Abstract
Lung Cancer remains the leading cause of cancer deaths in the USA and worldwide. Non-small cell lung cancer (NSCLC) harbors high transcriptomic intratumor heterogeneity (RNA-ITH) that limits the reproducibility of expression-based prognostic models. In this study, we used multiregional RNA-seq data (880 tumor samples from 350 individuals) from both public (TRACERx) and internal (MDAMPLC) cohorts to investigate the effect of RNA-ITH on prognosis in localized NSCLC at the gene, signature, and tumor microenvironment levels. At the gene level, the maximal expression of hazardous genes (expression negatively associated with survival) but the minimal expression of protective genes (expression positively associated with survival) across different regions within a tumor were more prognostic than the average expression. Following that, we examined whether multiregional expression profiling can improve the performance of prognostic signatures. We investigated 11 gene signatures collected from previous publications and one signature developed in this study. For all of them, the prognostic prediction accuracy can be significantly improved by converting the regional expression of signature genes into sample-specific expression with a simple function—taking the maximal expression of hazardous genes and the minimal expression of protective genes. In the tumor microenvironment, we found a similar rule also seems applicable to immune ITH. We calculated the infiltration levels of major immune cell types in each region of a sample based on expression deconvolution. Prognostic analysis indicated that the region with the lowest infiltration level of protective or highest infiltration level of hazardous immune cells determined the prognosis of NSCLC patients. Our study highlighted the impact of RNA-ITH on the prognostication of NSCLC, which should be taken into consideration to optimize the design and application of expression-based prognostic biomarkers and models. Multiregional assays have the great potential to significantly improve their applications to prognostic stratification.
Subject terms: Non-small-cell lung cancer, Prognostic markers, Tumour heterogeneity, High-throughput screening
Introduction
Lung cancer is the leading cause of cancer deaths in the USA1 and worldwide2. Approximately 80% to 85% of lung cancers are non-small cell lung cancer (NSCLC)3. With the wide implementation of CT-guided lung cancer screening, there has been a drastic increase in the detection of localized NSCLCs4. Although NSCLC is potentially curable if detected early, even for stage I NSCLC, ~30% of patients still recur and succumb to this disease after surgical resection with curative intent5. Therefore, there is an urgent need to accurately predict the recurrence risk and treatment sensitivity to improve personalized adjuvant therapy.
Over the past years, many efforts have been made to identify molecular features associated with postsurgical recurrence and develop biomarkers to select high-risk NSCLC patients for adjuvant therapy. Gene expression-based signatures have been scrutinized as gene expression may be reliable, reflecting cancer biology and clinical behavior6–24. Although some signatures have demonstrated potential clinical applications8,12,18,20–22, they have yet to be widely adopted in clinical practices due to poor reproducibility when applied to independent data25–27. One of the major hurdles originates from the high intratumor heterogeneity (ITH) of the lung cancer samples28–32, which poses challenges for successful treatment and is associated with an increased recurrence risk for lung cancer29,33,34. Transcriptomic intratumor heterogeneity (RNA-ITH) in lung cancer has been reported to impede the clinical application of gene signatures33,35,36. The majority of existing prognostic signatures were developed based on the expression profiling of a single tumor sample of each individual. Due to the RNA-ITH, it is arguable how much a single sample reflects the panoply of the whole tumor characteristics. As evidenced by the multiple-regional transcriptomic studies, the expression profiles from different regions within the same tumors varied substantially35,37,38. Applying the same gene signature to assess risk resulted in discordant risk scores among different regions35.
In this study, we leveraged publicly available and newly generated multiregional RNA-sequencing data to identify molecular features associated with postsurgical recurrence in the context of RNA ITH. Using the multiregional RNA-seq from the published TRACERx cohort32,39 and our internal MDAMPLC (MD Anderson Cancer Center Multiregional Profiling in Lung Cancer) cohort, we first investigated the effect of RNA-ITH on the prognostic association of individual genes. We revealed the strong correlation between intratumor and intertumor diversity at the gene expression level and found that the maximal expression of hazardous genes (indicative of shorter survival) and minimal expression of protective genes (indicative of longer survival) was more prognostic than their average expression in NSCLC. Furthermore, we demonstrated that by considering RNA-ITH, the prediction accuracy of 11 existing prognostic signatures could be significantly improved. Based on these findings, we developed a new gene signature, PACEG (Prognosis-Associated Clonally Expressed Genes), and proposed to adopt a multiregional assay to boost its prognostic performance in NSCLC. Besides, we deconvoluted the transcriptomic data and found that the prognostic impact of the tumor immune microenvironment ITH was consistent with the findings at the gene and signature levels, namely, the maximal/minimal infiltration of anti-/pro-tumor immune cells showed the highest prognostic association. In summary, our analyses indicated that RNA-ITH might provide unique insights into the development, optimization, and clinical application of gene signatures.
Results
Multiregional RNA-seq reveals transcriptomic intratumor heterogeneity
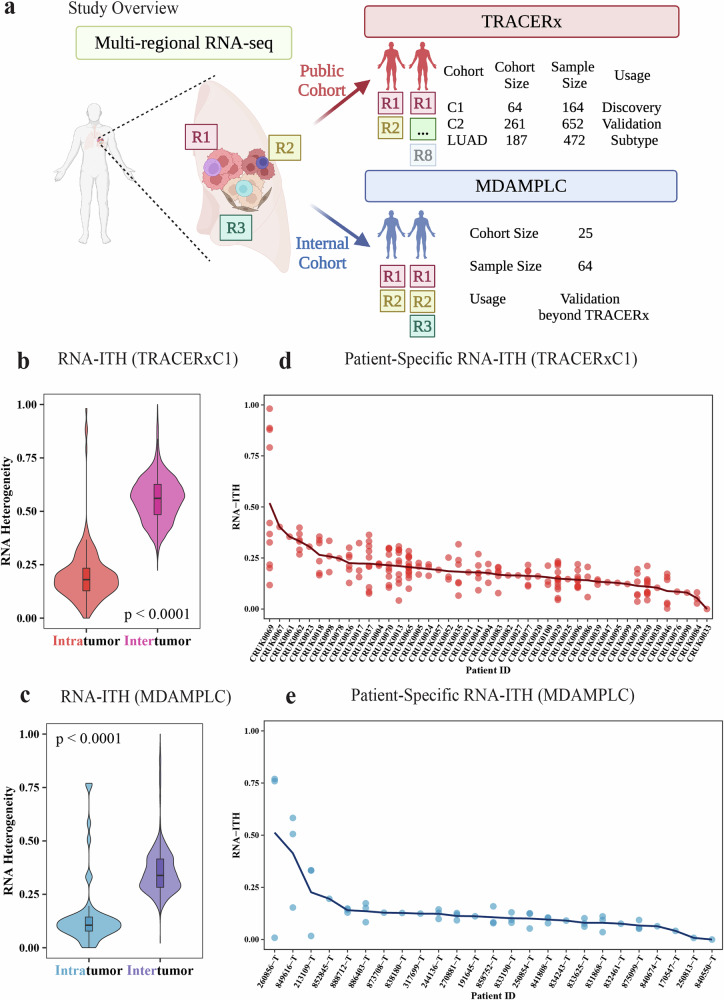
To investigate the transcriptomic intratumor heterogeneity (RNA-ITH) and its effect on patient prognosis in NSCLC, we analyzed the multiregional RNA-seq data from the TRACERx (TRAcking non-small cell lung Cancer Evolution through therapy [Rx]) project32,35,39,40. In the pursuit of robust and meaningful insights from the dataset and considering the data was collected and published over distinct timeframes, we partitioned the data into three cohorts, each serving a unique purpose (Fig. 1a, Supplementary Table 1). The first cohort of TRACERx (TRACERxC1) was released in 2019 (64 patients, 164 samples)35. We studied the RNA-seq data from this cohort, aiming to explore the intricacies of RNA-ITH and identify how to integrate RNA-ITH to improve the development and application of transcriptome-based signatures. Also, our results from this cohort are potentially comparable to previous studies, since it is widely investigated. The second cohort (TRACERxC2) we analyzed was published recently in 202340. To ensure it is an independent validation of our findings, we excluded the TRACERxC1 cohort from the latest dataset (261 patients, 652 samples). To control for histological subtypes that may confound our analysis, our third group exclusively focused on lung adenocarcinoma (LUAD) samples, the predominant histologic subtype in NSCLC and TRACERx project (TRACERxLUAD: 187 patients, 472 samples). To further confirm our findings, we also generated multiregional RNA sequencing data of 64 tumor regions from 25 NSCLC patients (MDAMPLC, MD Anderson Cancer Center Multiregional Profiling in Lung Cancer) (Fig. 1a, Supplementary Table 1).
Fig. 1. Transcriptomic intratumor heterogeneity (RNA-ITH) in NSCLC.
a Overview of the study. Multiregional RNA-seq data from two cohorts were used in this study. The main findings were identified in the TRACERxC1 cohort and confirmed in the TRACERxC2, TRACERxLUAD, and MDAMPLC cohorts. b, c Violin plot showing that RNA-ITH is lower than intertumor heterogeneity in both b TRACERxC1 and c MDAMPLC cohorts. d, e Patient-specific RNA-ITH in the d TRACERxC1 and e MDAMPLC cohorts. Each dot represents the paired tumor region from the patient. The curve indicates the patient-specific RNA-ITH, calculated by averaging the RNA-ITH of all region pairs. Significance is determined by the Wilcoxon Rank-Sum one-sided test.
For each pair of samples, we calculated the transcriptomic heterogeneity by comparing their expression profiles. The divergencies for within-patient region pairs reflect the RNA-ITH of individuals, which are compared with the intertumor heterogeneity, namely, divergencies for between-patient sample pairs (see Methods for details). As expected, the RNA-ITH is significantly lower than the intertumor heterogeneity in both the TRACERxC1 cohort (Fig. 1b) and the MDAMPLC cohort (Fig. 1c), which is consistent with the previous report35.
