Abstract
Some Basidiomycete fungi are important plant pathogens, and certain species have been associated with the grapevine trunk disease esca. We present the genomes of 4 species associated with esca: Fomitiporia mediterranea, Fomitiporia polymorpha, Tropicoporus texanus, and Inonotus vitis. We generated high-quality phased genome assemblies using long-read sequencing. The genomic and functional comparisons identified potential virulence factors, suggesting their roles in disease development. Similar to other white-rot fungi known for their ability to degrade lignocellulosic substrates, these 4 genomes encoded a variety of lignin peroxidases and carbohydrate-active enzymes (CAZymes) such as CBM1, AA9, and AA2. The analysis of gene family expansion and contraction revealed dynamic evolutionary patterns, particularly in genes related to secondary metabolite production, plant cell wall decomposition, and xenobiotic degradation. The availability of these genomes will serve as a reference for further studies of diversity and evolution of virulence factors and their roles in esca symptoms and host resistance.
Keywords: plant cell wall degradation, fungal secondary metabolism, comparative genomics, gene family evolution, Fomitiporia mediterranea, Fomitiporia polymorpha, Inonotus vitis, Tropicoporus texanus
Introduction
The majority of wood-colonizing species that cause grapevine trunk diseases belong to the Ascomycota phylum, with only a few Basidiomycete species confirmed as pathogens primarily of esca (Fischer 2000, 2002; Fischer and González García 2015; Brown et al. 2020; Del Frari et al. 2021; Martín et al. 2022). Whereas the primary pathogens of esca were long thought to be Ascomycetes Phaeomoniella chlamydospora and Phaeoacremonium minimum, various members of the Hymenochaetaceae family, including species like Fomitiporia mediterranea and Inonotus spp., have been isolated in esca-symptomatic vines in vineyards around the world (Fischer 2002; Cloete, Fischer, et al. 2015; Cloete, Mostert, et al. 2015; Fischer and González García 2015; Guerin-Dubrana et al. 2019; Brown et al. 2020; Mundy et al. 2020; Moretti et al. 2021; Pacetti et al. 2022). Brown et al. (2020) isolated multiple Basidiomycete species from vineyards with esca in California and Texas, and described novel species Fomitiporia ignea, Inonotus vitis, and Tropicoporus texanus. Tropicoporus texanus was capable of causing symptoms independently, without coinoculation with P. chlamydospora. Moreover, leaf symptoms resembling those of esca were more prevalent when Fomitiporia polymorpha was coinoculated with P. chlamydospora than when either was inoculated alone (Brown et al. 2020).
The ability of these fungi to colonize the wood and influence disease progression is determined by their functional arsenal. Wood-rotting Basidiomycetes are commonly categorized as either white- or brown-rot fungi, based in part on the type of residue they leave after degrading their host’s wood (Worrall et al. 1997; Sista Kameshwar and Qin 2018; Eichlerová and Baldrian 2020). White-rot species can degrade all of the main components of the wood (cellulose, hemicellulose, and lignin), whereas brown-rot species degrade cellulose and hemicellulose, leaving a modified lignin residue of brown color (Worrall et al. 1997; Zhou et al. 2014; Sista Kameshwar and Qin 2018; Eichlerová and Baldrian 2020; Moretti et al. 2021). These differences are attributed to the plant cell wall degrading enzymes (CWDEs) they produce, to the activities of the encoded enzymes in planta, to their nonenzymatic wood-decay capabilities, and also to the condition of the woody substrate they colonize. White-rot fungi are characterized by encoding lignin-oxidizing and lignin-degrading enzymes, which are grouped into the Auxiliary Activity (AA) family of the carbohydrate-active enzymes (CAZymes) (Sista Kameshwar and Qin 2018). Additionally, white-rot fungi secrete a more diverse array of glycoside hydrolases (GHs) than brown-rot fungi. To possibly compensate for the lower diversity of GH, brown-rot fungi secrete their GHs in greater abundance than their white-rot counterparts (Presley et al. 2018).
In addition to CAZymes for the breakdown of polysaccharides in the plant cell wall (Kameshwar and Qin 2016; Sista Kameshwar and Qin 2018), Basidiomycetes utilize other enzyme groups. Among them, peroxidases are used for decomposing structural components like lignin (Manavalan et al. 2015; Kameshwar and Qin 2016; Ayuso-Fernández et al. 2019). Cytochrome P450 enzymes support multiple primary and secondary metabolism, including the degradation of xenobiotic compounds (Syed et al. 2014; Morel-Rouhier 2021) and competition with other organisms (Riley et al. 2014; Castaño et al. 2022). Lastly, transporters facilitate the intake and secretion of various molecules and enzymes (Kovalchuk and Driessen 2010; Kovalchuk et al. 2013; Víglaš and Olejníková 2021). Additionally, some white-rot fungi have the ability to use nonenzymatic processes to aid in wood degradation (Moretti et al. 2023).
Regardless, not all wood-rotting Basidiomycetes can be categorized as strictly “white-rot fungi” or “brown-rot fungi,” based on the enzymes encoded by their genomes (or not) or results of various in vitro assays of wood degradation or enzyme activities (Riley et al. 2014). For example, the type of wood decay caused by F. mediterranea is characterized as a white rot, based originally on visibly descriptions of wood decay (Larignon and Dubos 1997), with further confirmation from in vitro assays of its enzymatic and nonenzymatic lignin-degrading properties (Cloete, Mostert, et al. 2015; Moretti et al. 2023). In contrast, F. polymorpha has characteristics of both white-rot fungi (production of lignin-degrading enzymes) and brown-rot fungi (persistence of lignin in inoculated wood) (Galarneau et al., in review).
To advance our study of Basidiomycetes that cause esca in grapevines, we have assembled high-quality genomes of the species F. mediterranea, F. polymorpha, T. texanus, and I. vitis. The type of wood decay caused by the latter 2 species has not been characterized. Therefore, with the aim to describe the functional arsenal encoded by these sets of species, we performed functional annotation and comparative analysis, focusing on putative virulence factors in these newly sequenced genomes against those found in known species causing brown rot and white rot, in addition to ascomycete wood-decay fungi. Additionally, we identified a set of putative virulence factors undergoing expansion and contraction in these fungi, highlighting potential avenues for further research.
Materials and methods
Sample collection
Fomitiporia polymorpha, I. vitis, and T. texanus were previously isolated in 2019 from grapevines (specifically Vitis vinifera L. ‘Chardonnay’ and V. vinifera ‘Cabernet-Sauvignon’) with the leaf symptoms of esca, in California, United States (Brown et al. 2020). Fomitiporia mediterranea was previously isolated in 1996 from grapevines (specifically V. vinifera ‘Ugni blanc’) with the leaf symptoms of esca, in the Charente wine region (France), and stored at the mycological collection at UMR SAVE (INRAE Nouvelle-Aquitaine Bordeaux) (Laveau et al. 2009).
DNA extraction and sequencing
A pure isolate of F. mediterranea was grown in liquid malt medium (cristomalt 15 g/L) at 100 rpm for 22 days. Pure fungal isolates of F. polymorpha, I. vitis, and T. texanus were grown in potato dextrose broth at 110 rpm for 7 days. The mycelium was filtered and washed twice with sterile water using vacuum filtration. The dry mycelium was frozen in liquid nitrogen and ground with a TissueLyser II (Qiagen) using a 50-mL stainless steel jar (Retch). High molecular DNA extraction was based on the method of Chin et al. (2016), using 5 g of ground mycelium. To confirm identity, we amplified and sequenced ITS (using primers ITS1–ITS4), elongation factor-1-alpha (using primers EF1-983F and EF1-1567R), and RPB2 (using primers bRPB2-6F and bRPB2-7.1R); the amplicons were aligned against NCBI nucleotide collection using blastn.
Prior to the preparation of the HiFi sequencing library of F. mediterranea, a preliminary step of short read elimination (SRE, Pacific Biosciences, Menlo Park, CA, United States) was carried out. This process ensured the overall quality and integrity of the HMW gDNA before library prep. SRE pretreated gDNA was sheared with a mode size of 15–18 kb using Diagenode's Megaruptor (Diagenode LLC, Denville, NJ, United States). The HiFi SMRTbell template was prepared with 5 µg of sheared DNA using the SMRTbell Prep Kit 3.0 (Pacific Biosciences, Menlo Park, CA, United States) following the manufacturer's instructions. AMpure PB bead size selection step was performed on HiFi SMRTbell template to deplete DNA fragments shorter than 5 kb using a 0.35X dilution of AMPure beads. Concentration and final size distribution of the library were evaluated using a Qubit 1X dsDNA HS Assay Kit (Thermo Fisher, Waltham, MA, United States) and Femto Pulse System (Agilent, Santa Clara, CA, United States), respectively. HiFi library was sequenced using a Revio sequencer (DNA Technology Core Facility, University of California, Davis).
The molecular weight of the DNA extracted from F. polymorpha, I. vitis, and T. texanus was evaluated with pulse field electrophoresis (Pippin pulse, Sage Science). Two hundred microliters of HMW gDNA at a concentration of 100 ng/µL was fragmented using a 26G blunt needle (SAI Infusion Technologies). gDNA shearing was accomplished by aspirating the entire volume and passing the sample through the 26G blunt needle 10 times. After shearing, the sample was cleaned and concentrated using 0.45X AMPure PB beads and the size distribution of the sheared gDNA fragments was assessed using pulse field gel electrophoresis (Pippin pulse, Sage Science, Beverly, MA, United States) prior to library preparation. SMRTbell templates were prepared with 10 µg of sheared DNA using SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, United States) following the manufacturer's instructions. The Blue Pippin instrument (Sage Science, Beverly, MA, United States) was used to size select SMRTbell templates using a cutoff size of 20–80 kb. Size-selected libraries were cleaned with 1x AMPure PB beads, and their concentration and final size distribution were evaluated using a Qubit HS Assay Kit (Thermo Scientific, Hanover Park, IL, United States) and pulse field gel electrophoresis (Pippin pulse, Sage Science), respectively. Finished libraries were sequenced on a Sequel II sequencer (DNA Technologies Core Facility, University of California, Davis).
