Abstract
Objectives
To develop and validate an open-source artificial intelligence (AI) algorithm to accurately detect contrast phases in abdominal CT scans.
Materials and methods
Retrospective study aimed to develop an AI algorithm trained on 739 abdominal CT exams from 2016 to 2021, from 200 unique patients, covering 1545 axial series. We performed segmentation of five key anatomic structures—aorta, portal vein, inferior vena cava, renal parenchyma, and renal pelvis—using TotalSegmentator, a deep learning-based tool for multi-organ segmentation, and a rule-based approach to extract the renal pelvis. Radiomics features were extracted from the anatomical structures for use in a gradient-boosting classifier to identify four contrast phases: non-contrast, arterial, venous, and delayed. Internal and external validation was performed using the F1 score and other classification metrics, on the external dataset “VinDr-Multiphase CT”.
Results
The training dataset consisted of 172 patients (mean age, 70 years ± 8, 22% women), and the internal test set included 28 patients (mean age, 68 years ± 8, 14% women). In internal validation, the classifier achieved an accuracy of 92.3%, with an average F1 score of 90.7%. During external validation, the algorithm maintained an accuracy of 90.1%, with an average F1 score of 82.6%. Shapley feature attribution analysis indicated that renal and vascular radiodensity values were the most important for phase classification.
Conclusion
An open-source and interpretable AI algorithm accurately detects contrast phases in abdominal CT scans, with high accuracy and F1 scores in internal and external validation, confirming its generalization capability.
Clinical relevance statement
Contrast phase detection in abdominal CT scans is a critical step for downstream AI applications, deploying algorithms in the clinical setting, and for quantifying imaging biomarkers, ultimately allowing for better diagnostics and increased access to diagnostic imaging.
Keywords: Contrast media, Abdomen, Machine learning, Artificial intelligence, Radiomics
Introduction
Abdominal computed tomography (CT) scans are commonly utilized to assess internal organs and structures. CT exams can be performed by scanning subjects in different phases related to the use of intravascular contrast agents, which enhance the radiodensity of blood vessels and vascularized internal organs facilitating the radiological differentiation of various tissues and structures [1, 2].
Accurate identification of contrast phases in abdominal CT scans is particularly critical for various downstream artificial intelligence (AI) applications and the reliable quantification of imaging biomarkers. For instance, an algorithm designed to differentiate hepatic lesions necessitates awareness of the specific contrast phase or, alternatively, must receive only a distinct phase as an input. This is due to the integral role that contrast characteristics play in the process of lesion characterization [3, 4]. Kidney tumors provide another prototypical example where lesions necessitate the interpretation of contrast phase properties for effective discrimination [5, 6]. In the field of opportunistic imaging and radiomics, when extracting measurements and biomarkers from organs, the phase of the contrast may significantly affect these measurements. For example, it has been demonstrated that measurements of bone and muscle attenuations as biomarkers of osteoporosis and sarcopenia are susceptible to the presence and phase of the contrast agent [7–10].
Currently, Digital Imaging and Communications in Medicine (DICOM) tags are widely used for identifying the contrast phases in abdominal CT scans. However, the contrast details can often be incomplete and unreliable due to various factors such as human error, inconsistency in entry, omission, or compatibility issues across different imaging systems. These inaccuracies represent a huge limitation for the quantification of imaging biomarkers in the field of radiomics and opportunistic imaging. As well as for the development and deployment of abdominal AI algorithms in the clinical setting [4, 11–14].
In the last few years, deep learning algorithms have demonstrated success in analyzing medical imaging. Especially with techniques such as convolutional neural networks that mimic the human ability to discern intricate patterns within images [15]. By making use of these algorithms, we can potentially automate the classification of the contrast phase in abdominal CT scans, solving the limitations and inaccuracies of DICOM tags.
We aimed to develop an AI-based algorithm to automate the detection of the contrast phases in abdominal CT scans. We employed an approach of mimicking how radiologists visually assess the contrast enhancement patterns by looking at key anatomical structures. We evaluated the algorithm on an external dataset to test for generalizability. In addition, we aimed to facilitate the integration of this tool into clinical workflows by making it available through an easy-to-use AI inference system: https://github.com/StanfordMIMI/Comp2Comp [16, 17].
Methods
This retrospective study was approved by the institutional review board and compliant with the Health Insurance Portability and Accountability Act.
Data preparation
We obtained 739 abdominal CT exams from Stanford Hospital from 200 unique patients undergoing cystectomy between June 2016 and August 2021, yielding a total of 1545 axial series. These series were partitioned into two datasets: 1183 series from 172 patients were used for training, and the remaining 362 from 28 patients formed the held-out test set. We ensured each patient’s data was exclusively assigned to one of these sets to maintain patient-wise separation (Fig. 1).
