CONSPECTUS
Membrane proteins mediate a plethora of cellular functions and represent important targets for drug development. Unlike soluble proteins, membrane proteins require native-like environments to fold correctly and be active. Therefore, modern structural biology techniques have aimed to determine the structure and dynamics of these membrane proteins at physiological temperature and in liquid crystalline lipid bilayers. With the flourishing of new NMR methodologies and improvements in sample preparations, magic angle spinning (MAS) and oriented sample solid-state NMR (OS-ssNMR) spectroscopy of membrane proteins is experiencing a new renaissance. Born as antagonistic approaches, these techniques nowadays offer complementary information on the structural topology and dynamics of membrane proteins reconstituted in lipid membranes. By spinning biosolid samples at the magic angle (θ = 54.7°), MAS NMR experiments remove the intrinsic anisotropy of the NMR interactions, increasing spectral resolution. Internuclear spin interactions (spin exchange) is reintroduced by RF pulses, providing distances and torsion angles to determine secondary, tertiary as well as quaternary structures of membrane proteins. OS-ssNMR, on the other hand, directly detects anisotropic NMR parameters such as dipolar couplings (DC) and anisotropic chemical shifts (CS), providing orientational constraints to determine the architecture (i.e., topology) of membrane proteins relative to the lipid membrane. Defining the orientation of membrane proteins and their interactions with lipid membranes is of paramount importance since lipid-protein interactions can shape membrane protein conformations and ultimately define their functional states. In this Accounts, we report selected studies from our group integrating MAS and OS-ssNMR techniques to give a comprehensive view of the biological processes occurring at cellular membranes. We focus on the main experiments for both techniques, with an emphasis on new implementation to increase both sensitivity and spectral resolution. We also describe how the structural constraints derived from both isotropic and anisotropic NMR parameters are integrated into dynamic structural modeling using replica-averaged orientational-restrained molecular dynamics simulations (RAOR-MD). We showcase small membrane proteins that are involved in Ca2+ transport and regulate cardiac and skeletal muscle contractility: phospholamban (PLN, 6 kDa), sarcolipin (SLN, 4 kDa), and DWORF (4 kDa). We summarize our results for the structures of these polypeptides free and in complex with the sarcoplasmic reticulum (SR) Ca2+-ATPase (SERCA, 110 kDa). Additionally, we illustrate the progress toward the determination of the structural topology of a six transmembrane protein associated with succinate and acetate transport (SaTP, hexamer 120 kDa). From these examples, the integrated MAS and OS-ssNMR approach, in combination with modern computational methods, emerges as a way to overcome the challenges posed by studying large membrane protein systems.
Graphical Abstract
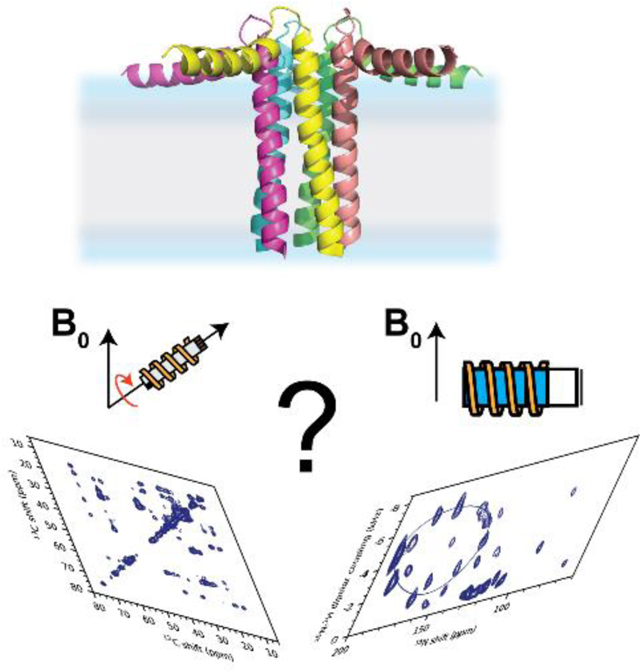
Introduction
Membrane proteins are involved in vital cellular events, mediating intra- and inter-cellular communication.5 Their structure and function are regulated by diverse lipid membranes that constitute various cellular compartments. The heterogeneous membrane environment is a significant barrier for classical structural biology methods, and the characterization of membrane proteins at atomic resolution remains an outstanding challenge. Currently, X-ray crystallography is the method of choice for determining the structural features of membrane proteins in their ground states. The majority of membrane protein structures deposited in the protein data bank (PDB) have been determined by X-ray in detergent preparations. However, detergents are a rough approximation of native membranes and are somewhat problematic, as they introduce structural distortions and deviations from a proteins’ native state.6,7 The outstanding progress in cryogenic electron microscopy (cryo-EM) has facilitated the structure determination of relatively large membrane proteins.8 However, both X-ray and cryo-EM fall short in the characterization of lipid-protein interactions and cannot probe the timescale of the functional dynamics of membrane proteins.
Solid-state NMR (ssNMR) methods are now reaching a level of sophistication, enabling the characterization of membrane proteins’ structure, dynamics, and interactions in fully hydrated lipid membranes.9 Therefore, ssNMR can be used as a tool to validate X-ray and cryo-EM structures, which are typically determined in the absence of a native-like environment. In the past, ssNMR of membrane proteins relied on two distinct techniques: magic angle spinning (MAS)10 and oriented sample (OS)11 ssNMR. While the first approach removes the anisotropy from the NMR physical observable by spinning at the magic angle to obtain high-resolution spectra,12 the latter exploits anisotropic NMR parameters to obtain the orientation of membrane proteins’ helical or β-sheet domains.13 While MAS NMR techniques are ideal for measuring distances and torsion angles of a protein’s backbone, OS-ssNMR directly measures the orientation of amide groups relative to the membrane bilayers. Specifically, this method provides tilt and rotation angles of membrane protein domains with respect to the bilayer normal. A significant advantage of ssNMR spectroscopy over X-ray and cryo-EM is the site-specific characterization of a protein’s motion, including the timescale at which these motions occur, as well as the depiction of the different energetic and functional states. Inspired by solution-state NMR,14 ssNMR is emerging as an atomic resolution technique suited for detecting conformationally excited states in lipid membranes.3,15–17 These high-energy conformations exemplify intermediates of protein folding reactions, active and inactive states, or alternate conformations that could be targeted by more specific allosteric drugs.
In the following synopsis, we describe the milestones that our group has reached in the past decade. By all means, this survey is not exhaustive and does not cover many breakthroughs achieved by other research groups in the study of membrane proteins.
Membrane mimetic systems for high-resolution ssNMR spectroscopy
The functional reconstitution of membrane proteins in membrane mimetic systems is an essential step for the structural and functional characterization by ssNMR. Detailed protocols have been outlined by Das et al.18 For MAS, we reconstitute recombinant membrane proteins in lipid vesicles via detergent-mediated preparations. For OS-ssNMR studies, we have been using two main procedures, involving either mechanically or magnetically aligned membrane preparations.18 Both reconstitution protocols have their merits and limitations. Mechanically aligned systems are prepared by spreading the lipid-protein mixtures on solid supports (typically glass plates), and with iterative hydration/dehydration cycles, both lipids and proteins align in a lasagna-like stacking of phospholipid and protein layers. Although with low hydration levels, these preparations are detergent-free and can be obtained using mixed lipid compositions to approximate native membranes.19 Magnetically oriented preparations include lipid bicelles,20 and more recently, nano- and macro-discs.21 Lipid bicelles were among the first systems to be utilized for magnetic alignment of membrane proteins.22 Bicelles are formed by one or more lipid types (long-chain component) and a detergent that solubilize the lipids (short-chain component). Depending on the ratio between the long- and short-chain components, anisotropic bicelles adopt a discoidal shape or a Swiss cheese phase.23 Unlike mechanically aligned membrane systems, the composition of bicelles has more restrictions as many lipids prevent the formation of stably aligned phases for NMR measurements.24 Also, the presence of proteins modifies the phase diagram of bicelles, and the conditions to obtain uniform orientation are often very narrow.
