Abstract
Considerable evidence suggests that breast cancer development and metastasis are driven by cancer stem-like cells (CSCs). Due to their unique role in tumor initiation, the interaction between CSCs and stromal cells is especially critical. In this work, we developed a platform to reliably isolate single cells in suspension and grow single-cell-derived spheres for functional enrichment of CSCs. The platform also allows adherent culture of stromal cells for cancer-stromal interaction. As a proof of concept, we grew SUM149 breast cancer cells and successfully formed single-cell-derived spheres. Cancer-associated fibroblasts (CAFs) as stromal cells were found to significantly enhance the formation and growth of cancer spheres, indicating elevated tumor-initiation potential. After on-chip culture for 14 days, we retrieved single-cell derived spheres with and without CAF co-culture for single-cell transcriptome sequencing. Whole transcriptome analysis highlights that CAF co-culture can boost cancer stemness especially ALDHhigh CSCs and alter epithelial/mesenchymal status. Single-cell resolution allows identification of individual CSCs and investigation of cancer cellular heterogeneity. Incorporating whole transcriptome sequencing data with public patient database, we discovered novel genes associated with cancer-CAF interaction and critical to patient survival. The preliminary works demonstrated a reliable platform for enrichment of CSCs and studies of cancer-stromal interaction.
Keywords: cell co-culture, RNA-Seq, single cell, tumor sphere, cancer-associated fibroblast
Insight Box
A focus of cancer biology is to understand cancer-stromal interaction in cancer niche, and cancer stem-like cells (CSCs) play a critical role. In this work, we enriched CSCs functionally and co-cultured them with cancer-associated fibroblasts. We found that the interaction elevated tumor-initiation potential indicated by high sphere formation rate. Furthermore, we performed single-cell RNA-sequencing to dissect the altered cancer stemness genes and epithelial/mesenchymal status. Single-cell resolution allows identification of individual CSCs and investigation of cancer cellular heterogeneity. Incorporating sequencing data with patient database, we discovered novel genes associated with cancer-stromal interaction and critical to patient survival as potential therapeutic targets.
INTRODUCTION
Considerable evidence suggests that breast cancer tumors can be initiated from a small sub-population of stem-like cells, called cancer stem-like cells (CSCs) [1, 2]. In addition to tumor initiation, CSCs are known to play critical roles in metastasis and acquire resistance to radiation and chemotherapies [3, 4]. Malignant features of CSC make it a critical target for curing cancer, yet there are controversies in defining it. In recent decades, various markers of CSC, including membrane proteins (e.g. CD44, CD24, CD90 and CD133), enzymatic markers (e.g. aldehyde dehydrogenase (ALDH)) and transcription factors (e.g. NOTCH, NANOG, octamer-binding transcription factor 4 and sex determining region Y box 2) have been identified [5–9]. However, markers are inconsistent across different malignancies and even within a single type of cancer. Multiple marker systems may isolate different sub-sets of CSCs and create controversies in this field [10]. One cause of this confusion is that the markers may not directly link to the function of tumor initiation and metastasis, so it is hard to judge and compare between marker sets [11]. As compared to marker-based approaches, functional isolation of CSC presents a new opportunity to overcome the ambiguity caused by complicated marker systems [12]. Clonal tumor sphere formation from single suspended cancer cell has been validated as a functional feature of CSCs in breast cancer [13]. It was reported that for differentiated bulk cells, disruption of cell adhesion to substrate/extracellular matrix leads to anoikis, a form of programmed cell death [14]. Stem-like CSCs are able to survive and proliferate to form tumor spheres. The unique function to resist harsh suspension environment is tightly correlated with their capability to survive in the circulatory system and metastasize in a distant site. As such, single-cell suspension culture is a useful approach to enrich CSCs for the investigation of this critical sub-population.
Given the importance of isolating single cells for suspension culture, deployment and monitoring of them remain challenging. Fluorescence-activated cell sorting (FACS) machine provides a function of single-cell deployment, yet there are chances of deploying zero or multiple cells in well-plates. Given that single cells can be deployed by FACS into well-plates, it is labor-intensive to monitor them in well-plates. More importantly, the throughput of 96 or 384 well-plates is low for enriching CSCs representing only a few percent of all cells, causing difficulty in drawing statistically significant conclusion. Growing a small number of cells in low-attachment plate/dish is even worse because cell aggregation can happen in an uncontrollable manner. Thus, there is no way to distinguish truly single-cell-derived tumor spheres or spheres formed by aggregation of multiple cells. It is far easier for cell aggregation than single cell to survive and grow, so both the quantification of sphere number and CSC enrichment condition are less reliable. Also, it would be difficult to monitor the growth of individual spheres in plate/dish. Due to the difficulty in single-cell suspension culture using conventional approaches, people have developed microfluidic approaches using hydrodynamic cell isolation, droplet platform and micro-wells [15–18]. The unique capability of microfluidics to precisely handle cells and create suspension micro-environment makes them an ideal approach for single-cell-derived tumor sphere formation.
Complicated cell–cell interaction in tumor microenvironment represents the other challenge in understanding tumor initiation and metastasis process [19]. Tumor niche includes fibroblasts, mesenchymal stem cells and cells of the immune and vascular systems, actively inducing tumor development and metastasis. Interestingly, the fibroblasts interacting with cancer cells are modified to cancer-associated fibroblasts (CAFs) and especially supportive to the dissemination of cancer [20]. Inhibition of this malignant interaction proposes a new strategy to stop tumor growth and reduce drug resistance and metastasis [21]. There are many cell–cell interaction mechanisms, including direct cell contact signaling, engulfment, communication by extracellular vesicles (e.g. exosomes) and protein secretion [22–25]. In this work, we focus on the cancer-stromal interaction through secreted growth factors (e.g. vascular endothelial growth factor, platelet-derived growth factor and fibroblast growth factor) and chemokines [22]. As mutual interaction is critical in this process, one-way interaction such as feeding conditioned media from stromal cell culture to cancer cells is less effective [26]. Mutual communication is recommended for mimicking tumor niche.