Fig. 1.
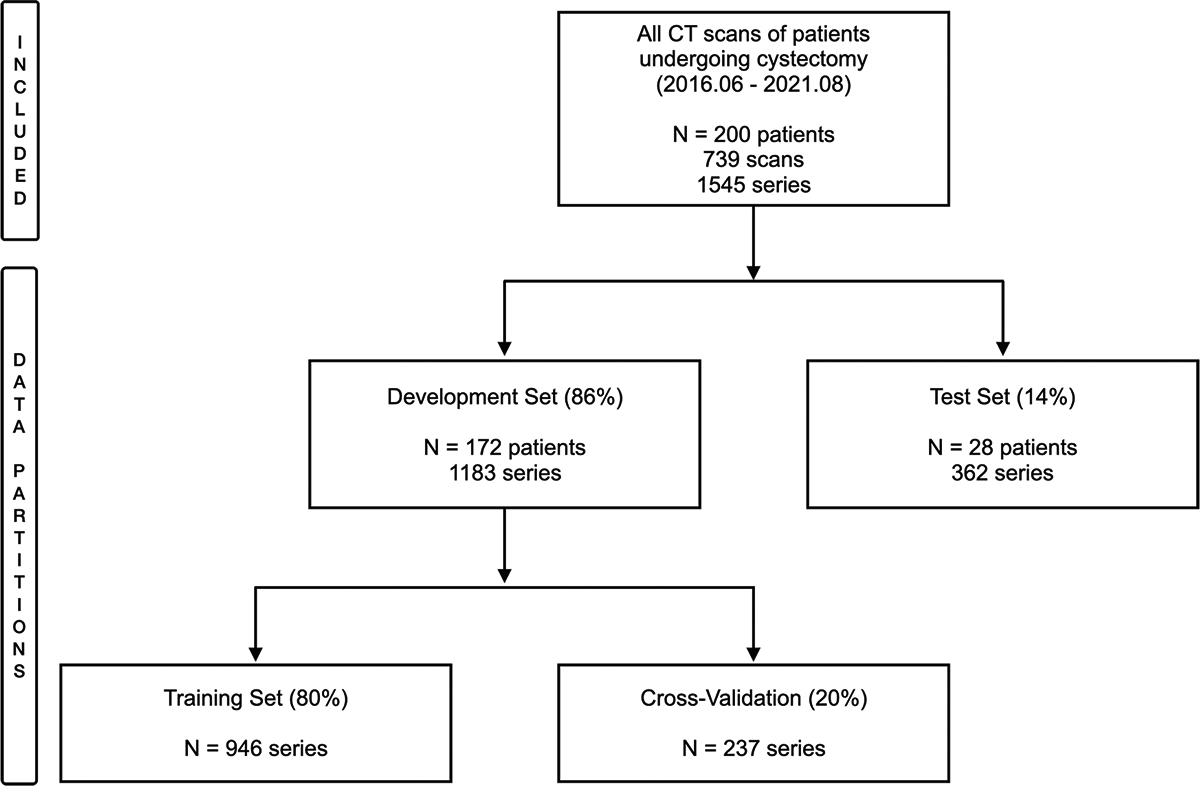
Flowchart depicting the included dataset and data partitioning for the algorithm development
Each series was initially weakly labeled into one of four classes: non-contrast, arterial, venous, or delayed. This preliminary classification was accomplished using Regex patterns to match words in the “Series Description” DICOM tag that indicated one of the categories, such as “arte”, “portal”, “venous”, “nephro”, “non con”, “w/o”, “delay” etc. Despite the known limitations of DICOM tags, this provided a useful first-pass labeling that would later undergo rigorous confirmation and correction when necessary [18, 19].
A board-certified radiologist with 5 years of experience reviewed each series to confirm or correct the initial labels when necessary. To facilitate this process, we segmented the organs using TotalSegmentator [20] and exported a JPEG image of two slices of each series in two strategic regions of the abdomen. The first was extracted on the level of the right adrenal, providing a good assessment of the aorta, inferior vena cava (IVC), portal vein, and hepatic artery. The second slice was extracted on the level of the left kidney which allowed a good assessment of the renal parenchyma and renal pelvis. The images were assessed on a standard monitor, and no special software was utilized to display the images. This dual-method process, the automated Regex and the human labeler was selected as a strategy to maximize the accuracy and reliability of our labeling (Fig. 2).
Fig. 2.