Genome assembly and polishing
The HiFi reads of F. mediterranea were assembled using Hifiasm v.0.19.5-r587 (Cheng et al. 2021). First, reads between 15 and 25 kb were selected. Multiple parameters with different values were tested to obtain the least fragmented genome. The optimized options were “-a 3 -k 41 -w 51 -f 0 -r 5 -s 0.8 -D 10 -N 10 -n 7 -m 10000000,” which generated the primary assembly and the haplotigs.
The subreads from the CLR sequencing of F. polymorpha, I. vitis, and T. texanus were randomly subsampled using bamsieve within the PacBio SMRT link v.8.0. using the option “--percentage 25” to keep only 25% of the reads. Canu v.2.2 (Koren et al. 2017) was used with the options “corOutCoverage=200 ‘batOptions=-dg 3 -db 3 -dr 1 -ca 50 -cp 50’ genomeSize=60m -pacbio” to assemble the genomes.
The pbmm2 v.0.8.1 align (https://github.com/PacificBiosciences/pbmm2) was used to map the subreads to the raw assembly using the options “--preset SUBREAD --sort -j 32 -J 8”. These alignments were used as input for gccp v2.02 (https://github.com/PacificBiosciences/gcpp) with the options “--algorithm=arrow -j 40” to polish the raw assembly. After confirming the duplication levels using BUSCO v.5.6.1 (Manni et al. 2021) with the basidiomycota_odb10 database (created on 2024 January 11), the genomes were split into primary scaffolds and the haplotigs. Inonotus vitis and T. texanus genomes were split using the pipeline purge_haplotigs (Roach et al. 2018) following the instruction at https://bitbucket.org/mroachawri/purge_haplotigs/src/master/. The purged genomes were evaluated with Benchmark of Universal Single-Copy Orthologs (BUSCO) to confirm their completeness and duplication levels.
The genome of F. polymorpha was tested further before continuing because purge_haplotigs yielded inconsistent results. First, the assembly was evaluated for external contamination using ContScout (Bálint et al. 2024) with the options “-d uniprotKB -a diamond -q 251363”. The taxonomy ID 251363 belonged to F. polymorpha. This analysis revealed no external contamination in the genome. Additionally, in silico PCR of the ITS sequences in the genome was performed with in_silico_pcr.pl (https://github.com/egonozer/in_silico_pcr) with the primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS-4 (5′-TCCTCCGCTTATTGATATGC-3′). The amplicons were blasted against the NCBI nucleotide collection, and they mapped to F. polymorpha. Additional in silico PCR was done with various bacterial markers; no amplificons were obtained. Next, the total predicted proteins of the species (see Repeat and gene annotation) were used with OrthoFinder v.2.5.4 (Emms and Kelly 2019) with default parameters. This was done to obtain a gene copy number profile. The profile was compared to the profile of the diploid assembly of F. mediterranea, and the results suggested an incomplete diploid assembly or a possible aneuploidy of F. polymorpha. Therefore, the primary assembly of the species was manually curated.
For the manual curation of F. polymorpha, the gene alignments were generated with blastall within BLAST+ 2.15.0 (Camacho et al. 2009) with the options “-p blastp -e 1e-10 -b 5 -v 5 -m 8 -a 8”. The alignments were used with the gff file of the genes to obtain the collinearity percentages using MCScanX (Wang et al. 2012) with default parameters. If more than 70% of the total genes in a scaffold were in collinearity with another scaffold, then it was moved to the haplotigs. This process allowed the curating of a primary assembly, retaining high completeness and low duplication based on the BUSCO analysis.
The contigs of all assembles were evaluated using TelFinder (Sun et al. 2023) to identify terminal telomeric repeats. The tool was used with default parameters, checking both ends of the contigs separately, and only considering telomeric repeats with scores ≥50 (Supplementary Table 1 in Supplementary File 1).
Repeat and gene annotation
The primary assembly of the species was used to annotate and mask repeats. First, repeat model libraries were predicted for each genome using RepeatModeler v.1.0.8 (Smit et al. 2015a) with default parameters. RepeatMasker v.4.06 (Smit et al. 2015b) was used with default parameter and custom repeat libraries to annotate and mask the repeats. The custom library contained the predicted models and the repbase library 20160829-2023 (Bao et al. 2015). The repeat annotation was used to extract the transposable elements (TEs) for further analysis. These TEs were classified into Class I and Class II.
The tool maskFastaFromBed (Quinlan 2014) was used with the option “-soft”, the repeat annotation, and the primary assembly to softmask the repeats. The genes were annotated using Braker v2.1.6 (Brůna et al. 2021). The soft-masked genomes with the OrthoDB11 database of fungi (Kuznetsov et al. 2023) were used with the options “--fungus --softmasking 1” to obtain the raw gene annotation. This annotation was further cleaned to remove proteins with internal stop codons and without stop or end codons.
To calculate the gene density, the tool makewindows from BEDtools v.2.29.1 (Quinlan 2014) was used to obtain windows of 10 kb from the primary assemblies. Then, the coverage tool from the same package was used with the windows and the gene and repeat annotation to obtain the density of the feature per 10 kb of genomic regions. These results were organized and plotted using ggplot2 v.3.5.0 (Wilkinson 2011) in R v.4.2.2 (R Core Team 2022). Similarly, in the TE composition plot, the elements that represented less than 1% of the total TE content in the species were grouped into the category others. The organized data were plotted with ggplot2.
Functional annotation
The general annotation of the predicted proteins was assigned using Pfamscan (https://www.ebi.ac.uk/jdispatcher/pfa/pfamscan) against the Pfam-A database using an E-value of 0.001. CAZymes were annotated with the dbCAN3 at https://bcb.unl.edu/dbCAN2/blast.php (Zheng et al. 2023), selecting the options “HMMER: dbCAN (E-Value < 1e-15, coverage > 0.35)”, “DIAMOND: CAZy (E-Value < 1e-102)”, and “HMMER: dbCAN-sub (E-Value < 1e-15, coverage > 0.35)”. The annotation was kept only when the genes were annotated with at least 2 algorithms. The signal peptides were assigned using SignalP 5.0 (Almagro Armenteros et al. 2019). The proteins with annotation in both databases (SignalP5 and dbCAN3) were annotated as secreted CAZymes. Secondary metabolite clusters were annotated using antiSMASH v.6.0 at https://fungismash.secondarymetabolites.org with default parameters (Blin et al. 2021). The genes in each cluster were given annotations of the cluster to which they belonged. The peroxidases were annotated using a specialized database for fungi fPoxDB (Choi et al. 2014) using hmmsearch within HMMER v.3.1b2 (Eddy 2011) with the option “-E 1e-5”. The cytochrome P450 proteins were annotated locally by intersecting the results of 3 methods, Pfam, blastp within Diamond v2.1.8.162 (Buchfink et al. 2021) with options “--id 60 --outfmt 6 --evalue 0.0001” against CYPED 6.0 database (Fischer et al. 2007), and phmmer (Eddy 2011) with options “-E 0.001 --incE 0.001 --noali” against CYPED 6.0 database. Genes with annotation with at least 2 methods were kept as P450. Last, the proteins involved in transportation functions were annotated using Diamond blastp with the option “-evalue 1e-10” against the TCDB database (Saier et al. 2021). The R package tidyheatmap v 0.2.1 (Mangiola and Papenfuss 2020) was used to build the heatmaps.
Phylogenetic analysis
Genomes and gene annotations from the 4 Basidiomycete species were compared to those of 8 Basidiomycete species and 2 Ascomycete species (Supplementary Table 2 in Supplementary File 1). As the 4 species are wood-colonizing fungi, we included for comparative purposes a combination of species that cause different types of wood decay, namely Basidiomycetes that cause brown rot or white rot and Ascomycetes that cause soft rot. White-rot fungi were Pleurotus ostreatus, Stereum hirsutum, and Trametes versicolor (Supplementary Table 2 in Supplementary File 1). Brown-rot fungi were Daedalea quercina (aka Fomitopsis quercina), Fomitopsis schrenkii, Gloeophyllum trabeum, Postia placenta, and Serpula lacrymans. Ascomycetes that cause the grapevine trunk disease Botryosphaeria dieback Neofusicoccum parvum (soft-rot fungus; Galarneau et al., in review) and Botryosphaeria dothidea were also included. Additionally, other nonpathogenic and pathogenic Ascomycetes known for colonizing wood and other organs of the plant were included for phylogenetic reference only (Supplementary Table 2 in Supplementary File 1).
The predicted proteins of all the genomes were used as input for OrthoFinder v.2.5.4 (Emms and Kelly 2019) with default parameters. The resulting single-copy orthologs were aligned using MUSCLE v.5.1 (Edgar 2004) with the option “-maxiters 16”. The alignments were concatenated and parsed with Gblocks v.0.91b (Castresana 2000) with default parameters. ModelTest-NG v.0.1.7 (Darriba et al. 2020) was used to optimize the evolutionary model. The maximum likelihood (ML) tree was obtained with RAxML-NG v.0.9.0 (Kozlov et al. 2019), the parsed alignment, and the evolutionary model “LG+I+G4” with the options “--tree pars{10} --bs-trees 100”. The clock-calibrated tree was constructed using BEAST v.2.7.6 (Bouckaert et al. 2019). The parsed alignment of single-copy orthologs was prepared with BEAUti v2.7.6 (Bouckaert et al. 2019). Calibration points were set for the Ascomycete crown to 539 million years ago (Mya; Prieto and Wedin 2013) with a normal distribution, and the Polyporales group set to 142 Mya (Ji et al. 2022) with a normal distribution. Five different Markov chain Monte Carlo chains of 1,000,000 generations were set. The LG substitution model with 4 gamma categories, a strict clock, and the birth–death model was used. Sampling was performed every 1,000 generations. The resulting log and tree files were combined using LogCombiner v.2.7.6 (Bouckaert et al. 2019), and the maximum clade credibility tree was generated using TreeAnnotator v2.7.6 (Bouckaert et al. 2019) with a burn-in of 10,000 generations. Figtree (Rambaut 2018) was used to plot the phylogenetic trees.