While there are microfluidic methods reported in cell–cell interaction, it is generally limited to the studies of many cells [27–30]. Thus, the approaches would not allow enrichment of critical CSCs using single-cell sphere formation. Droplet-based technology is great in single-cell isolation, suspension culture and high-throughput, yet its difficulty in media exchange makes it not suitable for weekslong tumor sphere assay [31]. We previously reported cell–cell interaction in single-cell level, yet it was limited to small-scale demonstration and by complicated fabrication process [32]. In this work, we present a simple and reliable method to combine single-cell suspension culture for enriching CSCs and CAF co-culture to understand how they behave in tumor niche. We demonstrated successful sphere formation using SUM149 breast cancer cells, and CAFs can significantly boost the formation and growth of tumor sphere. The large number of micro-wells, easy media exchange scheme and easy sphere retrieval facilitate downstream analysis using single-cell RNA sequencing. This cutting-edge analytic method detects gene expression alteration of whole transcriptome, making it suitable both for hypothesis-driven studies to understand the CAF-cancer interaction mechanism and also discovery of novel regulators. More importantly, single-cell resolution allows investigation of cellular heterogeneity that will be otherwise neglected in bulk analysis. As a proof concept, we successfully demonstrated CAF co-culture boosted CSC populations in breast cancer cells and altered epithelial/mesenchymal status of tumor cells. Our research also revealed novel gene regulators correlated with tumor-CAF interaction and critical for patient survival.
MATERIALS AND METHODS
Device design and fabrication
The micro-well device was composed of a single layer of PDMS (polydimethylsiloxane, Sylgard 184, Dow Corning), which was fabricated on a silicon substrate by standard soft lithography. One device contained 10 000 micro-wells (200 μm diameter for each micro-well, making an array of 100 by 100 micro-wells). The SU-8 (Microchem) mold used for soft-lithography was created by a photolithography process with a 100 μm layer for micro-wells, following the protocol described in the previous work [16]. The pattern was designed using a computer-aided design software (AutoCAD 2018, Autodesk®), and the mask was made by Desktop Based Lithography μPG 101 tool (Heidelberg instruments). The SU-8 mold was treated by vaporized trichloro(3,3,3-trifluoropropyl) silane (452807 Aldrich) under vacuum for 2 hours to promote the release of cured PDMS. PDMS was prepared by mixing with 10(elastomer):1(curing agent) (w/w) ratio, poured on SU-8 molds and cured at 100 °C overnight before peeling. After peeling the PDMS device, it was then bonded to a 3 by 2 inch glass slide for easy handling using oxygen plasma (80 W for 60 seconds). The device after bonding was heated at 80°C overnight to enhance bonding quality. The device was sterilized by UV radiation prior to use. Pluronic® F-108 (BASF, CAS 9003-11-6) solution (5% in DI water) was loaded to the device 12 hours before cell loading to create non-adherent PDMS substrate [16]. The device was then rinsed with PBS (Gibco 10010) for one hour to remove residual F-108 solution.
Cell culture
We cultured SUM149 cells in F-12 (Gibco 11765) media supplemented with 5% fetal bovine serum (Gibco 10082), 1% Pen/Strep (Gibco 15070), 1% GlutaMAX (Gibco 35050), 1 μg/mL hydrocortisone (Sigma H4001) and 5 μg/mL insulin (Sigma I6634). CAFs were harvested as described in the supplementary method and cultured in fibroblast medium (ScienCell 2301). All cells were cultured in regular polystyrene culture dishes and passaged at or before cells reached 80% confluency. We maintained all cells at 37°C in a humidified incubator with 5% CO2.
CAF co-cultured single-cell-derived sphere experiment
CAF cells were first harvested from a petri-dish with 0.05% Trypsin/EDTA (Gibco 25200) for 5 minutes, centrifuged at 100 × g for 5 minutes and re-suspended at 1 × 10 [6] cells/mL in CAF culture media. Then, 1 × 10 [4] CAFs were seeded in a permeable transwell insert (Corning #3450, 24 mm Transwell® with 0.4 μm Pore Polyester Membrane Insert) and cultured using CAF culture media for 2 days to completely adhere. At this time, SUM149 cancer cells were stained with 1 μM of Green Cell Tracker (Life Technologies, C2925) for 15 minutes, harvested from a petri-dish with 0.05% Trypsin/EDTA for 6 minutes, centrifuged at 100 × g for 5 minutes and re-suspended at 1 × 10 [6] cells/mL in regular SUM149 culture media. Then, 1500 SUM149 cells were seeded in the PDMS micro-wells. After 5 minutes, SUM149 cells settled in the micro-wells and then transwell insert was placed on the micro-wells for co-culture. After loading of the cancer cells, the culture media was replaced with standard tumor sphere assay media as described in the literature [12]. Media was exchanged every other day. When exchanging media, we first took out all the residual media in the transwell insert and then added 2 mL of fresh sphere media into transwell insert. On day 14, spheres were stained with 5 μM of LIVE staining (Life Technologies, L3224) for 30 minutes to identify live tumor cells/spheres in the micro-wells. The tumor sphere that had grown to at least 100 μm in diameter was scored as a sphere [33].
Single-cell-derived sphere retrieval and dissociation
After 2 weeks of culture, tumor spheres formed from a small portion of stem-like SUM149 cells in micro-wells. We removed 6-well insert (culturing CAFs) and then flipped PDMS micro-wells (containing tumor spheres) to retrieve spheres. The retrieved spheres were spun down and then re-suspended in 1 mL of 0.05% Trypsin/EDTA. The suspension was incubated at 37 °C for 4 minutes, pipetted up and down 20 times, incubated for 4 minutes and pipetted up and down 20 times again to enhance the dissociation of the spheres to single cells [15]. Dish-cultured cells were retrieved by regular 6-minute trypsinization process.
