Abstract
DNA methylation, by regulating the transcription of genes, is a major modifier of the eukaryotic genome. DNA methyltransferases (DNMTs) are responsible for both maintenance and de novo methylation. We have reported that human immunodeficiency virus type 1 (HIV-1) infection increases DNMT1 expression and de novo methylation of genes such as the gamma interferon gene in CD4+ cells. Here, we examined the mechanism(s) by which HIV-1 infection increases the cellular capacity to methylate genes. While the RNAs and proteins of all three DNMTs (1, 3a, and 3b) were detected in Hut 78 lymphoid cells, only the expression of DNMT1 was significantly increased 3 to 5 days postinfection. This increase was observed with either wild-type HIV-1 or an integrase (IN) mutant, which renders HIV replication defective, due to the inability of the provirus to integrate into the host genome. Unintegrated viral DNA is a common feature of many retroviral infections and is thought to play a role in pathogenesis. These results indicate another mechanism by which unintegrated viral DNA affects the host. In addition to the increase in overall genomic methylation, hypermethylation and reduced expression of the p16INK4A gene, one of the most commonly altered genes in human cancer, were seen in cells infected with both wild-type and IN-defective HIV-1. Thus, infection of lymphoid cells with integration-defective HIV-1 can increase the methylation of CpG islands in the promoters of genes such as the p16INK4A gene, silencing their expression.
One function widely ascribed to DNA methylation is that of a genome defense mechanism against foreign invaders. Retroviruses integrate randomly in the host genome, where they are susceptible to silencing through methylation, which depends on the local DNA environment (6, 26, 33, 87). However, methylation of proviral DNA, which retains the ability to be reactivated, leading to productive infection, allows the virus to survive by escaping immune surveillance. DNA methylation is a major modifier of the eukaryotic genome (8, 85). In mammals, DNA methylation is essential for normal embryonic development, as it plays an important role in the regulation of gene expression, X chromosome inactivation, genomic imprinting, chromatin modification, and silencing of endogenous retroviruses (2, 42, 58, 77). Exactly how DNA methylation recognizes methylated and nonmethylated sequences, transcriptionally represses genes, and stably maintains these patterns during cell replication is not clear. The packaging of the DNA template within chromatin largely controls the transcriptional activity of a gene (85). In 1998, two groups (37, 57) showed that a methyl-binding protein (MeCP2) forms a complex with histone deacetylase (HDAC) and a transcriptional repressor (Sin-3). This linked DNA methylation with histone deacetylation, a universal mechanism for gene silencing (34). To achieve stable repression, the chromatin around the inactive gene becomes more densely methylated and more condensed through histone deacetylation, which can be involved in oncogenic transformation and other pathogenic states (36).
Three enzymes responsible for DNA methylation, known as DNA methyltransferases (DNMTs), have been identified. The carboxy-terminal domain of DNMT1 catalyzes the methylation of DNA containing hemimethylated CpG dinucleotides more efficiently than that of unmethylated DNA in vitro (6). DNMT1 is the enzyme mainly responsible for maintaining DNA methylation patterns in adult mammalian tissues and also participates in de novo methylation on C-type CpG islands in human carcinogenesis (4, 38). It has recently been shown that the noncatalytic N-terminal domain of DNMT1 can act as a transcriptional repressor binding directly to HDAC2 and a new corepressor, DMAP1, to form a complex at the replication foci (73). DNMT1 also forms a complex with Rb, E2F1, and HDAC1 to repress transcription of E2F-responsive promoters (71), suggesting that DNMT1 has activities other than the enzymatic function of a methyltransferase. Two other forms of DNMTs have been isolated in mammals (62, 84). The recently identified DNMT3a and DNMT3b are essential for de novo methylation and for mouse development (64).
During retroviral infection, methylation is increased throughout the viral genome, particularly the viral long terminal repeats (LTR), suggesting that methylation can be a mechanism of suppression of viral expression and latency for human immunodeficiency virus type 1 (HIV-1) and human T-cell leukemia virus type 1 (HTLV-1) (20, 39, 51, 74, 75). Previously, we showed that acute infection of cells with HIV-1 results in an increase in DNMT1 expression and activity, an overall increase in methylated genomic DNA in infected cells, and the de novo methylation of a single CpG dinucleotide in the gamma interferon (IFN-γ) gene promoter, which subsequently down-regulated the expression of interferon (53). However, little is known about the mechanism(s) of the methylation changes seen after HIV-1 infection. As unintegrated circular retroviral DNA has been implicated in the pathogenesis of several retroviral infections, we asked whether replication of HIV-1 is necessary for the increases in DNMT activity and the increased capacity of infected cells to methylate genes de novo. Here, we report that integration-defective HIV-1 could also induce increased DNMT1 expression and activity, resulting in hypermethylation and reduced expression of the tumor suppressor gene p16INK4A following acute HIV-1 infection in lymphoid cell lines. This study further implicates aberrant methylation as a mechanism of pathogenesis during HIV-1 infection and suggests the methylation machinery as a novel target for AIDS therapy.
MATERIALS AND METHODS
Cell culture.
Hut 78, a mature CD4+ T-cell line derived from a T-cell lymphoma, was cultured at 37°C and 5% CO2, in RPMI 1640. Dulbecco's modified Eagle Medium (DMEM) was used to culture 293 T cells. Media (Biowhittaker, Walkersville, Md.) were supplemented with 10% heat-inactivated fetal calf serum (HyClone, Logan, Utah), 300 μg of l-glutamine/ml, 100 μg of penicillin/ml, and 100 μg of streptomycin/ml.
Plasmids and viral stocks.
All viral stocks were generated by transient transfection of 293 T cells using a calcium phosphate transfection system (Life Technologies, Inc., Gaithersburg, Md.). Cells were plated in 100-mm tissue culture dishes 24 h before transfection. Cells were refed fresh, complete DMEM 3 h before transfection. The precipitate was kept on cells for 24 h prior to replacement with fresh, complete medium and culture for an additional 24 h. Supernatants were filtered through a 0.45-μm-pore-size filter, which collected all of the medium cell-free. The amount of viral p24 antigen was determined using an enzyme-linked immunosorbent assay kit (Cellular Products, Buffalo, N.Y.) with a sensitivity of 10 pg/ml. The strains used included WT NL4–3 (a full-length molecular clone of HIV-1 [1]), D116N (containing a mutation in the catalytic core domain; kindly provided by Alan Engelmann, Dana-Farber Cancer Institute, Boston, Mass. [18, 19]), and a reverse transcription (RT)-negative mutant (kindly provided by Robert Gorelick, AIDS Vaccine Program, SAIC—Frederick, Frederick, Md. [23]).
Infection of Hut 78 cells with HIV-1.
Hut 78 cells were infected at a concentration of 107 cells in 1 ml of total volume in a 50-ml conical centrifuge tube to which 10 to 30 ng of p24 antigen from mutant or wild-type HIV-1 was added. After incubation in a shaking water bath for 2 h at 37°C, cells were washed twice, resuspended in 30 ml of RPMI complete medium in a T75 flask, and incubated at 37°C. At various times postinfection (p.i.), DNA, RNA, and nuclear proteins were isolated. In some experiments, Hut 78 cells were pretreated overnight with the hypomethylating agent 5-azacytidine (5-AzaC) before infection. Cells (5 × 106) were seeded in a 100-mm dish 12 to 24 h before treatment and were then exposed to 1 to 10 μM 5-AzaC or 5-deoxy-AzaC for 24 h. Control cultures (mock treated) were treated with the same volume of phosphate-buffered saline (PBS). Twenty-four hours after addition of 5-AzaC, the culture medium was replaced with drug-free medium, and the cells were infected using D116N or WT NL4–3.
Isolation and PCR analysis of HIV-1 DNA.
