Abstract
The highly conserved coadapters CREB binding protein (CBP) and p300 form complexes with CREB as well as other DNA binding transcription factors to modulate chromatin remodeling and thus transcription. Human T-lymphotropic virus type 1 (HTLV-1) transcription is controlled, in part, by the CREB/ATF family of transcription factors which bind promoter sequences and function as complexes with the viral oncogenic protein Tax. We have reported that the nuclear localizing protein p30II of HTLV-1 functions as a transcription factor, differentially modulates CREB-responsive promoters, and is critical for maintenance of proviral loads in rabbits. In this study, we tested whether p30II directly interacts with CBP/p300 to modulate gene transcription. Gal4(BD)-p30II-mediated transactivation was enhanced following exogenous expression of p300 and was competitively repressed by the p300 binding protein, adenovirus E1A, and E1ACR2 (mutated for retinoblastoma binding but retaining p300 binding). In contrast, E1ACR1 (mutated for p300 binding) failed to alter Gal4(BD)-p30II-mediated transactivation. In addition, Gal4(BD)-p30II-mediated transactivation was competitively inhibited by the cotransfection of CMV-p30II-HA and CMV-Tax but could be rescued by exogenous p300. Importantly, we demonstrate that p30II colocalizes with p300 in cell nuclei and directly binds to CBP/p300 in cells. Deletion mutants of CBP/p300 were used to localize the site critical for binding p30II to a highly conserved KIX region. DNA binding assays confirmed the interference of p30II with the assembly of CREB-Tax-p300/CBP multiprotein complexes on 21-bp repeat oligonucleotides in vitro. Collectively, our results demonstrate that CBP/p300 is a cellular protein target for HTLV-1 p30II and mediates its transcriptional effects in vivo.
The coactivators CREB binding protein (CBP) and p300 mediate transcriptional control of various cellular and viral DNA binding transcription factors. These coactivators are highly similar in nucleotide sequence, are evolutionarily conserved, and are often referred to together as CBP/p300, despite evidence of divergent functions (10, 25). These proteins bridge transcription factors to relevant promoters, have intrinsic histone acetyltransferase (HAT) activity, and form complexes with proteins such as CBP/p300 binding protein-associated factor, which also exhibits HAT activity (26). Recent reviews provide a growing list of cellular and viral proteins that interact with either CBP or p300, including steroid and retinoid hormone receptors, CREB, c-Jun, c-Myb, Sap-1a, c-Fos, MyoD, p53, Stat-1/2, NF-κB, pp90rsk, TATA-binding protein, and TFIIB (4, 25, 29, 30). Among viral regulatory proteins, human T-lymphotropic virus type 1 (HTLV-1) Tax, adenovirus E1A, Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor protein, and simian virus 40 large T antigen also target and affect CBP and p300 functions (1–3, 17, 32, 38, 41, 52).
Complex retroviruses, like HTLV-1, must regulate their gene expression in cooperation with host cell transcription factors including CBP/p300. HTLV-1 encodes typical gag, pol, and env gene products as well as unique regulatory and accessory genes encoded in four open reading frames (ORFs) (pX ORFs I to IV) between the env gene and the 3′ long terminal repeat (54). ORFs IV and III of HTLV-1 encode the well-characterized Tax and Rex proteins, respectively. Tax, a 40-kDa nuclear localizing phosphoprotein, mediates multiple virus-cell interactions by increasing viral transcription and by influencing cell proliferation, apoptosis, DNA repair, and cell cycle control (9, 33, 43, 44, 49). Rex is a 27-kDa nucleolar localizing phosphoprotein that increases the cytoplasmic accumulation of nonspliced and singly spliced viral RNA (18, 37, 51).
A number of alternatively spliced mRNAs are expressed from the pX region of HTLV-1 and have been identified from infected cell lines and freshly isolated cells from HTLV-1-infected subjects (11, 15, 36). Recently, serum antibodies and cytotoxic CD8+ T cells from HTLV-1-infected individuals have been demonstrated to recognize pX ORF I- and II-derived proteins, indicating that these viral proteins are expressed in vivo (19, 48). The HTLV-1 pX ORF II mRNA is spliced from the first tax exon and encodes two proteins, p30II and p13II. The smaller protein, p13II, is derived from initiation at the first internal methionine codon in ORF II and represents the 87 carboxyl-terminal residues of p30II. The p30II and p13II proteins are localized to the nucleus and mitochondria, respectively (16, 35, 57). p30II contains serine- and threonine-rich regions with distant homology to transcription factors Oct-1 and -2, Pit-1, and POU-1 (15). It has recently been reported that mutations in a viral clone, which destroys the initiator methionine of the mRNA encoding p13II and inserts an artificial termination codon in the mRNA encoding p30II, reduce proviral copy numbers up to 100-fold in rabbits (8). We have subsequently demonstrated that HTLV-1 p30II functions as a transcription factor, has opposing effects compared to Tax, and differentially modulates CREB-responsive promoters (57).
In this study, we sought to identify protein-protein interactions that mediate the transcription effects of p30II. Our previous work demonstrated that minimal promoter units (independent of enhancer elements) are equally influenced by p30II, and thus, we hypothesized that CBP/p300 may interact with p30II. Our data using a Gal4(BD)-p30II-mediated transactivation assay indicated that exogenous p300 enhanced the effects of p30II in transcription, whereas the p300 binding protein, wild-type adenovirus 12SE1A, and its CR2 mutant (retinoblastoma [Rb] binding mutant), but not the CR1 mutant (p300 binding mutant), reduced the effects of p30II. Gal4(BD)-p30II-mediated transactivation was also competitively inhibited by the cotransfection of CMV-p30II-HA and CMV-Tax and was rescued by exogenously expressed p300. Using immunoprecipitation and glutathione S-transferase (GST) pull-down assays, direct physical interaction between p30II and CBP/p300 was demonstrated in cells. The p30II and CBP/p300 association was confirmed by immunohistochemistry. Deletion mutants of CBP/p300 in GST pull-down assays and in functional Gal4-mediated transcription assays were used to localize the binding site of CBP/p300 for p30II to a highly conserved KIX region. Furthermore, we demonstrated that p30II inhibited the ability of CREB to bind its DNA target using biotinylated DNA precipitation assays. Taken together, our data are the first to demonstrate that CBP/p300 is a cellular protein target for HTLV-1 p30II and that these important cellular coadapters mediate the viral protein transcriptional effects in cells.
MATERIALS AND METHODS
Cell lines.
All cultured cells (HEK 293 cells were obtained from the American Type Culture Collection [no. CRL-1573]; HEK 293T cells were obtained from G. Franchini [National Cancer Institute, Bethesda, Md.]) were grown in 10-cm-diameter tissue culture dishes in Dulbecco's minimal essential medium containing 10% fetal bovine serum and 1% streptomycin and penicillin at 37°C. Cells were split and cultured in six-well plates to 50% confluence 16 h before transfection according to the manufacturer's protocol (Lipofectamine Plus; Gibco BRL).
Plasmids.
The luciferase reporter plasmid p5XG-E1b-Luc was a kind gift of Y. Shi (Harvard Medical School, Cambridge, Mass.) and contains five tandem Gal4 DNA binding sequences upstream of a TATA box derived from plasmid pE1b-CAT (50). The plasmids pCRE-Luc, pTRE-Luc, and pRSV-β-Gal-Luc have been described previously (57). The Gal4 effector plasmid pCMV-Gal4 (DNA binding domain, amino acids [aa] 1 to 147)-p30II [Gal4(BD)-p30II] was constructed by subcloning the p30II-encoding sequence into a pCMV-Gal4(BD) vector (Stratagene). The p30II-encoding sequence was synthesized by PCR amplification with 5′ primer ATATGAATTCATGGCACTATGCTGTTTCGCC(5′-A) and 3′ primer TATACTGCAGTAGAGGTTCTCGGGTG (3′-A) from the HTLV-1 molecular clone ACH (34), including the 5′ EcoRI and 3′ PstI restriction sites (underlined). ACH was also used as a template to synthesize p30II-encoding sequences for all subsequent p30II expression constructs using appropriate restriction sites as indicated. pBC-p30II, a GST-p30II fusion protein expression vector, was constructed by subcloning the same p30II-encoding sequence into the vector pBC (12) using NdeI and NheI restriction sites. pBluescript-p30II (pBS-p30II), a T7 and T3 promoter-driven p30II expression vector, was constructed by subcloning the p30II-encoding sequence into the pBS vector (Stratagene) using EcoRI and PstI restriction sites. All of the p30II expression vectors were sequenced to confirm correct reading frames. pCMV-p30II-HA, a p30II-HA expression vector, was a kind gift of G. Franchini (National Cancer Institute). Adenovirus E1A expression vectors including wild-type 12SE1A, pRSV-12SE1A, and two deletion mutants, pRSV-12SE1AΔCR1 and pRSV-12SE1AΔCR2, were kindly provided by T. Kouzarides (University of Cambridge, Cambridge, United Kingdom). pCMV-Tax expresses the HTLV-1 Tax protein and has been described previously (45). pCMV-p300 expresses the full-length p300 protein and was kindly provided by A. Leiter (Tufts University School of Medicine, Boston, Mass.). All bacterial GST-p300 and GST-CBP expression vectors have been described previously (20). pRSV-KIX (aa 379 to 654)/p300 expresses the KIX domain of p300 and was a gift of G. Louis (CNRS-University Claude Bernard, Lyon, France).
