Abstract
Helper-dependent minimal adenovirus vectors are promising tools for gene transfer and therapy because of their high capacity and the absence of immunostimulatory or cytotoxic viral genes. In order to characterize this new vector system with respect to its integrative properties, the integration pattern of a minimal adenovirus vector with a neor gene inserted centrally into a noncoding 27.4-kb genomic stuffer element derived from the human X chromosome after infection of a sex chromosome aneuploid (X0) human glioblastoma cell line was studied. Our results indicate that even extensive homologies and abundant chromosomal repeat elements present in the vector did not lead to integration of the vector via homologous or homology-mediated mechanisms. Instead, integration occurred primarily by insertion of a monomer with no or little loss of sequences at the vector ends, apparently at random sites, which is very similar to E1 deletion adenovirus vectors. It is therefore unlikely that the incorporation of stuffer elements derived from human genomic DNA, which were shown to allow long-term transgene expression in vivo in a number of studies, leads to an enhanced risk of insertional mutagenesis. Furthermore, our findings indicate that the potential of minimal adenovirus vectors as tools for targeted insertion and gene targeting is limited despite the possibility of incorporating long stretches of homologous sequences. However, we found an enhanced efficiency of stable neor transduction of the minimal adenovirus vector compared to an E1 deletion adenovirus vector, possibly caused by the absence of potential growth-inhibitory viral genes. Complete integration of the vector and tolerance of the integrated vector sequences by the cell might indicate a potential use of these vectors as tools for stable transfer of (large) genes.
Recombinant adenovirus vectors are promising vehicles for many gene delivery applications in molecular biology and medicine because of their high cellular transduction efficiency. First-generation adenovirus vectors based on human adenovirus serotype 5 (Ad5) are rendered replication deficient by deletion of the E1 region and can accommodate inserts of up to 8 kb (reviewed in reference 3). The potential of these vectors for gene therapy applications is limited by the rapid loss of transgene expression in vivo. Residual expression of adenovirus genes was shown to induce a cytotoxic-T-lymphocyte-mediated immune response, resulting in inflammation and loss of vector-transduced cells (11, 71, 72, 73). To overcome these limitations, we and others have developed helper-dependent adenovirus vectors with all viral coding sequences deleted (20, 26, 29, 33, 35, 40, 52, 56). These vectors—termed minimal adenovirus vectors herein—were shown to mediate prolonged transgene expression and to exhibit reduced tissue toxicity in vivo compared to E1 deletion adenovirus vectors in a number of recent studies (8, 9, 42, 43, 44, 57, 76).
Minimal adenovirus vectors containing only a heterologous expression unit and the cis-acting adenovirus signal elements (inverted terminal repeats [ITRs] and packaging signal) would usually be much smaller than wild-type adenovirus. Vectors of this size amplify less efficiently or multimerize (20, 25, 44). As this phenomenon was correlated to a lower packaging limit of Ad5 (1, 50), stuffer sequences are added to provide a packageable genome size. It has been shown that nature of the stuffer has a substantial influence on transgene expression in vivo. Long-term gene expression was observed with stuffers composed of noncoding human genomic sequences (8, 42, 44, 56, 57). In contrast, rapid loss of transgene expression was observed with a stuffer derived from phage lambda and could be correlated with a stuffer-directed cellular immune response (51). We have recently developed a cosmid-based system for simple and efficient construction of minimal adenovirus vectors. As a stuffer for transgene insertion a noncoding genomic fragment from the human X chromosome was used (29). Minimal adenovirus vectors with stuffer elements derived from human chromosomal DNA differ from E1 deletion adenovirus vectors not only by the absence of viral genes but also by the presence of homologies to the genome of human target cells. It has been suspected that the latter might lead to a more frequent integration into the host cell chromosome by homologous or homology-related recombination mechanisms (56).
In nonpermissive cells, wild-type adenovirus persists as an episome in the nucleus and can occasionally integrate into the cellular genome via an as-yet-unknown mechanism. In permissive cells (productive infection), recombinations between viral and cellular DNA can also occur, as evidenced by the emergence of recombinant viruses that contain cellular DNA (for a review see reference 17). Adenoviruses encoding a temperature-sensitive DNA-binding protein yield stable clones at the nonpermissive temperature (21). E1 deletion adenovirus vectors carrying selectable genes were shown to integrate at frequencies of ∼10−3 to 10−5 into cultured mammalian cells (27, 32, 62). The integration of E1 deletion adenovirus vectors was found to primarily occur as monomers with no or little loss of sequences at the vector end, apparently at random sites (27, 31, 61, 62). Occasionally, multiple copies of vector DNA arranged in tandem arrays were also observed (27, 62). These integrative properties have been exploited to generate cell lines that stably express nonselectable transgenes (31) or to produce transgenic mice by adenovirus-mediated gene transfer into fertilized eggs (61). Furthermore, targeted correction of mutant neor genes with E1+ or ΔE1 adenovirus vectors carrying an intact gene has also been reported (22, 41, 66).
The influence on integration behavior of extensive homologies shared by adenovirus vectors and the target cell genome has not been studied before. In the present study, the integration pattern of a helper-dependent minimal adenovirus vector with a neor gene inserted centrally into a 27.4-kb continous noncoding genomic stuffer derived from the human X chromosome was investigated. We were interested to evaluate whether the incorporation of genomic stuffer elements could lead to an enhanced risk of insertional mutagenesis. Furthermore, we wanted to study the potential of minimal adenovirus vectors as tools for gene targeting strategies, as the high capacity of these vectors allows the insertion of long stretches of homologous sequences. Finally, we also wanted to evaluate the potential of minimal adenovirus vectors as tools for stable transfer of large genes or complex genetic elements. In order to clarify whether extensive homologies lead to an enhanced integration rate and/or an altered integration pattern, the efficiency of the minimal adenovirus vector in stably transducing the neor gene after infection of a sex chromosome aneuploid (X0) human glioblastoma cell line was compared to that of an E1 deletion adenovirus vector and the integrated minimal adenovirus vector sequences were characterized.
MATERIALS AND METHODS
Cells and viruses.
The E1-transformed human embryonic kidney cell line 293 (24), the human hepatoma cell line Huh7 (45), and the human glioblastoma cell line U87-MG (54) were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL Life Technologies, Paisley, Scotland, United Kingdom) supplemented with 2 mM glutamine (Sigma, Deisenhofen, Germany), antibiotics (penicillin and streptomycin), and 10% fetal calf serum (Roche Diagnostics, Mannheim, Germany). The cultures were incubated at 37°C in a humidified atmosphere with 5% CO2. The helper-dependent minimal adenovirus MVX-lacZneo was constructed using a cosmid-based cloning system and was amplified with a ΔE1 helper virus containing a packaging signal flanked by loxP sites on a Cre recombinase-expressing packaging cell line derived from 293 cells as described in detail elsewhere (29). MVX-lacZneo was partially purified from helper virus by CsCl density gradient centrifugation. The final preparation contained 1.65 × 1011 viral particles/ml as determined by measurement of the optical density at 260 nm (37). Contamination with helper virus was ∼25% as judged by restriction analysis of DNA extracted from purified virus. AdΔE1neo and AdRSVβgal (59) are first-generation (ΔE1) adenovirus vectors that contain the same neor cassette as MVX-lacZneo or Rous sarcoma virus promoter-driven Escherichia coli lacZ expression cassette, respectively, inserted into the E1 region. AdΔE1neo was constructed using pΔE1sp1A (4) as a shuttle vector for the insertion into the E1 region and was rescued by cotransfection with pJM17 (38) onto 293 cells. The ΔE1 viruses were amplified on 293 cells and purified by CsCl banding as described elsewhere (30), and titers were determined by dilution endpoint assay on 293 cells. Contamination with replication-competent adenovirus was determined for MVX-lacZneo and AdΔE1neo as described previously (28) and was below 1/107 infectious particles (IP) for both viruses.
Comparative Southern blot-based titration of MVX-lacZneo.
Huh7 cells were infected with various multiplicities of infection (MOIs) of AdΔE1neo (ranging from 10 to 300 IP/cell as determined by dilution endpoint assay) and MVX-lacZneo (ranging from 300 to 9,000 VP/cell) for 1 h in phosphate-buffered saline (PBS)–1 mM MgCl2, thoroughly washed with PBS, and replenished with medium. Total DNA was extracted from the cells at 6 h postinfection and digested with NcoI, which releases an 894-bp fragment from the neor cassette contained in both viruses (see Fig. 1A). Southern blot analysis was performed using a radiolabeled probe specific for this fragment, and signal intensities were quantitated. This method was shown previously to detect only DNA from infectious virus that has been transported to the nucleus (6). Signal intensities generated with a given MOI of AdΔE1neo (3 × 109 IP/ml) correlated with signal intensities generated after infection with a nine-fold-higher number of viral particles/cell of MVX-lacZneo (1.65 × 1011 VP/ml), indicating a titer of 1.8 × 1010 IP/ml for the latter.
