Abstract
Pigs are permissive to both human and avian influenza viruses and have been proposed to be an intermediate host for the genesis of pandemic influenza viruses through reassortment or adaptation of avian viruses. Prospective virological surveillance carried out between March 1998 and June 2000 in Hong Kong, Special Administrative Region, People's Republic of China, on pigs imported from southeastern China, provides the first evidence of interspecies transmission of avian H9N2 viruses to pigs and documents their cocirculation with contemporary human H3N2 (A/Sydney/5/97-like, Sydney97-like) viruses. All gene segments of the porcine H9N2 viruses were closely related to viruses similar to chicken/Beijing/1/94 (H9N2), duck/Hong Kong/Y280/97 (H9N2), and the descendants of the latter virus lineage. Phylogenetic analysis suggested that repeated interspecies transmission events had occurred from the avian host to pigs. The Sydney97-like (H3N2) viruses isolated from pigs were related closely to contemporary human H3N2 viruses in all gene segments and had not undergone genetic reassortment. Cocirculation of avian H9N2 and human H3N2 viruses in pigs provides an opportunity for genetic reassortment leading to the emergence of viruses with pandemic potential.
Human infection and mortality in Hong Kong in 1997 associated with the avian influenza virus H5N1 (H5N1/97) focused global attention on the role of avian influenza viruses as a cause of human disease (7, 34, 37). Subsequently, human disease associated with H9N2 viruses was documented, suggesting that other avian viruses can also cross the species barrier to humans (20, 21, 25). In both instances, there was little evidence of human-to-human transmission, each human infection seemingly being an independent transmission event from the avian host (17, 25). The pandemic influenza viruses of 1957 and 1968 emerged through genetic reassortment of avian viruses with the prevailing human viruses (18, 27). It may be speculated that the poor human-to-human transmissibility of the H5N1/97 viruses was because these purely avian viruses had not reassorted with human influenza viruses.
It has been proposed that pigs can serve as mixing vessels for the reassortment of human and avian influenza viruses (28). Pigs are susceptible to experimental infection with a range of avian and human influenza viruses (19). However, in nature, interspecies transmission of avian viruses to pigs is not often documented. Avian H1N1 viruses have been transmitted to pigs in Europe (26) and in China (9). Recently a purely avian, i.e., nonreassorted, H4N6 influenza virus caused a disease outbreak in pigs in Canada (16).
Porcine tracheal cells have receptors for both human and avian viruses, and this provides a biological basis for the susceptibility of pigs to both avian and human influenza viruses and facilitates reassortment between them. There are instances of reassortment between avian and human viruses occurring in pigs in nature (2, 3, 5). However, direct evidence that genetic reassortment in pigs played a role in the genesis of a human pandemic virus is still lacking.
A number of influenza viruses have been isolated previously from pigs in the south China region. These include classical (9, 30) and avian-like (9) swine H1N1 viruses and H3N2 viruses similar to human A/Hong Kong/2/68 (A/HK/8/68) and A/Victoria/3/75 viruses (29, 31). Some of these early human H3N2 viruses have undergone reassortment with classical swine (H1N1) influenza viruses in southern China (23, 33) and with avian-like swine H1N1 viruses in Europe (5). More recently, triple-reassortant viruses with surface antigens similar to contemporary human H3N2 (Sydney97-like) virus but with other gene segments from avian and classical swine influenza viruses have been reported in the United Kingdom (3) and the United States (38). These recent North American porcine H3N2 viruses have acquired their polymerase gene segments from avian viruses of the American lineage, and at least three separate introductions of the human H3 hemagglutinin (HA) gene appear to have occurred (35).
Although the pathogenic H5N1/97 virus has not been detected since the poultry slaughter in Hong Kong in December 1997, its probable precursors are present in poultry in south China (6, 10, 14), a region regarded as an epicenter for the emergence of pandemic influenza viruses (32). Furthermore, different lineages of H9N2 viruses are now widespread in poultry in China (11), central Asia, and Europe (4). It is therefore important to examine whether these avian viruses infect pigs and cocirculate with human viruses in the hypothetical mixing vessel for influenza virus reassortment. In this paper, we show that avian H9N2 viruses cocirculate with contemporary human H3N2 Sydney-like viruses in pigs in the southeastern China region.
MATERIALS AND METHODS
Sampling of pigs.
Tracheal swabs were collected on a monthly basis from pigs slaughtered at an abattoir in Hong Kong between March 1998 and June 2000. Serum specimens were also collected during the visits: 117 in 1998, 46 in 1999, and 294 between January and June 2000. Currently, around 25% of pigs slaughtered in Hong Kong originate from the neighboring Guangdong Province, 60% originate from provinces in southeastern China (predominantly Hunan, Jiangxi, Hubei, and Henan and a few from Fujian, Zhejiang, and Guangxi), and the rest are raised in Hong Kong. Those imported from the mainland of China arrive in the Special Administrative Region within a day of slaughter. Tracheal swabs were collected into transport medium (9) and kept at 4°C until transported to the laboratory.
Virus isolation and identification.
The swab eluate was inoculated into embryonated chicken eggs by the allantoic route (36) and into Madin-Darby canine kidney (MDCK) cell cultures in Eagle's minimum essential medium with trypsin (2 μg/ml). The cell culture tubes were incubated for up to 7 days and examined for cytopathic effect. Virus isolates were passaged and identified using hemagglutination inhibition (HAI) tests and neuraminidase inhibition tests using a panel of reference sera (National Institute of Allergy and Infectious Diseases Resources for Influenza Research, National Institutes of Health).
Serology tests.
Sera were treated with receptor-destroying enzyme and used for HAI tests using turkey erythrocytes (36). Neutralization tests were carried out by mixing 100 50% tissue culture infective doses of the virus with serial dilutions of serum and incubating for 2 h followed by inoculation onto MDCK cells grown in 96-well microtiter plates. After adsorption of the virus-serum mixture for 2 h, the inoculum was removed and fresh serum-free tissue culture medium containing trypsin (2 μg/ml) was added. Complete neutralization of cytopathic effect (read under an inverted microscope) was considered evidence of neutralizing antibody.