In Fig. D and E, we displayed the normalized expression divergencies of all within-patient region pairs for each patient and used the average value (curve) to quantify the RNA-ITH at the patient level. In both cohorts, we observed substantial variations in the RNA-ITH among patients, which may offer critical information on patient prognosis that was rarely utilized by previous RNA-seq-based prognostic signatures or models.
Integrating gene-level RNA-ITH may improve biomarker design
Biomarker selection is one of the determinants of the performance of gene expression-based models. To further investigate the RNA-ITH in individual gene expression, we decomposed the total expression variance of each gene into between- and within-patient variances, which reflected its intertumor and intratumor expression diversity, respectively. As shown in Fig. 2a, the intratumor variation is highly correlated with the intertumor variation (R = 0.732, p < 0.001), indicating that genes informative for distinguishing patients (i.e., potential biomarkers) also tend to have high diversity among tumor regions within the same tumors, and vice versa indicating that leveraging RNA-ITH may improve biomarker design.
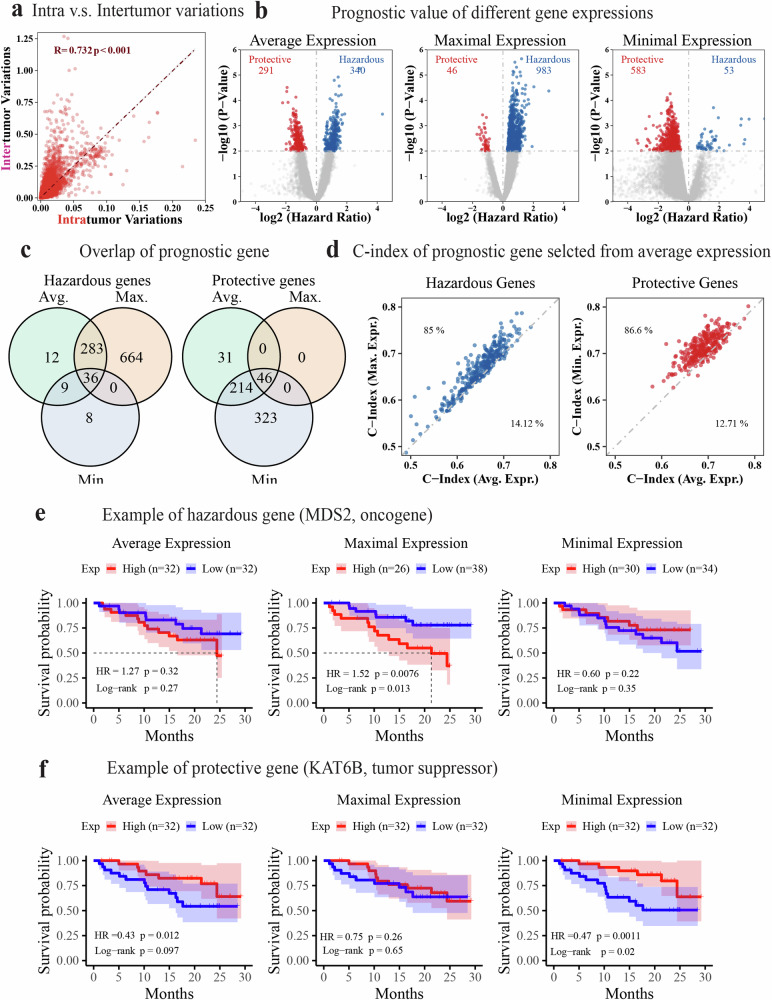
Fig. 2. Association of regional gene expression with patient survival in the TRACERxC1 cohort.
a Scatter plot showing a strong correlation between transcriptomic intra and intertumor variance at the gene expression level. The Pearson correlation coefficient (R) and the p-value (p) are shown in the figure with red text. b Volcano plots demonstrating the survival association of genes when their average, maximal, or minimal expression across all regions was used to represent its patient-specific expression level. The hazard ratio and p-value were calculated using the univariate Cox regression that fitted the expression of each gene as a continuous variable. c Venn diagram showing the numbers and overlap of survival-associated genes based on their average, maximal, and minimal expression, respectively. d Scatter plots comparing the C-index of selected prognostic genes using different representative expressions. Hazardous (HR > 1, p-value < 0.01) and protective genes (HR < 1, p-value < 0.01) were selected based on average expression and compared with results using maximal and minimal expression separately. The text labels the percentage of genes in that area. e, f Kaplan–Meier curve showing the recurrence-free survival of patients with high(red) or low(blue) expression of MDS2 (e) or KAT6B (f) using the median as a cutoff. The maximal expression of the hazardous gene MDS2 and the minimal expression of the protective gene KAT6B is more prognostic. The shadow represents the 95% confidence interval. The survival analysis measures disease-free survival.
To investigate the impact of RNA-ITH on the prognostic association at the gene level, we utilized the average, maximal, and minimal expression of each gene across all tumor regions within the same tumors to represent a patient-level expression. The prognostic association of resultant average, maximal, and minimal expression values was examined using univariate Cox regression models (Fig. 2b). Based on average expression, we identified a total of 631 prognostic genes (p < 0.01), including 340 hazardous (Hazard Ratio, HR > 1 for disease-free survival, DFS) and 291 protective (HR < 1) genes. The use of the maximal expression was more sensitive to identifying hazardous genes, resulting in 983 hazardous but only 46 protective genes (Fig. 2b). It captured the largest number of hazardous genes, including the majority identified using the average expression approach (Fig. 2c). In contrast, the use of the minimal expression was more sensitive to identifying protective genes, resulting in 53 hazardous and 583 protective genes (Fig. 2b). This approach had the advantage of protective gene identification over than average expression approach (Fig. 2c). Therefore, taking advantage of multiregional RNA-seq in the context of RNA-ITH may offer more prognostic biomarker candidates and improve the development of prognostic models.
Furthermore, the hazardous genes selected based on average expression demonstrated higher prognostic association when their maximal expression was used for survival analysis, as indicated by higher concordance indices and more significant p-values (Fig. 2d, Supplementary Fig. 1). In contrast, protective genes identified by average expression were more prognostic when their minimal expression was used in the survival analysis (Fig. 2d, Supplementary Fig. 1). As an example shown in Fig. 2e, f, the maximal expression of proto-oncogene MDS241,42 (hazardous gene, HR > 1) and the minimal expression of the tumor suppressor KAT6B41,42 (protective gene, HR < 1) achieved the best prognostic stratification of patients. Likely, the tumor region with the extreme expression of the prognostic gene represents the most aggressive or resistant (to treatments) region within the tumor and thus poses the strongest prognostic effects on patient survival. Taken together, these findings suggest that the performance of single region-derived signatures may be greatly improved in the application when integrated with RNA-ITH through multiregional profiling assays.
Improve performance of prognostic signatures with RNA-ITH
Since gene level- RNA-ITH has demonstrated the potential to improve prognostic biomarker selection and application, we next examined whether consideration of RNA-ITH can improve existing prognostic signatures.
To this end, we adopted two methods to calculate patient-level risk scores from multiregional expression data (Fig. 3a).
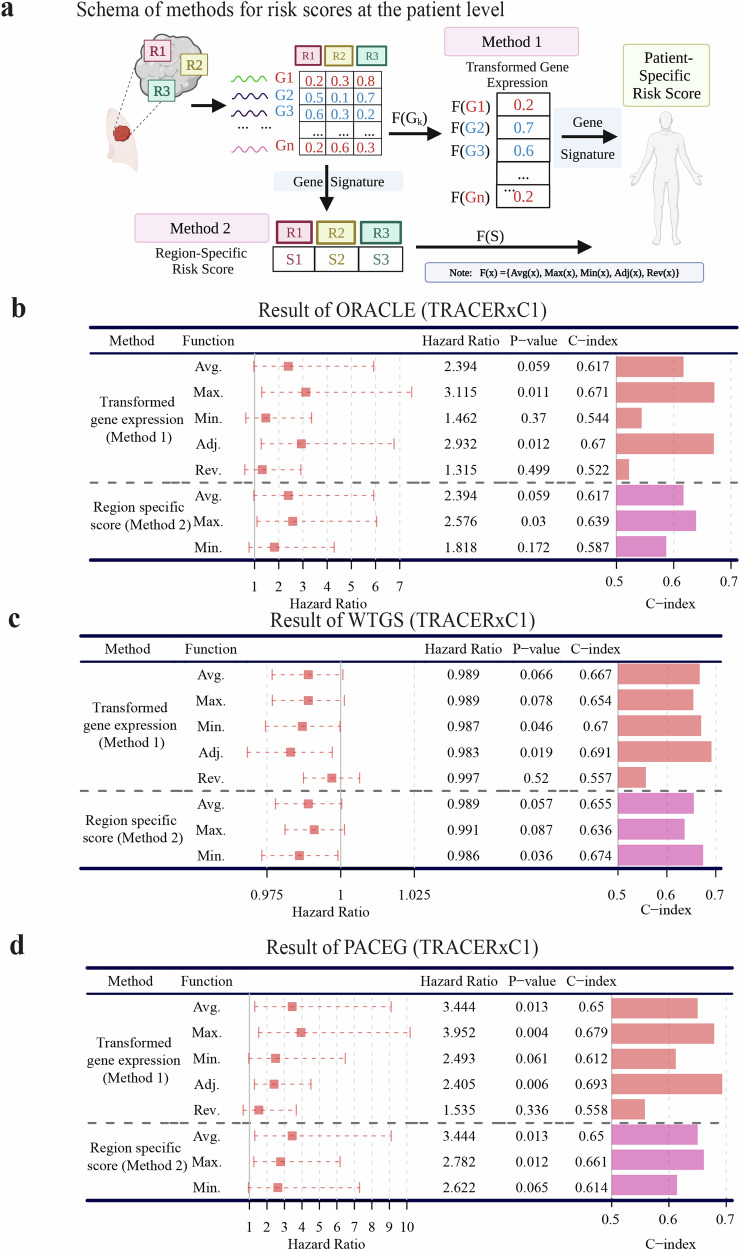
Fig. 3. Calculation of patient-specific risk scores using multiregional RNA-seq data.
a Two methods for calculating patient risk scores based on gene signatures. Method 1: Transform the expression of signature genes into patient-specific values and then compute the risk score of patients based on the signature. The transformation function calculated the average(Avg.)/maximal (Max.) (Adj.)/minimal (Min.) gene expression across all regions of each individual or summarized the maximal expression of hazardous genes and the minimal expression of protective genes (Adjusted, Adj.) or the reverse calculation (Rev.). Method 2: Apply the gene signature to all regions of a patient to obtain region-specific risk scores and then transform them into individual-level risk scores. The transformation function calculated the average(Avg.)/maximal (Max.) (Adj.)/minimal (Min.) region-specific scores of each individual. b–d Performance of three different prognostic signatures: b ORACLE, c WTGS, and d PACEG applied with eight functions from two methods of quantifying patient-specific risk score in the TRACERxC1 cohort. In the Hazard Ratio column, a 95% confidence interval was shown as a dotted line. The survival analysis measures disease-free survival.
