Abstract
The monomeric red fluorescent protein DsRed (mDsRed) is an optical probe widely used in multicolor applications in flow cytometry and fluorescence microscopy. Although the crystal structure of monomeric DsRed has been determined, its molecular dynamics have not been fully elucidated. To better understand its molecular flexibility, the crystal structure of mDsRed was recently determined, and its structure and temperature factors were analyzed. Solvent-accessible hole connected with the mDsRed chromophore was observed on the mDsRed surface structure. Electron density map analysis showed the tyrosine-ring group of the mDsRed chromophore in a cis-conformation, exhibiting flexibility with a nonplanar configuration between the tyrosine and imidazoline rings of the chromophore. Temperature factor analysis indicated that the top and bottom of the β-barrel are relatively flexible. These structural findings extended our understanding of the molecular flexibility of mDsRed. The detailed data collection and structure determination reported in this study can be used for future structural analyses.
Keywords: Substrate binding protein, Single domain, Structural flexibility, X-ray diffraction data, Crystal structure
Specifications Table
| Subject | Biological science |
| Specific subject area | Structural Biology |
| Type of data | Processed |
| Data collection | Synchrotron: Pohang Light Source II (PLS-II) Beamline: 7C Detector: Q270 CCD X-ray detector (ADSC, USA) Data collection energy: 14.82 keV Data collection temperature: 100 K Data processing program: HKL2000 |
| Data source location | Institution: Kookmin University City/Town/Region: Seoul Country: Republic of Korea |
| Data accessibility | Structure factor and coordinates Repository name: Protein Data Bank Data identification number: https://doi.org/10.2210/pdb8WGP/pdb Direct URL to data: https://www.rcsb.org/structure/8WGP Instructions for accessing these data: Coordinate and structure factor can be download without permission. |
| Related research article | K.H. Nam, Structural Flexibility of the Monomeric Red Fluorescent Protein DsRed. Crystals 14 (1), 62; https://doi.org/10.3390/cryst14010062 [1]. |
1. Value of The Data
-
•
These data provide the coordinates and structure factors of mDsRed to aid in understanding its molecular function.
-
•
These data offer useful information about the flexibility of the mDsRed chromophore in its nonplanar configuration, which contributes to the understanding of the molecular dynamics of fluorescent proteins (FPs).
-
•
These data show distinct temperature factors compared to the previously determined mDsRed structure, providing insights into the molecular flexibility of mDsRed.
-
•
These data can be used to engineer FPs to improve their fluorescence properties.
2. Background
Fluorescent proteins are widely used as optical probes in molecular and cellular biology and as biosensor probes for detecting metal ions and pH levels [[2], [3], [4], [5], [6]]. The original tetrameric DsRed, derived from the reef coral Dendronephthya sp., was engineered into monomeric DsRed (mDsRed) to avoid slow maturation and challenges in FRET applications [[7], [8], [9], [10]]. mDsRed is widely used in multicolor applications such as flow cytometry and fluorescence microscopy [11]. The overall structure of mDsRed has been reported, providing useful information for understanding the structural properties of mDsRed [10]. However, the structural dynamics of mDsRed have not been fully elucidated. To understand the fundamental properties of mDsRed, recent studies have analyzed the structural dynamics of mDsRed using new crystal structures [1]. In this study, the detailed data collection and structure determination of mDsRed are reported for future utilization of the mDsRed structure.