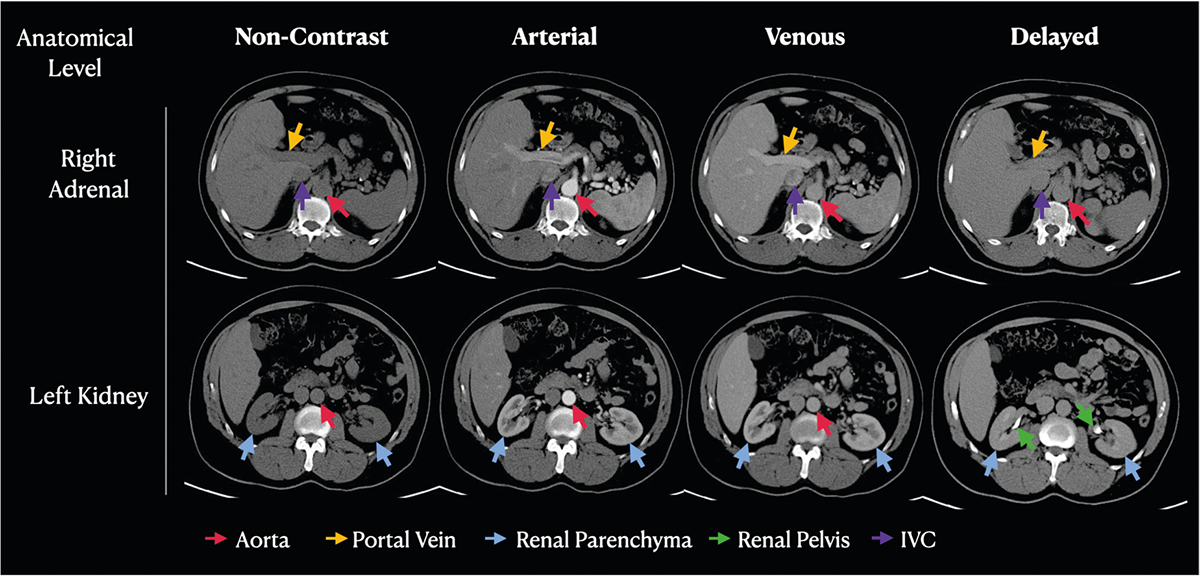
Example abdominal CT images at the anatomical level of the right adrenal gland and the left kidney, representing each of the four classes. Observe the variations in pixel intensity across the phases in the key structures: aorta (red arrows), portal vein (yellow arrows), renal parenchyma (blue arrows), renal pelvis (green arrows), and IVC (purple arrows)
Algorithm development
Our study methodology comprises a three-step process for training a contrast phase algorithm: segmentation of organs, feature extraction, and classification.
Stage 1—organ segmentation
We isolated the key anatomical structures of the aorta, IVC, portal vein, renal parenchyma, and renal pelvis. To perform the segmentation, we used our open-source toolbox, which included TotalSegmentator [20], a deep learning-based tool for multi-organ segmentation, and a rule-based approach to extract the renal pelvis. We apply a 3-pixel erosion to the segmented masks to avoid pixel intensity contamination from adjacent structures, such as fat or atherosclerotic calcification that could introduce bias to the pixel intensity measurements.
Stage 2—feature extraction
Once the organs were segmented, we computed 48 quantitative low-level radiomics features that characterize the radiodensity statistics of these structures. This process began by applying the segmentation masks from the previous step to isolate the organs of interest: aorta, IVC, portal vein, renal parenchyma, and renal pelvis. For each isolated organ, we applied binary erosion to refine the segmentation boundaries, ensuring precision in our feature extraction process.
Subsequent steps involved calculating radiodensity statistics such as maximum value, minimum value, mean, median, standard deviation, and variance of the pixel intensities within each organ’s segmented region. Additionally, we employed the SciPy library’s ConvexHull function to compute and fill the convex hull of the renal pelvis, enhancing our analysis of its radiodensity features. The statistical computations were performed using NumPy for basic statistics, and SciPy Multidimensional Image Processing—“scipy.ndimage”—for more complex spatial measurements. We then calculated the comparative radiodensity differences between the vascular structures (aorta, IVC, and portal vein). The calculated features were then saved into a NumPy array for subsequent model training [21, 22].
Stage 3—algorithm training
Data partitioning was performed initially between the development set (train + validation set) and the test set in an 85%/15% proportion on a patient level. Subsequently, the development set was split into training and validation sets in an 80%/20% proportion (Fig. 1).
