Abstract
Aminopeptidase N (APN), a 150-kDa metalloprotease also called CD13, serves as a receptor for serologically related coronaviruses of humans (human coronavirus 229E [HCoV-229E]), pigs, and cats. These virus-receptor interactions can be highly species specific; for example, the human coronavirus can use human APN (hAPN) but not porcine APN (pAPN) as its cellular receptor, and porcine coronaviruses can use pAPN but not hAPN. Substitution of pAPN amino acids 283 to 290 into hAPN for the corresponding amino acids 288 to 295 introduced an N-glycosylation sequon at amino acids 291 to 293 that blocked HCoV-229E receptor activity of hAPN. Substitution of two amino acids that inserted an N-glycosylation site at amino acid 291 also resulted in a mutant hAPN that lacked receptor activity because it failed to bind HCoV-229E. Single amino acid revertants that removed this sequon at amino acids 291 to 293 but had one or five pAPN amino acid substitution(s) in this region all regained HCoV-229E binding and receptor activities. To determine if other N-linked glycosylation differences between hAPN, feline APN (fAPN), and pAPN account for receptor specificity of pig and cat coronaviruses, a mutant hAPN protein that, like fAPN and pAPN, lacked a glycosylation sequon at 818 to 820 was studied. This sequon is within the region that determines receptor activity for porcine and feline coronaviruses. Mutant hAPN lacking the sequon at amino acids 818 to 820 maintained HCoV-229E receptor activity but did not gain receptor activity for porcine or feline coronaviruses. Thus, certain differences in glycosylation between coronavirus receptors from different species are critical determinants in the species specificity of infection.
Human coronaviruses (HCoV) in two serogroups represented by HCoV-229E (serogroup 1) and HCoV-OC43 (serogroup 2) are an important cause of upper respiratory tract infection. Serological studies suggest that they cause from 15 to 30% of human upper respiratory infections or colds (25). Although there is no animal model for the pathogenesis of HCoV respiratory infections, HCoV-229E and HCoV-OC43 have been administered intranasally to human volunteers, and this has shown that people can be repeatedly infected by the same strain (5). There is also in vitro and in vivo evidence for infection of cells of the central nervous system (CNS) by HCoV (39, 51, 68, 69). Coronaviruses have a lipid membrane that has large peplomers that protrude from the virion and give it the appearance of a corona. These peplomers or spike glycoproteins are the viral attachment proteins. The 27-kb genomic RNA of HCoV-229E is positive sense, has a 5′ cap, is polyadenylated at the 3′ terminus, and is infectious when transfected into cells of a wide variety of species (25). In contrast, infection by HCoV-229E virions is limited to cells of human or feline origin in vitro, demonstrating that the receptor for HCoV-229E is a major determinant in species specificity (25).
The cellular receptor for HCoV-229E is human aminopeptidase N (hAPN). HCoV-229E infection is blocked by monoclonal antibodies (MAbs) to hAPN, and murine fibroblasts that are nonpermissive for HCoV-229E become susceptible after transfection with hAPN expression plasmids (77). Additionally, infection of human neural cells by HCoV-229E requires hAPN (39). hAPN is also involved in human cytomegalovirus (CMV) infection (41, 57, 63). CMV incorporates hAPN into its envelope, and hAPN is implicated in autoimmunity induced by CMV in immunocompromised patients (21, 64, 65).
APN, also called CD13 (EC 3.4.11.2), is a 150- to 160-kDa type II glycoprotein that is a membrane peptidase (42). It is a metalloprotease that binds zinc through a highly conserved HELAH amino acid motif, and deletions or mutations in this motif inhibit enzymatic activity (42, 54). APN expressed on the cell surface is heavily glycosylated, and carbohydrates account for approximately 30% of the molecular mass (8, 60). APN is expressed as a dimer that protrudes 10.5 nm from the cell surface of epithelial cells of the kidney, intestine, and respiratory tract; granulocytes; monocytes; fibroblasts; endothelial cells; cerebral pericytes at the blood-brain barrier; and synaptic membranes in the CNS (4, 29, 38, 42, 48, 54, 60, 62, 66). APN has a wide range of biological activities, including the removal of individual amino acids from N termini of small peptides in the lumen of the small intestine, cleaving peptides bound to major histocompatibility complex class II molecules of antigen presenting cells, and degrading neurotransmitters at synaptic junctions (40, 48, 54). hAPN is a marker for acute myeloid leukemia and plays a role in tumor invasion (19, 31, 36, 50, 61, 67).
APN also serves as the major receptor for other serogroup 1 coronaviruses that infect other species, including transmissible gastroenteritis virus (TGEV) of swine, porcine respiratory coronavirus, feline infectious peritonitis virus (FIPV), and feline enteric coronavirus (FECoV) (10, 11, 71). The hAPN, feline APN (fAPN), and porcine APN (pAPN) proteins have strong sequence identity and similarity. For example, pAPN and fAPN both have 78% amino acid identity and 86% similarity to hAPN with the introduction of seven and five gaps, respectively, using BLASTP 2.0.9. (1). All three proteins have numerous N-linked glycosylation sequons; fAPN has 8, hAPN has 11, and pAPN has 13. In general, the APN receptors are used by serogroup 1 coronaviruses in a species-specific manner; i.e., hAPN serves as a receptor for the human virus (HCoV-229E) but not the porcine coronaviruses, and pAPN serves as a receptor for the porcine coronaviruses but not HCoV-229E (9, 35). However, fAPN serves as a functional receptor for many serogroup 1 coronaviruses, including feline (FECoV and FIPV), human (HCoV-229E), porcine (TGEV), and canine coronaviruses (71).
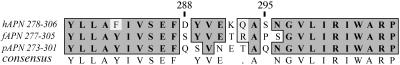
Studies of chimeric hAPN, fAPN, and pAPN glycoproteins identified two large regions that are important in receptor activity for various serogroup 1 coronaviruses. Chimeras between hAPN and pAPN showed that amino acids 717 to 813 of the pAPN are required for TGEV receptor activity (9). Chimeras between hAPN and fAPN showed that amino acids 670 to 840 of fAPN are required for receptor activity for feline and porcine coronaviruses (22). When amino acids 260 to 353 of hAPN or amino acids 135 to 297 of fAPN are substituted into pAPN for amino acids 255 to 348 or 132 to 295, respectively, the resulting chimeras have HCoV-229E and TGEV receptor activity (34, 35). Alignment of hAPN, fAPN, and pAPN around this region shows strong amino acid conservation, except at amino acids 288 to 295 of hAPN (Fig. 1). In pAPN substitution of amino acids 283 to 290 with amino acids 288 to 295 of hAPN resulted in a protein that serves as a receptor for TGEV and HCoV-229E (34). This suggests two possibilities: (i) that amino acids 288 to 295 of hAPN directly bind to the spike glycoprotein of HCoV-229E or (ii) that these substitutions alter pAPN conformation and expose a binding site that is evolutionarily conserved. This site must be more accessible in fAPN, which can act as a receptor for serogroup 1 coronaviruses of cats, pigs, dogs, and humans.
FIG. 1.
Amino acid alignment of a small region of hAPN, fAPN, and pAPN. The predicted amino acid sequences of hAPN, fAPN, and pAPN were aligned using Clustal W (version 1.4) and the Blosum similarity matrix. hAPN amino acids 278 to 306, aligned with fAPN amino acids 277 to 305 and pAPN amino acids 273 to 301, are shown. The numbers above the sequence correspond to the hAPN glycoprotein, and the consensus sequence is shown on the bottom line. Identical amino acids are shown on dark gray background, similar amino acids are shown on a light gray background, and dissimilar amino acids are shown on a white background.
To better understand the interaction of HCoV-229E spike and hAPN we extended previous studies, using sequence analysis of APN proteins from many species to identify and mutate specific amino acids in hAPN that had the potential to influence receptor activity for serogroup 1 coronaviruses. The coronavirus receptor activities of wild-type hAPN and nine mutant hAPN proteins expressed in nonpermissive cells were analyzed to determine the specific amino acids of hAPN that interact with HCoV-229E and to elucidate species-specific differences in APN that influence the receptor activity for different coronaviruses. The data demonstrate that the addition of a single N-linked glycosylation site at amino acid 291 of hAPN abrogates HCoV-229E receptor function and that it does this by directly inhibiting virus binding. Furthermore, the data strongly suggest that HCoV-229E does not bind directly to amino acids 288 to 295 of hAPN but that substitution of these amino acids for the analogous amino acids of pAPN (amino acids 283 to 290) inserts a glycosylation site that blocks the binding of HCoV-229E to a different region of APN that may be evolutionarily conserved in hAPN, pAPN, and fAPN.
Many chimeras and mutants have been generated between fAPN, pAPN, and hAPN to study how specific amino acid changes influence receptor activity and species specificity of serogroup 1 coronaviruses of cats, pigs, and people. However, this is the first study that demonstrates the influence that N-linked glycosylation of APN has on coronavirus receptor activity and species specificity. This study also augments the limited understanding of the influence N-linked glycosylation of viral receptors has on virus-receptor interactions.
MATERIALS AND METHODS
Cells and viruses.
All cells were maintained in the appropriate medium supplemented with 2% Antibiotic-Antimycotic (GIBCO BRL, Grand Island, N.Y.) at 37°C and 5% CO2 in Falcon tissue culture treated flasks or plates (Becton Dickinson and Company, Franklin Lakes, N.J.). Human diploid lung fibroblasts, MRC5 cells (American Type Culture Collection [ATCC], Manassas, Va.), were grown from passage 19 to 27 in Eagle's minimal essential medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1 mM sodium pyruvate (GIBCO BRL), and 1 mM nonessential amino acids (GIBCO BRL). The BHK-21 line of baby hamster kidney fibroblasts (from Robert Garcea, University of Colorado Health Sciences Center, Denver), CMT93 cells (ATCC), swine testicle cells (from David Brian, University of Tennessee, Knoxville), and Felis catus whole fetus (FCWF) cells (from Niels Pedersen, University of California at Davis) were grown in Dulbecco's modified MEM (DMEM) supplemented with 10% heat-inactivated FBS (GIBCO BRL).
