Abstract
Potato virus A (PVA) particles were bombarded with thermally activated tritium atoms, and the intramolecular distribution of the label in the amino acids of the coat protein was determined to assess their in situ steric accessibility. This method revealed that the N-terminal 15 amino acids of the PVA coat protein and a region comprising amino acids 27 to 50 are the most accessible at the particle surface to labeling with tritium atoms. A model of the spatial arrangement of the PVA coat protein polypeptide chain within the virus particle was derived from the experimental data obtained by tritium bombardment combined with predictions of secondary-structure elements and the principles of packing α-helices and β-structures in proteins. The model predicts three regions of tertiary structure: (i) the surface-exposed N-terminal region, comprising an unstructured N terminus of 8 amino acids and two β-strands, (ii) a C-terminal region including two α-helices, as well as three β-strands that form a two-layer structure called an abCd unit, and (iii) a central region comprising a bundle of four α-helices in a fold similar to that found in tobacco mosaic virus coat protein. This is the first model of the three-dimensional structure of a potyvirus coat protein.
Potato virus A (PVA) is a filamentous virus which belongs to the genus Potyvirus (family Potyviridae), the largest and one of the most economically important groups of plant viruses. PVA is serologically related to Potato virus Y (PVY), the type member of the genus (8). The potyvirus coat protein is a multifunctional protein which plays a role in virus transmission by vector aphids (4), virion formation, and virus movement (9).
The PVA coat protein comprises 269 amino acids and has a calculated Mr of 30,257 (26). PVA has 73 to 78% amino acid sequence homology to the coat proteins of other potyviruses (25, 26); the greatest differences are found in the N-terminal region, as among potyviruses in general (32, 33). Although detailed information on the genome organization and the deduced amino acid sequences of the proteins encoded by many potyviruses is available (22, 27), little is known about the conformation of the coat protein subunits and their packing arrangement in the virion.
Both the N- and C-terminal residues of potyviruses are thought to be surface located. When potyvirus particles are treated briefly with trypsin, 20 to 30 amino acids are removed from the N and C termini of the coat protein subunit, leaving infective trypsin-resistant core particles that have coat protein subunits trimmed to 216 to 218 amino acid residues (33). Epitope-mapping studies with monoclonal antibodies (MAbs) against PVA have shown that the N-terminal octapeptide of the coat protein is immunodominant (2), as has been found for other potyviruses (31, 33). Moreover, the N-terminal triplet DAG is implicated in transmissibility by vector aphids (3, 5, 21).
To assess the in situ spatial organization of the PVA coat protein, we subjected intact PVA particles to tritium bombardment. This approach has been used successfully to probe directly the accessible surfaces of proteins in macromolecular assemblies such as those of the plant viruses tobacco mosaic virus (TMV) (genus Tobamovirus) (16) and potato virus X (PVX) (genus Potexvirus) (7), the ribosome (1), and membrane structures (30, 34).
In this method, a specially prepared target, e.g., the virus particle preparation, is maintained at the temperature of liquid nitrogen and exposed to a beam of tritium atoms produced by catalytic dissociation of molecular tritium at the surface of a tungsten filament heated to 2,000 K. The initial energy of incident particles is just sufficient for abstraction of hydrogen atoms from the target molecule and substitution of tritium for hydrogen. Even a single unreactive collision instantly reduces the energy of the tritium atom below the threshold required for displacing hydrogen. Thus, the label is incorporated at the first impact or is not incorporated at all. The distribution of label along the polypeptide chain characterizes the steric accessibility to the bombarding tritium atoms; thus, the most-exposed parts of the macromolecule are the most intensely labeled (29). The comprehensive information on the accessible surfaces of macromolecules obtained by this method, in conjunction with theoretical approaches for predicting the topology of a protein, allows the construction of a three-dimensional model for the structure of the protein. It should be noted that a free globular protein (for example, myoglobin) which is randomly oriented in the target is uniformly irradiated in all directions, whereas a protein in an intact virus particle is effectively irradiated only in the region that is exposed at the outer surface of the virus particle.
The aims of this work were to determine the distribution of tritium label in the coat protein molecules in assembled PVA particles and to combine this information with stereochemical analysis for predicted topology to produce a three-dimensional model for the tertiary structure of the PVA coat protein subunit.
MATERIALS AND METHODS
Reagents.
Trypsin treated with l-1-tosylamide-2-phenylchloromethylketone (TPCK-trypsin) and sodium diethyldithiocarbamate (DIECA) were obtained from Sigma (St. Louis, Mo.); diisopropylfluorophosphate (DFP) was obtained from Merck (Darmstadt, Germany); and reagents for preparing gels were from Bio-Rad Laboratories (Hercules, Calif.). Water was obtained using a Milli-Q System (Millipore, Bedford, Mass.). All other chemicals were of analytical grade.
Virus purification.