Single-cell RNA-Seq and data analysis
For each SUM149 cell population (dish cultured control, mono-cultured single-cell derived sphere and CAF co-cultured single-cell derived sphere), we sequenced around 300 single cells for representative sampling. We performed high-throughput single-cell barcoding transcriptome sequencing using Hydro-Seq platform [34, 35]. After barcode beads captured mRNA from cells, we performed RT (Thermofisher Maxima RT kit), PCR (Kapa HiFi Hotstart PCR Readymix) and library preparation (Illumina Nextera XT Library Prep Kit). We obtained approximately 30 million reads (paired-end: one side 25 base pairs for barcode and the other side 100 base pairs for mRNA quantification) for each population (1 × 10 [5] reads per cell). Reads were aligned using STAR and processed by the standard flow suggested by Dropseq [34]. Then, we used open-source SEURAT kit to analyze single-cell sequencing data. We first identified significantly altered genes defined by logarithmic fold change as 0.25 and minimal portion of expressing cells as 10%. For significantly altered genes, we used a publicly available database, kmplot.com [36], to determine correlations with patient prognosis in breast cancer. To analyze the prognostic value of a particular gene, patient samples were split into two groups based on expression of that gene and compared by a Kaplan–Meier curve to calculate the hazard ratio with 95% confidence intervals and log rank P value. We selected genes for further evaluation based on concordance between changes in expression of CAF co-cultured condition and worse survival in breast cancer.
RESULTS AND DISCUSSION
Single-cell-derived sphere and CAF co-culture experiment
The presented co-culture platform is composed of a non-adherent substrate having 10 000 micro-wells for single-cell-derived tumor sphere formation and a porous permeable transwell insert for CAF adherent co-culture (Fig. 1(a)). For co-culture experiment, we first load CAFs in a transwell insert for adherent culture and let CAFs adhere to monolayer. Then, we load 1500 cancer cells onto the micro-well device. Since Poisson’s distribution determines the seeding process, 13.1% of 10 000 micro-wells isolate single cells for tumor sphere assay (Fig. 1b–d). By tracking the location of each micro-well, a small number of micro-wells trapping multiple cells can be excluded from sphere formation rate measurement. After cell seeding, the transwell insert culturing CAFs is placed onto the substrate. As the size of proteins is typically around 1–5 nm, pores (0.4 μm) on the transwell insert allow secreted proteins to diffuse through for cellular interaction while preventing cells from migrating to the other side [37, 38]. Physical separation between two cell types guarantees simple cell retrieval after tumorsphere assays for further downstream analysis without contamination. In addition, media can be simply exchanged by aspirating and replenishing media in the transwell insert, so the isolated single cells will not be disturbed. The media exchange process is easier and more reliable than conventional tumorsphere assays in low-attachment plate and hanging droplet platform.
Figure 1 .
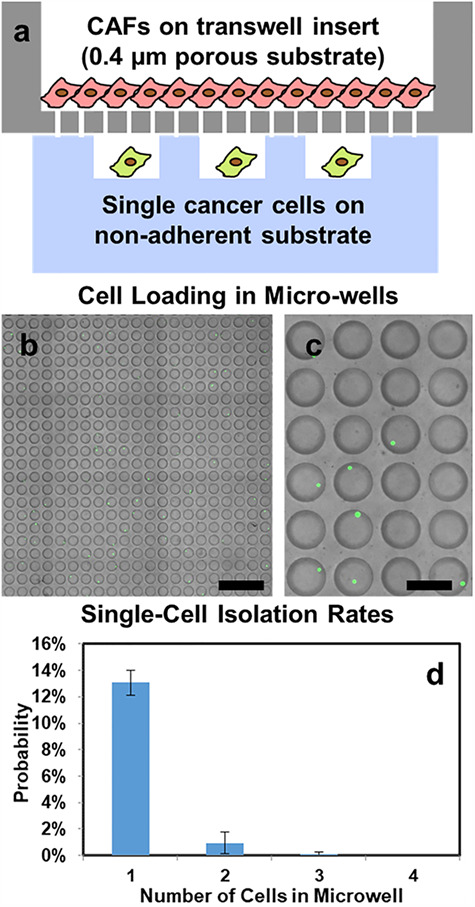
Schematics and cell loading of vertical adhesion-suspension co-culture platforms. (a) Single cancer cells are seeded in non-adherent micro-wells randomly (following Poisson’s distribution), and CAFs are cultured on a transwell insert. The 0.4 μm pores on transwell insert allow secreted proteins to pass through for cellular interactions between CAFs and cancer cells. (b) Micro-wells in a 20 by 20 array right after cell loading. SUM149 cancer cells were labeled with green fluorescence for counting (scale bar: 1 mm). (c) Enlarged view of micro-wells in a 4 by 6 array right after cell loading. (8 out of 24 wells captured single cells) (scale bar: 250 μm) (d) Distribution of the number of captured cancer cells per micro-well when loading 1500 cells into a 10 000-well device. (Error bar represents standard error of the mean (SEM), n = 3 devices).
As the capability to derive tumor spheres from single cells represents a unique capability of CSCs, non-stem-like cancer cells are likely to die after being cultured in suspension for 14 days. Compared with initial (right after cell loading) micro-well image (Fig. 1b and c), day 14 images (Fig. 2a and b) show fewer green fluorescent positive cells (labeled by LIVE staining), yet the surviving ones grow to large spheres. In this manner, stem-like cancer cells are enriched in the presented culture method. To assess cancer-stromal interaction effect, we quantified the number of single-cell-derived spheres in micro-well array using a custom MATLAB program. Compared with mono-cultured SUM149 single cells, CAF co-cultured ones have significantly higher sphere formation rate (Fig. 2a–c), indicating an increased tumorigenic potential with CAF support. In addition, CAF co-cultured spheres were significantly larger than mono-cultured ones (Fig. 2d), indicating a higher cancer cell proliferation rate boosted by cancer-stromal interaction. This experiment demonstrates the critical phenotypic differences of cancer cells with and without stromal support.
