Abstract
JCV, a small DNA virus of the polyomavirus family, has been shown to infect glial cells of the central nervous system, hematopoietic progenitor cells, and immune system lymphocytes. A family of DNA binding proteins called nuclear factor-1 (NF-1) has been linked with site-coding specific transcription of cellular and viral genes and replication of some viruses, including JC virus (JCV). It is unclear which NF-1 gene product must be expressed by cells to promote JCV multiplication. Previously, it was shown that elevated levels of NF-1 class D mRNA were expressed by human brain cells that are highly susceptible to JCV infection but not by JCV nonpermissive HeLa cells. Recently, we reported that CD34+ precursor cells of the KG-1 line, when treated with the phorbol ester phorbol 12-myristate 13-acetate (PMA), differentiated to cells with macrophage-like characteristics and lost susceptibility to JCV infection. These studies have now been extended by asking whether loss of JCV susceptibility by PMA-treated KG-1 cells is linked with alterations in levels of NF-1 class D expression. Using reverse transcription-PCR, we have found that PMA-treated KG-1 cells express mRNA that codes for all four classes of NF-1 proteins, although different levels of RNA expression were observed in the hematopoietic cells differentiated into macrophages. Northern hybridization confirms that the expression of NF-1 class D gene is lower in JCV nonpermissive PMA-treated KG-1 cells compared with non-PMA-treated cells. Further, using gel mobility shift assays, we were able to show the induction of specific NF-1–DNA complexes in KG-1 cells undergoing PMA treatment. The binding increases in direct relation to the duration of PMA treatment. These results suggest that the binding pattern of NF-1 class members may change in hematopoietic precursor cells, such as KG-1, as they undergo differentiation to macrophage-like cells. Transfection of PMA-treated KG-1 cells with an NF-1 class D expression vector restored the susceptibility of these cells to JCV infection, while the transfection of PMA-treated KG-1 cells with NF-1 class A, B, and C vectors was not able to restore JCV susceptibility. These data collectively suggest that selective expression of NF-1 class D has a regulatory role in JCV multiplication.
The nuclear factor-1 (NF-1) family of DNA binding proteins is encoded by four genes (NF-1 class A, B, C, and X [also known as NF-1 class D]) that are highly conserved from chickens to humans (17, 25, 26, 32, 41, 42). This protein family has been implicated in transcription of cellular genes (4, 9, 19, 23, 38, 40) and replication of several viruses (8, 13, 18, 20, 22, 37, 43, 46, 47), including JC virus (JCV) (1). NF-1 proteins were first isolated from HeLa cells and shown to contribute to adenovirus DNA replication in these cells (35, 36). All NF-1 family members have highly conserved N-terminal binding domains by which they bind to DNA, promote protein dimerization, and assist in virus replication (14, 32, 33). The C-terminal domains of NF-1 members vary considerably and function in transcriptional activation. A role for NF-1 proteins in cell type-specific gene expression and cellular differentiation also has been proposed (27, 48).
NF-1 proteins bind to a consensus sequence, 5′-TGG(A/C)N5GCCAA-3′, found within the promoter regions of cellular genes and those of several viruses (10, 15, 16, 28, 31). Different NF-1 family members bind to this sequence with equal affinity (26). Several NF-1 binding sites have been found within the promoter-enhancer region of JCV and are believed to be important for its replication in glial cells (1, 2, 47).
The screening of two human fetal brain cDNA libraries demonstrated that all NF-1 family members were expressed. NF-1 class D, however, was expressed at higher levels than those members of classes A, B, and C (45). Other studies have shown levels of NF-1 class D to vary in different cell types, with diminished expression in kidney and epithelial cells (5).
In a previous report, we provided evidence that JCV, a human polyomavirus, can infect cell types other than glial cells, including hematopoietic precursor cells and immune system lymphocytes (34). Additionally, we showed that the JCV-susceptible, undifferentiated progenitor cell line KG-1, when treated with the phorbol ester phorbol 12-myristate 13-acetate (PMA), differentiated into macrophage-like cells that lost JCV susceptibility (34). Although JCV binds to the surface of many cell types (49), it is unknown whether its tropism is due to the presence or absence of specific transcription factors and/or of a specific cellular receptor. Given that NF-1 expression has been implicated in JCV replication and cellular differentiation, we asked whether expression levels of NF-1 family members in undifferentiated KG-1 cells or PMA-treated KG-1 cells that have differentiated to macrophages correlate with the susceptibility of either cell type to JCV infection. Using reverse transcription (RT)-PCR, Northern blot, and gel mobility shift assays we have obtained evidence that, in hematopoietic cells, NF-1 class D expression is essential for JCV early transcription, which initiates viral multiplication.
MATERIALS AND METHODS
Hematopoietic progenitor cell lines.
The KG-1 and KG-1a cells lines, originating from the bone marrow of a patient with acute myelogenous leukemia, were purchased from the American Type Culture Collection (Manassas, Va.). When KG-1 cells were cultured in RPMI 1640 medium with 20% fetal bovine serum and PMA (2.5 μg/ml) added, they differentiated to cells with macrophage-like characteristics (24). However, KG-1a cells treated identically fail to differentiate.
Human primary stromal cells.
Human tonsillar stromal cells (HTSC) were obtained and processed by methods described previously (29).
Primary human astrocytes and SVG cell line.
All procedures involving human fetal tissue followed National Institutes of Health guidelines. Tissues from human fetal brain (gestational age, 7 to 10 weeks) were dissected, trypsinized, and resuspended in minimum essential medium and 10% fetal bovine serum (EMEM-10). The cell suspension was seeded into 162-cm2 flasks coated with collagen (100 μg/ml; Calbiochem, La Jolla, Calif.) and incubated at 37°C in a humidified air atmosphere containing 5% CO2. Ten to fifteen days after the cells were plated, microglial cells were detached from cultures by rotary shaking at 350 rpm for 90 min and removed from cultures. The remaining adherent cells were released by trypsinization, resuspended in EMEM-10, seeded into culture flasks, and passaged two to four times to obtain purified astrocytes (21).
Cells of the SVG line, established by immortalization of human fetal brain cells with an origin-defective mutant of simian virus 40 T protein (30), were cultured in EMEM-10 as described previously (30).
Isolation of cellular RNA.
Total cellular RNA was extracted from all cell types studied with an RNeasy Mini kit (Qiagen, Valencia, Calif.) by the procedure specified by the manufacturer. Briefly, samples were lysed and homogenized in the presence of denaturing guanidine isothiocyanate buffer. Ethanol was then added to the samples, and they were applied to a Qiagen RNeasy mini spin column. Sample RNA that bound to the column membrane was eluted in 30 μl of diethyl pyrocarbonate-treated distilled water.
RT-PCR and Southern blot analysis.
