Abstract
Further development of direct-acting antiviral agents against human SARS-CoV-2 infections remains a public health priority. Here, we report that an antisense peptide-conjugated morpholino oligomer (PPMO) named 5′END-2, targeting a highly conserved sequence in the 5′ UTR of SARS-CoV-2 genomic RNA, potently suppressed SARS-CoV-2 growth in vitro and in vivo. In HeLa-ACE 2 cells, 5′END-2 produced IC50 values of between 40 nM and 1.15 μM in challenges using six genetically disparate strains of SARS-CoV-2, including JN.1. In vivo, using K18-hACE2 mice and the WA-1/2020 virus isolate, two doses of 5′END-2 at 10 mg/kg, administered intranasally on the day before and the day after infection, produced approximately 1.4 log10 virus titer reduction in lung tissue at 3 days post-infection. Under a similar dosing schedule, intratracheal administration of 1.0–2.0 mg/kg 5′END-2 produced over 3.5 log10 virus growth suppression in mouse lungs. Electrophoretic mobility shift assays characterized specific binding of 5′END-2 to its complementary target RNA. Furthermore, using reporter constructs containing SARS-CoV-2 5′ UTR leader sequence, in an in-cell system, we observed that 5′END-2 could interfere with translation in a sequence-specific manner. The results demonstrate that direct pulmonary delivery of 5′END-2 PPMO is a promising antiviral strategy against SARS-CoV-2 infections and warrants further development.
Keywords: MT: Oligonucleotides: Therapies and Applications, peptide-conjugated morpholino oligomer, PPMO, antisense, antiviral, SARS-CoV-2, K-18-hACE2 mouse model, RNA virus, intratracheal delivery, intranasal delivery
Graphical abstract
Sakai and colleagues document pre-clinical development of a novel antiviral agent against SARS-CoV-2. An antisense peptide-conjugated morpholino oligomer called 5′END-2 targets highly conserved sequences in SARS-CoV-2 genomic RNA. 5′END-2 inhibited several SARS-CoV-2 strains in cell cultures, and when delivered via intratracheal administration, profoundly suppressed SARS-CoV-2 growth in the lungs of mice.
Introduction
COVID-19 is caused by the severe acute respiratory syndrome coronavirus 2 SARS-CoV-2 virus,1,2 and as of Feburary 2024, it has been the cause of over 7 million human deaths worldwide. As of December 2023, over 1 million new cases and 9,000 deaths were being reported monthly from the World Health Organization’s (WHO’s) six regions. Furthermore, at least two major epidemiologic studies conclude that mortality caused by COVID-19 has been underestimated.3,4
Despite US Food and Drug Administration (FDA) approval or Emergency Use Authorization of several therapeutics for the treatment of COVID-19, the need for the development of additional compounds to address SARS-CoV-2 infections continues to be a public health priority.5,6 Direct-acting antiviral agents (DAAs) target a physical component of the virus directly and are typically designed to interfere with the virus replication cycle. Several DAAs, including nucleotide (nt) analogs (e.g., remdesivir and molnupiravir), protease inhibitors (e.g., Paxlovid), and numerous monoclonal antibodies, when administered soon after diagnosis, can dramatically reduce the hospitalization rate of COVID-19 patients. However, drawbacks to current DAAs include, variously, the requirement for intravenous administration in a healthcare setting, side effects, interactions with other medications, cost, and the selection over time of drug-resistant virus variants.7,8,9,10 The development of additional DAAs, especially those that can be administered orally or by inhalation, is needed to improve medicinal strategies and to decrease global production of the virus and the generation of SARS-CoV-2 variants. The results observed to date with current DAAs suggest that the several-day overlap between the appearance of disease symptoms and the replication of SARS-CoV-2 in the respiratory tract of humans represents a period of time in which a pharmaceutical capable of direct interference in viral replication can be effective at reducing disease severity. Furthermore, combination treatments consisting of two or more DAAs may reduce the frequency of escape variant generation and improve clinical outcomes.11,12
Over the past few years, a number of sequence-specific RNA-targeted therapeutic compounds have been FDA approved and commercialized, including at least seven single-stranded oligonucleotide-type agents.13,14,15,16 Strategies targeting SARS-CoV-2 RNA, including small interfering RNA siRNA17,18,19 and antisense oligonucleotide mixmers,20,21 have demonstrated in vivo antiviral efficacy in mouse models of SARS-CoV-2 infection. A SARS-CoV-2-specific siRNA-peptide dendrimer formulation was evaluated in a single clinical trial in Russia in 2021 (this study was registered at ClinicalTrials.gov [NCT05184127]). The compound was reported as safe and produced clinical improvement in patients hospitalized with moderate COVID-19.22 However, there have been no further human clinical trials with this composition or any other compounds designed to target SARS-CoV-2 RNA in a sequence-specific manner.
Phosphorodiamidate morpholino oligomers (PMOs), also known as morpholinos, are single-stranded nucleic acid analogs containing the same bases as DNA, but having a non-natural backbone in place of the sugar-phosphate backbone of nucleic acids.23 PMOs feature high sequence specificity and biological compatibility and exert their antisense activity through steric blockade, as their hybridization to complementary single-stranded RNA does not form a substrate for RNase H activity.24,25 Four PMOs have been FDA approved for treatment of Duchenne’s muscular dystrophy.16,26,27,28 To improve entry into cells and subsequent intracellular distribution, PMOs can be conjugated to a cell-penetrating peptide to produce peptide-conjugated PMOs (PPMOs).29,30,31,32 PPMOs are water soluble, nuclease resistant, and non-toxic at effective concentrations across a range of in vitro and in vivo applications.33,34 At least three PPMOs are currently in human clinical trials (ClinicalTrials.gov: NCT06204809, NCT06079736, and NCT06185764). PPMOs have been documented to readily enter numerous cell types in vitro and in vivo, including primary airway epithelial, without toxicity.35,36,37,38 In studies designed to evaluate antiviral efficacy and specificity, PPMOs targeting viral sequences have demonstrated a considerable ability to suppress the growth of an array of RNA and DNA viruses in cell cultures and animal models (reviewed by Stein, Nan and Zhang, and Warren et al.39,40,41).
The SARS-CoV-2 genome comprises an approximately 29.9-kb single-stranded RNA of positive polarity featuring a 5′ m7G cap and 3′ polyadenylation.42,43,44 The genomic 5′ and 3′ UTRs are approximately 265 and 228 nt long, respectively.45,46 The antiviral efficacy and specificity of PPMO targeting the distal region of ancestral SARS-CoV-2 (strain WA-1/2020) 5′ UTR RNA have been demonstrated in Vero cell cultures.47 Here, we extend those observations by evaluating PPMO antiviral efficacy in vitro, using a human-derived cell line infected with several different strains of SARS-CoV-2, and in vivo, using a mouse model to compare the antiviral activity of PPMO delivered by two different routes of administration, intranasal (IN) and intratracheal (IT).
In this study, we evaluated sequence conservation across the SARS-CoV-2 virome in the region of 5′ UTR targeted by an antiviral PPMO, named 5′END-2, and found it to be highly conserved. We report here that 5′END-2 was potently effective against several virus strains in cell cultures and was able to suppress virus growth in the lungs of mice, using moderate dosing. We also observed that IT administration of a low dose of PPMO suppressed the growth of SARS-CoV-2 in the lungs of mice more effectively than a higher dose administered IN under similar conditions. Furthermore, we demonstrate direct evidence of PPMO hybridization to its RNA target sequence and its ability to interfere with the process of viral RNA translation.
Results
PPMO design
SARS-CoV-2 is a member of the Coronaviridae family, within the order Nidovirales. Considerations in the design of the virus-RNA-targeted PMO sequence, constituting the antisense component of the virus-targeted PPMO in this study, included historical results that identified the 5′-terminal region of the 5′ UTR in the nidoviruses mouse hepatitis virus, equine arteritis virus, and porcine reproductive and respiratory syndrome virus as a highly sensitive site for PPMO intervention.48,49,50,51,52,53,54 In addition, the 5′-terminal region of the 5′ UTR of the genomes of other non-nidovirus positive-strand RNA viruses that utilize cap-dependent translation has been a productive antiviral PPMO target region in numerous studies.30,55,56,57,58,59 PPMO design for this study was specifically informed by a previous study in which PPMO produced multi-log reductions in the titer of SARS-CoV-2 strain WA-1/2020 in Vero-E6 cell cultures.47 In that study, four PPMO, with two targeting the 5′-terminal region (named 5′END-1 and 5′END-2) and two targeting the transcription regulatory site leader sequence (TRS-L) region (named TRS-1 and TRS-2) of SARS-CoV-2 genomic RNA, had high activity, whereas a PPMO targeting the AUG translation start site region of ORF1a/b (AUG-1, targeting nt 251–275) had only moderate antiviral activity. Another specific consideration in the choice of the PPMO target in the SARS-CoV-2 genome for this study involved reports published during the early stages of the pandemic, indicating that the first four nt of the SARS-CoV-2 genome were more variable than nt 5–30.60,61 Considering the various studies and reports above, we focused the PPMO targeting for this study on sequences in the 5′-terminal region of the 5′ UTR of SARS-CoV-2 genome RNA, specifically nt 5–29 (PPMO 5′END-2). This region includes a majority of the nt that comprise stem-loop 1 (SL1), a secondary-structure feature comprising nt 7–33 (of the SARS-CoV-2 GenBank Reference Sequence NC_045512.2).60,62 The PPMOs used in this study are defined in Table 1. The 5′END-2 PPMO was designed with the intention of interfering with events of the virus life cycle which involve the 5′-terminal region of the genome 5′ UTR, including pre-initiation of the translation of genomic and most subgenomic mRNAs, as well as regulation of translation by viral protein NSP1.61,63,64,65 Along with the 5′END-2 PPMO, we produced a negative control PPMO (NC705) which contains the same peptide ((RXR)4XB) as is present in the 5′END-2 PPMO, but conjugated to a nonsense PMO sequence having little agreement with any RNA virus or human or mouse transcript sequences, as determined by BLASTn. Thus, NC705 PPMO is designed to serve as a test for antisense specificity. The peptide component of the PPMO used in this study was chosen based on previous studies demonstrating its ability as a PMO transporter,32,66,67,68 along with the typically high aqueous solubility and low toxicity of PPMO made with this arginine-rich peptide.33,34,69
Table 1.
PPMO used in this study
| PPMO name | PPMO sequence (5′-3′) | PPMO target location in SARS-CoV-2 genomea |
|---|---|---|
| 5′END-2 | TGTTACCTGGGAAGGTATAAACCTT | nt 5-29 |
| NC705 | CCTCTTACCTCAGTTACAATTTATA | N/A |
Based on Wuhan-Hu-1, GenBank: NC_045512.