DNA was isolated according to the Hirt method (30). Briefly, cells were first washed in PBS without Ca2+ and Mg2+ and then centrifuged at 750 × g; the pellet was resuspended in 470 μl of 10 mM EDTA, pH 7.5, and transferred to a 1.5-ml Eppendorf style centrifuge tube. Thirty microliters of 10% (wt/vol) sodium dodecyl sulfate was added to this pellet and mixed by gentle inversion of the tube to prevent shearing of chromosomal DNA. Following a 20-min incubation at room temperature, 125 μl of 5 M NaCl was added and mixed again by gentle inversion. After incubation overnight at 4°C, the solution was centrifuged at 17,000 × g for 30 min at 4°C. The supernatant was phenol-chloroform extracted at least three times until no interface was observed. The aqueous layer was precipitated in 70% ethanol and resuspended in 25 to 50 μl of Tris-EDTA (pH 8.0). The viral DNA synthesis status was analyzed by PCR using primers which amplify sequences from the 2-long terminal repeat (2-LTR)-containing circles formed in the nuclei of infected cells. The primer sequences were 5′-GAG ATC CCT CAG ACC CTT TTA G-3′ (sense) and 5′-GTC AGT GGA TAT CTG ATC CCT G-3′ (antisense). Reaction components were mixed at room temperature and heated to 94°C prior to addition of Taq polymerase. We performed 34 cycles of denaturation at 94°C for 45 s, annealing at 60°C for 30 s, and polymerization at 72°C for 2 min, and 1 cycle at 72°C for 5 min. We then electrophoresed 20 μl of each reaction mixture in 2.0% agarose gels (Novex, San Diego, Calif.). Gels were stained with 0.5 μg of ethidium bromide per ml to visualize the DNA. The oligonucleotide primer pair M667 (sense)–AA55 (antisense) was used to determine the 5′ R-U5 region of the LTR in HIV-1 DNA (88). The sequence of M667 was 5′-GGC TAA CTA GGG AAC CCA CTG-3′, and the sequence of AA55 was 5′-CTG CTA GAG ATT TTC CAC ACT GAC-3′. The PCR program was 91°C for 1 min, 65°C for 2 min, 72°C for 1 min, and one cycle at 72°C for 5 min.
Detection of intracellular HIV cores by flow cytometry
Cells were cultured at 106/ml for 6 h in 10 μg of brefeldin A (Sigma, St. Louis, Mo.)/ml. After three washes in PBS, the cells were resuspended in 1 ml of freshly prepared 2% paraformaldehyde in PBS (pH 7.2) and incubated for 2 h at 4°C. The cells were then washed three times in PBS, resuspended in 1 ml of PBS containing 0.1% saponin (Sigma), and incubated for 10 to 30 min at 4°C for efficient permeabilization, which was tested using an anti-actin antibody (Sigma). A phycoerythrin-labeled anti-HIV core antibody (KC-57; Beckman-Coulter, Brea, Calif.) was added, and the cells were incubated for 45 min at 4°C in the dark. After being washed, the cells were left undisturbed for 10 min, resuspended, and immediately analyzed by flow cytometry using a FASCAN (Becton Dickinson, Mountain View, Calif.). Data were analyzed using Flow Jo software (Tri Star, Inc., San Carlos, Calif.).
RT-PCR for DNMTs.
Total cellular RNA from Hut 78 cells infected with HIV-1 or mock infected was extracted and purified with TRIzol Reagent (Life Technologies). RNA was resuspended in diethyl pyrocarbonate-treated water and quantitated by the optical density at 260 or 280 nm. An agarose gel using ethidium staining also verified the quantitation. The samples were treated with DNase I (Roche, Indianapolis, Ind.). RT reactions using 2.5 μg of total RNA in a total reaction volume of 20 μl were performed using Superscript II reverse transcriptase (Life Technologies). PCR mixtures containing 2.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (Boehringer Mannheim), 1 μM each primer, 2 μl of cDNA from the RT reaction, and 2.5 U of Taq DNA polymerase (Sigma) in a volume of 50 μl were amplified using the following PCR conditions taken from the work of Robertson et al. (DNMT3a and β-actin) (69) and Mizuno et al. (DNMT1 and DNMT3b) (54). For DNMT3a and β-actin, 35 and 25 cycles, respectively, of 94°C for 2 min, 94°C for 0.5 min, the transcript-specific annealing temperature (65°C for DNMT3a and 60°C for β-actin) for 1 min, and 72°C for 1 min (with one 322-bp fragment of β-actin cDNA used as a control) were carried out. For DNMT1 and DNMT3b, the PCR programs were 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. Primer sequences (Life Technologies) used were as follows: for DNMT1 (GenBank accession number XM017218), 5′-ACC GCT TCT ACT TCC TCG AGG CCT A-3′ (sense) and 5′-CCA CAG TGT TCA CAG AGG ACT GCA AC-3′ (antisense); for DNMT3a (GenBank accession number NM022552), 5′-GGG GAC GTC CGC AGC GTC ACA C-3′ (sense) and 5′-CAG GGT TGG ACT CGA GAA ATC GC-3′ (antisense); for DNMT3b (GenBank accession number XM009449), 5′-AAT GTG AAT CCA GCC AGG AAA GGC-3′ (sense) and 5′-ACT GGA TTA CAC TCC AGG AAC CGT-3′ (antisense); and for β-actin (GenBank accession number XM004814), 5′-GGA GTC CTG TGG CAT CCA CG-3′ (sense) and 5′-CTA GAA GCA TTT GCG GTG GA-3′ (antisense). The density of each band obtained by RT-PCR in each lane was normalized to the amount of total RNA as determined by the density of the band obtained by RT-PCR for β-actin; i.e., if the β-actin control value was 30,000 U (pixels of brightness), then the calculation used to normalize DNMTs to β-actin can be expressed as [30,000/(density of β-actin)] × (density of DNMTs). The RT-PCR analysis was done at least three times.
Product analysis using real-time PCR.
Quantitative RT-PCR (QRT-PCR) was performed on a Light Cycler (Roche Molecular Systems) by using syber green, which fluoresces upon binding to double-stranded DNA, according to the manufacturer's instructions, and results were normalized to the β-actin control as described above. To discriminate between specific products and nonspecific products such as primer dimers, DNA melting curves were generated (68). Fluorescence data were converted to melting peaks using Roche data analysis software, then plotted as the negative derivative of fluorescence with respect to temperature (−dF/dT versus T, where F is fluorescence and T is temperature). The area of the specific melting peak is directly proportional to the amount of intended product (65, 68). The area under the melting peak was determined using Gaussian Fit software (Roche Molecular Systems).
DNMT protein expression using Western blot analysis.
To examine DNMT protein levels following acute HIV-1 infection, Western blotting was carried out. Nuclear extracts were prepared from both infected and uninfected Hut 78 cells by washing cells with PBS and lysing in buffer A (10 mM HEPES [pH 7.4], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg of protease inhibitors/ml, 0.025% NP-40) for 15 min with rotation at 4°C. The nuclear pellet was resuspended in buffer B (20 mM HEPES [pH 7.4], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.5 mM DTT, 0.2 mM PMSF, 1 μg of protease inhibitors/ml) for 30 min. The soluble nuclear protein was collected by centrifugation. A 200-μg portion of each nuclear extract was boiled in loading buffer for 5 min and then loaded onto a sodium dodecyl sulfate–14% polyacryamide gel. After electrophoresis, proteins were electroeluted onto a polyvinylidene difluoride membrane. A rabbit polyclonal antibody against DNMT1 (diluted 1:500) was purchased from New England Biolabs (Newton, Mass.). The rabbit polyclonal antibody against DNMT3a was a gift from Keith Robertson (National Cancer Institute [NCI], Bethesda, Md.). A mouse monoclonal antibody against DNMT3b (diluted 1:200) was obtained from Alexis Biochemicals (San Diego, Calif.). An antibody against β-actin (Sigma) was used as a control for protein input.
Genomic DNA methylation analysis.
A modified methyl-accepting assay (86) was used to determine the methylation status of DNA isolated from infected and uninfected Hut 78 cells. DNA (200 ng) was incubated with 4 U of SssI CpG methylase (New England Biolabs) in the presence of 1.5 μM S-adenosyl-l-[methyl-3H]methionine (60 to 85 Ci/mmol; TRK 581; Amersham) and 1.5 μM nonradioactive S-adenosylmethionine (SAM) (New England Biolabs). The reaction mixtures (20 μl) were incubated at 37°C for 4 h in a buffer containing 10 mM Tris-HCl (pH 7.9), 50 mM NaCl, 10 mM MgCl2, and 1 mM DTT. The reactions were stopped by addition of 5 μl of 2.5 mM nonradioactive SAM, and reaction products were spotted on 2.4-cm2 Whatman GF/C filter disks, which were air dried for 15 min and washed with 6 ml of 5% (wt/vol) trichloroacetic acid and 70% (vol/vol) ethanol. Disks were counted in Econofluor in a Beckman liquid scintillation counter. Control reactions without DNA or enzyme added were included as background, and these results never exceeded 5% of those in the test samples. All samples were done in triplicate, and values were obtained as disintegrations per minute per nanogram of DNA.
