Abstract
Neoadjuvant chemoimmunotherapy (NACI) has significant implications for the treatment of esophageal cancer. However, its clinical efficacy varies considerably among patients, necessitating further investigation into the underlying mechanisms. The rapid advancement of single-cell RNA sequencing (scRNA-seq) technology facilitates the analysis of patient heterogeneity at the cellular level, particularly regarding treatment outcomes. In this study, we first analyzed scRNA-seq data of esophageal squamous cell carcinoma (ESCC) following NACI, obtained from the Gene Expression Omnibus (GEO) database. After performing dimensionality reduction, clustering, and annotation on the scRNA-seq data, we employed CellChat to investigate differences in cell–cell communication among samples from distinct efficacy groups. The results indicated that macrophages in the non-responder exhibited stronger cell communication intensity compared to those in responders, with SPP1 and GALECTIN signals showing the most significant differences between the two groups. This finding underscores the crucial role of macrophages in the efficacy of NACI. Subsequently, reclustering of macrophages revealed that Mac-SPP1 may be primarily responsible for treatment resistance, while Mac-C1QC appears to promote T cell activation. Finally, we conducted transcriptome sequencing on ESCC tissues obtained from 32 patients who underwent surgery following NACI. Utilizing CIBERSORT, CIBERSORTx, and WGCNA, we analyzed the heterogeneity of tumor microenvironment among different efficacy groups and validated the correlation between SPP1+ macrophages and resistance to NACI in ESCC using publicly available transcriptome sequencing datasets. These findings suggest that SPP1+ macrophages may represent a key factor contributing to resistance against NACI in ESCC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-024-03848-6.
Keywords: Esophageal squamous cell carcinoma, Neoadjuvant chemoimmunotherapy, Macrophages, Single-cell RNA sequencing, Bulk RNA sequencing
Introduction
Esophageal cancer is recognized as the sixth leading cause of cancer-related deaths worldwide, with more than 90% of cases identified as esophageal squamous cell carcinoma (ESCC) [1, 2]. In spite of advances in diagnostics and treatment, less than 20% of ESCC patients survive five years after diagnosis [3]. Neoadjuvant chemoradiotherapy is frequently necessary in cases of resectable locally advanced esophageal cancer [4]. However, this treatment confers benefits to only a subset of patients, achieving a complete pathological response rate of less than 50% [5]. Consequently, there is a pressing clinical need for the development of more effective treatment strategies to enhance patient outcomes.
Recent studies have shown immune checkpoint inhibitors to be a promising treatment for esophageal cancer [6]. Several recent clinical trials have demonstrated that neoadjuvant chemoimmunotherapy (NACI) is a safe and feasible approach that can significantly improve clinical outcomes for patients [7–10]. However, despite notable advancements in NACI regimens, several issues continue to persist. For instance, while a subset of patients exhibits a favorable response to treatment, a significant proportion still does not demonstrate substantial tumor regression after treatment. Compared to conventional neoadjuvant chemotherapy, immunotherapy can inhibit the PD-1/PD-L1 axis to ameliorate T cell exhaustion [11]. Combined positive score (CPS), a well-known biomarker for immunotherapy, is based on the expression of PD-L1; however, its predictive capacity for the efficacy of immunotherapy is still considered suboptimal [12]. Understanding the complex interplay between tumors and immune system, as well as identifying potential factors that contribute to treatment resistance, is crucial for broadening the population of patients who can benefit from these therapies.
The interactions among various immune cells within the tumor microenvironment (TME) are highly complex and represent a significant factor influencing the efficacy of neoadjuvant immunotherapy; however, the underlying mechanisms remain poorly understood [13, 14]. Single-cell RNA sequencing (scRNA-seq) technology is a powerful tool for analyzing cellular heterogeneity, allowing researchers to group and analyze cells in the TME based on the expression of specific genes. Recent studies have meticulously characterized the cellular landscape of the ESCC TME before and after NACI, uncovering novel biomarkers that may predict the efficacy of NACI and revealing the strong relationship between the TME and NACI outcomes [15, 16]. In this study, we utilized scRNA-seq data from ESCC patients who received NACI to identify potential cellular populations influencing NACI efficacy. Using the CellChat method, we observed a significant increase in communication intensity among macrophages in the non-responder, with signals such as SPP1 and GALECTIN primarily transmitted to CD8+ T cells. Considering the heterogeneity of tumor-associated macrophages (TAMs), we identified three macrophage subclusters characterized by high expression of C1QC, SPP1, and HSP, which exhibited notable metabolic and functional differences. Furthermore, the SPP1+ macrophages also exhibited elevated expression of LAGLS9, a member of the GALECTIN family, suggesting that SPP1+ macrophages may play a pivotal role in mediating resistance to NACI. Subsequently, we analyzed transcriptomic sequencing data from 32 ESCC patients who underwent NACI and correlated these findings with scRNA-seq data using techniques such as CIBERSORTx, thereby validating the results obtained in the single-cell analysis. Finally, we leveraged publicly available transcriptomic datasets to confirm the association between SPP1+ macrophages and resistance to immunotherapy in ESCC. Our research highlights the heterogeneity of macrophage subclusters underlying NACI treatment in esophageal cancer and identifies SPP1+ macrophages as a potentially key factor contributing to NACI resistance, providing new insights for future therapeutic strategies in ESCC.
Materials and methods
Patients and specimen collection
We enrolled a total of 32 patients diagnosed with ESCC, whose tumors were classified according to the 8th edition of the TNM system of the American Joint Committee on Cancer (AJCC). All patients had no prior history of anticancer treatment and subsequently underwent two cycles of NACI prior to surgery, which included camrelizumab, cisplatin, and nab-paclitaxel. Specimens of ESCC were obtained from these 32 patients following neoadjuvant therapy through surgical procedures for bulk RNA sequencing analysis. The response to NACI was evaluated using the Tumor Regression Grade (TRG) system, which is based on the guidelines established by the American Joint Committee on Cancer and the College of American Pathologists (AJCC/CAP). TRG scores ranging from 0 to 1 were classified as responders, while scores ranging from 2 to 3 were categorized as non-responders. Consequently, the study cohort was stratified into two groups: 13 responders and 19 non-responders. The Ethics Committee at the First Affiliated Hospital of Zhengzhou University granted ethical approval for this research, with the approval number L2021-Y346-004. Informed consent was obtained from all patients prior to the start of the study.
Data acquisition
The scRNA-seq data for the ESCC cohort, including datasets GSE203115 [17] and GSE196756, were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Additionally, the bulk transcriptome data, GSE53625 [18], were also obtained from the GEO database. Furthermore, gene expression information for ESCC patients was downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/).
ScRNA-seq data processing and clustering
The GSE203115 dataset includes tumor samples from ESCC patients who underwent two cycles of NACI, comprising non-responder (n = 1) and responders (n = 2). The GSE196756 dataset comprises three ESCC tumor samples and three adjacent normal samples; however, only the three tumor samples were selected for combined analysis with those from GSE203115. ScRNA-seq samples were examined for quality control using their raw count matrix. Cells exhibiting more than 20% mitochondrial genes and fewer than 300 or exceeding 7000 expressed genes were considered not to meet the research requirements and were subsequently excluded from further analysis. Data normalization, identification of variable genes, scaling (utilizing the NormalizeData, FindVariableFeatures, and ScaleData functions), principal component analysis (RunPCA), dimensionality reduction (RunUMAP), and unsupervised graph-based clustering (FindNeighbors & FindClusters) were performed using Seurat (version 5.0.3) [19]. Harmony (version 1.2.0) [20] was employed to remove batch effects (RunHarmony). A total of 13 clusters were identified through UMAP clustering analysis, and differentially expressed genes (DEGs) for each cluster were ascertained using FindAllMarkers. Cell types were ultimately annotated by integrating previously reported and most significantly altered marker genes.
Cell–cell communication analysis
To explore the intricacies of cell–cell communication, a cell-to-cell communication network was constructed and visualized using CellChat (version 1.6.1) [21]. It simulates intercellular communication by evaluating the interactions among ligands, receptors, and their respective co-factors. We separately created CellChat objects for non-responder and responders, subsequently merging them for comparative analysis to identify cell clusters exhibiting differential functionality between the two groups and to uncover key ligand-receptor pairs and signaling pathways. The visualization methods employed include netVisual_circle, netVisual_bubble, and netVisual_chord_gene.
