Abstract
The amphipathic α-helices located in the cytoplasmic tail of the envelope (Env) transmembrane glycoprotein gp41 of human immunodeficiency virus type 1 have been implicated in membrane association and cytopathicity. Deletion of the last 12 amino acids in the C terminus of this domain severely impairs infectivity. However, the nature of the involvement of the cytoplasmic tail in Env-membrane interactions in cells and the molecular basis for the defect in infectivity of this mutant virus are still poorly understood. In this study we examined the interaction of the cytoplasmic tail with membranes in living mammalian cells by expressing a recombinant cytoplasmic tail fragment and an Escherichia coli β-galactosidase/cytoplasmic tail fusion protein, both of them lacking gp120, the gp41 ectodomain, and the transmembrane region. We found through cell fractionation, in vivo membrane flotation, and confocal immunofluorescence studies that the cytoplasmic tail contained determinants to be routed to a perinuclear membrane region in cells. Further mapping showed that each of the three lentivirus lytic peptide (LLP-1, LLP-2, and LLP-3) sequences conferred this cellular membrane-targeting ability. Deletion of the last 12 amino acids from the C terminus abolished the ability of the LLP-1 motif to bind to membranes. High salt extraction, in vitro transcription and translation, and posttranslational membrane binding analyses indicated that the β-galactosidase/LLP fusion proteins were inserted into membranes via the LLP sequences. Subcellular fractionation and confocal microscopy studies revealed that each of the LLP motifs, acting in a position-independent manner, targeted non-endoplasmic reticulum (ER)-associated β-galactosidase and enhanced green fluorescence protein to the ER. Our study provides a basis for the involvement of the gp41 cytoplasmic tail during Env maturation and also supports the notion that the membrane apposition of the C-terminal cytoplasmic tail plays a crucial role in virus-host interaction.
The cytoplasmic domain of human immunodeficiency virus type 1 (HIV-1) envelope (Env) transmembrane (TM) glycoprotein gp41 has multiple functions in the virus life cycle. Mutations, deletions, and truncations in this region may affect virus replication, infectivity, cytopathicity, Env incorporation into virions, cell type-dependent Env stability, and interaction with the viral matrix (MA) protein. A deletion of 144 amino acids, which comprise most of the cytoplasmic tail, from the C terminus of gp41 does not affect virus infectivity, Env assembly into virions, and cytopathogenicity in MT-4 cells (66). The differential virus infectivity of this mutant in permissive cells (MT-4 and M8166) and nonpermissive cells (most T-cell lines and primary cells) can be attributed to the differential requirement of the cytoplasmic tail to incorporate gp120 into virions in different cell types (1, 46). We previously reported that an Env mutant of HIV-1 lacking the whole cytoplasmic tail and the last two amino acids in the TM region can trans-dominantly interfere with wild-type (wt) virus infectivity by forming a dysfunctional wt-mutant hetero-oligomer when coexpressed with the wt Env protein (11). Tat-induced expression of this cytoplasmic tail truncation Env confers dominant interference with virus transmission when the mutant env gene is targeted to HeLa-CD4 cells (13). Using procaryotic and eucaryotic expression systems, we demonstrated that the C-terminal two-thirds portion of the gp41 cytoplasmic tail, per se, possesses the potential to self-assemble into a high-ordered multimeric structure (39). These results provide a structural basis for the role of the cytoplasmic tail in the virus life cycle.
Although the gp41 cytoplasmic tail sequence does not reveal typical membrane binding sequences, HIV-1 isolates show a remarkable conservation of the amphipathic α-helical secondary structures. The unusual large helical hydrophobic moments of the three highly conserved amphipathic α-helical segments, located at residues 828 to 856, 770 to 795, and 789 to 815, termed lentivirus lytic peptide 1 (LLP-1), LLP-2, and LLP-3, respectively, suggest that these motifs have interactions with membranes (2, 23, 45, 61). Peptides representing these motifs interact with membranes, decrease bilayer stability, alter membrane ionic permeability, and induce cytolytic effects on both procaryotic and eucaryotic cells (14, 17, 25, 26, 34, 43, 44, 57). The membrane association feature of these LLPs has led to a hypothesis that a contiguous long sequence located in the cytoplasmic tail, beginning with the first palmitoylation site at Cys-764 (68) and ending at the C terminus, is embedded in membranes (34).
Till now only limited information has been available regarding the interaction of the HIV-1 gp41 cytoplasmic tail with membranes in virus-infected or Env-expressing cells. An in vitro transcription-coupled translation assay showed that a chimera containing the cytoplasmic tail fused to the signal peptide and the N-terminal 27 amino acids of the herpes simplex virus (HSV) Env glycoprotein gD-1 translocates across microsomal membranes (30). This HSV gD-1/cytoplasmic tail fusion protein is expressed on the cell surface and is released into culture medium when expressed in eucaryotic cells (29). These observations suggest that the gp41 cytoplasmic tail has a close association with cellular membranes. However, it is not clear whether the gp41 cytoplasmic tail by itself contains regions associated with cellular membranes or whether the exogenous sequences derived from the HSV gD-1 Env also contribute to cytoplasmic tail binding to cellular membranes. The endogenous reverse transcription activity of intact HIV-1 virions, measured by the permeability of the viral envelope to deoxyribonucleoside triphosphates, which are substrates of DNA polymerization, decreases when the LLP-1 and LLP-2 sequences in the cytoplasmic tail are deleted (71). This result suggests that LLP-1 and LLP-2 may bind to the viral envelope, resulting in decreased stability of the viral envelope. Collectively, these previous studies point out a plausible role of the gp41 cytoplasmic tail in cellular membrane association. However, direct evidence for binding of the gp41 cytoplasmic tail or LLP sequences to membranes within eucaryotic cells is lacking. Also, sequences in the cytoplasmic tail involved in cellular membrane binding and the nature of this binding are not yet clearly defined.
The C terminus of the gp41 cytoplasmic tail appears to play an important role in virus infectivity. A deletion of 12 amino acids from the C terminus of the cytoplasmic tail, mutant TM844, results in virus replication with a strikingly slower kinetic than that of the wt virus (70). However, the fusion ability of this mutant, assembly and release of virions, and incorporation of TM844 mutant Env into mature virions are normal compared to these processes in the wt virus (70). It was previously shown that this Env mutant is able to trans-dominantly interfere with wt Env-mediated virus infectivity (13). We also demonstrated that truncation of the last 12 amino acids from the C terminus abolishes the ability of the LLP-1 motif to self-associate (39). These results raise the possibility that cytoplasmic tail multimerization may play a crucial role in a step after gp41 ectodomain-mediated membrane fusion. However, the molecular basis for such a defect in infectivity of this mutant virus is still unclear.
HIV-1 Env precursor processing and intracellular transport in cells are very inefficient (38, 58, 67); the vast majority of the Env remains uncleaved and is retained in the endoplasmic reticulum (ER) or in a cis-Golgi compartment (19). This phenotype of the Env protein is consistent with the endoglycosidase H-sensitive state of the majority of the Env in cells (3, 67). Twenty to 90% of the intracellular Env, depending on the cell type, is degraded (22, 67), and intracellular degradation can occur in the ER (18). The reasons for this inefficient transport out of the ER are not fully understood. By examining an HSV gD-1/HIV Env chimera, Haffar et al. proposed that the segment between residues 751 to 856 of the Env may contain information that inhibits proteolytic processing and Env transport out of the ER (31). However, whether the gp41 cytoplasmic tail possesses a unique structural element that functions as an ER binding or retention signal remains elusive.
To understand the membrane localization basis for the role of the gp41 cytoplasmic tail in Env maturation and Env-mediated fusion, in the present study we examined gp41 cytoplasmic tail-mediated cellular membrane association. Further mapping showed that each of the three LLP sequences located in the cytoplasmic tail can be posttranslationally inserted into cellular membranes. We also showed that each of the LLP sequences contains ER-targeting signals. The results may provide a basis to address the structural and organizational role of the cytoplasmic tail in Env maturation, Env-mediated cytopathicity, and pore formation after receptor binding and membrane fusion.
MATERIALS AND METHODS
Cells and antibodies.
COS-1 cells were cultured in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum. Mouse monoclonal antibody (MAb) directed against Escherichia coli β-galactosidase was obtained from Promega (Madison, Wis.). Hybridoma 183 (clone H12-5C) produces a murine MAb reactive with HIV-1 capsid (CA) protein p24. Hybridoma Chessie 8 produces a murine MAb directed against residues 727 to 732 of the HIV-1 Env. Affinity-purified goat antibodies directed against the C termini of human calnexin and calreticulin and a rabbit antibody against the C terminus of rat Rab1B were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). MAbs specific for residues 701 to 715 of β-COP, a subunit of the coatomer of the COP-coated vesicles, and for the microtubule-binding peripheral Golgi membrane protein 58K were purchased from Sigma (St. Louis, Mo.). The Living Colors peptide antibody, a mixture of rabbit anti-peptide antibodies against green fluorescence protein (GFP), was purchased from Clontech Laboratories, Inc. (Palo Alto, Calif.). Fluorescein-conjugated wheat germ agglutinin (WGA) was obtained from Molecular Probes (Eugene, Oreg.). Affinity-purified fluorescein isothiocyanate (FITC)-conjugated and tetramethyl rhodamine isothiocyanate (TRITC)-conjugated secondary antibodies were purchased from Zymad Laboratories Inc. (South San Francisco, Calif.) and Kirkegaard & Perry Laboratories (Gaithersburg, Md.).
Construction of plasmids.
