Abstract
Enterovirus D68 (EV-D68) virus is a non-polio enterovirus that typically causes respiratory illness and, in severe cases, can lead to paralysis and death in children. There is currently no vaccine or antiviral for EV-D68. We previously discovered the viral 2A protease (2Apro) as a viable antiviral drug target and identified telaprevir as a 2Apro inhibitor. 2Apro is a vial cysteine protease that cleaves the viral VP1-2A polyprotein junction. In this study, we report the X-ray crystal structures of EV-D68 2Apro, wild-type, and the C107A mutant, and the structure-based lead optimization of telaprevir. Guided by the X-ray crystal structure, we predicted the binding pose of telaprevir in 2Apro using molecular dynamics simulations. We then utilized this model to inform structure-based optimization of the telaprevir’s reactive warhead and P1-P4 substitutions. These efforts led to the discovery of 2Apro inhibitors with improved antiviral activity than telaprevir. These compounds represent promising lead compounds for further development as EV-D68 antivirals.
Keywords: Enterovirus D68, EV-D68, antiviral, 2A protease, acute flaccid myelitis
Graphical Abstract
INTRODUCTION
The genus Enterovirus of Picornaviridae contains many significant pathogens related to human and mammalian diseases. This genus comprises fifteen species: four human enteroviruses (A-D),1 eight animal enteroviruses (E-L), and three rhinoviruses (A-C). The human Enterovirus D68 (EV-D68) was first identified and characterized in California in 1962.2 Only 26 sporadic confirmed cases of EV-D68 were reported from 1970 to 2005, according to the U.S. National Enterovirus Surveillance System (NESS) statistics.3 Although infections associated with EV-D68 were considered rare, there was an outbreak in 2014 in the United States when 1,153 cases with acute respiratory symptoms were confirmed to be EV-D68 infections.3 A similar statistic of EV-D68 infection was reported in Europe during the same period.4 Respiratory illness is the most common clinical signature of EV-D68 infections.5 Although EV-D68 is a non-polio enterovirus, it was linked to acute flaccid myelitis (AFM), a polio-like neurological disorder, with symptoms including dysgeusia and muscle weakness.6 Most patients had an onset of AFM between August and November, with increases in AFM cases every two years since 2014. This unique linkage suggests a possibility that EV-D68 may result in neurological diseases. Although EV-D68 is a significant human pathogen, no vaccine or antiviral is available.
The genome of EV-D68 contains a positive-sense single-stranded RNA of about 7.6 kb, encoding a precursor polyprotein that yields four structural proteins (VP1, VP2, VP3, and VP4) and seven non-structural proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D) after autocatalytic cleavage.7 Upon translation, the 2A protease (2Apro), 3Cpro, and 3CDpro cleaves the polyprotein to produce mature viral proteins. The 2Apro and 3Cpro comprise a catalytic cysteine nucleophile and adopt a chymotrypsin-like fold.8 The 2Apro cleaves at the VP1-2A junction on a conserved amino acid sequence,9 whereas 3Cpro cleaves at all other junctions, including the 3C N-terminal and C-terminal.10 Several compounds have been reported to inhibit EV-D68 in cell culture, with the majority repurposed from antivirals targeting other enteroviruses, including EV-A71, CV-B3, and rhinovirus.11, 12 Representative examples include the VP1 capsid inhibitor pleconaril, 2C inhibitors fluoxetine and dibucaine, 3C protease inhibitor rupintrivir, 3D polymerase inhibitor FNC, and host-targeting antivirals enviroxime and itraconazole.11 However, none of these compounds were specifically designed for EV-D68. Their lack of potency and selectivity, coupled with pharmacokinetic property liabilities, impede further development into clinical candidates.
We recently discovered EV-D68 2Apro is a viral cysteine protease and a viable antiviral drug target.9 We expressed 2Apro and showed that it explicitly cleaves a peptide sequence corresponding to the viral polyprotein VP1-2A junction but not other unrelated sequences. Using a FRET-based enzymatic assay, we screened the protease inhibitor library and identified telaprevir as an EV-D68 2Apro inhibitor with an IC50 of 0.2 μM. Telaprevir also inhibited multiple strains of EV-D68 in viral cytopathic effect assay and plaque assay with EC50 values from 0.4 to 1.9 μM. In this follow-up study, we aim to optimize the antiviral activity of telaprevir. Guided by the X-ray crystal structure of EV-D68 2Apro and the binding pose of telaprevir predicted by molecular dynamics simulations, we systematically examined the reactive warhead, P1, P2, P3, and P4 substitutions. Several lead compounds with EC50 values around 100 nM, and improved selectivity indexes were identified. These lead compounds represent promising drug candidates for EV-D68.
RESULTS
X-ray crystal structures of EV-D68 2Apro and 2AC107A
The asymmetric unit contains six EV-D68 2Apro molecules (A-F). The overall structures of the six molecules demonstrate high similarity with r.m.s.d. values ranging from 0.27-0.41 Å over 109-116 Cα atoms. EV-D68 2Apro adopts the classical 2A protease fold consisting of two domains. The N-terminal domain is comprised of a four-stranded sheet (bI2, cI, eI2, and fI) and two orthogonally oriented β-sheets (aII, bII, cII2, dII, eII, and fII) with a tightly bound zinc atom C-terminal domain (Figure 1A). Four conserved residues, Cys53, Cys55, Cys113, and His115, comprise the zinc ion-binding site (Figure 1A). Mutation of C107A does not significantly alter the overall structure of EV-D68 2AC107A with an r.m.s.d. of 0.43 Å over 134 Cα atoms (molecule B of EV-D68 2Apro compared to molecule E of EV-D68 2AC107A) (Figure 1B).
Figure 1.
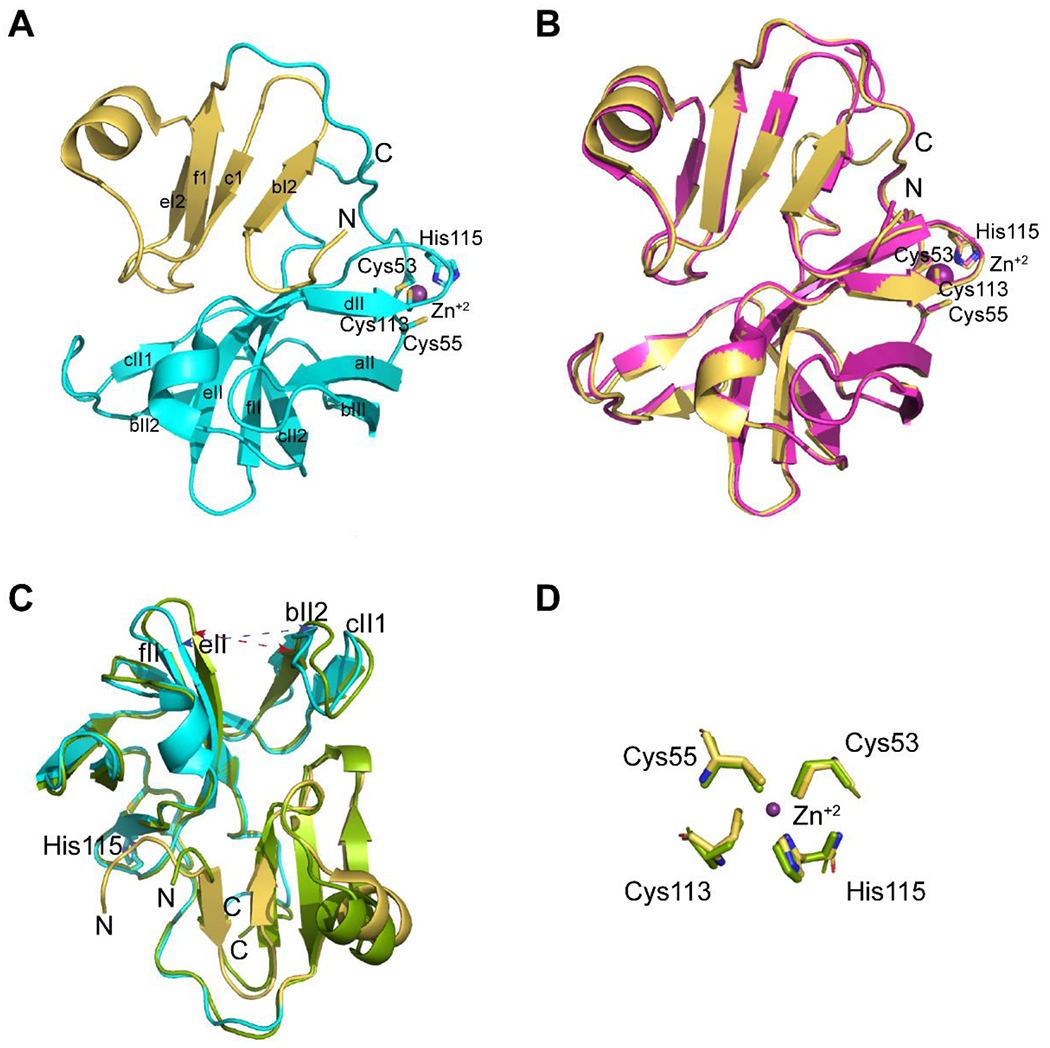
Crystal structures of EV-D68 2Apro and EV-D68 2AC107A and comparison of the crystal structures of EV-D68 2Apro (7MG0) and EV-71 2Apro (4FVB). (A) Crystal structure derived model of EV-D68 2Apro (7MG0) molecule B. The N-terminal domain is shown in gold, while the C-terminal domain is colored cyan. The four conserved residues coordinated to the bound Zn2+ are labeled. (B) Overlay of EV-D68 2Apro (gold, 7MG0) and EV-D68 2AC107A (magenta, 7JRE). Essential residues coordinating the bound Zn+2 atom are labeled. (C) Ribbon diagrams showing the overlay of Cα atoms EV-D68 2Apro (N-terminal domain, gold; C-terminal domain, cyan (7MG0) and EV-71 2Apro (green) (4FVB). The differences between the relative positions of the residues in the loops connecting β-strands bII2 and eII are shown by a blue dashed arrow for EV-D68 2Apro and a red dashed arrow for EV-71 2Apro. The molecules have been rotated ~180° around the x- and y-axis. (D) The coordination of the Zn+2 cation by Cys53, Cys55, Cys113, and His115 is highly spatially conserved. All model figures were generated using Pymol.
The structures of EV-D68 2Apro (7MG0) and EV-71 2Apro (4FVB)13 are overall very similar with an r.m.s.d. of 0.74 Å over 116 Cα atoms. However, closer inspection finds loop eII-fII and loop bII2-cII1 have shifted in the apo EV-D68 2Apro structure, widening the cavity between the loops from 10.1 Å (Val81 Cα to Gly 128 Cα, red dashed line) to 13.8 Å (Ile81 Cα to Gly 125 Cα, blue dashed line) (Figure 1C). The positions of the residues coordinating the Zn+2 cation binding are highly conserved (Figure 1D).
Molecular dynamics simulations of telaprevir in complex with EV-D68 2Apro
To gain insights into the binding pose of telaprevir in complex with EV-D68 2Apro, we produced with induced fit docking using the structure of the EV-D68 2Apro apo protein (PDB ID 7MG0), and the docking pose of telaprevir is shown in Figure 2. In the telaprevir complex, the ketoamide warhead was trapped in the H18-D106-C107 catalytic triad. The ketoamide oxygen sat in the oxyanion hole and can interact with close backbone amide nitrogens of protease residues G105 and D106. Meanwhile, the Nε nitrogen of H18 acts as the general base in the catalytic triad. In this docking pose, the C107 thiol is poised to form a covalent bond with the ketone from telaprevir.
Figure 2.
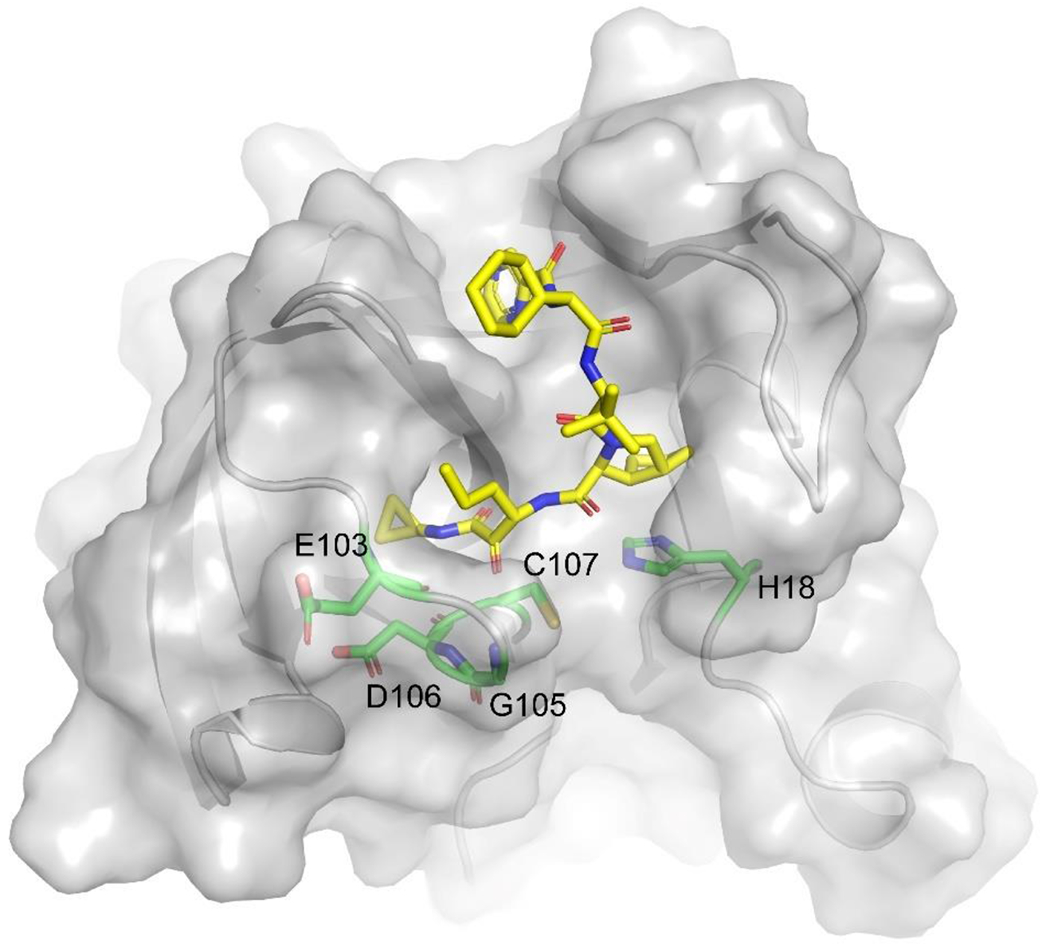
Docking pose of telaprevir produced with induced fit docking using the crystal structure of the EV-D68 2Apro (PDB ID 7MG0).
We then generated a covalently bound conformation with telaprevir in which a covalent bond was formed between the ketoamide warhead and the catalytic cysteine 107. Here, we generated an (S) configuration, which is typical for most protease inhibitors, although the (R) configuration has also been observed.14 We then performed a 500ns-MD simulation to optimize steric clashes between the ligand and the binding pocket and investigate possible conformational rearrangements. The final snapshot of the binding complex is shown in Figure 3A. The MD simulations showed that the 2Apro conformation was stabilized after ~ 50 ns while telaprevir retained the docking conformation during the trajectory (Figure 3B).
Figure 3.
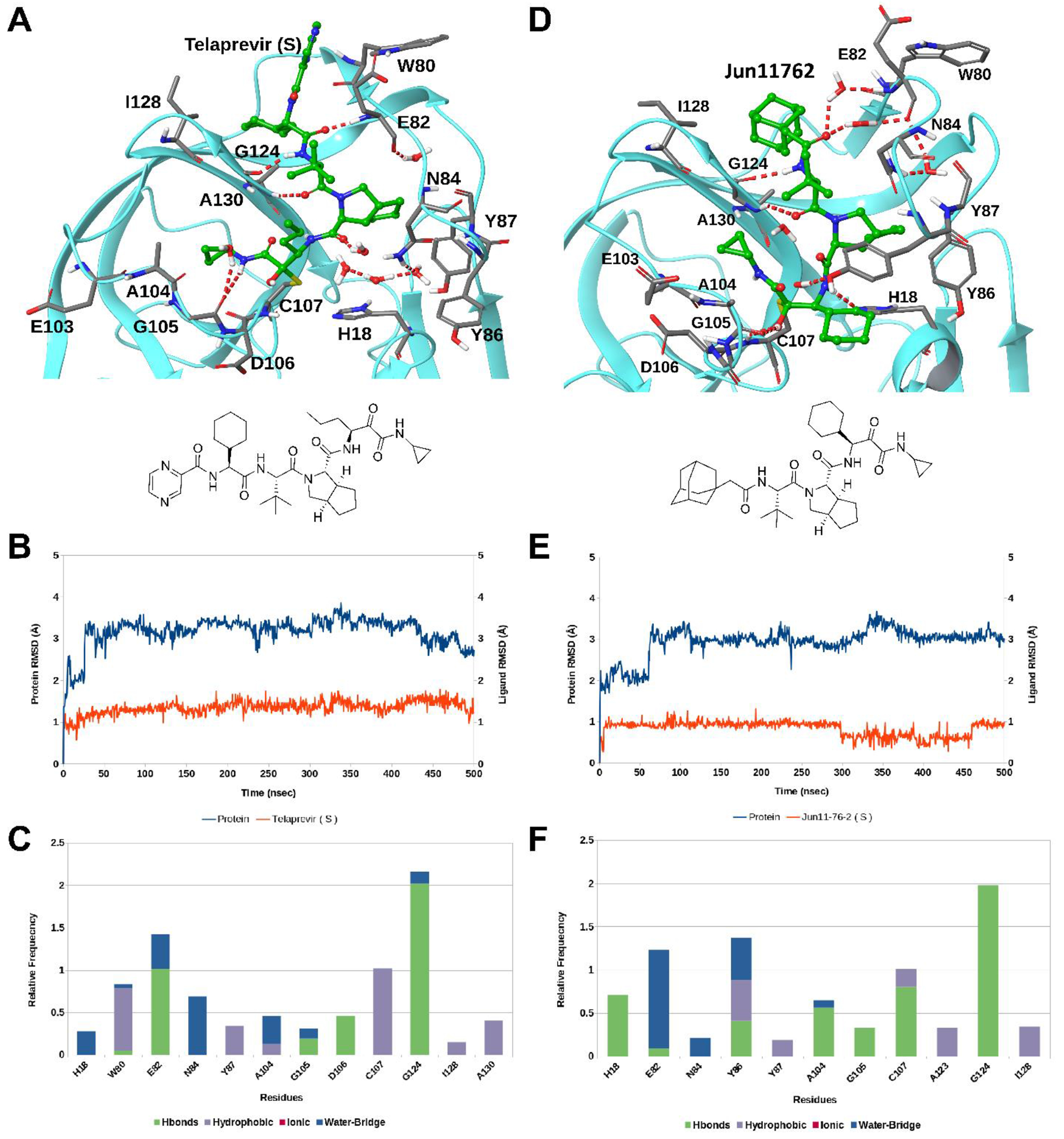
M.D. simulations of telaprevir and Jun11762 in complex with EV-D68 2Apro. Covalent docked poses of telaprevir (A) and Jun11762 (D) resulted from 500ns-MD simulations. Ligand carbons are shown in green, and protein in light blue, with important amino acid residues in grey sticks. Non-polar hydrogens were omitted for clarity. RMSD plots of Cα carbons of the protein (blue diagram) and telaprevir (B) and Jun11762 (E) (orange diagram). Stabilizing interactions inside the binding area of telaprevir (C) and Jun11762 (F); hydrogen bonding interactions bar is depicted in green, hydrophobic interactions in magenta, and water bridges in blue. They are considered important when the frequency bar is ≥ 0.2.