FIG. 1.
Structures and titration of AdΔE1neo and MVX-lacZneo. (A) Both viruses contain a neomycin resistance gene (neor) cassette controlled by the human CMV immediate early promotor and the bovine growth hormone polyadenylation signal (bGHpA). AdΔE1neo is a first-generation adenovirus vector that contains the neor cassette inserted into the ΔE1 region. Minimal adenovirus MVX-lacZneo has the neor cassette inserted centrally in a 27.4-kb noncoding genomic stuffer derived from the human X chromosome. Furthermore, the vector contains a lacZ expression cassette controlled by the Rous sarcoma virus promoter and the SV40 polyadenylation signal (SV40pA) at the 3′ end. Repeat elements in the X-chromosomal backbone are indicated (Table 1). The Ad5 ITRs and the packaging signal (Ψ) are also shown. (B) Verification of infectious titers as determined by a comparative Southern blot-based titration protocol. DNA extracted from Huh7 cells after infection with the viruses at the indicated MOIs was digested with NcoI, and the 894-bp fragment generated from the neor cassette was detected (PC, positive controls [purified NcoI fragment from the neor cassette]; NC, negative control [DNA from mock-infected Huh7 cells]).
Selection and analysis of G418 resistant clones.
U87-MG cells (106) in 25-cm2 cell culture flasks were infected with the viruses at the various MOIs in a total volume of 1 ml of PBS–1 mM MgCl for 1.5 h at room temperature, washed with PBS, and replenished with medium. At 2 days postinfection G418 (Gibco-BRL Life Technologies) was added at a final concentration of 250 μg/ml. Medium was changed twice weekly, and clones were allowed to grow for ∼5 weeks. For clone counts cells were stained with methylene blue. For characterization of MVX-lacZneo integration pattern a total of 200 clones were generated under conditions that allowed single clones to be isolated with a minimal risk of cross-contamination (infection with an MOI of 0.01 IP/cell, resulting in ∼10 to 20 clones per cell culture flask) and expanded individually under G418 selection. For PCR or Southern blot analyses, genomic DNA was extracted from one confluent 150-mm-diameter dish each. To analyze β-galactosidase activity, cells were plated into six-well plates and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) following standard protocols.
PCR analysis.
For each clone, 100 ng of genomic DNA were used for PCR with primers PNeo1H (5′-TTCTCACTGCTGCTGTCCTA-3′) and PNeo1R (5′-GGCAACTTGCTGGCACTGTA-3′), which bind 470 bp upstream or 272 bp downstream, respectively, of the SnaBI site at the X-chromosomal target region (see Fig. 4A), into which the neor cassette had been inserted in MVX-lacZneo (29). As controls for the unmodified target region DNA from original U87-MG cells and pMVX-lacZ, a cosmid that contains the unmodified X-chromosomal target region (29) was used. As a control for the modified target region pMVX-lacZneo was used, the cosmid vector from which minimal adenovirus MVX-lacZneo had been derived (29). The reaction was performed in a total volume of 25 μl with 35 cycles of 45 s at 94°C, 45 s at 55°C, and 40 s at 72°C in a PCT-100 thermocycler (MJ Research) using Platinum Taq (Gibco-BRL Life Technologies) with 2 mM MgCl2, 5% dimethyl sulfoxide, 0.2 mM deoxyribonucleotide triphosphates and primers at 1 μM each. As a number of unspecific bands arose from this first amplification, 1 μl of the product was used for a second (nested) PCR carried out under the same conditions to unequivocally visualize a specific product. Primers PNeo2H (5′-CCTCTGCCTTGACATGACCT-3′) and PNeo2R (5′-GGACAATGGAGATCAATGCC-3′) bind 281 bp upstream and 248 bp downstream of the SnaBI site, respectively, and give rise to a 529-bp product from the unmodified target region. The 2,063-bp product resulting from the modified target region with the neor cassette inserted into the SnaBI site was not amplified under these conditions. Therefore, the protocol served only to detect presence or absence of the unmodified target region. Analysis was done in duplicate in two seperate experiments for each clone.
FIG. 4.
Analyses for targeted insertion of MVX-lacZneo. (A) The neor cassette in MVX-lacZneo is flanked with 13.8 and 13.6 kb of sequences on the 5′ and 3′ sides respectively, which are homologous to the target site on the single X chromosome in U87-MG cells. Site-specific integration would lead to loss of the original structure of the target site, which would be detected by the absence of the 529-bp product generated by nested PCR over the SnaBI site, into which the neor cassette had been inserted (29), or by the absence of the 1,958-bp BglII/BamHI fragment hybridizing with the X probe. (B) Results of the nested PCR using genomic DNA from U87-MGneor clones 1 through 16 as a template. Cosmids pMVX-lacZ and pMVX-lacZneo, which contain the unmodified target region and the target region with an inserted neor cassette, respectively, and DNA from original U87MG cells were used as controls. Note that the 2,093-bp product characteristic of the modified target region (and of unspecific integration of MVX-lacZneo) was not generated under the PCR conditions used. (C) Results of the Southern blot analysis with genomic DNA from U87-MGneor clones 1 through 16. Besides integrated MVX-lacZneo sequences (MVX), the unmodified X-chromosomal target region (X) was present in all clones. BglII/BamHI-digested cosmid pMVX-lacZneo was used as a positive control (PC; one and three copies/cell); DNA from original U87-MG cells was used as a negative control (NC).
Southern blot analysis.
Digested DNAs (15 μg each) were seperated over 1% agarose gels and blotted onto Hybond-N+ membranes (Amersham). Radiolabeling of probes, hybridization, and washing were performed following standard procedures. An 894-bp NcoI fragment from the neor cassette (see Fig. 1A) was used as a probe for comparative Southern blot titration and for the detection of the neor cassette in U87-MGneor clones. A 473-bp XhoI fragment from pMVX-lacZneo (29) was used to detect X-chromosomal sequences as indicated in Fig. 4A (X probe). A 341-bp fragment containing bp 189 to 526 from the adenovirus 5′ end (Ψ probe) and a 415-bp fragment containing part of the simian virus 40 (SV40) polyadenylation signal (SV40pA probe) were used as probes for the detection of integrated minimal adenovirus 5′ and 3′ ends, respectively (see Fig. 5). Hybridizing bands were visualised by exposure to Kodak Biomax MR films, and band intensities were measured using a Fujix BAS 2000 phosphorimager (Fuji, Tokyo, Japan).
FIG. 5.
Southern blot analysis for the presence of the 3′ and 5′ ends of MVX-lacZneo in U87-MGneor clones 1 through 16. (A) Genomic DNA was digested with BglII and BamHI which cut at the indicated positions at the 3′ and 5′ ends of MVX-lacZneo, respectively (A). For hybridization, probes specific for the packaging signal at the 3′ end (Ψ probe) and the SV40 polyadenylation signal at the 5′ end of MVX-lacZneo (SV40pA probe) were used. (B and C) Results with the Ψ probe and the SV40pA probe, respectively. Fragments with similar sizes recognized by both probes which may indicate joined vector ends are marked by asterisks. Note that BglII/BamHI-digested pMVX-lacZneo, which was used as a positive control (PC; one and three copies/cell, respectively), generates fragments with sizes of 6,901 and 596 bp that hybridize with the Ψ probe and the SV40pA probe, respectively (arrows). DNA from original U87-MG cells was used as a negative control (NC).
Localization of repeat elements.
The X-chromosomal backbone in MVX-lacZneo was analyzed for the presence of chromosomal repeat elements using RepeatMasker version 5/99 programmed by A.F.A. Smit and P. Green at default settings with Repbase version 3.04 at http://repeatmasker.genome.washington.edu/cgi-bin.RM2_req.pl. and with BLAST2.0 search (2) using public sequence databases at http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST.
FISH.