The RT of cellular template RNA to cDNA was done at 42°C in 20 μl of reaction mixture prepared according to the manufacturer's instruction (Perkin-Elmer, Foster City, Calif.). Reverse transcriptase enzyme was then denatured by incubating samples at 95°C for 5 min, and cDNA was amplified by PCR in 100 μl of reaction mixture containing 2.5 U of Taq polymerase and 25 pmol of each primer. Reaction products were amplified for 30 cycles, by use of the following program: 1 min at 94°C, 2 min at 50°C, and 3 min at 72°C. Amplification was completed with a 7-min extension period at 72°C.
The PCR primers used for NF-1 class A were derived from the NF-1 class L cDNA clone (39), primers for NF-1 class B were derived from the NF-1/Red-1 cDNA clone (12), primers for NF-1 class C were derived from the NF-1/CTF1 cDNA clone (42), and those for NF-1 class D were derived from the NF-1/AT1 cDNA (45). After blotting samples to membranes, each membrane was probed with 32P-labeled oligonucleotide fragments specific for the different NF-1 classes.
RT-PCR analysis was also used in KG-1 and KG-1 PMA-treated cells transfected with the plasmid containing NF-1 class D and subsequently incubated with JCV. The primer sets used were specific for the conserved region of the JCV genome coding for T protein and were previously described (34). PCR products were analyzed by use of a specific JCV 32P-labeled pM1Tc DNA probe (11).
Hybridization was carried out at 42°C for 20 h. The filters were washed twice in 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–0.1% sodium dodecyl sulfate (SDS), 1× SSPE–0.5% SDS, and 0.1× SSPE–0.5% SDS at room temperature for 20 min each. After washing, filters were exposed to Kodak BioMAX-MS film for several days at −80°C.
Northern blot and RNA probes.
Total cellular RNA, extracted as described above, was analyzed by Northern hybridization with a NorthernMax kit (Ambion Inc., Austin, Tex.) by the procedure specified by the manufacturer. Briefly, 20 μg of RNA was electrophoresed on a 1% agarose-formaldehyde gel and RNA was transferred to a positively charged nylon membrane. The membrane containing transferred RNA was prehybridized for 30 min at 68°C and then hybridized overnight at 68°C with an NF-1 class D RNA probe (107 cpm) (Lofstrand, Inc. Gaithersburg, Md.). After hybridization, the membrane was washed twice for 5 min at room temperature with low-stringency 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) wash solution and twice for 15 min at 68°C with high-stringency 0.1× SSC wash solution.
After autoradiography, the membrane was rehybridized with a human GAPDH probe to serve as an RNA control. Densitometric data analysis was performed to quantify NF-1 class D and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA signals using ImageQuant software. Relative NF-1 class D mRNA in the different cell types was determined as the ratio of NF-1 class D to GAPDH.
RNA probe was generated by RT-PCR from the 3′ region of the human NF-1 class D gene. To accomplish this, the PCR primers, previously described for RT-PCR analysis, were used to amplify specific sequences in total RNA from human fetal brain cells. After PCR amplification, the specific sequences were TA cloned into the pCR2.1 vector (Invitrogen, Carlsbad, Calif.) and then sequenced. The plasmid was linearized with HindIII before synthesis of the sense strand of the 32P-labeled RNA probe with the T7 promoter.
Northern analysis was also used in KG-1 PMA-treated cells transfected with the plasmids containing NF-1 class A, B, C, and D, respectively, and subsequently incubated with JCV. mRNA was analyzed by use of a specific JCV 32P-labeled DNA probe (11).
Preparation of nuclear extracts.
Untreated and PMA-treated KG-1 cells were washed three times with phosphate-buffered saline (PBS) (4°C), and whole-cell extracts were prepared by a modification of the procedure of Andrews and Faller (3). Briefly, cell pellets were rapidly frozen in dry ice and thawed at 27°C three times. Two volumes of cold buffer C (20 mM Tris-HCl, pH 7.9; 1.5 mM MgCl2; 420 mM NaCl; 0.2 M EDTA; 25% glycerol) containing protease inhibitors (dithiothreitol, 0.5 mM; phenylmethylsulfonyl fluoride, 0.5 mM; antipain, 5 mg/ml; leupeptin, 5 mg/ml; aprotinin, 5 mg/ml; pepstatin A, 5 mg/ml; chymostatin, 5 mg/ml) were added to each sample of disrupted cells. Samples were then centrifuged at 9,000 × g for 5 min, and supernatant fractions containing DNA binding proteins were harvested and stored in small aliquots at −80°C. Protein concentrations were determined by the method of Bradford (6).
Electrophoretic mobility shift assays.
Oligonucleotides with the nucleotide sequence of the intact NF-1 binding site (5′-ATGGCTGCCAGCCAAG-3) or a mutated version (5′-ATTACTGCCAGCTGAG-3; mutated NF-1 residues are shown in boldface type) were synthesized by Life Technologies, Inc. (Invitrogen). Oligonucleotides with sequences complementary to those given above were also synthesized. The oligonucleotides for both DNA strands of the authentic or mutated binding sites were annealed to form double-stranded structures, labeled with [γ-32P]ATP for 30 min at 37°C, and centrifuged through a Bio-Rad Biospin column at 3,000 rpm for 5 min. The labeled probe (200,000 cpm; 0.5 ng/ml) was then incubated with 10 μg of nuclear extract from either human fetal brain cells (HFBC), untreated KG-1 cells, or PMA-treated KG-1 cells in the presence or absence of a 250-fold excess of either unlabeled authentic oligonucleotide or unlabeled mutant oligonucleotide. The reaction was incubated at room temperature for 30 min and electrophoresed on a 6% polyacrylamide–Tris-glycine gel. The gel was dried, and samples were visualized by autoradiography with Kodak BioMAX-MS film.
Transfection and infection with JCV.
KG-1 and KG-1 PMA-treated cells were transfected in triplicate with pAT1 (44) (plasmid containing NF-1 class D) or calf thymus DNA (each 7 μg/0.5 × 106 cells), using N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP) cationic liposome-mediated transfection reagent (Roche Molecular Biochemicals, Indianapolis, Ind.) and 1,2-dimyristyloxypropyl-3-dimethyl-hydroxy ethyl ammonium bromide and cholesterol (DMRIE-C) cationic liposome-mediated transfection reagent (Invitrogen). KG-1 PMA-treated cells were also transfected with the NF-1 plasmids pCHAmNF1-A1.1, pCHAmNF1-B2, pCHAmNF1-C2, and pCHAmNF1-X2 (this NF1-X2 plasmid expresses what will be referred to NF-1 class D protein), which were a generous gift from R. Gronostajski. Each plasmid contains the cDNA coding region of one of the four classes of murine NF-1 (A, B, C, or D). Each NF-1 cDNA was subcloned into the pCHA vector to form a fusion protein with an N-terminal hemagglutinin tag. This tag has shown no effect on NF-1 DNA-binding or transactivation functions. The pCHA vector was derived from pCMVβ, which contains the cytomegalovirus immediate-early promoter minus the LacZ gene excised from flanking NotI restriction enzyme sites (7).