PVA isolate B11 (provided by F. Rabenstein of the Institute of Plant Breeding Research and Pathogen Diagnostics, Aschersleben, Germany) was purified from systemically infected leaves of Nicotiana tabacum cv. Samsun harvested 3 to 4 weeks after inoculation. Leaves were pulverized in a Waring blender with 0.05 M phosphate buffer, pH 8.0, containing 0.01 M DIECA, 0.005 M EDTA, and 1% sodium sulfite (2 ml of buffer per g of leaf tissue). After low-speed centrifugation (LSC) (8,000 × g, 8°C, 20 min), the supernatant was stirred for 3 h at 4°C with Triton X-100 (1%, vol/vol); then 50 g of polyethylene glycol 6000 and 12 g of NaCl per liter were added, and the mixture was stirred at 4°C for 1.5 h. The precipitate was sedimented by LSC and resuspended with 0.1 M borate buffer, pH 8.0. After a further LSC to remove insoluble material, the preparation was subjected to high-speed centrifugation through a 20% (vol/vol) sucrose cushion (150,000 × g, 5°C, 2.5 h). The pellet was resuspended in 0.05 M borate buffer, pH 8.0, and subjected to a further cycle of differential centrifugation. Then the pellet was resuspended in 0.05 M borate buffer, pH 8.0. Virus concentration was estimated spectrophotometrically, assuming an A260 (0.1%; 1-cm path length) of 2.8. The purity of the preparations was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (19) and immunoblotting tests with the PVA-specific MAbs A5B6 and A3H4 as described previously (2).
PVA tritium bombardment.
PVA labeling with hot tritium atoms was performed as described previously (6, 16). Briefly, the tritium label was introduced by means of bombardment with a beam of tritium atoms heated to 2,000 K (13). A PVA suspension (1.0 ml at 1 to 2 mg/ml) in 0.05 M borate buffer, pH 8.0, was spread as a thin film on the inner surface of a glass vessel that had been precooled with liquid nitrogen. Liquid nitrogen was also used for cooling the vessel during the process of irradiation. Labeling conditions were three 15-s exposures to tritium gas (T2) carried out by filling the vessel with T2 to a pressure of 0.5 Pa each time. These conditions yielded virus preparations with a specific radioactivity of ∼500 Bq/μg of protein. After labeling, the samples were concentrated by centrifugation (105,000 × g, 5°C, 100 min).
Isolation of the labeled PVA coat protein.
Protein was isolated from labeled PVA by the LiCl method (17). The purity of the labeled protein samples was checked by SDS-PAGE (19).
CD spectroscopy.
Circular dichroism (CD) spectra were measured in a modified Jobin-Yvon (Longiomeau, France) Mark V dichrograph interfaced with an IBM-compatible computer. Measurements were made over 200 to 250 nm at 25°C in a 1-mm quartz-Suprasil cuvette (Hellma, Müllheim, Germany). PVA coat protein was diluted in 10 mM ammonium-bicarbonate buffer, pH 7.8, to concentrations of 90 and 180 μg/ml. The PVA coat protein absorption coefficient (A280 [0.1%; 1-cm path length]), calculated according to the work of Gill and von Hippel (15), was found to be 1.0 optical unit.
Tryptic digestion of the labeled PVA coat protein.
Tryptic digestion was done at an enzyme/substrate ratio of 1:50 (wt/wt) for 4 h at 37°C in 0.01 M ammonium bicarbonate buffer, pH 7.8. Samples (1 to 2 μg) were removed before and after the reaction to evaluate the degree of hydrolysis. The reaction was stopped by addition of 1.33 μg of trypsin inhibitor (7.2 nmol of DFP).
Reverse-phase high-performance liquid chromatography (HPLC) of tryptic peptides.
Samples dissolved in the starting eluent (0.1% trifluoroacetic acid [TFA]) in water) were separated in a Beckman 344 chromatographic system with a 165 UV-VIS dual-channel spectrophotometric detector combined with a Gilson 201 fraction collector on an Ultrasphere octyldecyl silane (ODS) column (particle diameter, 5 μm; column height, 250 mm; inside diameter, 4.6 mm; Beckman). All separations were performed at room temperature (20 ± 3°C) using linear gradients of acetonitrile in water containing 0.1% TFA (0 to 60% in 60 min and 60 to 80% in 10 min).
Analytical methods.
Tryptic peptides were hydrolyzed as described by Tsugita and Scheffler (35), and amino acid analysis was carried out on a Hitachi-835 analyzer (Tokyo, Japan) in the standard mode for protein hydrolysate analysis with cation-exchange separation and ninhydrin postcolumn derivatization. A Packard Radiomatic 150TR flow scintillation analyzer with an online amino acid analyzer was used for simultaneous analysis of amino acid content and levels of tritium radioactivity. The LSC cocktail Hionic-Fluor was used for counting. In our experiments using a model system, 150 dpm could be detected routinely in amino acid peaks with this cocktail, corresponding to a molar radioactivity of about 0.2 Bq/nmol. Optical densities at 570 and 440 nm and radioactive counts were measured and processed simultaneously using MultiChrom for Windows software (Ampersand Ltd., St. Petersburg, Russia). The specific radioactivity of each given amino acid residue was normalized for the probability of including the label (as described by Gedrovich and Badun [14]). Results are given as specific radioactivity for each amino acid, obtained by dividing the total radioactivity by the number of residues of that amino acid present in the peptide. The short N-terminal amino acid sequences of the tryptic peptides were determined by Edman degradation in combination with mass spectrometry. A matrix-assisted laser desorption ionization (MALDI) linear time-of-flight mass spectrometer (model REFLEX II; Bruker-Franzen Analytic GmbH, Bremen, Germany) was used throughout the study to check the identity and purity of the HPLC fractions, determine mass spectra, and select the desired fragment for subsequent N-terminal amino acid sequencing analysis.