Using the extracted radiomic features, we employed extreme gradient boosting (XGBoost) to train a classifier to categorize the CT images into the four contrast phases [23]. XGBoost is a machine learning technique that uses decision tree ensembles. We designed a multi-class classification problem aimed at categorizing the CT images into the four contrast phases. The model was trained to learn the distinct radiomic patterns associated with each contrast phase from our training dataset. Therefore, the inputs of the model were the 48 selected features, and the output was a one-hot-encoding vector representing one of the four classes. We developed the algorithm in Python, utilizing the open-source software “dmlc-XGBoost” [23], as well as other popular Python libraries such as Scikit-Learn, Numpy, Pickle, Pandas, and Matplotlib during the training process. The model was trained from “scratch”, with no special initialization or pre-training. We fit the model XGBoost classifier (class “XGBClassifier”) of type “tree-based model”, in the learning objective “multi:softprob”, which means multi-class with softmax returning the probability of each class. We performed a grid search to optimize the following parameters: n_estimators of 50, max_depth of 5, the learning rate of 0.1, and “min_child_weight” = 3. The other parameters were based on their demonstrated efficacy in the literature, and therefore set as default in the XGBoost library. We didn’t use any ensemble of models or data augmentation.
Evaluation
To ensure the generalizability of our model, we conducted internal and external validation of our algorithm with an internal test set and an external validation set from a different country, which was never exposed to the model during training.
For internal validation, we utilized our held-out test set of 362 CT scan series from 28 patients not included in the training. For external validation, we used 864 CT scan series from the “VinDr-Multiphase CT” [24] benchmark, which is a public dataset that includes non-contrast, arterial, and venous series. Notably, the external dataset includes a fourth class labeled “other”, which is distinct from the “delayed” category used in our dataset to specifically denote the delayed phase in abdominal CT scans. A radiologist performed an assessment of the VinDr dataset to ensure that the categories were compatible with our labeling schema, especially to assess if “other” corresponded to “delayed”, and if “venous” were free from misclassification of “delayed” cases.
All cases in “other” and a 50-case sample of “venous” were assessed. It was determined that most cases (77%) in “other” in fact represented the ‘delayed’ phase, therefore partially compatible with our label “delayed”. And almost all the cases in “venous” (96%) were correctly categorized, with only two instances (4%) being more fittingly described as ‘delayed’. We then excluded the incompatible instances of “other” in the VinDr dataset, making this category compatible with our “delayed” class. We refer to this category as “adjusted other/delayed” throughout the text.
The performance of the model was evaluated using a number of metrics: F1 score as the primary metric, accuracy, precision, sensitivity, specificity, and their 95% confidence interval [25], as well as the area under the receiver operating characteristic curve (AUROC), and area under the precision-recall curve (AUPRC). The overall performance of the algorithm was assessed through the F1 score and the other metrics using the macro-averaged approach, providing a balanced evaluation across all classes.
We utilized Shapley additive explanations (SHAP) plots to evaluate feature importance in our model. SHAP plots offer an interpretable summary of feature influence on model prediction for each class.
Results
Our study population consisted of adult patients with ages ranging from 21 to 90 years. The distribution of the contrast phases in our training and test sets was imbalanced (Table 1), with a low prevalence of the “arterial” class (4% of the cases in the training set and 6% in the test set) and a higher prevalence of the venous class (43% of the cases in the training set, and 38% in the test set). The distribution of the external dataset had a reduced number of non-contrast, and “adjusted other/delayed” classes, respectively accounting for 13.4% and 8.9% of the cases, while arterial and venous phases had a prevalence of 35.4% and 42.2%, respectively (Table 2).
Table 1.
Patient characteristics for the internal training and test sets, and the external validation set
| Characteristic | Training set | Internal test set | External validation |
|---|---|---|---|
| No. of patients | 172 | 28 | - |
| No. of scans | 364 | 87 | 209 |
| No. of series | 1183 | 362 | 864 |
| Female patients | 22% | 14% | 23.7% |
| Age (y) | |||
| Mean ± SD | 70 ± 8 | 68 ± 8 | – |
| Range | 21−85 | 44–90 | – |
Table 2.