Method 1 (M1)
Transformed gene expression. For each signature gene, the expression values across all regions from a patient were combined using transformation functions to obtain patient-level expression, which was then used to calculate the risk score of that patient. We tested five different transformation functions: (1) Average (Avg.), (2) Maximal (Max.), (3) Minimal (Min.) Function calculated average/maximal/minimal gene expression across all regions within each tumor, respectively; (4) Adjusted Function (Adj.) selected the maximal expression of hazardous genes (positive coefficients) and the minimal expression of protective genes (negative coefficients) across multiple regions within the same tumor, which was supposed to achieve the best performance based on the gene-level results; and (5) Reverse Function (Rev.) calculated maximal expression of protective genes and minimal expression of hazardous genes, which was anticipated to get the worst accuracy.
Method 2 (M2)
Region-specific risk score. The signature scores were calculated for each tumor region, and then the transformation function was applied to gain the patient-level scores based on the region-specific scores. The transformed functions could be Average (Avg.)/Maximal (Max.)/Minimal (Min.) Function that calculated average/maximal/minimal region scores across all regions of each individual, respectively.
ORACLE (outcome risk associated clonal lung expression) signature was developed in NSCLC from genes with low intra-tumor but high intertumor diversity using TRACERxC1 multiregional RNA-seq and TCGA LUAD RNA-seq data35. It was a hazardous signature for which higher scores denoted higher risks. We first applied the above-mentioned two methods to this signature to assess whether its prediction accuracy could be improved when RNA-ITH was taken into consideration. As shown in Fig. 3b, the signature score is based on the Adj. and Max. Functions of Method 1(M1-Adj and M1-Max) achieve the best performance in the TRACERxC1 cohort, as indicated by greater C-Indices and more significant p-values. The improved performance of M1-Adj (p-value = 0.012, C-index = 0.67) is in line with the prognostic results of individual genes, supporting our hypothesis that integrating RNA-ITH improves the existing signature application. In addition, the M1-Max achieves a comparable accuracy as the M1-Adj (p-value = 0.011, C-index = 0.671), presumably because most of the ORACLE signature genes (19 out of 23) are hazardous, defined by the positive coefficients. As for Method 2, the results of M2-Max achieve a more significant prognostic association (p-value = 0.03, C-index = 0.639), while the scores based on M2-Avg and M2-Min are not significant (Fig. 3b).
To validate our findings, we extended our analysis to the TRACERxC2 cohort, exclusively consisting of independent patients to the TRACERxC1 cohort to ensure the independence of the analysis (Supplementary Fig. 2a). With a larger cohort size and enhanced statistical power, our investigation consistently highlighted the superior performance of the M1-Adj (p-value = 0.0001, C-index = 0.61) and the M2-Max (p-value = 0.0007, C-index = 0.603).
To control the potential impact of histology on our analysis, we sought to perform the analysis within the same histology. As the ORACLE signature was originally developed for lung adenocarcinoma, we conducted survival analysis within the TRACERxLUAD cohort (Supplementary Fig. 2b). In this context, our results revealed that both the M1-Adj (p-value = 3 × 10−5, C-index = 0.627) and the M2-Max (p-value = 0.0003, C-index = 0.616) outperformed the M2-Avg, demonstrating the significant enhancement in prognostic capabilities achieved by incorporating RNA-ITH into the ORACLE signature.
We also tested the ORACLE signature integrated with the two methods above in our MDAMPLC cohort. Although the p-value was not significant due to the small sample size, we observed the same trends that the Max. Function of Method 1 and 2 enable prediction improvement compared to Avg. Function (Supplementary Fig. 2c). In summary, even though the ORACLE signature is selected from genes with low ITH, it can be improved when considering RNA-ITH in its application. In addition to ORACLE, the finding that integrating RNA-ITH will improve the performance of expression-based signatures was also supported by nine public hazardous signatures evaluated in the same way (Table 1, Supplementary Table 2, Supplementary Table 3).
Table 1.
Nine public signatures improved by multiregional RNA-seq data
| Cohort | TRACERxC1 | MDAMPLC | TRACERxC1 | MDAMPLC | TRACERxC1 | MDAMPLC | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Function | HR | p-Value | C-index | C-index | HR | p-Value | C-index | C-index | HR | p-Value | C-index | C-index |
| Signature | Boutros et al., 2008 (N = 5) | Krzystanek et al., 2016 (N = 6) | Bianchi et al., 2007 (N = 10) | ||||||||||
| Transformed gene expression (Method 1) | Avg. | 2.588 | 0.023 | 0.673 | 0.481 | 2.787 | 0.065 | 0.627 | 0.505 | 1.927 | 0.178 | 0.6 | 0.542 |
| Max. | 2.541 | 0.018 | 0.693 | 0.486 | 2.824 | 0.05 | 0.625 | 0.557 | 2.293 | 0.076 | 0.645 | 0.58 | |
| Min. | 2.529 | 0.037 | 0.67 | 0.476 | 2.419 | 0.089 | 0.622 | 0.495 | 1.507 | 0.396 | 0.578 | 0.481 | |
| Adj. | 2.304 | 0.012 | 0.706 | 0.618 | 1.998 | 0.018 | 0.685 | 0.608 | 2.071 | 0.035 | 0.667 | 0.575 | |
| Rev. | 1.549 | 0.315 | 0.579 | 0.439 | 1.114 | 0.776 | 0.537 | 0.462 | 0.961 | 0.923 | 0.486 | 0.458 | |
| Region-specific (Method 2) | Avg. | 2.588 | 0.023 | 0.673 | 0.481 | 2.787 | 0.065 | 0.627 | 0.505 | 1.927 | 0.178 | 0.6 | 0.542 |
| Max. | 2.624 | 0.008 | 0.698 | 0.509 | 3.021 | 0.045 | 0.628 | 0.524 | 1.981 | 0.142 | 0.62 | 0.557 | |
| Min. | 2.289 | 0.073 | 0.651 | 0.448 | 2.166 | 0.129 | 0.617 | 0.476 | 1.723 | 0.254 | 0.594 | 0.486 | |
| Signature | Kratz et al., 2012 (N = 11) | Zhu et al., 2010 (N = 15) | Garber et al., 2001 (N = 24) | ||||||||||
| Transformed gene expression (Method 1) | Avg. | 1.926 | 0.14 | 0.59 | 0.557 | 1.638 | 0.294 | 0.562 | 0.547 | 1.813 | 0.09 | 0.638 | 0.5 |
| Max. | 2.012 | 0.095 | 0.603 | 0.599 | 2.563 | 0.034 | 0.641 | 0.59 | 2.284 | 0.011 | 0.669 | 0.519 | |
| Min. | 1.861 | 0.141 | 0.613 | 0.472 | 0.862 | 0.734 | 0.456 | 0.538 | 1.281 | 0.471 | 0.576 | 0.434 | |
| Adj. | 2.031 | 0.035 | 0.646 | 0.608 | 2.586 | 0.021 | 0.657 | 0.58 | 2.404 | 0.004 | 0.708 | 0.538 | |
| Rev. | 1.281 | 0.558 | 0.518 | 0.472 | 0.756 | 0.462 | 0.41 | 0.538 | 0.919 | 0.77 | 0.492 | 0.467 | |
| Region-specific score (Method 2) | Avg. | 1.926 | 0.14 | 0.59 | 0.557 | 1.638 | 0.294 | 0.562 | 0.547 | 1.813 | 0.09 | 0.638 | 0.5 |
| Max. | 2.183 | 0.054 | 0.629 | 0.59 | 2.231 | 0.079 | 0.621 | 0.594 | 2.023 | 0.041 | 0.646 | 0.519 | |
| Min. | 1.587 | 0.307 | 0.558 | 0.467 | 1.083 | 0.855 | 0.496 | 0.533 | 1.485 | 0.243 | 0.6 | 0.439 | |
| Signature | Wistuba et al., 2013 (N = 30) | Raz et al., 2008 (N = 54) | Beer et al., 2002 (N = 92) | ||||||||||
| Transformed gene expression (Method 1) | Avg. | 2.086 | 0.006 | 0.696 | 0.533 | 1.386 | 0.15 | 0.581 | 0.514 | 1.163 | 0.258 | 0.591 | 0.524 |
| Max. | 2.256 | 0.003 | 0.711 | 0.59 | 1.622 | 0.015 | 0.654 | 0.58 | 1.301 | 0.045 | 0.63 | 0.552 | |
| Min. | 1.838 | 0.019 | 0.679 | 0.481 | 1.055 | 0.79 | 0.502 | 0.439 | 1.018 | 0.876 | 0.519 | 0.443 | |
| Adj. | 1.24 | 0.013 | 0.714 | 0.618 | 1.245 | 0.017 | 0.693 | 0.594 | 1.094 | 0.013 | 0.711 | 0.575 | |
| Rev. | 0.973 | 0.834 | 0.481 | 0.425 | 0.926 | 0.415 | 0.385 | 0.448 | 0.943 | 0.102 | 0.341 | 0.462 | |
| Region-specific score (Method 2) | Avg. | 2.086 | 0.006 | 0.696 | 0.533 | 1.386 | 0.15 | 0.581 | 0.514 | 1.163 | 0.258 | 0.591 | 0.524 |
| Max. | 2.08 | 0.004 | 0.7 | 0.599 | 1.354 | 0.127 | 0.598 | 0.542 | 1.286 | 0.062 | 0.637 | 0.533 | |
| Min. | 1.966 | 0.013 | 0.683 | 0.476 | 1.209 | 0.367 | 0.549 | 0.472 | 1.075 | 0.533 | 0.564 | 0.462 | |
As ORACLE and the other nine existing signatures only harbored a small subset of genes, the impact of RNA-ITH in prognostic signature might not be fully captured. Therefore, we next tested another signature called whole-transcriptomic gene signature (WTGS), which was a protective signature (higher score indicating longer survival) using all genes for the prognostication43. In this signature, each gene was assigned a weight based on its prognostic significance43. As shown in Fig. 3c, the survival analysis with the five transformation functions of Method 1 was performed in the TRACERxC1 cohort. Again, the best performance is achieved by the M1-Adj (p-value = 0.019, C-index = 0.691). Signature scores based on M1-Avg, M1-Max, and M1-Min result in weak prognostic associations, while the score based on M1-Rev is not prognostic (Fig. 3c). Using Method 2, the best performance is obtained by M2-Min (p-value = 0.036, C-index = 0.674) in line with the fact that WTGS is a protective signature (Fig. 3c).