Methylation-specific PCR (MSP-PCR) for p16INK4A.
We used the bisulfite treatment method of Clark et al. (15) with some modifications as follows. Two micrograms of total genomic DNA (from at least two independent infections corresponding to RT-PCR experiments) was isolated with the QIAamp DNA Blood Mini Kit (Qiagen Inc.) and then denatured with NaOH and modified with a freshly prepared sodium bisulfite solution (2.35 M) containing hydroquinone (0.04 M). The bisulfite-treated DNA was desalted using the Wizard DNA Clean Up kit (Promega). To amplify the p16INK4A promoter, we used a 0.1-μg aliquot of the converted DNA. Methylation of the 5′ CpG island in the p16INK4A gene was also determined in samples from Hut 78 cells infected with wild-type or mutant HIV-1, as well as those treated with 5-AzaC. Bisulfite-treated DNA was amplified by PCR using primers specific (GenBank accession number X94154) for the methylated (sense, 5′-TTA TTA GAG GGT GGG GCG GAT CGC-3′; antisense, 5′-GAC CCC GAA CCG CGA CCG TAA-3′) or unmethylated (sense, 5′-TTA TTA GAG GGT GGG GTG GAT TGT-3′; antisense, 5′-CAA CCC CAA ACC ACA ACC ATA A-3′ [28, 29]) CpG island. The thermocycler program was 95°C for 5 min, 5 cycles of 95°C for 1 min, 65°C (methylated) or 60°C (unmethylated) for 2 min, and 72°C for 3 min, and then 35 cycles of 95°C for 30 s, 65 or 60°C for 30 s, and 72°C for 30 s in a 50-μl volume containing 100 ng of bisulfite-treated DNA, 0.1 mM deoxynucleoside triphosphates, 2.0 mM MgCl2, and 0.5 μM primers. The PCR product was directly loaded onto 3% agarose gels and electrophoresed. The gel was stained with ethidium bromide and directly visualized under UV illumination. Oligonucleotide primers for a stretch of the MYOD1 gene completely devoid of CpG dinucleotides were used for a control reaction for equal loading and amplification of bisulfite-treated DNA (17). The sense primer was 5′-CCA ACT CCA AAT CCC CTC TCT AT-3′, and the antisense primer was 5′-TGA TTA ATT TAG ATT GGG TTT AGA GAA GGA-3′. The PCR program for MYOD1 was 30 s at 94 °C, 1 min at 55°C, and 72°C for 1 min. Futhermore, wild-type p16INK4A primers were used to ensure that complete conversion of DNA was obtained in the bisulfite reaction. A positive control for complete methylation was also amplified. In this control, DNA isolated from Hut 78 cells was treated with SssI methylase (New England Biolabs) prior to bisulfite treatment. This enzyme methylates all CpG dinucleotides.
RT-PCR of p16INK4A.
mRNA expression of the p16INK4A gene was determined by RT-PCR of RNA (using the same samples with which the RT-PCR analysis of DNMTs was done) from Hut 78 cells infected with D116N or WT NL4–3 with or without 5-AzaC treatment. The sequences (56) of the primers (GenBank accession number L27211) used were 5′-CCC GCT TTC GTA GTT TTC AT-3′ (sense) and 5′-TTA TTT GAG CTT TGG TTC TG-3′ (antisense). In PCRs, the concentration of each primer was 0.5 μM in 50 μl. PCR amplification was run for 35 cycles, with each amplification cycle consisting of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min. The size of the PCR product was 355 bp. The primer sequence and PCR program for β-actin cDNA amplification were as described for the DNMT RT-PCR. The PCR product was visualized on 2% agarose gels.
RESULTS
Increased expression of DNMT1 in lymphoid cells infected with integration-defective HIV-1.
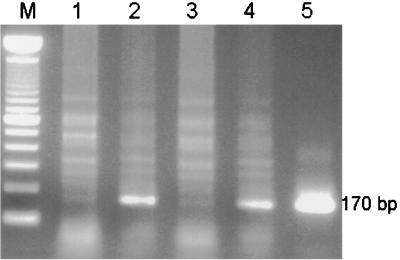
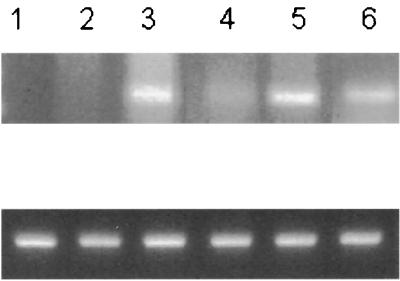
We previously reported that acute HIV infection of primary T cells and lymphoid cell lines results in up-regulation of the expression and activity of DNMT1 and that, consequently, infected cells have an increased capacity to methylate genes (53). In considering mechanisms for this increased DNMT1 expression, the presence of large amounts of unintegrated viral DNA during HIV infection (5, 50, 78) suggested that viral integration was not needed. Therefore, DNMT expression was examined in lymphoid cells at various time points following acute infection with the wild type HIV-1 strain NL4–3 (1), a mutant incapable of integration (D116N [18, 19]), and a mutant incapable of RT (RT negative [23, 24]). Since these mutant proviral clones produced virus particles at lower levels than the wild type when transfected into 293 T cells, in our studies, infections were normalized to the p24 core antigen (CA) content of the virus stock and carried out at a multiplicity of infection which would limit the spread of the wild-type virus. The presence of unintegrated 2-LTR circles demonstrates the nuclear presence of viral intermediates. D116N and WT NL4–3 showed similar levels of 2-LTR circles, while the RT-negative mutant showed no 2-LTR circle formation (Fig. 1). To compare the number of proviral DNA molecules in infected-cell populations, we performed PCR with primers specific for HIV-1 LTR. To estimate viral DNA amounts in this assay, we used DNA isolated from the ACH2 cell line as the positive control (Fig. 2, lane 5). This cell line contains 1 to 2 copies per cell. Similar levels of viral DNA were seen for the D116N and WT NL4–3 viruses, while the RT-negative mutant was essentially negative for viral DNA formation (Fig. 2). β-Actin was used to ensure equal loading. Despite the fact that the D116N mutant completely abolished detectable integrase (IN) activity (reference 18; also data not shown), unintegrated D116N DNA scored 30% positive in MAGI infectivity assays. The lack of integration is further supported by the absence of viral production as determined by cocultivation in Hut 78 cells (data not shown). Furthermore, we examined viral production at the single-cell level. In cultures infected with WT NL4–3, 43 to 52% of the cells had detectable intracellular HIV cores by day 5, while in cultures infected with D116N, no positive cells could be detected.
FIG. 1.
2-LTR circle formation in Hut 78 cells following HIV infection. Lane M, 100-bp DNA ladder; lane 1, uninfected cells; lanes 2 through 4, cells infected with the IN-negative mutant D116N, the RT-negative mutant, and WT NL4–3, respectively; lane 5, a positive control from plasmid pBR 541–14. Virus DNA was extracted 24 h p.i. by the Hirt method.
FIG. 2.
Presence of the R/U5 region of HIV-1 LTR DNA in Hut 78 cells infected with HIV-1. Genomic DNA was extracted from Hut 78 cells at day 3 p.i. (Top) HIV-1 LTR (140 bp). Lane 1, negative control; lane 2, uninfected cells; lanes 3 through 5, cells infected with D116N, the RT-negative mutant, and WT NL4–3, respectively; lane 6, positive-control DNA from ACH2 cells. (Bottom) β-Actin.
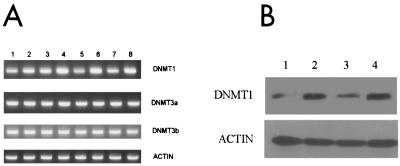
Next, the expression of DNMT1 and the recently discovered DNMT3a and DNMT3b (63) was examined in the human T-cell line Hut 78 at various time points following infection with wild-type and mutant viruses using RT-PCR, QRT-PCR, and Western analysis. Both cells infected with WT NL43 and cells infected with D116N showed increased levels of DNMT1 RNA (Fig. 3A), while DNMT3a and -3b showed little or no significant increase in expression in infected cells. The increased expression of DNMT1 was also observed at the protein level, as Western analysis showed increased DNMT1 protein levels in infected cells. (Fig. 3B, lanes 2 and 4). Western blotting for β-actin demonstrates that equivalent amounts of nuclear extracts were used in this experiment. Consistent with the RT-PCR results, no significant difference between DNMT3a and DNMT3b protein levels was seen after HIV-1 infection (data not shown).