PPMO target in the SARS-CoV-2 5′ UTR has high sequence conservation
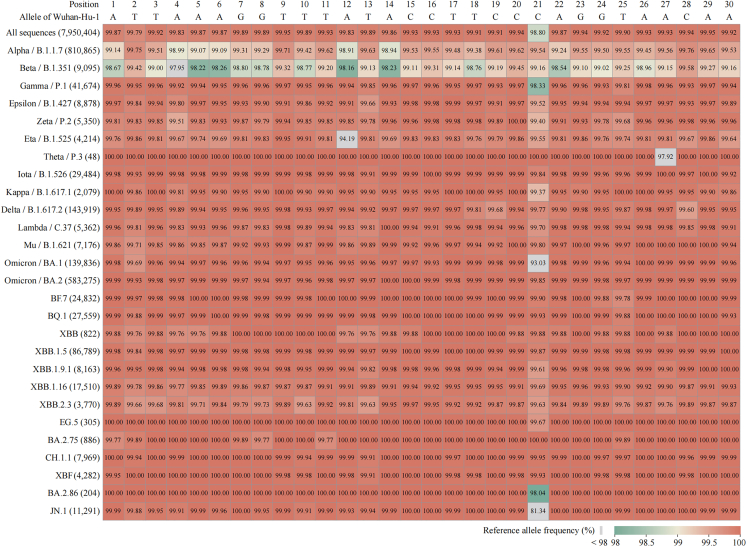
Another major factor in PPMO design for this study was PMO target-site sequence conservation across the SARS-CoV-2 virome. We evaluated sequence conservation in the 5′-terminal region (nt 1–30) of the 5′ UTR across virus strains representing the SARS-CoV-2 viromic spectrum. We carried out a comprehensive bioinformatic mutational analysis of the 5′-terminal region of the 5′ UTR to define the degree of complementary sequence agreement between the 5′END-2 PPMO and its target across the breadth of the SARS-CoV-2 virome. Nearly 8 million high-quality SARS-CoV-2 sequences, collected from January 2020 through January 2024 and representing an in-depth sampling of all lineages in the SARS-CoV-2 virome, were analyzed for mutations in the 5′END-2 PPMO target sequence region. At least 98.8% of all strains analyzed have perfect agreement between the 5′END-2 PPMO and its target site in the SARS-CoV-2 genome (nt 5–29) (Figure 1), with less than 0.3% having more than a single base mismatch between 5′END-2 and its target (Table 2). Interestingly, our analysis revealed that the SARS-CoV-2 Beta lineage (B.1.351) has overall around 1% less sequence conservation at nt 1–30 in relation to the ancestral Wuhan-Hu-1 lineage, compared to other SARS-CoV-2 lineages, including numerous lineages that did not appear until after Beta in the pandemic. Nevertheless, 5′END-2 PPMO has antisense sequence agreement with at least 98% of each of the 9,095 isolates of Beta SARS-CoV-2 analyzed at each target residue (Figure 1).
Figure 1.
Heatmap of reference allele frequency in the 5′END-2 PPMO target region of different SARS-CoV-2 lineages
The reference allele (GenBank: NC_045512) frequency at nt positions 1–30 of the SARS-CoV-2 genome was calculated from a dataset of complete and high-quality human-origin genome sequences, and visually represented in a heatmap. The reference allele sequence for nt 1–30 is shown in the top horizontal row. This terminal region of the viral 5′ UTR includes the target of the 5′END-2 PPMO, located from nt 5–29. As of January 8, 2024, the WHO has defined 27 variants of concern, variants of interest, and variants under monitoring. The vertical axis labels are indicative of the lineage names, which appear in order of their chronological appearance during the pandemic, with the accompanying numbers in parentheses representing the tally of genome sequences specific that were analyzed for each specific lineage.
Table 2.
Bioinformatic analysis of sequence agreement between 5′END-2 PPMO and its target in a wide array of SARS-CoV-2-genomes (January 2020–January 2024)
| No. of SARS-CoV-2 genome sequences analyzed | % With perfect agreement between 5′END-2 and SARS-CoV-2 | % With 1 mismatch between 5′END-2 and SARS-CoV-2 | % With 2 mismatches between 5′END-2 and SARS-CoV-2 | % With 3 mismatches between 5′END-2 and SARS-CoV-2 | % With 4 or more mismatches between 5′END-2 and SARS-CoV-2 |
|---|---|---|---|---|---|
| 7,950,404 | 98.31 | 1.41 | 0.07 | 0.02 | 0.19 |
Since mid-2022, strains of the Omicron lineage have become the dominant circulating SARS-CoV-2 worldwide. We therefore also carried out an Omicron-focused sequence conservation analysis on full-length high-quality sequences available through the Global Initiative on Sharing All Influenza Data (GISAID) from an 18-month period from April 2022 to October 2023. The mutational profiles in separate but contiguous 6-month windows appear in Table S1. We analyzed over 330,000 genomic sequences of Omicron SARS-CoV-2 in total and determined that for this 18-month period, over 97% of the sequences exhibit perfect agreement between the 5′END-2 PPMO and its target site in the SARS-CoV-2 genome, while approximately 2% have a single mispair of disagreement between 5′END-2 PPMO and its complementary target. Less than 0.2% of the sequences analyzed have more than 1 base of disagreement between 5′END-2 and its target.
Together, these analyses provide extensive validation of high sequence conservation at the 5′END-2 PPMO target site across the SARS-CoV-2 virome, and they suggest that this region of SARS-CoV-2 genomic sequence is not substantially increasing in variability over time.
Electrophoretic mobility shift assays confirm that 5′END-2 PMO and PPMO bind to target RNA specifically and with high affinity
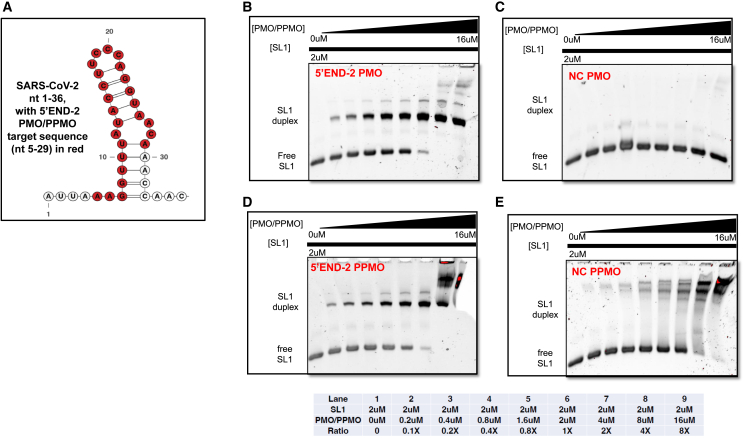
We wished to characterize the sequence-specific hybridization behavior of the PMO antisense portion of 5′END-2 PPMO with its RNA target (nt 5–29 of SARS-CoV-2 genomic RNA) and to validate that duplexing of 5′END-2 PPMO with its target RNA occurs in a sequence-specific manner, through Watson-Crick complementary base pairing. Furthermore, we sought to investigate whether the presence of the P7 peptide in the 5′END-2 PPMO had an effect on direct binding of the 5′END-2 PMO to its target RNA. We performed electrophoretic mobility shift assays (EMSAs), using the 5′END-2 PMO or PPMO and an RNA analyte comprising nt 1–36 of SARS-CoV-2 RNA (named SL1 RNA). As a control, we also ran identical assays but replaced the 5′END-2 PMO or PPMO with the NC705 PMO or PPMO. For these assays, the concentration of SL1 RNA was fixed at 2 μM per sample while the PMO or PPMO was titrated from 0 to 16 μM in serial reactions (Figure 2). On the gels running these reactions, the SL1 RNA (Figure 2A) is visualized as the lower band, whereas SL1 RNA duplexed with PMO or PPMO appears as a discrete upper band. Because PMO itself has little ionic charge, it does not appear as a discrete band on the gels. We observed that 5′END-2 PMO or PPMO behaved similarly (Figures 2B and 2D), producing a noticeable shift, as evidenced by the appearance of the higher-molecular-weight species (the discrete upper band) in a gradual concentration-dependent manner. The upper band becomes initially visible when the 5′END2 PMO or PPMO is present at only a 0.1× concentration in relation to its SL1 target RNA (lane 2 of Figures 2B and 2D). As the concentration of 5′END-2 PMO or PPMO increases, a gradual increase in the intensity of the upper band (duplexed material) and decrease in the intensity of the lower band (free SL1 RNA) is evident. Nearly complete duplexing is apparent when 5′END-2 PMO/PPMO was present at 2× the concentration of the target RNA, and apparently complete duplexing when present at a 4× concentration to that of the target RNA (lane 8 of Figures 2B and 2D). The NC705 PMO (Figure 2C) did not produce any specific electrophoretic shift. The reactions containing increasing amounts of NC705 PPMO exhibited increasing intensity of non-distinct higher-molecular-weight bands (Figure 2E), which migrate well above the SL1 duplexed species. Since PPMO is positively charged, due mostly to the presence of arginine residues in the P7 peptide, we speculate that these bands represent non-duplexed PPMO. In addition, a series of non-distinct high-molecular-weight bands, likely representing unduplexed PPMO, are also apparent in lane 8 of the 5′END-2 PPMO gel (Figure 2D). These results demonstrate that 5′END-2 PPMO is capable of annealing specifically and avidly to its RNA target sequence and that the presence of the peptide portion of the 5′END-2 PPMO does not alter fundamental antisense duplexing behavior.
Figure 2.
5′END-2 PMO and PPMO duplex specifically and efficiently with target RNA
(A) Schematic diagram of nt 1–36 of SARS-CoV-2 genome. This RNA (SL1) was produced by in vitro transcription and served as the analyte for the experiments of (B)–(E). The secondary structure of the diagram was produced by “RNAStructure” and the residues representing the 5′END-2 PMO or PPMO target site are shown in red. (B–E) EMSA gels of reactions containing SL1 and PMO/PPMO. A fixed concentration of SL1 RNA (2 μM) was used with titrations of the indicated PMO (B and C) or PPMO (D and E) from 0 to 16 μM, as described in detail in materials and methods. The reactions were run on native PAGE comprising 8% acylamide. A table indicating the overall lane-by-lane composition of the reactions is present below the gels. NC, negative control.
5′END-2 PPMO inhibits the growth of several SARS-CoV-2 variants in vitro
To evaluate antiviral activity, we first sought to determine whether PPMO 5′END-2, which had been identified as potently antiviral against the ancestral Wuhan-Hu-1-like human SARS-CoV-2 (WA-1/2020) in Vero-E6 cells, also had substantial antiviral activity against other SARS-CoV-2 strains, including representative variants of concern, in HeLa-ACE2 cells.70 In these antiviral and cytotoxicity assays, we included a positive control compound, nirmatelvir (Paxlovid), known to have potent antiviral activity against coronaviruses in vitro.71 To test the level of antiviral activity, each compound was applied in a 6- or 8-point dose response, using 3-fold serial dilutions from the highest concentration of 50 or 80 μM PPMO or 5 μM nirmatelvir. The compounds were added for 5 h, after which the drug-containing medium was removed before an infection period. The presence of the drug was omitted during the infection period to preclude possible direct extracellular interaction between the drug and the inoculating viral particles. After infection, the cells were incubated in the absence of the test compound for 24 h before virus quantification assays. The two PPMOs and the positive control nirmatelvir were tested against six strains of virus: the ancestral Wuhan-Hu-1-like USA WA/2020 (first isolated in the United States in January 2020), a strain from the Delta (B.1.617.2) lineage, and four strains representing disparate sublineages within the Omicron clade (BA.1, BA.5, XBB1.5, and JN.1). To evaluate the cytotoxicity of the PPMO, we used an MTT-based assay and a similar experimental format as above, but we omitted virus infection. The results of both assays are shown in the charts of Figure S1, and Table 3 summarizes the results from these experiments, showing the cytotoxic concentration 50% (CC50), the half-maximal inhibitory concentration (IC50), and the 90% inhibition concentration (IC90) for each compound against each virus. It was not possible to calculate meaningful selectivity index values for the compounds, as the CC50 values were almost all in excess of the highest concentration of the compounds used in this set of experiments. None of the PPMO produced significant cytotoxicity at any of the concentrations tested. Likewise, the positive control compound, as assessed by CC50, was benign even at its highest concentration. The 5′END-2 PPMO generated IC50 values from approximately 0.040–1.14 μM. The data indicate that the 5′END-2 PPMO had high antiviral efficacy against all the virus strains and that the activity was sequence specific and noncytotoxic. Overall, these results suggest that 5′END-2 PPMO has the potential to suppress the growth of a spectrum of SARS-CoV-2 strains. The favorable profile of the considerable antiviral efficacy of 5′END-2 PPMO against a genetic diversity of SARS-CoV-2 strains, with minimal impact on cell viability in a human cell line, provided rationale for further evaluation in an in vivo setting.
Table 3.