Gene family expansion and contraction analysis
The predicted proteins in all the genomes were concatenated in a single file and blasted to themselves using blastp within Diamond v2.1.8.162 (Buchfink et al. 2021) with the options “--evalue 1e-6 --very-sensitive --outfmt 6”. The proteins were grouped in families using Markov clustering with MCL v.14-137 (Enright et al. 2002). To do the clustering, first mcxload with the options “--stream-mirror --stream-neg-log10 -stream-tf ‘ceil(200)’” was used with the blast result file. Next, the output from mcxload was used as input for mcl with the option “-I 3”. Then, mcxdump was used with the output from the previous step to obtain a preliminary file needed to run CAFE v.5.0 (Mendes et al. 2021). The script cafetutorial_mcl2rawcafe.py at https://github.com/hahnlab/cafe_tutorial/tree/main/python_scripts was used to prepare the file. Computational analysis of gene family evolution (CAFE) was run with the option “-P 0.0100”, an estimated lambda value of 0.0011169463915058, the previously prepared input, and the clock-calibrated tree. The resulting file was used to extract the families with significant rates of gain or loss of genes (P < 0.01). The number of families expanding and contracting per phylogenetic node was incorporated into the clock-calibrated tree using CafePlotter (https://github.com/moshi4/CafePlotter). A Fisher's exact test was used to obtain the functions that were significantly enriched in the expanded and contracted families of the species of interest.
Results
Genome assemblies of F. mediterranea, F. polymorpha, I. vitis, and T. texanus
The isolates of the 4 Basidiomycete species described in this study were obtained from esca-symptomatic plants in vineyards in US states California and Texas (Brown et al. 2020), and in Charente in France (Laveau et al. 2009). The total genome assembly sizes were 148.9 Mb for F. polymorpha, 126.2 Mb for F. mediterranea, 71.7 Mb for I. vitis, and 68.3 Mb for T. texanus. The genome coverage ranged from 40X in F. polymorpha to 148X in F. mediterranea. Except for F. polymorpha, all genomes were divided into a primary assembly and haplotigs, each constituting roughly half the size of the total genome assembly (Table 1). This even distribution between the primary assembly and haplotigs suggests the complete diploid representation of the genomes. Conversely, the F. polymorpha assembly underwent manual curation, resulting in a primary assembly of 85.6 Mb and a set of haplotigs totaling 63.2 Mb. After ruling out the possibility of contamination with DNA from another species in the sequencing reads, a gene copy number analysis for F. polymorpha and F. mediterranea revealed a higher proportion of odd copy numbers in F. polymorpha compared to the diploid genome of F. mediterranea (Supplementary Fig. 1 in Supplementary File 1). This finding suggests that either the diploid assembly of the F. polymorpha genome was incomplete or aneuploid.
Table 1.
Assembly and annotation statistics of the 4 Basidiomycete species genomes.
| Species | Fomitiporia mediterranea (PHCO36) | Fomitiporia polymorpha (WFB1) | Inonotus vitis (OC1) | Tropicoporus texanus (TX9) |
|---|---|---|---|---|
| Region | Charente, France | Davis, CA, United States | Napa, CA, United States | Hays, TX, United States |
| Haploid assembly size (Mb) | 64.2 | 85.6 | 35.3 | 34.0 |
| Coverage | 148X | 40X | 82X | 86X |
| No. of scaffolds | 58 | 32 | 54 | 41 |
| N50 (Mb) | 5.4 | 5.4 | 0.9 | 1.5 |
| L50 (scaffold no.) | 4.0 | 6.0 | 13.0 | 7.0 |
| BUSCOs in assembly (%)a | 96.3 | 96.0 | 96.1 | 96.1 |
| Repeats masked (Mb) | 33.5 | 51.9 | 5.7 | 3.3 |
| Repeats masked (%) | 52.2 | 60.6 | 16.1 | 9.6 |
| No. of CDS | 12,550 | 11,018 | 9,935 | 10,851 |
| Mean protein size | 444 | 484 | 507 | 509 |
| BUSCOs in gene prediction (%)b | 99.2 | 97.6 | 98.0 | 98.4 |
| Mean gene density (gene/10 kb) | 2.3 | 1.5 | 3.3 | 3.8 |
| SEM | 0.03 | 0.02 | 0.03 | 0.03 |
a Percentage of complete BUSCO peptides found in the primary assembly.
b Percentage of complete BUSCO peptides found in the predicted genes of the primary assembly.
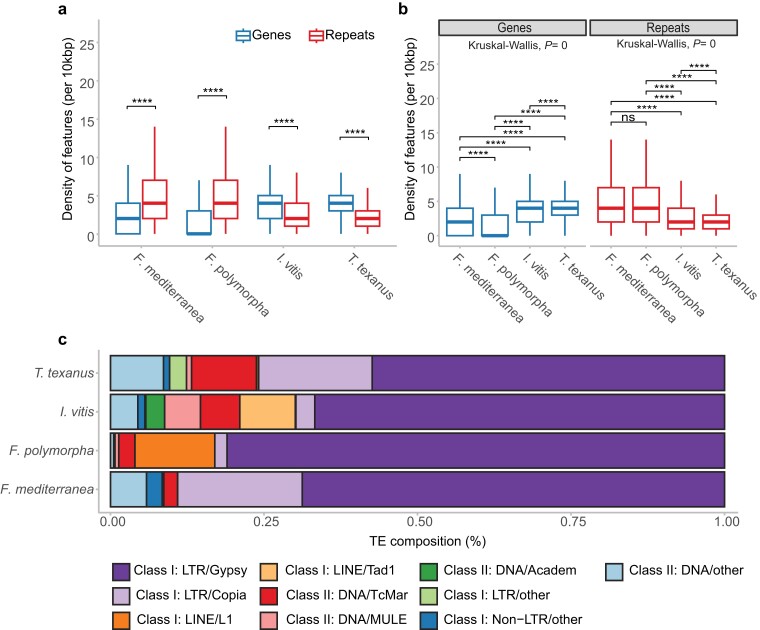
The statistics presented henceforth pertain to the primary assembly (i.e. the more contiguous haploid versions) of the genomes. The number of scaffolds in the haploid genomes varied from 32 in F. polymorpha to 58 in F. mediterranea (Table 1). The BUSCO analysis showed that the assemblies contain over 96% of the complete single-copy ortholog sequences curated for the Basidiomycete group (Table 1). Similarly, between 97.6 and 99.2% of the single-copy ortholog sequences curated for the Basidiomycetes were found after gene annotation (Table 1). Additionally, telomeric repeats were detected in some of the contigs of the genome assemblies (Supplementary Table 1 in Supplementary File 1). The highest repeat content was observed in F. polymorpha, constituting more than 60% of the genome size, followed by 52.2% in F. mediterranea, 16.1% in I. vitis, and 9.6% in T. texanus. This distribution of repeat content aligns with gene density, with the highest density observed in T. texanus and the lowest in F. polymorpha (Table 1). Comparing the density of genes and repeats within species revealed that repeats were significantly denser in F. polymorpha and F. mediterranea. Conversely, in I. vitis and T. texanus, the genes were significantly denser than the repeats (Fig. 1a). Additionally, a comparison of feature density between species showed that I. vitis and T. texanus had significantly higher gene density compared to both Fomitiporia. The reverse was true for repeat content, where both Fomitiporia species had higher repeat content density than I. vitis and T. texanus (Fig. 1b).
Fig. 1.
Gene and repeat density in the genome of the species in study. a) Density of genes and repeats per 10 kb of genome space with statistical comparisons within species. Significant differences were determined using the Mann–Whitney U test (****P ≤ 0.0001). b) Density of genes and repeats per 10 kb of genome space with statistical comparisons between species. Significant differences were determined using the Dunn test after a significant Kruskal–Wallis test (****P ≤ 0.0001). c) TE types as a proportion of the total TE length in the genome. Categories representing < 1% of the repeat content were classed as “other.”
A significant proportion of total repeats were TEs, with 48% of the total repeat content in T. texanus, 68% in F. mediterranea, 71% in I. vitis, and 75% in F. polymorpha. Within the TEs, the LTR/Gypsy covered an average of 68.5 ± 4.9%, the most significant proportion of the TEs. Fomitiporia polymorpha presents the largest proportion, with 81.0% of their total TE sequence being LTR/Gypsy (Fig. 1c). Next, the LTR/Copia elements covered a large proportion of TEs in F. mediterranea (20.3%) and T. texanus (18.5%). Moreover, the elements LINE/Tad1 and DNA/MULE were almost exclusive to I. vitis, whereas the LINE/L1 was almost exclusive to F. polymorpha (Fig. 1c).
Gene annotation focused on putative virulence factors
We focused the functional annotation of the protein-coding genes of 3 species on the potential virulence factors such as CAZymes, fungal peroxidases, cytochrome P450s, biosynthetic gene clusters (BGCs), and cellular transporters. The variation in gene numbers among putative virulence factors can indicate the virulence strategies adopted by fungi. For example, in previous studies among the species that cause Botryosphaeria dieback, the more virulent Neofusicoccum species have a larger repertoire of CAZymes compared to those of less virulent Dothiorella spp. and Diplodia spp. (Garcia et al. 2021). Similarly, our observations reveal that grapevine trunk pathogens like Eutypa lata and P. minimum exhibit a higher number of BGCs than N. parvum (Garcia et al. 2024). These observations are supported by evidence of recent evolutionary changes like gene family expansions in these virulence factor groups (Garcia et al. 2021, 2024).
In this study between the Basidiomycete species, both Fomitiporia genomes encoded a high number of fungal peroxidases, similar to those of other white-rot species, such as T. versicolor. Tropicoporus texanus encoded a high number of secreted CAZymes, whereas I. vitis appeared to be on the medium or lower end of counts per functional category when compared with all the species (Table 2).
Table 2.