Single‑cell trajectory analysis
Pseudotime analysis was performed on macrophage subsets using CytoTRACE2 (version 1.0.0) [22] and Monocle (version 2.30.1) [23]. CytoTRACE2 facilitated the determination of the differentiation status of each cell sub cluster. Subsequently, Monocle was utilized to conduct pseudotime analysis based on the differential genes among the cell subsets, followed by dimensionality reduction using the reduce dimension function. Visualization was subsequently achieved using functions such as plot_cell_trajectory and plot_pseudotime_heatmap.
Enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted utilizing the R package clusterProfiler (version 4.10.1) [24], a widely employed algorithm for identifying statistically enriched pathways in sorted gene lists. Gene Set Variation Analysis (GSVA) was performed utilizing the Hallmark gene sets from the MSigDB database to uncover differential pathways. Additionally, specific gene set scores were calculated for single-cell and bulk transcriptomic data utilizing the AUCell function from the Seurat package and ssGSEA, respectively, which included the scoring of macrophage subgroups such as M1 and M2 [25]. We also employed the scMetabolism [26] package to compare metabolic differences among various macrophage subsets. The STRING database was used to construct protein–protein interaction (PPI) networks.
DEGs analysis and weighted gene co-expression network analysis (WGCNA)
The R package DESeq2 [27] was used to analyze DEGs, identifying genes as DEGs if they met the criteria of p value < 0.05 and |log2FC|> 1.0. The R package WGCNA [28] was utilized to analyze the bulk transcriptomic data of clinical esophageal cancer. Gene co-expression networks were constructed based on the interaction patterns of genes. Finally, hierarchical clustering and the dynamic tree cutting algorithm were utilized to identify 38 gene modules.
Immune infiltration analysis
We conducted deconvolution analysis on bulk RNA-seq data utilizing the CIBERSORT algorithm and CIBERSORTx [29]. Firstly, CIBERSORT analysis, which correlated with reference mRNA expression values (LM22), was employed to compare the relative infiltration levels of 22 immune cell types between the non-responders and responders. Subsequently, three distinct macrophage subtypes derived from the ESCC single-cell transcriptome dataset were uploaded to the CIBERSORTx database, and the “Create Feature Matrix” module was utilized to construct feature genes for each cell subtype. Finally, the proportions of the three macrophage subtypes were calculated for the clinical ESCC bulk transcriptome dataset.
Statistical analysis
R software (version 4.3.0) was used for all statistical analyses. T-tests were utilized to compare the mean gene expression and gene features between two groups of cells. The Wilcoxon test was used to analyze data that were not normally distributed. Spearman's rank correlation coefficient was used for correlation analysis. Statistical significance was defined as a p value < 0.05 (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
Results
Cell–Cell communication analysis reveals macrophage heterogeneity between the non-responders and responders in NACI-treated ESCC patients
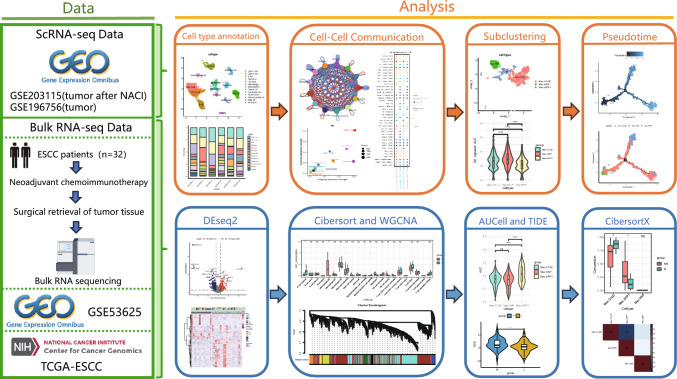
The experimental process is shown schematically in Fig. 1.
Fig. 1.
Study overview
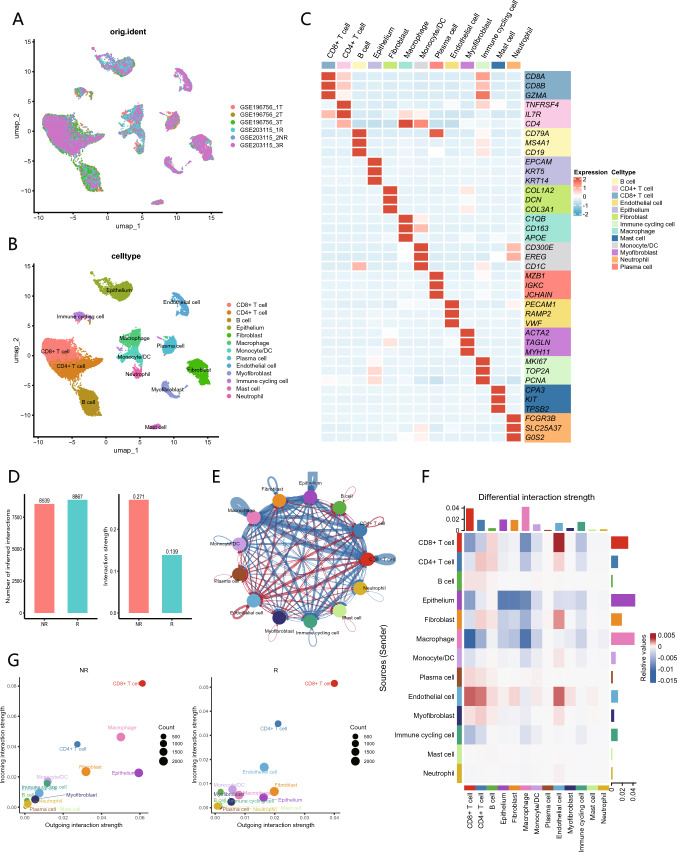
This study included six single-cell samples from patients with ESCC, consisting of three untreated samples and three samples obtained after NACI treatment, which included one non-responder and two responders. After rigorous quality control and filtering of the scRNA-seq data, a total of 24,325 cells were obtained from these samples. By employing unsupervised clustering, these cells were mapped onto a two-dimensional UMAP plane, leading to the identification of 13 distinct cell populations (Fig. 2A). These clusters included CD8+ T cells, CD4+ T cells, B cells, epithelial cells, fibroblasts, macrophages, monocytes/DC, plasma cells, endothelial cells, myofibroblasts, immune cycling cells, mast cells, and neutrophils. The distribution of these cell clusters across the samples was relatively uniform, with no evident sample-specific characteristics shown (Fig. 2B). The marker genes and expression patterns for each cell type are shown in a heatmap (Fig. 2C). Additionally, UMAP visualizations illustrating the relationship between treatment status and different efficacy groups, as well as the proportions of various cell types across different samples and groups, are shown (Fig. S1A-E).
Fig. 2.
Single-cell landscape of tumor microenvironment of ESCC and analysis of cell–cell communication between NR and R. A UMAP plot illustrates the cellular distribution across six samples. B UMAP plot displays the distribution of 13 cell populations within the tumor microenvironment, distinguished by different colors. C Heatmap presents the average expression levels of characteristic marker genes used to distinguish among cell populations. D Bar graph illustrates the total number (left) and strength (right) of ligand-receptor interactions between NR and R. E Network diagram compares the differences in the intensity of cell communication between NR and R, with the thickness of lines representing the levels of strength. The blue lines indicate stronger communication in NR, while the red lines represent stronger communication in R. F Heatmap illustrates the communication differences between different cell populations in NR, with vertical axis representing ligand cells and horizontal axis representing receptor cells. G Comparison of the incoming and outgoing interaction intensities among cell populations in NR and R
To investigate the differences in intercellular communication between the non-responder and responders, we employed the human CellChat database to analyze the intercellular interactions among the 13 identified cell clusters. Initially, we compared the number and strength of communications across all cell populations between the non-responder and responders, revealing that the non-responder exhibited a greater communication strength (Fig. 2D). Next, we employed network diagrams to compare the communication intensities among different cell populations across the groups, demonstrating that the communication strength between macrophages and CD8+ T cells, CD4+ T cells, and epithelial cells was significantly enhanced in the non-responder (Fig. 2E). The constructed intercellular communication heatmap indicated that macrophages ranked prominently in both signal reception and transmission strength, suggesting that they significantly influence the functional states of other cell types (Fig. 2F). Furthermore, we identified variations in the signaling intensities of different cell populations between the groups, again showing that macrophages represented the cell population with the most pronounced differences (Fig. 2G). These results indicate that macrophages within the TME play a pivotal role in shaping the outcomes of NACI.