After validation using the TRACERxC2 cohort, M1-Adj (p-value = 5 × 10−7., C-index = 0.65) and M2-Min (p-value = 5 × 10−5, C-index = 0.623) demonstrated great improvements compared to the corresponding Avg. Function (Supplementary Fig. 3a). When narrowing down to the adenocarcinoma subtype, both M1-Adj (p-value = 3 × 10−7, C-index = 0.687) and M2-Min (p-value = 5 × 10−5, C-index = 0.65) continued to exhibit superior prognostic value in the TRACERxLUAD cohort (Supplementary Fig. 3b).
Consistently, the M1-Adj and the M2-Min achieve the highest C-index in our MDAMPLC cohort (Supplementary Fig. 3c). Taken together, these results indicate that the RNA-ITH has a remarkable impact on the performance of prognostic signatures, and when multiregional expression data is available, M1-Adj achieves the best prognostic performance.
The PACEG gene signature
Biswas et al. utilized stably expressed genes with high intertumor but low intratumor diversity, as revealed by the TRACERxC1 multiregional expression data (defined as Q4 genes), and developed the ORACLE signature35. Although these genes are, in principle, less impacted by RNA-ITH, we observed improved performance using the Max. and Adj. Functions of Method 1 (Fig. 3B). Considering the high correlation between intertumor and intratumor variations at the gene level (Fig. 2a), the informative genes for patient stratification had probably been missed by the filter used in ORACLE. Indeed, only ~6.6% of genes were identified as the Q4 genes for the development of ORACLE, with many prognostic genes likely excluded35.
To improve ORACLE and further demonstrate the potential of integrating RNA-ITH in the signature application, we developed a new gene signature called PACEG (Prognosis-Associated Clonally Expressed Genes) using a similar procedure as ORACLE (see “Methods” for details). A critical finding of ORACLE is that Q4 genes were more likely to be clonal genes whose expression was driven by their copy numbers in the dominant clone ORACLE35. Instead of focusing on Q4 genes, PACEG selected signature genes from clonal genes identified from paired RNA-seq and copy number variation (CNV) data of LUAD (see Methods for details). As such, PACEG was developed fully based on LUAD data, which ensures its independent and unbiased application in the multiregional cohorts. In total, the signature contains 26 genes non-overlapped with ORACLE, including sixteen hazardous and ten protective genes (Supplementary Table 4).
Consistent with the previous 11 signatures, integrating PACEG with M1-Adj achieves the best performance (Fig. 3d, p-value = 0.006, C-index = 0.693) in the TRACERxC1 cohort. The M1-Max results in a slightly more significant p-value, but a smaller C-index. Similarly, the M2-Max demonstrates the highest performance (p-value = 0.012, C-index = 0.661) compared to M2-Avg and M2-Min (Fig. 3d). Within the TRACERxC2 cohort, it is noteworthy that the M1-Adj (p-value = 5 × 10−5, C-index = 0.614) and the M2-Max (p-value = 0.0002, C-index = 0.603) consistently exhibit the best performance (Supplementary Fig. 4a). Also confirmed in the MDAMPLC cohort, M1-Adj and M2-Max show the highest C-index among all transformation functions in the same methods (Supplementary Fig. 4b). Of note, when applied to the multiregional data, PACEG has a much better prediction accuracy than the ORACLE signature, especially with M1-Adj (Supplementary Table 5), possibly from the inclusion of more prognostically informative genes (non-Q4 genes).
Considering the signature was originally developed using TCGA LUAD data, we also assessed its performance within the adenocarcinoma subtype (TRACERxLUAD). Consistently, M1-Adj (p-value = 3 × 10−6, C-index = 0.642) and M2-Max (p-value = 8 × 10−5, C-index = 0.621) demonstrated the best results (Supplementary Fig. 4c). To examine whether PACEG provides independent prognostic values after considering established clinical factors, we performed multivariable Cox regression analysis, which includes variables such as the PACEG score calculated from the M1-Adj (PACEG-M1-Adj), smoking status, stage, age, and gender (Supplementary Table 6). As a result, PACEG-M1-Adj turned out to be the strongest predictor in the model (HR = 1.69, p-value = 0.0008), with a significant p-value after adjusting for those clinical factors. This result indicates that PACEG-M1-Adj offers valuable insights into survival predictions while complementing those important clinical factors. Integrating the expression-based signature with multiregional information not only improves the clinical utility of the signature itself but may surpass the predictive capability of several crucial clinical prognostic variables.
The prognostic impact of RNA-ITH in the tumor microenvironment
The infiltration level of immune cells in the tumor microenvironment has been reported to be associated with the prognosis of NSCLC patients. Our previous study has also demonstrated that a higher degree of T cell receptor (TCR) ITH was associated with an increased risk of postsurgical recurrence of NSCLC44. Next, we investigated the prognostic effect of tumor immune microenvironment ITH by computationally inferring the infiltration of major immune cell types from the multiregional RNA-seq45–47 (see “Methods” for details). After obtaining the immune infiltration scores in all tumor regions, we converted them into patient-specific infiltration scores by calculating the average, maximal, and minimal values and investigated their association with prognosis using Cox regression.
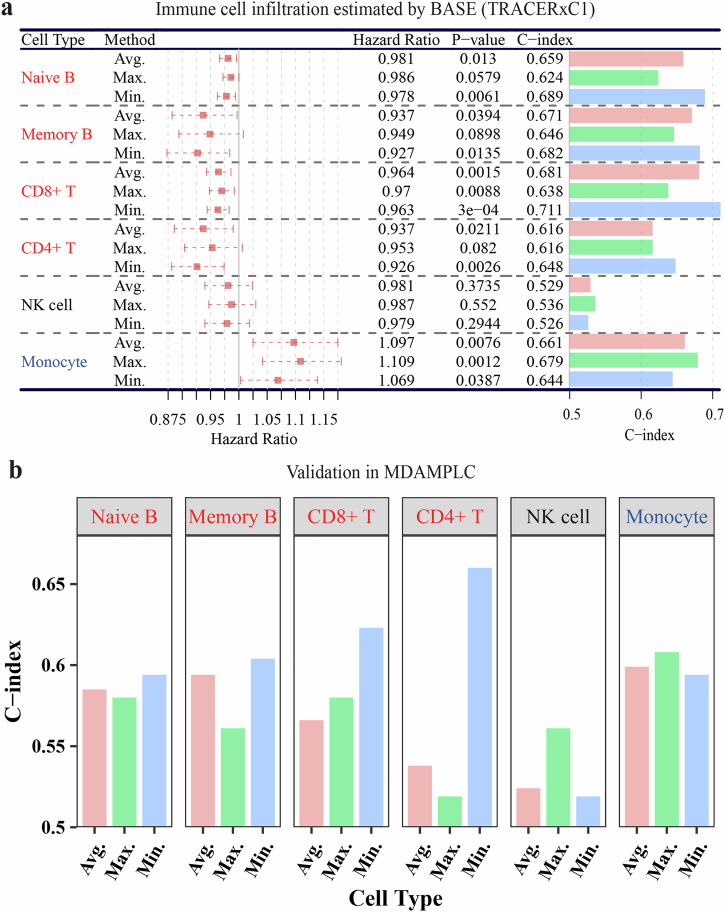
As shown in Fig. 4a, based on the average infiltration level, we found that Naïve B, Memory B, CD8+ T, and CD4+ T cells are protective immune cells with higher infiltration associated with longer survival, while Monocytes predominance is hazardous in NSCLC, consistent with previous studies47. Importantly, for protective immune cells, the more accurate prediction is achieved by using the minimal infiltration scores as indicated by both the p-values and C-indices, while for the hazardous immune cell, Monocytes, the best prognostic association is observed when the maximal infiltration level is used. Consistently in the MDAMPLC cohort, the highest C-indices of Naïve B, Memory B, CD8+ T, and CD4+ T cells are from using the minimal infiltration regions, while the highest C-index of Monocyte is obtained with the maximal infiltration regions (Fig. 4b). These results suggest that the tumor regions with the least favorable immune microenvironment contribute the most to the overall prognosis of patients.