RESULTS AND DISCUSSION
Characterization of PVA preparations.
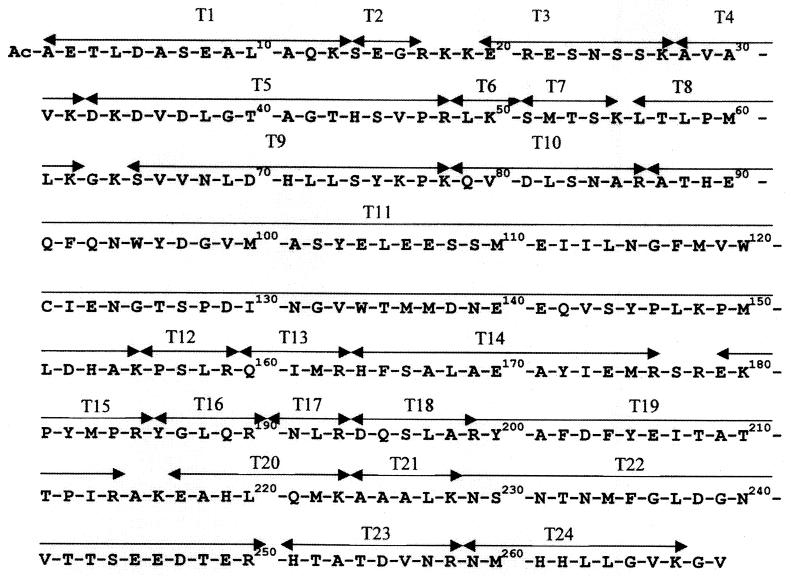
The coat protein from the intact PVA particles migrated as a single band in SDS–12% PAGE, gave one band with PVA-specific MAbs in an immunoblot, and reacted with anti-PVA MAb A5B6, which recognizes the N-terminal octapeptide of the PVA coat protein. The N-terminal sequence of the PVA coat protein was confirmed by sequence analysis and mass spectrometry. These tests showed that the N terminus of the coat protein was blocked and the C-terminal region was preserved. To determine the type of N-blocked group, the coat protein was hydrolyzed by trypsin, and the tryptic peptide T1 (Fig. 1) was isolated by HPLC. Analysis of peptide T1 (amino acids 1 to 13) by partial sequencing and mass spectrometry showed that the N-terminal alanine of the protein was acetylated.
FIG. 1.
Amino acid sequence of PVA coat protein according to Puurand et al. (26) (EMBL accession no. ZZ21670). T1 to T24 are tryptic peptides, marked by double-headed arrows.
Predictions of PVA coat protein-secondary structure elements.
The secondary structure of the PVA coat protein was predicted by a stereochemical method that takes into account the sequence patterns of standard structures in proteins. The analysis predicts that for amphiphilic β-strands the sequence periodicity of polar and nonpolar amino acids must be arranged with an alternating pattern (20), whereas for amphiphilic α-helices, the sequence periodicity must place a nonpolar residue every three or four positions (20, 28). Stereochemical analysis showed that there should be at least one hydrophobic residue per turn of the α-helix and that side chains of these residues should be arranged so as to form a continuous hydrophobic stripe on one side of the α-helix. Such minimal hydrophobic stripes can be of three types, those formed by hydrophobic residues in positions 1, 4, 8, 11, 15, 18, etc., positions 1, 4, 8, 12, 15, 19, 23, etc., or positions 1, 5, 9, 13, etc. (10). In practice, sequences coding for α-helices usually contain more hydrophobic residues than those necessary to form a minimal hydrophobic stripe.
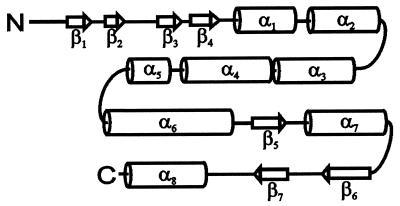
Analysis of the PVA coat protein amino acid sequence using this approach has led us to the conclusion that the protein should consist of three structural regions, the N-terminal region (amino acids 1 to 60), the central region (amino acids 61 to 180), and the C-terminal region (amino acids 181 to 269) (Fig. 2). The central region comprises five consecutive α-helices: α2 (amino acids 62 to 76), α3 (88 to 104), α4 (106 to 124), α5 (129 to 137), and α6 (150 to 176). The C-terminal region was predicted to consist of two α-helices, α7 (amino acids 194 to 210) and α8 (252 to 268), and three β-strands, β5 (182 to 188), β6 (218 to 227), and β7 (235 to 241). The first eight amino acids of the N-terminal region should be unstructured, followed by four consecutive β-strands: β1 (amino acids 9 to 13), β2 (17 to 20), β3 (28 to 32), and β4 (35 to 40) and α-helix α1 (45 to 57). From these measurements and the structural predictions, it follows that the PVA coat protein is highly ordered, with eight α-helical regions comprising 50%, and seven β-strands comprising 16%, of the protein sequence.
FIG. 2.