Cell transfection and reporter gene assay.
For each transfection, 0.3 to 0.9 μg of reporter vectors was cotransfected with various amounts of effector plasmids using Lipofectamine Plus (Gibco BRL) (57). As an internal control for transfection efficiency, 0.1 μg of pRSV-β-Gal (Gibco BRL) was also used in each transfection. pBluescript (Stratagene) was used as carrier DNA to equalize DNA concentrations for each transfection. Transfected cells were lysed with 1× lysis buffer (Promega) using 0.4 ml of buffer per well at room temperature for 25 min. Twenty microliters of each lysate was used to test luciferase reporter gene activity using an Enhanced Luciferase Assay kit (Promega). To normalize transfection experiments, 5 μl of each lysate was assayed for β-galactosidase activity according to the manufacturer's protocol (Lumigen). Results were expressed as mean fold increases or percentages of change ± standard deviations (SD) in arbitrary light units (ALU) of luciferase activity in four independent trials set up as duplicates in each experimental trial.
Western immunoblot assay.
Transiently transfected cells were lysed in RIPA buffer containing 1× phosphate-buffered saline (PBS), 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS). Cell lysates were prepared by centrifugation at 14,000 rpm (Beckman) for 20 min at 4°C. Equal amounts of proteins were mixed with Laemmli buffer (62.5 mM Tris [pH 6.8], 2% SDS, 10% glycerol, 0.2% bromophenol blue, 100 mM dithiothreitol [DTT]). After boiling for 5 min, samples were electrophoresed through 6 or 10% polyacrylamide gels. The fractionated proteins were transferred to nitrocellulose membranes (Amersham Pharmacia Biotechnology) at 100 V for 1 h at 4°C. Membranes were then blocked in Tris-buffered saline containing 5% nonfat milk and 0.1% Tween 20. Proteins were detected with the appropriate primary antibody followed by an anti-rabbit or anti-mouse (NEN Life Science) immunoglobulin G (IgG)-horseradish peroxidase-conjugated goat antibody. Blots were developed using an enhanced chemiluminescence detection system (NEN Life Science).
Biotin-labeled DNA pull-down assay.
To examine the effects of p30II on the recruitment of CREB and p300 by HTLV-1 Tax into a multiprotein complex bound to the HTLV-1 21-bp repeat DNA, we labeled 1.5 μg of the annealed oligonucleotides 5′-GATCTGGGCGTTGACGACAACCCCTCACCTCAAAAAACTTTC-3′ and 5′-TTTGAAAGTTTTTTGAGGTGAGGGGTTGTCGTCAACGCCCAGATC-3′ (the HTLV-1 21-bp repeat sequence is shown in bold) with biotin-14-dATP (Life Technologies, Inc.) using 10 units of Klenow (New England Biolabs) at 37°C for 30 min. Labeled oligonucleotides were electrophoresed and purified from a 7.5% Tris-borate-EDTA acrylamide gel, eluted in 250 μl of deionized distilled water, and quantified. About 40 ng of labeled DNA (20 μl) was added to 100 μl of nuclear lysate from 293T cells transfected with pCMV-Tax, pCMV-p300, and pCMV-p30II-HA, as indicated in the figure legends, in a total volume of 200 μl of binding buffer (25 mM HEPES [pH 7.9], 5 mM KCl, 0.5 mM MgCl2, 0.5 mM EDTA, 1 mg of bovine serum albumin per ml, 10% [vol/vol] glycerol, and 0.25 mM DTT). The nuclear mixtures with biotin-labeled oligonucleotide probes were preincubated on ice for 2 h with gentle agitation. Sixty microliters of a 50% slurry of washed streptavidin-agarose (Life Technologies) was added to the nuclear lysates and kept on ice for 1 h with gentle agitation. The bound matrices were collected, washed twice with 500 μl of binding buffer, pelleted by centrifugation, and resuspended in 40 μl of SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer. Each sample was heated at 95°C for 5 min and loaded onto a precast 4 to 20% Tris-glycine-acrylamide gel (Bio-Rad) for resolution of bound products. After electrophoresis, the gels were transferred to nitrocellulose membranes for immunoblotting to detect CREB, Tax, p30II-HA, and CBP/p300. CREB was detected with a polyclonal antibody against CREB (diluted 1:1,000; New England Biolabs), Tax was detected with a polyclonal anti-Tax antibody (diluted 1:500, Tax α-serum 6505; National Institutes of Health AIDS Research and Reference Reagents Program), and p30II-HA was detected by polyclonal anti-HA antibody (diluted 1:1,000; Babco). p300 was detected using a polyclonal anti-p300 antibody (diluted 1:1,000; Santa Cruz Biotechnology, Inc.). A horseradish peroxidase-conjugated anti-goat serum was used as secondary antibody (diluted 1:1,000; Santa Cruz Biotechnology, Inc.).
Coimmunoprecipitation of p30II with p300.
Sixty percent confluent 293T cells were cotransfected by pCMV-Gal4(BD)-p30II and pCMV-p300 or pCMV-p30II-HA and pCMV-p300 by calcium phosphate coprecipitation. After 48 h, the transfected cells were washed with PBS and resuspended in 400 μl of lysis buffer containing 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 μg of leupeptin per ml, and 1 μg of aprotinin per ml. Cell suspensions were incubated on ice for 20 min and then lysed by homogenization. The lysates were centrifuged at 14,000 rpm (Beckman) for 20 min at 4°C. Supernatants were precleared by incubation with 10 μl of control rabbit IgG antibody and 30 μl of 50% protein A-agarose slurry (Amersham Pharmacia Biotechnology) for 1 h at 4°C. After centrifugation at 3,000 rpm (Beckman) for 1 min, the cleared supernatants were incubated overnight at 4°C with 100 ng of a polyclonal antibody against Gal4(BD) (Santa Cruz Biotechnology, Inc.) or 100 ng of a polyclonal antibody against hemagglutinin (HA) (Babco). After adding 40 μl of 50% of protein A-agarose slurry, the mixture was incubated at 4°C for 1 h. The immunoprecipitated complexes were washed twice with 10 volumes of lysis buffer and three times with PBS buffer. The components of the complexes were resolved on SDS-polyacrylamide gels [either 10% for Gal4(BD)-p30II and p30II-HA or 6% for p300] and detected by Western immunoblot assay.
Colocalization of p30II and CBP/p300 by immunofluorescence assay.
To detect cellular colocalization of p30II and p300 by immunofluorescence, 293T cells were seeded in chamber slides (Fisher Scientific) at approximately 40% confluence 18 h prior to transfection. Transfection with 4 μg of pCMV-p30II-HA and 2 μg of pCMV-p300 was performed using Lipofectamine Plus (Gibco BRL). At 48 h posttransfection, media were removed and cells were fixed for 15 min using 4% paraformaldehyde at room temperature. Cells were then incubated with monoclonal anti-HA antiserum (diluted 1:500; Babco) and polyclonal p300 antiserum (diluted 1:1,000; Santa Cruz Biotechnology, Inc.) overnight at 4°C followed by incubation with indocarbocyanine-labeled anti-mouse immunoglobulin (diluted 1:1,000; Jackson Immunogen) and Alexa 488 goat anti-rabbit IgG (diluted 1:1,000; Molecular Probes) for 1 h at room temperature. The expression of p30II-HA and p300 was evaluated by immunofluorescence microscopy (Axioplan2; Zeiss). A digital camera (Diagnostic Instruments, Inc.) was used to produce standard light microscopic and immunofluorescent photomicrographs.