HCoV-229E was obtained from ATCC (VR740, lot W4) and was propagated in MRC5 cells at 34°C and 5% CO2. The stock virus was amplified for 24 h, plaque purified twice, and amplified by inoculation of a flask containing 2 × 10 7 MRC5 cells with a low multiplicity of infection (MOI ∼0.006) and incubation for 48 h to produce virus seed stock designated DW6-seed. Multiple T150 flasks were inoculated at an MOI of 0.002 and incubated for 70 h when the supernatant containing the working stock virus designated DW6-W was collected. The virus titer was determined by plaque and 50% tissue culture infective dose assays in MRC5 cells. FECoV 79-1683 was propagated in FCWF cells, and TGEV clone E (kindly provided by David Brian, University of Tennessee, Knoxville) was grown in porcine swine testicle cells as described (71).
hAPN plasmid construction, PCR mutagenesis, and sequence analysis.
The wild-type hAPN expression plasmid wt-hAPN (also designated E88A10) was produced by subcloning an hAPN cDNA fragment, accession number M22324, from pBSSK+ (Stratagene, La Jolla, Calif.) into the pCi-neo expression plasmid (Promega Corporation, Madison, Wis.) using XhoI and XbaI. To create p6-hAPN, two fragments of hAPN from nucleotides 111 to 990 and nucleotides 991 to 1216 were generated by PCR with Pfu; one fragment was produced with primers hAPN-111 and hAPN-990R, and the other was produced with h-pAPN-991 and DT22 (Table 1), according to the manufacturer's instructions (Stratagene, La Jolla, Calif.). The temperature cycle parameters used were 5 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 2 min and then 30 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min. These amplicons were mixed together and then digested with NcoI and ligated to NcoI-cut pCite2a (Novagen, Madison, Wis.) to produce E74B23. The naturally occurring NcoI sites at positions 121 and 1178 of hAPN ligated to the plasmid arms and blunt end ligation occurred between the PCR products from nucleotides 990 to 991. This changed D288YVEKQAS295 in hAPN to Q288SVNETAQ295 and created an HpaI site. This clone was sequenced to ensure that only the desired mutations were produced. An NcoI fragment from E74B23 was subcloned into an hAPN cDNA clone (E83A3) in pBSSK+ that lacks the wild-type NcoI fragment. This produced a full-length hAPN clone that was identical to wt-hAPN except for the desired substitutions (E84A4). This mutant hAPN cDNA was subcloned into the pCi-neo expression plasmid (Promega Corporation) using XhoI and XbaI to create p6-hAPN (laboratory designation E85A4). All the restriction enzymes used were purchased from New England Biolabs (Beverly, Mass.).
TABLE 1.
Oligonucleotides used for PCR, mutagenesis, and sequencing
| Primer | Sequence (5′-3′) |
|---|---|
| Forward | |
| hAPN-111 | CACCATCACCATGGCCAAGG |
| h-pAPN-991 | AACGAGACGGCACAAAATGGTGTCTTG |
| hAPN E291N-Q293T/F | GTTCGACTACGTTAACAAGACGGCATCCAATGG |
| m-hAPN N291E/F | TTCGACTACGTTGAGAAGACGGCATCC |
| m-hAPN T293Q/F | TACGTTAACAAGCAGGCTAGCAATGGTGTCTTG |
| hpAPN-N291E/F | CAATCCGTTGAGGAGACGGCACAA |
| hpAPN-T293Q/F | TCCGTTAATGAGCAGGCACAAAATGGT |
| hAPN-N818K/F | GAGCAGTTCCGGAAAGCCACACTGGTC |
| hAPN-T820E/F | CCGAAATGCCGAGCTCGTCAATGAGGC |
| DT-021 | CCCACTTCCAGAAGACCCC |
| Reverse | |
| hAPN-990R | AACGGATTGGAACTCACTGACAATGAA |
| DT-022 | AGTTCTCCCGGTAGGTCACC |
| hAPN E291N-Q293T/R | CCATTGGATGCCGTCTTGTTAACGTAGTCGAAC |
| m-hAPN N291E/R | GGATGCCGTCTTCTCAACGTAGTC |
| m-hAPN T293Q/R | CAAGACACCATTGCTAGCCTGCTTGTTAACGTA |
| hpAPN-N291E/R | TTGTGCCGTCTCCTCAACGGATTG |
| hpAPN-T293Q/R | ACCATTTTGTGCCTGCTCATTAACGGA |
| hAPN-N818K/R | GACCAGTGTGGCTTTCCGGAACTGCTC |
| hAPN-T820E/R | GCCTCATTGACGAGCTCGGCATTTCGG |
Site-directed mutagenesis was done by PCR amplifying various templates with Pfu polymerase using mutagenic primers (Table 1) with 18 temperature cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 18 min, according to the manufacturer's instructions (Stratagene). The products were digested with DpnI for 1 h at 37°C to remove the template and used to transform Escherichia coli DH5α (GIBCO BRL). Colonies were picked and clones were screened by restriction endonuclease digestion and/or PCR analysis. Each of the clones amplified and used for transfection was sequenced to ensure that it contained only the desired mutations. The hAPN/N291KT expression plasmid (laboratory designation, F1A1) was generated from wt-hAPN using primers hAPN E291N-Q293T/F and hAPN E291N-Q293T/R. Revertants hAPN/E291KT and hAPN/N291KQ (laboratory designations, F2A2 and F3A4) were generated from hAPN/N291KT using primers m-hAPN N291E/F and m-hAPN N291E/R or m-hAPN T293Q/F and m-hAPN T293Q/R, respectively. Expression plasmids p6-hAPN/N291E and p6-hAPN/T293Q (laboratory designations, E96A1 and E94A15) were generated using hpAPN-N291E/F and hpAPN-N291E/R or hpAPN-T293Q/F and hpAPN-T293Q/R. Plasmids hAPN/T820E or p6-hAPN/N818K and p6-hAPN/T820E (laboratory designations, E100A24 or E97A9 and E98A14) were produced from wt-hAPN or p6-hAPN using primers hAPN-T820E/F and hAPN-T820E/R or hAPN-N818K/F and hAPN-N818K/R. The plasmids were sequenced by the University of Colorado Cancer Center DNA Sequencing and Analysis Core Facility using ABI Prism kits from Applied Biosystems (Foster City, Calif.) containing AmpliTaq DNA Polymerase FS in one of the following: dRhodamine Terminator Cycle Sequencing Ready Reaction kit, BigDye Terminator Cycle Sequencing Ready Reaction kit, or dGTP BigDye Terminator Cycle Sequencing Ready Reaction kit. The standard sequencing thermocycling parameters were the following: denaturation for 5 min at 94°C, followed by 30 cycles of denaturation at 96°C for 10 s, annealing at 50°C for 5 s, and extension-termination at 60°C for 4 min, followed by incubation at 4°C until the samples were processed to remove the residual dye-labeled dideoxynucleotides (dye terminators). The reaction products were analyzed on either an ABI 373A or an ABI Prism 377 DNA fluorescent sequencer, both standard and XL versions (Applied Biosystems). The sequences were analyzed using Blast (1), MacVector 6.5.3 (Oxford Molecular Group, Campbell, Calif.), and Vector NTI suite (Informax, Bethesda, Md.). The hAPN, pAPN, and fAPN sequences were obtained from the GenBank database, accession numbers M22324, Z29522, and U58920, respectively.
Transfection and selection of cells.
BHK-21 cells were seeded at 2 × 105 per well in six-well dishes or on glass coverslips in six-well dishes; 18 to 20 h later, when the cells had reached 70 to 80% confluency, they were transfected with 1 μg of plasmid and 12 μl of Lipofectamine reagent (GIBCO BRL) per well, according to the manufacturer's instructions, and incubated for 5 h at 37°C and 5% CO2 in Optimem (GIBCO BRL). Then, the medium was doubled by adding 1 ml of DMEM containing 20% FBS. The cells were incubated for 16 to 18 h at 37°C and 5% CO2. The medium was changed to growth medium, and hAPN protein expression was determined by immunofluorescence or immunoblot and/or the cells were inoculated with HCoV-229E 24 to 48 h after transfection.
CMT93 cells were seeded at 3 × 105 per well in six-well dishes; 18 to 20 h later (at 60% confluency) the cells were transfected with 9 μg of plasmid and 15 μl of Pfx-2 reagent (Invitrogen) per well, according to the manufacturer's instructions. The cells were incubated for 6 h at 37°C and 5% CO2 in Optimem (GIBCO BRL), and then the transfection solution was replaced with growth medium and the cells were cultured for 20 h and split into 150-mm-diameter dishes in growth medium containing G418 (500 μg/ml; GIBCO BRL). The cells were grown to confluency, and this mixed population of G418-resistant cells was sorted two times by fluorescence-activated cell sorting (FACS) with MAb WM47, as described below, and tested for APN protein expression by immunofluorescence, FACS, immunoblot, and enzymatic activity assays (77). Limiting dilution was used to generate clones from the mixed population of hAPN-expressing cells.
Antibodies.
Anti hAPN MAbs WM47 (DAKO Corporation, Carpinteria, Calif.), MY7 (BioGenex, San Ramon, Calif.), and WM15 and SJ1D1 (Biodesign, Kennebunk, Maine) were purchased, and MY32 was kindly provided by C. I. Civin, Johns Hopkins Oncology Center, Baltimore, Md. Anti HCoV-229E spike glycoprotein MAb 5-11H.6 was kindly provided by Pierre Talbot, University of Quebec, Loval, Quebec, Canada. Polyclonal goat anti-HCoV-229E was produced from NP-40-dispersed purified HCoV-229E in collaboration with Lawrence Sturman (New York State Health Department, Albany), and polyclonal mouse anti hAPN was prepared by inoculating Swiss Webster mice with NIH 3T3 cells expressing hAPN (Zip cells) (77). FECoV and TGEV antigens were detected with polyclonal feline anti-FIPV serum (Biodesign, Kennebunk, Maine).