Cytotoxicity (CC) and antiviral (IC) values of nirmatrelvir and PPMO against six strains of SARS-CoV-2 in HeLa-ACE2 cell culture assays
| Drug | Virus | CC50, μM | IC50,μM | IC90, μM |
|---|---|---|---|---|
| Nirmatrelvir | WA-1/2020 | >5 | 0.11 | 0.21 |
| Nirmatrelvir | DELTA | >5 | 0.04 | 0.17 |
| Nirmatrelvir | BA.1 | >5 | 0.03 | 0.08 |
| Nirmatrelvir | BA.5 | >5 | 0.05 | 0.14 |
| Nirmatrelvir | XBB1.5 | >5 | 0.02 | 0.05 |
| Nirmatrelvir | JN.1 | >5 | 0.06 | 0.15 |
| NC705 | WA-1/2020 | >50 | >50 | >50 |
| NC705 | DELTA | >50 | >50 | >50 |
| NC705 | BA.1 | >50 | >50 | >50 |
| NC705 | BA.5 | >50 | >50 | >50 |
| NC705 | XBB1.5 | >50 | >50 | >50 |
| NC705 | JN.1 | >50 | 24.38 | >50 |
| 5′END-2 | WA-1/2020 | >50 | 0.17 | 2.23 |
| 5′END-2 | DELTA | >50 | 0.10 | 3.51 |
| 5′END-2 | BA.1 | >50 | 0.34 | 3.27 |
| 5′END-2 | BA.5 | >50 | 0.04 | 0.18 |
| 5′END-2 | XBB1.5 | >50 | 0.39 | 13.31 |
| 5′END-2 | JN.1 | >50 | 1.14 | 17.27 |
For methodologic details, see materials and methods section.
IN administration of 5′END-2 PPMO moderately inhibits SARS-CoV-2 growth in the lungs of K18-hACE2 mice
Dose regimen determination
In vitro and in vivo dose-dependent toxicity from P7-PMO is generally a function of the concentration of the P7 peptide component.30,72 In regard to the potential generic toxicity to mice from IN administration of the structural type of PPMO used in this study (P7-PMO), several previous studies using P7-PMO and a similar IN dosing regimen to that used here reported no apparent toxicity to uninfected animals.36,73,74,75 We limited the number of doses administered to the mice to two, occurring 1 day before and 1 day after infection. This regimen introduced minimal stress to the mice, as handling and anesthetization were limited to a total of three events. Furthermore, our dosing regimen, with PPMO administrations temporally separate from the virus infection event, precluded direct interactions between inoculation virus and drug in the airways on the day of virus inoculation.
IN delivery of treatments and viral titer determination
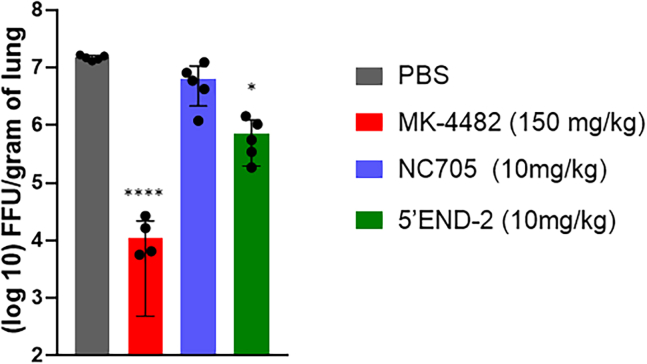
K18-hACE2 (human angiotensin-converting enzyme 2) mice are transgenic for hACE2 expression in epithelial cells and can grow SARS-CoV-2 to high titers in the lungs.76,77 To determine the effect of intranasal dosing of PPMO on SARS-CoV-2 growth, we dosed each group of five mice twice, with 10 mg/kg of PPMO or vehicle solution only (PBS) at 24 h before and 18 h after infection. The treatments were delivered via IN administration, as was the infection inoculum of 5,000 focus-forming units (ffu) of WA-1/2020 SARS-CoV-2. Along with the two PPMO tested, and a PBS-only (vehicle-control) group, we also included a positive control, the small-molecule antiviral compound MK-4482 (molnupiravir, an orally bioavailable nucleoside analog70), which was administered to the mice orally at a dose of 150 mg/kg, twice per day, starting at 1 day before infection and continuing through day 2 post-infection. At 3 days post-infection, all mice were euthanized and lung tissue collected, homogenized, and subject to viral titer determination by focus-forming assay. We observed that NC705 PPMO treatment produced virus titer of approximately 0.2 log10 ffu/g lung tissue below the PBS-treated control mice. The average titer reduction of 5′END-2 PPMO dosed at 10 mg/kg per dose was ∼1.4 log10 ffu/g tissue less than PBS-only-treated mice (Figure 3). Notably, the positive control compound MK448 lowered lung virus titer by approximately 3 log10 ffu/g tissue, thereby setting an existent standard of high activity in this model.
Figure 3.
5′END-2 PPMO administered IN limits virus growth
K18-hACE2 mice were treated with PBS, 5′END-2 PPMO, and NC705 (negative control) PPMO as indicated, via IN instillation at 24 h before and 18 h after IN administration of 5,000 ffu of WA-1/2020. MK-4482 (molnupravir), used as a positive control compound, was administered orally twice per day from 1 day before infection until 2 days post-infection. Viral load in the lungs was measured at 3 days post-infection by focus-forming assay, as described in materials and methods. Mean ± SD is shown (n = 5) and was analyzed by one-way ANOVA with Dunnett’s multiple comparisons (∗p < 0.05; ∗∗∗∗p < 0.0001).
IT administration of PPMO results in a greater amount of PPMO translocation to the lungs than IN administration
Results from previous studies using fluorescein-labeled P7-PMO (Fl-PPMO) administered via IN administration to uninfected mice suggested a distribution of signal having an overall gradated manner from upper to lower lung.36,37 In those studies, Fl-PPMO signal was most intense along the major bronchi, but signal was also detected in minor bronchial branches and was observed to enter epithelial cells surrounding alveoli.36
We hypothesized that IN administration of PPMO in the SARS-CoV-2 experiment described above may have resulted in loss of PPMO material in the upper respiratory tract, thus limiting antiviral efficacy against SARS-CoV-2 in the lungs. To gain insight into the relative efficiency with which IN and IT administration are able to deliver PPMO to the lungs, we compared the two routes of delivery with a lissamine-labeled 5′END-2 PPMO (PPMO-liss) in uninfected K18-hACE2 mice. The experiment had two arms, with one arm utilizing IN and the other arm utilizing IT administration. Each arm had two groups (n = 5) corresponding to dose levels of 0 and 10 mg/kg of PPMO-liss. A single dose was administered to the mice in the same manner as described for each delivery method in the antiviral experiments and the lungs collected 24 h later and bisected into upper and lower sections. Fluorescence of lung lysates was used for comparison between routes of administration and between upper and lower lung. Fluorescence was near background levels for the 0 mg/kg PPMO-liss samples (data not shown). While there was considerable variability within this experiment and differences between conditions were not statistically significant, results suggest a trend toward moderately higher PPMO concentrations in the lung following IT administration than with IN administration (Figures S2A and S2B). Furthermore, although this experiment did not address PPMO presence in specific cell types, the results indicate little difference between the amount of material found in the upper compared to the lower lung for each delivery method (Figures S2C and S2D), suggesting widespread PPMO distribution in the lungs with either route of administration.
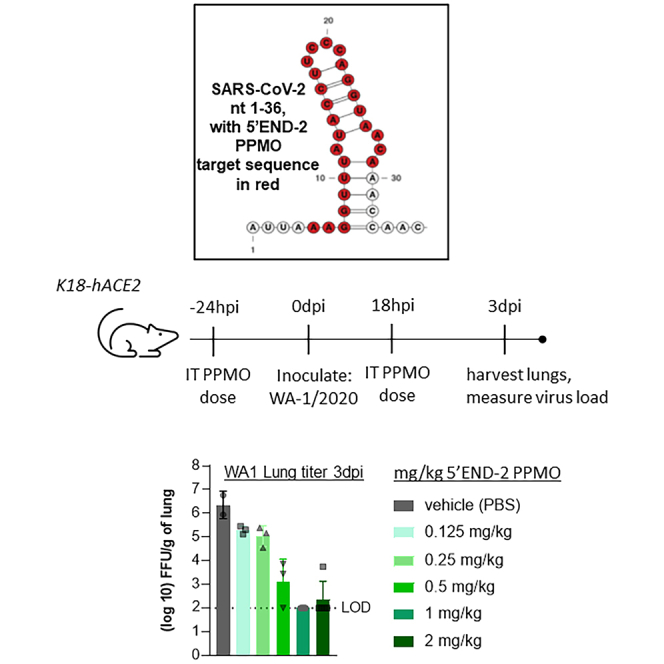
IT administration of 5′END-2 PPMO potently inhibits virus growth in the lungs of K18-hACE2 mice infected with WA-1/2020
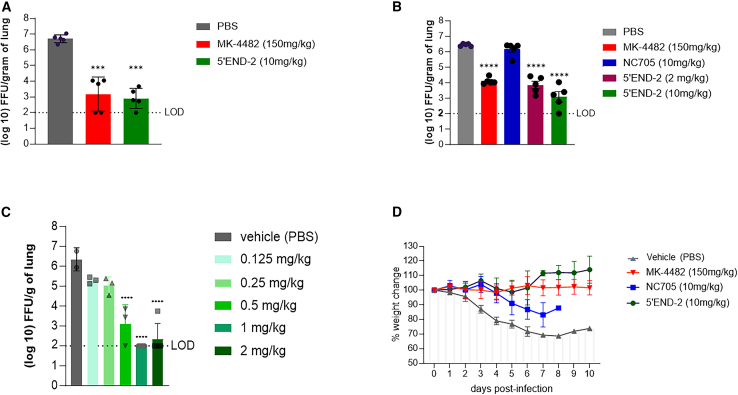
To promote greater PPMO delivery into the lungs, we chose to employ IT delivery of PPMOs. To evaluate virus titer in the lungs, we performed three experiments using IT administration of PPMO under similar conditions as described above for the evaluation of virus growth in the lungs after IN administration of PPMO. Our first experiment consisted of only three groups, with the mice treated with either PBS, a positive control (MK4482, described and administered as above), or 5′END-2 PPMO (Figure 4A). A second independent experiment included the same three groups and two additional groups, NC705 PPMO at a dose of 10 mg/kg and 5′END-2 PPMO at 2 mg/kg (Figure 4B). Overall, the second experiment was intended to serve as a bio-repeat of the first experiment, yet also include a negative control PPMO. The third independent experiment was a dose titration study with 5′END-2 PPMO at 2, 1, 0.5, 0.25, 0.125, and 0 mg/kg. All three experiments were performed under the same conditions and yielded the same central result: that 5′END-2 PPMO markedly suppressed virus growth in the lungs by over 3 log10 ffu/g of lung tissue. In the first two experiments, the level of virus reduction produced by 5′END-2 PPMO at 10 mg/kg was slightly greater than that observed with the positive control compound MK-4482. In the second experiment (Figure 4B), NC705 did not cause a reduction in virus growth, indicating that the antiviral effect of 5′END-2 was a function of sequence-specific binding of the PMO portion of the PPMO to viral RNA, and that the overall structural chemistry of P7-PMO was not a factor in the antiviral effect of 5′END-2 PPMO. In the third experiment, the antiviral effect of 5′END-2 was shown to be potent and dose responsive, as treatment with 1, 0.5, 0.25, and 0.125 mg/kg dosing produced approximately 4.2, 3.2, 1.2, and 1.0 log10 mean suppression of virus growth (Figure 4C), respectively. Under the conditions of this study, the minimum dose necessary to produce robust antiviral efficacy of at least 3 log10 ffu/g of lung tissue was 0.5 mg/kg. Overall, we observed a minor amount of experiment-to-experiment variation in the levels of virus growth inhibition by the various individual treatments.
Figure 4.
