Highlights
-
•
Analysis of residual host cell proteins (HCPs) is critical for biopharmaceuticals production and process development.
-
•
LC-MS is an orthogonal method for identifying and profiling HCPs in downstream streams.
-
•
HCP profile from intermediate crude extract to the final drug substance indicated successful removal of undesirable HCPs during the purification process.
-
•
LC-MS provides detailed insights into individual HCPs and guides process development efforts.
Keywords: Host cell proteins, Process-related impurities, LC-MS, Plant-produced biopharmaceuticals, Nicotiana benthamiana
Abstract
Host cell proteins (HCPs) are process-related impurities found in biopharmaceutical products that can impair their safety and efficacy. While ELISA has traditionally been employed to quantify HCPs, LC-MS emerges as a powerful alternative for precise identification of individual HCPs. In this study, we used LC-MS for profiling HCPs from Nicotiana benthamiana-derived biopharmaceuticals. Our approach involved rigorous false discovery rate control to ensure data integrity and reliability. Comprehensive analysis revealed a systematic reduction of HCPs following purification, demonstrating the efficiency of purification processes in removing non-essential proteins. Furthermore, LC-MS enabled the identification of potential contaminants, refining purification strategies and improving product purity and integrity. Our findings highlight the potential of LC-MS as an analytical tool for HCPs analysis in biopharmaceutical development and manufacturing. By providing detailed insights into HCPs profiles and contaminants, LC-MS facilitates informed decision-making in downstream processing steps, benefiting product quality, patient safety, and the biopharmaceutical sector.
1. Introduction
Biotherapeutics, also known as biopharmaceuticals or biological drugs, are pharmaceutical products derived from living organisms or their components. Unlike traditional small-molecule drugs that have been synthesized chemically, biotherapeutics are large, complex molecules typically produced through biotechnological processes [1]. These advanced therapeutic agents encompass various products, including proteins, peptides, antibodies, nucleic acids, and cell-based therapies [1]. They are designed to target specific molecules, pathways, or cells in the body to treat diseases at their root causes, offering novel approaches to addressing medical conditions ranging from cancer [2,3], autoimmune disorders [4,5], and infectious diseases [6,7].
Plant-based expression systems have been explored for the large-scale production of therapeutic proteins for >20 years. These systems can synthesize complex proteins with post-translational modifications and are highly scalable.
However, they face significant challenges, such as maintaining controlled growth conditions and the downstream processing of plant crude extracts. Achieving desirable purity is difficult because plants typically produce more debris than other expression systems, especially those in the tobacco family, which have significant secondary metabolite content [8]. As a result, clarifying plant crude extracts with a single round of filtering or one-step chromatography may not be sufficient, potentially leading to contaminated drug substances with host cell proteins.
The presence of residual HCPs in biological products is considered as a critical quality attribute due to their potential to impact product quality, efficacy, and safety [[9], [10], [11]]. These proteins may induce unwanted immunogenicity, alter product stability, or impair therapeutic efficacy, highlighting the importance of efficiently reducing their levels during downstream processing. Traditionally, ELISA has been the standard method for quantifying total HCPs levels in bioprocess samples [12,13]. However, with the recent emergence of plant expression platforms, the availability of commercial ELISA kits remains limited. Consequently, many plant-based pharmaceutical companies utilizing these platforms find themselves compelled to independently develop and validate their ELISA kits, a process that is both expensive and time-consuming. Meanwhile, LC-MS has emerged as an orthogonal method gaining importance for HCPs profiling and quantitation analysis. LC-MS offers distinct advantages over ELISA, notably providing deeper insights into the properties of individual HCPs and facilitating more precise identification and characterization [[13], [14], [15]].
Despite its advantages, the application of LC-MS in HCPs analysis poses challenges, particularly in achieving the wide dynamic range required to detect HCPs at low concentrations in the presence of dominant therapeutic proteins. Various strategies have been explored to solve this issue, including optimizing sample preparation techniques such as online or offline fractionation, depletion of monoclonal antibodies using affinity-based methods or molecular weight cutoff strategies, and employing enrichment methods for HCPs [11,[16], [17], [18]]. Furthermore, these challenges include ion suppression effects, matrix interference [19], and the need for robust data processing algorithms to accurately identify and quantify individual HCPs species. Addressing these challenges demands ongoing research and innovation in sample preparation, chromatographic separation, mass spectrometry instrumentation, and data analysis techniques.