Analysis of viral RNA.
Viral gene sequencing and analysis were carried out as described previously (11). In brief, viral RNA was directly extracted from infected allantoic fluids or cell culture using QIAamp ViralRNA MiniKit (Qiagen, Inc., Valencia, Calif.). Reverse transcription followed by PCR was performed using specific primers for each gene segment (primer sequences are available on request). PCR products were purified with the QIAquick PCR purification kit (Qiagen, Inc.) and sequenced using synthetic oligonucleotides. Reactions were performed with Big Dye-Terminator Cycle Sequencing Ready Reaction kits used with AmpliTaqDNA Polymerase FS (Perkin-Elmer/Applied Biosystems, Inc.). Samples were electrophoresed and analyzed on a model 377 DNA sequencer (Perkin-Elmer/Applied Biosystems, Inc.).
Sequence data were edited and analyzed using the Wisconsin Sequence Analysis Package (version 10.0; Genetics Computer Group, Madison, Wis.). Phylogenetic analyses were carried out using Phylogenetic Analysis Using Parsimony (version 4.0; David Swofford, Illinois Natural History Survey, Champaign).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained from this study are available from GenBank under accession numbers AF222810 through AF222825 and AF400752 through AF400791.
RESULTS
Virus isolation.
One hundred eighty-four influenza A viruses were isolated from 4,957 tracheal swabs collected from apparently healthy pigs from southeastern China between March 1998 and June 2000. Most viruses were isolated in both embryonated eggs and MDCK cells, but two (swine/Hong Kong/2106/98 [Sw/HK/2106/98] and Sw/HK/3297/98) of the four H9N2 and all the Sydney97-like H3N2 viruses (see below) were only isolated in MDCK cells and would have been missed if egg inoculation by the allantoic route had been the only method used.
Antigenic characterization.
Preliminary antigenic characterization of the virus isolates by HAI and neuraminidase inhibition tests identified these viruses to be H9N2 (n = 4), H3N2 (n = 28), and H1N1, classical swine like (n = 152). The H9N2 viruses were antigenically similar to chicken/Hong Kong/G9/97 (Ck/HK/G9/97), a virus of the duck/Hong Kong/Y280/97 (Dk/HK/Y280/97) (H9N2) lineage, and distinct from the quail/Hong Kong/G1/97 (Qa/HK/G1/97) (H9N2) virus (Table 1). A ferret antiserum to Ck/HK/G9/97 (H9N2) virus gave comparable HAI titers to the homologous virus and to the porcine isolates Sw/HK/2106/98 and Sw/HK/3297/98, but a four- to eightfold-lower reactivity with virus isolates Sw/HK/9/98 (H9N2) and Sw/HK/10/98 (H9N2), indicating some antigenic heterogeneity among the porcine H9N2 isolates.
TABLE 1.
Antigenic characterization of H9N2 isolates from pigs
| Virus | Result of HAI test using ferret antiseraa to:
|
||
|---|---|---|---|
| Qa/HK/G1/97b | Ck/HK/G9/97b | Sw/HK/9/98 | |
| Sw/HK/2106/98 | <40 | 2,560 | 80 |
| Sw/HK/9/98 | <40 | 640 | 640 |
| Sw/HK/10/98 | <40 | 640 | 320 |
| Sw/HK/3297/98 | 40 | 5,120 | 320 |
| Qa/HK/G1/97b | 640 | <40 | <40 |
| Ck/HK/G9/97b | <40 | 2,560 | 640 |
Provided by A. Hay, Mill Hill, London, United Kingdom.
Representatives of two main lineages of H9N2 viruses. Ck/HK/G9/97 is used as a representative of the Dk/HK/Y280/97 lineage.
Two antigenic groups of H3N2 viruses were distinguishable, with 13 being closely related to contemporary human (Sydney/5/97) viruses and the other 15 being related to A/Port Chalmers/1/73 and A/Victoria/3/75 viruses (data not shown). The antigenic characterization of three representative Sydney/5/97-related viruses is shown in Table 2.
TABLE 2.
Antigenic characterization of representative H3N2 isolates from pigs
| Virusa | Result of HAI test using ferret antisera tob:
|
A/PC/73 MAb 121/1c | ||||
|---|---|---|---|---|---|---|
| HK/8/68 | PC/1/73 | Vic/3/75 | Bk/1/79 | Syd/5/97 | ||
| A/Sw/HK/2422/98 | <40 | <40 | <40 | <40 | 5,120 | <400 |
| A/Sw/HK/2405/98 | <40 | <40 | <40 | <40 | 2,560 | <400 |
| A/Sw/HK/2329/98 | <40 | <40 | <40 | <40 | 5,120 | <400 |
| A/Sydney/5/97 | 80 | <40 | 40 | <40 | 1,280 | <400 |
The A/Sydney/5/97 antigen was an inactivated antigen provided as part of the World Health Organization reagent kit, while samples of untreated virus-infected egg allantoic fluid of the pig isolates were used as antigens in this test.
Abbreviations: PC, Port Chalmers; Vic, Victoria; Bk, Bangkok; Syd, Sydney.
Result of HAI test with monoclonal antibody (MAb).
The 13 Sydney97-like H3N2 viruses were isolated on eight sampling occasions between March 1998 and September 1999. The four H9N2 viruses were isolated on three sampling occasions during 1998, in March (Sw/HK/2106/98), April (Sw/HK/9/98 and Sw/HK/10/98), and October (Sw/HK/3297/98).
All four H9N2 isolates and three representative Sydney97-like H3N2 viruses were reisolated from the original swab specimen to confirm their validity. These were used in subsequent seroepidemiology, genetic characterization, and phylogenetic analysis.
Seroepidemiology.