Harvesting and chromosome preparation for molecular and cytogenetic analysis followed standard procedures. For X-chromosome-specific chromosome paint, fluorescence in situ hybridization (FISH) with an indirect labeled coatasome probe for chromosome X (Oncor) was used following the instructions of the manufacturer. The digoxigenin-labeled coatasome was detected by fluorescein isothiocyanate-conjugated antidigoxigenin antibody (Oncor). For the detection of integration sites of MVX-lacZneo, FISH with pMVX-lacZneo was performed. Probe DNA was labeled by nick translation with SpectrumOrange-dUTP (nick translation kit; VYSIS). Probe length after nick translation was 100 to 400 bp. To avoid background fluorescence, the slides were treated with RNase (stock solution: 20 mg of RNase A per ml, 10 mM Tris-HCl [pH 7.5], 15 mM NaCl) prior to hybridization. Chromosome preparations were equilibrated in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature and incubated with RNase (100 μg/ml) at 37°C for 1 h. After washing twice in 2× SSC at room temperature, slides were dehydrated through alcohol grades (70%, 85%, 2× absolute ethanol). Chromosomal DNA was denatured with 70% formamide–2× SSC at 70°C for 5 min and quickly transferred to ethanol grades. For each hybridization, 80 ng of labeled DNA, 2.5 μg of Cot-1 DNA, and glycogen were mixed and precipitated with sodium acetate (3 M) and isopropanol. Human highly repetitive DNA (Cot-1 DNA) was used to precipitate the small probe DNA, to block nonspecific sticking of probe DNA to cytoplasm, and to prevent repetitive sequences interspersed within the unique sequences of the probe from hybridizing to target DNA located all over in the human genome (53, 69). DNA was resuspended in 10 μl of hybridization mixture containing 50% formamide, 2× SSC, and 10% dextran sulfate, denatured at 73°C for 5 min, and hybridized to denatured target metaphase spreads. Slides were incubated at 37°C in a moist chamber overnight. Posthybridization washes were performed in 0.4× SSC–0.3% NP-40 at 73°C for 2 min. A second incubation was done in 2× SSC–0.1% NP-40 at room temperature for 30 s. Chromosomes were counterstained with 4,6-diamino-2-phenylindole (DAPI), and slides were mounted in Vectashield antifade solution (Vector). Hybridizations were analyzed using an epifluorescence microscope (Axioscope; Zeiss) equipped with a cooled charge coupled device camera (Hamamatsu). Image analysis was performed with an ISIS system (Metasystems, Altlussheim, Germany).
RESULTS
Integration frequency of MVX-lacZneo.
The helper-dependent minimal adenovirus vector MVX-lacZneo (29) contains the 5′-terminal 526 bp (5′ ITR and packaging signal), the 3′-terminal 157 bp (3′ ITR) of Ad5, and a human cytomegalovirus (CMV) immediate early promoter-driven neomycin resistance gene (neor) cassette centrally inserted into a 27.4-kb noncoding genomic stuffer derived from the human X chromosome. Furthermore, it contains an E. coli lacZ reporter gene cassette located at the 3′ end (Fig. 1A). As a model system to study chromosomal integration of MVX-lacZneo we chose U87-MG, a hypodiploid human glioblastoma cell line which is monosomic for the X chromosome (54). The presence of a single X chromosome in U87-MG cells was verified by X-chromosome-specific paint, and the presence of the homologous target region (the chromosomal fragment inserted into MVX-lacZneo) was verified by PCR (data not shown). Furthermore, U87-MG cells were found to be highly susceptible to adenovirus infection, as determined by staining of cells for β-galactosidase activity after infection with AdRSVβgal (∼60% or 95% transduced cells after infection with an MOI of 10 or 50 IP/cell, respectively).
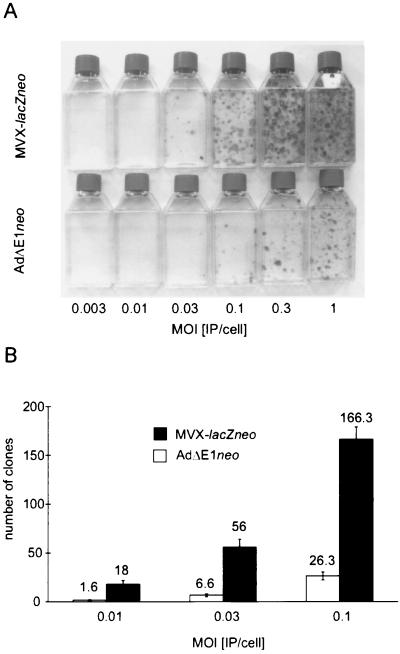
We were first interested to investigate integration frequencies of MVX-lacZneo compared to those of the first-generation adenovirus vector AdΔE1neo, which contains the same neor cassette as MVX-lacZneo inserted into the E1 region (Fig. 1A). In order to compare integration frequencies it was necessary to adjust infectious viral titers with respect to the efficiency for transducing DNA containing the neor cassette. To this end, a comparative Southern blot protocol was used, based on the quantification of transduced neor sequences after infection of cells with various MOIs of the viruses, as described in Materials and Methods. Titers were verified by repetition of the procedure with MOIs of 11, 33, 100, and 300 IP/cell for both viruses (Fig. 1B). To investigate integration frequencies, U87-MG cells were infected with the viruses at MOIs ranging from 0.003 to 1 IP/cell and selected for G418 resistance, which is conferred by the neor gene. No G418-resistant clones arose after mock infection or infection with AdRSVβgal in control experiments (data not shown). In contrast, G418-resistant clones (termed U87-MGneor clones herein) grew after infection with MVX-lacZneo and AdΔE1neo, indicating integration of the neor sequences. Clone numbers reflected the different MOIs used and were on average ∼10-fold higher with MVX-lacZneo (Fig. 2). Apparent integration frequencies were determined as the 100-fold ratio of the number of clones generated to the total number of infectious particles used for the infection and corresponded to ∼0.18% and ∼0.02% for MVX-lacZneo and AdΔE1neo, respectively.
FIG. 2.
Apparent chromosomal integration frequencies of AdΔE1neo and MVX-lacZneo. U87-MG cells (106) were infected with the viruses at the MOIs indicated and selected with G418. After 5 weeks, clones were stained and counted. Each experiment was done in triplicate. Representative results are shown (A). The average number of clones per 106 cells generated after infection with the viruses at MOIs of 0.01, 0.03, and 0.1 is also shown (B). Bars represent standard deviations.
Enhanced apparent integration frequency does not correlate with repeat-mediated integration of MVX-lacZneo sequences.
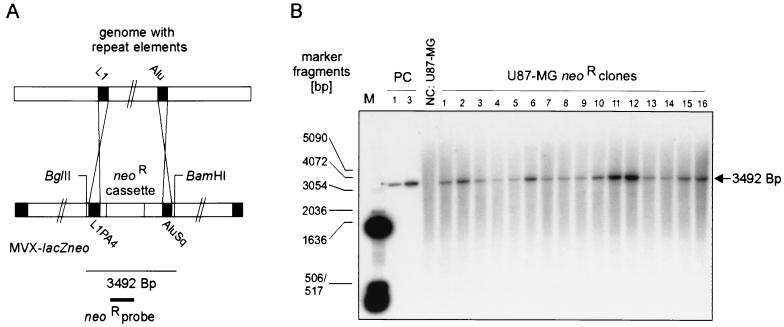
We first suspected that the enhanced apparent integration frequency of MVX-lacZneo might reflect a higher rate of chromosomal integration mediated by the homologies shared by the vector and the host cell genome. Alu and L1 elements constitute the majority of short and long interspersed repetitive elements (SINEs and LINEs), respectively, and account for approximately 5% of the human genome each (recently reviewed in references 39 and 48). Thus, a high number of potential targets might lead to integration by homology-related mechanisms, if the vector contains repeat elements. Therefore, the X-chromosomal backbone in MVX-lacZneo was screened for repeat elements that may contribute to its integration behavior (Table 1). Four complete Alu elements and two 5′-truncated L1 elements were found. Importantly, the neor cassette in MVX-lacZneo was found to be flanked by an L1 and an Alu repeat element. Integration of the neor cassette in MVX-lacZneo by homology-like recombination between the flanking L1 and/or Alu element and their abundant counterparts in the genome of U87-MG cells would lead to a loss of vector sequences, which are located upstream of the L1 and/or downstream of the AluSq element, respectively. This could be monitored by the presence or absence of the restriction sites for BglII and BamHI, which flank the L1 and the AluSq element in MVX-lacZneo, respectively (Fig. 3A). We therefore performed Southern blot analysis of BamHI/BglII-digested DNA from 16 U87-MGneor clones using a probe specific for the neor open reading frame (ORF) (Fig. 3B). In no case was loss of these sites observed. Instead, a fragment corresponding to the original fragment generated from MVX-lacZneo was detected in all clones. It was present at different copy numbers, as indicated by various signal intensities. Thus, the integration of the neor cassette had in no case been mediated by homology-related recombination events via the L1 or AluSq element.
TABLE 1.