5′END-2 PPMO administered via IT injection markedly limits virus growth in the lungs of K18-hACE2 mice and protects from weight loss
(A–C) K18-hACE2 mice were treated with PBS or the indicated PPMO via IT administration at 24 h before and 18 h after IN administration of 5,000 ffu of SARS-CoV-2 (strain WA-1/2020). MK-4482 (molnupravir), used as a positive control compound, was administered orally twice per day from 1 day before infection until 2 days post-infection. Virus load in the lungs was measured at 3 days post-infection by focus-forming assay, as described in materials and methods, and charted. The experiments represented by the graphs in (A)–(C) were carried out under the same conditions (see materials and methods), but at independent times. The limit of virus detection (LOD) in the focus-forming assay was 100 ffu/g tissue, as represented by a dotted line. n = 5, except the experiment of (C), which tested 5’END-2 at various concentrations, where each group contained 3 mice. (D) Mice were treated with PBS, MK-4482, or PPMO, as indicated, under the same experimental conditions as described above. Body weights were measured daily for 10 days post-infection and represented as a percentage of the animal weight on the day of infection. By 8 days post-infection, all the animals in the NC705 had lost greater than 35% of their original body weight and were considered moribund. n = 5/group. Data shown as mean ± SD and analyzed by one-way ANOVA with Dunnett’s multiple comparisons (∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
The moderately low dosing regimen of two treatments of 0.5–2.0 mg/kg of 5′END-2 PPMO delivered directly to the lungs via IT administration generated similar protection to eight doses of 150 mg/kg MK-4482 given orally. It is further noteworthy that while MK-4482 dosing included two doses on the day of infection, PPMO dosing did not occur on the same day as virus inoculation. In this study, we used the measurement of virus titer in the lungs as a primary indicator of the ability to limit virus replication. In addition, to investigate the ability of 5′END-2 PPMO to protect from virus-associated pathogenesis, we measured the ability of the PPMOs to protect mice from virus-induced weight loss, using the same experimental conditions as the virus-replication evaluations above. The mice were weighed daily for 10 days. Mice treated with NC-705 PPMO or PBS suffered 20%–35% body weight loss by days 5–6 and did not recover, whereas MK-4482 and 5′END-2 PPMO treatments completely protected the mice from body weight loss (Figure 4D). We note that the NC705 PPMO appeared to provide a minor protective effect against weight loss for several days, although it was not significant, and all of the mice in that group were moribund by day 9. These results demonstrate that IT-delivered 5′END-2 PPMO suppressed virus growth in the lungs and protected against virus-induced weight loss. Furthermore, the lack of weight loss in mice treated with 5′END-2 PPMO indicates that the dosing regimen did not cause PPMO-associated overt toxicity to the mice.
5′END-2 PPMO is capable of interfering with the process of translation
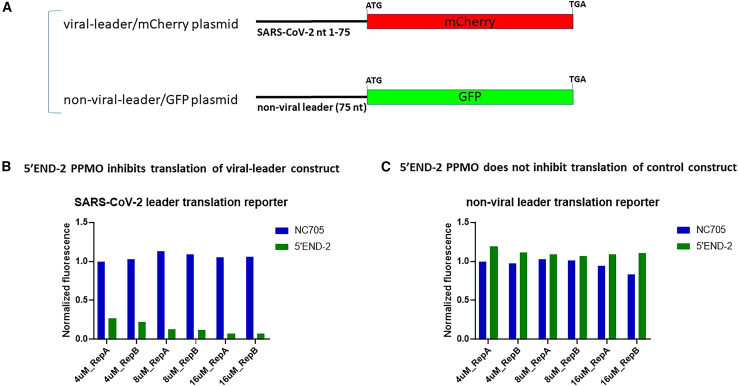
Finally, we sought to gain insight into which aspect of the virus replicative cycle the 5′END-2 PPMO was affecting, to exert its considerable inhibition of SARS-CoV-2 growth. To investigate whether 5′END-2 PPMO could specifically interfere with protein expression in a cellular milieu, we utilized two plasmid constructs, each containing different fluorescent-reporter coding sequences. In one construct (pSARS2-75/mCherry), the 5′-most 75 nt of the SARS-CoV-2 genome is followed by the mCherry coding sequence. A second construct (pRNDM-75/GFP) contains 75 nt of nonsense sequence followed by the GFP coding sequence (Figure 5A). HEK293 cells were treated with PPMO (5′END-2 or NC705) or PBS for 2.5 h and then were co-transfected with both plasmids. The contents of the cells were harvested 24 h later for reporter-specific fluorescence quantification by flow cytometry. The experiment was conducted twice, with similar results (Figures 5B and 5C). For the viral leader construct, the 5′END-2 PPMO interfered with the process of mCherry expression in a dose-dependent manner, while the NC705 PPMO generated little if any inhibitory effect on protein expression. The results from the non-viral leader/GFP construct, in which neither PPMO affected expression of the GFP reporter RNA in a significant manner, further validates the conclusion that 5′END-2 acted in a sequence-specific manner in its inhibitory effect on the translation of viral leader/mCherry RNA.
Figure 5.
The 5′END-2 PPMO restricts protein expression from a SARS-CoV-2 sequence-containing reporter construct, in a potent and specific manner
(A) Schematic of the two reporter constructs used in this experiment. One construct (pSARS2-75/mCherry) contains the first 75 nt of SARS-CoV-2 genomic sequence (viral leader) fused to mCherry coding sequence, while the second (pRNDM-75/GFP) contains 75 nt of non-viral sequence (non-viral leader) fused to GFP coding sequence. Both constructs contain a CMV promoter. (B and C) In-cell translation assays. HEK293 cells were treated with PBS or PBS containing PPMO at the indicated concentrations for 2.5 h, then co-transfected with 500 ng of each reporter construct. At 24 h post-transfection, the cells were assayed for their mCherry and GFP levels by flow cytometry. Translation of the construct containing SARS-CoV-2 leader RNA (B) was markedly limited by the 5′END-2 PPMO, whereas the NC705 PPMO had no effect. Neither PPMO produced appreciable inhibition of translation of the non-viral leader (NC) construct. These experiments were run twice independently, and both trials are shown.
Discussion
Despite significant advances in the development of drugs to treat infections with SARS-CoV-2, the magnitude and impact of the ongoing COVID19 public health threat necessitates continued efforts to develop additional DAAs against current and future virus variants. The study here provides a proof-of-principle demonstration that PPMOs, when administered IT, can generate the suppression of SARS-CoV-2 growth of over 99.9% in the lungs of mice. The level of antiviral activity by PPMO using IT delivery is comparable or superior to that reported for current drugs having FDA approval for the treatment of COVID-19 when those current drugs were tested in similar murine models using IN or oral administration.78,79,80
In vivo targeting of the 5′-terminal region of SARS-CoV-2 with antisense oligomers delivered IN has been reported in at least two other studies.20,21 Those two studies demonstrated that antisense oligomers delivered IN can markedly limit SARS-CoV-2 growth in the lungs of experimental animals. Furthermore, those studies helped characterize how the relationship between SL1 RNA and NSP1 protein regulates the translation of viral and cellular mRNAs in the infected cell. It is noteworthy (and remarkable) that the antisense oligomers in those studies, and in the present study, are apparently able to navigate the consortium of proteins typically associated with the 5′ terminal region of mRNA being readied for translation and access their target sequences. Although the SL1 region is highly ordered in its native state, it is unclear whether the various oligomers are invading SL1 secondary structure or rather accessing their target RNA once the SL is relaxed by helicase activity from a cellular initiation factor (e.g., eIF4A) associated with the 5′-terminal region of mRNA during the pre-initiation of translation. Surprising also is that the SL1-targeting oligomers in these various studies, by the nature of their targeting substantial stretches of residues on both sides of the SL, must themselves also contain substantial degree of self-complementarity under physiological conditions. Although the duplexing data presented here in Figure 2 was not obtained from an intact biological system, it suggests that PPMO is indeed capable of invading highly ordered secondary structure, since the duplexing titration reactions did not include any cellular translation pre-initiation factors. Data from studies with other RNA viruses in which effectively antiviral PPMOs were targeted against secondary structures not directly subject to the helicase activity associated with ribosomal processivity, such as the 3′ SL (3′SLT) of dengue virus55 and the distal panhandle formations present in influenza virus vRNA,35,81 further suggest that PPMOs can invade highly ordered regions of viral RNA.
The present study differs from the two previous SARS-CoV-2 studies targeting SL1 with antisense oligomers mentioned above in at least two important aspects. First, the previous studies utilized locked nucleic acid (LNA) mixmers, a hybrid structural type of oligomer composed of LNA residues in combination with phosphorothioate DNA (PSO) residues. Although the specific arrangement of the two different types of residues within the oligomers was not defined in either paper, most LNA mixmers contain several LNA residues at the distal regions (wings) of the oligomer, with alternating sections of several contiguous PSO residues and LNA residues interspersed in the interior region.82,83,84,85 LNA mixmers presumably exert their antisense activity primarily through RNase H-mediated cleavage of target-RNA in the interior of the mixmer, in regions where PSO is duplexed to RNA. In contrast, the present study employed PPMOs, which exert their antisense activity solely via steric blockade of complementary target RNA sequences by the PMO component of the PPMO, forming a duplex that does not constitute an RNase substrate. This structural and functional difference could have implications for the relative suitability for clinical development, as oligomers containing numerous PSO residues are more prone to off-target effects than oligomers composed entirely of PMO residues.24,86,87,88 The two previous LNA mixmer in vivo studies with SARS-CoV-2 utilized notably more aggressive IN dosing of 20 mg/kg/dose with a total of four or eight doses, respectively, in the two studies, compared to the only two doses of 10 mg/kg or less used in the study here. It is noteworthy that IT delivery of 5′END-2 PPMO produced considerably higher suppression in the titer of infectious virus in the lungs of mice than did IN delivery of PPMO or LNA mixmers using the same K18-hACE2 mouse model and WA-1/2020 virus.
The duplexing characteristics of 5′END-2 PPMO and its target RNA analyte in a gel shift assay (Figure 2), along with protein expression inhibition of an in-cell reporter construct (Figure 5), demonstrate that 5′END-2 PPMO can anneal to its intended target sequence and interfere with the expression of an RNA containing a 5′ UTR sequence of SARS-CoV-2. Although our experiments with dual-reporter constructs (Figure 5) do not directly demonstrate the inhibition of the process of translation, the results suggest that the reduction in reporter protein expression is due to sequence-specific inhibition of translation by the 5′END-2 PPMO. We speculate that the mechanism of action by which the 5′END-2 PPMO exerts antiviral inhibition against SARS-CoV-2 is primarily by interfering with events in the pre-initiation of the cap-dependent translation of ORF1a/b and/or subgenomic mRNAs, which contain the same 75 nt 5′-terminal leader sequence.44,89,90,91 However, it may also be possible that the duplexing of 5′END-2 PPMO with its target sequence also interferes in viral RNA capping, RNA decay, or binding of viral NS1 protein to SL1 RNA, which are thought to involve RNA sequences in this same terminal region of the SARS-CoV-2 5′ UTR. It has been established that the distal region of the 5′UTR of many plus-strand RNA viruses, including coronaviruses, contain highly-conserved sequences and structures involved in the processes of transcription and translation.92,93,94,95,96 Further experiments, preferably using authentic virus in an intact biological system, will be required to elucidate exactly which molecular events of the viral life cycle are being disrupted by 5′END-2. In any event, the present study demonstrates that through one or more mechanisms, the 5′END-2 PPMO is capable of potently inhibiting SARS-CoV-2 growth and pathology in vitro and in vivo.
A notable attribute of 5′END-2 is that it targets a sequence in the 5′-terminal region of the virus that is very highly conserved across the SARS-CoV-2 virome (Figure 1; Tables 2 and S1). Although our bioinformatics analysis suggests that a low percentage of the SARS-CoV-2 genomes may have a single base of disagreement within the 5′END-2 PPMO target site, previous studies have demonstrated that PPMOs having a single base mismatch with their target site retain approximately 90% of their activity compared to those having perfect agreement.50,81 Future studies should characterize the propensity for serial treatments of 5′END-2 PPMO to generate mutations in the SARS-CoV-2 genome. Because of current regulatory restrictions, those types of experiments were not possible for the present study. We note a previous study in which SARS-CoV-1 passaged 11 times in cells treated with 2–10 μM of a PPMO targeted to the TRS-L region produced virus that were mutated in the PPMO target site. The escape mutants exhibited significantly delayed growth kinetics in single-cycle growth curves and were described as partially resistant.97
Our in vitro data (Table 3; Figure S1) shows that PPMOs can potently suppress the growth of a broad spectrum of SARS-CoV-2 variants, including a representative ancestral Wuhan Hu-1 strain (USA WA/2020), a Delta variant of concern strain, and four Omicron strains, including JN.1, the predominant strain in the United States and globally as of early 2024 (CDC update, Jan 22, 2024) and the progenitor to several Omicron subvariants currently increasing in circulation as of mid-2024.98 None of the PPMOs caused significant cytotoxicity in our assays, as measured by CC50 values. There was modest variation in the antiviral efficacy of the SARS-CoV-2-targeted PPMOs against the various strains in vitro. This variation remains unexplained as sequencing of each strain used in the study showed that at least 24 of the 25 nt in the PPMO target region were complementary. We speculate that virus growth kinetics and cell culture conditions may have affected the results to a minor extent.