Most hydrogen bonding interactions observed in the docking pose in the non-covalent complex (Figure 2) were also observed in the covalent complex (Figure 3A, C). In particular, telaprevir formed three hydrogen bonds with the protease backbone: (1) the P1 ketoamide nitrogen with the carbonyl oxygen of G105 and D106, (2) the P2 carbonyl oxygen with the amide nitrogen of G124, and (3) the P4 amide carbonyl oxygen with the amide nitrogen of E82 (Figure 3A, C). As shown in Figure 3A, the hydrogen bond observed in the non-covalent complex between N84 carbonyl amide side chain and P2 amide nitrogen was disrupted by waters that entered this area concentrated between the H18 imidazole, N84 amide side chain, and C107. Indeed, in Figure 3B water bridges are shown between H18 and N84. Hydrophobic interactions were formed between W80 and P5 pyrazine ring, Y87 and P2 lactam cyclopentyl, C107 and P1 propyl, and P4 cyclohexyl and A130 (Figure 3).
We similarly performed M.D. simulations to gain insights into the binding mode of Jun11762, one of the most potent lead compounds from this study. The 500ns-MD simulations with Jun11762 complex are shown in Figures 3D, E, and F. The MD simulations showed that the 2Apro conformation was stabilized after ~ 60 ns while Jun11762 retained the docking conformation during the trajectory (Figure 3E). Jun11762 binds similarly to telaprevir except for the additional hydrogen bond between P1 amide N.H. and the H18 side chain imidazole (Figure 3D, F). The adamantyl group also contributes to more significant hydrophobic interactions with I128 (Figure 3F vs 3C).
Exploring P2, P3, P4 substitutions and the reactive warhead
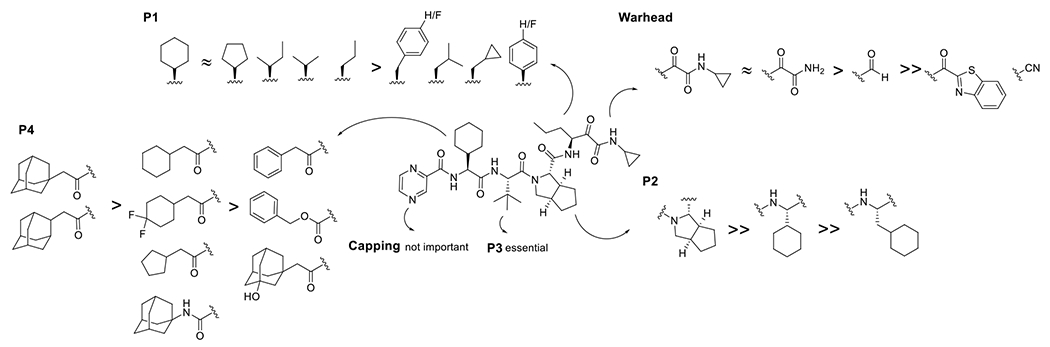
Guided by the docking pose of telaprevir with EV-D68 2Apro (Figure 3A), we performed structure-based lead optimization. Telaprevir inhibited EV-D68 2Apro and EV-D68 US/MO/14-18947 strain with IC50 of 0.18 μM and EC50 of 0.45 μM in the FRET enzymatic assay and CPE assay, respectively, and was included as a control. Replacing the pyrazine capping group in telaprevir with carboxybenzyl (Cbz) led to compound Jun7453 with slightly decreased antiviral potency (EC50 = 0.63 μM vs 0.45 μM) (Figure 4). To explore the importance of the P3 tert-butyl and P4 cyclohexyl substitutions in 2Apro inhibition, compounds Jun7424 and Jun8862 were designed. Compound Jun7424 with deletions in the P3 and P4 substitutions had drastically reduced enzymatic inhibition (IC50 = 4.35 μM vs 0.18 μM) and antiviral activity (EC50 = 25.89 μM vs 0.45 μM). Compound Jun8862 with a deletion in the P4 substitution also had reduced enzymatic inhibition (IC50 = 0.94 μM vs 0.18 μM) and antiviral activity (EC50 = 4.65 μM vs 0.45 μM). These results suggest that P3 and P4 substitutions are essential for 2Apro inhibition and antiviral activity. The SAR is consistent with the binding pose of telaprevir in 2Apro predicted by the M.D. simulations, which revealed that the P3 tert-butyl and P4 cyclohexyl substitutions form hydrophobic interactions with A130 and I128, respectively (Figure 3A).
Figure 4.
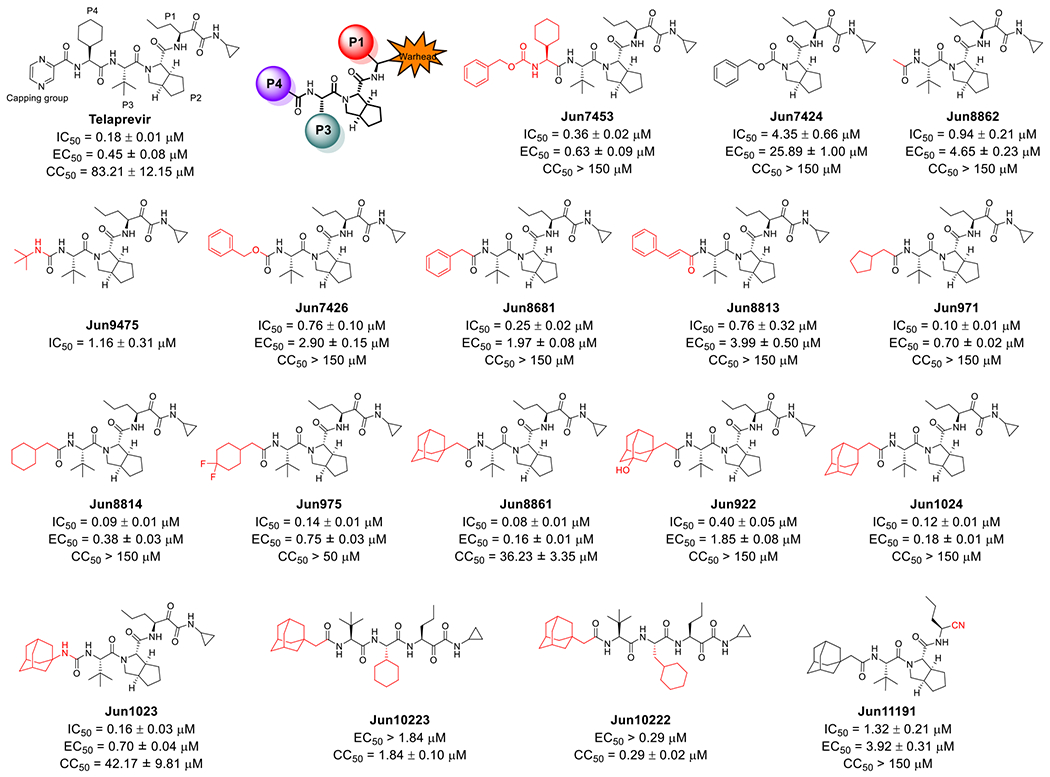
Telaprevir analogs with variations at the P2, P3, P4 substitutions and the reactive warhead. Key structural variations compared to telaprevir were highlighted in red. Enzymatic inhibitory activity IC50 value was determined in the FRET-based assay. Antiviral EC50 value was determined in the CPE assay with EV-D68 US/MO/14-18947 in R.D. cells. Cytotoxicity CC50 value was determined using the neutral red uptake method in R.D. cells. The results are the mean ± standard deviation of two independent experiments with three repeats in each experiment.
Next, a series of compounds were designed with various aromatic and hydrophobic substitutions as mimetics of the P4 cyclohexyl substitution in telaprevir. Jun9475 with the tert-butyl urea capping group had reduced enzymatic inhibition (IC50 = 1.16 μM vs 0.18 μM). Compounds Jun7426, Jun8681, and Jun8813 with aromatic capping groups at P4 position all had reduced enzymatic inhibition and antiviral activity compared to telaprevir. These results suggest that aromatic substitutions were not preferred at the P4 site. As the P4 cyclohexyl substitution at the P4 site of telaprevir forms hydrophobic interactions with I128 (Figure 3A), we hypothesized that aliphatic hydrophobic substitutions might be favored at this subsite. Indeed, compounds Jun971 (cyclopentyl), Jun8814 (cyclohexyl), Jun975 (difluorocyclohexyl), and Jun8861 (adamantly) all had comparable or improved enzymatic inhibition (IC50 = 0.08 to 0.14 μM) and antiviral activity (EC50 = 0.16 to 0.75 μM) compared to telaprevir. The most potent compound Jun8861 inhibited EV-D68 2Apro with an IC50 of 0.08 μM and the EV-D68 US/MO/14-18947 virus with an EC50 of 0.16 μM. Compound Jun922 with the 3-hydroxyadamantyl substitution had reduced antiviral activity compared to the adamantyl analog Jun8861 (EC50 = 1.85 μM vs 0.16 μM), suggesting hydrophilic substitution is not preferred. Compound 1024 with 2-adamantyl substitution had similar antiviral potency as Jun8861 (EC50 = 0.18 μM vs 0.16 μM). Replacing the amide linker in Jun1024 with a urea link led to compound Jun1023 with reduced enzymatic inhibition and antiviral activity (EC50 = 0.70 μM vs 0.16 μM) compared to Jun8861.
Compounds Jun10223 and Jun10222 were designed to explore the importance of the P2 cyclopentylproline substitution in telaprevir. However, both compounds were highly cytotoxic (CC50 < 2 μM) and were not further pursued.
Inspired by nirmatrelvir, which contains a nitrile reactive warhead,15–17 we designed the telaprevir analog Jun11191 with the nitrile warhead. However, Jun11191 had a significantly reduced enzymatic inhibition (IC50 = 1.32 vs 0.08 μM) and antiviral activity (EC50 = 3.92 vs 0.16 μM) compared to Jun8861.
In summary, our SAR results showed that the P2 cyclopentylproline, P3 tert-butyl, and the α-ketoamide reactive warhead in telaprevir are preferred for 2Apro inhibition, and aliphatic hydrophobic substitutions are preferred at the P4 position. The SAR results are consistent with the binding mode predicted by the M.D. simulations (Figure 3).
Exploring P1 substitution and the reactive warhead
Starting from the most potent lead compound Jun8861, we sought to explore the P1 substitution to further improve the antiviral potency (Figure 5). According to the binding pose predicted by the M.D. simulations, the P1 norvaline substitution in telaprevir forms hydrophobic interactions with A104 and intramolecular hydrophobic interactions with the P3 tert-butyl substitution (Figure 3A). We, therefore, hypothesized that other hydrophobic substitutions, including aromatic and aliphatic ones, might be tolerated at the P1 position. Compound Jun11385 with benzyl substitution at P1 position had similar enzymatic inhibition (IC50 = 0.09 vs 0.08 μM), antiviral activity (EC50 = 0.25 vs 0.16 μM), but increased cytotoxicity (CC50 = 17.35 vs 36.23 μM) compared to Jun8861. Compound Jun11763 with the 4-fluorobenzyl substitution had comparable enzymatic inhibition and antiviral activity as Jun11385. Replacing the amide linker in Jun11763 with a urea linker led to compound Jun11631 with similar enzymatic inhibition (IC50 = 0.16 vs 0.15 μM) and but reduced antiviral activity (EC50 = 0.35 μM vs 0.19 μM). Next, we examined the reactive warhead. Replacing the α-ketoamide with aldehyde led to compound Jun11707 with reduced enzymatic inhibition (IC50 = 1.82 vs 0.15 μM) and antiviral activity (EC50 = 1.30 μM vs 0.19 μM). Compound Jun11705 with the ketobenzothiazole warhead was inactive (IC50 > 20 μM). Compound Jun11671 with the 4-(tert-butoxy)phenyl substitution showed reduced enzymatic inhibition (IC50 = 0.18 vs 0.09 μM) and antiviral activity (EC50 = 1.73 μM vs 0.25 μM) compared to Jun11385, suggesting bulky substitutions are not preferred at P1 position. Jun11818 and Jun11862 with the thienyl and indole substitutions at P1 position had comparable enzymatic inhibition and antiviral activity as Jun11385. Notably, Jun11862 was not cytotoxic (CC50 > 150 μM). Jun11672 with the methoxymethyl P1 substitution was not active as Jun8861 with the propyl substitution, suggesting hydrophilic substitution is not preferred at P1 position. Compounds Jun11761 and Jun11389 with the cyclopropylmethyl and isobutyl substitutions also failed to improve the antiviral potency compared to Jun8861. Next, we examined compounds with β-branched substitutions at P1 position. It was found that compounds Jun11762 (cyclohexyl), Jun11808 (cyclopentyl), Jun11718 (isopropyl), and Jun11756 (isobutyl) had comparable enzymatic inhibition and antiviral activity as Jun8861 but had improved cytotoxicity (CC50 > 150 μM). In comparison, Jun11817 and Jun11861 with the benzyl substitutions were less active. Jun11724 with the tert-butyl substitution was inactive (IC50 > 20 μM). Similarly, compounds Jun11806 and Jun11805 with α-branched substitutions were also less active compared to Jun11762. Collectively, the structure-activity relationship studies revealed Jun11762 as the most potent lead with improved enzymatic inhibition (IC50 = 0.09 vs 0.18 μM), antiviral activity (EC50 = 0.13 vs 0.45 μM), cellular toxicity (CC50 >150 vs 83.21 μM) compared to telaprevir.
Figure 5.
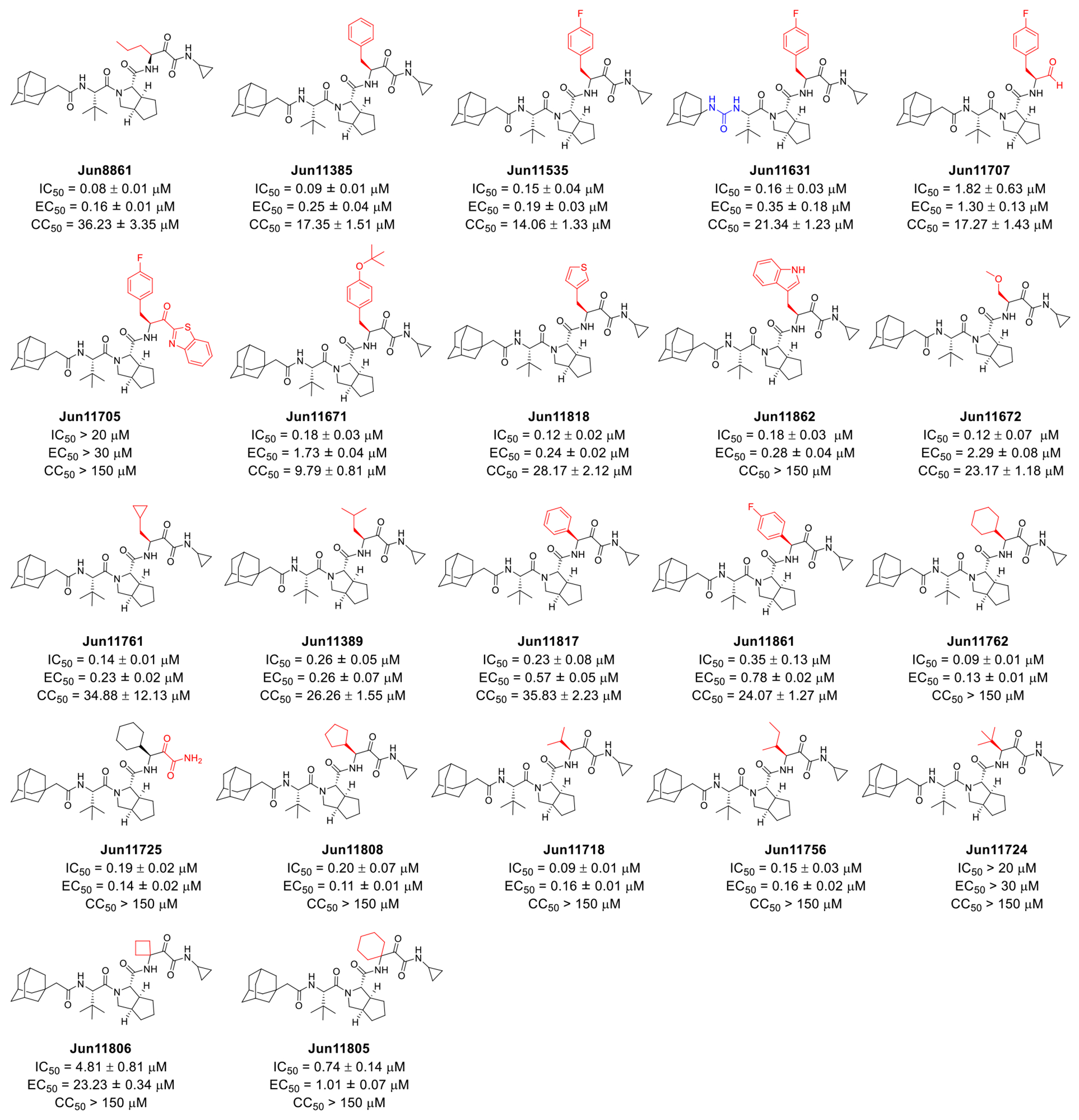
Telaprevir analogs with variations at the P1 substitutions and the reactive warhead. Key structural variations compared to Jun8861 were highlighted in red. Enzymatic inhibitory activity IC50 value was determined in the FRET-based assay. Antiviral EC50 value was determined in the CPE assay with EV-D68 US/MO/14-18947 in R.D. cells. Cytotoxicity CC50 value was determined using the neutral red uptake method in R.D. cells. The results are the mean ± standard deviation of two independent experiments with three repeats in each experiment.
Next, we selected four representative inhibitors, Jun11762, Jun11808, Jun11707, and Jun8861, for the enzyme kinetic studies. Telaprevir was included as a control. Fitting the kinetic curve of telaprevir revealed a two-phase inhibition mechanism (Figure 6): an initial reversible binding with a Ki of 823.9 nM, and a second step inactivation with a kinact of 0.001456 s−1. The overall kinact/Ki value is 1767.2 M−1s−1. The results indicate telaprevir is an irreversible covalent inhibitor of EV-D68 2Apro. In comparison, the four lead compounds Jun11762 (Ki = 51.4 nM), Jun11808 (Ki = 111.8 nM), Jun11707 (Ki = 275.8 nM), and Jun8861 (Ki = 58.6 nM) all showed reversible binding to EV-D68 2Apro (Figure 6). The Ki values suggested that Jun11762 and Jun8861 had more potent binding than Jun11808 and Jun11707, consistent with their corresponding IC50 values. Although Jun11762, Jun11808, and Jun8861 contain the same ketoamide reactive warhead as telaprevir, the mechanism of action appears to be different. Similar phenomena were also observed for SARS-CoV-2 main protease (Mpro) inhibitors, in which compounds containing the same warhead showed either reversible covalent or irreversible covalent binding.14, 18
Figure 6.
