Abstract
Background
Cellular senescence is pivotal in the occurrence and progression of atrial fibrillation (AF). This study aimed to identify senescence-related genes that could be potential therapeutic biomarkers for AF.
Methods
AF-related differentially expressed genes (DEGs) were identified using the Gene Expression Omnibus dataset. Weighted gene co-expression network analysis (WGCNA) was used to analyze important modules and potential hub genes. Integrating senescence-related genes, potential biomarkers were identified. Their differential expression levels were then validated in human atrial tissue, HL-1 cells, and Angiotensin II-infused mice. Finally, molecular docking analysis was conducted to predict potential interactions between potential biomarkers and the senolytic drug Navitoclax.
Results
We identified seven genes common to AF-related DEGs and senescence-related genes. Three significant modules were selected from WGCNA analysis. Taken together, three senescence-related genes (ETS1, SP1, and WT1) were found to be significantly associated with AF. Protein-protein interaction network analysis revealed biological connections among the predicted target genes of ETS1, SP1, and WT1. Notably, ETS1, SP1, and WT1 exhibited significant differential expression in clinical samples as well as in vitro and in vivo models. Molecular docking revealed favorable binding affinity between senolytic Navitoclax and these potential biomarkers.
Conclusions
This study highlights ETS1, SP1, and WT1 as crucial senescence-related genes associated with AF, offering potential therapeutic targets, with supportive evidence of binding affinity with senolytic Navitoclax. These findings provide novel insights into AF pathogenesis from a senescence perspective.
Keywords: Atrial fibrillation, Senescence, Gene analysis, Molecular docking, Navitoclax
1. Introduction
Atrial fibrillation (AF), the most common sustained heart rhythm disorder, is characterized by disordered electrical activity in the left atrium, causing irregular contractions. Globally, approximately 0.51 % of the population, 37.5 million people, have been diagnosed with AF [1]. Notably, AF is widely recognized as an age-related disease because age increases its prevalence, which reaches 10 % by the age of 65 [2]. Among the numerous risk factors for AF, aging is the most significant [3]. There are four types of AF: first-diagnosed, paroxysmal, persistent, and permanent [4]. Some patients with AF experience symptoms of varying severity, while others are asymptomatic. AF induces thrombus development and other major adverse cardiovascular events, such as heart failure, which may increase the mortality rate [5]. Although there are several theories, including the ectopic firing theory, AF re-entry, and atrial remodeling, which may contribute to AF progression, the causative molecular mechanisms of AF remain unclear. Therefore, a better and more definitive understanding of the mechanisms of AF is urgently needed to stratify patients and provide effective targeted therapeutics.
Cellular senescence is a state of steady and lasting growth arrest and is one of the key processes in aging. Naturally occurring senescent cells could lead to cardiac dysfunction [6]. Manipulating the biological process of cellular senescence can alleviate or delay many age-related diseases, such as AF. AF is closely related to aging and emerging research has shown that cellular senescence plays a major role. Senescence sparks and regulates, such as mitochondrial damage, shortened telomeres, and oxidative stress, contribute to AF progression [7]. Navitoclax (ABT-263), as an inhibitor of the B-cell lymphoma-2 (BCL-2) family, has been identified as a potential senolytic drug with the ability to selectively eliminate senescent cells. Initially explored in cancer treatment, Navitoclax has gained attention in recent years for its potential applications in cardiovascular diseases linked to senescence, such as pulmonary arterial hypertension and cardiac ischemia-reperfusion injury [8]. Our preliminary research findings indicate that Navitoclax can ameliorate angiotensin II-induced cardiac remodeling by mitigating cardiac fibrosis and inflammation. Notably, Navitoclax also demonstrates an improvement in cardiac electrophysiological characteristics, manifested by a decreased susceptibility to ventricular tachyarrhythmia induced by programmed electrical stimulation. These results suggest a beneficial impact of Navitoclax in the context of pre-existing senescence states during cardiac remodeling, emphasizing its potential to clear senescent cells. Despite senescence being implicated in the initiation and progression of AF, the mechanisms underlying senescence-induced AF remain unclear.
Due to the rapid advancement in high-throughput sequencing, investigating the pathogenesis of diseases at the molecular level is feasible. Bioinformatics analysis is a powerful tool, which has been widely used in basic research. However, little research has focused on the inter-linkages of key genes between senescence and AF. In this study, potential key genes involved in senescence and AF were identified using a sequencing database of AF and a set of senescence-related genes. These potential biomarkers were further validated for their crucial role in human atrial tissue and their interaction with Navitoclax using molecular docking. Our investigation integrates bioinformatics, histological validation, and molecular docking analysis to identify senescence-related genes that significantly correlate with AF, offering novel insights into how cellular senescence may impact AF, which may aid in individualized therapeutic strategies.
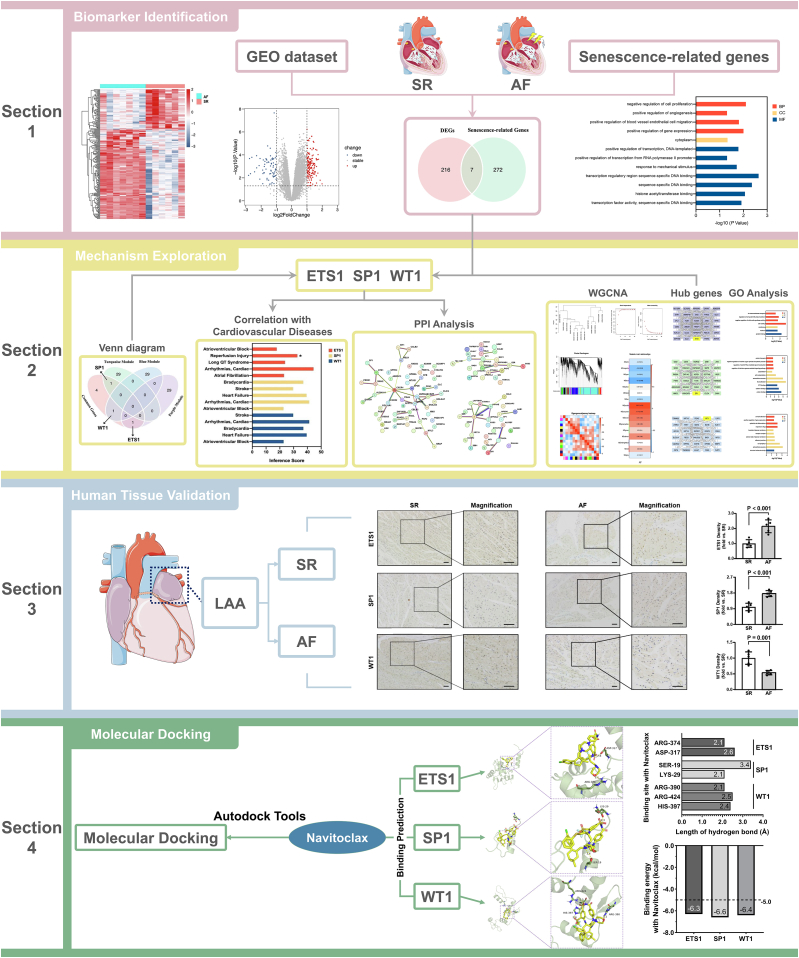
The flowchart of the present research has been illustrated in Fig. 1.
Fig. 1.
Flowchart of the research of identification of senescence-related genes for potential biomarkers of atrial fibrillation.