Another hurdle is represented by detergents (e.g., 1,2-dihexanoyl-sn-glycero-3-phosphocholine, DHPC) that may interact with membrane proteins competing out lipids and causing the disruption of the bicellar phase.7 Additional limitation of bicelles have been extensively discussed by Salnikov et al. 25–27 Nonetheless, membrane proteins reconstituted in bicelle preparations possess more favorable conditions for ssNMR spectroscopic analysis compared to mechanically aligned systems.28 The dynamics of bicelles lengthen the transverse spin relaxation (T2) of proteins, resulting in sharper and more intense resonances.29 As a consequence, it is possible to obtain highly-resolved two-dimensional (2D) separated local field (SLF) spectra. More importantly, membrane proteins reconstituted in bicellar preparations make possible the acquisition of 3D SLF experiments, i.e., polarization inversion spin exchange at the magic angle (PISEMA) and SAMPI4, and residue-specific sequential assignments via proton driven spin diffusion (PDSD).
To spin or not to spin?
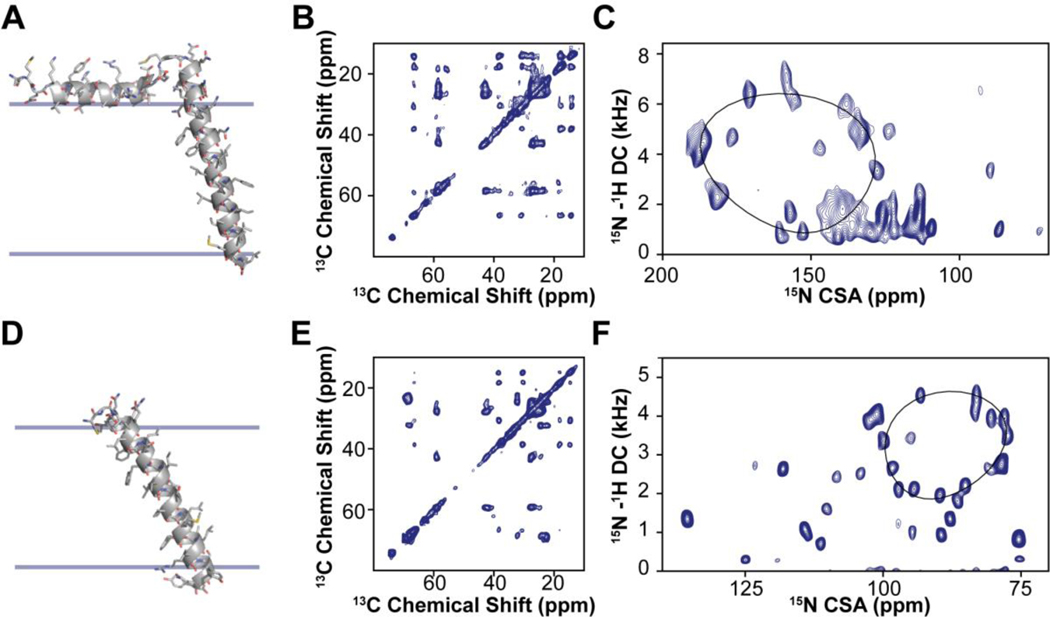
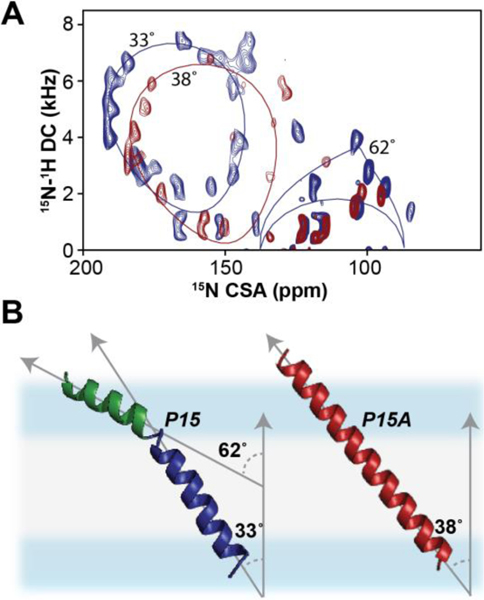
Since its inception, ssNMR of membrane proteins has been carried out using MAS techniques.30 OS-ssNMR has been less practiced than MAS NMR due to the more demanding sample preparations. However, the redundancy of the primary sequences of helical membrane proteins and the inherent conformational heterogeneity hampers the complete sequential assignments and structure determination. To date, the majority of the structures solved by NMR of membrane proteins are backbone structures of small and medium-sized membrane proteins obtained by OS-ssNMR.9 A few research groups have pioneered the combination of the two techniques to determine distances, torsion angles as well as orientational restraints.31–36 The marriage of these techniques, often performed in similar membrane preparations, is very powerful. Both isotropic and anisotropic parameters can be combined to describe the structural dynamics of membrane proteins that transition from one structural state to another (Fig.1). MAS techniques easily capture changes in the secondary structures. However, topological changes are more difficult to identify using isotropic NMR parameters. Transmembrane (TM) helix signaling occurs via intramembrane topological changes of helical domains, such as translation, piston, pivot (tilting), or rotation motions.37,38 While translation or piston-like motions can be monitored using inter-helical distances, tilt and rotation motions are often silent to the MAS analysis, but can be easily mapped using OS-ssNMR techniques. An example is the effects of a single mutation on the TM helix of DWarf Open Reading Frame (DWORF) (Fig.2). The proline to alanine mutation of this small protein affects not only the helical content, but also its tilt and rotation angles relative to the lipid membrane. These topological changes are silent to MAS techniques, but they can be readily characterized by SLF experiments (Fig.2). Although rotationally aligned MAS experiments for determining the topology of membrane proteins have been developed, they require fast rotational diffusion of proteins within lipid membranes and are often challenging for membrane proteins interacting strongly with lipids.39,40
Figure 1: MAS and OS-ssNMR studies of single pass membrane proteins PLN and SLN.
A. Structure and membrane orientation of PLN obtained from a combination of isotropic and anisotropic restraints. B. 13C,13C-DARR spectrum of PLN in lipid vesicles. C. SE-SAMPI4 spectrum of PLN in oriented lipid bicelles. D. Structure and orientation of SLN in lipid membranes calculated using both MAS and OS-ssNMR data. E. 13C,13C-DARR spectrum of SLN in lipid vesicles. F. SE-SAMPI4 spectrum of PLN in oriented lipid bicelles. Note that the oriented spectrum of SLN was obtained using paramagnetic doping and in the absence of Yb3+ (unflipped bicelles).
Figure 2: Effects of single mutation on the topology of DWORF in lipid membranes.
A. SE-SAMPI4 spectra of wild-type DWORF (blue) and P15A mutant (red) reconstituted into flipped 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)/1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) bicelles. B. Structural models of DWORF backbone obtained from replica-averaged orientational restrained molecular dynamics (RAOR-MD). Distinctive helical domains associated with the N-terminus (green) and C-terminus (blue) are fitted to PISA models.