Performance metrics and 95% confidence intervals on the internal test set and external validation dataset
| Metric | Non-contrast | Arterial | Venous | Delayed | Total/macro-average |
|---|---|---|---|---|---|
| No. training examples | 285 (24.0%) | 47 (4.1%) | 503 (42.5%) | 346 (29.2%) | 1181 |
| No. internal validation examples | 76 (20.9%) | 22 (6.0%) | 139 (38.4%) | 125 (34.5%) | 362 |
| F1 score | 96.6% | 78.9% | 92.2% | 95.0% | 90.7% |
| Precision | 100.0% (94.9%–100.0%) | 93.7% (67.5%–99.0%) | 87.1% (81.7%–91.1%) | 97.4% (92.6%–99.1%) | 94.6% |
| Sensitivity | 93.4% (85.3%–97.8%) | 68.1% (45.1%–86.1%) | 97.8% (93.8%–99.6%) | 92.8% (86.8%–96.7%) | 88.1% |
| Specificity | 100.0% (98.7%–100.0%) | 99.7% (98.4%–99.9%) | 91.0% (86.5%–94.4%) | 98.7% (96.4%–99.7%) | 97.4% |
| AUROC | 98.0% | 99.4% | 98.6% | 99.6% | 98.9% |
| AUPRC | 98.1% | 84.3% | 97.6% | 99.4% | 94.9% |
| Accuracy | 98.6% (96.6%–99.5%) | 97.8% (95.5%–99.0%) | 93.6% (90.6%–95.9%) | 96.7% (94.1%–98.2%) | 92.3% |
| No. external validation examples | 116 (13.4%) | 306 (35.4%) | 365 (42.2%) | 77 (8.9%) | 864 |
| F1 score | 97.0% | 85.8% | 75.4% | 59.9% | 79.5% |
| Precision | 96.6% (91.3%–98.7%) | 99.7% (97.7%–99.9%) | 73.2% (69.3%–76.7%) | 43.5% (38.8%–48.3%) | 78.3% |
| Sensitivity | 97.4% (92.6%–99.5%) | 75.3% (70.9%–79.4%) | 77.8% (73.1%–82.1%) | 96.1% (89.0%–99.2%) | 86.7% |
| Specificity | 99.5% (98.8%–99.9%) | 99.8% (99.0%–100.0%) | 84.1% (80.9%–86.9%) | 88.9% (86.6%–90.9%) | 93.1% |
| AUROC | 99.9% | 94.3% | 87.3% | 87.1% | 92.2% |
| AUPRC | 98.5% | 95.5% | 77.7% | 60.9% | 83.2% |
| Accuracy | 99.3% (98.5%–99.7%) | 89.5% (87.4%–91.4%) | 81.9% (79.3%–84.3%) | 89.5% (87.3%–91.4%) | 90.1% |
Distribution for the training, test, and external validation datasets
The class adjusted other/delayed is an adjusted version of the original “other” category in the VinDr dataset to exclusively include “delayed” cases
In total, 874 labels across all classes underwent manual labeling with the following breakdown: 550 “venous”, 200 “non-contrast”, 67 “delayed”, and 57 “arterial” labels required manual assignment. Before the manual inspection, the initial automatic labeling assigned labels to the 713 series, while the 832 series remained unlabeled. Of the 713 initially labeled series, 413 were “delayed”, of which 9 needed manual correction; 161 were “non-contrast” series, none of which required correction; 122 were “venous”, with 30 requiring correction; and 17 were “arterial”, with 3 requiring correction. In the external validation dataset, the assessment of the class “other”, found that 25 instances did not correspond to the ‘delayed’ phase and were consequently excluded.
Table 2 provides a detailed breakdown of the performance metrics across both internal and external validations, including AUROC, AUPRC, precision, recall, specificity, and their 95% confidence intervals. In internal validation (held-out test set), the classifier produced F1 scores of 96.6% for non-contrast, 78.9% for arterial, 92.2% for venous, and 95.0% for the delayed phase, with overall accuracy of 92.3%. Precision ranged from 87.1% to 100%, sensitivity from 68.1% to 97.8%, and specificity from 91.0% to 100.0%. The AUROC and AUPRC values were consistently high across all phases, contributing to an overall AUROC of 98.9% and AUPRC of 94.8%.
For the external validation conducted on the publicly available “VinDr-Multiphase CT” dataset, consisting of 864 abdominal CT scan series, the algorithm maintained high performance. The classifier achieved F1 scores of 97.0% for non-contrast, 85.8% for arterial, 75.4% for venous, and 59.9% for “adjusted other/delayed” phases. With an overall accuracy of 90.1%. Precision values varied from 43.5% to 96.6%, sensitivity from 75.3% to 97.4%, and specificity from 84.1% to 99.8%. The overall AUROC was 92.1 and AUPRC 83.2%.
The feature importance analysis demonstrated that the radiodensity statistics of certain anatomical structures played a significant role in the classification of the different classes. For the non-contrast phase, the most significant features were the renal parenchyma and renal pelvis (Fig. 3a). For the arterial phase, relative radiodensity values between the aorta and portal vein and between the aorta and IVC were the most relevant features (Fig. 3b). For the venous phase, the portal vein’s radiodensity was the most important feature (Fig. 3c). And for the delayed phase, the most significant features were the relative radiodensity values between the aorta and IVC and the radiodensity measurements from the kidney pelvis (Fig. 3d).
