol 10, 664–674 (2010). [DOI] [PubMed] [Google Scholar]
- 125.Li Y et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147, 629–640 (2011). [DOI] [PubMed] [Google Scholar]
- 126.Kiss EA et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334, 1561–1565 (2011). [DOI] [PubMed] [Google Scholar]
- 127.Lee JS et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol 13, 144–151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qiu J et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36, 92–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chieppa M, Rescigno M, Huang AY & Germain RN Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J. Exp. Med 203, 2841–2852 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hadis U et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 34, 237–246 (2011). [DOI] [PubMed] [Google Scholar]
- 131.Satpathy AT et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol 14, 937–948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zaghouani H, Hoeman CM & Adkins B Neonatal immunity: faulty T-helpers and the shortcomings of dendritic cells. Trends Immunol. 30, 585–591 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jaensson E et al. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J. Exp. Med 205, 2139–2149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Iwata M et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity 21, 527–538 (2004). [DOI] [PubMed] [Google Scholar]
- 135.Mora JR et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science 314, 1157–1160 (2006). [DOI] [PubMed] [Google Scholar]
- 136.Sommer A Vitamin A deficiency and clinical disease: an historical overview. J. Nutr 138, 1835–1839 (2008). [DOI] [PubMed] [Google Scholar]
- 137.Cheroutre H, Lambolez F & Mucida D The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol 11, 445–456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ooi JH, Chen J & Cantorna MT Vitamin D regulation of immune function in the gut: why do T cells have vitamin D receptors? Mol. Aspects Med 33, 77–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yu S, Bruce D, Froicu M, Weaver V & Cantorna MT Failure of T cell homing, reduced CD4/CD8αα intraepithelial lymphocytes, and inflammation in the gut of vitamin D receptor KO mice. Proc. Natl Acad. Sci. USA 105, 20834–20839 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gaboriau-Routhiau V et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009). [DOI] [PubMed] [Google Scholar]
- 141.Ivanov II et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huber S, Gagliani N & Flavell RA Life, death, and miracles: Th17 cells in the intestine. Eur. J. Immunol 42, 2238–2245 (2012). [DOI] [PubMed] [Google Scholar]
- 143.Fujino S et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut 52, 65–70 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Seiderer J et al. Role of the novel Th17 cytokine IL-17F in inflammatory bowel disease (IBD): upregulated colonic IL-17F expression in active Crohn’s disease and analysis of the IL17F p.His161Arg polymorphism in IBD. Inflamm. Bowel Dis 14, 437–445 (2008). [DOI] [PubMed] [Google Scholar]
- 145.Duerr RH et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Elson CO et al. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology 132, 2359–2370 (2007). [DOI] [PubMed] [Google Scholar]
- 147.Lee YK et al. Late developmental plasticity in the T helper 17 lineage. Immunity 30, 92–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tanoue T & Honda K Induction of Treg cells in the mouse colonic mucosa: a central mechanism to maintain host-microbiota homeostasis. Semin. Immunol 24, 50–57 (2012). [DOI] [PubMed] [Google Scholar]
- 149.Huber S et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3− and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity 34, 554–565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rubtsov YP et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008). [DOI] [PubMed] [Google Scholar]
- 151.Franke A et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat. Genet 40, 1319–1323 (2008). [DOI] [PubMed] [Google Scholar]
- 152.Glocker EO, Kotlarz D, Klein C, Shah N & Grimbacher B IL-10 and IL-10 receptor defects in humans. Ann. NY Acad. Sci 1246, 102–107 (2011). [DOI] [PubMed] [Google Scholar]
- 153.Khor B, Gardet A & Xavier RJ Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Josefowicz SZ et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482, 395–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mucida D et al. Oral tolerance in the absence of naturally occurring Tregs. J. Clin. Invest 115, 1923–1933 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cha HR et al. Downregulation of Th17 cells in the small intestine by disruption of gut flora in the absence of retinoic acid. J. Immunol 184, 6799–6806 (2010). [DOI] [PubMed] [Google Scholar]
- 157.Wang C, Kang SG, Hogen Esch H, Love PE & Kim CH Retinoic acid determines the precise tissue tropism of inflammatory Th17 cells in the intestine. J. Immunol 184, 5519–5526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Smith PM et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Arpaia N et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Furusawa Y et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 161.Geuking MB, McCoy KD & Macpherson AJ Metabolites from intestinal microbes shape Treg. Cell Res. 23, 1339–1340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Xu W & Di Santo JP Taming the beast within: regulation of innate lymphoid cell homeostasis and function. J. Immunol 191, 4489–4496 (2013). [DOI] [PubMed] [Google Scholar]
- 163.Spencer SP et al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science 343, 432–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van de Pavert SA et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature 508, 123–127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Aagaard K et al. The placenta harbors a unique microbiome. Sci. Transl. Med 6, 237ra65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mold JE & McCune JM Immunological tolerance during fetal development: from mouse to man. Adv. Immunol 115, 73–111 (2012). [DOI] [PubMed] [Google Scholar]
- 167.Gareau MG, Sherman PM & Walker WA Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol 7, 503–514 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rautava S, Luoto R, Salminen S & Isolauri E Microbial contact during pregnancy, intestinal colonization and human disease. Nat. Rev. Gastroenterol. Hepatol 9, 565–576 (2012). [DOI] [PubMed] [Google Scholar]