Schematic drawing of the coat protein subunit secondary structure of PVA, showing the distribution of α-helices (indicated by cylinders) and β-strands (indicated by arrows).
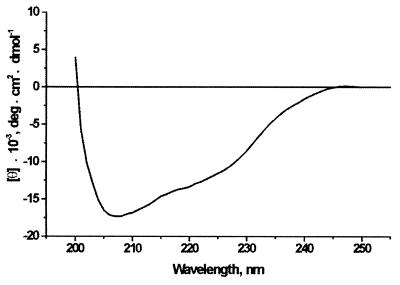
To support these predictions we measured the CD spectrum of PVA coat protein in the region of 200 to 250 nm (Fig. 3). Since the aggregation state of PVA coat protein in 10 mM ammonium bicarbonate buffer is not known, and the sample absorption spectra had a small amount of turbidity at a λ of >310 nm, only the α-helical content of the protein could be reliably determined using the Greenfield-Fasman equation (18); it was found to be 46%, which is in good agreement with the predicted value (50%).
FIG. 3.
CD spectrum of PVA coat protein. The spectrum of LiCl-purified protein (90 μg/ml) in 10 mM ammonium bicarbonate buffer, pH 7.8, was measured in a 1-mm cuvette at 25°C.
It is noteworthy that the borders of the predicted secondary-structure elements either coincide exactly with the sites of trypsin hydrolysis or are located in close proximity to these sites (Fig. 1 and 2): for example, peptide T1 (amino acids 1 to 13) and the β1-strand (9 to 13), peptide T4 (amino acids 28 to 32) and the β3-strand (28 to 32), peptide T9 (amino acids 64 to 78) and the α2-helix (62 to 76), the trypsin-resistant core peptide T11 (amino acids 87 to 155) and the α-helical block (α3-, α4-, and α5-helices and loops, amino acids 88 to 149), and finally, the two peptides T23 and T24 (amino acids 251 to 267) and the α8-helix (252 to 268). This interesting coincidence of the hydrolysis sites with the borders of the predicted secondary-structure elements may testify to the importance of the specific location of some amino acid residues and the protein sequence segments for PVA coat protein tertiary-structure formation.
Tritium bombardment of PVA particles and analysis of label distribution in the protein.
Freshly prepared, rapidly frozen, vitrified aqueous suspensions of virus particles were bombarded with a beam of hot tritium atoms. To determine the label distribution within the tritiated protein, two approaches may be used, each with its merits and demerits. Direct sequencing of the tritiated peptides may give specific labeling of each amino acid residue. In this case, reverse phase HPLC analysis of the amino acid phenylthiohydantoin (PTH) derivatives with simultaneous analysis of the level of tritium radioactivity is necessary. However, since there are tritium labels 3 orders of magnitude more labile (exchangeable) (bonds O-3H, N-3H, and S-3H) than the informative label (bonds C-3H), its use is questionable. Moreover, it is impossible to eliminate all exchangeable label by dialysis or chromatography of a protein. A second approach is fragmentation of the protein into relatively short peptides, each of which has a minimum number of duplicated amino acids (large peptides may be further enzymatically cleaved using an enzyme with a different specificity), followed by amino acid analysis with simultaneous radioactivity measurements. This approach obviates the problem of removing labile label, since under the conditions of acid hydrolysis used, it will be removed in full. In this work, we used this approach, which has been widely adopted in a number of recent studies (6, 16, 30). Here we present the results of the tryptic digestion of tritium-labeled PVA protein. The peptides obtained by trypsin hydrolysis are mainly rather short: most have fewer than 10 residues, six peptides have 13 to 22 residues, and only one peptide (peptide T11, amino acids 87 to 155) consists of 69 residues. These results may be considered a first approximation, but they are sufficient for modeling the three-dimensional structure of the protein. They may be further refined by using proteases with different specificities to improve the resolution.
Using HPLC, we isolated 24 peptides (Fig. 1) comprising approximately 92% of the entire coat protein sequence. Three tryptophan residues, cysteine 121, arginine 176, and five dipeptides, KK (amino acids 18 and 19), GK (63 and 64), SR (177 and 178), AK (215 and 216) and GV (268 and 269), were not identified. In addition, the specific radioactivity values for histidines at positions 71, 89, 153, 164, 261, and 262 could not be obtained because of very high levels of ammonium radioactivity in this region of the chromatograms.