The data generated from our study can be utilized in strategic planning for optimizing purification processes and enhancing HCP clearance in biologics manufacturing.
2. Material and methods
2.1. HCPs library preparation
Leaves of N. benthamiana were ground and lysed in a solution containing 0.2 % sodium dodecyl sulfate (SDS) (Amresco, USA), 20 mM dithiothreitol (DTT) (USB, USA), 100 mM NaCl (Bio basic, USA), and 50 mM Tris–HCl, pH 8.0 (Bio basic, USA). The proteins were precipitated in a cold acetone solution at a 1:5 (v/v) ratio and resolubilized with 0.25 % Rapidgest SF (Waters, USA) in 20 mM ammonium bicarbonate (Sigma Aldrich, Germany). Protein concentrations of the lysates were determined using the Bradford Reagent protein assay (Sigma Aldrich, Germany).
Protein sample (25 μg) was reduced with 4 mM DTT at 72 °C for 30 min, then alkylated with 12 mM iodoacetamide (IAA) (GE Healthcare, UK) at room temperature in the dark for 30 min. The sample was desalted using a Zeba spin desalting column (Thermo Scientific, Sweden) before digestion with trypsin (Thermo Scientific, Lithuania) at a protein-to-trypsin ratio of 1:50 (w/w) at 37 °C overnight. The solution was further dried and reconstituted with 0.1 % formic acid (Sigma Aldrich, Germany) in LC-MS grade water (Supelco®, Germany).
2.2. Intermediate crude extract and Drug substance preparation
The recombinant SARS-CoV-2 RBD-Fc protein was previously generated and produced in plants [20]. Briefly, the receptor binding domain (RBD) of SARS-CoV-2 was fused with the Fc region of human immunoglobulin G1 (IgG1) (GenBank accession number: 4CDH_A), and then cloned into a plant expression geminiviral vector [21]. This construct was subsequently introduced into N. benthamiana plants via transient expression method. After expression, the recombinant RBD-Fc protein was extracted from the plants and clarified through filtration and centrifugation, resulting in an intermediate crude extract (ICE). The crude extract was purified using protein A affinity resin MabSelectSURE® (Cytiva, Buckinghamshire, UK) to obtain the drug substance (DS), which was subsequently formulated into the drug product without additional purification steps. Both the ICE and the DS were digested with trypsin, following the protocol described above. The recombinant SARS-CoV-2 RBD-Fc protein utilized in this study serves both as a model for evaluating downstream processing in plant-based biopharmaceutical production and as a COVID-19 vaccine which currently undergoing human clinical trials (NCT04953078 and NCT05197712).
2.3. LC-MS and SWATH analysis
Protein sample (1 µg) was loaded into a nanoLC system (Thermo Scientific, Germany) onto a trap column (300 µm i.d. x 5 mm, packed with 5 µm C18 100 Å PepMap™) (Thermo Scientific, Germany), where desalting occurred with 2 % acetonitrile (ACN) (VWR, France) and 0.05 % trifluoroacetic acid (TFA) (Sigma Aldrich, Germany) at a flow rate of 10 µl/min for 3 min. Subsequently, peptides were separated using an analytical column (75 µm i.d. x 15 cm, packed with Acclaim PepMap™ C18) (Thermo Scientific, Germany) at a flow rate of 300 nl/min.
Peptide elution was achieved with a linear gradient of 3–35 % buffer B in buffer A over 92 min (A: 0.1 % formic acid (FA) in water; B: 0.1 % FA in 80 % ACN). The eluted peptides were then analyzed on a 6600+ TripleTOF LC-MS/MS system (AB SCIEX, Germany). The mass spectrometry data acquisition was set from gradient time zero to 120 min, with MS1 spectra collected in the mass range of 400 – 1500 m/z for 250 ms in "high sensitivity" mode. Each MS1 spectrum was further fragmented using up to 30 precursors per cycle. Switch criteria included a charge of 2+ to 5+, a 500 cps intensity threshold, and dynamic exclusion for 15 s. All of the samples were analyzed in triplicate.
2.4. Data processing
The spectral library was processed using ProteinPilot™ Software 5.0.2 (AB SCIEX, Germany) against the Nicotiana tabacum database from UniProtKB, which is currently available, unlike N. benthamiana, which lacks a comprehensive database. Proteins identified by LC-MS/MS data in each pooled tobacco sample with unused scores above 0.05 (indicating > 95 % confidence) and a false discovery rate (FDR) below 1 % were considered significant and included in the next analysis.