Figure 2 .
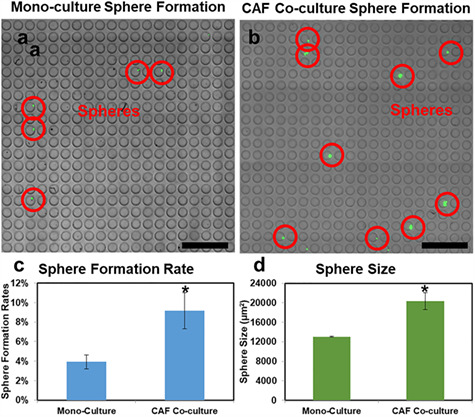
Single-cell-derived sphere formation with and without CAF co-culture. (a, b) Micro-wells in a 20 by 20 array after 14 days of (a) cancer mono-culture and (b) with CAF co-culture on-chip. Most non-stem-like single cells were dead due to culturing in suspension, yet a small number of stem-like SUM149 breast cancer cells grew to large tumor spheres indicated by green fluorescence and red circles. (scale bar: 1 mm). (c) Sphere formation rate of mono-cultured and CAF co-cultured SUM149 cells after 14-day. (Error bar represents SEM, n = 3 devices), *P < 0.05. (d) Average size of mono-cultured and CAF co-cultured SUM149 spheres derived from single cells (Error bar represents SEM, n = 3 devices), *P < 0.05.
Distinct transcriptome profiles of dish-cultured and single-cell suspension-cultured SUM149 breast cancer cells
In addition to the phenotypic observation of single-cell-derived spheres, we aim to investigate the gene expression differences between dish-cultured spheres (control), mono-cultured spheres (CSC enriched) and CAF co-cultured spheres (CSC enriched and regulated by CAF). The alteration can help us identify critical genes/pathways associated with CSC and CAF regulation. As demonstrated in Supplementary Fig. 1(a), the presented platform allows easy retrieval of spheres for downstream transcriptome analysis. After flipping PDMS micro-wells upside down, spheres will get out of the micro-wells by gravity. After 5 minutes, around 90% of the spheres can be retrieved. Using a combination of chemical (trypsinization) and mechanical (pipetting) method, tumor spheres were dissociated into single cells (Supplementary Fig. 1). The combination allows rapid sphere dissociation and maintains good cell viability [15, 32]. As shown in Supplementary Fig. 1(b), the dissociated cells preserved good viability indicated by sharp phase contrast of intact cell membrane. More importantly, no doublet or cluster of cells was observed after dissociation. After the dissociation process, we successfully collected around 2000 dissociated single cells from mono-cultured spheres and around 10 000 cells from co-cultured spheres. The collected single cells were then processed by Hydro-Seq for single-cell whole transcriptome sequencing using the mRNA barcoding technique, which allows all mRNAs from one cell to be uniquely labeled and identified after sequencing [34, 35]. Hydro-Seq microfluidic bead-cell pairing scheme can precisely isolate single cells without contamination from multiple cell capture. As non-stem-like cancer cells are likely to die in suspension environment, those dead cells can be excluded by a low number of transcripts in data processing, even if being sequenced. In this manner, stem-like cancer cells can be enriched for investigation.
We first visualized single-cell data using principal component analysis (PCA) and t-distributed stochastic neighbor embedding (tSNE) clustering methods. We found obvious separation between dish-cultured, single-cell suspension mono-cultured and single-cell suspension CAF co-cultured cells, indicating distinct transcriptome profiles (Fig. 3a and b). We first examined the differences between dish-cultured and single-cell suspension mono-cultured conditions. To define key pathways associated with single-cell suspension culture, we selected significantly altered genes defined by thresholds of fold change and portion of expressing cells and applied pathway analysis using the NCI-Nature 2016 pathway database (Table 1). The pathway analysis identifies critical genetic networks in the sphere formation process and provides mechanistic and functional implications. We first identified cell cycle-related pathways in the top-ranked list, including ‘PLK1 signaling events,’ ‘Validated targets of C-MYC transcriptional activation,’ and ‘Aurora B signaling.’ [39–41] We further examined critical genes related to cell cycle such as MKI67, PCNA, CCND1 and CDK1 and c-Myc pathway such as NPM1, NME1, HMGA1 and THBS1 (Fig. 3c and d) [42, 43]. It is obvious that single-cell suspension culture cells were far more dormant as compared to dish-cultured ones, and dormancy is indeed a marker of stem cells [44]. The phenotypic dormancy matched well with the transcriptome regulation in cell cycle. More interestingly, single-cell suspension-cultured cells showed down-regulation of proteasome genes (Fig. 3e), matching well with recent discovery that low proteasome activity is a marker for CSCs[45]. These observations collectively strengthen the link between single-cell suspension culture and stem-like characteristic.
Figure 3 .