Following transfection, the cells were grown and selected in medium containing G418 (500 μg/ml) for 1 to 3 weeks. PMA was added to the medium each time it was changed.
After the selection, 106 cells were incubated with 4,000 hemagglutination units of JCV (MAD-4 strain). On various days (5, 16, 23, and 44 days) postinfection the cells were harvested, seeded onto glass coverslips, and fixed for immunofluorescence analysis. RNA was extracted from aliquots of these cells and used for RT-PCR amplification and Northern analysis of JCV T-antigen expression.
Immunofluorescence analysis of T and V antigens.
Mock (negative control)- and pAT-1-transfected KG-1 or PMA-treated KG-1 cells were incubated with JCV, placed on coverslips, treated with antibody to simian virus 40 T-antigen (PAB 416; Oncogene Science Inc., Boston, Mass.), diluted 1:10 in PBS, and incubated secondarily in the dark for 30 min at 4°C with fluorescein isothiocyanate-conjugated goat anti-mouse antibody (Jackson Immunoresearch, West Grove, Pa.), diluted 1:50 in PBS.
Cells were also tested for JCV virion (V) antigen by incubation overnight at 4°C with a mouse monoclonal antibody specific for its capsid protein (Novo Castra-Vector Laboratories, Burlingame, Calif.), diluted 1:20 in PBS. The secondary antibody and its usage were described above. The coverslips were washed twice in PBS and mounted on glass slides with 2% antifade (1,4-diazabicyclo-[2.2.2] octane; Sigma, St. Louis, Mo.) in 90% glycerol. Sample fluorescence was analyzed with a Zeiss ICM 405 epifluorescence microscope.
RESULTS
Expression of different NF-1 classes of mRNA in hematopoietic cells.
Several studies provided evidence that NF-1 binding sites in the promoter-enhancer region of the JCV genome are linked with JCV multiplication in glial cells (1, 2, 47). The NF-1 class D protein was implicated specifically in JCV expression in human brain-derived cells (44). Previously, we showed that the human hematopoietic precursor cell line, KG-1, when treated with PMA, lost its susceptibility to JCV infection (34). Furthermore, this loss of susceptibility to JCV in PMA-treated KG-1 cells correlated with the cell phenotype of increased adherence of cells to culture flask surfaces and increased expression of the monocyte surface marker, CD11b (data not shown), indicating differentiation of PMA-treated KG-1 cells to cells with macrophage-like characteristics.
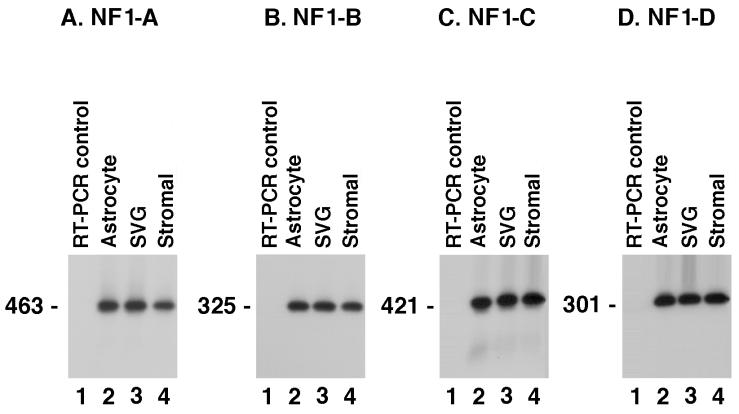
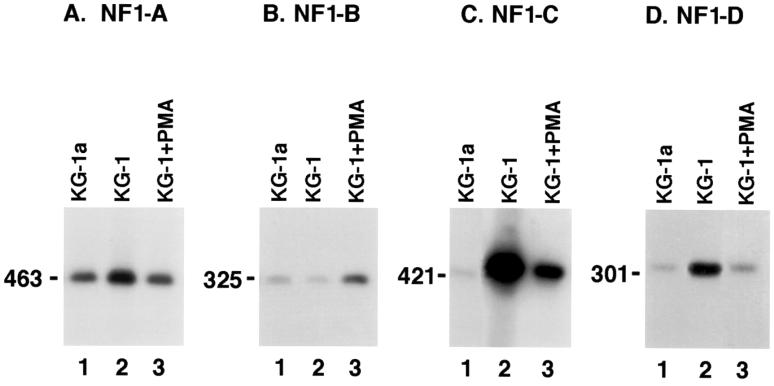
Total cellular RNA was extracted from HFBC, primary HTSC, and SVG cells, all of which are highly susceptible to JCV infection and therefore were used as positive controls for this study (Fig. 1). Total cellular RNA was also isolated from untreated KG-1 cells, PMA-treated KG-1 cells, and parental KG-1a cells to determine whether these cell types synthesized mRNA coding for the various classes of NF-1 protein (Fig. 2). Specific primers were used to identify sequences from each of the four NF-1 class proteins. The RT-PCR products obtained were Southern blotted and hybridized with probes to detect nucleotide sequences specific to each of the four NF-1 class members. Results of this work showed that the respective NF-1 class probes were specific for RT-PCR products generated by the various NF-1 specific primers (Fig. 1 and 2).
FIG. 1.
RT-PCR amplification and Southern blot analysis of human fetal brain cells (lanes 2), SVG cell line (lanes 3), and HTSC (lanes 4) using primers specific for NF-1 classes A, B, C, and D (panels A to D, respectively). All cell types examined expressed all four NF-1 classes at comparable levels. The cell types examined are those most highly susceptible to JCV infection. In each panel, lane 1 contains the negative control that corresponds to RT-PCR amplification without template. Results included are representative of three independent experiments.
FIG. 2.
RT-PCR amplification and Southern blot analysis of KG-1a (lanes 1), KG-1 (lanes 2), and PMA-treated KG-1 (30 days of treatment) (lanes 3) cells using primers for different NF-1 classes A, B, C, and D (panels A to D, respectively). Class-specific probes showed that the RT-PCR products were specific for their respective NF-1 class. All cell types examined expressed the four classes of NF-1, but at different levels. PMA-treated KG-1 cells (lanes 3) showed a downregulation of class A, C, and D and an increase in the expression of NF-1 class B relative to untreated KG-1 control cells. Results included are representative of three independent experiments.
