Abstract
Hepatocellular Carcinoma (HCC) is a serious primary solid tumor that is prevalent worldwide. Due to its high mortality rate, it is crucial to explore both early diagnosis and advanced treatment for HCC. In recent years, multi-omics approaches have emerged as promising tools to identify biomarkers and investigate molecular mechanisms of biological processes and diseases. In this study, we performed proteomics, phosphoproteomics, metabolomics, and lipidomics to reveal the molecular features of early- and advanced-stage HCC. The data obtained from these omics were analyzed separately and then integrated to provide a comprehensive understanding of the disease. The multi-omics results unveiled intricate biological pathways and interaction networks underlying the initiation and progression of HCC. Moreover, we proposed specific potential biomarker panels for both early- and advanced-stage HCC by overlapping our data with CPTAC database for HCC diagnosis, and deduced novel insights and mechanisms related to HCC origination and development, such as glucose depletion during tumor progression, ROCK1 deactivation and GSK3A activation.
Keywords: Hepatocellular carcinoma, Proteomics and phosphoproteomics, Metabolomics and lipidomics, Multi-omics analysis and integration
Highlights
-
•
Multi-omics approaches were used to characterize the molecular features of early- and advanced-stage HCC.
-
•
The integrated and comprehensive analysis of multi-omics data were performed.
-
•
The biomarker panels for both early- and advanced-stage HCC were proposed for cancer diagnosis and therapeutic intervention.
1. Introduction
Hepatocellular Carcinoma (HCC) is the most common form of liver cancer and originates in hepatocytes. It is often associated with various liver diseases, such as cirrhosis or chronic hepatitis B or C infection [[1], [2], [3]]. HCC is now one of the most lethal cancer-induced diseases worldwide [4,5], and patients in developing countries have an average survival time of only a few months after clinical diagnosis via traditional medical imaging strategies, although there has been significant improvement in HCC survival rates in recent years [6]. Due to its heterogeneity, complexity, and malignancy, both early diagnosis and intervention for HCC remain challenging. Although molecular characterization of HCC tissue is increasingly important, alpha-fetoprotein (AFP), a glycoprotein produced by the fetal liver, is the only commonly used serum biomarker for the early screening test [7,8]. The elevated levels of the isoform AFP-L3 are more specifically associated with hepatocellular carcinoma (HCC) compared to total AFP levels [9]. In addition to the AFP family, PIVKA-II (Protein Induced by Vitamin K Absence or Antagonist-II), also known as Des-gamma carboxy prothrombin, serves as an additional diagnostic tool for the diagnosis and monitoring of HCC [10]. Other current diagnostic methods for HCC include traditional imaging techniques such as CT scans, ultrasound, and MRI [2,11], as well as the GALAD scoring system, which incorporates clinical parameters to assess the risk of HCC [12]. Also, there is an overuse of the current diagnostic testing, which may lead to potential harm from unnecessary follow-up testing or treatment, and patient anxiety or stress due to false positive results. In the same vein, different studies use their own definitions of overuse, making it challenging to directly compare assessments of similar procedures. In contrast to diagnosis, the intervention methods for HCC often depend on the disease stage, which is classified as stages I to IV using the TNM (Tumor, Nodule & Metastasis) cancer staging system [4,11,13]. The cancer stage is a crucial parameter used to describe the tumor's growth and spreading.
Surgical resection and liver transplantation are commonly used to treat early-stage HCC, while radio ablation and systemic therapy are frequently used to treat advanced-stage HCC [3,4,[14], [15], [16], [17]]. For example, Sorafenib, the first multikinase inhibitor agent approved by the Food and Drug Administration (FDA), has significantly improved the survival of HCC patients [17,18]. However, its efficacy is limited by side effects, low response rates, and the development of drug resistance [18,19]. Another pitfall is the lack of international agreement on low-value service definitions and standardized assessment methods, which hinders consistent evaluation and comparison of treatments across studies. Although many other targeted and combinational therapies are in rapid development and Phase III clinical trials [20,21], a better understanding of the molecular mechanisms underlying HCC origin and progression is still needed to guide interventions.
Multi-omics integration is an emerging approach that combines high-throughput data from genomics, transcriptomics, proteomics, metabolomics, and lipidomics to provide a comprehensive understanding of HCC. Genomic and transcriptomic studies, which are based on whole-genome sequencing and RNA-seq or microarray assays, have identified numerous genetic alterations and dysregulated gene expression patterns associated with HCC [22,23]. Mass spectrometry-based proteomics can identify differentially expressed proteins in HCC-related cells or tissues compared to normal cells or tissues. Early proteomic work mostly focused on identifying prognostic biomarkers in serum or tumor tissue samples [24], while recent research has employed proteomics, phosphoproteomics, and AI-based data interpretation to uncover more mechanisms underlying HCC development and progression. These cutting-edge approaches have facilitated the identification of novel therapeutic targets, including Sterol O-acyltransferase 1 (SOAT1), Src family kinases, Proliferating Cell Nuclear Antigen (PCNA), Cyclin-Dependent Kinase 1 (CDK1), Fibroblast Growth Factor Receptor 4 (FGFR4), and Microfibril Associated Protein 5 (MFAP5), offering promising avenues for targeted intervention in HCC treatment [[25], [26], [27], [28], [29], [30]].
Proteogenomic characterization is a multi-omics strategy that integrates genomic, transcriptomic, and proteomic data. It has emerged as a valuable approach in cancer research, providing biological and diagnostic insights [31]. For example, Gao et al. performed a proteogenomic study using 159 paired HBV-related HCC tissues and identified PYCR2 and ADH1A as two prognostic biomarkers. Similarly, Ng et al. used proteogenomic analysis to reveal dysregulation of Wnt-β-catenin, AKT/mTOR, and Notch pathways in HCC [32,33]. In addition to genomics, transcriptomics and proteomics, metabolomics and lipidomics are alternative tools for investigating biomarkers and therapeutic targets of HCC at the level of the final products of genes and proteins.
Compared to lipidomics, the metabolomic approach primarily focuses on hydrophilic metabolites, particularly those involved in central metabolism pathways. In addition to the well-known Warburg effect, which is a general characteristic of cancer cell metabolism, HCC exhibits a distinct metabolite profile compared to other cancers. Previous studies have shown that metabolic molecules involved in glycolysis, the TCA cycle, amino acid metabolism, and bile acid metabolism are disrupted in association with HCC [[34], [35], [36]]. Lipidomics is a complementary approach to metabolomics that primarily profiles endogenous hydrophobic compounds. Numerous studies have shown that alterations in phospholipid, sphingolipid, β-oxidation, and ketone body biosynthesis in HCC cells or tissues contribute to the initiation and development of the disease [37,38]. Recently, more sets of combinational biomarkers for premalignant liver disease have been identified and summarized using metabolomic and lipidomic approaches [39].
Multi-omics studies offer considerable advantages over traditional methodologies by addressing complex biological systems and providing a holistic understanding of disease pathogenesis. However, these approaches are not without their challenges. A primary concern is the underrepresentation of racial and ethnic diversity in multi-omics datasets, often due to the logistical difficulties associated with global sample collection. Consequently, the findings may not be generalizable across all populations. Additionally, the requirement for large sample sizes increases the risk of false positives, a concern that is particularly pronounced in studies involving early-stage tissues, where standardized assessment protocols are lacking. Such limitations can lead to the inadvertent exclusion of dysregulated molecules during statistical analysis. To mitigate these issues, it is crucial to implement strategies that include comprehensive assessment of sample provenance, refinement of data preprocessing techniques, and benchmarking against centralized data repositories like Clinical Proteomic Tumor Analysis Consortium (CPTAC). These measures are essential for enhancing the robustness and applicability of multi-omics research findings.
Numerous studies have been conducted to identify specific patterns associated with hepatocellular carcinoma (HCC) disease using single or multiple omics approaches. However, most studies have focused on early diagnosis and treatment, and research on advanced-stage HCC remains insufficient. To address this gap, we employed a multi-omics approach, including quantitative proteomics, phosphoproteomics, metabolomics, and lipidomics, to investigate differential patterns between early- and advanced-stage HCC tumors. We compared our results with CPTAC database and integrated the multi-omics data using QIAGEN Ingenuity Pathway Analysis (IPA), revealing the complexity of the pathway network during tumor progression. These findings not only provide valuable insights into liver cancer biology but also demonstrate the feasibility of integrating multi-dimensional information from macro biomolecules, protein translational modification, and small compounds.