Antibody to H9N2 virus and Sydney97-like H3N2 viruses was detected in porcine sera by HAI and neutralization tests, providing independent evidence of the activity of these viruses (Table 3). In the HAI test, antibody to H9N2 viruses (Dk/HK/Y280/97, Sw/HK/9/98, and/or Ck/HK/G9/97) was present at low prevalence in sera collected in each year of the study. Some of these sera also had HAI antibody to an H9N7 reassortant virus [Ck/HK/G9/97 (H9N2) × A/seal/Massachusetts/1/80 (H7N7)] (data not shown), confirming that the observed immunological reactivity was to the HA (H9) rather than the steric effects resulting from antibody binding to the neuraminidase (N2). Three of these serum samples (collected in the year 2000) had neutralizing activity, one to Dk/HK/Y280/97, one to Sw/HK/9/98, and one to both these viruses. No serological evidence of infection with Qa/HK/G1/97 or Dk/HK/439/97 (“Korean”) lineage H9N2 viruses was documented. Neutralizing antibody to the porcine Sw/HK/2422/98 (Sydney97-like) isolate was detected in all 3 years, with prevalence ranging from 4 to 29%.
TABLE 3.
Seroprevalencea to influenza A subtype H3N2 and H9N2 viruses in pigs
| Viral antigen | Serological testb | No. of sera (%) reactive in yrc
|
||
|---|---|---|---|---|
| 1998 (n = 117) | 1999 (n = 46) | 2000d(n = 294) | ||
| Dk/HK/Y280/97 (H9N2) | NT | 0 (0) | 0 (0) | 2 (0.7) [20–40] |
| Sw/HK/9/98 (H9N2) | NT | 0 (0) | 0 (0) | 2 (0.7) [20] |
| Qa/HK/G1/97 (H9N2) | NT | 0 (0) | 0 (0) | NDe |
| Dk/HK/Y280/98 (H9N2) | HAI | 2 (2) [80] | 2 (4) [160] | 16 (5) [80–640] |
| Sw/HK/9/98 (H9N2) | HAI | 1 (1) [80] | 1 (2) [80] | 8 (3) [80–320] |
| Ck/HK/G9/97 (H9N2) | HAI | 1 (1) [160] | 5 (11) [80–320] | 8 (3) [80–160] |
| Qa/HK/G1/97 (H9N2) | HAI | 0 (0) | 0 (0) | 0 (0) |
| Dk/HK/Y439/97 (H9N2) | HAI | 0 (0) | 0 (0) | 0 (0) |
| Sw/HK/2422/98 (H3N2) | NT | 34 (29) [20–>320] | 2 (4) [160, 320] | 43 (15) [20–>320] |
| Sw/HK/2422/98 (H3N2) | HAI | 18 (15) [80–1,280] | 2 (4) [320–2,560] | 57 (19) [80–1,280] |
HAI positives were taken as titers of 1/80 or more. Neutralization positives were taken as titers of 1/20 or more.
NT, neutralization test.
Values in brackets show the range of the antibody titers.
January to June.
ND, not determined..
Genotyping of porcine H3N2 and H9N2 influenza viruses.
The four H9N2 pig isolates and three representative Sydney97-like H3N2 viruses were selected for genetic analysis. The gene segments of the H3N2 and H9N2 viruses were sequenced in part or completely, and their homologies were determined by comparison with sequences available in GenBank and between viruses from the same group.
Sw/HK/9/98 (H9N2) was closely related to H9N2 viruses of the Dk/HK/Y280/97 (H9N2) lineage in all eight gene segments, with homologies ranging from 97 to 98% (Table 4). The HA, PB1, and NS genes had greater homology with viruses circulating in chicken in 1994 than to more recently isolated viruses of the Dk/HK/Y280/97 lineage. The H3N2 viruses represented by Sw/HK/2405/98 were closely related to H3N2 Sydney97-like viruses currently circulating in humans. The homology was >98% in each of the eight gene segments, strongly suggesting that the virus is of human origin and was probably introduced into pigs recently. The close homology of the PB2, PB1, and PA genes to H3N2 viruses isolated in 1996 reflects the lack of sequence data in GenBank for these gene segments of Sydney97-like viruses. Homology within the H3N2 pig virus isolates was 99 to 100% (results not shown).
TABLE 4.
Homology of swine influenza viruses isolated in Hong Kong
| Gene | Sw/HK/2045/98 (H3N2)a
|
Sw/HK/9/98 (H9N2)b
|
||
|---|---|---|---|---|
| Homologous virus | % Homology | Homologous virus | % Homology | |
| PB2 | Fukushima/140/96 | 99 | Dk/HK/Y280/97 | 97 |
| PB1 | Fukushima/140/96 | 99 | Ck/HK/739/94 | 97 |
| PA | Fukushima/140/96 | 99 | Dk/HK/Y280/97 | 98 |
| HA | Nagasaki/93/98 | 99 | Ck/Bei/1/94 | 97 |
| NP | Nagasaki/76/98 | 99 | Ck/HK/G23/97 | 98 |
| NA | Shiga/25/97 | 99 | Dk/HK/Y280/97 | 98 |
| M | Hong Kong/498/97 | 99 | Pg/HK/Y233/97 | 98 |
| NS | Hong Kong/497/97 | 98 | Ck/Bei/1/94 | 98 |
Ranges of sequences (in base pairs) used to carry out homology search are as follows: PB2, 4 to 2,269; PB1, 286 to 2,256; PA, 1,442 to 2,251; HA, 78 to 1,725; NP, 17 to 1,513; NA, 725 to 1,410; M, 125 to 1,023; NS, 9 to 888).
Ranges of sequences (in base pairs) used to carry out homology search are as follows: PB2, 1 to 2,246; PB1, 1 to 2,237; PA, 499 to 2,127; HA, 12 to 1,650; NP, 7 to 1,482; NA, 1 to 1,368; M, 1 to 993; NS, 1 to 831.
Thus, primary genotyping of these isolates confirmed the antigenic characterization that avian H9N2 viruses similar to Dk/HK/Y280/97 and contemporary human Sydney97-like H3N2 cocirculated in southeastern China. Reassortants of these viruses were not detected.
Phylogenetic relationships of porcine influenza viruses from southeastern China.