In Fig. 2, all of the cell types examined expressed mRNA coding for the four classes of NF-1 protein, but class-specific levels varied in some cell types. Interestingly, compared to expression levels in untreated KG-1 cells, PMA-treated KG-1 cells expressed lower levels of NF-1 class A, C, and D proteins (Fig. 2A, C, and D, lanes 3) and an elevated level of NF-1 class B protein (Fig. 2B, lane 3). In untreated KG-1 cells, mRNA for NF-1 class C protein was expressed at a higher level than any of the other three NF-1 class members (Fig. 2C, lane 2).
Differential expression of NF-1 class D protein in PMA-treated KG-1 cells.
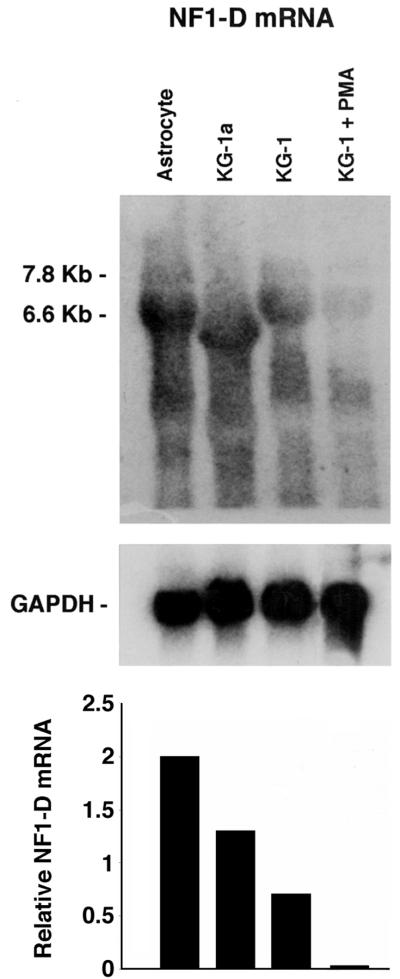
To further examine NF-1 class D expression in the hematopoietic cell and astrocytes derived from HFBC, Northern hybridization analysis was performed. Total RNA was extracted from the different cell types, and Northern blot analysis was accomplished with a 32P-labeled RNA probe for NF-1 class D. Figure 3 shows a difference in the amount of RNA from KG-1 PMA-treated cells and the other cell types (untreated KG-1, KG-1a and astrocytes derived from HFBC; all these cell types are susceptible to JCV infection) hybridizing to the NF-1 class D labeled probe. At least two related species (7.8 and 6.6 kb) of NF-1 class D RNA can be seen in the astrocytes derived from HFBC, KG-1 cells, and PMA-treated KG-1 cells, as previously described (7, 45).
FIG. 3.
Northern analysis of NF-1 class D mRNA expression in hematopoietic cell lines. Total RNA was extracted from primary astrocytes and KG-1a, untreated KG-1, and PMA-treated KG-1 (30 days of treatment) cells. A labeled specific NF-1 class D RNA probe was hybridized to each RNA sample. The marker on the left of the panel indicates the mRNA species. Hybridization signals were quantitated using ImageQuant as described in Materials and Methods and normalized for the intensity of the GAPDH signal. The relative RNA levels were then calculated and plotted.
The results of the Northern hybridization analysis suggest that the gene for NF-1 class D was more highly expressed in JCV susceptible cells than in PMA-treated KG-1 cells that had lost JCV susceptibility.
Induction of NF-1 binding in PMA-treated KG-1 cells.
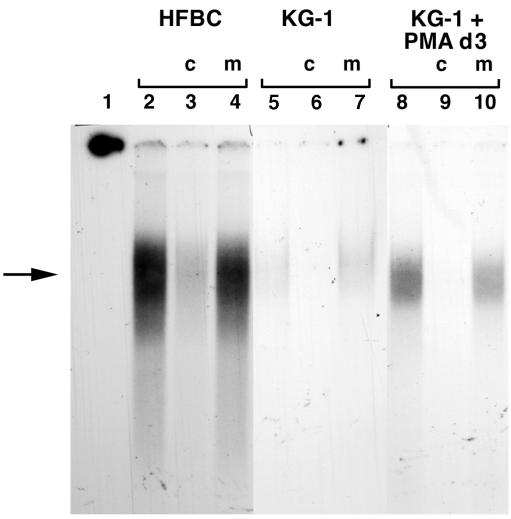
Electrophoretic mobility shift assays were performed to determine if there are differences in the NF-1 binding proteins of KG-1 cells and HFBC. Nuclear proteins were extracted at different times following KG-1 cell treatment with PMA, as described in Materials and Methods, from HFBC, untreated KG-1 cells, and PMA-treated KG-1 cells, and competitive gel shift experiments were performed. A gel-shifted band was detected when extracts from either HFBC or PMA-treated KG-1 cells were incubated with a probe containing the binding sequences specific for NF-1 proteins (Fig. 4, lanes 2 and 8). This shifted band depicts NF-1 protein bound specifically to its consensus binding site sequence. When subjected to competition with excess unlabeled NF-1 competitor, this band disappeared (Fig. 4, lanes 3, 6, and 9), but it remained when subjected to competition with excess mutant NF-1 competitor (Fig. 4, lanes 4, 7, and 10). A detectable gel-shifted band was present when extracts from untreated KG-1 cells were incubated with site-specific probes (Fig. 4, lanes 5 to 7).
FIG. 4.
Competitive gel shift analysis of the binding of nuclear proteins from HFBC (lanes 2 to 4), untreated KG-1 (lanes 5 to 7), and PMA-treated KG-1 (lanes 8 to 10) cells to a radiolabeled oligonucleotide containing an NF-1 binding site. Competitors were either unlabeled cold homologous oligonucleotide (c) or unlabeled mutant oligonucleotide (m). Lane 1 contains the probe without any added nuclear extract and shows migration off the gel. The arrow indicates the position of a specific gel-shifted band present in HFBC and induced in KG-1 cell extracts by PMA treatment, compared to the band from extracts from untreated KG-1 cells.
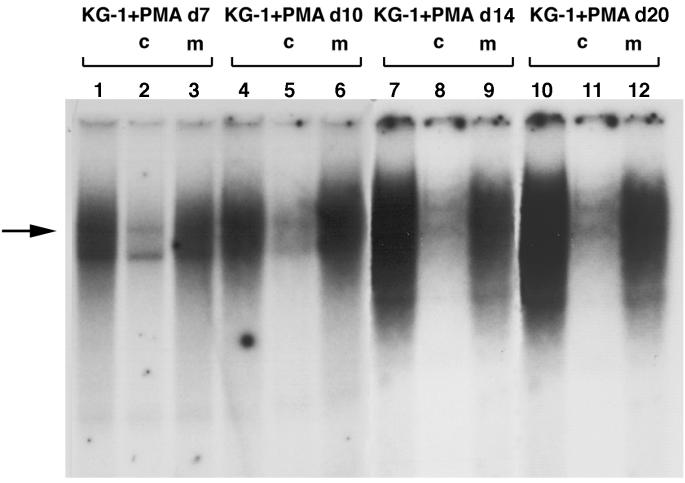
As the time interval of continuous PMA stimulation was increased, specific binding of NF-1 proteins also increased (Fig. 5, lanes 1, 4, 7, and 10). These results show that PMA treatment of KG-1 cells induces increased binding of NF-1 proteins to their specific DNA binding sequence. However, without specific antibodies for each of the four NF-1 classes, it is not possible to determine the precise composition of these NF-1–DNA complexes.