2. Materials and methods
2.1. Clinical sample collection and study cohort
All patients involved in this study provided informed consent for biobank recruitment at Shulan (Hangzhou) Hospital. Both tumor and normal tissues adjacent to the tumor (NAT) samples were procured from hepatocellular carcinoma (HCC) patients during surgical procedures. These samples were carefully sectioned into pieces ranging from 0.3 to 0.8 cm3 using a scalpel. Following extraction, the fresh samples were promptly frozen and preserved in liquid nitrogen prior to sample processing. Ethical clearance was granted by the Research Ethics Committee at Shulan (Hangzhou) Hospital.
The diagnosis of HCC was confirmed either through histopathological examination or by adhering to established imaging criteria. The clinical stage of the disease was subsequently ascertained based on the TNM cancer staging system. The study encompassed 14 pairs of HCC and corresponding non-tumor liver tissue samples. Half of these pairs were derived from patients diagonosed with early-stage (TNM stage I, characterized by a solitary tumor of ≤2 cm with no vascular invasion), while the remining pairs were from patients with advanced-stage (TNM stage III & IV, marked by a solitary tumor of ≥2 cm or multiple tumors, vascular invasion, and in certain instances, distant metastasis). The statistical characteristics of patients with early-stage and advanced-stage HCC are depicted in Table 1.
Tables 1.
Characteristics of the cohorts.
| Early-stage | Advanced-stage | ||
|---|---|---|---|
| Age | Mean ± SD | 60.3 ± 11.9 | 46.4 ± 13.5 |
| Gender | Male% | 57.1 | 42.8 |
| Smoking | Yes% | 28.6 | 28.6 |
| No% | 71.4 | 71.4 | |
| Alcohol | Yes% | 0 | 28.6 |
| No% | 100 | 71.4 | |
| Cirrhosis | Yes% | 14.3 | 14.3 |
| Cancer grade | Low% | 14.3 | 57.1 |
| Medium% | 85.7 | 42.9 | |
| Tumor size | Mean ± SD | 110.5 ± 146.2 cm3 | 684.1 ± 582.9 cm3 |
| Invasion | Yes% | 0 | 71 |
| Metastasis | Yes% | 0 | 57.1 |
All tissue samples were ground and homogenized using a cryogenic grinder (SPEX 6875 Freezer/Mill) and divided into four parts by weight: 3 mg for proteomic experiments, 80 mg for phosphoproteomic experiments, 50 mg for metabolite extraction, and 50 mg for lipid extraction.
2.2. Reagents, protein, metabolite, lipid extraction and phosphopeptide enrichment
All the detailed information was provided in Supplementary Information (SI) part. Briefly, all the main reagents used in this study were from Sigma, Promega, Thermo Fisher, etc. Protein, metabolite and lipid were exacted by RIPA buffer, 80 % methanol and Methyl tert-butyl ether (MTBE) from the ground tissue, respectively. Protein was digested by trypsin overnight and further labeled by TMT regent for proteomics, phosphopeptide was enriched by IMAC Fe-NTA and TiO2.
2.3. LC-MS/MS methods and data pretreatment
The TMT-labeled peptides were separated using a Thermo Vanquish Neo integrated nano-HPLC system directly interfaced with a Thermo Exploris 480 mass spectrometer equipped with FAIMS Pro. The enriched phosphopeptides were analyzed using a Bruker Nano Elute HPLC system directly interfaced with a Bruker timsTOF Pro 2 mass spectrometer. Both metabolomic and lipidomic analyses were performed using an Agilent 1290 Infinity HPLC system coupled to an Agilent 6545 Q-tof mass spectrometer with an electrospray ion source operating in both positive and negative ion modes. The detailed information can be found in SI part.
Quality control (QC) runs were collected using a pooled mixture derived from all samples and several parameters including peak width, total ion current chromatogram, and precursor mass accuracy were examined to assess data quality of each sample. Samples exhibiting significantly low TIC values or signal to noise were omitted from subsequent analysis to maintain data quality. Each acquired MS raw data was searched against the proper database (protein, metabolite and lipid) to generate the identification lists, which were further quantified by reporter ion intensity, precursor ion intensity and extract ion area for TMT-labeled proteins, phosphopeptides and small molecules, respectively. All the quantified omics data were normalized before further comparison and analysis. The TMT-labeled proteomics data were normalized by the total intensities of the reporter ions from the QC channel, the label-free phosphoproteomics data were normalized by the total abundances of the identified peptides, the metabolite and lipid scaling were preformed based on the total ion area of the sample.
2.4. Bioinformatic analysis
After database searching and quantification procedures, proteomic data were analyzed using, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome assays. GO enrichment analysis was conducted on the website (http://geneontology.org/) by importing the gene IDs of up- or down-regulated proteins [40,41], and a cut-off less than 0.05 was used for GO category analysis after multiple test correction. KEGG pathway enrichment was performed on the website (http://www.genome.jp/kegg/kaas/) by importing the gene IDs of up- or down-regulated proteins with a 0.01 cut-off for p values [42]. For Reactome pathway identifier mapping, IDs of dysregulated proteins were submitted to Analysis Tools in Reactome (https://reactome.org/) [43]. IntAct interactors were included to increase the analysis background and all none-human identifiers were converted to their human equivalents. Reactome pathways with adjusted p values < 0.05 were considered statistically over-representation.
A kinase-substrate relation was established using the KSEA app (casecpb.shinyapps.io/ksea/) based on phosphoproteomic data [44]. Only significantly changed (p < 0.05) phosphopeptides were used for the assay, and the filtered output contained kinase predictions with a z-score of ≥2.0.
Venn diagram was generated by calculating the intersections of list of elements on the website (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Survival analysis was performed on the GEPIA2 website (http://gepia2.cancer-pku.cn/#survival) based on a multi-gene signature and plot a Kaplan-Meier curve [45]. The default setting of GEPIA2 was used, which first calculated and ranked all samples based on the expression levels of the genes in the signature, then defined the top 50 % of the samples as high and bottom 50 % of samples as low before running standard Kaplan-Meier curves.
To perform multi-omics integrative analysis, Ingenuity Pathway Analysis (IPA) was used. Significant up- or down-regulated proteins, metabolites, and lipids were combined into a single sheet, which was uploaded to IPA for core analysis. Multiple parameters for pathway enrichment and network building were tested and optimized as shown in Supplementary Information. The data of dysregulated phosphopeptides were uploaded separately for phosphorylation analysis. Comparison analysis was used to summarize the enriched pathways from the protein, metabolite, lipid, and phosphorylation assays for both early and advanced-stage HCC.
2.5. Statistics
Data were presented following the analysis of variance (ANOVA). To calculate the statistical significance between groups, Student's t-test, assuming two-tailed distributions, or the nonparametric Mann-Whitney test was used, without assuming that the variance was similar between the groups being statistically compared. Principal component analysis (PCA) was performed for cluster analysis of proteomic data for all samples using Omicsolution analysis platform (https://www.omicsolution.com/wkomics/pca/) [46]. For the TMT-labeled proteomics data, the absolute values of fold change ≥1.5 were considered as the dysregulated threshold. For the label-free phosphoproteomics, metabolomics and lipidomics data, the absolute values of fold change ≥2 were deemed to be differentially existed. A p-value of <0.05 was considered statistically significant.
3. Results
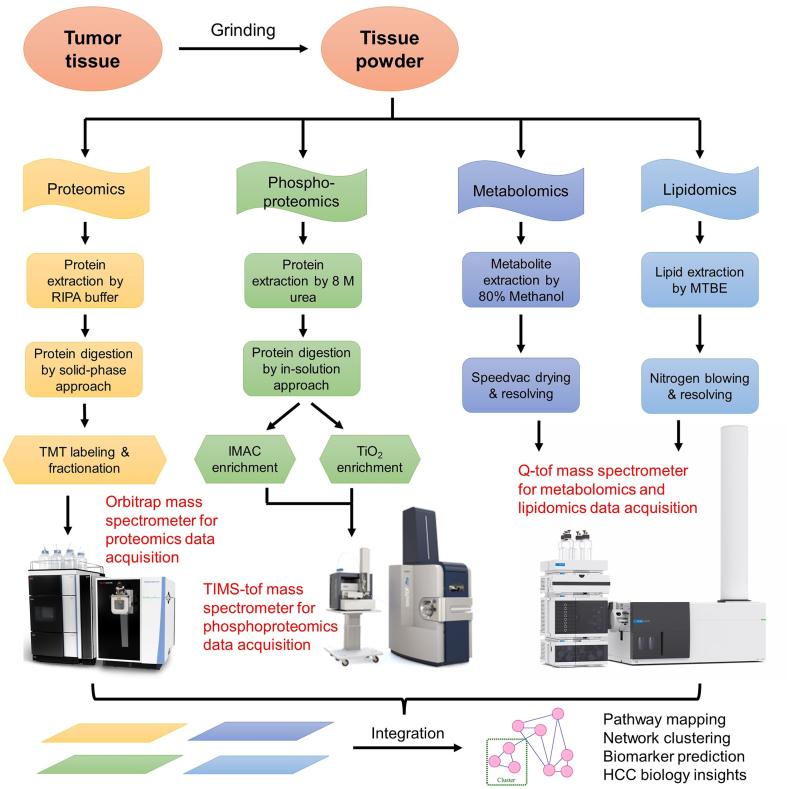
LC-MS/MS data of TMT-labeled quantitative proteomics, phosphoproteomics, metabolomics, and lipidomics for the same HCC tumor or normal tissues adjacent to the tumor (NAT) were acquired in parallel from three different instruments (Fig. 1). Each omics dataset was analyzed and annotated separately. Dysregulated proteins, phosphopeptides, metabolites, and lipids were selected and summarized for both early stage and advanced stage HCC, respectively.