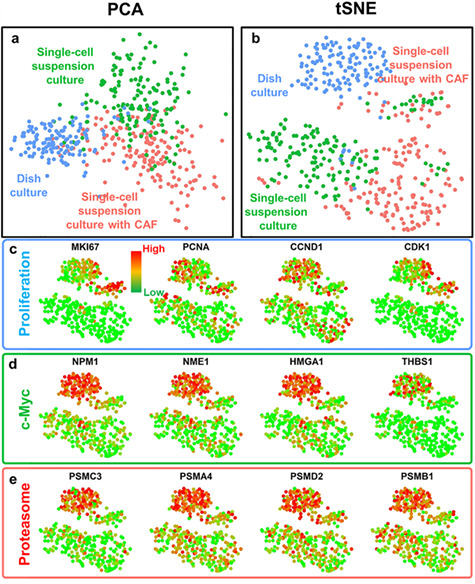
Distinct gene expression profiles of SUM149 breast cancer cells from different culture conditions. (a, b) Principal component analysis (PCA) and t-distributed Stochastic Neighbor Embedding (tSNE) plots of single-cell transcriptome analysis for single-cell suspension cultured with CAFs (red color), single-cell suspension cultured without CAFs (green color) and dish cultured (blue color) cancer cells. Three populations of the same cell line are clearly separated, indicating distinct gene expression profiles under different culture conditions. (c–e) Gene expression and clustering of SUM149 cells from different culture conditions. Each dot represents one cell. Red color represents high (90th percentile) expression of a gene, and green color represents low (10th percentile) expression of a gene. The expression is logarithmically normalized. (c) The expression of proliferation-related genes: proliferation marker protein Ki-67 (MKI67), proliferating cell nuclear antigen (PCNA), cyclin D1 (CCND1) and cyclin-dependent kinase 1 (CDK1). (d) The expression of c-Myc pathway-related genes: nucleophosmin (NPM1), nucleoside diphosphate kinase A (NME1), high mobility group AT-hook 1 (HMGA1), and thrombospondin 1 (THBS1). (e) The expression of proteasome genes: proteasome 26S Subunit ATPase 3 (PSMC3), proteasome subunit alpha 4 (PSMA4), proteasome 26S subunit, non-ATPase 2 (PSMD2) and proteasome subunit beta 1 (PSMB1). Dish-cultured cells have elevated proliferative, c-Myc-related and proteasome genes.
Table 1 .
Top-ranked altered pathways (NCI-Nature pathway database) comparing the differences between dish-cultured and single-cell suspension-cultured SUM149 cells.
| Top-ranked pathways | P-value |
|---|---|
| PLK1 signaling events | 7.57E-10 |
| Validated targets of C-MYC transcriptional activation | 1.51E-09 |
| a6b1 and a6b4 integrin signaling | 8.77E-07 |
| Validated transcriptional targets of deltaNp63 isoforms | 8.77E-07 |
| Aurora B signaling | 4.92E-07 |
| FoxO family signaling | 1.205E-05 |
| HIF-1-alpha transcription factor network | 2.073E-05 |
| ATR signaling pathway | 1.608E-05 |
| Validated targets of C-MYC transcriptional repression | 0.0001353 |
| FOXM1 transcription factor network | 0.0001057 |
CAF co-culture boosted CSC-associated genes and altered epithelial/mesenchymal status
After comparing the dish and single-cell suspension culture conditions, we further investigated the effects induced by CAF co-culture (Fig. 4a and b). As demonstrated in Table 2, we found pathways associated with epithelial/mesenchymal status as top ranked ones: ‘E-cadherin signaling in the nascent adherens junction’ and ‘HIF-1-alpha transcription factor network.’ The regulation of epithelial/mesenchymal status is critical for tumor development and metastasis. Then, we further examined individual genes differentially expressed by mono-culture and CAF co-culture conditions. We first noticed a prominent activation of pan-aldehyde dehydrogenase isoforms (Pan-ALDH), especially ALDH1A3 in co-cultured cells (Fig. 4c). As an established marker of CSC, ALDHhigh cells demonstrate elevated tumorigenic and metastatic capability.[46] While not as significant as ALDH, another well-accepted CSC marker CD44 was also up-regulated (P-value < 0.01) in co-cultured SUM149 cells. The up-regulation of CSC-associated genes matches well with the correlation between cancer-CAF interaction and poor prognosis reported in the literature [20–22]. We also examined established epithelial and mesenchymal regulators. Interestingly, we found that the CAF co-cultured cells were more epithelial-like indicated by up-regulation of EpCAM, CDH1, KRT8 and KRT16 and also down-regulation of VIM, HIF1A, SNAI1 and NNMT (Fig. 4d and e).[47] The results follow the same trend with enrichment of epithelial ALDHhigh CSCs in CAF co-culture condition.
Figure 4 .
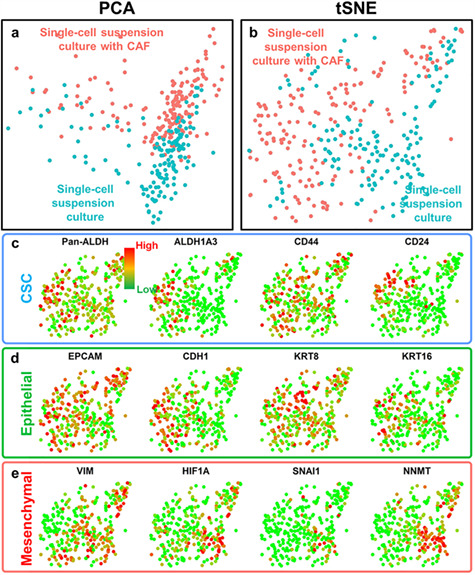
CAF co-culture-altered gene expression profile of SUM149 breast cancer cells. (a, b) PCA and tSNE plots of single-cell transcriptome analysis for single-cell suspension-cultured cells with CAFs (red color) and single-cell suspension-cultured cells without CAFs (blue color). Two populations of the same cell line are clearly separated, indicating CAF co-culture-altered gene expression profile. (c–e) Gene expression and clustering of SUM149 cells from different culture conditions. Each dot represents one cell. Red color represents high (90th percentile) expression of a gene, and green color represents low (10th percentile) expression of a gene. The expression is logarithmically normalized. (c) The expression of CSC-related genes: pan-aldehyde dehydrogenase isoforms (Pan-ALDH), aldehyde dehydrogenase 1 family member A3 (ALDH1A3), CD44 and CD24. (d) The expression of epithelial genes: epithelial cell adhesion molecule (EpCAM), cadherin 1 (CDH1), keratin 8 (KRT8) and keratin 16 (KRT16). (e) The expression of mesenchymal genes: vimentin (VIM), hypoxia inducible factor 1 alpha (HIF1A), snail transcriptional repressor 1 (SNAI1) and nicotinamide N-methyltransferase (NNMT). The p-values comparing the expression of Pan-ALDH, ALDH1A3, CD44, CD24, VIM, EpCAM, CDH1, HIF1A, SNAI1 and NNMT between two cell populations are lower than 0.01. CAF-co-cultured cells have elevated CSC and epithelial genes and down-regulated mesenchymal genes.