Advanced MAS and OS-ssNMR techniques
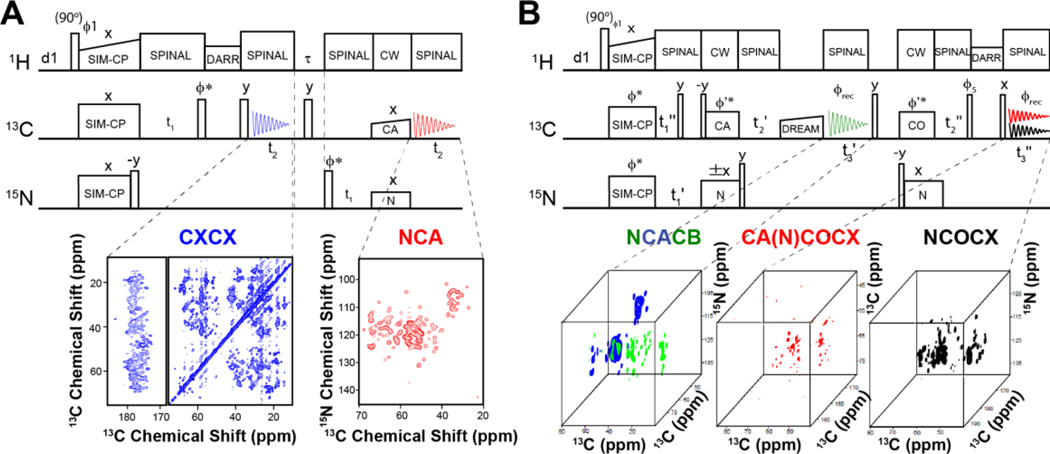
In the past decades, there have been several breakthroughs in MAS of membrane proteins. The first and most significant advancement involves the use of dynamic nuclear polarization (DNP)41, which enhances the nuclear polarization via dipolar interactions with unpaired spin electrons, giving rise to high sensitivity spectra. In a few cases, this technique has dramatically improved the NMR spectra, and for selected membrane proteins, DNP has accessed information that was difficult to achieve using classical spectroscopic methods or enabled the direct analysis of peptides in cell.42,43 Also, protein perdeuteration44 and paramagnetic doping45 have contributed to speeding up NMR data acquisition. Our contribution has been in the development of novel pulse sequences with higher sensitivity for both MAS and OS-ssNMR.4,46 Our strategy is to recover orphan spin operators that are discarded during the execution of conventional pulse programs. We called these experiments POE for Polarization Optimized Experiments (Fig.3).4 The essential element of POE is the simultaneous cross-polarization (SIM-CP) that enables the transfer of polarization from the 1H bath to two (or more) heteronuclei 13C and 15N. Dual acquisition MAS spectroscopy (DUMAS)47,48 was the first implementation of POE. In the DUMAS scheme, 13C- and 15N-edited 2D (or 3D) experiments are simultaneously registered in a single experiment using two 13C acquisition periods per scan. In our laboratory, 2D CC and NC correlation spectra are routinely acquired using DUMAS-based CXCX-NCA and Double Quantum Single Quantum (DQSQ)-NCO pulse sequences. After an initial analysis of CC and NC fingerprints, one can move on to more robust sequential assignment protocols, e.g., a 3D DUMAS NCACX-CANCO experiment.
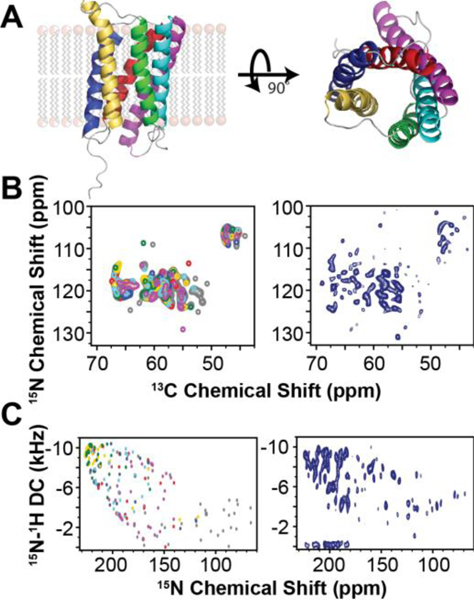
Figure 3: MAS Polarization optimized experiments (POE) on the six transmembrane helices SaTP protein transporter reconstituted in lipid membranes.
A. 2D DUMAS experiments for the simultaneous acquisition of DARR (200 ms) and NCA experiments. B. MEIOSIS experiment with the simultaneous acquisition of three 3D experiments: NCACB, CA(N)COCX, and NCOCX.
The sequential walk of carbonyl chemical shifts is obtained by matching the NCA planes of NCACX and CANCO data sets. In 3D DUMAS, a bidirectional SPECFIC-CP enables the polarization transfer from 15N to 13Cα and vice versa. For selecting the NC bidirectional transfer, we use a four-step phase cycle on 15N (+x, -x, +x, -x) and 13C (+x, +x, -x, -x) radio frequency (RF) spin-locks.49 This phase cycle selects the N to Cα or C to N transferred polarization and eliminates the 13C and 15N residual polarization pathways. To recover both transferred and residual polarization pathways of SPECIFIC-CP, we have developed the Multiple ExperIments via Orphan SpIn operatorS (MEIOSIS) approach that records four 2D spectra using two acquisitions per scan.50 The RF phases of SPECIFIC-CP spin-lock pulses are Hadamard-encoded, which enables the decoding of both transferred and residual polarization pathways leading to simultaneous acquisition of four multidimensional spectra. Similarly, a 3D MEIOSIS pulse sequence was developed for acquiring 3D CCC or CA(N)COCX correlation together with two other 3D spectra, NCACB and NCOCX, as shown for succinate and acetate transport membrane protein (SatP) (Fig.3). We have also exploited residual polarization to concatenate up to eight two-dimensional experiments using Multiple acquisitions via sequential transfer of orphan spin polarization (MAeSTOSO) approach.51 The MAeSTOSO approach can be very useful for acquiring 2D CXCX and N(C)C spectra with Dipolar Assisted Rotational Resonance (DARR) mixing periods for both long- and short-range correlations. All of these pulse sequences were tested with both crystalline preparations of globular proteins and single and/or multi-span membrane proteins such as sarcolipin (SLN), phospholamban (PLN), and SatP.52 POE can also take advantage of the afterglow phenomenon, introduced by Traaseth and co-workers,53 which can be incorporated into Transferred Echo DOuble Resonance (TEDOR)-NCX-based pulse sequences.52 The 3D version of TEDOR-NCACX-NCOCX enables the simultaneous measurement of CC DARR restraints in the 1st acquisition and the TEDOR NC distance restraints in the 2nd acquisition. This subtype of POE enables one to record two different experiments, the first for resonance assignment and the second for distance measurements. More recently, POE were developed to include fast MAS experiments for the acquisition of ten experiments simultaneously.54,55
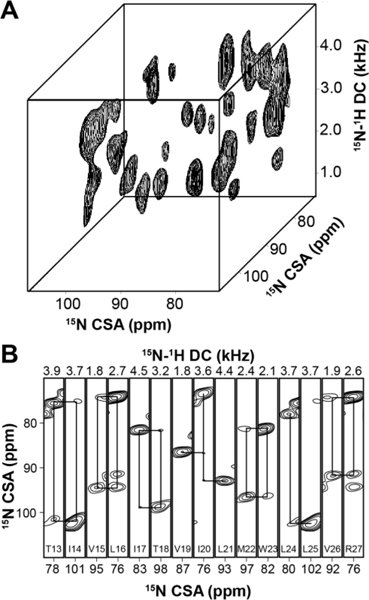
Parallel to the improvements of MAS spectroscopy, our group has developed sensitivity enhancement (SE) SLF and heteronuclear correlation (HETCOR) experiments.46 SE-SLF experiments increase the signal-to-noise ratio (S/N) by .56,57 We routinely use SE-SLF for 2D experiments to determine the topology of membrane protein backbones with respect to the bilayer normal. Recently, our laboratory has utilized paramagnetic relaxation enhancement (PRE) for the fast acquisition of SE-SLF experiments. By doping bicelles with 5% 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-diethylenetriaminepentaacetic acid copper salt lipid (Cu2+-DMPE-DTPA), we found that it is possible to accelerate the acquisition of multidimensional SE-SLF experiments up to 3 times. The improvements in sample stability combined with SE techniques and PRE made it possible to acquire 3D experiments for sequential residues assignments in magnetically oriented bicelle samples (Fig. 4).2 A significant drawback of the application of paramagnetic doping to multidimensional SE-SLF pulse sequences is the temperature variations during the experiments, which is due to to the relatively short pulse delay (~ 1 s). These temperature oscillations not only affect the thermal stability of membrane proteins, but also disturb the magnetic alignment of the bicellar system. To address this issue, we designed heat-compensated SE-SLF pulse sequences (hcSE-SLF).58 By removing the heterogeneity of the resonances in the spectra, the hcSE-SLF pulse sequences provide approximately 20% increase in sensitivity relative to SE-SLF experiments. The use of PRE is now being combined with Dual Acquisition orIented ssNMR spectroscopy (DAISY), a technique that records 2D SLF and SLF-Proton Driven Spin Diffusion (PDSD) OS-ssNMR spectra, simultaneously.49
Figure 4: Example of 3D SLF experiments performed on SLN aligned in lipid bicelles.