Repeat elements in MVX-lacZneoa
| Positionb (bp) in MVX-lacZneo | Length (bp) | Repeat element | Repeat class and family | Matching position (bp) in consensusc | % bp diversity |
|---|---|---|---|---|---|
| 1077–3492 (+) | 2,415 | Tigger1 | DNA, MER2 type | 4–2418 (0) | 15.4 |
| 10135–11376 (C) | 1,241 | L2 | LINE, L2 | (955) 2358–931 | 27.4 |
| 13636–13861 (+) | 225 | L1PA4 | LINE, L1 | 5893–6122 (24) | 2.5 |
| 16345–16641 (+) | 296 | AluSq | SINE, Alu | 1–312 (1) | 8.4 |
| 18031–18352 (+) | 321 | LIMB1 | LINE, L1 | 5840–6159 (11) | 21.4 |
| 18463–18668 (+) | 205 | AluSp | SINE, Alu | 1–207 (106) | 9.7 |
| 19240–19521 (C) | 281 | AluY | SINE, Alu | (28) 283–1 | 7.1 |
| 20343–20588 (+) | 245 | MLT2D | LTR, retroviral | 2–256 (297) | 21.9 |
| 20589–20883 (C) | 294 | AluSg | SINE, Alu | (14) 296–1 | 10.8 |
| 20884–21206 (+) | 322 | MLT2D | LTR, retroviral | 256–548 (5) | 21.9 |
| 23795–24323 (C) | 528 | MLT2B | LTR, retroviral | (40) 408–1 | 18.6 |
| 25904–26962 (+) | 1,058 | MER11C | LTR, retroviral | 1–1036 (35) | 8.7 |
| 27879–29028 (+) | 1,149 | L1PA3 | LINE, L1 | 4999–6145 (1) | 1.7 |
Repeat elements were detected by analysis of the MVX-lacZneo sequence by RepeatMasker version 5/99 with Repbase version 3.04 with default settings. Lower-complexity sequences and simple repeats (total, 1.9% of the sequence) and repeat elements with a Smith-Waterman score of <1,000 are not included. Repeat elements with >200-bp lengths and <20% bp diversity from consensus are in bold. LTR, long terminal repeat.
“+” and “C” (complementary strand) indicate the orientation of the repeat in MVX-lacZneo.
Numbers in parentheses show the number of base pairs missing to the 3′ end of the consensus.
FIG. 3.
Southern blot analysis for the involvement of L1 and AluSq elements in MVX-lacZneo integration. (A) The integration of MVX-lacZneo via homology-mediated recombination involving the L1 and/or the AluSq element flanking the neor cassette and their abundant cellular counterparts would lead to loss of the BgIII and/or BamHI site flanking these elements, resulting in BglII/BamHI fragments containing neor sequences with sizes different from the 3,492 bp generated from the complete vector. (B) Genomic DNA from U87MGneor clones 1 through 16 was digested with BglII and BamHI and hybridized with the 894-bp NcoI fragment (Fig. 1A) to detect neor sequences. As positive controls (PC), BglII/BamHI-digested pMVX-lacZneo corresponding to one and three copies/cell was used. DNA from original U87-MG cells was used as a negative control (NC).
Lack of homologous targeting of MVX-lacZneo sequences to the X-chromosomal target site.
The previous results could have been indicative of (i) unspecific integration of a large proportion of the vector or (ii) targeted insertion into the X chromosome via a double-crossover event. The neor cassette in MVX-lacZneo is flanked by 13.8 and 13.6 kb of sequences homologous to the target region in the X chromosome (Fig. 4A). Gene targeting is most efficient using linearized replacement type vectors which contain the transgene flanked on both sides with sequences that are homologous the target region (for a recent review see reference 70). Furthermore, the frequency of targeted insertion by a double-crossover event depends on the length of the flanking homologous sequences and was shown to be saturated at a length of 14 kb (14). Thus, MVX-lacZneo would represent a replacement-type vector for gene targeting with optimal length of flanking homologous DNA sequences. We therefore wanted to analyze whether the higher apparent integration frequency of MVX-lacZneo might be correlated with targeted insertion into the homologous target region in the single X chromosome of U87-MG cells. To this end, genomic DNA from U87-MGneor clones 1 through 16 was analyzed by PCR over the X-chromosomal target site (Fig. 4B). The characteristic product for the unmodified X-chromosomal target region was present in all clones, indicating that a homologous targeting to the X-chromosomal site had not occurred. These results were verified by Southern blot analysis using BamHI/BglII-digested DNA from U87-MG clones 1 through 16 and a probe specific for the X-chromosomal sequence between the neor cassette and the AluSq element. Two hybridizing bands were detected in all clones (Fig. 4C): the signal from the integrated MVX-lacZneo sequences and a signal from the unmodified target site on the X chromosome. It was thus clear that homologous targeting was not the reason for the enhanced apparent integration frequency of MVX-lacZneo. However, we wanted to further examine the potential of minimal adenovirus vectors to target a specific chromosomal site. Therefore, another 184 U87-MGneor clones were analyzed by PCR over the X-chromosomal target site. Again, with all clones the product characteristic of the unmodified target site was detected (data not shown). Thus, despite the long homologous stretches on both sides of the neor cassette in MVX-lacZneo, no homologous targeting event could be detected in a total of 200 clones tested.
MVX-lacZneo integrates completely, with no or little loss of sequences at the vector ends.
In the previous experiments no evidence for the involvement of homologous or homology-mediated mechanisms involving specific sequences of the X-chromosomal backbone in MVX-lacZneo integration was found. We therefore suspected that the enhanced number of clones obtained after infection with MVX-lacZneo compared to AdΔE1neo might be due not to different integration behavior but results from other mechanisms. We wanted to test whether MVX-lacZneo, like AE1 adenovirus vectors (27, 31, 61, 62), integrates at full length. To this end, Southern hybridization experiments were performed with probes for the Ad5 packaging signal (5′ end, Ψ probe) (Fig. 5B) and the SV40 poly(A) signal (3′ end, SV40pA probe) (Fig. 5C), using BamHI/BglII-digested DNA from U87-MGneor clones 1 through 16. In 8 of 16 clones (50%) both termini were detected (clones 2, 3, 4, 9, 10, 12, 15, and 16). Five of the remaining clones were positive only with the Ψ probe (clones 1, 6, 8, 11, and 13), two were positive only with the SV40pA probe (clones 5 and 7), and only in clone 14 was no signal detected with either probes. These data were confirmed by analysis for β-galactosidase activity of these clones and an additional 15 clones (total, 31). In 25 clones (80.6%), β-galactosidase activity was clearly detected, indicating integration of the lacZ cassette located at the 3′ end of MVX lacZneo. Table 2 summarizes the results for U87-MGneor clones 1 through 16. As expected, all clones that had been positive with the SV40pA probe in the Southern analysis expressed β-galactosidase. Interestingly, β-galactosidase activity was also detected in three of six clones that were negative with the SV40pA probe (clones 6, 13, and 14), indicating that in these clones, integration involved loss of 3′-terminal sequences after the lacZ ORF. Thus, in 13 of 16 clones, MVX-lacZneo obviously had integrated completely with no or little loss of sequences at both ends. The sizes of the terminal fragments detected by Southern blot varied, indicating insertion at different chromosomal sites. In two clones (clones 2 and 4) (Fig. 5B and C, lanes 6 and 8, respectively), a fragment of similar apparent size hybridized with both probes. This might indicate joining of vector ends prior to integration, especially in clone 4, where the size of the fragment (slightly above 2 kb) would be consistent with a direct head-to-tail junction (1,515 bp plus 596 bp from the 5′- and 3′-terminal fragments, respectively). There was a discrepancy between the signal intensities and the number of signals after hybridization with the Ψ probe and the SV40pA probe. With the SV40pA probe, only one signal was detected in positive clones. Furthermore, signal intensities were very similar and may correspond to ∼1 copy per genome as determined by comparison with the positive controls. With the Ψ probe, signal intensities strongly varied. Intense signals correlated with a high copy number of integrated neor cassettes (Fig. 4B), especially in clones 11 and 12 Fig. 4B and 5B, lanes 15 and 16, respectively). Furthermore, more than one band hybridizing with the Ψ probe was present in five clones (clones 2, 6, 11, 12, and 16). Thus, although both ends of MVX-lacZneo were detected in most clones, sequences from the 3′ end were underrepresented in many clones compared to sequences from the 5′ end and the neor cassette.
TABLE 2.
β-Galactosidase activity of U87-MGneor clones 1 through 16
| Hybridizing probe(s)a | No. of clones | No. of clones with β-galactosidase activityb
|
|||
|---|---|---|---|---|---|
| − | + | ++ | +++ | ||
| None | 1 | 1 | |||
| Ψ | 5 | 3 | 1 | 1 | |
| SV40pA | 2 | 1 | 1 | ||
| Ψ + SV40pA | 8 | 2 | 3 | 3 | |
As determined by Southern blot analysis (Fig. 5). Ψ, probe for Ad5 packaging signal (5′ end of MVX-lacZneo).
β-Galactosidase activity was determined by the percentage of cells turning blue after X-Gal staining (−, 0%; +, <10%; ++, 10 to 50%; +++, 50 to 100%).
Integrated MVX-lacZneo sequences are located at a single chromosomal site.