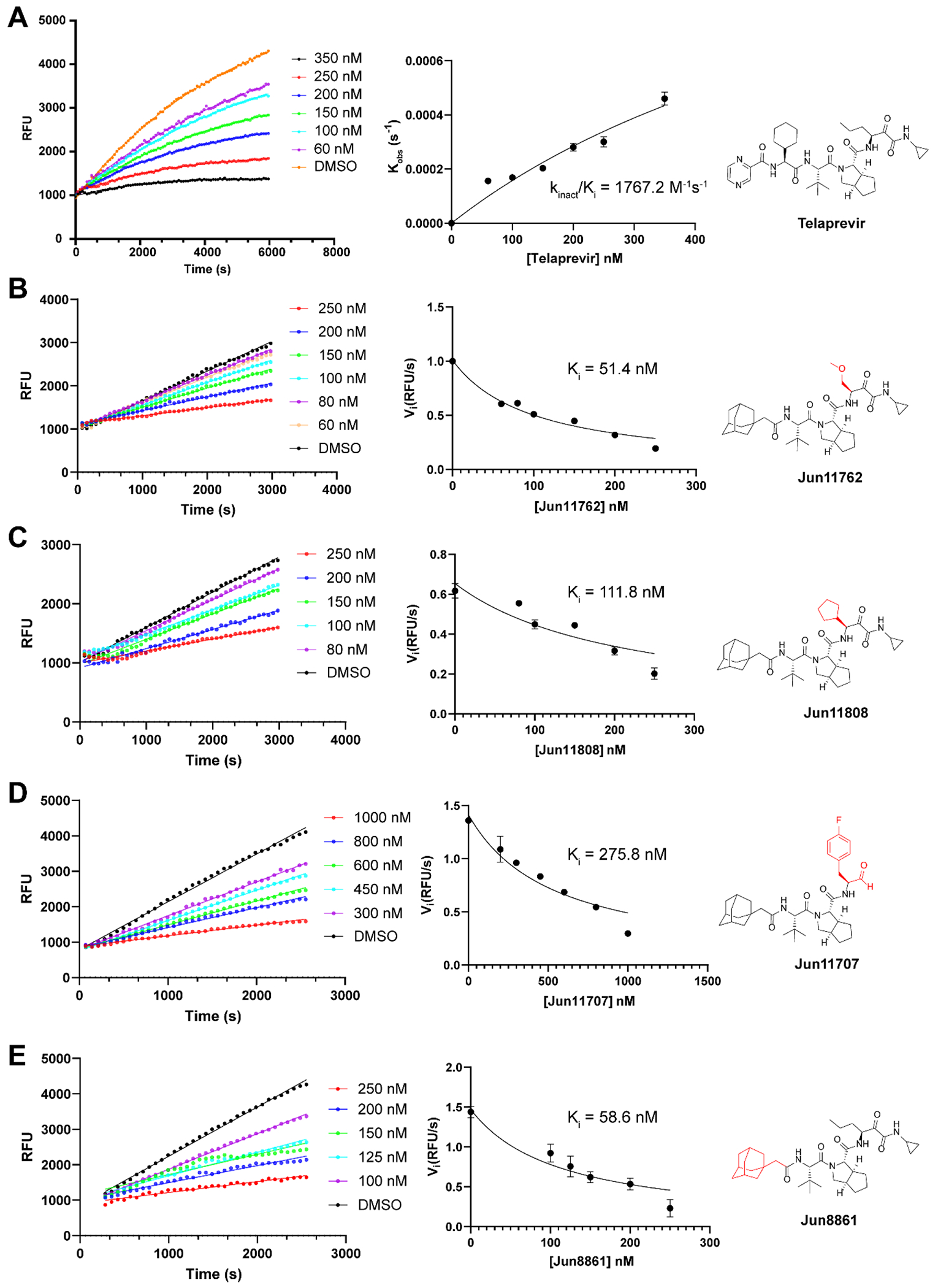
Enzyme kinetic studies of EV-D68 2Apro inhibitors. In the kinetic studies, 0.4 μM EV-D68 2Apro was added to a solution containing various concentrations of protease inhibitors and 40 μM FRET substrate to initiate the reaction. The reaction was then monitored for up to 2 h. The left column shows the reaction progression curves used for curve fitting to generate the plot shown in the middle column. Detailed methods were described in the “Materials and methods” section. Compound structures were shown in the right column. Telaprevir (A); Jun11762 (B); Jun11808 (C); Jun11707 (D); Jun8861 (E).
Broad-spectrum antiviral activity of lead compounds
We chose the three most potent hits, Jun11762, Jun12808, and Jun11756, based on the CPE assay results and tested them in the secondary plaque assay. Telaprevir was included as a control. Jun11762 (EC50 = 15.89 nM), Jun12808 (EC50 = 15.92 nM), and Jun11756 (EC50 = 13.52 nM) showed 3 to 4-fold improvement compared to telaprevir (EC50 = 52.12 nM) (Figure 7), which is consistent with the CPE assay results.
Figure 7.
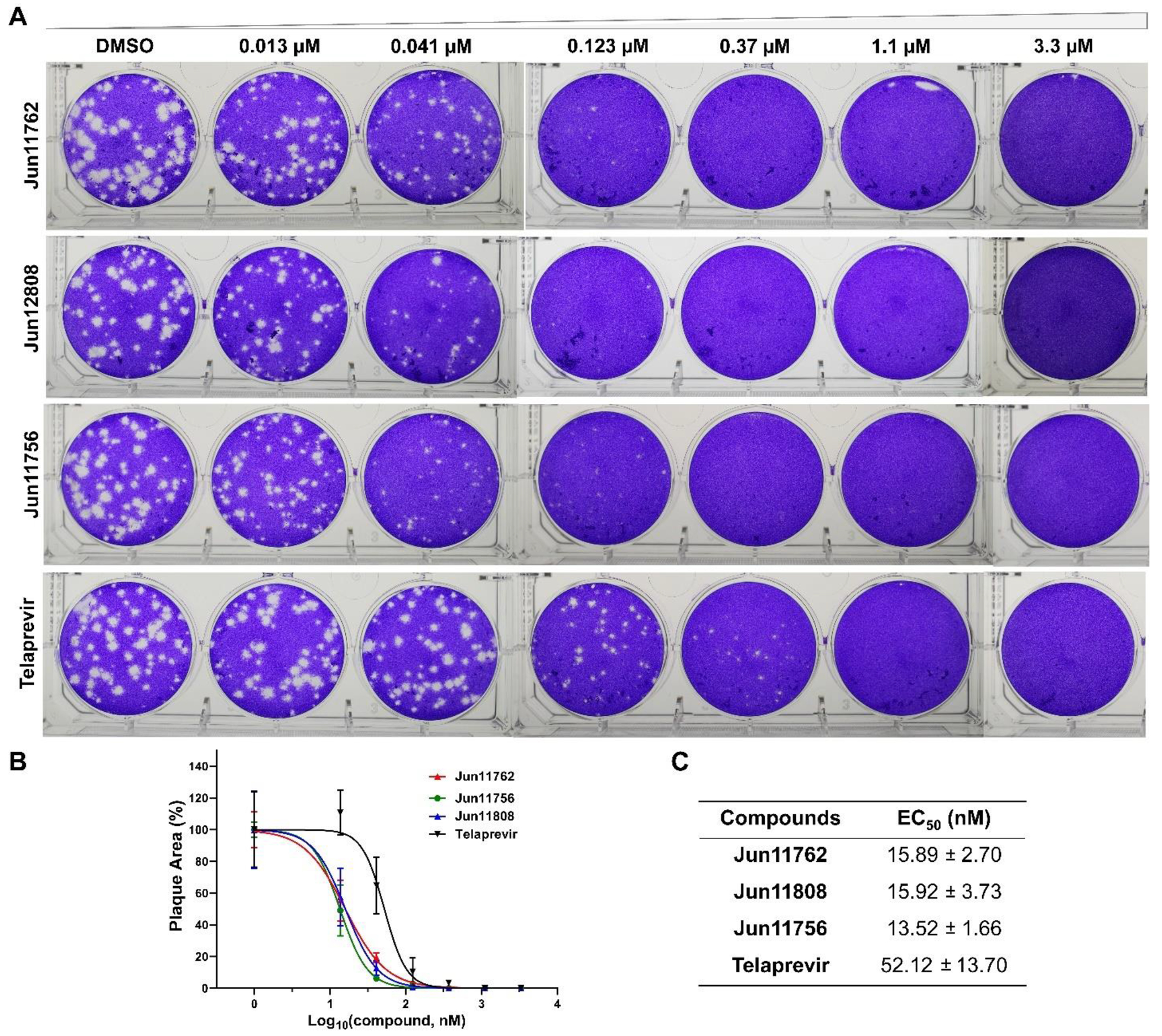
Plaque assay results of telaprevir analogs in inhibiting EV-D68 US/MO/14-18947. (A) Plaque images. The images are representatives of two repeats. (B) Curve fitting. (C) EC50 values. The results are the mean ± standard deviation of two repeats.
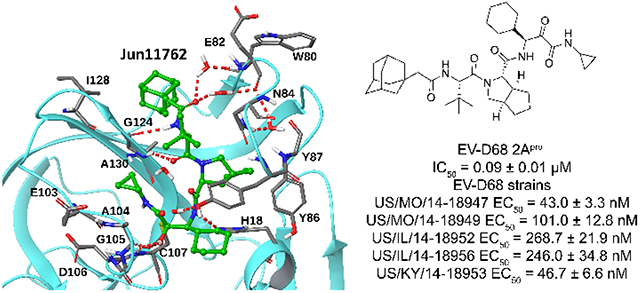
As the 2Apro is conserved among EV-D68, 2Apro inhibitors are expected to have broad-spectrum antiviral activity. To test this hypothesis, we tested Jun11762 against four additional EV-D68 strains in the B1, B2, and D clades. It was found Jun11762 inhibited all five strains with EC50 values from 43.0 to 268.7 nM, corresponding to a 3.1-4.6-fold improvement compared to telaprevir (Table 1).
Table 1.
Broad-spectrum antiviral activity of Jun11762 against five strains of EV-D68.
| EV-D68 viruses | EC50 (nM) | |
|---|---|---|
| Jun11762 | Telaprevir | |
| US/MO/14-18947 (clade B1) | 43.0 ± 3.3 | 199.1 ± 29.1 |
| US/MO/14-18949 (clade B1) | 101.0 ± 12.8 | 368.4 ± 34.4 |
| US/IL/14-18952 (clade B2) | 268.7 ± 21.9 | 828.4 ± 106.6 |
| US/IL/14-18956 (clade B2) | 246.0 ± 34.8 | 836.0 ± 68.5 |
| US/KY/14-18953 (clade D) | 46.7 ± 6.6 | 209.4 ± 37.1 |
Summary of the structure-activity relationship studies
The structure-activity relationship of telaprevir in inhibiting EV-D68 2Apro is summarized in Figure 8. P1 to P4 substitutions and a reactive warhead are essential for enzymatic inhibition and antiviral activity. The terminal capping group is not essential. For the reactive warhead, α-ketoamide is preferred over aldehyde. Ketobenzothiazole and nitrile are not tolerated, although both were shown to be preferred warheads for SARS-CoV-2 main protease inhibition.15, 17 For the P1 substitution, cyclohexyl, cyclopentyl, isopropyl, propyl, and isobutyl are preferred over phenyl, cyclopropyl, and benzyl. For the P2 substutiton, cyclopentylproline is preferred over cyclohexyl and cyclohexylmethyl. The P3 substitution is essential, and its deletion leads to a significant decrease of enzymatic inhibition. For the P4 substitution, 1, and 2-adamantylmethyl are preferred over cyclohexylmethyl, cyclopentylmethyl, benzyl, Cbz, and (3-hydroxyl)adamantylmehtyl.
Figure 8.
Summary of the structure-activity relationships of telaprevir in inhibiting EV-D68 2Apro.
In vivo pharmacokinetic (P.K.) and toxicity studies in mice
We selected the lead compound Jun11762 for the preliminary in vivo P.K. and toxicity studies in mice. We first tested Jun11762 against five CYP isoforms, and the results were shown in Table S2. It was found that Jun11762 did not inhibit CYP1A2, CYP2C9, CYP2C19, and CYP2D6 (IC50 > 50 μM). However, it had potent inhibition of CYP3A-M with an IC50 of 0.57 μM, which needs to be optimized in the following lead optimization. For the in vivo P.K., male C57BL/6J mice (3 per group) were dosed by intraperitoneal injection (i.p.) of 50 mg/kg Jun11762 in 10% DMSO and 90% corn oil. The results showed that the plasma concentration of Jun11762 was above 4-fold of EC50 value (0.13 μM) for at least 5 h (Figure 9A). We further evaluated the in vivo toxicity of Jun11762 in BALB/c mice. Mice (3 per group) were dosed at 100 mg/kg by i.p. injection at day 1 followed by a second injection of 200 mg/kg at day 3, a third injection of 300mg/kg at day 7, and a fourth injection of 400mg/kg at day 12. Body weight, survival, and clinical signs were monitored daily up to day 16. It was found that Jun11762 was tolerated at up to 400mg/kg, and no mortality or significant body weight loss was observed (Figure 9B, C).
Figure 9. In vivo snap P.K. and toxicity of Jun11762 in mice.
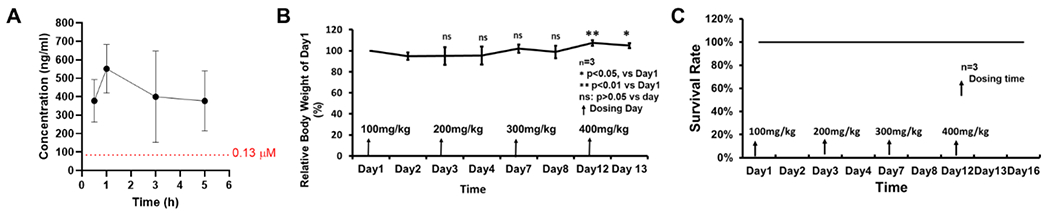
(A) In vivo P.K. of Jun11762 in C57BL/6J mice (n=3). Jun11762 was formulated in 10% DMSO and 90% corn oil and was dosed by intraperitoneal injection at 50mg/kg. Blood samples were collected at 30min, 1h, 3h, and 5h and drug concentration was quantified by LCMS. (B, C) In vivo toxicity of Jun11762 in BALB/c mice. Dosage was sequentially escalated from 100mg/kg to 400mg/kg in the increment of 100mg/kg each time by i.p. injection. Body weight (B) and survival (C) were monitored daily.
Chemistry
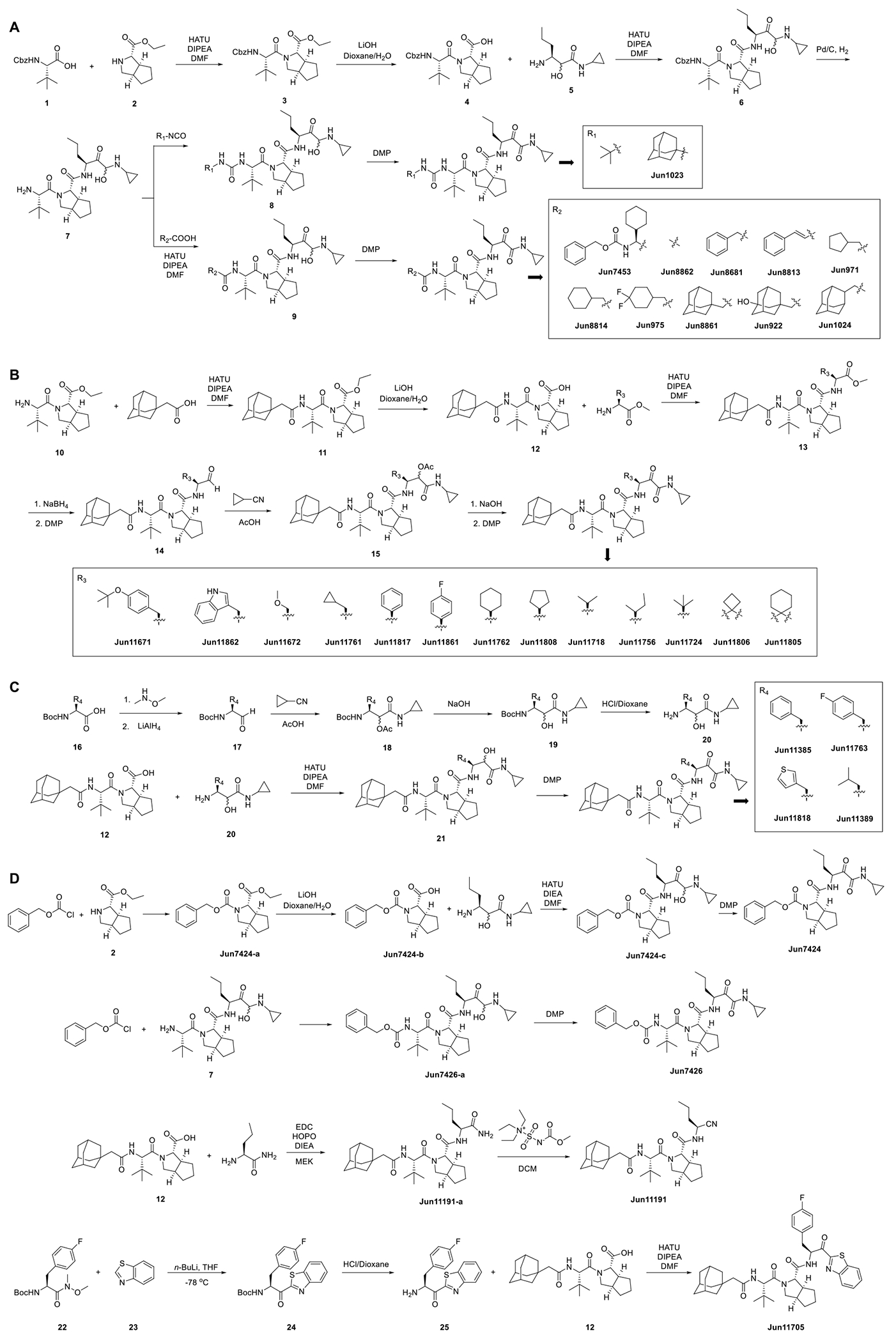
The 2Apro inhibitors were assembled using standard solution phase peptide chemistry (Scheme 1). For compounds with different terminal urea or amide capping groups (Scheme 1A), we first prepared the advanced intermediate 7, which subsequently reacted with isocyanate or coupled with carboxylic acid to give the intermediates 8 and 9, respectively. Final step oxidation using Dess-Martin periodinane (DMP) gave the final products Jun1023, Jun7453, Jun8862, Jun8681, Jun8813, Jun971, Jun8814, Jun975, Jun8861, Jun922, and Jun1024.
Scheme 1.
Synthesis routes for the 2Apro inhibitors.
For compounds with various R3 substitutions at the P1 position, two synthesis routes were designed (Scheme 1B and C), depending on the availability of starting materials. For amino acids available in the ester form, intermediate 13 was prepared by coupling 12 with the amino ester (Scheme 1B). The ester in intermediate 13 was reduced to hydroxyl by NaBH4, which was then oxidized to aldehyde 14 by DMP. Next, Passerini reaction involving aldehyde 14, cyclopropyl isocyanide, and acetic acid gave intermediate 15, which was subjected to ester hydrolysis and DMP oxidation to give final products Jun11671, Jun11862, Jun11672, Jun11761, Jun11817, Jun11861, Jun11762, Jun11808, Jun11718, Jun11756, Jun11724, Jun11806, and Jun11805. The amino acids available in the Boc-protected form were first converted to Weinreb amides, which were reduced to aldehydes 17 by LiAlH4 (Scheme 1C). Next, Passerini reaction by reacting aldehyde 17 with cyclopropyl isocyanide and acetic acid gave the ester intermediate 18, which was converted to the amino alcohol 20 by ester hydrolysis and Boc deprotection. Coupling of acid 12 with the amino alcohol 20 gave intermediate 21, which was oxidized by DMP to give the final products Jun11385, Jun11763, Jun11818, and Jun11389. Jun7424, Jun7426, Jun11191, and Jun11705 were synthesized (Scheme 1D) using the advanced intermediates shown in Schemes 1A, B, and C.
Conclusion
EV-D68 is one of the most important non-polio enteroviruses.19 Studies have shown that contemporary circulating EV-D68 strains have acquired the capacity to infect human neuronal cells,20, 21 corroborating with increased cases of AFM. Since 2014, there has been a biennial EV-D68 outbreak pattern in the U.S. and Europe, coinciding with increasing numbers of severe respiratory illnesses and neurological complications such as AFM, meningitis, and encephalitis.6, 22–25 EV-D68 is transmitted by the respiratory route, which is unusual for an enterovirus. Despite the disease severity, EV-D68 antiviral development is far left behind.11 No drug-like small molecule has been reported with in vivo antiviral efficacy against EV-D68 in animal models. Given their promising in vitro antiviral activity, enviroxime, pirodavir, pleconaril, ribavirin, and rupintrivir have been tested in the EV-D68 respiratory and neurological disease mouse models.26 However, none of these antivirals showed therapeutic efficacy,26 possibly due to the low potency or pharmacokinetic liabilities.