A. 3D SE-SAMPI4-PDSD spectrum of [U-15N]-SLN in bicelles doped with 5% Cu2+-chelated lipids (DMPE-DTPA-Cu2+). B. 3D strip plots at specific dipolar coupling values from the 3D SE-SAMPI4-PDSD spectrum. The solid lines indicate [i, i+1] cross peaks.
Structure calculations using ssNMR restraints
There are several approaches to implement isotropic and orientational restraints in the structure determination of membrane proteins. MAS experiments provide structural restraints, primarily isotropic chemical shifts that are converted into torsion angle restrains using TALOS+,59 and distances derived from DARR (or PDSD), chemical shift perturbation (CSP), and PREs, similarly to solution-state NMR. In most of the calculation protocols distances, angles, and chemical shift index (CSI) are implemented as harmonic restraints.60 However, distances and angular restraints for membrane proteins are often very sparse and may result in ill-defined structures, with the register of TM helices poorly defined. This problem is common to solution NMR structures of membrane proteins in detergent micelles.6 For OS-ssNMR, Opella and Nevzorov introduced structural fitting procedures that would offer the best fit of chemical shift anisotropy (CSA) and dipolar coupling (DC) to calculate backbone orientations.61 A more efficient procedure was introduced by Marassi and co-workers,62 where the orientational restraints were treated as harmonic restraints and minimized using a simulated annealing algorithm. Together with the Cross, Marassi, Opella, and Hong groups, we recognized the importance of using a hybrid approach that would include not only the physical parameters obtained by MAS or solution NMR, but also orientation dependent parameters obtained by OS-ssNMR.1,33 In our original calculations, we included in the force field a depth of insertion potential developed by DeGrado’s group that restrains the conformational freedom of membrane proteins within a low dielectric slab. This limits their conformational space to more physical minima defined by the hydrophobicity and electrostatics of membrane proteins. Using this protocol, we were able to determine the high-resolution structures of SLN and PLN (monomeric and pentameric assembly).1,33,35,36 Later on, we implemented distance, angular, and orientational restraints in a simulation system with explicit water and lipid environments (DMPC and POPC bilayers).63,64 The explicit environment provides an improved description and a more accurate search of the membrane protein conformational phase space.63,64 Restrained molecular dynamics samplings have been shown to be applicable to a many biological systems, ranging from relatively rigid structural states to heterogeneous conformational ensembles of proteins, including those adopting multiple topological states. Given the ensemble-averaged nature of the experimental ssNMR data, the latter case is accounted for by imposing the structural restraints as an average over independent simulations (replicas) evolving simultaneously in the so-called replica-averaged approach. These restrained simulations are able to give a view on the collective protein backbone motions at picosecond-to-millisecond timescales. To this extent, we were able to accurately describe the heterogeneous conformational ensembles of PLN both in its monomeric63 and pentameric64 states. The conformational equilibrium of PLN matched with the distinct states that were observed by both MAS and OS-ssNMR.
Topological allostery: transmembrane signaling via dynamic interactions.
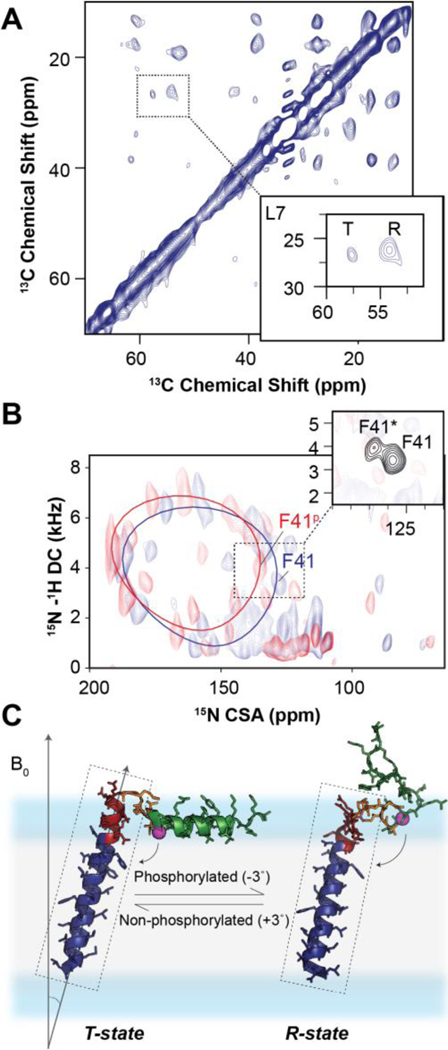
Unlike their soluble counterparts, membrane proteins are embedded in lipid bilayers where hydrophobicity is no longer a dominant force stabilizing ternary and quaternary structure.65 Concomitant with the allostery occurring throughout an extensive network of interactions, signal transduction throughout TM domains must be communicated throughout a structure held together by weaker van der Waals forces, side-chain packing motifs (i.e., leucine zippers), specific interactions with lipids and physical constraints imposed by the dimensions of lipid bilayers.66,67 Indeed, hydrogen bonding plays a significant role in stabilizing larger α-helical bundles,68 but how external signals are transduced throughout transmembrane helices is still unknown. As mentioned above, rigid-body motions of the helical domains (i.e., topological changes) are among the possible structural transitions that characterize TM dynamic signaling. In the current literature, there are many examples of rigid-body transitions such as those occurring in mechanosensitive channels, G protein-coupled receptors (GPCRs), etc.65 Distance and torsion angle restraints are insufficient to define these global topological changes. In contrast, SLF experiments are extremely sensitive to small TM changes in rotation and tilt that propagate from one to the opposite leaflet of the lipid membranes. Among the most remarkable examples are the topological changes mapped by Traaseth and co-workers for EmrE using OS-ssNMR.69 In this case, the topological changes of EmrE were so pronounced to enable the detection of exchanging topologies in the SLF spectra. Another example is the topological transitions of PLN in lipid membranes. (Fig.5) PLN possesses two major regions: a hydrophobic TM domain that inhibits the sarcoplasmic reticulum Ca2+-ATPase (SERCA), and a cytoplasmic domain that tunes the extent of inhibition via phosphorylation by protein kinase A (PKA) and calmodulin-dependent protein kinase II (CaMKII).70 Our original topology of monomeric and pentameric PLN were obtained in mechanically aligned lipid bilayers.1,33 Under these conditions, the backbone resonances in the SLF spectra were so broad that we could not fully appreciate the presence of two different populations of resonances for the TM domain. In contrast, the spectra in lipid bicelles display a major and a minor population with slightly different tilt and rotation angles (Fig.5), mirroring the two populations (R and T states) identified for the cytoplasmic region by MAS spectroscopy (Fig.5A).
Figure 5: Two-state topological equilibrium of monomeric PLN.
A. Observation by MAS of two conformational states of PLN: the ground T-state and the excited R-state. B. 2D [15N-1H] SE-SAMPI4 spectrum of PLNAFA with (blue) and without (red) phosphorylation in flipped DMPC/POPC bicelles. The insets show the two-state equilibrium for a selective 15N-Phe labeled PLNAFA. C. T-state (PDB 2KB7) and R-state (PDB 2LPF) of PLNAFA.