2. Materials and methods
2.1. Data source and processing
Gene expression profiles associated with AF (GSE79768) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) [9]. We selected seven left atrial samples with AF and six with sinus rhythm (SR) for subsequent analysis. Patients with AF in this study had persistent AF continuously for more than 6 months, while those with SR showed no clinical evidence of AF and were not taking antiarrhythmic medications [10]. The annotation packages in Bioconductor (https://www.bioconductor.org) and hgu133a.db were used to convert the probe ID to a gene symbol. Additionally, 279 genes associated with cellular senescence, obtained from CellAge (https://genomics.senescence.info/cells/), were included in this study [11].
2.2. Identification of differentially expressed genes (DEGs)
DEGs in the left atrial samples of patients with AF compared with those of patients with SR were identified using the R package “limma”. The false discovery rate process was used to calculate fold-changes (FC). Applying FC and p-value filters, DEGs were identified between the AF and SR samples (| logFC | >1 and P < 0.05) [12,13].
2.3. Gene ontology (GO) enrichment analysis
Database for annotation, visualization, and integrated discovery (DAVID) (https://david.ncifcrf.gov/home.jsp) was used to perform the gene ontology (GO) enrichment analysis [14]. GO is a database containing a series of GO terms that describes and defines the functions of gene in various species, continually updated with new research [15]. The enriched GO terms include biological process (BP), cellular component (CC), and molecular function (MF) ontologies. The significant enrichment for GO threshold was P < 0.05, and the count was ≥2.
2.4. Weighted gene co-expression network analysis (WGCNA) analysis
We used the WGCNA R package to build co-expression network [16]. Genes with an upper 25 % variation across samples in the dataset were imported into WGCNA. Cluster the samples to detect the presence of outliers. Co-expression networks were then constructed using automatic network construction. The soft thresholding power β was calculated using the pick-Soft-Threshold function. Modules were obtained via hierarchical clustering and the dynamic tree cut function. Correlations between the clinical traits and each module were calculated. Gene information corresponding to the modules was used for further analysis.
2.5. Analysis of hub genes
Edges with upper 1000 wt from the corresponding module were introduced into Cytoscape, a free visualization software. The Cytoscape cytoHubba plugin was used to recognize the hub genes in the network. The top 30 hub genes in each module were detected through the maximal clique centrality (MCC) method.
2.6. Identification of genes associated with disease
The correlation between AF and relevant gene expression levels was evaluated by Spearman's correlation analysis. The Comparative Toxicogenomics Database (CTD) (http://ctdbase.org/) was used to identify comprehensive gene-disease interactions, predict possible associations, and generate a network of systems [17]. The relationships between relevant genes and diseases of the cardiovascular system were analyzed using the database.
2.7. Protein-protein interaction (PPI) network analysis of predicted target genes
We obtained sets of predicted target genes for the three transcription factors by intersecting the results of the TRANSFAC database analysis with AF-DEGs, followed by PPI analysis [18]. The PPI interaction network of these predicted target genes was constructed with a confidence level of 0.4 (medium confidence = 0.4) using the online Search Tool for the Retrieval of Interacting Genes (STRING) (https://string-db.org/) [[19], [20], [21]].
2.8. Human left atrial tissue samples and immunohistochemistry
Human left atrial appendage (LAA) samples were obtained from patients undergoing mitral valve surgery at the Shanghai Ruijin Hospital and divided into two groups: sinus rhythm (SR) and persistent atrial fibrillation (AF). There were no significant differences between groups in terms of age, gender, left ventricular ejection fraction (LVEF), and left atrial diameter. The study received approval by Institutional Review Board of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine (approval no. RJH201656). Written informed consents were obtained from all participants.
The expression level of ETS1, SP1, and WT1 in human LAA tissues were assessed through immunohistochemical staining. Briefly, after fixation in 4 % paraformaldehyde, samples were embedded in paraffin and sectioned consecutively (thickness: 4 μm). Sections were incubated overnight at 4 °C with anti-ETS1 antibody (CST: #14069S), anti-SP1 antibody (Abcam: #ab13370) and anti-WT1 antibody (Abcam: #ab89901) followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (CST: #3900S) at room temperature for 1 h. Images of all sections were captured using a Zeiss (Axio Scope. A1) microscope and quantified using Image J.
2.9. Culture of HL-1 cells
The HL-1 cells (mouse atrial myocytes) were obtained from the Cell Bank of the Chinese Academy of Sciences. The cells were cultured in DMEM containing 10 % fetal bovine serum (Gibco, USA), 0.1 mM norepinephrine, 2 mM L-glutamine, and 1 % penicillin/streptomycin solution at 37 °C in a 5 % CO2 environment. Prior to stimulation with angiotensin II (Ang II) at 1 μM (Sigma, USA), HL-1 cells were incubated in a serum-free medium overnight.
2.10. Animal experiments
All animal experiments were conducted in compliance with the National Institutes of Health Guidelines on the Care and Use of Laboratory Animals and approved by the Animal Ethics Committee of Shanghai Jiao Tong University School of Medicine. Male C57BL/6J mice aged 6–8 weeks were infused with either saline or Ang II (1 mg/kg/min; Merck Millipore: #05230101) with osmotic minipumps for 4 weeks before being euthanized. Mouse hearts were analyzed using real-time quantitative PCR (RT-qPCR) and western blotting.
2.11. RNA Isolation and RT-qPCR
RT-qPCR was conducted to verify the expression levels of ETS1, SP1, and WT1 in human LAA, HL-1, and mouse hearts. Total RNA extraction was performed using TRIzol Reagent (Thermo Fisher Scientific: #15596026CN), followed by reverse transcription (Takara: #RR036A). RT-qPCR was conducted using a TB Green RT-qPCR Kit (Takara: #RR820A) to measure mRNA levels of ETS1, SP1, and WT1, with GAPDH serving as the housekeeping gene. The primer sequences are presented in Table 1.
Table 1.
Information of PCR primers.
| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| GAPDH (human) | Forward | AAGGTGAAGGTCGGAGTCAAC |
| Reverse | CTTCCCGTTCTCAGCCATGTA | |
| ETS1 (human) | Forward | GATAGTTGTGATCGCCTCACC |
| Reverse | GTCCTCTGAGTCGAAGCTGTC | |
| SP1 (human) | Forward | AGTTCCAGACCGTTGATGGG |
| Reverse | GTTTGCACCTGGTATGATCTGT | |
| WT1 (human) | Forward | CACAGCACAGGGTACGAGAG |
| Reverse | CAAGAGTCGGGGCTACTCCA | |
| GAPDH (mouse) | Forward | AGGTCGGTGTGAACGGATTTG |
| Reverse | TGTAGACCATGTAGTTGAGGTCA | |
| ETS1 (mouse) | Forward | CGACTCTCACCATCATCAAGACA |
| Reverse | GAGAACTCTGAGGGAGGAACACA | |
| SP1 (mouse) | Forward | TGCAAACCAACAGATCATCCC |
| Reverse | TGACAGGTAGCAAGGTGATGT | |
| WT1 (mouse) | Forward | TCTTCCGAGGCATTCAGGATG |
| Reverse | TGCACACATGAAAGGACGTTT |
2.12. Western blot analysis
Human LAA, HL-1 cells, and mouse hearts were lysed using RIPA solution, and protein concentrations were quantified using the Bradford protein assay (Thermo Fisher Scientific: #23227). Protein samples underwent gel electrophoresis and were then transferred to polyvinylidene difluoride membranes (Millipore: #IPVH00010). The membranes were blocked in 5 % nonfat milk solution for 1 h, followed by overnight incubation at 4 °C with primary antibodies. After washing three times with PBS, the proteins were incubated with HRP-conjugated secondary antibodies. Protein bands were identified using an electrochemiluminescence system. The primary antibodies used were as follows: anti-GAPDH (Proteintech: #60004); anti-ETS1 (CST: #14069S); anti-SP1 (Proteintech: #21962); anti-WT1 (Abcam: #ab89901).
