Abstract
The life cycle of human papillomaviruses (HPVs) is tightly linked to the differentiation status of the host cell. While early genes are expressed during the initial stages of viral infection, late gene expression occurs in the suprabasal layers of the cervical epithelium. Late genes encode E1^E4, a cytosolic protein, and capsid proteins L1 and L2. We have mapped over 30 initiation sites for late transcripts and show that the transcripts initiate in a 200-nucleotide region within the E7 open reading frame. The mechanisms regulating the activation of late gene expression, however, are not yet understood. DNase I hypersensitivity analysis of HPV-31 chromatin in cell lines that maintain viral genomes extrachromosomally indicates that a major shift in nuclease digestion occurs upon differentiation. In undifferentiated cells, hypersensitive regions exist in the upstream regulatory region proximal to the E6 open reading frame. Upon differentiation, a region between nucleotides 659 and 811 in the E7 open reading frame becomes accessible to DNase I. These results indicate that the late transcript initiation region becomes accessible to transcription factor binding upon differentiation. Several complexes mediate chromatin rearrangement, and we tested whether histone acetylation was sufficient for late transcript activation. Treatment with the histone deacetylase inhibitor trichostatin A was found to be insufficient to activate late gene expression in undifferentiated cells. However, it did activate expression of early transcripts. These results suggest that chromatin remodeling around the late promoter occurs upon epithelial differentiation and that mechanisms in addition to histone deacetylation contribute to activation of late gene expression.
Papillomaviruses (PVs) are a family of small DNA viruses that infect epithelial tissue at various anatomical locations. Over 85 PV types that are specific for humans (HPVs) have been identified, and they show a high degree of sequence and functional conservation. A subset of HPVs infect the genital epithelia and are broadly divided into two categories: low-risk viruses that cause benign hyperproliferative lesions, and high-risk types that can potentially progress to neoplastic transformation (23).
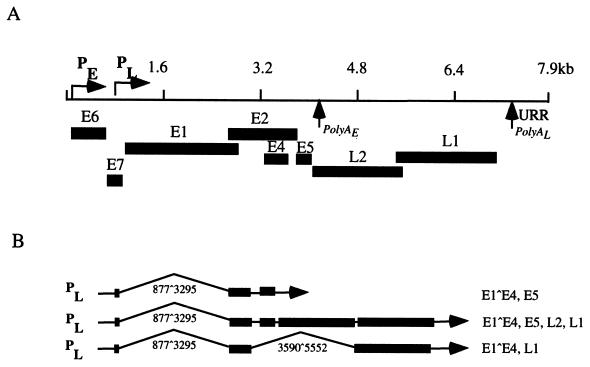
High-risk HPV types include HPV-16, -18, -31, -33, -45, and -54 (23, 35). During the initial stages of HPV infection, viral DNA is established as episomes in the basal layer of stratified epithelia, with a copy number of 50 to 100 copies per cell. Early transcripts are expressed from a promoter upstream of E6 and, in HPV-31, initiate at approximately nucleotide 97 (p97) (Fig. 1A) (25). Similar promoters have been identified in HPV-16 and -18 (4, 7). Early transcripts are polycistronic messages that encode a number of viral proteins, including E1, E2, E6, and E7 (25, 47). The early promoter is regulated by sequences within a noncoding upstream regulatory region (URR) containing the origin of viral replication as well as binding sites for viral and cellular proteins (23, 24, 28, 34, 54). Copy number and early gene expression are tightly regulated at this initial stage by viral proteins E1 and E2 (13, 14, 17, 36). As basal cells divide and daughter cells migrate from basal strata, they begin to differentiate. As differentiation occurs, two viral processes take place: viral genome amplification and late gene induction (6, 18). Late genes consist of polycistronic messages that include E1^E4, a cytoplasmic protein thought to facilitate viral egress from infected cells, as well as L1 and L2, the two viral capsid proteins (Fig. 1B) (15, 21, 25, 26, 40, 43, 44). Keratinocyte differentiation is therefore required for a productive viral infection.
FIG. 1.
(A) Linear representation of the HPV-31 genome. Open reading frames are indicated underneath the map. PE refers to the early promoter (p97); PL refers to the late promoter. PolyA indicates polyadenylation sites. (B) Cartoon depicting late HPV-31 transcripts. PL designates the late promoter. Splice sites are indicated by the numbers below each transcript. Open reading frames are specified to the right of each message.
Past work from Meyers et al. used organotypic raft culture to mimic the complete HPV-31 life cycle in vitro using the CIN 612 cell line, which was established from a cervical biopsy that maintained episomal copies of HPV-31b (38). The raft system successfully induced epithelial differentiation, viral genome amplification, induction of late genes, and assembly of infectious virions. Additional studies identified late transcripts that appeared upon differentiation of CIN 612 cells and initiated in a region around nucleotide 742 (Fig. 1) (25, 26). Differentiation-induced transcripts also appear in HPV-16 and HPV-6; furthermore, these transcripts initiate in a similar region of the E7 open reading frame (ORF) (20, 22, 29, 39). While some of the initiation sites of the late promoter have been mapped, a detailed analysis of the extent of these initiation sites is incomplete.
The mechanisms that mediate the switch from early to late promoter usage have not been elucidated, and the role of chromatin in this regulation has not been established. Nuclear DNA is condensed into chromatin and, in this state, is generally inaccessible to the transcriptional machinery (59). In contrast, transcriptionally active regions of DNA are readily accessible to transcription factors. The basic unit of chromatin is the nucleosome, a complex of approximately 200 nucleotides tightly wound around a core octamer of histones H2A, H2B, H3, and H4 and one linker histone. Nucleosomes are assembled into higher-order structures of increased compaction. It is generally agreed that chromatin remodeling must occur for the transcriptional machinery to gain access to DNA (59). Two cellular processes facilitate gene expression via chromatin modification: ATP-dependent chromatin remodeling and histone acetylation (8, 30, 33). ATP-dependent remodeling is thought to be a catalytic process that shifts the thermodynamic equilibrium between different nucleosomal conformations to favor the relaxed state. A second process is mediated by histone acetyltransferases (HATs) and covalently modifies histones. HATs add acetyl moieties to lysine residues in histone tails; as a result, acetylated histones are unable to bind DNA tightly. The process can be reversed by histone deacetylases.