FISH analyses with spread metaphase chromosomes using the complete MVX-lacZneo sequence as a probe were performed with three U87-MGneor clones (clones 2, 11, and 15) that exhibited overrepresentation of sequences from the 5′ terminus and the neor cassette and/or more than one specific fragment detected with the Ψ probe. Several metaphases analyzed for each clone gave identical signal patterns, confirming clonality of the cell populations. Representative results are shown in Fig. 6. As expected, in all clones a signal was detected at the distal part of the q arm of the X chromosome, the region from which the stuffer sequences in MVX-lacZneo were derived. Only one additional signal representing the integration site of MVX-lacZneo sequences was detected for each clone. The integration site was located on different chromosomes as judged by chromosome morphology. Interestingly, the signal resulting from integrated MVX-lacZneo sequences had an intensity similar to that of the signal at Xq in clones 2 and 15, whereas in clone 11 it was clearly severalfold more intense. Thus, signal intensities corresponded to the estimated number of integrated neor sequences and multiple copies of MVX-lacZneo sequences in clone 11 colocalized at a single integration site.
FIG. 6.

FISH analysis for integrated MVX-lacZneo sequences. pMVX-lacZneo, which contains the genome of MVX-lacZneo, was labeled with SpectrumOrange-dUTP and used as a probe to detect integrated MVX-lacZneo sequences on spread metaphase chromosomes of U87MG-neor clones 2, 11, and 15. For each clone two signals were detected. X, X chromosome with the probe hybridizing to the distal part of the q arm, the region from where the X-chromosomal backbone in the vector was derived; MVX, integration site of MVX-lacZneo sequences.
DISCUSSION
MVX-lacZneo and U87-MG cells were particularly suited to analysis of the influence of homologies shared by minimal adenovirus vectors and the target cell genome on vector integration pattern for the following reasons: (i) the neor cassette in the vector was inserted centrally into the 27.4-kb stuffer derived from human noncoding genomic DNA from chromosome X, resulting in ∼14 kb of sequences on both sides of the marker which are homologous to the target site on human chromosome X; (ii) the neor cassette in the vector was furthermore flanked by an L1 and an Alu repeat element, which have more than 105 related sequences each in the human genome; and (iii) U87-MG cells are monosomic for the X chromosome (54), which facilitates the detection of targeting events. However, no homology-mediated integration events of MVX-lacZneo sequences could be detected. Instead, our data indicate that the minimal adenovirus vector had integrated completely, with no or little loss of sequences at the vector ends. The sizes of fragments hybridizing with probes for the vector termini were different, indicating insertion at different chromosomal sites. The pattern of hybridizing fragments was consistent with a head-to-tail junction for 2 of 16 clones.
Thus, the integration pattern of MVX-lacZneo resembles that of E1 deletion adenovirus vectors carrying a selectable gene, which in a number of previous studies was shown to involve in most cases the insertion of a single viral genome into the host cell genome with no or little loss of sequences at the vector ends, apparently at random sites (27, 31, 61, 62). Occasionally, multiple copies of vector DNA arranged in tandem arrays have been found (27, 62), which is also a common phenomenon with wild-type Ad12 integration into nonpermissive hamster cells (15). Multimeric insertion of viral DNA with linkage of the left- and right-hand ends of the viral genomes, either by direct linkage of the viral ends or with short stretches of intervening cellular DNA, has been observed upon integration of Ad2 and Ad5 (63, 64, 65). The organization of the integrated MVX-lacZneo sequences clearly indicates that the homologies of the X-chromosomal backbone of the vector to the host cell genome were not the cause of the enhanced number of G418-resistant clones generated after infection with this vector, compared to the E1 deletion vector AdΔE1neo, which shared no homologies with the target cell genome. From the structure of the integrated MVX-lacZneo sequences and considering the low MOI (0.01 IP/cell) used for the infections, we speculate that the typical initial event was the unspecific integration of a single complete linear monomer. Given the low MOI, it is unlikely that the possible presence of a head-to-tail junction of the vector termini in 2 of 16 clones was indicative of a concatemeric insertion of more than one vector genome. Instead, in these clones, circularization of the vector genome prior to integration might have occurred, since wild-type adenovirus does circularize, at low efficiency, during productive infection (55), and similar events might also have occurred to some extent after transduction with the minimal vector. Lack of viral coding genes in the minimal vector might have played a role in this process, as the E4 region was shown to antagonize joining of viral ends (68).
The reason for the enhanced apparent integration frequency of the minimal vector remains speculative. Different integration frequencies could, in theory, be explained by loss of terminal vectors sequences during the integration event, as the neor cassette was located at the 5′ end in the E1 deletion vector, whereas it was located centrally in the minimal vector. As mentioned above, however, loss of larger parts of the vector termini is obviously a rare event during integration of E1 deletion or minimal adenovirus vectors. Furthermore, in 80% of the neor clones generated after infection with the minimal vector, the lacZ expression cassette, which is located at the 3′ terminus of the minimal vector and which was not selected for, was present (Table 2). Thus, whereas loss of terminal sequences could explain minor differences in the integration frequency, it cannot account for a 10-fold-reduced integration frequency of the E1 deletion vector compared to the minimal vector. Interestingly, our results are very similar to those obtained by Harui et al. (27) with a minimal adenovirus vector containing a selectable β-geo gene, which in various mammalian cells exhibited an up-to-50-fold-higher integration frequency than a ΔE1 adenovirus vector with the same expression cassette inserted into the E1 region, reaching more than 1% integration frequency in CHO cells. Importantly, the β-geo expression cassette was located at the vector ends in both vectors. The minimal vector furthermore differed from the one in the present study in that it contained a stuffer which was composed entirely of repeated bacterial sequences from plasmid pBR and not of human chromosomal DNA. The similarity of their results and ours therefore adds further proof for our conclusions that (i) loss of terminal sequences during the integration process cannot be the reason for the reduced number of neor clones generated after infection with the E1 deletion vector and (ii) homologies between the vector and the host cell genome (i.e., chromosomal stuffer elements) per se do not influence the integration pattern of minimal adenovirus vectors.
The enhanced efficiency of minimal adenovirus vectors to stably transduce selectable genes might have been mediated by the absence of viral genes. These are expressed at low levels from first-generation adenovirus vectors (11, 71, 72, 73) and may inhibit normal cellular machinery upon vector integration. Transient adenovirus gene expression might also have occurred in some cells after infection with the minimal vector, due to the contamination with the E1 deletion helper virus. However, nonintegrated helper virus genomes will be diluted out after some cell divisions. Furthermore, integration of both helper virus and minimal virus into the genome of a given cell can practically be excluded, considering the low MOIs used and the integration rates of the E1 deletion and minimal vectors determined in our study. The reduced number of stably transduced clones after infection of first-generation adenovirus vectors might therefore reflect not a reduced integration frequency but rather the need for a balanced expression of the adenovirus genes and the selectable gene. However, other factors may also contribute. Adenovirus E4orf3 and E4orf6 can act to inhibit recombination and double strand break (DSB) repair under replicative conditions (5, 46). These viral genes are present in the E1 deletion vector but not in the minimal vector. Thus, although U87-MG cells do not allow replication of the E1 deletion vector, it cannot be excluded that specific viral functions contribute to reduce the integration frequency of the E1 deletion vector.
These conclusions imply that the incorporation into minimal adenovirus vectors of stuffer elements derived from noncoding human genomic DNA most probably does not lead to an enhanced risk of insertional mutagenesis by homologous or homology-related recombination events. This is highly relevant for the design of minimal adenovirus vectors as tools for in vivo gene transfer, as minimal adenovirus vectors with stuffer elements composed of human chromosomal DNA were shown to allow long-term gene expression in vivo in a number of studies (8, 42, 44, 51, 56, 57), whereas the incorporation of a stuffer element derived from phage lambda led to rapid loss of transduced cells by a stuffer-directed immune response (51). This phenomenon was suspected to be related to the presence of immunostimulatory CpG elements (reviewed in reference 34) in bacterial and phage DNA and the suppression of these elements in mammalian genomic DNA.
Although much work has been dedicated to the issue, no specific sites or mechanisms for adenovirus integration have been found (16, 19, 23). However, a few base pairs of homology between the vector and the host cell chromosome were occasionally present at the integration site (16, 19, 23). This is very similar to the products generated by nonhomologous end joining (NHEJ), which in mammalian cells is the most prominent repair mechanism for DNA DSBs and was recently found to be also involved in retrovirus DNA integration (12). NHEJ results in ligation of nonhomologous DNA ends without nucleotide loss or generates products in which only a few nucleotides are deleted where ligation has taken place at short regions of microhomology between the two recombining ends (for a recent review see reference 18). It is therefore tempting to speculate that adenovirus vectors are recognized as substrates for NHEJ, leading to integration of the complete vector at sites of DSBs, which arise by errors in DNA metabolism (36). Interestingly, the integration frequency of adenovirus vectors was shown to dramatically increase upon treatment of cells with ionizing radiation, which induces DSBs (75). In this context it should also be mentioned that in U87-MGneor clone 11, the MVX-lacZneo sequences were located in a deviative chromosome which had resulted from an interchromosomal translocation, with the insertion site being the junction site (data not shown).