3. Data Description
The indexing results of X-ray diffraction (XRD) experiments showed that rod-shaped mDsRed crystals belonged to the monoclinic space group P21. The unit cell dimensions of mDsRed were a = 39.56 Å, b = 107.00 Å, c = 50.02 Å, and β = 104.86° (Table 1). Matthews coefficient analysis showed that two mDsRed molecules occupied an asymmetric unit, with a VM of 2.11 ų/Da and a solvent content of 41.84 %. XRD data were processed in the range of 50.0–2.9 Å resolution, with 8,136 unique reflections. The overall completeness, redundancy, and I/σ of mDsRed were 91.5 %, 2.8, and 12.62, respectively. The overall I/σ, Rmerge, Rmeas, Rpim, CC1/2, and CC* values of mDsRed XRD data were 12.62, 0.157, 0.152, 0.188, 0.929, and 0.980, respectively (Fig. 1). At low (50.00–7.86 Å) resolution, the I/σ, Rmerge, Rmeas, Rpim, CC1/2, and CC* values were 30.16, 0.090, 0.107, 0.056, 0.990, and 0.998, respectively. At high (2.95–2.90 Å) resolution, the I/σ, Rmerge, Rmeas, Rpim, CC1/2, and CC* values were 4.04, 0.354, 0.443, 0.261, 0.646, and 0.886, respectively.
Table 1.
Data collection statistics for mDsRed
| Data collection | mDsRed |
|---|---|
| X-ray energy (eV) | 14820 |
| Space group | P21 |
| Cell dimension a, b, c (Å) α, β, γ (°) |
39.56, 107.00, 50.02 90.00, 104.86, 90.00 |
| Resolution (Å) | 50.0–2.90 (2.95–2.90) |
| Unique reflections | 8136 (362) |
| Completeness (%) | 91.5 (80.6) |
| Multiplicity | 2.8 (2.1) |
| I/sigma | 36.47 (5.63) |
| Rmerge | 0.157 (0.354) |
| Rmeas | 0.152 (0.441) |
| Rpim | 0.261 (0.102) |
| CC1/2 | 0.929 (0.646) |
| CC* | 0.980 (0.886) |
Values for the outer shell are given in parentheses.
Fig. 1.
Data processing results of mDsRed for (A) I/σ and Rmerge, (B) Rmeas and Rpim, and (C) CC1/2 and CC*.
The phasing problem of mDsRed was solved using the molecular replacement method. The electron density map was suitable for model building for all residues. The resolution of the final refinement of the mDsRed structure was 26.10 to 2.90 Å, with a completeness of 90.7 %. A total of 7,733 reflections were used in refinement, with a free R-value test set count of 388. The Rwork (working + test set), Rwork (working set), and Rfree values of the final structure of mDsRed were 0.199, 0.197, and 0.242, respectively (Table 2). A total of 3,510 protein atoms without hydrogen and 44 water molecules were defined. The correlation coefficients Fo-Fc and Fo-Fc free were 0.903 and 0.856, respectively. The RMS deviation of bond lengths and angles were 0.011 and 1.680, respectively.
Table 2.
Refinement statistics for mDsRed.
| Refinement | mDsRed |
|---|---|
| Resolution (Å) | 26.10–2.90 |
| Rworka | 0.197 |
| Rfreeb | 0.242 |
| RMS deviations | |
| Bonds (Å) | 0.011 |
| Angles (°) | 1.680 |
| B factors (Å2) | |
| Protein | 13.50 |
| Chromophore | 25.53 |
| Ramachandran plot | |
| Favored (%) | 90.5 |
| Allowed (%) | 7.9 |
| Disallowed (%) | 1.6 |
Rwork = Σ||Fobs| Σ |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
Rfree was calculated as Rwork using a randomly selected subset of unique reflections not used for structural refinement.
Ramachandran analysis of the deposited mDsRed showed that 388 and 33 of 427 residues were in the favored (90.9 %) and allowed (7.7 %) regions, respectively (Fig. 2). Meanwhile, Lys50 (phi, psi: 157.6, 114.8), Pro190 (−64.5, −136.7), and Asn192 (−59.2, 97.1) from chain A and Glu5 (−35.0, −31.6), Glu34 (170.1, 147.5), and Lys50 (166.4, 126.2) from chain B were in the outlier (1.4 %) region (Fig. 2).
Fig. 2.
Ramachandran plot of the deposited crystal structure of mDsRed.