To characterize the genetic relationships of the H9N2 and H3N2 viruses more precisely, phylogenetic analyses were carried out of the HA1 region of the HA and part of the M gene sequence of each of the H9N2 and Sydney97-like H3N2 viruses.
(i) H9 HA tree.
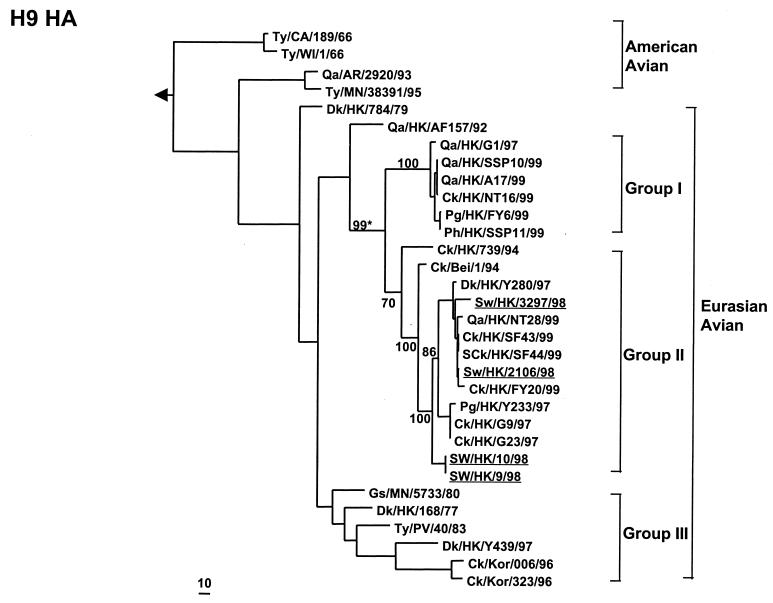
The H9 HA1 tree separated into two distinct lineages, one including viruses from North America and the other comprising viruses isolated in Asia, especially from China, in recent years (Fig. 1). All four porcine H9N2 viruses isolated in the present study clustered into a sublineage formed by recent avian viruses from land-based poultry in southeastern China. Two H9N2 viruses, Sw/HK/2106/98 and Sw/HK/3297/98, isolated in March and October 1998, respectively, appear closely related to contemporary viruses of the Dk/HK/Y280/97 lineage isolated from chicken. The others (Sw/HK/9/98 and Sw/HK/10/98) isolated in April 1998 were more closely related to early chicken isolates (Ck/HK/739/94 or Ck/Beijing/1/94 [Ck/Bei/1/94]) of the same sublineage and exhibit an “outgroup” relationship to contemporary avian Dk/HK/Y280/97-like viruses. Since these four porcine H9N2 viruses do not form a direct parent-descendant relationship, the findings imply that there were at least two independent introductions from an avian host to pigs.
FIG. 1.
Phylogenetic tree for the H9 HA1 gene of influenza A viruses. The nucleotide sequences of the HA1 gene (960 bp from position 55 to 1014) were analyzed with the Phylogenetic Analysis Using Parsimony program using a maximum-parsimony algorithm. The HA1 phylogenetic tree is rooted to A/Dk/Alberta/60/76 (H12N5). The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes. Vertical lines are for spacing branches and labels. The bootstrap value (∗) for 1,000 replications was computed using MEGA software (version 1.0; Pennsylvania State University, University Park), and the results for key branches in the tree are depicted. Abbreviations used in virus designations not used in the text are as follows: Gs, goose; Pg, pigeon; Ph, pheasant; SCk, silkie chicken; Ty, turkey; Kor, Korea; CA, California; WI, Wisconsin; AR, Arkansas; MN, Minnesota. Viruses isolated in this study are underlined. Group I, Qa/HK/G1/97 (H9N2) lineage; group II, Dk/HK/Y280//97 (H9N2) lineage; group III, Ck/Kor/006/96 (H9N2) lineage.
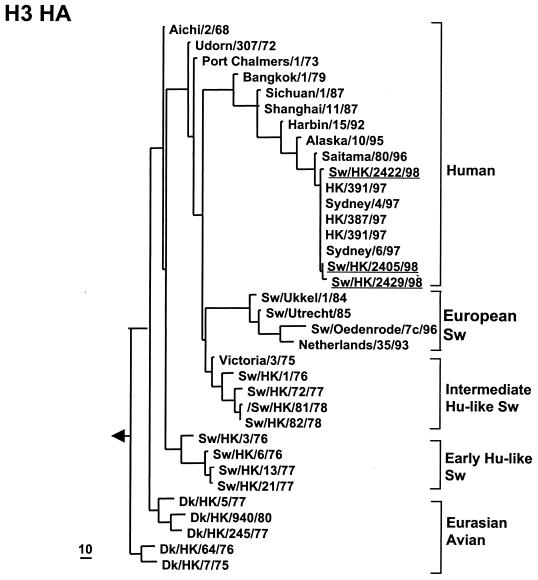
(ii) H3 HA1 tree.
The H3 HA1 tree (Fig. 2) shows that three groups of human H3N2 viruses have transmitted to and persisted in pigs for various periods of time. Viruses related to the initial H3N2 pandemic virus of 1968 (Aichi/2/68) persisted in pigs (early human like) until 1977. Subsequently, A/Victoria/3/75-like viruses transmitted to pigs (intermediate human like) in China and viruses of this sublineage continued to be isolated in China at least until 1978 (Fig. 2). A/Victoria/3/75-like viruses also entered pigs independently in Europe and have persisted as avian-like swine H3N2 (European swine) viruses. The H3N2 viruses Sw/HK/2405/99 and Sw/HK/2429/99 were directly derived from recent Sydney97-like human H3N2 viruses.
FIG. 2.
Phylogenetic tree of the partial H3 HA1 gene (778 bp from position 33 to 810) of influenza A viruses. The method used and abbreviations are as given in the legend to Fig. 1. Viruses isolated in this study are underlined. The tree is rooted at Dk/Czechoslovakia/56 (H4N6).