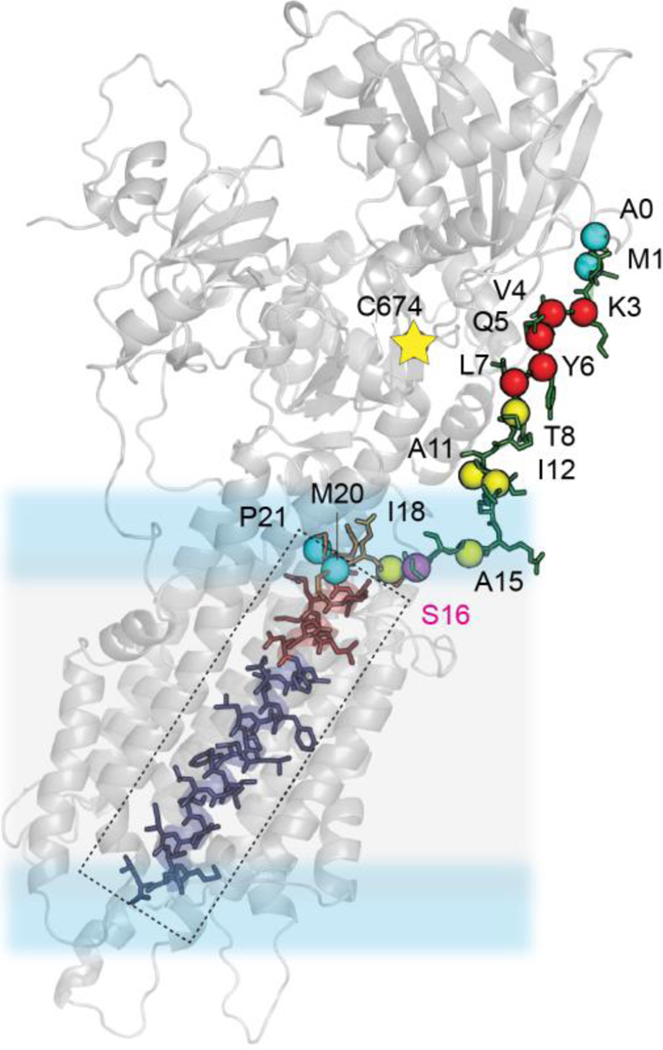
Upon phosphorylation of Ser16 by PKA, we observed a shift of the ground population toward the lowest populated state, which becomes the dominating state. These changes are very pronounced for Asn30 and Asn34, whose hydrogen bond interactions stabilize the E2 (Ca2+-free) state of SERCA. Since the inhibition of SERCA by PLN occurs via allosteric interactions between the TM domain of the regulator and the Ca2+ binding site of the ATPase (Fig.6), the topological changes of the TM domain allosterically modulate the extent of inhibition of the enzyme (topological allostery).71 It is possible that different SERCA modulators expressed in non-cardiac cells, i.e., regulins, might display different topologies reflecting their various biological activities. Therefore, the topological and allosteric diversity in regulins may modulate the ATPase affinity for Ca2+ ions, thereby differentially affecting calcium cycling.72
Figure 6: Structural determination of the SERCA-PLN complex by ssNMR.
Mapping the binding between the dynamic N-terminus was achieved previously by PRE experiments followed by MAS [13C-1H] rINEPT and [13C-13C] DARR measurements (Data from ref. 70). Residues marked by red spheres were found to experience unambiguous PRE-induced line broadening, yellow spheres ambiguous and blue sphere unambiguously no-PRE quenched.
Characterization of motions via ssNMR
The characterization of the site-specific motions and more importantly the timescale of conformational changes are among the most important contributions of NMR to structural biology.73 However, the analysis of the dynamics of membrane proteins in lipid bilayers is still a significant challenge. Hong and co-workers are using the dipolar-coupling chemical-shift correlation (DIPSHIFT) as a means to determine the dynamics of proteins and peptides.30 Although it is possible to characterize the backbone dynamics for a few selected cases, the heterogeneity of membrane proteins spectra represents a significant hurdle. Our group has been relying on a semi-quantitative analysis of the segmental motions using a combination of CP and refocused Insensitive Nuclei Enhanced by Polarization Transfer (rINEPT) experiments.3 Since the intensity of polarization obtained from these experiments depends on the dynamics of the proteins, the comparison of these spectra gives an overall view of the complex motions of membrane proteins. In particular, the membrane-associated protein domains are mapped via CP based experiments that rely on DCs, whereas the dynamic residues that undergo fast dynamics average out anisotropic interactions and retain longer T2 relaxation properties, which enables the mapping of these residues via J-coupling based INEPT transfer experiments.74 We typically start with 1D 15N CP and rINEPT experiments to look at the extent of dynamics by comparing the backbone spectral intensities. Due to the presence of intense lipid signals, 1D 13C rINEPT experiments are usually less informative for a semi-quantitative analysis of mobile residues. However, a more detailed characterization can be made by using 13C-detected 2D 13C-13C DARR and INEPT-TOtal through Bond correlation SpectroscopY (TOBSY) experiments that map immobile and dynamic residues, respectively. To these experiments, we recently added 1H-detected sensitivity enhanced refocused INEPT heteronuclear single quantum correlation (RI-HSQC) experiments for probing sparse conformational states with populations as low as 1%.75 These experiments can be hybridized into new pulse sequences to map rigid and dynamic domains simultaneously.76
Interactions of intrinsically disordered proteins (IDPs) with lipid membranes.
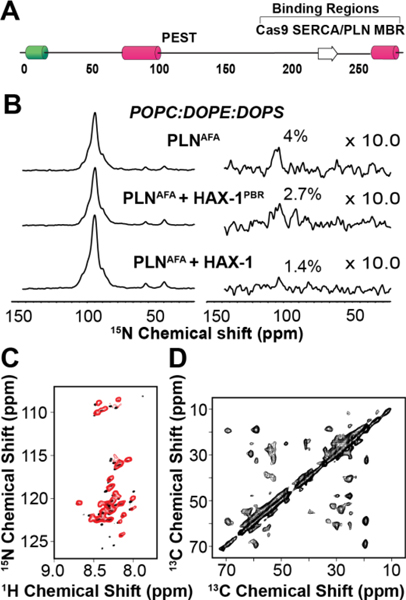
Although both MAS and OS-ssNMR have been extensively used to study dynamic small peptides interacting with membranes, studies on the interactions between IDPs and the membrane bilayer are still limited. In collaboration with the De Simone laboratory, our group has carried out a series of studies on the interactions between α-synuclein (α−Syn) and membrane model systems.64,77,78 α−Syn in solution exists as a disordered polypeptide, adopting a random coil ensemble of conformers. Upon interactions with lipid membranes, α−Syn adopts a helical conformation.79 However, the membrane-binding region has been virtually invisible to solution-state NMR analysis. We used INEPT-based experiments to image the ‘dark side’ of α−Syn and identify the residues directly involved with the membrane interactions and possibly responsible for the pre-fibrillar aggregates that have been hypothesized to constitute the toxic species. More recently, we used a combination of CP and rINEPT experiments to determine the interaction of hematopoietic-substrate-1 associated protein X-1 or HAX-1 with PLN (Fig.7).80 HAX-1 is a 279 amino acid intrinsically disordered protein (IDP) that is thought to interact with PLN, modulating the inhibition of SERCA. Indeed, our data confirm this hypothesis, but further suggest these interactions occur via an amphipathic helix located on the C-terminus of HAX-1 that interacts with lipid membranes (Fig.7). This helix may be crucial for localizing HAX-1 near the SERCA/PLN complex as well as promoting a rigid, more inhibitory state of PLN.
Figure 7: MAS ssNMR experiments on HAX-1, an intrinsically disordered protein of 279 amino acids.
A. Schematic of the predicted structural and functional domains of HAX-1. B. rINEPT spectra (right) normalized to CP spectra (left) of PLNAFA in complex with HAX-1203−245, the PLN binding region (PBR) of HAX-1 and full length HAX-1. C. Overlay of IDP HAX-1 spectra, obtained from MAS RI-HSQC (red), and solution NMR [1H,15N]-HSQC (black). D. 13C-13C DARR experiments of HAX-1 in lipid vesicles.