Fig. 3.
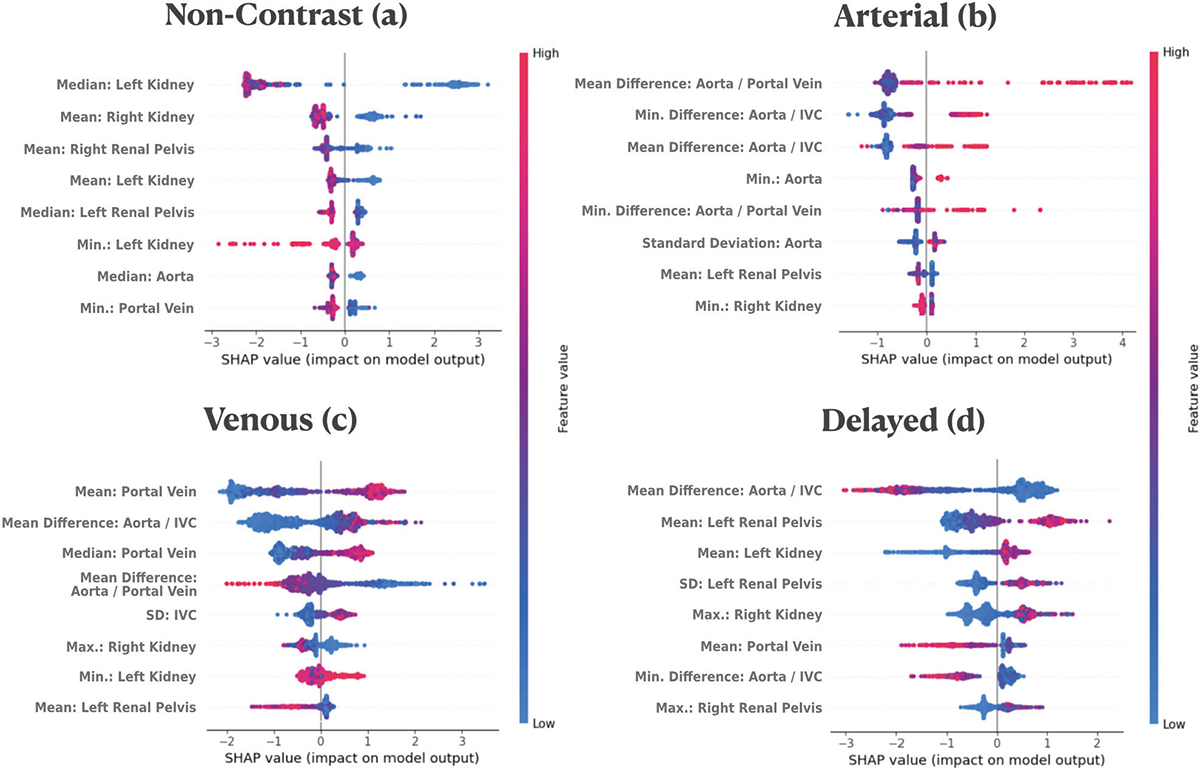
SHAP importance plot for the four contrast phases. This plot shows the top eight most influential features for each contrast phase: non-contrast (a), arterial (b), venous (c), and delayed (d). Each dot represents a feature instance. Features are arranged on the y-axis, while their impact on the model’s output is displayed along the x-axis. Dots on the right of the 0-line increase the model’s output, while those on the left decrease it. Color intensity represents the feature value: high values in red and low values in blue. The plot elucidates how each feature instance affects the model’s prediction, providing insights into the decision-making process across different contrast phases
Discussion
Our study addresses the problem of identifying contrast phases in abdominal CT scans. To tackle this, we developed an interpretable algorithm that first uses a deep learning-based tissue segmentation to extract radiomic radiodensity features, which are passed into an XGBoost model to detect the contrast phase for each series. This algorithm demonstrated a high F1 score and other performance metrics on internal and external validation sets, demonstrating a robust generalizability of our model.