While it has been observed that HPV late transcript expression requires epithelial differentiation, the process by which late gene expression is activated remains unknown. It is possible that differentiated cells express cellular factors that are required for late gene expression or that viral genome amplification results in viral chromatin rearrangement to make it more accessible to the transcriptional machinery. In this study, we have mapped in detail the initiation sites for late transcripts and examined chromatin remodeling around the HPV-31 late promoter at different stages of the viral life cycle.
Late transcripts initiate at various sites within the E7 ORF.
The HPV-31 life cycle includes a temporal pattern of expression of early and late genes. Late gene expression requires epithelial differentiation, and late transcripts have been observed to initiate at heterogeneous sites (20, 22, 29, 39). Previous studies induced differentiation by means of organotypic raft cultures, which provided a mix of both undifferentiated and differentiated cells. We have recently determined that differentiation induced by suspension in 1.5% methylcellulose for 24 h, in contrast to raft culture, resulted in a relatively uniform degree of differentiation in all cells and induced viral DNA amplification and late transcript expression (45, 46).
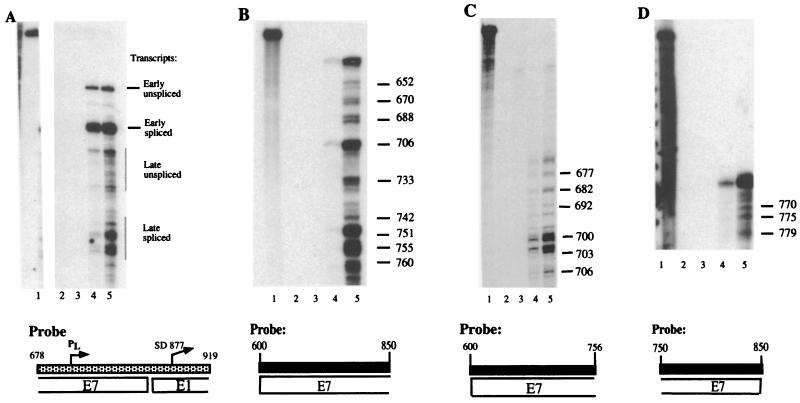
An RNase protection assay is shown in Fig. 2A, in which we compared initiation site usage between early and late genes. RNA samples from an undifferentiated population of CIN 612 cells (lane 4) and from CIN 612 cells differentiated in methylcellulose for 24 h (lane 5) were hybridized to a probe (pRP-p742) that detects early and late transcripts (51). Two bands representing early transcripts are seen in both differentiated and undifferentiated cells. These bands correspond to spliced and unspliced messages initiated at the early promoter. In contrast, late transcripts are induced upon differentiation and consist of several bands corresponding to a number of initiation sites (compare lane 4 with lane 5). The pattern of initiation among unspliced and spliced late messages appears to differ; however, we have not determined whether splicing events affect initiation site usage among late transcripts. Previous work from Hummel et al. and Ozbun and Meyers indicated that a number of late messages initiate at nucleotides other than nucleotide 742 (25, 26, 40, 41). We therefore performed a more extensive analysis to map the differentiation-dependent transcripts by RNase protection assay on RNA isolated from the CIN 612 cell line.
FIG. 2.
RNase protection assays. Total RNA was isolated from monolayer or differentiated cells using the Trizol reagent (Invitrogen, Carlsbad, Calif.) per the manufacturer's instructions. Antisense riboprobes labeled with 32P were synthesized using the Riboprobe transcription system from Promega. Probes were purified following gel electrophoresis in 6% acrylamide. RNase protection assay was performed as described by Stubenrauch et al. (50), with the following exceptions: 15 μg of RNA was used for each hybridization to 1.5 × 105 cpm of probe, and the RNA was digested with 12-μg/ml RNase A and 60-U/ml RNase T1 (Roche Molecular Biochemicals, Indianapolis, Ind.). Labeled 100-bp ladder (Invitrogen) and a DNA sequencing ladder were run with samples as size markers. Lanes contain hybridization samples as follows: 1, undigested probe; 2, no RNA; 3, yeast tRNA; 4, RNA from monolayer CIN 612 cells; 5, RNA from CIN 612 cells induced to differentiate in methylcellulose for 24 h. (A) The pRP-p742 probe, which detects both early and late messages, has been described previously (50). The probe is illustrated by a stippled rectangle below the autoradiograph. Corresponding open reading frames are specified under the probe. PL designates the late promoter, while SD877 represents a splice donor site at nucleotide 877. Early and late transcripts are marked to the right of the bands. (B to D) RNase protection assay for mapping of late transcripts. All probes are specified below each autoradiograph. The probe used in panel B spans nucleotides 600 to 850; the probe in panel C spans nucleotides 600 to 756; the probe in panel D includes nucleotides 750 to 850. The numbers to the right of each band indicate the nucleotide location of each initiation site. The slowest-migrating band in lanes 4 and 5 in each panel corresponds to the fully protected probe.
We mapped transcript initiation sites for the late promoter in the E7 ORF using five overlapping probes that spanned approximately 300 nucleotides within the E7 ORF. To ensure that our studies identified real start sites, we required that the putative sites be detected in analysis with at least two overlapping probes. None of these probes spanned a known splice donor or acceptor site. The transcripts were mapped by size comparison with a DNA sequencing ladder, and three representative experiments are shown in Fig. 2B to 2D. Because RNA has a lower mobility in denaturing polyacrylamide gels than DNA of the same size, transcript initiation sites were mapped within a 5-nucleotide margin of error.