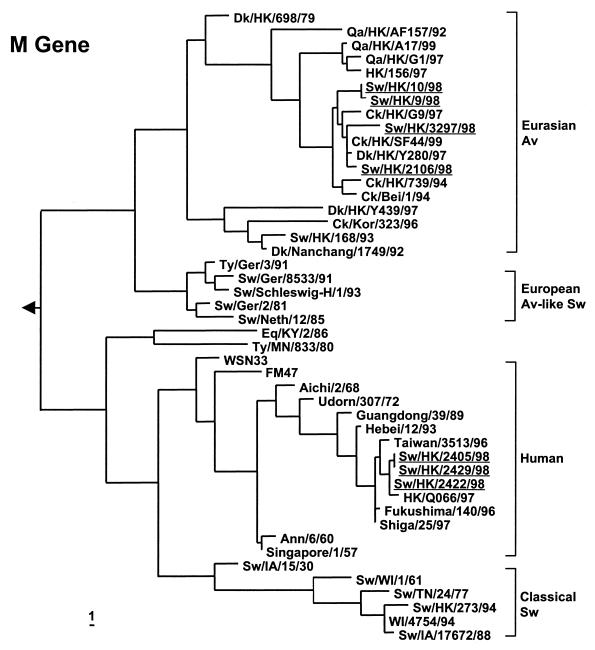
(iii) M gene tree.
The H9N2 pig isolates were derived from the avian H9N2 (Y280-like) lineage from China, but the Sw/HK/9/98 and Sw/HK/10/98 (H9N2) viruses retain their outgroup relationship with recent H9N2 isolates from poultry and the other two H9N2 pig virus isolates. The Sydney/5/97-related H3N2 viruses Sw/HK/2405/99 and Sw/HK/2429/99 are closely related to contemporary human H3N2 virus isolates (Fig. 3).
FIG. 3.
Phylogenetic trees for the M gene (residues 41 to 671) of swine influenza viruses from southeastern China. The method used and abbreviations are as given in the legend to Fig. 1. Viruses isolated in this study are underlined. The tree is rooted at A/Equine/Prague/1/56 (H7N7). Abbreviations used in virus designations not used in the text are as follows: Ger, Germany; IA, Iowa; KY, Kentucky; Neth, The Netherlands.
Molecular analysis of HA and NA genes of the pig H9N2 isolates.
Alignment of the deduced amino acid sequences of the HA1 region of the four H9N2 isolates with those of other avian isolates did not reveal common amino acid changes associated with transmission to pigs. All four pig isolates have amino acid L at position 226 (H3 numbering) and G at position 228 within the receptor-binding pocket (Table 5). Amino acid L at position 226 is also found in recent H9N2 viruses of the Dk/HK/Y280/97 lineage isolated from poultry but differs from that (226 Q) in the earliest chicken virus isolates of this lineage, viz., Ck/Bei/1/94 and Ck/HK/739/94. Both Sw/HK/9/98 and Sw/HK/10/98 H9N2 viruses are uniquely different from other avian viruses and the other two pig isolates (Sw/HK/2106/98 and Sw/HK/3297/98) in containing an amino acid residue H at position 227 within the receptor-binding region. Furthermore, Sw/HK/9/98 and Sw/HK/10/98 viruses differ from Sw/HK/2106/98 and Sw/HK/3297/98 and many other recent avian H9N2 isolates in having V at amino acid residue 190. An early chicken virus (Ck/Bei/94) (H9N2) and a contemporary quail isolate (Qa/HK/NT/298/99) (H9N2) also possess V at this position. In common with many other contemporary H9N2 isolates of the Dk/HK/Y280/97 lineage, the pig H9N2 isolates Sw/HK/2106/98 and Sw/HK/3297/98 have amino acid A at residue 190, though Dk/HK/Y280/97 (H9N2), the prototype virus of this phylogenetic lineage, has T at this position. The HA1 genes of Sw/HK/9/98 and Sw/HK/10/98 viruses are similar to early chicken H9N2 viruses (Ck/HK/739/94; Ck/Bei/94) and differ from contemporary H9N2 viruses at a number of other amino acid residues unconnected with receptor binding, viz., positions 32, 174, and 205. These findings suggest that Sw/HK/9/98 and Sw/Hk/10/98 are more related to Ck/HK/739/94 and Ck/Bei/94 than to contemporary Ck/HK/Y280/97-like H9N2 viruses. The Sw/HK/3297/98 virus differs from other avian and porcine H9N2 viruses at residues 274, 279, and 286 (Table 5). Compared to other H9N2 viruses of this lineage, the potential glycosylation sites on the HA1 region in the four pig isolates remain unaltered.
TABLE 5.
Comparison of amino acid sequences of pig and avian H9N2 HAs
| Virus | Amino acid residue relevant in receptor for binding
|
Other amino acid change
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 183 | 190 | 226 | 227 | 228 | 32 | 174 | 205 | 274 | 279 | 286 | |
| Ck/Beijing/1/94 | N | V | Q | Q | G | T | S | T | N | V | H |
| Ck/HK/739/94 | H | A | •a | • | • | • | • | • | S | • | • |
| Sw/HK/9/98 | • | • | L | H | • | • | • | • | • | • | • |
| Sw/HK/10/98 | • | • | L | H | • | • | • | • | • | • | • |
| Qa/HK/NT/28/99 | • | • | L | • | • | N | N | A | • | • | • |
| Ck/HK/G9/97 | • | A | L | • | • | N | • | A | • | • | • |
| Dk/HK/Y280/97 | • | T | L | • | • | N | N | A | • | • | • |
| Ck/HK/FY20/99 | • | A | L | • | • | N | N | A | • | • | • |
| SCk/HK/SF43/99 | • | A | L | • | • | N | N | A | • | • | • |
| Sw/HK/2106/98 | • | A | L | • | • | N | N | A | • | • | • |
| Sw/HK/3297/98 | • | A | L | • | • | N | N | A | K | A | Q |
•, amino acid is conserved relative to Ck/Beijing/1/94.