Genes annotated per functional category.
| Species | Total predicted genes | Signal peptide | CAZymes | Secreted CAZymes | Peroxidases | P450 | BGC genes | Transporters |
|---|---|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | 6,716 | 352 | 137 | 41 | 21 | 3 | 31 | 1,670 |
| Botryosphaeria dothidea | 12,424 | 1,341 | 611 | 348 | 56 | 262 | 926 | 3,219 |
| Neofusicoccum parvum | 13,067 | 1,429 | 642 | 390 | 58 | 272 | 930 | 3,363 |
| Fomitiporia mediterranea | 12,550 | 888 | 408 | 233 | 51 | 137 | 330 | 2,597 |
| Fomitiporia polymorpha | 11,018 | 911 | 429 | 247 | 55 | 154 | 305 | 2,736 |
| Inonotus vitis | 9,935 | 801 | 377 | 220 | 47 | 118 | 299 | 2,357 |
| Tropicoporus texanus | 10,851 | 925 | 440 | 268 | 49 | 114 | 336 | 2,488 |
| Stereum hirsutum | 14,072 | 1,059 | 521 | 294 | 53 | 211 | 408 | 2,711 |
| Gloeophyllum trabeum | 11,846 | 779 | 345 | 180 | 32 | 123 | 480 | 2,417 |
| Pleurotus ostreatus | 12,330 | 1,094 | 507 | 283 | 54 | 151 | 295 | 2,450 |
| Serpula lacrymans | 12,789 | 686 | 297 | 144 | 35 | 159 | 609 | 2,339 |
| Trametes versicolor | 14,296 | 1,084 | 450 | 257 | 68 | 188 | 428 | 2,499 |
| Postia placenta | 12,541 | 737 | 276 | 114 | 37 | 215 | 597 | 2,359 |
| Daedalea quercina | 12,199 | 790 | 301 | 152 | 34 | 138 | 389 | 2,386 |
| Fomitopsis schrenkii | 13,885 | 893 | 351 | 177 | 37 | 182 | 297 | 2,509 |
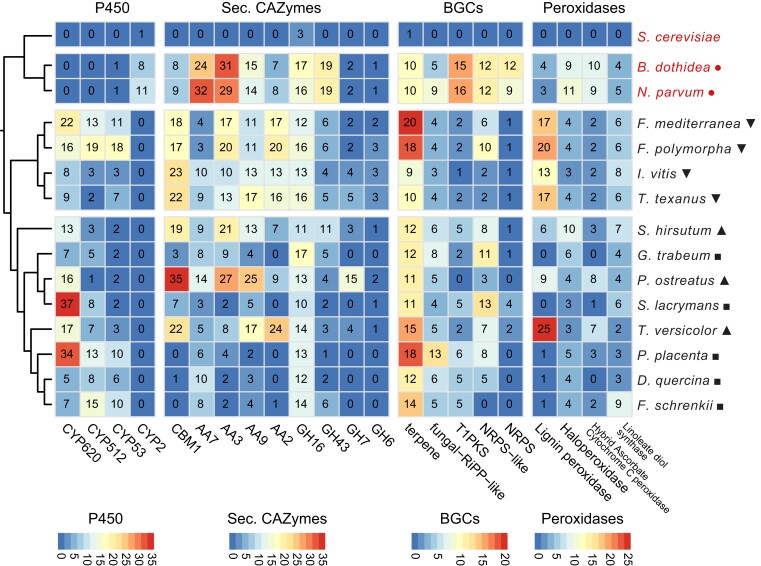
The abundance patterns of specific families within the functional categories varied between the species analyzed. Among the families of the cytochrome P450 enzymes, the activity can range from degrading xenobiotic molecules and some members can be involved in synthesizing secondary metabolites, which generally contribute to adaptation to different environmental conditions (Crešnar and Petrič 2011; Moktali et al. 2012; Chen et al. 2014). Interestingly, the families CYP620 and CYP512 appeared to be exclusive to Basidiomycete species. Some members of these families have been reported to be involved in polyketide biosynthesis and triterpenoid biosynthesis (Nazmul Hussain Nazir et al. 2010; Chen et al. 2012; Syed et al. 2014; Yu et al. 2020). The number of genes annotated as CYP512 was particularly high in F. polymorpha. Similarly, CYP53, with some member involved in detoxifying benzoate residues (Faber et al. 2001), was highly abundant in F. polymorpha (Fig. 2).
Fig. 2.
Number of protein-coding genes annotated as putative P450, secreted CAZymes, BGC, and peroxidases. The specific functions per functional category were selected based on the highest number of genes across all genomes. The first 3 species are Ascomycetes and the rest are Basidiomycetes. Species marked with inverted triangles are the fungi of interest in this study, triangles are known white-rot species, squares are known brown-rot species, and circles are known soft-rot species.
The successful establishment of these fungi in their hosts relies heavily on their ability to degrade the various polysaccharides that constitute the plant cell wall (Morales-Cruz et al. 2015). CAZymes are primarily responsible for the modification of these host cell wall polysaccharides (Kubicek et al. 2014). The identification of CAZymes with signal peptides for secretion is a common method for studying the potential array of enzymes capable of degrading the plant cell wall (Blanco-Ulate et al. 2014; Morales-Cruz et al. 2015; Garcia et al. 2021). Compared to white-rotters S. hirsutum and T. versicolor, the species F. mediterranea, F. polymorpha, I. vitis, and T. texanus were characterized by similarly high number of genes encoding carbohydrate-binding module 1 (CBM1), which is associated with cellulose-binding activity (Lehtiö et al. 2003). In contrast, CMB1 was absent from brown-rotters P. placenta and F. schrenkii (Fig. 2). The AA Families 7, 3, and 9 are gluco-oligosaccharide oxidases, cellobiose dehydrogenase, and polysaccharide monooxygenases, respectively. AA7 and AA3 were more prevalent in the pathogens B. dothidea and N. parvum, whereas AA3 also showed high gene counts in P. ostreatus, S. hirsutum, F. polymorpha, and F. mediterranea. Conversely, AA9 was more abundant in species like P. ostreatus, T. texanus, and T. versicolor (Fig. 2). The members of the GH family 16 (GH16) are widely distributed across different taxonomic groups. Their primary functions include the degradation or remodeling of cell wall polysaccharides, such as those involving endoglucanases, transglycosylases, and chitin-glucosyltransferase (Viborg et al. 2019). This gene family is consistently large across all the species studied, except for Saccharomyces cerevisiae, where only 3 genes have been annotated as GH16 (Fig. 2). Additionally, members of GH43, known for their xylosidase and arabinofuranosidase activities (Flipphi et al. 1993; Shallom et al. 2005), were found to be more abundant in B. dothidea and N. parvum. The GH families GH7 and GH6, with cellobiose hydrolase functions (Riley et al. 2014), were almost absent from the brown-rot species. On the other hand, the F. mediterranea, F. polymorpha, I. vitis, and T. texanus have similar number of GH7 and GH6 than those of the white-rot species.
BGCs produce secondary metabolites critical for fungal development and interactions with their plant host and other organisms (Keller 2019). Fomitiporia mediterranea, F. polymorpha, and P. placenta showed the highest number of terpene BGCs. Compared to all brown-rotters and all white-rotters, F. mediterranea, F. polymorpha, I. vitis, and T. texanus were characterized by similarly low numbers of polyketide synthases (T1PKS) and nonribosomal peptide synthetases (NRPSs). In contrast, T1PKS and NRPS were most numerous in soft-rotter N. parvum and related species B. dothidea.
The capacity to fully degrade lignin significantly distinguishes white-rot from brown-rot Basidiomycetes. In this study, white-rot species T. versicolor, P. ostreatus, and S. hirsutum, along with F. mediterranea, possessed a significantly higher number of lignin peroxidases compared to brown-rot species (P = 0.049; Fig. 2). However, the new genomes of F. polymorpha, I. vitis, and T. texanus showed no significant difference with the aforementioned white-rot fungi (P = 0.015).
Gene family expansion and contraction
Next, we tested if the differences in putative virulence factor repertoires reflected accelerated processes of gene family expansion and contraction. We implemented the CAFE approach (Mendes et al. 2021). CAFE computes the birth and death rates of genes within gene families, identifies families with accelerated rates of gain and loss, and pinpoints the specific lineages responsible for significant changes in family size. CAFE requires a clock-calibrated phylogenetic tree and the sizes of gene families as input.
To establish the phylogenetic relationships of the species under study, we obtained a set of single-copy orthologs for all species (Supplementary Table 2 in Supplementary File 1) using OrthoFinder. These orthologs were used to build a ML tree (Supplementary Fig. 2 in Supplementary File 1), with most tree nodes showing a bootstrap value of 100, indicating high topological accuracy. The newly presented genomes in this study form a single clade, clustering the Fomitiporia species and I. vitis with T. texanus. Upon confirming the ML tree’s topology, we used the set of orthologous genes to generate a clock-calibrated tree through a Bayesian approach with BEAST (Bouckaert et al. 2019). The crown ages of the Ascomycota and the Polyporales groups served as calibration points (Fig. 3a). The topology of the Bayesian tree aligns with that of the ML tree, and the clock calibration suggests that the 4 studied species shared a common ancestor approximately 80 Mya. The Fomitiporia species diverged around 8 Mya, while I. vitis and T. texanus diverged approximately 55 Mya.
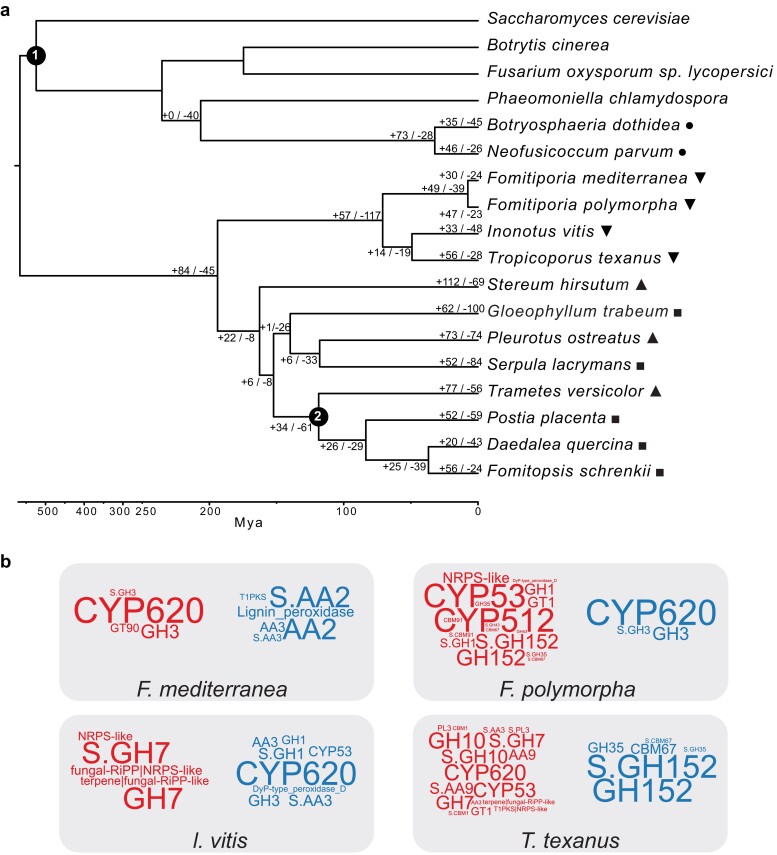
Fig. 3.