The SPP1 and GALECTIN signaling pathways in macrophages may contribute to resistance to NACI in ESCC patients
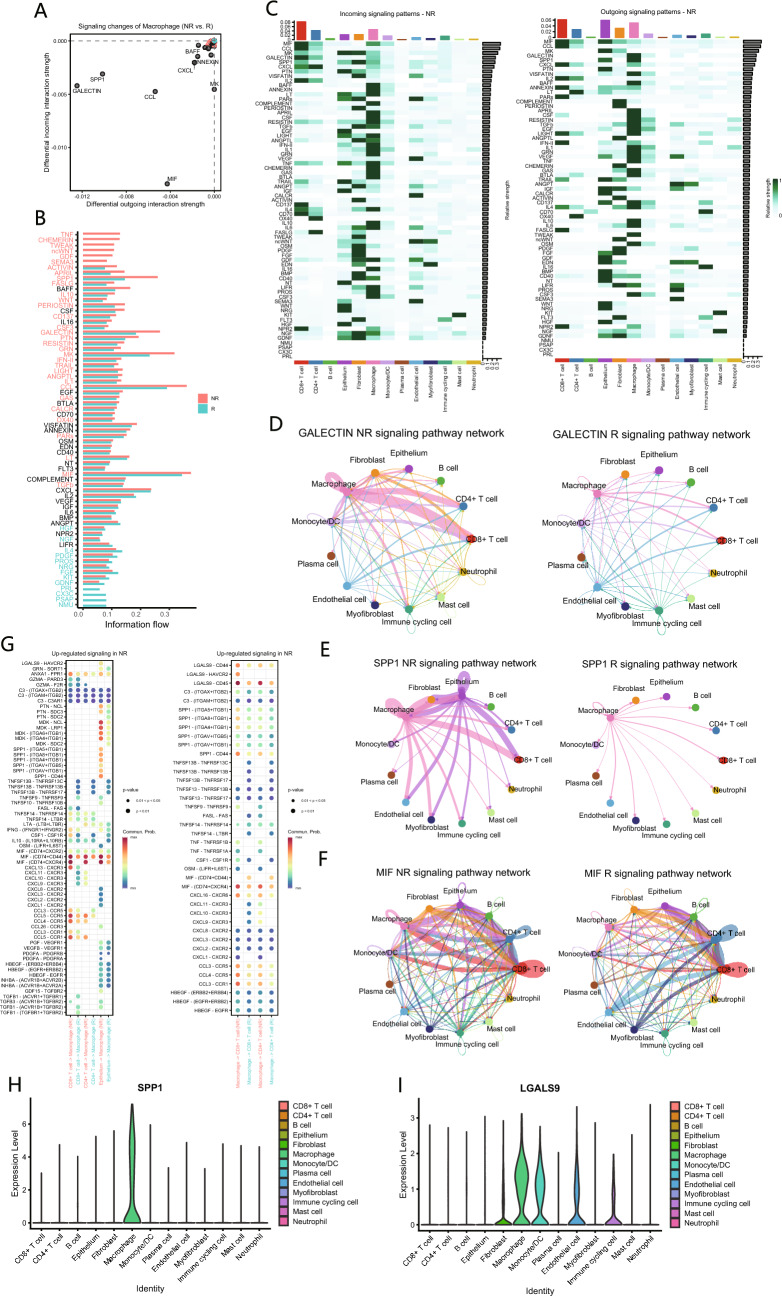
We further investigated the specific signaling changes in macrophages between the non-responder and responders, discovering that macrophages in the non-responder exhibited elevated levels of SPP1 and GALECTIN signaling, while also receiving enhanced MIF signals (Fig. 3A). At the global signaling pathway level, the predominant signaling patterns in the non-responder encompassed SPP1, GALECTIN, CCL, MK, and MIF pathways, with SPP1 demonstrating significant differences between the two groups (Fig. 3B). A heatmap illustrating the incoming and outgoing signaling patterns across various cell populations revealed that macrophages served as the primary senders of SPP1 and GALECTIN signals (Fig. 3C). To elucidate the sending and receiving relationships, a network diagram was employed to compare the strength of various signaling pathways between the two groups. The results indicated that macrophages primarily influenced CD8+ T cells through the SPP1 and GALECTIN pathways (Fig. 3D and E), suggesting their potential significance in NACI efficacy. Additionally, macrophages received enhanced MIF signals from epithelial cells and CD8+ T cells, which may contribute to alterations in macrophage function (Fig. 3F).
Fig. 3.
Cell–cell communication between macrophages and other cell populations. A Differences in various signaling pathways in macrophages between NR and R. B Differences in the overall signaling pathway between NR and R. C Heatmap illustrates the incoming and outgoing signaling patterns of NR and R, with color intensity representing relative strength of different signaling pathways. D-F Network diagram displays GALECTIN(D), SPP1(E) and MIF(F) signaling pathway network in NR and R. G Communication probabilities of important ligand-receptor pairs between macrophages and T cells in NR. The color of dots represents the communication probability, while the size of dots indicates p value. H, I Comparison of the expression levels of SPP1(H) and LGALS9(I) among different cell populations
Finally, we delineated the interaction pathways between macrophages and T cells, discovering that the SPP1 signals emitted by macrophages in the non-responder are predominantly received by CD8+ T cells via the receptors CD44 and ITGB1. In the context of GALECTIN signaling, the LGALS9 emitted by macrophages is primarily received by CD8+ T cells through receptors CD44, CD45, and HAVCR2, as well as by CD4+ T cells via receptors CD44 and CD45. Additionally, macrophages predominantly receive MIF signals via receptors CD74, CD44, and CXCR4 (Fig. 3G). Furthermore, we observed that, in comparison to LGALS9, SPP1 demonstrates a characteristic of specific expression in macrophages (Fig. 3H and I).
SPP1+ macrophages are the primary macrophage subcluster contributing to resistance to NACI in ESCC patients
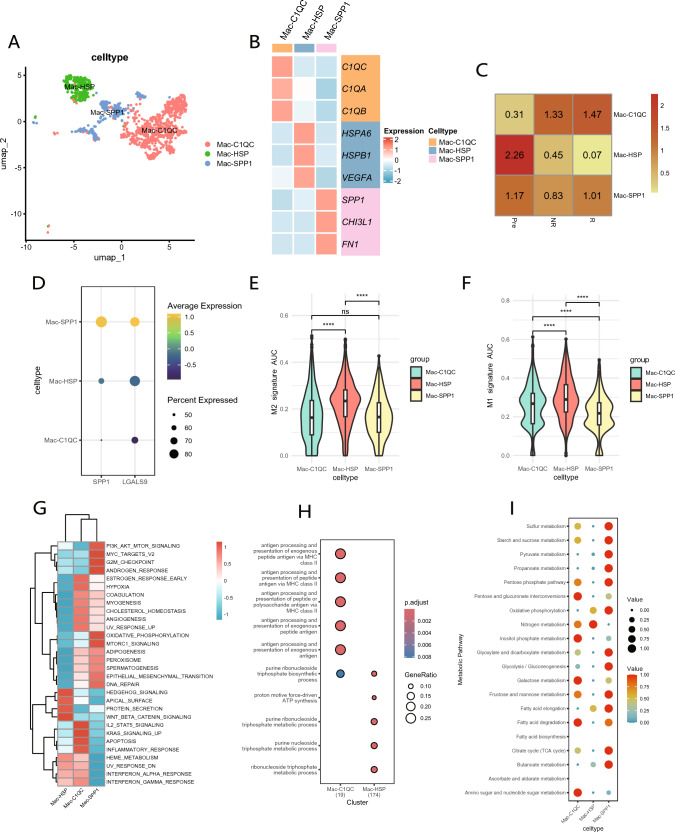
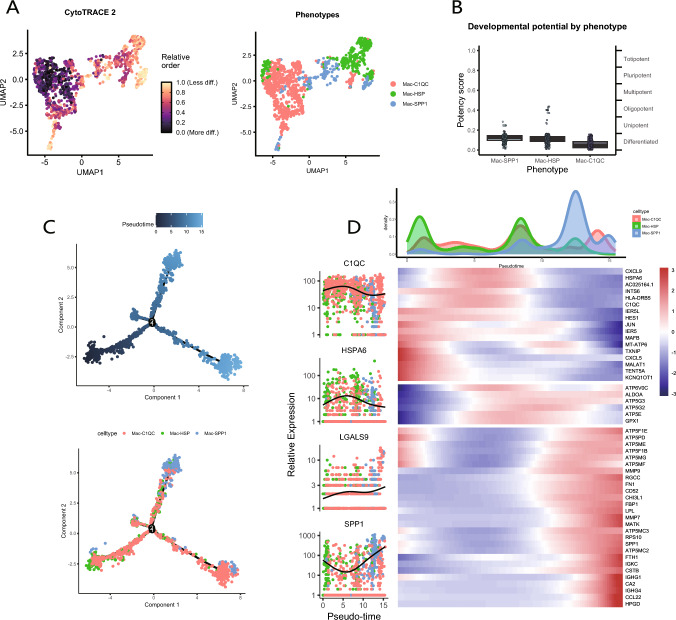
Evidently, macrophages exhibit a crucial role in intercellular communication within the non-responder. To further elucidate their characteristics, we classified them into three distinct cell clusters based on unique gene markers, designated as Mac-C1QC, Mac-HSP, and Mac-SPP1 (Fig. 4A and B). The ratio of observed over expected cell numbers (Ro/e) was employed to illustrate the tissue preferences of macrophage subclusters, revealing that Mac-HSP predominantly resided in pre-treatment samples, while Mac-C1QC was primarily identified in post-treatment samples. The proportion of Mac-SPP1 exhibited minimal variation before and after treatment (Fig. 4C). Notably, in addition to SPP1, Mac-SPP1 demonstrated high expression of LGALS9 (Fig. 4D), suggesting that Mac-SPP1 may be a critical factor contributing to resistance to NACI. Mac-C1QC exhibited elevated levels of inflammatory cytokines such as CXCL1, CXCL2, CXCL3, and CXCL8, potentially facilitating the recruitment and activation of immune cells (Fig. S2A).