2.13. Molecular docking
The structure of Navitoclax was retrieved from the PubChem database. Corresponding protein structures for ETS1, SP1, and WT1 were obtained from the Protein Data Bank. Molecular docking of these key targets with Navitoclax was conducted using AutoDock Tools (version 1.5.6). The binding affinities between Navitoclax and ETS1, SP1, and WT1 were assessed through docking energy scores by using PyMOL software 2.3.0.
3. Results
3.1. DEGs and functional enrichment of common genes
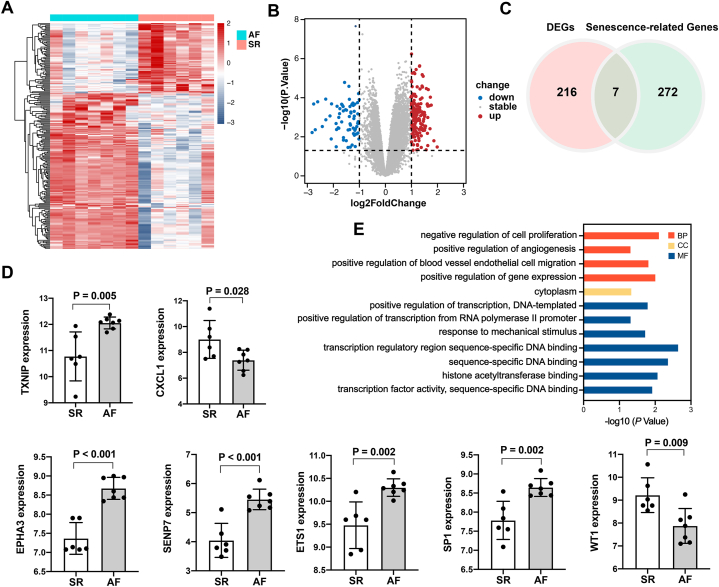
In this study, we identified 54,674 probes corresponding to 20,862 genes in the GSE79768 dataset. Comparing patients with SR to those with AF, we identified 222 DEGs in left atrial specimens from patients with AF, including 153 up-regulated and 69 down-regulated genes. DEGs are displayed as a heatmap (Fig. 2A) and volcano map (Fig. 2B). As shown in Fig. 2C, among the 222 DEGs, seven genes overlapped with 279 cellular senescence-related genes downloaded from CellAge (ETS1, EPHA3, SENP7, TXNIP, SP1, WT1, and CXCL1). Differential expression levels of the seven genes between SR and AF were shown in Fig. 2D. The DAVID database was used to confirm the GO term enrichment related to biological processes, cellular components, and molecular functions, and the seven common genes were associated with various processes, as indicated in Fig. 2E. The seven common genes were mainly involved in BP terms such as “negative regulation of cell proliferation,” “positive regulation of angiogenesis,” “positive regulation of blood vessel endothelial cell migration”, and “positive regulation of gene expression”.
Fig. 2.
DEGs and functional enrichment analysis of genes common to senescence-related genes. (A) Rows represent DEGs, and columns represent samples. DEGs that are upregulated or downregulated are represented by red and blue, respectively. (B) Upregulated DEGs are represented by red plot points, while downregulated DEGs are represented by blue plot points. (C) A venn diagram illustrating 7 overlapping genes in DEGs and senescence-related genes. (D) Differential expression levels of 7 common genes between SR and AF. (E) Analysis of 7 common genes using Gene Ontology (GO). Biological processes (BP) are represented by the red bars, cellular components (CC) by the yellow bars, and molecular functions (MF) by the blue bars. DEG, differentially expressed gene; AF, atrial fibrillation; SR, sinus rhythm.
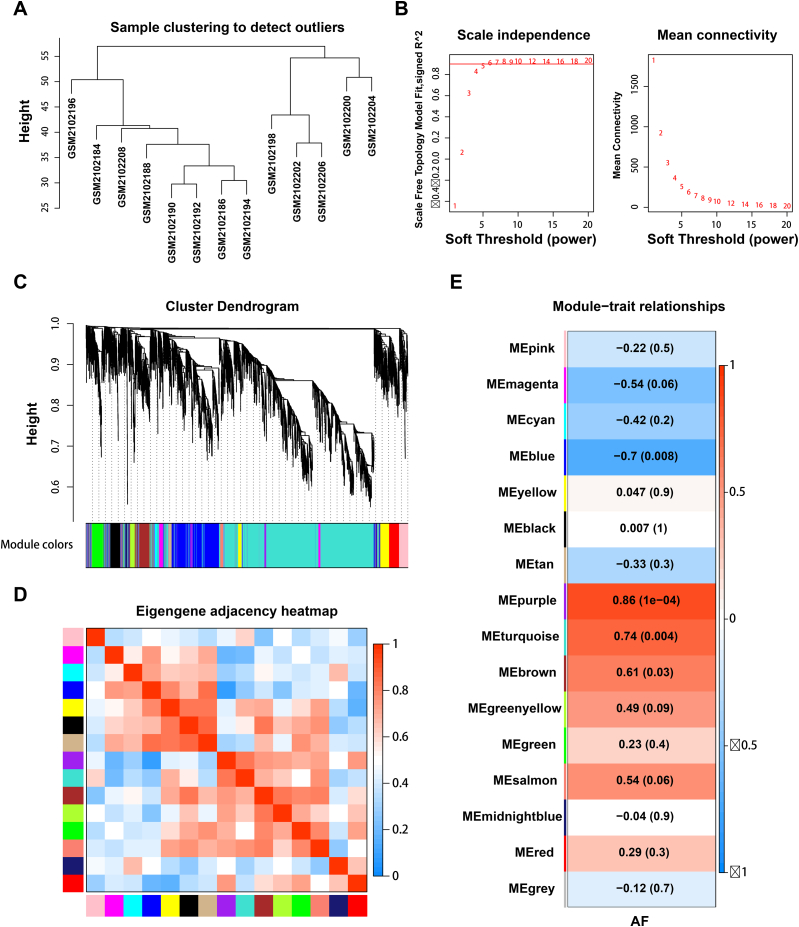
3.2. Identification of gene co-expression networks and modules
The top 25 % variance (5216) genes in 13 left atrial appendage samples from seven patients with AF and six patients with SR were selected for analysis. The sample cluster analysis results are depicted in Fig. 3A. Before constructing the weighted co-expression network, we carefully selected a suitable soft-thresholding power value to ensure a balance between scale independence and the mean connectivity [16]. Utilizing the pickSoftThreshold function in our analysis, we determined that a power value of 8 yielded a scale-free topology fit index exceeding 0.90 and a higher mean connectivity, indicating a robust network structure (Fig. 3B). Fig. 3C shows 16 modules derived from genes with comparable co-expression characteristics. We also analyzed the interaction associations between these modules. The eigengene adjacency heatmap showed that the 16 modules could be divided into several smaller groups, suggesting the existence of co-expression clusters with distinct functions in the genetic network of AF (Fig. 3D). Furthermore, module‐trait association analysis showed that the purple and turquoise modules exhibited a high positive correlation (r = 0.86, P < 0.001; r = 0.74, P = 0.004), while the blue module showed the highest negative correlation (r = −0.70, P = 0.008) (Fig. 3E). Subsequently, these three modules were selected as critical modules for AF and were included in further analyses.
Fig. 3.