GST-p30II fusion protein pull-down assay.
Eighty percent confluent 293T cells were transfected with 15 μg each of pBC-p30II and pCMV-p300 by calcium-phosphate coprecipitation. At 48 h posttransfection, cells were washed and resuspended in 400 μl of GST binding buffer containing 50 mM Tris-HCl, 150 mM NaCl, 0.5% NP-40, 5% glycerol, 1 mM PMSF, 0.5 μg of leupeptin per ml, and 1 μg of aprotinin per ml. After being kept on ice for 30 min, the cells were lysed by homogenization. Cell lysates were cleared by centrifugation at 14,000 rpm (Beckman) for 20 min at 4°C. Fifty microliters of a 50% glutathione-Sepharose slurry (Amersham Pharmacia) was added to the cell lysates, and the mixture was incubated for 1.5 h at 4°C. The GST beads were washed four times using GST washing buffer (50 mM Tris-HCl, 250 mM NaCl, 1.0% NP-40, 1 mM PMSF, 1 mM DTT, and 5% glycerol) and eluted in Laemmli buffer by boiling for 4 min. Proteins were electrophoresed in 4 to 20% gradient polyacrylamide gels and analyzed by Western immunoblot assay with anti-p300 antibody as the primary antibody (Santa Cruz Biotechnology, Inc.)
GST-CBP/p300 fusion protein pull-down assay.
To determine the binding region of CBP/p300 with p30II, GST fusion proteins containing defined regions of each protein were expressed in Escherichia coli BL21 cells using plasmids expressing either GST, GST-p300 (aa 1 to 159), GST-p300 (aa 744 to 1540), GST-p300 (aa 1540 to 2368), GST-CBP (aa 1 to 250), GST-CBP (aa 251 to 450), GST-CBP (aa 451 to 682), or GST-CBP (aa 1 to 717) (20). Protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h during the exponential growth phase of the bacterial culture. The bacteria were harvested, pelleted, and resuspended in 25 ml of PBS, pH 7.4, containing 1 mg of lysozyme per ml and 1 mM PMSF. After an incubation for 20 min at 4°C with gentle shaking, cells were ruptured by mild sonication. After centrifugation to remove cell debris, 2 ml of GST fusion protein-containing supernatant was mixed with 100 μl of 50% glutathione-Sepharose bead slurry (Amersham Pharmacia Biotechnology) for 2 h at 4°C. After extensive washing five times in lysis buffer, the glutathione-agarose beads were resuspended in 200 μl of GST binding buffer and mixed with 35 μl of reaction mixture containing 35S-labeled p30II synthesized by the T3-driven pBS-p30II vector using an in vitro transcription and translation kit (Promega). After incubation at 4°C for 2 h with gentle agitation, glutathione-agarose beads were collected by centrifugation at 3,000 rpm (Beckman) for 1 min. After washing three times with GST binding buffer, the GST-agarose beads were resuspended in Laemmli buffer and subjected to SDS–10% PAGE. Gels were dried at 80°C for 1 h, and 35S labeled p30II was detected by autoradiography.
RESULTS
Exogenous p300 enhances Gal4(BD)-p30II-mediated transactivation.
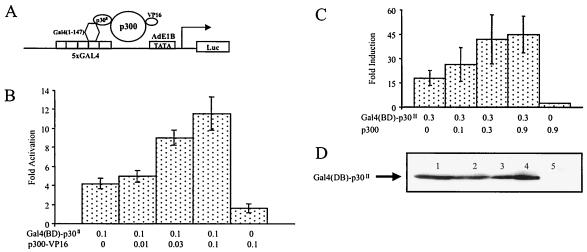
It has been reported that p30II functions as a transcription factor by using both Gal4-driven and CREB-responsive element (CRE)-driven reporter gene assays (57). When fused with Gal4(BD), p30II enhances Gal4-driven reporter gene expression by promoting transcription at core TATA box sites. One potential mechanism by which Gal4(BD)-p30II may modulate transcription is through interaction with common coadapters of transcription, such as CBP/p300. To test this possibility, we used a mammalian two-hybrid assay to determine potential synergistic activation between Gal4(BD)-p30II and p300. This functional assay was designed to show novel physical interactions between proteins regulating adenovirus virus transcription in vivo (21). We reasoned that, if p30II and p300 physically interact with each other, Gal4(BD)-p30II-mediated Gal4 reporter gene activities using the adenovirus E1B promoter system would also be stimulated in the presence of the chimeric protein p300-Vp16 (Fig. 1A) because the interaction of p30II and p300 would recruit Vp16 to the promoter region of the reporter gene. Vp16 as a chimeric protein with p300 is a potent stimulator of transcription and has been useful for identifying proteins that stabilize the transcription complex. Our data using this assay indicated that the Gal4 reporter gene activities were clearly increased by cotransfection of the p300-Vp16 vector and Gal4(BD)-p30II (Fig. 1B).
FIG. 1.
p300 enhances Gal4(BD)-p30II-dependent transactivation through protein-protein interactions. (A) Schematic representation of the p300-Vp16 mammalian two-hybrid system. For the transient transfection, 293T cells were transfected with 0.3 μg of the p5XG-E1b-Luc reporter plasmid together with the indicated quantities (in micrograms) of pCMV-Gal4(BD)-p30II plus increasing amounts of pRSV-p300-Vp16 (B) or pCMV-p300 plasmid (C). Thirty-six hours after transfection, cells were collected and an aliquot of cell extract was tested for luciferase reporter gene activity while another aliquot of cell extract was analyzed by Western blotting for the Gal4(BD)-p30II fusion protein (D). The luciferase activity of cells transfected with p5XG-E1b-Luc alone was used as the reference for basal reporter activity. Results are expressed as fold activation of basal luciferase reporter activity (measured in ALU) in the presence of indicated effector plasmids. Data represent the mean values ± SD derived from four independent experiments performed in duplicate. The basal luciferase activity of cells transfected with p5XG-E1b-Luc plasmid alone is 600 ALU.
We then tested the influence of p300 independently by cotransfecting pCMV-Gal4(BD)-p30II with pCMV-p300 in the same Gal4 reporter gene assay. Our data indicated that Gal4(BD)-p30II-dependent reporter gene activities were stimulated by exogenously expressed p300 in a dose-dependent manner (Fig. 1C). This stimulation was Gal4(BD)-p30II dependent, as it was not observed in the absence of pGal4(BD)-p30II (Fig. 1C). Gal4(BD)-p30II was expressed in similar amounts regardless of whether pCMV-p300 was coexpressed (Fig. 1D, lanes 1 to 4), which suggests that p300 participates in Gal4(BD)-p30II-dependent transactivation as a coactivator instead of inducing Gal4(BD)-p30II expression. Taken together, our results supported the tenet that p300 might be a coactivator in Gal4(BD)-p30II-modulated transcriptional events.
12SE1A protein competitively inhibits Gal4(BD)-p30II-dependent transactivation.