Fig. 4.
Single-cell RNA-seq reveals the characteristics of macrophage subclusters. A UMAP plot displays three distinct subclusters of macrophages. B Heatmap shows the average expression levels of characteristic marker genes used to distinguish different macrophage subclusters. C Heatmap displays the Ro/e index to compare the tissue distribution preferences of the three macrophage subclusters. D Comparison of the expression levels of SPP1 and LGALS9 among different macrophage subclusters. E–F Comparison of M2(E) and M1(F) polarization signature scores among the three macrophage subclusters. G Heatmap illustrates the different pathways that are enriched in the DEGs of three macrophage subclusters. H GO enrichment analysis of differentially expressed genes among macrophage subclusters. I Comparison of metabolic pathways among three macrophage subclusters
Subsequently, we utilized M1 and M2-related gene sets to compute AUC scores for the various macrophage subclusters. The results indicated that Mac-HSP, predominantly identified in pre-treatment tissues, exhibited the highest M2 score, thereby confirming its classification as classic TAMs (Fig. 4E). Among the two cell populations primarily present in post-treatment samples, Mac-C1QC demonstrated a higher M1 score, indicating its beneficial role in NACI (Fig. 4F). Furthermore, the M2 scores of pre-treatment samples were significantly higher than those observed in post-treatment samples, but no significant difference was found in M2 scores between the non-responder and responders. (Fig. S2B). This suggests that NACI reverses the M2 polarization of macrophages, resulting in a shift towards the anti-tumor M1 phenotype. No significant difference was observed in M1 scores between the pre-treatment patients and post-treatment non-responder; however, both were higher than those in responders, indicating that a high M1 score may not be adequate to predict a favorable response to NACI (Fig. S2C).
To further elucidate the differences among various macrophage subclusters, we conducted GSVA on DEGs (Fig. 4G). The results indicated that Mac-SPP1 exhibited enrichment in pathways such as PI3K_AKT_MTOR_SIGNALING, OXIDATIVE_PHOSPHORYLATION, and MTORC1_SIGNALING. Oxidative phosphorylation represents a primary metabolic pathway in M2 macrophages, suggesting that Mac-SPP1 may exhibit characteristics typical of M2 macrophages. Mac-C1QC exhibited enrichment in pathways such as INFLAMMATORY_RESPONSE, KRAS_SIGNALING_UP, and IL2_STAT5_SIGNALING, which indicate its involvement in anti-tumor immune responses. Mac-HSP exhibited enrichment in pathways associated with HEDGEHOG_SIGNALING and WNT_BETA_CATENIN_SIGNALING, indicating that these cells play significant roles in tissue repair and regeneration, consistent with its predominant presence in pre-treatment samples.
In GO enrichment analysis (Fig. 4H), Mac-C1QC was enriched in gene sets related to the processing and presentation of exogenous peptide antigens through MHC class II, underscoring its pivotal role in antigen processing and presentation, specifically regarding MHC class II-mediated presentation of exogenous peptide antigens. This suggests that Mac-C1QC is likely to play a crucial role in T cell activation and the initiation of adaptive immune responses. Mac-HSP was enriched in gene sets associated with proton motive force-driven ATP synthesis, purine ribonucleoside triphosphate, and purine nucleoside triphosphate metabolic processes. The proton motive force-driven ATP synthesis is a key step in mitochondrial oxidative phosphorylation, indicating that Mac-HSP is likely to be highly active in nucleic acid synthesis and cell signaling.
Lastly, we conducted a comprehensive exploration of metabolic differences among the three macrophage subclusters utilizing scMetabolism package (Fig. 4I). The results indicated that Mac-SPP1 exhibited the activation of multiple metabolic pathways, including citrate cycle (TCA cycle), pentose phosphate pathway, and glycolysis/gluconeogenesis. These metabolic characteristics imply that Mac-SPP1 cells constitute a metabolically active macrophage subcluster, potentially engaged in complex energy demands and metabolic reprogramming.
To investigate the characteristics of macrophage subtype differentiation, we employed the CytoTRACE tool to analyze the cellular stemness among various macrophage subtypes (Fig. 5A). The results indicated that Mac-C1QC exhibited a higher level of cellular differentiation compared to two other subtypes, representing the final stage of macrophage differentiation (Fig. 5B). Subsequently, we utilized the Monocle tool to perform pseudotime analysis of the macrophage subtypes (Fig. 5C), which shown the distribution of the three macrophage subclusters (Fig. S2D). As the pseudotime trajectory progressed, the expression changes of markers associated with the macrophage subtypes (C1QC, HSPA6, SPP1) were shown, as shown in this figure (Fig. 5D). Notably, C1QC exhibited a similar expression pattern to HSPA6, while SPP1 displayed a similar pattern to LGALS9, with contrasting trends between the two pairs. The pseudotime trajectory plot, based on the expression levels of SPP1 and LGALS9, demonstrated a remarkably similar distribution of high expression regions for both genes (Fig. S2E). Furthermore, Mac-C1QC and Mac-SPP1 were identified as the most predominant macrophage subclusters in post-treatment samples. Collectively, these findings suggest that Mac-SPP1 may be associated with resistance to NACI, while Mac-C1QC may hold greater significance in the context of NACI treatment.
Fig. 5.