Fig. 1.
Schematic representation of multi-omic experiments including proteomics, phosphoproteomics, metabolomics and lipidomics.
3.1. Overview of proteomic, metabolomic, and lipidomic results
For the TMT-labeled proteomic study, a total of 12,152 protein groups (Table S1) and 166,260 unique peptides were identified from 742,816 PSMs. Unsupervised PCA was performed on the proteomic data and the leading principal components of the global proteomic data separated tumor from NAT samples (Fig. S1). Among these proteins, 233 upregulated and 303 downregulated proteins were detected in early-stage HCC tissues compared to NAT (Fig. 2A). In contrast, 299 upregulated and 632 downregulated proteins were identified in advanced-stage HCC (Fig. 2B). As expected, more dysregulated proteins were present during disease progression. GO analysis was used to cluster dysregulated proteins involved in metabolic processes, cell cycle and organization, nucleic acid and protein metabolism, transport, signal transduction, and development in both early and advanced stages (Fig. S2). Analyzed by KEGG, cell cycle, DNA replication, and repair pathways were enriched in upregulated proteins, while various metabolism pathways were enriched in downregulated proteins (Fig. S3).
Fig. 2.
Proteomics, metabolomics and lipidomics of early and advanced-stage HCC. (A) and (B) Volcano plot analysis of TMT-labeled proteomics on early (A) and advanced (B) stage HCC. (C) The numbers of differentially accumulated metabolites and lipids quantified between early or advanced stage tumor and the normal tissue adjacent to the tumor. (D) and (E) Volcano plot analysis of metabolites quantified in early (D) or advanced (E) stage HCC. (F) and (G) Volcano plot analysis of lipids quantified in early (F) or advanced (G) stage HCC. The Statistically significantly (p-value<0.05, Student's t-test) up- (Fold change>1.5) and down-regulated (Fold change<0.67) molecules were showed in purple and red dots, respectively.
To gain deeper understanding of tumor genesis and procession, we subjected the dysregulated proteins identified in our study to Reactome for pathway and network analysis. Consistent with KEGG analysis, Reactome search indicated an enrichment of cell cycle pathways among upregulated proteins, while metabolic pathways were predominantly associated with downregulated proteins. Notably, early-stage HCC exhibited a distinct pattern of upregulated pathways compared to advanced-stage HCC (Fig. S4). Within the hierarchical arrangement of Reactome pathways, the three most significantly enriched top-level pathways in early-stage HCC were related to the cell cycle, DNA replication, and chromatin organization. In contrast, DNA repair pathways were the most enriched in advanced-stage HCC, although cell cycle and DNA replication pathways were also significantly represented. The divergent pathway patterns observed between the two HCC stages highlight distinct biological processes, offering potential insights for the development of biomarkers to monitor cancer progression.
For metabolomics and lipidomics, 251 metabolites and 330 lipids were annotated from 1201 metabolite and 3354 lipid features under positive mode; simultaneously, 245 metabolites and 395 lipids were identified from 1197 metabolite and 2698 lipid features under negative mode (Table S2). The numbers of upregulated and downregulated metabolites and lipids under positive/negative mode for both early-stage and advanced-stage HCC were summarized in Fig. 2C. Combining the compounds detected from positive and negative modes, a total of 58 metabolites and 72 lipids were dysregulated in early-stage HCC, and 86 metabolites and 102 lipids were dysregulated in advanced-stage HCC (Fig. 2D–G). Consistent with the trend observed in proteomic data, the higher the tumor stage, the greater the number of significantly changed compounds discovered.
3.2. Phosphoproteomics and kinase-substrates relationship
For label-free quantitative phosphoproteomics, 25,071 unique phosphopeptides containing 35,574 unique phosphorylation sites were profiled on 6686 proteins (Table S3). As illustrated in Fig. 3A, 49.4 % of phosphopeptides were identified simultaneously under both conditions, while 45.3 % and 5.3 % were profiled exclusively by IMAC-Fc and TiO2, respectively. The distribution of single-, double-, and multiple-phosphorylation sites on each phosphopeptide was shown in Fig. 3B, with nearly 90 % of peptides being mono-phosphorylated. The ratios of phosphorylation sites on Ser, Thr, and Tyr were exhibited in Fig. 3C; consistent with basic biochemical knowledge, the ratio of detected phosphoTyosine was less than 3 %. Only peptides with a single phosphorylation site were used for further quantitative variance analysis. The numbers of up- and down-regulated phosphopeptides with unique mono-phosphorylation sites in early and advanced-stage HCC were calculated and displayed on the volcano plots (Table 2 & Fig. 3D-E).
Fig. 3.
Phosphopeptide enrichment of early and advanced-stage HCC using the IMAC Fe-NTA and TiO2 strategies. (A) Overlap between all phosphopeptides quantified by IMAC Fe-NTA or TiO2 phosphopeptides enrichment method. (B) The composition of the mono- (1P), double- (2P), and multi- (3P) phosphorylated peptides quantified. (C) The percentage of phosphorylated serine (p-ser), threonine (p-thr), and tyrosine (p-tyr) in the quantified phosphopeptides. (D) Volcano plot analysis of quantified mono-phosphorylated peptides between early-stage tumor and the normal tissue adjacent to the tumor. (E) Volcano plot analysis of quantified mono-phosphorylated peptides between advanced-stage tumor and the normal tissue adjacent to the tumor. Statistically significantly (p-value<0.05, Student's t-test) up- (Fold change>2) and down-regulated (Fold change<0.5) phosphopeptides with unique mono-phosphorylation sites were showed in purple and red dots, respectively.
Table 2.
The numbers of dysregulated phosphopeptides.
| Up-regulation | Down-regulation | |
|---|---|---|
| Early-stage | 1145 | 825 |
| Advanced-stage | 1253 | 945 |
The kinase-substrate relationship assay was performed using the KSEA App to predict altered kinase activities in early and advanced-stage HCC from phosphoproteomic data. The enriched kinase activities in early and advanced-stage HCC are displayed in Fig. 4A and B, respectively. Kinases represented by red bars (z-score ≥2) indicate hyperactive kinase activity, while kinases denoted by blue bars (z-score ≤ −2) represent deactivated kinase activity. On one hand, most hyperactive kinase families were shared in both early and advanced-stage HCC. The high activation of cyclin-dependent kinase (CDK), mitogen-activated protein kinase (MAPK), and Aurora kinase (AURK) was expected, as they are all related to the cell cycle, cell proliferation, and mitosis, leading to tumorigenesis. On the other hand, apart from AMP-activated protein kinase (AMPK), deactivated kinases exhibited some degree of diversity, indicating distinct signal transduction pathways in different stages of HCC. For example, Rho-associated coiled-coil-containing protein kinase 1 (ROCK1), whose substrates include RDN3 and NOS3, was dramatically deactivated in the advanced-staged samples compared to the early-staged samples. In contrast, the activity of the isozyme ROCK2 remained unchanged, indicating differential regulatory mechanisms, which will be discussed later.
Fig. 4.
Kinase-substrate enrichment analysis of phosphoproteomic data of early- or advanced-stage HCC. (A) and (B) The enriched kinase identified in early (A) or advanced (B) stage tumor tissues when compared with the normal tissue adjacent to the tumor. Significantly hyperactivated and deactivated kinase are illustrated by red or blue, respectively. Significance thresholds were set to a z-score cutoff of 2, p value cutoff of 0.05, and substrate count cutoff of 5.