For SWATH-MS data analysis, PeakView 2.2 software (AB SCIEX, Germany) was employed. The library spectra generated were used as the database for SWATH analysis. Data processing entailed utilizing an extracted ion chromatogram (XIC) extraction window of 5 min and an XIC width of 75 ppm. Peak areas from peptides exhibiting >95 % confidence and <1 % global FDR were extracted using MarkerView v1.3.0 (AB SCIEX, Germany).
3. Results
3.1. Generation of host cell proteins spectral library
We used the crude extract of whole leaf samples, where total proteins were digested and then analyzed using the LC-MS/MS technique. To generate the library search, results of peptides were searched against the NCBI database for N. tabacum. We also employed SWATH DIA, which has higher sensitivity compared to conventional data-dependent acquisition (DDA) [15,22,23]. The processing of spectral library aimed to generate comprehensive lists of high-confidence proteins, which are crucial for avoiding biased interpretations and ensuring the reliability of downstream analyses. Effective FDR control is imperative to maintain the integrity and validity of data obtained from proteomic experiments [24]. In this study, a stringent FDR threshold of <0.01 (1 %) was applied, thereby defining confidently identified peptides and proteins for subsequent analysis. Supplementary Table S1 shows the identification of 318 peptides with an FDR <1 %. Data that met these stringent criteria underwent further statistical analysis, including Venn diagram analysis to gain a profound understanding of the underlying patterns and relationships within the dataset.
3.2. Matching ICE and DS to the HCP library
The Venn diagram analysis provided valuable information about the distribution of identified proteins across the experimental groups. Among the shared proteins, 163 were consistently detected in both groups, indicating robust identification across experimental conditions. However, the presence of 12 unique proteins solely in the high confidence proteins group suggests that there may be discrepancies in the extraction or classification methods used for this group (Fig. 1 and Table 1).
Fig. 1.
(A) Venn diagram illustrating the numbers of proteins identified in host cell proteins (HCPs), intermediate crude extract (ICE), and drug substance (DS). (B) Distribution of protein numbers identified.
Table 1.
List of 12 proteins in the high confidence proteins group.
| UniProtKB | Protein names | p-value |
|---|---|---|
| Q8S950 | Kinesin-like protein NACK1 OS=Nicotiana tabacum OX=4097 GN NACK1 PE=1 SV=1 | 0.001 |
| P93354 | Histone H2B OS=Nicotiana tabacum OX=4097 GN HIS2B PE=2 SV=3 | 0.003 |
| Q7FNT3 | Photosystem II protein D1 OS=Atropa belladonna OX=33,113 GN=psbA PE=3 SV=1 | 0.005 |
| Q3C1G3 | Photosystem II protein D1 OS=Nicotiana sylvestris OX=4096 GN=psbA PE=3 SV=1 | 0.005 |
| Q33C59 | Photosystem II protein D1 OS=Nicotiana tomentosiformis OX=4098 GN=psbA PE=3 SV=1 | 0.005 |
| P69556 | Photosystem II protein D1 OS=Nicotiana tabacum OX=4097 GN=psbA PE=3 SV=2 | 0.005 |
| Q76MV0 | Histone H3.2 OS=Nicotiana tabacum OX=4097 GN=B34 PE=1 SV=1 | 0.008 |
| Q76N23 | Histone H3.3 OS=Nicotiana tabacum OX=4097 GN H3 PE=1 SV=1 | 0.008 |
| Q3C1G9 | DNA-directed RNA polymerase subunit beta OS=Nicotiana sylvestris OX=4096 GN=rpoC2 PE=3 SV=1 | 0.017 |