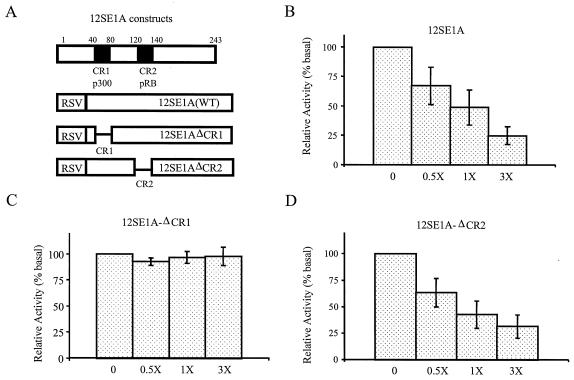
Adenovirus E1A protein is a multifunctional viral protein that modulates gene transcription by interacting with several key cellular proteins. It is well documented that adenovirus 12SE1A protein can inactivate p300-mediated gene transactivation by directly binding p300 (55). Since our results suggested that p300 stimulated Gal4(BD)-p30II-mediated transactivation, we tested the possibility that 12SE1A could competitively inhibit Gal4(BD)-p30II-dependent transactivation. We cotransfected vectors that expressed either wild-type E1A (12SE1A) or two deletion mutants which lack the ability to bind p300 (12SE1AΔCR1) or pRb, a cell cycle regulatory protein (12SE1AΔCR2), with CMV-Gal4(BD)-p30II in our Gal4-Luc reporter gene system (Fig. 2A). Our data indicated that Gal4(BD)-p30II-mediated transactivation of our Gal4-Luc reporter gene activities was inhibited in the presence of increasing concentrations of a pRSV-12SE1A expression vector (Fig. 2B), whereas the control RSV vector had no effect on Gal4(BD)-p30II transactivation (data not shown). In contrast, RSV-12SE1AΔCR1, which lacks N-terminal amino acid residues (aa 40 to 80) necessary for its interaction with p300, did not inhibit Gal4(BD)-p30II-mediated transactivation when transfected at levels comparable to that of the wild-type 12SE1A expression vector (Fig. 2C). Cotransfection with 12SE1AΔCR2, which lacks C-terminal amino acid residues (aa 120 to 140) corresponding to the region that interacts with pRb, resulted in repression of Gal4(BD)-p30II-mediated transactivation similar to that of wild-type 12SE1A (Fig. 2D). These data are consistent with previous reports of 12SE1A inhibition of CBP/p300-dependent transcription (1), and the data further implicated p300 as an essential member of a functional complex with p30II in transcriptional regulation.
FIG. 2.
In vivo competition of Gal4(BD)-p30II transactivation by E1A-expressing plasmids. (A) Schematic diagram representing the structure of E1A-expressing plasmids. Top bar depicts 243-aa wild-type E1A with conserved regions. CR1, site of p300 binding; CR2, site of pRb binding. 293T cells were transfected with 0.3 μg of p5XG-E1b-Luc reporter plasmid together with increasing amounts of competitor pRSV plasmid containing either wild-type 12SE1A (B) or mutated 12SE1AΔCR1 (C) or 12SE1AΔCR2 (D) E1A plasmid in fold excess relative to Gal4(BD)-p30II (0.3 μg). A relative luciferase activity value of 100% was assigned to cells that received no competitor plasmid DNA. Data represent the mean values ± SD derived from four independent experiments performed in duplicate. The basal luciferase activity of cells transfected with p5XG-E1b-Luc plasmid alone was 550 ALU.
Transactivation by Gal4(BD)-p30II through p300.
Our data suggested that Gal4(BD)-p30II-mediated transactivation is dependent upon p300. CBP/p300 has been shown to interact with a variety of transcriptional activators, including Tax of HTLV-1 (23, 38). In this context, CBP/p300 serves as a coadapter between these activators and the RNA polymerase II transcriptional machinery. We reasoned, therefore, that if CBP/p300 does participate in Gal4(BD)-p30II-dependent transactivation, overexpression of those transcription factors should competitively repress the transactivation by Gal4(BD)-p30II in our Gal4-Luc reporter gene assay by reducing the available CBP/p300 pool. In contrast, by adding exogenous p300 back to the cell using transient transfection, the repression should be reversed.
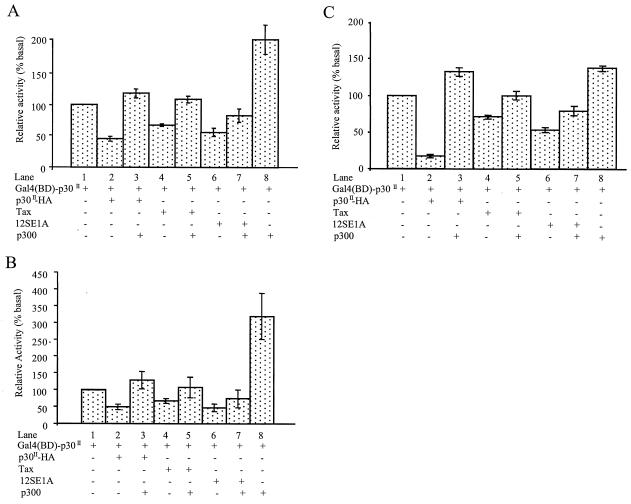
We tested the necessity of p300 in Gal4(BD)-p30II-mediated transactivation using in vivo competition assays as previously described (13, 22). In this assay, saturating concentrations of p30II-HA, Tax, and E1A expression vectors were introduced by cotransfection with the Gal4-p30II expression vector in our Gal4-Luc reporter gene assay. As predicted, transactivation by Gal4(BD)-p30II was repressed by cotransfection with p30II-HA, Tax, and E1A (Fig. 3A, lanes 2, 4, and 6). However, the repression by each of the expression vectors was restored by transfection with pCMV-p300 (Fig. 3A, lanes 3, 5, and 7). Consistent with our previous data, the cotransfection of pCMV-p300 with pGal4(BD)-p30II markedly enhanced Gal4-mediated transcription (Fig. 3A, lane 8). Similar results were obtained in parallel assays performed using 293 cells (Fig. 3B), which indicated that T-antigen expression in 293T cells did not influence the interaction of p30II with p300. Importantly, our results were also confirmed using Jurkat T cells (Fig. 3C). Collectively, these data indicated that p300 actively participates in Gal4(BD)-p30II-mediated transactivation.
FIG. 3.
In vivo competition assays of transactivation by p30II. 293T cells (A) were cotransfected with 0.3 μg of pGal4(BD)-p30II (lane 1) and 0.3 μg of pCMV-p30II-HA (lanes 2 and 3), pCMV-Tax (lanes 4 and 5), or pCMV-12SE1A (lanes 6 and 7) with (+) (lanes 3, 5, 7, and 8) or without (−) (lanes 1, 2, 4, and 6) 0.3 μg of pCMV-p300. All transfections contained 0.3 μg of p5XG-E1b-Luc as a reporter. Similar transfections using the same sets of plasmids were performed in 293 (B) and Jurkat T (C) cells. The relative luciferase activity value of 100% was assigned to cells that received only pGal4(BD)-p30II (lane 1) as the effector plasmid: 15,000, 22,000, and 14,000 ALU for 293T, 293, and Jurkat T cells, respectively. Data represent the mean values ± SD derived from four independent experiments performed in duplicate.
p300 expression reverses p30II inhibition of CRE- and TRE-driven gene transcription.
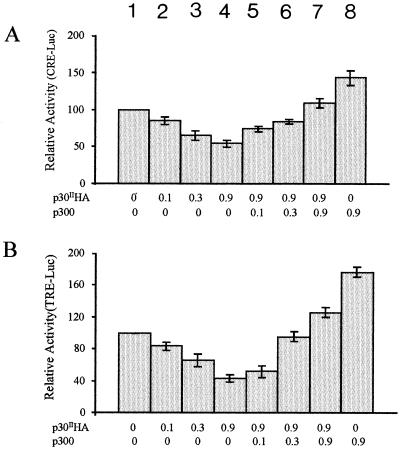
We have reported that p30II, in contrast to Tax, repressed cellular CRE-driven reporter gene activity and only at lower concentrations enhanced the HTLV-1 long terminal repeat Tax-responsive element (TRE)-driven reporter gene activity (57). Our results suggested that p30II and Tax serve divergent roles in the regulation of transcription. The coadapters CBP and p300 have been demonstrated to be integral components of HTLV-1 Tax transactivation. Since our data indicated that p30II interacts with p300, we postulated that p30II might repress basal CRE- and TRE-driven reporter gene activities as a consequence of competition for limited basal quantities of CBP/p300. In this scenario, overexpression of p300 should prevent p30II-mediated repression of CRE- and TRE-driven reporter gene expression. To test this possibility, we performed cotransfection studies using luciferase reporter plasmids driven by either the CRE (CRE-Luc) or the TRE (TRE-Luc) promoter in the presence of increasing concentrations of p30II. As has been reported (57), both reporter gene activities were repressed by p30II in a dose-dependent manner. Then we attempted to reverse the p30II repressive effects by cotransfection with increasing concentrations of the pCMV-p300 expression vector. A fixed concentration of pCMV-p30II-HA expression vector (0.9 μg) was cotransfected with increasing amounts of pCMV-p300 in the presence of pCMV-CRE-Luc (Fig. 4A) or pTRE-Luc vector (Fig. 4B). As has been reported (57), our CMV empty vector at identical concentrations did not influence either CRE- or TRE-mediated transcription. As predicted, p30II-dependent repression of both CRE- and TRE-responsive promoters was completely restored by increasing amounts of exogenous p300. These data further support our hypothesis that p300 mediates p30II-dependent transcriptional activity.