3.3. Multi-omic characterization of early and advanced-staged HCC
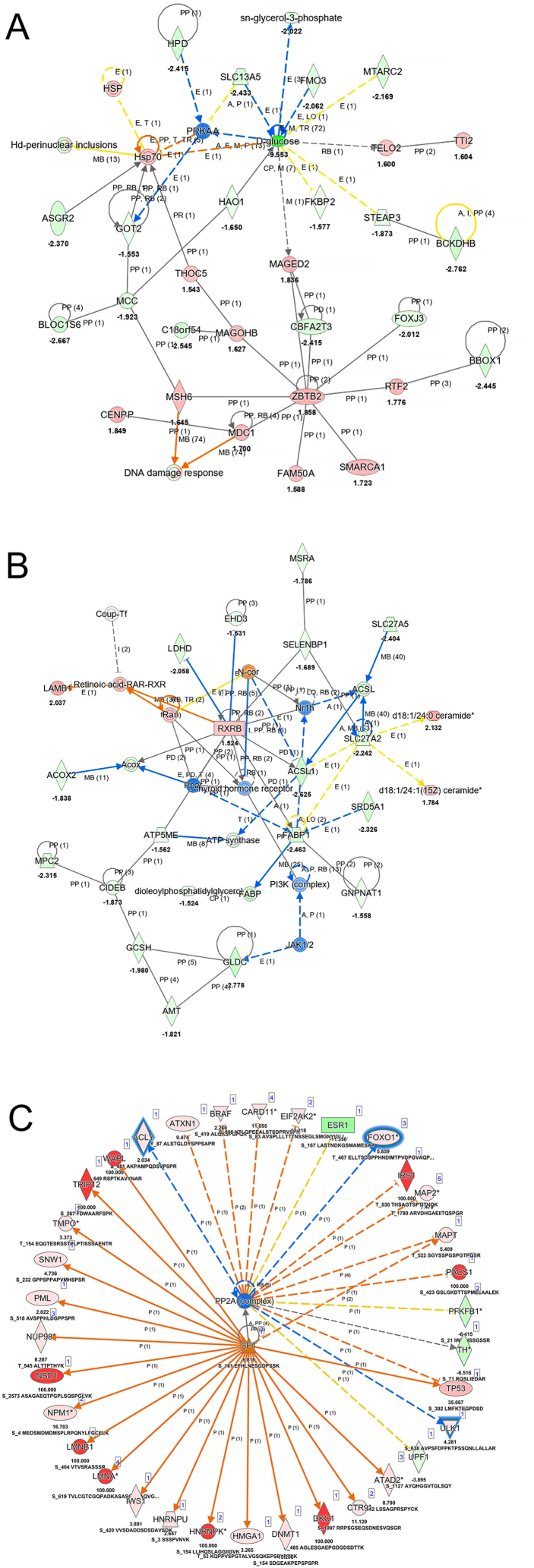
The lists of dysregulated proteins, metabolites, and lipids were combined and subjected to IPA for core analysis, abbreviated as PML (Protein, Metabolite and Lipid) analysis. Compared to advanced-stage HCC, a limited number of pathways were enriched in early-stage HCC, indicating that a few universal tumorigenesis processes and pathways were activated or inhibited, including DNA replication, cell proliferation, transcription factor activities, and p53 function (Fig. S5). As HCC progressed to advanced stage, an increasing number of bioprocesses and pathways contributing to tumor growth were activated (Fig. S6A), and various metabolism pathways, particularly those involved in anabolism, were inhibited (Fig. S6B), consistent with findings from separate proteomic and metabolomic data. Two selected networks of the multi-omics assay in advanced-stage HCC are exhibited in Fig. 5A & B: one depicts a network from glucose depletion to triggering DNA damage response, while the other mostly describes how retinoid X receptor influences lipid metabolism. Although these networks were reconstructed based on the current database containing diverse information from different cell lines, tissues, and species, these findings could help researchers understand connections between individual changes in proteins or compounds and reveal the complexity of the biological system.
Fig. 5.
Interaction networks based on PML analysis (A&B) and phosphorylation analysis (C) using IPA. (A) A network of glucose depletion triggering DNA damage response. (B) Interaction networks of retinoid X and lipid metabolism. (C) Upstream analysis of SET and PP2A. Orange and blue solid lines indicate direct activation and inhibition, respectively. Yellow lines present findings were not consistent with state of downstream molecules. Dashed lines indicate indirect relationship while gray lines signify effect was not predicted. Shapes filled with red and green presents increased and decreased measurement, respectively.
Phosphorylation assay was submitted to IPA as a separate analysis since IPA permits the integration of protein-level data but lacks the capability to incorporate phosphorylation events directly with metabolomic and lipidomic profiles. In contrast to the results of PML analysis, several kinases signaling pathways were prone to activation for tumor initiation and metastasis in early-stage HCC (Fig. S7A), suggesting that post-translational modification (PTM) signaling is more sensitive than protein and metabolite changes. As expected, the bioprocesses and pathways enriched in advanced-stage HCC demonstrated a higher degree of malignancy (Fig. S7B). Fig. S8 displays a complex phosphorylation interaction network predicted from the phosphoproteomic data, revealing multiple protein phosphorylation factors affecting ATPase.
The upstream regulator analysis was also performed using upregulated phosphoprotein lists (Fig. S9). Multiple factors including kinase, chemical reagent/drug, cytokine, growth factor and phosphatase were disclosed for both early- and advanced-stage HCC. Intriguingly, the analysis revealed uniform inhibition of all phosphatases and concurrent activation of all kinases, along with SET, which serves as a potent inhibitor of protein phosphatase 2A (PP2A). Given that PP2A acts as a tumor suppressor with the capability to deactivate multiple signaling pathways, its inhibition via SET activation is a critical event that may contribute to tumorigenesis [47]. Phosphorylation of Ser161, located within the earmuff domain, was significantly elevated in our phosphoproteomic data (Table S3). The earmuff domain has been shown to facilitate SET protein translocation from the cytoplasm to the nucleus and participate in nucleosome remolding [48,49]. While SET exhibits specificity for unwrapped nucleosomes [50], it is plausible that post-translational modifications, such as Ser161 phosphorylation, modulate its interaction with intact nucleosomes. This study's phosphoproteomic analysis further illuminates the intricate interplay between SET, PP2A, and their associated proteins (Fig. 5C). Notably, several nuclear proteins, including Lamin A/C (LMNA), Thymopoietin (TMPO), Nucleoporin 98 (NUP98), and NUP210, exhibited increased phosphorylation in tumor tissues. Given that phosphorylation of Lamin A/C influences nuclear lamina assembly, a process in which PP2A is also implicated [51,52], these findings suggest that these nuclear proteins may function as downstream effectors of SET signaling, potentially impacting tumor cell proliferation. Although phosphorylation data could not be merged with other omics data in IPA, the comparison assay provided an alternative way to integrate observations from the four omics under the current study.
3.4. Comparison of advanced and early-stage HCC
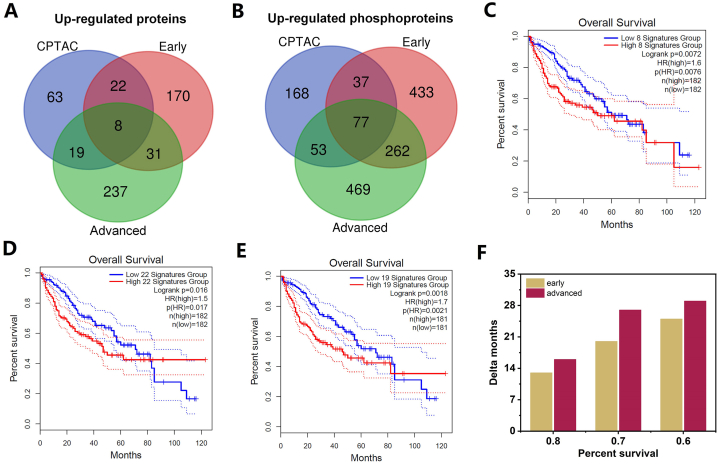
To identify the most clinically relevant targets from dysregulated multi-omics data, we conducted a comparison of up- and down-regulated proteins and phosphoproteins using the CPTAC database [25,32,53], which offers a repository for multidimensional proteomic data across diverse cancer types. As a well-established resource, it can be leveraged as a benchmark for validating our omics data findings. Such validation can lead to the identification of novel biomarker panels, potentially accelerating research and enriching our understanding of the complexities inherent in cancer biology. Fig. 6A and B illustrate that approximately 50 % of the up-regulated proteins and phosphoproteins from the CPTAC database were detected in either early or advanced-stage samples. Notably, 8 up-regulated proteins appeared in all three groups (Fig. 6A), designating them as the shared prominent biomarker panel. Furthermore, 22 up-regulated proteins, which are the specific intersection of CPTAC database and our early-stage HCC omics data, were identified as specific early-stage HCC biomarker panels; while 19 up-regulated proteins, the specific intersection of CPTAC database and our advanced-stage HCC omics data, were unique to advanced-stage HCC (shown in Table 3).