Fig. 4. Association of immune cell infiltration with patient survival.
a Forest plot showing the survival analysis results of six immune cells in TRACERxC1 cohort. The average (Avg.), maximal (Max.), and minimal (Min.) infiltration values of each immune cell were used to obtain the patient-level immune infiltration and then were analyzed with univariate Cox regression. In the Hazard Ratio column, a 95% confidence interval was shown as a dotted line. b The bar graph of the C-index evaluating the same immune cells in the MDAMPLC cohort. The infiltration level of six immune cell types was calculated in all regions of each patient. The minimal infiltration level of Naïve B, Memory B, CD8+ T, and CD4+ T cells (protective) but the maximal infiltration level of Monocyte (hazardous) achieves the highest prognostic association. The survival analysis measures disease-free survival.
Prognostic risk variation across different regions and signatures
Herein, we revealed the impact of RNA-ITH at the gene expression level, gene signature level and tumor microenvironment level (Supplementary Fig. 5). For both tumor-intrinsic gene signatures (ORACLE, WTGS, and PACEG) and tumor immune microenvironment signatures (immune cell infiltration), we have shown that tumor regions with the least favorable signatures scores (i.e., maximal for hazardous and minimal for protective signatures) are more informative for prognostication. We next sought to investigate whether the same regions within each tumor would be defined as the least favorable by different signatures. We constructed univariate Cox regression models for the three tumor-intrinsic prognostic signatures and five immune cell signatures. The models were trained based on the maximal scores from the three hazardous signatures (ORACLE, PACEG, and Monocyte) and the minimum scores for the five protective signatures (WTGS, Naïve B, Memory B, CD8+ T, and CD4+ T cells). Then all models were applied to the expression data of all tumor regions to calculate the region-specific risk scores. After normalization to make the risk scores comparable, patients with more than three tumor regions were included for further analysis (Supplementary Fig. 6).
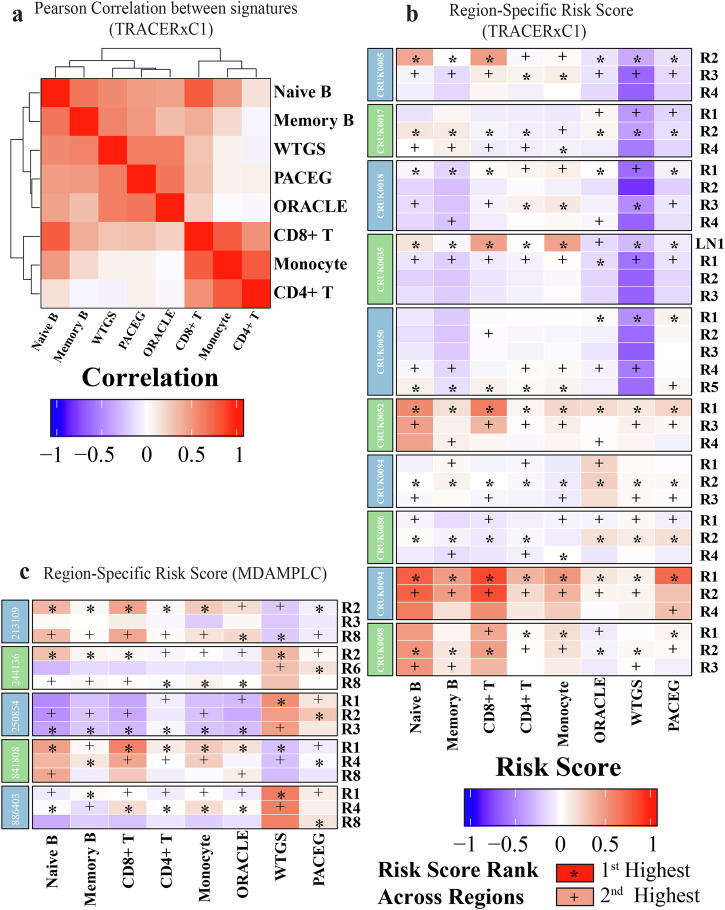
As shown in Fig. 5a, overall, the risk scores from the three tumor-intrinsic gene signatures (ORACLE, PACEG, and WTGS) are highly correlated with each other. However, the immune cell signatures fall into two groups: CD8+ T, CD4+ T, and Monocyte signatures are correlated, while Memory and Naïve B cell signatures make similar predictions with tumor-intrinsic signatures. For each gene signature, the predicted prognostic risk scores from different regions within the same tumors overall are similar, with some exceptions whereby substantial variations among different tumor regions are observed (e.g., CRUK0035 by the CD8+ T cell signature) (Supplementary Fig. 6). Of note, the WTGS signature, which uses the whole transcriptome for risk prediction, is the least influenced by RNA-ITH.
Fig. 5. Region-specific risk scores predicted by different gene signatures.
a Heatmap showing the correlation between risk scores predicted by the eight signatures. b, c Heatmap displaying the risk scores of different regions from selected patients in b TRACERxC1 and c MDAMPLC cohorts predicted by eight gene signatures. The risk score was normalized by subtracting the median value and scaled to the (−1, 1) interval“*” represents the highest risk scores across different regions of the same patients. “+” shows the second highest risk scores.
More interestingly, we found that the eight signatures identified the same tumor regions to carry the highest risks for recurrence in some patients (Fig. 5b, c). When we divided the signatures into intrinsic gene signatures (ORACLE, PACEG, and WTGS) and the tumor immune microenvironment signatures (Monocyte, Naïve B, Memory B, CD8+ T, and CD4+ T cells), there was more consistency within the two groups of signatures (Supplementary Fig. 6). For the three tumor-intrinsic gene signatures, the same regions in 75% (21 out of 28) TRACERxC1 cohort and 71% (10 out of 14) MDAMPLC cohort are predicted as the top two high-risk regions within the same tumors (e.g., CRUK0050), which suggests that these regions may be sensitive to chemotherapy. The five tumor immune microenvironment signatures identify the same regions as the top two high-risk regions in 43% (12 out of 28) TRACERxC1 cohort and 86% (12 out of 14) MDAMPLC cohort (e.g., CRUK0050), which indicates that these regions may be sensitive to immunotherapy. These findings suggest that the most aggressive and/or the least immune-infiltrated tumor regions may have the most prognostic impact.
Discussion
The adjuvant therapeutic paradigm for localized NSCLC has significantly changed recently by the addition of targeted therapy and immunotherapy to patients with EGFR mutations or positive PD-L1 expression48,49. However, not all patients benefit from these revolutionary therapies. A considerable proportion of patients still suffer from inevitable postsurgical recurrence or severe toxicities. In addition, the associated high cost continues to put additional pressure on the current health system. Therefore, studies are still needed to understand the mechanisms underlying postsurgical recurrence and develop reliable biomarkers for personalized adjuvant therapy.
Extensive efforts have been made to identify molecular features associated with postsurgical recurrence. One potential risk factor is ITH, namely, tumors are composed of cancer cells, stromal cells, and immune cells with distinct molecular and phenotypic features28–34. In NSCLC, a series of studies from our group and others have demonstrated ITH that has emerged at precancerous stages50,51 and continues to evolve along with local invasion31,38,52, metastatic spread53, and upon treatment54–57 and increased molecular ITH was associated with impaired T cell response and increased risk of postsurgical recurrence44.
In line with the profound impact on cancer biology, ITH also has a significant impact on the performance of prognostic signatures. In this study, we investigated the impact of RNA-ITH on the prognostic association of individual genes, gene signatures, and tumor immune environment using the multiregional RNA-seq in localized NSCLCs. At the gene level, we found that the maximal expression of hazardous genes and the minimal expression of protective genes were more prognostic than the average expression across all regions from the same tumors. Though the average expression enables selecting both hazardous and protective genes, the numbers are smaller, highlighting the limitation of previous studies in which biomarker design relied on single-sample gene expression. If only one biopsy per tumor is used, the potential sampling bias may impede the biomarker selection (lower ability to identify protective genes by maximal expression approach and lower ability to identify hazardous genes by minimal expression approach). On the other hand, if the whole tumor is sequenced as a bulk sample, it may lose valuable biomarker candidates (e.g., the limited prognostic genes identified by average expression). Therefore, taking advantage of multiregional RNA-seq in the context of RNA-ITH may improve the biomarker design and prognostic model performance.
Motivated by the observations at the gene level, we then investigated how to leverage multiregional gene expression data to improve the prognostication capability of existing prognostic signatures. We tested two different strategies. First, region-specific expression of all genes in a signature was transformed into patient-specific expression by taking transformation functions (Avg./Max./Min./Adj./Rev.), which were then used to calculate patient-level risk scores. In Method 1, we found that the best prognostic performance was achieved by applying the Adj. Function, a combination of max/min function for hazardous/protective genes, respectively. Second, a gene signature was applied to all tumor regions, and the maximal, minimal, and average scores were used for patient-specific scores. By this approach, the maximal scores for hazardous signatures and the minimum scores for protective signatures achieved the best performance, consistent with results at the gene level. These results were consistently observed in the publicly available TRACERx dataset for eleven widely studied NSCLC gene signatures and our newly developed signature PACEG. Those results were further supported by multiregional transcriptomic data of the MDAMPLC cohort, an independent dataset generated by our group. Although the Cox regression results did not reach statistical significance in the MDAMPLC cohort, likely due to the small sample size, the C-indexes demonstrated the same trends.
It is also worth noting that different prognostic signatures are in overall agreement when identifying the tumor regions associated with the highest risks of recurrence within the same tumors, suggesting that certain tumor regions may carry the most aggressive cancer cell subclones that may have driven the overall prognosis of that particular patient. These findings are reminiscent of oncologic knowledge and clinical practice that in patients with lung cancers of mixed histology, the prognosis and treatment are usually determined by the most aggressive histology58. Together with previous studies, our results underscored the importance of ITH in the prognostication of localized NSCLS and suggested that transcriptomic ITH should be considered when developing gene expression-based prognostic biomarkers.