| Q33C48 | DNA-directed RNA polymerase subunit beta OS=Nicotiana tomentosiformis OX=4098 GN=rpoC2 PE=3 SV=1 | 0.017 |
| P38550 | DNA-directed RNA polymerase subunit beta OS=Nicotiana tabacum OX=4097 GN=rpoC2 PE=3 SV=2 | 0.017 |
| Q8S8Y1 | DNA-directed RNA polymerase subunit beta'' OS=Atropa belladonna OX=33,113 GN=rpoC2 PE=3 SV=1 | 0.017 |
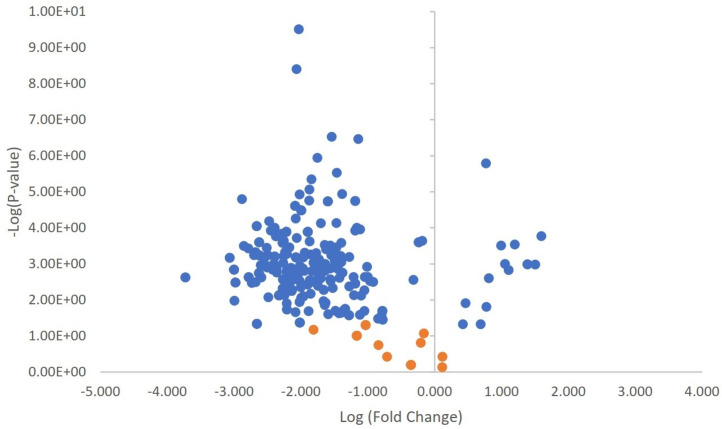
Expanding our analysis, a comprehensive investigation into the HCPs profiles of ICE showed a consistent trend across multiple datasets. Notably, the majority of proteins (n = 286) identified in a less stringent set with a p-value < 0.05 showed a reducing pattern (Fig. 2). These findings suggest a decrease in the levels of these HCPs in the ICE samples. In total, we identified 305 proteins from the ICE sample. We then purified the ICE sample using a single-step protein A chromatography process, which effectively eliminated 142 proteins. Moreover, we did not observe any proteins that were uniquely present in both groups. Despite the lack of unique protein identifications in the DS groups, the presence of recognized proteins in the ICE group underscores the efficiency of the purification process in removing a significant amount of impurity proteins, thereby improving the overall quality and purity of the drug substance.
Fig. 2.
Volcano plot of quantitative ICE data. The plot denotes statistical significance plotted against protein ratio intensity on a log2 scale. The X-axis represents log-transformed fold change in protein abundance, with positive values indicating increased levels and negative values indicating decreased levels. The Y-axis shows the negative logarithm of the p-value, where higher values indicate greater statistical significance. Proteins with p-values < 0.05 are depicted in blue, while those with p-values > 0.05 are shown in orange.
After the DS analysis, 163 proteins were identified, and their relative amount compared to ICE were uniformly downregulated (Fig. 3 and Supplementary Table S2). This consistent reducing pattern across various datasets underscores the reliability and robustness of our findings. In pharmaceutical purification processes, proteins can nonspecifically bind to various components of the purification system. In addition, prior studies have found that the co-elution of HCPs is primarily caused by their association with the monoclonal antibody on agarose-based protein A resins, rather than a direct interaction with the resins [[25], [26], [27], [28]]. Such interactions can prevent the target protein from being isolated, potentially affecting downstream applications. From this data, we were able to identify the top 10 high intensity non-specifically co-eluting proteins, as well as the fold changes in intensity when compared to ICE (Table 2).
Fig. 3.
Volcano plot of quantitative DS data. Data statistical significance is plotted in function of the protein ratio intensity in a log2 scale. The X-axis represents log-transformed fold change in protein abundance, with positive values indicating increased levels and negative values indicating decreased levels. The Y-axis shows the negative logarithm of the p-value, where higher values indicate greater statistical significance. Proteins with p-values 〈 0.05 are depicted in blue, while those with p-values 〉 0.05 are shown in orange.
Table 2.
Identification of the Top 10 Proteins Co-eluting during protein A Chromatography.