Conclusions and Perspectives
As for the entire field of structural biology,81,82 NMR is relying on multiple approaches to fully characterize the structure and dynamics of membrane proteins. Neither MAS nor OS-ssNMR possess the silver bullet; instead, the synergistic nature of these different approaches is emerging. Both MAS and OS-ssNMR are compatible with lipid compositions and ratios similar to physiological membranes in liquid crystalline phases, which represents a significant advantage over the other structural biology approaches. Under these conditions, membrane proteins, if properly reconstituted, are active and undergo conformational transitions that mimic their cellular function. From a spectroscopic viewpoint, MAS NMR methods are progressing rapidly. However, multidimensional spectroscopy of OS-ssNMR is still in its infancy. Our group is able to run double resonance 2D and 3D [1H,15N] OS-ssNMR experiments routinely on selected samples. The quantum leap will be possible when triple resonance 3D [1H,13C,15N] experiments will become available for the unambiguous assignments of membrane proteins. The next frontier is the conjugation of these ssNMR methodologies to determine the structure of multi-span membrane proteins. Fig.8 shows the application of MAS and OS-ssNMR to a six TM protein (SaTP) involved in the transport of succinate and acetate. Both the simulated spectra and the experimental fingerprints for both MAS and OS-ssNMR are rather promising, and the sensitivity of these experiments supports the feasibility of the structure determination at physiological temperature with lipids in a liquid crystalline phase. These results suggest that the future of ssNMR is bright. Although improvements in sample preparations, hardware, and pulse sequences must be in place, critical information on challenging biological systems can be obtained for the time being.
Figure 8:
Integrated MAS and OS-ssNMR spectroscopy of SaTP in lipid membranes. A. X-ray structure of SaTP (PDB 5ZUG). B. Predicted MAS NCA spectrum (left) using the ShiftX2 Server versus experimental NCA spectrum (right). C. Predicted SE-PISEMA spectrum obtained from MD simulation versus experimental SE-PISEMA spectrum (right).
Funding
This work is supported by the National Institute of Health to G.V. (GM64742 and HL 144130).
Biographies
Gianluigi Veglia obtained his Ph.D. in Chemistry from the University of Rome, La Sapienza, under the direction of Prof. M. Delfini and Prof. A. Di Nola. In 1993, he was a visiting fellow at SUNY Stonybrook, NY, under the supervision of Prof. M. Eisenberg and Prof. G. Prestwich. In 1995, he joined the laboratory of Prof. S. Opella as a Postdoctoral Associate. In 2000, he joined the University of Minnesota and is currently a Professor of Chemistry and Biochemistry. His research interests span from the structural characterization of soluble and membrane-associated proteins using both solution and solid-state NMR methods.
Daniel Weber is an American Heart Association Postdoctoral Fellow at the University of Minnesota, Minneapolis. He received his B.Sc. (Hons.) degree in Chemistry from James Cook University, Australia, and his Ph.D. in Chemistry from the University of Melbourne under the supervision of Prof. Frances Separovic. Daniel was a scientist at IBM Research before joining the Veglia Lab in 2017. His research interests include the application and development of solid-state NMR and computational modeling approaches for studying the structure and dynamics of membrane proteins.
Songlin Wang received his B.Sc. degree in Chemistry from Peking University, China, and his Ph.D. in Chemistry from the University of Illinois at Chicago under the direction of Prof. Yoshitaka Ishii. In 2016, he joined the laboratory of Prof. Gianluigi Veglia at the University of Minnesota as a postdoctoral associate. His research interests include characterizing the structures and interactions of the membrane proteins and developing novel NMR methodology.
Erik Larsen received his undergraduate degree in Chemistry from the University of Minnesota, Twin Cities, in 2015. Currently, he is graduate student the University of Minnesota, Twin Cities, working under the direction of Prof. Jiali Gao in the Department of Chemistry. His current research focuses on calcium cycling regulation in cardiomyocytes towards drug development for heart disease.
T. Gopinath is a solid-state NMR scientist at Minnesota NMR Center, University of Minnesota, Minneapolis. In 2007, he received his Ph.D. in NMR Quantum Computing from the Indian Institute of Science, Bangalore, India, under the supervision of Prof. Anil Kumar. From 2008–2012, he worked as a postdoctoral scientist in the Veglia group and contributed to novel solid-state NMR methodological developments to speed-up the data collection using multi-acquisition polarization experiments. His current research focuses on further developments and applications of ssNMR methods to various biomacromolecules.
REFERENCES
- (1).Verardi R; Shi L; Traaseth NJ; Walsh N; Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc Natl Acad Sci U S A 2011, 108, 9101–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wang S; Gopinath T; Veglia G. Application of paramagnetic relaxation enhancements to accelerate the acquisition of 2D and 3D solid-state NMR spectra of oriented membrane proteins. Methods 2018, 138–139, 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gopinath T; Veglia G. Probing membrane protein ground and conformationally excited states using dipolar- and J-coupling mediated MAS solid state NMR experiments. Methods 2018, 148, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gopinath T; Veglia G. Experimental Aspects of Polarization Optimized Experiments (POE) for Magic Angle Spinning Solid-State NMR of Microcrystalline and Membrane-Bound Proteins. Methods Mol Biol 2018, 1688, 37–53. [DOI] [PubMed] [Google Scholar]
- (5).The Structure of Biological Membranes; Second ed.; CRC Press, 2005. [Google Scholar]
- (6).Chipot C; Dehez F; Schnell JR; Zitzmann N; Pebay-Peyroula E; Catoire LJ; Miroux B; Kunji ERS; Veglia G; Cross TA; Schanda P. Perturbations of Native Membrane Protein Structure in Alkyl Phosphocholine Detergents: A Critical Assessment of NMR and Biophysical Studies. Chem Rev 2018, 118, 3559–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhou HX; Cross TA Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys 2013, 42, 361–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ognjenovic J; Grisshammer R; Subramaniam S. Frontiers in Cryo Electron Microscopy of Complex Macromolecular Assemblies. Annu Rev Biomed Eng 2019. [DOI] [PubMed] [Google Scholar]
- (9).Ladizhansky V. Applications of solid-state NMR to membrane proteins. Biochim Biophys Acta Proteins Proteom 2017, 1865, 1577–1586. [DOI] [PubMed] [Google Scholar]
- (10).McDermott A. Structure and dynamics of membrane proteins by magic angle spinning solid-state NMR. Annu Rev Biophys 2009, 38, 385–403. [DOI] [PubMed] [Google Scholar]
- (11).Gong XM; Franzin CM; Thai K; Yu J; Marassi FM Nuclear magnetic resonance structural studies of membrane proteins in micelles and bilayers. Methods Mol Biol 2007, 400, 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Griffin RG Dipolar recoupling in MAS spectra of biological solids. Nat Struct Biol 1998, 5 Suppl, 508–512. [DOI] [PubMed] [Google Scholar]
- (13).Opella SJ; Marassi FM Structure determination of membrane proteins by NMR spectroscopy. Chem Rev 2004, 104, 3587–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Mulder FA; Mittermaier A; Hon B; Dahlquist FW; Kay LE Studying excited states of proteins by NMR spectroscopy. Nat Struct Biol 2001, 8, 932–935. [DOI] [PubMed] [Google Scholar]
- (15).Gustavsson M; Traaseth NJ; Veglia G. Probing ground and excited states of phospholamban in model and native lipid membranes by magic angle spinning NMR spectroscopy. Biochim Biophys Acta 2012, 1818, 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Traaseth NJ; Veglia G. Probing excited states and activation energy for the integral membrane protein phospholamban by NMR CPMG relaxation dispersion experiments. Biochim Biophys Acta 2010, 1798, 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Solid-State NMR. In Applications in biomembrane structure; Separovic F, Sani M-A, Eds.; IOP Publishing, 2020. [Google Scholar]