FIG. 3.
Expression of DNMTs in Hut 78 cells following HIV infection. (A) The DNMT1 transcription level is up-regulated. RT-PCR analysis was performed as described in Materials and Methods. The sizes for DNMT1, DNMT3a, and DNMT3b are 335, 280, and 191 bp, respectively. Lanes 1 to 4 show results from day 3 p.i., and lanes 5 to 8 show results from day 5 p.i. Lanes 1 and 5, uninfected cells; lanes 2 and 6, D116N-infected cells; lanes 3 and 7, RT-negative mutant-infected cells; lanes 4 and 8, WT NL4–3-infected cells. All samples were normalized to β-actin as described in Materials and Methods. (B) DNMT1 protein is induced by HIV-1 infection on Hut 78 cells. Western blot analysis was carried out using an antibody against DNMT1 (diluted 1:200) and a 1:5,000-diluted, peroxidase-conjugated secondary antibody (anti-rabbit). Protein was isolated from Hut 78 cells on day 3 p.i. Lane 1, uninfected cells; lanes 2 through 4, cells infected with D116N, the RT-negative mutant, and WT NL4–3, respectively.
QRT-PCR has the advantage that PCR amplification and product analysis can occur simultaneously, conferring a higher level of specificity on quantitation. To discriminate between specific products and nonspecific products such as primer dimers, DNA melting curves were generated. Fluorescence data were converted to melting peaks using the manufacturer's software, then plotted as the negative derivative of fluorescence with respect to temperature (−dF/dT versus T) (65, 68). The area of the specific melting peak is directly proportional to the amount of specific product (Table 1). Using QRT-PCR to quantify the changes in DNMT expression that we observed by semiquantitative RT-PCR and Western analysis, we showed that by day 5 p.i., the amounts of β-actin RNA present were remarkably similar in control and virus-infected cultures. Both DNMT3a (Table 1) and DNMT3b (data not shown) had less RNA in infected than in control cultures. In contrast, by day 5, DNMT1 RNA amounts were 22% greater in D116N-infected cultures and 44% greater in WT NL4–3-infected cultures (Table 1).
TABLE 1.
Increased levels of DNMT1 expression following HIV-1 infection
| Cell infectiona | Specific melting curve area ± SDb (specific Tm) at the following time p.i.:
|
|
|---|---|---|
| Day 3 | Day 5 | |
| β-Actin | ||
| Uninfected | 34.37 ± 0.58 (89.84) | 30.39 ± 0.56 (89.79) |
| IN/D116N | 31.37 ± 0.54 (89.77) | 30.38 ± 0.60 (89.78) |
| WT NL4-3 | 30.77 ± 0.54 (89.76) | 29.81 ± 0.55 (89.79) |
| RT negative | 34.18 ± 0.54 (89.85) | 31.52 ± 0.62 (89.78) |
| DNMT3a | ||
| Uninfected | 36.25 ± 0.60 (89.86) | 36.07 ± 0.64 (89.87) |
| IN/D116N | 33.48 ± 0.59 (89.81) | 33.51 ± 0.64 (89.90) |
| WT NL4-3 | 32.11 ± 0.65 (89.81) | 31.80 ± 0.67 (89.91) |
| RT negative | 33.67 ± 0.58 (89.85) | 33.92 ± 0.66 (89.91) |
| DNMT1 | ||
| Uninfected | 21.89 ± 0.58 (91.06) | 18.76 ± 0.62 (90.98) |
| IN/D116N | 23.65 ± 0.58 (91.09) | 22.15 ± 0.62 (90.99) |
| WT NL4-3 | 25.35 ± 0.56 (91.03) | 26.74 ± 0.67 (91.01) |
| RT negative | 22.05 ± 0.56 (91.13) | 19.83 ± 0.58 (90.99) |
Cells were either not infected or infected with wild-type HIV-1 (WT NL4-3), an IN-defective mutant (IN/D116N), or an RT-negative mutant.
DNMT1 levels are expressed as area under a specific melting curve as determined by real-time PCR using Syber Green as described in Materials and Methods. Values are representative of two experiments.
Increased overall genomic methylation in lymphoid cells acutely infected with integration-defective HIV-1.
To determine the functional consequences of increased DNMT1 expression in the lymphoid cell line, we examined the overall methylation status of genomic DNA using a methyl acceptor assay as described in Materials and Methods. The assay takes advantage of the ability of bacterial SssI methylase to methylate all unmethylated CpG dinucleotides. By using radiolabeled SAM as a substrate, a quantitative measure of overall genomic methylation was obtained. If the DNA is more methylated in infected cell lines, then less SAM will be incorporated. Data were expressed relative to SAM uptake in uninfected Hut 78 cells. As shown in Table 2, the uptake of radiolabeled SAM in WT NL-43-infected cells was 55% of that for the control at day 3 and 80% at day 7, demonstrating significantly more methylation of CpG dinucleotides in the genomic DNA of infected cells than in that of uninfected cells. Viral infection of Hut 78 cells with the D116N mutant showed smaller but still significant increases in overall genomic methylation (20 to 25% [Table 2]), while infection with the RT-negative virus showed no significant change. These results correlated with the increases seen in DNMT expression. Since knowledge of the genes that either are regulated by methylation or contain CpG islands in their promoters has been steadily increasing, particularly with sequencing of the human genome and array technology using CpG islands (32), it seems likely that numerous genes that have altered methylation status following HIV-1 infection will be identified.
TABLE 2.
Increased levels of genomic methylation following HIV-1 infection
| Cell infectiona | Mean DNA methylation level ± SDb (% of control) at the following time p.i.:
|
|
|---|---|---|
| Day 3 | Day 7 | |
| Uninfected | 19,207 ± 242 (100) | 20,082 ± 135 (100) |
| IN/D116N | 15,242 ± 700 (79) | 15,262 ± 150 (76) |
| WT NL4-3 | 10,711 ± 600 (56) | 16,073 ± 700 (80) |
| RT negative | 17,922 ± 400 (93) | 18,817 ± 250 (94) |
Cells were either not infected or infected with either wild-type HIV-1 (WT NL4-3), an IN-defective mutant (IN/D116N), or an RT-negative mutant.
DNA methylation levels are expressed in disintegrations per minute per nanogram of DNA and as percent increases over the methylation levels of mock-infected matched control cultures at the indicated times. Genomic methylation was measured by the methyl acceptor assay as described in Materials and Methods. Values are representative of three experiments.
Increased methylation in the p16INK4A promoter in lymphoid cell lines acutely infected with HIV-1 and mutant viruses.
The cyclin-dependent kinase inhibitor p16 normally inhibits the phosphorylation of RB by cyclin D and cyclin-dependent kinases 4 and 6. This gene is frequently altered in neoplasia, including hematological malignancies, which often result from homozygous deletion or promoter region hypermethylation (29). Since hypermethylation of p16INK4A has recently been demonstrated in HTLV-1-infected cell lines (79) and since p16INK4A is frequently methylated in non-Hodgkin's lymphoma (29), commonly seen in AIDS patients, we examined the methylation status of p16INK4A following acute infection with wild-type HIV-1 and mutant viruses using MSP-PCR (28). Bisulfite treatment converts the cytosine residues in the genomic DNA to uracil, which are amplified as thymine during subsequent PCR.
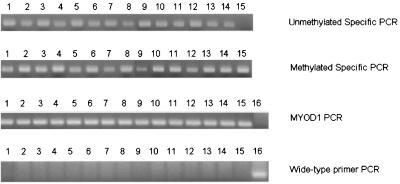
As shown in Fig. 4, Hut 78 cells showed positive 150- and 151-bp bands for methylated and unmethylated specific primer sets for p16INK4A, respectively, indicating that the p16INK4A gene is partially methylated in this cell line. The methylated bands for the p16INK4A gene in D116N- and WT NL4–3-infected Hut 78 cells were consistently stronger than the products of uninfected Hut 78 cells or of Hut 78 cells infected with RT-negative mutant virus (Fig. 4 and Table 3). As a control for equal loading, a sequence from the MYOD1 gene lacking methylatable CpG dinucleotides (17) was used for normalization instead of β-actin as described in Materials and Methods. Thus, unmethylated product levels were significantly lower, and methylated product levels were correspondingly higher, in HIV-infected cells. Further, bisulfite genomic sequencing of eight CpG islands in the p16INK4A gene promoter revealed 80 to 90% methylation in Hut 78 cells infected with WT NL4–3 or D116N. Partial (30 to 40%) but not complete methylation of the p16INK4A gene promoter was detected in uninfected Hut 78 cells and those infected with the RT-negative mutant (data not shown).