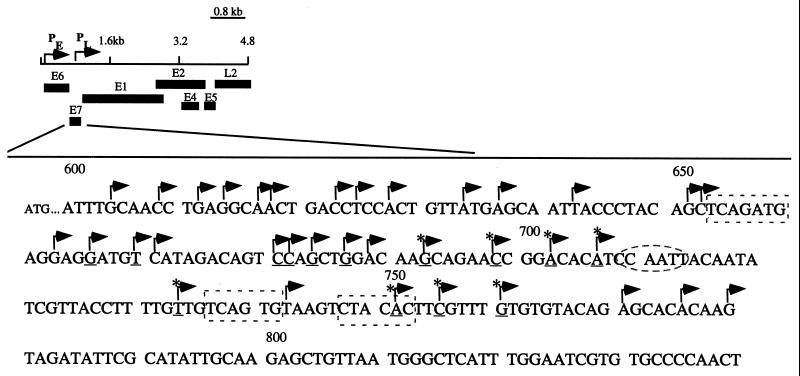
We observed a set of protected bands in differentiated cells that corresponded to approximately 35 different initiation sites within the E7 ORF. Three representative RNase protection assays, which use three different probes that detect 18 initiation sites, are shown on Fig. 2B to 2D. The frequency of initiation site usage varied among transcripts, as judged by band intensity on RNase protection assay. Interestingly, several protected bands were observed to initiate in the E7 ORF in undifferentiated cells as well as in differentiated cells. These transcripts therefore appear to be expressed constitutively throughout the life cycle of the virus (Fig. 2B and 2C). An example of these messages included a transcript initiating at nucleotide 706 that is expressed in monolayer CIN 612 cells and is upregulated in cells that have undergone differentiation (Fig. 2B). The region of differentiation-dependent transcript initiation that we identified spans approximately 200 nucleotides within the E7 ORF. Figure 3 summarizes initiation site usage within the E7 ORF. The 35 transcripts that we mapped include initiation sites at nucleotides 737, 742, 750, and 767, consistent with previous reports mapping some of these sites (40, 41).
FIG. 3.
Partial linear map of HPV-31 with corresponding open reading frames at the bottom. The portion of the E7 ORF from nucleotides 600 to 850 has been magnified to mark with arrows the initiation sites that were identified in this study. Initiation sites that show a high frequency of usage are underlined. Transcripts that are constitutively expressed are indicated by asterisks. Sequences in dashed boxes are initiator elements, and the CCAAT box is indicated by a dashed oval.
A change in chromatin architecture around the late initiation region coincides with epithelial differentiation.
The mechanism by which late genes are induced remains unclear. PV genomes are associated with histone proteins (16), and chromatin remodeling has been shown to affect gene expression (5, 9, 55, 58, 60, 61). We therefore examined the state of chromatin throughout the life cycle of HPV-31 in order to observe any changes in the architecture of viral chromatin. The DNase I hypersensitivity assay was used, as it allows us to observe patterns of permeability to nuclease digestion throughout the genome at different stages of the viral life cycle. This assay is based on the fact that nucleosomal arrays are resistant to nuclease cleavage, while regions associated with nonhistone proteins are sensitive to DNase I (10). Furthermore, the pattern of DNase I sensitivity has been shown to correlate with the binding of transcriptional regulators to hypersensitive sites.
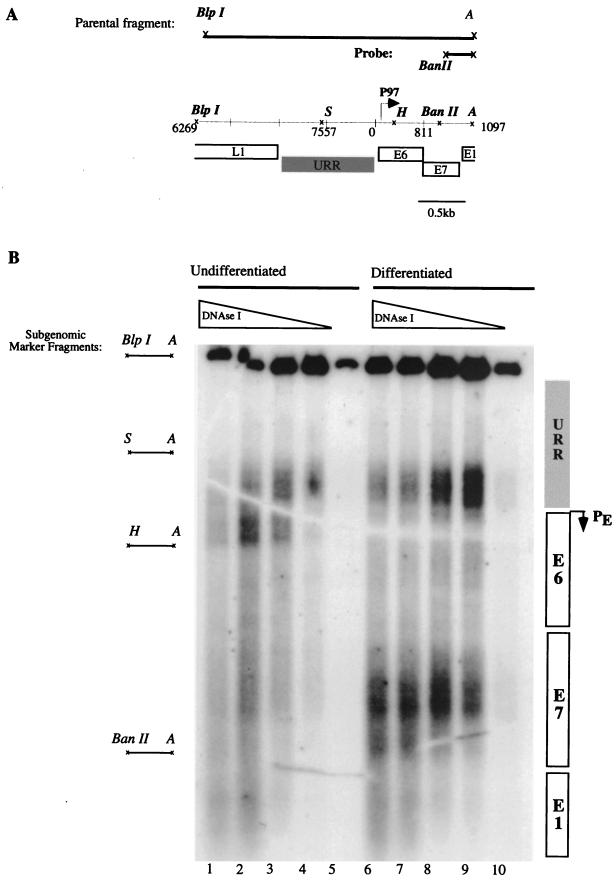
For this study, we isolated nuclei from undifferentiated and methylcellulose-differentiated populations of CIN 612 cells by sucrose gradient centrifugation. Following isolation, the nuclei were lightly treated with various concentrations of DNase I. DNA extracts from treated nuclei were prepared and digested with BlpI and AlwNI, each of which cleaves the HPV-31 genome at a single site. Digestion with these two enzymes yields a subgenomic restriction fragment from nucleotides 6269 to 1097, which spans the 3′ half of the L1 ORF to the 5′ half of the E1 ORF (Fig. 4A). Because the nuclei were lightly treated with DNase I, a portion of the HPV-31 molecules escaped nuclease digestion. This full-length BlpI-AlwNI restriction fragment ran as a 2.7-kb “parental” band (Fig. 4A). The digested DNA was examined by Southern analysis using a subgenomic probe positioned at the 3′ end of the BlpI-AlwNI restriction fragment. The sites of nuclease digestion were mapped by their distance from the 3′ end of the probe.