Table 2 .
Top-ranked altered pathways (NCI-Nature pathway database) comparing the differences between mono-cultured and CAF co-cultured SUM149 cells.
| Top-ranked pathways | P-value |
|---|---|
| E-cadherin signaling in the nascent adherens junction | 2.01E-07 |
| HIF-1-alpha transcription factor network | 5.06E-07 |
| RAC1 signaling pathway | 0.000008934 |
| Validated transcriptional targets of AP1 family members Fra1 and Fra2 | 0.000004479 |
| AP-1 transcription factor network | 0.0000255 |
| ErbB1 downstream signaling | 0.00004934 |
| Validated targets of C-MYC transcriptional activation | 0.00002596 |
| a6b1 and a6b4 Integrin signaling | 0.00004988 |
| VEGFR1 specific signals | 0.00003069 |
| Validated targets of C-MYC transcriptional repression | 0.0002151 |
CSCs and cellular heterogeneity based on transcriptome profiles
While ALDH and CD44 expression levels were overall up-regulated in CAF co-cultured cells, there was a significant cellular heterogeneity among single cells. Using single-cell resolution, we could pinpoint those more stem-like cells. It is clear that all ALDHhigh cells were from CAF co-culture condition, while CD44high cells distribute more uniformly in the plot. More interestingly, we also identified cells simultaneously expressing ALDHhigh and CD44high markers.[48] These dual positive cells may be endowed with the highest degree of cellular plasticity and metastatic potential (Fig. 5a). In addition to CSC markers, we re-visited cell proliferation-related genes. Given that single-cell suspension-cultured cells are generally more dormant, there is a highly proliferative sub-population clustered in the upper right corner (Fig. 5b). The cells in this cluster have enhanced expression of cell proliferation indexes (MKI67 and PCNA) and also cyclin and related kinase (CCND1 and CDK1). Without single-cell resolution, we will not be able to identify dual positive CSCs and highly proliferative cells. The capability to monitor cellular heterogeneity represents the unique value of single-cell analysis.
Figure 5 .
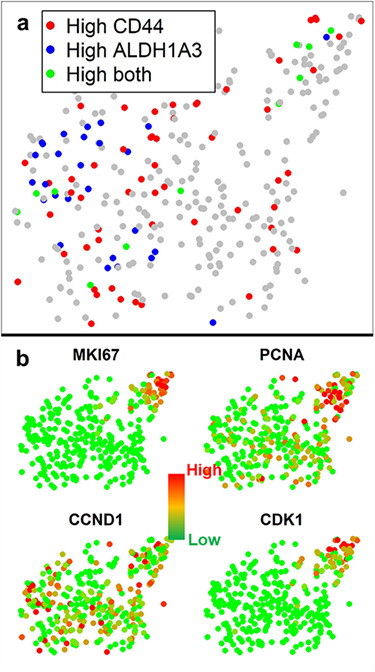
tSNE clustering demonstrates the cellular heterogeneity of CAF co-cultured and mono-cultured SUM149 breast cancer cells. (a) Mesenchymal-like CSCs (CD44 high), epithelial-like CSCs (ALDH1A3 high) and dual positive CSCs (both CD44 and ALDH1A3 high) on the tSNE plot. Epithelial-like CSCs are enriched in CAF co-cultured group, and mesenchymal-like CSCs are distributed more uniformly. (b) Gene expression of CAF co-cultured and mono-cultured SUM149 breast cancer cells. Each dot represents one cell. Red color represents high (90th percentile) expression of a gene, and green color represents low (10th percentile) expression of a gene. The expression is logarithmically normalized. Upper right cluster having elevated proliferation-related genes (MKI67, PCNA, CCND1 and CDK1) represents the fast proliferating cells in single-cell derived spheres.
Identification of novel regulators induced by CAF co-culture
In addition to known cell-cycle, epithelial, mesenchymal and CSC-related genes, our data provide an opportunity to discover novel gene regulators related to CAF co-culture as potential prognostic markers and/or drug targets. We compared significantly altered genes by statistics and identified 552 genes up-regulated and 460 genes down-regulated in the CAF co-cultured cells as compared to mono-cultured ones. Among potential candidates, we used a publicly available database (kmplot.com) to determine correlations with prognosis in breast cancer. Combining transcriptome analysis of migratory cells and data mining for patient outcomes, we successfully identified top-ranked candidate genes with significant differences in survival of patients with breast cancer. As examples, we highlight schlafen family member 5 (SLFN5) up-regulated in CAF co-cultured cancer cells (Fig. 6a) and up-regulation of this gene correlated with bad patient prognosis (Fig. 6b). We also identified dual specificity phosphatase 4 (DUSP4) down-regulated in CAF co-cultured cells (Fig. 6a) and down-regulation of this gene correlated with bad patient prognosis (Fig. 6c). The potential to discover new prognostic biomarkers and/or drug targets related to CAF co-culture demonstrates the value of presented co-culture platform with simple sample retrieval capability for single-cell whole transcriptome sequencing.
Figure 6 .