| UniProtKB | Protein names | p-value | Fold Change |
|---|---|---|---|
| P09042 | Pathogenesis-related protein 1C OS=Nicotiana tabacum OX=4097 PE=2 SV=3 | 0.012 | 0.0392 |
| P08299 | Pathogenesis-related protein 1A OS=Nicotiana tabacum OX=4097 PE=1 SV=1 | 0.004 | 0.0297 |
| P07053 | Pathogenesis-related protein 1B OS=Nicotiana tabacum OX=4097 PE=2 SV=1 | 0.004 | 0.0297 |
| P07052 | Pathogenesis-related protein R minor form OS=Nicotiana tabacum OX=4097 PE=2 SV=1 | 0.012 | 0.0235 |
| Q3C1J3 | Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, chloroplastic OS=Nicotiana sylvestris OX=4096 GN=accD PE=3 SV=2 | 0.044 | 0.0234 |
| Q33C25 | Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, chloroplastic OS=Nicotiana tomentosiformis OX=4098 GN=accD PE=3 SV=2 | 0.044 | 0.0234 |
| P12219 | Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, chloroplastic OS=Nicotiana tabacum OX=4097 GN=accD PE=1 SV=2 | 0.044 | 0.0234 |
| P15797 | Glucan endo-1,3-beta-glucosidase, basic vacuolar isoform OS=Nicotiana tabacum OX=4097 PE=1 SV=2 | 0.001 | 0.0221 |
| P23546 | Glucan endo-1,3-beta-glucosidase, basic vacuolar isoform GGIB50 OS=Nicotiana tabacum OX=4097 PE=1 SV=1 | 0.001 | 0.0221 |
| P27666 | Glucan endo-1,3-beta-glucosidase, basic vacuolar isoform GLB OS=Nicotiana tabacum OX=4097 PE=2 SV=1 | 0.001 | 0.0221 |
Most proteins identified in the DS belong to the photosynthesis-related and coenzyme groups.
Hydrolytic degradation is frequently linked to residual HCPs impurities originating from the upstream process [[29], [30], [31], [32]]. These HCPs can enzymatically hydrolyze the ester bonds in polysorbate, leading to the release of free fatty acids that accumulate during drug product storage. This enzymatic activity can degrade polysorbate to a degree where it fails to adequately protect the protein, potentially causing particle formation or other adverse effects [33,34]. Interestingly, no lipase, esterase, and proteases were found, which are enzymes known to affect the stability of the DS and final product, including excipients and some stabilizers [11,29,35,36]. This is important because it implies that the DS may be more stable, as it lacks enzymes that could cause unwanted reactions or degrade the final product. However, it is imperative to conduct additional comprehensive stability studies to ensure the reliability and consistency of the results. Further method development for sample preparation and instrument conditions could broaden the coverage of the HCPs library, maximizing the detection of potential impurities and increasing the sensitivity and accuracy of the analysis. This enhancement allows the identification and resolution of a wider spectrum of HCP-related challenges during the purification process, leading to the production of biopharmaceuticals with higher purity, stability, and safety profiles.
4. Discussion
Reducing HCPs to an acceptable level in biotherapeutic products is a core goal in downstream purification processes to guarantee safety and efficacy of final product. While ELISA is commonly utilized to quantify total HCPs levels, LC-MS analysis provides a more in-depth understanding by allowing for the identification and quantification of individual HCP. The HCPs profiling method and Venn diagram analysis presented a visual representation of the overlap and distinctiveness of proteins identified across three experimental groups. This graphical approach simplified the identification of proteins shared by multiple groups, as well as those unique in specific conditions.
In this study, we successfully identified and profiled HCPs from N. benthamiana by using N. tabacum as a spectral reference, given its close relationship to N. benthamiana and ready availability in the database. We demonstrated that relative quantification is useful in designing the purification process, but absolute levels are critical to ensure the safety of human therapeutics. We comprehensively investigated and interpreted the proteomic data by integrating multiple analytical approaches, including stringent FDR control. FDR analysis in proteomic results is critical for ensuring the quality and accuracy of protein identifications. Typically, FDR assessment is conducted using a target-decoy search strategy, where each peptide-spectrum match is assigned a score based on the frequency of false discoveries in peptide identifications [37,38]. After the HCPs library was established, peptides from the ICE and DS were searched against the HCPs library.
The information obtained from this analysis is important for determining experiment design. These purification steps played a pivotal role in enhancing the purity and specificity of the target protein, thereby reducing sample complexity and improving overall product quality. Depending on the extent of non-specific binding observed, additional purification steps may be necessary to ensure target protein purity and integrity. Conversely, if non-specific binding is minimal and meets the required standards for intended use, it may be feasible to proceed without further purification, thus saving time and resources. One of our examples is the Baiya SARS-CoV-2 vaccine, which was developed by Baiya Phytopharm – a Thailand-based biotech company. It utilized a single-step purification process based on comprehensive data analysis in its manufacturing process. The vaccine has shown safety and high efficacy in preclinical studies [39,40] and is currently in phase 1 clinical trials (NCT04953078 and NCT05197712).