FIG. 5.
Competitive gel shift analysis at different time points (lanes 1 to 12) of the binding of nuclear proteins from PMA-treated cells to a radiolabeled oligonucleotide containing an NF-1 binding site. The arrow indicates the position of a specific gel-shifted band in KG-1 cells treated with PMA. The intensity of the band increased in nuclear extracts from cells exposed to PMA for a longer time.
Restoration of JCV susceptibility in PMA-treated KG-1 cells transfected with a plasmid containing NF-1 class D.
To determine whether expression of NF-1 class D protein has functional significance for JCV infectibility of cells, JCV-nonsusceptible PMA-treated KG-1 cells were transfected with an expression vector containing the NF-1 class D cDNA AT-1 sequence (44). After transfection and selection of cells expressing NF-1 class D using G418, PMA-treated KG-1 cells were adsorbed with JC virions. The presence of JC virus-positive cells was detected by immunofluorescence techniques at 5, 16, 23, and 44 days postincubation. In the PMA-treated cultures not transfected with pAT-1 class D expression vector, no immunofluorescence-positive cells were detected at any time point (Table 1). However, cells positive for both JCV T and V antigens were detected in cultures transfected with the pAT-1 expression vector for NF-1 class D at 23 and 44 days postincubation (Table 1).
TABLE 1.
Summary of the correlation between NF-1 class D expression and JCV susceptibility
| Cell type | NF-1 class Dab | JCV T- and V-Ag proteinac | Reference(s) |
|---|---|---|---|
| HFBC | +++ | +++ | 34; this study |
| HTSC | +++ | ++ | 34; this study |
| SVG | +++ | +++ | 30; this study |
| KG-1a | + | + | 34 |
| KG-1 | + | + | This study |
| KG-1 + PMA | ± | − | This study |
| KG-1 + PMA + NF-1 class D expression vector | ++ | + | This study |
+++, >50% of cells expressing; +, ∼20% of cells expressing; ±, barely detectable.
Indicates expression of mRNA as determined by RT-PCR and Northern blotting.
Indicates degree of immunocytochemical positivity of JCV antigen expression.
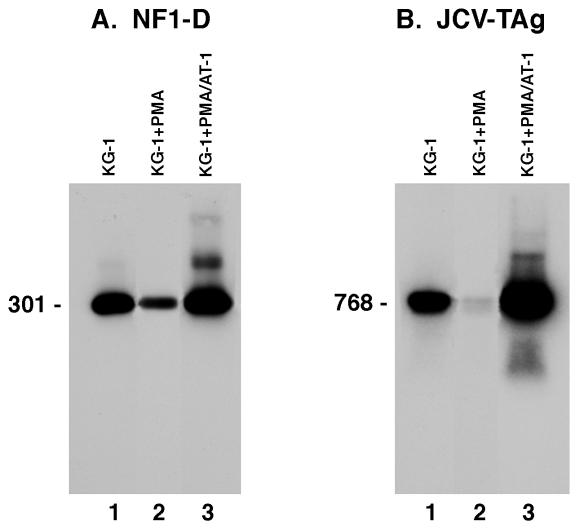
To further verify that susceptibility to JCV infection can be restored by transfection of NF-1 class D protein into nonsusceptible, PMA-treated KG-1 cells, these cells were transfected with the plasmid AT-1, which contains NF-1 class D cDNA, and then were adsorbed with JCV. RNA was extracted from both the NF-1 class D- or calf thymus DNA-transfected cultures, and mRNA sequences specific for NF-1 class D were amplified by RT-PCR with primer pairs specific for a conserved 301-bp sequence and subjected to Southern blot analysis. Figure 6A shows that the calf thymus control DNA-transfected cultures (lanes 1 and 2) demonstrated only endogenous levels of NF-1 class D, but the cells transfected with pAT-1 class D expression vector (lane 3) showed elevated levels of the 301-bp NF-1 class D sequence. The same cellular RNA samples were also amplified by RT-PCR with primer pairs coding for a 768-bp conserved region of the JCV genome coding for its T protein (Fig. 6B). When tested with a probe specific for this JCV T-antigen sequence, results showed that the JCV-infected, NF-1 class D-transfected, PMA-treated KG-1 cells were highly positive for the specific JCV T sequence (Fig. 6B, lane 3), while the JCV-infected, calf thymus DNA-transfected PMA-treated KG-1 cells showed only a barely detectable JCV T-protein mRNA band (Fig. 6B, lane 2). In these cells, however, there were no evidence of either JCV T- or V-antigen expression.
FIG. 6.
Comparative expression of nucleotide sequences specific to NF-1 class D or JCV T antigen in KG-1 or PMA-treated KG-1 cells. Cultures of KG-1 or PMA-treated KG-1 cells were transfected with either calf thymus DNA or a plasmid containing the NF-1 class D gene and subsequently adsorbed with JCV. RNA was extracted from these cells, and specific nucleotide sequences for NF-1 class D or JCV T antigen were reverse transcribed and amplified by PCR. (A) Results of expression of the NF-1 class D-specific sequence. Lanes 1 and 2 show the respective results from KG-1 or PMA-treated KG-1 cells transfected with calf thymus DNA and adsorbed with JCV; lane 3 shows the results from PMA-treated KG-1 cells transfected with the NF-1 class D containing plasmid and adsorbed with JCV. (B) Results of expression of the JCV T-antigen (TAg)-specific sequence. The order of the samples is the same as that in panel A.
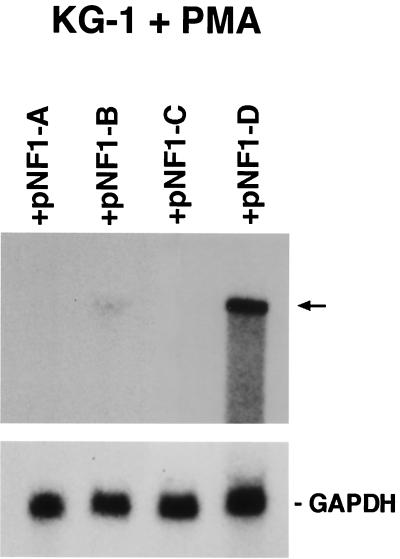
Furthermore, to verify that the observed effect is specific to the NF-1 class D factor, PMA-treated KG-1 cells were transfected with specific plasmids coding for the four NF-1 different classes (pCHAmNF1-A1.1, pCHAmNF1-B2, pCHAmNF1-C2, and pCHAmNF1-D2) and then adsorbed with JCV. RNA was extracted from all four transfected cultures, and Northern hybridization analysis, using a specific JCV DNA probe, was performed. The result confirmed the expression of JCV T-antigen proteins only in those cells that were transfected with NF-1 class D. Moreover, the experiment of control transfection using vectors that express NF-1 class A, NF-1 class B, and NF-1 class C confirmed that only the PMA-treated KG-1 cells that overexpress NF-1 class D protein were expressing specific JCV T-antigen proteins (Fig. 7).