Pseudotime analysis of macrophage subclusters. A, B CytoTRACE detects differences in the degree of differentiation among macrophage subclusters. C Differences in the changes and distribution of pseudotime trajectories among the three macrophage subclusters. D The waveform plot illustrates the density changes of the three macrophage subclusters during the pseudotime process. The heatmap displays the differential gene expression changes and clustering of the three macrophage subclusters during the pseudotime process. The left panel shows the expression patterns of marker genes and LGALS9 during the pseudotime process
Bulk transcriptome sequencing analysis confirms the association between macrophage subset and resistance to NACI in ESCC
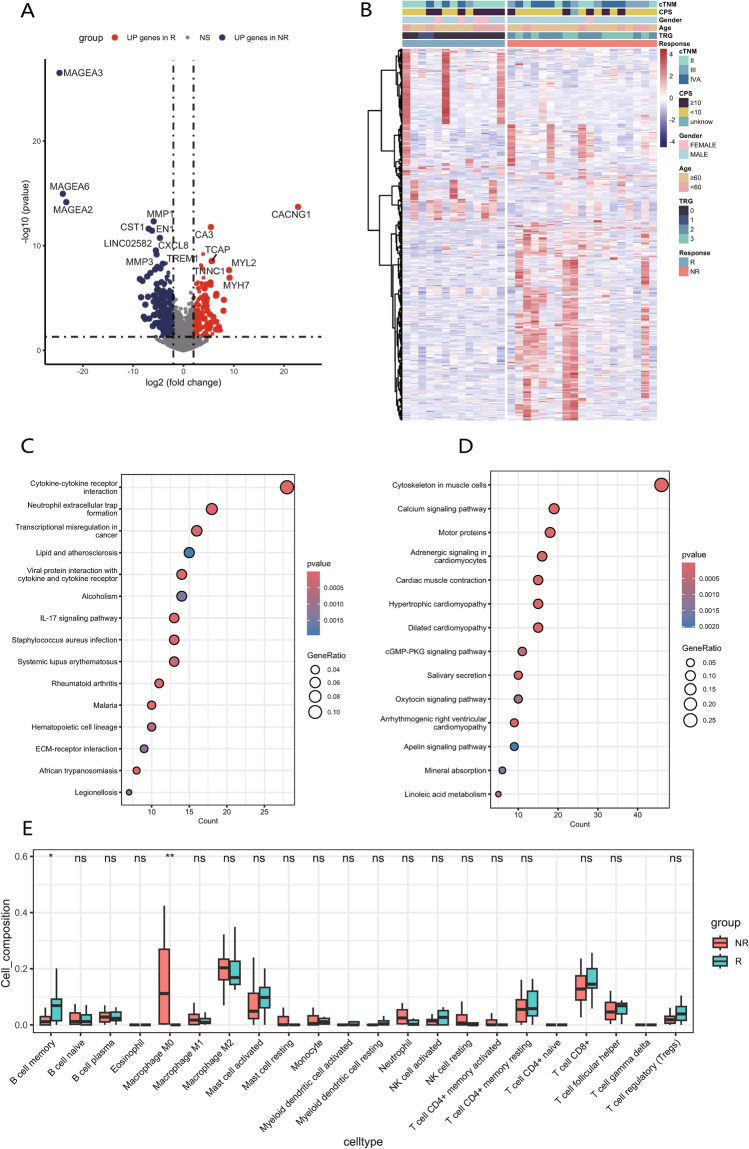
We conducted transcriptome sequencing on tumor specimens obtained from 32 patients diagnosed with ESCC who underwent NACI, with their clinical information shown in Table 1. Based on pathological TRG scores of postoperative specimens, samples with scores of 0 and 1 were categorized into responders, whereas those with scores of 2 and 3 were categorized into non-responders. Utilizing our transcriptome sequencing data, we identified a total of 1,346 DEGs between non-responders and responders, with 511 genes upregulated in responders and 835 genes upregulated in non-responders (Fig. 6A). The heatmap of DEGs demonstrated distinct clustering of samples from non-responders and responders (Fig. 6B). We subsequently performed KEGG enrichment analysis on identified DEGs. Genes that were highly expressed in non-responders (Fig. 6C) were enriched in pathways including cytokine-cytokine receptor interaction, transcriptional misregulation in cancer, and viral protein interaction with cytokine and cytokine receptor. In contrast, genes that were highly expressed in responders (Fig. 6D) were enriched in pathways such as cytoskeleton in muscle cells and calcium signaling pathway. These findings suggest that non-responders exhibit significant inflammatory responses, abnormal immune system activation, and dysregulation of metabolic and transcriptional processes.
Table 1.
Clinical characteristics of ESCC patients (N = 32)
| Characteristics | N (%) |
|---|---|
| Sex | |
| Female | 5(15.6) |
| Male | 27(84.4) |
| Age | |
| < 60 | 11(34.4) |
| ≥ 60 | 21(65.6) |
| cTNM | |
| II | 14(43.7) |
| III | 7(21.9) |
| IV | 11(34.4) |
| CPS | |
| ≥ 10 | 12(37.5) |
| < 10 | 19(59.4) |
| Unknown | 1(3.1) |
| TRG score | |
| 0 | 11(34.4) |
| 1 | 2(6.2) |
| 2 | 10(31.3) |
| 3 | 9(28.1) |
| Pathological response | |
| NR | 19(59.4) |
| R | 13(40.6) |
CPS combined positive score, TRG Tumor Regression Grade, NR non-responders, R responders
Fig. 6.
Differential analysis of clinical bulk transcriptome sequencing samples. A Volcano plot illustrates the differentially expressed genes between NR and R. B Heatmap displays the expression patterns of the 200 genes with the largest fold changes between NR and R, along with their correlations with clinical traits. C, D KEGG analysis of genes that are highly expressed in NR(C) and R(D). E Box plot illustrates the differences in the proportions of 22 immune cell types between NR and R as assessed by CIBERSORT. The box represents the interquartile range (IQR), showing the middle 50% of the data, with a line for the median. The whiskers extend to the smallest and largest values within 1.5 times the IQR from the quartiles
Next, CIBERSORT deconvolution algorithm was employed to analyze the immune microenvironment between non-responders and responders (Fig. 6E). There were no statistically significant differences in the proportions of the majority of immune cell types between two groups. However, non-responders exhibited a significantly higher composition of “Macrophage M0” compared to responders, suggesting that genes associated with “Macrophage M0” may correlate with resistance to NACI in ESCC.
The characteristic gene set for SPP1+ macrophages is identified by integrating bulk and single‑cell RNA-seq data
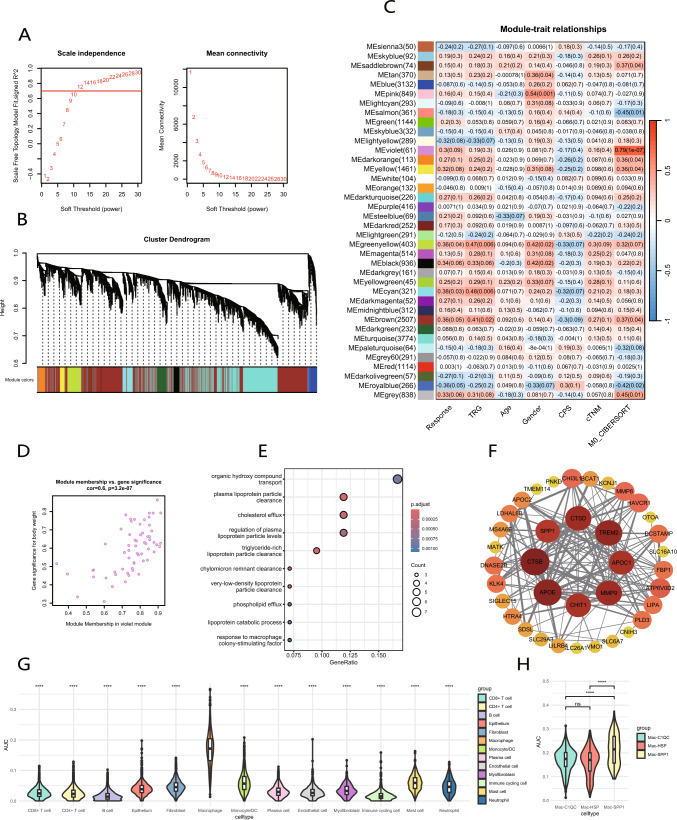
The WGCNA algorithm was employed to modularize gene expression across all patients to identify the core gene set most closely associated with “Macrophage M0”. A soft threshold of 12 was selected (Fig. 7A), and the clustering dendrogram revealed a total of 38 gene modules (Fig. 7B). The violet module, which contains 61 genes, exhibited a strong correlation with “Macrophage M0” (Fig. 7C). Subsequently, we calculated and visualized the correlation matrix between the violet module and individual genes, along with the phenotypic correlation. It was shown that genes highly correlated with the traits also played critical roles within the key module (Fig. 7D). Based on the significant p values of gene-trait relationships, 58 hub genes were identified from the 61 genes in the violet module. Subsequently, we conducted GO enrichment analysis on these 58 genes (Fig. 7E), identifying the enriched terms such as response to macrophage colony-stimulating factor, lipoprotein catabolic process, and plasma lipoprotein particle clearance. Thus, this module is likely associated with alterations in lipid metabolism and macrophage activity. Furthermore, the PPI network analysis highlighted TREM2, APOE, MMP9, SPP1, CTSB, CDST, CHIT1, and APOC1 as hub genes, indicating their potential significance (Fig. 7F).
Fig. 7.