We aim to explore new drug targets in designing EV-D68 antiviral to address this unmet medical need. Our previous study validated the 2Apro as a viable antiviral drug target and discovered telaprevir as the first 2Apro inhibitor.9, 27 In this follow-up study, we solved the X-ray crystal structures of 2Apro wild-type and the C107A mutant. Unfortunately, our efforts in co-crystallizing telaprevir with 2Apro have not been successful, mainly due to compound precipitation at high drug concentrations (mM). We, therefore, conducted molecular dynamics simulations to predict the binding poses of telaprevir in 2Apro, which was used to guide the lead optimization. We employed a stepwise optimization strategy to identify favorable P1 to P4 substitutions. In addition, we also examined several reactive warheads, including aldehyde, nitrile, and ketobenzothiazole. These efforts led to the discovery of Jun11762 with improved antiviral activity than telaprevir. Unlike the structure-activity relationship of SARS-CoV-2 Mpro inhibitors,16, 17 ketoamide is the preferred warhead for EV-D68 2Apro. Jun11762 showed consistent antiviral potency against five contemporary strains of EV-D68, suggesting 2Apro inhibitors are promising antiviral drug candidates for combating current and possibly future EV-D68 variants. Preliminary in vivo P.K. profiling showed that Jun11762 had acceptable drug exposure with plasma concentration above 4-fold of EC50 for at least 5 hours when dosed at 50 mg/kg by i.p. administration. Jun11762 was also tolerated at up to 400 mg/kg in BALB/c mice. Further studies are warranted to optimize the antiviral potency and pharmacokinetic properties of these 2Apro inhibitors and advance them to the in vivo mouse model studies. Furthermore, it is desired to develop 2Apro inhibitors with a high genetic barrier to drug resistance. The substrate envelop hypothesis will be considered in designing 2Apro inhibitors.28
Materials and Methods
Cell lines and viruses.
Human rhabdomyosarcoma (R.D., ATCC, CCL-136) cells were maintained at 37 °C in a 5% CO2 atmosphere and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin antibiotics. The following reagents were obtained through BEI Resources, NIAID, NIH: Human Enterovirus D68, US/MO/14-18949, NR-49130; Enterovirus D68, US/MO/14-18947, NR-49129; Enterovirus D68, US/IL/14-18952, NR-49131; Enterovirus D68, US/KY/14-18953, NR-49132; Enterovirus D68, US/IL/14-18956, NR-49133.
Antiviral assays.
Cells for antiviral assays were seeded and grown overnight at 37 °C in a 5% CO2 atmosphere for ~90% confluency the next day. For all infections, cells were washed with PBS containing magnesium and calcium and infected with virus diluted in DMEM with 2% FBS and 30 mM MgCl2. Viruses were incubated for at least 1h at 33 °C in a 5% CO2 atmosphere, followed by the addition of compound as well as 1% penicillin-streptomycin. For CC50 measurements, the experiment was performed similarly but excluded viral infection. For cytopathic effect (CPE) assays, cells were stained with 66 μg/mL neutral red for 2h and neutral red uptake was measured at absorbance 540 nm using a Multiskan FC Microplate Photometer (ThermoFisher Scientific). The EC50 and CC50 values were calculated from best-fit dose response curves using GraphPad Prism. For plaque reduction assays, an 1.2% avicel microcystalline cellulose (FMC BioPolymer, Philadelphia, PA) overlay was used and cells were stained after 3 days as previously described 29.
Protein expression and purification.
EV-D68 2Apro gene from strain US/KY/14-18953 was ordered from Genscript (Piscataway, NJ) in pET28b(+) vector with E. coli codon optimization. 2Apro gene was subcloned into pE-SUMOstar vector according to the manufacturer’s protocol. Plasmid encoding WT-2Apro was transformed into BL21(DE3) competent E. coli cells, and a single colony was picked and used to inoculate 10 mL of L.B. grown supplemented with 50 μg/mL kanamycine at 37 °C at 250 rpm. The 10 mL inoculant was added to 1 L of L.B. with 50 μg/mL kanamycine and grown to an OD600 of 0.8-1.0, cooled to 18 °C, then induced using 0.5 mM IPTG. Induced cultures were incubated at 18 °C for an additional 24h, then harvested and resuspended in lysis buffer (25 mM Tris (pH 7.5); 750 mM NaCl, 2 mM DTT with 0.5 mg/mL lysozyme, 0.5 mM PMSF and 0.02 mg/mL DNase I) and lysed with alternating sonication and French press cycles. The cell debris were removed by centrifugation at 12,000 × g for 45 min (20% amplitude, 1 s on/off). The supernatant was incubated with Ni-NTA resin for over 2h at 4 °C on a rotator. The Ni-NTA resin was thoroughly washed with 50 mM imidazole in wash buffer (50 mM Tris (pH 7.5), 150 mM NaCl, and 2 mM DTT); for the SUMO-tagged protein, the protein was directly eluted with 300 mM imidazole; for the nontagged 2Apro Ni-NTA resin was treated with SUMO Protease 1 to remove the SUMO tag, then 2Apro was washed off from the column with 10 mM imidazole in wash buffer. The imidazole was removed via dialysis or 10 K molecular weight cut-off centrifugal concentrator spin column. The purity of the protein was confirmed with SDS-PAGE. The protein concentration was determined by classic BCA assay using bovine serum albumin as a standard. EV-D68 3Cpro in pET28b(+) vector was expressed as previously described 10.
Protein crystallization.
Crystallization of EV-D68 2Apro protein (10 mg/ml) was carried out using hanging-drop vapor diffusion at 20 °C. Crystals of EV-D68 2Apro protein were obtained using a well solution containing 0.05 M sodium cacodylate (pH 6.0), 15% (v/v) 2-propanol, 0.05 M MgCl2, 0.002 M CaCl2, and 0.001 M Spermine. The EV-D68 2AC107A (13mg/ml) protein crystallized under similar conditions to the wild-type protein. Crystals were generated by hanging-drop vapor diffusion using a well solution containing 0.05 M sodium cacodylate (pH 6.5), 10% (v/v) 2-propanol, 0.005 M MgCl2 and 0.005 M Spermine at 20 °C. Crystals were incubated for 10 minutes in a mother liquor solution containing 40% glycerol as a cryoprotectant before flash freezing in liquid nitrogen.
Data Collection and Processing.
X-ray diffraction data were collected remotely at the National Synchrotron Light Source II beamline 17-ID-2 using an Eiger 16M detector. Single crystal diffraction data were processed by X.D.,30 and the space group was determined to belong to P4. Partial twining was detected by Xtriage in Phenix.31 Data collection and processing statistics are given in Table 1.
Structure Solution and Refinement.
The phase of EV-D68 2AC107A was determined using molecular replacement (M.R.) by the program BALBES on the CCP4 online server32 using the structure of human rhinovirus 2 (HRV2) 2Apro (PDB code: 2HRV) as the search model. To achieve a model with better statistics and geometry, a second round of molecular replacement was done using the initial M.R. model (Phaser-MR in Phenix), along with a poly-Alanine model to reduce bias. The model was further refined by multiple rounds of refinement, followed by Rosetta refinement (Phenix) (twin operator applied with default setting) and Buster refinement.33 Ramachandran and rotamer outliers were manually fixed using KING.34 The structure of EV-D68 2Apro was solved in the same way as the described above using EV-D68 2AC107A as the search model. The statistics for refinement are given in Table 1.
Peptide synthesis.
The FRET substrate of EV-D68 2Apro, Dabcyl-KIRIVNT/GPGFGGE-Edans, was synthesized using the Fmoc solid phase synthesis strategy.35
Enzymatic assays.
The reaction buffer contains 50 mM Tris (pH7.0), 150 mM NaCl, 10% Glycerol, and 2 mM DTT, and the reaction was carried out at 30 °C in Cytation 5 imaging reader (Fisher Scientific) with EX360/40-EM460/40 filter. Reactions were monitored every 90 sec. For telaprevir IC50 measurements, 1.0 μM 2Apro was incubated with varying concentrations of telaprevir at 30 °C for 1h in reaction buffer. The reaction was initiated by adding 20 μM of FRET substrate, and the reaction was monitored for 2h and initial velocity was calculated for the first 30 min via linear regression. The IC50 was calculated by plotting the initial velocity against various concentrations of telaprevir with the dose-response curve in Prism 5.0.
The enzyme kinetic progression curve measurements with 2Apro inhibitors were carried out as follows: 2Apro FRET substrate was diluted to a final concentration of 40 μM in 190 μL of reaction buffer containing 50 mM Tris (pH 7.0), 100 mM NaCl, 2 mM DTT, and 10% glycerol. Various concentrations of inhibitors were then added to the substrate-buffer mixture. The proteolytic reaction was initiated by adding 10 μL of 2Apro to a final concentration of 0.4 μM. The reaction was monitored at 30°C, every 71 seconds, for 2 hours in the Cytation 5 imaging reader (Thermo Fisher Scientific) with the excitation of 360/40 nm and emission of 460/40 nm. The initial velocity was calculated using linear regression with the first 2500-3000 seconds of fluorescence signal. The kinetic curve of telaprevir was fitted using the equations described previously.9 The kinetic curves of Jun11762, Jun11808, Jun11707, and Jun8861 were fitted using the Morrison equotation as described previously.36
Molecular docking calculations.
The structures of the telaprevir and Jun11762 were generated using Maestro (Schrödinger Release 2020-4: Maestro, Schrödinger, LLC, New York, NY, 2020; Schrödinger Release 2020-4: LigPrep, Schrödinger, LLC, New York, NY, 2020), their protonation state was inspected using Epik (Schrödinger Release 2020-4: Epik, Schrödinger, LLC, New York, NY, 2020) at pH 7.4 and were subsequently minimized using Macromodel (Schrödinger Release 2020-4: MacroModel, Schrödinger, LLC, New York, NY, 2020) and the MMFF94 force field 37, 38 using the conjugate gradient (CG) method and a distance-dependent dielectric constant of 4.0 until a convergence value of 0.0001 kJ Å−1 mol−1 was reached.
For the docking calculations, we used the structure of the apo protein (PDB ID 7JRE) and the structure of its complex with a peptide substrate (PDB ID 4FVD).13 The two structures were superimposed and then the protein structure of PDB ID 4FVD was deleted. The most favored protonation states of ionizable residues at pH 7 in the complex between PDB ID 7JRE and peptide substrate were assigned using Epik. All hydrogen atoms of the protein complex were minimized using Maestro/Macromodel and OPLS-2005,39, 40 using a distance-dependent dielectric constant of 4.0. The molecular mechanics minimization was performed with a CG method, and a root mean square of the energy gradient (threshold) value of 0.005 kJ Å−1 mol−1 was used as the convergence criterion. The structures of the apo-protein and the ligand were saved separately for the docking calculations.
The docking of telaprevir and Jun11762 was carried out with Glide program 6 (Schrödinger Release 2020-4: Glide, Schrödinger, LLC, New York, NY, 2020) with the induced fit module and Glide XP using default settings allowing the generation of 30 docking solutions for each ligand using as a criterion that the docking poses should differ by an RMSD of 1.5 Ǻ.41 The highest docking poses were visually inspected using the UCSF Chimera package.42 The RMSD of the highest in score docking pose of peptide substrate from the crystallographic structure was 1.2 Å, suggesting that the docking calculations method was reliable for calculating the docking poses of telaprevir and Jun11762. All hydrogens atoms of the generated protein complexes were minimized with the OPLS-2005 force field using Maestro/Macromodel using a distance-dependent dielectric constant of 4.0. These complexes consist of c.a. 315 amino acid residues and 4,951 atoms. We then generated a covalent bond between the ligands and the Cys107.
MD simulations.
MD simulations were carried out to the covalently bound telaprevir and Jun11762 with EV-D68 2Apro prepared as described above. Each complex was solvated using the TIP3P 43 water model. Using the “System Builder” utility of Schrodinger Desmond v.11.1 each complex was embedded in an orthorhombic water box extending beyond the solute 10 Å in x,y,z direction leading to 14500 waters. Na+ and Cl− ions were placed in the water phase to neutralize the systems and to reach the experimental salt concentration of 0.150 M NaCl. The total number of atoms was c.a. 48000.
The OPLS-2005 force field was used to model all protein and ligand interactions and lipids. The particle mesh Ewald method (PME) 44, 45 was employed to calculate long-range electrostatic interactions with a grid spacing of 0.8 Å. Van der Waals and short-range electrostatic interactions were smoothly truncated at 9.0 Å. The Langevin thermostat 46 was utilized to maintain a constant temperature in all simulations, and the Berendsen barostat 47 was used to control the pressure. Periodic boundary conditions were applied (73×102×65) Ǻ3. The equations of motion were integrated using the multistep RESPA integrator 48 with an inner time step of 2 fs for bonded interactions and non-bonded interactions within a cutoff of 9 Å. An outer time step of 6.0 fs was used for non-bonded interactions beyond the cut-off. Each system was equilibrated in MD simulations with a default protocol for water-soluble proteins provided in Desmond, which consists of a series of restrained MD simulations designed to relax the system while not deviating substantially from the initial coordinates.
The first simulation was a Brownian dynamics run for 100 ps at a temperature of 10 K in the NVT (constant number of particles, volume, and temperature) ensemble with solute heavy atoms restrained with a force constant of 50 kcal mol Å−2. The Langevin thermostat 46 was applied in the NVT ensemble and a MD simulation for 12 ps with solute heavy atoms restrained with a force constant of 50 kcal mol Å−2. The velocities were randomized and MD simulation for 12 ps was performed in the NPT (constant number of particles, pressure, and temperature) ensemble and a Berendsen barostat 47 with solute heavy atoms equally restrained at 10 K and another one at 300 K. The velocities were again randomized and unrestrained MD simulation for 24 ps was performed in the NPT ensemble. The above-mentioned equilibration was followed by 500ns simulation without restrains. Two simulations were performed in a workstation with GTX 970. The visualization of the produced trajectories and structures was performed using Maestro or programs Chimera 42 and VMD.49
Animals and treatments for the in vivo toxicity study.
Female BALB/c mice (around 19g) were purchased from Taconic Biosciences, Inc (Germantown, NY). All mice were housed under conditions of controlled temperature (22°C) with on–off light cycle, with food and water provided ad libitum. For the in vivo toxicity study of Jun11762, 3 female BALB/c mice were given the starting dose of 100mg/kg (in corn oil containing 10% DMSO) by intraperitoneal injection (i.p.). After enough time of compound elimination in vivo, the next dose of 200mg/kg was administered to the same animals, and the dosages were sequentially escalated to 300mg/kg and 400mg/kg. Bodyweights were measured, and the general health status of the animals was observed during the study period. The Rutgers University Animal Care and Use Committee approved all animal studies.
General Chemical Methods
All chemicals were purchased from commercial vendors and used without further purification unless otherwise noted. 1H and 13C NMR spectra were recorded on a Bruker-400 or −500 NMR spectrometer. Chemical shifts are reported in parts per million referenced with respect to residual solvent (CD3OD) 3.31 ppm, (DMSO-d6) 2.50 ppm, and (CDCl3) 7.26 ppm or from internal standard tetramethylsilane (TMS) 0.00 ppm. The following abbreviations were used in reporting spectra: s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet; dd, doublet of doublets; ddd, doublet of doublet of doublets. All reactions were carried out under N2 atmosphere, unless otherwise stated. HPLC-grade solvents were used for all reactions. Flash column chromatography was performed using silica gel (230-400 mesh, Merck). High resolution mass spectra were obtained using an OrbitrapTM for all the compounds, obtained in an Ion Cyclotron Resonance (ICR) spectrometer. The purity was assessed by using Shimadzu UPLC with Shimdazu C18-AQ column (4.6 × 150 mm P/N #227-30767-05) at a flow rate of 1 mL/min; λ = 254 and 220 nm; mobile phase A, 0.1% trifluoroacetic acid in H2O, and mobile phase B, 0.1% trifluoroacetic acid in 90% CH3CN and 10% H2O. The gradients are 0-2 min 10% B, 2-15 min 10%-100% B, 15-18 min, 100% B, 18.1-20 min 10% B. All compounds submitted for testing were confirmed to be >95.0% purity by HPLC traces. All final products were characterized by proton and carbon NMR, HPLC, and HRMS.
General amide coupling procedure:
Corresponding acid (1 eq.), HATU (1.2 eq.) were dissolved in DMF and stirred for 30-60 mins. Then corresponding amine (1-1.2 eq.), DIEA (4 eq.) were added. The reaction mixture was stirred at ambient temperature for 20 hours. Then E.A. and water were added. The organic phase was washed by water, 1 M HCl (aq.), saturated NaHCO3 (aq.), brine, dried by Na2SO4, filtered, removed the solvent to give the crude product which can be purified with silica gel chromatography.
General ester hydrolysis procedure:
Corresponding ester was dissolved in THF/MeOH/H2O(1:1:1). LiOH (2 eq.) was added. After 12 hours stirring at ambient temperature, the solvent was partly removed in vacuo and 1 N HCl (aq.) was added till the pH=2. The aqueous phase was extracted by DCM 3 times, dried by dried by Na2SO4. The solvent was removed in vacuo to give the crude product without purification.
General ester reduction procedure:
Corresponding ester was dissolved in MeOH. NaBH4 (10 eq.) was added slowly at 0 °C. After stirring at ambient temperature for 1 hour, the solvent was partly removed in vacuo and 1 N HCl (aq.) was added. After 30 min stirring, E.A. was added. The organic phase was separated and dried with NaSO4. The solvent was removed in vacuo and purified with silica gel chromatography or without further purification.
General Passerini reaction procedure:
Corresponding aldehyde was dissolved in E.A. Corresponding isocyanide (1.2 eq.), and acetic acid (1.2 eq.) were added at 0 °C. The reaction mixture was stirred at ambient temperature for 12 hours till the starting martial disappeared. After removed the solvent in vacuo, the crude product was purified with silica gel chromatography or without further purification.
General Dess-Martin periodinane oxidation procedure:
Corresponding alcohol was dissolved in anhydrous DCM and Dess-Martin periodinane (1.5 eq.) was added at 0 °C. The reaction mixture was stirred at ambient temperature for 1 hour then the solvent was removed in vacuo. 10% Na2S2O3 (aq.) was added and stirred for 30 min then E.A. was added. The organic phase was washed by 10% Na2S2O3 (aq.), saturated NaHCO3 (aq.), brine, dried by Na2SO4. The solvent was removed in vacuo and purified with silica gel chromatography.
General Cbz deprotection procedure:
Corresponding material (1 eq.), wet palladium/carbon (5% based on palladium) were dissolved in MeOH. The reaction mixture was stirred at ambient temperature under H2 (1 atm, balloon) for 12 hours. The solid was filtered and the solvent was removed in vacuo to give the product without further purification.
General Boc deprotection procedure:
Corresponding material (1 eq.) was added 4 M HCl in dioxane (1 mL/mmol) at 0 °C under N2. The reaction mixture was stirred at ambient temperature for 1 hour. The solvent was removed in vacuo to give the product without further purification.
ethyl (1S,3aR,6aS)-2-((S)-2-(((benzyloxy)carbonyl)amino)-3,3-dimethylbutanoyl)octahydrocyclopenta[c]pyrrole-1-carboxylate (3).
Compound 3 was synthesized from starting materials 1 and 2 using the general amide coupling procedure. Yield: 68%. 1H NMR (400 MHz, CDCl3) δ 7.41 – 7.21 (m, 5H), 5.46 (d, J = 9.8 Hz, 1H), 5.11 – 5.00 (m, 2H), 4.38 – 4.29 (m, 2H), 4.16 (m, 2H), 3.85 (m, 1H), 3.69 (m, 1H), 2.77 – 2.58 (m, 2H), 1.89 (m, 2H), 1.74 (m, 3H), 1.45 (m, 1H), 1.24 (t, J = 7.1 Hz, 3H), 1.02 (s, 9H).