ORACLE is a pioneer prognostic signature considering RNA-ITH in its development for localized NSCLC35. It is developed by selecting genes with high intertumor heterogeneity but low-ITH (Q4 genes) to overcome tumor sampling bias. However, the increase in reproducibility across independent datasets may be at the cost of reduced prognostication performance, as 93.4% of genes, including many prognostic indicators, are excluded. Based on the findings about clonal transcriptomic biomarkers in Biswas et al.‘s study, we developed a 26-gene signature, PACEG, by intentionally preserving all clonally expressed prognostic genes, rather than restricted to the Q4 genes, since we found that the transcriptomic intratumor and intertumor heterogeneity were highly correlated (R = 0.733, p < 0.001), in order to improve the prognostic performance. Indeed, our results from the multiregional transcriptomic data demonstrated that the PACEG signature achieved better prognostic performance than ORACLE and many other signatures, especially when using Adj. Function of Method 1 or Max. Function of Method 2.
If validated, multiregional transcriptomic profiling followed by the Adjusted Transformation for recurrence risk evaluation may provide improved insight for prognostication of patients with localized NSCLC (Supplementary Fig. 7). For resected NSCLC, multiregional sampling is not a barrier. From technology perspective, whole exome sequencing and whole transcriptomics profiling have already made into clinical practice as CLIA-certified assays to guide treatment decision59–61. Novel and cheaper transcriptomic technologies are making the cost associated with these tests acceptable, particularly taking into account the money/resources saved from resulting in better patient selection for personalized adjuvant therapy. Multiregional sequencing of small gene panels such as PACEG will further drive down the cost. Furthermore, the development of spatial transcriptomic technologies makes it even more practical for multiregional gene expression analysis from one single pathologic slide.
The therapeutic landscape of perioperative oncology is rapidly evolving, with an increasing number of patients now receiving neoadjuvant therapy. In our current study, it is important to note that none of the patients included had undergone neoadjuvant therapy. As a result, the gene signatures we developed did not account for the potential effects of such treatments. Neoadjuvant therapy can profoundly influence the tumor microenvironment and alter the expression levels of various genes within the tumor. These changes can, in turn, impact the performance and applicability of our gene signatures. Therefore, for patients who receive neoadjuvant therapy, it will be necessary to develop new or updated gene signatures tailored to the specific therapeutic context. Nonetheless, we believe that the principle of multiregional profiling, which helps to mitigate sampling bias and improve prognostic performance, would still be relevant and beneficial in the neoadjuvant setting.
Intratumor heterogeneity is a universal phenomenon across various cancer types. Our strategic approach, aimed at enhancing the clinical applicability and precision of expression-based signatures through the integration of multiregional RNA-seq, theoretically, holds the potential for broader application in heterogeneous cancer categories. Although we were unable to validate our concept within multiregional RNA-seq datasets from other cancer types due to limited data availabilities, we fervently aspire to stimulate further initiatives in multiregional RNA-seq data generation and exploration within diverse cancer types through our research.
Methods
TRACERx multiregional RNA-seq dataset
TRACERxC1 included tumor samples, and clinical details came from the first 100 patients enrolled in the TRACERx lung cancer study (TRACERx100)32. Multiregional RNA-seq data were downloaded as FASTQ files from the European Genome-phenome Archive (EGAS00001003458)39. It included 164 samples from 64 patients with NSCLC (Supplementary Table 1). Out of these patients, 45 of them had multiregional RNA-seq profiles accounting for a total of 145 tumor regions. For each patient, 2–6 samples were collected from different regions of the same tumor of a patient. The cohort had 41 male and 23 female patients with NSCLC, with a median age of 67.5. The majority were localized NSCLC: IA (n = 12), IB (n = 25), IIA (n = 7), IIB (n = 9), IIIA (n = 10), and IIIB (n = 1). The subtype of the cohort was predominantly adenocarcinoma (n = 41) and squamous cell carcinoma (n = 16). The rest were adenosquamous carcinoma (n = 3), carcinosarcoma (n = 2), large cell carcinoma (n = 1), and large cell neuroendocrine carcinoma (n = 1). Forty-four had no adjuvant treatment, and 20 had adjuvant therapy.
TRACERxC2 included tumor samples, and clinical details came from the first 421 patients enrolled in the TRACERx lung cancer study (TRACERx421)40. Multiregional RNA-seq data were preprocessed as described by Martínez-Ruiz et al.40 and downloaded from Zenodo (https://zenodo.org/record/7819449/). For additional validation, we excluded the TRACERxC1 cohort from the latest TRACERx421 cohort (TRACERxC2). TRACERxC2 included 652 samples from 261 patients with NSCLC. Out of these patients, 208 of them had multiregional RNA-seq profiles accounting for a total of 599 tumor regions. For each patient, 2–8 samples were collected from different regions of the same tumor of a patient. More clinical characteristics were summarized in Supplementary Table 1.
TRACERxLUAD included 472 samples from 187 patients with adenocarcinoma subtype from the TRACERx421 datasets. Out of these patients, 152 of them had multiregional RNA-seq profiles accounting for a total of 437 tumor regions. For each patient, 2–8 samples were collected from different regions of the same tumor of a patient. More clinical characteristics were summarized in Supplementary Table 1.
MDAMPLC cohorts and samples
A total of 64 tumor regions from 25 patients were included in this cohort (MD Anderson Cancer Center Multiregional Profiling in Lung Cancer, MDAMPLC). For each tumor, 2–3 tumor regions were subjected to RNA-seq (Supplementary Table 1). There were 13 male and 12 female patients with a median age of 62. The cohort was dominatingly localized NSCLC: IA (n = 2), IB (n = 9), IIA (n = 6), IIB (n = 4), IIIA (n = 3), and IV (n = 1). The histologic subtypes include adenocarcinoma (n = 13), squamous cell carcinoma (n = 8), and neuroendocrine (n = 4). Written informed consent for sample collection and analysis was obtained from all patients. The study protocols adhered to the Declaration of Helsinki and were approved by the Institutional Review Board at the University of Texas MD Anderson Cancer Center. No individual person’s data in any form (including any individual details, images, or videos) was used in this study.
MDAMPLC RNA-seq
Multi-site frozen tumor tissues were collected by Core Needle Biopsy (CNB) and were trimmed thoroughly away embedded OCT around tissues on dry ice. Tissues were then transferred to 2 ml vials (Qiagen 990381) containing Bead (Qiagen 69989) precooled at −20 °C. Vials were added with Qiagen Lysis Buffer QIAzol (Qiagen 79306) and put into the precooled insert of the Adapter of TissueLyser LT (Qiagen 85600) and followed the manual of TissueLyser LT for Tissue Disruption & Homogenization. RNA was extracted following the Qiagen kit Protocol from Animal tissue of miRNeasy Mini Handbook (Qiagen 217004). The samples were qualified by the Agilent Bioanalyzer. RNA was considered high quality if both ribosomal peaks were present on the Agilent Bioanalyzer trace and had an RIN of greater than 7.5.
Purified double-stranded cDNA was constructed using the NuGEN Ovation RNA-Seq protocol from total RNA, which was amplified using both 3’ poly(A) selection and random priming throughout the transcriptome. Next, all cDNA was quantified using the Invitrogen Qubit 2.0 DNA quantitation assay qualified by the Agilent Bioanalyzer HS-DNA chip, and if required, sheared using the Covaris S2 focused-ultrasonicator following the NuGEN Encore NGS Library System 1 protocol.
RNA-seq preprocessing
FASTQ data of TRACERx100 and MDAMPLC RNA-seq underwent quality control. Against the hg19 reference genome, RSEM (v1.2.3) with the option “—bowtie2(v2.2.3)” was used to calculate FPKM (Fragments Per Kilobase of transcript per Million mapped reads) from FASTQ62,63. The preprocessed RNA-seq data of TRACERx421 was downloaded from Zenodo and directly used.
TCGA LUAD RNAseq and CNV data
The Cancer Genome Atlas (TCGA) clinical information, RNA-seq data, and Copy Number Variation (CNV) segment files for lung adenocarcinoma (LUAD) were downloaded from Firehose (https://gdac.broadinstitute.org/). This RNA-seq dataset consisted of RSEM-normalized gene expression data for 20,501 genes. The details of the preprocessing of the CNV segment files have been previously described64. The CNV of genes was determined based on mapping the gene to the significant CNV segments provided in the CNV segment files. If a gene was mapped to multiple consecutive segments, the weighted mean was used, where the weight was calculated based on the fraction mapped to the corresponding segment.
Calculation of transcriptomic heterogeneity
After filtering out expression genes and performing log transformation, we calculated the Euclidean Distance of gene expression for each pair of tumor regions to quantify the transcriptomic heterogeneity based on the multiregional RNA-seq. Specifically, the transcriptomic divergence between tumor regions i and j () was quantified as:
| 1 |
Where ek,· is the expression value of gene k, and m is the total number of genes.
If tumor regions i and j were from different tumors, the divergencies for between-patient sample pairs represented intertumor heterogeneity. If tumor regions i and j were from the same tumors, the divergencies for within-patient sample pairs reflected the RNA-ITH in individuals. The average value of all within-tumor region pairs () was calculated to represent the RNA-ITH of the patient.
Calculation of ORACLE signature scores
The ORACLE (outcome risk associated clonal lung expression) signature is a 23-gene signature defined by Biswas et al.35. This signature was developed by selecting clonally expressed prognostic genes with low intratumor but high intertumor heterogeneity. Given the RNA-seq profile of a lung cancer sample, the ORACLE risk score was calculated as the weighted sum of the log2 expression values of the 23 signature genes. The weights of signature genes were downloaded from the ref. 35.
Calculation of nine public signature scores
Nine NSCLC signatures were selected from the literature because they are hazardous signatures composed of no less than five genes. The signature genes were collected from their publications6,7,11–13,15,18,21,24. Given that the weights were unavailable in most signatures, the coefficients of each signature were trained in the TCGA LUAD RNA-seq data as weights, respectively, through the multivariate Cox regression, to make it comparable among all signatures mentioned in this study. The risk score was calculated as the weighted sum of the log2 expression values of genes of each signature.