FIG. 7.
Northern analysis of mRNA expression for JCV T antigen in PMA-treated KG-1 cells transfected with the plasmids containing NF-1 class A, B, C, and D, respectively. (The NF-1 plasmids pCHAmNF1-A1.1, pCHAmNF1-B2, pCHAmNF1-C2, and pCHAmNF1-X2 were described in Materials and Methods.) Specific radiolabeled probes for JCV and GAPDH were used to hybridize each RNA sample. The arrow on the right indicates the mRNA species for JCV T antigen.
DISCUSSION
It has been known for over 30 years that glial cells derived from human fetal brain are susceptible to the neurotropic polyomavirus JCV. Recently, we also showed that JCV can infect hematopoietic progenitor cells and cells of the immune system (34). We further showed that cells of the progenitor cell line KG-1 were susceptible to JCV infection but, when treated with the phorbol ester PMA, lost susceptibility and differentiated to cells with macrophage-like characteristics. The NF-1 class D protein has been associated with JCV's ability to infect certain cell types (45). Human fetal glial cells, the cells in which JCV replicates most efficiently, have been reported to express higher levels of NF-1 class D protein than NF-1 class C protein, while HeLa cells, nonsusceptible to JCV, were found to express higher levels of class C than class D proteins (45). Moreover, transfection of an expression clone of NF-1 class D, AT-1, in HeLa cells was able to activate the JCV early promoter in those cells that are normally nonpermissive to infection (44). Our findings that PMA-treated KG-1 cells lost susceptibility to infection by JCV prompted us to initiate work to determine whether this loss of JCV susceptibility might also be associated with reduced levels of NF-1 class D expression.
We have shown, by RT-PCR, that transcription levels of mRNA in the untreated KG-1 cells or PMA-treated KG-1 cells differed for all four classes of NF-1. In the untreated KG-1 controls, NF-1 class B transcripts were expressed at the lowest level, NF-1 class D and A transcripts were expressed at an intermediate level, and NF-1 class C transcripts were expressed at the highest level.
In PMA-treated KG-1 cells, mRNA levels of NF-1 classes A, C, and D decreased, while those of class B increased. Moreover, Northern analysis showed a lower expression of NF-1 class D in these cells. Together, these findings suggest that levels of NF-1 class D mRNA differ in specific cell types and that these differences may correlate with their susceptibility to JCV infection. The lower expression level of NF-1 class D protein is also observed in other cell types that are not permissive to JCV infection, such as primary human T lymphocytes and microglial cells (M. C. G. Monaco and E. O. Major, unpublished data). Other authors observed a low expression of NF-1 class D in human kidney, a site thought to be commonly infected by JCV since virus can be excreted in the urine (5). However, the exact cell types in the kidney that are susceptible to JCV infection have not been clearly identified. Also, the viral regulatory sequences of virion particles from the urine universally display the archetype arrangement of single, not repeat, nucleotides, which is the arrangement found most frequently in pathological tissues such as the brain (34, 44). Virions with the archetype arrangement of the regulatory region do not produce the early mRNA without the T protein provided in trans, nor are they infectious in kidney or glial cells in culture. It remains unknown what governs JCV excretion in the urine, as the virion found isolated in the urine cannot propagate in cell culture.
To determine whether JCV susceptibility could be restored in the nonsusceptible PMA-treated KG-1 cells, we transfected them with a plasmid containing the NF-1 class D cDNA sequence. Elevated expression of the transfected NF-1 class D gene was detected in the PMA-treated KG-1 cells (Fig. 6A, lane 3), and their susceptibility to JCV infection was restored as shown by immunostaining of JCV T antigen or capsid protein by specific antibodies (Table 1). Further evidence for restoration of JCV susceptibility in PMA-treated KG-1 cells was obtained by RT-PCR. A 768-bp sequence specific to mRNA of the JCV T antigen was found in extracts from NF-1-transfected, PMA-treated KG-1 cells (Fig. 6B, lane 3). In KG-1 control cells transfected with only calf thymus DNA as a control and treated with PMA and JCV, the expression of JCV T antigen was virtually undetectable by RT-PCR (Fig. 6B, lane 2). T antigen was never detected by immunofluorescence assay in non-NF-1 class D-transfected cells (data not shown). Northern analysis of the control experiments, using PMA-treated KG-1 cells transfected independently with NF-1 class A, B, C, or D, clearly demonstrated the detection of mRNA expression for JCV T antigen only in those cells that were transfected with NF-1 class D (Fig. 7).
By use of competitive gel shift experiments, we have shown that nuclear protein extracts from PMA-treated KG-1 cells contain a binding protein(s) that complexes specifically with an oligonucleotide NF-1 protein recognition site probe. However, nuclear protein extracts from untreated KG-1 cells complexed at lower levels to the DNA probe. Although we do not know the precise composition of the protein-probe complexes, this is the first evidence that binding patterns of the NF-1 family of proteins may change in hematopoietic progenitor cells as they differentiate to macrophage-like cells as a result of PMA treatment. The duration of PMA treatment may also modulate KG-1 cell differentiation and their NF-1 class protein binding patterns.
Our results also are consistent with those of other authors who detected altered expression of the NF-1 gene family members after phorbol ester-induced differentiation in cell lines from several leukemia patients (27). These authors also described the presence in nuclear extracts from these leukemic cell lines of a faster-migrating band consisting of NF-1–DNA complex, a result that is suggestive of hematopoietic differentiation and implies a possible role for NF-1 family members in mammalian development (27).
We cannot state unequivocally why PMA-treated KG-1 cells lose susceptibility to JCV infection. However, their loss of susceptibility may be linked to the quantity and/or quality of NF-1 class D protein they produce. Interestingly, JCV cell binding experiments demonstrated that KG-1 cells treated with PMA bind significantly more JCV than untreated KG-1 cells (W. J. Atwood, personal communication), indicating that virion-cell attachment is not a factor.
Our results have shown that untreated KG-1 cells have low susceptibility to JCV infection (34) and express a lower level of NF-1 class D protein than highly JCV-susceptible human fetal brain-derived glial cells. This already-low level of NF-1 class D protein in KG-1 cells was further reduced by PMA treatment that caused these cells to differentiate to cells with macrophage-like characteristics and lose JCV susceptibility. Perhaps PMA treatment of KG-1 cells reduced NF-1 class D protein expression below the threshold level required for JCV infection.