FACS.
After G418 selection, the cells were trypsinized with trypsin-EDTA (GIBCO BRL) and washed three times in 1 ml of wash buffer (0.1% [wt/vol] bovine albumin Fraction V [Sigma, St. Louis, Mo.] in phosphate-buffered saline [PBS]). One million cells were incubated on ice in 100 μl of a 1:100 dilution of WM47 for 1 h at 4°C. The cells were washed three times with 1 ml of wash buffer and incubated for 30 min at 4°C in 100 μl of a 1:100 dilution of affinity-purified, phycoerythrin-conjugated goat F(ab′)2 anti-mouse immunoglobulin (DAKO Corporation). The cells were washed three times with 1 ml of wash buffer and resuspended in 500 μl of wash buffer and sorted into G418 selection medium by flow cytometry at the University of Colorado Health Sciences Center Flow Cytometry Core Facility in a Coulter XL with System II software (Beckman Coulter, Hialeah, Fla.). These cells were expanded, sorted again, and grown for one to nine passages. Each cell line was shown by FACS with WM47 to express similar levels of hAPN and had mean fluorescence intensity that ranged from 5.4 (hAPN/N291KQ) to 9.9 (p6-hAPN). FACS of the mutant hAPN cell lines by a panel of anti hAPN MAbs was done, as described above, with WM47, WM15, MY7, MY32, or SJ1D1. However, after the last wash, the cells were fixed in 500 μl of fix buffer (1.6% paraformaldehyde [Ted Pella, Inc., Redding, Calif.] in PBS).
IFA.
Immunofluorescence assays (IFA) for expression of hAPN and HCoV-229E spike glycoprotein were done with 1:100 dilutions of WM47 and MAb 5-11H.6, respectively. Cells on glass coverslips were inoculated with HCoV-229E (DW6-W) at an MOI of 1, incubated for 20 to 22 h, washed with PBS, and fixed with cold 100% acetone. Antigens were detected by immunofluorescence with 1:100 dilutions of the various MAbs, followed by fluorescein-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories, West Grove, Pa.) (71). The coverslips were analyzed using a Zeiss Axioplan 2 microscope at a magnification of ×40 (Carl Zeiss, Inc., New York, N.Y.).
Immunoblots.
Six-well dishes of the transiently transfected BHK-21 cells or the CMT93 cell lines were inoculated with HCoV-229E (DW6-W) at an MOI of 1 to 2, incubated at 34°C and 5% CO2 for 1 h, and washed three times with 3 ml of PBS, and then 1 ml of growth medium was added and the cells were incubated overnight at 34°C and 5% CO2. At 20 to 22 h postinoculation the wells were washed once with 5 ml of PBS, and then cell lysates were prepared by adding 500 μl of ice-cold cell lysis buffer (58). Immunoblots were done using sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) to separate 20 μl of the cell lysates and transferred to Immobilon-P as described (78). The blots were probed for HCoV-229E with a 1:4,000 dilution of polyclonal goat antiserum to HCoV-229E and for hAPN with a 1:1,250 dilution of polyclonal mouse antiserum to hAPN. These blots were incubated for 1 h at 20 to 25°C, washed, and incubated for 30 min to 1 h with a 1:4,000 dilution of horseradish peroxidase conjugated rabbit anti-goat (Cappel, Durham, N.C.) or goat anti-mouse (Sigma) antibody. Bands were detected with Renaissance Chemiluminescence reagent and exposure of autoradiography film (Dupont/NEN, Boston, Mass).
HCoV-229E virus binding assay.
To prepare radiolabeled virus, [3H]-uridine (50 Ci/mmol; final concentration = 20 μCi/ml; NEN) was added to the growth medium 1 h postinoculation with HCoV-229E (DW6-W) at an MOI of 1 and incubated overnight. Supernatant was centrifuged at 3,000 × g for 5 min to remove cell debris and then layered over 6 ml of buffered 30% sucrose and centrifuged at 100,000 × g for 2 h at 4°C. The virus pellet was resuspended in MEM containing 5% FBS (GIBCO BRL), flash frozen, and stored at −80°C.
CMT93 cell lines transfected with wild-type hAPN, p6-hAPN, hAPN/N291KT, hAPN/E291KT, and hAPN/N291KQ that were sorted for similar levels of hAPN expression by FACS or cells transfected with pCi-neo were trypsinized and counted. A total of 106 cells was mixed with 3.1 × 105 cpm of 3H-labeled virus at an MOI of 0.5 in 500 μl of medium, in triplicate, and rocked at 4°C for 2 h. The cells were washed three times with 1 ml of medium at 4°C, resuspended in 200 μl of cell lysis buffer (58), and then transferred to scintillation vials containing 5 ml of EcoScint (Life Science Products Inc., Denver, Colo.) and counted in a model LS 6500 scintillation counter (Beckman Coulter, Inc., Fullerton, Calif.). The average number of counts per minute bound by the wild-type hAPN was defined as 100% binding, and the average number of counts per minute bound by the cells transfected with empty pCi-neo was defined as 0% binding. The average of each of the other cell lines was reported as a percentage of wild-type binding activity, and the error bars were determined by the standard deviation of the mean of the three replicates.
RESULTS
Substitution of hAPN amino acids 288 to 295 with pAPN amino acids 283 to 290 blocked infection by HCoV-229E.
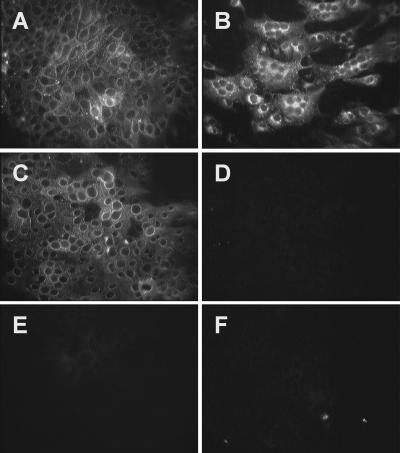
To determine if six amino acid substitutions in an eight-amino-acid stretch, from amino acids 288 to 295, of hAPN alters its function as a receptor for HCoV-229E, we used primer-based PCR mutagenesis to change the amino acids in this region of hAPN to the analogous pAPN residues, creating the p6-hAPN construct. Murine CMT93 cells, which are nonpermissive for HCoV-229E, were transfected with plasmids expressing wt-hAPN, a mutant hAPN that has six substitutions to the porcine amino acids at positions 288, 289, 291, 292, 293, and 295, (p6-hAPN), or the empty expression plasmid (pCi-neo). These cells were selected for G418 resistance and sorted for similar hAPN expression levels using FACS with MAb WM47. The cells transfected with wt-hAPN and p6-hAPN expressed equivalent levels of APN as shown by indirect IFA with MAb WM47 (Fig. 2A and C). Inoculation of cells transfected with wt-hAPN, p6-hAPN, or the pCi-neo expression plasmid resulted in HCoV-229E infection of cells that expressed wild-type hAPN as shown by indirect IFA with anti-HCoV-229E spike MAb 511H.6 (Fig. 2B). In contrast, transfection with the mutant hAPN p6-hAPN or the plasmid control pCi-neo resulted in no HCoV-229E receptor activity (Fig. 2D and F, respectively).
FIG. 2.
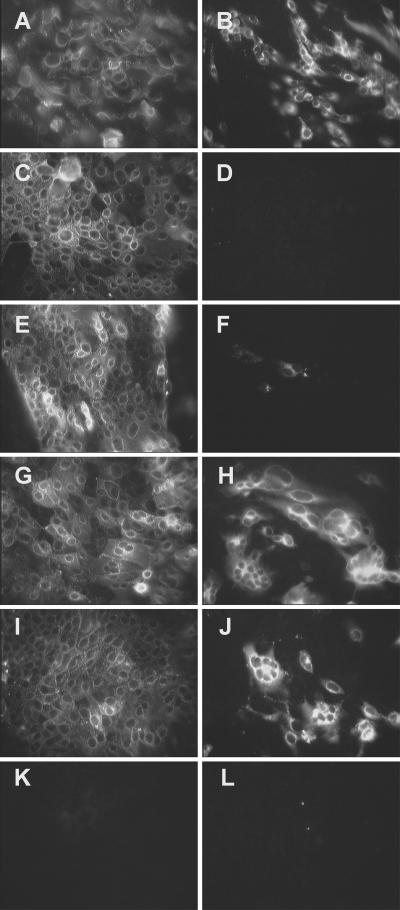
Mutations in amino acids 288 to 295 of hAPN blocked HCoV-229E infection. CMT93 cell lines were generated by transfection with wt-hAPN (A and B), p6-hAPN (C and D), or pCi-neo (E and F), and the hAPN-expressing lines (A to D) were sorted by FACS for similar expression levels of hAPN protein. hAPN protein was shown by indirect IFA with MAb WM47 (A, C, and E) and infection by HCoV-229E was shown by indirect IFA, 22 h postinoculation, with anti-spike glycoprotein MAb 511H.6 (B, D, and F).