- (18).Das N; Murray DT; Cross TA Lipid bilayer preparations of membrane proteins for oriented and magic-angle spinning solid-state NMR samples. Nat Protoc 2013, 8, 2256–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Aisenbrey C; Michalek M; Salnikov ES; Bechinger B: Solid-State NMR Approaches to Study Protein Structure and Protein–Lipid Interactions. In Lipid-Protein Interactions: Methods and Protocols; Kleinschmidt JH, Ed.; Humana Press: Totowa, NJ, 2013; pp 357–387. [DOI] [PubMed] [Google Scholar]
- (20).Durr UH; Gildenberg M; Ramamoorthy A. The magic of bicelles lights up membrane protein structure. Chem Rev 2012, 112, 6054–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Radoicic J; Park SH; Opella SJ Macrodiscs Comprising SMALPs for Oriented Sample Solid-State NMR Spectroscopy of Membrane Proteins. Biophys J 2018, 115, 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Howard KP; Opella SJ High-resolution solid-state NMR spectra of integral membrane proteins reconstituted into magnetically oriented phospholipid bilayers. Journal of Magnetic Resonance Series B 1996, 112, 91–94. [DOI] [PubMed] [Google Scholar]
- (23).Durr UHN; Soong R; Ramamoorthy A. When detergent meets bilayer: Birth and coming of age of lipid bicelles. Progress in Nuclear Magnetic Resonance Spectroscopy 2013, 69, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yamamoto K; Soong R; Ramamoorthy A. Comprehensive Analysis of Lipid Dynamics Variation with Lipid Composition and Hydration of Bicelles Using Nuclear Magnetic Resonance (NMR) Spectroscopy. Langmuir 2009, 25, 7010–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Salnikov ES; Aisenbrey C; Anantharamaiah GM; Bechinger B. Solid-state NMR structural investigations of peptide-based nanodiscs and of transmembrane helices in bicellar arrangements. Chemistry and Physics of Lipids 2019, 219, 58–71. [DOI] [PubMed] [Google Scholar]
- (26).Aisenbrey C; Salnikov ES; Bechinger B. Solid-State NMR Investigations of the MHC II Transmembrane Domains: Topological Equilibria and Lipid Interactions. The Journal of membrane biology 2019, 252, 371–384. [DOI] [PubMed] [Google Scholar]
- (27).Salnikov ES; Aisenbrey C; Pokrandt B; Brügger B; Bechinger B. Structure, Topology, and Dynamics of Membrane-Inserted Polypeptides and Lipids by Solid-State NMR Spectroscopy: Investigations of the Transmembrane Domains of the DQ Beta-1 Subunit of the MHC II Receptor and of the COP I Protein p24. Frontiers in Molecular Biosciences 2019, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Veglia G; Traaseth NJ; Shi L; Verardi R; Gopinath T; Gustavsson M. The Hybrid Solution/Solid-State NMR Method for Membrane Protein Structure Determination. Comprehensive Biophysics, Vol 1: Biophysical Techniques for Structural Characterization of Macromolecules 2011, 182–198. [Google Scholar]
- (29).Salnikov ES; Aisenbrey C; Raya J; Bechinger B: CHAPTER 12 Investigations of the Structure, Topology and Dynamics of Membrane-Associated Polypeptides by Solid-State NMR Spectroscopy. In Advances in Biological Solid-State NMR: Proteins and Membrane-Active Peptides; The Royal Society of Chemistry, 2014; pp 214–234. [Google Scholar]
- (30).Hong M. Structure, topology, and dynamics of membrane peptides and proteins from solid-state NMR spectroscopy. J Phys Chem B 2007, 111, 10340–10351. [DOI] [PubMed] [Google Scholar]
- (31).Hu F; Luo W; Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science 2010, 330, 505–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sharma M; Yi MG; Dong H; Qin HJ; Peterson E; Busath DD; Zhou HX; Cross TA Insight into the Mechanism of the Influenza A Proton Channel from a Structure in a Lipid Bilayer. Science 2010, 330, 509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Traaseth NJ; Shi L; Verardi R; Mullen DG; Barany G; Veglia G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc Natl Acad Sci U S A 2009, 106, 10165–10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Verardi R; Traaseth NJ; Shi L; Porcelli F; Monfregola L; De Luca S; Amodeo P; Veglia G; Scaloni A. Probing membrane topology of the antimicrobial peptide distinctin by solid-state NMR spectroscopy in zwitterionic and charged lipid bilayers. Biochim Biophys Acta 2011, 1808, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Vostrikov VV; Mote KR; Verardi R; Veglia G. Structural dynamics and topology of phosphorylated phospholamban homopentamer reveal its role in the regulation of calcium transport. Structure 2013, 21, 2119–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Mote KR; Gopinath T; Veglia G. Determination of structural topology of a membrane protein in lipid bilayers using polarization optimized experiments (POE) for static and MAS solid state NMR spectroscopy. J Biomol NMR 2013, 57, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hulko M; Berndt F; Gruber M; Linder JU; Truffault V; Schultz A; Martin J; Schultz JE; Lupas AN; Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 2006, 126, 929–940. [DOI] [PubMed] [Google Scholar]
- (38).Matthews EE; Zoonens M; Engelman DM Dynamic helix interactions in transmembrane signaling. Cell 2006, 127, 447–450. [DOI] [PubMed] [Google Scholar]
- (39).Cady SD; Goodman C; Tatko CD; DeGrado WF; Hong M. Determining the orientation of uniaxially rotating membrane proteins using unoriented samples: A (2)H, (13)C, and (15)N solid-state NMR investigation of the dynamics and orientation of a transmembrane helical bundle. Journal of the American Chemical Society 2007, 129, 5719–5729. [DOI] [PubMed] [Google Scholar]
- (40).Das BB; Nothnagel HJ; Lu GJ; Son WS; Tian Y; Marassi FM; Opella SJ Structure determination of a membrane protein in proteoliposomes. J Am Chem Soc 2012, 134, 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Su Y; Andreas L; Griffin RG Magic angle spinning NMR of proteins: high-frequency dynamic nuclear polarization and (1)H detection. Annu Rev Biochem 2015, 84, 465–497. [DOI] [PubMed] [Google Scholar]
- (42).Ong YS; Lakatos A; Becker-Baldus J; Pos KM; Glaubitz C. Detecting substrates bound to the secondary multidrug efflux pump EmrE by DNP-enhanced solid-state NMR. J Am Chem Soc 2013, 135, 15754–15762. [DOI] [PubMed] [Google Scholar]
- (43).Sani M-A; Zhu S; Hofferek V; Separovic F. Nitroxide spin–labeled peptides for DNP-NMR in-cell studies. The FASEB Journal 2019, 33, 11021–11027. [DOI] [PubMed] [Google Scholar]
- (44).Reif B. Deuterated peptides and proteins: structure and dynamics studies by MAS solid-state NMR. Methods Mol Biol 2012, 831, 279–301. [DOI] [PubMed] [Google Scholar]
- (45).Wickramasinghe NP; Parthasarathy S; Jones CR; Bhardwaj C; Long F; Kotecha M; Mehboob S; Fung LW; Past J; Samoson A; Ishii Y. Nanomole-scale protein solid-state NMR by breaking intrinsic 1HT1 boundaries. Nat Methods 2009, 6, 215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Gopinath T; Mote KR; Veglia G. Sensitivity and resolution enhancement of oriented solid-state NMR: application to membrane proteins. Prog Nucl Magn Reson Spectrosc 2013, 75, 50–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Gopinath T; Veglia G. 3D DUMAS: simultaneous acquisition of three-dimensional magic angle spinning solid-state NMR experiments of proteins. J Magn Reson 2012, 220, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Gopinath T; Veglia G. Dual acquisition magic-angle spinning solid-state NMR-spectroscopy: simultaneous acquisition of multidimensional spectra of biomacromolecules. Angew Chem Int Ed Engl 2012, 51, 2731–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Gopinath T; Mote KR; Veglia G. Simultaneous acquisition of 2D and 3D solid-state NMR experiments for sequential assignment of oriented membrane protein samples. J Biomol NMR 2015, 62, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Gopinath T; Veglia G. Orphan spin operators enable the acquisition of multiple 2D and 3D magic angle spinning solid-state NMR spectra. The Journal of chemical physics 2013, 138, 184201–184201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Gopinath T; Veglia G. Multiple acquisitions via sequential transfer of orphan spin polarization (MAeSTOSO): How far can we push residual spin polarization in solid-state NMR? J