While accurate identification of co-eluting proteins provides invaluable information into the purification process and aids informed decision-making regarding subsequent HCPs clearance strategies, the use of a triple quadrupole mass detector (QqQ) in conjunction with isotope-labeled peptides [41] or isobaric tags [42] could enhance the validation and confirmation of proteins identified through proteomic profiling, given its high sensitivity and accuracy [43,44], especially for absolute quantification purposes. However, implementing this technique requires time for optimization and validation to ensure the reliability of results.
5. Conclusion
In conclusion, the LC-MS profiling methods presented in this study have high sensitivity and exhibit promise for becoming routine analytical techniques in a wide range of biopharmaceutical applications. Their sensitivity and versatility make them valuable tools for characterizing and monitoring the quality of biopharmaceutical products, contributing to advancements in drug development and manufacturing processes.
CRediT authorship contribution statement
Chalisa Panapitakkul: Writing – review & editing, Writing – original draft, Investigation, Conceptualization. Christine Joy I. Bulaon: Writing – original draft, Writing – review & editing. Nuttapat Pisuttinusart: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Waranyoo Phoolcharoen: Writing – review & editing, Writing – original draft, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Waranyoo Phoolcharoen from Chulalongkorn University is a founder/shareholder of Baiya Phytopharm Co., Ltd., Thailand. Chalisa Panapitakkul, Christine Joy I. Bulaon, and Nuttapat Pisuttinusart are employees of Baiya Phytopharm Co., Ltd.
Acknowledgments
Acknowledgments
We appreciate the technical assistance provided by the technicians and staff during the experimental study.
Funding
This project is funded by National Research Council of Thailand (NRCT) and Chulalonkorn University (Grant No. N42A670577).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.btre.2024.e00856.
Contributor Information
Nuttapat Pisuttinusart, Email: nuttapat.nut@gmail.com.
Waranyoo Phoolcharoen, Email: waranyoo.p@chula.ac.th.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.Morrow T., Felcone L.H. Defining the difference: What Makes Biologics Unique. Biotechnol. Healthc. 2004;1(4):24–29. [PMC free article] [PubMed] [Google Scholar]
- 2.Sahu M., Suryawanshi H. Immunotherapy: The future of cancer treatment. J. Oral Maxillofac. Pathol. 2021;25(2):371. doi: 10.4103/0973-029X.325257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esfahani K., et al. A review of cancer immunotherapy: from the past, to the present, to the future. Curr. Oncol. 2020;27(Suppl 2):S87–s97. doi: 10.3747/co.27.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raychaudhuri S.P., Raychaudhuri S.K. Biologics: target-specific treatment of systemic and cutaneous autoimmune diseases. Indian J. Dermatol. 2009;54(2):100–109. doi: 10.4103/0019-5154.53175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yasunaga M. Antibody therapeutics and immunoregulation in cancer and autoimmune disease. Semin. Cancer Biol. 2020;64:1–12. doi: 10.1016/j.semcancer.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Hammitt L.L., et al. Nirsevimab for Prevention of RSV in Healthy Late-Preterm and Term Infants. N. Engl. J. Med. 2022;386(9):837–846. doi: 10.1056/NEJMoa2110275. [DOI] [PubMed] [Google Scholar]
- 7.Waickman A.T., et al. Biologics for dengue prevention: up-to-date. Expert. Opin. Biol. Ther. 2023;23(1):73–87. doi: 10.1080/14712598.2022.2151837. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q., Davis K.R. The potential of plants as a system for the development and production of human biologics. F1000Res. 2016:5. doi: 10.12688/f1000research.8010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Hunter A.K., Mozier N.M. Host cell proteins in biologics development: Identification, quantitation and risk assessment. Biotechnol. Bioeng. 2009;103(3):446–458. doi: 10.1002/bit.22304. [DOI] [PubMed] [Google Scholar]
- 10.Goey C.H., Alhuthali S., Kontoravdi C. Host cell protein removal from biopharmaceutical preparations: Towards the implementation of quality by design. Biotechnol. Adv. 2018;36(4):1223–1237. doi: 10.1016/j.biotechadv.2018.03.021. [DOI] [PubMed] [Google Scholar]