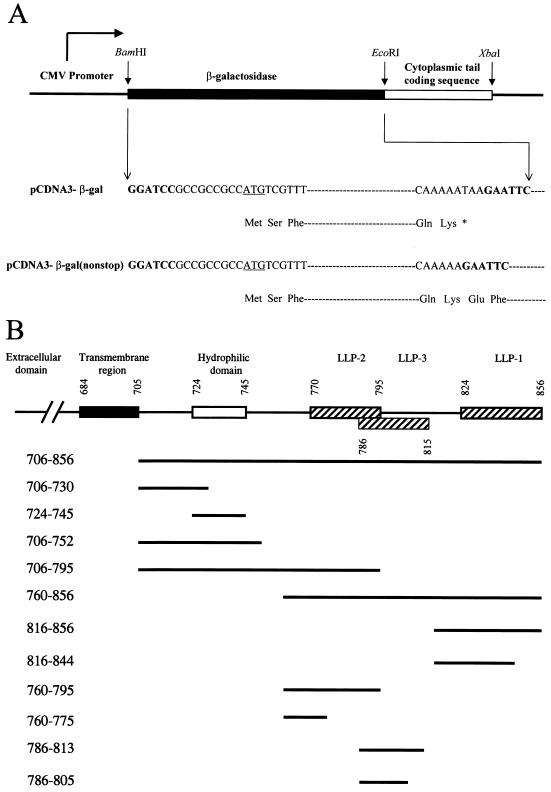
For construction of a gp41 cytoplasmic tail expression plasmid, the coding sequence of the entire cytoplasmic tail spanning residues 706 to 856 of the Env of the HXB2 strain was PCR amplified. This was performed using wt pSVE7-puro (13) as the template, oligonucleotide 706fEcoRI(Met) (39) as the forward primer, and 856rXbaI (5′-GCTCTAGATTATAGCAAAATCCTTTCCA-3′; nucleotides 8794 to 8775 of the HXB2 provirus) as the reverse primer. The nucleotides underlined indicate the recognition sequence of the restriction enzyme shown in the name of the oligonucleotide. The EcoRI-XbaI fragment was inserted in the corresponding sites in pCDNA3 (Fig. 1A), a cytomegalovirus (CMV) enhancer/promoter-driven plasmid (Invitrogen, Carlsbad, Calif.). To construct pCDNA3-gal, which encodes a stop codon following the C-terminal lysine residue of E. coli β-galactosidase (Fig. 1A), the entire 3.1-kB E. coli β-galactosidase-coding sequence was cloned by PCR using pCMVβ (Clontech) as the template and oligonucleotides β-gal-f-BamHI (5′-CGCGGATCCGCCGCCGCCATGTCGTTTACTTTGACCAAC-3′; nucleotides 1 to 21 of the β-galactosidase gene) and β-gal-r-EcoRI (5′-CCCGAATTCTTATTTTTGACACCAGACCAACTG-3′; nucleotides 3144 to 3121) as the forward and reverse primers, respectively. To construct pCDNA3-β-gal(nonstop) (Fig. 1A), β-gal-f-BamHI and β-gal-r-EcoRI(nonstop) (5′-CCCGAATTCTTTTTGACACCAGACCAACTG-3′; 3141 to 3121 of the β-galactosidase gene) were used as the primers. For construction of pCDNA3-β-gal plasmids that encode residues 706 to 856, 706 to 795, and 706 to 752 (Fig. 1B), 706fEcoRI (39) and 856rXbaI were used as the primers, and wt, TM795, and TM752, respectively, of pSVE7-puro (13) were used as the templates. For construction of pCDNA3-β-gal plasmids that encode residues 760 to 856, 760 to 795, and 760 to 775 (Fig. 1B), oligonucleotides 760fEcoRI (39) and 856rXbaI were used as primers, and wt, TM795, and TM775, respectively, of pSVE7-puro (13) were used as the templates in PCR. Oligonucleotides 816fEcoRI (39) and 856rXbaI were used as the primers, and wt and TM844, respectively, of pSVE7-puro (13) were used as the templates to clone the coding sequences for residues 816 to 856 and 816 to 844 (Fig. 1B). 706fEcoRI and 730rXbaI (5′-GCTCTAGATTAGGGCCTGTCGGGTCCCCTCGG-3′; 8413 to 8393 of the HXB2 provirus) were primed on wt pSVE7-puro to generate the 706-to-730 coding sequence (Fig. 1B). 724fEcoRI (5′-CCGAATTCCCGAGGGGACCCGACAGG-3′; 8393 to 8410) and 745rXbaI (5′-CCTCTAGATTAGGATCTGTCTCTGTCTCTCTC-3′; 8458 to 8438) were primed on wt pSVE7-puro to obtain the 724-to-745 coding sequence (Fig. 1B). For construction of cytoplasmic tail fragments tagged with enhanced GFP (EGFP) at the C terminus, DNA fragments corresponding to residues 706 to 856, 816 to 856, 760 to 795, and 786 to 813 with EcoRI and SalI sites flanked at the 5′ and 3′ ends, respectively, were generated by PCR using wt pSVE7-puro as the template and were then inserted in pEGFP(N2) (Clontech). Oligonucleotides 5′-CGCGGATCCGCCGCCGCCATGAACCGGGGAGTCCCTTTTAGG-3′ and 5′-CGGAATTCTCAAATGGGGCTACATGTCTTCTGAAACCG-3′ were used as sense and anti-sense primers, respectively, to clone the CD4 gene from a human CD4 expression plasmid, pBS-CD4, into the pCDNA3 vector.
FIG. 1.
Plasmid construction. (A) Construction of β-galactosidase expression plasmids. An E. coli β-galactosidase-coding sequence with a stop codon immediately after the codon for the C-terminal lysine residue was generated by PCR and inserted in the BamHI and EcoRI sites in pCDNA3 to yield pCDNA3-β-gal. Nucleotide sequences in bold indicate the restriction enzyme linker sequences, and the asterisk indicates the stop codon. A coding sequence of β-galactosidase without a stop codon after the C-terminal lysine was also inserted in pCDNA3 to yield pCDNA3-β-gal(nonstop). (B) Schematic diagram of recombinant β-galactosidase/cytoplasmic tail chimeras. pCDNA3-β-gal constructs that encode different regions of the cytoplasmic tail fused to the C terminus of β-galactosidase as depicted were constructed by PCR and inserted in the EcoRI and XbaI sites of pCDNA3-β-gal(nonstop).
PCR amplifications and DNA sequencing
All PCRs were performed using Vent DNA polymerase (New England BioLabs, Beverly, Mass.) according to the PCR amplification program previously described (39). For amplification of the β-galactosidase and human CD4 genes, a final concentration of 4 mM MgSO4 and 5% dimethyl sulfoxide was included in the Vent polymerase buffer. The amplification was subjected to 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 3 min. All pCDNA3-β-galactosidase/cytoplasmic tail constructs were autosequenced using oligonucleotide 5′-GGGGATTGGTGGCGACG-3′ (nucleotides 3045 to 3061 of the β-galactosidase gene) as the primer to confirm the cytoplasmic tail-coding sequences. Oligonucleotide 5′-CGTCGCCGTCCAGCTCGACCAG-3′ was used to sequence the coding sequences of cytoplasmic tail segments cloned in pEGFP(N2) chimeras.
Plasmid transfection, cell fractionation, and membrane flotation assay.
COS-1 cells were transfected with plasmid DNA by the DEAE-dextran method as previously described (10, 12). For cell fractionation, the washed transfected cells were sonicated twice on ice in 0.5 ml of hypotonic TE buffer (10 mM Tris-HCl, pH 7.5, containing 1 mM EDTA and complete protease inhibitor cocktail [Roche Molecular Biochemicals, Mannheim, Germany]) using an Ultrasonic Processor model W-375 (Ultrasonics, Inc., Farmingdale, N.Y.) with an output power of 20 to 30%, each time for 15 s. After centrifugation at 1,000 × g at 4°C for 10 min, postnuclear supernatants were centrifuged in a Beckman TLA100.2 rotor at 100,000 × g for 1 h. The membrane pellet fractions were resuspended with 0.5 ml of 12 mM phosphate buffer, pH 7.4, containing 3.2 mM KCl and 137 mM NaCl (referred to as PBS hereafter). Equal volumes of soluble and membrane fractions were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by Western blotting. For membrane flotation, the washed cells were sonicated twice in 0.5 ml of hypotonic TE buffer supplemented with 10% (wt/vol) sucrose on ice as described above. Membrane flotation assay was performed as previously described (32).
Salt extraction of membrane-bound LLP fusion proteins.
Washed transfected cells were sonicated in TE buffer, and after ultracentrifugation the P100 fractions were incubated with 0.5 ml of phosphate buffer containing 0.15 M NaCl or 1 M NaCl at 4°C for 30 min, with constant agitation. The mixtures were then ultracentrifuged and equal volumes of soluble and membrane fractions were subjected to Western blotting analysis.
Subcellular fractionation.
The subcellular fractionation procedure as previously described by Yang et al. (69) was performed, with minor modifications. The cells were sonicated in homogenization buffer containing the complete protease inhibitor cocktail twice on ice, each time for 15 s to prepare postnuclear fractions. Aliquots (0.5 ml) of the postnuclear fractions were loaded on the top of the preformed linear 0 to 26% Optiprep (Life Technologies, Inc.) gradients prepared according to the protocol provided by the manufacturer. The gradients were centrifuged at 40,000 rpm in a Beckman SW41 rotor at 4°C for 2 h. After fractionation, fractions were diluted and membranes in each fraction were pelleted by ultracentrifugation in a Beckman TLA100.2 rotor at 47,000 rpm for 1 h. The pelleted materials were then analyzed by Western blotting.
Laser scanning immunofluorescence microscopy and image analysis
COS-1 cells were transfected with 2.5 μg of DNA plasmids and then seeded onto glass coverslips placed in 24-well culture plates. Two days posttransfection cells were fixed, permeabilized with 0.25% Triton X-100, and processed for immunostaining according to procedures previously described (13). The concentrations of primary antibodies used were the following: Chessie 8 ascitic fluid, 1:200; β-galactosidase MAb, 1:500; anti-calnexin, anti-calreticulin, anti-Rab1B, and anti-EGFP, all 1:100; and anti-Golgi 58K protein, 1:50. The concentration of FITC-conjugated and TRITC-conjugated secondary antibodies used was 1:100. The concentration of fluorescein-labeled WGA was 1:500. Immunostained cells were analyzed under a Bio-Rad MRC 1000 confocal immunofluorescence microscope (Hercules, Calif.) using an oil object lens. Images were then analyzed by Confocal Assistant (Bio-Rad), and composite files were generated.