WGCNA identifies the “M0 macrophage” module, and AUCell reveals its association with Mac-SPP1. A Selecting a soft threshold to construct a gene co-expression network. B The clustering diagram illustrates the distribution of different gene modules. C Heatmap reveals the correlation between gene modules and sample traits. D Correlation between gene significance and module membership in violet module. E GO enrichment analysis of genes in violet module. F PPI network constructed using genes from violet module. G, H AUC scores of violet module genes across different cell clusters and three macrophage subclusters in scRNA-seq data
To investigate which cell population is associated with “Macrophage M0” characteristic gene set, we utilized AUCell to evaluate different cell populations derived from the aforementioned scRNA-seq data (Fig. 7G). The AUC score for macrophage cell population was significantly higher than that of the other cell populations. More importantly, Mac-SPP1 score was significantly higher than that of both Mac-C1QC and Mac-HSP (Fig. 7H), whereas no significant difference was observed in the AUC values between Mac-C1QC and Mac-HSP. This indicates that the gene set associated with “Macrophage M0” is characteristic of Mac-SPP1.
Public database validation confirms that SPP1+ macrophages are associated with immunotherapy resistance in ESCC
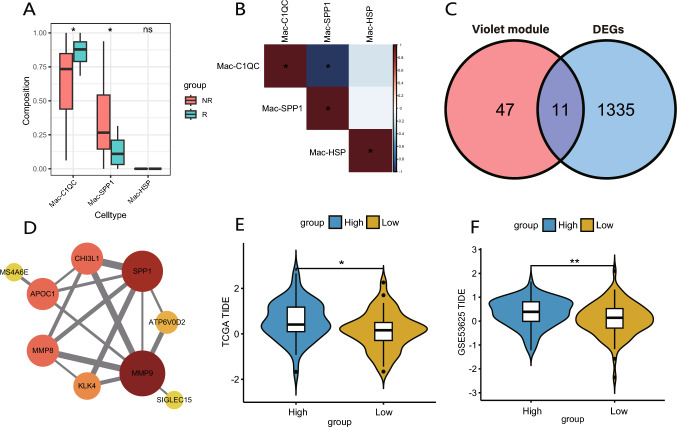
To investigate whether there were differences in the proportions of Mac-SPP1, Mac-C1QC, and Mac-HSP between non-responders and responders in the clinical bulk transcriptome data, we employed the deconvolution algorithm CIBERSORTx to generate a macrophage profile from the previously mentioned scRNA-seq data (Fig. 8A). Non-responders exhibited a higher proportion of Mac-SPP1, whereas responders demonstrated a greater proportion of Mac-C1QC, with negligible presence of Mac-HSP in both groups. Due to the limited availability of single-cell samples from ESCC following NACI treatment, a similar distribution of macrophage subsets as in bulk RNA-seq was not found in the scRNA-seq data. Nevertheless, the markedly low abundance of Mac-HSP in post-treatment ESCC samples was observed in both scRNA-seq data and our transcriptomic findings. Correlation analysis revealed a significant negative correlation between the proportions of Mac-SPP1 and Mac-C1QC (Fig. 8B, Fig. S2F), which aligns with the conclusions derived from the pseudotime analysis of macrophage subsets.
Fig. 8.
Validation of the association between Mac-SPP1 characteristics and immune therapy resistance using public transcriptome databases. A Using CIBERSORTx to infer the proportions of three macrophage subclusters in clinical bulk transcriptome sequencing data. The box represents the interquartile range (IQR), showing the middle 50% of the data, with a line for the median. The whiskers extend to the smallest and largest values within 1.5 times the IQR from the quartiles. B Heatmap displays the correlations between the proportions of three macrophage subclusters in bulk transcriptome sequencing data. C Venn diagram shows the intersection between the genes in violet module and the differentially expressed genes between NR and R. D PPI network of the intersecting genes. E, F In TCGA(E) and GEO(F) datasets, groups with higher scores for the intersecting genes have higher TIDE scores, indicating poorer outcomes for immune therapy
Finally, we aim to further validate our findings in public databases. We identified 12 genes through the intersection of the DEGs between non-responders and responders in bulk transcriptomic data and the genes within the violet module (Fig. 8C), among which SPP1 and MMP9 are pivotal genes (Fig. 8D). Based on expression characteristics of T cell dysfunction and exclusion, Tumor Immune Dysfunction and Exclusion (TIDE) analysis predicts immunotherapy responses. In the TCGA cohort, ESCC patients were divided into two groups according to the median score based on the intersected genes and then subjected to TIDE analysis. The group with higher scores had elevated TIDE scores, suggesting a greater likelihood of developing resistance to immunotherapy (Fig. 8E). Subsequently, we conducted a similar analysis on GSE53625 dataset, yielding consistent conclusions with those from TCGA database (Fig. 8F).
Discussion
Increasing evidence indicates that immunosuppressive components within the TME play a pivotal role in conferring resistance to immunotherapy [30]. The TME of ESCC constitutes a highly immunosuppressive environment, presenting substantial challenges to the eradication of cancer cells and the induction of antitumor immunity [31]. TAMs, as a principal component of TME, exhibit considerable plasticity and heterogeneity, and can facilitate cancer cell survival, induce angiogenesis, and suppress CD8+ T cell function through cell–cell interactions, inhibitory cytokine secretion, and upregulation of immune checkpoints, thereby playing a significant role in treatment resistance [32]. Nevertheless, the roles of distinct macrophage subsets in the NACI treatment of ESCC remain ambiguous. Single-cell technologies have empowered researchers to delineate cell populations based on unique transcriptomic characteristics, thereby revealing their diverse biological significance.
Our analysis revealed that in the non-responder receiving NACI, the GALECTIN signaling pathway in macrophages was significantly enhanced, interacting with CD8+ T cells and CD4+ T cells through receptors such as CD44, CD45, and HAVCR2. Similarly, SPP1 signaling was also markedly pronounced, interacting with CD8+ T cells via receptors including CD44, ITGB1, ITGAV, and ITGA5. SPP1, also known as osteopontin, is a critical extracellular matrix protein that plays a significant role in various biological processes, including cell adhesion, migration, and proliferation [33]. Recent studies have demonstrated that SPP1 plays a crucial role in the initiation and progression of many cancers [34–36]. High levels of SPP1 have been detected in various types of cancer and are associated with poor prognosis, acting as a potential biomarker and therapeutic target [37–39]. SPP1 has also been shown to be involved in the regulation of the TME [40]. The interaction between SPP1 and the cell surface receptor CD44 can inhibit T cell activation and proliferation, thereby promoting tumor immune evasion [41]. Likewise, LGALS9, the gene encoding galectin-9, has been documented to be implicated in tumor progression [42]. The LGALS9-CD44 interaction has been demonstrated to enhance the differentiation and maintenance of suppressive regulatory T cells [43]. Thus, it may provide a potential strategy to reverse the immunosuppression in ESCC by attenuating these signaling pathways through the downregulation of CD44 expression.
Furthermore, we observed that in the non-responder, macrophages received more robust MIF signals from CD8+ T cells and epithelial cells via CD74, CD44, and CXCR4. MIF is a multifunctional molecule that functions as a pro-inflammatory factor and chemokine, regulating cellular proliferation, angiogenesis, and fibrosis [44]. Research has demonstrated that MIF can enhance tumor cell proliferation by influencing the tumor suppressor gene p53 [45]. MIF secreted by tumor cells can promote the formation of myeloid-derived suppressor cells (MDSCs), which subsequently inhibit the proliferation and antitumor functions of CD8+ T cells [46]. Additionally, MIF may contribute to chemotherapy resistance [47]. Therefore, targeting MIF not only directly inhibits tumor cell growth but also suppresses the generation of MDSCs, potentially reversing the tumor immunosuppressive microenvironment and enhancing the efficacy of NACI in ESCC.