FIG. 4.
DNase I hypersensitivity analysis of HPV-31. (A) Schematic representation of the parental fragment, probe, and corresponding regions in HPV-31. P97, early promoter. URR and open reading frames are indicated by boxes. Restriction sites are indicated by italics: BlpI; BanII; A, AlwNI; S, SpeI; H, HpaI. (B) Autoradiograph corresponding to a DNase I hypersensitivity assay. Lanes 1 to 5, nuclei from undifferentiated CIN 612 cells. Lanes 6 to 10, nuclei from CIN 612 cells that were induced to differentiate in semisolid medium for 24 h. Nuclei were treated with a titration of DNase I corresponding to 10, 5, 2.5, 1.25, and 0 U/μl. Genomic DNA was digested with BlpI and AlwNI restriction endonucleases to yield a 2.7-kb parental fragment, and areas of hypersensitivity were observed by Southern blot analysis with a probe corresponding to the BanII-AlwNI fragment of HPV-31. Subgenomic marker fragments used for mapping areas of nuclease digestion are indicated at the left. Regions of the HPV-31 genome that show hypersensitivity are indicated to the right of the autoradiograph.
Several species of higher mobility were detected in nuclei treated with DNase I. An area of hypersensitivity was observed in the vicinity of the SpeI-AlwNI marker in undifferentiated CIN 612 cells, between nucleotides 7557 and 215 (Fig. 4B, lanes 1 to 4). This region corresponds to sequences within the URR and proximal to the early promoter. This pattern of hypersensitivity is absent in nuclei that were not treated with DNase I (lane 5), and its intensity increases proportionally to the concentration of DNase I that was used. The location of these DNase I-hypersensitive sites is in accordance with published studies of HPV-31 promoter usage in undifferentiated cells.
We detected a major shift in the pattern of hypersensitivity in cells that had undergone differentiation in methylcellulose. An area of sensitivity mapping close to marker fragment BanII-AlwNI appeared in differentiated CIN 612 cells (Fig. 4B, lanes 6 to 9). This area of nuclease digestion occurs within the region between nucleotides 659 and 811. The observed digestion pattern was absent from untreated nuclei and was not seen in nuclei isolated from undifferentiated cells at similar concentrations of DNase I. The area of hypersensitivity maps within the E7 ORF and corresponds to the region where we mapped a number of initiation sites for differentiation-dependent transcripts. We have observed the same pattern of hypersensitivity in at least three independent experiments using CIN 612 cells at different passage. The pattern of nuclease digestion suggests that DNA accessibility around the E7 ORF changes with differentiation so that the DNA becomes associated with nonhistone proteins, which could potentially recruit the transcriptional machinery to the region. We also observed a decrease in accessibility to DNase I in the proximity of the early promoter upon differentiation (Fig. 4B, lanes 6 to 9).
Histone hyperacetylation is insufficient to activate late transcript expression.
Because histone acetylation is one mechanism that mediates chromatin remodeling, we tested whether it could induce an altered chromatin configuration similar to that seen upon epithelial differentiation. If changes in histone acetylation were solely responsible for altered DNase I hypersensitivity, we would expect that treatment of undifferentiated cells with an inhibitor of histone deacetylases would lead to the same pattern of sensitivity seen in differentiated cells. Undifferentiated monolayer cultures of CIN 612 cells were therefore treated with various concentrations of trichostatin A (TSA), a noncompetitive inhibitor of histone deacetylases (62, 63), and harvested after 24 h. We then performed DNase I hypersensitivity analysis and observed that TSA treatment of monolayer cells did not induce a significant degree of hypersensitivity in the E7 ORF (data not shown).
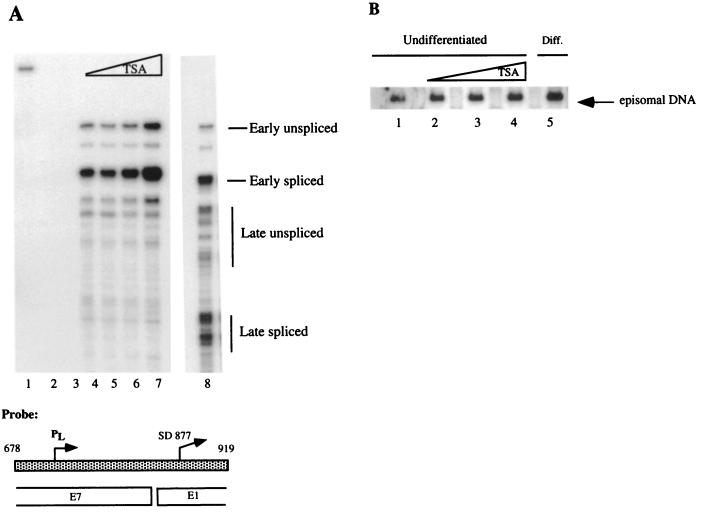
It is possible that any chromatin alterations induced by histone hyperacetylation in the TSA-treated cells occur in only a subset of cells or are too subtle to be detected by Southern blotting. We therefore performed RNase protection assays as a functional test for a relationship between histone hyperacetylation and late gene expression. Antisense riboprobe pRP-p742, which detects both early and late transcripts, was hybridized to RNA harvested after treatment with TSA, and an RNase protection assay was performed (Fig. 5A) (51). As shown in Fig. 5A, histone hyperacetylation does not induce late transcript activation in monolayer cells (lanes 5 to 7). This is in contrast to the strong activation of late gene expression in cells that were induced to differentiate in methylcellulose (lane 8). These results suggest that late gene expression is linked to a change that occurs upon epithelial differentiation and that histone acetylation alone is not responsible for late gene induction.