Fig. 6.
Potential biomarker panels with clinical prognostic significance. Venn diagram showing the overlap among the up-regulated proteins (A) and phosphoproteins (B) analyzed from CPTAC database, early- and advanced stage HCC data in current study. Kaplan–Meier analysis for the overall survival of the 8- (C), 22- (D) and 19- (E) gene signatures. The comparisons of the reduced month time of the high expression between early- and advanced-stage specific gene signatures (F).
Table 3.
The specific biomarker panels of early- and advanced-stage HCC.
| Early-stage | Advanced-stage |
|---|---|
| SQSTM1, JTB, RACGAP1, HELLS, ASNS, MTHFD1L, SULT1C2, SMC4, COA7, MSI1, CEP131, SCAMP3, CPD, GPATCH4, ROBO1, PLVAP, PODXL, HKDC1, UBD, MSTO1, ACSL4, PARP1 | IGF2BP3, LAMB1, ATAD2, JMJD6, MSH6, CDK1, FABP5, MDK, MCM3, VCAN, MKI67, PCNA, PYCR1, LAMA4, PEG10, TFRC, MAGED2, RFC4, KIF23 |
Next, to evaluate the specificity of the selected biomarker panels for different stages of hepatocellular carcinoma (HCC), we conducted survival analyses using the Kaplan-Meier method, which connects gene signatures with patient survival rates. These analyses were performed using the GEPIA2 program, which is based on RNA expression data. In the HCC CPTAC database, RNA and protein levels demonstrated over 90 % positive correlation [32]. Our identified gene signatures from the shared, specific early-stage, and specific advanced-stage biomarker panels showed nearly 100 %, 70 %, and 80 % presence, respectively, and exhibited strong correlation with the CPTAC RNA data (Table S5). As illustrated in Fig. 6C, higher expression of the 8 up-regulated gene signatures (shared group) was significantly associated with shorter survival. Intriguingly, both specific early-stage and specific advanced-stage groups demonstrated higher expression correlated with shorter survival, but the 19-gene signature group exhibited a longer time distance between high and low gene expression survival curves compared to the 22-gene signature group (Fig. 6D & E). High expression of the advanced-group panel indicated a longer time distance (Fig. 6F), representing lower survival time compared to the high expression of the early group panel, highlighting the clinical relevance of our specific biomarker panels. A similar trend was also observed for down-regulated proteins (Fig. S10). These agreements also revealed a direct relationship between transcriptional and translational regulation in the current HCC samples.
By utilizing the comparison assay, the enriched a hundred pathways either activated or inhibited from both phosphorylation analysis and PML analysis were summarized in a single graphical representation (Fig. 7A). Most of the pathway changing trends were consistent with each other, and more significant activation or inhibition (darker colors) was observed in advanced-stages and CPTAC data than in early-stages. Some of the top changing pathways were obviously to understand. For instance, the top two enriched pathways were related to the dysregulation of xenobiotic metabolism, leading to the loss of detoxification function of liver [54]. Serotonin degradation (the third line in Fig. 7A), have been widely reported to be strongly inhibited, leading to serotonin accumulation, and promoting tumor growth in HCC [55]. As mentioned previously, the PML analysis in early stages only revealed some top changing pathways. In contrast, the phosphorylation analysis in early stages exposed most of the changing pathways observed in advanced stages. This phenomenon suggests that protein phosphorylation is an upstream signal of protein and small compound changes. Concordant findings emerged from the Reactome pathway analysis, which revealed an overrepresentation of not only cell cycle and DNA replication pathways but also post-translational modification pathways, such as phosphorylation and methylation, in early-stage HCC. EML4 and NUDC in mitotic spindle formation, polo-like kinase mediated events, phosphorylation of Emi1 and activation of NIMA kinases NEK6, 7, and 9 were significantly enriched in early-stage HCC alone (Table S4). These elements play a critical role in cellular reprogramming via phosphorylation, thereby facilitating tumorigenesis. This comparison assay list provides an overall perspective of biological pathways in HCC initiation and progression.
Fig. 7.
The summarized enriched pathways and an illustrated mechanism of HCC progression. (A) The comparison analysis of enriched pathways for PML assay in early-stage HCC (Group I), PML assay in advanced-stage HCC (Group II), proteome assay of CPTAC database (Group III), phosphorylation assay in early-stage HCC (Group IV), phosphorylation assay in advanced-stage HCC (Group V) and phosphorylation assay of CPTAC database (Group VI). The orange color and blue color in the heatmap represent pathway activation and inhibition, respectively. The darker color is related to larger z-score, which means the higher degree of activation or inhibition. (B) The summarized characterization of early- and advanced-stage HCC from the current omics study.
4. Discussion
Hepatocellular carcinoma (HCC) is one of the most malignant tumors worldwide, posing a severe threat to human health. Although surgical resection and liver transplantation can efficiently treat early-stage HCC, early diagnosing and intervening in advanced-stage HCC patients remain challenging. Multi-omics technologies offer systematic insights into the complex biological processes related to disease initiation and development, which are helpful for developing biomarkers for diagnosis and drug targets for systemic therapy. Several points related to this study warrant discussion.
Firstly, at the technical level, multi-omics and their applications are still challenging in today's research. Simultaneously acquiring multi-omics data from the same tissue requires an adequate sample amount and diverse instrument support, especially for PTM-omics, which typically need substantial proteins and sophisticated experimental workflows, such as enrichment and fractionation. As shown in Fig. 6, protein phosphorylation exhibited higher sensitivity for signal alteration than downstream proteins and compounds in early-stage HCC, suggesting that it may be more suitable for biomarker selection in early diagnosis theoretically [56,57]. Although phosphopeptides, ordinary tryptic peptides, metabolites, and lipids are all detected by LC-MS/MS, at a practical level, phosphoproteomics is considerably more complex and challenging to implement for clinical samples than other omics. Previous studies have identified over 20,000 phosphopeptides or phosphorylation sites in HCC tumors using high-pH reverse phase fractionation (one sample, six injections) and TiO2 enrichment [27]. Taking advantage of the high sensitivity and speed of the timsTOF Pro 2 mass spectrometer and the complementary results of IMAC Fe-NTA and TiO2 profiling [[58], [59], [60]], a comparable number of phosphopeptides has been obtained using a much simpler approach (one sample, two injections; routine DDA data acquisition without project-specific proteomics libraries). Additionally, multi-omic data have traditionally been difficult to integrate due to their complexity and heterogeneity [61]. With the assistance of the IPA platform, all the omic data generated from proteins, including PTMs, to small compounds in the current study have been recognized and integrated for analysis, revealing enriched pathways and biomarkers in different stages of HCC. All reagents, instruments, database search and analysis engines used in this research are commercial or open source, reflecting the lowered threshold for multi-omics studies.
Next, the down-regulated proteins and metabolites from proteomics and metabolomics data were found to be quite consistent, demonstrating a global attenuation of cell metabolism that prominently occurred in advanced-stage HCC. Unlike the phenotypes obtained from cell lines or mouse models, both the enzymes and metabolites in glycolysis and anaerobic respiration in these clinical samples showed significant decreases, seemingly opposite to the Warburg effect. However, MAPK deactivation from phosphoproteomic data definitively supported the activation of aerobic glycolysis [62,63]. The paradox can be explained from several perspectives. As mentioned earlier, proteome and metabolome changes are the downstream consequence of cell signaling, and the tumor tissue may already be in a low-glucose equilibrium if insufficient materials are supplied, which may be caused by therapy-induced metabolic alterations for the advanced-stage samples. Additionally, the traditional Warburg effect has been increasingly challenged due to different cancer types and diverse tumor heterogeneity and microenvironments, which play a significant role in shaping the metabolic profile of cancer cells, e.g., tumor stromal cells including cancer-associated fibroblasts can act as metabolic supporters that can fuel the growth of HCC cells, resulting in the observation of a reverse Warburg effect [[64], [65], [66]]. Moreover, glutamate was significantly increased in advanced-stage HCC, indicating the activation of an alternative energy generation pathway called glutaminolysis, which can not only produce energy from α-ketoglutarate entering TCA cycle, but also directly provide building blocks to synthesize nucleotides and lipids that are essential for tumor proliferation [67]. Although the glutaminolysis pathway can compensate for some energy supply, it still seemed that global catabolism decreases during tumor progression. This viewpoint has been recently demonstrated by a Flux work that indicates solid tumors generally produce ATP at a slower rate than normal [68].