Calculation of WTGS signature scores
The WTGS signature scores of lung cancer samples were calculated using the statistical framework called WTSPP (whole transcriptome signatures for prognostic prediction)43. Instead of selecting a small set of prognostic genes, WTSPP used all genes for prognostic prediction by assigning a weight for each gene based on its prognostic association in training data. In this study, we used the published signature previously defined based on the TCGA LUAD data43. This signature was applied to the multiregional RNA-seq data to calculate the risk scores of samples based on a rank-based function provided by the WTSPP framework.
The development of the PACEG signature and calculation of risk scores
The PACEG (Prognosis-Associated Clonally Expressed Genes) signature was defined by using the TCGA LUAD RNA-seq data, CNV data, and clinical information in the following steps. First, from all genes, we selected 10,250 genes with expression levels above the median expression. Lowly expressed genes were generally associated with a low signal/noise ratio due to technical reasons and, therefore, were excluded at this step. Second, genes associated with overall patient survival (false discovery rate <0.01 in Univariate Cox regression models) were selected, resulting in a set of 736 genes. Third, from them, we further selected 124 genes with clonal expression in lung cancer—genes with expression correlated with their copy numbers (Pearson correlation coefficient >0.5). The expression of these genes was largely driven by their copy numbers and therefore, they were more likely to be clonally expressed compared to randomly selected genes. Finally, we applied a Cox regression model with L1 regularization (LASSO) and selected 26 genes (out of the 124 genes) to form the PACEG signature. In contrast to the procedure to define the ORACLE signature, we did not restrict genes to Q4 genes (genes with high intertumor but low intratumor diversity)35, which excludes nearly 93.4% of genes, including most prognostic genes.
For a lung cancer sample, the PACEG signature was applied to calculate the risk score as a linear combination of signature gene expression values, weighted by the model coefficients fitted in the TCGA cohort.
Inference of immune cell infiltration
We applied our previously developed binding association with sorted expression (BASE) algorithm to infer immune cell infiltration based on tumor gene expression profiles45–47. Full details on the BASE algorithm45 and the weight profile calculation of six immune cell types (Naïve B, Memory B, CD8+ T, CD4+ T, NK cells, and monocyte)46 have been described previously. Given the lung cancer gene expression data and pre-defined immune cell-specific reference profiles, the method outputs the inferred infiltration scores of different immune cells in each of the samples. The effectiveness of computational inference for these cell types has been validated by comparing them with flow cytometry data in both human peripheral blood mononuclear cells (PBMC) and lung tumor samples47.
Prediction of region-specific risk scores
First, we constructed univariate Cox regression models for eight prognostic signatures, respectively. The models were trained based on the maximal scores for the three hazardous signatures (ORACLE, PACEG, and Monocyte) and the minimum scores for the five protective signatures (WTGS, Naïve B, Memory B, CD8+ T, and CD4+ T cells). Second, all models were applied to the expression data of all tumor regions to calculate the region-specific risk scores. Third, the risk scores obtained from each signature were normalized by subtracting the median of scores and rescaled to the [0,1] interval by dividing the difference between maximum and minimum. Last, the patients with at least three regions were displayed, and the top two highest-risk scores of each tumor were highlighted.
Statistical analyses
Statistical analyses were performed with the R platform (v4.1.0) and visualized with ggplot2 R package (v3.3.5)65, ggpubr R package (v0.4.0)66, ggvenn R package (v0.1.9)67, forestplot R package (v1.10.1)68, ComplexHeatmap R package (v2.10.0)69, and circlize R package (v0.4.13)70. Survival analyses, specifically measuring disease-free survival, were performed using survival R package (v3.2-11)71. Specifically, The “coxph” and “survreg” functions were applied to build univariate and multivariate Cox regression models for evaluating the association of signature scores with patient overall or relapse-free survival. The “survdiff” function was used to compare the survival time between two patient groups. The “survfit” function was used to create Kaplan–Meier survival curves which were visualized by survminer R package (v0.4.9)72. Other survival analyses such as log-rank tests were also performed using functions included in the survival and survminer R packages. The glmnet R package (v4.1-2)73,74 was used to implement multivariate Cox regression models with L1 regularization (LASSO). The Student t-test and Wilcoxon Rank-Sum test were used to compare the interested metrics between two sample groups with the R function “t.test” and “wilcox.test”, respectively. Correlation analysis between two variables was conducted by using the R functton “cor.test”. For all analyses, if applicable, the p-values from statistical tests and models were adjusted by the Benjamini–Hochberg method to correct for multiple testing.
Supplementary information
Acknowledgements
This study was supported by the National Cancer Institute of the National Institute of Health Research Project Grant (R01CA269764 to C. Cheng, R01CA234629 to Jianjun Zhang), the AACR-Johnson & Johnson Lung Cancer Innovation Science Grant (18-90-52-ZHAN to Jianjun Zhang), the MD Anderson Lung Cancer Moon Shot Program, the Cancer Prevention and Research Institute of Texas Multi-Investigator Research Award grant (RP160668 to Jianjun Zhang) and the UT Lung Specialized Programs of Research Excellence Grant (P50CA70907 to Jianjun Zhang), Rydin Family Research Fund (Jianjun Zhang) and the Cancer Prevention Research Institute of Texas (CPRIT) (RR180061 to C. Cheng). C. Cheng is a CPRIT Scholar in Cancer Research. This study used the data generated by The TRAcking Non-small Cell Lung Cancer Evolution Through Therapy (Rx) (TRACERx) Consortium and provided by the UCL Cancer Institute and The Francis Crick Institute. The TRACERx study is sponsored by University College London, funded by Cancer Research UK, and coordinated through the Cancer Research UK and UCL Cancer Trials Center. The authors would like to acknowledge the support of the High-Performance Computing for Research facility at the University of Texas MD Anderson Cancer Center for providing computational resources that have contributed to the research results reported in this paper. Figure 1a, Fig. 3a, and Supplementary Fig. 7 were created with BioRender.com.
Author contributions
Conceptualization: C. Li and C. Cheng; Methodology: C. Li and C. Cheng; Software: C. Li and C. Cheng; Validation: C. Li; Formal Analysis: C. Li and C. Cheng; Investigation, C. Li, C. Cheng, and T. T. Nguyen; Resources: Jianjun Zhang, I. I. Wistuba, and A. P. Futreal; Data Curation: C. Li, J. Li, X. Song, S. M. Hubert, C. B. Chow, J. Fujimoto, L. Little, C. Gumb, and Jianhua Zhang; Writing—Original Draft: C. Li; Writing—Review & Editing: C. Cheng, Jianjun Zhang, C. B. Chow, J. Fujimoto, L. Little, C. Gumb, C. I. Amos, J. Wu, T. T. Nguyen, J. Li, X. Song, S. M. Hubert, J. V. Heymach and Jianhua Zhang, I. Wistuba, and A.P. Futreal; Visualization, C. Li; Supervision: Jianjun Zhang and C. Cheng; Funding Acquisition: Jianjun Zhang and C. Cheng.
Data availability
The TRACERx100 RNA-seq data (EGAS00001003458), TRACERx421 RNA-seq (https://zenodo.org/record/7819449/) and the TCGA LUAD data (https://gdac.broadinstitute.org/) are publicly available. The preprocessed RNA-seq and de-identified clinical information of the MDAMPLC cohort are deposited on GitHub: https://github.com/CSkylarL/MSofRNAITH.
Code availability
All codes and intermediate results in this study are deposited on GitHub: https://github.com/CSkylarL/MSofRNAITH.
Competing interests
Jianjun Zhang reports grants from Merck, grants and personal fees from Johnson and Johnson and Novartis, and personal fees from Bristol Myers Squibb, AstraZeneca, GenePlus, Innovent, and Hengrui outside the submitted work. Ignacio Wistuba has provided consulting or advisory roles for AstraZeneca/MedImmune, Bayer, Bristol-Myers Squibb, Genentech/Roche, GlaxoSmithKline, Guardant Health, HTG Molecular Diagnostics, Merck, MSD Oncology, OncoCyte, Jansen, Novartis, Flame Inc, Regeneron, and Pfizer; has received grants and personal fees from Genentech/Roche, Bristol Myers Squibb, AstraZeneca/MedImmune, HTG Molecular, Merck, and Guardant Health; has received personal fees from GlaxoSmithKline and Oncocyte, Daiichi-Sankyo, Roche, Astra Zeneca, Regeneron, Sanofi, Pfizer, and Bayer; has received research funding to his institution from 4D Molecular Therapeutics, Adaptimmune, Adaptive Biotechnologies, Akoya Biosciences, Amgen, Bayer, EMD Serono, Genentech, Guardant Health, HTG Molecular Diagnostics, Iovance Biotherapeutics, Johnson & Johnson, Karus Therapeutics, MedImmune, Merck, Novartis, OncoPlex Diagnostics, Pfizer, Takeda, and Novartis. The remaining authors declare no potential conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Jianjun Zhang, Chao Cheng.
Contributor Information
Jianjun Zhang, Email: JZhang20@mdanderson.org.
Chao Cheng, Email: Chao.Cheng@bcm.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00680-0.
References
- 1.Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin.72, 7–33 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Araujo, L. H. et al. in Abeloff’s Clinical Oncology 1108–1158.e1116 (Elsevier, 2020).