FIG. 4.
p300 reverses the inhibitory effects of p30II on CRE- and TRE-driven reporter gene activity. 293T cells were transfected with 0.3 μg of a reporter gene plasmid, pCRE-Luc (A) or pTRE-Luc (B), and the indicated quantities of pCMV-p30II-HA (in micrograms) (bars 2 to 7) together with increasing quantities of pCMV-p300 (in micrograms) (bars 5 to 8). The relative luciferase activity shown in bar 1 (without competitor plasmids) was established as 100%. Data represent the mean values ± SD derived from four independent experiments performed in duplicate. The basal luciferase activities of cells transfected with the CRE-Luc and TRE-Luc reporter plasmids alone are 7,500 and 850 ALU, respectively.
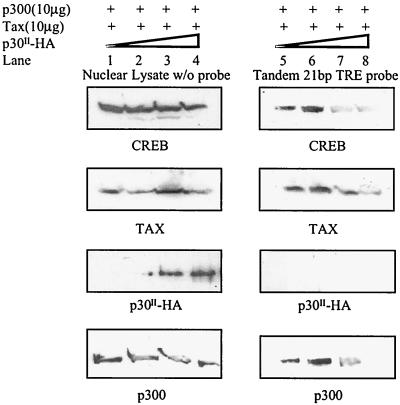
p30II disrupts the assembly of CREB-Tax-p300 complexes on TRE probes.
As a coadapter CBP/p300 functions to stabilize the multiple protein complex that includes CREB, Tax, TATA binding protein, and other transcription machinery molecules on CRE and TRE promoters, thereby promoting transcription (23, 38, 40). To test the direct effect of p30II on the assembly of these complexes, we expressed Tax and p300 in the presence of increasing amounts of pCMV-p30II-HA. Nuclear lysates from transfected cells were tested for the presence of multiprotein complexes formed on biotin-labeled HTLV-1 21-bp repeat oligonucleotides (Fig. 5). Increasing amounts of expressed p30II had no effect on the expression of CREB, Tax, or p300 (Fig. 5, lanes 1 to 4). However, increasing concentrations of p30II dramatically reduced the binding of CREB, Tax, and p300 to the biotin-labeled 21-bp repeat oligonucleotide (Fig. 5, lanes 5, 6, 7, and 8). These data are consistent with the sequestration of p300 by p30II and suggest this tenet as a mechanism for the repression of basal TRE reporter gene activities by p30II. p30II did not bind directly to CRE and TRE cis elements in electrophoretic mobility shift assays using probes containing tandem repeats of consensus CRE or HTLV-1 21-bp repeat TRE sequences (data not shown). Taken together, our data indicated that p30II negatively influences the assembly of multiprotein complexes at the HTLV-1 21-bp repeat TRE site.
FIG. 5.
p30II inhibits the assembly of Tax-p300-CREB multiprotein complexes on TRE oligonucleotides. Exogenous Tax, p300, and p30II-HA were introduced into 293T cells by transfection using expression plasmids. At 48 h posttransfection, nuclear lysates of transfected cells were incubated with the biotin-labeled TRE (HTLV-1 21-bp repeat) DNA probe as described in Materials and Methods. Streptavidin-agarose was used to isolate bound components. After extensive washing, bound proteins (right panel) were resolved using SDS-PAGE (4 to 20% gradient acrylamide) gels and detected by Western immunoblot assay. In parallel, equal amounts of nuclear lysates used as input were examined by Western immunoblot assay to determine protein expression levels of each component in transfected cells (left panel).
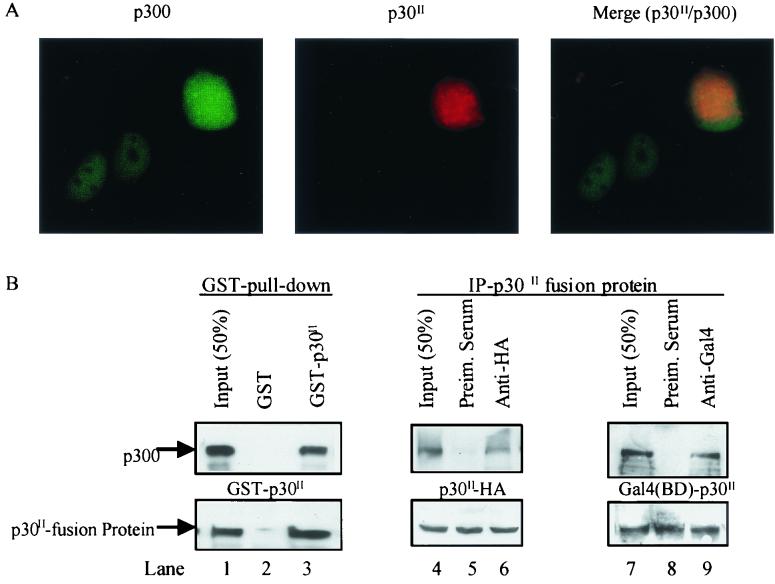
p30II directly binds p300/CBP in vivo.
Our data strongly suggested that p30II-mediated transcription is dependent, at least in part, upon CBP/p300. To test the cellular localization of each protein, we performed immunofluorescence microscopy following the cotransfection of pCMV-p30II-HA and pCMV-p300 in 293T cells. As expected, p30II-HA was expressed in the nucleus of cells and overlapped with p300 expression in cells that expressed both proteins (Fig. 6A). To test if p30II physically interacted with CBP/p300 in vivo, we cotransfected 293T cells with pBC-p30II, pCMV-p30II-HA, or pCMV-Gal4(BD)-p30II together with pCMV-p300. Whole-cell extracts were prepared and used in GST pull-down or coimmunoprecipitation assays. Following binding to glutathione-Sepharose beads, GST-p30II effectively pulled down p300 from cellular lysates, unlike the GST-vector control (Fig. 6B, compare lanes 2 and 3). p300, when coexpressed with pCMV-p30II-HA or pCMV-Gal4(BD)-p30II, was immunoprecipitated as a complex by using both anti-HA (Fig. 6B, lane 6) and anti-Gal4(BD) antibodies (Fig. 6B, lane 9). In contrast, p300 could not be detected in the complexes precipitated by using preimmune serum (Fig. 6B, lanes 5 and 8). All three p30II fusion proteins could be detected from their respective cellular lysates (Fig. 6B, lower panel, lanes 1 and 3 to 9). In addition, p30II failed to bind CREB directly when tested in immunoprecipitation and Western blot assays (data not shown).
FIG. 6.
p30II colocalizes and forms physical complexes with p300 in cells. (A) Immunofluorescence analysis of 293T cells transfected with pCMV-p30II-HA and pCMV-p300 were performed 16 h posttransfection as described in Materials and Methods. p30II-HA (red) and p300 (green) were detected using monoclonal anti-HA antiserum and polyclonal anti-p300 antiserum, respectively. Images were merged to detect (in cells expressing p30II) the overlap between p30II and p300 expression (yellow). (B) To determine physical binding between p30II and p300, 293T cells were cotransfected with 10 μg of pCMV-p300 together with 10 μg of each of the following plasmids, as indicated: pBC (empty vector), pBC-p30II, pCMV-p30II-HA, and pCMV-Gal4(BD)-p30II. At 48 h posttransfection, GST pull-down or immunoprecipitation assays were performed as described in Materials and Methods. p300 pulled down by GST-p30II (lane 3) or precipitated with p30II-HA (lane 6) or Gal4(BD)-p30II (lane 9) was detected by Western immunoblot assay using anti-p300 antibody. pBC (GST control plasmid)-transfected cell extract was used as a negative control for the GST pull-down assay (lane 2). Normal rabbit IgG was used as a negative control in immunoprecipitation assays (lanes 5 and 8). Equal amounts of input cell extracts were processed to determine the expression of p30II fusion proteins by Western immunoblot assay (lower panel, lanes 1, 4, and 7).