In this study, we identified three macrophage subsets in ESCC; among these, Mac-SPP1 exhibited elevated levels of SPP1 and LGALS9 and was metabolically active through various pathways. Previous research has indicated that Mac-SPP1 is abundant in cancer, displaying increased M2 signaling, and is generally associated with a poor prognosis [48]. However, in our study, we did not observe characteristics of M2 polarization in Mac-SPP1, which may be attributed to the reversal of TAM M2 polarization induced by NACI. In contrast to Mac-SPP1, although Mac-C1QC has been implicated in predicting poor prognosis in various cancers, there is also evidence suggesting that Mac-C1QC represents an antitumor immune cell population [49, 50]. In our study, Mac-C1QC exhibited higher expression levels of inflammatory cytokines, including IL1B, CXCL1, CXCL2, CXCL3, CXCL8, and CCL3. CXCL1, CXCL2, and CXCL3 bind to the chemokine receptor CXCR3, inducing the recruitment of granulocytes to sites of injury or infection, thereby triggering inflammatory responses [51]. CCL3 can bind to CCR1, CCR3, and CCR5, which are involved in recruiting monocytes and lymphocytes during inflammation [52]. Due to this, Mac-C1QC may play an active role in recruiting and regulating immune cells during tumor-associated inflammation. Additionally, enrichment analysis of Mac-C1QC indicated its efficient antigen-presenting function, suggesting that it is a primary macrophage subset responsible for clearing apoptotic cells. Previous studies have also confirmed that the C1Q molecule can enhance the phagocytic capacity of macrophages [53]. Our findings suggest that Mac-C1QC is an important antigen-presenting macrophage subset that promotes T cell activation in esophageal cancer, revealing its significant antitumor role within the TME. Understanding the mechanisms by which Mac-C1QC regulates tumor immune responses represents a promising avenue for future research. Mac-HSP is primarily found in untreated ESCC samples and exhibits high levels of M2 polarization, which is characteristic of classical TAMs cells. Studies have shown that Mac-HSP is present in a higher proportion in ESCC compared to normal tissues and is negatively correlated with survival outcomes [54].
CIBERSORT analysis of clinical samples revealed a significantly higher proportion of “M0 macrophages” in non-responders. Given the heterogeneity of tumors, WGCNA was used to identify the most relevant gene set; additionally, we observed that this gene set was significantly enriched in Mac-SPP1. These findings suggest a greater abundance of Mac-SPP1 in non-responders, as indicated by transcriptomic sequencing data. Furthermore, CIBERSORTx analysis demonstrated that responders exhibited elevated levels of Mac-C1QC, and a significant negative correlation was observed between the proportions of Mac-C1QC and Mac-SPP1. This finding further supports the positive role of Mac-C1QC in the response to NACI. Previous studies have classified patients into distinct groups based on the gene markers of Mac-C1QC and Mac-SPP1, revealing differences in prognoses, tumor stages, and levels of immune cell infiltration [55]. This underscores the functional heterogeneity exhibited by these two macrophage subtypes. Finally, TIDE analysis of the GEO and TCGA cohorts further validated that the presence of Mac-SPP1 is associated with unfavorable therapeutic outcomes.
Our study presents several limitations. For instance, the limited availability of single-cell samples of ESCC treated with NACI in public databases complicates the dissection of highly heterogeneous cell clusters at a finer resolution, which may lead to the oversight of other significant features. Future research will necessitate larger datasets to investigate the relationship between various cell subclusters and clinical outcomes. Furthermore, our conclusions are predominantly derived from data mining and have yet to be validated through additional experimental studies. Future experimental research is essential to explore the specific mechanisms that various macrophage subclusters exert their functions and to identify more precise targets for enhancing clinical outcomes.
As a result, we identify the distinct roles of macrophage subclusters in NACI-treated esophageal cancer. Mac-SPP1 may contribute to NACI resistance through the secretion of SPP1 and LAGLS9, thereby inhibiting T cell response activity. Conversely, Mac-C1QC may play a crucial role in the recruitment and regulation of immune cells. Our research offers novel insights for future therapeutic strategies in ESCC.
Supplementary Information
Below is the link to the electronic supplementary material.
Fig. S1 A-B The UMAP plot illustrates the distribution of 13 cell clusters before and after treatment, as well as among different efficacy groups. C-E Bar chart displays the proportions of 13 cell clusters within each sample, categorized by pre-treatment and post-treatment groups, as well as by different efficacy groups.
Fig. S2 A Comparison of inflammatory cytokine expression levels across three macrophage clusters. B-C Comparison of M2 (B) and M1 (C) polarization characteristic scores of macrophages between the pre-treatment group and different efficacy groups. D The distribution of three macrophage subclusters on the trajectory plot. E The distribution of SPP1 and LGALS9 expression levels on the trajectory plot. F The scatter plot depicts the correlation between the proportions of Mac-C1QC to Mac-SPP1 in clinical ESCC bulk transcriptome data.
Author contributions
L.H. and Y.Q. designed the research framework and supervised the research procedures. Z.G., F.L. and Z.Y. participated in data organization and analysis. B.L. and Y.X. collected the clinical data of the patients. B.W., Y.S. and P.Y. participated in the clinical work and collected surgical specimens. Y.Q. contributed to funding acquisition. Z.G. and F.L participated in the writing of the article.
Funding
This work was supported by the funds from the Cultivation Project of Henan Health Science and Technology Innovation Talents (Grant No. YXKC2022014).
Data availability
The single-cell transcriptome data (GSE203115) and bulk transcriptome data (GSE53625) can be obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). Furthermore, gene expression information for ESCC patients was downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). No datasets were generated or analyzed during the current study.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University (No. L2021-Y346-004).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhenyang Geng, Feng Li and Zhichang Yang have equally contributed to this work.
Contributor Information
Lan Huang, Email: lanhuang@zzu.edu.cn.
Yu Qi, Email: qiyu_zzu@126.com.
References
- 1.Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin 71:209–249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Abnet CC, Arnold M, Wei W-Q (2018) Epidemiology of esophageal squamous cell carcinoma. Gastroenterology 154:360–373. 10.1053/j.gastro.2017.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Napier KJ (2014) Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. WJGO 6:112. 10.4251/wjgo.v6.i5.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ajani JA, D’Amico TA, Bentrem DJ et al (2023) Esophageal and esophagogastric junction cancers, version 2.2023, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 21:393–422. 10.6004/jnccn.2023.0019 [DOI] [PubMed] [Google Scholar]
- 5.Yang H, Liu H, Chen Y et al (2018) Neoadjuvant chemoradiotherapy followed by surgery versus surgery alone for locally advanced squamous cell carcinoma of the esophagus (NEOCRTEC5010): a phase III multicenter, randomized, open-label clinical trial. JCO 36:2796–2803. 10.1200/JCO.2018.79.1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kono K, Mimura K, Yamada R et al (2018) Current status of cancer immunotherapy for esophageal squamous cell carcinoma. Esophagus 15:1–9. 10.1007/s10388-017-0596-2 [DOI] [PubMed] [Google Scholar]
- 7.Sun J-M, Shen L, Shah MA et al (2021) Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): a randomised, placebo-controlled, phase 3 study. The Lancet 398:759–771. 10.1016/S0140-6736(21)01234-4 [DOI] [PubMed] [Google Scholar]
- 8.Sihag S, Ku GY, Tan KS et al (2021) Safety and feasibility of esophagectomy following combined immunotherapy and chemoradiotherapy for esophageal cancer. J Thorac Cardiovasc Surg 161:836-843.e1. 10.1016/j.jtcvs.2020.11.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Li J, Lin W et al (2022) Neoadjuvant camrelizumab plus chemotherapy for resectable, locally advanced esophageal squamous cell carcinoma ( NIC-ESCC2019): a multicenter, phase 2 study. Int J Cancer 151:128–137. 10.1002/ijc.33976 [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Yang Y, Liu Z et al (2022) Multicenter, single-arm, phase II trial of camrelizumab and chemotherapy as neoadjuvant treatment for locally advanced esophageal squamous cell carcinoma. J Immunother Cancer 10:e004291. 10.1136/jitc-2021-004291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLane LM, Abdel-Hakeem MS, Wherry EJ (2019) CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol 37:457–495. 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- 12.Paydary K, Reizine N, Catenacci DVT (2021) Immune-checkpoint inhibition in the treatment of gastro-esophageal cancer: a closer look at the emerging evidence. Cancers 13:5929. 10.3390/cancers13235929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D, Coussens LM (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21:309–322. 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- 14.Sanmamed MF, Chen L (2018) A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell 175:313–326. 10.1016/j.cell.2018.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji G, Yang Q, Wang S et al (2024) Single-cell profiling of response to neoadjuvant chemo-immunotherapy in surgically resectable esophageal squamous cell carcinoma. Genome Med 16:49. 10.1186/s13073-024-01320-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, Zhang Y, Ma N et al (2023) Progenitor-like exhausted SPRY1+CD8+ T cells potentiate responsiveness to neoadjuvant PD-1 blockade in esophageal squamous cell carcinoma. Cancer Cell 41:1852-1870.e9. 10.1016/j.ccell.2023.09.011 [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Leng X, Liu Q et al (2023) Intratumoral microbiota composition regulates chemoimmunotherapy response in esophageal squamous cell carcinoma. Can Res 83:3131–3144. 10.1158/0008-5472.CAN-22-2593 [DOI] [PubMed] [Google Scholar]