- 11.Ji Q., et al. A highly sensitive and robust LC-MS platform for host cell protein characterization in biotherapeutics. Biologicals. 2023;82 doi: 10.1016/j.biologicals.2023.101675. [DOI] [PubMed] [Google Scholar]
- 12.Seisenberger C., et al. Questioning coverage values determined by 2D western blots: A critical study on the characterization of anti-HCP ELISA reagents. Biotechnol. Bioeng. 2021;118(3):1116–1126. doi: 10.1002/bit.27635. [DOI] [PubMed] [Google Scholar]
- 13.Guo J., et al. Technical advancement and practical considerations of LC-MS/MS-based methods for host cell protein identification and quantitation to support process development. MAbs. 2023;15(1) doi: 10.1080/19420862.2023.2213365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trauchessec M., et al. An innovative standard for LC-MS-based HCP profiling and accurate quantity assessment: Application to batch consistency in viral vaccine samples. Proteomics. 2021;21(5) doi: 10.1002/pmic.202000152. [DOI] [PubMed] [Google Scholar]
- 15.Kreimer S., et al. Host Cell Protein Profiling by Targeted and Untargeted Analysis of Data Independent Acquisition Mass Spectrometry Data with Parallel Reaction Monitoring Verification. Anal. Chem. 2017;89(10):5294–5302. doi: 10.1021/acs.analchem.6b04892. [DOI] [PubMed] [Google Scholar]
- 16.Doneanu C., et al. Analysis of host-cell proteins in biotherapeutic proteins by comprehensive online two-dimensional liquid chromatography/mass spectrometry. MAbs. 2012;4(1):24–44. doi: 10.4161/mabs.4.1.18748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schenauer M.R., Flynn G.C., Goetze A.M. Identification and quantification of host cell protein impurities in biotherapeutics using mass spectrometry. Anal. Biochem. 2012;428(2):150–157. doi: 10.1016/j.ab.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Doneanu C.E., et al. Enhanced detection of low-abundance host cell protein impurities in high-purity monoclonal antibodies down to 1 ppm using ion mobility mass spectrometry coupled with multidimensional liquid chromatography. Anal. Chem. 2015;87(20):10283–10291. doi: 10.1021/acs.analchem.5b02103. [DOI] [PubMed] [Google Scholar]
- 19.Furey A., et al. Ion suppression; A critical review on causes, evaluation, prevention and applications. Talanta. 2013;115:104–122. doi: 10.1016/j.talanta.2013.03.048. [DOI] [PubMed] [Google Scholar]
- 20.Siriwattananon K., et al. Plant-Produced Receptor-Binding Domain of SARS-CoV-2 Elicits Potent Neutralizing Responses in Mice and Non-human Primates. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.682953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Q., et al. Geminiviral vectors based on bean yellow dwarf virus for production of vaccine antigens and monoclonal antibodies in plants. Hum. Vaccin. 2011;7(3):331–338. doi: 10.4161/hv.7.3.14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heissel S., et al. Evaluation of spectral libraries and sample preparation for DIA-LC-MS analysis of host cell proteins: A case study of a bacterially expressed recombinant biopharmaceutical protein. Protein Expr. Purif. 2018;147:69–77. doi: 10.1016/j.pep.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Gillet L.C., et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell Proteomics. 2012;(6):11. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aggarwal S., Yadav A.K. False Discovery Rate Estimation in Proteomics. Methods Mol. Biol. 2016;1362:119–128. doi: 10.1007/978-1-4939-3106-4_7. [DOI] [PubMed] [Google Scholar]
- 25.Tarrant R.D., et al. Host cell protein adsorption characteristics during protein A chromatography. Biotechnol. Prog. 2012;28(4):1037–1044. doi: 10.1002/btpr.1581. [DOI] [PubMed] [Google Scholar]
- 26.Shukla A.A., Hinckley P. Host cell protein clearance during protein A chromatography: development of an improved column wash step. Biotechnol. Prog. 2008;24(5):1115–1121. doi: 10.1002/btpr.50. [DOI] [PubMed] [Google Scholar]
- 27.Nogal B., Chhiba K., Emery J.C. Select host cell proteins coelute with monoclonal antibodies in protein a chromatography. Biotechnol. Prog. 2012;28(2):454–458. doi: 10.1002/btpr.1514. [DOI] [PubMed] [Google Scholar]