Third, the multi-omics integrated assay revealed comprehensive signal networks, providing new insights into the mechanisms of HCC disease. As described above, most reported biological processes in HCC, including epithelial-mesenchymal transition (EMT), abnormal tumor microenvironment formation, and senescence bypass, emerged through the multi-omics data. EMT is considered one of the common features of cancer development [69], and in HCC, besides transcription factor promotion, signaling pathways such as AURK, MAPK, and NF-κB also play important roles in the EMT process [[70], [71], [72]]. Consistent with previous studies, these canonical kinase pathways were hyperactive from early to advanced-stage HCC. Our data also contained some differing results that seemed contradictory to previous reports; for example, Rho-associated coiled-coil-containing protein kinase 1 (ROCK1) was significantly deactivated in advanced-stage HCC. Although ROCK2 was observed as an overexpressed kinase leading to tumor malignancy both by proteomic profiling and biology studies [27,73], ROCK1 might contribute to HCC progression through an alternative way involving the peritumoral microenvironment (PME), a new concept describing the tissue surrounding the tumor that influences cancer development [74]. Existing evidence showed that hepatic stellate cells (HSCs) accumulating in HCC PME with an increase in VEGF were able to induce angiogenesis [75], and ROCK1 was reported as a negative regulator of VEGF-driven angiogenic activation [76]. We hypothesize that deactivated ROCK1 would appear in HSCs and further promote angiogenesis. Due to the heterogeneity of tumor tissue, omics studies in this research could not identify specific cell types. Although this limitation might be partially addressed by bioinformatic assays [29], elaborate biological experiments are needed for validation. Additionally, glycogen synthase kinase 3α (GSK3A) was hyperactivated in advanced-stage HCC, which is responsible for glycogen accumulation and tumorigenesis by the suppression of Hippo signaling through glycogen-induced phase separation [77]. The simultaneous discovery of ROCK1 deactivation and GSK3A activation only in advanced-stage HCC adds new insights to the HCC progression landscape.
Finally, the clinical significance of early- and advanced-stage HCC were further scrutinized using the CPTAC dataset, which provided a valuable 22-gene signature and a 19-gene signature as the biomarker panels. These findings not only enhanced our understanding of HCC initiation and progression, but also offered new perspective on tumor diagnosis and potential therapeutic approaches. The insights gained from this study would be pivotal in guiding future therapeutic interventions and improving patient outcomes in HCC disease.
While the application of multi-omics technologies in this study has yielded novel insights into HCC progression, the realization of its full potential is hindered by several challenges. These include, but are not limited to, the limitations of current bioinformatics tools in analyzing PTM data in conjunction with metabolomic and lipidomic profiles, constraints imposed by a limited sample size, and the absence of validation for the proposed biomarker panels. Specifically, the integration of phosphoproteomic data with metabolomics and lipidomics within the IPA platform is currently not feasible, an issue that substantially limits the capacity for comprehensive multi-omic analysis. PTMs such as phosphorylation provide a more sensitive gauge of cellular signaling dynamics than changes in protein abundance or metabolite concentrations alone. The ability to analyze phosphoproteomic data alongside metabolomic and lipidomic data would significantly enhance our understanding of the interconnected pathways and regulatory networks underpinning biological processes. Furthermore, a broader sample cohort is anticipated to enrich the understanding of tumorigenesis. Despite the limited sample size in the current study, rigorous evaluation of the samples was conducted to minimize false-positive results, and the parameters in each group, e.g. gender, alcohol drinking, etc., were closely matched [78]. The biomarker panels identified herein are preliminary and have not undergone experimental validation. For the translation of these findings into clinical practice, it is imperative to validate the biomarker panels through extensive biological and clinical studies across diverse patient cohorts. After confirming their accuracy and reliability, targeted MS workflow or ELISA procedure would be developed to improve the sensitivity and specificity of the method, reduce the cost of the detection, fitting the clinical diagnosis demands. Finally, the optimized biomarker panels would be incorporated into clinical decision-making and continuously monitored the outcomes for the further adjustment.
In conclusion, performing multi-omic profiling from proteins to small compounds provided vast amounts of information in early and advanced-stage HCC. We demonstrated a feasible and comprehensive workflow for multi-omic integration studies, encompassing experimental design, sample preparation, data acquisition, and analysis. We also established a correlation network of HCC initiation and development, proposing new mechanisms for HCC, including reversed hypermetabolism and ROCK1 deactivation. Our results can also be verified by CPTAC data to some extent, and the specific biomarker panels, including a 22-gene signature and a 19-gene signature, of early- and advanced-stage HCC were also be identified, respectively (summarized in Fig. 7B). These insights can broaden our knowledge pool and provide potential gene signatures or biomarkers for HCC diagnosis and therapeutic interventions.
Ethics statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Research Ethics Committee of Shulan (Hangzhou) Hospital (Reference Number: KY2023033).
Data availability statement
The datasets used and/or analyzed during the current study are available at the Massive website (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with the ID number of MSV000091782.
CRediT authorship contribution statement
Mingzhu Fan: Writing – review & editing, Methodology, Investigation, Formal analysis. Jin Hu: Methodology, Investigation, Formal analysis. Xiaoyan Xu: Investigation, Formal analysis. Jia Chen: Investigation, Data curation. Wenwen Zhang: Writing – review & editing, Investigation. Xiaoping Zheng: Investigation. Jinheng Pan: Investigation. Wei Xu: Methodology, Conceptualization. Shan Feng: Writing – review & editing, Writing – original draft, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Yalin Wang, the director of the Biomedical Research Core Facility at Westlake University, China for supporting this research. We also thank Beibei Ma and Xiaoxian Du from Bruker Corporation for the database searching and quantification of the MS data acquired by the timsTOF Pro2 mass spectrometer.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e38182.
Contributor Information
Wei Xu, Email: 202211010211013@zcmu.edu.cn.