In vitro protein synthesis, protease protection assay, and posttranslational membrane binding.
In vitro coupled transcription/translation was performed using the reticulocyte-based TNT quick coupled transcription/translation kit (Promega). For analysis of protein synthesis in the presence of membranes, 2 μl of canine pancreatic microsomal membrane (Promega) was added per 25 μl of reaction mixture. After incubation at 30°C for 90 min, excess unlabeled methionine was added into the reaction to stop [35S]methionine incorporation into newly synthesized proteins. For protease protection, protein samples synthesized in the presence of membranes were divided into three portions. One portion received no treatment. The second portion was treated with 0.1 mg of proteinase K/ml as previously described (30). After incubation on ice for 1 h, samples were added with a final concentration of 5 mM phenylmethylsulfonyl fluoride (dissolved in dimethyl sulfoxide) and incubated on ice for 10 min. The samples were then boiled at 95°C for 10 min to inactivate proteinase K activity. The third portion was solubilized with a final concentration of 1% Triton X-100 before proteinase K treatment. All samples were then placed on ice, solubilized with detergents, and analyzed by SDS-PAGE. For posttranslational membrane binding analysis, cycloheximide at a final concentration of 0.2 mg/ml was added into the protein samples synthesized in vitro in the absence of membranes. The mixtures were spun at 40,000 rpm in a TLA45 rotor at 4°C for 1 h to remove insoluble materials. Aliquots (25 μl) of supernatants were added with 3 μl of microsomal membranes, incubated at 30°C for 30 min, and ultracentrifuged to pellet membranes. The membrane fractions were washed once with 100 μl of PBS and then ultracentrifuged. The supernatants from the second ultracentrifugation were combined with the first supernatants. Equal volumes of supernatant and membrane fractions were analyzed by SDS-PAGE.
RESULTS
The HIV-1 gp41 cytoplasmic tail possesses membrane-binding ability as determined by cell fractionation
To determine whether the HIV-1 gp41 cytoplasmic tail per se is able to bind cellular membranes, the coding sequence of the entire cytoplasmic tail, spanning residues 706 to 856, was cloned by PCR and inserted in pCDNA3 to yield pCDNA3/706-856. The expressed cytoplasmic tail recombinant protein contained a Met initiation residue attached N terminal to the Asn residue at position 706. Since neither the Env signal peptide nor the TM region was fused to the cytoplasmic tail, this recombinant protein was presumably synthesized by the free ribosome protein synthesis machinery, not by membrane-associated ribosomes.
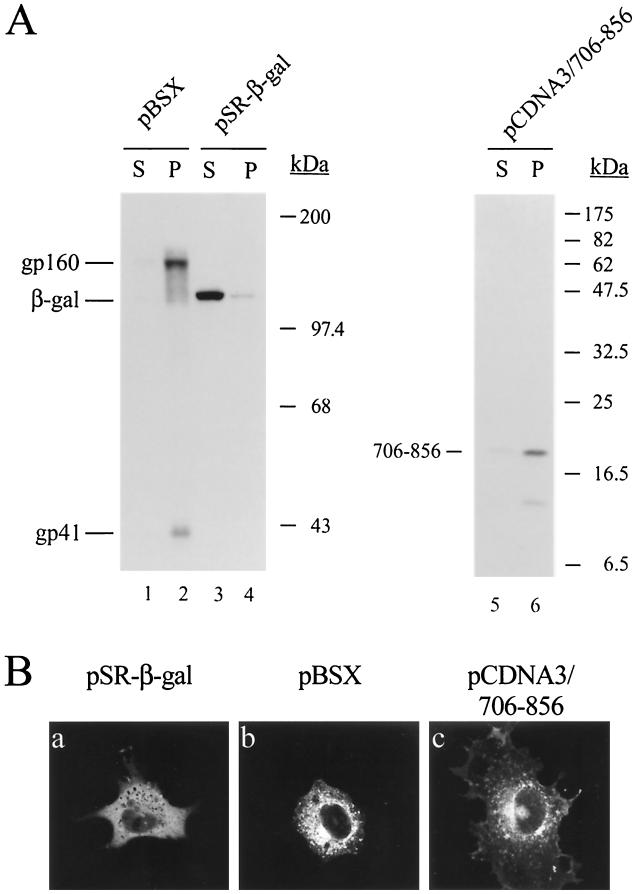
The intracellular localization of the 706-856 recombinant protein was then determined by cell fractionation. To determine the fidelity of the cell fractionation in separating membrane proteins from cytosolic proteins, COS-1 cells were also transfected in parallel with pSR-β-gal or pBSX. pSR-β-gal encodes an E. coli β-galactosidase gene driven by the SRα promoter composed of the simian virus 40 (SV40) promoter and the R-U5 segment of the human T-cell leukemia virus type 1 (HTLV-1) long terminal repeat (59). pBSX is an SV40 late promoter-driven HXB2 strain Env expression plasmid (11). gp160 and gp41 were exclusively distributed to the membrane fraction (Fig. 2A, lane 2), indicating the membrane-associated nature of these Env species. β-Galactosidase predominantly located to the soluble fraction (Fig. 2A, lane 3), confirming that β-galactosidase is a cytoplasmic soluble protein. The cytoplasmic tail recombinant protein predominantly located to the membrane fraction (Fig. 2A, lane 6).
FIG. 2.
Membrane association of a gp41 cytoplasmic tail recombinant protein. (A) Cell fractionation of a cytoplasmic domain fragment expressed in the absence of other Env segments. COS-1 cells were transfected with 5 μg (each) of pBSX, pSR-β-gal, or pCDNA3/706-856 by the DEAE-dextran method. Two days after transfection, postnuclear supernatants were prepared and ultracentrifuged to resolve into soluble (S) and membrane pellet (P) fractions. Equal portions of soluble and pellet fractions were separated by SDS–7.5% PAGE (lanes 1 to 4) or SDS–15% PAGE (lanes 5 and 6) followed by Western blotting using Chessie 8 (lanes 1, 2, 5, and 6) and β-galactosidase (lanes 3 and 4) MAbs. (B) Examination of subcellular localization of the cytoplasmic tail recombinant protein by confocal immunofluorescence. COS-1 cells transfected with pSR-β-gal, pBSX, or pCDNA3/706-856 were grown on coverslips. Transfected cells were fixed, permeabilized, and immunostained with β-galactosidase MAb (panel a) or Chessie 8 MAb (panels b and c) followed by incubation with FITC-conjugated anti-mouse immunoglobulin G. The slides were viewed under a confocal immunofluorescence microscope.
Examination of subcellular localization of the gp41 cytoplasmic tail recombinant protein by confocal microscopy.
The intracellular localization of β-galactosidase, HIV-1 Env, and the 706-856 recombinant protein was examined under a confocal immunofluorescence microscope. β-Galactosidase exhibited the homogeneous staining characteristic of a cytoplasmic location, and the plasma membrane was not stained (Fig. 2B, panel a). There were intense signals in the periphery of the nuclei of cells expressing HIV-1 Env (Fig. 2B, panel b). These sites were presumably associated with membranes. Although staining of HIV-1 Env expression was mostly in the perinuclear area and in peripheric dots in the cytoplasm, cell surface speckles were also evident (Fig. 2B, panel b). The cytoplasmic tail recombinant protein was perinuclearly stained in the cytoplasm, and speckles of fluorescence were also observed on the peripheral plasma membrane (Fig. 2B, panel c).
The C-terminal, but not the N-terminal, segment of the cytoplasmic tail contains membrane-targeting signals.
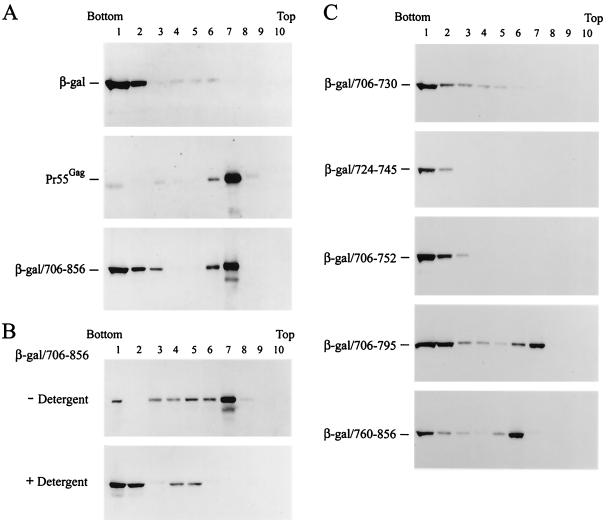
To determine whether the cytoplasmic tail contains membrane-targeting signals, the ability of the cytoplasmic tail to target the heterologous, cytosolic E. coli β-galactosidase to membranes was examined by an in vivo protein-membrane interaction assay (47, 55). This assay was previously used to study the membrane-binding ability of vesicular stomatitis virus M protein and myristylated HIV-1 Gag proteins (15, 16, 32, 56). COS-1 cells were transfected with pCDNA3-β-gal, a pCDNA3-based vector that encodes the E. coli β-galactosidase (Fig. 1A), with pCMV-Gag, a CMV promoter-driven Gag expression plasmid, or with pCDNA3-β-gal/706-856, which encodes a fusion protein with the gp41 cytoplasmic tail attached at the C terminus of β-galactosidase (Fig. 1B). β-Galactosidase exclusively located to the bottom fractions (fractions 1 and 2) of the gradient (Fig. 3A, top panel). The Gag precursor Pr55Gag exclusively floated to membrane fractions 6 and 7 (Fig. 3A, middle panel), which correspond to the 6 to 65% sucrose interface. This observation was consistent with the previous finding that Pr55Gag is predominantly associated with membranes (55). About 60% of the total β-galactosidae/706-856 fusion protein synthesized floated to membrane fractions 6 and 7 (Fig. 3A, bottom panel). These observations indicated that although the cytoplasmic tail recombinant protein is designed to be synthesized by free ribosomes, it is routed to cellular membranes.