The amino acid sequence at the HA cleavage site is highly conserved in all the H9N2 viruses, including the pig isolates described here, and no change in the multiple basic amino acids associated with highly pathogenic avian influenza viruses was observed.
The four pig H9N2 isolates share the same three-amino-acid deletion in the NA stalk region found in other Dk/HK/Y280/97-like viruses and are distinct from Ck/Bei/1/94 and Ck/HK/G9/97 in this regard. The potential glycosylation sites on the NA are also similar to those found in Dk/HK/Y280/97-like viruses. Alignment of the gene sequences of the internal gene segments of the four pig H9N2 viruses fail to reveal common amino acid changes associated with interspecies transmission of avian viruses into pigs. However, three silent nucleotide substitutions at positions 1273 (C→T), 1323 (C→T) and 1423 (T→C) in the PA gene were common to the four pig H9N2 viruses and were not seen in other avian H9N2 viruses sequenced to date. In the other internal genes, there were nucleotide signatures that were shared by Sw/HK/9/98 and Sw/HK/10/98 (H9N2) that were distinct from that of Ck/Bei/1/94, as well as from those of contemporary avian Dk/HK/Y280/97-like viruses.
DISCUSSION
Avian H9N2 viruses cocirculate with contemporary human Sydney97-like H3N2 viruses in pigs in southeastern China, a hypothetical epicenter for the genesis of influenza pandemics (32).
The four H9N2 viruses belonged to the Dk/HK/Y280/97 lineage in all eight gene segments; i.e., they are not reassortants. They were isolated on three separate occasions in 1998 and confirmed by independent reisolation from the original clinical specimens. The Sw/HK/3297/98 (H9N2) and Sw/HK/2106/98 (H9N2) viruses were antigenically and genetically similar to contemporary H9N2 isolates from chicken. In contrast, Sw/HK/9/98 (H9N2) and Sw/HK/10/98 (H9N2) viruses are clearly distinct and more closely related to earlier H9N2 viruses of this lineage, viz., Ck/Bei/1/94 and Ck/HK/739/94. These avian viruses were not handled in the laboratory during the period of the study, and this argues against Sw/HK/9/98 (H9N2) and Sw/HK/10/98 (H9N2) being laboratory contaminants. Furthermore, serological data provide independent evidence of H9N2 virus infection in pigs and indicate that infection with a similar virus continued in subsequent years (Table 3).
That influenza A viruses can seemingly persist in pigs with minimal antigenic and genetic change has been shown by previous studies of human H3N2 viruses isolated from pigs in the 1970s (1, 29, 31). Thus, one explanation for finding viruses similar to early avian Ck/Bei/94 (H9N2) viruses in pigs is that they were introduced to pigs some years ago and were preserved antigenically and genetically in the porcine host. In order to confirm this hypothesis, it will be necessary to demonstrate that similar viruses continue to be isolated from pigs and form a distinct evolutionary lineage. An alternative (and more likely) explanation is that H9N2 viruses similar to Ck/Bei/94 continue to circulate in poultry in regions of China from which sequence data of avian viruses are presently scarce. Land-based poultry sampled in Hong Kong only comes from the immediately adjacent area of Guangdong, but pigs are imported from much farther afield and may well acquire infection with H9N2 viruses different from those currently isolated in Hong Kong and Guangdong. In any event, the low rate of H9N2 virus isolation, low seroprevalence, and lack of a direct precursor-descendant relationship among these four isolates indicate that there were at least two separate interspecies transmission events.
All four pig H9N2 virus isolates have amino acid residue L at position 226 within the receptor-binding pocket, an amino acid residue associated with a preferential binding of the virus to “human” α2,6- rather than “avian” α2,3-NeuAcGal receptors (8, 15, 22). The importance of this amino acid residue in the binding of HA to human α2,6-NeuAcGal receptors has been confirmed by recent data on the crystal structure of the Sw/HK/9/98 virus (12). In conjunction with amino acid residue L at 226, position 190 is reported to influence the affinity of the binding to the α2,6-NeuAcGal receptor, the binding affinity being highest with V at position 190, intermediate with T at this position, and weakest with A at this position (22). It is predicted, therefore, that Sw/HK/9/98 (H9N2) and Sw/HK/10/98 (H9N2) viruses have high-affinity binding to the α2,6-NeuAcGal receptor found in human cells. Experimentally, this has been found to be true with the related H9N2 virus Qa/HK/NT28/99, which has a similar amino acid motif at the receptor binding site (22). The other two pig H9N2 isolates, Sw/HK/2106/98 and Sw/HK/3297/98, have A at position 190 and, while still showing preference for the α2,6-NeuAcGal human receptor, would be predicted to have a lower-affinity interaction with this receptor. These predictions need to be confirmed by experimental studies of the receptor specificity of these viruses. It is interesting that two of the pig H9N2 viruses were isolated in MDCK cells rather than in the allantoic cavity of embryonated eggs, possibly a reflection of their affinity for α2,6-NeuAcGal binding.
It is important to note that the Q→L change at amino acid position 226 in the HA1 region of H9N2 viruses occurred in the avian host and preceded their introduction to pigs rather than being an adaptive change following the interspecies transmission event. When compared to related H9N2 viruses isolated from avian hosts, the only unique amino acid changes found in the HA1 of the pig isolates were at residue 227 within the receptor-binding pocket found in Sw/HK/9/98 (H9N2) and Sw/HK/10/98 (H9N2) and in residues 274, 279, and 286 outside the receptor-binding area found in Sw/HK/3297/98 (H9N2). The significance of these changes remains to be elucidated.
In the gene regions sequenced, there were no unique amino acid substitutions common to all four pig H9N2 viruses that may be markers of interspecies transmission. Three silent nucleotide substitutions in the PA gene were shared by all four pig viruses and distinguished them from other avian viruses sequenced to date, but these presumably have no functional significance.