Expression of wt-hAPN and p6-hAPN in clones obtained from this mixed population of cells was determined by FACS analysis using MAb WM47. FACS analysis showed that the wild type hAPN clone F4G9 and two p6-hAPN clones, F4C10 and C5F8, expressed hAPN levels with mean fluorescence intensity values of 2.95, 2.22, and 9.4, respectively. Only the clone expressing wild-type hAPN was permissive to HCoV-229E (data not shown). p6-hAPN clone C5F8 expressed approximately three times more hAPN than did wild-type hAPN F4G9, yet lacked HCoV-229E receptor activity (data not shown). Thus, the lack of HCoV-229E receptor activity of p6-hAPN was not due to a low level of the mutant receptor on the cell surface. Additionally, FACS analysis of these clones using MAb SJ1D1, MY32, MY7, or WM15 illustrated that this panel of MAbs recognized both the wild-type and mutant hAPN glycoproteins (data not shown). These MAbs recognize four different epitopes of hAPN: WM15 blocks HCoV-229E infection and APN enzymatic activity, but does not bind a deletion mutant lacking the HELAH motif; MY7 also blocks infection, partially inhibits enzymatic activity, and binds to the HELAH deletion mutant; MY32 blocks infection, has no effect on enzymatic activity, and binds to the deletion mutant; SJ1D1 has no effect on infection or APN activity and binds to the deletion mutant (2). All of these MAbs bound hAPN and p6-hAPN; however, MY7 showed a slight decrease in binding to the p6-hAPN mutant (data not shown). Furthermore, cells expressing the p6-hAPN mutant had enzymatic activity equal to or greater than cells expressing wt-hAPN (data not shown). The observed reactivity with this panel of MAbs and the aminopeptidase activity indicate that the six amino acid substitutions in this region of hAPN did not result in a dramatic change in the protein conformation and show that substitution of these six amino acids of hAPN with pAPN blocks HCoV-229E infection.
Removal of a potential N-linked glycosylation site within the substituted region of hAPN restored HCoV-229E receptor activity.
Analysis of the amino acid sequence within the substituted region showed that this change inserted a putative N-linked glycosylation signal (NET) at amino acids 291 to 293 of p6-hAPN that is not conserved in hAPN or fAPN (Fig. 1). To investigate the influence of this glycosylation signal on HCoV-229E receptor activity, we used site-directed mutagenesis to remove this glycosylation signal from the p6-hAPN construct that did not serve as a functional receptor. Reversion of asparagine 291 to glutamic acid (N291E) encoded by wild-type hAPN or threonine 293 to glutamine (T293Q) created two separate expression plasmids, p6-hAPN/N291E and p6-hAPN/T293Q, respectively. Transient transfection of BHK-21 cells with wt-hAPN, p6-hAPN, p6-hAPN/N291E, p6-hAPN/T293Q, or pCi-neo resulted in expression of hAPN in all of the cells except those transfected by vector alone or untransfected BHK-21 cells as demonstrated by immunoblotting with polyclonal mouse antiserum to hAPN (Fig. 3A, lanes 1 to 5, respectively). HCoV-229E inoculation of BHK-21 cells transiently transfected with these expression constructs resulted in infection of cells that expressed the wild-type hAPN and cells that expressed the revertants p6-hAPN/N291E and p6-hAPN/T293Q (Fig. 3B, lanes 1, 3, and 4, respectively). In contrast, cells that expressed p6-hAPN, pCi-neo-transfected cells, and untransfected cells were not susceptible to HCoV-229E (Fig. 3B, lanes 2, 5, and 6, respectively).
FIG. 3.
Single amino acid reversions that remove an N291 glycosylation signal in p6-hAPN result in HCoV-229E receptor activity. Immunoblots show hAPN expression (A) and HCoV-229E nucleocapsid expression (B) in transiently transfected BHK-21 cells. Cell lysates were isolated from BHK-21 cells transiently transfected with wt-hAPN (lane 1), p6-hAPN (lane 2), p6-hAPN/N291E (lane 3), p6-hAPN/T293Q (lane 4), pCi-neo (lane 5), or untransfected BHK-21 cells (lane 6). The cells were inoculated at an MOI of 1 with HCoV-229E 24 h after transfection, and cell lysates were prepared 20 h postinoculation. The proteins were separated by SDS–10% PAGE and transferred to Immobilon-P. (A) hAPN protein expression (arrow) was identified by reactivity with polyclonal mouse antiserum to hAPN; (B) HCoV-229E nucleocapsid (N) protein expression (arrow), was shown by polyclonal goat antiserum to HCoV-229E.
Another way to analyze receptor function is to determine the titer of virus released postinoculation. We found that there was a 100- to 1,000-fold increase in virus titer in supernatants of cells transfected with wt-hAPN or either of the two single-amino-acid revertants but no increase in virus titer from cells transfected with the p6-hAPN construct. None of these constructs had receptor activity for the porcine coronavirus TGEV (data not shown). The data showed that hAPN proteins with five amino acid substitutions that changed this region from D288YVEKQAS295 to Q288SVEETAQ295 or Q288SVNEQAQ295 served as functional HCoV-229E receptors; however, the glycosylation signal, underlined, in Q288SVNETAQ295 blocked receptor activity.
Glycosylation of hAPN at amino acid 291 blocked infection by HCoV-229E.
We used site-directed mutagenesis to insert the glycosylation signal into wild-type hAPN to determine if glycosylation of N291 was the only change required to block receptor activity of hAPN for HCoV-229E. When this clone (hAPN/N291KT) had been characterized, we made single amino acid reversions, changing N291E or T293Q and creating two new hAPN expression constructs, hAPN/E291KT and hAPN/N291KQ, respectively. These wild-type and mutant hAPN expression constructs, as well as the pCi-neo expression plasmid, were used to transfect nonpermissive murine CMT93 cells in order to create stable cell lines that expressed similar levels of hAPN. The transfected cells were selected for G418 resistance, and those transfected with hAPN expression plasmids were sorted twice for hAPN expression using FACS with MAb WM47. The stably transfected cell lines, except those transfected by the pCi-neo expression plasmid, all expressed similar levels of hAPN as demonstrated by FACS using MAb WM47 (data not shown). Ensuring that the cell lines expressed similar levels of hAPN showed that glycosylation of hAPN at N291 did not disrupt intracellular transport or expression at the cell membrane.
The cell lines were inoculated with HCoV-229E for 1 h, and cell lysates were isolated 20 h postinoculation. Immunoblot analysis of the cell lysates with polyclonal goat antiserum to HCoV-229E clearly demonstrated infection of cells expressing the wild-type hAPN or the revertants (hAPN/E291KT or hAPN/N291KQ) that remove the glycosylation signal at N291 (Fig. 4, lanes 1, 4, and 5, respectively). In contrast, cells that expressed p6-hAPN and hAPN with the glycosylation sequon at amino acids 291 to 293 (hAPN/N291KT) lacked HCoV-229E receptor activity as shown by immunoblotting (Fig. 4, lanes 2 and 3). Expression of wild-type and mutant hAPN proteins in these various cell lines was also shown by IFA using MAb WM47 (Fig. 5A, C, E, G, and I). Analysis of HCoV-229E infection of the transfected cells after a 20-h inoculation showed a high level of receptor activity in the cell lines that expressed wt-hAPN (Fig. 5B) and the two single-amino-acid revertants (hAPN/E291KT or hAPN/N291KQ) that remove the N291-glycosylation signal (Fig. 5H and J). However, the cell line that expressed hAPN with a glycosylation sequon at amino acids 291 to 293 (hAPN/N291KT) showed a very low level of HCoV-229E receptor activity (Fig. 5F). Data from the immunoblots was slightly different from IFA data in that cells expressing p6-hAPN or hAPN/N291KT completely lacked receptor activity as shown by immunoblotting (Fig. 4, lanes 2 and 3), whereas very few cells were positive by IFA (Fig. 5F). These differences are likely because immunoblots are less sensitive than IFA when very few cells are infected and due to the extreme virus challenge used in the IFA experiment. Comparison of titers of virus released by the various cell lines showed a 100- to 1,000-fold increase when the cells expressed the wild-type hAPN or the revertants (hAPN/E291KT or hAPN/N291KQ), but no increase in titer from cells expressing p6-hAPN, hAPN/N291KT or pCi-neo. In summary, immunoblotting, IFA, and release of virus from stably transfected cell lines demonstrated that N-linked glycosylation of hAPN at amino acid 291 alone blocked infection by HCoV-229E.
FIG. 4.
Addition of an N-linked glycosylation site at amino acid 291 of hAPN blocked HCoV-229E receptor activity. Cell lysates isolated from stably transfected CMT93 cell lines that were sorted by FACS to express similar levels of hAPN were analyzed by immunoblotting. Cell lysates were isolated from cells expressing wt-hAPN (lane 1), p6-hAPN (lane 2), hAPN/N291KT (lane 3), hAPN/E291KT (lane 4), hAPN/N291KQ (lane 5), or a line created with the empty expression vector pCi-neo (lane 6). The cells were inoculated with HCoV-229E at an MOI of 2, and the cell lysates were prepared 21.5 h postinoculation. The proteins were separated by SDS–10% PAGE and transferred to Immobilon-P, and HCoV-229E nucleocapsid (N) protein expression (arrow) was shown using polyclonal goat antiserum to HCoV-229E.
FIG. 5.
Wild-type and mutant hAPN glycoproteins expressed at similar levels on the plasma membrane are differentially infected by HCoV-229E. Cell lines that expressed similar levels of wt-hAPN (A and B), p6-hAPN (C and D), hAPN/N291KT (E and F), hAPN/E291KT (G and H), hAPN/N291KQ (I and J), or a line created by transfection with pCi-neo (K and L) were inoculated with HCoV-229E at an MOI of 1. hAPN expression was identified by indirect IFA with MAb WM47 (A, C, E, G, I, and K), and HCoV-229E spike glycoprotein expression was identified with anti-spike glycoprotein MAb 511H.6 (B, D, F, H, J, and L).
Glycosylation of hAPN at amino acid 291 blocked infection by inhibiting binding of HCoV-229E.