FIG. 3.
Determination of membrane-targeting signals by membrane flotation assay. (A) Membrane-targeting ability of the gp41 cytoplasmic tail. The postnuclear fractions prepared from COS-1 cells transfected with pCDNA3-β-gal, pCMV-Gag, or pCDNA3-β-gal/706-856 were subjected to equilibrium flotation centrifugation and analyzed by Western blotting using β-galactosidase MAb (top panel), CA MAb (middle panel), or Chessie 8 MAb (bottom panel). (B) Effect of detergent on membrane binding of β-galactosidase/706-856. The postnuclear fraction prepared from COS-1 cells expressing β-galactosidase/706-856 was divided into two portions. One portion was directly analyzed by membrane flotation assay and another portion was treated with 1% SDS prior to flotation analysis. After centrifugation and SDS-PAGE, the samples were analyzed by Western blotting using β-galactosidase MAb. (C) Mapping of membrane-targeting elements located in the gp41 cytoplasmic tail. The postnuclear fractions obtained from COS-1 cells transfected with pCDNA3-β-gal constructs that encoded various subdomains of the cytoplasmic tail as indicated were analyzed by membrane flotation assay followed by Western blotting using β-galactosidase MAb.
To confirm that β-galactosidase/706-856, which floated to fractions 6 and 7, was membrane associated, the postnuclear fraction obtained from cells expressing the fusion protein was either treated with 1% SDS or left untreated prior to membrane flotation assay. The β-galactosidase/706-856 fusion protein predominantly located to the bottom cytoplasmic fractions after SDS treatment (Fig. 3B), indicating that the fusion protein which floated to fractions 6 and 7 is indeed membrane associated and that the interaction of the 706-856 fusion protein with the membrane is disrupted by detergent treatment.
To map regions in the cytoplasmic tail crucial for membrane targeting, the ability of the N-terminal and C-terminal segments of the cytoplasmic tail (Fig. 1B) to direct β-galactosidase to cellular membranes was assessed by membrane flotation assay. β-Galactosidase fusion proteins containing residues 706 to 730, 724 to 745, and 706 to 752 all located to the cytosolic fractions, whereas a fraction of the 706-795 fusion proteins floated to membrane fractions 6 and 7 (Fig. 3C). A fraction of the C-terminal fusion protein 760-856 also floated to the membrane fractions (Fig. 3C). These observations showed that the C-terminal two-thirds, but not the N-terminal one-third, segment of the cytoplasmic tail contains membrane-targeting signals.
Membrane targeting by LLP sequences
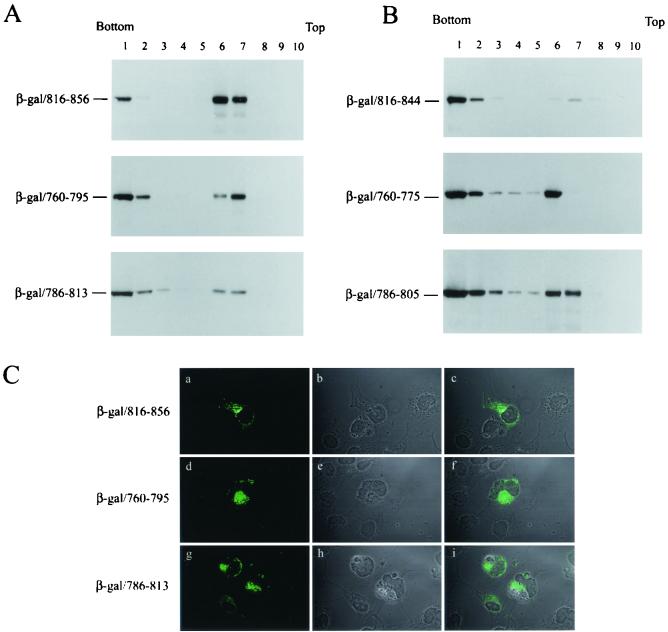
To assess whether each of the three LLP α-helices located in the cytoplasmic tail encodes membrane-targeting signals, β-galactosidase fusion proteins encoding residues 816 to 856, 760 to 795, and 786 to 813, which span LLP-1, LLP-2, and LLP-3, respectively, were analyzed by membrane flotation. All these β-galactosidase/LLP fusion proteins were able to float to membrane fractions 6 and 7 (Fig. 4A). To further determine the role of LLP motifs in membrane binding, the effect of deletions in the C termini of these LLP motifs on membrane association was examined. The 816-844 fusion protein predominantly located to the cytosolic fraction (Fig. 4B, top panel), indicating that an intact LLP-1 sequence is critical for membrane association. The 760-775 and 786-805 fusion proteins located to both the cytosolic and membrane fractions (Fig. 4B, middle and bottom panels, respectively), indicating that residues 760 to 775 and 786 to 805 possess the membrane association property. When COS-1 cells expressing each of the β-galactosidase/LLP fusion proteins were examined by confocal microscopy, all of these fusion proteins predominantly located to a juxtanuclear compartment (Fig. 4C), suggestive of an ER and/or Golgi localization of these fusion proteins.
FIG. 4.
Each of the three LLP motifs confers membrane-targeting ability. (A) The membrane-targeting ability of the LLP sequences determined by membrane flotation assay. pCDNA3-β-gal chimeras each encoding sequences encompassing the LLP-1, LLP-2, or LLP-3 motif as indicated were assessed by membrane flotation assay. (B) Effects of deletions in the C terminus of the LLP motifs on membrane binding. Postnuclear supernatants obtained from COS-1 cells expressing sequences containing deletions in the C terminus of LLP-1, LLP-2, and LLP-3, respectively, were analyzed by sucrose gradient equilibrium centrifugation. (C) Subcellular localization of the β-galactosidase/LLP fusion proteins. COS-1 cells expressing β-galactosidase/LLP fusion proteins were immunostained with β-galactosidase MAb and FITC-conjugated anti-mouse immunoglobulin G and then were examined by confocal microscopy as shown in panels a, d, and g, respectively. The phase-contrast images for the fields examined are shown in panels b, e, and h, respectively. The immunofluorescence staining and phase-contrast images were superimposed, and the images are shown in panels c, f, and i, respectively.
Apposition of cytoplasmic tail fusion proteins to membranes.
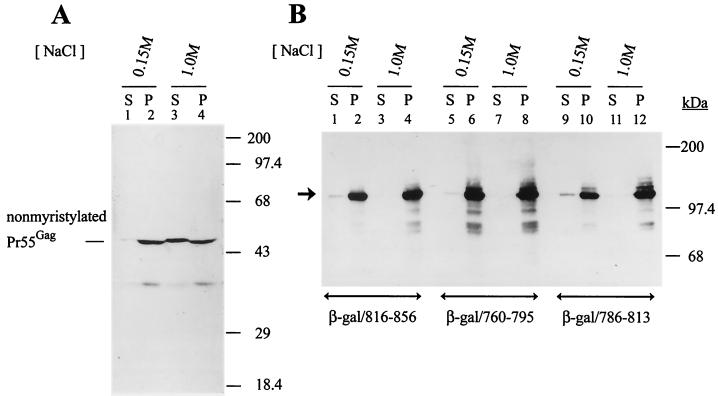
To study the nature of the interaction of the LLP motifs with membranes, membrane pellets obtained from COS-1 cells expressing each of the LLP fusion proteins were extracted with phosphate buffer containing 0.15 or 1 M NaCl. High-salt treatment extracts nonintegral membrane proteins from membranes (4, 56). Membranes obtained from cells transfected with HIVgptmyr− (63) were also extracted with low or high salt. HIVgptmyr− encodes a glycine-to-alanine substitution at residue 2 of MA, which abolishes the Pr55Gag myristylation signal and severely impairs Gag membrane binding (7, 47, 55, 72). High-salt conditions significantly extracted nonmyristylated Pr55Gag from the membrane fraction into the soluble fraction (Fig. 5A), which was consistent with the previous observation that nonmyristylated Pr55Gag binds to membranes under physiologic salt conditions (0.15 M NaCl) but dissociates from membranes at 1 M NaCl (7). In contrast, each of the β-galactosidase/LLP fusion proteins was still associated with membranes upon high-salt treatment (Fig. 5B), indicating that each of these LLP motifs may embed into cellular membranes when interacting with membranes.
FIG. 5.
Resistance of membrane-bound β-galactosidase/LLP fusion proteins to high-salt extraction. (A) High-salt extraction of nonmyristylated Pr55Gag from membranes. The postnuclear supernatant prepared from COS-1 cells transfected with HIVgptmyr− was divided into two portions and then ultracentrifuged to obtain the membranes. The pelleted membranes were incubated with phosphate buffer, pH 7.4, supplemented with 0.15 or 1 M NaCl as indicated, and the resultant mixtures were ultracentrifuged to yield soluble (S) and membrane-bound (P) fractions. Equal volumes of the soluble and membrane fractions were analyzed by Western blotting using CA MAb. (B) Resistance of the β-galactosidase/LLP fusion proteins to high-salt extraction. Membrane pellets prepared from COS-1 cells expressing various β-galactosidase/LLP fusion proteins were extracted with low or high salt. The extracted mixtures were ultracentrifuged to resolve them into soluble and membrane fractions and then were analyzed by Western blotting using the β-galactosidase MAb. The arrowhead indicates the migration of the β-galactosidase/LLP fusion proteins.
Insertion of in vitro synthesized β-galactosidase/LLP fusion proteins into membranes.