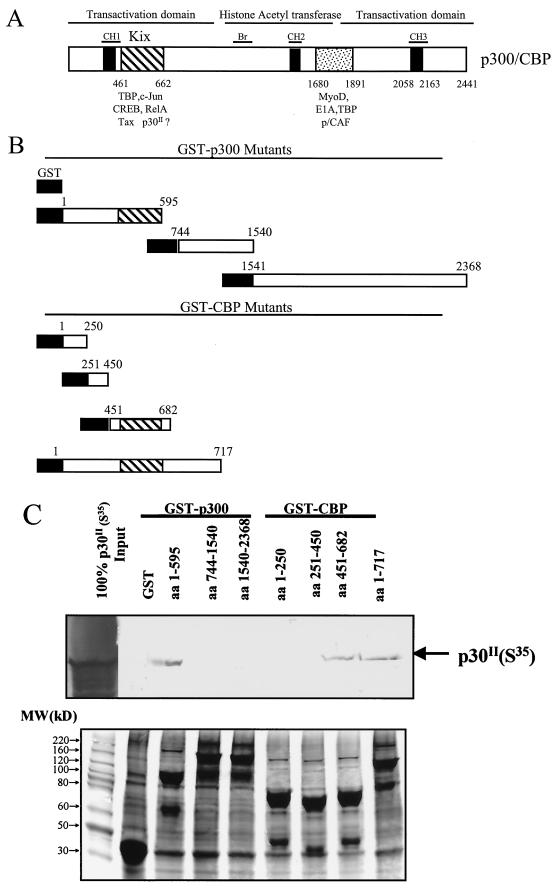
p30II interacts with CBP/p300 through the KIX domain.
We next tested which domains of CBP/p300 bind with p30II by expressing eight GST-CBP/p300 fusion proteins containing defined portions of p300 or CBP (Fig. 7A and B). To improve the sensitivity for detection of p30II, we synthesized and labeled p30II with [35S]methionine using an in vitro transcription and translation system (46). After incubation of glutathione-Sepharose beads bound with GST fusion proteins with comparable amounts of in vitro-synthesized 35S-labeled p30II, bound proteins were separated by SDS-PAGE and detected by autoradiography. Among the proteins pulled down by GST-Sepharose beads, 35S-labeled p30II could only be detected in GST-p300, aa 1 to 595; GST-CBP, aa 451 to 682; and GST-CBP, aa 1 to 717 (Fig. 7C). These data suggested that the region located between aa 461 and 662, corresponding to the KIX domain, of CBP/p300 contributed to the interaction between p30II and CBP/p300.
FIG. 7.
p30II directly binds the KIX domain of CBP/p300 in vitro. (A) Schematic diagram of binding motifs and functional domains of CBP and p300. (B) Diagram representing the structure of GST-CBP- and GST-p300-expressing plasmids used in the GST pull-down assay. (C) GST fusion proteins containing defined residues of CBP and p300 were expressed and coupled to glutathione-agarose beads. After extensive washing, glutathione-agarose beads coupled with GST-CBP or GST-p300 fusion protein were incubated with 35S-labeled p30II synthesized by in vitro transcription and translation as described in Materials and Methods. 35S-labeled p30II pulled down by glutathione-agarose beads coupled with GST-CBP or GST-p300 fusion protein (upper panel) was visualized by autoradiography after being resolved in SDS-PAGE gels. Coomassie-stained SDS-PAGE gel for each of the corresponding GST-fusion proteins used in pull-down assay (upper panel). The major protein in each lane was of the expected molecular mass (lane 1, molecular mass marker) (lower panel).
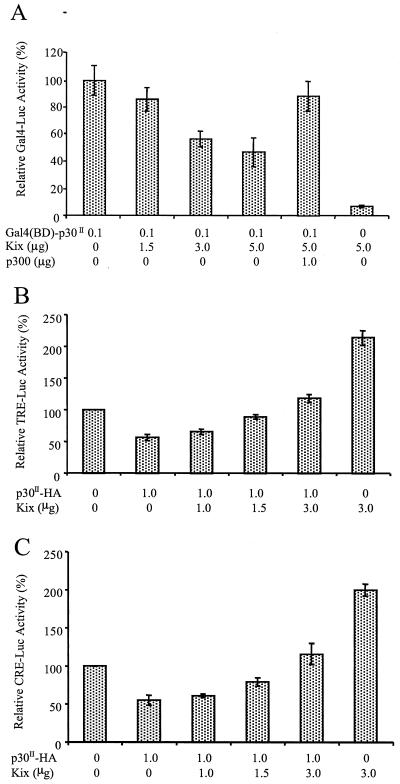
To further verify the ability of p30II to interact with the KIX domain, we performed a functional analysis of the ability of KIX domain-containing proteins to competitively inhibit p30II-mediated transcription. This analysis was based on the premise that a vector expressing a protein representing the isolated KIX domain would interfere with the physical interaction between CBP/p300 and p30II. As predicted, the overexpression of KIX domain molecules effectively reduced the transactivation of Gal4 reporter gene activities by Gal4(BD)-p30II in a dose-dependent manner (Fig. 8A). More importantly, this repression could be released by elevating exogenous p300 levels (Fig. 8A, compare lanes 5 and 6). In reciprocal studies, we also tested if this same plasmid (pRSV-KIX [aa 379 to 654]/p300) would alleviate the repression of both TRE- and CRE-mediated transcription induced by p30II. As KIX domain expression was increased, the ability of p30II-mediated repression was diminished, which resulted in enhanced luciferase reporter gene expression (Fig. 8B and C). Taken together, our data strongly support an interaction between HTLV-1 p30II and the CBP/p300 KIX domain, which would support the role of this unique viral protein in regulation of transcription during HTLV-1 replication.
FIG. 8.
KIX domain of p300 diminishes p30II-mediated transcription. (A) Using the Gal4-dependent transcription assay, 293T cells were cotransfected with 0.3 μg of the reporter plasmid p5XG-E1b-Luc and 0.1 μg of the effector plasmid pGal4(BD)-p30II and increasing amounts of pRSV-KIX (aa 379 to 654)/p300 (in micrograms). The wild-type p300 expression plasmid eliminated KIX domain suppression, whereas KIX domain alone failed to mediate transcription without p30II. Effects of KIX domain expression on p30II repression of pCRE-Luc (B) and pTRE-Luc (C) reporter plasmids are shown. pCRE-Luc or pTRE-Luc reporter plasmid and fixed amounts p30II-HA were transfected in the presence of increasing amounts of pRSV-KIX (aa 379 to 654)/p300 (in micrograms). Thirty-six hours after transfection, the cells were assayed for luciferase reporter gene activity. The relative luciferase activity from cells that received only pGal4(BD)-p30II (A) or no effector plasmid (B and C) was established as 100%. Data represent the mean values ± SD derived from four independent experiments performed in duplicate.
DISCUSSION
It is well documented that CBP and p300, as coactivators, participate in the activation of gene expression by bridging upstream transcription factor complexes with the general transcription machinery. Furthermore, CBP/p300 has intrinsic histone acetylase activity (7) to potentially increase the state of nucleosome acetylation, which is essential for the initiation of gene transcription. In addition, CBP and p300 are general integrators of signal-dependent transcription, and a diverse array of enhancer binding factors (e.g., CREB) utilizes these coactivators for transcriptional activation in response to extracellular signals (24, 28, 39).
Previous studies have demonstrated that p30II is a transcription factor and differentially modulates CREB-responsive gene activity (57). As a Gal4 fusion protein, p30II transactivates Gal4 reporter gene activity. However, unlike Tax, p30II could also repress CRE and TRE reporter gene activity in a dose-dependent manner. We hypothesized that p30II might modify target gene activities by interacting with common coactivators like CBP and p300. Our data, herein, demonstrate that p300 enhances p30II-dependent transcription in three different cell culture models, including Jurkat T cells. Like Tax, p30II physically interacted in cells with CBP and p300 through the KIX domain, as shown by our protein binding and transcription assay data. These data are consistent with our hypothesis that p30II modulates gene expression by interacting with CBP/p300, and the data provide a mechanism to explain the p30II-dependent modulation of gene transcription.