Our algorithm mirrored clinical intuition by prioritizing the radiodensity features of anatomical structures that are typically considered most representative of each specific contrast phase. For instance, the feature importance plot (Fig. 2) shows that the aorta, and its comparison with other vascular structures, were the most important features for the arterial phase. Similarly, the portal vein was the most important feature for the venous phase, and the renal pelvis was the second most important feature in delayed, while the other five high-ranking features were derived from renal structures. Interestingly the most important features of the non-contrast phase were dominated primarily by renal structures, accounting for the six most important features. Our hypothesis is that it is due to the homogeneity of the renal structures on non-contrast versus more heterogeneous after contrast. This explainable nature of the algorithm enables this parallelism with clinical knowledge, making the algorithm more transparent and easier to be trusted by clinicians, both in success and failure modes.
Our results revealed that the algorithm’s performance in identifying the arterial phase improved during external validation, reinforcing the generalization capability. The success in generalizing implies that the model has learned the underlying patterns associated with the different contrast phases, including the arterial phase, despite the limited number of examples on the training set. In contrast, we observed a decrease in performance for the venous and “adjusted other/delayed” classes.
The limitations of the study encompass the labeling discrepancy between our label “delayed” and the external dataset label “other”. To address labeling discrepancies, we conducted a thorough review of the “other” and “venous” categories. This ensured the dataset’s appropriateness for testing our algorithm by confirming the “venous” category was free from incorrectly labeled “delayed” cases. We also adjusted the “other” category to exclusively include “delayed” cases, what we called “adjusted other/delayed”. When selecting the external validation dataset, we focused on its accessibility and representativeness, opting for the ‘VinDr-Multiphase CT’ due to its size, multiphase nature, and public availability, enabling independent verification and benchmarking by the research community. Future work includes expanding the algorithm to other anatomical regions, once the same underlying method carries the potential to be trained in other modalities and body parts. The algorithm’s availability through the Comp2Comp Inference Pipeline is intended to facilitate this expansion by the broader research community.
In summary, the presented algorithm offers an effective and precise method for identifying contrast phases in abdominal CT scans, independent of the analysis of the DICOM tags. The accurate identification of the contrast phase in abdominal CT scans holds substantial utility for downstream AI applications, particularly those algorithms trained to function on specific series. By automating and standardizing the detection process, it reduces the potential for human error, and speeds up workflow. We hope that by supporting the integration of AI algorithms in the clinical setting and the consistent quantification of imaging biomarkers this algorithm can ultimately contribute to better and faster patient care, and broader access to diagnostic imaging. The algorithm’s public availability through the Comp2Comp Inference Pipeline, hosted on the GitHub repository “https://github.com/StanfordMIMI/Comp2Comp”, encourages broader use and exploration of this AI tool.
Key Points.
Digital Imaging and Communications in Medicine labels are inaccurate for determining the abdominal CT scan phase.
AI provides great help in accurately discriminating the contrast phase.
Accurate contrast phase determination aids downstream AI applications and biomarker quantification.
Acknowledgements
We would like to acknowledge the team behind the TotalSegmentator open-source project, for their work on Abdominal CT segmentations. We would like to acknowledge funding from the NIH, Stanford Precision Health and Integrated Diagnostics Seed Grant, Stanford Human-Centered AI, and Center for AI in Medicine and Image Seed Grant.
Funding
This study has received funding from NIH NHLBI R01 HL167974, Stanford Precision Health and Integrated Diagnostics Seed Grant; Stanford Human-Centered AI, and Center for AI in Medicine and Image Seed Grant.
Abbreviations
- AI
Artificial intelligence
- AUPRC
Area under precision-recall curve
- AUROC
Area under the receiver operating characteristic curve
- CT
Computed tomography
- DICOM
Digital Imaging and Communications in Medicine
- IVC
Inferior vena cava
- SHAP
Shapley additive explanations
- XGBoost
Extreme gradient boosting
Footnotes
Guarantor
The scientific guarantor of this publication is Eduardo Pontes Reis.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board (Stanford University School of Medicine) approval was obtained.
Study subjects or cohorts overlap
The study subjects or cohorts have not been previously reported.