Weighted gene co-expression network analysis. (A) Sample dendrogram. (B) Process for selecting soft thresholds. (C) Cluster dendrogram. Co-expression modules are represented by specific colors. (D) Different gene co-expression modules adjacency heatmap. (E) Heatmap showing the correlation between AF and module eigengenes.
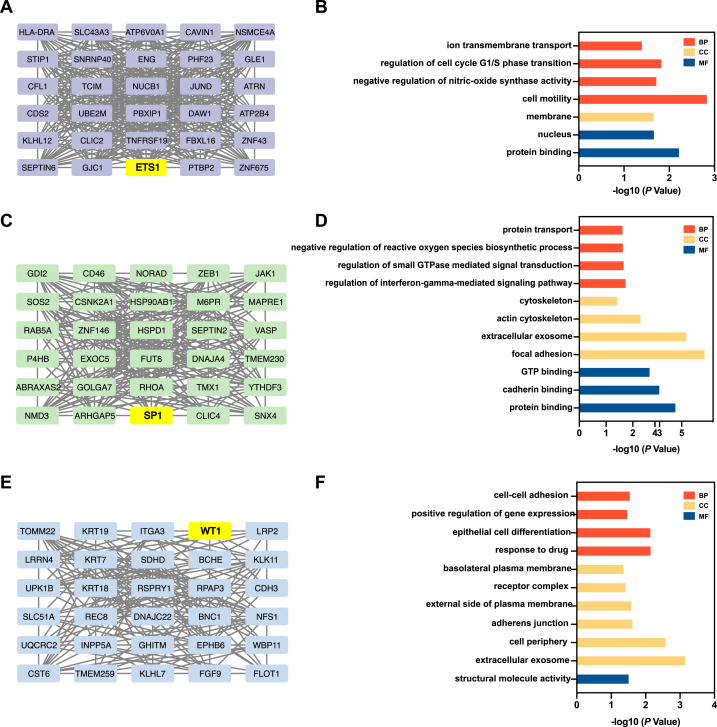
3.3. Hub genes selection and enrichment analysis
The top 30 hub genes in the purple, turquoise, and blue modules were determined using cytoHubba via the MCC method, as shown in Fig. 4. Meanwhile, corresponding functional enrichment analyses were conducted. In the purple module, which exhibited the strongest correlation, these 30 hub genes were enriched in the BP terms “ion transmembrane transport”, “regulation of cell cycle G1/S phase transition”, “negative regulation of nitric-oxide synthase activity”, and “cell motility”.
Fig. 4.
The selection and enrichment analysis of hub genes. The top 30 hub genes in purple module (A), turquoise module (C) and blue module (E). Gene Ontology (GO) analysis of hub genes in purple module (B), turquoise module (D) and blue module (F). Biological processes (BP) are represented by the red bars, cellular components (CC) by the yellow bars, and molecular functions (MF) by the blue bars.
3.4. Correlation between key genes and AF or other cardiovascular diseases
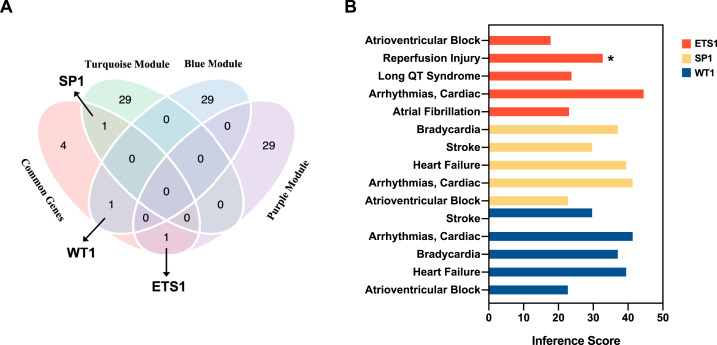
Among the hub genes identified in the three modules, three genes—ETS1, SP1, and WT1—overlapped with the seven DEGs mentioned above and shared senescence-related genes, as indicated in the Venn diagram (Fig. 5A). Notably, the Spearman correlation analyses revealed significant correlations between ETS1 (ρ = 0.866, P < 0.001), SP1 (ρ = 0.784, P = 0.002), and WT1 (ρ = −0.701, P = 0.008) and AF. We then validated the three senescence-related genes with external GEO datasets GSE41177. The data of left atrial appendage (LAA) from 16 AF patients and 3 controls in GSE41177 dataset was selected for validation (Fig. S1). Furthermore, the CTD database showed that ETS1, SP1, and WT1 target several other cardiovascular diseases (Fig. 5B).
Fig. 5.
Identification of ETS1, SP1 and WT1 and correlation with cardiovascular diseases. (A) Venn diagram showing 3 overlapping genes (ETS1, SP1, WT1) in DEGs and hub genes of three modules. (B) Relationship to cardiovascular diseases related to ETS1, SP1 and WT1 based on the CTD database. ETS1 are represented by the red bars, SP1 by the yellow bars, and WT1 by the blue bars. * Evidence of a marker in the disease.
3.5. PPI network of predicted target genes
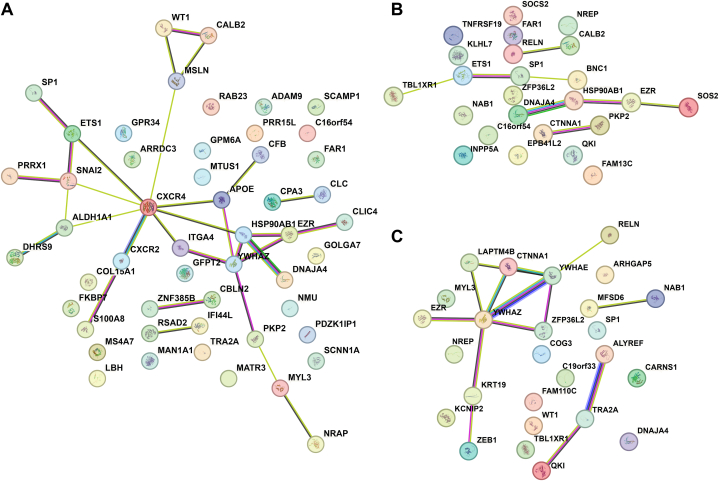
By intersecting TRANSFAC-predicted target genes and DEGs, we obtained a total of 52, 24, and 26 predicted target genes of ETS1, SP1, and WT1, respectively, involved in AF. A PPI network of three sets of predicted target genes was constructed (Fig. 6). The PPI enrichment p-values were 0.005, 0.029, and 0.001, respectively, indicating that the proteins were partially biologically connected.
Fig. 6.
Protein–protein interaction network analysis of predicted target genes. Protein–protein interaction network analysis of predicted target genes of ETS1 (A), SP1 (B) and WT1 (C).