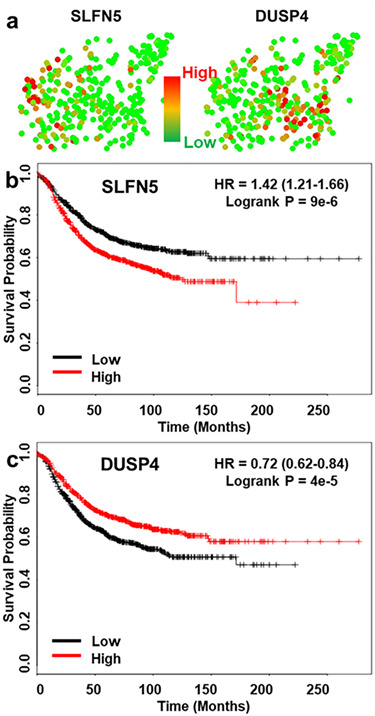
Sequencing of CAF co-cultured and mono-cultured breast cancer cells identifies new prognostic markers and therapeutic targets. (a) Gene expression of CAF co-cultured and mono-cultured SUM149 breast cancer cells. Each dot represents one cell. Red color represents high (90th percentile) expression of a gene, and green color represents low (10th percentile) expression of a gene. The expression is logarithmically normalized. Left cluster (CAF co-cultured) has up-regulated schlafen family member 5 (SLFN5) and down-regulated dual specificity phosphatase 4 (DUSP4) as compared to mono-cultured right cluster. (b, c) The Kaplan–Meier plots show that high expression levels of SLFN5 and low expression level of DUSP4 correlate with reduced relapse-free survival (RFS) in breast cancer.
CONCLUSION
We have successfully demonstrated a cell co-culture platform incorporating single-cell suspension culture of cancer cells with adherent culture of stromal cells. Single-cell suspension culture functionally enriches critical stem-like cancer cell population driving tumor initiation and metastasis. Adherent culture on transwell insert is favorable for the survival of stromal cells. The co-culture platform allows them to communicate with each other to mimic tumor microenvironment. As a proof of concept, we tested SUM149 breast cancer cells and primary CAFs as stromal cells. Single cancer cells successfully formed single-cell-derived spheres on-chip, and CAFs can significantly boost sphere formation and growth. The presented method allows simple and reliable media exchange which is critical for 2-week sphere formation process. With reliable cell retrieval capability, we harvested single-cell- derived spheres and dissociated them to single cells for downstream transcriptome analysis. Using cutting-edge single-cell RNA sequencing platform to analyze dish-cultured and single-cell suspension-cultured SUM149 breast cancer cells, we first exhibited that the single-cell-derived spheres are overall more quiescent, matching well with dormant stem-like capability. Then, we further investigated the differences between CAF co-cultured and mono-cultured cancer cells. Remarkably, we found CAF co-culture boosted CSCs especially ALDHhigh subtype. The CAF co-cultured cells were also more epithelial-like as compared to mono-cultured ones. While the presented method is limited to secretion-based cell-cell interaction, significant transcriptome alteration was observed in the experiment. With single-cell resolution, we could pinpoint individual CSCs of different sub-types rather than merely measure average gene expression of many cells. More interestingly, we found while single-cell suspension-cultured cells are generally more quiescent, there is a small sub-population composed of highly proliferative cells. The cellular heterogeneity information will otherwise be lost without single-cell resolution. Combining whole transcriptome sequencing data and publicly available database, we identified novel regulators associated with CAF co-culture. Those markers are pivotal in patient’s prognosis and can potentially be developed to predictive and/or therapeutic targets. In this preliminary study, we performed a successful cancer-CAF co-culture study and demonstrated the capability of CSC enrichment, CAF co-culture, reliable cell retrieval and single-cell RNA sequencing. In this manner, we verified boosting CSCs by CAF co-culture and identified novel gene targets for future biological investigation.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institute of Health to E.Y. (R01 CA 203810 and R21 CA 195016). Y.-C. Chen acknowledges the support from Forbes Institute for Cancer Discovery. We thank the Lurie Nanofabrication Facility of the University of Michigan (Ann Arbor, MI) for device fabrication and Dr. Jason Cong’s lab in UCLA for computation of sequencing read alignment.
Competing financial interests
The authors declare no competing financial interests.
Supporting Information
Supporting information includes: [1] detailed methods for harvesting cancer associated fibroblast cells, image acquisition and image analysis. [2] Images support successful sphere dissociation. [3] Pathway analysis to compare cell populations.