- 28.Pinto N.D., et al. Immunoglobulin G elution in protein A chromatography employing the method of chromatofocusing for reducing the co-elution of impurities. Biotechnol. Bioeng. 2017;114(1):154–162. doi: 10.1002/bit.26053. [DOI] [PubMed] [Google Scholar]
- 29.Hall T., et al. Polysorbates 20 and 80 Degradation by Group XV Lysosomal Phospholipase A2 Isomer X1 in monoclonal antibody formulations. J. Pharm. Sci. 2016;105(5):1633–1642. doi: 10.1016/j.xphs.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 30.Dixit N., et al. Residual host cell protein promotes polysorbate 20 degradation in a sulfatase drug product leading to free fatty acid particles. J. Pharm. Sci. 2016;105(5):1657–1666. doi: 10.1016/j.xphs.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 31.Labrenz S.R. Ester hydrolysis of polysorbate 80 in mAb drug product: Evidence in support of the hypothesized risk after the observation of visible particulate in mAb formulations. J. Pharm. Sci. 2014;103(8):2268–2277. doi: 10.1002/jps.24054. [DOI] [PubMed] [Google Scholar]
- 32.Kovner D., et al. Characterization of recombinantly-expressed hydrolytic enzymes from chinese hamster ovary cells: Identification of host cell proteins that degrade polysorbate. J. Pharm. Sci. 2023;112(5):1351–1363. doi: 10.1016/j.xphs.2023.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Kannan A., et al. A Mechanistic Understanding of Monoclonal Antibody Interfacial Protection by Hydrolytically Degraded Polysorbate 20 and 80 under IV Bag Conditions. Pharm. Res. 2022;39(3):563–575. doi: 10.1007/s11095-022-03217-x. [DOI] [PubMed] [Google Scholar]
- 34.Doshi N., Martin J., Tomlinson A. Improving prediction of free fatty acid particle formation in biopharmaceutical drug products: Incorporating ester distribution during polysorbate 20 degradation. Mol. Pharm. 2020;17(11):4354–4363. doi: 10.1021/acs.molpharmaceut.0c00794. [DOI] [PubMed] [Google Scholar]
- 35.Li X., et al. The measurement and control of high-risk host cell proteins for polysorbate degradation in biologics formulation. Antib. Ther. 2022;5(1):42–54. doi: 10.1093/abt/tbac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X., et al. Profiling active enzymes for polysorbate degradation in biotherapeutics by activity-based protein profiling. Anal. Chem. 2021;93(23):8161–8169. doi: 10.1021/acs.analchem.1c00042. [DOI] [PubMed] [Google Scholar]
- 37.Lee S., Park H., Kim H. False discovery rate estimation using candidate peptides for each spectrum. BMC. Bioinformatics. 2022;23(1):454. doi: 10.1186/s12859-022-05002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta N., et al. Target-decoy approach and false discovery rate: when things may go wrong. J. Am. Soc. Mass Spectrom. 2011;22(7):1111–1120. doi: 10.1007/s13361-011-0139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phoolcharoen W., et al. Preclinical evaluation of immunogenicity, efficacy and safety of a recombinant plant-based SARS-CoV-2 RBD vaccine formulated with 3M-052-Alum adjuvant. Vaccine. 2023;41(17):2781–2792. doi: 10.1016/j.vaccine.2023.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shanmugaraj B., et al. Preclinical evaluation of a plant-derived SARS-CoV-2 subunit vaccine: Protective efficacy, immunogenicity, safety, and toxicity. Vaccine. 2022;40(32):4440–4452. doi: 10.1016/j.vaccine.2022.05.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brun V., et al. Isotope-labeled protein standards: Toward absolute quantitative proteomics*. Mol. Cell Proteomics. 2007;6(12):2139–2149. doi: 10.1074/mcp.M700163-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Philip L.R., et al. Multiplexed Protein Quantitation in Saccharomyces cerevisiae Using Amine-reactive Isobaric Tagging Reagents*. Mol. Cell Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Dillen L., et al. Comparison of triple quadrupole and high-resolution TOF-MS for quantification of peptides. Bioanal.. 2012;4(5):565–579. doi: 10.4155/bio.12.3. [DOI] [PubMed] [Google Scholar]
- 44.Gautam S.S., et al. Relative quantitation of endogenous proteins by quadrupole-time of flight and tandem mass spectrometry. J. Chromatogr. B. 2019:11–18. doi: 10.1016/j.jchromb.2018.12.023. 1106-1107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.