As shown by our electrophoretic mobility experiments, nuclear extracts from PMA-treated KG-1 cells bound specifically to an NF-1 nucleotide binding sequence probe and formed a complex with retarded electrophoretic mobility. Nuclear extracts from JCV-susceptible, non-PMA-treated KG-1 cells subjected to the same procedure, however, showed reduced levels of binding to the probe.
Coupled with previous data (34), our results showing restoration of JCV susceptibility to nonsusceptible PMA-treated KG-1 cells following their transfection with an NF-1 class D plasmid suggest that NF-1 class D protein levels strongly influence JCV infectibility of specific cell types.
ACKNOWLEDGMENTS
We gratefully acknowledge Maneth Gravell for critical review of the manuscript. We thank Diane Lawrence, Nazila Janabi, and Peter Jensen for insightful discussions. We also thank Jean Hou and Conrad Messam for providing technical assistance with the RNA probe and Janet Stephens for assistance with the figures. The NF-1 expression plasmids were generously supplied by R. Gronostajski (Department of Cancer Biology, Research Institute, Cleveland Clinic Foundation and Department of Biochemistry, Case Western Reserve University, Cleveland, Ohio).
REFERENCES
- 1.Amemiya K, Traub R, Durham L, Major E O. Interaction of a nuclear factor-1-like protein with the regulatory region of the human polyomavirus JC virus. J Biol Chem. 1989;264:7025–7032. [PubMed] [Google Scholar]
- 2.Amemiya K, Traub R, Durham L, Major E O. Adjacent nuclear factor-1 and activator protein binding sites in the enhancer of the neurotropic JC virus. J Biol Chem. 1992;267:14204–14211. [PubMed] [Google Scholar]
- 3.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cell. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoyama A, Tamura T, Mikoshiba K. Regulation of brain-specific transcription of the mouse myelin basic protein gene: function of the NF-1-binding site in the distal promoter. Biochem Biophys Res Commun. 1990;167:648–653. doi: 10.1016/0006-291x(90)92074-a. [DOI] [PubMed] [Google Scholar]
- 5.Apt D, Liu Y, Bernard H U. Cloning and functional analysis of spliced isoforms of human nuclear factor I-X: interference with transcriptional activation by NFI/CTF in a cell-type specific manner. Nucleic Acids Res. 1994;22:3825–3833. doi: 10.1093/nar/22.19.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhry A Z, Lyons G E, Gronostajski R M. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 8.Chong T, Apt D, Gloss B, Isa M, Bernard H. The enhancer of human papillomavirus type 16: binding sites for the ubiquitous transcription factors oct 1, NFA, TEF-2, NF-1 and AP-1 participate in epithelial cell-specific expression. J Virol. 1991;65:5933–5943. doi: 10.1128/jvi.65.11.5933-5943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtois S J, Lafontaine D A, Lemaigre F P, Durviaux S M, Rousseau G G. Nuclear factor-1 and activator protein-2 bind in a mutually exclusive way to overlapping promoter sequences and trans-activate the human growth hormone gene. Nucleic Acids Res. 1990;18:57–64. doi: 10.1093/nar/18.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeVries E, Van Driel W, Tromp M, Van Boom J, Van der Vliet P C. Adenovirus DNA replication in vitro: site-directed mutagenesis of the nuclear factor I binding site of the Ad 2 origin. Nucleic Acids Res. 1985;13:4935–4952. doi: 10.1093/nar/13.13.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frisque R J, Bream G L, Cannella M T. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil G, Smith J R, Goldstein J L, Slaughter C A, Orth K, Brown M S, Osborne T F. Multiple genes encode nuclear factor 1-like proteins that bind to the promoter for 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci USA. 1988;91:192–196. doi: 10.1073/pnas.85.23.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gloss B, Yeo-Gloss M, Meisterernst M, Rogge L, Winnacker E L, Bernard H U. Clusters of nuclear factor I binding sites identify enhancers of several papillomaviruses but alone are not sufficient for enhancer function. Nucleic Acids Res. 1989;17:3517–3532. doi: 10.1093/nar/17.9.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gounari F, De Francesco R, Schmitt J, van der Vliet P, Cortese R, Stunnenberg H. Amino-terminal domain of NF1 binds to DNA as a dimmer and activates adenovirus DNA replication. EMBO J. 1990;9:559–566. doi: 10.1002/j.1460-2075.1990.tb08143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gronostajski R. Analysis of nuclear factor I binding to DNA using degenerate oligonucleotides. Nucleic Acids Res. 1986;14:9117–9132. doi: 10.1093/nar/14.22.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gronostajski R, Adhya S, Nagata K, Guggenheimer R A, Hurwitz J. Site-specific DNA binding of nuclear factor I. Analysis of cellular binding sites. Mol Cell Biol. 1985;5:964–971. doi: 10.1128/mcb.5.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gronostajski R M. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 18.Hay R T. The origin of adenovirus DNA replication: minimal DNA sequence requirement in vivo. EMBO J. 1985;4:421–426. doi: 10.1002/j.1460-2075.1985.tb03645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hennighausen L, Siebenlist U, Danner D, Leder P, Rawlins D, Rosenfeld P, Kelly T J. High-affinity binding site for a specific nuclear protein in the human IgM gene. Nature. 1985;314:289–292. doi: 10.1038/314289a0. [DOI] [PubMed] [Google Scholar]
- 20.Hennighausen L, Fleckenstein B. Nuclear factor 1 interacts with five DNA elements in the promoter region of the human cytomegalovirus major immediate early gene. EMBO J. 1986;5:1367–1371. doi: 10.1002/j.1460-2075.1986.tb04368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janabi N, Chabrier S, Tardieu M. Endogenous nitric oxide activates prostaglandin F2 alpha production in human microglial cells but not in astrocytes: a study of interactions between eicosanoids, nitric oxide, and superoxide anion (O2−) regulatory pathways. J Immunol. 1996;157:2129–2135. [PubMed] [Google Scholar]
- 22.Jeang K-T, Rawlins D R, Rosenfeld P J, Shero J H, Kelly T J, Hayward G S. Multiple tandemly repeated binding sites for cellular nuclear factor 1 that surround the major immediate-early promoters of simian and human cytomegalovirus. J Virol. 1987;61:1559–1570. doi: 10.1128/jvi.61.5.1559-1570.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones K A, Kadonaga T K, Rosenfeld P J, Kelly T J, Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987;4:79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- 24.Koeffler H P, Bar-Eli M, Territo M C. Phorbol ester effect on differentiation of human myeloid leukemia cell lines blocked at different stages of maturation. Cancer Res. 1981;41:919–926. [PubMed] [Google Scholar]