3.6. Validation of ETS1, SP1, and WT1 in human LAA and in vitro and in vivo experiments
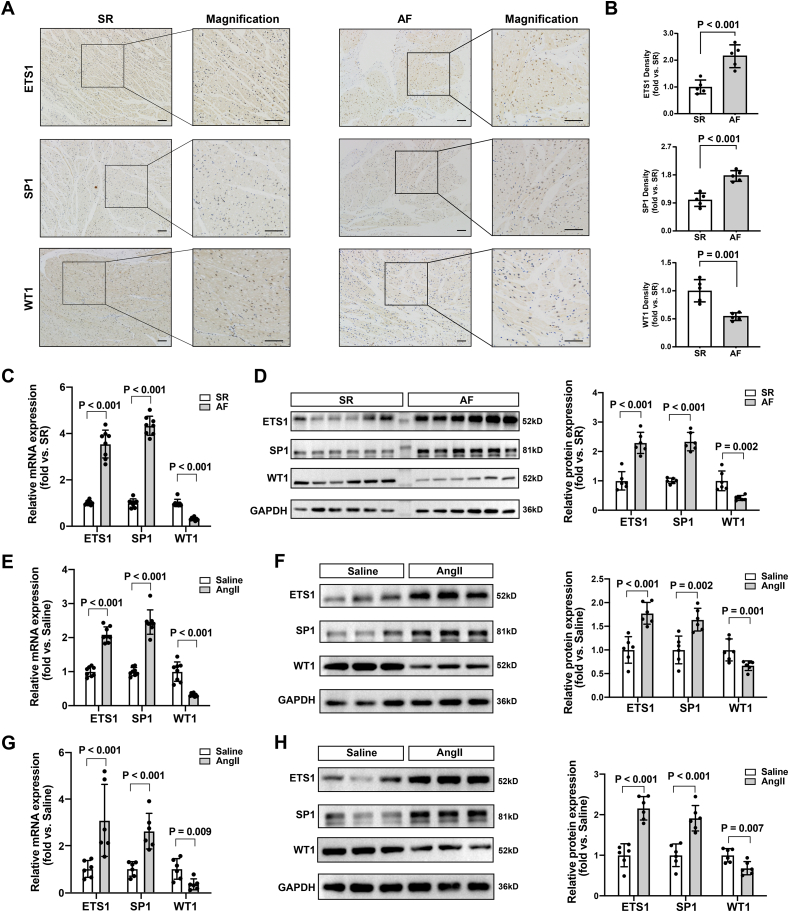
To further validate the significance of ETS1, SP1, and WT1 in AF, we performed immunohistochemical staining of LAA tissue which revealed significantly higher levels of ETS1 and SP1 in AF group compared to the SR group and lower levels of WT1 in the AF group than in the SR group (Fig. 7A and B). These differential expression results were further validated in human LAA using RT-qPCR and western blotting (Fig. 7C and D). Ang II stimulation significantly upregulated the expression levels of ETS1 and SP1 and downregulated the expression level of WT1 in HL-1 cells (Fig. 7E and F). Further in vivo experiments confirmed changes in the expression levels of ETS1, SP1, and WT1 in the LAA of mice administered Ang II (Fig. 7G and H).
Fig. 7.
Validation of ETS1, SP1, and WT1 in human LAA and in vitro and in vivo experiments. Representative immunohistochemical staining images of ETS1, SP1and WT1 (A) and quantification (B). Data are expressed as mean ± SD (n = 5 per group). scale bar = 100 μm. (C). Relative mRNA expression levels of ETS1, SP1 and WT1 in human LAA. Data are expressed as mean ± SD (n = 8 per group). (D). Western blot analyses of ETS1, SP1 and WT1 in human LAA and statistical plot. Data are expressed as mean ± SD (n = 6 per group) (E). Relative mRNA expression levels of ETS1, SP1 and WT1 in HL-1 cells treated with saline or Ang II. Data are expressed as mean ± SD (n = 8 per group). (F). Western blot analyses of ETS1, SP1 and WT1 in HL-1 cells treated with saline or Ang II and statistical plot. Data are expressed as mean ± SD (n = 6 per group). (G). Relative mRNA expression levels of ETS1, SP1 and WT1 in LAA of mice administered with saline or Ang II for 4 weeks. Data are expressed as mean ± SD (n = 6 per group). (H). Western blot analyses of ETS1, SP1 and WT1 in LAA of mice administered with saline or Ang II for 4 weeks. Data are expressed as mean ± SD (n = 6 per group).
3.7. Molecular docking
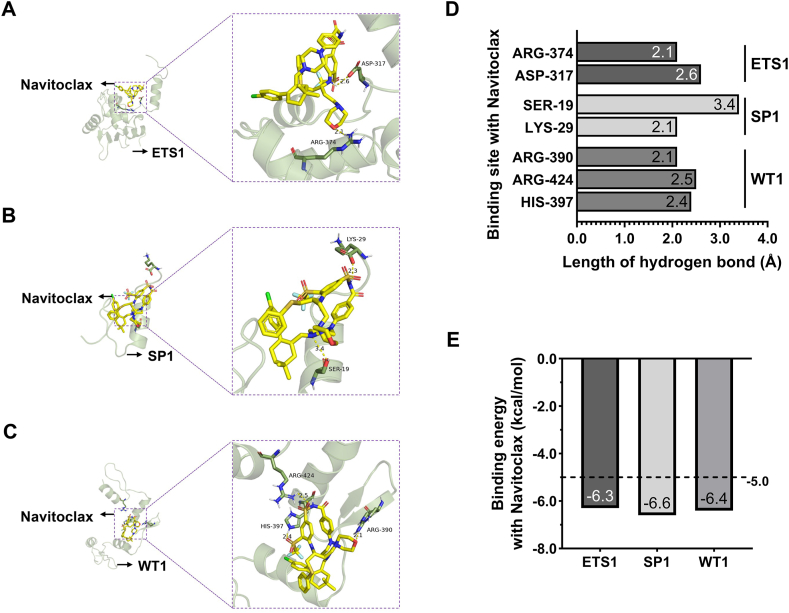
To investigate the potential interactions between Navitoclax and the identified potential AF biomarkers: ETS1, SP1, and WT1, molecular docking analysis was conducted. The results revealed affinity values of −6.3, −6.6, and −6.4 for the interactions of Navitoclax with ETS1, SP1, and WT1, respectively. Generally, binding energies below −5.0 indicates a favorable binding potential. Therefore, it is suggested that the senolytic agent Navitoclax likely interacts with ETS1, SP1, and WT1 (Fig. 8).
Fig. 8.
Molecular docking results of Navitoclax interaction with ETS1, SP1 and WT1. Molecular docking conformation of Navitoclax interaction with ETS1 (A), SP1 (B) and WT1 (C). (D). The hydrogen bonding sites and lengths formed by the interaction between Navitoclax and ETS1, SP1, WT1. (E). Navitoclax's affinity values for ETS1, SP1, and WT1.
4. Discussion
In the present study, we investigated senescence-related genes those could be potential therapeutic biomarkers of AF using bioinformatics, human atrial tissue, and then conducted molecular docking analysis to investigate the interaction between these biomarkers and Navitoclax, a senolytic agent. Overall, 153 upregulated and 69 downregulated genes were found in the dataset of patients with AF, of which seven genes overlapped with senescence-related genes: ETS1, EPHA3, SENP7, TXNIP, SP1, WT1, and CXCL1. Subsequently, we further screened AF-related modules using WGCNA. As a comprehensive network module-based analysis method, WGCNA could uncover new targets for functional studies and offer deeper insights into the pathogenesis of AF [22,23]. Hub genes of the three modules that were highly correlated with AF were identified. By intersecting the hub genes with the above seven common genes, we found that ETS1, SP1, and WT1 were key senescence-related transcription factors in AF. Furthermore, their association with AF and other cardiovascular diseases was verified using Spearman correlation and the CTD database. The PPI network of their predicted target genes was constructed. Importantly, immunohistochemical staining, RT-qPCR, and Western blot were performed on LAA tissues in the SR and AF groups, confirming the differential expression of three aging-related transcription factors between the two groups: ETS1, SP1, and WT1. This indicates their potential as biomarkers in AF. Molecular docking analysis revealed favorable binding potential of the senolytic agent Navitoclax for ETS1, SP1, and WT1, indicating that these factors may be crucial targets for understanding and treating AF in relation to senescence. To the best of our knowledge, this is the first study investigating senescence-related genes in patients with AF and identifying potential biomarkers and targets of senolytic agents. These findings enhance our understanding of AF pathogenesis through cellular senescence and offer potential therapeutic insights.