A major conclusion from this study is that, using the same dosing regimen and timing of doses, IT instillation of 5′END-2 PPMO is superior to IN instillation at suppressing virus growth in the lungs in this mouse model of experimental disease. In experiments employing either route of drug administration, the PPMO behaved in a sequence-specific and nontoxic manner at doses that may be meaningful for clinical application. Future studies using IT administration of 5′END-2 PPMO will include evaluations of the effect of the timing of post-infection doses only, to determine the kinetics of therapeutic inhibition of virus growth in vivo. Furthermore, it will be of interest to determine whether direct pulmonary delivery can produce improved antiviral activity of PPMOs designed against influenza virus,35 respiratory syncytial virus,36 or other respiratory virus infections in experimental animal models as compared to that previously produced by IN delivery of the same PPMO.
Although SARS-CoV-2 has tropism for numerous cell types and regions of the respiratory tract, it is generally accepted that in humans the primary site of pathology is the lungs.99,100 Within the lung, alveolar epithelial cells are impactful sites of virus infection and replication.101,102,103 Both the antiviral data (Figures 3 and 4) and the detection of PPMO-fluorochrome data (Figure S2) suggest that IT delivery resulted in a higher percentage of PPMOs having access to respiratory epithelia in the lungs than did PPMO delivered IN. These results support further preclinical development of this technology against viral infections of the lower respiratory tract. Numerous safe, effective, approved drugs are administered through inhalation.104,105 Notably, the 5′END-2 PPMO is water soluble at relatively high concentrations (up to at least 20 mg/mL in saline; data not shown) without the need for any excipient. Considering the high antiviral efficacy of 5′END-2 PPMO in vivo at a dose of 1–2 mg/kg, along with its water solubility, an aerosol mist formulation of this compound, dispensed through a nebulizer for inhalation, may be well suited for further preclinical development. However, direct pulmonary drug administration through inhalation presents many challenges,106 and additional biological, mechanical, pharmacokinetic, formulation, and toxicity issues will need to be addressed in future preclinical studies of inhalable PPMOs designed to prevent or treat SARS-CoV-2 infection or other respiratory disorders.
In summary, 5′END-2 targets RNA sequence that is highly conserved across the SARS-CoV-2 virome, and IT administration of this oligomer generates potent suppression of SARS-CoV-2 growth and pathology in vivo. Considering the structural similarity of PPMOs to other FDA-approved drugs, 5′END-2 can be considered a promising candidate for further preclinical development as a pre- or post-exposure intervention designed to reduce the pathogenic burden caused by SARS-CoV-2 infection.
Materials and Methods
PPMO synthesis
PPMO were synthesized by covalently conjugating the peptide (RXR)4XB (where R is arginine, X is 6-aminohexanoic acid, and B is β-alanine) to PMO (purchased from Gene Tools, Philomath, OR) at the 3′ end through a noncleavable linker, using methods described previously.107 The lab name of (RXR)4XB peptide is P7, and the type of PPMO used in this study is sometimes referred to as P7-PMO. PPMO-liss was produced from PMO-liss (purchased from Gene Tools),108 consisting of lissamine conjugated to the 3′ end of PMO, and containing a primary amine at the 5′ end for the conjugation of P7 peptide following the PPMO synthesis methods outlined above. PPMO compounds were analyzed at the Mass Spectrometry Facility at Oregon State University, Corvallis. All PPMO compounds were solubilized in sterile PBS (vehicle) unless otherwise noted.
Bioinformatics analysis
A comprehensive, non-redundant collection of 17,097,968 SARS-CoV-2 genome sequences was obtained on January 8, 2024 from RCoV19,109 maintained by the National Genomics Data Center,110 China National Center for Bioinformation.111 To ensure accuracy in our mutational analysis, we analyzed only the 7,950,404 entries judged to be complete and high-quality human-origin genome sequences. For this sub-dataset, a sequence was deemed “complete” if its length exceeded 29,000 bp and encompassed all the protein-coding regions of SARS-CoV-2 (nt 266–29,674 of GenBank: NC_045512.2). A sequence was deemed high-quality if it contained 15 or fewer unknown bases (Ns) and no more than 50 degenerate bases, which are positions that may have multiple base types. The alignment of genome sequences was performed using MUSCLE (version 3.8.425),112 with reference to the SARS-CoV-2 GenBank Reference Sequence (NC_045512.2). Mutations in the sequences were directly identified using an in-house Perl program. We analyzed sequence conservation in the target region of 5′END-2 PPMO, corresponding to nt 5–29 of the SARS-CoV-2 genome (GenBank: NC_045512.2), by performing statistical calculations on sequence mutations. We determined the percentage of sequences exhibiting perfect complementarity with the 5′END-2 PMO sequence. Subsequently, we counted the instances of mutation within the 5′END-2 PMO-target region for each sequence, considering only SNPs for this analysis. We calculated the proportion of sequences with a single base discrepancy (i.e., not complementary with the 5′END-2 PMO sequence). We extended this approach to include the quantification of sequences with two-base disagreements, three-base mismatches, and ultimately, those with four or more base differences. To interrogate sequence conservation of the 5′END-2 target site specifically across Omicron strains of SARS-CoV-2, sequences collected between April 1, 2022 and October 1, 2023 were downloaded from the GISAID EpiCoV database. The methods and parameters for the Omicron-specific analysis were similar to those described above. Briefly, only sequences with a length greater than 29,000 bp and with less than 1% Ns were retained. Makeblastdb (version 2.10.1+) was used to build an nt database from these sequences, then queried using BLASTn with the following parameters: word_size: 7; evalue: 6 million; penalty: −1; reward: +2. The data were visualized with custom Python scripts.
EMSA
RNA representing nt 1–36 of SARS-COV-2 was transcribed in vitro using synthetic DNA oligos (Integrated DNA Technologies) and protonated nucleoside triphosphates (NTPs) (Sigma-Aldrich, St. Louis, MO), incubating at 37°C with gentle rocking for 3 h. Transcription condition was optimized using in-house purified wild-type T7 RNA polymerase following published protocols with a customized NTP ratio.113,114 The transcribed RNA, called SL1, was purified to homogeneity by 15% urea-PAGE (National Diagnostics, Atlanta, GA) and electro-eluted in 1× Tris-borate-EDTA (TBE) buffer (Sigma-Aldrich) post-transcription. SL1 RNA was desalted and diluted to 20 μM before reannealing in the RNA refolding buffer (10 mM Tris-HCl, 50 mm KCl, pH 6.5) by incubating the sample at 95°C for 5 min followed by flash cooling on ice for more than 15 min. The annealed sample was concentrated using a centrifugation filtration system (Amicon, Burlington, MA) and stored in RNA stock buffer (10 mM Tris-HCl, 50 mM KCl, 1 mM MgCl2, pH 6.5). For these experiments, refolded SL1 was freshly prepared and kept at 4°C for short-term use.
EMSA analysis was performed on the PAGE electrophoresis Mini-PROTEAN system (Bio-Rad, Hercules, CA), using pre-chilled 1× TBE buffer (with the addition of 0.1% Triton X-100) at 4°C with 120 V for 30 min Native PAGE 8% was self-prepared using 30% acrylamide/bis solution, 29:1 (Bio-Rad), with a final 1× TBE and 0.5% w/w glycerol. The mini-gels were post-stained with 1× SYBR Gold (Invitrogen, Waltham, MA) in 1× TBE buffer and imaged with GelDoc system (Bio-Rad). Nine titration samples for each group were prepared for EMSA. In each group, the concentration of SL1 was fixed at 2 μM. The 5′END-2 PMO or PPMO, or NC705 PMO or PPMO at room temperature, were individually titrated into SL1 with a final concentration of 0, 0.2, 0.4, 0.8, 1.6, 2, 4, 8, and 16 μM PMO/PPMO. Titration samples were preincubated in EMSA buffer (10 mM Tris-HCl, 50 mM KCl, 1 mM MgCl2, 10% glycerol, 0.1% Triton X-100, pH 6.5) at 4°C for 30 min before loading onto the gels. A total of 15 μL of titration sample per lane was loaded onto the 8% native PAGE for electrophoresis, without addition of loading dye.
Cells and viruses
HeLa-ACE2 cells (BPS Bioscience, San Diego, CA), were maintained in DMEM (Corning, Corning, NY) supplemented with 10% fetal bovine serum (FBS), 0.5 μg/mL puromycin, and penicillin-streptomycin (Corning) at 37°C and 5% CO2.
All cell lines used in this study were regularly screened for mycoplasma contamination using the MycoStrip Mycoplasma Detection Kit (rep-mys-20; InvivoGen, San Diego, CA). Cells were infected with SARS-CoV-2, isolate USA-WA/2020 (NR-52281; BEI Resources, Manassas, VA) or representative isolates of variants Delta (B.1.617.2) and Omicron (BA.1, BA.5, XBB1.5, and JN.1). SARS-Cov-2 variants were collected from nasopharyngeal swab specimens as part of the routine SARS-CoV-2 surveillance conducted by the Mount Sinai Pathogen Surveillance program (institutional review board approved, HS#13-00981). Viruses were grown in Vero-TMPRSS2 cells (BPS Bioscience) for 4–6 days; the supernatant was clarified by centrifugation at 4,000 × g for 5 min and aliquots were frozen at −80°C for long-term use. Expanded viral stocks were sequence verified to be the identified SARS-CoV-2 variant and titered on Vero-TMPRSS2 cells before use in antiviral assays. Infections with viruses were performed under Biosafety Level 3 (BSL3) containment in accordance with the biosafety protocols developed by the Icahn School of Medicine at Mount Sinai and/or Scripps Institute.
In vitro antiviral and cytotoxicity assays
Four thousand HeLa-ACE2 cells (BPS Bioscience) per well were seeded into 96-well plates in DMEM (10% FBS) and incubated for 24 h at 37°C, 5% CO2. Five hours before infection, the medium was replaced with 100 μL of DMEM (2% FBS) containing PBS, nirmatrelvir (Paxlovid), or PPMO. The highest final concentrations for nirmatrelvir and PPMO in the media were 5 μM and 50 or 80 μM, respectively. Plates were then transferred into the BSL3 facility, the drug-containing medium removed, and 100 PFU (MOI 0.025) of virus were added in 50 μL DMEM (2% FBS). Plates were then incubated for 24 h at 37°C, after which supernatants were removed and cells fixed with 4% formaldehyde for 24 h prior to being removed from the BSL3 facility. The cells were then immunostained for the viral N protein (with an Icahn School of Medicine at Mount Sinai in-housmonoclonal antibody, 1C7, provided by Dr. Andrew Duty, andrew.duty@mssm.edu) along with a DAPI counterstain. Infected cells (488 nm) and total cells (DAPI) were quantified using the Cytation 1 (BioTek, Winooski, VT) imaging cytometer. Infectivity was measured by the accumulation of viral N protein (fluorescence accumulation). Percent infection was quantified as ((Infected cells/Total cells) − Background) × 100, and the PBS control was then set to 100% infection for analysis. Data from 6- or 8-point dose-response curves were used to calculate IC50s and IC90s by nonlinear regression for each experiment using GraphPad Prism version 10.0.0 (San Diego, CA). Cytotoxicity evaluation was performed using the MTT assay (Roche, Indianapolis, IN), according to the manufacturer’s instructions. Cytotoxicity evaluations were performed in uninfected cells using the same cell culture conditions and compound dilutions as the viral replication assays and were carried out concurrently with the viral replication assays. All assays were performed in biologically independent triplicate, and the means are reported.