Analysis of gene family expansion and contraction in the species of interest. a) Clock-calibrated phylogenetic tree constructed with single-copy gene orthologs. The branches represent divergence times in million years. Calibration point (1) at Ascomycete crown set to ∼539 Mya. Calibration point (2) at the Polyporales group set to ∼142 Mya. Positive and negative numbers indicated on the branches represent expansions and contractions, respectively, as determined by gene family evolution analysis CAFE. Species marked with inverted triangles are the fungi of interest in this study, triangles are known white-rot species, squares are known brown-rot species, and circles are soft-rot Ascomycete species. The unmarked species were included for phylogenetic reference only. b) Word clouds representing the genes enriched in the rapidly evolving families of the species F. mediterranea, F. polymorpha, I. vitis, and T. texanus. The word size represents the enrichment’s strength based on the P value. The red color (left side) represents the enriched genes within expanding families, and the blue (right side) represents enriched genes within contracting families.
The clock-calibrated tree, along with the gene family sizes for all species, served as input for CAFE. Fomitiporia polymorpha, F. mediterranea, and T. texanus showed an overall expansion of gene families. Conversely, in I. vitis, the majority of the gene families are contracting (Fig. 3a). An enrichment test conducted on the genes from families with accelerated gain or loss rates of gain or loss identified specific functions associated with these rapidly evolving families (Fig. 3b).
Genes annotated as CYP620 were highly enriched in the expanded gene families of F. mediterranea (Fig. 3b), whereas lignin peroxidases and AA2 were enriched in the contracting families. In F. polymorpha, CYP512, CYP53, and secreted GH152 showed high enrichment in expanding families (Fig. 3b). Similarly, in I. vitis, genes of the BGC fungal-RiPP|NPRS-like and the secreted GH7 were enriched in expanding families (Fig. 3b), whereas CYP620 was highly enriched in the contracting families. Lastly, in T. texanus, genes of the secreted CAZymes (GH10 and GH7) and P450s (CYP620 and CYP53) were enriched in the expanding families, while GH152 genes were highly enriched in the contracting families (Fig. 3b).
Discussion
Only a few Basidiomycete species have been associated with grapevine diseases and predominantly with esca. Traditionally, these species have been considered secondary pathogens, as they seemingly lack the capability to independently cause disease symptoms. However, researchers have observed a reduction of esca leaf symptoms after removing the white rot through vine surgery (Lecomte et al. 2022). More recently, researchers in California have demonstrated that some species can, in fact, trigger and exacerbate esca-related symptoms (Brown et al. 2020). In our study, we have assembled the genomes of Hymenochaetaceae family members associated with esca across various viticulture regions worldwide. With the exception of F. mediterranea (Floudas et al. 2012), these genome assemblies are the first to be published for each species.
The haploid genome sizes of previously published genomes in the Hymenochaetaceae family range from approximately 28 to 63 Mb. This range encompasses the assembly sizes of I. vitis and T. texanus presented in this study. The manually purged assembly size of F. polymorpha exceeds that of its close relative, F. mediterranea, by roughly 20 Mb. Nonetheless, the completeness and duplication levels, assessed with BUSCO, are comparable to those of the other assemblies. Identifying more than 96% of the Basidiomycete orthologs in our assemblies aligns with studies on close relatives, where completeness varied from 84.1 to 94.9% (Zhao, Dai, et al. 2023). Additionally, the low number of scaffolds (from 32 to 58), the low L50 values (from 4 to 13), and the high N50 values (from 0.9 to 5.4 Mb) of our assemblies indicate highly contiguous genomes, suggesting the presence of complete and nearly complete chromosomes. It is worth noting that the sequencing technology and assembly software likely had an effect on the coverage differences observed between the species, where F. mediterranea, the only species sequenced with HiFi, presented higher coverage.
The repetitive content in fungal genomes directly affects genome plasticity, including both structural and functional alterations (Castanera et al. 2017). Repeat coverage values exceeding 41% have been documented for F. mediterranea (Floudas et al. 2012) and 21% Inonotus obliquus (Duan et al. 2022), aligning with the findings for our genomes. As expected, the high repetitive content observed in Fomitiporia species corresponds to a sparser gene space compared to other genomes analyzed in this study. Among the identified repetitive sequences in the species of interest, LTR/Gypsy and LTR/Copia elements were the most prevalent, a finding that is consistent with other fungal studies (Floudas et al. 2012; Castanera et al. 2016, 2017). Moreover, the presence and coverage of elements such as LINE/L1 and LINE/Tad1 in F. polymorpha and I. vitis are of particular interest. Despite various studies indicating that these elements are not uncommon in fungi, they are typically present in small quantities (Daboussi and Capy 2003; Castanera et al. 2016, 2017; Qian et al. 2023).
Annotation of the genes in the newly assembled genomes is comparable with those of other fungal pathogens, as reported in Table 2, ranging from ∼10k to ∼12k genes. Besides the widely known Ascomycete pathogenic species N. parvum and B. dothidea, the new genomes of F. mediterranea, F. polymorpha, and T. texanus present a high number of genes annotated as putative virulence factors, which support the idea of their role as important wood-decay microorganisms. More specifically, the P450 gene family CYP53 is highly abundant in F. polymorpha. Some members of this family confer abilities related to the degradation of benzoate residues, which is a common plant defense compound (Podobnik et al. 2008; Lah et al. 2011; Jawallapersand et al. 2014). Similarly, the CBM1, AA9, and AA2 CAZymes, higher in white-rot fungi and the newly assembled genomes, are involved in lignocellulose substrate degradation (Lehtiö et al. 2003; Liu et al. 2017; Garrido et al. 2020), suggesting these enzymes may play a crucial role in the activity of the white-rot fungi, consistent with previous reports by other researchers (Levasseur et al. 2014; Liu et al. 2017; Li et al. 2019; Floudas et al. 2020; Miyauchi et al. 2020; Wu et al. 2020). Additionally, researchers have reported high correlation between white-rot species and the families CBM1, GH7, and GH6 (Floudas et al. 2012, 2015; Riley et al. 2014). This group of CAZymes was also observed in the newly assembled genomes in this study. The same applies to lignin peroxidases observed in the white-rot pathogens and the newly assembled genomes. Moreover, besides species like B. dothidea and N. parvum encoding more polyketide synthase clusters, more terpene clusters were annotated in the Fomitiporia species. The production of terpene metabolites like frustulosin and dihydroactinolide have been reported in F. mediterranea (Fischer and Thines 2017), and multiple terpene synthases have been annotated in the close relative I. obliquus (Duan et al. 2022).
We assessed the relationship between the species presented in this study through ML and Bayesian phylogenetic trees of single-copy orthologs. The separation of the Ascomycetes from the Basidiomycetes, the clustering of the members of the Hymenochaetaceae family (F. mediterranea, F. polymorpha, I. vitis, and T. texanus), and the grouping of the members of the order Polyporales are consistent with the literature (Chen et al. 2015; Song and Cui 2017; Ji et al. 2022; Liu et al. 2023; Zhao, Vlasák, et al. 2023). Additionally, the clock-calibrated tree estimation for the split between the Fomitiporia species is consistent with other studies (Chen et al. 2015; Liu et al. 2023).
The clock-calibrated tree allowed us to study patterns of gene family expansion and contraction in the newly assembled genomes. Identifying gene families with higher rates of gain and loss can help elucidate how these fungi may be adapting to their environments (Hahn et al. 2005). As mentioned before, some members of the CYP620 family can be associated with the production of polyketides. Therefore, its enrichment in the expanding families of F. mediterranea may suggest that the production of polyketides, a class of secondary metabolites with a wide range of activities, may play an important role in this species (Macheleidt et al. 2016). Also, the lignin peroxidases are enriched in the contracting families of F. mediterranea, an unexpected result based on their known ability to degrade lignin (Schilling et al. 2022). This result indicates that the common ancestor probably encoded a larger set of lignin peroxidases, an idea supported by the fact that the species F. polymorpha (phylogenetically closer to the common ancestor; Alves-Silva et al. 2020) encodes more proteins with this function than F. mediterranea.
Multiple gene families in F. polymorpha are undergoing expansion, and genes like CYP512, CYP53, and secreted GH152 are enriched in these families. As mentioned earlier, these P450 genes may be involved in producing secondary metabolites needed for interaction with the environment and the degradation of benzoate produced by plants, improving fungal defense mechanisms. On the other hand, GH152 presents endo-β-1,3-glucanase activity, which targets fungal cell walls (Grenier et al. 2000; Gavande and Goyal 2023; Matsumoto et al. 2023). The role of this enzyme in fungi is not fully understood, but studies have suggested activity in modifying fungal cell walls and senescence of fruiting bodies (Grenier et al. 2000; Sakamoto et al. 2006), which could be related to competition with other fungi.