- 18.Li J, Chen Z, Tian L et al (2014) LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut 63:1700–1710. 10.1136/gutjnl-2013-305806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart T, Butler A, Hoffman P et al (2019) Comprehensive integration of single-cell data. Cell 177:1888-1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korsunsky I, Millard N, Fan J et al (2019) Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16:1289–1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin S, Guerrero-Juarez CF, Zhang L et al (2021) Inference and analysis of cell-cell communication using cell chat. Nat Commun 12:1088. 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulati GS, Sikandar SS, Wesche DJ et al (2020) Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367:405–411. 10.1126/science.aax0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Den Berge K, Roux De Bézieux H, Street K et al (2020) Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun 11:1201. 10.1038/s41467-020-14766-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu T, Hu E, Xu S et al (2021) clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2:100141. 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Saeed AFUH, Liu Q et al (2023) Macrophages in immunoregulation and therapeutics. Sig Transduct Target Ther 8:207. 10.1038/s41392-023-01452-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y, Yang S, Ma J et al (2022) Spatiotemporal Immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov 12:134–153. 10.1158/2159-8290.CD-21-0316 [DOI] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinf 9:559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman AM, Steen CB, Liu CL et al (2019) Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 37:773–782. 10.1038/s41587-019-0114-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Visser KE, Joyce JA (2023) The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell 41:374–403. 10.1016/j.ccell.2023.02.016 [DOI] [PubMed] [Google Scholar]
- 31.Davern M, Donlon NE, Power R et al (2021) The tumour immune microenvironment in oesophageal cancer. Br J Cancer 125:479–494. 10.1038/s41416-021-01331-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mantovani A, Marchesi F, Malesci A et al (2017) Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 14:399–416. 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kariya Y, Kariya Y (2022) Osteopontin in Cancer: Mechanisms and Therapeutic Targets. IJTM 2:419–447. 10.3390/ijtm2030033 [Google Scholar]
- 34.Wu Q, Li L, Miao C et al (2022) Osteopontin promotes hepatocellular carcinoma progression through inducing JAK2/STAT3/NOX1-mediated ROS production. Cell Death Dis 13:341. 10.1038/s41419-022-04806-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao J, Li J, Sun L et al (2019) Hypoxia-driven paracrine osteopontin/integrin αvβ3 signaling promotes pancreatic cancer cell epithelial–mesenchymal transition and cancer stem cell-like properties by modulating forkhead box protein M1. Mol Oncol 13:228–245. 10.1002/1878-0261.12399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo X, Ruhland MK, Pazolli E et al (2011) Osteopontin stimulates preneoplastic cellular proliferation through activation of the MAPK pathway. Mol Cancer Res 9:1018–1029. 10.1158/1541-7786.MCR-10-0472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng B, Zhou M, Wu H, Xiong Z (2018) SPP1 promotes ovarian cancer progression via Integrin β1/FAK/Akt signaling pathway. OTT 11:1333–1343. 10.2147/OTT.S154215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao H, Chen Q, Alam A et al (2018) The role of osteopontin in the progression of solid organ tumour. Cell Death Dis 9:356. 10.1038/s41419-018-0391-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi L, Wang X (2017) Role of osteopontin in lung cancer evolution and heterogeneity. Semin Cell Dev Biol 64:40–47. 10.1016/j.semcdb.2016.08.032 [DOI] [PubMed] [Google Scholar]
- 40.Matsubara E, Yano H, Pan C et al (2023) The significance of SPP1 in lung cancers and its impact as a marker for protumor tumor-associated macrophages. Cancers 15:2250. 10.3390/cancers15082250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klement JD, Paschall AV, Redd PS et al (2018) An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J Clin Investig 128:5549–5560. 10.1172/JCI123360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daley D, Mani VR, Mohan N et al (2017) Dectin 1 activation on macrophages by galectin 9 promotes pancreatic carcinoma and peritumoral immune tolerance. Nat Med 23:556–567. 10.1038/nm.4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C, Thalhamer T, Franca RF et al (2014) Galectin-9-CD44 interaction enhances stability and function of adaptive regulatory T cells. Immunity 41:270–282. 10.1016/j.immuni.2014.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez-Niño MD, Sanz AB, Ruiz-Andres O et al (2013) MIF, CD74 and other partners in kidney disease: tales of a promiscuous couple. Cytokine Growth Factor Rev 24:23–40. 10.1016/j.cytogfr.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 45.Kong F, Deng X, Kong X et al (2018) ZFPM2-AS1, a novel lncRNA, attenuates the p53 pathway and promotes gastric carcinogenesis by stabilizing MIF. Oncogene 37:5982–5996. 10.1038/s41388-018-0387-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson KD, Templeton DJ, Cross JV (2012) Macrophage migration inhibitory factor promotes tumor growth and metastasis by inducing myeloid-derived suppressor cells in the tumor microenvironment. J Immunol 189:5533–5540. 10.4049/jimmunol.1201161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ha W, Sevim-Nalkiran H, Zaman AM et al (2019) Ibudilast sensitizes glioblastoma to temozolomide by targeting Macrophage Migration Inhibitory Factor (MIF). Sci Rep 9:2905. 10.1038/s41598-019-39427-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi J, Sun H, Zhang Y et al (2022) Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and SPP1+ macrophages in colorectal cancer. Nat Commun 13:1742. 10.1038/s41467-022-29366-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tu J, Wang D, Zheng X, Liu B (2023) Single-cell RNA datasets and bulk RNA datasets analysis demonstrated C1Q+ tumor-associated macrophage as a major and antitumor immune cell population in osteosarcoma. Front Immunol 14:911368. 10.3389/fimmu.2023.911368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Revel M, Sautès-Fridman C, Fridman W-H, Roumenina LT (2022) C1q+ macrophages: passengers or drivers of cancer progression. Trends Cancer 8:517–526. 10.1016/j.trecan.2022.02.006 [DOI] [PubMed] [Google Scholar]
- 51.Griffith JW, Sokol CL, Luster AD (2014) Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol 32:659–702. 10.1146/annurev-immunol-032713-120145 [DOI] [PubMed] [Google Scholar]
- 52.Nagarsheth N, Wicha MS, Zou W (2017) Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol 17:559–572. 10.1038/nri.2017.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galvan MD, Foreman DB, Zeng E et al (2012) Complement component C1q regulates macrophage expression of mer tyrosine kinase to promote clearance of apoptotic cells. J Immunol 188:3716–3723. 10.4049/jimmunol.1102920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li P, Gao X, Huang D, Gu X (2024) Identification and characterization of prognosticmacrophage subpopulations for humanesophagealcarcinoma. CMC. 10.2174/0109298673284207240108105724 [DOI] [PubMed] [Google Scholar]
- 55.Li X, Zhang Q, Chen G, Luo D (2021) Multi-omics analysis showed the clinical value of gene signatures of C1QC+ and SPP1+ TAMs in cervical cancer. Front Immunol 12:694801. 10.3389/fimmu.2021.694801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 A-B The UMAP plot illustrates the distribution of 13 cell clusters before and after treatment, as well as among different efficacy groups. C-E Bar chart displays the proportions of 13 cell clusters within each sample, categorized by pre-treatment and post-treatment groups, as well as by different efficacy groups.
Fig. S2 A Comparison of inflammatory cytokine expression levels across three macrophage clusters. B-C Comparison of M2 (B) and M1 (C) polarization characteristic scores of macrophages between the pre-treatment group and different efficacy groups. D The distribution of three macrophage subclusters on the trajectory plot. E The distribution of SPP1 and LGALS9 expression levels on the trajectory plot. F The scatter plot depicts the correlation between the proportions of Mac-C1QC to Mac-SPP1 in clinical ESCC bulk transcriptome data.
Data Availability Statement
The single-cell transcriptome data (GSE203115) and bulk transcriptome data (GSE53625) can be obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). Furthermore, gene expression information for ESCC patients was downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). No datasets were generated or analyzed during the current study.