Animal experiments
All infection studies were carried out under BSL3 conditions at Scripps Research and were conducted in accordance with guidelines and approval of the Institutional Animal Care and Use Committee of Scripps Research Institute. Heterozygous K18-hACE2 mice (strain B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from The Jackson Laboratory. Male or female mice aged 7–9 weeks were administered 50 μL PBS or PPMO (5′END-2 or NC705) in 50 μL PBS, at 24 h pre-infection and 18 h post-infection. Mice were anesthetized with isoflurane, and the compounds were administered IN or IT. MK-4482 (molnupravir) was administered orally twice daily for 4 days, from 1 day before infection to 2 days post-infection, at a dose of 150 mg/kg per dose, to maintain consistency with previous projects using this compound.70,115 All virus infections were carried out on anesthetized mice via IN administration, using 5000 ffu WA-1/2020 SARS-CoV-2. For experiments using IN delivery of test compounds, doses were administered in a dropwise manner. For IT delivery, PBS or PPMO-containing solution was administered through the Endotracheal Tube Introducer (Hallowell, Pittsfield, MA). To quantify infectious virus in the lung, lung tissue was harvested 3 days post-infection (the day of peak virus titer in the lungs in this model77,116), homogenized, and titrated by focus-forming assay, as described previously.117 Body weights of the mice were measured from 0 DPI for 10 days, or until the day of euthanasia. Mice losing greater than 35% of their body weight were considered moribund and were humanely euthanized.
Mouse lung fluorimetry
Mice (n = 5) were treated with P7-5′END2-liss at 0 or 10 mg/kg by either IN or IT instillation (as described above) and euthanized 24 h following treatment. Lungs were separated into upper and lower portions and homogenized in Qiagen RLT lysis buffer at 100 mg/mL using bead tubes and agitating at 5.65 m/s for 45 s for two cycles with a 30-s delay. Lysate fluorescence signal (excitation 520 nm/emission 580–640 nm) was measured using a fluorescence plate reader and used to interpolate PPMO concentration based on a standard curve (R2 = 0.9995) prepared using P7-5′END2-liss-spiked untreated mouse lung lysate. Data were analyzed using unpaired two-tailed Student’s t tests.
In-cell reporter constructs and assay
To measure the ability of a SARS-CoV-2-sequence-directed PPMO to interfere with the process of gene expression, we co-transfected two fluorescent reporter plasmids into HEK293 cells. One plasmid contained the first 75 nt of the SARS-CoV-2 genome (pSARS2-75/mCherry) followed by mCherry coding sequence, while the other contained a non-viral sequence followed by the coding sequence for GFP (pRNDM-75/GFP). Each plasmid contained a cytomegalovirus (CMV) promoter. For the reporter gene translation assays, HEK293 cells were grown to near-confluence in 24-well plates. The growth media (consisting of DMEM [Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS (Seradigm Premium Grade HI FBS, VWR, Radnor, PA), 1× penicillin-streptomycin (Gibco), 1× MEM non-essential amino acids (Gibco), and 1 mM sodium pyruvate (Gibco) was removed and replaced with 450 μL growth media and 50 μL PBS or PPMO-containing-PBS (producing final concentrations of 0, 4, 8, and 16 μM PPMO in the wells) for 2.5 h. The cells were then transfected with 500 ng of each plasmid using BioT transfection reaction (BioLand Scientific, Cerritos, CA), following the manufacturer’s protocol. At 24 h after transfection, the cells were trypsinized and prepared for flow cytometry analysis, as previously described.118
Data and code availability
All data collected for this study are available upon request from the corresponding authors.
Acknowledgments
This work was supported partially through the National Institute of Allergy and Infectious Diseases (NIAID) Antiviral Drug Discovery program, award no. U19AI171443, along with grants NIH/NIAID R01AI160706 and NIH/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01DK130425 to M.S. The authors thank Dr. Randy Albrecht for BSL3 lab management at the Icahn School of Medicine at Mount Sinai and Dr. John Adelman (Oregon Health & Science University) for a critical reading of the manuscript.
Author contributions
D.A.S., H.M.M., J.R.T., A. Chatterjee, S.C., A.G.-S., and M.S. conceptualized the study. M.K. and H.M.M. produced the PPMO. A.S., G.S., S.B., C.V.L., R.D.-T., P.W., K.W., L.L.L., M.B., A. Chow, J.H., C.L., J.R.T., M.S., H.M.M., and D.A.S contributed to data production and analysis. K.W., B.T., M.G., S.M.-S., Y.B., A. Chatterjee, J.R.T., H.M.M., and D.A.S. supervised various aspects of the project. D.A.S. wrote the initial draft and revisions of the paper. All authors reviewed, offered comments on, and approved the manuscript. Funding for this project was acquired by S.C., M.S., and A.G.-S.
Declaration of interests
D.A.S. and H.M.M are listed as inventors on a US patent application, which includes a PPMO developed in this study, filed by Oregon State University. The H.M.M. laboratory has received research support from Radiation Control Technology, BriSight Biosciences, and Autoimmunity BioSolutions. The A.G.-S. laboratory has received research support from GSK, Pfizer, Senhwa Biosciences, Kenall Manufacturing, Blade Therapeutics, Avimex, Johnson & Johnson, Dynavax, 7 Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma, Applied Biological Laboratories, and Merck outside of the reported work. A.G.-S. has consulting agreements with the following companies involving cash and/or stock: Castlevax, Amovir, Vivaldi Biosciences, Contrafect, 7 Hills Pharma, Avimex, Pagoda, Accurius, Esperovax, Applied Biological Laboratories, Pharmamar, CureLab Oncology, CureLab Veterinary, Synairgen, Paratus, Pfizer, and Prosetta outside of the reported work. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott and Astrazeneca. A.G.-S. is listed as an inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work. None of the support defined above is directly related to the research described in this paper.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2024.102331.
Contributor Information
Hong M. Moulton, Email: hong.moulton@oregonstate.edu.
David A. Stein, Email: dave.stein@oregonstate.edu.
Supplemental information
References
- 1.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Msemburi W., Karlinsky A., Knutson V., Aleshin-Guendel S., Chatterji S., Wakefield J. The WHO estimates of excess mortality associated with the COVID-19 pandemic. Nature. 2023;613:130–137. doi: 10.1038/s41586-022-05522-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.COVID-19 Excess Mortality Collaborators Estimating excess mortality due to the COVID-19 pandemic: a systematic analysis of COVID-19-related mortality, 2020-21. Lancet. 2022;399:1513–1536. doi: 10.1016/S0140-6736(21)02796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Delft A., Hall M.D., Kwong A.D., Purcell L.A., Saikatendu K.S., Schmitz U., Tallarico J.A., Lee A.A. Accelerating antiviral drug discovery: lessons from COVID-19. Nat. Rev. Drug Discov. 2023;22:585–603. doi: 10.1038/s41573-023-00692-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh A.K., Mishevich J.L., Mesecar A., Mitsuya H. Recent Drug Development and Medicinal Chemistry Approaches for the Treatment of SARS-CoV-2 Infection and COVID-19. ChemMedChem. 2022;17 doi: 10.1002/cmdc.202200440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards A.M., Baric R.S., Saphire E.O., Ulmer J.B. Stopping pandemics before they start: Lessons learned from SARS-CoV-2. Science. 2022;375:1133–1139. doi: 10.1126/science.abn1900. [DOI] [PubMed] [Google Scholar]
- 8.Li G., Hilgenfeld R., Whitley R., De Clercq E. Therapeutic strategies for COVID-19: progress and lessons learned. Nat. Rev. Drug Discov. 2023;22:449–475. doi: 10.1038/s41573-023-00672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh M., de Wit E. Antiviral agents for the treatment of COVID-19: Progress and challenges. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanderson T., Hisner R., Donovan-Banfield I., Hartman H., Lochen A., Peacock T.P., Ruis C. A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes. Nature. 2023;623:594–600. doi: 10.1038/s41586-023-06649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenke K., Lewis M.C., Feldmann F., Bohrnsen E., Schwarz B., Okumura A., Bohler W.F., Callison J., Shaia C., Bosio C.M., et al. Combined molnupiravir-nirmatrelvir treatment improves the inhibitory effect on SARS-CoV-2 in macaques. JCI Insight. 2023;8 doi: 10.1172/jci.insight.166485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shyr Z.A., Cheng Y.S., Lo D.C., Zheng W. Drug combination therapy for emerging viral diseases. Drug Discov. Today. 2021;26:2367–2376. doi: 10.1016/j.drudis.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moumne L., Marie A.C., Crouvezier N. Oligonucleotide Therapeutics: From Discovery and Development to Patentability. Pharmaceutics. 2022;14 doi: 10.3390/pharmaceutics14020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond S.M., Aartsma-Rus A., Alves S., Borgos S.E., Buijsen R.A.M., Collin R.W.J., Covello G., Denti M.A., Desviat L.R., Echevarría L., et al. Delivery of oligonucleotide-based therapeutics: challenges and opportunities. EMBO Mol. Med. 2021;13 doi: 10.15252/emmm.202013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul S., Caruthers M.H. Synthesis of Backbone-Modified Morpholino Oligonucleotides Using Phosphoramidite Chemistry. Molecules. 2023;28 doi: 10.3390/molecules28145380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egli M., Manoharan M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 2023;51:2529–2573. doi: 10.1093/nar/gkad067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Idris A., Supramaniam A., Tayyar Y., Kelly G., McMillan N.A.J., Morris K.V. An intranasally delivered ultra-conserved siRNA prophylactically represses SARS-CoV-2 infection in the lung and nasal cavity. Antivir. Res. 2024;222 doi: 10.1016/j.antiviral.2024.105815. [DOI] [PubMed] [Google Scholar]
- 18.Hariharan V.N., Shin M., Chang C.W., O'Reilly D., Biscans A., Yamada K., Guo Z., Somasundaran M., Tang Q., Monopoli K., et al. Divalent siRNAs are bioavailable in the lung and efficiently block SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2219523120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Idris A., Davis A., Supramaniam A., Acharya D., Kelly G., Tayyar Y., West N., Zhang P., McMillan C.L.D., Soemardy C., et al. A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Ther. 2021;29:2219–2226. doi: 10.1016/j.ymthe.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vora S.M., Fontana P., Mao T., Leger V., Zhang Y., Fu T.M., Lieberman J., Gehrke L., Shi M., Wang L., et al. Targeting stem-loop 1 of the SARS-CoV-2 5' UTR to suppress viral translation and Nsp1 evasion. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2117198119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu C., Lee J.Y., Woo J.Z., Xu L., Nguyenla X., Yamashiro L.H., Ji F., Biering S.B., Van Dis E., Gonzalez F., et al. An intranasal ASO therapeutic targeting SARS-CoV-2. Nat. Commun. 2022;13:4503. doi: 10.1038/s41467-022-32216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khaitov M., Nikonova A., Kofiadi I., Shilovskiy I., Smirnov V., Elisytina O., Maerle A., Shatilov A., Shatilova A., Andreev S., et al. Treatment of COVID-19 patients with a SARS-CoV-2-specific siRNA-peptide dendrimer formulation. Allergy. 2023;78:1639–1653. doi: 10.1111/all.15663. [DOI] [PubMed] [Google Scholar]