FIG. 4.
Methylation status of the p16INK4A gene promoter in Hut 78 cells following HIV infection. Lanes 1 to 4, uninfected cells and cells infected with D116N, the RT-negative mutant, and WT NL4–3, respectively, shown at day 3; lanes 5 to 8, uninfected cells and cells infected with D116N, the RT-negative mutant, and WT NL4–3, respectively, shown at day 5; lanes 9 to 11, 5-AzaC-treated Hut 78 cells that were uninfected or infected with D116N or WT NL4–3, respectively, shown at day 3; lanes 12 to 14, 5-AzaC-treated Hut 78 cells that were uninfected or infected with D116N or WT NL4–3, respectively, shown at day 5. Hut 78 cells were first treated with 1 μM 5-AzaC for 24 h and then infected with D116N or WT NL4–3. Lane 15, DNA that was completely methylated using SssI methylase, as a control for methylation level. Lane 16, unmodified DNA and wild-type primers as a positive control for bisulfite DNA treatment. MSP-PCR was performed with the specific primers described in Materials and Methods.
TABLE 3.
HIV-1 infection induces the hypermethylation of the p16INK4A promoter
| Samplea | Resultb for HIV-1 infected Hut 78 cells obtained by:
|
|
|---|---|---|
| Methylated PCR (% of methylated control) | Unmethylated PCR | |
| Uninfected, day 3 | 18,428.87 (43.00) | 26,646.78 |
| D116N, day 3 | 33,415.30 (77.96) | 18,732.93 |
| RT−, day 3 | 26,805.28 (62.54) | 24,308.68 |
| NL4-3, day 3 | 36,021.73 (84.04) | 18,506.52 |
| Uninfected, day 5 | 17,875.43 (41.71) | 22,352.07 |
| D116N, day 5 | 24,772.55 (57.80) | 18,384.99 |
| RT−, day 5 | 15,329.25 (35.76) | 21,502.76 |
| NL4-3, day 5 | 26,108.31 (60.91) | 16,554.79 |
| 5-AzaC, day 3 | 13,314.00 (31.06) | 36,351.70 |
| 5-AzaC + D116N, day 3 | 30,038.79 (70.08) | 27,717.58 |
| 5-AzaC + NL4-3, day 3 | 28,631.32 (66.80) | 23,715.96 |
| 5-AzaC, day 5 | 18,708.91 (43.65) | 30,460.06 |
| 5-AzaC + D116N, day 5 | 35,469.48 (82.75) | 22,962.43 |
| 5-AzaC + NL4-3, day 5 | 40,988.65 (95.63) | 24,714.90 |
| SssI treated | 42,861.24 (100.00) | No band |
Hut 78 cells were either uninfected or infected with wild-type (NL4-3), IN-defective (D116N), or RT-negative (RT−) HIV-1 and were either not treated or treated with 5-AzaC or SssI. SssI treatment leads to complete methylation.
Expressed as the band density determined on the indicated day (day 3 or day 5) p.i. as described in Materials and Methods. The density of each band from MSP-PCR was normalized to the amount of bisulfite-treated input DNA as determined by the density of the band from PCR for MYOD1, which lacks methylatable CpGs. Data shown are representative of three separate experiments.
These results suggest that the p16INK4A gene is a target of the increased DNMT activity in HIV-1-infected Hut 78 cells. To further examine this, we decreased methylation of the p16INK4A gene using 5-AzaC. Three days after treatment of uninfected Hut 78 cells with 1 μM 5-AzaC, MSP-PCR revealed a significant increase in the amount of unmethylated product (Fig. 4, lane 9), while a significantly more intense methylated 150-bp band was seen for DNA treated with 5-AzaC and infected with either D116N or WT NL4–3 (Fig. 4, lanes 10 and 11). By day 5, there was partial remethylation of this gene (Fig. 4, lane 12) that was markedly accelerated by HIV-1 infection (Fig. 4, lanes 13 and 14).
Decreased expression of the p16INK4A gene in lymphoid cells acutely infected with integration-defective HIV-1.
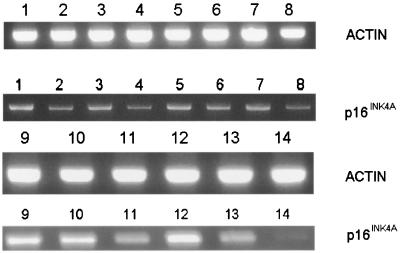
To correlate increased methylation of the p16INK4A gene promoter with expression of the gene, we examined the expression of p16INK4A RNA in Hut 78 cells, using semiquantitative RT-PCR (Fig. 5; Table 4). Decreased levels of p16INK4A expression were seen in Hut 78 cells infected with D116N or WT NL4–3 (Fig. 5, lanes 2 and 4; Table 4) but not in cells infected with the RT-negative mutant (Fig. 5, lane 3; Table 4). Hut 78 cells treated with 5-AzaC had a four- to fivefold increase in p16INK4A expression (Fig. 5, lanes 9 and 12; Table 4). Furthermore, infection of Hut 78 cells previously treated with 5-AzaC for 24 h, using either D116N or WT NL4–3, markedly decreased expression (50 and 90%, respectively) at 5 days p.i. (Fig. 5, lanes 12 to 14; Table 4). In addition, we found that p16INK4A mRNA levels were decreased in Hut 78 cells chronically infected with HIV-1 strain 3B, and hypermethylation in the p16INK4A promoter was present (data not shown). These data suggest that methylation of the p16INK4A gene is one of the mechanisms for silencing of p16INK4A expression in Hut 78 cells and that HIV-1 infection can modulate the methylation status of the CpG island in the promoter of p16INK4A, leading to decreased transcription.
FIG. 5.
Expression of p16INK4A mRNA in Hut 78 cells following HIV infection. RT-PCR was performed as described in Materials and Methods. Lanes 1 to 4, uninfected cells and cells infected with D116N, the RT-negative mutant, and WT NL4–3, respectively, shown at day 3 p.i.; lanes 5 to 8, uninfected cells and cells infected with D116N, the RT-negative mutant, and WT NL4–3, respectively, shown at day 5 p.i.; lanes 9 to 11, Hut 78 cells treated with 5-AzaC and either left uninfected or infected with D116N or WT NL4–3, respectively, shown at day 3 p.i.; lanes 12 to 14, Hut 78 cells treated with 5-AzaC and either left uninfected or infected with D116N or WT NL4–3, respectively, shown at day 5 p.i. β-Actin was used as a loading and amplification control.
TABLE 4.
Decreased p16INK4A gene expression in Hut 78 cells following HIV infection
| Cell condition |
p16INK4A mRNA levela at the following time p.i.:
|
|
|---|---|---|
| Day 3 | Day 5 | |
| Untreated | ||
| Uninfected | 36,267 | 29,421 |
| D116N | 13,640 | 16,989 |
| RT-negative mutant | 25,299 | 21,058 |
| WT NL4-3 | 9,854 | 12,145 |
| Treated with 5-AzaC | ||
| Uninfected | 148,124 | 163,208 |
| D116N | 136,478 | 80,105 |
| WT NL4-3 | 88,629 | 1,320 |
The density of each band from RT-PCR in each lane of Fig. 5 was normalized to the amount of total RNA as determined by the density of band in RT-PCR for β-actin. Assuming that the control value for β-actin was 30,000 U (pixels of brightness), the p16 band would be normalized to the β-actin band by the calculation [30,000/(density of actin)] × (density of p16). Data shown are representative of three separate experiments.