FIG. 5.
(A) RNase protection assay. Probe pRP-p742 was used to examine whether histone hyperacetylation induced late gene expression after treatment of undifferentiated CIN 612 cells with TSA (Sigma-Aldrich, St. Louis, Mo.). TSA was dissolved in ethanol and diluted directly into culture media to a final concentration of 100, 300, or 900 nM. The same volume of ethanol was added to the untreated samples. Monolayer cells were treated for 24 h and used directly to isolate RNA or induced to differentiate in semisolid medium for 24 h. Lane 1, undigested probe; lane 2, no RNA; lane 3, yeast tRNA; lane 4, untreated monolayer CIN 612 cells; lanes 5 to 7, undifferentiated CIN 612 cells treated with TSA at 100 nM (lane 5), 300 nM (lane 6), or 900 nM (lane 7). Lane 8 shows the pattern of late gene expression induced following suspension in methylcellulose for 24 h in the absence of TSA. Transcripts are labeled to the right of bands. The low levels of late transcript initiation in undifferentiated cells are due to a small percentage of cells that differentiate spontaneously (43). (B) Southern blot analysis for episome copy number after treatment with TSA. The Southern blot assay was performed as previously described (24). Briefly, 10 μg of sheared genomic DNA was digested with BglII (New England Biolabs), an enzyme that does not cut the HPV-31 genome, or with HpaI (New England Biolabs), a single cutter in HPV-31. The digests were separated in 0.8% agarose, transferred to a ZetaProbe GT membrane (Bio-Rad), and hybridized to an HPV-31 genomic probe at 42°C. Lanes: 1, untreated monolayer CIN 612 cells; 2 to 4, monolayer CIN 612 cells treated with 100, 300, and 900 nM TSA, respectively; 5, untreated CIN 612 cells that were induced to differentiate in semisolid medium for 24 h. All bands were quantified by PhosphorImager analysis.
Interestingly, our studies suggest that histone acetylation may play a role in regulation of early transcript expression. PhosphorImager analysis indicates that early transcript expression is upregulated approximately threefold in samples treated with 900 nM TSA (data not shown). This number is an average of two different experiments. Because histone acetylation also plays a role in DNA synthesis (1, 12, 32), we decided to investigate whether the increase in early transcripts was correlated with an increase in viral template number after TSA treatment. Undifferentiated monolayer cultures of CIN 612 cells were treated with TSA for 24 h, DNA extracts were prepared, and viral copy number was determined by Southern blot analysis. PhosphorImager analysis revealed that episomal DNA was increased in undifferentiated samples treated with 900 nM TSA by 1.8-fold relative to the untreated cells (Fig. 5B). This increase in copy number after TSA treatment may partly account for increased early transcripts in undifferentiated cells.
Western blot analysis was performed to detect the extent of histone acetylation after treatment with TSA. Undifferentiated CIN 612 cells were treated with TSA for 24 h, and protein extracts were prepared for Western blotting. We observed a dose-dependent increase in the levels of acetylated H4 following TSA treatment (data not shown). These results indicate that, while TSA treatment does induce histone hyperacetylation, this effect is insufficient to activate late gene expression.
The HPV life cycle is tightly linked to the differentiation status of the host cell. Two sets of viral genes follow a temporal pattern of expression: early genes are expressed throughout the life cycle, while late genes require epithelial differentiation for expression. In this study, we have performed RNase protection assays to map HPV-31 transcripts that initiate at various sequences within the E7 ORF. Previous studies mapped transcript initiation sites at nucleotides 737, 742, 750, and 767. In this study, we mapped approximately 35 additional start sites that span 200 nucleotides within the E7 ORF. These 5′ termini include start sites at nucleotides 626, 642, 651, 680, 733, and 751. In addition, we observed that a number of transcripts that initiate in the E7 ORF are expressed constitutively throughout the life cycle. For example, a transcript initiating at nucleotide 706 was expressed in monolayer cells, although at lower levels than observed when cells underwent differentiation. These constitutive transcripts could provide low-level expression of the replication proteins E1 and E2 (31). The promiscuous initiation site usage that we observed among HPV-31 late transcripts is similar to that of other DNA tumor viruses such as simian virus 40 (SV40). In the case of SV40, approximately two dozen initiation sites for late transcripts have been identified, and they span a 300-nucleotide region of the genome (49).
No consensus TATA box (TATAAA) is present in the E7 ORF of HPV-31, HPV-6, HPV-16, or bovine PV type 1 (3, 20, 22, 29, 39). In the case of most TATA-less promoters, an initiator element overlaps the initiation site and performs an analogous function to the TATA box: it directs assembly of the basal transcriptional machinery at the correct initiation site (10). Sequence analysis indicates that initiator elements may be present at nucleotides 653 to 659, 736 to 742, and 747 to 753 of HPV-31. Some of the start sites that we mapped overlap these putative elements. The rest of the transcripts, however, lack both a TATA box and initiator element. Transcription from TATA-less and initiatorless promoters appears to be mediated by transcription factors, such as Sp1, that presumably direct the transcriptional machinery to particular initiation sites. Initiation from this type of promoter has been shown to occur in a promiscuous manner over hundreds of nucleotides throughout the promoter (48).