AF is the most common sustained arrhythmia worldwide. Some people with AF are asymptomatic or have unnoticed mild symptoms. However, transient episodes of AF, if frequent enough, can be devastating. Therefore, it is imperative to understand the mechanisms of AF and make a prompt diagnosis. Despite current multifaceted treatments for AF, such as rate control, cardioversion, and catheter ablation, none offer a complete cure, and some are associated with side effects or recurrence [24,25]. A deeper understanding of the molecular mechanisms underlying AF will provide opportunities for the development of more effective targeted therapies. The incidence and prevalence of AF increase with age, highlighting a strong association between AF and aging currently under intense investigation. However, the molecular mechanisms involved still need to be elucidated. Cellular senescence is a characteristic of aging at the cellular level. Many studies have examined the key genes and modules related to AF through using bioinformatics. However, the correlation between senescence and AF has not been fully investigated to date.
Among the seven DEGs we initially identified, EPHA3 was reported to play a crucial role in heart development. SENP7, a member of the SENPs family, is a key enzyme regulating SUMOylation, a transient post-translational modification, in cardiovascular disease [26,27]. TXNIP has been identified as a regulator of the myocardial metabolic switch, and its deletion prevents cardiac dysfunction in response to pressure overload. Additionally, the TXNIP/NLRP3 inflammasome signaling pathway contributes to atrial fibrosis in AF [28]. Meanwhile, CXCL1, a pro-inflammatory factor, promotes cardiac remodeling and fibrosis, with patients with AF showing higher circulating blood CXCL1 levels than those with SR [29]. Numerous bioinformatics studies have also identified CXCL1 as a key gene in the development of AF, which is consistent with our findings [30,31]. Intriguingly, in the GO biological process analysis of these seven common genes, “negative regulation of cell proliferation” corresponds with cellular senescence, whereas “positive regulation of blood vessel endothelial cell migration” and “positive regulation of angiogenesis” were related to AF, as a previous study revealed that endothelial cell activation concurs with extracellular matrix remodeling in AF [32]. Using WGCNA, we identified 16 gene modules, with the purple module exhibiting the highest correlation with AF. Following functional enrichment analysis of the key genes from the purple module, we found that, in addition to “ion transmembrane transport,” which underlies the pathophysiology of AF, GO terms were also involved in the regulation of the cell cycle G1/S transition, which is linked to senescence characteristics. This finding further confirmed the strong connection between AF and senescence. To validate our findings, we intersected the hub genes from significant modules with the seven above-mentioned genes, identifying three key senescence-related genes in AF.
ETS1, a member of the E26 transformation-specific transcription factor family, induces the transcription of downstream target genes, many of which are involved in cell proliferation, invasion, and angiogenesis [33]. Abundant evidence has shown that ETS1 plays a critical role in almost all carcinomas by regulating the key cancer-related events [34]. ETS1 also mediates tissue remodeling by regulating the expression of genes encoding the extracellular matrix (ECM) and the enzymes involved in matrix degradation. It regulates Ang II-induced fibroblast activation and renal fibrosis and is involved in airway remodeling by upregulating TNF-α-induced expression of matrix metalloproteinase 9 (MMP9). ETS1 upregulation mediates Ang II -related cardiac fibrosis [35]. Taken together, these studies underscore the role of ETS1 as a mediator in the activation of profibrotic and pro-inflammatory pathways. Our study identified ETS1 as a key senescence-related gene in AF, consistent with the knowledge that extracellular matrix proteins, including MMPs and angiotensin II, have a role in senescence-induced AF [36].
SP1 is a member of a transcription factor family and plays a central role in cell cycle regulation and development. It functions as a well-characterized transcriptional activator that binds to the GC-rich sequences that are required for the expression and regulation of various genes. Initially recognized as a constitutive transcriptional activator for housekeeping and other TATA-free genes, SP1 has since been found to regulate genes involved in apoptosis and senescence [37]. In human diploid fibroblasts, SP1 is essential for inducing CDNK2A expression, a key driver of cellular senescence [38]. SP1 promotes ECM gene expression and activates the TGF-β1/Smad signaling pathway, which is considered a potential mechanism of senescence-induced AF [39,40]. Consistent with this, our identification of SP1 confirmed its significant role in senescence and AF.
WT1, which acts as a transcriptional regulator, influences cell differentiation, growth, apoptosis, and metabolism [41]. It is essential for the development of organs and tissues, including the heart. WT1 is recognized as pivotal in cardiovascular development and is involved in crucial processes such as coronary vessel formation and cardiac autonomic nervous system regulation [42]. Furthermore, WT1 has been implicated in cardiac disease and is believed to play a role in cardiac repair and regeneration, owing to its strong re-expression in the epicardium, endothelium, and cardiomyocytes after myocardial infarction. Vicent et al. also identified WT1 as a critical regulator of senescence and proliferation [43]. Our findings revealed that WT1 is a key gene concurrently expressed in both senescence and AF, indicating that it could be a therapeutic target for this disease.
Clearing senescent cells through pharmacological and genetic engineering strategies may ameliorate senescence-associated conditions. Senolytic compounds have proven effective in eliminating senescent cells and delaying senescence-related diseases [44]. In a previous study conducted by our team, Navitoclax, a small-molecule compound known for its senolytic activity, was found to reduces susceptibility to ventricular tachyarrhythmias induced by programmed electrical stimulation [45]. Many believe that senolytic drugs possess great potential, as senescence plays a role in the pathogenesis of AF [36]. The molecular docking results of this study showed that Navitoclax may interact with the three identified transcription factors: ETS1, SP1 and WT1. We speculate that Navitoclax may affect the activity or transcriptional regulatory function of these three proteins through binding, thereby exerting its senolytic and protective effects in AF, that is, ETS1, SP1 and WT1 could be potential targets for Navitoclax. In addition, studies have reported that the sensitivity of patients with acute myeloid leukemia to Navitoclax is related to genetic aberrations of WT1, and that Navitoclax can specifically eliminate senescent cells mediated by the WT1-ATF2-TGFb2 axis, which also partly support our speculation [46,47]. The identification of senescence-related biomarkers offers fresh insights into understanding AF pathophysiology, suggesting that targeting these genes with specific senolytic compounds that eliminate senescent cells could be a potential treatment approach for AF.
4.1. Limitations
This study inevitably has some limitations. First, our analysis relied on a single dataset due to the limited availability of sequencing data of the left atria of paired patients with AF and SR. Therefore, future studies should consider large-scale samples from multiple sources. Second, comprehensive in vivo and in vitro assays are necessary to validate the mechanisms of the identified senescence-related genes in the development of AF. Third, further validation of the interactions between Navitoclax and these three proteins, as well as their functional implications in AF, requires more in-depth basic research in the future.
5. Conclusions
ETS1, SP1, and WT1 were identified as potential senescence-related biomarkers in AF. using bioinformatics. Validation in human tissues and molecular docking analysis further confirmed that these genes—ETS1, SP1, and WT1—could serve as therapeutic biomarkers associated with AF due to their strong binding affinity with the senolytic drug Navitoclax.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81900290, 81870250).
Ethics approval and consent to participate
This research and involved experimental procedures were received approval by Institutional Review Board of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine (approval no. RJH201656). Written informed consents were obtained from all participants.
Consent for publication
All authors agree to publish.