- Retrospective
- Diagnostic study
- Performed at one institution
References
- 1.Kammerer S, Höink AJ, Wessling J et al. (2015) Abdominal and pelvic CT: Is positive enteric contrast still necessary? Results of a retrospective observational study. Eur Radiol 25:669–678 [DOI] [PubMed] [Google Scholar]
- 2.Shirkhoda A (1991) Diagnostic pitfalls in abdominal CT. Radiographics 11:969–1002 [DOI] [PubMed] [Google Scholar]
- 3.Radiya K, Joakimsen HL, Mikalsen KØ, Aahlin EK, Lindsetmo R-O, Mortensen KE (2023) Performance and clinical applicability of machine learning in liver computed tomography imaging: a systematic review. Eur Radiol. 10.1007/s00330-023-09609-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocha BA, Ferreira LC, Vianna LGR et al. (2022) Contrast phase recognition in liver computer tomography using deep learning. Sci Rep 12:20315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schieda N, Nguyen K, Thornhill RE, McInnes MDF, Wu M, James N (2020) Importance of phase enhancement for machine learning classification of solid renal masses using texture analysis features at multi-phasic CT. Abdom Radiol (NY) 45:2786–2796 [DOI] [PubMed] [Google Scholar]
- 6.Han S, Hwang SI, Lee HJ (2019) The classification of renal cancer in 3-phase CT images using a deep learning method. J Digit Imaging 32: 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boutin RD, Kaptuch JM, Bateni CP, Chalfant JS, Yao L (2016) Influence of IV contrast administration on CT measures of muscle and bone attenuation: implications for sarcopenia and osteoporosis evaluation. AJR Am J Roentgenol 207:1046–1054 [DOI] [PubMed] [Google Scholar]
- 8.Pickhardt PJ, Lauder T, Pooler BD et al. (2016) Effect of IV contrast on lumbar trabecular attenuation at routine abdominal CT: correlation with DXA and implications for opportunistic osteoporosis screening. Osteoporos Int 27:147–152 [DOI] [PubMed] [Google Scholar]
- 9.Rühling S, Navarro F, Sekuboyina A et al. (2022) Automated detection of the contrast phase in MDCT by an artificial neural network improves the accuracy of opportunistic bone mineral density measurements. Eur Radiol 32:1465–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang S, Graffy PM, Ziemlewicz TJ, Lee SJ, Summers RM, Pickhardt PJ (2019) Opportunistic osteoporosis screening at routine abdominal and thoracic CT: normative L1 trabecular attenuation values in more than 20 000 adults. Radiology 291:360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye Z, Qian JM, Hosny A et al. (2022) Deep learning-based detection of intravenous contrast enhancement on CT scans. Radiol Artif Intell 4:e210285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Na S, Sung YS, Ko Y et al. (2022) Development and validation of an ensemble artificial intelligence model for comprehensive imaging quality check to classify body parts and contrast enhancement. BMC Med Imaging 22:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muhamedrahimov R, Bar A, Laserson J, Akselrod-Ballin A, Elnekave E (2022) Using machine learning to identify intravenous contrast phases on computed tomography. Comput Methods Programs Biomed 215:106603 [DOI] [PubMed] [Google Scholar]
- 14.Blankemeier L, Yao L, Long J et al. (2024) Skeletal muscle area on CT: determination of an optimal height scaling power and testing for mortality risk prediction. AJR Am J Roentgenol 222:e2329889. [DOI] [PubMed] [Google Scholar]
- 15.Chartrand G, Cheng PM, Vorontsov E et al. (2017) Deep learning: a primer for radiologists. Radiographics 37:2113–2131 [DOI] [PubMed] [Google Scholar]
- 16.Fuentes-Orrego JM, Pinho D, Kulkarni NM, Agrawal M, Ghoshhajra BB, Sahani DV (2014) New and evolving concepts in CT for abdominal vascular imaging. Radiographics 34:1363–1384 [DOI] [PubMed] [Google Scholar]
- 17.Blankemeier L, Desai A, Chaves JMZ et al. (2023) Comp2Comp: open-source body composition assessment on computed tomography. Preprint at 10.48550/ARXIV.2302.06568 [DOI] [Google Scholar]
- 18.Gauriau R, Bridge C, Chen L et al. (2020) Using DICOM metadata for radiological image series categorization: a feasibility study on large clinical brain MRI datasets. J Digit Imaging 33:747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reis EP, De Paiva JPQ, Da Silva MCB et al. (2022) BRAX, Brazilian labeled chest x-ray dataset. Sci Data 9:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wasserthal J, Breit H-C, Meyer MT et al. (2023) TotalSegmentator: robust segmentation of 104 anatomic structures in CT images. Radiol Artif Intell 5:e230024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdhary CL, Acharjya DP (2020) Segmentation and feature extraction in medical imaging: a systematic review. Procedia Comput Sci 167:26–36 [Google Scholar]
- 22.Van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B (2020) Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging 11:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen T, Guestrin C (2016) XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, San Francisco, 13–17 [Google Scholar]
- 24.Dao BT, Nguyen TV, Pham HH, Nguyen HQ (2022) Phase recognition in contrast-enhanced CT scans based on deep learning and random sampling. Med Phys 49:4518–4528 [DOI] [PubMed] [Google Scholar]
- 25.Clopper CJ, Pearson ES (1934) The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26:404–413 [Google Scholar]