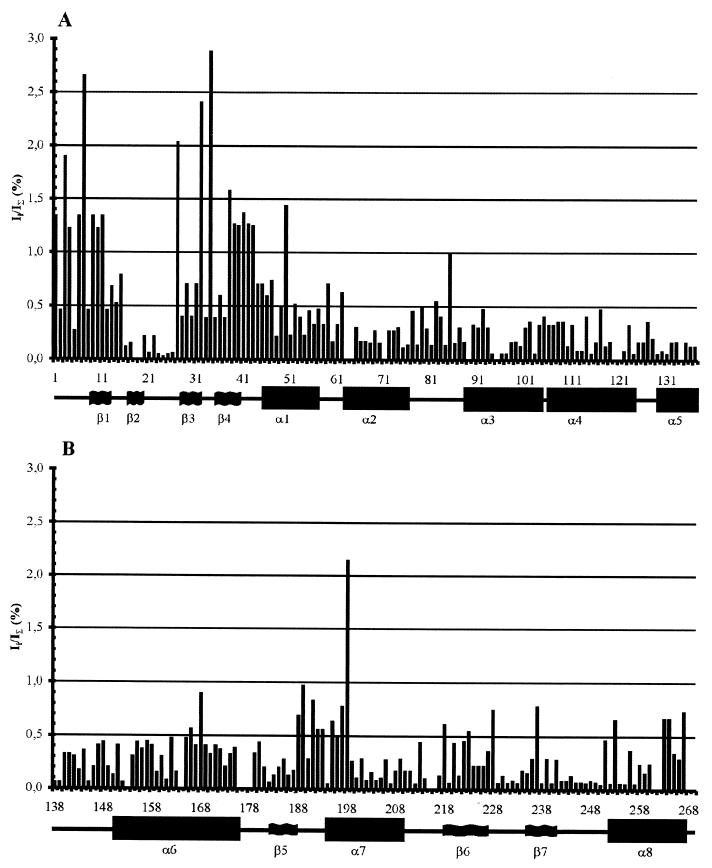
Figure 4 shows the tritium label distribution along the polypeptide chain. The experimental data are the average values of tritium labeling for each type of amino acid residue in each tryptic peptide. Thus, for example, peptide T1 (amino acids 1 to 13) contains four alanine residues (amino acids 1, 6, 9, and 11 [Fig. 1]), and the specific radioactivity values are considered equal, each constituting one-quarter of the total value of the alanine tritium label in peptide T1. Therefore, the contribution of an individual residue cannot be determined unless it is the only one of its kind in the peptide. It is evident from Fig. 4 that the profile of accessibility for tritium labeling of the coat protein amino acids has a number of prominent features. The N-terminal region (the first 50 amino acids) contains about 40% of the total stable tritium label incorporated into the protein. In this region, the most accessible parts were amino acid residues 1 to 15 (the T1 peptide and part of T2) and amino acid residues 27 to 50 (the C-terminal lysine of the T3 peptide and peptides T4 to T6). The trypsin-resistant core peptide T11 (amino acids 87 to 159) from the central part of the protein contains the lowest level of tritium labeling. In addition, surprisingly, the C-terminal region of the coat protein had a relatively low level of tritium labeling. Also, Fig. 4 shows that, with the exception of A85 and L168, the long central part of the coat protein molecule (residues 60 to 180) contains rather small and uniform amounts of the label. Average specific radioactivity values in α-helices α2 to α5 (residues 62 to 76, 88 to 104, 106 to 124, and 129 to 137, respectively) are significantly lower than those for other parts of the molecule (Table 1). Similarly low levels of the label were incorporated into the α6-helix (residues 150 to 176) (excluding L168), indicating that these segments of PVA coat protein are located in the internal part of the subunit in the virion.
FIG. 4.
Distribution of the tritium labels in the coat protein (A and B) upon tritium bombardment of PVA particles. Ii/IΣ (%), ratio of the specific radioactivity of the given amino acid (Ii) to the total specific radioactivity of the protein (IΣ) expressed as a percentage. The number of the amino acid residue (from the N terminus) is given along the abscissa, and the predicted α-helices and β-strands are indicated below.
TABLE 1.
Average specific radioactivity values for amino acids in the predicted secondary structure elements
| Unit | Region | ΣIi/na |
|---|---|---|
| β1 | A9–K13 | 19.7 |
| β2 | R17–E20 | 3.7 |
| β3 | A28–K32 | 18.0 |
| β4 | D35–T40 | 17.8 |
| α1 | S45–T57 | 10.3 |
| α2 | K62–K76 | 3.9 |
| α3 | T88–E104 | 3.9 |
| α4 | E106–N124 | 4.1 |
| α5 | D129–M137 | 2.2 |
| α6 | M150–R176 | 6.5 |
| β5 | Y182–L188 | 4.7 |
| α7 | D194–T210 | 7.1 |
| β6 | A218–L227 | 6.4 |
| β7 | F235–V241 | 5.4 |
| α8 | T252–G268 | 6.5 |
ΣIi, the sum of specific radioactivities of amino acid residues in the secondary-structure element; n, number of amino acids in the secondary-structure element.
Spatial reconstruction.
To model the PVA coat protein three-dimensional structure, we used an approach based on the known principles of protein structure (for a review, see, e.g., references 10 and 12). In proteins, the packing of α-helices and/or β-sheets is restricted by both the principle of close packing and the chemical nature of the side chains. In globular proteins, α-helices and β-strands are usually amphiphilic and pack so that their hydrophobic faces are buried in a hydrophobic core and the polar side chains are accessible to the solvent. The principle of close packing restricts possible packing angles (Ω) between α-helices and/or β-strands to predetermined values. The three-dimensional arrangement of α-helices and/or β-strands depends also on the length and conformation of the loops between them. Another important structural feature used in our prediction studies is that in proteins the α-helices and/or β-strands adjacent along the chain often form a few well-defined types of super-secondary structures or structural motifs, for example, the four-α-helix bundles and abCd-units. The three-dimensional structural modeling of PVA coat protein also included data on the topography of the protein. The label-accessible surface of the coat protein obtained by tritium planigraphy was used as a criterion for choosing the optimal packing of α-helices and β-strands to ensure that the tritium-accessible parts of the protein were surface located in the model and the inaccessible ones were in the interior of the protein.