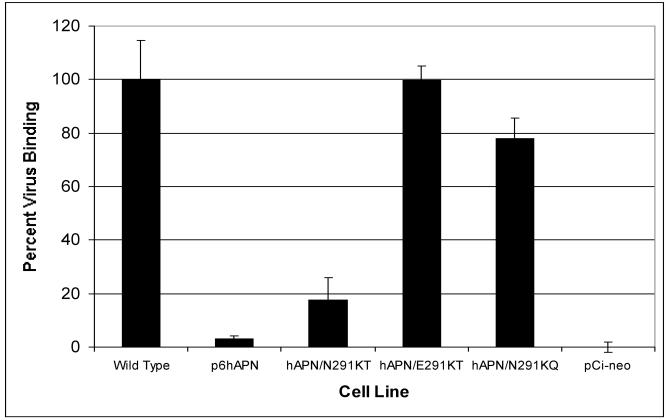
Glycosylation of N291 may inhibit HCoV-229E receptor activity of hAPN either by blocking virus binding or by inhibiting a postbinding step required for virus entry, such as a conformational change in APN or viral S glycoproteins. To determine if N291 glycosylation influenced virus binding, we tested the ability of HCoV-229E to bind to stably transfected, FACS-sorted, CMT93 cell lines. The cell lines expressed wild-type hAPN, mutants containing the glycosylation sequon at amino acids 291 to 293, p6-hAPN (which has six amino acid substitutions) or hAPN/N291KT (which has two amino acid substitutions), the two single amino acid revertants (hAPN/E291KT, hAPN/N291KQ) or the expression vector (pCi-neo). All of the hAPN cell lines expressed similar levels of hAPN at the cell surface. HCoV-229E containing RNA labeled with [3H]-uridine was incubated with the various cell lines at 4°C for 2 h, the cells were washed, and virus binding was quantitated by scintillation counting. Cell lines expressing wild-type hAPN, hAPN/E291KT, or hAPN/N291KQ, which lacked the glycosylation sequon at amino acids 291 to 293, bound 100, 99.8, and 78.0% of the virus, respectively (Fig. 6). In contrast, cells that expressed p6-hAPN or hAPN/N291KT, which had the glycosylation sequon at amino acids 291 to 293, bound 3.1 and 17.4% of the virus, respectively (Fig. 6). These experiments illustrate that N291 glycosylation of hAPN directly inhibited binding of HCoV-229E to the receptor on the plasma membrane.
FIG. 6.
Binding of HCoV-229E was inhibited by the addition of a glycosylation signal at amino acid 291 of hAPN. CMT93 cell lines that had similar levels of hAPN surface expression were mixed with 3H-labeled virus, the cells were washed and lysed, and 3H was counted as described in Materials and Methods. The average 3H-labeled virus bound by the wt-hAPN was 1.38 × 104 cpm and was defined as 100% binding. The average total counts bound by cells transfected with pCi-neo was defined as 0% binding. The average of the other cell lines was reported as a percentage of wild-type binding activity, and the error bars represent the standard deviation of the three replicates.
Removal of a glycosylation signal at N818 of hAPN did not change HCoV-229E receptor activity or allow infection by TGEV or FECoV.
The results described above proved that glycosylation of hAPN at N291 blocked the HCoV-229E receptor activity of hAPN. Furthermore, fAPN, which serves as a receptor for feline and porcine coronavirus and HCoV, is less glycosylated than hAPN or pAPN. To determine if glycosylation sites in hAPN that are not conserved in pAPN or fAPN were responsible for the species-specific receptor interaction observed in serogroup 1 coronaviruses, we removed a potential glycosylation site from wt-hAPN and p6-hAPN. Sequence analysis of APN glycoproteins identified a potential glycosylation sequon at amino acids 818 to 820 of hAPN that is not conserved by pAPN or fAPN. N818 corresponds with Q820 of pAPN and is within amino acids 670 to 840 of fAPN, which are required for TGEV and FIPV receptor activity (22).
To determine if the removal of this N-linked glycosylation sequon converted hAPN into a functional receptor for porcine or feline coronaviruses, we used site-directed mutagenesis to remove this sequon from wild-type hAPN and p6-hAPN, creating hAPN/N818E and p6-hAPN/N818E or p6-hAPN/T820E. These new expression plasmids were transfected into CMT93 cells, selected for G418 resistance, and sorted twice for hAPN expression using FACS with MAb WM47. The stably transfected CMT93 cells and FCWF cells were inoculated with HCoV-229E, TGEV, or FECoV 79-1683 for 1 h. Cell lysates were isolated 22 h postinoculation, separated by SDS-PAGE and immunoblotted with goat polyclonal anti-HCoV-229E serum (Fig. 7A) or feline polyclonal anti-FIPV serum (Fig. 7B and C). The FCWF cells express fAPN and are permissive to HCoV-229E, TGEV, and FECoV or FIPV (71). FCWF cells were used as a positive control for virus infection (Fig. 7, lanes 1). The results indicated that removal of the potential glycosylation site at N818 of hAPN did not alter the ability of HCoV-229E to use the protein as a receptor (Fig. 7A, lane 4). Although this protein has a glycosylation pattern more similar to fAPN, it lacked receptor activity for TGEV and FECoV (Fig. 7B and C, lanes 4). Removal of the glycosylation sequon at amino acids 818 to 820 from p6-hAPN results in a protein that should have a glycosylation pattern that is similar to that of pAPN. However, cells that expressed p6-hAPN/N818E or p6-hAPN/T820E proteins also lacked receptor activity for TGEV and FECoV (Fig. 7B and C, lanes 5 and 6). The results from these experiments suggest that glycosylation of N818 of APN is not the sole discriminator of species specificity.
FIG. 7.
The influence of a glycosylation signal at N818 of hAPN and p6-hAPN on receptor activity for serogroup 1 coronaviruses of humans, pigs, and cats. Nucleocapsid (N) proteins (arrows) were identified by immunoblotting with goat polyclonal antisera to HCoV-229E (A) or feline polyclonal antisera to FIPV that cross-reacts with TGEV (B and C). FCWF (lane 1) or CMT93 cell lines that express wt-hAPN (lane 2), p6-hAPN (lane 3), hAPN/N818E (lane 4), p6-hAPN/N818E (lane 5), and p6-hAPN/T820E (lane 6) or cell lines created with the empty expression vector pCi-neo (lane 7) were inoculated with HCoV-229E (A), TGEV (B), or FECoV 79-1863 (C).
DISCUSSION
A major determinant of the species specificity and tissue tropism of viruses is the presence of a specific receptor(s) on the plasma membrane of the host cells. This is true for many viruses, including human immunodeficiency virus (HIV) (CD4 and CXCR4 or CCR5), Epstein-Barr virus (CD21), major rhinoviruses (ICAM-1), poliovirus (PVR), measles virus (CD46), mouse hepatitis virus (murine CEACAM1), and serogroup 1 coronaviruses (APN or CD13) (14, 15, 18, 72, 77). Many viral attachment proteins and cellular receptors have posttranslational modifications that affect their interaction. These include proteolytic processing, protein folding, and glycosylation. Glycosylation of the hemagglutinin of influenza A and envelope of lentiviruses can have dramatic effects on cleavage activation, immune escape, virulence, and interactions with cellular receptors (3, 6, 7, 12, 13, 16, 27, 47, 53, 55, 56, 76). In contrast to the ample information regarding effects of glycosylation on virus attachment proteins is the limited knowledge of the effects of receptor glycosylation on virus-receptor interactions. The glycosylation of the cellular receptor impacts virus receptor interactions of measles, HIV, and murine leukemia viruses (MuLVs) (17, 45, 59). Studies of the measles virus receptor (CD46) demonstrate that N-glycan addition increases receptor activity (26, 43–45). In contrast, removal of two N-linked glycosylation sites in CXCR4, an HIV coreceptor, increases fusion and entry of HIV type 2 strain ROD/B, illustrating that glycosylation of virus receptors may weaken their interaction with viral attachment proteins (59).
The data presented in this work show that glycosylation of hAPN at amino acid 291 blocks infection by HCoV-229E. The initial hAPN mutant (p6-hAPN), which has pAPN amino acids 283 to 290 substituted for amino acids 288 to 295 of hAPN, completely lacks receptor activity for HCoV-229E. One possible reason for the lack of receptor activity could be poor surface expression of the mutant receptors. To control for this we used cell lines sorted for similar levels of wild-type and mutant hAPN surface expression as well as clones that express p6-hAPN at similar levels and levels three times higher than wild-type hAPN on the plasma membrane. Mixed cell lines and clones that express relatively normal levels of p6-hAPN and a clone that has high-level expression of p6-hAPN lacked receptor activity. This mutant contains six amino acid substitutions, of which two confer a potential glycosylation signal. Glycosylation of N291 could alter APN's folding in the endoplasmic reticulum and Golgi and its conformation on the cell surface and indirectly cause a loss of HCoV-229E receptor activity (23, 52). However, APN enzymatic assay, FACS selection with WM47, and further analysis with a panel of MAbs that bind four other epitopes of wt-hAPN indicate that the amino acid substitutions have little effect on five different epitopes or the aminopeptidase activity (2). These data indicate that the protein is folded normally and expressed correctly on the cell surface. This demonstrates that the loss of HCoV-229E receptor function is due to a change in the interaction of hAPN with the spike glycoprotein. The most dramatic change caused by these mutations is the introduction of a potential N-linked glycosylation sequon at amino acids 291 to 293.