Compound 4 was synthesized from compound 3 by following the general ester hydrolysis procedure and was used for the next step without further purification.
Compound 6 was synthesized from starting materials 4 and 5 following the general amide coupling procedure and used for the next step without further purification.
Compound 7 was synthesized from compound 6 following the general Cbz deprotection procedure and used for the next step without further purification.
Jun9475 and Jun1023 were synthesized from compound 7 through a two-step procedure. In the first step, compound 7 (1 eq), triethylamine (1.2 eq), and isocyanate (1 eq) were added to anhydrous DCM at 0oC. The solution was warmed to room temperature and stirred overnight. The solution was extacted with 1N HCl and brine to give intermediate 8. In the second step, compound 8 was oxidaized by following the general DMP oxidation procedure to give the final products Jun9475 and Jun1023.
(1S,3aR,6aS)-2-((S)-2-(3-(tert-butyl)ureido)-3,3-dimethylbutanoyl)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun9475).
White solid. Yield: 56%. 1H NMR (400 MHz, CDCl3) δ 7.42 (d, J = 6.8 Hz, 1H), 6.94 (d, J = 3.6 Hz, 1H), 5.34-5.21 (m, 1H), 4.98 (d, J = 9.6 Hz, 1H), 4.55-4.43 (m, 3H), 3.85 (dd, J = 10.8, 4.0 Hz, 1H), 3.74 (dd, J = 10.4, 8.0 Hz, 1H), 3.51 (d, J = 3.2 Hz, 1H), 2.98-2.89 (m, 1H), 2.85-2.76 (m, 2H), 1.96-1.81 (m, 3H), 1.65-1.56 (m, 3H), 1.48-1.36 (m, 3H), 1.33 (s, 9H), 1.01 (s, 9H), 0.93 (t, J = 7.2 Hz, 3H), 0.89-0.84 (m, 2H), 0.66-0.58 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 196.6, 172.9, 171.3, 160.5, 157.0, 66.0, 57.3, 54.2, 54.2, 50.4, 45.0, 43.2, 35.2, 33.5, 32.1, 31.6, 29.4, 26.5, 25.4, 22.4, 19.1, 13.6, 6.5, 6.5. C28H47N5O5 ESI-MS: m/z (M + H+): 534.7 (calculated), 534.4 (found).
(1S)-2-((S)-2-(3-((3R,5R,7R)-adamantan-1-yl)ureido)-3,3-dimethylbutanoyl)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun1023).
White solid. Yield: 71%. 1H NMR (400 MHz, CDCl3) δ 7.82 (d, J = 7.1 Hz, 1H), 7.07 (d, J = 4.1 Hz, 1H), 5.50 (d, J = 9.8 Hz, 1H), 5.20-5.15 (m, 1H), 4.86 (s, 1H), 4.55 (d, J = 9.7 Hz, 1H), 4.44 (d, J = 3.2 Hz, 1H), 3.83-3.77 (m, 2H), 2.84-2.71 (m, 4H), 2.04 (s, 4H), 1.93 (s, 6H), 1.88-1.84 (m, 3H), 1.65 (s, 7H), 1.39 (d, J = 7.3 Hz, 2H), 0.97 (s, 10H), 0.90 (t, J = 7.3 Hz, 4H), 0.84 (d, J = 7.4 Hz, 2H), 0.63-0.60 (m, 2H).13C NMR (101 MHz, CDCl3) δ 197.25, 172.71, 171.85, 160.84, 156.71, 65.99, 56.99, 54.49, 54.27, 50.67, 46.16, 43.23, 42.43, 36.43, 35.55, 33.29, 32.07, 31.52, 29.51, 26.51, 25.43, 22.50, 19.09, 13.54, 6.46, 6.42. C34H53N5O5, HRMS calculated for m/z [M+H]+: 612.4125 (calculated), 612.4128 (found).
The following compounds Jun7453, Jun8862, Jun8681, Jun8813, Jun971, Jun8814, Jun975, Jun8861, Jun922, and Jun1024 were synthesized from compound 7 through a two-step procedure. In the first step, compound 9 was synthesized from compound 7 by following the general amide coupling reaction. In the second step, compound 9 was oxidaized by following the general DMP oxidation procedure to give the final product.
Benzyl ((S)-1-cyclohexyl-2-(((S)-1-((1S,3aR,6aS)-1-(((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)carbamoyl)hexahydrocyclopenta[c]pyrrol-2(1H)-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-2-oxoethyl)carbamate (Jun7453)
White solid. Yield: 51%. 1H NMR (400 MHz, CDCl3) δ 7.90 (dd, J = 21.4, 6.7 Hz, 2H), 7.62 (d, J = 9.5 Hz, 1H), 7.28 (m, 5H), 5.60 (d, J = 9.3 Hz, 1H), 5.47 (d, J = 9.6 Hz, 1H), 5.12 – 5.03 (m, 2H), 4.77 (d, J = 9.6 Hz, 1H), 4.61 (s, 1H), 4.32 (m, 1H), 3.93 (m, 1H), 3.57 (m, 1H), 2.95 – 2.70 (m, 3H), 2.59 (m, 1H), 1.75 (m, 6H), 1.55 (t, J = 12.2 Hz, 4H), 1.41 (m, 6H), 1.23 – 0.98 (m, 5H), 0.95 (s, 1H), 0.87 (s, 9H), 0.74 (m, 2H), 0.69 – 0.61 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 198.52, 171.36, 171.33, 170.21, 160.29, 156.22, 136.28, 128.45, 128.05, 127.66, 66.78, 65.72, 59.67, 56.39, 55.14, 53.17, 46.18, 42.84, 40.43, 36.75, 33.44, 32.82, 32.53, 29.36, 28.78, 26.36, 25.99, 25.78, 22.72, 19.17, 13.19, 6.07. C39H57N5O7, HRMS calculated for m/z [M+H]+: 708.6336 (calculated), 708.5358 (found).
(1S,3aR,6aS)-2-((S)-2-acetamido-3,3-dimethylbutanoyl)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun8862).
White solid. Yield: 56%. 1H NMR (400 MHz, DMSO-d6) δ 9.23 (d, J = 8.2 Hz, 1H), 8.74 – 8.65 (m, 1H), 8.30 – 8.18 (m, 1H), 5.03 – 4.92 (m, 1H), 4.50 (d, J = 8.3 Hz, 1H), 4.28 (dd, J = 6.7, 3.8 Hz, 1H), 3.82 – 3.67 (m, 1H), 3.65 – 3.55 (m, 1H), 2.80 – 2.71 (m, 1H), 2.68 – 2.61 (m, 1H), 2.51 – 2.46 (m, 1H), 1.87 – 1.50 (m, 7H), 1.50 – 1.33 (m, 6H), 1.03 – 0.97 (m, 6H), 0.97 – 0.83 (m, 7H), 0.72 – 0.61 (m, 2H), 0.61 – 0.53 (m, 2H). 13C NMR (101 MHz, DMSO-d6) δ 197.48, 172.17, 167.98, 162.57, 162.47, 65.53, 58.56, 54.46, 53.83, 47.77, 43.04, 35.28, 32.12, 31.75, 28.55, 26.76, 26.65, 25.12, 22.92, 19.17, 13.93, 13.89, 5.87, 5.82. C25H40N4O5, HRMS calculated for m/z [M+H]+: 477.3077 (calculated), 477.3188 (found).
(1S,3aR,6aS)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)-2-((S)-3,3-dimethyl-2-(2-phenylacetamido)butanoyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun8681).
White solid. Yield: 72%. 1H NMR (400 MHz, CDCl3) δ 7.42 – 7.25 (m, 5H), 7.13 (d, J = 7.1 Hz, 1H), 6.93 (d, J = 3.9 Hz, 1H), 6.15 (d, J = 9.3 Hz, 1H), 5.31 – 5.21 (m, 1H), 4.64 (d, J = 9.3 Hz, 1H), 4.44 (d, J = 2.7 Hz, 1H), 3.75 (d, J = 6.1 Hz, 2H), 2.98 – 2.88 (m, 1H), 2.87 – 2.74 (m, 2H), 1.97 – 1.81 (m, 3H), 1.76 – 1.66 (m, 1H), 1.65 – 1.52 (m, 2H), 1.55 – 1.32 (m, 4H), 1.00 – 0.81 (m, 15H), 0.67 – 0.57 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 196.22, 170.98, 170.96, 170.62, 160.38, 134.80, 129.21, 128.97, 127.34, 77.34, 77.02, 76.70, 65.99, 56.67, 54.37, 54.09, 45.05, 43.71, 43.13, 35.52, 33.50, 32.14, 31.80, 26.26, 25.54, 22.42, 19.09, 13.58, 6.50, 6.41. C31H44N4O5, HRMS calculated for m/z [M+H]+: 553.3390 (calculated), 553.4110 (found).
(1S,3aR,6aS)-2-((S)-2-cinnamamido-3,3-dimethylbutanoyl)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun8813).
White solid. Yield: 68%. 1H NMR (400 MHz, DMSO-d6) δ 8.71 (d, J = 5.2 Hz, 1H), 8.24 (d, J = 6.9 Hz, 1H), 8.06 (d, J = 8.7 Hz, 1H), 7.58 – 7.51 (m, 2H), 7.45 – 7.36 (m, 3H), 7.36 – 7.29 (m, 1H), 7.01 – 6.90 (m, 1H), 5.02 – 4.92 (m, 1H), 4.56 (dd, J = 9.2, 3.5 Hz, 1H), 4.25 (d, J = 3.9 Hz, 1H), 3.83 – 3.74 (m, 1H), 3.74 – 3.67 (m, 1H), 2.80 – 2.55 (m, 2H), 1.87 – 1.29 (m, 11H), 1.03 – 0.95 (m, 9H), 0.90 – 0.84 (m, 3H), 0.70 – 0.48 (m, 4H). 13C NMR (101 MHz, DMSO-d6) δ 197.52, 172.34, 169.75, 165.44, 162.57, 139.51, 135.46, 129.88, 129.37, 127.95, 122.49, 65.37, 57.37, 54.38, 53.84, 47.76, 43.04, 40.55, 40.34, 40.13, 39.92, 39.71, 39.50, 39.30, 36.36, 35.13, 32.15, 32.11, 31.83, 26.91, 25.19, 22.94, 19.17, 13.93, 5.89, 5.84. C32H44N4O5, HRMS calculated for m/z [M+H]+: 565.3390 (calculated), 565.4113 (found).
(1S,3aR,6aS)-2-((S)-2-(2-cyclopentylacetamido)-3,3-dimethylbutanoyl)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun971).
White solid. Yield: 61%. 1H NMR (500 MHz, DMSO-d6) δ 8.70 (d, J = 5.2 Hz, 1H), 8.21 (d, J = 7.0 Hz, 1H), 7.75 (d, J = 9.1 Hz, 1H), 5.00 – 4.92 (m, 1H), 4.47 (d, J = 9.1 Hz, 1H), 4.25 (d, J = 3.7 Hz, 1H), 3.77 – 3.69 (m, 1H), 3.70 – 3.63 (m, 1H), 2.79 – 2.68 (m, 1H), 2.67 – 2.57 (m, 1H), 2.23 – 2.02 (m, 3H), 1.85 – 1.30 (m, 16H), 1.19 – 1.06 (m, 2H), 0.91 (d, J = 19.8 Hz, 10H), 0.72 – 0.58 (m, 2H), 0.61 – 0.52 (m, 2H). 13C NMR (126 MHz, DMSO-d6) δ 197.50, 172.50, 172.35, 170.00, 162.58, 65.25, 56.73, 55.36, 54.33, 53.85, 47.69, 42.91, 41.31, 40.48, 37.55, 34.86, 32.27, 32.20, 32.18, 32.16, 31.92, 26.91, 25.11, 24.98, 24.97, 22.93, 19.18, 13.94, 5.88, 5.82. C30H48N4O5, HRMS calculated for m/z [M+H]+: 545.3703 (calculated), 545.4003 (found).
(1S,3aR,6aS)-2-((S)-2-(2-cyclohexylacetamido)-3,3-dimethylbutanoyl)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun8814).
White solid. Yield: 76%. 1H NMR (400 MHz, DMSO-d6) δ 8.71 (d, J = 5.2 Hz, 1H), 8.22 (d, J = 6.9 Hz, 1H), 7.76 (d, J = 9.0 Hz, 1H), 5.01 – 4.91 (m, 1H), 4.48 (d, J = 9.1 Hz, 1H), 4.25 (d, J = 3.6 Hz, 1H), 3.79 – 3.64 (m, 2H), 2.81 – 2.69 (m, 1H), 2.69 – 2.53 (m, 1H), 2.13 – 2.02 (m, 2H), 1.81 – 1.34 (m, 15H), 1.19 – 1.10 (m, 2H), 0.96 – 0.87 (m, 14H), 0.71 – 0.60 (m, 2H), 0.63 – 0.52 (m, 2H). 13C NMR (101 MHz, DMSO-d6) δ 197.46, 172.33, 172.15, 169.99, 162.55, 65.23, 56.72, 55.34, 54.34, 53.84, 47.66, 43.04, 42.86, 35.58, 34.77, 32.85, 32.83, 32.30, 32.13, 31.95, 26.89, 26.35, 26.13, 26.08, 25.09, 22.93, 19.18, 13.93, 5.87, 5.82. C31H50N4O5, HRMS calculated for m/z [M+H]+: 559.3859 (calculated), 559.4583 (found).
(1S,3aR,6aS)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)-2-((S)-2-(2-(4,4-difluorocyclohexyl)acetamido)-3,3-dimethylbutanoyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun975).
White solid. Yield: 71%. 1H NMR (500 MHz, DMSO-d6) δ 8.70 (d, J = 5.2 Hz, 1H), 8.21 (d, J = 6.9 Hz, 1H), 7.83 (d, J = 8.9 Hz, 1H), 5.00 – 4.92 (m, 1H), 4.46 (d, J = 9.0 Hz, 1H), 4.25 (d, J = 3.7 Hz, 1H), 3.77 – 3.70 (m, 1H), 3.70 – 3.64 (m, 1H), 2.79 – 2.70 (m, 1H), 2.67 – 2.57 (m, 1H), 2.25 – 2.05 (m, 2H), 1.99 – 1.91 (m, 2H), 1.83 – 1.35 (m, 6H), 1.24 – 1.15 (m, 2H), 0.96 – 0.87 (m, 12H), 0.70 – 0.61 (m, 2H), 0.64 – 0.53 (m, 2H). 13C NMR (126 MHz, DMSO-d6) δ 197.51, 172.33, 171.78, 169.92, 162.57, 65.26, 56.88, 55.36, 54.37, 53.85, 47.67, 42.90, 34.78, 32.96, 32.89, 32.28, 32.17, 31.95, 28.58, 28.51, 26.90, 25.10, 22.93, 19.18, 13.93, 5.88, 5.82. C31H48F2N4O5, HRMS calculated for m/z [M+H]+: 595.3671 (calculated), 595.4499 (found).
(1S,3aR,6aS)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)-2-((S)-3,3-dimethyl-2-(2-phenylacetamido)butanoyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun8861).
White solid. Yield: 63%. 1H NMR (400 MHz, DMSO-d6) δ 8.70 (d, J = 5.2 Hz, 1H), 8.21 (d, J = 6.8 Hz, 1H), 7.60 (d, J = 9.2 Hz, 1H), 5.01 – 4.91 (m, 1H), 4.51 (d, J = 9.2 Hz, 1H), 4.26 (d, J = 3.5 Hz, 1H), 3.77 – 3.69 (m, 2H), 2.81 – 2.69 (m, 1H), 2.68 – 2.58 (m, 2H), 2.07 – 1.99 (m, 1H), 1.94 – 1.86 (m, 4H), 1.84 – 1.70 (m, 1H), 1.69 – 1.28 (m, 21H), 1.04 – 0.82 (m, 12H), 0.72 – 0.62 (m, 2H), 0.61 – 0.55 (m, 2H). 13C NMR (101 MHz, DMSO-d6) δ 197.43, 172.33, 170.78, 170.08, 162.55, 65.17, 56.44, 54.46, 53.86, 49.49, 47.65, 42.80, 42.48, 42.33, 36.92, 34.70, 32.98, 32.46, 32.13, 28.58, 26.91, 25.16, 22.93, 19.19, 13.93, 5.87, 5.82. C35H54N4O5, HRMS calculated for m/z [M+H]+: 611.4276 (calculated), 611.5035 (found).
(1S,3aR,6aS)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)-2-((S)-2-(2-((1r,3R,5R,7S)-3-hydroxyadamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun922).
White solid. Yield: 57%. 1H NMR (400 MHz, DMSO-d6) δ 8.70 (d, J = 5.2 Hz, 1H), 8.21 (d, J = 6.9 Hz, 1H), 7.63 (d, J = 9.2 Hz, 1H), 5.01 – 4.91 (m, 1H), 4.53 – 4.43 (m, 1H), 4.31 (s, 1H), 4.26 (d, J = 3.6 Hz, 1H), 3.79 – 3.67 (m, 2H), 2.81 – 2.69 (m, 1H), 2.66 – 2.57 (m, 1H), 2.11 – 1.92 (m, 4H), 1.87 – 1.24 (m, 22H), 1.04 – 0.81 (m, 12H), 0.71 – 0.60 (m, 2H), 0.63 – 0.54 (m, 2H). 13C NMR (101 MHz, DMSO-d6) δ 197.47, 172.35, 170.73, 170.06, 162.57, 67.17, 65.19, 56.52, 55.35, 54.41, 53.86, 50.87, 48.67, 47.66, 45.07, 45.01, 42.84, 41.35, 41.13, 36.13, 35.75, 34.71, 32.40, 32.14, 32.09, 30.57, 26.94, 25.19, 22.93, 19.19, 13.94, 5.87, 5.82. C35H54N4O6, HRMS calculated for m/z [M+H]+: 627.4121 (calculated), 627.4238 (found).
(1S)-2-((S)-2-(2-((1S,3S,5S,7S)-adamantan-2-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 1024).
White solid. Yield: 72%. 1H NMR (400 MHz, CDCl3) δ 7.64 (d, J = 7.6 Hz, 1H), 7.08 (d, J = 4.1 Hz, 1H), 6.65 (d, J = 9.6 Hz, 1H), 4.74 (d, J = 9.6 Hz, 1H), 4.46 (d, J = 2.8 Hz, 1H), 3.86-3.75 (m, 2H), 2.86-2.74 (m, 3H), 2.49-2.42 (m, 1H), 2.31-2.23 (m, 2H), 1.95-1.79 (m, 13H), 1.72 (d, J = 4.6 Hz, 4H), 1.66 (dd, J = 7.9, 4.9 Hz, 2H), 1.55 (q, J = 6.1, 5.3 Hz, 4H), 1.48-1.34 (m, 5H), 0.99 (s, 9H), 0.91 (t, J = 7.3 Hz, 4H), 0.84 (d, J = 7.2 Hz, 2H), 0.61 (dd, J = 8.9, 3.8 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 196.98, 172.52, 171.43, 171.22, 160.47, 66.10, 56.39, 54.46, 53.88, 53.43, 45.75, 43.11, 41.79, 40.08, 38.82, 38.80, 38.13, 35.33, 33.46, 32.28, 32.00, 31.83, 31.66, 31.47, 31.45, 27.92, 27.79, 26.40, 25.51, 22.42, 19.02, 13.43, 6.36, 6.32. C35H54N4O5, HRMS calculated for m/z [M+H]+: 612.4172 (calculated), 612.4175 (found).
(S)-3-((S)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanamido)-2-cyclohexylacetamido)-N-cyclopropyl-2-oxohexanamide (Jun10223).