Taking into account these structural features, the very low level of tritium labeling of α-helices 2 to 6 (Table 1), and the volume occupied by 1 protein subunit in the PVA virion, we suggest that four long α-helices (α2 [amino acids 62 to 76], α3 [88 to 104], α4 [106 to 124], and α6 [150 to 176]) located in the central part of the PVA coat protein are packed into a four-α-helical bundle at packing angles (Ω) of ≅20° (Fig. 5). The distribution of these α-helices along the sequence and their lengths are very similar to those of the TMV coat protein. So it looks very likely that the three-dimensional arrangement is also similar to that of the TMV coat protein (23). This region contains also the short α5-helix (amino acids 129 to 137). This domain may play an important role in oligomerization of the PVA coat protein.
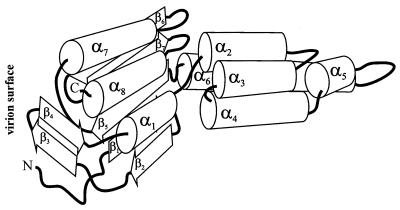
FIG. 5.
Schematic representation of the overall fold of the PVA coat protein. α-Helices are shown as cylinders, and β-strands are shown as angled open arrows, with thick solid lines indicating interconnecting loops and the N and C termini of the polypeptide chain.
The C-terminal region has been predicted to consist of two α-helices, α7 (amino acids 194 to 200) and α8 (252 to 268), and three β-strands, β5 (182 to 188), β6 (218 to 227), and β7 (235 to 241), organized in βαββα order. In proteins, polypeptide chains having such a topology are folded into the so-called abCd unit (11). Thus, we conclude that the C-terminal domain is a two-layer structure in which one layer is formed by three β-strands and the other by two α-helices. Similar structural units have been found in many RNA-binding proteins.
In contrast to predictions of the central and C-terminal regions, modeling of the N-terminal part was rather difficult because of the long connecting loops. In order to predict the three-dimensional arrangement of the secondary structural elements in the N-terminal part, we used the C-terminal two-layer α/β structure as a template, since it is very likely that the C-terminal and N-terminal regions interact, because the N and C ends of the central four-α-helix bundle are very close to each other (see Fig. 5). Therefore, in accordance with the structural features of two-layer α/β proteins (11), α-helix α1 (amino acids 45 to 57) should be packed in the α-helical layer and β-strands β1 (9 to 13) and β2 (17 to 20) should be packed into the β layer of the common two-layer structure (Fig. 5). Another possible interpretation is that the N-terminal residues form a separate domain, but this model was rejected because the tritium labeling of the β2-strand (amino acids 17 to 20) is rather low and it does not fit well with structural predictions. The predicted overall fold of the N-terminal region is reminiscent of that of a ββα unit. This unit has a long loop region (residues 21 to 44) which connects helix α1 and the β-hairpin formed by β-strands β1 (residues 9 to 13) and β2 (17 to 20). We suggest that the regions of residues 28 to 32 and 35 to 40 (the β3- and β4-strands) of this loop can also form a short β-hairpin.
At first sight, the proposed location of the β1-strand (residues 9 to 13) in the model (Fig. 5) seems contrary to the high level of labeling (Table 1). However, it should be noted that this region includes part of the highly labeled tryptic peptide T1 (residues 1 to 13) and contains amino acids (A, L, E/Q) which are also present among the first eight amino acids. As stated above, it is not possible to separate the individual contributions made by residues represented more than once in the sequence, and we suppose that the label is mainly present in the first eight amino acids. In the future, site-directed mutagenesis may be used to determine the contributions of the individual residues and increase the reliability of the data.
The C-terminal abCd unit and the N-terminal ββα unit should form the second structural domain of the PVA coat protein. Since the hinge regions comprising amino acids 58 to 61 and 177 to 181 are rather flexible, the orientation of the central α-helical domain and the second domain may be different in different functional states of the coat protein.
This model of PVA coat protein is supported by the results of in situ degradation of the coat protein in potyvirus particles, which occurs at K32 and K34. According to our model the first eight amino acids, the beginning of the β1-strand (from before residue 9 to residue 13), and the β-hairpin, formed by the two β-strands β3 (residues 28 to 32) and β4 (35 to 40), which are located in the principal site of trypsin proteolysis, are exposed on the surface of the virion.
The proposed spatial organization of the PVA coat protein in the virus particles differs from the schematic models suggested for a related potyvirus—PVY, in which both the N and C termini of the coat protein molecule are surface located (32, 33). According to our model, the 17 amino acids at the C terminus of the PVA coat protein form the α8-helix (252 to 268), which is oriented with the C-terminal end of the helix toward the outer surface of the virion. We have shown that it remains intact after mild proteolysis by TPCK-trypsin.