Inonotus vitis and T. texanus showed a similar pattern of expansion and contraction when compared to the Fomitiporia species. The significant enrichment in secondary metabolite biosynthetic genes, GHs, and P450s in the expanding families suggests that the repertoire of virulence factors of these pathogens has recently evolved, possibly in response to pressure imposed by the plant hosts, the environment, or microbial competitors, and antagonists (Gladieux et al. 2014; Sacristán et al. 2021). Intraspecific genome comparisons would help determine whether these gene families are still evolving. The expansion of secreted GH7 and GH10 suggests a potential increase in cellulose and hemicellulose degradation capabilities. Similarly, the expansion of CYP53 could imply evolution driven by the pressure imposed by plant defense compounds. In summary, these 4 Basidiomycetes are actively evolving various functions of their putative virulence arsenal, some of which could improve their ability to thrive in their role as wood decomposers.
Supplementary Material
Contributor Information
Jadran F Garcia, Department of Viticulture and Enology, University of California, Davis, Davis, CA 95616, USA.
Rosa Figueroa-Balderas, Department of Viticulture and Enology, University of California, Davis, Davis, CA 95616, USA.
Gwenaëlle Comont, INRAE, Bordeaux Sciences Agro, ISVV, SAVE, 33140 Villenave d’Ornon, France.
Chloé E L Delmas, INRAE, Bordeaux Sciences Agro, ISVV, SAVE, 33140 Villenave d’Ornon, France.
Kendra Baumgartner, Crops Pathology and Genetics Research Unit, United States Department of Agriculture—Agricultural Research Service, Davis, CA 95616, USA.
Dario Cantu, Department of Viticulture and Enology, University of California, Davis, Davis, CA 95616, USA; Genome Center, University of California, Davis, Davis, CA 95616, USA.
Data availability
The sequencing data generated for this project are available at NCBI (PRJNA1099290; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1099290). The references for previously published genomic data used in this study can be seen in Supplementary Table 2 of Supplementary File 1. The genome assemblies and gene models produced in this study are publicly available at Zenodo (https://doi.org/10.5281/zenodo.10957629) and NCBI (PRJNA1136927; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1136927). Dedicated genome browsers and BLAST tools are available at https://www.grapegenomics.com. The code used in this project is displayed in Supplementary File 2.
Supplemental material available at G3 online.
Funding
This work was supported by the USDA National Institute of Food and Agriculture Specialty Crop Research Initiative (grant 2012-51181-19954). DC was also partially supported by the Louis P. Martini Endowment in Viticulture. DC, KB, and CELD by the Champion program framework INRAE—UC Davis. We thank the DNA Technologies Core at the Genome Center (UC Davis), Andrea Minio (UC Davis), Nathalie Ferrer (INRAE), and Olivier Fabreguettes (INRAE) for their technical support.
Literature cited
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. Signalp 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 37(4):420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Alves-Silva G, Drechsler-Santos ER, da Silveira RMB. 2020. Bambusicolous Fomitiporia revisited: multilocus phylogeny reveals a clade of host-exclusive species. Mycologia. 112(3):633–648. doi: 10.1080/00275514.2020.1741316. [DOI] [PubMed] [Google Scholar]
- Ayuso-Fernández I, Rencoret J, Gutiérrez A, Ruiz-Dueñas FJ, Martínez AT. 2019. Peroxidase evolution in white-rot fungi follows wood lignin evolution in plants. Proc Natl Acad Sci U S A. 116(36):17900–17905. doi: 10.1073/pnas.1905040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bálint B, Merényi Z, Hegedüs B, Grigoriev IV, Hou Z, Földi C, Nagy LG. 2024. ContScout: sensitive detection and removal of contamination from annotated genomes. Nat Commun. 15(1):936. doi: 10.1038/s41467-024-45024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ulate B, Morales-Cruz A, Amrine KC, Labavitch JM, Powell AL, Cantu D. 2014. Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front Plant Sci. 5:435. doi: 10.3389/fpls.2014.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T. 2021. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49(W1):W29–W35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, et al. 2019. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 15(4):e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AA, Lawrence DP, Baumgartner K. 2020. Role of basidiomycete fungi in the grapevine trunk disease esca. Plant Pathol. 69(2):205–220. doi: 10.1111/ppa.13116. [DOI] [Google Scholar]
- Brůna T, Hoff KJ, Lomsadze A, Stanke M, Borodovsky M. 2021. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom Bioinform. 3(1):lqaa108. doi: 10.1093/nargab/lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Reuter K, Drost HG. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 18(4):366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanera R, Borgognone A, Pisabarro AG, Ramírez L. 2017. Biology, dynamics, and applications of transposable elements in basidiomycete fungi. Appl Microbiol Biotechnol. 101(4):1337–1350. doi: 10.1007/s00253-017-8097-8. [DOI] [PubMed] [Google Scholar]
- Castanera R, López-Varas L, Borgognone A, LaButti K, Lapidus A, Schmutz J, Grimwood J, Pérez G, Pisabarro AG, Grigoriev IV, et al. 2016. Transposable elements versus the fungal genome: impact on whole-genome architecture and transcriptional profiles. PLoS Genet. 12(6):e1006108. doi: 10.1371/journal.pgen.1006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño JD, Muñoz-Muñoz N, Kim YM, Liu J, Yang L, Schilling JS. 2022. Metabolomics highlights different life history strategies of white and brown rot wood-degrading fungi. mSphere. 7(6):e0054522. doi: 10.1128/msphere.00545-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Cui BK, Zhou LW, Korhonen K, Dai YC. 2015. Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers. 71:185–200. doi: 10.1007/s13225-014-0317-2. [DOI] [Google Scholar]
- Chen S, Xu J, Liu C, Zhu Y, Nelson DR, Zhou S, Li C, Wang L, Guo X, Sun Y, et al. 2012. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat Commun. 3(1):913. doi: 10.1038/ncomms1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lee MK, Jefcoate C, Kim SC, Chen F, Yu JH. 2014. Fungal cytochrome P450 monooxygenases: their distribution, structure, functions, family expansion, and evolutionary origin. Genome Biol Evol. 6(7):1620–1634. doi: 10.1093/gbe/evu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 18(2):170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 13(12):1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Détry N, Kim KT, Asiegbu FO, Valkonen JP, Lee YH. 2014. fPoxDB: fungal peroxidase database for comparative genomics. BMC Microbiol. 14:117. doi: 10.1186/1471-2180-14-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloete M, Fischer M, Mostert L, Halleen F. 2015. Hymenochaetales associated with esca-related wood rots on grapevine with a special emphasis on the status of esca in South African vineyards. Phytopathol Mediterr. 54(2):299–312. doi: 10.14601/Phytopathol_Mediterr-16364. [DOI] [Google Scholar]
- Cloete M, Mostert L, Fischer M, Halleen F. 2015. Pathogenicity of South African hymenochaetales taxa isolated from esca-infected grapevines. Phytopathol Mediterr. 54(2):368–379. doi: 10.14601/Phytopathol_Mediterr-16237. [DOI] [Google Scholar]
- Crešnar B, Petrič Š. 2011. Cytochrome P450 enzymes in the fungal kingdom. Biochim Biophys Acta. 1814(1):29–35. doi: 10.1016/j.bbapap.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Daboussi MJ, Capy P. 2003. Transposable elements in filamentous fungi. Annu Rev Microbiol. 57(1):275–299. doi: 10.1146/annurev.micro.57.030502.091029. [DOI] [PubMed] [Google Scholar]
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Frari G, Oliveira H, Boavida Ferreira R. 2021. White rot fungi (Hymenochaetales) and esca of grapevine: insights from recent microbiome studies. J Fungi (Basel). 7(9):770. doi: 10.3390/jof7090770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Han H, Qi J, Gao JM, Xu Z, Wang P, Zhang J, Liu C. 2022. Genome sequencing of Inonotus obliquus reveals insights into candidate genes involved in secondary metabolite biosynthesis. BMC Genomics. 23(1):314. doi: 10.1186/s12864-022-08511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichlerová I, Baldrian P. 2020. Ligninolytic enzyme production and decolorization capacity of synthetic dyes by saprotrophic white rot, brown rot, and litter decomposing basidiomycetes. J Fungi (Basel). 6(4):301. doi: 10.3390/jof6040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30(7):1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber BW, van Gorcom RF, Duine JA. 2001. Purification and characterization of benzoate-para-hydroxylase, a cytochrome P450 (CYP53A1), from Aspergillus niger. Arch Biochem Biophys. 394(2):245–254. doi: 10.1006/abbi.2001.2534. [DOI] [PubMed] [Google Scholar]
- Fischer J, Thines E. 2017. Secondary metabolites of fungal vine pathogens. In: König H, Unden G, Fröhlich J, editors. Biology of Microorganisms on Grapes, in Must and in Wine. Cham: Springer International Publishing. p. 165–185. [Google Scholar]
- Fischer M. 2000. Grapevine wood decay and lignicolous basidiomycetes. Phytopathol Mediterr. 39(1):100–106. doi: 10.1400/57826. [DOI] [Google Scholar]
- Fischer M. 2002. A new wood-decaying basidiomycete species associated with esca of grapevine: Fomitiporia mediterranea (Hymenochaetales). Mycol Prog. 1(3):315–324. doi: 10.1007/s11557-006-0029-4. [DOI] [Google Scholar]
- Fischer M, González García V. 2015. An annotated checklist of European basidiomycetes related to white rot of grapevine (Vitis vinifera). Phytopathol Mediterr. 54(2):281–298. doi: 10.14601/Phytopathol_Mediterr-16293. [DOI] [Google Scholar]