To assess the consequence of the introduction of the glycosylation signal at N291, two new mutants generated from p6-hAPN that revert N291E or T293Q but contain five other amino acid changes were tested (Fig. 8). The two single amino acid revertants that eliminate the glycosylation signal regain receptor activity for HCoV-229E. Additionally, fAPN also has amino acid differences from hAPN at D288, K292, Q293, and S295, suggesting that the block in receptor activity by the substitution of six amino acids in hAPN to those of pAPN is due to glycosylation of N291 rather than the individual amino acid substitutions (Fig. 8). Some of the amino acids in p6-hAPN/N291E and p6-hAPN/T293Q have different properties from those of hAPN. For example, there are side chain structural differences (Y289S) and conversion of basic amino acids to acidic amino acids (K292E). These observations suggest that S of HCoV-229E may not interact directly with any of amino acids 288 to 295 of hAPN, except perhaps V290 or A294 that are conserved in hAPN, pAPN, and fAPN proteins. Given this information one might predict that the removal of the glycosylation sequon N286–288 in pAPN would convert this protein into a functional receptor for HCoV-229E and TGEV. However, previous studies showed that when this glycosylation sequon in pAPN was removed (plasmids AP36 and AP37) changing three or one amino acid(s) of pAPN, respectively, the resulting proteins maintained TGEV receptor activity but did not gain receptor activity for HCoV-229E (33). Thus, in the context of hAPN, glycosylation of N291 blocks infection by HCoV-229E, but the removal of the analogous glycosylation site from pAPN does not convert it into a functional HCoV-229E receptor. This shows that glycosylation of pAPN at N286 is not the only block of HCoV-229E receptor activity in pAPN.
FIG. 8.
Amino acid sequence alignment from amino acids 288 to 295 of the hAPN expression plasmids summarizes the consequence of an N291 glycosylation sequon on HCoV-229E receptor activity. The predicted amino acids of hAPN expression plasmids were aligned using Clustal W (version 1.4) and the Blosum similarity matrix. The numbers above the sequences correspond to amino acids of the hAPN glycoprotein. Identical amino acids are shown on a dark gray background.
A new set of hAPN expression plasmids was generated to demonstrate that glycosylation of N291 of hAPN is the key block of receptor function of p6-hAPN. The mutants used have substitutions that encode the glycosylation sequon (hAPN/N291KT) and two revertants that restore N291E (hAPN/E291KT) or T293Q (hAPN/N291KQ) and remove the glycosylation signal (Fig. 8). Immunoblots of cell lysates from the CMT93 cell lines and virus titration of supernatants after inoculation show no evidence of HCoV-229E receptor activity from either the hAPN/N291KT cell line or the p6-hAPN cell line. Additionally, the data from virus binding experiments using wild-type hAPN, p6-hAPN, hAPN/N291KT and the revertants hAPN/E291KT or hAPN/N291KQ demonstrate that glycosylation of N291 blocks receptor activity by directly inhibiting virus-receptor interaction. However, IFA shows a small difference when the inoculum is incubated with the cell lines for 20 h. IFA of p6-hAPN cell lines directly correlates with immunoblot and virus titration data and shows no HCoV-229E receptor activity. In contrast, IFA data from hAPN/N291KT cell lines show a few HCoV-229E positive cells. The few infected cells likely result from incomplete N291 glycosylation of hAPN. Glycosylation of asparagine residues is more complex than a minimal NX(S/T) sequon. The specific amino acids in the X position that follow the S/T position and the cell type dramatically influence complete glycosylation of the protein (20, 24, 28, 49). The sequence of this mutant hAPN protein is N291KTA, and the sequence of p6-hAPN is N291ETA. The effects of K or E in the second position are not well understood. However, the presence of the threonine residue in the third position confers strong glycosylation properties to rabies virus glycoprotein, as well as 38 other characterized glycoproteins, and minimizes the effect of the amino acid in the second position, with the exception of proline (28). Other studies with rabies virus glycoprotein show that the NLTA sequon is efficiently glycosylated, demonstrating that alanine in the fourth position, as in hAPN/N291KT, does not inhibit glycosylation (49). Our data suggest that the amino acids surrounding the p6-hAPN glycosylation sequon confer more efficient asparagine glycosylation than that of hAPN/N291KT. Alternatively, the amino acid changes that accompany p6-hAPN may be synergistic to the effect of glycosylation, although they do not change spike-receptor interaction enough to completely block receptor activity in the glycosylation revertants p6-hAPN/N291E and p6-hAPN/T293Q. If this were the case, then we would expect to see some reduction in receptor activity of p6-hAPN/N291E and p6-hAPN/T293Q. Yet, similar levels of infectious virus and viral proteins (N and S) are produced from cells expressing wild-type hAPN, p6-hAPN/N291E, and p6-hAPN/T293Q.
Glycosylation of cellular receptors also inhibits infection by reticuloendotheliosis virus, B-lymphotropic papovaviruses and MuLVs (17, 30, 37). The virus receptor interaction that is most analogous to APN and HCoV-229E S is that of the MuLV ecotropic retroviruses. MuLVs are subgrouped into five different categories based on receptor interference characteristics and host range. Ecotropic MuLVs infect rodent cells but not human cells, while amphotropic MuLVs infect cells of rodents and primates. The murine transporter for cationic amino acids (CAT-1) is the cell surface receptor for ecotropic MuLVs (32, 73). CAT-1 expressed by Mus dunni tail fibroblasts is a functional receptor for all ecotropic MuLVs except Moloney-MuLV. However, the removal of the N-linked glycosylation sequon at amino acids 229 to 231 of M. dunni tail fibroblast CAT-1 converts the glycoprotein into a functional receptor for all ecotropic viruses, including Moloney MuLV (17). This is also illustrated by a 220-fold increase in transduction efficiency of an ecotropic MuLV vector upon inhibition of cellular glycosylation (70). Furthermore, N-linked glycosylation of hamster CAT-1 protein inhibits its receptor activity for ecotropic MuLVs and contributes to the resistance of hamsters to these viruses (17, 74, 75). This is very similar to our observations with the substitution of a glycosylation sequon encoded by pAPN into hAPN that blocks HCoV-229E receptor activity. The spike of TGEV likely evolved to use its cognate receptor that contains this glycosylation site in a manner similar to that seen by MuLV strain PVC-211, which is a neurotropic variant of Friend MuLV that has an expanded host range because of changes in the env gene (46).
We also evaluated the ability of the mutant hAPN proteins to serve as receptors for TGEV. Our initial prediction was that none of the substitutions from 288 to 295 would confer TGEV receptor activity because the previous hAPN-pAPN chimeras had demonstrated that a region much closer to the carboxyl terminus of APN is required for TGEV entry (9). It is possible that these two regions of APN that are separated by more than 400 amino acids are close together in the three-dimensional structure of APN, and evidence of FIPV receptor function by an hAPN-pAPN (AP33) chimera suggests that it is likely (33). Additionally, it is possible that the N-linked glycosylation site that blocks receptor activity for HCoV-229E may enhance TGEV receptor function as is true for measles virus and CD46 (26, 43, 44). However, none of the hAPN constructs with mutations from amino acids 288 to 295 showed any receptor activity for TGEV.
We also identified other potential glycosylation sites in hAPN that are not conserved in pAPN or fAPN in the region previously shown to be a critical determinant of receptor activity for porcine, feline, and canine viruses (amino acids 670 to 840). Glycosylation of hAPN in this region may block porcine and feline viruses in the same manner that glycosylation of N291 blocks HCoV-229E. A potential glycosylation sequon from amino acids 818 to 820 of hAPN that is not conserved in pAPN or fAPN was identified, and mutant expression plasmids that no longer have this glycosylation sequon were generated from hAPN or p6-hAPN. hAPN/N818E, a mutant that has a glycosylation pattern more similar to fAPN, has receptor activity for HCoV-229E but not TGEV or FECoV. p6-hAPN/N818E, a mutant that has a glycosylation pattern more similar to pAPN, lacks receptor activity for HCoV-229E, TGEV, and FECoV. Thus, it appears glycosylation of APN is not the sole discriminator of the species specificity in the serogroup 1 coronaviruses. All of the data on the differences observed between hAPN and pAPN from amino acids 717 to 813 of pAPN illustrate that none of the amino acid changes made individually or in combination convert hAPN into a functional receptor for TGEV (9). Thus, simultaneous amino acid substitutions and glycosylation changes may be required for hAPN to function as a receptor for porcine and feline coronaviruses.
In this study we see how expression of the glycosylation signal, normally present in pAPN, blocks infection in LAPN, and the other studies mentioned demonstrate how glycan addition to receptors may aid or inhibit interaction with virus. Thus, species-, tissue-, and cell-specific glycosylation of receptors may play a role in tropism in vivo. We believe that glycosylation of amino acid 291 of hAPN blocks the interaction of HCoV-229E spike glycoprotein with a region of hAPN outside of amino acids 288 to 295 and that the region the virus binds to is conserved among APN glycoproteins of many species. It is likely that the progenitor of all serogroup 1 coronaviruses bound APN in this conserved region. As APN glycoproteins evolved in different species, the viral spike glycoproteins evolved to use certain receptors more efficiently and in doing so became more species specific.
ACKNOWLEDGMENTS
We thank Pierre Talbot for the anti-spike MAb 5-11H.6, Justin Hagee for his technical assistance, Karen Helm for FACS analysis at the University of Colorado Cancer Center Flow Cytometry Core, and Christopher Korch for sequencing the DNA samples at the University of Colorado Cancer Center DNA Sequencing and Analysis Core Facility. We also thank Bruce Zelus, Dianna Blau, Ken Tyler, and Bruce Banfield for critiques of the manuscript.