Sequence analysis of the E7 ORF using the TRANSFAC database (57) reveals that a number of potential transcription factor-binding sites occur in this region. In preliminary studies, we observed bandshifts in electrophoretic mobility shift assays using probes that spanned the E7 ORF and identified in vitro binding of Oct-1, SOX5, and SRY (L. Peña, unpublished data). It is therefore possible that these binding sites function as transcriptional cis elements to direct expression of late transcripts. It is also possible that sequences outside the E7 ORF, such as the URR, play a role in late gene expression.
One model for late gene activation involves transcription factors that are expressed exclusively upon differentiation and direct late transcript initiation within the E7 ORF. Alternatively, the viral replication that occurs at suprabasal strata may play an important role in activating late gene expression through one of two mechanisms. One possibility is that viral DNA replication leads to increased template number, so that low levels of constitutive late transcript expression can now be detected. A second possibility is that viral genome amplification titrates a repressor of late gene expression in a manner similar to that in SV40 (56). Both models require episomal templates for activation of late gene expression and are consistent with two previous observations. First, Frattini et al. showed that HPV-31 must exist as an episome for late gene expression to be activated (19). Second, CIN 612 cells that spontaneously amplify viral DNA in monolayer cultures also express late genes (43).
It is generally accepted that the state of chromatin can have a major impact on gene expression. DNA that is densely packaged into nucleosomes is inaccessible to the transcriptional machinery, and expression of a particular gene is therefore contingent on modification of the chromatin in the region. In our study, we used the DNase I hypersensitivity assay to observe the configuration of viral chromatin throughout the life cycle of HPV-31 and observed a major shift in nuclease hypersensitivity upon epithelial differentiation. Differentiated cells show increased permeability to DNase I digestion in the vicinity of nucleotides 659 and 811. This region is located within the E7 ORF and encompasses the late transcript initiation sites that we mapped by RNase protection assay. The observed shift in hypersensitivity suggests that transcription factor binding occurs in the E7 ORF upon epithelial differentiation.
SV40 gene expression is a prominent example of the role of chromatin remodeling in transcription. A series of studies indicate that nucleosome positioning along the SV40 origin of replication, the early promoter, and the late promoter affects the activity of these elements (2, 27, 42, 53). Cereghini et al. have observed that a DNase I-hypersensitive site along the late transcript initiation region appears during the switch to late gene expression (11). Chromatin remodeling therefore leads to increased accessibility to the SV40 late promoter and facilitates late gene expression.
We have identified a change in the in vivo chromatin structure of HPV-31 in the vicinity of the early promoter and the late transcript initiation region. Our data suggest that a change in differentiated cells coincides with chromatin remodeling in the E7 ORF, which is in turn associated with late gene expression. Studies by Stünkel and Bernard provide evidence that chromatin rearrangement plays a role in HPV gene expression (52). In vivo studies of CaSki cells, which carry approximately 500 copies of integrated HPV-16, indicate that nucleosome positioning in the URR occurs over the viral enhancer, the origin of viral replication, and the early promoter. The presence of a nucleosome over the HPV-16 early promoter has a repressive effect on promoter activity. In vitro studies also showed that a nucleosome assembles over the early promoter of HPV-18 at approximately the same site as in HPV-16. These results suggest that viral chromatin organization may be dictated by the DNA sequence and that it regulates viral gene expression.
Several cellular complexes remodel chromatin so that the DNA template can be accessed for transcription. These include histone acetylation and ATP-dependent remodeling complexes. Although they differ in function, both complexes increase DNA accessibility to the transcriptional machinery. Histone acetylation decreases the affinity of histone protein binding to DNA and leads to transcriptional activation of a number of promoters. Our analysis of TSA-treated CIN 612 cells indicates that histone acetylation is not sufficient to activate late gene expression in undifferentiated cells. It is possible that the altered architecture in differentiated cells is induced by another chromatin remodeling complex, such as SWI/SNF. Interestingly, human SNF5 was recently reported to interact with the viral protein E1 and participate in viral DNA replication (37). Whether this interaction plays a role in late promoter activation remains to be seen.
Our studies suggest that histone acetylation may play a role in HPV early gene expression from episomal templates. Zhao et al. have also reported increased transcription from the URR-E6 promoter of HPV-11 after TSA treatment (64). In addition, we observed a 1.8-fold increase in episome copy number in undifferentiated cells treated with TSA. In contrast, differentiation induces, on average, a threefold increase in viral copy number. The increase in copy number that was observed in TSA-treated cells may therefore be insufficient to activate late gene expression. It is also possible that, while copy number may play an integral role in late gene induction, other processes may also be at work. Additional studies examining the sequences that regulate induction of the late promoter will be required to further understand the mechanisms that regulate differentiation-dependent viral gene expression.
Acknowledgments
This work was supported by grants from the National Cancer Institute to L. Peña (1F31CA80673-01) and to L. Laimins (CA59655).
We thank members of the Laimins laboratory for technical advice and A. Aiyar and K. Rundell for helpful comments on the manuscript.