Contemporary human H3N2 viruses antigenically and genetically similar to Sydney97-like viruses were isolated repeatedly from pigs in southeastern China. Virus isolation and seroepidemiology suggest that these viruses continued to circulate throughout the period of the study. Unlike the recent reassortant H3N2 viruses isolated from pigs in the North American continent, viruses isolated in the present study have not undergone reassortment and are similar in all gene segments to contemporary human H3N2 viruses. Seroprevalence to this virus appears higher than would be expected from discrete introductions of the virus into the pig population, and it is likely that the H3N2 Sydney-like virus in pigs is now being maintained by transmission within pigs. While it is known that human viruses readily infect pigs in an experimental setting (19) and have repeatedly crossed into humans (13), only a few have established themselves in the pig population in nature. The early A/HK/68-like H3N2 viruses established themselves in pigs in Asia (29) and persisted in this host till at least 1977 (Fig. 2) without reassortment with other avian or swine viruses (33). Victoria75-like human viruses have been detected in pigs in China (31), Europe (24), and Canada (1). In Europe, the virus acquired internal genes from avian-like H1N1 viruses through reassortment and persists to this day, viz., avian-like swine H3N2 viruses (Fig. 3). However, other antigenic variants of human H3N2 viruses have not established themselves in pigs until recently, when the Sydney97-like virus was found to have contributed its HA gene to reassortants causing disease outbreaks in pigs in the United States (35, 38).
We now report the isolation of unreassorted H3N2 Sydney-like viruses from pigs in southeastern China. These findings appear to indicate that, like Victoria/3/75, the H3N2 Sydney97 variant may have a greater propensity to cross the species barrier and establish itself in pigs. There are sera with evidence of antibody to both viruses, providing evidence of cocirculation of avian H9N2 and human H3N2 viruses in pigs in southeastern China. These human H3N2 viruses have not yet undergone reassortment with porcine viruses. The H9N2 viruses are still in the process rapid evolution in the avian host (11) and now have crossed to a new host—the pig. Taking these results together, it may be predicted that both viruses have an increased propensity to reassort.
In southern China, pigs are reared in abundance and, being the major source of protein for an increasingly affluent population, are raised in increasing numbers. While some of this pig husbandry is carried out in large-scale farms, small-holder raising of animals with close interaction between humans, poultry, and pigs continues, providing the opportunity for interspecies transmission of influenza viruses (32). H9N2 viruses are widespread in poultry in this region (11), and interspecies transmission to pigs is now documented. Repeated introductions of avian H9N2 viruses into pigs which may be coinfected with human H3N2 Sydney-like viruses provide the opportunity for the emergence of reassortants containing an H9 HA and internal genes adapted to replication in human cells. Unlike H5N1/97 viruses, the HA of these H9N2 viruses would be predicted to already have affinity to bind to the sialyl-oligosaccharides found on human cells. In the context of a human population immunologically naïve to the H9 antigen, such a virus would pose a significant pandemic threat.
ACKNOWLEDGMENTS
This study was supported by Wellcome Trust grant 057476/2/99/Z, National Institute of Allergy and Infectious Diseases grant NO1-A1-05357, Cancer Center support CORE grant CA-21765, and the American Lebanese Syrian Associated Charities.
The assistance of T. M. Ellis and K. M. Dyrting of the Department of Agriculture, Fisheries, and Conservation and H. C. Sit of the Department of Food and Environmental Hygiene of the Hong Kong Special Administrative Region is acknowledged with thanks. We thank A. Hay and N. J. Cox for providing reference antisera. We acknowledge the excellent technical assistance of C. Y. Cheung, S. K. Ma, S. Krauss, and L. J. Zhang.
REFERENCES
- 1.Bikour M H, Frost E H, Deslandes S, Talbot B, Weber J M, Elazhary Y. Recent H3N2 swine influenza virus with haemagglutinin and nucleoprotein genes similar to 1975 human strains. J Gen Virol. 1995;76:697–703. doi: 10.1099/0022-1317-76-3-697. [DOI] [PubMed] [Google Scholar]
- 2.Brown I H, Alexander D J, Chakraverty P, Harris P A, Manvell R J. Isolation of an influenza A virus of unusual subtype (H1N7) from pigs in England, and the subsequent experimental transmission from pig to pig. Vet Microbiol. 1994;39:125–134. doi: 10.1016/0378-1135(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 3.Brown I H, Harris P A, McCauley J W, Alexander D J. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J Gen Virol. 1998;79:2947–2955. doi: 10.1099/0022-1317-79-12-2947. [DOI] [PubMed] [Google Scholar]
- 4.Cameron K R, Gregory V, Banks J, Brown I H, Alexander D J, Hay A J, Lin Y P. H9N2 subtype influenza A viruses in poultry in Pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology. 2000;278:36–41. doi: 10.1006/viro.2000.0585. [DOI] [PubMed] [Google Scholar]
- 5.Castrucci M R, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster R G. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 6.Cauthen A N, Swayne D E, Shultz-Cherry S, Perdue M L, Suarez D L. Continued circulation in China of highly pathogenic avian influenza viruses encoding the hemagglutinin gene associated with the 1997 H5N1 outbreak in poultry and humans. J Virol. 2000;74:6592–6599. doi: 10.1128/jvi.74.14.6592-6599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claas E C J, Kawaoka Y, de Jong J C, Masurel N, Webster R G. Infection of children with avian-human reassortant influenza virus from pigs in Europe. Virology. 1994;204:453–457. doi: 10.1006/viro.1994.1553. [DOI] [PubMed] [Google Scholar]
- 8.Connor R J, Kawaoka Y, Webster R G, Paulson J C. Receptor specificity in human, avian and equine H2 and H3 influenza isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 9.Guan Y, Shortridge K F, Krauss S, Li P H, Kawaoka Y, Webster R G. Emergence of avian H1N1 influenza viruses in pigs in China. J Virol. 1996;70:8041–8046. doi: 10.1128/jvi.70.11.8041-8046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan Y, Shortridge K F, Krauss S, Webster R G. Molecular characterisation of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan Y, Shortridge K F, Krauss S, Chin P S, Dyrting K C, Ellis T M, Webster R G, Peiris M. H9N2 influenza viruses possessing H5N1-like internal genes continue to circulate in poultry in southeastern China. J Virol. 2000;74:9372–9380. doi: 10.1128/jvi.74.20.9372-9380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ha, Y., J. J. Skehel, and D. C. Wiley. High resolution crystal structure of hemagglutinin from two recently emerged influenza A viruses. Proceedings of the Options for the Control of Influenza IV, in press.