The crystal structure of mDsRed exhibited the β-barrel fold (Fig. 3A). The mDsRed chromophore, generated by post-translational modification of the Gln66-Tyr67-Gly68 tripeptide, is almost at the center of the β-barrel fold (Fig. 3B). The surface structure of mDsRed showed that the mDsRed-B molecule exhibited a solvent-accessible hole with ∼3.80 Å diameter between the β7 and β10 strands, which connects with the chromophore, whereas no accessible hole was observed in the mDsRed-A molecule, indicating that the residues near the β7 and β10 strands were flexible. The observed hole in mDsRed-B is ∼8.16 Å away from the hydroxyl group of the tyrosine ring in the chromophore.
Fig. 3.
Crystal structure of mDsRed. (A) Cartoon representation of mDsRed containing the chromophore comprising the tripeptide Gln66-Tyr67-Gly68. (B) Surface structure of mDsRed showing the hole between the β7 and β10 strands of mDsRed-B.
The electron density map of the β-barrel region containing the residues near the chromophore was observed, whereas the electron density map of the tyrosine-ring group in the chromophore was relatively poor (Fig. 4A), indicating the conformational flexibility of the mDsRed chromophore. The two mDsRed chromophores are in cis-configuration between the tyrosine- and imidazoline-ring groups. The side view of the chromophore showed a nonplanar configuration between the tyrosine- and imidazoline-ring groups in the mDsRed chromophore (Fig. 4B). The position of the tyrosine ring of mDsRed-A was moved and rotated to the bottom direction by 2.0 Å and 30°, respectively. The position of the tyrosine ring of mDsRed-B was moved and rotated to the bottom direction by 0.7 Å and 30°, respectively (Fig. 4B). mDsRed chromophores interacted with Pro63, Arg95, Ser146, His163, Ser197, and Glu215 (Table 3).
Fig. 4.
Conformation of the mDsRed chromophore. (A) 2mFo-DFc (blue mesh, 0.8 σ) and mFo-DFc (green mesh, 3.0 σ and red mesh, −3.0 σ) electron density maps of the mDsRed chromophore. (B) Analysis of the configuration of the mDsRed chromophore. The figure was obtained from [1] and modified.
Table 3.
Interactions between the chromophore and its neighboring residues.
| Chromophore (atom) | Residue (atom) | Distance (Å) |
|
|---|---|---|---|
| mDsRed-A | mDsRed-B | ||
| CRQ66 (OH) | Ser146 (OG) | 2.49 | 2.29 |
| His163 (NE2) | 3.89 | 3.26 | |
| Ser197 (O) | 3.49 | 3.44 | |
| CRQ66 (N2) | Pro63 (O) | 3.49 | 3.79 |
| Glu215 (OE1) | 3.18 | 3.26 | |
| CRQ66 (O2) | Arg95 (NH2) | 3.14 | 2.99 |
| CRQ66 (OE1) | Gln213 (NE2) | 3.27 | 3.01 |
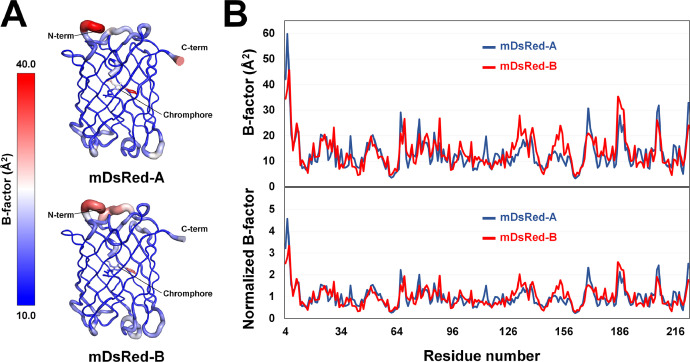
The B-factor putty representation showed that the β-barrel fold of mDsRed was rigid, whereas the N- and C-terminal regions exhibited high flexibility (Fig. 5A). B-factor analysis indicated that the loop containing Asp78 and Asp207 in mDsRed-A and that containing Pro186, Gln188, and Asp132 in mDsRed-B were relatively more flexible compared to β-barrel regions (Fig. 5B). Comparison of the B-factor and normalized B-factor values for mDsRed-A and mDsRed-B revealed distinct molecular flexibility, which may be attributed to the influence of crystal packing.