Shan Feng, Email: fengshan@westlake.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Hartke J., Johnson M., Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin. Diagn. Pathol. 2017;34:153–159. doi: 10.1053/j.semdp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 4.Ganesan P., Kulik L.M. Hepatocellular carcinoma: new developments. Clin. Liver Dis. 2023;27:85–102. doi: 10.1016/j.cld.2022.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 6.Ding J., Wen Z. Survival improvement and prognosis for hepatocellular carcinoma: analysis of the SEER database. BMC Cancer. 2021;21:1157. doi: 10.1186/s12885-021-08904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piñero F., Dirchwolf M., Pessôa M.G. Biomarkers in hepatocellular carcinoma diagnosis, prognosis and treatment response assessment. Cells. 2020;9:1370. doi: 10.3390/cells9061370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galle P.R., Foerster F., Kudo M., Chan S.L., Llovet J., Qin S., Schelman W., Chintharlapalli S., Abada P., Sherman M., et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019;39:2214–2229. doi: 10.1111/liv.14223. [DOI] [PubMed] [Google Scholar]
- 9.Li D., Mallory T., Satomura S. AFP-L3: a new generation of tumor marker for hepatocellular carcinoma. Clin. Chim. Acta. 2001;313:15–19. doi: 10.1016/s0009-8981(01)00644-1. [DOI] [PubMed] [Google Scholar]
- 10.Liebman H.A., Furie B.C., Tong M.J., Blanchard R.A., Lo K.J., Lee S.D., Coleman M.S., Furie B. Des-gamma-carboxy (abnormal) prothrombin as a serum marker of primary hepatocellular carcinoma. N. Engl. J. Med. 1984;310:1427–1431. doi: 10.1056/NEJM198405313102204. [DOI] [PubMed] [Google Scholar]
- 11.Grandhi M.S., Kim A.K., Ronnekleiv-Kelly S.M., Kamel I.R., Ghasebeh M.A., Pawlik T.M. Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol. 2016;25:74–85. doi: 10.1016/j.suronc.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Johnson P.J., Pirrie S.J., Cox T.F., Berhane S., Teng M., Palmer D., Morse J., Hull D., Patman G., Kagebayashi C., et al. The detection of hepatocellular carcinoma using a prospectively developed and validated model based on serological biomarkers. Cancer Epidemiol. Biomarkers Prev. 2014;23:144–153. doi: 10.1158/1055-9965.EPI-13-0870. [DOI] [PubMed] [Google Scholar]
- 13.Tellapuri S., Sutphin P.D., Beg M.S., Singal A.G., Kalva S.P. Staging systems of hepatocellular carcinoma: a review. Indian J. Gastroenterol. 2018;37:481–491. doi: 10.1007/s12664-018-0915-0. [DOI] [PubMed] [Google Scholar]
- 14.Marrero J.A., Kulik L.M., Sirlin C.B., Zhu A.X., Finn R.S., Abecassis M.M., Roberts L.R., Heimbach J.K. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American association for the study of liver diseases. Hepatology. 2018;68:723–750. doi: 10.1002/hep.29913. [DOI] [PubMed] [Google Scholar]
- 15.Fan J., Yang G.S., Fu Z.R., Peng Z.H., Xia Q., Peng C.H., Qian J.M., Zhou J., Xu Y., Qiu S.J., et al. Liver transplantation outcomes in 1,078 hepatocellular carcinoma patients: a multi-center experience in Shanghai, China. J. Cancer Res. Clin. Oncol. 2009;135:1403–1412. doi: 10.1007/s00432-009-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tateishi R., Shiina S., Teratani T., Obi S., Sato S., Koike Y., Fujishima T., Yoshida H., Kawabe T., Omata M. Percutaneous radiofrequency ablation for hepatocellular carcinoma. An analysis of 1000 cases. Cancer. 2005;103:1201–1209. doi: 10.1002/cncr.20892. [DOI] [PubMed] [Google Scholar]
- 17.Llovet J.M., Castet F., Heikenwalder M., Maini M.K., Mazzaferro V., Pinato D.J., Pikarsky E., Zhu A.X., Finn R.S. Immunotherapies for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2022;19:151–172. doi: 10.1038/s41571-021-00573-2. [DOI] [PubMed] [Google Scholar]
- 18.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., de Oliveira A.C., Santoro A., Raoul J.L., Forner A., et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 19.Keating M.G., Santoro A. Sorafenib: a review of its use in advanced hepatocellular carcinoma. Drugs. 2009;69:223–240. doi: 10.2165/00003495-200969020-00006. [DOI] [PubMed] [Google Scholar]
- 20.Yarchoan M., Agarwal P., Villanueva A., Rao S., Dawson L.A., Llovet J.M., Finn R.S., Groopman J.D., El-Serag H.B., Monga S.P., et al. Recent developments and therapeutic strategies against hepatocellular carcinoma. Cancer Res. 2019;79:4326–4330. doi: 10.1158/0008-5472.CAN-19-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn R.S., Qin S., Ikeda M., Galle P.R., Ducreux M., Kim T.Y., Kudo M., Breder V., Merle P., Kaseb A.O., et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 2020;382:1894–1905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 22.Schulze K., Nault J.C., Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J. Hepatol. 2016;65:1031–1042. doi: 10.1016/j.jhep.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 23.Jin Y., Lee W.Y., Toh S.T., Tennakoon C., Toh H.C., Chow P.K., Chung A.Y., Chong S.S., Ooi L.L., Sung W.K., et al. Comprehensive analysis of transcriptome profiles in hepatocellular carcinoma. J. Transl. Med. 2019;17:273. doi: 10.1186/s12967-019-2025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Megger D.A., Naboulsi W., Meyer H.E., Sitek B. Proteome analyses of hepatocellular carcinoma. J Clin Transl Hepatol. 2014;2:23–30. doi: 10.14218/JCTH.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z.H., Yang D.L. Identification of a protein signature for predicting overall survival of hepatocellular carcinoma: a study based on data mining. BMC Cancer. 2020;20:720. doi: 10.1186/s12885-020-07229-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren L., Li C., Wang Y., Teng Y., Sun H., Xing B., Yang X., Jiang Y., He F. In vivo phosphoproteome analysis reveals kinome reprogramming in hepatocellular carcinoma. Mol. Cell. Proteomics. 2018;17:1067–1083. doi: 10.1074/mcp.RA117.000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Y., Sun A., Zhao Y., Ying W., Sun H., Yang X., Xing B., Sun W., Ren L., Hu B., et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567:257–261. doi: 10.1038/s41586-019-0987-8. [DOI] [PubMed] [Google Scholar]
- 28.Gu Y., Guo Y., Gao N., Fang Y., Xu C., Hu G., Guo M., Ma Y., Zhang Y., Zhou J., et al. The proteomic characterization of the peritumor microenvironment in human hepatocellular carcinoma. Oncogene. 2022;41:2480–2491. doi: 10.1038/s41388-022-02264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X., Xing X., Dowlut D., Zeng Y., Liu J., Liu X. Integrating phosphoproteomics into kinase-targeted cancer therapies in precision medicine. J. Proteonomics. 2019;191:68–79. doi: 10.1016/j.jprot.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 30.Moldogazieva N.T., Mokhosoev I.M., Zavadskiy S.P., Terentiev A.A. Proteomic profiling and artificial intelligence for hepatocellular carcinoma translational medicine. Biomedicines. 2021;9:159. doi: 10.3390/biomedicines9020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mani D.R., Krug K., Zhang B., Satpathy S., Clauser K.R., Ding L., Ellis M., Gillette M.A., Carr S.A. Cancer proteogenomics: current impact and future prospects. Nat. Rev. Cancer. 2022;22:298–313. doi: 10.1038/s41568-022-00446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Q., Zhu H., Dong L., Shi W., Chen R., Song Z., Huang C., Li J., Dong X., Zhou Y., et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179:561–577. doi: 10.1016/j.cell.2019.08.052. [DOI] [PubMed] [Google Scholar]
- 33.Ng C.K.Y., Dazert E., Boldanova T., Coto-Llerena M., Nuciforo S., Ercan C., Suslov A., Meier M.A., Bock T., Schmidt A., et al. Integrative proteogenomic characterization of hepatocellular carcinoma across etiologies and stages. Nat. Commun. 2022;13:2436. doi: 10.1038/s41467-022-29960-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khalil A., Elfert A., Ghanem S., Helal M., Abdelsattar S., Elgedawy G., Obada M., Abdel-Samiee M., El-Said H. The role of metabolomics in hepatocellular carcinoma. Egyptian Liver J. 2021;11:41. [Google Scholar]
- 35.Feng N., Yu F., Yu F., Feng Y., Zhu X., Xie Z., Zhai Y. Metabolomic biomarkers for hepatocellular carcinoma: a systematic review. Medicine (Baltim.) 2022;101 doi: 10.1097/MD.0000000000028510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrarini A., Di Poto C., He S., Tu C., Varghese R.S., Kara Balla A., Jayatilake M., Li Z., Ghaffari K., Fan Z., et al. Metabolomic analysis of liver tissues for characterization of hepatocellular carcinoma. J. Proteome Res. 2019;18:3067–3076. doi: 10.1021/acs.jproteome.9b00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sangineto M., Villani R., Cavallone F., Romano A., Loizzi D., Serviddio G. Lipid metabolism in development and progression of hepatocellular carcinoma. Cancers. 2020;12:1419. doi: 10.3390/cancers12061419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan S.L.W., Israeli E., Ericksen R.E., Chow P.K.H., Han W. The altered lipidome of hepatocellular carcinoma. Semin. Cancer Biol. 2022;86:445–456. doi: 10.1016/j.semcancer.2022.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Beyoğlu D., Idle J.R. Metabolomic and lipidomic biomarkers for premalignant liver disease diagnosis and therapy. Metabolites. 2020;10:50. doi: 10.3390/metabo10020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabregat A., Sidiropoulos K., Viteri G., Marin-Garcia P., Ping P., Stein L., D'Eustachio P., Hermjakob H. Reactome diagram viewer: data structures and strategies to boost performance. Bioinformatics. 2018;34:1208–1214. doi: 10.1093/bioinformatics/btx752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiredja D.D., Koyutürk M., Chance M.R. The KSEA App: a web-based tool for kinase activity inference from quantitative phosphoproteomics. Bioinformatics. 2017;33:3489–3491. doi: 10.1093/bioinformatics/btx415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S., Zheng W., Hu L., Gong M., Yang H. MixProTool: a powerful and comprehensive web tool for analyzing and visualizing multigroup proteomics data. J. Comput. Biol. 2018;25:1123–1127. doi: 10.1089/cmb.2018.0050. [DOI] [PubMed] [Google Scholar]