It is of interest to look at the general fold of the polypeptide chain in the model (Fig. 5). The ends of all the α-helices are directed toward the outer surface of the virus particle. This spatial packing of α-helices can explain both their low level of tritium labeling and the unexpected labeling of some amino acids located far from the virion surface, for example, A85, L168, and A198. A similar phenomenon was observed in tritium planigraphy studies of TMV virions (16), where several amino acid residues that are located far from the virion surface and are in contact with the RNA (according to the X-ray data) were labeled. It is likely that the small size of tritium atoms (0.9 Å) allows them, without any loss of energy, to penetrate into the virion depth through channels at the junctures between three protein subunits (16). Channels of this type have also been observed by Neiman et al. (24) in their study of the structure of the MS2 phage.
Finally, the results of PVA coat protein structure modeling suggest that the subunit should have an elongated shape with a total length of about 75 to 80 Å, which is larger than the radius of the PVA virion. However, changing the mutual orientation of the α-helices of the central and peripheral domains may reduce the total length of the subunit in the virion. In addition, it is possible that in the virion, the coat protein subunits are oriented at a significant angle to the virus particle axis, which would have the effect of reducing their projection on the virion radius. We plan to use the proposed model and the results of some future experiments to predict the mutual orientation of PVA coat protein subunits in the virion and the possible location of the RNA.
ACKNOWLEDGMENTS
We thank Igor V. Nazimov and Stanislav E. Esipov of the Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, for technical assistance and helpful discussions.
This study was supported by EU INCO Copernicus grant ERBIC 15CT970900 and Estonian Science Foundation grant 4224. The Scottish Crop Research Institute receives grant aid from the Scottish Executive Rural Affairs Department.
REFERENCES
- 1.Agafonov D E, Kolb V A, Spirin A S. Protein on ribosome surface: measurement of protein exposure by hot tritium bombardment technique. Proc Natl Acad Sci USA. 1997;94:12892–12897. doi: 10.1073/pnas.94.24.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrejeva J, Järvekülg L, Rabenstein F, Torrance L, Harrison B D, Saarma M. Antigenic analysis of potato virus A particles and coat protein. Ann Appl Biol. 1994;125:337–348. [Google Scholar]
- 3.Andrejeva J, Puurand Ü, Merits A, Rabenstein F, Järvekülg L, Valkonen J. Potyvirus helper component-proteinase and coat protein (CP) have co-ordinated functions in virus-host interactions and the same CP motif affects virus transmission and accumulation. J Gen Virol. 1999;80:1133–1139. doi: 10.1099/0022-1317-80-5-1133. [DOI] [PubMed] [Google Scholar]
- 4.Atreya C D, Raccah B, Pirone T P. A point mutation in the coat protein abolishes aphid transmission of a potyvirus. Virology. 1990;178:161–165. doi: 10.1016/0042-6822(90)90389-9. [DOI] [PubMed] [Google Scholar]
- 5.Atreya P L, Lopez-Moya J J, Chu M, Atreya C D, Pirone T P. Mutational analysis of the coat protein N-terminal amino acids involved in potyvirus transmission by aphids. J Gen Virol. 1995;76:265–270. doi: 10.1099/0022-1317-76-2-265. [DOI] [PubMed] [Google Scholar]
- 6.Baratova L A, Grebenshchikov N I, Shishkov A V, Kashirin I A, Radavsky J L, Järvekülg L, Saarma M. The topography of the surface of potato virus X: tritium planigraphy and immunological analysis. J Gen Virol. 1992;73:229–235. doi: 10.1099/0022-1317-73-2-229. [DOI] [PubMed] [Google Scholar]
- 7.Baratova L A, Grebenshchikov N I, Dobrov E N, Gedrovich A V, Kashirin I A, Shishkov A V, Efimov A V, Järvekülg L, Radavsky J L, Saarma M. The organization of potato virus X coat proteins in virus particles studied by tritium planigraphy and model building. Virology. 1992;188:175–180. doi: 10.1016/0042-6822(92)90747-d. [DOI] [PubMed] [Google Scholar]
- 8.Berger P, Barnett O W, Brunt A A, et al. Family Potyviridae. In: van Regenmortel M H V, et al., editors. Virus taxonomy. New York, N.Y: Springer-Verlag; 2000. pp. 703–724. [Google Scholar]
- 9.Dolja V V, Haldeman R, Robertson N L, Dougherty W G, Carrington J C. Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J. 1994;13:1482–1491. doi: 10.1002/j.1460-2075.1994.tb06403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efimov A V. Standard structures in proteins. Prog Biophys Mol Biol. 1993;60:201–239. doi: 10.1016/0079-6107(93)90015-c. [DOI] [PubMed] [Google Scholar]
- 11.Efimov A V. Structural similarity between two-layer α/β and β-proteins. J Mol Biol. 1995;245:402–415. doi: 10.1006/jmbi.1994.0033. [DOI] [PubMed] [Google Scholar]
- 12.Efimov A V. Complementary packing of α-helices in proteins. FEBS Lett. 1999;463:3–6. doi: 10.1016/s0014-5793(99)01507-0. [DOI] [PubMed] [Google Scholar]