DISCUSSION
Insertion of foreign invaders into the eukaryotic genome can occur naturally, e.g., during viral infections or under experimental conditions such as microinjection or transfection of DNA for immunization purposes. It has long been suggested that de novo methylation is one mechanism by which the cell or genome is protected from expression of foreign DNA such as viruses (7, 33). Retroviruses, which are essentially movable genetic elements, integrate randomly in the host genome and, depending on the local DNA environment, are susceptible to DNA methylation. A relationship among DNA methylation, retroviral replication, and pathophysiology was first shown for murine leukemia virus (MuLV) (26). The MuLV LTRs became hypermethylated, silencing viral expression that could be reactivated to produce active virus by several means. Methylation has been shown to be a mechanism of suppression of viral expression and latency for both disease-causing human retroviruses, HIV-1 and HTLV-1 (52, 53, 74, 75). The ability of the provirus to become latent through methylation and escape the immune response is a two-edged sword for the host that is clinically relevant to human disease and therapy, e.g., HIV latency after highly active antiretroviral therapy (14, 20). It is possible that primary retroviral virulence (direct viral replicative pathogenesis) is inversely related to the number of proviral CpG dinucleotides available for nuclear methylation and silencing by the vertebrate host. If true, the most pathogenic retroviruses would be those with the lowest frequency of CpGs (39). Interestingly, as in the human genome, CpG dinucleotides are underrepresented in retroviral genomes. To counteract the host defense system of methylation and silencing, HIV-1 evolved or is evolving to decrease the number of methylatable CpGs in its genome, thereby escaping the host's capacity to inactivate viral DNA through methylation. The methylatable-CpG content in the HIV-1 genome is lower than that encountered in the many viruses elsewhere in the animal kingdom (61).
In addition to de novo methylation of foreign viral DNA, the methylation patterns of cellular genes and DNA structures can be profoundly altered (27, 65) by viral infection. These virus-induced changes are likely a reflection of more general alterations in chromatin structure. Compelling evidence for the role of methylation in chromatin structure and vice versa has been published (42, 48, 67). Exactly how DNA methylation silences gene expression has been further elucidated by the observation that methyl binding proteins form complexes with other proteins, such as HDAC, that affect chromatin structure and gene regulation, and complexes such as Sin-3, which are transcriptional repressors (37, 57, 58, 82, 83). There seem to be several levels of stable gene silencing. The chromatin of an inactive gene can be densely methylated by de novo methylation, and additional chromatin condensation can occur through histone deacetylation (46).
Previous studies from our laboratory showed that acute infection of cells with HIV-1 results in increased DNMT1 expression and activity, an overall increase in methylation DNA in infected cells, and de novo methylation of a CpG dinucleotide in the IFN-γ gene promoter, resulting in the subsequent down-regulation of expression of this cytokine (53). However, little is known about the mechanism(s) of increased methylation and DNMT1 activity during HIV infection. DNMT1 is the chief enzyme responsible for maintaining methylation patterns in adult mammalian cells. Disruption of the Dnmt1 gene results in embryonic lethality (45). DNMT1 is a large enzyme (193.5 kDa) composed of a C-terminal catalytic domain, which transfers methyl groups from SAM to cytosines in CpG nucleotides, and a large N-terminal regulatory domain with several functions, including targeting to replication foci (13, 44). Forced overexpression of DNMT1 or cleavage between the N-terminal regulatory domain and the C-terminal catalytic domain has been shown to result in increased de novo methylation activity (6, 80) and cellular transformation (86). Other forms of DNMTs have been isolated in mammals (62, 84), and two recently identified DNA methyltransferases, DNMT3a and DNMT3b, are thought to be essential for de novo methylation (31, 64, 69). Moreover, it has recently been shown that in addition to its capacity to methylate CpG sites, DNMT1 can play other roles in transcriptional regulation. DNMT1 can bind HDAC2 and novel corepressors to form a complex at replication foci (73). DNMT1 also forms a complex with Rb, E2F1, and HDAC1, repressing transcription from E2F-responsive promoters (71).
Is HIV-1 replication necessary for increases in DNMTs and genomic methylation? The integration process is the keystone of retroviral replication. Once integrated into the chromosome, the provirus will remain stable throughout the life span of the target cell (12). Integration of retroviral DNA into the host cell genome is required for virus replication and is mediated by viral IN (11). IN function is essential for HIV-1 replication in T-cell lines (9, 43, 76). Mutational analyses of HIV-1 IN indicate that the protein consists of three functional domains: the N-terminal, core (catalytic domain), and C-terminal domains (18, 19). A mutation in the catalytic domain (present in the D116N mutant used in this study) completely abolishes 3′ processing, DNA strand transfer, and disintegration in vitro (18). Although the D116N mutant shows a significant titer in a CD4+ indicator cell assay, it is clearly integration and replication defective (19). As reported for other tissues (69), DNMT1 was expressed at the highest level of the three DNMTs in lymphoid cells. QRT-PCR and Western analysis also showed that acute infection of Hut 78 cells with wild-type or integration-negative HIV-1 markedly up-regulates DNMT1 mRNA and protein expression, respectively. The RT-negative mutant showed no significant ability to regulate DNMT1 expression and no detectable effect on methylation. In contrast to DNMT1, DNMT3a and -3b showed no significant increase in expression following acute HIV-1 infection.
In addition to our previous demonstration that HIV infection can result in the aberrant methylation of single CpG dinucleotides in the IFN-γ promoter (53), we show here that genes which have CpG-rich regions of 1 kb of DNA, termed “CpG islands,” and are usually hypomethylated can be aberrantly methylated in HIV-1-infected cells. In malignant cells, these CpG island regions become methylated and expression of the associated gene is silenced (16, 35). The p16INK4A gene, which encodes a specific inhibitor of cyclin-dependent kinases 4 and 6, is located at the 9p2/1 chromosomal region. Loss of p16INK4A gene expression is a frequent molecular alteration involved in tumorigenesis. Recently, changes of expression and methylation status of the p16INK4A gene have been demonstrated in tumor cell lines and a variety of cancers (3, 21, 22, 25, 40, 49, 54, 60, 81). The hypothesis that expression of the p16INK4A gene may be regulated in part by changes in the methylation status of this CpG island has been substantiated in several tumor models (21, 47, 59). Partial or complete promoter methylation rather than deletion of the p16INK4A gene has been observed in some HTLV-1-infected T-cell lines (79) and here in Hut 78 cells. However, by using completely methylated and unmethylated controls, increased methylation of the p16INK4A gene in HIV-1-infected cells was shown. The p16INK4A gene is frequently methylated in non-Hodgkin's leukemia, a malignancy common in AIDS patients (29), suggesting that HIV-induced aberrant methylation could play a role in disease development.
What is the mechanism of increased DNMT1 expression and hypermethylation which results from infection with integration-defective HIV-1? Infection with the HIV-1 mutant D116N results in accumulation of unintegrated 2-LTR circles in the nucleus, a sensitive indicator of a recent infection. In contrast to wild-type HIV-1, infection with D116N resulted in more 2-LTR circle formation in Hut 78 cells, as previously reported (18, 19). This increased DNMT1 expression is not caused by the mere presence of foreign DNA (RT-negative HIV had no effect). Unintegrated DNA can serve as a template for HIV tat expression (18, 19), a transactivator of many genes, which could stimulate DNMT1 expression in trans. Moreover, it has been shown that 2-LTR circles produce large amounts of tat, which stimulates the β-galactosidase readout in the MAGI assay and leads to p24 production (10). On the other hand, since the N terminus of DNMT1 has recently been shown to form complexes with HDAC, transcription factors, and corepressors to silence transcription of specific genes (70, 71, 73), the effect of HIV-1 on DNMT1 may be due to an as yet unknown effect of viral proteins on a non-DNA methyltransferase function of DNMT1. For example viral proteins, such as tat, could interrupt the normal complex formation. We are currently studying this and other hypotheses to determine how HIV infection might alter these DNMT1 functions.
As nonintegrated circular DNA has long been implicated in the pathogenesis of retroviral infections, the ability to aberrantly methylate genes and/or alter chromatin structure could play a role in such pathogenesis. Examples of such pathogenesis are the association of cell killing with nonintegrated spleen necrosis virus (41), disease-specific production of unintegrated feline leukemia virus DNA in feline AIDS (55), and avian leukosis-virus induced-osteoporosis (72). In AIDS, circular forms of unintegrated HIV have been associated with dementia and giant cell formation (78) and may play a role in neuropathogenesis. Although unintegrated viral DNA has been linked to cell killing during HIV infection, it is not always associated with cytopathology (5, 50). Regardless of the mechanism, this study shows that integration-defective HIV-1 can alter the DNA methylation patterns of infected cells, further implicating aberrant methylation as a mechanism of pathogenesis in AIDS and AIDS-associated malignancies and suggesting that the methylation machinery can be a novel target for AIDS therapy.