- 4.Flores, R., Patel, P., Alpert, N., Pyenson, B. & Taioli, E. Association of stage shift and population mortality among patients with non–small cell lung cancer. JAMA Netw. Open4, e2137508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uramoto, H. & Tanaka, F. Recurrence after surgery in patients with NSCLC. Transl. Lung Cancer Res.3, 242–249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beer, D. G. et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med.8, 816–824 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Bianchi, F. et al. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J. Clin. Investig.117, 3436–3444 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bueno, R. et al. Validation of a molecular and pathological model for five-year mortality risk in patients with early stage lung adenocarcinoma. J. Thorac. Oncol.10, 67–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H.-Y. et al. A five-gene signature and clinical outcome in non–small-cell lung cancer. N. Engl. J. Med.356, 11–20 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Eguchi, T. et al. Cell cycle progression score is a marker for five-year lung cancer-specific mortality risk in patients with resected stage I lung adenocarcinoma. Oncotarget7, 35241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garber, M. E. et al. Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl Acad. Sci. USA98, 13784–13789 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kratz, J. R. et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet379, 823–832 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krzystanek, M., Moldvay, J., Szüts, D., Szallasi, Z. & Eklund, A. C. A robust prognostic gene expression signature for early stage lung adenocarcinoma. Biomark. Res.4, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, B., Cui, Y., Diehn, M. & Li, R. Development and validation of an individualized immune prognostic signature in early-stage nonsquamous non–small cell lung cancer. JAMA Oncol.3, 1529–1537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raz, D. J. et al. A multigene assay is prognostic of survival in patients with early-stage lung adenocarcinoma. Clin. Cancer Res.14, 5565–5570 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Shukla, S. et al. Development of a RNA-Seq based prognostic signature in lung adenocarcinoma. J. Natl Cancer Inst.109, djw200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki, K. et al. Prognostic immune markers in non-small cell lung cancer. Clin. Cancer Res.17, 5247–5256 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Wistuba, I. I. et al. Validation of a proliferation-based expression signature as prognostic marker in early stage lung adenocarcinoma. Clin. Cancer Res.19, 6261–6271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang, H. et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non–small cell lung cancer patients. Clin. Cancer Res.19, 1577–1586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Laar, R. K. Genomic signatures for predicting survival and adjuvant chemotherapy benefit in patients with non-small-cell lung cancer. BMC Med. Genomics5, 30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu, C.-Q. et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non–small-cell lung cancer. J. Clin. Oncol.28, 4417 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Director’s Challenge Consortium for the Molecular Classification of Lung, A. et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat. Med.14, 822–827 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau, S. K. et al. Three-gene prognostic classifier for early-stage non–small-cell lung cancer. J. Clin. Oncol.25, 5562–5569 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Boutros, P. C. et al. Prognostic gene signatures for non-small-cell lung cancer. Proc. Natl Acad. Sci. USA106, 2824–2828 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian, J. & Simon, R. Gene expression–based prognostic signatures in lung cancer: ready for clinical use? J. Natl Cancer Inst.102, 464–474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vargas, A. J. & Harris, C. C. Biomarker development in the precision medicine era: lung cancer as a case study. Nat. Rev. Cancer16, 525–537 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu, C. Q. & Tsao, M. S. Prognostic markers in lung cancer: is it ready for prime time? Transl. Lung Cancer Res.3, 149–158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Sousa, V. M. L. & Carvalho, L. Heterogeneity in lung cancer. Pathobiology85, 96–107 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Marino, F. Z. et al. Molecular heterogeneity in lung cancer: from mechanisms of origin to clinical implications. Int. J. Med. Sci.16, 981 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senosain, M.-F. & Massion, P. P. Intratumor heterogeneity in early lung adenocarcinoma. Front. Oncol.10, 349 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, J. et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science346, 256–259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jamal-Hanjani, M. et al. Tracking the evolution of non–small-cell lung cancer. N. Engl. J. Med.376, 2109–2121 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Ramón y Cajal, S. et al. Clinical implications of intratumor heterogeneity: challenges and opportunities. J. Mol. Med.98, 161–177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamal-Hanjani, M., Quezada, S. A., Larkin, J. & Swanton, C. Translational implications of tumor heterogeneity. Clin. Cancer Res.21, 1258–1266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biswas, D. et al. A clonal expression biomarker associates with lung cancer mortality. Nat. Med.25, 1540–1548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz-Cano, S. J. Tumor heterogeneity: mechanisms and bases for a reliable application of molecular marker design. Int. J. Mol. Sci.13, 1951–2011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gyanchandani, R. et al. Intratumor heterogeneity affects gene expression profile test prognostic risk stratification in early breast cancerintratumor heterogeneity in GEP test risk stratification. Clin. Cancer Res.22, 5362–5369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, W.-C. et al. Multiregion gene expression profiling reveals heterogeneity in molecular subtypes and immunotherapy response signatures in lung cancer. Mod. Pathol.31, 947–955 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Rosenthal, R. et al. Neoantigen-directed immune escape in lung cancer evolution. Nature567, 479–485 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martínez-Ruiz, C. et al. Genomic–transcriptomic evolution in lung cancer and metastasis. Nature616, 1–10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consortium, A. P. G. et al. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov.7, 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sondka, Z. et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer18, 696–705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaafsma, E. et al. Whole transcriptome signature for prognostic prediction (WTSPP): application of whole transcriptome signature for prognostic prediction in cancer. Lab. Investig.100, 1356–1366 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuben, A. et al. TCR repertoire intratumor heterogeneity in localized lung adenocarcinomas: an association with predicted neoantigen heterogeneity and postsurgical recurrenceTCR intratumor heterogeneity and relapse in lung cancer. Cancer Discov.7, 1088–1097 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varn, F. S., Andrews, E. H., Mullins, D. W. & Cheng, C. Integrative analysis of breast cancer reveals prognostic haematopoietic activity and patient-specific immune response profiles. Nat. Commun.7, 10248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varn, F. S., Wang, Y., Mullins, D. W., Fiering, S. & Cheng, C. Systematic pan-cancer analysis reveals immune cell interactions in the tumor microenvironment. Cancer Res.77, 1271–1282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varn, F. S., Tafe, L. J., Amos, C. I. & Cheng, C. Computational immune profiling in lung adenocarcinoma reveals reproducible prognostic associations with implications for immunotherapy. Oncoimmunology7, e1431084 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, Y. L. et al. Osimertinib in resected EGFR-mutated non-small-cell lung cancer. N. Engl. J. Med.383, 1711–1723 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Felip, E. et al. Adjuvant atezolizumab after adjuvant chemotherapy in resected stage IB-IIIA non-small-cell lung cancer (IMpower010): a randomised, multicentre, open-label, phase 3 trial. Lancet398, 1344–1357 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Hu, X. et al. Multi-region exome sequencing reveals genomic evolution from preneoplasia to lung adenocarcinoma. Nat. Commun.10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu, X. et al. Evolution of DNA methylome from precancerous lesions to invasive lung adenocarcinomas. Nat. Commun.12, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quek, K. et al. DNA methylation intratumor heterogeneity in localized lung adenocarcinomas. Oncotarget8, 21994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee, W.-C. et al. Multiomics profiling of primary lung cancers and distant metastases reveals immunosuppression as a common characteristic of tumor cells with metastatic plasticity. Genome Biol.21, 1–21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nong, J. et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat. Commun.9, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le, X. et al. Landscape of EGFR-dependent and-independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLCOsimertinib resistance landscape. Clin. Cancer Res.24, 6195–6203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin, Y. et al. Distinct co-acquired alterations and genomic evolution during TKI treatment in non-small-cell lung cancer patients with or without acquired T790M mutation. Oncogene39, 1846–1859 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Chen, R. et al. Evolution of genomic and T-cell repertoire heterogeneity of malignant pleural mesothelioma under dasatinib treatmentImmunogenomic ITH evolution of MPM. Clin. Cancer Res.26, 5477–5486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruffini, E. et al. Lung tumors with mixed histologic pattern. Clinico-pathologic characteristics and prognostic significance. Eur. J. Cardiothorac. Surg.22, 701–707 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Cuppen, E. et al. Implementation of whole-genome and transcriptome sequencing into clinical cancer care. JCO Precis. Oncol.6, e2200245 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.George, B. et al. Transcriptomic-based microenvironment classification reveals precision medicine strategies for PDAC. Gastroenterology166, 859–871.e3 (2024). [DOI] [PubMed] [Google Scholar]
- 61.Heeke, S. et al. Tumor-and circulating-free DNA methylation identifies clinically relevant small cell lung cancer subtypes. Cancer Cell42, 225–237. e225 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma.12, 1–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, B., Yao, K., Zhou, E., Zhang, L. & Cheng, C. Chr20q amplification defines a distinct molecular subtype of microsatellite stable colorectal cancer. Cancer Res.81, 1977–1987 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wickham, H. Package ‘ggplot2’: elegant graphics for data analysis. Springe -Verl. N. Y. doi10, 978–970 (2016). [Google Scholar]
- 66.Kassambara, A. ggpubr:“ggplot2” based publication ready plots. (2020).
- 67.Yan, L. ggvenn: Draw Venn Diagram by ‘ggplot2’. R Package Version19 (2021).
- 68.Gordon, M., Lumley, T. & Gordon, M. M. Package ‘forestplot’. Advanced forest plot using ‘grid’graphics. The Comprehensive R Archive Network, Vienna (2019).
- 69.Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Therneau, T. M. Survival Analysis [R package survival version 2.42-6]. (2015).
- 72.Kassambara, A., Kosinski, M., Biecek, P. & Fabian, S. Survminer: Drawing Survival Curves Using Ggplot2. https://CRAN.R-project.org/package=survminer. R package version 0.49 (2021).
- 73.Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw.33, 1 (2010). [PMC free article] [PubMed] [Google Scholar]
- 74.Simon, N., Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J. Stat. Softw.39, 1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The TRACERx100 RNA-seq data (EGAS00001003458), TRACERx421 RNA-seq (https://zenodo.org/record/7819449/) and the TCGA LUAD data (https://gdac.broadinstitute.org/) are publicly available. The preprocessed RNA-seq and de-identified clinical information of the MDAMPLC cohort are deposited on GitHub: https://github.com/CSkylarL/MSofRNAITH.
All codes and intermediate results in this study are deposited on GitHub: https://github.com/CSkylarL/MSofRNAITH.