Fig. 5.
Flexibility of mDsRed. (A) B-factor putty representation of mDsRed-A and mDsRed-B. (B) Profile of the B-factor and normalized B-factor for mDsRed-A (blue) and mDsRed-B (red).
The coordinates and structure factors of mDsRed can be downloaded from the Protein Data Bank (PDB; https://www.rcsb.org/structure/8WGP) in the following formats: structure coordinates (PDBx/mmCIF, PDBML, PDB), XRD data (PDBx/mmCIF), and map coefficients (MTZ format). The validation report (PDF, mmCIF) for the deposited mDsRed is also available for download. Additionally, information on data collection and structure determination has been deposited in the Protein Data Bank.
4. Experimental Design, Materials and Methods
The sample preparation, XRD data collection, and structure determination were reported previously [1]. Briefly, the pDsRed-Monomer vector containing mDsRed (catalog no. 632467; Takara, Shiga, Japan) was transformed into Escherichia coli BL21 (DE3). Cells were grown in an LB broth medium containing ampicillin (0.1 µg/mL) at 37°C until an OD600 of 0.5–0.6 was reached. Protein expression was induced by adding isopropyl β-D-thiogalactopyranoside. Cells were incubated at 20°C overnight and stored at 4°C for 1 day. After cell disruption by sonication, the mDsRed supernatant was purified by heat treatment and size exclusion chromatography. mDsRed (purity: 70–80 %) was crystallized using the sitting-drop vapor diffusion method at 18°C. mDsRed crystals were obtained using a crystallization solution containing 0.1 M bis-Tris (pH 5.5), 0.2 M magnesium chloride, and 25 % (w/v) polyethylene glycol 3,350. Rod-shaped mDsRed crystals (10 × 25 × 200 µm³) were obtained within 2 months and used for XRD data collection at the 7C beamline at the Pohang Light Source II (Republic of Korea) [12]. A mDsRed crystal was cryoprotected with a crystallization solution supplemented with 20 % (v/v) glycerol for 10 s. XRD data were collected at 100 K, recorded on a Pilatus 6M detector (DECTRIS, Baden, Switzerland), and processed with the HKL2000 program [12]. The electron density map was obtained using the molecular replacement method with MOLREP (version 11.2.08) [13] implemented in CCP4 [14]. Structure building was performed using COOT (version 0.9.6) [15]. The model structure was refined with REFMAC5 (version 5.8.0267) [16] implemented in CCP4 [14]. The model structure was evaluated with MolProbity [17]. The structure factor and coordinate were deposited in the PDB (http://rcsb.org) under accession code 8WGP.
Limitations
Not applicable.
Ethics Statement
This work meets the ethical requirements for publication in this journal. This work does not involve human subjects, animal experiments, or any data collected from social media.
CRediT authorship contribution statement
Ki Hyun Nam: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft.
Acknowledgements
We would like to thank the beamline staffs at the 11C beamline at the Pohang Light Source II for their assistance with data collection. This work was funded by the National Research Foundation of Korea (NRF) (NRF-2021R1I1A1A01050838).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability
Crystal structure of DsRed-Monomer (Original data) (Protein Data Bank).