ACKNOWLEDGMENTS
J.-Y.F. and J.A.M. contributed equally to this report.
We thank Weimin Zhu, Paula Roberts, Angela Brennan, and Dorjbal Dorjsuren for technical assistance. We also thank Robert Gorelick for providing HIV-1 wild-type and RT mutant plasmids and for useful discussions and Howard Young for review of the manuscript.
The NIH Intramural AIDS Targeted Antiviral Program provided support for this study. Portions of this work were also supported by funds from the NCI under contract NO1-CO-56000.
REFERENCES
- 1.Adachi A, Gendelman H, Koenig S, Folks T, Willey R, Rabson A, Martin M. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartolomei M S, Tilghman S M. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 3.Baur A, Shaw P, Burri N, Delacretaz F, Bosman F, Chaubert P. Frequent methylation silencing of p15INK4B (MTS2) and p16INK4A (MTS1) in B-cell and T-cell lymphomas. Blood. 1999;94:1773–1781. [PubMed] [Google Scholar]
- 4.Baylin S. Tying it all together: epigenetics, genetics, cell cycle, and cancer. Science. 1997;277:1948–1949. doi: 10.1126/science.277.5334.1948. [DOI] [PubMed] [Google Scholar]
- 5.Bergeron L, Sodroski J. Dissociation of unintegrated viral DNA accumulation from single-cell lysis induced by human immunodeficiency virus type 1. J Virol. 1992;66:5777–5787. doi: 10.1128/jvi.66.10.5777-5787.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bestor T. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bestor T, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 8.Bird A. DNA methylation de novo. Science. 1999;286:2287–2288. doi: 10.1126/science.286.5448.2287. [DOI] [PubMed] [Google Scholar]
- 9.Cannon P, Wilson W, Byles E, Kingsman S, Kingsman A. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol. 1994;68:4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cara A, Cereseto A, Lori F, Reitz M S., Jr HIV-1 protein expression from synthetic circles of DNA mimicking the extrachromosomal forms of viral DNA. J Biol Chem. 1996;27:5393–5397. doi: 10.1074/jbc.271.10.5393. [DOI] [PubMed] [Google Scholar]
- 11.Chen J C, Krucinski J, Miercke L J, Finer-Moore J, Tang A, Leavitt A, Stroud R. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci USA. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherepanov P, Pluymers W, Claeys A, Proost P, Clercq E, Debyser Z. High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 2000;14:1389–1399. doi: 10.1096/fj.14.10.1389. [DOI] [PubMed] [Google Scholar]
- 13.Chuang L, Ian H, Koh T, Ng H, Xu G, Li B. Human DNA (cytosine-5)-methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 14.Chun T, Davey R, Jr, Ostrowski M, Shawn J, Engel D, Mullins J, Fauci A. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat Med. 2000;6:757–761. doi: 10.1038/77481. [DOI] [PubMed] [Google Scholar]
- 15.Clark S, Harrison J, Paul C, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Counts J, Goodman J. Alterations in DNA methylation may play a variety of roles in carcinogenesis. Cell. 1995;83:13–15. doi: 10.1016/0092-8674(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 17.Eads C A, Danenberg K D, Kawakami K, Saltz L B, Blake C, Shibata D, Danenberg P V, Laird P W. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:e32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman A, Englund G, Orenstein J, Martin M, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finzi D, Hermankova M, Pierson T, Carruth L, Buck C, Chaisson R, Quinn T, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D, Richman D, Siliciano R. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 21.Fueyo J, Gomez-Manzano C, Bruner J, Saito Y, Zhang B, Zhang W, Levin V, Yung W, Kyritsis A. Hypermethylation of the CpG island of p16/CDKN2 correlates with gene inactivation in gliomas. Oncogene. 1996;13:1615–1619. [PubMed] [Google Scholar]
- 22.Garcia J, Villuendas R, Algara P, Saez A, Sanchez-Verde L, Martinez-Montero J, Martinez P, Piris M. Loss of p16 protein expression associated with methylation of the p16INK4A gene is a frequent finding in Hodgkin's disease. Lab Investig. 1999;79:1453–1459. [PubMed] [Google Scholar]
- 23.Gorelick R, Gagliardi T, Bosche W, Wiltrout T, Coren L, Chabot D, Lifson J D, Henderson L, Arthur L. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology. 1999;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- 24.Gorelick R, Fu W, Gagliardi T, Bosche W, Rein A, Henderson L, Arthur L O. Characterization of the lock-in replication of nucleocapsid protein zinc finger mutants from Moloney murine leukemia virus. J Virol. 1999;73:8185–8195. doi: 10.1128/jvi.73.10.8185-8195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo S X, Taki T, Ohnishi H, Piao H Y, Yabuchi K, Bessho F, Hanada R, Yanagisawa M, Hayashi Y. Hypermethylation of p16 and p15 genes and RB protein expression in acute leukemia. Leuk Res. 2000;24:39–46. doi: 10.1016/s0145-2126(99)00158-7. [DOI] [PubMed] [Google Scholar]
- 26.Harbers K, Schnieke A, Stuhlmann H, Jahner D, Jaenisch R. DNA methylation and gene expression: endogenous retroviral genome becomes infectious after molecular cloning. Proc Natl Acad Sci USA. 1981;78:7609–7613. doi: 10.1073/pnas.78.12.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heller H, Kammer C, Wilgenbus P, Doerfler W. Chromosomal insertion of foreign (adenovirus type 12, plasmid, or bacteriophage lambda) DNA is associated with enhanced methylation of cellular DNA segments. Proc Natl Acad Sci USA. 1995;92:5515–5519. doi: 10.1073/pnas.92.12.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herman J G, Civin C, Issa J, Collector M, Sharkis S, Baylin S. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997;57:837–841. [PubMed] [Google Scholar]
- 30.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh C L. In vivo activity of murine de novo methyltransferases DNMT3a and DNMT3b. Mol Cell Biol. 1999;19:8211–8218. doi: 10.1128/mcb.19.12.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang T H M, Perry M R, Laux D E. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 33.Jahner D, Stuhlmann H, Stewart C L, Harbers K, Lohler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 34.Johnson C A, Turner B M. Histone deacetylases: complex transducers of nuclear signals. Semin Cell Dev Biol. 1999;10:179–188. doi: 10.1006/scdb.1999.0299. [DOI] [PubMed] [Google Scholar]
- 35.Jones P A. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 36.Jones P A, Laird P W. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–166. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 37.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 38.Kanai Y, Ushijima S, Kondo Y, Nakanishi Y, Hirohashi S. DNA methyltransferase expression and DNA methylation of CpG islands and pericentromeric satellite region in human colorectal and stomach cancers. Int J Cancer. 2001;91:205–212. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1040>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Karlin S, Doerfler W, Cardon L R. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J Virol. 1994;68:2889–2897. doi: 10.1128/jvi.68.5.2889-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawamura M, Ohnishi H, Guo S X, Sheng X M, Minegishi M, Hanada R, Horibe K, Hongo T, Kaneko Y, Bessho F, Yanagisawa M, Sekiya T, Hayashi Y. Alterations of the p53, p21, p16, p15 and RAS genes in childhood T-cell acute lymphoblastic leukemia. Leuk Res. 1999;23:115–126. doi: 10.1016/s0145-2126(98)00146-5. [DOI] [PubMed] [Google Scholar]
- 41.Keshet E, Temin H M. Cell killing by spleen necrosis virus is correlated with a transient accumulation of spleen necrosis virus DNA. J Virol. 1979;31:376–388. doi: 10.1128/jvi.31.2.376-388.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 43.Lafemina R L, Schneider C L, Robbins H L, Callahan P L, LeGrow K, Roth E, Schleif W A, Emini E A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehmann A, Arlett C, Harcourt S, Steingrimsdottir H, Gebara M. Mutagenic treatments result in inactivation of expression of a transfected bacterial gene integrated into a human cell line. Mutat Res. 1989;220:255–262. doi: 10.1016/0165-1110(89)90029-8. [DOI] [PubMed] [Google Scholar]
- 45.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 46.Li E. The mojo of methylation. Nat Genet. 1999;23:5–6. doi: 10.1038/12595. [DOI] [PubMed] [Google Scholar]