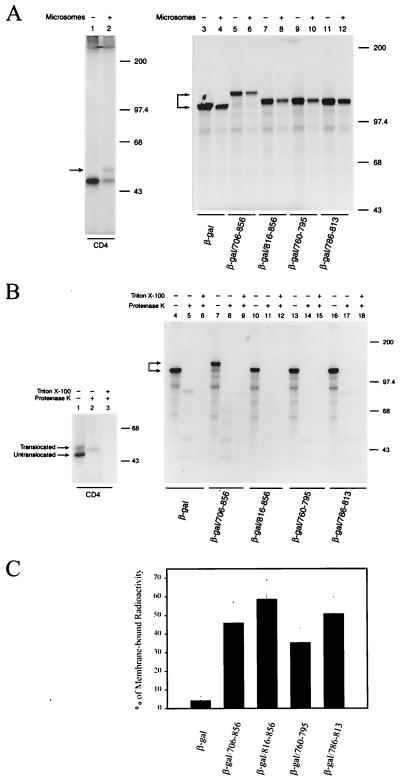
To further define the mode of binding of β-galactosidase/LLP fusion proteins to membranes, 35S-labeled β-galactosidase and β-galactosidase/LLP fusion proteins were synthesized by in vitro coupled transcription/translation in the presence or absence of microsomal membranes. Synthesis of human CD4 encoded by pCDNA3/CD4 in the presence and absence of microsomes was also performed as a control. In addition to the lower-molecular-weight form of CD4, which was also seen in the absence of membranes during in vitro synthesis, a higher-molecular-weight form of CD4, as marked by the arrowhead, was detected in the presence of membranes (Fig. 6A, lane 2). This higher-molecular-weight form represented the CD4 molecule translocated across membranes and modified by posttranslational glycosylation. Although the presence of membranes reduced the synthesis of β-galactosidase and β-galactosidase/LLP fusion proteins, the presence of membranes did not apparently affect the mobility of these proteins on SDS-PAGE (Fig. 6A, lanes 3 to 12), suggesting that these β-galactosidase/LLP fusion proteins are not translocated across membranes through the β-galactosidase moiety.
FIG. 6.
Characterization of in vitro synthesized β-galactosidase fusion proteins. (A) In vitro synthesis of β-galactosidase/LLP fusion proteins. [35S]methionine-labeled human CD4 and β-galactosidase/LLP fusion proteins synthesized in vitro in the presence or absence of canine pancreatic microsomal membranes were analyzed by SDS-PAGE. (B) Treatment of in vitro synthesized β-galactosidase/LLP fusion proteins with proteinase K. In vitro transcription/translation-synthesized 35S-labeled CD4, β-galactosidase, and β-galactosidase/LLP fusion proteins were treated with or without proteinase K or were first solubilized with 1% Triton X-100 before proteinase K digestion and then analyzed by SDS-PAGE. (C) Posttranslational binding of β-galactosidase/cytoplasmic tail proteins to membranes. 35S-labeled β-galactosidase and β-galactosidase/LLP fusion protein synthesized in vitro in the absence of membranes were incubated with canine pancreatic microsomal membranes at 30°C for 30 min. The reaction mixtures were ultracentrifuged to resolve them into the soluble and membrane fractions and then were analyzed by SDS-PAGE. The radioactivity of bands corresponding to the β-galactosidase and β-galactosidase/LLP fusion proteins partitioned in each fraction was quantitated by an Instant Imager (Packard Instrument Company, Meriden, Conn.). The ratio of the radioactivity in the membrane fraction to the total radioactivity is expressed as a percentage. The diagram represents the percentage of membrane-bound β-galactosidase and each β-galactosidase/LLP fusion protein from four experiments (mean average ± standard deviation).
A protease protection assay was then performed to determine whether these β-galactosidase/LLP fusion proteins were translocated across membranes during in vitro synthesis. Proteinase K treatment completely digested the untranslocated form of CD4 but converted the membrane-translocated form to a faster-migrating form (Fig. 6B, lane 2), indicating that the translocated form is protected from protease digestion. The increase in mobility of the protected form indicated that the CD4 cytoplasmic tail, which faces the cytoplasmic side, is digested by proteinase K. As expected, solubilization of membranes by Triton X-100 before protease treatment completely destroyed the CD4 molecule (Fig. 6B, lane 3). Under the same conditions, proteinase K treatment destroyed β-galactosidase and β-galactosidase/LLP fusion proteins, although some nonspecific bands were seen (Fig. 6B, lanes 5, 8, 11, 14, and 17, respectively). These nonspecific bands were also observed in samples disrupted by Triton X-100 prior to proteinase K treatment (Fig. 6B, lanes 6, 9, 12, 15, and 18, respectively). Taken together, these studies indicated that most of the β-galactosidase/LLP molecule faces the cytoplasm.
To provide evidence that the synthesized β-galactosidase/LLP fusion protein can posttranslationally anchor to membranes via the LLP sequences, 35S-labeled β-galactosidase/LLP fusion proteins synthesized in vitro in the absence of membranes were incubated with microsomes. The mixtures were then ultracentrifuged to resolve them into the soluble and membrane fractions. β-Galactosidase was predominantly partitioned into the soluble fraction, whereas 35 to 58% of the β-galactosidase/LLP fusion proteins were bound to membranes (Fig. 6C). The differential membrane binding among these LLP fusion proteins may represent the differential intrinsic characteristic of these LLP sequences to insert into membranes under the in vitro membrane binding condition.
Subcellular fractionation of the LLP fusion proteins.
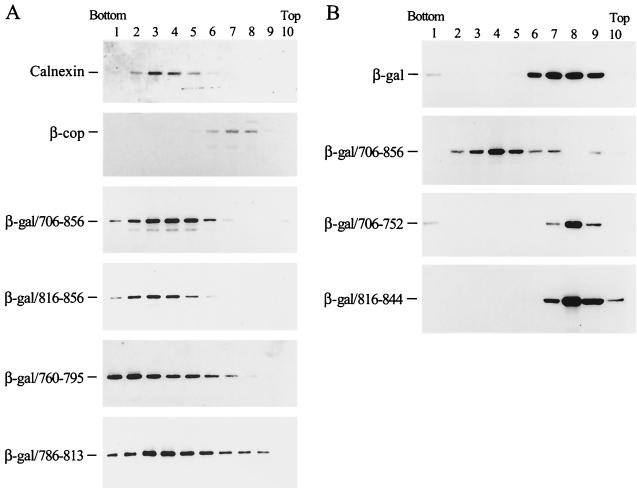
To evaluate the localization of the LLP fusion proteins to the ER or the Golgi apparatus when expressed in living cells, subcellular fractionation using iodixanol-based linear density gradient centrifugation was performed. Antibodies raised against calnexin, an ER resident protein (21), and β-COP, a Golgi marker, were also used to probe the ER and Golgi membrane distributions in untransfected cells. The membrane distribution of these LLP fusion proteins corresponded with that of calnexin to the denser ER membranes, which was distinct from the distribution of β-COP to the less dense Golgi membranes (Fig. 7A). This finding indicated a predominant ER localization of these β-galactosidase/LLP fusion proteins in intact cells. To assess the specificity of the binding of these LLP fusion proteins to the ER, subcellular fractionation of β-galactosidase fusion proteins containing 706-to-752 and 816-to-844 segments was performed. These two fusion proteins sedimented to the top fractions of the gradient where the cytosolic β-galactosidase protein also sedimented (Fig. 7B), showing that the 706-to-752 and 816-to-844 segments do not confer ER-targeting ability on β-galactosidase.
FIG. 7.
Subcellular fractionation of the β-galactosidase fusion proteins. (A) LLP fusion proteins. Postnuclear fractions obtained from COS-1 expressing β-galactosidase fusion proteins as indicated were analyzed by iodixanol-based linear density gradient centrifugation. After fractionation, membranes in each fraction were pelleted by ultracentrifugation and then analyzed by immunoblotting using β-galactosidase MAb. Subcellular fractionation of untransfected COS-1 was also performed, and aliquots of fractionated samples were analyzed by immunoblotting using antibodies directed against calnexin (ER marker) or β-COP (Golgi marker). (B) 706-752 and 816-844 fusion proteins. Postnuclear supernatants obtained from COS-1 expressing 706-752 or 816-844 fusion proteins were analyzed by subcellular fractionation followed by Western blotting using β-galactosidase MAb.
Localization of LLP sequences to subcellular organelles examined by confocal microscopy.
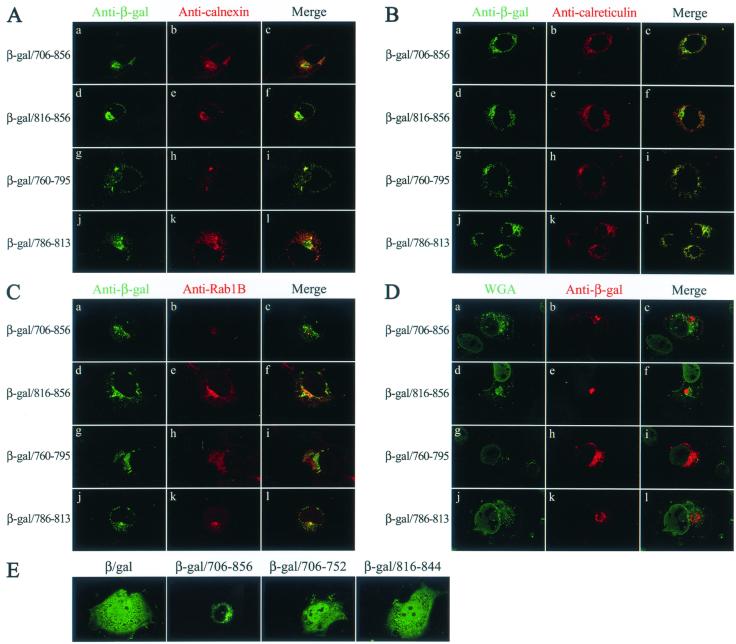
To confirm that these LLP fusion proteins are transported to the ER, colocalization of β-galactosidase/LLP fusion proteins with calnexin was examined. β-Galactosidase MAb staining of the cytoplasmic tail and each of the LLP fusion proteins revealed a large reticular network typical of the ER, together with intense staining in the perinuclear region (Fig. 8A, column 1). Anti-calnexin also revealed a distinct punctate staining located in the perinuclear region of the cell (Fig. 8A, column 2). A superimposition of the green and red fluorescence images showed that each of the fusion proteins substantially localized to the region stained by anti-calnexin (Fig. 8A, column 3). Colocalization of these fusion proteins with calreticulin, an ER luminal protein (36, 42), was also examined. These fusion proteins located to a perinuclear membrane compartment where calreticulin also colocalized (Fig. 8B). Colocalization of these proteins with Rab1B, which locates to the ER/cis-Golgi intermediate compartment or to the cis-Golgi (28, 54), was also performed. Although some of the green and red fluorescence images appeared to be slightly superimposed (Fig. 8C), the degree of superimposition was much less than that seen in Fig. 8A and B. Moreover, these fusion proteins did not colocalize with WGA (Fig. 8D), a trans-Golgi network marker (27, 62).