This work was supported by NIH grant AI26075, and D.E.W. was supported by NIH Neurovirology-Molecular Biology Training Grant-T32 NS07321. FACS analysis at the University of Colorado Cancer Center Flow Cytometry Core was supported by NIH grant 2 P30 CA 46934-09, and sequencing of DNA samples at the University of Colorado Cancer Center DNA Sequencing and Analysis Core Facility was supported by an NIH/NCI Cancer Core Support grant (CA46934).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashmun R A, Holmes K V, Shapiro L H, Favaloro E J, Razak K, de Crom R P G, Howard C J, Look A T. M3 CD13 (aminopeptidase N) cluster workshop report. 1. Leucocyte typing V. In: Schlossman S F, Boumsell L, Gilks W, Harlan J M, Kishimoto T, Morimoto C, Ritz J, Shaw S, Silverstein R, Springer T, Tedder T F, Todd R F, editors. White cell differentiation antigens. Proceedings of the Fifth International Workshop and Conference. Oxford, United Kingdom: Oxford University Press; 1995. pp. 771–775. [Google Scholar]
- 3.Bandres J C, Wang Q F, O'Leary J, Baleaux F, Amara A, Hoxie J A, Zolla-Pazner S, Gorny M K. Human immunodeficiency virus (HIV) envelope binds to CXCR4 independently of CD4, and binding can be enhanced by interaction with soluble CD4 or by HIV envelope deglycosylation. J Virol. 1998;72:2500–2504. doi: 10.1128/jvi.72.3.2500-2504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes K, Kenny A J, Turner A J. Localization of aminopeptidase N and dipeptidyl peptidase IV in pig striatum and in neuronal and glial cell cultures. Eur J Neurosci. 1994;6:531–537. doi: 10.1111/j.1460-9568.1994.tb00297.x. [DOI] [PubMed] [Google Scholar]
- 5.Callow K A, Parry H F, Sergeant M, Tyrrell D A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chackerian B, Rudensey L M, Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J Virol. 1997;71:7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng-Mayer C, Brown A, Harouse J, Luciw P A, Mayer A J. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J Virol. 1999;73:5294–5300. doi: 10.1128/jvi.73.7.5294-5300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danielsen E M, Noren O, Sjostrom H. Co- and post-translational events in the biogenesis of pig small intestinal aminopeptidase N. Tokai J Exp Clin Med. 1982;7(Suppl.):135–140. [PubMed] [Google Scholar]
- 9.Delmas B, Gelfi J, Kut E, Sjostrom H, Noren O, Laude H. Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase-N that is distinct from the enzymatic site. J Virol. 1994;68:5216–5224. doi: 10.1128/jvi.68.8.5216-5224.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delmas B, Gelfi J, L'Haridon R, Vogel L K, Sjostrom H, Noren O, Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmas B, Gelfi J, Sjostrom H, Noren O, Laude H. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv Exp Med Biol. 1993;342:293–298. doi: 10.1007/978-1-4615-2996-5_45. [DOI] [PubMed] [Google Scholar]
- 12.Deshpande K L, Fried V A, Ando M, Webster R G. Glycosylation affects cleavage of an H5N2 influenza virus hemagglutinin and regulates virulence. Proc Natl Acad Sci USA. 1987;84:36–40. doi: 10.1073/pnas.84.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desrosiers R C. Strategies used by human immunodeficiency virus that allow persistent viral replication. Nat Med. 1999;5:723–725. doi: 10.1038/10439. [DOI] [PubMed] [Google Scholar]
- 14.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 15.Dveksler G S, Pensiero M N, Cardellichio C B, Williams R K, Jiang G S, Holmes K V, Dieffenbach C W. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edmonson P, Murphey-Corb M, Martin L N, Delahunty C, Heeney J, Kornfeld, Donahue P R, Learn G H, Hood L, Mullins J I. Evolution of a simian immunodeficiency virus pathogen. J Virol. 1998;72:405–414. doi: 10.1128/jvi.72.1.405-414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eiden M V, Farrell K, Wilson C A. Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J Virol. 1994;68:626–631. doi: 10.1128/jvi.68.2.626-631.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 19.Fujii H, Nakajima M, Saiki I, Yoneda J, Azuma I, Tsuruo T. Human melanoma invasion and metastasis enhancement by high expression of aminopeptidase N/CD13. Clin Exp Metastasis. 1995;13:337–344. doi: 10.1007/BF00121910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Beato R, Martinez I, Franci C, Real F X, Garcia-Barreno B, Melero J A. Host cell effect upon glycosylation and antigenicity of human respiratory syncytial virus G glycoprotein. Virology. 1996;221:301–309. doi: 10.1006/viro.1996.0379. [DOI] [PubMed] [Google Scholar]
- 21.Giugni T D, Soderberg C, Ham D J, Bautista R M, Hedlund K O, Moller E, Zaia J A. Neutralization of human cytomegalovirus by human CD13-specific antibodies. J Infect Dis. 1996;173:1062–1071. doi: 10.1093/infdis/173.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hegyi A, Kolb A F. Characterization of determinants involved in the feline infectious peritonitis virus receptor function of feline aminopeptidase N. J Gen Virol. 1998;79:1387–1391. doi: 10.1099/0022-1317-79-6-1387. [DOI] [PubMed] [Google Scholar]
- 23.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 24.Hollsberg P, Scholz C, Anderson D E, Greenfield E A, Kuchroo V K, Freeman G J, Hafler D A. Expression of a hypoglycosylated form of CD86 (B7-2) on human T cells with altered binding properties to CD28 and CTLA-4. J Immunol. 1997;159:4799–4805. [PubMed] [Google Scholar]
- 25.Holmes K V, Lai M M. Coronaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1075–1094. [Google Scholar]
- 26.Hsu E C, Dorig R E, Sarangi F, Marcil A, Iorio C, Richardson C D. Artificial mutations and natural variations in the CD46 molecules from human and monkey cells define regions important for measles virus binding. J Virol. 1997;71:6144–6154. doi: 10.1128/jvi.71.8.6144-6154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson D C, Drummer H E, Urge L, Otvos L J, Brown L E. Glycosylation of a synthetic peptide representing a T-cell determinant of influenza virus hemagglutinin results in loss of recognition by CD4+ T-cell clones. Virology. 1994;199:422–430. doi: 10.1006/viro.1994.1140. [DOI] [PubMed] [Google Scholar]
- 28.Kasturi L, Chen H, Shakin-Eshleman S H. Regulation of N-linked core glycosylation: use of a site-directed mutagenesis approach to identify Asn-Xaa-Ser/Thr sequons that are poor oligosaccharide acceptors. Biochem J. 1997;323:415–419. doi: 10.1042/bj3230415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny A J, Maroux S. Topology of microvillar membrance hydrolases of kidney and intestine. Physiol Rev. 1982;62:91–128. doi: 10.1152/physrev.1982.62.1.91. [DOI] [PubMed] [Google Scholar]
- 30.Keppler O T, Herrmann M, Oppenlander M, Meschede W, Pawlita M. Regulation of susceptibility and cell surface receptor for the B-lymphotropic papovavirus by N glycosylation. J Virol. 1994;68:6933–6939. doi: 10.1128/jvi.68.11.6933-6939.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khalidi H S, Medeiros L J, Chang K L, Brynes R K, Slovak M L, Arber D A. The immunophenotype of adult acute myeloid leukemia: high frequency of lymphoid antigen expression and comparison of immunophenotype, French-American-British classification, and karyotypic abnormalities. Am J Clin Pathol. 1998;109:211–220. doi: 10.1093/ajcp/109.2.211. [DOI] [PubMed] [Google Scholar]
- 32.Kim J W, Closs E I, Albritton L M, Cunningham J M. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature. 1991;352:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 33.Kolb A F, Hegyi A, Maile J, Heister A, Hagemann M, Siddell S G. Molecular analysis of the coronavirus-receptor function of aminopeptidase N. Adv Exp Med Biol. 1998;440:61–67. doi: 10.1007/978-1-4615-5331-1_8. [DOI] [PubMed] [Google Scholar]
- 34.Kolb A F, Hegyi A, Siddell S G. Identification of residues critical for the human coronavirus 229E receptor function of human aminopeptidase N. J Gen Virol. 1997;78:2795–2802. doi: 10.1099/0022-1317-78-11-2795. [DOI] [PubMed] [Google Scholar]
- 35.Kolb A F, Maile J, Heister A, Siddell S G. Characterization of functional domains in the human coronavirus HCV 229E receptor. J Gen Virol. 1996;77:2515–2521. doi: 10.1099/0022-1317-77-10-2515. [DOI] [PubMed] [Google Scholar]
- 36.Konikova E, Glasova M, Kusenda J, Babusikova O. Intracellular markers in acute myeloid leukemia diagnosis. Neoplasma. 1998;45:282–291. [PubMed] [Google Scholar]
- 37.Koo H M, Parthasarathi S, Ron Y, Dougherty J P. Pseudotyped REV/SRV retroviruses reveal restrictions to infection and host range within members of the same receptor interference group. Virology. 1994;205:345–351. doi: 10.1006/viro.1994.1651. [DOI] [PubMed] [Google Scholar]
- 38.Kunz J, Krause D, Kremer M, Dermietzel R. The 140-kDa protein of blood-brain barrier-associated pericytes is identical to aminopeptidase N. J Neurochem. 1994;62:2375–2386. doi: 10.1046/j.1471-4159.1994.62062375.x. [DOI] [PubMed] [Google Scholar]
- 39.Lachance C, Arbour N, Cashman N R, Talbot P J. Involvement of aminopeptidase N (CD13) in infection of human neural cells by human coronavirus 229E. J Virol. 1998;72:6511–6519. doi: 10.1128/jvi.72.8.6511-6519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsen S L, Pedersen L O, Buus S, Stryhn A. T cell responses affected by aminopeptidase N (CD13)-mediated trimming of major histocompatibility complex class II-bound peptides. J Exp Med. 1996;184:183–189. doi: 10.1084/jem.184.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsson S, Soderberg-Naucler C, Moller E. Productive cytomegalovirus (CMV) infection exclusively in CD13-positive peripheral blood mononuclear cells from CMV-infected individuals: implications for prevention of CMV transmission. Transplantation. 1998;65:411–415. doi: 10.1097/00007890-199802150-00021. [DOI] [PubMed] [Google Scholar]