White solid. Yield: 81%. 1H NMR (500 MHz, DMSO-d6) δ 8.73 (d, J = 5.5 Hz, 1H), 8.17 (d, J = 6.5 Hz, 1H), 7.85 (d, J = 9.0 Hz, 1H), 7.46 (d, J = 9.5 Hz, 1H), 5.04-4.96 (m, 1H), 4.32 (d, J = 9.5 Hz, 1H), 4.22 (t, J = 8.5 Hz, 1H), 2.81-2.71 (m, 1H), 2.04 (d, J = 12.55 Hz, 1H), 1.93-1.84 (m, 4H), 1.72-1.26 (m, 24H), 1.19-0.81 (m, 15H), 0.70-0.55 (m, 4H). 13C NMR (125 MHz, DMSO-d6) δ 196.9, 171.2, 170.6, 170.4, 162.3, 59.8, 56.9, 53.6, 49.9, 42.5, 37.0, 34.5, 33.0, 31.7, 29.4, 28.6, 27.2, 26.4, 26.0, 23.0, 19.2, 13.8, 5.9, 5.8. C35H56N4O5 HRMS calculated for m/z [M+H]+: 613.4329 (calculated), 613.4400 (found).
(S)-3-((S)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanamido)-3-cyclohexylpropanamido)-N-cyclopropyl-2-oxohexanamide (Jun10222).
White solid. Yield: 74%. 1H NMR (500 MHz, DMSO-d6) δ 8.70 (d, J = 5.0 Hz, 1H), 8.07 (d, J = 7.0 Hz, 1H), 7.95 (d, J = 8.0 Hz, 1H), 7.50 (d, J = 9.0 Hz, 1H), 5.01-4.95 (m, 1H), 4.44-4.35 (m, 1H), 4.25 (d, J = 9.0 Hz, 1H), 2.80-2.71 (m, 1H), 1.97 (dd, J = 23.0, 12.5 Hz, 2H), 1.89 (s, 3H), 1.72-1.07 (m, 27H), 0.96-0.78 (m, 14H), 0.69-0.54 (m, 4H). 13C NMR (125 MHz, DMSO-d6) δ 197.1, 172.5, 170.5, 170.4, 162.4, 60.0, 53.7, 50.1, 49.9, 42.5, 37.0, 34.5, 33.7, 33.6, 32.9, 32.3, 31.9, 28.6, 27.2, 26.6, 26.3, 26.0, 23.0, 19.2, 13.9, 5.9, 5.9. C36H58N4O5 HRMS calculated for m/z [M+H]+: 627.4485 (calculated), 627.4589 (found).
Synthesis procedure for Jun7424
(1S,3aR,6aS)-Octahydrocyclopenta[c]pyrrole-1-carboxylic acid ethyl ester hydrochloride (2) and triethylamine (TEA) were dissolved in dichloromethane (DCM). CbzCl was added dropwise at 0 °C. After stirring 12 hours, water was added. The aqueous phase was extracted by DCM 3 times, and dried with Na2SO4. The solvent was removed in vacuo to give the crude product, which was purified by silica gel chromatography to give Jun7424-a (90% yield) 1H NMR (400 MHz, CDCl3) δ 7.46 – 7.12 (m, 5H), 5.20 – 4.96 (m, 2H), 4.23 – 3.95 (m, 3H), 3.75 (m, 1H), 3.35 (m, 1H), 2.73 – 2.60 (m, 2H), 2.01 – 1.91 (m, 1H), 1.88 – 1.68 (m, 2H), 1.62 – 1.55 (m, 2H), 1.52 – 1.39 (m, 1H), 1.29 – 1.08 (m, 3H).
Jun7424-b was synthesized from Jun7424-a following the general ester hydrolysis procedure and used for the next step without further purification.
Jun7424-c was synthesized from Jun7424-b following the general amide coupling procedure and used for the next step without further purification.
Benzyl (1S,3aR,6aS)-1-(((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate (Jun7424).
Jun7424 was synthesized from Jun7424-c by following the general DMP oxidation procedure. White solid. Yield: 60%. 1H NMR (400 MHz, DMSO-d6) δ 8.71 (d, J = 5.1 Hz, 1H), 8.33 (d, J = 6.7 Hz, 1H), 7.42 – 7.25 (m, 5H), 5.08 – 4.96 (m, 2H), 4.91 (m, 1H), 4.12 (dd, J = 8.9, 2.8 Hz, 1H), 3.64 (m, 1H), 3.22 (m, 1H), 2.76 (m, 1H), 2.59 (m, 1H), 1.89 (s, 1H), 1.78 – 1.51 (m, 5H), 1.42 (m, 2H), 1.29 (s, 1H), 0.90 (m, 1H), 0.81 (m, 2H), 0.67 (m, 2H), 0.59 (m, 2H). 13C NMR (101 MHz, DMSO-d6) δ 197.40 , 172.85 , 162.52 , 128.83 , 128.64 , 128.00 , 127.78 , 127.39 , 66.28 , 65.70 , 54.03 , 53.42 , 49.93 , 48.67 , 42.22 , 41.19 , 32.72 , 32.65 , 32.30 , 32.05 , 31.98 , 25.45 , 22.93 , 19.38 , 14.01 , 5.87. C25H33N3O5 HRMS calculated for m/z [M+H]+: 456.2498 (calculated), 456.3018 (found).
Synthesis procedure for Jun7426
Benzyl ((S)-1-((1S,3aR,6aS)-1-(((S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl)carbamoyl)hexahydrocyclopenta[c]pyrrol-2(1H)-yl)-3,3-dimethyl-1-oxobutan-2-yl)carbamate (Jun7426).
Jun7426 was synthesized by following a similar procedure as Jun7424. White solid. Yield: 40%. 1H NMR (400 MHz, DMSO-d6) δ 8.70 (d, J = 5.2 Hz, 1H), 8.23 (d, J = 6.8 Hz, 1H), 7.38 – 7.31 (m, 6H), 7.26 – 7.17 (m, 1H), 5.10 – 4.96 (m, 3H), 5.00 – 4.92 (m, 1H), 4.28 (d, J = 3.8 Hz, 1H), 4.17 (d, J = 8.7 Hz, 1H), 3.79 – 3.70 (m, 1H), 3.70 – 3.62 (m, 1H), 2.79 – 2.72 (m, 1H), 1.86 – 1.67 (m, 3H), 1.68 – 1.52 (m, 3H), 1.46 – 1.32 (m, 3H), 1.00 – 0.84 (m, 14H), 0.70 – 0.63 (m, 2H), 0.62 – 0.56 (m, 2H). 13C NMR (101 MHz, DMSO-d6) δ 197.53, 172.35, 169.97, 162.59, 156.86, 137.57, 128.75, 128.20, 128.08, 65.87, 65.36, 59.51, 54.33, 53.87, 47.65, 43.01, 40.61, 40.40, 40.20, 39.99, 39.78, 39.57, 39.36, 38.71, 34.90, 32.17, 31.86, 26.84, 25.17, 22.93, 19.19, 13.94, 5.88, 5.82. C31H44N4O6 HRMS calculated for m/z [M+H]+: 569.3339 (calculated), 569.4084 (found).
Synthesis procedure for Jun11191
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl) acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-amino-1-oxopentan-2-yl) octahydrocyclopenta [c] pyrrole-1-carboxamide (Jun11191-a).
Under nitrogen, 2-Hydroxypyridine 1-oxide (HOPO) (22.8 mg, 0.25 eq) was added to a solution of (1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl) octahydrocyclopenta [c] pyrrole-1-carboxylic acid (12) (368 mg, 0.82 mmol) and (S)-2-aminopentanamide HCl salt (141 mg, 0.93 mmol) in butan-2-one (MEK) (4 mL), and the mixture was cooled to 0 °C. DIPEA (435 μL, 2.5 mmol) was then added, followed by the addition of EDCI (189 mg, 0.98 mmol). The reaction mixture was stirred at 25 °C overnight. The solution was diluted with E.A. The separated organic layer was washed with water and saturated aqueous sodium chloride solution, dried over Na2SO4, filtered, and concentrated to afford Jun11191-a as a yellow solid (336.7 mg, 75% yield). 1H NMR (400 MHz, CDCl3) δ 7.12 (d, J = 8.0 Hz, 1H), 6.53-6.47 (m, 1H), 6.37 (d, J = 12.0 Hz, 1H), 5.85-5.80 (m, 1H), 4.71 (d, J = 8.0 Hz, 1H), 4.41-4.36 (m, 2H), 3.80 (s, 1H), 2.81 (s, 2H), 1.96-1.95 (m, 6H), 1.70-1.55(m, 18H), 1.51-1.42 (m, 3H), 1.00 (s, 9H), 0.92-0.87 (m, 3H). C31H50N4O4 ESI-MS calculated for m/z [M+H]+: 543.8 (calculated), 543.4 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl) acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-cyanobutyl) octahydro-cyclopenta [c] pyrrole-1-carboxamide (Jun11191).
Burgess reagent (320 mg, 1.25 mmol) was added to a solution of the Jun11191-a (271 mg, 0.5 mmol) in dichloromethane (6 mL). The reaction mixture was stirred at 25 °C overnight before being diluted with DCM (10 mL). The reaction mixture was quenched by sat. NaHCO3 (40 mL), washed by 1M HCl and brine (40 mL). The separated organic phase was concentrated to get a residue which was purified by flash column chromatography (E.A./hexane=1/4-1/2) to give Jun11191 as a white solid (60 mg, 23% yield). 1H NMR (400 MHz, CDCl3) δ 7.65 (d, J = 8.0 Hz, 1H), 6.08 (d, J = 12.0 Hz, 1H), 4.84 (q, J = 8.0 Hz, 1H), 4.67 (d, J = 8.0 Hz, 1H), 4.32 (s, 1H), 3.83-3.74 (m, 2H), 2.94-2.85 (m, 8H), 1.97-1.92 (m, 1H), 1.70-1.60 (m, 18H), 1.51-1.46 (m, 3H), 0.99 (s, 9H), 0.94 (d, J = 10.0 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 172.12, 171.09, 170.91, 119.12, 66.41, 56.76, 54.70, 51.64, 45.29, 43.38, 42.81, 40.30, 36.90, 35.36, 35.25, 33.13, 32.41, 31.90, 28.81, 26.70, 25.68, 18.81, 13.33. C31H48N4O3 HRMS calculated for m/z [M+H]+: 525.3805 (calculated), 525.4193 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-4-(cyclopropylamino)-3,4-dioxo-1-phenylbutan-2-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun11385).
White solid. Yield: 78%. 1H NMR (400 MHz, CDCl3) δ 7.28 (s, 3H), 7.18 (dd, J = 14.6, 7.3 Hz, 2H), 6.86 (d, J = 7.0 Hz, 1H), 6.02 (d, J = 10.1 Hz, 1H), 4.72 (t, J = 11.0 Hz, 1H), 4.46 – 4.31 (m, 1H), 3.78 (dd, J = 10.9, 4.9 Hz, 1H), 3.72 – 3.62 (m, 1H), 3.39 – 3.25 (m, 1H), 3.20 (dd, J = 14.3, 6.9 Hz, 1H), 2.82 (ddt, J = 26.3, 14.1, 6.4 Hz, 3H), 1.99 (s, 4H), 1.89 (dt, J = 12.9, 6.6 Hz, 2H), 1.73 (d, J = 12.1 Hz, 3H), 1.66 (s, 6H), 1.56 – 1.32 (m, 4H), 1.02 (d, J = 8.6 Hz, 3H), 1.01 – 0.89 (m, 9H), 0.88 (d, J = 4.2 Hz, 3H), 0.62 (d, J = 8.4 Hz, 2H). 13C NMR (101 MHz, DMSO) δ 196.55, 172.43, 170.79, 170.12, 162.37, 137.68, 129.43, 128.72, 126.95, 65.10, 56.40, 55.61, 54.43, 49.47, 47.78, 42.46, 36.91, 34.65, 32.98, 32.56, 32.17, 28.57, 26.94, 25.17, 22.95, 5.87, 5.83. C39H54N4O5, HRMS calculated for m/z [M+H]+: 659.4172 (calculated), 659.3376 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-4-(cyclopropylamino)-1-(4-fluorophenyl)-3,4-dioxobutan-2-yl)octahydrocyclo-penta[c]pyrrole-1-carboxamide (Jun11763).
White solid. Yield: 76%. 1H NMR (400 MHz, CDCl3) δ 7.17 – 7.05 (m, 2H), 7.01 – 6.88 (m, 4H), 6.08 (d, J = 9.6 Hz, 1H), 5.52 (td, J = 6.7, 5.3 Hz, 1H), 4.68 (d, J = 9.6 Hz, 1H), 4.37 (d, J = 2.8 Hz, 1H), 3.80 – 3.64 (m, 2H), 3.25 (dd, J = 14.3, 5.4 Hz, 1H), 3.11 (dd, J = 14.3, 6.9 Hz, 1H), 2.78 (dtt, J = 16.6, 8.6, 3.6 Hz, 3H), 1.95 (d, J = 3.3 Hz, 5H), 1.85 (dd, J = 13.4, 7.0 Hz, 2H), 1.74 – 1.65 (m, 4H), 1.60 (d, J = 12.4 Hz, 9H), 1.49 – 1.42 (m, 1H), 1.37 (dt, J = 13.4, 6.6 Hz, 1H), 1.00 (d, J = 7.8 Hz, 1H), 0.94 (s, 9H), 0.86 (dt, J = 6.8, 2.0 Hz, 2H), 0.60 (dd, J = 5.8, 3.1 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 195.18, 171.54, 171.04, 170.80, 160.35, 131.33, 130.81, 115.62, 66.03, 56.30, 55.35, 54.32, 51.71, 45.67, 42.99, 42.64, 36.73, 35.13, 32.97, 32.23, 31.78, 28.63, 26.46, 25.38, 22.47, 6.52, 6.44. C39H53FN4O5, HRMS calculated for m/z [M+H]+: 677.4078 (calculated), 677.4344 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-(4-fluorophenyl)-3-oxopropan-2-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun11-70-7).
White solid. Yield: 65%. 1H NMR (400 MHz, CDCl3) δ 9.60 (d, J = 2.8 Hz, 1H), 7.13 (td, J = 5.6, 2.8 Hz, 2H), 6.98 (dd, J = 6.8, 2.7 Hz, 3H), 4.75 – 4.66 (m, 2H), 4.33 (q, J = 3.5, 2.7 Hz, 1H), 3.81 (dt, J = 7.0, 3.0 Hz, 2H), 3.14 (dd, J = 6.5, 2.5 Hz, 2H), 2.81 (s, 2H), 2.39 – 2.31 (m, 1H), 2.05 (d, J = 2.7 Hz, 2H), 1.96 (s, 4H), 1.68 (s, 2H), 1.59 (s, 4H), 1.28 – 1.22 (m, 6H), 1.02 (s, 1H), 0.98 (d, J = 2.8 Hz, 9H), 0.94 (d, J = 4.0 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 198.51, 171.66, 171.61, 170.94, 160.71, 139.88 – 133.10 (m), 130.84, 115.47, 66.20, 59.88, 56.40, 54.40, 51.55, 45.99, 43.11, 42.63, 42.48, 36.73, 35.17, 32.96, 32.24, 31.75, 28.63, 26.49, 25.38. C35H48FN3O4, HRMS calculated for m/z [M+H]+: 594.3707 (calculated), 594.2794 (found).
(1S,3aR,6aS)-2-((S)-2-(3-((3R,5R,7R)-adamantan-1-yl)ureido)-3,3-dimethylbutanoyl)-N-((S)-4-(cyclopropylamino)-1-(4-fluorophenyl)-3,4-dioxobutan-2-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun11631).
White solid. Yield: 79%. 1H NMR (400 MHz, CDCl3) δ 7.20 (d, J = 7.0 Hz, 1H), 7.10 (dd, J = 8.4, 5.3 Hz, 2H), 6.93 (t, J = 8.6 Hz, 3H), 5.49 (q, J = 6.4 Hz, 1H), 5.21 (d, J = 9.8 Hz, 1H), 4.58 – 4.44 (m, 2H), 4.40 (d, J = 2.6 Hz, 1H), 3.81 (dd, J = 10.8, 3.6 Hz, 1H), 3.71 (dd, J = 10.7, 7.1 Hz, 1H), 3.47 (s, 1H), 3.23 (dd, J = 14.2, 5.8 Hz, 1H), 3.10 (dd, J = 14.2, 6.7 Hz, 1H), 2.75 (dtq, J = 14.9, 8.1, 3.9 Hz, 3H), 2.06 – 2.02 (m, 3H), 1.92 (s, 5H), 1.82 (q, J = 6.5 Hz, 2H), 1.65 (d, J = 6.3 Hz, 5H), 1.51 (ddd, J = 11.1, 8.1, 4.7 Hz, 2H), 1.44 – 1.38 (m, 1H), 0.98 (d, J = 8.1 Hz, 1H), 0.93 (s, 9H), 0.85 (d, J = 7.4 Hz, 2H), 0.64 – 0.52 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 195.43, 172.96, 171.43, 160.73, 160.50, 156.70, 131.42, 130.88, 115.56, 66.09, 57.15, 55.43, 54.12, 50.86, 46.12, 43.10, 42.43, 36.41, 35.19, 32.04, 31.43, 29.53, 26.51, 25.25, 22.48, 6.51, 6.44. C38H52FN5O5, HRMS calculated for m/z [M+H]+: 678.4031 (calculated), 678.3202 (found).
Synthesis of Jun11705.
To a solution of benzothiazole 23 (5.4 g, 40 mmol) in 100 mL anhydrous tetrahydrofuran was added n-butyl lithium (2.5 M solution in hexane, 40 mmol) dropwise at −78 °C in 5 min with stirring under nitrogen. Upon completion of the addition, the reaction mixture was stirred for 30 min at −78 °C. A solution of tert-butyl (S)-(3-(4-fluorophenyl)-1-(methoxy(methyl)amino)-1-oxopropan-2-yl)carbamate 22 (3.26 g, 10 mmol) in tetrahydrofuran (50 mL) was added at a rate to maintain the reaction temperature below −70 °C. The resulting mixture was then slowly warmed to room temperature and quenched with saturated aqueous NH4Cl (50 mL). The resulting organic layer was separated, washed with water. The organic extract was dried over Na2SO4, filtered, and concentrated under reduced pressure. The crude material was purified by chromatography to afford tert-butyl (S)-(1-(benzo[d]thiazol-2-yl)-3-(4-fluorophenyl)-1-oxopropan-2-yl)carbamate 24 (3.2 g, 80%).
To a solution of 24 (2.0 g, 5 mmol) in 20 mL dichloromethane was added 5M, HCl in dioxane (4.0 mL, 20 mmol) The reaction mixture was stirred at room temperature for 2 h and the solvent was removed under reduced pressure to give crude (S)-2-amino-1-(benzo[d]thiazol-2-yl)-3-(4-fluorophenyl)propan-1-one 25. The resulting solid was used directly for the next step reaction without further purification. A mixture of substituted of carboxylic acid 12 (2.0 g, 4.6 mmol) and the resulting amine 25 (1.4 g, 4.6 mmol) in 10 mL DMF was added HATU (1.7 g, 4.6 mmol) followed by DIPEA (1.7 mL, 9.2 mmol). The reaction mixture was stirred at room temperature for 6 h, then diluted with water (30 mL) and extracted with EtOAc (40 mL). The organics were dried with Na2SO4 and concentrated in vacuo to give (1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-4-(benzo[d]thiazol-2-yl)-1-(4-fluorophenyl)-3,4-dioxobutan-2-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide Jun11705 (2.8 g, 80%) which were purified by flash chromatography.
(S)-2-amino-1-(benzo[d]thiazol-2-yl)-3-(4-fluorophenyl)propan-1-one (25).