- 13.Haesebrouck F, Biront P, Pensaert M B, Leunen J. Epizootics of respiratory tract disease in swine in Belgium due to H3N2 influenza viruses and experimental reproduction of disease. Am J Vet Res. 1985;46:1926–1928. [PubMed] [Google Scholar]
- 14.Hoffmann E, Stech J, Leneva I, Krauss S, Scholtissek C, Chin P S, Peiris M, Shortridge K F, Webster R G. Characterization of the influenza A virus gene pool in avian species in southern China: was H6N1 a derivative or a precursor of H5N1? J Virol. 2000;74:6309–6315. doi: 10.1128/jvi.74.14.6309-6315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito T, Couceiro J S, Kelm S, Baum L G, Krauss S, Castrucci M R, Donatelli I, Kida H, Paulson J C, Webster R G, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karasin A I, Brown I H, Carman S, Olsen C W. Isolation and characterization of H4N6 avian influenza virus from pigs with pneumonia in Canada. J Virol. 2000;74:9322–9327. doi: 10.1128/jvi.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katz J M, Lim W, Buxton Bridges C, Rowe T, Hu-Primmer J, Lu X, Abernathy R A, Clarke M, Conn L, Kwong H, Lee M, Au G, Ho Y Y, Mak K H, Cox N J, Fukuda K. Antibody response in individuals infected with avian influenza A (H5N1) viruses and detection of anti-H5 antibody among household and social contacts. J Infect Dis. 1999;180:1763–1770. doi: 10.1086/315137. [DOI] [PubMed] [Google Scholar]
- 18.Kawaoka Y, Krauss S, Webster R G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge K F, Kawaoka Y, Webster R G. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75:2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y P, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. Avian-to-human transmission of H9N2 subtype influenza viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci USA. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, Y. P., W. Lim, V. Gregory, K. Cameron, M. Bennett, A. Klimov, K. Subbarao, M. Shaw, K. Shortridge, R. G. Webster, N. Cox, and A. Hay. Recent examples of human infection by animal and avian influenza viruses in Hong Kong. Proceedings of the Options for the Control of Influenza IV, in press.
- 22.Matrosovich M N, Krauss S, Webster R G. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281:156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 23.Nerome K, Kanegae Y, Shortridge K F, Sugita S, Ishida M. Genetic analysis of procine H3N2 viruses originating in southern China. J Gen Virol. 1995;76:613–624. doi: 10.1099/0022-1317-76-3-613. [DOI] [PubMed] [Google Scholar]
- 24.Otis K, Sidoli L, Bachman P A, Webster R G, Kaplan M M. Human influenza viruses in pigs: isolation of a H3N2 strain antigenically related to A/England/42/72 and evidence for continuous circulation of human viruses in the pig population. Arch Virol. 1982;73:103–108. doi: 10.1007/BF01314719. [DOI] [PubMed] [Google Scholar]
- 25.Peiris M, Yuen K Y, Leung C W, Chan K H, Ip P L S, Lai R W M, Orr W K, Shortridge K F. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 26.Pensaert M, Otis K, Vandeputte J, Kaplan M M, Bachmann P A. Evidence for the natural transmission of influenza A viruses from wild ducks to swine and its potential importance for man. Bull W H O. 1981;59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 27.Scholtissek C, Rhode W, von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 28.Scholtissek C, Burger H, Kistner O, Shortridge K. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 29.Shortridge K F, Webster R G, Butterfield W K, Campbell C H. Persistence of Hong Kong influenza virus variants in pigs. Science. 1977;196:1454–1455. doi: 10.1126/science.867041. [DOI] [PubMed] [Google Scholar]
- 30.Shortridge K F, Webster R G. Geographical distribution of swine (Hsw1N1) and Hong Kong (H3N2) influenza virus variants in pigs in southeast Asia. Intervirology. 1979;11:9–15. doi: 10.1159/000149006. [DOI] [PubMed] [Google Scholar]
- 31.Shortridge K F, Cherry A, Kendal A P. Further studies on the antigenic properties of H3N2 strains of influenza A viruses isolated from swine in southeast Asia. J Gen Virol. 1979;44:251–254. doi: 10.1099/0022-1317-44-1-251. [DOI] [PubMed] [Google Scholar]
- 32.Shortridge K F, Stuart-Harris C H. An influenza epicentre? Lancet. 1982;ii:812–813. doi: 10.1016/s0140-6736(82)92693-9. [DOI] [PubMed] [Google Scholar]
- 33.Shu L L, Lin Y P, Wright S M, Shortridge K F, Webster R G. Evidence for interspecies transmission and reassortment of influenza A viruses in pigs in southern China. Virology. 1994;202:825–833. doi: 10.1006/viro.1994.1404. [DOI] [PubMed] [Google Scholar]
- 34.Subbarao K, Klimov A, Katz J, Regenery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterisation of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 35.Webby R J, Swenson S L, Krauss S L, Gerrish P J, Goyal S M, Webster R G. Evolution of swine H3N2 influenza viruses in the United States. J Virol. 2000;74:8243–8251. doi: 10.1128/jvi.74.18.8243-8251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization Collaborating Centers for Reference and Research on Influenza. Concepts and procedures for laboratory based influenza surveillance, p. B-17–B-44. Atlanta, Ga: World Health Organization Collaborating Centers for Reference and Research in Influenza, Centers for Disease Control; 1982. [Google Scholar]
- 37.Yuen K Y, Chan P K S, Peiris M, Tsang D N C, Que T L, Shortridge K F, Cheung P T, To W K, Ho E T F, Sung R, Cheng A F B. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 38.Zhou N N, Senne D A, Landgraf J S, Swenson S L, Erikson G, Rossow K, Liu L, Yoon K J, Krauss S, Webster R G. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–8856. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]