Magn Reson 2016, 267, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Gopinath T; Wang S; Lee J; Aihara H; Veglia G. Hybridization of TEDOR and NCX MAS solid-state NMR experiments for simultaneous acquisition of heteronuclear correlation spectra and distance measurements. J Biomol NMR 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Banigan JR; Traaseth NJ Utilizing afterglow magnetization from cross-polarization magic-angle-spinning solid-state NMR spectroscopy to obtain simultaneous heteronuclear multidimensional spectra. J Phys Chem B 2012, 116, 7138–7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Gopinath T; Weber DK; Veglia G. Multi-receiver solid-state NMR using polarization optimized experiments (POE) at ultrafast magic angle spinning. J Biomol NMR 2020, 74, 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Gopinath T; Veglia G. Proton-detected polarization optimized experiments (POE) using ultrafast magic angle spinning solid-state NMR: Multi-acquisition of membrane protein spectra. J Magn Reson 2020, 310, 106664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Gopinath T; Traaseth NJ; Mote K; Veglia G. Sensitivity enhanced heteronuclear correlation spectroscopy in multidimensional solid-state NMR of oriented systems via chemical shift coherences. J Am Chem Soc 2010, 132, 5357–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Gopinath T; Veglia G. Sensitivity enhancement in static solid-state NMR experiments via single- and multiple-quantum dipolar coherences. J Am Chem Soc 2009, 131, 5754–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Wang S; Gopinath T; Veglia G. Improving the quality of oriented membrane protein spectra using heat-compensated separated local field experiments. J Biomol NMR 2019, 73, 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Shen Y; Delaglio F; Cornilescu G; Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 2009, 44, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Robustelli P; Kohlhoff K; Cavalli A; Vendruscolo M. Using NMR Chemical Shifts as Structural Restraints in Molecular Dynamics Simulations of Proteins. Structure 2010, 18, 923–933. [DOI] [PubMed] [Google Scholar]
- (61).Nevzorov AA; Opella SJ Structural fitting of PISEMA spectra of aligned proteins. J Magn Reson 2003, 160, 33–39. [DOI] [PubMed] [Google Scholar]
- (62).Tian Y; Schwieters CD; Opella SJ; Marassi FM A Practical Implicit Membrane Potential for NMR Structure Calculations of Membrane Proteins. Biophys J 2015, 109, 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).De Simone A; Mote KR; Veglia G. Structural Dynamics and Conformational Equilibria of SERCA Regulatory Proteins in Membranes by Solid-State NMR Restrained Simulations. Biophysical Journal 2014, 106, 2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Fusco G; De Simone A; Arosio P; Vendruscolo M; Veglia G; Dobson CM Structural Ensembles of Membrane-bound alpha-Synuclein Reveal the Molecular Determinants of Synaptic Vesicle Affinity. Sci Rep 2016, 6, 27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Phillips R; Ursell T; Wiggins P; Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature 2009, 459, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Popot JL Do Membrane Proteins Need to Fold in a “Membrane-Mimetic” Environment in Order to Reach a Functional 3D Structure? Biophysical Journal 2013, 104, 194a–194a. [Google Scholar]
- (67).Lee AG Biological membranes: the importance of molecular detail. Trends Biochem Sci 2011, 36, 493–500. [DOI] [PubMed] [Google Scholar]
- (68).Bowie JU Membrane protein folding: how important are hydrogen bonds? Current Opinion in Structural Biology 2011, 21, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Gayen A; Banigan JR; Traaseth NJ Ligand-induced conformational changes of the multidrug resistance transporter EmrE probed by oriented solid-state NMR spectroscopy. Angew Chem Int Ed Engl 2013, 52, 10321–10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Gustavsson M; Verardi R; Mullen DG; Mote KR; Traaseth NJ; Gopinath T; Veglia G. Allosteric regulation of SERCA by phosphorylation-mediated conformational shift of phospholamban. Proc Natl Acad Sci U S A 2013, 110, 17338–17343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Weber DK; Sanz-Hernandez M; Venkateswara Reddy U; Wang S; Larsen EK; Gopinath T; Gustavsson M; Cornea RL; Thomas DD; De Simone A; Veglia G. A Bitopic Miniprotein Regulates a Membrane-Embedded Enzyme via Topological Allostery. Biorxv 2020, doi: 10.1101/2020.08.28.271940. [DOI] [Google Scholar]
- (72).Akin BL; Hurley TD; Chen Z; Jones LR The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum. J Biol Chem 2013, 288, 30181–30191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Sekhar A; Kay LE An NMR View of Protein Dynamics in Health and Disease. Annu Rev Biophys 2019, 48, 297–319. [DOI] [PubMed] [Google Scholar]
- (74).Andronesi OC; Becker S; Seidel K; Heise H; Young HS; Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc 2005, 127, 12965–12974. [DOI] [PubMed] [Google Scholar]
- (75).Gopinath T; Nelson SED; Veglia G. (1)H-detected MAS solid-state NMR experiments enable the simultaneous mapping of rigid and dynamic domains of membrane proteins. J Magn Reson 2017, 285, 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Gopinath T; Nelson SED; Soller KJ; Veglia G. Probing the Conformationally Excited States of Membrane Proteins via (1)H-Detected MAS Solid-State NMR Spectroscopy. J Phys Chem B 2017, 121, 4456–4465. [DOI] [PubMed] [Google Scholar]
- (77).Fusco G; De Simone A; Gopinath T; Vostrikov V; Vendruscolo M; Dobson CM; Veglia G. Direct observation of the three regions in alpha-synuclein that determine its membrane-bound behaviour. Nat Commun 2014, 5, 3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Fusco G; Pape T; Stephens AD; Mahou P; Costa AR; Kaminski CF; Kaminski Schierle GS; Vendruscolo M; Veglia G; Dobson CM; De Simone A. Structural basis of synaptic vesicle assembly promoted by alpha-synuclein. Nat Commun 2016, 7, 12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Bodner CR; Maltsev AS; Dobson CM; Bax A. Differential phospholipid binding of alpha-synuclein variants implicated in Parkinson’s disease revealed by solution NMR spectroscopy. Biochemistry 2010, 49, 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Larsen EK; Weber DK; Wang S; Gopinath T; Blackwell DJ; Dalton MP; Robia SL; Gao J; Veglia G. Intrinsically disordered HAX-1 regulates Ca(2+) cycling by interacting with lipid membranes and the phospholamban cytoplasmic region. Biochim Biophys Acta Biomembr 2020, 1862, 183034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Robinson CV; Sali A; Baumeister W. The molecular sociology of the cell. Nature 2007, 450, 973–982. [DOI] [PubMed] [Google Scholar]
- (82).Koukos PI; Bonvin AMJJ Integrative Modelling of Biomolecular Complexes. Journal of Molecular Biology 2020, 432, 2861–2881. [DOI] [PubMed] [Google Scholar]
Key References
- Verardi R, Shi L, Traaseth NJ, Walsh N. & Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc Natl Acad Sci U S A 108, 9101–6 (2011).1 This paper illustrates how a combination of isotropic and anisotropic NMR parameters makes it possible to determine the structural topology of membrane proteins in lipid bilayers.
- Wang S, Gopinath T. & Veglia G. Application of paramagnetic relaxation enhancements to accelerate the acquisition of 2D and 3D solid-state NMR spectra of oriented membrane proteins. Methods 138–139, 54–61 (2018).2 This paper reports sample preparations and NMR pulse sequences to obtain 2D and 3D oriented solid-state NMR spectra for membrane protein structure determination.
- Gopinath T. & Veglia G. Probing membrane protein ground and conformationally excited states using dipolar- and J-coupling mediated MAS solid state NMR experiments. Methods 148, 115–122 (2018).3 A survey of the available methods to characterize conformationally excited states of membrane proteins.
- Gopinath T. & Veglia G. Experimental Aspects of Polarization Optimized Experiments (POE) for Magic Angle Spinning Solid-State NMR of Microcrystalline and Membrane-Bound Proteins. Methods Mol Biol 1688, 37–53 (2018).4 A review of the newly developed methods for multi-acquisition experiments in solid-state NMR spectroscopy.