White solid. Yield: 82%. 1H NMR (400 MHz, MeOD) δ 8.22 – 8.08 (m, 1H), 8.06 (dd, J = 7.3, 1.9 Hz, 1H), 7.57 (pd, J = 7.2, 1.5 Hz, 2H), 7.21 (dd, J = 8.4, 5.3 Hz, 2H), 6.93 (t, J = 8.7 Hz, 2H), 5.44 (dd, J = 8.0, 5.4 Hz, 1H), 3.59 – 3.43 (m, 1H), 3.26 (dd, J = 14.5, 8.0 Hz, 1H). 13C NMR (101 MHz, MeOD) δ 189.47, 162.46, 153.25, 137.18, 131.17, 131.09, 128.64, 127.57, 125.41, 122.57, 115.53, 57.16, 47.41, 47.20, 46.98, 36.00. C16H14FN2OS, LCMS calculated for m/z [M+H]+: 301.4 (calculated), 301.0 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((R)-4-(benzo[d]thiazol-2-yl)-1-(4-fluorophenyl)-3,4-dioxobutan-2-yl)octahydrocyclo-penta[c]pyrrole-1-carboxamide (Jun11705).
White solid. Yield: 81%. 1H NMR (400 MHz, CDCl3) δ 8.21 (t, J = 8.3 Hz, 1H), 8.00 (d, J = 7.9 Hz, 1H), 7.64 – 7.49 (m, 2H), 7.11 – 7.07 (m, 2H), 6.92 (dt, J = 13.9, 8.6 Hz, 2H), 6.16 – 5.95 (m, 2H), 4.71 (d, J = 9.6 Hz, 1H), 4.43 (dd, J = 11.1, 2.8 Hz, 1H), 3.76 (d, J = 5.8 Hz, 2H), 3.44 (ddd, J = 26.5, 14.1, 5.5 Hz, 1H), 3.24 (ddd, J = 43.2, 14.2, 6.9 Hz, 1H), 2.98 – 2.84 (m, 1H), 2.77 (dddd, J = 29.1, 11.9, 6.8, 3.2 Hz, 2H), 1.97 (d, J = 7.7 Hz, 5H), 1.89 – 1.81 (m, 2H), 1.70 (d, J = 12.1 Hz, 3H), 1.61 (d, J = 16.4 Hz, 8H), 1.51 – 1.46 (m, 1H), 1.41 – 1.36 (m, 1H), 1.26 (d, J = 3.1 Hz, 2H), 0.99 (s, 9H), 0.87 (d, J = 7.0 Hz, 1H). 13C NMR (101 MHz, CDCl3) δ 192.30, 191.90, 171.76, 171.10, 170.75, 170.66, 163.72, 153.54, 137.27, 131.05, 128.15, 127.24, 127.21, 125.86, 122.40, 115.24, 66.33, 56.38, 54.39, 51.79, 46.04, 43.03, 42.65, 36.76, 35.27, 32.99, 32.38, 31.90, 28.66, 26.53, 25.43, 22.64, 14.10. C42H51FN4O4S, HRMS calculated for m/z [M+H]+: 727.3693 (calculated), 727.3794 (found).
(1S)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N ((S)-1-(4-(tert-butoxy)phenyl)-4-(cyclopropylamino)-3,4-dioxobutan-2 yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11671).
White solid. Yield: 61%. 1H NMR (400 MHz, CDCl3) δ 7.01 (d, J = 8.4 Hz, 2H), 6.96 (dd, J = 12.8, 5.4 Hz, 2H), 6.87 (d, J = 8.3 Hz, 2H), 6.16 (d, J = 9.5 Hz, 1H), 5.52 (q, J = 6.4 Hz, 1H), 4.69 (d, J = 9.6 Hz, 1H), 4.41 (d, J = 3.0 Hz, 1H), 3.79-3.67 (m, 2H), 3.24-3.10 (m, 2H), 2.82-2.71 (m, 3H), 1.97 (s, 5H), 1.90-1.80 (m, 2H), 1.71-1.56 (m, 16H), 1.32 (s, 8H), 1.00 (s, 2H), 0.97 (s, 8H), 0.83 (d, J = 6.5 Hz, 2H), 0.62-0.55 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 195.33, 171.42, 170.98, 170.72, 160.42, 154.48, 130.26, 129.71, 124.09, 124.07, 78.27, 66.03, 56.32, 55.27, 54.27, 51.62, 45.89, 42.93, 42.60, 36.70, 36.66, 35.17, 32.92, 32.25, 31.76, 28.81, 28.59, 26.52, 25.36, 22.41, 6.45, 6.38. C43H62N4O6, HRMS calculated for m/z [M+H]+: 731.4748 (calculated), 731.4737 (found).
(1S)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((2S)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-(thiophen-3-yl)butan-2 yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun11818).
White solid. Yield: 63%. 1H NMR (400 MHz, CDCl3) δ 7.11 (d, J = 5.1 Hz, 1H), 6.93-6.86 (m, 3H), 6.82 (s, 1H), 6.11 (d, J = 9.6 Hz, 1H), 5.50 (q, J = 5.8 Hz, 1H), 4.70 (d, J = 9.6 Hz, 1H), 4.39 (d, J = 2.9 Hz, 1H), 3.79-3.73 (m, 2H), 3.59 (dd, J = 15.3, 5.8 Hz, 1H), 3.50-3.46 (m, 1H), 2.84 (dd, J = 9.6, 4.1 Hz, 1H), 2.80-2.74 (m, 2H), 1.96 (s, 2H), 1.95-1.93 (m, 3H), 1.90-1.83 (m, 2H), 1.70-1.6 (m, 4H), 1.59 (dd, J = 13.0, 2.8 Hz, 9H), 1.52-1.35 (m, 3H), 0.97 (s, 9H), 0.85 (q, J = 7.2, 6.7 Hz, 2H), 0.60 (t, J = 4.4 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 194.75, 171.62, 171.05, 170.83, 160.24, 137.15, 127.24, 127.02, 124.82, 66.26, 60.37, 56.37, 55.54, 54.34, 53.41, 51.70, 45.99, 42.97, 42.65, 36.74, 35.21, 32.98, 32.31, 31.77, 31.49, 28.64, 26.75, 26.55, 26.51, 25.38, 22.45, 14.18, 6.55, 6.44. C37H52N4O5S, HRMS calculated for m/z [M+H]+: 665.3737 (calculated), 665.3740 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-4-(cyclopropylamino)-1-(1H-indol-3-yl)-3,4-dioxobutan-2-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11862).
White solid. Yield: 51%. 1H NMR (400 MHz, CDCl3) δ 8.47 (s, 1H), 7.50 (d, J = 7.9 Hz, 1H), 7.30 (d, J = 8.0 Hz, 1H), 7.15 (d, J = 10.2 Hz, 2H), 7.06 (d, J = 7.5 Hz, 1H), 6.99 (d, J = 6.6 Hz, 1H), 6.83 (d, J = 3.9 Hz, 1H), 6.11 (d, J = 9.6 Hz, 1H), 5.63 (q, J = 6.1 Hz, 1H), 4.65 (d, J = 9.6 Hz, 1H), 4.39 (d, J = 2.8 Hz, 1H), 3.72-3.67 (m, 2H), 3.41 (dd, J = 5.9, 2.6 Hz, 2H), 2.80-2.71 (m, 4H), 1.94 (d, J = 5.5 Hz, 8H), 1.59 (d, J = 11.8 Hz, 11H), 0.88 (s, 10H), 0.82 (d, J = 5.2 Hz, 2H), 0.53 (d, J = 4.1 Hz, 2H).13C NMR (101 MHz, DMSO-d6) δ 195.47, 171.40, 171.03, 170.83, 160.55, 136.15, 127.32, 123.58, 122.12, 119.52, 118.55, 111.23, 109.25, 66.04, 56.40, 54.80, 54.39, 51.68, 45.90, 42.99, 42.64, 36.73, 35.14, 32.96, 32.34, 31.87, 28.63, 27.06, 26.53, 26.39, 25.41, 22.38, 6.42, 6.30. C41H55N5O5, HRMS calculated for m/z [M+H]+: 698.4281 (calculated), 698.4259 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-4-(cyclopropylamino)-1-methoxy-3,4-dioxobutan-2-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11672).
White solid. Yield: 63%. 1H NMR (400 MHz, CDCl3) δ 7.36 (dd, J = 18.7, 7.5 Hz, 1H), 7.01 (dd, J = 21.2, 4.0 Hz, 1H), 6.19 (d, J = 9.6 Hz, 1H), 5.47-5.41 (m, 1H), 4.74 (t, J = 9.1 Hz, 1H), 4.52 (dd, J = 13.0, 2.8 Hz, 1H), 4.17-4.11 (m, 1H), 3.82-3.69 (m, 3H), 3.26 (d, J = 6.8 Hz, 3H), 2.97 (td, J = 8.1, 4.1 Hz, 1H), 2.82-2.75 (m, 2H), 1.95 (d, J = 4.1 Hz, 6H), 1.88 (dd, J = 12.6, 7.1 Hz, 2H), 1.68 (s, 3H), 1.60 (d, J = 12.7 Hz, 10H), 1.52-1.39 (m, 3H), 1.02 (d, J = 6.3 Hz, 9H), 0.85 (d, J = 5.8 Hz, 2H), 0.61 (d, J = 3.3 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 193.60, 193.25, 171.66, 171.40, 170.98, 170.91, 170.83, 170.79, 160.29, 160.27, 71.88, 71.86, 65.80, 59.15, 59.11, 56.36, 56.34, 55.53, 55.45, 54.28, 54.20, 51.66, 45.02, 44.99, 42.95, 42.83, 42.61, 36.70, 35.34, 35.29, 32.94, 32.31, 32.24, 31.94, 31.88, 28.60, 26.45, 26.43, 25.50, 25.48, 22.37, 22.34, 6.41, 6.35. C34H52N4O6, HRMS calculated for m/z [M+H]+: 613.3965 (calculated), 613.3887 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-cyclopropyl-4-(cyclopropylamino)-3,4-dioxobutan-2-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11761).
White solid. Yield: 53%. 1H NMR (400 MHz, CDCl3) δ 7.21 (d, J = 6.7 Hz, 1H), 6.91 (s, 1H), 6.01 (d, J = 9.6 Hz, 1H), 5.37 (q, J = 6.3 Hz, 1H), 4.71 (d, J = 9.5 Hz, 1H), 4.46 (d, J = 2.6 Hz, 1H), 3.79- 3.6 (m, 3H), 2.96 (d, J = 8.1 Hz, 1H), 2.84-2.74 (m, 3H), 1.96 (s, 8H), 1.86 (ddd, J = 26.9, 13.5, 6.7 Hz, 4H), 1.70 (d, J = 12.8 Hz, 6H), 1.62 (s, 7H), 1.55 (s, 2H), 0.99 (s, 8H), 0.85 (d, J = 7.5 Hz, 2H), 0.59 (dd, J = 8.5, 3.7 Hz, 2H), 0.44 (d, J = 8.1 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 196.47, 171.42, 170.91, 170.78, 160.47, 65.99, 56.36, 54.85, 54.43, 53.42, 51.55, 45.43, 43.06, 42.64, 42.61, 36.75, 36.71, 35.27, 32.91, 32.30, 31.89, 28.60, 26.49, 25.51, 22.38, 7.48, 6.44, 6.36, 5.27, 4.60. C36H54N4O5, HRMS calculated for m/z [M+H]+: 623.4172 (calculated), 623.4183 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethyl-butanoyl)-N-((S)-1-(cyclopropylamino)-5-methyl-1,2-dioxohexan-3-yl)octahydro-cyclopenta[c]pyrrole-1-carboxamide (Jun11-38-9).
White solid. Yield: 80%. 1H NMR (400 MHz, CDCl3) δ 7.04 (d, J = 7.0 Hz, 1H), 6.91 (d, J = 3.9 Hz, 1H), 6.03 (d, J = 9.6 Hz, 1H), 5.30 (s, 1H), 4.72 (d, J = 9.5 Hz, 1H), 4.44 (d, J = 2.5 Hz, 1H), 3.81 – 3.61 (m, 2H), 2.96 (q, J = 7.8 Hz, 1H), 2.79 (ddq, J = 14.9, 7.4, 4.0 Hz, 2H), 1.95 (s, 4H), 1.91 – 1.85 (m, 2H), 1.69 (dt, J = 13.0, 6.2 Hz, 7H), 1.62 (s, 4H), 1.58 (d, J = 4.3 Hz, 4H), 1.56 – 1.44 (m, 4H), 1.38 (dt, J = 20.6, 8.1 Hz, 2H), 0.99 (d, J = 7.8 Hz, 11H), 0.92 (d, J = 5.7 Hz, 3H), 0.89 – 0.87 (m, 1H), 0.87 – 0.81 (m, 3H), 0.59 (dt, J = 11.5, 6.0 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 196.34, 171.66, 170.93, 170.83, 160.54, 65.99, 56.40, 54.42, 52.90, 51.80, 44.74, 43.08, 42.72, 40.47, 36.79, 35.31, 33.04, 32.33, 28.70, 26.49, 25.57, 25.17, 23.28, 22.43, 21.34, 6.43. C36H56N4O5, HRMS calculated for m/z [M+Na]+: 647.4148 (calculated), 647.3351 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-3-(cyclopropylamino)-2,3-dioxo-1-phenylpropyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11817).
White solid. Yield: 62%. 1H NMR (400 MHz, CDCl3) δ 7.73 (d, J = 6.5 Hz, 1H), 7.37 (d, J = 8.6 Hz, 3H), 7.30 (s, 2H), 7.03 (d, J = 4.0 Hz, 1H), 6.42 (d, J = 6.4 Hz, 2H), 6.21 (d, J = 9.6 Hz, 1H), 4.66 (d, J = 9.5 Hz, 1H), 4.57 (d, J = 2.6 Hz, 1H), 3.77-3.71 (m, 2H), 3.64 (dd, J = 10.8, 7.9 Hz, 1H), 3.03-2.96 (m, 2H), 2.83-2.75 (m, 2H), 2.70-2.65 (m, 2H), 1.95 (s, 7H), 1.67 (s, 5H), 1.05 (s, 5H), 0.80 (s, 10H), 0.60-0.53 (m, 2H), 0.51-0.45 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 192.63, 192.26, 171.85, 171.66, 170.87, 170.35, 170.24, 159.97, 133.78, 133.68, 129.11, 129.08, 128.72, 128.66, 128.46, 128.43, 65.76, 65.72, 57.94, 57.92, 56.41, 56.29, 54.32, 51.57, 51.54, 44.35, 44.32, 42.93, 42.91, 42.60, 36.70, 35.30, 35.10, 32.97, 32.94, 32.30, 32.24, 32.03, 31.92, 28.60, 26.51, 26.30, 25.58, 25.55, 22.42, 22.39, 6.35, 6.31. C38H52N4O5, HRMS calculated for m/z [M+H]+: 645.4016 (calculated), 645.4038 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-3-(cyclopropylamino)-1-(4-fluorophenyl)-2,3-dioxopropyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun11861).
White solid. Yield: 71%. 1H NMR (400 MHz, CDCl3) δ 7.79 (dd, J = 17.4, 6.4 Hz, 1H), 7.40-7.35 (m, 2H), 7.01 (d, J = 8.9 Hz, 2H), 6.40 (t, J = 6.8 Hz, 1H), 6.19 (dd, J = 27.0, 9.5 Hz, 1H), 4.70 (dd, J = 40.8, 9.5 Hz, 1H), 4.51 (dd, J = 23.9, 2.7 Hz, 1H), 3.73 (dddd, J = 45.0, 26.4, 10.8, 5.9 Hz, 3H), 3.02-2.94 (m, 1H), 2.83-2,75 (m, 1H), 2.72-2.66 (m, 1H), 1.97 (d, J = 8.0 Hz, 6H), 1.86 (dt, J = 14.3, 7.1 Hz, 2H), 1.70-1.57 (m, 16H), 1.43 (ddt, J = 33.1, 13.0, 6.6 Hz, 3H), 1.05 (s, 3H), 0.82 (s, 4H), 0.59-0.48 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 192.41, 192.09, 171.93, 171.74, 170.91, 170.38, 170.29, 159.90, 159.84, 130.36, 130.28, 129.69, 116.20, 115.99, 65.80, 57.23, 57.19, 56.42, 56.30, 54.34, 51.62, 51.59, 44.32, 42.96, 42.92, 42.61, 36.69, 35.29, 35.09, 32.98, 32.96, 32.28, 32.22, 31.98, 31.88, 28.60, 26.50, 26.29, 25.56, 25.52, 22.45, 22.42, 6.39, 6.36. C38H51FN4O5, HRMS calculated for m/z [M+H]+: 663.3922 (calculated), 663.3927 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-cyclohexyl-3-(cyclopropylamino)-2,3-dioxopropyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11762).
White solid. Yield: 40%. 1H NMR (400 MHz, DMSO-d6) δ 8.69 (d, J = 5.1 Hz, 1H), 8.11 (d, J = 7.5 Hz, 1H), 7.63 (d, J = 9.2 Hz, 1H), 4.96 (t, J = 6.6 Hz, 1H), 4.51 (d, J = 9.1 Hz, 1H), 4.33 (d, J = 3.5 Hz, 1H), 4.03 (q, J = 7.1 Hz, 1H), 3.72 (d, J = 7.1 Hz, 2H), 2.74 (td, J = 7.4, 3.7 Hz, 1H), 2.63 (tq, J = 7.8, 4.0, 3.2 Hz, 1H), 1.88 (s, 5H), 1.66-1.53 (m, 19H), 1.19-1.04 (m, 8H), 0.92 (s, 11H), 0.65 (d, J = 6.3 Hz, 2H), 0.56 (d, J = 3.6 Hz, 2H). 13C NMR (101 MHz, DMSO-d6) δ 197.63, 172.44, 170.77, 170.74, 170.16, 162.91, 65.10, 60.20, 58.22, 56.42, 54.45, 49.48, 47.43, 42.84, 42.49, 39.25, 36.94, 34.74, 32.97, 32.31, 32.11, 29.72, 28.58, 28.24, 26.91, 26.15, 26.05, 25.98, 25.14, 22.97, 21.20, 14.54, 5.89, 5.85. C38H58N4O5, HRMS calculated for m/z [M+H]+: 651.4485 (calculated), 651.4490 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-3-amino-1-cyclohexyl-2,3-dioxopropyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11725).
White solid. Yield: 62%. 1H NMR (400 MHz, CDCl3) δ 7.33 (d, J = 7.5 Hz, 1H), 6.94 (d, J = 3.1 Hz, 1H), 6.55 (d, J = 3.3 Hz, 1H), 6.38 (d, J = 9.7 Hz, 1H), 5.17 (t, J = 6.7 Hz, 1H), 4.75 (d, J = 9.6 Hz, 2H), 4.46 (d, J = 2.7 Hz, 1H), 3.80 (dt, J = 10.8, 3.8 Hz, 2H), 3.75-3.68 (m, 2H), 2.96-2.92 (m, 1H), 2.82 (ddt, J = 12.1, 7.9, 4.3 Hz, 2H), 1.96 (s, 9H), 1.91-1.82 (m, 5H), 1.69 (d, J = 12.5 Hz, 12H), 1.02 (s, 13H). 13C NMR (101 MHz, CDCl3) δ 196.19, 171.73, 171.06, 170.95, 161.56, 66.05, 58.40, 56.37, 54.45, 51.53, 44.82, 43.00, 42.63, 39.91, 36.73, 35.26, 32.98, 32.22, 31.87, 30.03, 28.63, 28.29, 26.47, 25.88, 25.80, 25.75, 25.49. C35H54N4O5, HRMS calculated for m/z [M+H]+: 611.4172 (calculated), 611.4176 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-cyclopentyl-3-(cyclopropylamino)-2,3-dioxopropyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11808).