- 25.Kruse U, Qian F, Sippel A E. Identification of a fourth nuclear factor I gene in chicken by cDNA cloning: NFI-X. Nucleic Acids Res. 1991;19:6641. doi: 10.1093/nar/19.23.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruse U, Sippel A E. Transcription factor nuclear factor I proteins from stable homo- and heterodimers. FEBS Lett. 1994;348:46–50. doi: 10.1016/0014-5793(94)00585-0. [DOI] [PubMed] [Google Scholar]
- 27.Kulkarni S, Gronostajski R M. Altered expression of the developmentally regulated NFI gene family during phorbol ester-induced differentiation of human leukemic cells. Cell Growth Differ. 1996;7:501–510. [PubMed] [Google Scholar]
- 28.Leegwater P A, Van Driel W, Van der Vliet P C. Recognition site of nuclear factor I, a sequence-specific DNA-binding protein from HeLa cells that stimulates adenovirus DNA replication. EMBO J. 1985;4:1515–1521. doi: 10.1002/j.1460-2075.1985.tb03811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lisignoli G, Monaco M C G, Facchini A, Toneguzzi S, Cattini L, Hilbert D M, Lavaroni S, Belvedere O, Degrassi A. In vitro cultured stromal cells from human tonsils display a distinct phenotype and induce B cell adhesion and proliferation. Eur J Immunol. 1996;26:17–27. doi: 10.1002/eji.1830260104. [DOI] [PubMed] [Google Scholar]
- 30.Major E O, Miller A E, Mourrain P, Troub R, De Widt E, Sever J. Establishment of a line of human glial cells that supports JC virus multiplication. Proc Natl Acad Sci USA. 1985;82:1257–1261. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meisterernst M, Gander I, Rogge L, Winnacker E L. A quantitative analysis of nuclear factor I/DNA interactions. Nucleic Acids Res. 1988;16:4419–4435. doi: 10.1093/nar/16.10.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meisterernst M, Rogge L, Foeckler R, Karaghiosoff M, Winnacker E L. Structural and functional organization of a porcine gene coding for nuclear factor I. Biochemistry. 1989;28:8191–8200. doi: 10.1021/bi00446a034. [DOI] [PubMed] [Google Scholar]
- 33.Mermod N, O'Neill E A, Kelly T J, Tjian R. The proline-rich transcriptional activator of CTF/NF-1 is distinct from the replication and DNA binding domain. Cell. 1989;58:741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 34.Monaco M C G, Atwood W, Gravell M, Tornatore C, Major E O. JC Virus infection of hematopoietic progenitor cells, primary B lymphocytes, and tonsillar stromal cells: implications for viral latency. J Virol. 1996;70:7004–7012. doi: 10.1128/jvi.70.10.7004-7012.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagata K, Guggenheimer R A, Hurwitz J. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc Natl Acad Sci USA. 1983;80:6177–6181. doi: 10.1073/pnas.80.20.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagata K, Guggenheimer R, Enomoto T, Lichy J, Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci USA. 1982;79:6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsson P, Hallberg B, Thornell A, Grundstroem T. Mutant analysis of protein interactions with a nuclear factor I binding site in the SL3-3 virus enhancer. Nucleic Acids Res. 1989;17:4061–4075. doi: 10.1093/nar/17.11.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowock J, Borgmeyer U, Peuschel A W, Rupp R A W, Sippel A E. The TGGCA-binding protein binds to the MMTV-LTR, the adenovirus origin of replication, and the BK virus enhancer. Nucleic Acids Res. 1985;13:2045–2061. doi: 10.1093/nar/13.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paonessa G, Gaunari F, Frank R, Cortese R. Purification of a NF-1-like DNA binding protein from rat liver and cloning the corresponding cDNA. EMBO J. 1988;7:3115–3123. doi: 10.1002/j.1460-2075.1988.tb03178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi P, Karsenty G, Roberts A B, Roche N S, Sporn M B, De Crombrugghe B. A nuclear factor 1 binding site mediates the transcriptional activation of a type I collagen promoter by transforming growth factor-beta. Cell. 1988;52:405–414. doi: 10.1016/s0092-8674(88)80033-3. [DOI] [PubMed] [Google Scholar]
- 41.Rupp R, Kruse U, Multhaup G, Gobel U, Beyreuther K, Sippel A. Chicken NFI/TGGCA proteins are encoded by at least three independent genes, NFI-A, NFI-B, NFI-C with homologues in mammalian genomes. Nucleic Acids Res. 1990;18:2607–2616. doi: 10.1093/nar/18.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santoro C, Mermod N, Andrews P, Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988;334:218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- 43.Shaul Y, Ben-Levy R, De Medina T. The high affinity binding site for nuclear factor I next to the hepatitis B virus S gene promoter. EMBO J. 1986;5:1967–1971. doi: 10.1002/j.1460-2075.1986.tb04451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinohara T, Nagashima K, Major E O. Propagation of the human polyomavirus, JCV, in human neuroblastoma cell lines. Virology. 1997;228:269–277. doi: 10.1006/viro.1996.8409. [DOI] [PubMed] [Google Scholar]
- 45.Sumner C, Shinohara T, Durham L, Traub R, Major E O, Amemiya K. Expression of multiple classes of the nuclear factor-1 family in the developing human brain: differential expression of two classes of NFI genes. J Neurovirol. 1996;2:87–100. doi: 10.3109/13550289609146542. [DOI] [PubMed] [Google Scholar]
- 46.Sundsfjord A, Johansen T, Flaegstad T, Moens U, Villand P, Subramani S, Traavik T. At least two types of control regions can be found among naturally occurring BK virus strains. J Virol. 1990;64:3864–3871. doi: 10.1128/jvi.64.8.3864-3871.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura T, Inoue T, Nagata K, Mikoshiba K. Enhancer of human polyoma JC virus contains nuclear factor-1-binding sequences: analysis using mouse brain nuclear extracts. Biochem Biophys Res Commun. 1988;157:419–425. doi: 10.1016/s0006-291x(88)80265-1. [DOI] [PubMed] [Google Scholar]
- 48.Ways D K, Qin W, Garris T O, Chen J, Hao E, Cooper D R, Usala S J, Parker P J, Cook P P. Effects of chronic phorbol ester treatment on protein kinase C activity, content, and gene expression in the human monoblastoid U937 cell. Cell Growth Differ. 1994;5:161–169. [PubMed] [Google Scholar]
- 49.Wei G, Liu C K, Atwood W J. JC virus binds to primary human glial cells, tonsillar stromal cells, and B-lymphocytes, but not to T lymphocytes. J Neurovirol. 2000;6:127–136. doi: 10.3109/13550280009013156. [DOI] [PubMed] [Google Scholar]