FIG. 8.
Subcellular localization of β-galactosidase/LLP fusion proteins. (A) Colocalization of β-galactosidase/LLP fusion proteins with calnexin. After fixation and permeabilization, COS-1 cells expressing β-galactosidase fusion proteins as indicated were successively incubated with mouse anti-β-galactosidase, goat anti-calnexin, and appropriate FITC- and TRITC-conjugated secondary antibodies. The cells were analyzed by confocal microscopy for localization of β-galactosidase fusion proteins (panels a, d, g, and j) and calnexin (panels b, e, h, and k). The green and red fluorescence images were merged and are shown in panels c, f, i, and l. (B) Colocalization of β-galactosidase fusion proteins with calreticulin. Transfected cells were immunostained with mouse anti-β-galactosidase and goat anti-calreticulin and then were examined by confocal microscopy. (C) Subcellular localization of β-galactosidase/LLP fusion proteins to a compartment distinct from where Rab1B localizes. Transfected cells were immunostained with β-galactosidase MAb and rabbit anti-Rab1B and then were visualized by immunofluorescence microscopy. (D) Localization of β-galactosidase/cytoplasmic tail fusion proteins to a region distinct from the trans-Golgi network. Transfected cells were successively incubated with mouse anti-β-galactosidase, TRITC-conjugated anti-mouse immunoglobulin G, and fluorescence-labeled WGA and then were examined under confocal microscopy. (E) Subcellular localization of 706-752 and 816-844 fusion proteins. Cells expressing β-galactosidase and β-galactosidase fusion proteins as indicated were examined by confocal microscopy using β-galactosidase MAb.
When subcellular localization of β-galactosidase fused to 706-to-752 and 816-to-844 segments was examined by confocal microscopy, these two fusion proteins, along with β-galactosidase, were distributed in the cytoplasm and nucleus (Fig. 8E). Unlike the pSR-β-gal plasmid examined in Fig. 2B, which encodes a weak SV40 and HTLV-1 hybrid promoter/enhancer, pCDNA3-β-gal encodes a strong CMV promoter for protein expression. Thus, a fraction of overexpressed β-galactosidase is also transported to the nucleus. This observation together with the membrane flotation and subcellular fractionation results (Fig. 3C, 4B, and 7B) indicated that the 706-to-752 and 816-to-844 segments do not contain an ER-targeting signal.
Subcellular localization of EGFP-tagged LLP sequences to the ER
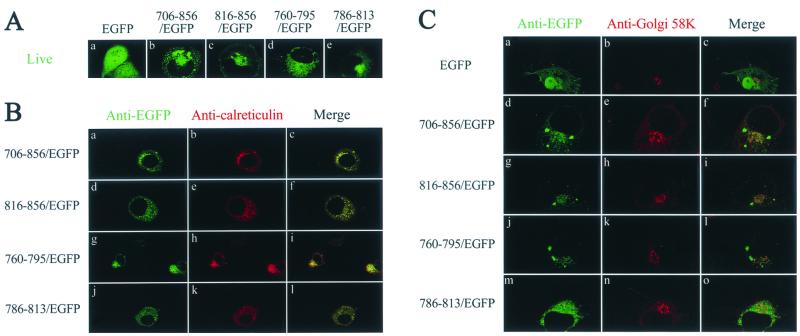
To examine whether the ER-targeting ability of the LLP sequences is dependent on its N-terminal or C-terminal location in a fusion protein, the entire cytoplasmic tail and each of the LLP motifs were individually fused to the N terminus of heterologous protein EGFP from the jellyfish Aequorea victoria. To avoid artifacts that may arise during the fixation and permeabilization process, live cells were examined by confocal microscopy. EGFP, whose expression is driven by a strong CMV immediate-early promoter, was distributed in the cytoplasm as well as in the nucleus (Fig. 9A). Fusion at the N terminus of EGFP with either the cytoplasmic tail or any of the LLP sequences redirected EGFP from its cytoplasmic and nucleus locations to a perinuclear region (Fig. 9A), where calreticulin also coresided (Fig. 9B). These LLP/EGFP fusion proteins were not transported to the Golgi apparatus, as judged by the poor colocalization of these proteins with Golgi 58K protein (Fig. 9C), which localizes to the cytoplasmic face of the Golgi apparatus (5).
FIG. 9.
Subcellular localization of cytoplasmic tail subdomains fused to the N terminus of EGFP. (A) Examination in a living condition. COS-1 cells were transfected with pEGFP(N2) or a pEGFP(N2) chimera that encoded the cytoplasmic tail or each of the LLP sequences as indicated. One day after transfection cells were directly examined under a confocal microscope. (B) Colocalization with calreticulin. COS-1 cells expressing EGFP or EGFP/LLP fusion proteins were fixed, permeabilized, and successively incubated with rabbit anti-EGFP, FITC-conjugated anti-rabbit immunoglobulin G, goat anti-calreticulin, and TRITC-conjugated anti-goat immunoglobulin G. (C) Localization of EGFP fusion proteins to a compartment distinct from the Golgi apparatus. COS-1 cells expressing EGFP or EGFP/LLP fusion proteins were successively incubated with rabbit anti-EGFP, FITC-conjugated anti-rabbit immunoglobulin G, mouse anti-Golgi 58K, and TRITC-conjugated anti-mouse immunoglobulin G.
DISCUSSION
Although the interaction of the HIV-1 gp41 cytoplasmic tail with lipid bilayers has been extensively studied using peptide modeling, the molecular basis for the interaction of this domain with cellular membranes in HIV-1-infected or Env-expressing cells is still poorly understood. In the present study, we dissected the role of the gp41 cytoplasmic tail in cellular membrane interaction from that of the signal peptide and the TM domain of the Env by examining intracellular localization of cytoplasmic tail recombinant proteins expressed in the absence of other Env segments in mammalian cells. We demonstrated a direct binding of a gp41 cytoplasmic tail recombinant protein to cellular membranes (Fig. 2). We also found that the C-terminal two-thirds segment of the cytoplasmic tail, residues 760 to 856, contains information to direct a cytosolic E. coli β-galactosidase to cellular membranes (Fig. 3C). Our study provides a model system to assess the membrane-binding potential in eucaryotic cells of various proteins or domains of interest. Since each of the three α-helical LLP sequences by itself possesses membrane-targeting ability (Fig. 4A and C), it is unlikely that the membrane binding ability of the cytoplasmic tail is via association of the LLP motifs with other membrane-bound proteins. Association with cellular membranes is a rather unique and intrinsic feature of these α-helices.
The observation that β-galactosidase/LLP fusion proteins are resistant to high-salt extraction (Fig. 5) implies that these LLP sequences are embedded in membranes when interacting with cellular membranes. This result is consistent with the previous finding that an antibody directed against the C terminus of the gp41 cytoplasmic tail fails to gain access to the target sequence when the HSV gD-1/cytoplasmic tail chimera is bound to microsomes (30). In addition, carbonate treatment cannot extract this cytoplasmic tail fusion protein from membranes (30), and the 789-815 peptide is protected from proteolysis when it is in a membrane-bound state (34). The membrane-targeting property of the gp41 cytoplasmic tail is also supported by the ability of the β-galactosidase/LLP fusion proteins to posttranslationally bind to membranes in vitro (Fig. 6C). Also, a protease protection assay showed that most of the β-galactosidase/LLP molecule faces the cytoplasm, resulting in sensitivity to protease digestion (Fig. 6B). Collectively, our study demonstrates that these β-galactosidase/LLP fusion proteins insert into membranes via the LLP sequences.
There are precedents that show this mode of tail-anchored posttranslational membrane insertion. A class of eucaryotic integral membrane proteins translocates posttranslationally across membranes via a C-terminal hydrophobic anchor sequence (52). A short C-terminal hydrophobic sequence of synaptobrevin can posttranslationally anchor the molecule to the ER, resulting in a trans-membrane orientation of the hydrophobic tail (65). This type of membrane insertion is mechanistically different from protein translocation with signal peptides (37). Vaccinia virus H3L Env protein, lacking signal peptide cleavage and N glycosylation, is exposed on the surfaces of intracellular immature virions after synthesis in the cytoplasm. This protein posttranslationally inserts into membranes via its C-terminal, hydrophobic anchor sequence, leaving most of the molecule facing the cytoplasm (20).