- 47.Lo K W, Chung S T, Leung S F, van Hasselt A, Tsang Y S, Mak K F, Chung Y F, Woo J K, Lee J C, Huang D P. Hypermethylation of the p16 gene in nasopharyngeal carcinoma. Cancer Res. 1996;56:2721–2725. [PubMed] [Google Scholar]
- 48.Lubbert M, Miller C W, Koeffler H P. Changes of DNA methylation and chromatin structure in the human myeloperoxidase gene during myeloid differentiation. Blood. 1991;78:345–356. [PubMed] [Google Scholar]
- 49.Melki J R, Vincent P C, Clark S J. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999;59:3730–3740. [PubMed] [Google Scholar]
- 50.Mercure L, Phaneuf D, Wainberg M A. Detection of unintegrated human immunodeficiency virus type 1 DNA in persistently infected CD8+ cells. J Gen Virol. 1993;74:2077–2083. doi: 10.1099/0022-1317-74-10-2077. [DOI] [PubMed] [Google Scholar]
- 51.Mikovits J A, Raziuddin M, Gonda M, Ruta M, Lohrey N, Kung H, Ruscetti F. Negative regulation of HIV replication in monocytes: distinctions between restricted and latent expression in THP-1 cells. J Exp Med. 1990;171:1705–1720. doi: 10.1084/jem.171.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mikovits J A, Lohrey N C, Schuloff R, Courtless J, Ruscetti F W. Immune activation of HIV expression from latently infected monocytes from asymptomatic seropositive patients. J Clin Investig. 1992;90:1486–1491. doi: 10.1172/JCI116016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikovits J A, Young H, Vertino P, Issa J, Pitha P, Turcoski-Corrales S, Taub D, Petrow C, Baylin S, Ruscetti F. Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the gamma interferon (IFN-γ) promoter and subsequent downregulation of IFN-γ production. Mol Cell Biol. 1998;18:5166–5177. doi: 10.1128/mcb.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–1179. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 55.Mullins J I, Chen C S, Hoover E A. Disease-specific and tissue-specific production of unintegrated feline leukaemia virus variant DNA in feline AIDS. Nature. 1986;319:333–336. doi: 10.1038/319333a0. [DOI] [PubMed] [Google Scholar]
- 56.Nakashima R, Fujita M, Enomoto T, Haba T, Yoshino K, Wada H, Kurachi H, Sasaki M, Wakasa K, Inoue M, Buzard G, Murata Y. Alteration of p16 and p15 genes in human uterine tumors. Br J Cancer Res. 1999;59:458–467. doi: 10.1038/sj.bjc.6690379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 58.Ng H H, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 59.Ng M H, Chung Y F, Lo K W, Wickham N W, Lee J C, Huang D P. Frequent hypermethylation of p16 and p15 genes in multiple myeloma. Blood. 1997;89:2500–2506. [PubMed] [Google Scholar]
- 60.Nuovo G J, Plaia T W, Belinsky S A, Baylin S B, Herman J G. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc Natl Acad Sci USA. 1999;96:12754–12759. doi: 10.1073/pnas.96.22.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nyce J W. CpG suppression in HIV-1 versus HIV-2: correlation with pathogenicity and possible implications for the design of antiretroviral vaccines. In: Russo V, Martienssen R, Riggs A, editors. Epigenetic mechanisms of gene regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 561–572. [Google Scholar]
- 62.Okano M, Xie S, Li E. DNMT2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26:2536–2540. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 64.Okano M, Bell D W, Haber D A, Li E. DNA methyltransferases DNMT3a and DNMT3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 65.Rasmussen R, Morrison T, Herrman M, Wittwer C. Quantitative PCR by fluorescence monitoring of a double strand DNA specific binding dye. Biochemica. 1998;2:8–11. [Google Scholar]
- 66.Remus R, Kammer C, Heller H, Schmitz B, Schell G, Doerfler W. Insertion of foreign DNA into an established mammalian genome can alter the methylation of cellular DNA sequences. J Virol. 1999;73:1010–1022. doi: 10.1128/jvi.73.2.1010-1022.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rideout W M, III, Eversole-Cire P, Spruck III C H, Hustad C M, Coetzee G A, Gonzales F A, Jones P A. Progressive increases in the methylation status and heterochromatinization of the myoD CpG island during oncogenic transformation. Mol Cell Biol. 1994;14:6143–6152. doi: 10.1128/mcb.14.9.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ririe K M, Rasmussen R P, Wittner C T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 69.Robertson K D, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales F A, Jones P A. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robertson K D, Jones P A. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 71.Robertson K D, Ait-Si-Ali S, Yokochi T, Wade P A, Jones P L, Wolffe A P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 72.Robinson H L, Miles B D. Avian leukosis virus-induced osteopetrosis is associated with the persistent synthesis of viral DNA. Virology. 1985;141:130–143. doi: 10.1016/0042-6822(85)90189-8. [DOI] [PubMed] [Google Scholar]
- 73.Roundtree M, Bachman K, Baylin S. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 74.Saggioro D, Panozzo M, Chieco-Bianchi L. Human T-lymphotropic virus type I transcriptional regulation by methylation. Cancer Res. 1990;50:4968–4973. [PubMed] [Google Scholar]
- 75.Saggioro D, Forino M, Chieco-Bianchi L. Transcriptional block of HTLV-1 LTR by sequence-specific methylation. Virology. 1991;182:68–75. doi: 10.1016/0042-6822(91)90649-v. [DOI] [PubMed] [Google Scholar]
- 76.Shin C, Taddeo B, Haseltine W A, Farnet C M. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J Virol. 1994;68:1633–1642. doi: 10.1128/jvi.68.3.1633-1642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Surani M A. Imprinting and the initiation of gene silencing in the germline. Cell. 1998;93:309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 78.Teo I, Veryard C, Barnes H, An S, Jones M, Lantos P, Luthert P, Shaunak S. Circular forms of unintegrated human immunodeficiency virus type 1 DNA and high levels of viral protein expression: association with dementia and multinucleated giant cells in the brains of patients with AIDS. J Virol. 1997;71:2928–2933. doi: 10.1128/jvi.71.4.2928-2933.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trovato R, Cereseto A, Takemoto S, Gessain A, Watanabe T, Waldmann T, Franchini G. Deletion of the p16INK4A gene in ex vivo acute adult T cell lymphoma/leukemia cells and methylation of the p16INK4A promoter in HTLV type I-infected T cell lines. AIDS Res Hum Retrovir. 2000;16:709–713. doi: 10.1089/088922200308701. [DOI] [PubMed] [Google Scholar]
- 80.Vertino P M, Yen R W C, Gao J, Baylin S B. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5)-methyltransferase. Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vonlanthen S, Heighway J, Tschan M, Borner M, Altermatt H, Kappeler A, Tobler A, Fey M, Thatcher N, Yarbrough W, Betticher D. Expression of p16INK4a/p16α and p19ARF/p16β is frequently altered in non-small cell lung cancer and correlates with p53 overexpression. Oncogene. 1998;17:2779–2785. doi: 10.1038/sj.onc.1202501. [DOI] [PubMed] [Google Scholar]
- 82.Wade P, Jones P, Vermaak D, Veenstra G, Imhof A, Sera T, Tse C, Ge H, Shi Y B, Hansen J, Wolffe A. Histone deacetylase directs the dominant silencing of transcription in chromatin: association with MeCP2 and the Mi-2 chromodomain SWI/SNF ATPase. Cold Spring Harbor Symp Quant Biol. 1998;63:435–445. doi: 10.1101/sqb.1998.63.435. [DOI] [PubMed] [Google Scholar]
- 83.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 84.Wilkinson C R, Bartlett R, Nurse P, Bird A P. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res. 1995;23:203–210. doi: 10.1093/nar/23.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wolffe A P, Jones P L, Wade P A. DNA demethylation. Proc Natl Acad Sci USA. 1999;96:5894–5896. doi: 10.1073/pnas.96.11.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu J, Issa J P J, Herman J, Bassett D E, Nelkin B D, Baylin S B. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:8891–8895. doi: 10.1073/pnas.90.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoder J A, Walsh C, Bestor T H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 88.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]