REFERENCES
- 1.Ait-Si-Ali S, Polesskaya A, Filleur S, Ferreira R, Duquet A, Robin P, Vervish A, Trouche D, Cabon F, Harel-Bellan A. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene. 2000;19:2430–2437. doi: 10.1038/sj.onc.1203562. [DOI] [PubMed] [Google Scholar]
- 2.Alexiadis V, Varga-Weisz P D, Bonte E, Becker P B, Gruss C. In vitro chromatin remodelling by chromatin accessibility complex (CHRAC) at the SV40 origin of DNA replication. EMBO J. 1998;17:3428–3438. doi: 10.1093/emboj/17.12.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker C C, Howley P M. Differential promoter utilization by the bovine papillomavirus in transformed cells and productively infected wart tissues. EMBO J. 1987;6:1027–1035. doi: 10.1002/j.1460-2075.1987.tb04855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker C C, Phelps W C, Lindgren V, Braun M J, Gonda M A, Howley P M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker P, Renkawitz R, Schutz G. Tissue-specific DNaseI hypersensitive sites in the 5′-flanking sequences of the tryptophan oxygenase and the tyrosine aminotransferase genes. EMBO J. 1984;3:2015–2020. doi: 10.1002/j.1460-2075.1984.tb02084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedell M A, Hudson J B, Golub T R, Turyk M E, Hosken M, Wilbanks G D, Laimins L A. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol. 1991;65:2254–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedell M A, Jones K H, Grossman S R, Laimins L A. Identification of human papillomavirus type 18 transforming genes in immortalized and primary cells. J Virol. 1989;63:1247–1255. doi: 10.1128/jvi.63.3.1247-1255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger S L. Gene regulation; local or global? Nature. 2000;408:412–415. doi: 10.1038/35044160. [DOI] [PubMed] [Google Scholar]
- 9.Bryan P N, Olah J, Birnstiel M L. Major changes in the 5′ and 3′ chromatin structure of sea urchin histone genes accompany their activation and inactivation in development. Cell. 1983;33:843–848. doi: 10.1016/0092-8674(83)90026-0. [DOI] [PubMed] [Google Scholar]
- 10.Carey M, Smale S T. Transcriptional regulation in eukaryotes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 11.Cereghini S, Saragosti S, Yaniv M, Hamer D H. SV40-alpha-globulin hybrid minichromosomes. Differences in DNase I hypersensitivity of promoter and enhancer sequences. Eur J Biochem. 1984;144:545–553. doi: 10.1111/j.1432-1033.1984.tb08500.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Tini M, Evans R M. HATs on and beyond chromatin. Curr Opin Cell Biol. 2001;13:218–224. doi: 10.1016/s0955-0674(00)00200-3. [DOI] [PubMed] [Google Scholar]
- 13.Del Vecchio A M, Romanczuk H, Howley P M, Baker C C. Transient replication of human papillomavirus DNAs. J Virol. 1992;66:5949–5958. doi: 10.1128/jvi.66.10.5949-5958.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demeret C, Le Moal M, Yaniv M, Thierry F. Control of HPV 18 DNA replication by cellular and viral transcription factors. Nucleic Acids Res. 1995;23:4777–4784. doi: 10.1093/nar/23.23.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egawa K, Iftner A, Doorbar J, Honda Y, Iftner T. Synthesis of viral DNA and late capsid protein L1 in parabasal spinous cell layers of naturally occurring benign warts infected with human papillomavirus type 1. Virology. 2000;268:281–293. doi: 10.1006/viro.1999.0174. [DOI] [PubMed] [Google Scholar]
- 16.Favre M, Breitburd F, Croissant O, Orth G. Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J Virol. 1977;21:1205–1209. doi: 10.1128/jvi.21.3.1205-1209.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frattini M G, Laimins L A. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology. 1994;204:799–804. doi: 10.1006/viro.1994.1596. [DOI] [PubMed] [Google Scholar]
- 18.Frattini M G, Lim H B, Doorbar J, Laimins L A. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J Virol. 1997;71:7068–7072. doi: 10.1128/jvi.71.9.7068-7072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frattini M G, Lim H B, Laimins L A. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc Natl Acad Sci USA. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grassmann K, Rapp B, Maschek H, Petry K U, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J Virol. 1996;70:2339–2349. doi: 10.1128/jvi.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haller K, Stubenrauch F, Pfister H. Differentiation-dependent transcription of the epidermodysplasia verruciformis-associated human papillomavirus type 5 in benign lesions. Virology. 1995;214:245–255. doi: 10.1006/viro.1995.0028. [DOI] [PubMed] [Google Scholar]
- 22.Higgins G D, Uzelin D M, Phillips G E, McEvoy P, Marin R, Burrell C J. Transcription patterns of human papillomavirus type 16 in genital intraepithelial neoplasia: evidence for promoter usage within the E7 open reading frame during epithelial differentiation. J Gen Virol. 1992;73:2047–2057. doi: 10.1099/0022-1317-73-8-2047. [DOI] [PubMed] [Google Scholar]
- 23.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 947–978. [Google Scholar]
- 24.Hubert W G, Kanaya T, Laimins L A. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5′ of the minimal origin. J Virol. 1999;73:1835–1845. doi: 10.1128/jvi.73.3.1835-1845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hummel M, Hudson J B, Laimins L A. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hummel M, Lim H B, Laimins L A. Human papillomavirus type 31b late gene expression is regulated through protein kinase C-mediated changes in RNA processing. J Virol. 1995;69:3381–3388. doi: 10.1128/jvi.69.6.3381-3388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jongstra J, Reudelhuber T L, Oudet P, Benoist C, Chae C B, Jeltsch J M, Mathis D J, Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984;307:708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- 28.Kanaya T, Kyo S, Laimins L A. The 5′ region of the human papillomavirus type 31 upstream regulatory region acts as an enhancer which augments viral early expression through the action of YY1. Virology. 1997;237:159–169. doi: 10.1006/viro.1997.8771. [DOI] [PubMed] [Google Scholar]
- 29.Karlen S, Offord E A, Beard P. Functional promoters in the genome of human papillomavirus type 6b. J Gen Virol. 1996;77:11–16. doi: 10.1099/0022-1317-77-1-11. [DOI] [PubMed] [Google Scholar]