Data availability statement
The datasets analyzed during the current study are available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/). These accession numbers for the datasets are GSE79768 and GSE41177. The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
CRediT authorship contribution statement
Jingmeng Liu: Writing – original draft, Visualization, Validation, Software, Resources, Investigation, Formal analysis, Conceptualization. Taojie Zhou: Writing – review & editing, Validation, Software, Resources, Investigation, Data curation, Conceptualization. Yangyang Bao: Methodology, Data curation. Changjian Lin: Methodology. Qiujing Chen: Methodology. Yang Dai: Methodology. Ning Zhang: Methodology. Wenqi Pan: Methodology, Investigation. Qi Jin: Validation, Supervision. Lin Lu: Supervision. Qiang Zhao: Supervision. Tianyou Ling: Writing – review & editing, Supervision, Project administration. Liqun Wu: Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors have no competing interests to declare.
Acknowledgements
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e37366.
Contributor Information
Tianyou Ling, Email: lty11498@rjh.com.cn.
Liqun Wu, Email: wlq10583@rjh.com.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lippi G., Sanchis-Gomar F., Cervellin G. Global epidemiology of atrial fibrillation: an increasing epidemic and public health challenge. Int. J. Stroke. 2021;16(2):217–221. doi: 10.1177/1747493019897870. [DOI] [PubMed] [Google Scholar]
- 2.Zou R., Zhang D., Lv L., Shi W., Song Z., Yi B., Lai B., Chen Q., Yang S., Hua P. Bioinformatic gene analysis for potential biomarkers and therapeutic targets of atrial fibrillation-related stroke. J. Transl. Med. 2019;17(1):45. doi: 10.1186/s12967-019-1790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chugh S.S., Havmoeller R., Narayanan K., Singh D., Rienstra M., Benjamin E.J., Gillum R.F., Kim Y.H., McAnulty J.H., Jr., Zheng Z.J., Forouzanfar M.H., Naghavi M., Mensah G.A., Ezzati M., Murray C.J. Worldwide epidemiology of atrial fibrillation: a Global Burden of disease 2010 study. Circulation. 2014;129(8):837–847. doi: 10.1161/CIRCULATIONAHA.113.005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirchhof P., Benussi S., Kotecha D., Ahlsson A., Atar D., Casadei B., Castella M., Diener H.C., Heidbuchel H., Hendriks J., Hindricks G., Manolis A.S., Oldgren J., Popescu B.A., Schotten U., Van Putte B., Vardas P., Group E.S.C.S.D. 2016 ESC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Eur. Heart J. 2016;37(38):2893–2962. doi: 10.1093/eurheartj/ehw210. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Cai W., Gu L., Ji X., Shen Q. Comprehensive analysis of pertinent genes and pathways in atrial fibrillation. Comput. Math. Methods Med. 2021;2021 doi: 10.1155/2021/4530180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McHugh D., Gil J. Senescence and aging: causes, consequences, and therapeutic avenues. J. Cell Biol. 2018;217(1):65–77. doi: 10.1083/jcb.201708092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin P.H., Lee S.H., Su C.P., Wei Y.H. Oxidative damage to mitochondrial DNA in atrial muscle of patients with atrial fibrillation. Free Radic. Biol. Med. 2003;35(10):1310–1318. doi: 10.1016/j.freeradbiomed.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Dookun E., Walaszczyk A., Redgrave R., Palmowski P., Tual-Chalot S., Suwana A., Chapman J., Jirkovsky E., Donastorg Sosa L., Gill E., Yausep O.E., Santin Y., Mialet-Perez J., Andrew Owens W., Grieve D., Spyridopoulos I., Taggart M., Arthur H.M., Passos J.F., Richardson G.D. Clearance of senescent cells during cardiac ischemia-reperfusion injury improves recovery. Aging Cell. 2020;19(10) doi: 10.1111/acel.13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai F.C., Lin Y.C., Chang S.H., Chang G.J., Hsu Y.J., Lin Y.M., Lee Y.S., Wang C.L., Yeh Y.H. Differential left-to-right atria gene expression ratio in human sinus rhythm and atrial fibrillation: implications for arrhythmogenesis and thrombogenesis. Int. J. Cardiol. 2016;222:104–112. doi: 10.1016/j.ijcard.2016.07.103. [DOI] [PubMed] [Google Scholar]
- 11.Avelar R.A., Ortega J.G., Tacutu R., Tyler E.J., Bennett D., Binetti P., Budovsky A., Chatsirisupachai K., Johnson E., Murray A., Shields S., Tejada-Martinez D., Thornton D., Fraifeld V.E., Bishop C.L., de Magalhaes J.P. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 2020;21(1):91. doi: 10.1186/s13059-020-01990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Song Z., Wu R., Kong X., Zhang H., Li S., Gong X., Gong S., Cheng J., Yuan F., Wu H., Wang S., Yuan Z. PRRC2B modulates oligodendrocyte progenitor cell development and myelination by stabilizing Sox2 mRNA. Cell Rep. 2024;43(3) doi: 10.1016/j.celrep.2024.113930. [DOI] [PubMed] [Google Scholar]
- 13.Zhang M., Xu G., Xi C., Yu E. Identification of immune-related tumor antigens and immune subtypes in osteosarcoma. Heliyon. 2024;10(11) doi: 10.1016/j.heliyon.2024.e32231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., King B.L., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J. The comparative Toxicogenomics database: update 2017. Nucleic Acids Res. 2017;45(D1):D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28(1):316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subbarayan K., Ulagappan K., Wickenhauser C., Bachmann M., Seliger B. Immune interaction map of human SARS-CoV-2 target genes: implications for therapeutic avenues. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.597399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H., Yu M., Li Z., Zhang E., Ma H. Identification and analysis of mitochondria-related key genes of heart failure. J. Transl. Med. 2022;20(1):410. doi: 10.1186/s12967-022-03605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan N., Chung M.K., Smith J.D., Hsu J., Serre D., Newton D.W., Castel L., Soltesz E., Pettersson G., Gillinov A.M., Van Wagoner D.R., Barnard J. Weighted gene coexpression network analysis of human left atrial tissue identifies gene modules associated with atrial fibrillation. Circ Cardiovasc Genet. 2013;6(4):362–371. doi: 10.1161/CIRCGENETICS.113.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wass S.Y., Offerman E.J., Sun H., Hsu J., Rennison J.H., Cantlay C.C., McHale M.L., Gillinov A.M., Moravec C., Smith J.D., Van Wagoner D.R., Barnard J., Chung M.K. Novel functional atrial fibrillation risk genes and pathways identified from coexpression analyses in human left atria. Heart Rhythm. 2023;20(9):1219–1226. doi: 10.1016/j.hrthm.2023.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchhof P., Benussi S., Kotecha D., Ahlsson A., Atar D., Casadei B., Castella M., Diener H.C., Heidbuchel H., Hendriks J., Hindricks G., Manolis A.S., Oldgren J., Popescu B.A., Schotten U., Van Putte B., Vardas P., Agewall S., Camm J., Baron Esquivias G., Budts W., Carerj S., Casselman F., Coca A., De Caterina R., Deftereos S., Dobrev D., Ferro J.M., Filippatos G., Fitzsimons D., Gorenek B., Guenoun M., Hohnloser S.H., Kolh P., Lip G.Y., Manolis A., McMurray J., Ponikowski P., Rosenhek R., Ruschitzka F., Savelieva I., Sharma S., Suwalski P., Tamargo J.L., Taylor C.J., Van Gelder I.C., Voors A.A., Windecker S., Zamorano J.L., Zeppenfeld K. 2016 ESC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Europace. 2016;18(11):1609–1678. doi: 10.1093/europace/euw295. [DOI] [PubMed] [Google Scholar]