- Fischer M, Knoll M, Sirim D, Wagner F, Funke S, Pleiss J. 2007. The cytochrome P450 engineering database: a navigation and prediction tool for the cytochrome P450 protein family. Bioinformatics. 23(15):2015–2017. doi: 10.1093/bioinformatics/btm268. [DOI] [PubMed] [Google Scholar]
- Flipphi MJ, Visser J, van der Veen P, de Graaff LH. 1993. Cloning of the Aspergillus niger gene encoding α-l-arabinofuranosidase A. Appl Microbiol Biotechnol. 39(3):335–340. doi: 10.1007/BF00192088. [DOI] [PubMed] [Google Scholar]
- Floudas D, Bentzer J, Ahrén D, Johansson T, Persson P, Tunlid A. 2020. Uncovering the hidden diversity of litter-decomposition mechanisms in mushroom-forming fungi. ISME J. 14(8):2046–2059. doi: 10.1038/s41396-020-0667-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, et al. 2012. The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 336(6089):1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- Floudas D, Held BW, Riley R, Nagy LG, Koehler G, Ransdell AS, Younus H, Chow J, Chiniquy J, Lipzen A, et al. 2015. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genet Biol. 76:78–92. doi: 10.1016/j.fgb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JF, Lawrence DP, Morales-Cruz A, Travadon R, Minio A, Hernandez-Martinez R, Rolshausen PE, Baumgartner K, Cantu D. 2021. Phylogenomics of plant-associated Botryosphaeriaceae species. Front Microbiol. 12:652802. doi: 10.3389/FMICB.2021.652802/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JF, Morales-Cruz A, Cochetel N, Minio A, Figueroa-Balderas R, Rolshausen PE, Baumgartner K, Cantu D. 2024. Comparative pangenomic insights into the distinct evolution of virulence factors among grapevine trunk pathogens. Mol Plant Microbe Interact. 37(2):127–142. doi: 10.1094/MPMI-09-23-0129-R. [DOI] [PubMed] [Google Scholar]
- Garrido MM, Landoni M, Sabbadin F, Valacco MP, Couto A, Bruce NC, Wirth SA, Campos E. 2020. PsAA9A, a C1-specific AA9 lytic polysaccharide monooxygenase from the white-rot basidiomycete Pycnoporus sanguineus. Appl Microbiol Biotechnol. 104(22):9631–9643. doi: 10.1007/s00253-020-10911-6. [DOI] [PubMed] [Google Scholar]
- Gavande PV, Goyal A. 2023. Chapter 6—Endo-β-1,3-glucanase. In: Goyal A, Sharma K, editors. Glycoside Hydrolases. Biochemistry, Biophysics, and Biotechnology. A volume in Foundations and Frontiers in Enzymology. Cambridge (MA): Academic Press. p. 121–133. [Google Scholar]
- Gladieux P, Ropars J, Badouin H, Branca A, Aguileta G, de Vienne DM, Rodríguez de la Vega RC, Branco S, Giraud T. 2014. Fungal evolutionary genomics provides insight into the mechanisms of adaptive divergence in eukaryotes. Mol Ecol. 23(4):753–773. doi: 10.1111/mec.12631. [DOI] [PubMed] [Google Scholar]
- Grenier J, Potvin C, Asselin A. 2000. Some fungi express β-1,3-glucanases similar to thaumatin-like proteins. Mycologia. 92(5):841–848. doi: 10.1080/00275514.2000.12061228. [DOI] [Google Scholar]
- Guerin-Dubrana L, Fontaine F, Mugnai L. 2019. Grapevine trunk disease in European and Mediterranean vineyards: occurrence, distribution and associated disease-affecting cultural factors. Phytopathol Mediterr. 58(1):49–71. doi: 10.14601/Phytopathol_Mediterr-25153. [DOI] [Google Scholar]
- Hahn MW, De Bie T, Stajich JE, Nguyen C, Cristianini N. 2005. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 15(8):1153–1160. doi: 10.1101/gr.3567505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawallapersand P, Mashele SS, Kovačič L, Stojan J, Komel R, Pakala SB, Kraševec N, Syed K. 2014. Cytochrome P450 monooxygenase CYP53 family in fungi: comparative structural and evolutionary analysis and its role as a common alternative anti-fungal drug target. PLoS One. 9(9):e107209. doi: 10.1371/journal.pone.0107209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zhou JL, Song CG, Xu TM, Wu DM, Cui BK. 2022. Taxonomy, phylogeny and divergence times of Polyporus (Basidiomycota) and related genera. [cited 2024 Apr 1]. Available from https://www.cabidigitallibrary.org/doi/full/10.5555/20230042657.
- Kameshwar AKS, Qin W. 2016. Lignin degrading fungal enzymes. In: Fang Z, Smith Jr Richard L, editors. Production of Biofuels and Chemicals from Lignin. Singapore: Springer. p. 81–130. [Google Scholar]
- Keller NP. 2019. Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol. 17(3):167–180. doi: 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk A, Driessen AJ. 2010. Phylogenetic analysis of fungal ABC transporters. BMC Genomics. 11:177. doi: 10.1186/1471-2164-11-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk A, Lee YH, Asiegbu FO. 2013. Diversity and evolution of ABC proteins in basidiomycetes. Mycologia. 105(6):1456–1470. doi: 10.3852/13-001. [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek CP, Starr TL, Glass NL. 2014. Plant cell wall–degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol. 52(1):427–451. doi: 10.1146/annurev-phyto-102313-045831. [DOI] [PubMed] [Google Scholar]
- Kuznetsov D, Tegenfeldt F, Manni M, Seppey M, Berkeley M, Kriventseva EV, Zdobnov EM. 2023. OrthoDB v11: annotation of orthologs in the widest sampling of organismal diversity. Nucleic Acids Res. 51(D1):D445–D451. doi: 10.1093/nar/gkac998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lah L, Podobnik B, Novak M, Korošec B, Berne S, Vogelsang M, Kraševec N, Zupanec N, Stojan J, Bohlmann J, et al. 2011. The versatility of the fungal cytochrome P450 monooxygenase system is instrumental in xenobiotic detoxification. Mol Microbiol. 81(5):1374–1389. doi: 10.1111/j.1365-2958.2011.07772.x. [DOI] [PubMed] [Google Scholar]
- Larignon P, Dubos B. 1997. Fungi associated with esca disease in grapevine. Eur J Plant Pathol. 103(2):147–157. doi: 10.1023/A:1008638409410. [DOI] [Google Scholar]
- Laveau C, Letouze A, Louvet G, Bastien S, Guérin-Dubrana L. 2009. Differential aggressiveness of fungi implicated in esca and associated diseases of grapevine in France. Phytopathol Mediterr. 48(1):32–46. doi: 10.14601/Phytopathol_Mediterr-2873. [DOI] [Google Scholar]
- Lecomte P, Cholet C, Bruez E, Martignon T, Giudici M, Simonit M, Ugaglia AA, Forget D, Miramon J, Arroyo M, et al. 2022. Recovery after curettage of grapevines with esca leaf symptoms. Phytopathol Mediterr. 61(3):473–489. doi: 10.36253/phyto-13357. [DOI] [Google Scholar]
- Lehtiö J, Sugiyama J, Gustavsson M, Fransson L, Linder M, Teeri TT. 2003. The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc Natl Acad Sci U S A. 100(2):484–489. doi: 10.1073/pnas.212651999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur A, Lomascolo A, Chabrol O, Ruiz-Dueñas FJ, Boukhris-Uzan E, Piumi F, Kües U, Ram AF, Murat C, Haon M, et al. 2014. The genome of the white-rot fungus Pycnoporus cinnabarinus: a basidiomycete model with a versatile arsenal for lignocellulosic biomass breakdown. BMC Genomics. 15(1):486. doi: 10.1186/1471-2164-15-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ma F, Zhao H, Zhang S, Wang L, Zhang X, Yu H. 2019. A lytic polysaccharide monooxygenase from a white-rot fungus drives the degradation of lignin by a versatile peroxidase. Appl Environ Microbiol. 85(9):e02803-18. doi: 10.1128/AEM.02803-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Olson Å, Wu M, Broberg A, Sandgren M. 2017. Biochemical studies of two lytic polysaccharide monooxygenases from the white-rot fungus Heterobasidion irregulare and their roles in lignocellulose degradation. PLoS One. 12(12):e0189479. doi: 10.1371/journal.pone.0189479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Shen LL, Xu TM, Song CG, Gao N, Wu DM, Sun YF, Cui BK. 2023. Global diversity, molecular phylogeny and divergence times of the brown-rot fungi within the Polyporales. Mycosphere. 14(1):1564–1664. doi: 10.5943/mycosphere/14/1/18. [DOI] [Google Scholar]
- Macheleidt J, Mattern DJ, Fischer J, Netzker T, Weber J, Schroeckh V, Valiante V, Brakhage AA. 2016. Regulation and role of fungal secondary metabolites. Annu Rev Genet. 50(1):371–392. doi: 10.1146/annurev-genet-120215-035203. [DOI] [PubMed] [Google Scholar]
- Manavalan T, Manavalan A, Heese K. 2015. Characterization of lignocellulolytic enzymes from white-rot fungi. Curr Microbiol. 70(4):485–498. doi: 10.1007/s00284-014-0743-0. [DOI] [PubMed] [Google Scholar]
- Mangiola S, Papenfuss A. 2020. tidyHeatmap: an R package for modular heatmap production based on tidy principles. J Open Source Softw. 5(52):2472. doi: 10.21105/joss.02472. [DOI] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 38(10):4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín L, García-García B, Alguacil MDM. 2022. Interactions of the fungal community in the complex patho-system of esca, a grapevine trunk disease. Int J Mol Sci. 23(23):14726. doi: 10.3390/ijms232314726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto R, Mehjabin JJ, Noguchi H, Miyamoto T, Takasuka TE, Hori C. 2023. Genomic and secretomic analyses of the newly isolated fungus Perenniporia fraxinea SS3 identified CAZymes potentially related to a serious pathogenesis of hardwood trees. Appl Environ Microbiol. 89(5):e0027223. doi: 10.1128/aem.00272-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes FK, Vanderpool D, Fulton B, Hahn MW. 2021. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics. 36(22–23):5516–5518. doi: 10.1093/bioinformatics/btaa1022. [DOI] [PubMed] [Google Scholar]