The gp41 cytoplasmic tail has a tyrosine-based motif located in its membrane-proximal region. This motif is involved in the trafficking and endocytosis of the HIV-1 Env via clathrin-associated AP-1 and AP-2 adapter complexes (6, 53). Also, this membrane proximal, tyrosine-based signal targets the HIV-1 Env to the basolateral membrane in polarized cells, resulting in basolateral virus budding in the polarized cells (40, 41, 48). We demonstrated in the present study that each of the LLP motifs contains a novel, position-independent targeting signal that is sufficient to target non-ER-binding proteins, such as β-galactosidase and EGFP, to the ER (Fig. 7, 8, and 9). In contrast, the N-terminal portion of the cytoplasmic tail and an LLP-1 sequence lacking the last 12 amino acids in the C terminus do not confer ER-binding ability on β-galactosidase (Fig. 7B and 8E). Although these findings provide a basis for the previous observation that the presence of the gp41 C-terminal segment in the Env hinders the Env from transporting out of the ER and the subsequent precursor cleavage (31), the precise role of LLP-mediated ER-targeting during Env maturation remains to be determined. It is likely that these multiple subcellular sorting signals, including the ER-targeting signal examined here, may govern the intracellular transport of the HIV-1 Env to different locations at different stages during the virus replication cycle.
In the present study, we also noted a correlation between the membrane-binding ability and multimerization potential of subdomains in the cytoplasmic tail (Table 1). In peptide modeling studies, binding of LLP-1 peptides to membrane bilayers is often accompanied by conformational changes of the peptides from being less structured in solution to a predominant helical structure in the membrane-bound state (26, 35, 57). The conformational alterations also include transitions of the peptides from a monomeric state in solution to an oligomeric state in the membrane environment, which results in hydrophilic pore formation and leads to osmotic disintegrity (17, 43, 44).
TABLE 1.
Correlation of the membrane-binding ability and multimerization potentials of subdomains located in the HIV-1 gp41 cytoplasmic tail
| Segment | Membrane associationa | Multimerization potentialb |
|---|---|---|
| 706–856 | + | Multimer |
| 706–752 | − | Monomer |
| 706–795 | + | Multimer |
| 760–856 | + | Multimer |
| 816–856 | + | Multimer |
| 760–795 | + | Multimer |
| 786–813 | + | ND |
| 816–844 | − | Monomer |
| 760–775 | + | Low-ordered multimer and monomer |
| 786–805 | + | ND |
The membrane association ability of subdomains in the gp41 cytoplasmic tail was assessed by membrane flotation assay as described in the text.
The results for the multimerization potential of the cytoplasmic tail subdomains, as assessed by gel filtration analysis of the maltose-binding protein fused to these subdomains, were extracted from a previous paper (39). ND, not determined.
HIV-1 Env-mediated membrane fusion is a complicated cascade process; different domains of gp41 are involved in a series of concerted protein-protein and host cell-membrane interactions. Previous studies showed that peptides representing the hydrophobic fusion domain, which is located at the N terminus of gp41, oligomerize in a membrane-mimetic environment (33, 51). These peptides can destabilize the cellular membranes by inserting into membranes in an oblique orientation (9, 33, 49). An Env mutant with a point mutation in the fusion domain was shown to possess dominant interference with infectious virus production, indicating that an oligomeric state of the fusion domain is required for membrane fusion (24). Another region in gp41 crucial for virus infectivity is the leucine heptad repeat sequence, which is adjacent to the fusion domain. The conformation of the HIV-1 gp41 core required for membrane fusion is believed to be the six-stranded coiled-coil heterodimer formed by the leucine heptad repeat sequence and the C-terminal α-helix located proximally to the TM region (8, 60, 64). A peptide that mimics the α-helical heptad repeat sequence has been shown to bind to membranes (50). Results from these studies illustrate that multimerization of the fusion domain and leucine heptad repeat sequence, and the interactions of these two regions with cellular membranes, play critical roles in virus infectivity and membrane fusion.
The correlation of membrane binding ability and the multimerization potential of subdomains of the cytoplasmic tail (Table 1) also has implications for understanding the role of the cytoplasmic tail in virus entry into host cells. It is likely that the C-terminal gp41 cytoplasmic tail is in proximity to or interacts with membranes in a step during or after virus entry into host cells. A previous study showed that virus encoding an Env with deletion of the last 12 amino acids in the C-terminal cytoplasmic tail displayed an impaired infectivity phenotype (70). Our present study showed that a deletion of these 12 amino acids from the C terminus of LLP-1 results in the loss of LLP-1-mediated membrane binding (Fig. 4B and 8E). This same deletion also causes the LLP-1 to lose its self-assembly potential (39). In addition, this mutant is able to interfere in trans with the wt Env function (13). Collectively, the present and previous studies show that the multimerization and membrane-binding abilities of the LLP-1 sequence are critical to virus infectivity. It is likely that the C-terminal segment of the cytoplasmic tail may exert two functions at a step after TM ectodomain core-mediated fusion by (i) organizing into an extended coiled coil and by (ii) providing a hydrophobic face that binds and/or inserts into cellular or viral membranes. Knowledge regarding cytoplasmic tail multimerization and membrane association will lead to refinement of our understanding of the molecular mechanisms involving gp41 during virus-host cell interaction and LLP-mediated permeable pathways.
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Council (NSC 89-2320-B-001-047) and the Institute of Biomedical Sciences at Academia Sinica, Taipei, Taiwan, Republic of China.
We are indebted to Shi-Lan Hong for technical assistance and to Kuan-Yu Chou for assistance in confocal immunofluorescence microscopy.
REFERENCES
- 1.Akari H, Fukumori T, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J Virol. 2000;74:4891–4893. doi: 10.1128/jvi.74.10.4891-4893.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreassen H, Bohr H, Bohr J, Brunak S, Bugge T, Cotterill R M, Jacobsen C, Kusk P, Lautrup B, Petersen S B, Saermark T, Ulrich K. Analysis of the secondary structure of the human immunodeficiency virus (HIV) proteins p17, gp120, and gp41 by computer modeling based on neural network methods. J Acquir Immune Defic Syndr. 1990;3:615–622. [PubMed] [Google Scholar]
- 3.Berman P W, Nunes W M, Haffar O K. Expression of membrane-associated and secreted variants of gp160 of human immunodeficiency virus type 1 in vitro and in continuous cell lines. J Virol. 1988;62:3135–3142. doi: 10.1128/jvi.62.9.3135-3142.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthiaume L, Resh M D. Biochemical characterization of a palmitoyl acyltransferase activity that palmitoylates myristoylated proteins. J Biol Chem. 1995;270:22399–22405. doi: 10.1074/jbc.270.38.22399. [DOI] [PubMed] [Google Scholar]
- 5.Bloom G S, Brashear T A. A novel 58-kDa protein associates with the Golgi apparatus and microtubules. J Biol Chem. 1989;264:16083–16092. [PubMed] [Google Scholar]
- 6.Boge M, Wyss S, Bonifacino J S, Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 7.Bryant M, Ratner L. Myristylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 9.Chang D K, Cheng S F, Chien W J. The amino-terminal fusion domain peptide of human immunodeficiency virus type 1 gp41 inserts into the sodium dodecyl sulfate micelle primarily as a helix with a conserved glycine at the micelle-water interface. J Virol. 1997;71:6593–6602. doi: 10.1128/jvi.71.9.6593-6602.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S S-L. Functional role of the zipper motif region of human immunodeficiency virus type 1 transmembrane protein gp4. J Virol. 1994;68:2002–2010. doi: 10.1128/jvi.68.3.2002-2010.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S S-L, Ferrante A A, Terwilliger E F. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology. 1996;226:260–268. doi: 10.1006/viro.1996.0654. [DOI] [PubMed] [Google Scholar]
- 12.Chen S S-L, Lee C-N, Lee W-R, McIntosh K, Lee T-H. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane glycoprotein. J Virol. 1993;67:3615–3619. doi: 10.1128/jvi.67.6.3615-3619.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S S-L, Lee S-F, Chuang C-K, Raj V S. trans-dominant interference with human immunodeficiency virus type 1 replication and transmission in CD4+ cells by an envelope double mutant. J Virol. 1999;73:8290–8302. doi: 10.1128/jvi.73.10.8290-8302.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chernomordik L, Chanturiya A N, Suss-Toby E, Nora E, Zimmerberg J. An amphipathic peptide from the C-terminal region of the human immunodeficiency virus envelope glycoprotein causes pore formation in membranes. J Virol. 1994;68:7115–7123. doi: 10.1128/jvi.68.11.7115-7123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chong L D, Rose J K. Interaction of normal and mutant vesicular stomatitis virus matrix proteins with the plasma membrane and nucleocapsids. J Virol. 1994;68:441–447. doi: 10.1128/jvi.68.1.441-447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong L D, Rose J K. Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J Virol. 1993;67:407–414. doi: 10.1128/jvi.67.1.407-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comardelle A M, Norris C H, Plymale D R, Gatti P J, Choi B, Fermin C D, Haislip A M, Tencza S B, Mietzner T A, Montelaro R C, Garry R F. A synthetic peptide corresponding to the carboxy terminus of human immunodeficiency virus type 1 transmembrane glycoprotein induces alterations in the ionic permeability of Xenopus laevis oocytes. AIDS Res Hum Retrovir. 1997;13:1525–1532. doi: 10.1089/aid.1997.13.1525. [DOI] [PubMed] [Google Scholar]
- 18.Courageot J, Fenouillet E, Bastiani P, Miquelis R. Intracellular degradation of the HIV-1 envelope glycoprotein. Evidence for, and some characteristics of, an endoplasmic reticulum degradation pathway. Eur J Biochem. 1999;260:482–489. doi: 10.1046/j.1432-1327.1999.00193.x. [DOI] [PubMed] [Google Scholar]