White solid. Yield: 57%. 1H NMR (400 MHz, CDCl3) δ 7.43 (d, J = 7.0 Hz, 1H), 7.16 (d, J = 4.2 Hz, 1H), 6.33 (d, J = 9.6 Hz, 1H), 5.15 (t, J = 7.2 Hz, 1H), 4.76 (d, J = 9.5 Hz, 1H), 4.50 (d, J = 2.0 Hz, 1H), 3.78 (d, J = 5.1 Hz, 2H), 2.82 (d, J = 3.7 Hz, 3H), 2.35 (q, J = 8.2 Hz, 1H), 2.05-1.96 (m, 5H), 1.85 (dt, J = 13.7, 7.0 Hz, 2H), 1.71-1.56 (m, 18H), 1.54-1.46 (m, 4H), 1.36 (dt, J = 13.0, 6.4 Hz, 2H), 1.30-1.23 (m, 1H), 1.01 (s, 8H), 0.84 (d, J = 7.6 Hz, 2H), 0.62 (d, J = 8.9 Hz, 2H).13C NMR (101 MHz, CDCl3) δ 196.09, 171.41, 171.20, 170.80, 160.91, 65.74, 57.04, 56.31, 54.46, 51.46, 45.37, 42.93, 42.60, 41.44, 36.69, 35.22, 32.91, 32.20, 31.83, 28.92, 28.83, 28.58, 26.48, 25.45, 24.95, 24.55, 22.50, 6.43, 6.30. C37H56N4O5, HRMS calculated for m/z [M+H]+: 637.4329 (calculated), 637.4333 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-(cyclopropylamino)-4-methyl-1,2-dioxopentan-3-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11718).
White solid. Yield: 42%. 1H NMR (400 MHz, CDCl3) δ 7.19 (d, J = 7.7 Hz, 1H), 6.94 (d, J = 3.9 Hz, 1H), 6.14 (d, J = 9.6 Hz, 1H), 5.24 (dd, J = 7.6, 5.0 Hz, 1H), 4.74 (d, J = 9.6 Hz, 1H), 4.45 (d, J = 2.6 Hz, 1H), 3.77-3.75 (m, 2H), 2.90-2.85 (m, 1H), 2.84-2.75(m, 2H), 2.40-2.32 (m, 1H), 1.96 (s, 4H), 1.86 (dd, J = 13.3, 6.8 Hz, 2H), 1.68 (t, J = 10.7 Hz, 6H), 1.63 (s, 5H), 1.59 (s, 4H), 1.56 (s, 1H), 1.53-1.34 (m, 3H), 1.01 (d, J = 2.5 Hz, 10H), 0.85 (d, J = 7.0 Hz, 5H), 0.60 (dt, J = 6.7, 3.0 Hz, 2H).13C NMR (101 MHz, CDCl3) δ 196.38, 171.62, 171.26, 170.79, 160.60, 66.15, 58.80, 56.38, 54.44, 51.75, 45.25, 43.07, 42.69, 36.77, 35.34, 33.00, 32.25, 31.84, 30.21, 28.67, 26.55, 25.50, 22.49, 19.91, 17.53, 6.53, 6.46. C35H54N4O5, HRMS calculated for m/z [M+H]+: 611.4172 (calculated), 611.4175 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((3S,4S)-1-(cyclopropylamino)-4-methyl-1,2-dioxohexan-3-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun11756).
White solid. Yield: 74%. 1H NMR (400 MHz, CDCl3) δ 7.26 (d, J = 7.7 Hz, 1H), 7.02 (d, J = 4.0 Hz, 1H), 6.20 (d, J = 9.6 Hz, 1H), 5.24 (dd, J = 7.7, 5.5 Hz, 1H), 4.74 (d, J = 9.7 Hz, 1H), 4.46 (d, J = 2.6 Hz, 1H), 3.76 (d, J = 6.9 Hz, 2H), 2.91-2.76 (m, 4H), 2.13-2.05 (m, 1H), 1.97 (s, 5H), 1.90-1.82 (m, 3H), 1.68 (t, J = 11.6 Hz, 6H), 1.62 (s, 5H), 1.52-1.45 (m, 2H), 1.41-1.31 (m, 3H), 1.01 (s, 9H), 0.96 (d, J = 6.8 Hz, 3H), 0.85 (t, J = 7.3 Hz, 5H), 0.62-0.59 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 196.54, 171.53, 171.16, 170.75, 160.70, 66.03, 60.34, 58.34, 56.33, 54.41, 51.68, 51.63, 45.17, 43.01, 42.64, 42.62, 36.86, 36.71, 35.28, 32.95, 32.21, 31.83, 28.61, 26.48, 25.47, 24.52, 22.45, 16.05, 14.16, 14.10, 11.25, 6.48, 6.40. C36H56N4O5, HRMS calculated for m/z [M+H]+: 625.4329 (calculated), 625.4330 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-(cyclopropylamino)-4,4-dimethyl-1,2-dioxopentan-3-yl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun11724).
White solid. Yield: 50%. 1H NMR (400 MHz, CDCl3) δ 7.52 (dd, J = 32.5, 7.2 Hz, 1H), 6.98 (dd, J = 51.0, 4.0 Hz, 1H), 6.16 (dd, J = 30.5, 9.6 Hz, 1H), 5.37 (dd, J = 7.2, 3.1 Hz, 1H), 4.74 (d, J = 9.6 Hz, 1H), 4.48 (dd, J = 5.4, 2.2 Hz, 1H), 3.76 (d, J = 5.2 Hz, 1H), 2.85-2.73 (m, 3H), 1.96 (d, J = 2.9 Hz, 5H), 1.89-1.82 (m, 2H), 1.71-1.56 (m, 14H), 1.51-1.31 (m, 3H), 1.03-0.98 (m, 18H), 0.84 (ddd, J = 7.4, 4.6, 3.1 Hz, 2H), 0.65-0.53 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 197.56, 171.61, 170.93, 170.74, 161.21, 65.83, 65.62, 58.76, 58.68, 56.35, 56.33, 54.43, 54.25, 51.68, 51.61, 45.09, 44.21, 42.97, 42.90, 42.63, 42.61, 36.70, 35.22, 35.05, 34.77, 32.96, 32.94, 32.15, 32.08, 31.84, 31.71, 28.60, 26.67, 26.51, 26.43, 25.54, 25.44, 22.54, 22.46, 6.60, 6.56, 6.36, 6.32. C36H56N4O5, HRMS calculated for m/z [M+H]+: 625.4329 (calculated), 625.4425 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-(1-(2-(cyclopropylamino)-2 oxoacetyl)cyclobutyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11-80-6).
White solid. Yield: 68%. 1H NMR (400 MHz, CDCl3) δ 8.33 (s, 1H), 6.95 (d, J = 4.3 Hz, 1H), 6.46 (d, J = 9.6 Hz, 1H), 4.75 (d, J = 9.6 Hz, 1H), 4.39 (d, J = 3.6 Hz, 1H), 3.87-3.77 (m, 2H), 2.97-2.90 (m, 1H), 2.79-2.73 (m, 2H), 2.71-2.65 (m, 1H), 2.61-2.53 (m, 1H), 2.43 (dt, J = 12.3, 8.8 Hz, 1H), 2.15-2.07 (m, 2H), 1.98-1.95 (m, 5H), 1.84 (ddd, J = 13.9, 8.3, 6.1 Hz, 3H), 1.70 (d, J = 11.8 Hz, 4H), 1.64 (s, 8H), 1.60-1.57 (m, 2H), 1.57 (d, J = 6.6 Hz, 1H), 1.46-1.40 (m, 2H), 1.02 (s, 9H), 0.79 (d, J = 6.5 Hz, 2H), 0.58 (dd, J = 15.8, 3.5 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 195.15, 171.46, 171.27, 170.93, 162.13, 65.69, 61.03, 56.47, 54.48, 51.27, 46.26, 43.17, 42.59, 36.72, 35.27, 32.90, 32.02, 31.59, 31.32, 30.59, 28.60, 26.68, 25.33, 22.36, 14.74, 6.36, 6.22. C35H52N4O5, HRMS calculated for m/z [M+H]+: 609.4016 (calculated), 609.4020 (found).
(1S,3aR,6aS)-2-((S)-2-(2-((3S,5S,7S)-adamantan-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-(1-(2-(cyclopropylamino)-2-oxoacetyl)cyclohexyl)octahydrocyclopenta[c]pyrrole-1-carboxamide (Jun 11-80-5).
White solid. Yield: 51%. 1H NMR (400 MHz, CDCl3) δ 7.43 (s, 1H), 6.76 (d, J = 3.8 Hz, 1H), 6.13 (d, J = 9.6 Hz, 1H), 4.75 (d, J = 9.6 Hz, 1H), 4.46 (d, J = 2.6 Hz, 1H), 3.76 (d, J = 6.2 Hz, 2H), 2.93-2.86 (m, 1H), 2.82-2.76 (m,1H), 2.70-2.64 (m, 1H), 2.18 (s, 1H), 1.97-1.95 (m, 5H), 1.91-1.78 (m, 5H), 1.70 (d, J = 12.9 Hz, 8H), 1.63-1.55 (m, 8H), 1.53-1.45 (m, 4H), 1.38 (dd, J = 13.0, 6.6 Hz, 1H), 1.03 (s, 10H), 0.77 (d, J = 5.5 Hz, 2H), 0.54 (d, J = 3.7 Hz, 2H).13C NMR (101 MHz, CDCl3) δ 198.10, 171.76, 170.97, 170.72, 162.69, 65.90, 61.40, 56.34, 54.34, 51.65, 44.61, 42.90, 42.62, 36.71, 35.25, 32.96, 32.18, 31.83, 31.72, 31.33, 28.61, 26.57, 26.46, 25.49, 24.93, 22.15, 21.41, 21.28, 6.34, 6.24. C37H56N4O5, HRMS calculated for m/z [M+H]+: 637.4329 (calculated), 637.4334 (found).
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the National Institutes of Health (NIH) grants AI157046 and AI147325 to J. W.
ABBREVIATIONS
- AFM
acute flaccid myelitis
- EV-A71
enterovirus A71
- EV-D68
enterovirus D68
- 2Apro
2A protease
- MD
molecular dynamics simulations
Footnotes
Competing interests
Jun Wang is an inventor of a filed patent claiming the use of Jun11762 and related compounds as potential EV-D68 antiviral drugs.
Supporting information: Tables S1; HNMR and CNMR spectra for final products; HPLC traces of selected compound; Molecular Formula Strings.
REFERENCES
- (1).Sun J; Hu XY; Yu XF, Current Understanding of Human Enterovirus D68. Viruses 2019, 11, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Schieble JH; Fox VL; Lennette EH, A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol 1967, 85, 297–310. [DOI] [PubMed] [Google Scholar]
- (3).Midgley CM; Watson JT; Nix WA; Curns AT; Rogers SL; Brown BA; Conover C; Dominguez SR; Feikin DR; Gray S, et al. , Severe respiratory illness associated with a nationwide outbreak of enterovirus D68 in the USA (2014): a descriptive epidemiological investigation. Lancet Respir Med 2015, 3, 879–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Poelman R; Schuffenecker I; Van Leer-Buter C; Josset L; Niesters HG; Lina B; group E.-E. E.-D. s., European surveillance for enterovirus D68 during the emerging North-American outbreak in 2014. J Clin Virol 2015, 71, 1–9. [DOI] [PubMed] [Google Scholar]
- (5).Oberste MS; Maher K; Schnurr D; Flemister MR; Lovchik JC; Peters H; Sessions W; Kirk C; Chatterjee N; Fuller S, et al. , Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J Gen Virol 2004, 85, 2577–2584. [DOI] [PubMed] [Google Scholar]
- (6).Messacar K; Asturias EJ; Hixon AM; Van Leer-Buter C; Niesters HGM; Tyler KL; Abzug MJ; Dominguez SR, Enterovirus D68 and acute flaccid myelitis-evaluating the evidence for causality. Lancet Infect Dis 2018, 18, e239–e247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Opanda SM; Wamunyokoli F; Khamadi S; Coldren R; Bulimo WD, Genetic diversity of human enterovirus 68 strains isolated in Kenya using the hypervariable 3’-end of VP1 gene. PLoS One 2014, 9, e102866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Agol VI, [Various problems in the molecular biology of picornaviruses]. Mol Biol (Mosk) 1977, 11, 1213–9. [PubMed] [Google Scholar]
- (9).Musharrafieh R; Ma C; Zhang J; Hu Y; Diesing JM; Marty MT; Wang J, Validating Enterovirus D68–2A(pro) as an Antiviral Drug Target and the Discovery of Telaprevir as a Potent D68–2A(pro) Inhibitor. J Virol 2019, 93, e02221–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Tan J; George S; Kusov Y; Perbandt M; Anemuller S; Mesters JR; Norder H; Coutard B; Lacroix C; Leyssen P, et al. , 3C protease of enterovirus 68: structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J Virol 2013, 87, 4339–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hu Y; Musharrafieh R; Zheng M; Wang J, Enterovirus D68 Antivirals: Past, Present, and Future. ACS Infect Dis 2020, 6, 1572–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Baggen J; Thibaut HJ; Strating J; van Kuppeveld FJM, The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol 2018, 16, 368–381. [DOI] [PubMed] [Google Scholar]
- (13).Cai Q; Yameen M; Liu W; Gao Z; Li Y; Peng X; Cai Y; Wu C; Zheng Q; Li J, et al. , Conformational plasticity of the 2A proteinase from enterovirus 71. J Virol 2013, 87, 7348–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ma C; Sacco MD; Hurst B; Townsend JA; Hu Y; Szeto T; Zhang X; Tarbet B; Marty MT; Chen Y, et al. , Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res 2020, 30, 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tan B; Joyce R; Tan H; Hu Y; Wang J, SARS-CoV-2 Main Protease Drug Design, Assay Development, and Drug Resistance Studies. Acc. Chem. Res 2023, 56, 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Owen DR; Allerton CMN; Anderson AS; Aschenbrenner L; Avery M; Berritt S; Boras B; Cardin RD; Carlo A; Coffman KJ, et al. , An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [DOI] [PubMed] [Google Scholar]
- (17).Joyce RP; Hu VW; Wang J, The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): an orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations. Med Chem Res 2022, 31, 1637–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hu Y; Ma C; Szeto T; Hurst B; Tarbet B; Wang J, Boceprevir, Calpain Inhibitors II and XII, and GC-376 Have Broad-Spectrum Antiviral Activity against Coronaviruses. ACS Infect Dis 2021, 7, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Elrick MJ; Pekosz A; Duggal P, Enterovirus D68 molecular and cellular biology and pathogenesis. J Biol Chem 2021, 296, 100317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Brown DM; Hixon AM; Oldfield LM; Zhang Y; Novotny M; Wang W; Das SR; Shabman RS; Tyler KL; Scheuermann RH, Contemporary Circulating Enterovirus D68 Strains Have Acquired the Capacity for Viral Entry and Replication in Human Neuronal Cells. mBio 2018, 9, e01954–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zhang Y; Cao J; Zhang S; Lee AJ; Sun G; Larsen CN; Zhao H; Gu Z; He S; Klem EB, et al. , Genetic changes found in a distinct clade of Enterovirus D68 associated with paralysis during the 2014 outbreak. Virus Evol 2016, 2, vew015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Morens DM; Folkers GK; Fauci AS, Acute Flaccid Myelitis: Something Old and Something New. mBio 2019, 10, e00521–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Murphy OC; Pardo CA, Acute Flaccid Myelitis: A Clinical Review. Semin Neurol 2020, 40, 211–218. [DOI] [PubMed] [Google Scholar]
- (24).Dyda A; Stelzer-Braid S; Adam D; Chughtai AA; MacIntyre CR, The association between acute flaccid myelitis (AFM) and Enterovirus D68 (EV-D68) - what is the evidence for causation? Euro Surveill 2018, 23, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hixon AM; Frost J; Rudy MJ; Messacar K; Clarke P; Tyler KL, Understanding Enterovirus D68-Induced Neurologic Disease: A Basic Science Review. Viruses 2019, 11, 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hurst BL; Evans WJ; Smee DF; Van Wettere AJ; Tarbet EB, Evaluation of antiviral therapies in respiratory and neurological disease models of Enterovirus D68 infection in mice. Virology 2019, 526, 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Frost J; Rudy MJ; Leser JS; Tan H; Hu Y; Wang J; Clarke P; Tyler KL, Telaprevir Treatment Reduces Paralysis in a Mouse Model of Enterovirus D68 Acute Flaccid Myelitis. J. Virol 2023, 97, e0015623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Matthew AN; Leidner F; Lockbaum GJ; Henes M; Zephyr J; Hou S; Rao DN; Timm J; Rusere LN; Ragland DA, et al. , Drug Design Strategies to Avoid Resistance in Direct-Acting Antivirals and Beyond. Chem Rev 2021, 121, 3238–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Musharrafieh R; Ma C; Wang J, Profiling the in vitro drug-resistance mechanism of influenza A viruses towards the AM2-S31N proton channel blockers. Antiviral Res 2018, 153, 10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kabsch W, XDS. Acta Crystallogr D Biol Crystallogr 2010, 66, 125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Adams PD; Afonine PV; Bunkóczi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung LW; Kapral GJ; Grosse-Kunstleve RW, et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010, 66, 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Krissinel E; Uski V; Lebedev A; Winn M; Ballard C, Distributed computing for macromolecular crystallography. Acta Crystallogr D Struct Biol 2018, 74, 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Smart OS; Womack TO; Flensburg C; Keller P; Paciorek W; Sharff A; Vonrhein C; Bricogne G, Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr 2012, 68, 368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Chen VB; Davis IW; Richardson DC, KING (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci 2009, 18, 2403–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Cady SD; Wang J; Wu Y; DeGrado WF; Hong M, Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy. J Am Chem Soc 2011, 133, 4274–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Tan B; Sacco M; Tan H; Li K; Joyce R; Zhang X; Chen Y; Wang J, Exploring diverse reactive warheads for the design of SARS-CoV-2 main protease inhibitors. Eur J Med Chem 2023, 259, 115667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Halgren TA, MMFF VII. Characterization of MMFF94, MMFF94s, and other widely available force fields for conformational energies and for intermolecular-interaction energies and geometries. J Comput Chem 1999, 20, 730–748. [DOI] [PubMed] [Google Scholar]
- (38).Halgren TA, Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 1996, 17, 490–519. [Google Scholar]
- (39).Jorgensen WL; Maxwell DS; TiradoRives J, Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 1996, 118, 11225–11236. [Google Scholar]
- (40).Kaminski GA; Friesner RA; Tirado-Rives J; Jorgensen WL, Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B 2001, 105, 6474–6487. [Google Scholar]
- (41).Friesner RA; Murphy RB; Repasky MP; Frye LL; Greenwood JR; Halgren TA; Sanschagrin PC; Mainz DT, Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 2006, 49, 6177–96. [DOI] [PubMed] [Google Scholar]
- (42).Pettersen EF; Goddard TD; Huang CC; Couch GS; Greenblatt DM; Meng EC; Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004, 25, 1605–12. [DOI] [PubMed] [Google Scholar]
- (43).Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML, Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys 1983, 79, 926–935. [Google Scholar]
- (44).Darden T; York D; Pedersen L, Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J Chem Phys 1993, 98, 10089–10092. [Google Scholar]
- (45).Essmann U; Perera L; Berkowitz ML; Darden T; Lee H; Pedersen LG, A Smooth Particle Mesh Ewald Method. J Chem Phys 1995, 103, 8577–8593. [Google Scholar]
- (46).Izaguirre JA; Catarello DP; Wozniak JM; Skeel RD, Langevin stabilization of molecular dynamics. J Chem Phys 2001, 114, 2090–2098. [Google Scholar]
- (47).Feller SE; Zhang YH; Pastor RW; Brooks BR, Constant-Pressure Molecular-Dynamics Simulation - the Langevin Piston Method. J Chem Phys 1995, 103, 4613–4621. [Google Scholar]
- (48).Humphreys DD; Friesner RA; Berne BJ, A Multiple-Time-Step Molecular-Dynamics Algorithm for Macromolecules. J Phys Chem 1994, 98, 6885–6892. [Google Scholar]
- (49).Isgro TP, C. J; Sotomayor M; Villa E VMD NAMD Tutorial. VMD Tutor 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