- 23.Summerton J., Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 24.Stein D., Foster E., Huang S.B., Weller D., Summerton J. A specificity comparison of four antisense types: morpholino, 2'-O-methyl RNA, DNA, and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev. 1997;7:151–157. doi: 10.1089/oli.1.1997.7.151. [DOI] [PubMed] [Google Scholar]
- 25.Moulton J.D. Using Morpholinos to Control Gene Expression. Curr. Protoc. Nucleic Acid Chem. 2017;68:4.30.1–4.30.29. doi: 10.1002/cpnc.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filonova G., Aartsma-Rus A. Next steps for the optimization of exon therapy for Duchenne muscular dystrophy. Expet Opin. Biol. Ther. 2023;23:133–143. doi: 10.1080/14712598.2023.2169070. [DOI] [PubMed] [Google Scholar]
- 27.Roberts T.C., Wood M.J.A., Davies K.E. Therapeutic approaches for Duchenne muscular dystrophy. Nat. Rev. Drug Discov. 2023;22:917–934. doi: 10.1038/s41573-023-00775-6. [DOI] [PubMed] [Google Scholar]
- 28.Wilton-Clark H., Yokota T. Recent Trends in Antisense Therapies for Duchenne Muscular Dystrophy. Pharmaceutics. 2023;15 doi: 10.3390/pharmaceutics15030778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moulton H.M., Nelson M.H., Hatlevig S.A., Reddy M.T., Iversen P.L. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjugate Chem. 2004;15:290–299. doi: 10.1021/bc034221g. [DOI] [PubMed] [Google Scholar]
- 30.Deas T.S., Binduga-Gajewska I., Tilgner M., Ren P., Stein D.A., Moulton H.M., Iversen P.L., Kauffman E.B., Kramer L.D., Shi P.Y. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 2005;79:4599–4609. doi: 10.1128/JVI.79.8.4599-4609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moulton H.M., Hase M.C., Smith K.M., Iversen P.L. HIV Tat peptide enhances cellular delivery of antisense morpholino oligomers. Antisense Nucleic Acid Drug Dev. 2003;13:31–43. doi: 10.1089/108729003764097322. [DOI] [PubMed] [Google Scholar]
- 32.Marshall N.B., Oda S.K., London C.A., Moulton H.M., Iversen P.L., Kerkvliet N.I., Mourich D.V. Arginine-rich cell-penetrating peptides facilitate delivery of antisense oligomers into murine leukocytes and alter pre-mRNA splicing. J. Immunol. Methods. 2007;325:114–126. doi: 10.1016/j.jim.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Moulton H.M., Moulton J.D. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim. Biophys. Acta. 2010;1798:2296–2303. doi: 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 34.Moulton H.M. In vivo delivery of morpholino oligos by cell-penetrating peptides. Curr. Pharmaceut. Des. 2013;19:2963–2969. doi: 10.2174/1381612811319160010. [DOI] [PubMed] [Google Scholar]
- 35.Gabriel G., Nordmann A., Stein D.A., Iversen P.L., Klenk H.D. Morpholino oligomers targeting the PB1 and NP genes enhance the survival of mice infected with highly pathogenic influenza A H7N7 virus. J. Gen. Virol. 2008;89:939–948. doi: 10.1099/vir.0.83449-0. [DOI] [PubMed] [Google Scholar]
- 36.Lai S.H., Stein D.A., Guerrero-Plata A., Liao S.L., Ivanciuc T., Hong C., Iversen P.L., Casola A., Garofalo R.P. Inhibition of respiratory syncytial virus infections with morpholino oligomers in cell cultures and in mice. Mol. Ther. 2008;16:1120–1128. doi: 10.1038/mt.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lupfer C., Stein D.A., Mourich D.V., Tepper S.E., Iversen P.L., Pastey M. Inhibition of influenza A H3N8 virus infections in mice by morpholino oligomers. Arch. Virol. 2008;153:929–937. doi: 10.1007/s00705-008-0067-0. [DOI] [PubMed] [Google Scholar]
- 38.Limburg H., Harbig A., Bestle D., Stein D.A., Moulton H.M., Jaeger J., Janga H., Hardes K., Koepke J., Schulte L., et al. TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes. J. Virol. 2019;93:10–1128. doi: 10.1128/JVI.00649-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein D.A. Inhibition of RNA virus infections with peptide-conjugated morpholino oligomers. Curr. Pharmaceut. Des. 2008;14:2619–2634. doi: 10.2174/138161208786071290. [DOI] [PubMed] [Google Scholar]
- 40.Nan Y., Zhang Y.J. Antisense Phosphorodiamidate Morpholino Oligomers as Novel Antiviral Compounds. Front. Microbiol. 2018;9:750. doi: 10.3389/fmicb.2018.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren T.K., Shurtleff A.C., Bavari S. Advanced morpholino oligomers: a novel approach to antiviral therapy. Antivir. Res. 2012;94:80–88. doi: 10.1016/j.antiviral.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park G.J., Osinski A., Hernandez G., Eitson J.L., Majumdar A., Tonelli M., Henzler-Wildman K., Pawłowski K., Chen Z., Li Y., et al. The mechanism of RNA capping by SARS-CoV-2. Nature. 2022;609:793–800. doi: 10.1038/s41586-022-05185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steiner S., Kratzel A., Barut G.T., Lang R.M., Aguiar Moreira E., Thomann L., Kelly J.N., Thiel V. SARS-CoV-2 biology and host interactions. Nat. Rev. Microbiol. 2024;22:206–225. doi: 10.1038/s41579-023-01003-z. [DOI] [PubMed] [Google Scholar]
- 44.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma R., Saha S., Kumar S., Mani S., Maiti T.K., Surjit M. RNA-Protein Interaction Analysis of SARS-CoV-2 5' and 3' Untranslated Regions Reveals a Role of Lysosome-Associated Membrane Protein-2a during Viral Infection. mSystems. 2021;6 doi: 10.1128/mSystems.00643-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao C., Cai Z., Xiao X., Rao J., Chen J., Hu N., Yang M., Xing X., Wang Y., Li M., et al. The architecture of the SARS-CoV-2 RNA genome inside virion. Nat. Commun. 2021;12:3917. doi: 10.1038/s41467-021-22785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenke K., Leventhal S., Moulton H.M., Hatlevig S., Hawman D., Feldmann H., Stein D.A. Inhibition of SARS-CoV-2 in Vero cell cultures by peptide-conjugated morpholino oligomers. J. Antimicrob. Chemother. 2021;76:413–417. doi: 10.1093/jac/dkaa460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Born E., Stein D.A., Iversen P.L., Snijder E.J. Antiviral activity of morpholino oligomers designed to block various aspects of Equine arteritis virus amplification in cell culture. J. Gen. Virol. 2005;86:3081–3090. doi: 10.1099/vir.0.81158-0. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J., Stein D.A., Timoney P.J., Balasuriya U.B.R. Curing of HeLa cells persistently infected with equine arteritis virus by a peptide-conjugated morpholino oligomer. Virus Res. 2010;150:138–142. doi: 10.1016/j.virusres.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y.J., Stein D.A., Fan S.M., Wang K.Y., Kroeker A.D., Meng X.J., Iversen P.L., Matson D.O. Suppression of porcine reproductive and respiratory syndrome virus replication by morpholino antisense oligomers. Vet. Microbiol. 2006;117:117–129. doi: 10.1016/j.vetmic.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burrer R., Neuman B.W., Ting J.P.C., Stein D.A., Moulton H.M., Iversen P.L., Kuhn P., Buchmeier M.J. Antiviral effects of antisense morpholino oligomers in murine coronavirus infection models. J. Virol. 2007;81:5637–5648. doi: 10.1128/JVI.02360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel D., Opriessnig T., Stein D.A., Halbur P.G., Meng X.J., Iversen P.L., Zhang Y.J. Peptide-conjugated morpholino oligomers inhibit porcine reproductive and respiratory syndrome virus replication. Antivir. Res. 2008;77:95–107. doi: 10.1016/j.antiviral.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Opriessnig T., Patel D., Wang R., Halbur P.G., Meng X.J., Stein D.A., Zhang Y.J. Inhibition of porcine reproductive and respiratory syndrome virus infection in piglets by a peptide-conjugated morpholino oligomer. Antivir. Res. 2011;91:36–42. doi: 10.1016/j.antiviral.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 54.Patel D., Stein D.A., Zhang Y.J. Morpholino oligomer-mediated protection of porcine pulmonary alveolar macrophages from arterivirus-induced cell death. Antivir. Ther. 2009;14:899–909. doi: 10.3851/IMP1409. [DOI] [PubMed] [Google Scholar]
- 55.Holden K.L., Stein D.A., Pierson T.C., Ahmed A.A., Clyde K., Iversen P.L., Harris E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3' stem-loop structure. Virology. 2006;344:439–452. doi: 10.1016/j.virol.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 56.Paessler S., Rijnbrand R., Stein D.A., Ni H., Yun N.E., Dziuba N., Borisevich V., Seregin A., Ma Y., Blouch R., et al. Inhibition of alphavirus infection in cell culture and in mice with antisense morpholino oligomers. Virology. 2008;376:357–370. doi: 10.1016/j.virol.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stein D.A., Huang C.Y.H., Silengo S., Amantana A., Crumley S., Blouch R.E., Iversen P.L., Kinney R.M. Treatment of AG129 mice with antisense morpholino oligomers increases survival time following challenge with dengue 2 virus. J. Antimicrob. Chemother. 2008;62:555–565. doi: 10.1093/jac/dkn221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stein D.A., Skilling D.E., Iversen P.L., Smith A.W. Inhibition of Vesivirus infections in mammalian tissue culture with antisense morpholino oligomers. Antisense Nucleic Acid Drug Dev. 2001;11:317–325. doi: 10.1089/108729001753231696. [DOI] [PubMed] [Google Scholar]
- 59.Kinney R.M., Huang C.Y.H., Rose B.C., Kroeker A.D., Dreher T.W., Iversen P.L., Stein D.A. Inhibition of dengue virus serotypes 1 to 4 in vero cell cultures with morpholino oligomers. J. Virol. 2005;79:5116–5128. doi: 10.1128/JVI.79.8.5116-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miao Z., Tidu A., Eriani G., Martin F. Secondary structure of the SARS-CoV-2 5'-UTR. RNA Biol. 2021;18:447–456. doi: 10.1080/15476286.2020.1814556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sosnowski P., Tidu A., Eriani G., Westhof E., Martin F. Correlated sequence signatures are present within the genomic 5'UTR RNA and NSP1 protein in coronaviruses. RNA. 2022;28:729–741. doi: 10.1261/rna.078972.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wacker A., Weigand J.E., Akabayov S.R., Altincekic N., Bains J.K., Banijamali E., Binas O., Castillo-Martinez J., Cetiner E., Ceylan B., et al. Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy. Nucleic Acids Res. 2020;48:12415–12435. doi: 10.1093/nar/gkaa1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eriani G., Martin F. Viral and cellular translation during SARS-CoV-2 infection. FEBS Open Bio. 2022;12:1584–1601. doi: 10.1002/2211-5463.13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tidu A., Janvier A., Schaeffer L., Sosnowski P., Kuhn L., Hammann P., Westhof E., Eriani G., Martin F. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA. 2020;27:253–264. doi: 10.1261/rna.078121.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bujanic L., Shevchuk O., von Kügelgen N., Kalinina A., Ludwik K., Koppstein D., Zerna N., Sickmann A., Chekulaeva M. The key features of SARS-CoV-2 leader and NSP1 required for viral escape of NSP1-mediated repression. RNA. 2022;28:766–779. doi: 10.1261/rna.079086.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swenson D.L., Warfield K.L., Warren T.K., Lovejoy C., Hassinger J.N., Ruthel G., Blouch R.E., Moulton H.M., Weller D.D., Iversen P.L., Bavari S. Chemical modifications of antisense morpholino oligomers enhance their efficacy against Ebola virus infection. Antimicrob. Agents Chemother. 2009;53:2089–2099. doi: 10.1128/AAC.00936-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lebleu B., Moulton H.M., Abes R., Ivanova G.D., Abes S., Stein D.A., Iversen P.L., Arzumanov A.A., Gait M.J. Cell penetrating peptide conjugates of steric block oligonucleotides. Adv. Drug Deliv. Rev. 2008;60:517–529. doi: 10.1016/j.addr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abes R., Moulton H.M., Clair P., Yang S.T., Abes S., Melikov K., Prevot P., Youngblood D.S., Iversen P.L., Chernomordik L.V., Lebleu B. Delivery of steric block morpholino oligomers by (R-X-R)4 peptides: structure-activity studies. Nucleic Acids Res. 2008;36:6343–6354. doi: 10.1093/nar/gkn541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amantana A., Moulton H.M., Cate M.L., Reddy M.T., Whitehead T., Hassinger J.N., Youngblood D.S., Iversen P.L. Pharmacokinetics, biodistribution, stability and toxicity of a cell-penetrating peptide-morpholino oligomer conjugate. Bioconjugate Chem. 2007;18:1325–1331. doi: 10.1021/bc070060v. [DOI] [PubMed] [Google Scholar]