References
- 1. Al-Hajj M, Wicha MS, Benito-Hernandez Aet al. . Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100:3983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ye X, Tam WL, Shibue Tet al. . Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 2015;525:256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li X, Lewis MT, Huang Jet al. . Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst 2008;100:672–9. [DOI] [PubMed] [Google Scholar]
- 4. Diehn M, Cho RW, Lobo NAet al. . Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 2009;458:780–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karsten U, Goletz S. What makes cancer stem cell markers different? Springerplus 2013;2:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tomita H, Tanaka K, Tanaka Tet al. . Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 2016;7:11018–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takebe N, Miele L, Harris PJet al. . Notch, hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol 2015;12:445–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jeter CR, Yang T, Wang Jet al. . Concise review: NANOG in cancer stem cells and tumor development: an update and outstanding questions. Stem Cells 2015;33:2381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang D, Lu P, Zhang H. Oct-4 and Nanog promote the epithelial-mesenchymal transition of breast cancer stem cells and are associated with poor prognosis in breast cancer patients. Oncotarget 2014;5:10803–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prestegarden L, Svendsen A, Wang Jet al. . Glioma cell populations grouped by different cell type markers drive brain tumor growth. Cancer Res 2010;70:4274–9. [DOI] [PubMed] [Google Scholar]
- 11. Liu Y, Nenutil R, Appleyard MVet al. . Lack of correlation of stem cell markers in breast cancer stem cells. Br J Cancer 2014;110:2063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dontu G, Abdallah WM, Foley JMet al. . In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 2003;17:1253–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu S, Dontu G, Mantle IDet al. . Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res 2006;66:6063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer 2006;6:449–58. [DOI] [PubMed] [Google Scholar]
- 15. Chen YC, Ingram PN, Fouladdel Set al. . High-throughput single-cell derived sphere formation for cancer stem-like cell identification and analysis. Sci Rep 2016;6:27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng YH, Chen YC, Brien Ret al. . Scaling and automation of a high-throughput single-cell-derived tumor sphere assay chip. Lab Chip 2016;16:3708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brouzes E. Droplet microfluidics for single-cell analysis. Methods Mol Biol 2012;853:105–39. [DOI] [PubMed] [Google Scholar]
- 18. Rettig JR, Folch A. Large-scale single-cell trapping and imaging using microwell arrays. Anal Chem 2005;77:5628–34. [DOI] [PubMed] [Google Scholar]
- 19. Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008;27:5904–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shiga K, Hara M, Nagasaki Tet al. . Cancer-associated fibroblasts: their characteristics and their roles in tumor growth. Cancers (Basel) 2015;7:2443–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barnett RM, Vilar E. Targeted therapy for cancer-associated fibroblasts: are we there yet? J Natl Cancer Inst 2018;110:11–3. [DOI] [PubMed] [Google Scholar]
- 22. Weber CE, Kuo PC. The tumor microenvironment. Surg Oncol 2012;21:172–7. [DOI] [PubMed] [Google Scholar]
- 23. Mayor R, Carmona-Fontaine C. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 2010;20:319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen YC, Gonzalez ME, Burman B, Zhao X, Anwar T, Tran M, Medhora N, Hiziroglu AB, Lee W, Cheng YH, Choi Y, Yoon E, Kleer CG. Mesenchymal Stem/Stromal Cell Engulfment Reveals Metastatic Advantage in Breast Cancer. Cell Rep. 2019;27:3916–3926.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maia J, Caja S, Strano Moraes MC, Couto N, Costa-Silva B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front Cell Dev Biol. 2018;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Melzer C, Yang Y, Hass R.. Interaction of MSC with tumor cells. Cell Commun Signal 2016;14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jeong SY, Lee JH, Shin Y, Chung S, Kuh HJ. Co-Culture of Tumor Spheroids and Fibroblasts in a Collagen Matrix-Incorporated Microfluidic Chip Mimics Reciprocal Activation in Solid Tumor Microenvironment. PLoS One 2016;11:e0159013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Menon NV, Chuah YJ, Cao Bet al. . A microfluidic co-culture system to monitor tumor-stromal interactions on a chip. Biomicrofluidics 2014;8:064118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu T, Lin B, Qin J. Carcinoma-associated fibroblasts promoted tumor spheroid invasion on a microfluidic 3D co-culture device. Lab Chip 2010;10:1671–7. [DOI] [PubMed] [Google Scholar]
- 30. Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol 2011;21:745–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frimat JP, Becker M, Chiang YYet al. . A microfluidic array with cellular valving for single cell co-culture. Lab Chip 2011;11:231–7. [DOI] [PubMed] [Google Scholar]
- 32. Chen YC, Zhang Z, Fouladdel Set al. . Single cell dual adherent-suspension co-culture micro-environment for studying tumor-stromal interactions with functionally selected cancer stem-like cells. Lab Chip 2016;16:2935–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Faustino GM, Gattass M, Rehen Set al. . Automatic embryonic stem cells detection and counting method in fluorescence microscopy images. In: Proc. IEEE Int. Symp. Biomed. Imag, From Nano to Macro, 2009, 799–802.
- 34. Macosko EZ, Basu A, Satija Ret al. . Highly parallel genome-wide expression profiling of individual cells using Nanoliter droplets. Cell 2015;161:1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng YH, Chen YC, Lin Eet al. . Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells. Nat Commun 2163;10:2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Györffy B, Lanczky A, Eklund ACet al. . An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725–31. [DOI] [PubMed] [Google Scholar]
- 37. Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced 2009;11:32–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. David MS, Kelly E, Zoellner H. Opposite cytokine synthesis by fibroblasts in contact co-culture with osteosarcoma cells compared with transwell co-cultures. Cytokine 2013;62:48–51. [DOI] [PubMed] [Google Scholar]
- 39. Lee SY, Jang C, Lee KA. Polo-like kinases (plks), a key regulator of cell cycle and new potential target for cancer therapy. Dev Reprod 2014;18:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bretones G, Delgado MD, León J. Myc and cell cycle control. Biochim Biophys Acta 2015;1849:506–16. [DOI] [PubMed] [Google Scholar]
- 41. Goldenson B, Crispino JD. The aurora kinases in cell cycle and leukemia. Oncogene 2015;34:537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sobecki M, Mrouj K, Colinge Jet al. . Cell-cycle regulation accounts for variability in Ki-67 expression levels. Cancer Res 2017;77:2722–34. [DOI] [PubMed] [Google Scholar]
- 43. Strzalka W, Ziemienowicz A. Proliferating cell nuclear antigen (PCNA): A key factor in DNA replication and cell cycle regulation. Ann Bot 2011;107:1127–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kleffel S, Schatton T. Tumor dormancy and cancer stem cells: Two sides of the same coin? Adv Exp Med Biol 2013;734:145–79. [DOI] [PubMed] [Google Scholar]
- 45. Lenos KJ, Vermeulen L. Cancer stem cells don't waste their time cleaning-low proteasome activity, a marker for cancer stem cell function. Ann Transl Med 2016;4:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ginestier C, Hur MH, Charafe-Jauffret Eet al. . ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007;1:555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 2009;119:1429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu S, Cong Y, Wang Det al. . Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep 2013;2:78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