- 25.Rienstra M., Lubitz S.A., Mahida S., Magnani J.W., Fontes J.D., Sinner M.F., Van Gelder I.C., Ellinor P.T., Benjamin E.J. Symptoms and functional status of patients with atrial fibrillation: state of the art and future research opportunities. Circulation. 2012;125(23):2933–2943. doi: 10.1161/CIRCULATIONAHA.111.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephen L.J., Fawkes A.L., Verhoeve A., Lemke G., Brown A. A critical role for the EphA3 receptor tyrosine kinase in heart development. Dev. Biol. 2007;302(1):66–79. doi: 10.1016/j.ydbio.2006.08.058. [DOI] [PubMed] [Google Scholar]
- 27.Hotz P.W., Muller S., Mendler L. SUMO-Specific Isopeptidases Tuning cardiac SUMOylation in health and disease. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.786136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu H., Liu W., Lan T., Pan W., Chen X., Wu H., Xu D. Salvianolate reduces atrial fibrillation through suppressing atrial interstitial fibrosis by inhibiting TGF-beta1/Smad2/3 and TXNIP/NLRP3 inflammasome signaling pathways in post-MI rats. Phytomedicine. 2018;51:255–265. doi: 10.1016/j.phymed.2018.09.238. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y.L., Cao H.J., Han X., Teng F., Chen C., Yang J., Yan X., Li P.B., Liu Y., Xia Y.L., Guo S.B., Li H.H. Chemokine receptor CXCR-2 Initiates atrial fibrillation by Triggering Monocyte Mobilization in mice. Hypertension. 2020;76(2):381–392. doi: 10.1161/HYPERTENSIONAHA.120.14698. [DOI] [PubMed] [Google Scholar]
- 30.Qu Q., Sun J.Y., Zhang Z.Y., Su Y., Li S.S., Li F., Wang R.X. Hub microRNAs and genes in the development of atrial fibrillation identified by weighted gene co-expression network analysis. BMC Med. Genom. 2021;14(1):271. doi: 10.1186/s12920-021-01124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan G., Wei J. Identification of potential novel biomarkers and therapeutic targets involved in human atrial fibrillation based on bioinformatics analysis. Kardiol. Pol. 2020;78(7–8):694–702. doi: 10.33963/KP.15339. [DOI] [PubMed] [Google Scholar]
- 32.van den Berg N.W.E., Kawasaki M., Fabrizi B., Nariswari F.A., Verduijn A.C., Neefs J., Wesselink R., Al-Shama R.F.M., van der Wal A.C., de Boer O.J., Aten J., Driessen A.H.G., Jongejan A., de Groot J.R. Epicardial and endothelial cell activation concurs with extracellular matrix remodeling in atrial fibrillation. Clin. Transl. Med. 2021;11(11):e558. doi: 10.1002/ctm2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westermarck J., Seth A., Kahari V.M. Differential regulation of interstitial collagenase (MMP-1) gene expression by ETS transcription factors. Oncogene. 1997;14(22):2651–2660. doi: 10.1038/sj.onc.1201111. [DOI] [PubMed] [Google Scholar]
- 34.Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin. Cancer Biol. 2015;35:20–38. doi: 10.1016/j.semcancer.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Hao G., Han Z., Meng Z., Wei J., Gao D., Zhang H., Wang N. Ets-1 upregulation mediates angiotensin II-related cardiac fibrosis. Int. J. Clin. Exp. Pathol. 2015;8(9):10216–10227. [PMC free article] [PubMed] [Google Scholar]
- 36.Guo G., Watterson S., Zhang S.D., Bjourson A., McGilligan V., Peace A., Rai T.S. The role of senescence in the pathogenesis of atrial fibrillation: a target process for health improvement and drug development. Ageing Res. Rev. 2021;69 doi: 10.1016/j.arr.2021.101363. [DOI] [PubMed] [Google Scholar]
- 37.Torabi B., Flashner S., Beishline K., Sowash A., Donovan K., Bassett G., Azizkhan-Clifford J. Caspase cleavage of transcription factor Sp1 enhances apoptosis. Apoptosis. 2018;23(1):65–78. doi: 10.1007/s10495-017-1437-4. [DOI] [PubMed] [Google Scholar]
- 38.Wu J., Xue L., Weng M., Sun Y., Zhang Z., Wang W., Tong T. Sp1 is essential for p16 expression in human diploid fibroblasts during senescence. PLoS One. 2007;2(1):e164. doi: 10.1371/journal.pone.0000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verrecchia F., Rossert J., Mauviel A. Blocking sp1 transcription factor broadly inhibits extracellular matrix gene expression in vitro and in vivo: implications for the treatment of tissue fibrosis. J. Invest. Dermatol. 2001;116(5):755–763. doi: 10.1046/j.1523-1747.2001.01326.x. [DOI] [PubMed] [Google Scholar]
- 40.Cao F., Li Z., Ding W.M., Yan L., Zhao Q.Y. LncRNA PVT1 regulates atrial fibrosis via miR-128-3p-SP1-TGF-beta1-Smad axis in atrial fibrillation. Mol. Med. 2019;25(1):7. doi: 10.1186/s10020-019-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugiyama H. Wilms' tumor gene WT1: its oncogenic function and clinical application. Int. J. Hematol. 2001;73(2):177–187. doi: 10.1007/BF02981935. [DOI] [PubMed] [Google Scholar]
- 42.Wagner N., Wagner K.D. Every Beat You Take-the Wilms' tumor Suppressor WT1 and the heart. Int. J. Mol. Sci. 2021;22(14) doi: 10.3390/ijms22147675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vicent S., Chen R., Sayles L.C., Lin C., Walker R.G., Gillespie A.K., Subramanian A., Hinkle G., Yang X., Saif S., Root D.E., Huff V., Hahn W.C., Sweet-Cordero E.A. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J. Clin. Invest. 2010;120(11):3940–3952. doi: 10.1172/JCI44165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang J., Wang Y., Shao L., Laberge R.M., Demaria M., Campisi J., Janakiraman K., Sharpless N.E., Ding S., Feng W., Luo Y., Wang X., Aykin-Burns N., Krager K., Ponnappan U., Hauer-Jensen M., Meng A., Zhou D. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22(1):78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia K., Dai Y., Liu A., Li X., Wu L., Lu L., Bao Y., Jin Q. Senolytic agent Navitoclax inhibits angiotensin II-induced heart failure in mice. J. Cardiovasc. Pharmacol. 2020;76(4):452–460. doi: 10.1097/FJC.0000000000000878. [DOI] [PubMed] [Google Scholar]
- 46.Kontro M., Kumar A., Majumder M.M., Eldfors S., Parsons A., Pemovska T., Saarela J., Yadav B., Malani D., Floisand Y., Hoglund M., Remes K., Gjertsen B.T., Kallioniemi O., Wennerberg K., Heckman C.A., Porkka K. HOX gene expression predicts response to BCL-2 inhibition in acute myeloid leukemia. Leukemia. 2017;31(2):301–309. doi: 10.1038/leu.2016.222. [DOI] [PubMed] [Google Scholar]
- 47.Zheng H., Liu M., Shi S., Huang H., Yang X., Luo Z., Song Y., Xu Q., Li T., Xue L., Lu F., Wang J. MAP4K4 and WT1 mediate SOX6-induced cellular senescence by synergistically activating the ATF2-TGFbeta2-Smad2/3 signaling pathway in cervical cancer. Mol. Oncol. 2024;18(5):1327–1346. doi: 10.1002/1878-0261.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/). These accession numbers for the datasets are GSE79768 and GSE41177. The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.