- 13.Gedrovich A V, Goldanskii V I, Rumjantsev Y M, Unukovich M S, Shishkov A V. Obtaining of the labeled polypeptides and proteins by means of thermally activated tritium atoms. Radiokchimija. 1984;26:483–494. [Google Scholar]
- 14.Gedrovich A V, Badun G A. Study of the spatial structure of globular proteins by tritium planigraphy. Short peptides as a model of a fully extended polypeptide chain. Molekularnaja Biologija. 1992;26:558–564. [PubMed] [Google Scholar]
- 15.Gill S C, von Hippel P H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 16.Goldanskii V I, Kashirin I A, Shishkov A V, Baratova L A, Grebenshchikov N I. The use of thermally activated tritium atoms for structural-biological investigations: the topography of the TMV protein-accessible surface of the virus. J Mol Biol. 1988;201:567–574. doi: 10.1016/0022-2836(88)90638-9. [DOI] [PubMed] [Google Scholar]
- 17.Goodman R M. Reconstruction of potato virus X in vitro. I. Properties of the dissociation protein structural subunits. Virology. 1975;68:287–298. doi: 10.1016/0042-6822(75)90272-x. [DOI] [PubMed] [Google Scholar]
- 18.Greenfield N, Fasman G D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;10:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lim V I. Structural principles of the globular organization of protein chains. A stereochemical theory of globular protein secondary structure. J Mol Biol. 1974;88:857–872. doi: 10.1016/0022-2836(74)90404-5. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Moya J J, Wang R Y, Pirone T P. Context of the coat protein DAG motif affects potyvirus transmissibility by aphids. J Gen Virol. 1999;80:3281–3288. doi: 10.1099/0022-1317-80-12-3281. [DOI] [PubMed] [Google Scholar]
- 22.Morozov S Y, Solovyev A G. Genome organization in RNA viruses. In: Mandahar C L, editor. Molecular biology of plant viruses. Boston, Mass: Kluwer; 1999. pp. 47–98. [Google Scholar]
- 23.Namba K, Pattanayek R, Stubbs G. Visualization of protein-nucleic acid interactions in a virus. Refined structure of intact tobacco mosaic virus at 2.9 Å resolution by X-ray fiber diffraction. J Mol Biol. 1989;208:307–325. doi: 10.1016/0022-2836(89)90391-4. [DOI] [PubMed] [Google Scholar]
- 24.Neiman L A, Andropova L P, Zalesskaya M A, Budowsky E I. Tritium labeling of phage: MS2 RNA and protein. Bioorganicheskaja Ximija. 1986;12:1070–1072. [PubMed] [Google Scholar]
- 25.Puurand Ü, Mäkinen K, Paulin L, Saarma M. The complete nucleotide sequence of potato virus A genomic RNA and its sequence similarities with other potyviruses. J Gen Virol. 1994;75:457–461. doi: 10.1099/0022-1317-75-2-457. [DOI] [PubMed] [Google Scholar]
- 26.Puurand Ü, Mäkinen K, Baumann M, Saarma M. Nucleotide sequence of the 3′-terminal region of potato virus A RNA. Virus Res. 1992;23:99–105. doi: 10.1016/0168-1702(92)90070-p. [DOI] [PubMed] [Google Scholar]
- 27.Riechmann J L, Lain S, Garcia J A. Highlights and prospects of potyvirus molecular biology. J Gen Virol. 1992;73:1–16. doi: 10.1099/0022-1317-73-1-1. [DOI] [PubMed] [Google Scholar]
- 28.Schiffer M, Edmundson A B. Use of helical wheels to represent the structures of protein and to identify segments with helical potential. Biophys J. 1967;7:121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shishkov A V, Baratova L A. Tritium planigraphy of biological systems. Russian Chem Rev. 1994;63:781–796. [Google Scholar]
- 30.Shishkov A V, Goldanskii V I, Baratova L A, Fedorova N V, Ksenofontov A L, Zhirnov O P, Galkin A V. The in situ spatial arrangement of the influenza A virus matrix protein MI assessed by tritium bombardment. Proc Natl Acad Sci USA. 1999;96:7827–7830. doi: 10.1073/pnas.96.14.7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shukla D D, Tribbick G, Mason T J, Hewish D R, Geysen H M, Ward C M. Localization of virus-specific and group-specific epitopes of plant potyviruses by systematic immunochemical analysis of overlapping peptide fragments. Proc Natl Acad Sci USA. 1989;86:8192–8196. doi: 10.1073/pnas.86.21.8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shukla D D, Ward C M. Structure of potyvirus coat proteins and its application in the taxonomy of the potyvirus group. Adv Virus Res. 1989;36:273–314. doi: 10.1016/s0065-3527(08)60588-6. [DOI] [PubMed] [Google Scholar]
- 33.Shukla D D, Strike P M, Tracy S L, Gough K H, Ward C M. The N and C termini of the coat proteins of potyviruses are surface-located and the N terminus contains major virus-specific epitopes. J Gen Virol. 1988;69:1497–1508. [Google Scholar]
- 34.Tsetlin V I, Alynycheva T N, Shemyakin V V, Neiman L A, Ivanov V T. Tritium thermal activation study of bacteriorhodopsin topography. Eur J Biochem. 1988;178:123–129. doi: 10.1111/j.1432-1033.1988.tb14437.x. [DOI] [PubMed] [Google Scholar]
- 35.Tsugita A, Scheffler J-J. A rapid method for acid hydrolysis of protein with a mixture of trifluoroacetic acid and hydrochloric acid. Eur J Biochem. 1982;124:585–588. doi: 10.1111/j.1432-1033.1982.tb06634.x. [DOI] [PubMed] [Google Scholar]