- 47.Dacol E.C., Wang S., Chen Y., Lepique A.P. The interaction of SET and protein phosphatase 2A as target for cancer therapy. Biochim. Biophys. Acta Rev. Canc. 2021;1876 doi: 10.1016/j.bbcan.2021.188578. [DOI] [PubMed] [Google Scholar]
- 48.Muto S., Senda M., Akai Y., Sato L., Suzuki T., Nagai R., Senda T., Horikoshi M. Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:4285–4290. doi: 10.1073/pnas.0603762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bayarkhangai B., Noureldin S., Yu L., Zhao N., Gu Y., Xu H., Guo C. A comprehensive and perspective view of oncoprotein SET in cancer. Cancer Med. 2018;7:3084–3094. doi: 10.1002/cam4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buzón P., Velázquez-Cruz A., Corrales-Guerrero L., Díaz-Quintana A., Díaz-Moreno I., Roos W.H. The histone chaperones SET/TAF-1β and NPM1 exhibit conserved functionality in nucleosome remodeling and histone eviction in a cytochrome c-dependent manner. Adv. Sci. 2023;10 doi: 10.1002/advs.202301859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmitz M., Held M., Janssens V., Hutchins J., Hudecz O., Ivanova E., Goris J., Trinkle-Mulcahy L., Lamond A., Poser I., et al. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 2010;12:886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon D., Wilson K. Partners and post-translational modifications of nuclear lamins. Chromosoma. 2013;122:13–31. doi: 10.1007/s00412-013-0399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edwards N.J., Oberti M., Thangudu R.R., Cai S., McGarvey P.B., Jacob S., Madhavan S., Ketchum K.A. The CPTAC data portal: a resource for cancer proteomics research. J. Proteome Res. 2015;14:2707–2713. doi: 10.1021/pr501254j. [DOI] [PubMed] [Google Scholar]
- 54.Timsit Y.E., Negishi M. CAR and PXR: the xenobiotic-sensing receptors. Steroids. 2007;72:231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soll C., Jang J.H., Riener M.O., Moritz W., Wild P.J., Graf R., Clavien P.A. Serotonin promotes tumor growth in human hepatocellular cancer. Hepatology. 2010;51:1244–1254. doi: 10.1002/hep.23441. [DOI] [PubMed] [Google Scholar]
- 56.Carter A.M., Tan C., Pozo K., Telange R., Molinaro R., Guo A., De Rosa E., Martinez J.O., Zhang S., Kumar N., et al. Phosphoprotein-based biomarkers as predictors for cancer therapy. Proc Nal Acad Sci. 2020;117:18401–18411. doi: 10.1073/pnas.2010103117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henriques A.G., Müller T., Oliveira J.M., Cova M., da Cruz E Silva C.B., da Cruz E Silva O.A. Altered protein phosphorylation as a resource for potential AD biomarkers. Sci. Rep. 2016;6 doi: 10.1038/srep30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meier F., Brunner A.D., Koch S., Koch H., Lubeck M., Krause M., Goedecke N., Decker J., Kosinski T., Park M.A., et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics. 2018;17:2534–2545. doi: 10.1074/mcp.TIR118.000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliinyk D., Meier F. Ion mobility-resolved phosphoproteomics with dia-PASEF and short gradients. Proteomics. 2023;23 doi: 10.1002/pmic.202200032. [DOI] [PubMed] [Google Scholar]
- 60.Yue X., Schunter A., Hummon A.B. Comparing multistep immobilized metal affinity chromatography and multistep TiO2 methods for phosphopeptide enrichment. Anal. Chem. 2015;87:8837–8844. doi: 10.1021/acs.analchem.5b01833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian I., Verma S., Kumar S., Jere A., Anamika K. Multi-omics data integration, interpretation, and its application. Bioinform Biol Insights. 2020;14 doi: 10.1177/1177932219899051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinberg G.R., Carling D. AMP-activated protein kinase: the current landscape for drug development. Nat. Rev. Drug Discov. 2019;18:527–551. doi: 10.1038/s41573-019-0019-2. [DOI] [PubMed] [Google Scholar]
- 63.Meng S.S., Gu H.W., Zhang T., Li Y.S., Tang H.B. Gradual deterioration of fatty liver disease to liver cancer via inhibition of AMPK signaling pathways involved in energy-dependent disorders, cellular aging, and chronic inflammation. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1099624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee M., Yoon J.H. Metabolic interplay between glycolysis and mitochondrial oxidation: the reverse Warburg effect and its therapeutic implication. World J. Biol. Chem. 2015;6:148–161. doi: 10.4331/wjbc.v6.i3.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen H., Wu Q., Peng L., Cao T., Deng M., Liu Y., Huang J., Hu Y., Fu N., Zhou K., et al. Mechanism, clinical significance, and treatment strategy of Warburg effect in hepatocellular carcinoma. J. Nanomater. 2021;2021 [Google Scholar]
- 66.Feng J., Li J., Wu L., Yu Q., Ji J., Wu J., Dai W., Guo C. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2020;39:126. doi: 10.1186/s13046-020-01629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang L., Venneti S., Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu. Rev. Biomed. Eng. 2017;19:163–194. doi: 10.1146/annurev-bioeng-071516-044546. [DOI] [PubMed] [Google Scholar]
- 68.Bartman C.R., Weilandt D.R., Shen Y., Lee W.D., Han Y., TeSlaa T., Jankowski C.S.R., Samarah L., Park N.R., da Silva-Diz V., et al. Slow TCA flux and ATP production in primary solid tumours but not metastases. Nature. 2023;614:349–357. doi: 10.1038/s41586-022-05661-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ye X., Weinberg R.A. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du R., Huang C., Liu K., Li X., Dong Z. Targeting AURKA in Cancer: molecular mechanisms and opportunities for Cancer therapy. Mol. Cancer. 2021;20:15. doi: 10.1186/s12943-020-01305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen L., Guo P., He Y., Chen Z., Chen L., Luo Y., Qi L., Liu Y., Wu Q., Cui Y., et al. HCC-derived exosomes elicit HCC progression and recurrence by epithelial-mesenchymal transition through MAPK/ERK signaling pathway. Cell Death Dis. 2018;9:513. doi: 10.1038/s41419-018-0534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi C., Chen Y., Chen Y., Yang Y., Bing W., Qi J. CD4+ CD25+ regulatory T cells promote hepatocellular carcinoma invasion via TGF-β1-induced epithelial-mesenchymal transition. OncoTargets Ther. 2018;12:279–289. doi: 10.2147/OTT.S172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wong C.C., Wong C.M., Tung E.K., Man K., Ng I.O. Rho-kinase 2 is frequently overexpressed in hepatocellular carcinoma and involved in tumor invasion. Hepatology. 2009;49:1583–1594. doi: 10.1002/hep.22836. [DOI] [PubMed] [Google Scholar]
- 74.McGranahan N., Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 75.Lu Y., Lin N., Chen Z., Xu R. Hypoxia-induced secretion of platelet-derived growth factor-BB by hepatocellular carcinoma cells increases activated hepatic stellate cell proliferation, migration and expression of vascular endothelial growth factor-A. Mol. Med. Rep. 2015;11:691–697. doi: 10.3892/mmr.2014.2689. [DOI] [PubMed] [Google Scholar]
- 76.Kroll J., Epting D., Kern K., Dietz C.T., Feng Y., Hammes H.P., Wieland T., Augustin H.G. Inhibition of Rho-dependent kinases ROCK I/II activates VEGF-driven retinal neovascularization and sprouting angiogenesis. Am. J. Physiol. Heart Circ. Physiol. 2009;296:H893–H899. doi: 10.1152/ajpheart.01038.2008. [DOI] [PubMed] [Google Scholar]
- 77.Liu Q., Li J., Zhang W., Xiao C., Zhang S., Nian C., Li J., Su D., Chen L., Zhao Q., et al. Glycogen accumulation and phase separation drives liver tumor initiation. Cell. 2021;184:5559–5576.e19. doi: 10.1016/j.cell.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 78.Li H., Rong Z., Wang H., Zhang N., Pu C., Zhao Y., Zheng X., Lei C., Liu Y., Luo X., et al. Wang J. Proteomic analysis revealed common, unique and systemic signatures in gender-dependent hepatocarcinogenesis. Biol. Sex Differ. 2020;11:46. doi: 10.1186/s13293-020-00316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available at the Massive website (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with the ID number of MSV000091782.