It is widely assumed that adenovirus vectors integrate at low frequency, which is regarded as an advantage for in vivo gene transfer because it minimizes the risk of insertional mutagenesis of cancer-related genes. Our study and previous findings (27, 31, 62), however, indicate that the ability of adenovirus vectors to integrate into host cell chromosomes is actually high compared to that of naked plasmid DNA, although the end is protected by the terminal protein. The apparent integration frequency of AdΔE1neo in U87-MG cells (0.02%) was in good agreement with earlier studies, which involved E1 deletion adenovirus vectors carrying a neor gene and various mammalian cell lines and were performed under very similar experimental conditions (27, 62). As discussed above, the severalfold-enhanced apparent integration frequency of the minimal adenovirus vector possibly gives a better estimate of the integration frequency of adenovirus vectors in general, which might therefore be higher than suspected so far. Furthermore, even this integration frequency may be an underestimate, since under the conditions used not every infectious particle present actually infects a target cell (47) and not every integration event would be expected to lead to the generation of stably transduced clone, as local effects at the integration site (e.g., silencers or higher-order silencing chromatin structures) may repress sufficient marker gene expression in some cases. In contrast to wild-type Ad12, the integration of which in hamster cells has been extensively studied as a model system for in vivo oncogenesis (17), the integration of recombinant adenovirus vectors in vivo is poorly investigated. There are reports that suggest that adenovirus vectors do integrate after in vivo gene transfer into animals (7, 49), although no transmission into the germ line was observed in mice after administration of a ΔE1/ΔE4 adenovirus (74). Considering the high viral doses that are usually envisaged for gene therapy applications, the risk of insertional mutagenesis after in vivo gene transfer by adenoviral vectors is an essentially unsolved issue that will need direct examination.
The high efficiency of adenovirus vector integration at random sites is probably sufficient to explain the fact that in none of 200 clones tested could a targeted insertion of MVX-lacZneo sequences into the X-chromosomal target site via homologous recombination be detected in the present study. This finding is consistent with the notion that homologous targeting with viral or plasmid vectors is generally a rare event in mammalian cells, with an average ratio of random to targeted insertion of around 1,000:1 (for a recent review, see reference 36). Thus, the potential of minimal adenovirus vectors for gene targeting strategies seems to be limited, although they can accommodate long stretches of homologous sequences. Interestingly, the limited targeting potential of most viral and nonviral vector systems including the minimal adenovirus vector in the present study is in striking contrast to the observation that E1+ and ΔE1 adenovirus vectors carrying an intact neor gene are capable of correcting a mutant neor gene inserted into a host cell's chromosome with an extraordinarily high ratio (up to 40%) of gene targeting versus random integration of the complete vector (22, 41, 66). The high targeting rates obtained with these vectors may be explained by the aforementioned assumption that the integration of adenovirus coding regions is not tolerated by the host cell in most cases. If this was true, first generation adenovirus vectors, unlike minimal adenovirus vectors, would resemble replacement-type vectors for positive and negative selection that promote homologous targeting because it leads to loss of the adenovirus coding sequences. This assumption would also be in agreement with the finding that the absolute targeting frequencies of the E1+ and ΔE1 vectors were not enhanced compared to those of other vector systems (22, 41, 66). Possibly, the targeting rates of minimal adenovirus vectors could be improved to some minor extent by incorporation of flanking homologous sequences which were isolated from the cells to be infected (i.e., which are isogenic), as targeting rates can be reduced up to fivefold if sequence polymorphisms are present (14). This approach, however, does not seem very promising, as it would require the laborious isolation and insertion into the vector of isogenic sequences for each individual cell line to be infected.
Watson and coworkers have shown that linear plasmid-derived replacement vectors containing a neor cassette flanked by two complete L1 repeats or by one complete L1 repeat on one side and an Alu repeat on the other efficiently integrate via repeat-mediated homology-like recombination with chromosomal counterparts of these repeat elements in up to 20% of the clones (67). The difference between their results and ours might be related to the different sizes of the L1 elements: the L1 repeat before the neor cassette in MVX-lacZneo was severely 5′ truncated (total of 225 bp, matching bp 5893 to 6122 of the 6,146-bp consensus) (Table 1). As most genomic L1 elements are also 5′ truncated, retaining only the 100 to 1,000 3′ terminal bp of the 6-kb full-length element (48), this did not reduce the high number of related sequences in the genome. However, the L1 element (and the 296-bp Alu element at the other side of the neor cassette) might have been too short to promote homology-like recombination at a significant rate. Interestingly, in the study by Watson and coworkers (67), a vector carrying the neor gene flanked by two repeats of the Alu family (< 300 bp in length) did not exhibit repeat-mediated integration. Their and our data are therefore in agreement with the assumption that ∼300 bp of imperfect homology does not suffice to mediate homologous recombination of targeting vectors in mammalian cells (58, 60).
During clonal growth under selection, rearrangements of integrated MVX-lacZneo sequences seem to have occurred in some clones that led to amplification of the neor gene, as visualized by the band intensities in Fig. 4B. Growth under G418 selection could have been the driving force for these rearrangements despite the neor gene being driven by the strong CMV promoter, because its activity could have been reduced to various extents by silencing cis-active elements at the integration sites or by trans-acting silencing mechanisms (10). Interestingly, sequences from the 5′ end of the vector were also overrepresented in most clones containing multiple copies of the neo gene, whereas sequences from the 3′ end of the vector were underrepresented in these clones, being either absent or present only at ∼1 copy/cell. This phenomenon was most pronounced in U87-MGneor clone 11, where multiple copies of the neor gene and the packaging signal (5′ end) were present but no sequences from the 3′ end of the vector were detected. These multiple sequences all colocalized to a single chromosomal site (Fig. 6). Interestingly, the density of chromosomal repeat elements in the backbone of MVX-lacZneo was significantly higher downstream of the neor cassette, with 11 of total 15 elements and namely all four Alu elements being located in this region (Table 1). Alu elements occur at an average of one per 3 to 6 kb in the human genome but are completely absent in MVX-lacZneo upstream of the neor cassette. Thus, recombination between Alu elements located 5′ of the MVX-lacZneo integration site and the Alu elements in the 3′ terminal part of the vector which would result in amplification of the neor gene by unequal crossover events would, at the same time, amplify the complete 5′ sequences of pMVX-lacZneo, whereas sequences in the 3′ part of the vector would not be amplified. It therefore seems likely that growth under selection led to the amplification of the neor cassette by repeat-mediated intrachromosomal rearragements. In fact, chromosomal rearrangements due to recombination between Alu elements represent a major source of genome plasticity and have been related to a variety of deseases in humans (reviewed in reference 13). Similarly, rearrangements of integrated minimal adenovirus vector sequences were also reported by Harui and colleagues (27) and were attributed to the repetitive nature of the backbone in their vector.
These conclusions have important implications for the potential use of minimal adenovirus vectors as tools for the generation of stable cell lines and transgenic animals, which is a promising strategy in cases where transduction of the cells by conventional transfection techniques is too inefficient. Minimal adenovirus vectors would be particularly interesting for these purposes for a number of reasons: (i) as adenovirus vectors, they infect a large variety of cell types at high efficiency; (ii) they obviously integrate at high efficiency, primarily as one complete monomer per cell; (iii) the high capacity of these vectors could enable stable transfer of genes together with complex regulatory elements to achieve regulated or tissue-specific expression; and (iv) adverse effects mediated by residual adenovirus gene expression are excluded. From our data we conclude that in the design of vectors for these purposes it will be important to exclude chromosomal repeat elements or internally redundant sequences from the vector in order to maintain vector structure upon integration.
ACKNOWLEDGMENTS
We acknowledge the excellent technical assistance of Heidrun Peter and Uta Fischer with the cell culture work and of Antje Gerlach with the FISH experiments. We thank David Bauer for help with the computer analyses and Frank Schnieders for helpful discussions. Finally, we thank Gary S. Jennings, Peter Löser, and Christian Hofmann for critical reading of the manuscript.