- 42.Look A T, Ashmun R A, Shapiro L H, Peiper S C. Human myeloid plasma membrane glycoprotein CD13 (gp150) is identical to aminopeptidase N. J Clin Investig. 1989;83:1299–1307. doi: 10.1172/JCI114015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maisner A, Alvarez J, Liszewski M K, Atkinson D J, Atkinson J P, Herrler G. The N-glycan of the SCR 2 region is essential for membrane cofactor protein (CD46) to function as a measles virus receptor. J Virol. 1996;70:4973–4977. doi: 10.1128/jvi.70.8.4973-4977.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maisner A, Herrler G. Membrane cofactor protein with different types of N-glycans can serve as measles virus receptor. Virology. 1995;210:479–481. doi: 10.1006/viro.1995.1365. [DOI] [PubMed] [Google Scholar]
- 45.Manchester M, Gairin J E, Patterson J B, Alvarez J, Liszewski M K, Eto D S, Atkinson J P, Oldstone M B. Measles virus recognizes its receptor, CD46, via two distinct binding domains within SCR1-2. Virology. 1997;233:174–184. doi: 10.1006/viro.1997.8581. [DOI] [PubMed] [Google Scholar]
- 46.Masuda M, Hanson C A, Hoffman P M, Ruscetti S K. Analysis of the unique hamster cell tropism of ecotropic murine leukemia virus PVC-211. J Virol. 1996;70:8534–8539. doi: 10.1128/jvi.70.12.8534-8539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsas R, Stephenson S L, Hryszko J, Kenny A J, Turner A J. The metabolism of neuropeptides. Phase separation of synaptic membrane preparations with Triton X-114 reveals the presence of aminopeptidase N. Biochem J. 1985;231:445–449. doi: 10.1042/bj2310445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mellquist J L, Kasturi L, Spitalnik S L, Shakin-Eshleman S H. The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry. 1998;37:6833–6837. doi: 10.1021/bi972217k. [DOI] [PubMed] [Google Scholar]
- 50.Menrad A, Speicher D, Wacker J, Herlyn M. Biochemical and functional characterization of aminopeptidase N expressed by human melanoma cells. Cancer Res. 1993;53:1450–1455. [PubMed] [Google Scholar]
- 51.Murray R S, Brown B, Brian D, Cabirac G F. Detection of coronavirus RNA and antigen in multiple sclerosis brain. Ann Neurol. 1992;31:525–533. doi: 10.1002/ana.410310511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naim H Y, Joberty G, Alfalah M, Jacob R. Temporal association of the N- and O-linked glycosylation events and their implication in the polarized sorting of intestinal brush border sucrase-isomaltase, aminopeptidase N, and dipeptidyl peptidase IV. J Biol Chem. 1999;274:17961–17967. doi: 10.1074/jbc.274.25.17961. [DOI] [PubMed] [Google Scholar]
- 53.Nakayama E E, Shioda T, Tatsumi M, Xin X, Yu D, Ohgimoto S, Kato A, Sakai Y, Ohnishi Y, Nagai Y. Importance of the N-glycan in the V3 loop of HIV-1 envelope protein for CXCR-4- but not CCR-5-dependent fusion. FEBS Lett. 1998;426:367–372. doi: 10.1016/s0014-5793(98)00375-5. [DOI] [PubMed] [Google Scholar]
- 54.Noren O, Sjostrom H, Olsen J. Aminopeptidase N. In: Kenney A J, Boustead C M, editors. Cell-surface peptidases in health and disease. Oxford, United Kingdom: BIOS Scientific Publishers; 1997. pp. 175–191. [Google Scholar]
- 55.Olsen C W, McGregor M W, Cooley A J, Schantz B, Hotze B, Hinshaw V S. Antigenic and genetic analysis of a recently isolated H1N1 swine influenza virus. Am J Vet Res. 1993;54:1630–1636. [PubMed] [Google Scholar]
- 56.Perdue M L, Latimer J W, Crawford J M. A novel carbohydrate addition site on the hemagglutinin protein of a highly pathogenic H7 subtype avian influenza virus. Virology. 1995;213:276–281. doi: 10.1006/viro.1995.1571. [DOI] [PubMed] [Google Scholar]
- 57.Phillips A J, Tomasec P, Wang E Y, Wilkinson G G, Borysiewicz L K. Human cytomegalovirus infection downregulates expression of the cellular aminopeptidases CD10 and CD13. Virology. 1998;250:350–358. doi: 10.1006/viro.1998.9385. [DOI] [PubMed] [Google Scholar]
- 58.Philpott M S, Easterday B C, Hinshaw V S. Antigenic and phenotypic variants of a virulent avian influenza virus selected during replication in ducks. J Wildl Dis. 1989;25:507–513. doi: 10.7589/0090-3558-25.4.507. [DOI] [PubMed] [Google Scholar]
- 59.Potempa S, Picard L, Reeves J D, Wilkinson D, Weiss R A, Talbot S J. CD4-independent infection by human immunodeficiency virus type 2 strain ROD/B: the role of the N-terminal domain of CXCR-4 in fusion and entry. J Virol. 1997;71:4419–4424. doi: 10.1128/jvi.71.6.4419-4424.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riemann D, Kehlen A, Langner J. CD13 —not just a marker in leukemia typing. Immunol Today. 1999;20:83–88. doi: 10.1016/S0167-5699(98)01398-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saiki I, Fujii H, Yoneda J, Abe F, Nakajima M, Tsuruo T, Azuma I. Role of aminopeptidase N (CD13) in tumor-cell invasion and extracellular matrix degradation. Int J Cancer. 1993;54:137–143. doi: 10.1002/ijc.2910540122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Semenza G. Anchoring and biosynthesis of stalked brush border membrane proteins: glycosidases and peptidases of enterocytes and renal tubuli. Annu Rev Cell Biol. 1986;2:255–313. doi: 10.1146/annurev.cb.02.110186.001351. [DOI] [PubMed] [Google Scholar]
- 63.Soderberg C, Giugni T D, Zaia J A, Larsson S, Wahlberg J M, Moller E. CD13 (human aminopeptidase N) mediates human cytomegalovirus infection. J Virol. 1993;67:6576–6585. doi: 10.1128/jvi.67.11.6576-6585.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soderberg C, Larsson S, Rozell B L, Sumitran-Karuppan S, Ljungman P, Moller E. Cytomegalovirus-induced CD13-specific autoimmunity—a possible cause of chronic graft-vs-host disease. Transplantation. 1996;61:600–609. doi: 10.1097/00007890-199602270-00015. [DOI] [PubMed] [Google Scholar]
- 65.Soderberg C, Sumitran-Karuppan S, Ljungman P, Moller E. CD13-specific autoimmunity in cytomegalovirus-infected immunocompromised patients. Transplantation. 1996;61:594–600. doi: 10.1097/00007890-199602270-00014. [DOI] [PubMed] [Google Scholar]
- 66.Stange T, Kettmann U, Holzhausen H J. Immunoelectron microscopic single and double labelling of aminopeptidase N (CD 13) and dipeptidyl peptidase IV (CD 26) Acta Histochem. 1996;98:323–331. doi: 10.1016/S0065-1281(96)80025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stasi R, Del P G, Venditti A, Bruno A, Suppo G, Aronica G, Di C, Papa G. Lineage identification of acute leukemias: relevance of immunologic and ultrastructural techniques. Hematol Pathol. 1995;9:79–94. [PubMed] [Google Scholar]
- 68.Stewart J N, Mounir S, Talbot P J. Human coronavirus gene expression in the brains of multiple sclerosis patients. Virology. 1992;191:502–505. doi: 10.1016/0042-6822(92)90220-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Talbot P J, Ekande S, Cashman N R, Mounir S, Stewart J N. Neurotropism of human coronavirus 229E. Adv Exp Med Biol. 1993;342:339–346. doi: 10.1007/978-1-4615-2996-5_52. [DOI] [PubMed] [Google Scholar]
- 70.Tavoloni N, Rudenholz A. Variable transduction efficiency of murine leukemia retroviral vector on mammalian cells: role of cellular glycosylation. Virology. 1997;229:49–56. doi: 10.1006/viro.1996.8412. [DOI] [PubMed] [Google Scholar]
- 71.Tresnan D B, Levis R, Holmes K V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J Virol. 1996;70:8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tyler K L, Fields B N. Pathogenesis of viral infections. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 173–218. [Google Scholar]
- 73.Wang H, Kavanaugh M P, North R A, Kabat D. Cell-surface receptor for ecotropic murine retroviruses is a basic amino-acid transporter. Nature. 1991;352:729–731. doi: 10.1038/352729a0. [DOI] [PubMed] [Google Scholar]
- 74.Wang H, Klamo E, Kuhmann S E, Kozak S L, Kavanaugh M P, Kabat D. Modulation of ecotropic murine retroviruses by N-linked glycosylation of the cell surface receptor/amino acid transporter. J Virol. 1996;70:6884–6891. doi: 10.1128/jvi.70.10.6884-6891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson C A, Eiden M V. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J Virol. 1991;65:5975–5982. doi: 10.1128/jvi.65.11.5975-5982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu Z, Kayman S C, Honnen W, Revesz K, Chen H, Vijh-Warrier S, Tilley S A, McKeating J, Shotton C, Pinter A. Characterization of neutralization epitopes in the V2 region of human immunodeficiency virus type 1 gp120: role of glycosylation in the correct folding of the V1/V2 domain. J Virol. 1995;69:2271–2278. doi: 10.1128/jvi.69.4.2271-2278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeager C L, Ashmun R A, Williams R K, Cardellichio C B, Shapiro L H, Look A T, Holmes K V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zelus B D, Wessner D R, Williams R K, Pensiero M N, Phibbs F T, deSouza M, Dveksler G S, Holmes K V. Purified, soluble recombinant mouse hepatitis virus receptor, Bgp1(b), and Bgp2 murine coronavirus receptors differ in mouse hepatitis virus binding and neutralizing activities. J Virol. 1998;72:7237–7244. doi: 10.1128/jvi.72.9.7237-7244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]