- 30.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 31.Klumpp D J, Laimins L A. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology. 1999;257:239–246. doi: 10.1006/viro.1999.9636. [DOI] [PubMed] [Google Scholar]
- 32.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 33.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 34.Kyo S, Tam A, Laimins L A. Transcriptional activity of human papillomavirus type 31b enhancer is regulated through synergistic interaction of AP1 with two novel cellular factors. Virology. 1995;211:184–197. doi: 10.1006/viro.1995.1390. [DOI] [PubMed] [Google Scholar]
- 35.Laimins L A. The biology of human papillomaviruses: from warts to cancer. Infect Agents Dis. 1993;2:74–86. [PubMed] [Google Scholar]
- 36.Lambert P F. Papillomavirus DNA replication. J Virol. 1991;65:3417–3420. doi: 10.1128/jvi.65.7.3417-3420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee D, Sohn H, Kalpana G V, Choe J. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature. 1999;399:487–491. doi: 10.1038/20966. [DOI] [PubMed] [Google Scholar]
- 38.Meyers C, Frattini M G, Hudson J B, Laimins L A. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 39.Nilsson C H, Bakos E, Petry K U, Schneider A, Durst M. Promoter usage in the E7 ORF of HPV16 correlates with epithelial differentiation and is largely confined to low-grade genital neoplasia. Int J Cancer. 1996;65:6–12. doi: 10.1002/(SICI)1097-0215(19960103)65:1<6::AID-IJC2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 40.Ozbun M A, Meyers C. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J Virol. 1997;71:5161–5172. doi: 10.1128/jvi.71.7.5161-5172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozbun M A, Meyers C. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J Virol. 1998;72:2715–2722. doi: 10.1128/jvi.72.4.2715-2722.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powers J H, Bina M. In vitro assembly of a positioned nucleosome near the hypersensitive region in simian virus 40 chromatin. J Mol Biol. 1991;221:795–803. doi: 10.1016/0022-2836(91)80176-u. [DOI] [PubMed] [Google Scholar]
- 43.Pray T R, Laimins L A. Differentiation-dependent expression of E1–E4 proteins in cell lines maintaining episomes of human papillomavirus type 31b. Virology. 1995;206:679–685. doi: 10.1016/s0042-6822(95)80088-3. [DOI] [PubMed] [Google Scholar]
- 44.Rotenberg M O, Chow L T, Broker T R. Characterization of rare human papillomavirus type 11 mRNAs coding for regulatory and structural proteins, using the polymerase chain reaction. Virology. 1989;172:489–497. doi: 10.1016/0042-6822(89)90191-8. [DOI] [PubMed] [Google Scholar]
- 45.Ruesch M N, Laimins L A. Human papillomavirus oncoproteins alter differentiation-dependent cell cycle exit on suspension in semisolid medium. Virology. 1998;250:19–29. doi: 10.1006/viro.1998.9359. [DOI] [PubMed] [Google Scholar]
- 46.Ruesch M N, Stubenrauch F, Laimins L A. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J Virol. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seedorf K, Oltersdorf T, Krammer G, Rowekamp W. Identification of early proteins of the human papilloma viruses type 16 (HPV 16) and type 18 (HPV 18) in cervical carcinoma cells. EMBO J. 1987;6:139–144. doi: 10.1002/j.1460-2075.1987.tb04731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smale S T. Core promoter architecture for eukaryotic protein-coding genes. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. Vol. 3. New York, N.Y: Raven Press Ltd.; 1994. pp. 63–82. [Google Scholar]
- 49.Somasekhar M B, Mertz J E. Sequences involved in determining the locations of the 5′ ends of the late RNAs of simian virus 40. J Virol. 1985;56:1002–1013. doi: 10.1128/jvi.56.3.1002-1013.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stubenrauch F, Colbert A M, Laimins L A. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J Virol. 1998;72:8115–8123. doi: 10.1128/jvi.72.10.8115-8123.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stubenrauch F, Lim H B, Laimins L A. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J Virol. 1998;72:1071–1077. doi: 10.1128/jvi.72.2.1071-1077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stünkel W, Bernard H-U. The chromatin structure of the long control region of human papillomavirus type 16 represses viral oncoprotein expression. J Virol. 1999;73:1918–1930. doi: 10.1128/jvi.73.3.1918-1930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tack L C, Beard P. Both trans-acting factors and chromatin structure are involved in the regulation of transcription from the early and late promoters in simian virus 40 chromosomes. J Virol. 1985;54:207–218. doi: 10.1128/jvi.54.1.207-218.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turek L P. The structure, function, and regulation of papillomaviral genes in infection and cervical cancer. Adv Virus Res. 1994;44:305–356. doi: 10.1016/s0065-3527(08)60332-2. [DOI] [PubMed] [Google Scholar]
- 55.Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 56.Wiley S R, Kraus R J, Zuo F, Murray E E, Loritz K, Mertz J E. SV40 early-to-late switch involves titration of cellular transcriptional repressors. Genes Dev. 1993;7:2206–2219. doi: 10.1101/gad.7.11.2206. [DOI] [PubMed] [Google Scholar]
- 57.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 59.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 60.Wu C, Gilbert W. Tissue-specific exposure of chromatin structure at the 5′ terminus of the rat preproinsulin II gene. Proc Natl Acad Sci USA. 1981;78:1577–1580. doi: 10.1073/pnas.78.3.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu C, Wong Y C, Elgin S C. The chromatin structure of specific genes. II. Disruption of chromatin structure during gene activity. Cell. 1979;16:807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- 62.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida M, Hoshikawa Y, Koseki K, Mori K, Beppu T. Structural specificity for biological activity of trichostatin A, a specific inhibitor of mammalian cell cycle with potent differentiation-inducing activity in Friend leukemia cells. J Antibiot (Tokyo) 1990;43:1101–1106. doi: 10.7164/antibiotics.43.1101. [DOI] [PubMed] [Google Scholar]
- 64.Zhao W, Noya F, Chen W Y, Townes T M, Chow L T, Broker T R. Trichostatin A up-regulates human papillomavirus type 11 upstream regulatory region-E6 promoter activity in undifferentiated primary human keratinocytes. J Virol. 1999;73:5026–5033. doi: 10.1128/jvi.73.6.5026-5033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]