REFERENCES
- 1.Alemany R, Dai Y, Lou Y C, Sethui E, Prokopenko E, Josephs S F, Zhang W-W. Complementation of helper-dependent adenoviral vectors: size effects and titer fluctuations. J Virol Methods. 1997;68:147–159. [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benihoud K, Yeh P, Perricaudet M. Adenovirus vectors for gene delivery. Curr Opin Biotechnol. 1999;10:440–447. doi: 10.1016/s0958-1669(99)00007-5. [DOI] [PubMed] [Google Scholar]
- 4.Bett A J, Haddara W, Prevec L, Graham F L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer J, Rohleder K, Ketner G. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology. 1999;263:307–312. doi: 10.1006/viro.1999.9866. [DOI] [PubMed] [Google Scholar]
- 6.Brand K, Klocke R, Possling A, Paul D, Strauss M. Induction of apoptosis and G2/M arrest by infection with replication-deficient adenovirus at high multiplicity of infection. Gene Ther. 1999;6:1054–1063. doi: 10.1038/sj.gt.3300914. [DOI] [PubMed] [Google Scholar]
- 7.Brown G R, Thiele D L, Silva M, Beutler B. Adenoviral vectors given intravenously to immunocompromised mice yield stable transduction of the colonic epithelium. Gastroenterology. 1997;112:1586–1594. doi: 10.1016/s0016-5085(97)70040-4. [DOI] [PubMed] [Google Scholar]
- 8.Chen H-H, Mack L M, Kelly R, Ontell M, Kochanek S, Clemens P R. Persistence in muscle of an adenoviral vector that lacks all viral genes. Proc Natl Acad Sci USA. 1997;94:1645–1650. doi: 10.1073/pnas.94.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H-H, Mack L M, Choi S-Y, Ontell M, Kochanek S, Clemens P R. DNA from both high-capacity and first-generation adenoviral vectors remain intact in skeletal muscle. Hum Gene Ther. 1999;10:365–373. doi: 10.1089/10430349950018814. [DOI] [PubMed] [Google Scholar]
- 10.Chen W Y, Bailey E C, McCune S L, Dong J-Y, Townes T M. Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc Natl Acad Sci USA. 1997;94:5798–5803. doi: 10.1073/pnas.94.11.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai Y, Schwarz E M, Gu D, Zhang W W, Sarvetnick N, Verma I M. Cellular and humoral responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long term expression. Proc Natl Acad Sci USA. 1995;92:1401–1405. doi: 10.1073/pnas.92.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel R, Katz R A, Skalka A M. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 13.Deininger P L, Batzer M A. Alu repeats and human desease. Mol Gen Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 14.Deng C, Capecchi M R. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doerfler W. Uptake, fixation, and expression of foreign DNA in mammalian cells: the organization of integrated adenovirus DNA sequences. Curr Top Microbiol Immunol. 1982;101:127–194. doi: 10.1007/978-3-642-68654-2_6. [DOI] [PubMed] [Google Scholar]
- 16.Doerfler W. The integration of foreign DNA into mammalian genomes and its consequences: a concept for oncogenesis. Adv Cancer Res. 1995;66:313–344. doi: 10.1016/s0065-230x(08)60259-6. [DOI] [PubMed] [Google Scholar]
- 17.Doerfler W. A new concept in (adenoviral) oncogenesis: integration of foreign DNA and its consequences. Biochim Biophys Acta. 1996;1288:F79–F99. doi: 10.1016/0304-419x(96)00024-8. [DOI] [PubMed] [Google Scholar]
- 18.Featherstone C, Jackson S P. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 19.Fechteler K, Tatzelt J, Huppertz S, Wilgenbus P, Doerfler W. The mechanism of adenoviral DNA integration: studies in a cell-free system. Curr Top Microbiol Immunol. 1995;199:109–137. doi: 10.1007/978-3-642-79499-5_5. [DOI] [PubMed] [Google Scholar]
- 20.Fisher K J, Choi H, Burda J, Chen S-J, Wilson J M. Recombinant adenovirus deleted for all viral genes for gene therapy of cystic fibrosis. Virology. 1996;217:11–22. doi: 10.1006/viro.1996.0088. [DOI] [PubMed] [Google Scholar]
- 21.Fisher P B, Babiss L E, Weinstein I B, Ginsberg H S. Analysis of type 5 adenovirus transformation with a cloned rat embryo cell line (CREF) Proc Natl Acad Sci USA. 1982;79:3527–3531. doi: 10.1073/pnas.79.11.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita A, Sakagami K, Kanagae Y, Saito I, Kobayashi I. Gene targeting with a replication-defective adenovirus vector. J Virol. 1995;69:6180–6190. doi: 10.1128/jvi.69.10.6180-6190.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gahlmann R, Leisten R, Vardimon L, Doerfler W. Patch homologies and the integration of adenovirus DNA in mammalian cells. EMBO J. 1982;1:1101–1104. doi: 10.1002/j.1460-2075.1982.tb01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from a human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 25.Haecker S E, Stedman H H, Balice-Gordon R J, Smith D B J, Greelish J P, Mitchell M A, Wells A, Sweeney H L, Wilson J M. In vivo expression of full-length human dystrophin from adenoviral vectors deleted of all viral genes. Hum Gene Ther. 1996;7:1907–1914. doi: 10.1089/hum.1996.7.15-1907. [DOI] [PubMed] [Google Scholar]
- 26.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phillips L M. Construction of adenovirus vectors through Cre/lox-recombination. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harui A, Suzuki S, Kochanek S, Mitani K. Frequency and stability of chromosomal integration of adenovirus vectors. J Virol. 1999;73:6141–6146. doi: 10.1128/jvi.73.7.6141-6146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hehir K M, Armetano D, Cardoza L M, Choquette T L, Berthelette P B, White G A, Couture L A, Everton M B, Keegan J, Martin J M, Pratt D A, Smith M P, Smith A E, Wadsworth S C. Molecular characterization of replication-competent variants of adenovirus vectors and genome modifications to prevent their occurrence. J Virol. 1996;70:8459–8467. doi: 10.1128/jvi.70.12.8459-8467.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillgenberg M, Schnieders F, Löser P, Strauss M. System for efficient helper-dependent minimal adenovirus construction and rescue. Hum Gene Ther. 2001;12:643–657. doi: 10.1089/104303401300057342. [DOI] [PubMed] [Google Scholar]
- 30.Kanagae Y, Makimura M, Saito I. A simple and efficient method for purification of infectious recombinant adenovirus. Jpn J Med Sci Biol. 1994;47:157–166. doi: 10.7883/yoken1952.47.157. [DOI] [PubMed] [Google Scholar]
- 31.Karlsson S, Van Doren K, Schweiger S G, Nienhuis A W, Gluzman Y. Stable gene transfer and tissue-specific expression of a human globin gene using adenoviral vectors. EMBO J. 1986;5:2377–2385. doi: 10.1002/j.1460-2075.1986.tb04507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlsson S, Humphries R K, Gluzman Y, Nienhuis A W. Transfer of genes into hematopoietic cells using recombinant DNA viruses. Proc Natl Acad Sci USA. 1985;82:158–162. doi: 10.1073/pnas.82.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kochanek S, Clemens P R, Mitani K, Chen H-H, Chan S, Caskey C T. A new adenoviral vector: replacement of all viral coding sequences with 28 kb of DNA independently expressing both full-length dystrophin and β-galactosidase. Proc Natl Acad Sci USA. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krieg A M, Yi A-K, Hartmann G. Mechanisms and therapeutic applications of immune stimulatory CpG DNA. Pharmacol Ther. 1999;84:113–120. doi: 10.1016/s0163-7258(99)00023-6. [DOI] [PubMed] [Google Scholar]
- 35.Kumar-Singh R, Chamberlain J S. Encapsidated adenovirus minichromosomes allow delivery and expression of a 14 kb dystrophin cDNA to muscle cells. Hum Mol Genet. 1996;5:913–921. doi: 10.1093/hmg/5.7.913. [DOI] [PubMed] [Google Scholar]
- 36.Lanzov V A. Gene targeting for gene therapy: prospects. Mol Genet Metab. 1999;68:276–282. doi: 10.1006/mgme.1999.2910. [DOI] [PubMed] [Google Scholar]
- 37.Maizel J V, White D O, Scharff M D. The polypeptides of adenovirus. I. Evidence for multiple components in the virion and a comparison of types 2, 7A, and 12. Virology. 1968;36:115–125. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- 38.McGrory W J, Bautista D S, Graham F L. A simple technique for the rescue of early region 1 mutations into infectious human adenovirus type 5. Virology. 1988;163:614–617. doi: 10.1016/0042-6822(88)90302-9. [DOI] [PubMed] [Google Scholar]