References
- 1.Nam K.H. Structural flexibility of the monomeric red fluorescent protein DsRed. Crystals. 2024;14(1):62. doi: 10.3390/cryst14010062. [DOI] [Google Scholar]
- 2.Tsien R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 3.Bizzarri R., Arcangeli C., Arosio D., Ricci F., Faraci P., Cardarelli F., Beltram F. Development of a novel GFP-based ratiometric excitation and emission pH indicator for intracellular studies. Biophys. J. 2006;90(9):3300–3314. doi: 10.1529/biophysj.105.074708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim I.J., Kim S., Park J., Eom I., Kim S., Kim J.H., Ha S.C., Kim Y.G., Hwang K.Y., Nam K.H. Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development. FEBS Lett. 2016;590(17):2982–2990. doi: 10.1002/1873-3468.12316. [DOI] [PubMed] [Google Scholar]
- 5.Kim I.J., Xu Y., Nam K.H. Metal-induced fluorescence quenching of photoconvertible fluorescent protein DendFP. Molecules. 2022;27(9):2922. doi: 10.3390/molecules27092922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nam K.H. Fluorescent protein-based metal biosensors. Chemosensors. 2023;11(4):216. doi: 10.3390/chemosensors11040216. [DOI] [Google Scholar]
- 7.Matz M.V., Fradkov A.F., Labas Y.A., Savitsky A.P., Zaraisky A.G., Markelov M.L., Lukyanov S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999;17(10):969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 8.Bevis B.J., Glick B.S. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat. Biotechnol. 2002;20(1):83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 9.Ranganathan R., Wall M.A., Socolich M. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat. Struct. Biol. 2000;7(12):1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 10.Strongin D.E., Bevis B., Khuong N., Downing M.E., Strack R.L., Sundaram K., Glick B.S., Keenan R.J. Structural rearrangements near the chromophore influence the maturation speed and brightness of DsRed variants. Protein Eng. Des. Sel. 2007;20(11):525–534. doi: 10.1093/protein/gzm046. [DOI] [PubMed] [Google Scholar]
- 11.Hawley T.S., Telford W.G., Ramezani A., Hawley R.G. Four-color flow cytometric detection of retrovirally expressed red, yellow, green, and cyan fluorescent proteins. BioTechniques. 2001;30(5):1028–1034. doi: 10.2144/01305rr01. [DOI] [PubMed] [Google Scholar]
- 12.Park S.Y., Ha S.C., Kim Y.G. The protein crystallography beamlines at the pohang light source II. Biodesign. 2017;5(1):30–34. [Google Scholar]
- 13.Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66(Pt 1):22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 14.Agirre J., Atanasova M., Bagdonas H., Ballard C.B., Baslé A., Beilsten-Edmands J., Borges R.J., Brown D.G., Burgos-Mármol J.J., Berrisford J.M., Bond P.S., Caballero I., Catapano L., Chojnowski G., Cook A.G., Cowtan K.D., Croll T.I., Debreczeni J.É., Devenish N.E., Dodson E.J., Drevon T.R., Emsley P., Evans G., Evans P.R., Fando M., Foadi J., Fuentes-Montero L., Garman E.F., Gerstel M., Gildea R.J., Hatti K., Hekkelman M.L., Heuser P., Hoh S.W., Hough M.A., Jenkins H.T., Jiménez E., Joosten R.P., Keegan R.M., Keep N., Krissinel E.B., Kolenko P., Kovalevskiy O., Lamzin V.S., Lawson D.M., Lebedev A.A., Leslie A.G.W., Lohkamp B., Long F., Malý M., McCoy A.J., McNicholas S.J., Medina A., Millán C., Murray J.W., Murshudov G.N., Nicholls R.A., Noble M.E.M., Oeffner R., Pannu N.S., Parkhurst J.M., Pearce N., Pereira J., Perrakis A., Powell H.R., Read R.J., Rigden D.J., Rochira W., Sammito M., Sánchez Rodríguez F., Sheldrick G.M., Shelley K.L., Simkovic F., Simpkin A.J., Skubak P., Sobolev E., Steiner R.A., Stevenson K., Tews I., Thomas J.M.H., Thorn A., Valls J.T., Uski V., Usón I., Vagin A., Velankar S., Vollmar M., Walden H., Waterman D., Wilson K.S., Winn M.D., Winter G., Wojdyr M., Yamashita K. The CCP4 suite: integrative software for macromolecular crystallography. Acta Crystallogr. D Struct. Biol. 2023;79(6):449–461. doi: 10.1107/s2059798323003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casañal A., Lohkamp B., Emsley P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 2020;29(4):1055–1064. doi: 10.1002/pro.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., Jain S., Lewis S.M., Arendall W.B., 3rd, Snoeyink J., Adams P.D., Lovell S.C., Richardson J.S., Richardson D.C. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27(1):293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Crystal structure of DsRed-Monomer (Original data) (Protein Data Bank).