- 70.Bakowski M.A., Beutler N., Wolff K.C., Kirkpatrick M.G., Chen E., Nguyen T.T.H., Riva L., Shaabani N., Parren M., Ricketts J., et al. Drug repurposing screens identify chemical entities for the development of COVID-19 interventions. Nat. Commun. 2021;12:3309. doi: 10.1038/s41467-021-23328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 72.Deas T.S., Bennett C.J., Jones S.A., Tilgner M., Ren P., Behr M.J., Stein D.A., Iversen P.L., Kramer L.D., Bernard K.A., Shi P.Y. In Vitro Resistance Selection and In Vivo Efficacy of Morpholino Oligomers against West Nile Virus. Antimicrob. Agents Chemother. 2007;51:2470–2482. doi: 10.1128/AAC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soonthornvacharin S., Rodriguez-Frandsen A., Zhou Y., Galvez F., Huffmaster N.J., Tripathi S., Balasubramaniam V.R.M.T., Inoue A., de Castro E., Moulton H., et al. Systems-based analysis of RIG-I-dependent signalling identifies KHSRP as an inhibitor of RIG-I receptor activation. Nat. Microbiol. 2017;2:17022. doi: 10.1038/nmicrobiol.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tripathi S., Pohl M.O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D.A., Moulton H.M., DeJesus P., Che J., Mulder L.C.F., et al. Meta- and Orthogonal Integration of Influenza "OMICs" Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin-Sancho L., Tripathi S., Rodriguez-Frandsen A., Pache L., Sanchez-Aparicio M., McGregor M.J., Haas K.M., Swaney D.L., Nguyen T.T., Mamede J.I., et al. Restriction factor compendium for influenza A virus reveals a mechanism for evasion of autophagy. Nat. Microbiol. 2021;6:1319–1333. doi: 10.1038/s41564-021-00964-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fumagalli V., Di Lucia P., Ravà M., Marotta D., Bono E., Grassi S., Donnici L., Cannalire R., Stefanelli I., Ferraro A., et al. Nirmatrelvir treatment of SARS-CoV-2-infected mice blunts antiviral adaptive immune responses. EMBO Mol. Med. 2023;15 doi: 10.15252/emmm.202317580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y., Cao L., Li G., Cong F., Li Y., Sun J., Luo Y., Chen G., Li G., Wang P., et al. Remdesivir Metabolite GS-441524 Effectively Inhibits SARS-CoV-2 Infection in Mouse Models. J. Med. Chem. 2022;65:2785–2793. doi: 10.1021/acs.jmedchem.0c01929. [DOI] [PubMed] [Google Scholar]
- 80.Abdelnabi R., Foo C.S., Kaptein S.J.F., Boudewijns R., Vangeel L., De Jonghe S., Jochmans D., Weynand B., Neyts J. A SCID Mouse Model To Evaluate the Efficacy of Antivirals against SARS-CoV-2 Infection. J. Virol. 2022;96 doi: 10.1128/jvi.00758-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ge Q., Pastey M., Kobasa D., Puthavathana P., Lupfer C., Bestwick R.K., Iversen P.L., Chen J., Stein D.A. Inhibition of multiple subtypes of influenza a virus in cell cultures with morpholino oligomers. Antimicrob. Agents Chemother. 2006;50:3724–3733. doi: 10.1128/AAC.00644-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Okamoto S., Echigoya Y., Tago A., Segawa T., Sato Y., Itou T. Antiviral Efficacy of RNase H-Dependent Gapmer Antisense Oligonucleotides against Japanese Encephalitis Virus. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms241914846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dhorne-Pollet S., Fitzpatrick C., Da Costa B., Bourgon C., Eléouët J.F., Meunier N., Burzio V.A., Delmas B., Barrey E. Antisense oligonucleotides targeting ORF1b block replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.915202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hagey R.J., Elazar M., Pham E.A., Tian S., Ben-Avi L., Bernardin-Souibgui C., Yee M.F., Moreira F.R., Rabinovitch M.V., Meganck R.M., et al. Programmable antivirals targeting critical conserved viral RNA secondary structures from influenza A virus and SARS-CoV-2. Nat. Med. 2022;28:1944–1955. doi: 10.1038/s41591-022-01908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lulla V., Wandel M.P., Bandyra K.J., Ulferts R., Wu M., Dendooven T., Yang X., Doyle N., Oerum S., Beale R., et al. Targeting the Conserved Stem Loop 2 Motif in the SARS-CoV-2 Genome. J. Virol. 2021;95 doi: 10.1128/JVI.00663-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iannitti T., Morales-Medina J.C., Palmieri B. Phosphorothioate oligonucleotides: effectiveness and toxicity. Curr. Drug Targets. 2014;15:663–673. doi: 10.2174/1389450115666140321100304. [DOI] [PubMed] [Google Scholar]
- 87.Ferdinandi E.S., Vassilakos A., Lee Y., Lightfoot J., Fitsialos D., Wright J.A., Young A.H. Preclinical toxicity and toxicokinetics of GTI-2040, a phosphorothioate oligonucleotide targeting ribonucleotide reductase R2. Cancer Chemother. Pharmacol. 2011;68:193–205. doi: 10.1007/s00280-010-1473-z. [DOI] [PubMed] [Google Scholar]
- 88.Flynn L.L., Li R., Pitout I.L., Aung-Htut M.T., Larcher L.M., Cooper J.A.L., Greer K.L., Hubbard A., Griffiths L., Bond C.S., et al. Single Stranded Fully Modified-Phosphorothioate Oligonucleotides can Induce Structured Nuclear Inclusions, Alter Nuclear Protein Localization and Disturb the Transcriptome In Vitro. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.791416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang D., Jiang A., Feng J., Li G., Guo D., Sajid M., Wu K., Zhang Q., Ponty Y., Will S., et al. The SARS-CoV-2 subgenome landscape and its novel regulatory features. Mol. Cell. 2021;81:2135–2147. doi: 10.1016/j.molcel.2021.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Malone B., Urakova N., Snijder E.J., Campbell E.A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022;23:21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brinton M.A., Basu M. Functions of the 3' and 5' genome RNA regions of members of the genus Flavivirus. Virus Res. 2015;206:108–119. doi: 10.1016/j.virusres.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Madhugiri R., Karl N., Petersen D., Lamkiewicz K., Fricke M., Wend U., Scheuer R., Marz M., Ziebuhr J. Structural and functional conservation of cis-acting RNA elements in coronavirus 5'-terminal genome regions. Virology. 2018;517:44–55. doi: 10.1016/j.virol.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang D., Leibowitz J.L. The structure and functions of coronavirus genomic 3' and 5' ends. Virus Res. 2015;206:120–133. doi: 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ziv O., Price J., Shalamova L., Kamenova T., Goodfellow I., Weber F., Miska E.A. The Short- and Long-Range RNA-RNA Interactome of SARS-CoV-2. Mol. Cell. 2020;80:1067–1077.e5. doi: 10.1016/j.molcel.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hyde J.L., Chen R., Trobaugh D.W., Diamond M.S., Weaver S.C., Klimstra W.B., Wilusz J. The 5' and 3' ends of alphavirus RNAs--Non-coding is not non-functional. Virus Res. 2015;206:99–107. doi: 10.1016/j.virusres.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Neuman B.W., Stein D.A., Kroeker A.D., Churchill M.J., Kim A.M., Kuhn P., Dawson P., Moulton H.M., Bestwick R.K., Iversen P.L., Buchmeier M.J. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 2005;79:9665–9676. doi: 10.1128/JVI.79.15.9665-9676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li P., Faraone J.N., Hsu C.C., Chamblee M., Zheng Y.M., Carlin C., Bednash J.S., Horowitz J.C., Mallampalli R.K., Saif L.J., et al. Neutralization escape, infectivity, and membrane fusion of JN.1-derived SARS-CoV-2 SLip, FLiRT, and KP.2 variants. Cell Rep. 2024;43 doi: 10.1016/j.celrep.2024.114520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu J., Li Y., Liu Q., Yao Q., Wang X., Zhang H., Chen R., Ren L., Min J., Deng F., et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021;7:17. doi: 10.1038/s41421-021-00249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bösmüller H., Matter M., Fend F., Tzankov A. The pulmonary pathology of COVID-19. Virchows Arch. 2021;478:137–150. doi: 10.1007/s00428-021-03053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lamers M.M., Haagmans B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022;20:270–284. doi: 10.1038/s41579-022-00713-0. [DOI] [PubMed] [Google Scholar]
- 102.Bryce C., Grimes Z., Pujadas E., Ahuja S., Beasley M.B., Albrecht R., Hernandez T., Stock A., Zhao Z., AlRasheed M.R., et al. Pathophysiology of SARS-CoV-2: the Mount Sinai COVID-19 autopsy experience. Mod. Pathol. 2021;34:1456–1467. doi: 10.1038/s41379-021-00793-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao F., Ma Q., Yue Q., Chen H. SARS-CoV-2 Infection and Lung Regeneration. Clin. Microbiol. Rev. 2022;35 doi: 10.1128/cmr.00188-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ruge C.A., Kirch J., Lehr C.M. Pulmonary drug delivery: from generating aerosols to overcoming biological barriers-therapeutic possibilities and technological challenges. Lancet Respir. Med. 2013;1:402–413. doi: 10.1016/S2213-2600(13)70072-70079. [DOI] [PubMed] [Google Scholar]
- 105.Hickey A.J. Back to the future: inhaled drug products. J. Pharmacol. Sci. (Tokyo, Jpn.) 2013;102:1165–1172. doi: 10.1002/jps.23465. [DOI] [PubMed] [Google Scholar]
- 106.Forbes B., Asgharian B., Dailey L.A., Ferguson D., Gerde P., Gumbleton M., Gustavsson L., Hardy C., Hassall D., Jones R., et al. Challenges in inhaled product development and opportunities for open innovation. Adv. Drug Deliv. Rev. 2011;63:69–87. doi: 10.1016/j.addr.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 107.Abes S., Moulton H.M., Clair P., Prevot P., Youngblood D.S., Wu R.P., Iversen P.L., Lebleu B. Vectorization of morpholino oligomers by the (R-Ahx-R)(4) peptide allows efficient splicing correction in the absence of endosomolytic agents. J. Contr. Release. 2006;116:304–313. doi: 10.1016/j.jconrel.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 108.Li Y.F. End-Modifications on Morpholino Oligos. Methods Mol. Biol. 2017;1565:39–50. doi: 10.1007/978-1-4939-6817-6_4. [DOI] [PubMed] [Google Scholar]
- 109.Li C., Ma L., Zou D., Zhang R., Bai X., Li L., Wu G., Huang T., Zhao W., Jin E., et al. RCoV19: A One-stop Hub for SARS-CoV-2 Genome Data Integration, Variant Monitoring, and Risk Pre-warning. Dev. Reprod. Biol. 2023;21:1066–1079. doi: 10.1016/j.gpb.2023.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Members C.-N., Partners Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2024. Nucleic Acids Res. 2024;52:D18–D32. doi: 10.1093/nar/gkad1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bao Y., Xue Y. From BIG Data Center to China National Center for Bioinformation. Dev. Reprod. Biol. 2023;21:900–903. doi: 10.1016/j.gpb.2023.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Levengood J.D., Rollins C., Mishler C.H.J., Johnson C.A., Miner G., Rajan P., Znosko B.M., Tolbert B.S. Solution structure of the HIV-1 exon splicing silencer 3. J. Mol. Biol. 2012;415:680–698. doi: 10.1016/j.jmb.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 114.Luo L., Chiu L.Y., Sugarman A., Gupta P., Rouskin S., Tolbert B.S. HnRNP A1/A2 Proteins Assemble onto 7SK snRNA via Context Dependent Interactions. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.166885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., 3rd, Liu H., Madden V.J., Krzystek H.M., De C., White K.K. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 117.Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., et al. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell. 2020;183:1325–1339. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data collected for this study are available upon request from the corresponding authors.