- 19.Crise B, Buonocore L, Rose J K. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J Virol. 1990;64:5585–5593. doi: 10.1128/jvi.64.11.5585-5593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Fonseca F G, Wolffe E J, Weisberg A, Moss B. Characterization of the vaccinia virus H3L envelope protein: topology and posttranslational membrane insertion via the C-terminal hydrophobic tail. J Virol. 2000;74:7508–7517. doi: 10.1128/jvi.74.16.7508-7517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David V, Hochstenbach F, Rajagopalan S, Brenner M B. Interaction with newly synthesized and retained proteins in the endoplasmic reticulum suggests a chaperone function for human integral membrane protein IP90 (calnexin) J Biol Chem. 1993;268:9585–9592. [PubMed] [Google Scholar]
- 22.Earl P L, Moss B, Doms R W. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991;65:2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenberg D, Wesson M. The most amphiphilic α-helices include two amino acid segments in human immunodeficiency virus glycoprotein gp41. Biopolymers. 1990;29:171–177. doi: 10.1002/bip.360290122. [DOI] [PubMed] [Google Scholar]
- 24.Freed E O, Delwart E L, Buchschacher G L, Jr, Panganiban A T. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujii G, Horvath S, Woodward S, Eiserling F, Eisenberg D. A molecular model for membrane fusion based on solution studies of an amphiphilic peptide from HIV gp41. Protein Sci. 1992;1:1454–1464. doi: 10.1002/pro.5560011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gawrisch K, Han K-H, Yang J-S, Bergelson L D, Ferretti J A. Interaction of peptide fragment 828–848 of the envelope glycoprotein of human immunodeficiency virus type 1 with lipid bilayers. Biochemistry. 1993;32:3112–3118. doi: 10.1021/bi00063a024. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths G, Brands R, Burke B, Louvard D, Warren G. Viral membrane proteins acquire galactose in trans Golgi cisternae during intracellular transport. J Cell Biol. 1982;95:781–792. doi: 10.1083/jcb.95.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths G, Ericsson M, Krijnse-Locker J, Nilsson T, Goud B, Soling H D, Tang B L, Wong S H, Hong W. Localization of the Lys, Asp, Glu, Leu tetrapeptide receptor to the Golgi complex and the intermediate compartment in mammalian cells. J Cell Biol. 1994;127:1557–1574. doi: 10.1083/jcb.127.6.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haffar O K, Dowbenko D J, Berman P W. The cytoplasmic tail of HIV-1 gp160 contains regions that associate with cellular membranes. Virology. 1991;180:439–441. doi: 10.1016/0042-6822(91)90054-f. [DOI] [PubMed] [Google Scholar]
- 30.Haffar O K, Dowbenko D J, Berman P W. Topogenic analysis of the human immunodeficiency virus type 1 envelope glycoprotein, gp160: in microsomal membranes. J Cell Biol. 1988;107:1677–1687. doi: 10.1083/jcb.107.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haffar O K, Nakamura G R, Berman P W. The carboxy terminus of human immunodeficiency virus type 1 gp160 limits its proteolytic processing and transport in transfected cell lines. J Virol. 1990;64:3100–3103. doi: 10.1128/jvi.64.6.3100-3103.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiernan R E, Ono A, Freed E O. Reversion of a human immunodeficiency virus type 1 matrix mutation affecting Gag membrane binding, endogenous reverse transcriptase, and virus infectivity. J Virol. 1999;73:4728–4737. doi: 10.1128/jvi.73.6.4728-4737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kliger Y, Aharoni A, Rapaport D, Jones P, Blumenthal R, Shai Y. Fusion peptides derived from the HIV type 1 glycoprotein 41 associate within phospholipid membranes and inhibit cell-cell fusion. Structure-function study. J Biol Chem. 1997;272:13496–13505. doi: 10.1074/jbc.272.21.13496. [DOI] [PubMed] [Google Scholar]
- 34.Kliger Y, Shai Y. A leucine zipper-like sequence from the cytoplasmic tail of the HIV-1 envelope glycoprotein binds and perturbs lipid bilayers. Biochemistry. 1997;36:5157–5169. doi: 10.1021/bi962935r. [DOI] [PubMed] [Google Scholar]
- 35.Koenig B W, Bergelson L D, Gawrisch K, Ward J, Ferretti J A. Effect of the conformation of a peptide from gp41 on binding and domain formation in model membranes. Mol Membr Biol. 1995;12:77–82. doi: 10.3109/09687689509038499. [DOI] [PubMed] [Google Scholar]
- 36.Krause K H, Michalak M. Calreticulin. Cell. 1997;88:439–443. doi: 10.1016/s0092-8674(00)81884-x. [DOI] [PubMed] [Google Scholar]
- 37.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport T A. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasky L A, Groopman J E, Fennie C W, Benz P M, Capon D J, Dowbenko D J, Nakamura G R, Nunes W M, Renz M E, Berman P W. Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science. 1986;233:209–212. doi: 10.1126/science.3014647. [DOI] [PubMed] [Google Scholar]
- 39.Lee S-F, Wang C-T, Liang J Y-P, Hong S-L, Huang C-C, Chen S S-L. Multimerization potential of the cytoplasmic domain of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J Biol Chem. 2000;275:15809–15819. doi: 10.1074/jbc.M000601200. [DOI] [PubMed] [Google Scholar]
- 40.Lodge R, Götlinger H G, Gabuzda D, Cohen E A, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lodge R, Lalonde J-P, Lemay G, Cohen E A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michalak M, Baksh S, Opas M. Identification and immunolocalization of calreticulin in pancreatic cells: no evidence for “calciosomes.”. Exp Cell Res. 1991;197:91–99. doi: 10.1016/0014-4827(91)90484-c. [DOI] [PubMed] [Google Scholar]
- 43.Miller M A, Cloyd M W, Liebmann J, Rinaldo C R, Jr, Islam K R, Wang S Z S, Mietzner T A, Montelaro R C. Alterations in cell membrane permeability by the lentivirus lytic peptide (LLP-1) of HIV-1 transmembrane protein. Virology. 1993;196:89–100. doi: 10.1006/viro.1993.1457. [DOI] [PubMed] [Google Scholar]
- 44.Miller M A, Garry R F, Jaynes J M, Montelaro R C. A structural correlation between lentivirus transmembrane proteins and natural cytolytic peptides. AIDS Res Hum Retrovir. 1991;7:511–519. doi: 10.1089/aid.1991.7.511. [DOI] [PubMed] [Google Scholar]
- 45.Miller M A, Montelaro R C. Amphipathic helical segments of HIV-1 transmembrane (TM) proteins and their potential role in viral cytopathicity. In: Aloia R C, editor. Advances in membrane fluidity. Vol. 6. New York, N.Y: A. R. Liss; 1992. pp. 351–364. [Google Scholar]
- 46.Murakami T, Freed E O. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci USA. 2000;97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ono A, Freed E O. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Owens R J, Dubay J W, Hunter E, Compans R W. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pritsker M, Rucker J, Hoffman T L, Doms R W, Shai Y. Effect of nonpolar substitutions of the conserved Phe11 in the fusion peptide of HIV-1 gp41 on its function, structure, and organization in membranes. Biochemistry. 1999;38:11359–11371. doi: 10.1021/bi990232e. [DOI] [PubMed] [Google Scholar]
- 50.Rabenstein M, Shin Y-K. A peptide from the heptad repeat of human immunodeficiency virus gp41 shows both membrane binding and coiled-coil formation. Biochemistry. 1995;34:13390–13397. doi: 10.1021/bi00041a016. [DOI] [PubMed] [Google Scholar]
- 51.Rapaport D, Shai Y. Interaction of fluorescently labeled analogues of the amino-terminal fusion peptide of Sendai virus with phospholipid membranes. J Biol Chem. 1994;269:15124–15131. [PubMed] [Google Scholar]
- 52.Rapoport T A, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 53.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of endogenously synthesized HIV-1 envelope protein: mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 54.Schwaninger R, Plutner H, Bokoch G M, Balch W E. Multiple GTP-binding proteins regulate vesicular transport from the ER to Golgi membranes. J Cell Biol. 1992;119:1077–1096. doi: 10.1083/jcb.119.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spearman P, Wang J-J, Heyden N Y, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srinivas S K, Srinivas R V, Anantharamaiah G M, Segrest J P, Compans R W. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J Biol Chem. 1992;267:7121–7127. [PubMed] [Google Scholar]
- 58.Stein B S, Engleman E G. Intracellular processing of the gp160 HIV-1 envelope precursor. Endoproteolytic cleavage occurs in a cis or medial compartment of the Golgi complex. J Biol Chem. 1990;265:2640–2649. [PubMed] [Google Scholar]
- 59.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SR alpha promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan K, Liu J-H, Wang J-H, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venable R M, Pastor R W, Brooks B R, Carson F W. Theoretically determined three-dimensional structures for amphipathic segments of HIV-1 gp41 envelope protein. AIDS Res Hum Retrovir. 1989;5:7–22. doi: 10.1089/aid.1989.5.7. [DOI] [PubMed] [Google Scholar]
- 62.Virtanen I, Ekblom P, Laurila P. Subcellular compartmentalization of saccharide moieties in cultured normal and malignant cells. J Cell Biol. 1980;85:429–434. doi: 10.1083/jcb.85.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Willey D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 65.Whitley P, Grahn E, Kutay U, Rapoport T A, von Heijne G. A 12-residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J Biol Chem. 1996;271:7583–7586. doi: 10.1074/jbc.271.13.7583. [DOI] [PubMed] [Google Scholar]
- 66.Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 67.Willey R L, Bonifacino J S, Potts B J, Martin M A, Klausner R D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang C, Spies C P, Compans R W. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc Natl Acad Sci USA. 1995;92:9871–9875. doi: 10.1073/pnas.92.21.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang M, Ellenberg J, Bonifacino J S, Weissman A M. The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J Biol Chem. 1997;272:1970–1975. doi: 10.1074/jbc.272.3.1970. [DOI] [PubMed] [Google Scholar]
- 70.Yu X, Yuan X, McLane M F, Lee T-H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang H, Dornadula G, Alur P, Laughlin M A, Pomerantz R J. Amphipathic domains in the C terminus of the transmembrane protein (gp41) permeabilize HIV-1 virions: a molecular mechanism underlying natural endogenous reverse transcription. Proc Natl Acad Sci USA. 1996;93:12519–12524. doi: 10.1073/pnas.93.22.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]