There is precedence for a similar transcriptional repression by other viral regulatory proteins. CBP/p300 interactions are important for the biological effects of adenovirus oncoprotein E1A, including the regulation of transcription, suppression of differentiation, and immortalization of cells in culture (1, 3, 42). Simian virus T antigen regulates the expression of a group of cellular genes by modifying the HAT activity of CBP/p300 or by bridging the gap between DNA binding transcription factors and components of the general transcription machinery (4, 25). In the context of HTLV replication, Tax-mediated recruitment of CBP/p300 to the promoter has been shown to be necessary for efficient Tax-mediated transcription of HTLV-1 (23, 38, 53, 56). Our data indicate that p30II also modulates transcription in a unique manner compared to Tax but uses similar interactions with coadapter proteins important for RNA Pol II-mediated transcription.
Our results indicated that p30II acted as a repressor to down-regulate CRE- and TRE-driven reporter gene activities, which is in contrast to the role of HTLV-1 Tax. Tax is critical for high-level HTLV-1 transcription and propagation. To activate viral gene expression, Tax participates in a series of protein-protein and protein-DNA interactions, forming a stable nucleoprotein complex on the HTLV-1 promoter (56). Within this complex, Tax serves as a high-affinity binding site for the recruitment of CBP and p300 (23, 38, 56). Once associated with the viral promoter, CBP/p300 is believed to remodel chromatin and/or facilitate communication with the basal transcription machinery. To recruit CBP/p300, Tax has been shown to specifically bind to a region of CBP/p300 called the KIX domain (27, 32). This region of CBP/p300 also interacts with several other transcription factors, including Ser-133-phosphorylated CREB (14, 39). Tethering of CBP/p300 to the HTLV-1 transcriptional control region promotes the strong transcriptional activation associated with Tax (53). Interestingly, p30II shares this physical interaction with CBP/p300 through the same KIX domain. In addition, we also provide data demonstrating that p30II could destabilize Tax, p300, and CREB multiprotein complexes formed on 21-bp probes in vitro. We believe that the competitive CBP/p300 binding between p30II and Tax might explain how p30II could attenuate the formation of these multiprotein complexes and thereby repress transcription on CREB-responsive promoters. Similarly, Colgin and Nyborg (17) demonstrated that Tax expression interferes with the transcriptional activity of c-Myb and that the binding of Tax and c-Myb to the KIX domain of CBP is mutually exclusive. KIX expression, by itself, enhances the binding of Tax and CREB to CRE-containing oligonucleotides (17) and, as we have demonstrated, would be expected to enhance CRE- and TRE-mediated transcription. Additionally, Tax has been proposed to interfere with CBP-mediated transcription by binding the coactivator (53). Thus, p30II at higher concentrations may serve to promote viral persistence by reducing viral expression, thereby reducing immune elimination of virus-infected cells.
Alternatively, it has been shown that at low concentrations p30II acts to enhance TRE (viral)- over CRE (cellular)-mediated transcription (57). Thus, while promoting viral transcription, this viral protein may competitively repress CBP/p300-dependent cellular gene transcription (e.g., p53-dependent p21WAF1/CIP1 gene activity), promoting cell proliferation (5) and allowing efficient viral spread in vivo. This tenet would explain the data from the rabbit model of HTLV-1 infection, which demonstrated that proviral clones with selective mutations involving pX ORF II (p30II and p13II) fail to maintain normal viral loads in vivo (8). Like Tax and E1A, p30II appears to behave in a sequence-independent manner through sequestration of CBP/p300 (1, 25, 43, 52). Our present studies are focused on the functional significance of the interactions between p30II and CBP/p300, such as the requirement for acetyltransferase activity of CBP/p300 for the transcriptional effects of p30II.
It is noteworthy that the CBP/p300 protein is generally present at limiting concentrations within the cell nucleus, creating an environment for competition between coactivators and transcription factors, thus providing an additional layer of regulated gene expression (47, 52). Several recent studies suggest that a functional antagonism between transcription factors occurs as a consequence of direct competition for binding to common regions of CBP/p300 (6, 17, 31). HTLV-1 p30II, another accessory protein of HTLV-1, is critical for optimal viral loads and differentially modulates CREB-responsive promoters perhaps by the sequestration of cellular CBP/p300. By tightly controlling the level of viral expression in vivo, it is likely that HTLV-1 uses these accessory proteins to avoid the overexpression of viral proteins or to repress cellular genes necessary to maintain viral persistence. It will be important for future studies to define relevant target genes of p30II in addition to CREB-responsive genes to provide a more complete understanding of the role of p30II in the pathogenesis and replication of this important human pathogen.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant no. RR-14324 from the National Center for Research Resources and grant no. CA-70259 from the National Cancer Institute awarded through the Comprehensive Cancer Center of The Ohio State University. W. Zhang was supported by a David White Fellowship award from The Ohio State University. M. Lairmore was supported by an Independent Scientist Career Award from the National Institutes of Health (K02 AI01474).
We thank Tim Vojt for preparation of figures. We also thank S. Beebe, S. Yang, T. Kouzarides, G. Louis, P. Quinn, G. Franchini, and S. McKnight for valuable reagents. We thank Q. Xi for technical assistance. We also thank P. Green and K. Boris-Lawrie for critical reviews of the manuscript.
REFERENCES
- 1.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 2.Ariumi Y, Kaida A, Lin J Y, Hirota M, Masui O, Yamaoka S, Taya Y, Shimotohno K. HTLV-1 tax oncoprotein represses the p53-mediated trans-activation function through coactivator CBP sequestration. Oncogene. 2000;19:1491–1499. doi: 10.1038/sj.onc.1203450. [DOI] [PubMed] [Google Scholar]
- 3.Avantaggiati M L, Carbone M, Graessmann A, Nakatani Y, Howard B, Levine A S. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 1996;15:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- 4.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 5.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 6.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 8.Bartoe J T, Albrecht B, Collins N D, Robek M D, Ratner L, Green P L, Lairmore M D. Functional role of pX open reading frame II of human T-lymphotropic virus type 1 in maintenance of viral loads in vivo. J Virol. 2000;74:1094–1100. doi: 10.1128/jvi.74.3.1094-1100.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bex F, Gaynor R B. Regulation of gene expression by HTLV-I tax protein. Methods. 1998;16:83–94. doi: 10.1006/meth.1998.0646. [DOI] [PubMed] [Google Scholar]
- 10.Blobel G A. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- 11.Cereseto A, Berneman Z, Koralnik I, Vaughn J, Franchini G, Klotman M E. Differential expression of alternatively spliced pX mRNAs in HTLV-I-infected cell lines. Leukemia. 1997;11:866–870. doi: 10.1038/sj.leu.2400665. [DOI] [PubMed] [Google Scholar]
- 12.Chatton B, Bahr A, Acker J, Kedinger C. Eukaryotic GST fusion vector for the study of protein-protein associations in vivo: application to interaction of ATFa with Jun and Fos. BioTechniques. 1995;18:142–145. [PubMed] [Google Scholar]
- 13.Chen X, Bieker J J. Erythroid Kruppel-like factor (EKLF) contains a multifunctional transcriptional activation domain important for inter- and intramolecular interactions. EMBO J. 1996;15:5888–5896. [PMC free article] [PubMed] [Google Scholar]
- 14.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 15.Ciminale V, Pavlakis G N, Derse D, Cunningham C P, Felber B K. Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV type I. J Virol. 1992;66:1737–1745. doi: 10.1128/jvi.66.3.1737-1745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciminale V, Zotti L, Dagostino D M, Ferro T, Casareto L, Franchini G, Bernardi P, Chiecobianchi L. Mitochondrial targeting of the p13(II) protein coded by the x-II ORF of human T-cell leukemia/lymphotropic virus type I (HTLV-I) Oncogene. 1999;18:4505–4514. doi: 10.1038/sj.onc.1203047. [DOI] [PubMed] [Google Scholar]