- Miyauchi S, Hage H, Drula E, Lesage-Meessen L, Berrin JG, Navarro D, Favel A, Chaduli D, Grisel S, Haon M, et al. 2020. Conserved white-rot enzymatic mechanism for wood decay in the Basidiomycota genus Pycnoporus. DNA Res. 27(2):dsaa011. doi: 10.1093/dnares/dsaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moktali V, Park J, Fedorova-Abrams ND, Park B, Choi J, Lee YH, Kang S. 2012. Systematic and searchable classification of cytochrome P450 proteins encoded by fungal and oomycete genomes. BMC Genomics. 13:525. doi: 10.1186/1471-2164-13-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Cruz A, Amrine KC, Blanco-Ulate B, Lawrence DP, Travadon R, Rolshausen PE, Baumgartner K, Cantu D. 2015. Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens. BMC Genomics. 16(1):1–22. doi: 10.1186/S12864-015-1624-Z/FIGURES/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel-Rouhier M. 2021. Chapter 4—Wood as a hostile habitat for ligninolytic fungi. In: Morel-Rouhier M, Sormani R, editors. Advances in Botanical Research. Wood Degradation and Ligninolytic Fungi. Vol. 99. Cambridge (MA): Academic Press. p. 115–149. [Google Scholar]
- Moretti S, Goddard ML, Puca A, Lalevée J, Di Marco S, Mugnai L, Gelhaye E, Goodell B, Bertsch C, Farine S. 2023. First description of non-enzymatic radical-generating mechanisms adopted by Fomitiporia mediterranea: an unexplored pathway of the white rot agent of the esca complex of diseases. J Fungi (Basel). 9(4):498. doi: 10.3390/jof9040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti S, Pacetti A, Pierron R, Kassemeyer HH, Fischer M, Péros JP, Perez-Gonzalez G, Bieler E, Schilling M, Di Marco S, et al. 2021. Fomitiporia mediterranea M. Fisch., the historical esca agent: a comprehensive review on the main grapevine wood rot agent in Europe. Phytopathol Mediterr. 60(2):351–379. doi: 10.36253/phyto-13021. [DOI] [Google Scholar]
- Mundy DC, Brown A, Jacobo F, Tennakoon K, Woolley RH, Vanga B, Tyson J, Johnston P, Ridgway HJ, Bulman S. 2020. Pathogenic fungi isolated in association with grapevine trunk diseases in New Zealand. N Z J Crop Hortic Sci. 48(2):84–96. doi: 10.1080/01140671.2020.1716813. [DOI] [Google Scholar]
- Nazmul Hussain Nazir KH, Ichinose H, Wariishi H. 2010. Molecular characterization and isolation of cytochrome P450 genes from the filamentous fungus Aspergillus oryzae. Arch Microbiol. 192(5):395–408. doi: 10.1007/s00203-010-0562-z. [DOI] [PubMed] [Google Scholar]
- Pacetti A, Moretti S, Perrin C, Gelhaye E, Bieler E, Kassemeyer HH, Mugnai L, Farine S, Bertsch C. 2022. Grapevine wood-degrading activity of Fomitiporia mediterranea M. Fisch.: a focus on the enzymatic pathway regulation. Front Microbiol. 13:844264. doi: 10.3389/fmicb.2022.844264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podobnik B, Stojan J, Lah L, Kras̆evec N, Selis̆kar M, Riz̆ner TL, Rozman D, Komel R. 2008. CYP53A15 of Cochliobolus lunatus, a target for natural antifungal compounds. J Med Chem. 51(12):3480–3486. doi: 10.1021/jm800030e. [DOI] [PubMed] [Google Scholar]
- Presley GN, Panisko E, Purvine SO, Schilling JS. 2018. Coupling secretomics with enzyme activities to compare the temporal processes of wood metabolism among white and brown rot fungi. Appl Environ Microbiol. 84(16):e00159-18. doi: 10.1128/AEM.00159-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto M, Wedin M. 2013. Dating the diversification of the major lineages of Ascomycota (fungi). PLoS One. 8(6):e65576. doi: 10.1371/journal.pone.0065576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Ibrahim HMM, Erz M, Kümmel F, Panstruga R, Kusch S. 2023. Long noncoding RNAs emerge from transposon-derived antisense sequences and may contribute to infection stage-specific transposon regulation in a fungal phytopathogen. Mob DNA. 14(1):17. doi: 10.1186/s13100-023-00305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. 2014. BEDTools: the Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics. 47(1):11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2018. FigTree, A Graphical Viewer of Phylogenetic Trees (Version 1.4. 4). Edinburgh (Scotland): Institute of Evolutionary Biology, University of Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/.
- R Core Team . 2022. R: A Language and Environment for Statistical Computing. Vienna (Austria): R Foundation for Statistical Computing. https://www.R-project.org/.
- Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, et al. 2014. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A. 111(27):9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR. 2018. Purge haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 19(1):460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacristán S, Goss EM, Eves-van den Akker S. 2021. How do pathogens evolve novel virulence activities? Mol Plant Microbe Interact. 34(6):576–586. doi: 10.1094/MPMI-09-20-0258-IA. [DOI] [PubMed] [Google Scholar]
- Saier MH, Reddy VS, Moreno-Hagelsieb G, Hendargo KJ, Zhang Y, Iddamsetty V, Lam KJK, Tian N, Russum S, Wang J, et al. 2021. The transporter classification database (TCDB): 2021 update. Nucleic Acids Res. 49(D1):D461–D467. doi: 10.1093/nar/gkaa1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y, Watanabe H, Nagai M, Nakade K, Takahashi M, Sato T. 2006. Lentinula edodes tlg1 encodes a thaumatin-like protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol. 141(2):793–801. doi: 10.1104/pp.106.076679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling M, Maia-Grondard A, Baltenweck R, Robert E, Hugueney P, Bertsch C, Farine S, Gelhaye E. 2022. Wood degradation by Fomitiporia mediterranea M. Fischer: physiologic, metabolomic and proteomic approaches. Front Plant Sci. 13:988709. doi: 10.3389/fpls.2022.988709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shallom D, Leon M, Bravman T, Ben-David A, Zaide G, Belakhov V, Shoham G, Schomburg D, Baasov T, Shoham Y. 2005. Biochemical characterization and identification of the catalytic residues of a family 43 β-d-xylosidase from Geobacillus stearothermophilus T-6. Biochemistry. 44(1):387–397. doi: 10.1021/bi048059w. [DOI] [PubMed] [Google Scholar]
- Sista Kameshwar AK, Qin W. 2018. Comparative study of genome-wide plant biomass-degrading CAZymes in white rot, brown rot and soft rot fungi. Mycology. 9(2):93–105. doi: 10.1080/21501203.2017.1419296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2015a. RepeatModeler Open-1.0. 2008–2015. http://www.repeatmasker.org.
- Smit AFA, Hubley R, Green P. 2015b. RepeatMasker Open-4.0. 2013–2015.http://www.repeatmasker.org.
- Song J, Cui BK. 2017. Phylogeny, divergence time and historical biogeography of Laetiporus (Basidiomycota, Polyporales). BMC Evol Biol. 17(1):102. doi: 10.1186/s12862-017-0948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Wang H, Tao S, Xi X. 2023. Large-scale detection of telomeric motif sequences in genomic data using TelFinder. Microbiol Spectr. 11(2):e0392822. doi: 10.1128/spectrum.03928-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Shale K, Pagadala NS, Tuszynski J. 2014. Systematic identification and evolutionary analysis of catalytically versatile cytochrome p450 monooxygenase families enriched in model basidiomycete fungi. PLoS One. 9(1):e86683. doi: 10.1371/journal.pone.0086683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Víglaš J, Olejníková P. 2021. An update on ABC transporters of filamentous fungi—from physiological substrates to xenobiotics. Microbiol Res. 246:126684. doi: 10.1016/j.micres.2020.126684. [DOI] [PubMed] [Google Scholar]
- Viborg AH, Terrapon N, Lombard V, Michel G, Czjzek M, Henrissat B, Brumer H. 2019. A subfamily roadmap of the evolutionarily diverse glycoside hydrolase family 16 (GH16). J Biol Chem. 294(44):15973–15986. doi: 10.1074/jbc.RA119.010619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson L. 2011. Ggplot2: elegant graphics for data analysis by WICKHAM, H. Biometrics. 67(2):678–679. doi: 10.1111/j.1541-0420.2011.01616.x. [DOI] [Google Scholar]
- Worrall JJ, Anagnost SE, Zabel RA. 1997. Comparison of wood decay among diverse Lignicolous fungi. Mycologia. 89(2):199–219. doi: 10.1080/00275514.1997.12026772. [DOI] [Google Scholar]
- Wu H, Nakazawa T, Takenaka A, Kodera R, Morimoto R, Sakamoto M, Honda Y. 2020. Transcriptional shifts in delignification-defective mutants of the white-rot fungus Pleurotus ostreatus. FEBS Lett. 594(19):3182–3199. doi: 10.1002/1873-3468.13890. [DOI] [PubMed] [Google Scholar]
- Yu F, Song J, Liang J, Wang S, Lu J. 2020. Whole genome sequencing and genome annotation of the wild edible mushroom, Russula griseocarnosa. Genomics. 112(1):603–614. doi: 10.1016/j.ygeno.2019.04.012. [DOI] [PubMed] [Google Scholar]
- Zhao H, Dai YC, Wu F, Liu XY, Maurice S, Krutovsky KV, Pavlov IN, Lindner DL, Martin FM, Yuan Y. 2023. Insights into the ecological diversification of the hymenochaetales based on comparative genomics and phylogenomics with an emphasis on coltricia. Genome Biol Evol. 15(8):evad136. doi: 10.1093/gbe/evad136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Vlasák J, Yuan Y. 2023. Outline, phylogenetic and divergence times analyses of the genus Haploporus (Polyporales, Basidiomycota): two new species are proposed. MycoKeys. 98:233–252. doi: 10.3897/mycokeys.98.105684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Ge Q, Yan Y, Zhang X, Huang L, Yin Y. 2023. dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 51(W1):W115–W121. doi: 10.1093/nar/gkad328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LW, Wei YL, Dai YC. 2014. Phylogenetic analysis of ligninolytic peroxidases: preliminary insights into the alternation of white-rot and brown-rot fungi in their lineage. Mycology. 5(1):29–42. doi: 10.1080/21501203.2014.895784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated for this project are available at NCBI (PRJNA1099290; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1099290). The references for previously published genomic data used in this study can be seen in Supplementary Table 2 of Supplementary File 1. The genome assemblies and gene models produced in this study are publicly available at Zenodo (https://doi.org/10.5281/zenodo.10957629) and NCBI (PRJNA1136927; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1136927). Dedicated genome browsers and BLAST tools are available at https://www.grapegenomics.com. The code used in this project is displayed in Supplementary File 2.
Supplemental material available at G3 online.