- 39.Mighell A J, Markham A F, Robinson P A. Alu sequences. FEBS Lett. 1997;417:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 40.Mitani K, Graham F L, Caskey C T, Kochanek S. Rescue, propagation and partial purification of a helper-dependent adenovirus vector. Proc Natl Acad Sci USA. 1995;92:3854–3858. doi: 10.1073/pnas.92.9.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitani K, Wakamiya M, Hasty P, Graham F L, Bradley A, Caskey C T. Gene targeting in mouse embryonic stem cells with an adenoviral vector. Somat Cell Mol Genet. 1995;21:221–231. doi: 10.1007/BF02255777. [DOI] [PubMed] [Google Scholar]
- 42.Morral N, Parks R J, Zhou H, Langston C, Schiedner G, Quitones J, Graham F L, Kochanek S, Beaudet A L. High doses of a helper-dependent adenoviral vector yield supraphysiological levels of α1-antitrypsin with negligible toxicity. Hum Gene Ther. 1998;9:2709–2716. doi: 10.1089/hum.1998.9.18-2709. [DOI] [PubMed] [Google Scholar]
- 43.Morral N, O'Neal W, Rice K, Leland M, Kaplan J, Piedra P A, Zhou H, Parks R J, Velji R, Aguilar-Cordova E, Wadsworth S, Graham F L, Kochanek S, Carey K D, Beaudet A L. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proc Natl Acad Sci USA. 1999;96:12816–12821. doi: 10.1073/pnas.96.22.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morsy M A, Gu M C, Motzel S, Zhao J, Lin J, Su Q, Allen H, Franklin L, Parks R J, Graham F L, Kochanek S, Bett A J, Caskey T C. An adenoviral vector deleted for all viral coding sequences results in enhanced safety and extended expression of a leptin transgene. Proc Natl Acad Sci USA. 1998;95:7866–7871. doi: 10.1073/pnas.95.14.7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakabayashi H, Taketa K, Miyano K, Yamane T, Saito J. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 46.Nicolas A L, Munz P L, Falck-Pedersen E, Young C S H. Creation and repair of specific DNA double-strand breaks in vivo following infection with adenovirus vectors expressing Saccharomyces cerevisiae HO endonuclease. Virology. 2000;266:211–224. doi: 10.1006/viro.1999.0062. [DOI] [PubMed] [Google Scholar]
- 47.Nyberg-Hoffman C, Shabram P, Li W, Giroux D, Aguilar-Cordova E. Sensitivity and reproducibility of adenoviral infectious titer determination. Nat Med. 1997;3:808–811. doi: 10.1038/nm0797-808. [DOI] [PubMed] [Google Scholar]
- 48.Okada N, Hamada M, Okiwara I, Ohshima K. SINEs and LINEs share common 3′ sequences: a review. Gene. 1997;205:229–243. doi: 10.1016/s0378-1119(97)00409-5. [DOI] [PubMed] [Google Scholar]
- 49.Overturf K, Al-Dhalimy M, Ou C N, Finnegold M, Tanguay R, Lieber A, Kay M, Grompe M. Adenovirus-mediated gene therapy in a mouse model of hereditary tyrosinemia type 1. Hum Gene Ther. 1997;8:513–521. doi: 10.1089/hum.1997.8.5-513. [DOI] [PubMed] [Google Scholar]
- 50.Parks R J, Graham F L. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J Virol. 1997;71:3293–3298. doi: 10.1128/jvi.71.4.3293-3298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parks R J, Bramson J L, Wan Y, Addison C L, Graham F L. Effects of stuffer DNA on transgene expression from helper-dependent adenovirus vectors. J Virol. 1999;73:8027–8034. doi: 10.1128/jvi.73.10.8027-8034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parks R J, Chen L, Anton M, Sankar U, Rudnicki M A, Graham F L. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J. Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci USA. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ponten J, Macintyre E H. Long term culture of normal and neoplastic human glia. Acta Pathol Microbiol Scand. 1968;74:465–486. doi: 10.1111/j.1699-0463.1968.tb03502.x. [DOI] [PubMed] [Google Scholar]
- 55.Ruben M, Bacchetti S, Graham F. Covalently closed circles of adenovirus 5 DNA. Nature. 1983;301:172–174. doi: 10.1038/301172a0. [DOI] [PubMed] [Google Scholar]
- 56.Sandig V, Youil R, Bett A J, Franlin L L, Oshima M, Maione D, Wang F, Metzker K L, Savino R, Caskey T C. Optimization of the helper-dependent adenovirus system for production and potency in vivo. Proc Natl Acad Sci USA. 2000;97:1002–1007. doi: 10.1073/pnas.97.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schiedner G, Morral N, Parks R J, Wu Y, Koopmans S C, Langston C, Graham F L, Beaudet A, Kochanek S. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxixity. Nat Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 58.Shen M R, Deininger P L. An in vivo assay for measuring the recombination potential between DNA sequences in mammalian cells. Anal Biochem. 1992;205:83–89. doi: 10.1016/0003-2697(92)90582-r. [DOI] [PubMed] [Google Scholar]
- 59.Stratford-Perricaudet L D, Makeh I, Perricaudet M, Briand P. Widespread long-term gene transfer to mouse skeletal muscles and heart. J Clin Investig. 1992;90:626–630. doi: 10.1172/JCI115902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas K R, Deng C, Capecchi M R. High-fidelity gene targeting in embryonic stem cells by using sequence replacement vectors. Mol Cell Biol. 1992;12:2919–2923. doi: 10.1128/mcb.12.7.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsukui T, Kanagae Y, Saito I, Toyoda Y. Transgenesis by adenovirus-mediated gene transfer into zona-free eggs. Nat Biotechnol. 1996;14:982–985. doi: 10.1038/nbt0896-982. [DOI] [PubMed] [Google Scholar]
- 62.Van Doren K, Hanahan D, Gluzman Y. Infection of eucaryotic cells by helper-independent recombinant adenoviruses: early region 1 is not obligatory for integration of viral DNA. J Virol. 1984;50:606–614. doi: 10.1128/jvi.50.2.606-614.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Visser L, Reemet A C, van Mansfeld A D, Rozijn T H. Nucleotide sequence analysis of the linked left and right hand terminal regions of adenovirus type 5 DNA present in the transformed rat cell line 5RK20. Nucleic Acids Res. 1982;10:2189–2198. doi: 10.1093/nar/10.7.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Visser L, Wassenaar A T, van Maarschalkerweerd M W, Rozijn T H. Arrangement of integrated viral DNA sequences in cells transformed by adenovirus types 2 and 5. J Virol. 1981;39:684–693. doi: 10.1128/jvi.39.3.684-693.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Visser L, van Maarschalkerweerd M W, Rozijn T H, Wassenaar A T, Reemet A C, Sussenbach J S. Viral DNA sequences in adenovirus transformed cells. Cold Spring Harbor Symp Quant Biol. 1979;44:541–550. doi: 10.1101/sqb.1980.044.01.056. [DOI] [PubMed] [Google Scholar]
- 66.Wang Q, Taylor M W. Correction of a deletion mutant by gene targeting with an adenoviral vector. Mol Cell Biol. 1993;13:918–927. doi: 10.1128/mcb.13.2.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watson J E V, Slorach E M, Maule J, Lawson D, Porteous D J, Brookes A J. Human repeat-mediated integration of selectable markers into somatic cell hybrids. Genome Res. 1995;5:444–452. doi: 10.1101/gr.5.5.444. [DOI] [PubMed] [Google Scholar]
- 68.Weiden M D, Ginsberg H S. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc Natl Acad Sci USA. 1994;91:153–157. doi: 10.1073/pnas.91.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiner A M, Deininger P L, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 70.Yanez R J, Porter A C G. Therapeutic gene targeting. Gene Ther. 1998;5:149–159. doi: 10.1038/sj.gt.3300601. [DOI] [PubMed] [Google Scholar]
- 71.Yang Y, Ertl H C, Wilson J M. MHC class I restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1 deleted recombinant adenoviruses. Immunity. 1994;1:433–442. doi: 10.1016/1074-7613(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 72.Yang Y, Li Q, Ertl H C, Wilson J M. Cellular immunity to viral antigens limits E1-deleted recombinant adenoviruses for gene therapy. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang Y, Li Q, Ertl H C, Wilson J M. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J Virol. 1995;69:2004–2015. doi: 10.1128/jvi.69.4.2004-2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye X, Gao G P, Pabin C, Raper S E, Wilson J M. Evaluating the potential of germ line transmission after intravenous administration of recombinant adenovirus in the C3H mouse. Hum Gene Ther. 1998;9:2135–2142. doi: 10.1089/hum.1998.9.14-2135. [DOI] [PubMed] [Google Scholar]
- 75.Zeng M, Cerniglia G J, Eck S L, Stevens C W. High-efficiency stable gene transfer of adenovirus into mammalian cells using ionizing radiation. Hum Gene Ther. 1997;8:1025–1032. doi: 10.1089/hum.1997.8.9-1025. [DOI] [PubMed] [Google Scholar]
- 76.Zhang W-W, Josephs S F, Zhou J, Fang X, Alemany R, Belagué C, Dai Y, Ayares D, Prokopenko E, Lou Y-C, Sethi E, Hubert-Leslie D, Kennedy M, Ruiz L, Rockow-Magnone S. Development and application of a minimal-adenoviral vector system for gene therapy of hemophilia A. Thromb Haemostasis. 1999;82:562–571. [PubMed] [Google Scholar]