- 17.Colgin M A, Nyborg J K. The human T-cell leukemia virus type 1 oncoprotein Tax inhibits the transcriptional activity of c-Myb through competition for the CREB binding protein. J Virol. 1998;72:9396–9399. doi: 10.1128/jvi.72.11.9396-9399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Agostino D M, Ciminale V, Zotti L, Chieco-Bianchi L. Influence of Rex and intronic sequences on expression of spliced mRNAs produced by human T cell leukemia virus type I. AIDS Res Hum Retrovir. 1999;15:1351–1363. doi: 10.1089/088922299310061. [DOI] [PubMed] [Google Scholar]
- 19.Dekaban G A, Peters A A, Mulloy J C, Johnson J M, Trovato R, Rivadeneira E, Franchini G. The HTLV-I orfI protein is recognized by serum antibodies from naturally infected humans and experimentally infected rabbits. Virology. 2000;274:86–93. doi: 10.1006/viro.2000.0406. [DOI] [PubMed] [Google Scholar]
- 20.Deng L, de La F C, Fu P, Wang L, Donnelly R, Wade J D, Lambert P, Li H, Lee C G, Kashanchi F. Acetylation of HIV-1 tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones. Virology. 2000;277:278–295. doi: 10.1006/viro.2000.0593. [DOI] [PubMed] [Google Scholar]
- 21.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 22.Geiman D E, Ton-That H, Johnson J M, Yang V W. Transactivation and growth suppression by the gut-enriched Kruppel-like factor (Kruppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res. 2000;28:1106–1113. doi: 10.1093/nar/28.5.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giebler H A, Loring J E, van Orden K, Colgin M A, Garrus J E, Escudero K W, Brauweiler A, Nyborg J K. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol Cell Biol. 1997;17:5156–5164. doi: 10.1128/mcb.17.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giles R H, Peters D J, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 25.Goodman R H, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 26.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 27.Harrod R, Tang Y, Nicot C, Lu H S, Vassilev A, Nakatani Y, Giam C-Z. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol Cell Biol. 1998;18:5052–5061. doi: 10.1128/mcb.18.9.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 29.Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 30.Janknecht R, Hunter T. Transcriptional control: versatile molecular glue. Curr Biol. 1996;6:951–954. doi: 10.1016/s0960-9822(02)00636-x. [DOI] [PubMed] [Google Scholar]
- 31.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 32.Kashanchi F, Duvall J F, Kwok R P, Lundblad J R, Goodman R H, Brady J N. The coactivator CBP stimulates human T-cell lymphotrophic virus type I Tax transactivation in vitro. J Biol Chem. 1998;273:34646–34652. doi: 10.1074/jbc.273.51.34646. [DOI] [PubMed] [Google Scholar]
- 33.Kibler K V, Jeang K T. Taxing the cellular capacity for repair: human T-cell leukemia virus type 1, DNA damage, and adult T-cell leukemia. J Natl Cancer Inst. 1999;91:903–904. doi: 10.1093/jnci/91.11.903. [DOI] [PubMed] [Google Scholar]
- 34.Kimata J T, Wong F, Wang J, Ratner L. Construction and characterization of infectious human T-cell leukemia virus type 1 molecular clones. Virology. 1994;204:656–664. doi: 10.1006/viro.1994.1581. [DOI] [PubMed] [Google Scholar]
- 35.Koralnik I J, Fullen J, Franchini G. The p12I, p13I, and p30II proteins encoded by human T-cell leukemia/lymphotropic virus type 1 open reading frames I and II are localized in three different cellular compartments. J Virol. 1993;67:2360–2366. doi: 10.1128/jvi.67.4.2360-2366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koralnik I J, Gessain A, Klotman M E, Lo Monico A, Berneman Z N, Franchini G. Protein isoforms encoded by the pX region of human T-cell leukemia/lymphotropic virus type 1. Proc Natl Acad Sci USA. 1992;89:8813–8817. doi: 10.1073/pnas.89.18.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusuhara K, Anderson M, Pettiford S M, Green P L. Human T-cell leukemia virus type 2 Rex protein increases stability and promotes nuclear to cytoplasmic transport of gag/pol and env RNAs. J Virol. 1999;73:8112–8119. doi: 10.1128/jvi.73.10.8112-8119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwok R P, Laurance M E, Lundblad J R, Goldman P S, Shih H, Connor L M, Marriott S J, Goodman R H. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 39.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 40.Lenzmeier B A, Baird E E, Dervan P B, Nyborg J K. The Tax protein-DNA interaction is essential for HTLV-I transactivation in vitro. J Mol Biol. 1999;291:731–744. doi: 10.1006/jmbi.1999.2969. [DOI] [PubMed] [Google Scholar]
- 41.Li M, Damania B, Alvarez X, Ogryzko V, Ozato K, Jung J U. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol Cell Biol. 2000;20:8254–8263. doi: 10.1128/mcb.20.21.8254-8263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 43.Mesnard J M, Devaux C. Multiple control levels of cell proliferation by human T-cell leukemia virus type 1 Tax protein. Virology. 1999;257:277–284. doi: 10.1006/viro.1999.9685. [DOI] [PubMed] [Google Scholar]
- 44.Neuveut C, Jeang K T. HTLV-I Tax and cell cycle progression. Prog Cell Cycle Res. 2000;4:157–162. doi: 10.1007/978-1-4615-4253-7_14. [DOI] [PubMed] [Google Scholar]
- 45.Newbound G C, Andrews J M, O'Rourke J, Brady J N, Lairmore M D. Human T-cell lymphotropic virus type 1 Tax mediates enhanced transcription in CD4+ T lymphocytes. J Virol. 1996;70:2101–2106. doi: 10.1128/jvi.70.4.2101-2106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newbound G C, O'Rourke J P, Collins N D, DeWille J, Lairmore M D. Comparison of HTLV-I basal transcription and expression of CREB/ATF-1/CREM family members in peripheral blood mononuclear cells and Jurkat T cells. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:1–10. doi: 10.1097/00042560-199901010-00001. [DOI] [PubMed] [Google Scholar]
- 47.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C, Masuno M, Tommerup N, van Ommen G J, Goodman R H, Peters D J. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 48.Pique C, Dokhelar M C. In vivo production of rof and tof proteins of HTLV type 1: evidence from cytotoxic T lymphocytes. AIDS Res Hum Retrovir. 2000;16:1783–1786. doi: 10.1089/08892220050193317. [DOI] [PubMed] [Google Scholar]
- 49.Pise-Masison C A, Mahieux R, Radonovich M, Jiang H, Duvall J, Guillerm C, Brady J N. Insights into the molecular mechanism of p53 inhibition by HTLV type 1 Tax. AIDS Res Hum Retrovir. 2000;16:1669–1675. doi: 10.1089/08892220050193128. [DOI] [PubMed] [Google Scholar]
- 50.Potter J, Cheneval D, Dang C, Ressar L, Mezey E, Yang X. The upstream stimulatory factor binds to and activates the promoter of the rat class I alcohol dehydrogenase gene. J Biol Chem. 1991;266:15457–15463. [PubMed] [Google Scholar]
- 51.Reddy T R, Xu W D, Wong-Staal F. General effect of Sam68 on Rev/Rex regulated expression of complex retroviruses. Oncogene. 2000;19:4071–4074. doi: 10.1038/sj.onc.1203749. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki T, Uchidatoita M, Yoshida M. Tax protein of HTLV-1 inhibits CBP/p300-mediated transcription by interfering with recruitment of CBP/p300 onto DNA element of E-box or p53 binding site. Oncogene. 1999;18:4137–4143. doi: 10.1038/sj.onc.1202766. [DOI] [PubMed] [Google Scholar]
- 53.Van Orden K, Yan J P, Ulloa A, Nyborg J K. Binding of the human T-cell leukemia virus Tax protein to the coactivator CBP interferes with CBP-mediated transcriptional control. Oncogene. 1999;18:3766–3772. doi: 10.1038/sj.onc.1202703. [DOI] [PubMed] [Google Scholar]
- 54.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–70. [PubMed] [Google Scholar]
- 55.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 56.Yan J P, Garrus J E, Giebler H A, Stargell L A, Nyborg J K. Molecular interactions between the coactivator CBP and the human T-cell leukemia virus Tax protein. J Mol Biol. 1998;281:395–400. doi: 10.1006/jmbi.1998.1951. [DOI] [PubMed] [Google Scholar]
- 57.Zhang W, Nisbet J W, Bartoe J T, Ding W, Lairmore M D. Human T-lymphotropic virus type 1 p30II functions as a transcription factor and differentially modulates CREB-responsive promoters. J Virol. 2000;74:11270–11277. doi: 10.1128/jvi.74.23.11270-11277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]